Summary
Developmental stuttering is a speech disorder characterized by disruption in the forward movement of speech. This disruption includes part-word and single-syllable repetitions, prolongations, and involuntary tension that blocks syllables and words, and the disorder has a life-time prevalence of 6–12%. Within Vanderbilt’s electronic health record (EHR)-linked biorepository (BioVU), only 142 individuals out of 92,762 participants (0.15%) are identified with diagnostic ICD9/10 codes, suggesting a large portion of people who stutter do not have a record of diagnosis within the EHR. To identify individuals affected by stuttering within our EHR, we built a PheCode-driven Gini impurity-based classification and regression tree model, PheML, by using comorbidities enriched in individuals affected by stuttering as predicting features and imputing stuttering status as the outcome variable. Applying PheML in BioVU identified 9,239 genotyped affected individuals (a clinical prevalence of ∼10%) for downstream genetic analysis. Ancestry-stratified GWAS of PheML-imputed affected individuals and matched control individuals identified rs12613255, a variant near CYRIA on chromosome 2 (B = 0.323; p value = 1.31 × 10−8) in European-ancestry analysis and rs7837758 (B = 0.518; p value = 5.07 × 10−8), an intronic variant found within the ZMAT4 gene on chromosome 8, in African-ancestry analysis. Polygenic-risk prediction and concordance analysis in an independent clinically ascertained sample of developmental stuttering cases validate our GWAS findings in PheML-imputed affected and control individuals and demonstrate the clinical relevance of our population-based analysis for stuttering risk.
Introduction
Developmental stuttering is a speech disorder characterized by disruption in the forward movement of speech. This disruption includes part-word and single-syllable word repetitions, sound prolongations, and involuntary breaks in syllables and words.1 Previous population-based studies estimate that 6–12% of children aged 2-4 will develop a stutter and that 15–25% of these speech impediments will persist to adulthood, resulting in approximately 1% prevalence in the adult population.2 Risk factors for developmental stuttering include sex—males demonstrate increased risk—and a family history of stuttering.3 Elevated risk in males increases with age; the male-to-female ratio is approximately 2:1 (or lower) in children under 44,5 but rises to 5:1 in adolescents and adults,4 suggesting a higher rate of recovery in females, by age.6
The impact of stuttering across the lifespan is significant and well documented. Children who stutter, especially those in whom stuttering persists, experience decreased overall school performance, including social withdrawal and reduced classroom participation.7 In addition to the impact on their academic experiences, adolescents who stutter often experience a higher incidence of bullying.7 Adults who continue to stutter can also experience impaired career trajectories because stuttering can increase the risk of unemployment and reduce perceived job performance, both of which contribute to reducing socioeconomic status among people who stutter.8 Despite a clear social and vocational impact, no direct causes of developmental stuttering in populations have been previously identified. Given the observed enrichment in families, genetic studies offer a particularly promising approach to understanding underlying genetic causes and provide insight into potential biological mechanisms contributing to this phenotype.9
Heritability estimates of developmental stuttering have varied greatly across studies; they have ranged from 0.42 to 0.84 in the two largest twin studies, each comprising a sample size exceeding 20,000 individuals.10,11 Though heritability estimates vary, there is clear evidence that a genetic component for developmental stuttering exists, and consequently several linkage-based genetic analyses have sought to identify loci within potentially causative genes. These familial genetic studies identified significant hits within GNPTAB (MIM: 607840), GNPTG (MIM: 607838), NAGPA (MIM: 607985), and AP4E1 (MIM: 607244), although there is little concordance in identified loci across studies, indicating that results might be specific to the tested family.9,12,13 Follow-up studies have demonstrated that disruptions in GNPTAB resulted in deficits in astrocyte pathology in the corpus callosum and disruptions in mouse vocalization.14 The roles of these astrocytes in the onset of stuttering are not well characterized; however, recent studies have contributed to a growing body of evidence that dopamine receptor D2 blockers can impact stuttering behavior, perhaps because of increased astrocyte metabolism in the striatum.15 These studies suggest that dopamine projection from the basal ganglia might contribute to disturbances in speech and vocalization, and the findings potentially support pharmacological means for treatment.16,17 Recent evidence also implicates autoimmune reactions from group A beta-hemolytic streptococcus (GAS [MIM: 607395]) infections that target specific cell types within the basal ganglia as a potential cause of stuttering.18 Rheumatic fevers (MIM: 268240) and other sequelae resulting from GAS have been more generally linked with pediatric autoimmune neuropsychiatric disorders and historically have correlated strongly with stuttering in children.18
Still, to date, most genetic research has provided limited biological insight into potential mechanisms of action that contribute to the stuttering phenotype, and the lack of replicability across the linkage studies suggests that these genetic risk loci do not explain the genetic basis of stuttering at a population level.9
Genome-wide association studies (GWASs), an alternative method to linkage analysis for disease gene discovery, typically utilize genome-wide genetic data in large samples drawn from populations to identify common genetic variants that are associated with increased risk of a disease or trait. Prior to this study, no population-based genome-wide association study (GWAS) has successfully identified variants significantly associated with developmental stuttering. Stuttering has a high recovery rate and is frequently diagnosed outside of a hospital or clinical setting; therefore, one reason for the lack of genetic discoveries that explain the general prevalence of stuttering is the challenge of acquiring large numbers of developmental stuttering cases for GWAS approaches to be well powered. To address the issue of case acquisition, today, researchers are frequently turning to large-scale biobanks linked to electronic health records (EHRs) to efficiently and cost effectively develop studies well powered for genetic discovery.19 Cases for a particular phenotype are often identified in EHRs through the use of phenotyping algorithms based on ICD-9/10 billing codes, CPT procedural codes, and/or notes from clinical records.20 However, the billing codes traditionally used to assess patient status in the electronic health record are heavily underreported for developmental stuttering.21
In Vanderbilt University’s large HER-linked DNA database (BioVU), only 142 of the 92,762 (0.15%) patient samples genotyped on the Illumina Multi-Ethnic Genotyping Array (MEGAEX) had recorded billing codes denoting developmental stuttering (see Table S1), a proportion well below even the most stringent expected prevalence. The nature of this condition might shed some light on its underrepresentation in Vanderbilt’s EHR. In cases where patients do not have an overt stutter or in the event that the patient exhibits early recovery, doctors might simply overlook the condition and not record a diagnosis in the EHR. Even if a patient were to seek evaluation and treatment for developmental stuttering, speech evaluations are typically performed by speech-language pathologists, usually outside of a hospital context (e.g., schools or private clinics).21 There are also currently no FDA-approved medications or medical procedures to treat developmental stuttering, making it significantly less likely to be noted in a medical setting. Finally, although treatment exists for stuttering in the form of therapy and even though this is a chronic condition for many adults, this condition is not considered a parity diagnosis, and as such most government and private insurance plans do not cover treatment costs for stuttering. An inability to bill for these diagnoses makes it less likely for providers to include the ICD code for this diagnosis during a patient visit.
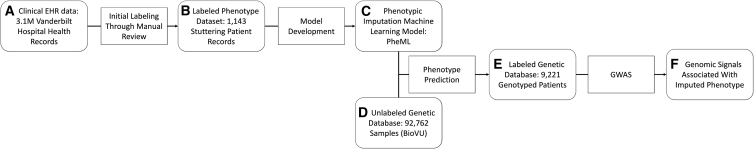
In our previous research, we identified individuals affected by stuttering by applying a phenotype-driven machine-learning algorithm (PheML) that uses commonly reported phenotypes significantly associated with clinically diagnosed developmental stuttering as predictor variables to impute a developmental stuttering phenotype in BioVU (individuals with this phenotype are predicted by PheML to be affected by stuttering).21 Our model takes a series of binary proxy parameters to impute developmental stuttering in a patient population by using a Gini impurity-based classification and regression tree classifier.22 The PheML algorithm was built and tested with an initial pool of manually reviewed records from individuals affected by stuttering in Vanderbilt University’s EHR (no subjects within BioVU were used) across all ancestries (Figure 1). Model validation testing in an independent dataset containing manually reviewed records resulted in a positive prediction rate of 83.3% (Table 1).21 Applying this model in BioVU resulted in a higher proportion of individuals with imputed developmental stuttering (∼10%) than were observed by diagnostic code (0.15%) or manual chart review.21
Figure 1.
Outline of PheML development and application
Within a set of 3.1 million deidentified electronic health records (A), we first identified a small pool of subjects (B) with developmental stuttering through expert manual review. We selected these patients and their demographically matched controls to identify comorbidities as predictive features and develop and test a machine-learning model (C) that would impute stuttering in BioVU (D), an independent EHR dataset linked to genetic data. We then performed a GWAS by using the imputed phenotype as the dependent variable in the labeled genetic dataset (E) to identify genetic variants associated with imputed stuttering (F).
Table 1.
Performance of PheML classification model
|
Predicted status |
|||
|---|---|---|---|
| Affected individuals | Control individuals | Total | |
| Classified as stuttering by manual review | 97 | 44 | 141 |
| No indications of stuttering | 19 | 665 | 684 |
| Total | 116 | 709 | |
To execute a well-powered GWAS aimed at identifying associated genetic loci for stuttering, we applied PheML in BioVU to impute a stuttering phenotype in patients with genetic data linked to their EHR. We then leveraged the imputed stuttering phenotype as the dependent variable in a GWAS to identify associated variants and generate a polygenic-risk-prediction model. We validated these results by comparing the concordance of our GWAS summary statistics to the GWAS results obtained from an independent clinically ascertained stuttering sample set acquired through the International Stuttering Project (ISP) and the polygenic-risk-prediction scores in the clinically ascertained cases versus matched population-based controls.23 This approach allowed us to impute a stuttering phenotype in a large patient set on the basis of the presence of a phenotypic profile that approximates a developmental stuttering phenotype and to amass statistical power from a large sample size to help accommodate the lack of clinical specificity. In doing so, we were able to perform a GWAS that identified genome-wide-significant variants associated with the clinical profile of developmental stuttering.
Subjects and methods
Model development and application to BioVU
We developed a model that classified patients as having a high probability of having developmental stuttering if they had one of the phenotypes (denoted as phecodes) associated with developmental stuttering; details are described in Pruett et al.21 Phecodes were mapped from ICD-9 codes clustered on the basis of a grouping system developed through the Phecode Map project.23 Features were selected on the basis of phecode enrichment in individuals with developmental stuttering as compared to matched controls.21 Patients with one or more instance of a phecode in their EHR were noted as positive for that feature, and they were notedas negative if they had no mentions of the phecode; only phecodes observed more often in the set of affected individuals than in 10,000 simulations of matched controls were carried forward into model building (corresponding to a p value of 0). A Gini impurity-based classification-and-regression-tree machine-learning model was developed from these features via scikit-learn tree regression software,22 and the model was tested in an independent set of 141 individuals with developmental stuttering and 684 matched controls in Vanderbilt University’s HER; phenotypic status of these individuals was confirmed by expert manual review.21 We then applied this model to a set of 92,762 individuals genotyped on the MEGAEX array with available ICD-9 records and resulting phecodes to impute developmental stuttering status for a downstream GWAS.
Genotyping, imputation, and quality control
All BioVU participants as well as the participants in the independent clinically ascertained developmental stuttering dataset were genotyped on Illumina’s Infinium Expanded Multi-Ethnic Genotyping Array (MEGAEX). Duplicate variants and indels were removed; for duplicate samples, the duplicate with a lower call rate was removed.
BioVU
Quality control was performed primarily with PLINK v. 1.90.24 Initial filtering thresholds for the entire BioVU sample included excluding variants with a call rate less than 98% and samples with a call rate less than 97%.25 Eigenvectors and eigenvalues were calculated through principal-component analysis (PCA) run in PLINKv1.90, and data were separated according to genetic principal components (eigenvectors) into five broad ancestry groups—European, African, South East Asian (EAS), EAS, and Hispanic (AMR) —with the 1000 Genomes reference for ancestry classification verification (Figure S1).26 Each ancestry subset was subsequently analyzed separately; a minor-allele filter of 1%, a variant-missingness filter of 5%, and a sample-missingness filter of 10% were applied, and checks for heterozygosity, sex, and variants that did not align with Hardy-Weinberg expectations (variants with a Hardy-Weinberg [hwe] statistic <1 × 10−10 were removed) were performed.25 Data were prepared for imputation according to specifications outlined on the Michigan server webpage; these included using a pre-imputation data-preparation toolkit (see McCarthy Group tools in the web resources).27 Imputation was performed for each ancestry cohort on the Michigan Imputation server through the use of EAGLE2 phasing, Minimac4 imputation, and the Haplo-type Reference Consortium (HRC) reference.27, 28, 29 Final post-imputation quality-control filtering included selecting variants with a minor-allele frequency above 1% within each ancestry group, as well as removing all variants with an R2 imputation info score of less than 0.4.
International Stuttering Project (ISP) dataset
As an independent reference dataset, we obtained 1,345 clinically ascertained developmental stuttering patients (965 male and 380 female) collected from Curtin University Stuttering Center in Perth, Australia, the SpeechMatters Clinic in Dublin, Ireland, the Stuttering Research Laboratory at the University of Pittsburgh, Dr. Shelly Jo Kraft’s research group at Wayne State University, and the Attadale Stuttering Treatment Facility in Australia and through a social-media outreach campaign led by Drs. Below and Kraft on reddit.com. We paired these with 7,019 demographically matched controls from BioVU (4,951 males and 2,068 females; selected as described below and with no overlap with the dataset used in our primary GWAS) (Table S2). A speech pathologist evaluated each participant to confirm their phenotypic status. We applied the methods described by Pluzhnikov et al. to identify possible plate or batch effects prior to merging unique batches of genotypes from affected individuals.30 No plate or batch effects were observed. Initial filtering for stuttering excluded variants and samples with a call rate less than 90%. Next, data for affected individuals were separately assessed for quality control according to broad ancestral groups (European, American/Hispanic, African American, and Asian) as defined by PCA in which the HAPMAP3 reference was used for ancestry classification.31 Each ancestry cohort was subsequently analyzed; analysis incorporated a minor-allele filter of 1%, a variant-missingness filter of 3%, and a sample-missingness filter of 5%, as well as checks for heterozygosity, sex, and variants that did not align with Hardy-Weinberg expectations (variants with a hwe statistic <1 × 10−15 were removed). Then, approximately five ancestry and sex-matched population-based controls per case were drawn from a quality-control filtered BioVU set (quality control for BioVU as described above). To select ancestry-matched controls, we calculated eigenvectors and eigenvalues through PCA generated by PLINK v.1.90. PCA was performed on the maximally unrelated set of affected indivdiuals and potential control individuals (as identified by PRIMUS32, 33, 34, 35) through use of a panel of SNPs in low linkage disequilibrium (LD); additional related affected individuals and potential control individuals were projected along each of the calculated eigenvectors. Data from affected individuals were merged with that from their selected matched controls (the control selection method is described below) for imputation according to standard protocols and specifications outlined for the TOPMed server; these included using the same pre-imputation data-preparation toolkit as above (see McCarthy Group tools in web resources).27 The autosomal region was imputed on the TOPMed server with EAGLE_v. 2.4 phasing, Minimac4 imputation, and the TOPMed reference.29,36,37 Post-imputation quality-control filtering included selecting variants with a minor-allele frequency above 1% and removing all variants with R2 imputation info score less than 0.4.
Genome-wide association studies
BioVU
For the GWASs, developmental stuttering patients were stratified by ancestry, with independent association analyses performed for each ancestry group: European, African, South EAS, EAS, and Hispanic. For each identified case, up to six controls were selected from the cohort of patients identified as controls by the PheML prediction model. Control individuals were matched by age (within 5 years of their matched affected individual) and sex. Additionally, control individuals were matched to the lowest genetic Euclidean pairwise case-control distance that met the previously mentioned criteria. Euclidean pairwise distance was calculated as the sum of the square of the difference in the eigenvectors scaled by their eigenvalue for each principal component calculated from our PCA.38 Any pairwise case-control distance not within two standard deviations of the mean distribution of all case-control pairwise distances were removed from the analysis. A logistic-regression model was used for the variant association analyses in SUGEN ,39 and corrections were made for sex, age, and ancestry (genetic ancestry captured by the first three principal components). To correct for multiple testing, we considered variants with an association p value below 5.0 × 10−8 to be significant. Manhattan plots were generated with the qqman R package.40 Loci and LD structure visualization and qq-plots plots were generated with the LocusZoom browser tool.41
International Stuttering Project (ISP) GWAS
Control individuals were selected from a sample set of individuals who were not identified as being affected by stuttering according to ICD9 and ICD10 codes (Table S1) or by the PheML prediction algorithm. These individuals were matched by sex, similarly to those described for the BioVu individuals described above. Genetic Euclicean pairwise distance was minimized, and any pairs of affected and control individuals not within two standard deviations of the mean distribution of all pairwise distances were removed. Individuals under 18 were also excluded as potential controls. Dates of birth for individuals in the ISP sample sets were not available, so subjects were not matched by age. A GWAS for the ISP stuttering sample set was performed with a frequency-based additive logistic model via SAIGE (scalable and accurate implementation of generalized mixed model), a method developed for biobank data in order to control for unbalanced case-control ratios and sample relatedness.42 The regression model accounted for population substructure by including the first six principal components as covariates.
Calculations of genetic heritability
Genome-wide SNP-based liability-scale heritability within our European ancestry (EUR) sample set was calculated through a genomic-relatedness-based restricted maximum-likelihood (GREML) approach implemented through GCTA software.43,|,44 Observed variance estimates from the observed scale were transformed to an expected underlying scale, for which an expected population prevalence was set to 0.1 on the basis of the observed frequency of predicted cases within BioVU. Heritability estimates included all variants tested in the GWASs (see Genotyping, imputation, and quality control in the methods section for exclusion criteria). We corrected for sex, age, and the first three principal components (see Genotyping, imputation, and quality control).
Variant-effect-size concordance analysis
For the concordance analysis, we compared summary statistics from the EUR GWAS to summary statistics produced from the ISP GWAS to determine whether the concordance rate between the two summary statistics was higher than expected. The concordance rate was calculated by the proportion of variants that had the same direction of effect over the total variants present in both GWAS analyses. 7,570,420 variants that passed previously described QC metrics were present in both GWAS analyses, aligned by strand and reference allele, and analyzed here. Additional concordance rates were calculated for variants with p values below 0.5, 0.05, and 0.005 thresholds in both GWASs. We performed a one-sample t test to determine whether each concordance rate was significantly higher than an expected concordance rate of 0.5.
Modeling of the polygenic risk score
We used the summary statistics resulting from our GWAS of PheML-defined EUR-imputed stuttering to develop a polygenic risk (PRS) model by using data from all 7,751,954 autosomal SNPs meeting the quality-control criteria outlined above. The PRS model was developed with PRScs python software, which creates a model that esimtaes genetic liability through a linear combination of the weight of SNP dosage on effect size and p values from the provided GWAS summary data.45 Our global shrinkage parameter (phi) was set to 1. This model was applied to the genetic data of the independent clinically ascertained developmental stuttering cohort as well as their matched controls. This analysis was restricted to only samples that were of EUR. Individual polygenic scores were calculated through PLINK v. 1.9.25 To assess the significance of the difference in genetic liability for stuttering between the individuals with clinically ascertained stuttering and the matched control individuals, we ran a two-sample t test comparing the overall score distribution between these two groups.
Results
Efficacy of PheML prediction
To test the efficacy of our model predicting PheML stuttering, we applied the model to a set of 825 patients (141 patients with developmental stuttering confirmed by manual review and 709 patients with no indications of stuttering in their health records). Of the 116 subjects that our prediction model scored as having developmental stuttering, 97 were among those manually reviewed as having developmental stuttering, and 19 had no indications of stuttering in their records, resulting in a positive prediction rate of ∼83% (Table 1). Of the 141 manually reviewed individuals with developmental stuttering in the testing set, 97 were predicted as having developmental stuttering by the PheML model, whereas 44 were not, suggesting that despite our high positive predictive values, 30% or more affected individuals might still be missed by our approach.21
PheML imputation of developmental stuttering identifies a large case sample
Of a set of 92,742 BioVU subjects, PheML labeled 9,239 as having a high likelihood for developmental stuttering (9.96% prevalence). Of this set, 5,732 affected individuals were female (62.0%; average age of 47.9 years). 6,639 (68.6%) were of EUR, 1,869 (20.2%) were of African ancestry (AFR), 398 (4.3%) were of Hispanic ancestry, 124 were of EAS ancestry (1.3%), and 41 (0.6%) were of South Asian ancestry (SAS) (Table 2). Broad ancestry groups were stratified via principal-component analysis (Figure S1).
Table 2.
Demographics of BioVU subjects classified by PheML algorithm
| Predicted to exhibit stuttering | Predicted not to stutter | |
|---|---|---|
| Total | 9,239 | 83,503 |
| Male | 3,507 (38.0%) | 36,140 (43.3%) |
| Female | 5,732 (62.0%) | 47,363 (56.7%) |
| Demographics | ||
| Mean age in years (SD) | 47.9 (24.9) | 55.2 (22.7) |
| Ancestry | ||
| European | 6,339 (68.6%) | 63,471 (76.0%) |
| African | 1,869 (20.2%) | 13,728 (16.4%) |
| East Asian | 124 (1.3%) | 772 (0.9%) |
| South Asian | 51 (0.6%) | 363 (0.4%) |
| Hispanic | 398 (4.3%) | 2,068 (2.5%) |
| Unknown/other | 158 (1.7%) | 3,101 (3.7%) |
Ancestry was determined through principal-component analysis. Testing set included 825 subjects (141 individuals confirmed to exhibit developmental stuttering and 684 subjects with no indication of stuttering in their health records). The positive-prediction rate for the model is ∼83%.
GWASs in the PheML prediction set identify genetic loci associated with developmental stuttering
The PheML predicted-stuttering sample set was stratified by PCA-based genetic ancestry (African, EAS, European, Hispanic, and South Asian) for genome-wide association studies (Table 3). We performed a separate GWAS for each ancestry group. Across analyses in the five ancestry groups, the European and AFR groups were the largest and best powered. One locus reached genome-wide significance in the EUR analysis and one locus reached near genome-wide significance in the AFR sample set (Table 4).
Table 3.
Demographics of BioVU subjects used in GWAS
| Individuals predicted to stutter | Predicted control individuals | |
|---|---|---|
| Total | 9221 | 45,793 |
| Male | 3,491 (37.9%) | 17,162 (37.5%) |
| Female | 5,730 (62.1%) | 28,631 (62.5%) |
| Demographics | ||
| Mean age in years (SD) | 47.7 (24.8) | 48.6 (24.2) |
| Ancestry | ||
| European | 6,339 (68.7%) | 33,172 (72.4%) |
| African | 1,853 (20.1%) | 8,372 (18.3%) |
| East Asian | 124 (1.3%) | 592 (1.3%) |
| South Asian | 51 (0.6%) | 228 (0.5%) |
| Hispanic | 397 (4.3%) | 1,395 (3.0%) |
Ancestry was determined through principal-component analysis. GWASs were performed with stratifications by ancestry.
Table 4.
Top hits identified in GWAS of PheML-imputed affected and control individuals
| rsID | Ancestry | Chr. | Position | Ref. | Alt. | Beta (95% CI) | p value | Nearest gene | Location | Replicating analysis (p < 0.05) | Replicating beta | Replicating p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12613255 | EUR | 2 | 16,628,186 | A | T | 0.323 (0.211, 0.434) | 1.31 × 10−8 | CYRIA | 113 kb 3′ | N/A | N/A | N/A |
| rs7837758 | AFR | 8 | 40,624,542 | C | T | 0.518 (0.331, 0.704) | 5.07 × 10−8 | ZMAT4 | intronic | N/A | N/A | N/A |
| rs10872381 | EAS | 6 | 131,706,657 | C | T | 0.803 (0.512, 1.093) | 6.38 × 10−8 | AKAP7 | 102 kb 3′ | N/A | N/A | N/A |
| rs115024493 | AFR | 12 | 91,973,678 | G | A | 0.376 (0.239, 0.512) | 6.85 × 10−8 | DCN | 397 kb 5′ | N/A | N/A | N/A |
| rs78072807 | AFR | 1 | 237,682,933 | T | C | 0.371 (0.235, 0.507) | 8.73 × 10−8 | RYR2 | intronic | N/A | N/A | N/A |
| rs10464899 | AFR | 8 | 60,209,436 | A | G | 0.216 (0.135, 0.296) | 1.51 × 10−7 | TOX | 178 kb 5′ | ISP | 0.138 (0.038, 0.238) | 7.21 × 10−3 |
| rs2997903 | AFR | 9 | 131,599,311 | G | A | 0.308 (0.188, 0.429) | 5.32 × 10−7 | KYAT1 | intronic | EUR | 0.086 (0.018, 0.015) | 1.27 × 10−2 |
| rs6981922 | AFR | 8 | 60,219,451 | T | A | 0.197 (0.119, 0.276) | 9.35 × 10−7 | TOX | 188 kb 5′ | ISP, EAS | 0.109 (0.099,0.119), 0.372 (0.021, 0.723) | 3.21 × 10−2, 3.79 × 10−2 |
| rs6415726 | HIS | 9 | 16,247,629 | G | A | 0.730 (0.438, 1.022) | 9.61 × 10−7 | C9orf92 | intronic | ISP | 0.223 (0.105, 0.341) | 2.17 × 10−4 |
| rs8013614 | EUR | 14 | 36,425,517 | C | G | −0.120 (−0.170, −0.071) | 1.59 × 10−6 | BRMS1L | 84 kb 5′ | AFR | −0.091 (−0.171, −0.011) | 2.62 × 10−2 |
| rs34456770 | AFR | 16 | 126,219 | G | A | 0.256 (0.114, 0.399) | 1.87 × 10−6 | MPG | 797 bp 5′ | ISP | 0.349 (0.070, 0.628) | 1.41 × 10−2 |
| rs10036373 | EUR | 5 | 23,909,919 | G | A | 0.097 (0.057, 0.137) | 2.26 × 10−6 | C5orf17 | 42 kb 5′ | ISP | 0.104 (0.007, 0.201) | 3.61 × 10−2 |
The table includes sentinel variants from loci that exceeded p values of 1 × 10−7, as well as any variants that exceeded 5 × 10−6 and that were replicated in a separate analysis. EUR, AFR, EAS, and HIS refer to association results from samples of European, African, East Asian, and Hispanic ancestry, respectively. ISP indicates variant results from the International Stuttering Project stuttering GWAS (see methods). "Ref." refers to the reference allel, and "Alt." refers to the alternative allele the association analysis was conditioned on.
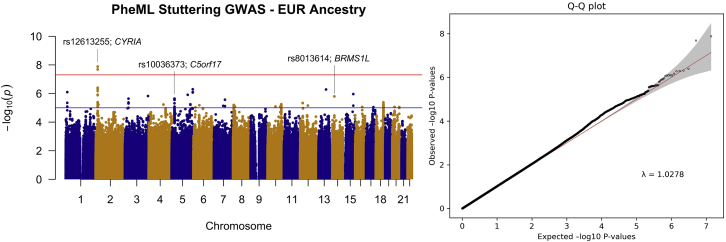
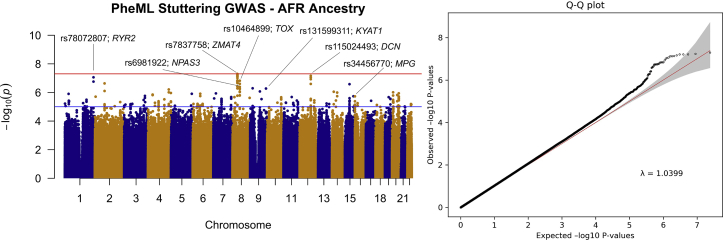
In the EUR case set, the GWAS included 6,339 predicted developmental stuttering cases and 33,172 ancestry and sex-matched controls and 7,751,954 imputed variants (Figure 2; see methods section). One statistically significant locus was identified, and the sentinel variant was determined to be at rs12613255 (beta = 0.323; p = 1.31 × 10−8), 113 kb 3¢ of CYFIP-related Rac1 interactor A (CYRIA) (MIM: 606322) (see Figure 3). The developmental-stuttering GWAS in subjects of AFR included 1,853 affected individuals, 8,402 ancestry- and sex-matched control individuals, and 13,636,593 variants (Figure 4; see methods). The top variant, rs7837758, reached near-genome-wide significance (beta = 0.518; p = 5.07 × 10−8). rs7837758 is found the third intron of ZMAT4, located on chromosome 8 (Figure 5). The GWAS performed on subjects of Hispanic ancestry included 397 affected individuals, 1,457 ancestry- and sex-matched control individuals, and 8,147,169 variants (Figure S2). For subjects of EAS ancestry, the GWAS included 124 affected individuals, 716 ancestry- and sex-matched controls, and 6,922,517 variants (Figure S3). For subjects of South Asian ancestry, the GWAS included 51 affected individuals, 279 ancestry- and sex-matched controls, and 7,058,354 variants (Figure S4). Most likely because of the reduced power in smaller sample sizes, association analyses in the Hispanic, South Asian, and EAS cohorts did not result in any significantly associated variants with our PheML prediction set (see Figures S2–S4 and Table S3). We also report several loci that exceeded a suggestive significance threshold of p = 5 × 10−6 (108 variants across all ancestry GWASs) and were replicated (p < 0.05) in one or more independent developmental stuttering GWASs (Table 4, Figures S5–S13). Our strongest replications across these studies include rs6415726 (HIS GWAS; beta = 0.730; p = 9.61 × 10−7), an intronic C9orf92 variant that replicated in the ISP GWAS (beta = 0.197; p = 6.29 × 10−4), and rs10464899 (AFR GWAS; beta = 0.216; p = 1.51 × 10−7; see Figure S13), a variant that is 178 kb 5′ of TOX [MIM: 606863] and replicated in the ISP GWAS (beta = 0.139; p = 6.88 × 10−3; see Figure S6).
Figure 2.
Manhattan plot and qq-plot of results from GWAS of European-ancestry individuals predicted by PheML to exhibit developmental stuttering
Analysis included 7,751,954 variants across chromosomes 1–22. One locus in chromosome 2 reached genome-wide significance (p < 5 × 10−8); the sentinel variant, rs12613255 (BETA = 0.323; p = 1.31 × 10−8), was 113 kb 3' of CYRIA (FAM49A is an alias for CYRIA). The red line indicates the threshold for genome-wide significance (5.0 × 10−8), and the blue line indicates the threshold for suggestive significance (1.0 × 10−5). Loci reported in Table 4 are labeled on the plot as well as the nearest gene.
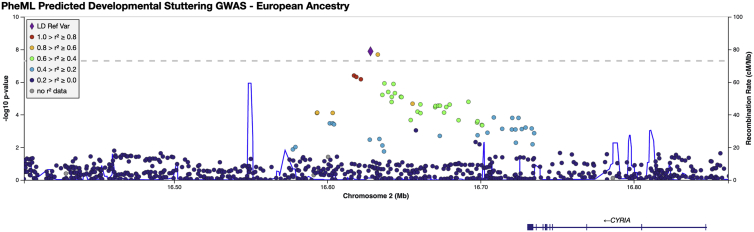
Figure 3.
LocusZoom plot for rs12613255 locus in EUR PheML stuttering GWAS
The lead variant (marked as a diamond) was found in chromosome 2, 113 kb 3' of CYRIA. A dashed line indicates the threshold for genome-wide significance (5.0 × 10−8).
Figure 4.
Manhattan plot and qq plot of results from GWAS of African-ancestry individuals predicted by PheML to exhibit developmental stuttering
Analysis included 13,643,593 variants across chromosomes 1–22. One variant, rs7837758, reached genome-wide significance (BETA = 0.518; p = 5.07 × 10−8), on chromosome 8 within the third intron of ZMAT4. The red line indicates the threshold for genome-wide significance (5.0 × 10−8), and the blue line indicates the threshold for suggestive significance (1.0 × 10−5). Loci reported in Table 4 are labeled on the plot as well as the nearest gene.
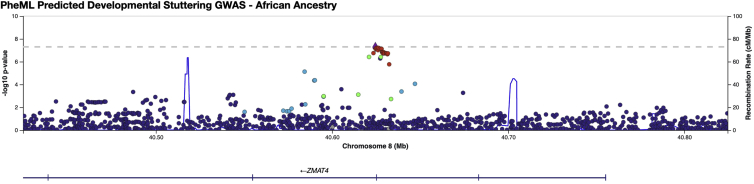
Figure 5.
LocusZoom plot for the rs7837758 locus in the AFR PheML Stuttering GWAS
The lead variant (marked as a diamond) was found on chromosome 8, within the third intron of ZMAT4. A dashed line indicates the threshold for genome-wide significance (5.0 × 10−8).
Genome-wide explained variance within the EUR PheML sample set
Genome-wide SNP-based liability-scale heritability within our EUR sample set was calculated through GCTA within the European-ancestry PheML sample set. The proportion of phenotypic variance explained by genetic factors was reported at 0.0232 (SE = 0.0083).43 Through GCTA we also transformed the explained variance estimates from the observed scale to the underlying liability scale to account for an expected prevalence of affected individuals of 0.1. The proportion of phenotypic variance (liability-scale heritability) was 0.0453 (SE = 0.016, p = 2.29 × 10−3).
Concordance analysis reveals genetic similarity between PheML-predicted stuttering individuals and those clinically ascertained by the ISP as having developmental stuttering
To ensure that the genetic profile of our PheML-predicted stuttering individuals properly recapitulated effects associated with clinical developmental stuttering, we compared the direction of effect estimated in a GWAS between our GWAS of EUR PheML-predicted stuttering individuals and a GWAS of an independent, largely European-ancestry, and clinically ascertained set of individuals with developmental stuttering (see Table S2); this latter set was also genotyped by the genotyping core facility at Vanderbilt University, VANTAGE, on the MEGAEX. 7,570,420 imputed variants were present and tested in both analyses by an approach similar to that described in the 2014 DIAGRAM paper.46
For all variants present in both GWASs, 50.41% of the variants were found to have the same direction of effect (3,816,091 of 7,570,420 variants; p = 6.86 × 10−112). For all variants that had a p value threshold below 0.5 in both GWASs, the concordance-of-effect rate was 50.86% (982,614 of 1,931,927 variants; p = 3.77 × 10−127).
Variants with a p value threshold below 0.05 in both GWASs had a concordance rate of 53.47% (10,830 of 20,255 variants; p = 2.84 × 10−23). Variants below a p value threshold of 0.005 in both GWASs had a concordance rate of 73.19% (121 of 171 variants; p = 6.25 × 10−10) (Table 5).
Table 5.
Results of variant-concordance analysis
| Concordant variants | Concordance rate (%) | Binomial p value | |
|---|---|---|---|
| All variants | 3,816,091/7,570,420 | 50.41 | 6.86 × 10−112 |
| p < 0.5 | 982,614/1,931,927 | 50.86 | 3.77 × 10−127 |
| p < 0.05 | 10,830/20,255 | 53.47 | 2.84 × 10−23 |
| p < 0.005 | 121/171 | 73.10 | 6.25 × 10−10 |
Results from analyses comparing summary statistics of the European PheML stuttering GWAS to those of the ISP stuttering GWAS. Concordant variants include any variants that were present in both analyses and had the same direction of effect.
Stuttering polygenic-risk-score models developed with results from the PheML stuttering GWAS show increased genetic liability within the ISP stuttering set
We developed a PRS-score model by using the summary statistics for 7,751,954 variants produced by the GWAS of EUR PheML-predicted stuttering individuals. We then applied this model to the genetic datasets of our ISP developmental-stuttering subjects and their matched control individuals (the same set used in the variant concordance analysis, although he sample set only included those of PCA-based European ancestry). Our ISP stuttering set scored significantly higher on the PRS model (mean = 8.56 × 10−8, SD = 1.13 × 10−6) than their matched control individuals (mean = −3.59 × 10−7, SD = 1.01 × 10−6; two-sample t test, t(1131) = 13.12, p = 6.83 × 10−39), providing compelling evidence that the genetic architecture identified in the model-imputed phenotyping discriminates the genetic liability for developmental stuttering in clinically ascertained cases and population-based controls (Figure S14).
Discussion
Stuttering classification model development and application to BioVU
We set out to utilize a phenotype-based machine learning algorithm, PheML, to identify unlabeled cases of developmental stuttering, an underdiagnosed phenotype, in a large EHR. Our model shows a positive prediction rate of 83.3%, though it’s important to note that while testing, those who were classified as “controls” during manual review may have had stuttering and simply did not have any mentions of it in their records. We expect that roughly 16.7% of patients in BioVU that were classified as a case are false positives, though this is likely an overestimate.
Applying PheML to BioVU resulted in a large population of patients that, even in the absence of a direct diagnosis of stuttering, exhibited a constellation of traits associated with stuttering; an underlying phenotypic signature that could be leveraged to predict developmental stuttering in a manner akin to imputation. Though 9.96% of the cohort was predicted to be a case by our PheML model, the modest sensitivity of the model (68.8%) suggests that this is likely an underestimation of the actual proportion of the true cases in our sample.
Interestingly, although males exhibit a higher prevalence of stuttering, more females were identified as exhibiting stuttering by our PheML prediction model (the female-to-male ratio was 1.6:1). Although there are more females in BioVU than males (1.3:1; female:male), this does not fully explain the discrepancy. There are several possible explanations for this imbalance. There might be sex imbalance in the rate or quality of diagnosis of selected predictive phenotypes; also, the ability to predict stuttering might be greater in women than in men. The positive-prediction rate for our model is >83.3%, and more women might be misspecified as affected by stuttering. A third possibility is that the prevalence of stuttering in women is higher than reported but less frequently diagnosed or detected because of a faster or higher rate of recovery. Prior evidence from Ambrose et al. supports this last potential explanation by suggesting that the male-to-female ratio for lifetime prevalence might be more balanced when mild cases of developmental stuttering and individuals who recover early are included.47 Future analyses of sex-stratified GWASs of developmental stuttering and its associated clinical phenome are needed if researchers are to further explore differential risk factors that might contribute to differences in age and rate of recovery between men and women.
Genetic discovery in predicted-stuttering cohort
PheML-imputed affected individuals and well-matched control individuals were stratified by ancestry for GWASs. Our estimated positive-prediction rate (83.3%, implying a false-positive rate of 16.7%) among cases is most likely markedly lower than the positive-prediction rate of samples acquired from affected individuals in treatment clinics, where speech and language pathologists confirm patient status. However, as with studies that leverage population-based controls, our power reduction resulting from imprecision in definitions of affected and control individuals is offset by the size of the dataset identified by our PheML model.
Controls for GWASs were selected from patients our model classified as having low likelihood for developmental stuttering (i.e., these patients were not predicted to stutter). Our sensitivity analysis indicates that the model is classifying 68.8% of manually reviewed developmental stuttering cases as being at high risk for stuttering, whereas 31.2% were classified as control individuals. Therefore, we expect some model-defined controls, roughly equal to one-third of the biobank prevalence for stuttering, to exhibit stuttering, potentially further reducing our power to discriminate allele-frequency differences between affected individuals and controls.
Despite the challenge of misclassification of affected and control individuals in the sample size attained with our phenotype-imputation approach in biobank-scale data, our analyses in EUR participants not only identified a genome-wide significant locus but also demonstrate that a significant portion of the variance of the trait captured by our model is heritable (h2 = 0.045, SE = 0.016, p = 2.29 × 10−3), indicating that there exists a common underlying genetic background among those who were classified as exhibiting developmental stuttering in our model. This heritability estimate is in line with other common complex neurological and psychological traits, such as PTSD in males and anxiety.48,49 We also demonstrate that this common genetic background is consistent with the genetic architecture identified in a clinically ascertained independent GWAS of stuttering.
Association analyses were separated by ancestry groups. For the analyses conducted in AFR, HIS, SAS, and EAS ancestry groups, no variants were observed to be significantly associated with our developmental stuttering cohort after genome-wide Bonferroni correction (p < 5 × 10−8) (Table S3), although two variants came close to reaching significance in the EAS sample (rs10872381, beta = 0.803; p = 6.4 × 10−8; see Figure S10) and AFR sample (rs7837758, beta = 0.518; p = 5.07 × 10−8, see Figure 5). These studies were markedly smaller in size than our EUR study (see Table 3) and were therefore likely insufficiently powered to discover variants of modest effect size.
GWAS in the European cohort revealed one significant locus; the top hit was at rs12613255 (beta = 0.323; p = 1.31 × 10−8); see Figure 3). The closest gene to this variant, which resides on chromosome 2, is CYRIA, also referred to in the literature as FAM49A. RNA expression data show that CYRIA is highly expressed in the central nervous system (specifically in the cerebral cortex, basal ganglia, and olfactory region) and is also highly expressed in the thyroid gland, granulocytes, and monocytes.50 CYRIA has not previously been implicated in developmental stuttering, although in Asian and Brazilian populations it has been reproducibly associated with cleft lip and palate,51, 52, 53, 54 a trait that was not used a predictor variable in our model.
In the AFR GWAS, variant rs7837758 nearly reached the genome-wide significance threshold of 5.0 × 10−8 (beta = 0.518; p = 5.07 × 10−8) (see Figures 4 and 5). This variant is located on chromosome 8 in the intron of ZMAT4 (Zinc finger matrin-type protein 4) (see Figure 5). Variants within this gene have previously been observed to be associated with myopia and fasting blood glucose in African Americans.55,56 Neither of these phenotypes have been previously associated with developmental stuttering, nor did these phenotypes serve as proxy variables in our prediction algorithm.21 ZMAT4 has been observed to be highly expressed in the central nervous system, especially in tissue types present in the cerebral cortex, cerebellum, and hippocampus and to be modestly expressed in the basal ganglia.50
Little is known about the neuronal basis of developmental stuttering, although imaging studies have demonstrated that patients who stutter show abnormal function in the form of overactivity in the cortical motor and pre-motor areas associated with speech, as well as disruptions in the basal ganglia and dopaminergic systems.57, 58, 59 Although the previous linkage analyses have identified candidate genes, including DRD2 (MIM: 126450), AP4E1, CYP17A1 (MIM: 609300), GNPTAB, GNPTG, and NAGPA, 13,60, 61, 62 the mechanisms of action remain uncertain, although both GNPTAB and GNPTG are active in lysosomal enzyme-targeting pathways and energy metabolism.63 We checked for replication within these genes but failed to demonstrate any significant findings (see Table S4). Although the mechanisms of action of our top associated variants on the clinical profile of developmental stuttering are not yet known, our approach enabled variant discovery, and future work will be needed to reproduce these findings and establish their functional role in the clinical profile of susceptibility to developmental stuttering.
Validation of GWAS results: concordance analysis
Using the summary statistics resulting from our GWAS of EUR individuals predicted by PheML to exhibit stuttering, we ran a variant concordance analysis that assessed how many variants had the same direction of effect as the variants tested in a GWAS run on a clinically ascertained developmental-stuttering sample set via the approach outlined in the 2014 DIAGRAM paper, in which concordance measures were used for assessing T2D risk alleles across various ancestry groups.46 Variants that were not well imputed (r2 > 0.4) in either GWAS were removed from the concordance analysis, and datasets were verified to have the same strand and reference-allele orientation. This was repeated with only variants that surpassed thresholds of p = 0.5, 0.05, and 0.005 in both association analyses. For example, for the group “p < 0.05,” only variants with a p value that was below 0.05 in both the PheML GWAS results and the ISP GWAS results were included in the analysis. Together, the four concordance analyses demonstrated that the proportion of variants with the same direction of effect was significantly greater than random. Additionally, as the significance threshold for variants included in the concordant analysis became more stringent, the proportion of variants with the same direction of effect increased, and 73.1% of all variants that surpassed a threshold of p = 0.005 in both GWAS have the same direction of effect (see Table 5). The clinically ascertained stuttering sample set was from a multi-ethnic analysis, although it was predominantly comprised of EUR participants (84.2%; see Figure S1 and Table S2). This remarkable finding provides compelling evidence that the GWAS using our PheML-imputed stuttering phenotype is capturing a portion of the genetic architecture of clinical developmental stuttering.
Validation of GWAS results: stuttering PRS model development
To further explore our genetic findings, we developed a PRS model built with summary statistics from the GWAS conducted in our EUR PheML stuttering sample. PRS models are used for summarizing variant effects and assessing genetic liability for a trait. We applied this model to the genetic dataset of our ISP developmental-stuttering EUR cohort, as well as their matched controls. The ISP stuttering set scored significantly higher than their matched controls (two-sample t test, t(1131) = 13.12, p = 6.83 × 10−39; see Figure S14), indicating that the PRS model developed from our imputed stuttering GWAS is significantly predictive of stuttering liability. This additional evidence strongly supports our conclusion that the PheML classifier captures a phenotype sufficiently similar to that of the ISP stuttering sample set to identify genetic risk factors relevant for stuttering. Although statistically significant, the difference observed in the score distributions for stuttering and control individuals suggests that this PRS model has limited clinical value for predicting developmental-stuttering status (receiver operating characteristic AUC = 0.601; see Figure S15). Ge et al.45 simulate the predictive performance of PRS across various sample sizes and genetic architectures. In these simulations they demonstrate that predictive performances for more polygenic traits benefitting from greater sample sizes, showing that PRS models developed from 50 to 100K sample sets have drastically better performance metrics than models with smaller sample size.45 Our PRS model was developed from a sample set of 39,511, which may be underpowered for developing a model that would be useful for predictive purposes.
We note that the developmental-stuttering comorbidities that formed the basis of our PheML model were determined in a dataset largely comprising European and African American individuals. As such, the underrepresentation of other racial and ethnic minority groups in our comorbidity detection and model building is a limitation. The effect of this population structure in our EHR might lead to our model’s missing some population-specific comorbidities, consequentially reducing model performance in these subgroups. Future research exploring comorbidities of stuttering and genetic architecture within and across populations is warranted.
Through our stuttering-prediction algorithm, we designed and conducted the largest GWAS for the clinical profile of developmental stuttering. Despite a lack of clinical evaluation and diagnosis of stuttering in our affected individuals and controls, our approach allowed use of an existing EHR-linked DNA databank of sufficient size and power to identify genome-wide significant loci through a population based genetic analysis of this important trait. These data provide insights into the genetic contributions to developmental stuttering in patients of African and European descent, demonstrating that genetic risk of this clinical profile is dominated by modest to low genetic effects, as well as providing a framework for studying underreported diseases in large-scale EHRs.
Acknowledgments
This work and investigators D.M.S, H.P., D.G.P., L.E.P., H.-H.C., R.M.J., S.J.K., and J.E.B. were supported by R03DC015329, R01DC017175, and R21DC016723 from the National Institute on Deafness and Other Communication Disorders (NIDCD), National Institutes of Health. D.M.S. is supported by NIDCD grant 5T32GM80178-13. Some of the datasets used for the described analyses were obtained from Vanderbilt University Medical Center’s BioVU, which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH-funded shared instrumentation grant S10RR025141 and Clinical and Translational Science Awards grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, and R01HD074711 and additional funding sources listed at https://victr.vumc.org/biovu-funding.
Declaration of interests
The authors declare no competing interests.
Published: December 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.11.004.
Data and code availability
All code used for developing the PheML model described in the methods section is available for download on github (see web resources). Genotyping data from BioVU are only available upon submission of a study proposal and approval through the BioVU Review Committee.
Web resources
LocusZoom GWAS visualization software, http://locuszoom.org/
McCarthy Group tools, www.well.ox.ac.uk/∼wrayner/tools/
PheML development code GitHub page, https://github.com/shawdm1/PheMLStutteringCART
PLINK v. 1.90 whole-genome data-analysis toolset, http://pngu.mgh.harvard.edu/purcell/plink/
PRScs Model GitHub page, https://github.com/getian107/PRScs
QQman R software packages, https://cran.r-project.org/web/packages/qqman/
Scikit-learn modeling software, https://github.com/scikit-learn/scikit-learn
Supplemental information
References
- 1.Wingate M.E. A standard definition of stuttering. J. Speech Hear. Disord. 1964;29:484–489. doi: 10.1044/jshd.2904.484. [DOI] [PubMed] [Google Scholar]
- 2.Yairi E., Ambrose N. Epidemiology of stuttering: 21st century advances. J. Fluency Disord. 2013;38:66–87. doi: 10.1016/j.jfludis.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajdacic-Gross V., Vetter S., Müller M., Kawohl W., Frey F., Lupi G., Blechschmidt A., Born C., Latal B., Rössler W. Risk factors for stuttering: A secondary analysis of a large data base. Eur. Arch. Psychiatry Clin. Neurosci. 2010;260:279–286. doi: 10.1007/s00406-009-0075-4. [DOI] [PubMed] [Google Scholar]
- 4.Yairi E. The onset of stuttering in two- and three-year-old children: a preliminary report. J. Speech Hear. Disord. 1983;48:171–177. doi: 10.1044/jshd.4802.171. [DOI] [PubMed] [Google Scholar]
- 5.Singer C.M., Hessling A., Kelly E.M., Singer L., Jones R.M. Clinical characteristics associated with stuttering persistence: A meta-analysis. J. Speech Lang. Hear. Res. 2020;63:2995–3018. doi: 10.1044/2020_JSLHR-20-00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seider R.A., Gladstien K.L., Kidd K.K. Recovery and persistence of stuttering among relatives of stutterers. J. Speech Hear. Disord. 1983;48:402–409. doi: 10.1044/jshd.4804.402. [DOI] [PubMed] [Google Scholar]
- 7.Daniels D.E., Gabel R.M., Hughes S. Recounting the K-12 school experiences of adults who stutter: A qualitative analysis. J. Fluency Disord. 2012;37:71–82. doi: 10.1016/j.jfludis.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 8.McAllister J., Collier J., Shepstone L. The impact of adolescent stuttering on educational and employment outcomes: Evidence from a birth cohort study. J. Fluency Disord. 2012;37:106–121. doi: 10.1016/j.jfludis.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Frigerio-Domingues C., Drayna D. Genetic contributions to stuttering: The current evidence. Mol. Genet. Genomic Med. 2017;5:95–102. doi: 10.1002/mgg3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Beijsterveldt C.E.M., Felsenfeld S., Boomsma D.I. Bivariate genetic analyses of stuttering and nonfluency in a large sample of 5-year-old twins. J. Speech Lang. Hear. Res. 2010;53:609–619. doi: 10.1044/1092-4388(2009/08-0202). [DOI] [PubMed] [Google Scholar]
- 11.Fagnani C., Fibiger S., Skytthe A., Hjelmborg J.V.B. Heritability and environmental effects for self-reported periods with stuttering: A twin study from Denmark. Logoped. Phoniatr. Vocol. 2011;36:114–120. doi: 10.3109/14015439.2010.534503. [DOI] [PubMed] [Google Scholar]
- 12.Kang C., Riazuddin S., Mundorff J., Krasnewich D., Friedman P., Mullikin J.C., Drayna D. Mutations in the lysosomal enzyme-targeting pathway and persistent stuttering. N. Engl. J. Med. 2010;362:677–685. doi: 10.1056/NEJMoa0902630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raza M.H., Mattera R., Morell R., Sainz E., Rahn R., Gutierrez J., Paris E., Root J., Solomon B., Brewer C., et al. Association between rare variants in AP4E1, a component of intracellular trafficking and persistent stuttering. Am. J. Hum. Genet. 2015;97:715–725. doi: 10.1016/j.ajhg.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han T.-U., Root J., Reyes L.D., Huchinson E.B., Hoffmann J.D., Lee W.S., Barnes T.D., Drayna D. Human GNPTAB stuttering mutations engineered into mice cause vocalization deficits and astrocyte pathology in the corpus callosum. Proc. Natl. Acad. Sci. USA. 2019;116:17515–17524. doi: 10.1073/pnas.1901480116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maguire G.A., Yoo B.R., SheikhBahaei S. Investigation of Riperidone treatment associated with enhanced brain activity in patients who stutter. Front. Neurosci. 2019;15:100. doi: 10.3389/fnins.2021.598949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turk A.Z., Mahsa L.M., Fritsch I., Maguire G.A., SheikhBahaei S. Dopamine, vocalization, and astrocytes. Front. Neurosci. 2019;15:100. doi: 10.1016/j.bandl.2021.104970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang S.-E., Guenther F.H. Involvement of the cortico-basal ganglia-thalamocortical loop in developmental stuttering. Front. Psychol. 2020;10:3088. doi: 10.3389/fpsyg.2019.03088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alm P.A. Streptococcal infection as a major historical cause of stuttering: Data, mechanisms, and current importance. Front. Hum. Neurosci. 2020;14:569519. doi: 10.3389/fnhum.2020.569519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohane I.S. Using electronic health records to drive discovery in disease genomics. Nat. Rev. Genet. 2011;12:417–428. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 20.Wei W.-Q., Teixeira P.L., Mo H., Cronin R.M., Warner J.L., Denny J.C. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J. Am. Med. Inform. Assoc. 2016;23(e1):e20–e27. doi: 10.1093/jamia/ocv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruett D.G., Shaw D.M., Chen H.-H., Petty L.E., Polikowsky H.G., Kraft S.J., Jones R.M., Below J.E. Identifying developmental stuttering and associated comorbidities in electronic health records and creating a phenome risk classifier. J. Fluency Disord. 2021;68:105847. doi: 10.1016/j.jfludis.2021.105847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedregosa F., Gael V., Gramfort A., Michel V., Thririon B. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 23.Polikowsky H.G., Shaw D.M., Petty L.E., Chen H.-H., Pruett D.G., Linklater J.P., Viljoen K.Z., Beilby J.M., Highland H.M., Levitt B., et al. Population-based genetic effects for developmental stuttering. HGG Advances. 2021;3 doi: 10.1016/j.xhgg.2021.100073. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denny J.C., Bastarache L., Ritchie M.D., Carroll R.J., Zink R., Mosley J.D., Field J.R., Pulley J.M., Ramirez A.H., Bowton E., et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., et al. Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R., et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pluzhnikov A., Below J.E., Konkashbaev A., Tikhomirov A., Kistner-Griffin E., Roe C.A., Nicolae D.L., Cox N.J. Spoiling the whole bunch: Quality control aimed at preserving the integrity of high-throughput genotyping. Am. J. Hum. Genet. 2010;87:123–128. doi: 10.1016/j.ajhg.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 32.Staples J., Qiao D., Cho M.H., Silverman E.K., Nickerson D.A., Below J.E., University of Washington Center for Mendelian Genomics PRIMUS: Rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staples J., Nickerson D.A., Below J.E. Utilizing graph theory to select the largest set of unrelated individuals for genetic analysis. Genet. Epidemiol. 2013;37:136–141. doi: 10.1002/gepi.21684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staples J., Witherspoon D.J., Jorde L.B., Nickerson D.A., Below J.E., Huff C.D., University of Washington Center for Mendelian Genomics PADRE: Pedigree-aware distant-relationship estimation. Am. J. Hum. Genet. 2016;99:154–162. doi: 10.1016/j.ajhg.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staples J., Ekunwe L., Lange E., Wilson J.G., Nickerson D.A., Below J.E. PRIMUS: improving pedigree reconstruction using mitochondrial and Y haplotypes. Bioinformatics. 2016;32:596–598. doi: 10.1093/bioinformatics/btv618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuchsberger C., Abecasis G.R., Hinds D.A. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luca D., Ringquist S., Klei L., Lee A.B., Gieger C., Wichmann H.E., Schreiber S., Krawczak M., Lu Y., Styche A., et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am. J. Hum. Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin D.-Y., Tao R., Kalsbeek W.D., Zeng D., Gonzalez F., 2nd, Fernández-Rhodes L., Graff M., Koch G.G., North K.E., Heiss G. Genetic association analysis under complex survey sampling: the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2014;95:675–688. doi: 10.1016/j.ajhg.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner S.D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. Journal of Open Source Software. 2018;3:731. [Google Scholar]
- 41.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A., et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge T., Chen C.-Y., Ni Y., Feng Y.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J., Ferreira T., Horikoshi M., Johnson A.D., Ng M.C., Prokopenko I., et al. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. Mexican American Type 2 Diabetes (MAT2D) Consortium. Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ambrose N.G., Cox N.J., Yairi E. The genetic basis of persistence and recovery in stuttering. J. Speech Lang. Hear. Res. 1997;40:567–580. doi: 10.1044/jslhr.4003.567. [DOI] [PubMed] [Google Scholar]
- 48.Nievergelt C.M., Maihofer A.X., Klengel T., Atkinson E.G., Chen C.-Y., Choi K.W., Coleman J.R.I., Dalvie S., Duncan L.E., Gelernter J., et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat. Commun. 2019;10:4558. doi: 10.1038/s41467-019-12576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trzaskowski M., Eley T.C., Davis O.S.P., Doherty S.J., Hanscombe K.B., Meaburn E.L., Haworth C.M., Price T., Plomin R. First genome-wide association study on anxiety-related behaviours in childhood. PLoS ONE. 2013;8:e58676. doi: 10.1371/journal.pone.0058676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 51.Leslie E.J., Carlson J.C., Shaffer J.R., Feingold E., Wehby G., Laurie C.A., Jain D., Laurie C.C., Doheny K.F., McHenry T., et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum. Mol. Genet. 2016;25:2862–2872. doi: 10.1093/hmg/ddw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang L., Jia Z., Shi Y., Du Q., Shi J., Wang Z., Mou Y., Wang Q., Zhang B., Wang Q., et al. Genetic factors define CPO and CLO subtypes of nonsyndromicorofacial cleft. PLoS Genet. 2019;15:e1008357. doi: 10.1371/journal.pgen.1008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y., Suzuki A., Iwata J., Jun G. Secondary genome-wide association study using novel analytical strategies disentangle genetic components of cleft lip and/or cleft palate in 1q32.2. Genes (Basel) 2020;11:1280. doi: 10.3390/genes11111280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Y., Zuo X., He M., Gao J., Fu Y., Qin C., Meng L., Wang W., Song Y., Cheng Y., et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramos E., Chen G., Shriner D., Doumatey A., Gerry N.P., Herbert A., Huang H., Zhou J., Christman M.F., Adeyemo A., et al. Replication of genome-wide association studies (GWAS) loci for fasting plasma glucose in African-Americans. Diabetologia. 2011;54:783–788. doi: 10.1007/s00125-010-2002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheong K.X., Yong R.Y.Y., Tan M.M.H., Tey F.L.K., Ang B.C.H. Association of VIPR2 and ZMAT4 with high myopia. Ophthalmic Genet. 2020;41:41–48. doi: 10.1080/13816810.2020.1737951. [DOI] [PubMed] [Google Scholar]
- 57.Watkins K.E., Smith S.M., Davis S., Howell P. Structural and functional abnormalities of the motor system in developmental stuttering. Brain. 2008;131:50–59. doi: 10.1093/brain/awm241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giraud A.-L., Neumann K., Bachoud-Levi A.-C., von Gudenberg A.W., Euler H.A., Lanfermann H., Preibisch C. Severity of dysfluency correlates with basal ganglia activity in persistent developmental stuttering. Brain Lang. 2008;104:190–199. doi: 10.1016/j.bandl.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 59.Shahed J., Jankovic J. Re-emergence of childhood stuttering in Parkinson’s disease: a hypothesis. Mov. Disord. 2001;16:114–118. doi: 10.1002/1531-8257(200101)16:1<114::aid-mds1004>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 60.Lan J., Song M., Pan C., Zhuang G., Wang Y., Ma W., Chu Q., Lai Q., Xu F., Li Y., et al. Association between dopaminergic genes (SLC6A3 and DRD2) and stuttering among Han Chinese. J. Hum. Genet. 2009;54:457–460. doi: 10.1038/jhg.2009.60. [DOI] [PubMed] [Google Scholar]
- 61.Mohammadi H., Joghataei M.T., Rahimi Z., Faghihi F., Khazaie H., Farhangdoost H., Mehrpour M. Sex steroid hormones and sex hormone binding globulin levels, CYP17 MSP AI (-34T:C) and CYP19 codon 39 (Trp:Arg) variants in children with developmental stuttering. Brain Lang. 2017;175:47–56. doi: 10.1016/j.bandl.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Kazemi N., Estiar M.A., Fazilaty H., Sakhinia E. Variants in GNPTAB, GNPTG and NAGPA genes are associated with stutterers. Gene. 2018;647:93–100. doi: 10.1016/j.gene.2017.12.054. [DOI] [PubMed] [Google Scholar]
- 63.Chow H.M., Garnett E.O., Li H., Etchell A., Sepulcre J., Drayna D., Chugani D., Chang S.-E. Linking lysosomal enzyme targeting genes and energy metabolism with altered gray matter volume in children with persistent stuttering. Neurobiol Lang. 2020;1:365–380. doi: 10.1162/nol_a_00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All code used for developing the PheML model described in the methods section is available for download on github (see web resources). Genotyping data from BioVU are only available upon submission of a study proposal and approval through the BioVU Review Committee.