Abstract
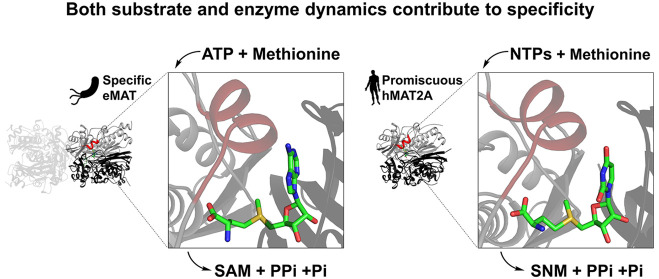
Protein conformational changes can facilitate the binding of noncognate substrates and underlying promiscuous activities. However, the contribution of substrate conformational dynamics to this process is comparatively poorly understood. Here, we analyze human (hMAT2A) and Escherichia coli (eMAT) methionine adenosyltransferases that have identical active sites but different substrate specificity. In the promiscuous hMAT2A, noncognate substrates bind in a stable conformation to allow catalysis. In contrast, noncognate substrates sample stable productive binding modes less frequently in eMAT owing to altered mobility in the enzyme active site. Different cellular concentrations of substrates likely drove the evolutionary divergence of substrate specificity in these orthologues. The observation of catalytic promiscuity in hMAT2A led to the detection of a new human metabolite, methyl thioguanosine, that is produced at elevated levels in a cancer cell line. This work establishes that identical active sites can result in different substrate specificity owing to the effects of substrate and enzyme dynamics.
Keywords: methionine adenosyltransferases, specificity, noncognate substrates, nonproductive binding, substrate dynamics, enzyme dynamics
Introduction
Enzymes can exhibit promiscuous activities with noncognate substrates that are not involved in the main physiological function of the enzyme.1 These promiscuous activities are often vestigial traits of a distant ancestor2 or have originated by chance through evolution.3−6 The importance of promiscuous enzymatic activities is becoming increasing evident, as they have been shown to contribute to evolvability,7 stress responses,8 and potentially, susceptibility to disease.8−10 Protein conformational sampling has been shown to play a role in substrate promiscuity,11−14 as conformational change can allow enzymes to occasionally sample alternative conformations with different charge preorganization, allowing different transition states to be stabilized.15 While the role of protein structural dynamics in this process has been described, the role of substrate conformational sampling is comparatively poorly understood.16 It has recently been reported that large active sites can accommodate multiple different productive substrate conformations without changing the conformation of the catalytic pocket17,18 and that in some cases new Michaelis complexes can be recognized.19
The methionine adenosyltransferases (MATs) are found in all kingdoms of life, and the product of their reaction, S-adenosyl-l-methionine (SAM), is a necessary metabolite in several essential cellular processes.20−22 Because of the physiological importance of SAM, dysfunction in the production of SAM by MATs can lead to disease.23,24 Mechanistically, the enzyme-catalyzed formation of S-adenosyl-l-methionine (SAM) from adenosine triphosphate (ATP) and methionine occurs in two steps:25 SAM is formed by SN2 attack by the sulfur of methionine at the C5′ carbon of ATP followed by hydrolysis of triphosphate (PPPi) into pyrophosphate (PPi) and orthophosphate (Pi)26 (Figure 1a). This second step is believed to provide the energy required for the conformational rearrangement of the enzyme necessary for product release.27 Two Mg2+ ions are involved in coordination of the triphosphate moiety of ATP, and K+ is known to enhance the reaction rate by allowing the active site to adopt the optimal conformation.21,28,29
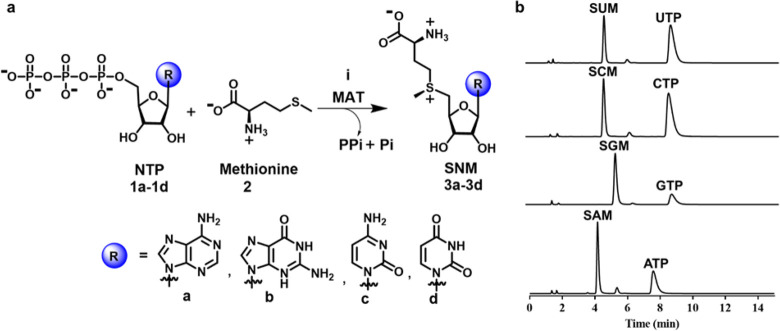
Figure 1.
SNM biochemical synthesis and identification of SNM analogues by UPLC. (a) Synthesis of S-nucleoside-l-methionine (SNM) analogues S-adenosyl-l-methionine (3a, SAM), S-guanosyl-l-methionine (3b, SGM), S-cytidyl-l-methionine (3c, SCM), and S-uridyl-l-methionine (3d, SUM) from different nucleotides (ATP, GTP, CTP, UTP) and methionine. (b) UPLC chromatograms of the reaction of NTPs (5 mM) and methionine (10 mM) in the presence of hMAT2A (20 μM) (1 h, 37 °C, details are in the Methods). Noted are the peaks corresponding to SAM (tR = 4.1 min), SCM (tR = 4.6 min), SUM (tR = 4.6 min), SGM (tR = 5.3 min), ATP (tR = 7.5 min), GTP (tR = 7.8 min), CTP (tR = 8.3 min), and UTP (tR = 8.5 min).
MATs are an excellent model system for the study of substrate promiscuity because the chemical reactivity of the cognate physiological nucleotide substrate, ATP, is independent from the nucleobase. The C5′ atom, which acts as an electrophile in the MAT-catalyzed reaction, belongs to the sugar moiety of the nucleotide and is therefore distant from the nucleobase.30,31 Moreover, SAM is not an intrinsically better methyl donor than the potential products (S-guanosyl-l-methionine (SGM), S-cytidyl-l-methionine (SCM), or S-uridyl-l-methionine (SUM)) from promiscuous reactions with noncognate substrates (GTP, CTP, UTP), since the nucleobase does not influence the sulfonium reactivity. It is worth noting that nucleobase change in SNM (S-(nucleoside)-l-methionine) analogues might influence the ability of methyltransferases or other downstream pathway enzymes to use these substrates, providing a biological basis for the potential evolution of MAT specificity. While E. coli MAT (eMAT) has been reported to display specificity for ATP in vitro,28 the mechanism by which eMAT gates substrate binding remains unknown. In addition, the substrate specificity of human MAT (hMAT2A) has not yet been systematically explored.
In this work, we have performed a systematic study of the substrate (ATP, GTP, CTP, UTP) promiscuities of human and E. coli MATs. Human MAT exits as a heterotetramer in the cell, consisting of an hMAT2A homodimer, which forms the catalytic unit, and two regulatory subunits (hMAT2β).32 Since the enzyme catalytic pocket is at the hMAT2A homodimer interface and the regulatory subunits are not required for catalysis,33 in this study we focus on the hMAT2A homodimer. We show that hMAT2A, unlike eMAT, exhibits substrate promiscuity toward other noncognate NTPs. Structural analysis reveals that eMAT specificity is a consequence of altered structural constraints on noncognate substrates in combination with increased active-site loop dynamics vs hMAT2A. The increased conformational freedom of the noncognate substrates results in eMAT sampling catalytically nonproductive states at higher frequency than the native substrate, ATP, providing a molecular explanation for the observed enzyme kinetics. We demonstrate that the substrate promiscuity of hMAT2A is relevant in vivo, and this knowledge allowed us to identify a new metabolite, methyl thioguanosine, a breakdown product of SGM, that is produced in a human liver cancer cell line but was not produced at detectable levels in a normal liver cell line.
Results
Catalytic Promiscuity of MATs
To probe the substrate specificity and promiscuity of eMAT and hMAT2A, we developed a sensitive and specific assay based on S-(nucleoside)-l-methionine product formation analysis using ultraperformance liquid chromatography (UPLC) (see the Methods). This method allowed us to analyze the catalytic efficiency of these enzymes when utilizing both the cognate substrate ATP and noncognate substrates GTP, CTP, and UTP, with confirmation of the respective reaction products via mass spectrometry (Figure 1b; Appendix). From these data kinetic parameters were derived (Table 1; Figure S1). Our assay validated the specificity eMAT for ATP, with kcat/KM measurements that were 61-fold, 8.5-fold, and 139-fold lower for GTP, CTP, and UTP, respectively, in comparison to ATP (Table 1; Figure S1).28 For hMAT2A, catalytic efficiency with ATP was comparable to that of eMAT, albeit with higher kcat and KM values. However, hMAT2A demonstrated considerably higher activity against noncognate substrates than eMAT, with kcat/KM values against noncognate substrates that were within 42–93% of ATP (Table 1). In addition, no spontaneous product formation was observed in no-enzyme controls (Figure S2a). Thus, while eMAT is comparatively specific for ATP, hMAT2A is catalytically promiscuous with various NTPs (Figure S2b).
Table 1. Kinetic Parametersa for SNM Analogue formation by hMAT2A and eMAT.
| enzyme:substrate | kcat (s–1) | KM (mM) | kcat/KM (M–1 s–1) |
|---|---|---|---|
| hMAT2A:ATP | 764 ± 61 | 0.27 ± 0.07 | 2.8 × 106 |
| hMAT2A:GTP | 3270 ± 600 | 1.26 ± 0.40 | 2.6 × 106 |
| hMAT2A:CTP | 100 ± 4.8 | 0.08 ± 0.02 | 1.3 × 106 |
| hMAT2A:UTP | 1180 ± 140 | 0.97 ± 0.2 | 1.2 × 106 |
| eMAT:ATP | 66 ± 4 | 0.06 ± 0.02 | 1.1 × 106 |
| eMAT:GTP | 18 ± 2 | 0.97 ± 0.30 | 1.8 × 104 |
| eMAT:CTP | 168 ± 12 | 1.3 ± 0.25 | 1.3 × 105 |
| eMAT:UTP | 23 ± 3 | 2.90 ± 0.7 | 7.9 × 103 |
Kinetic parameters for the SNM analogue formation by hMAT2A and eMAT using a concentration of ATP, GTP, CTP, and UTP in the range of 0.025–5 mM and a fixed saturating concentration of methionine (10 mM) in the presence of HEPES (100 mM), KCl (50 mM), and MgCl2 (10 mM), pH 8, at 37 °C. [hMAT2A] was 0.5 μM and [eMAT] was 0.5 μM for ATP, 5 μM for GTP and CTP, and 10 μM for UTP. Product formation was analyzed by UPLC and data fitted to the Michaelis–Menten equation using GraphPad Prism 7.02 (Figure S1).
Molecular Basis for MAT Specificity
Oligomeric State
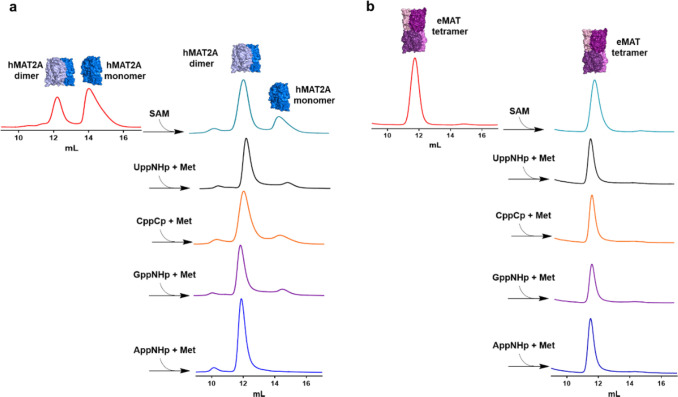
The active sites of both enzymes are located at the dimer interface.34,35 Accordingly, we investigated whether differences between the native oligomeric states of either eMAT or hMAT2A underlie their different substrate specificity. We used non-hydrolyzable NTP analogues (adenosine-5′-[(β,γ)-imido]triphosphate (AppNHp), guanosine-5′-[(β,γ)-imido]triphosphate (GppNHp), cytidine-5′-[(β,γ)-methyleno] triphosphate (CppCp), and uridine-5′-[(β,γ)-imido]triphosphate (UppNHp)]), which have nonhydrolyzable P–C–P or P–N–P β–γ phosphate linkages, to trap the enzymes in their product-bound state (at least for the time scale of these experiments).36 In this manner, we confirmed using size-exclusion chromatography that native apo-hMAT2A exists in equilibrium between monomeric (63%) and dimeric (37%) states (Figure 2a), whereas native apo-eMAT is tetrameric (Figure 2b). The oligomeric equilibrium of hMAT2A shifts almost entirely toward the dimeric state upon incubation with the non-hydrolyzable NTP analogues and methionine (Figure 2a). If any of a non-hydrolyzable NTP analogue, methionine, triphosphate, or SAM was added alone (i.e., if the ternary Michaelis complex is unable to form), no change in the oligomeric state was observed (Figure S3a). This result suggests that formation of the ternary Michaelis complex (enzyme:NTP:Met) drives dimer formation in the case of hMAT2A. In the case of eMAT, no change in oligomeric state was observed (Figure 2b and Figure S3b). In terms of the question of whether differences in oligomeric state underlie the differences in substrate specificity between hMAT2A and eMAT, we did not observe any differences between the cognate and noncognate analogues with either enzyme. Therefore, it can be concluded that the differences in substrate specificity are independent of the oligomeric state of the enzymes.
Figure 2.
Analysis of oligomeric state of hMAT2A and eMAT by size-exclusion chromatography. (a) hMAT2A (20 μM) is incubated with non-hydrolyzable NTPs (1 mM) adenosine-5′-[(β, γ)-imido]triphosphate (AppNHp), guanosine-5′-[(β, γ)-imido]triphosphate (GppNHp), cytidine-5′-[(β, γ)-methyleno] triphosphate (CppCp), and uridine-5′-[(β, γ)-imido]triphosphate (UppNHp)] together with methionine (Met, 10 mM) using reaction buffer (100 mM HEPES, 10 mM KCl, 10 mM MgCl2, pH 8, 37 °C for 1 h). hMAT2A is in an equilibrium of a monomer and dimer. When incubated with both substrates, the enzyme converts completely to a dimeric state. No change in oligomeric state is observed when incubated with SAM alone. (b) eMAT (20 μM) is incubated using the same conditions as used for hMAT2A. eMAT is in a tetrameric state, and no change in oligomeric state was observed after incubation with both substrates and SAM. Size-exclusion chromatography was performed using a GE Healthcare Life Sciences using Superdex 200 Increase 10/300 GL column.
Enzyme Active Site
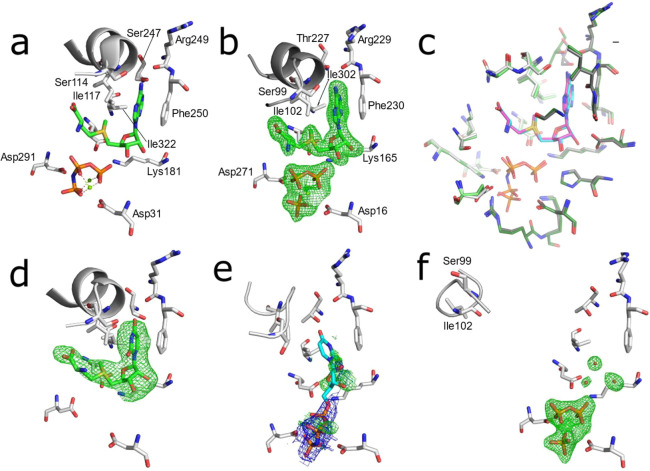
To investigate the structural basis for the observed differences in MAT substrate promiscuity, we then examined differences between the substrate binding cavities in eMAT and hMAT2A. The structure of hMAT2A in a complex with SAM has previously been reported35 (Figure 3a). Here, we solved a crystal structure of eMAT in the presence of ATP and methionine, which enabled us to capture the SAM product-bound state of eMAT at a resolution of 1.95 Å (Figure 3b). This allowed us to align the eMAT:SAM structure to the previously published hMAT2A:SAM structure (Figure 3c). The alignment of the residues within the active site is strikingly similar, both in terms of identity (20/21) and structure (RMSD 0.5 Å). Indeed, every amino acid side chain adopts the same rotamer, and the product is bound in an identical conformation (Figure 3c). We observe stabilizing interactions between the enzyme and adenine ring that include a π-stacking interaction (3.5 Å) with Phe230/250 (eMAT/hMAT2A numbering) and hydrogen bonds between the amine group of the adenine ring and the carbonyl oxygen of Arg229/249 and the N1 adenine nitrogen with the side chain of Thr227/Ser247. Closure of the active-site loop also brings Ile102′/117′ close to the adenine ring, forming van der Waals contacts. The only difference between the two structures is a conserved substitution at position 227/247 (Thr in eMAT vs Ser in hMAT2A; Figure S4). To investigate the effect of this substitution, we made Ser247Thr and Thr227Ser mutants in hMAT2A and eMAT, respectively. Neither mutation resulted in any significant change in substrate specificity in either enzyme (Figures S5). Thus, structures of the SAM-bound active sites of the two enzymes do not explain the observed differences in their substrate specificity.
Figure 3.
Substrate binding sites of eMAT and hMAT2A with bound substrates. Omit electron density (mF0–DFc) is shown in green mesh (3.0 σ), 2mF0–DFc density is shown as blue mesh (1.5σ). (a) Published structure of hMAT2A bound to the products SAM and PPNP (PDB ID 4NDN). (b) Structure of eMAT obtained via cocrystallization with ATP and methionine. (c) Superimposition of the substrate binding sites of eMAT (green, cyan SAM, PPi, and Pi) and hMAT2A (gray, magenta SAM, PPNP). The binding site is comprised of two chains within a homodimer; these are distinguished by light/dark coloring. (d) hMAT2A in complex with the product SUM after cocrystallization with UppNHp. (e) eMAT cocrystallized with UppNHp (2.24 Å); the PPNP group is included in the model and shown with 2mF0–DFc; ambiguous omit density potentially corresponding to disordered substrate/product is shown. A poorly fitting model of UppNHp is shown in a stick representation (cyan). (f) eMAT bound to the products PPi and Pi (1.89 Å), with the active-site loop captured in the “wide-open” conformation obtained via cocrystallization with CTP and methionine.
To understand the structural basis for specificity in hMAT2A, we next compared structures of hMAT2A:SAM and hMAT2A:SUM structures. hMAT2A displays similar turnover rates for both ATP and UTP, so we were interested in observing whether the products were likewise bound similarly. To obtain the hMAT2A:SUM complex (2.5 Å), we cocrystallized with an analogue of UTP (UppNHp). UppNHp is a substrate for the first methionine transfer step and can produce the product SUM in an identical fashion to UTP, but the imido linkage between the β–γ phosphate units prevents hydrolysis of the triphosphate moiety.36 Opening of the active-site loop (residues 113–131 in hMAT2A, 98–108 in eMAT) and product release are thus inhibited.29,35 In the resulting crystal structure, the product SUM was bound within the active site at high occupancy (Figure 3d) and with a similar active-site configuration to the structure of hMAT2A in complex with the cognate product SAM. The active-site loop was fully closed and interacted with SUM in the same manner as with SAM, and the π-stacking interaction with Phe250 was present. The main difference was that the hydrogen bond between the amine group of the adenine and the carbonyl oxygen of Arg249 is not present, although a hydrogen bond between the carbonyl group of the uridine ring and Ser247 is observed (Figure 3d). The close similarity between the hMAT2A:SAM and hMAT2A:SUM complexes is consistent with the similar rates of the UTP and ATP turnover observed in the enzyme kinetics (Table 1), suggesting both substrates are stable in catalytically competent configurations.
We then performed the same comparison, this time for eMAT, which displays a preference for ATP over UTP. To obtain an eMAT:SUM complex (2.24 Å), eMAT was cocrystallized with UppNHp in an identical fashion to hMAT2A. However, unlike the hMAT2A:SUM complex, the electron density observed in the nucleoside binding region of eMAT was weak and disordered, despite the presence of clear electron density for the triphosphate moiety. The poor electron density for the nucleoside groups in these structures could be due to either (or a combination of) disordered binding of the nucleoside moiety of the substrate or diffusion/disorder of the SUM product. The active-site loop was in a partially open, disordered conformation, suggesting a weaker interaction with the nucleoside than in the eMAT:SAM structure, in which this loop adopted a fully closed conformation. Thus, it appears that unlike the hMAT2A:SAM/SUM comparison, in which both product molecules were stable in the binding site, for eMAT, only SAM is stable, with the SUM complex being characterized by significant disorder, both of the product and the active-site loop. Even if the weaker nucleoside density is fully due to diffusion of SUM, this behavior is very different from the eMAT:SAM, hMAT2A:SAM, and hMAT2A:SUM structures in which the product was clearly stable within the active site. To investigate whether this was unique to UTP/UppNHp, we also cocrystallized with a second noncognate substrate analogue, GppNHP. Co-crystallization of eMAT with GppNHP yielded a 2.50 Å structure that displayed identical disordered electron density within the nucleoside binding site and disordered active-site loop as was observed in the eMAT:SUM structure (Figure S6).
Finally, we cocrystallized eMAT with the various noncognate NTPs (CTP, UTP, GTP) to confirm the results observed with the imido-NTP analogues with natural noncognate substrates. No electron density for any SNM product was observed in any of these structures. Within the active site of the 1.89 Å resolution CTP cocrystal, clear difference density for the PPi and Pi products was observed (Figure 3), although there is unambiguously no electron density for the SCM product. Ordered water molecules were observable, suggesting the SCM had fully diffused from the active site. The active-site loop, which was stable and closed in the eMAT:SAM structure, was instead observed in a “wide-open” conformation, which we believe is the first time this fully open conformation has been fully modeled. For the cocrystals of eMAT with UTP (2.25 Å) and GTP (2.39 Å), which crystallized in a different space group to CTP (Table S1), we again observe PPi and Pi in good electron density (Figure S6). Like the CTP cocrystal, we do not observe density for the products of the methionine transferase reaction. In these crystals the density in this region appears to correspond to a phosphate molecule, which presumably rebinds to the protein after hydrolysis.
Altogether, these structural studies suggest that the nonadenine-containing SNM products are less stable within the active site of eMAT than SAM, consistent with eMAT being selective for ATP. Thus, in contrast to hMAT2A, which hydrolyzes ATP/UTP at similar rates and interacts in an essentially identical manner with both SAM and SUM, suggesting they bind and are stabilized similarly within the active site, eMAT demonstrates significantly greater stabilization of SAM within the active site and unstable interactions with noncognate substrates containing uridine, guanine, and cytosine, which is likewise consistent with the enzyme kinetics (Table 1).
Differences in Protein and Substrate Dynamics
The crystallographic analysis of the eMAT:SUM and eMAT:SGM complexes suggested the poor density could be due, at least in part, to a disordered substrate binding mode. To examine this possibility in more detail, we performed molecular dynamics (MD) simulations of the eMAT tetramer and hMAT2A homodimer, each in complex with both ATP and UTP, to investigate whether there were significant differences between enzyme:substrate interactions across proteins that could explain their differing substrate specificities. In order to not bias these simulations, all four simulations began with a starting model in which the loop was fully closed over the active site; i.e., the eMAT:UTP bound structure was modeled on the stable SUM bound structure observed in hMAT2A to position the uridine moiety. However, during triplicate 1 μs MD simulations of each complex (Figure S7), the closed conformation was found to be unstable in the absence of bound methionine. Over the course of the simulations, nearly all active-site loops across all complexes and replicates transitioned to a dynamic open conformation. To avoid sampling bias arising from the variability in closed-to-open transition time points across domains and replicates, we used representative open-state structures at the end point of these trajectories as seed structures for open-state simulations. In addition, because the closed state is less relevant in the absence of methionine and loop opening motions are generally not well sampled on the time scale of MD simulations,37 we did not consider these initial trajectories during further analysis and turned our focus to the open state.
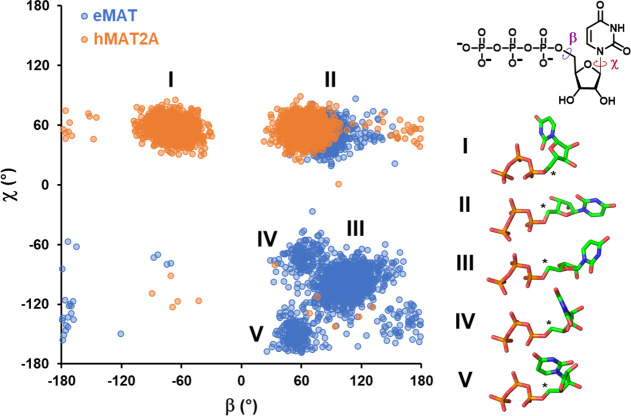
Triplicate 500 ns simulations of all four complexes (eMAT:UTP, eMAT:ATP, hMAT2A:ATP, hMAT2A:UTP) in the open state (Figure S8) show no clear differences in backbone dynamics between eMAT and hMAT2A (Figures S9 and S10), suggesting that conformational fluctuations in the protein backbone are not responsible for nucleotide discrimination in eMAT. However, during these simulations, changes in substrate positioning were observed (Figure S11). While the triphosphate moiety in both ATP and UTP-bound simulations remain stable, the sugar and purine/pyrimidine moieties adopt varied conformations, as the active-site loop open state lacks the stabilizing interaction with Ile102′ observed in the closed-state structures (Figures 3; Figure S12). The resulting substrate conformations are largely dictated by rotations around the β and χ dihedral angles (Figure 4).
Figure 4.
Conformational states populated by the UTP substrate bound to eMAT or hMAT2A. A plot of the UTP β vs χ dihedral angles (shown on the inset 2D representation of UTP) highlights the differences in conformational diversity exhibited by UTP in complex with eMAT or hMAT2A. Each data point represents a dihedral angle pair from one UTP molecule in one simulation frame, sampled every nanosecond over triplicate 500 ns trajectories. Dihedral angle measurements from different enzyme subunits were treated as independent data points. Major conformational clusters in the resulting landscape are shown as a stick representation with the electrophilic C5′ identified with an asterisk. The different conformational states adopted by UTP in eMAT and hMAT2A are indicative of differing enzyme–substrate interactions that constrain the UTP conformation and may contribute to enzyme specificity.
In hMAT2A, the β dihedral angle varies between gauche– (I) and gauche+ (II) conformations, while the χ dihedral angle is observed to predominantly adopt the favorable gauche+ (II) conformation. In eMAT however, interactions between Lys165 and the O4′/O5′ of UTP prevent the adoption of the β dihedral gauche– conformation. In several domain replicates of the eMAT:UTP complex, a strained eclipsed χ dihedral conformation (III) is observed that likely arises from electrostatic repulsion between UTP’s uracil moiety and the nearby Asp118′ side chain and Gly117′ backbone carbonyl (Figure S13). Dissociation of the nucleotide in the eMAT:UTP complex was never observed over the time scale of the simulations performed due to strongly favorable interactions between bound Mg2+ ions and the triphosphate moiety.38,39 However, the strained nucleotide conformations observed in the eMAT:UTP complex, which are absent in the hMAT2A:UTP complex, may indicate a weaker binding capacity for UTP in the open state for eMAT comparatively to hMAT2A. Interestingly, in one instance, this conformation also transitioned to an alternate conformation that rapidly fluctuated between gauche– (Figure 4, IV) and trans (Figure 4, V) χ dihedral conformations. The trans (V) χ dihedral conformation, which is observed only in the eMAT:UTP complex, positions the UTP pyrimidine ring such that it blocks binding and nucleophilic attack from methionine on C5′ of UTP (Figure 4). Thus, subtle substrate–enzyme interactions in eMAT that are not present in hMAT2A result in an altered UTP conformational landscape that destabilizes substrate binding and forces the adoption of nonproductive binding modes. These observations are consistent with the disorder in the active-site loop and the poor electron density for the nucleoside moieties in the substrates/products from cocrystallization with noncognate NTPs or imido-NTP analogues (Figure 3), as well as the observed substrate specificity of eMAT (Table 1). However, it is important to note that these results show that productive binding of noncognate NTPs can occur (consistent with slow turnover) but that it is less stable/frequent in eMAT than in hMAT2A.
Relevance of MAT Promiscuity In Vivo
Enzyme promiscuity sometimes reflects the physiological conditions under which these enzymes operate. Therefore, to better understand the biological implications of the differing eMAT and hMAT2A substrate specificities, we compared the physiological concentrations of NTPs in human40 and E. coli(41) cells (Table S2). In human cells, the concentration of ATP is ∼2.5 mM and the other NTPs (GTP 0.2 mM, CTP 0.08 mM, and UTP 0.2 mM) are almost 10-fold lower, whereas in E. coli the NTPs are all present at similar concentrations. This biological context supports the different promiscuities of eMAT and hMAT2A. In human cells, the lower concentration of noncognate NTPs in comparison to ATP limits competition for enzyme binding. In E. coli, however, this is not the case, and the higher enzyme specificity could contribute to limited noncognate NTPs concentrations.
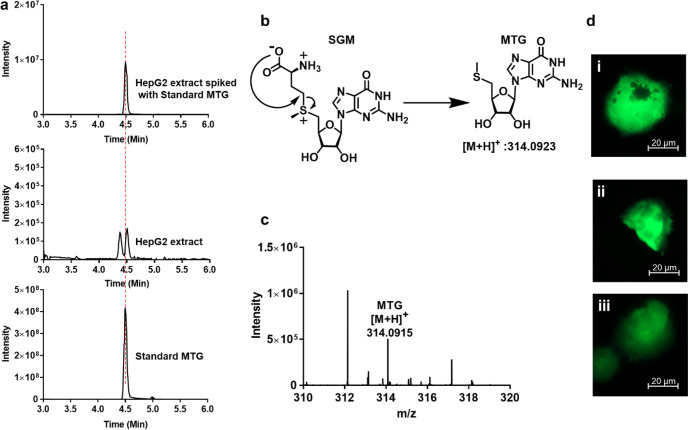
To validate these biological implications of hMAT2A promiscuity, we then investigated whether the promiscuous products of hMAT2A could be detected in vivo. We performed metabolite analysis of SNM abundance using liquid chromatography–mass spectrometry (LC–MS) of extracts from the normal human liver cell line THLE-2 and the hepatocarcinoma cell (HCC) line HepG2, in which hMAT2A is known to be upregulated.42−44 As a control, we could detect SAM and its breakdown product methionine thioadenosine (MTA) in both samples (Appendix). Notably, we also detected the breakdown product of SGM, methionine thioguanosine (MTG), in the cancer cell line extract (Figure 5a) and not in the normal THLE-2 cell line extract (Figure S14). The presence of MTG was confirmed by mass spectrometry analysis (Figure 5c). The presence of MTG only in the HepG2 cell line might be a combination of the following reasons. (i) The concentration of GTP is 2-fold higher in cancer cells than normal cells (Table S2). (ii) Overexpression of hMAT2A in cancer cells compared to normal cell line. It has been reported that hepatocellular carcinoma tissues have 6-fold higher hMAT2A mRNA content.45 (iii) The downstream pathway enzymes are unable to utilize MTG. To the best of our knowledge, there is no biological source of MTG other than from SGM breakdown (Figure 5b). While it is unclear whether MTG formed during the extraction procedure or is generated endogenously in the cells, SNM analogues were found to have comparable stability in aqueous buffer over the same period (Figure S15), suggesting SGM is not significantly less stable and more prone to degradation. Interestingly, even though the KM of hMAT2A with CTP (0.08 mM) is lower than for ATP (0.27 mM), no other SNMs or SNM breakdown products were detected in any of the cell lines within the sensitivity range of the experiment. This is most likely due to the lower CTP concentration within the cells (0.083 mM in normal cells and 0.4 mM in cancer cells).
Figure 5.
LC–MS analysis of metabolite and effect of SGM and SAM on HepG2. (a) Extracted chromatograms of the standard MTG, HepG2 cell extract, and cell extract samples spiked with the standard MTG. (b) Schematic representation of degradation of SGM into MTG after attack of carboxylate on the γ carbon atom of the methionine. (c) Mass spectrum of HepG2 extract showing the mass of MTG [M + H]+314.0915. Data was collected using a Q-Exactive HF mass spectrometer coupled with Waters UPLC ACQUITY M-Class liquid chromatography system. An analytical column (ACQUITY UPLC HSS T3 1.8 um, 1.0 × 150 mm) was used for sample chromatographic separation. (d) Fluorescence microscopy images showing no morphological effect of SGM and SAM on HepG2 cells. HepG2 cells electroporated with (i) pmaxGFP plasmid and with (ii) 1 mM SAM and (iii) 1 mM SGM. Imaging is done using a Celldiscover 7 microscope with 20× resolution with 2× magnification changer. Experiment was performed in biological triplicate.
After the inferred identification of SGM in liver cancer cells, we investigated whether increased SGM levels result in cellular toxicity or morphological changes to these cells. To overcome the low cell membrane permeability of SGM,46,47 we performed electroporation of HepG2 cells in the presence of three different concentrations of SGM (0.01, 0.1, and 1 mM). Electroporation was carried out along with a pmaxGFP plasmid to allow fluorescence microscopy observation. Cells were observed after an overnight incubation. The number of cells in the sample electroporated with SGM was comparable to the control (electroporation only with pmaxGFP plasmid) (Figure S16a), even at the highest SGM concentration, which indicates that the concentrations of SGM used do not affect cell survival. In addition, no microscopic effects on cell morphology could be detected (Figure 5d; Figure S16b,c). The same experiment was also performed using SAM (Figure 5d; Figure S16d,e) resulting in the same observations. Since SGM carries the same methyl transferring group as SAM, it is possible that SGM can neutrally substitute for SAM in the methylation or polyamine downstream pathways (Figure S17) or that it is simply inert. Overall, we have shown that hMAT2A promiscuity is maintained in vivo, allowing for the detectable production of SGM (and/or its breakdown product MTG) in the cancer cell line HepG2 in which hMAT2A is upregulated. The SNM products arising from promiscuous hMAT2A activity could therefore serve as potential biomarker targets for the detection of cancers.
Discussion
The enzyme kinetics and structural analysis suggest that the catalytic specificity of eMAT is a result of the noncognate substrates failing to adopt stable and catalytically competent binding modes. This leads to two questions: first, why are the unique substrate binding modes observed in the eMAT:UTP simulations not catalytically competent? The sulfur of methionine performs its nucleophilic substitution at the ribose C5′ atom; thus, the accessibility of this atom is of paramount importance. In the nonproductive states sampled by UTP throughout the simulation, the position of the C5′ atom is sterically occluded by the pyrimidine ring and methionine attack is sterically blocked (Figure 4). Clearly, UTP is a viable substrate for eMAT; indeed, we observe in the MD simulations that catalytically productive enzyme:substrate complexes are stable for hundreds of nanoseconds. Thus, the disorder observed here is best conceptualized as a partial depletion of catalytically productive substrate binding and weaker binding stability, compared with the cognate substrate, ATP. Second, what are the contributions of structural dynamics to hMAT2A catalysis with noncognate substrates in comparison to eMAT? The active sites are essentially identical (20/21 residues) and substitutions of Ser/Thr in either enzyme at the one variant position have no effect on specificity. However, there are many sequence differences between eMAT and hMAT2A in the second and third shells of the active-site loop (Figure S18). A plausible explanation is therefore that the crystallographic closed state of hMAT2A observed in the presence of noncognate substrates is promoted by additional stabilizing interactions in the second and third shells of the active-site loop, even though noncognate substrates make fewer stabilizing interactions with first shell residues. In contrast, eMAT cannot as easily sustain the closed active-site loop conformation without the additional stabilizing interactions from the adenine group, which are not present in the binding modes of the other noncognate nucleotides.
The selective pressure that drove the divergence in catalytic specificity between these orthologous enzymes most likely relates to the different cellular abundance of these molecules; i.e., there has been little selective pressure for hMAT2A to be specific because the other NTPs are not present at sufficiently high concentrations to compete with ATP. Indeed, the concentration of ATP is ∼10-fold higher than the KM, whereas for GTP/CTP/UTP the physiological concentrations are at or below the respective KM values (Table 1). In contrast, the concentrations of these NTPs in E. coli are more similar: ATP is 3.5 mM while GTP is 1.6 mM. Thus, eMAT likely evolved specificity owing to the selective pressure to discriminate between ATP and other nucleotides: the KM of eMAT for ATP is at least 16-fold lower than for any of the noncognate NTPs (Table 1).
Finally, in this work, we showed how MAT promiscuity is relevant in vivo as a putative example of “underground metabolism”.10 It is thought that promiscuous functions of enzymes are likely to be physiologically irrelevant.1 For instance, many promiscuous activities cannot occur at sufficiently high frequency to be relevant owing to the substrate concentrations encountered in physiological contexts,48 or the extremely low catalytic efficiency of many promiscuous activities making it irrelevant on biological time scales.49−51 This study is therefore a rare example where we could detect the promiscuous activity of hMAT2A for GTP in vivo. Moreover, we showed that it could be used as a biomarker to distinguish between normal and cancer cell lines.
In summary, these results show how enzyme dynamics have substantial effects on the conformational sampling of substrates within the active site of an enzyme, which can in turn result in large changes in catalytic specificity. The concept of nonproductive substrate binding is not new,52 nor is the notion that protein dynamics can affect substrate turnover,53,54 but this is an interesting example where the link between these two effects can be clearly seen. Moreover, because we have compared orthologous enzymes that have been on different evolutionary trajectories because of their distinct cellular environments, we have been able to show that the sequence differences controlling this specificity originate in the outer shells of the active site, which builds on a growing body of work that supports a model in which these outer-shell residues are critical for maintaining the optimum active-site architecture and controlling conformational changes that are important in the catalytic cycle.15 Consideration of these effects should aid enzyme engineers, evolutionists and synthetic chemists in the design and study of enzymes, substrates, and inhibitors. For example, we hope that this work will aid in the design of SAM analogues with unnatural bases;55,56 such analogues could show promise for reaching cellular bio-orthogonal probes or inhibitors of methyltransferases.
Materials
ATP, GTP, CTP, UTP, methionine, S-adenosylmethionine (SAM), HEPES, MgCl2, KCl, isopropyl-1-thio-β-d-galactopyranoside (IPTG), Tris–HCl, Na2HPO4, NaH2PO4, potassium phosphate, NaCl, imidazole, β-mercaptoethanol, dithiothreitol (DTT), kanamycin, glycerol, NaOH, HCl, ammonium acetate, bacto agar, bacto tryptone, bacto yeast extract, all other chemicals, and HPLC-grade solvents were purchased from commercial sources and used as supplied unless otherwise mentioned. PageRuler prestained protein ladder, 10–180 kDa and TrypLE Express Enzyme (1X), no phenol red, DMEM - Dulbecco’s Modified Eagle Medium, trypsin–EDTA, fetal bovine serum (FBS) PBS, and penicillin–streptomycin solution were purchased from ThermoFischer scientific. BL21 (DE3) competent cells and Q5 Site-Directed Mutagenesis Kit were purchased from New England Biolabs (NEB). RedTaq Ready Mix PCR reaction mix and benzonase, complete His-Tag Purification Resin (NiNTA), glass beads acid washed, and Bovine Collagen Solution Type I were purchased from Sigma-Aldrich. Protein inhibitor cocktail (PIC), bovine serum albumin (BSA), and lysozyme were purchased from Nacalai Tesque, Inc. 12% Mini-PROTEAN TGX Precast Protein Gels, 12-well, were purchased from Bio-Rad. Amicon centrifugal filters were purchased from Merck. Standards for size-exclusion chromatography were purchased from GE Healthcare. AppNHp, GppNHp, UppNHp, and CppCp were purchased from Jena Bioscience. THLE-2 cells were purchased from ATCC. A BEGM Bronchial Epithelial Cell Growth Medium Bullet Kit was purchased from Lonza. Fibronectin Human Protein were purchased from Life technologies. Phosphorylethanolamine was purchased from Funakoshi. The SF Cell Line 4D-Nucleofector X Kit and P1 Primary Cell 4D-Nucleofector X Kit S were purchased from Lonza. All of the experiments were performed using an ultrapure water purification system from a Milli-Q Integral MT10 type 1 (Millipore).
Methods
Protein Expression and Purification
The eMAT plasmid was a generous gift from Prof. Ronald E. Viola. E. coli BL21 (DE3) cells were transformed with the eMAT plasmid, and protein was expressed as reported previously.20 Cell pellets were resuspended in lysis buffer (40 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM imidazole) supplemented with 0.5 units of turbonuclease (T4330, Sigma-Aldrich), 0.3 mg·mL–1 lysozyme, 0.2 mM PMSF, and 5 mM DTT. Solubilized pellets were lysed by sonication and centrifuged at 30000g for 30 min. The soluble fraction was applied to a 5 mL HisTrap HP Ni2+-NTA IMAC column (GE Healthcare) pre-equilibrated with lysis buffer and washed with 50 mM imidazole. eMAT was eluted in lysis buffer supplemented with 400 mM imidazole and concentrated with an Amicon Ultra-15 spin concentrator (30 kDa MW cutoff, Millipore). eMAT was further purified by size-exclusion chromatography (SEC) using a HiLoad 26/600 Superdex 200 pg column (GE Healthcare) in SEC buffer A (50 mM Tris–HCl pH 8.0, 100 mM NaCl and 5 mM DTT). Analysis of MAT protein purity was verified with Coomassie SDS polyacrylamide gel electrophoresis, and protein concentrations were calculated using the molar extinction coefficient predicted by the ExPASY ProtParam server tool at A280. The hMAT2A plasmid was gift from Jon S. Thorson and purified as reported.57 hMAT2A pellets were processed in the same manner as eMAT, using sonication and Ni2+–NTA IMAC except for the composition of lysis buffer (50 mM Na2HPO4 pH 8.0, 300 mM NaCl, and 10 mM imidazole). hMAT2A elution was then incubated with 10 mM l-methionine, 10 mM MgCl2, and 100 μM UppNHp for 1 h on ice before purification in SEC buffer B (25 mM HEPES pH 7.6, 150 mM NaCl, 5 mM KCl, 5 mM DTT and 10% (v/v) glycerol) for crystallization.
Protein Crystallization, Data Collection, And Structure Determination
eMAT crystals were grown at 19 °C using the hanging-drop vapor diffusion method with reservoir solutions containing 0.1 M BIS–TRIS pH 6.5 and 10–20% (v/v) ethylene glycol while screening two different lengths of polyethylene glycol (PEG) at varying concentrations: PEG 8000 from 6 to 9% (w/v) and PEG 3350 from 16 to 22% (w/v). Drops were setup at 1:1 ratio and 1:2 ratio of reservoir to protein volume. Co-crystals formed within 2–4 days at 19 °C with various substrates. hMAT2A-UppNHp was concentrated to 10 mg·mL–1 for protein X-ray crystallographic studies. hMAT2A-UppNHp hanging drops were grown at 19 °C at a 1:1 and 1:2 ratio of reservoir to protein volume. The optimized screening matrix consisted of 0.1 M BIS–TRIS pH 6.5 and 10% (v/v) ethylene glycol while screening PEG 3350 at concentrations of 7–10% (w/v). Cubic diamond crystals formed within 2 days at 19 °C. The cocrystals were cryoprotected in solutions containing the mother liquor and increasing the concentrations of PEG 8000 or PEG 3350 to 25–35% (w/v) before being flash-frozen in liquid nitrogen. Diffraction data was collected on the macromolecular crystallography beamline (MX2) at the Australian Synchrotron using the Eiger X 6 M detector at a wavelength of 0.9537 Å.58 Data was processed using XDS59 and Aimless,60 and molecular replacement was performed using Phaser.61 Iterative cycles of manual model building and refinement were performed using Coot 0.9.362 and phenix.refine.63 Iterative cycles of manual model building and refinement were performed using Coot 0.9.3,62 and phenix.refine.63 TLS refinement was used in all cases, using TLS groups automatically selected by phenix.refine. Notably, chain B in the hexagonal space groups exhibited significant disorder in places. All crystallization conditions, data collection, and refinement details are provided in Table S1.
Molecular Dynamics Simulations
All molecular dynamics simulations were carried out using GROMACS 2018.3.64 Closed-state simulations were run using nucleotide-bound models derived from hMAT2A:SAM, hMAT2A:SUM, and eMAT:SAM crystal structure as starting points. For eMAT:UTP simulations, UTP was modeled in the eMAT:ATP model structure at the ATP position. Open-state simulations were run using the final frame from a randomly selected closed-state simulation replicate in which all domains had transitioned to the open state as starting points. Completed structures were solvated in a dodecahedral simulation box with a minimum distance of 10 Å from any protein atom to the box wall, followed by addition of roughly 50 mM NaCl into the aqueous phase, neutralizing the system charge. All systems were subjected to steepest-descent energy minimization followed by a 100 ps equilibration in the NVT ensemble with position restrains of 1000 kJ/mol/nm2 on all protein atoms, with velocities initializing from a Maxwell distribution at 300 K. All NVT equilibrated systems were then subjected to 100 ps equilibration in the NPT ensemble with position restraints of 1000 kJ/mol/nm2 on all protein atoms. Position restraints were released, and free simulation was performed at 300 K for 1 μs for each replicate. All simulations were performed using the CHARMM36-feb2021 force field.65 Water was explicitly modeled using the TIP3P model. Ionizable residues were set to their standard protonation state at pH 7. All equilibration and production simulations were conducted under periodic boundary conditions. Temperature was maintained close to the reference value of 300 K using V-rescale temperature coupling. Pressure was maintained close to the reference value of 1 atm using a Parinello-Rahman barostat with isotropic pressure coupling. The LINCS algorithm66 was used to constrain the lengths of all bonds to hydrogen. The Verlet cutoff scheme was used to evaluate the nonbonded interaction pair lists. van der Waals interactions were evaluated using a simple cut off scheme with a radius of 12 Å. Coulomb interactions were evaluated using the Particle Mesh Ewald (PME) method with a grid spacing of 1.6 Å. A 2 fs time-step was used for integrating the equations of motion. GROMACS tools64 were used for correction of periodic boundary conditions. Visual Molecular Dynamics (VMD)67 was used to view trajectories and for RMSD, RMSF, and dihedral angle calculations, and PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.) was used to produce figures.
Mutagenesis
Site-directed mutagenesis for Ser247Thr mutation on hMAT2A plasmid and Thr227Ser mutation on eMAT plasmid was carried out using Q5 Site-Directed Mutagenesis Kit (NEB) by following kit protocol, expressed, and purified as hMAT2A and eMAT, respectively. The primers used for mutagenesis are listed in Table S3.
Kinetics Assay for MATs
To observe the reaction efficiency of SNM product formation during catalysis with different substrates ATP/GTP/CTP/UTP (5 mM) and methionine (10 mM), HEPES (100 mM), MgCl2 (10 mM), KCl (50 mM), and hMAT2A/eMAT/Ser247Thr hMAT2A/Thr227Ser eMAT (20 μM) were mixed in water and the pH was adjusted to 8 with 10% NaOH. The reactions were incubated at 37 °C for 1 h. The reaction was quenched by acetonitrile followed by centrifugation at 12000 rpm for 5 min to precipitate the enzymes. Finally, the supernatant was filtered through 0.22 μm filter (Merck) and injected in UPLC for analysis (Waters UPLC Acquity H class). Diluted reaction aliquots were analyzed by using a HILIC column (SeQuant ZIC-cHILIC 3 μm,100 Å 150 × 2.1 mm PEEK coated HPLC column). An isocratic method was used with solvent A (100 mM ammonium acetate, pH 5.3) 35% and solvent B (acetonitrile) 65% for 15 min. Each injection was 3 μL with a flow rate of 0.3 mL/min and detected at 260 nm. Using this UPLC method, retention times for molecules were MTA 1.3 min, MTU 1.3 min, MTC 1.4 min, MTG 1.5 min, adenine 1.6 min, uracil 1.6 min, cytosine 1.8 min, guanine 2 min, SAM 4.1 min, SCM 4.6 min, SUM 4.6 min, SGM 5.3 min, ADP 5.3 min, UDP 6 min, CDP 6.1 min, GDP 6.3 min, ATP 7.5 min, GTP 7.8 min, CTP 8.3 min. Product formation was further confirmed by mass analysis (Appendix). SNM were purified using above-mentioned UPLC method and standard curves were plotted. For kinetic assay concentrations of the NTPs were in the range of 0.0250–5 mM and constant methionine concentration 10 mM were used. The kinetic parameters were determined using the Michaels-Menten equation using GraphPad Prism 7.02. The release of nucleotide bases from SNM analogues was also detected by UPLC (Figure S5a). SAM is prone to alkaline depurination,68 but release of nucleotide bases for pyrimidine ring in our reaction conditions might be due to deprotonation at C-5′ in basic conditions followed by the opening of the ribose ring which eliminates nucleotide base, further attack of water reforms ribose ring to give S-ribosylmethionine.69 Elimination of nucleotide bases was not observed from NTPs (Figure S2a) under the same conditions, which demonstrate that release of nucleotide base was from SNM analogues.
Analytical Size-Exclusion Chromatography
Size-exclusion chromatography was performed using GE Healthcare Life Sciences using Superdex 200 Increase 10/300 GL column. The injection volume was 100 μL, detection at 280 nm and flow rate was 0.5 mL/min. Nonhydrolyzable NTPs (1 mM), adenosine-5′-[(β,γ)-imido]triphosphate (AppNHp), guanosine-5′-[(β,γ)-imido]triphosphate (GppNHp), cytidine-5′-[(β,γ)-methyleno] triphosphate (CppCp), and uridine-5′-[(β,γ)-imido]triphosphate (UppNHp)] were incubated with methionine (l-Met) (10 mM) in HEPES (100 mM), KCl (50 mM), MgCl2 (10 mM), pH 8 at 37 °C for 1 h and then injected in the column.
Cell Culture and Extraction of Metabolites
HepG2 cell line was grown in DMEM medium containing 10% FBS and penicillin (100 U/mL) and streptomycin (100 mg/mL) by incubation in a 5% CO2 at 37 °C with 95% humidity. For routine maintenance, cells were trypsinized and split before becoming fully confluent. Cultured cells were washed with cold PBS (5 mL) twice. Cells (20 M) were harvested by trypsinization using TrypLE Express Enzyme (1X), no phenol red for 3 min at 37 °C in CO2 incubator. Centrifuged for 5 min at 100g. TrypLE was discarded, and the pellet was resuspended into cold PBS. The cell pellet was washed with cold PBS twice. Further extraction steps were performed on ice. Internal standards (10 nmol of HEPES and PIPES) were added to the sample. Cells were disrupted using 1 mL of cold acetonitrile, methanol, and water (40:40:20) with 0.1 M formic acid and glass beads acid washed by vortexing. Metabolites were collected by the centrifugation. Samples were concentrated using speed vac and finally dissolved in 100 μL of 10% acetonitrile with 0.1% formic acid and filtered through a 0.22 μm filter and injected into LC–MS.
LC–MS Method for Metabolite Analysis
Data were collected using Q-Exactive HF mass spectrometer (Thermo Fisher Scientific) coupled with Waters UPLC ACQUITY M-Class liquid chromatography system. An analytical column (ACQUITY UPLC HSS T3 1.8 um, 1.0 × 150 mm) was used for sample chromatographic separation. An injection volume of 2 μL was separated at flow rate of 50 μL/min using a gradient of 10–95% solvent B over 8 min, using water with 0.1% formic acid as solvent A and acetonitrile with 0.1% formic acid as solvent B. MS data were collected using Q-Exactive HF mass spectrometer (Thermo Fisher Scientific). The parameters are listed here: spray voltage, 3.0 kV; sheath gas, 16; auxiliary gas, 2; capillary temperature, 250 °C; aux gas heater temp, 150 °C; S-lens RF, 50; tuning method name, HESI; Spray interface, HESI, with metal needle for small flow (1 to 10 μL/min). The mass spectrometry method was set to acquire MS1 data for 14 min, positive mode, mass range 80 to 1000 m/z. Resolution was set at 60000. The maximum injection time was 30 ms. The auto gain was targeted to 500000 ions. Extracted ion chromatograms were done using a 5-ppm tolerance and smoothing with Boxcar method using 7 points.
Cell Electroporation with SGM, SAM, and pmaxGFP Plasmid
Cells were harvested by trypsinization, and 2 × 106 cells were pelleted by centrifugation at 100g for 3 min. Cells were resuspended in Nucleofector solution from Lonza. SF cell line 4d-Nucleofector X kit S (V4XC-2032) for HepG2 cells. Cells were electroporated with 0.4 μg of pmaxGFP plasmid and different concentrations of SGM and SAM (0.01, 0.1, 1 mM) using 4D-Nucleofector X Unit from Lonza. EH-100 program was used for HepG2 by following the manufactures protocol. Cells were incubated overnight in the incubator and observed under the fluorescence microscope. Cells were observed using Leica DMiL microscope using 10× objective. For higher magnification, cells were observed using a ZEISS Celldiscoverer 7 using a 20× objective with a 2× magnification changer.
Acknowledgments
We thank Yohsuke Moriyama for assistance with the cell electroporation experiment and Keiko Kono for sharing the fluorescent microscope in her unit. We thank Saacnicteh Toledo-Patino and Benjamin Clifton for insightful comments on this manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.1c00464.
Kinetic parameters, UPLC chromatograms, size exclusion chromatography, multiple sequence alignment, models and electron density, MD simulation, LC–MS analysis of metabolite, SNM degradation plots, results of electroporation of HepG2, schematics of SGM synthesis and possible downstream pathways, sequence differences, crystal data collection, NTPs concentration tables, plasmid sequences (PDF)
Mass spectrum of SAM using the Acquity UPLC H-Class system with an Acquity QDa detector (PDF)
Author Contributions
M.G. performed MAT kinetics assay (including protein purification), ran SEC analysis, performed all the cell experiments (including culturing), metabolite extractions, and imaging and processed and analyzed data. M.G. and A.V.B. ran LC–MS for metabolites and processed and analyzed data. L.L.T. expressed and purified protein for crystallography and collected crystallographic data. L.L.T., A.D., J.S.B., and C.J.J. processed, solved, and analyzed crystallographic data. A.M.D. and M.S. performed molecular dynamics simulations. P.L. and C.J J. analyzed data and wrote the paper with input from M.G. and A.M.D. P.L. and C.J.J. conceived and supervised the project.
Financial support from the Okinawa Institute of Science and Technology to P.L. is gratefully acknowledged. The Laurino lab is supported by a Kakenhi Grant (No. 90812256). This project was supported by a OIST Kick start-up grant. Funding by the Australian Research Council for the Centres of Excellence in Synthetic Biology and Innovation in Peptide and Protein Science is gratefully acknowledged. A.M.D. acknowledges funding from the Human Frontier Science Program (LT-000366/2020-C). This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of ANSTO, and made use of the Australian Cancer Research Foundation (ACRF) detector.
The authors declare no competing financial interest.
Supplementary Material
References
- Copley S. D. Shining a light on enzyme promiscuity. Curr. Opin. Struct. Biol. 2017, 47, 167–175. 10.1016/j.sbi.2017.11.001. [DOI] [PubMed] [Google Scholar]
- O’Brien P. J.; Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 1999, 6 (4), R91–R105. 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- Jensen R. A. Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 1976, 30, 409–25. 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- Khersonsky O.; Roodveldt C.; Tawfik D. S. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 2006, 10 (5), 498–508. 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Copley S. D. Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 2003, 7 (2), 265–72. 10.1016/S1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- Jeffery C. J. Moonlighting proteins. Trends Biochem. Sci. 1999, 24 (1), 8–11. 10.1016/S0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- Khersonsky O.; Tawfik D. S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010, 79, 471–505. 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Piedrafita G.; Keller M. A.; Ralser M. The Impact of Non-Enzymatic Reactions and Enzyme Promiscuity on Cellular Metabolism during (Oxidative) Stress Conditions. Biomolecules 2015, 5 (3), 2101–22. 10.3390/biom5032101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward P. S.; Patel J.; Wise D. R.; Abdel-Wahab O.; Bennett B. D.; Coller H. A.; Cross J. R.; Fantin V. R.; Hedvat C. V.; Perl A. E.; Rabinowitz J. D.; Carroll M.; Su S. M.; Sharp K. A.; Levine R. L.; Thompson C. B. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010, 17 (3), 225–34. 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ari R.; Casadesus J. Underground metabolism. BioEssays 1998, 20 (2), 181–6. . [DOI] [PubMed] [Google Scholar]
- Gora A.; Brezovsky J.; Damborsky J. Gates of enzymes. Chem. Rev. 2013, 113 (8), 5871–923. 10.1021/cr300384w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N.; Tawfik D. S. Protein dynamism and evolvability. Science 2009, 324 (5924), 203–7. 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- Maria-Solano M. A.; Serrano-Hervas E.; Romero-Rivera A.; Iglesias-Fernandez J.; Osuna S. Role of conformational dynamics in the evolution of novel enzyme function. Chem. Commun. (Cambridge, U. K.) 2018, 54 (50), 6622–6634. 10.1039/C8CC02426J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton B. E.; Jackson C. J. Ancestral Protein Reconstruction Yields Insights into Adaptive Evolution of Binding Specificity in Solute-Binding Proteins. Cell Chem. Biol. 2016, 23 (2), 236–245. 10.1016/j.chembiol.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Campbell E.; Kaltenbach M.; Correy G. J.; Carr P. D.; Porebski B. T.; Livingstone E. K.; Afriat-Jurnou L.; Buckle A. M.; Weik M.; Hollfelder F.; Tokuriki N.; Jackson C. J. The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol. 2016, 12 (11), 944–950. 10.1038/nchembio.2175. [DOI] [PubMed] [Google Scholar]
- Kaltenbach M.; Burke J. R.; Dindo M.; Pabis A.; Munsberg F. S.; Rabin A.; Kamerlin S. C. L.; Noel J. P.; Tawfik D. S. Evolution of chalcone isomerase from a noncatalytic ancestor. Nat. Chem. Biol. 2018, 14 (6), 548–555. 10.1038/s41589-018-0042-3. [DOI] [PubMed] [Google Scholar]
- Barrozo A.; Duarte F.; Bauer P.; Carvalho A. T.; Kamerlin S. C. Cooperative Electrostatic Interactions Drive Functional Evolution in the Alkaline Phosphatase Superfamily. J. Am. Chem. Soc. 2015, 137 (28), 9061–76. 10.1021/jacs.5b03945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. J.; Evans R. L. 3rd; Mylrea N. J.; Chaudhury D.; Luo C.; Guan B.; Pierce C. T.; Gordon W. R.; Wilmot C. M.; Kazlauskas R. J. Larger active site in an ancestral hydroxynitrile lyase increases catalytically promiscuous esterase activity. PLoS One 2020, 15 (6), e0235341 10.1371/journal.pone.0235341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miton C. M.; Jonas S.; Fischer G.; Duarte F.; Mohamed M. F.; van Loo B.; Kintses B.; Kamerlin S. C. L.; Tokuriki N.; Hyvonen M.; Hollfelder F. Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (31), E7293–E7302. 10.1073/pnas.1607817115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parungao G. G.; Zhao M.; Wang Q.; Zano S. P.; Viola R. E.; Blumenthal R. M. Complementation of a metK-deficient E. coli strain with heterologous AdoMet synthetase genes. Microbiology 2017, 163 (12), 1812–1821. 10.1099/mic.0.000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham G. D.; Pajares M. A. Structure-function relationships in methionine adenosyltransferases. Cell. Mol. Life Sci. 2009, 66 (4), 636–48. 10.1007/s00018-008-8516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merali S.; Clarkson A. B. Jr. S-adenosylmethionine and Pneumocystis. FEMS Microbiol. Lett. 2004, 237 (2), 179–86. 10.1111/j.1574-6968.2004.tb09694.x. [DOI] [PubMed] [Google Scholar]
- Lu S. C.; Mato J. M. S-adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 2012, 92 (4), 1515–42. 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola R. E.; Zhao M.; Blumenthal R. M.; Wijayasinghe Y. S.; Bhansali P. A surprising range of modified-methionyl S-adenosylmethionine analogues support bacterial growth. Microbiology 2015, 161, 674–82. 10.1099/mic.0.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni G. L. The Nature of the Active Methyl Donor Formed Enzymatically from L-Methionine and Adenosinetriphosphate. J. Am. Chem. Soc. 1952, 74 (11), 2942–2943. 10.1021/ja01131a519. [DOI] [Google Scholar]
- Mudd S. H.; Cantoni G. L. Activation of methionine for transmethylation. III. The methionine-activating enzyme of Bakers’ yeast. J. Biol. Chem. 1958, 231 (1), 481–92. 10.1016/S0021-9258(19)77322-0. [DOI] [PubMed] [Google Scholar]
- McQueney M. S.; Anderson K. S.; Markham G. D. Energetics of S-adenosylmethionine synthetase catalysis. Biochemistry 2000, 39 (15), 4443–54. 10.1021/bi992876s. [DOI] [PubMed] [Google Scholar]
- Lu Z. J.; Markham G. D. Enzymatic properties of S-adenosylmethionine synthetase from the archaeon Methanococcus jannaschii. J. Biol. Chem. 2002, 277 (19), 16624–31. 10.1074/jbc.M110456200. [DOI] [PubMed] [Google Scholar]
- Komoto J.; Yamada T.; Takata Y.; Markham G. D.; Takusagawa F. Crystal structure of the S-adenosylmethionine synthetase ternary complex: A novel catalytic mechanism of S-adenosylmethionine synthesis from ATP and Met. Biochemistry 2004, 43 (7), 1821–1831. 10.1021/bi035611t. [DOI] [PubMed] [Google Scholar]
- Markham G. D.; Parkin D. W.; Mentch F.; Schramm V. L. A Kinetic Isotope Effect Study and Transition-State Analysis of the S-Adenosylmethionine Synthetase Reaction. J. Biol. Chem. 1987, 262 (12), 5609–5615. 10.1016/S0021-9258(18)45617-7. [DOI] [PubMed] [Google Scholar]
- Parry R. J.; Minta A. Studies of enzyme stereochemistry. Elucidation of the stereochemistry of S-adenosylmethionine formation by yeast methionine adenosyltransferase. J. Am. Chem. Soc. 1982, 104 (3), 871–872. 10.1021/ja00367a048. [DOI] [Google Scholar]
- Kotb M.; Kredich N. M. S-Adenosylmethionine synthetase from human lymphocytes. Purification and characterization. J. Biol. Chem. 1985, 260 (7), 3923–30. 10.1016/S0021-9258(18)89210-9. [DOI] [PubMed] [Google Scholar]
- Halim A. B.; LeGros L.; Geller A.; Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J. Biol. Chem. 1999, 274 (42), 29720–5. 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- Takusagawa F.; Kamitori S.; Misaki S.; Markham G. D. Crystal structure of S-adenosylmethionine synthetase. J. Biol. Chem. 1996, 271 (1), 136–147. 10.1074/jbc.271.1.136. [DOI] [PubMed] [Google Scholar]
- Murray B.; Antonyuk S. V.; Marina A.; Lu S. C.; Mato J. M.; Hasnain S. S.; Rojas A. L. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (8), 2104–9. 10.1073/pnas.1510959113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham G. D.; Hafner E. W.; Tabor C. W.; Tabor H. S-Adenosylmethionine synthetase from Escherichia coli. J. Biol. Chem. 1980, 255 (19), 9082–92. 10.1016/S0021-9258(19)70530-4. [DOI] [PubMed] [Google Scholar]
- Liao Q.; Kulkarni Y.; Sengupta U.; Petrovic D.; Mulholland A. J.; van der Kamp M. W.; Strodel B.; Kamerlin S. C. L. Loop Motion in Triosephosphate Isomerase Is Not a Simple Open and Shut Case. J. Am. Chem. Soc. 2018, 140 (46), 15889–15903. 10.1021/jacs.8b09378. [DOI] [PubMed] [Google Scholar]
- Wilson J. E.; Chin A. Chelation of divalent cations by ATP, studied by titration calorimetry. Anal. Biochem. 1991, 193 (1), 16–9. 10.1016/0003-2697(91)90036-S. [DOI] [PubMed] [Google Scholar]
- Zea C. J.; Camci-Unal G.; Pohl N. L. Thermodynamics of binding of divalent magnesium and manganese to uridine phosphates: implications for diabetes-related hypomagnesaemia and carbohydrate biocatalysis. Chem. Cent. J. 2008, 2, 15. 10.1186/1752-153X-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut T. W. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994, 140 (1), 1–22. 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- Buckstein M. H.; He J.; Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 2008, 190 (2), 718–26. 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J.; Mao Z.; Hwang J. J.; Lu S. C. Differential expression of methionine adenosyltransferase genes influences the rate of growth of human hepatocellular carcinoma cells. Cancer Res. 1998, 58 (7), 1444–50. [PubMed] [Google Scholar]
- Chang C. F.; Liau M. C.; Becker F. F. Alteration of S-Adenosylmethionine Synthetases during Chemical Hepatocarcinogenesis and in Resulting Carcinomas. P Am. Assoc Canc Res. 1979, 20 (Mar), 13–13. [PubMed] [Google Scholar]
- Paneda C.; Gorospe I.; Herrera B.; Nakamura T.; Fabregat I.; Varela-Nieto I. Liver cell proliferation requires methionine adenosyltransferase 2A mRNA up-regulation. Hepatology 2002, 35 (6), 1381–91. 10.1053/jhep.2002.32538. [DOI] [PubMed] [Google Scholar]
- Avila M. A.; Berasain C.; Torres L.; Martin-Duce A.; Corrales F. J.; Yang H.; Prieto J.; Lu S. C.; Caballeria J.; Rodes J.; Mato J. M. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J. Hepatol. 2000, 33 (6), 907–14. 10.1016/S0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Wang R.; Zheng W.; Luo M. A sensitive mass spectrum assay to characterize engineered methionine adenosyltransferases with S-alkyl methionine analogues as substrates. Anal. Biochem. 2014, 450, 11–9. 10.1016/j.ab.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q.; Jiang F.; Schultz P. G.; Gray N. S. Design of allele-specific protein methyltransferase inhibitors. J. Am. Chem. Soc. 2001, 123 (47), 11608–13. 10.1021/ja011423j. [DOI] [PubMed] [Google Scholar]
- Bar-Even A.; Milo R.; Noor E.; Tawfik D. S. The Moderately Efficient Enzyme: Futile Encounters and Enzyme Floppiness. Biochemistry 2015, 54 (32), 4969–77. 10.1021/acs.biochem.5b00621. [DOI] [PubMed] [Google Scholar]
- Copley S. D. An evolutionary biochemist’s perspective on promiscuity. Trends Biochem. Sci. 2015, 40 (2), 72–8. 10.1016/j.tibs.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin S. Y.; Copley S. D. A compromise required by gene sharing enables survival: Implications for evolution of new enzyme activities. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (36), 13497–502. 10.1073/pnas.0804804105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal A.; Yu McLoughlin S.; Kershner J. P.; Copley S. D. Differential effects of a mutation on the normal and promiscuous activities of orthologs: implications for natural and directed evolution. Mol. Biol. Evol. 2015, 32 (1), 100–8. 10.1093/molbev/msu271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan S. K.; Sierks M. R. Minimizing nonproductive substrate binding: a new look at glucoamylase subsite affinities. Biochemistry 1997, 36 (48), 14946–55. 10.1021/bi970820x. [DOI] [PubMed] [Google Scholar]
- Agarwal P. K. Role of protein dynamics in reaction rate enhancement by enzymes. J. Am. Chem. Soc. 2005, 127 (43), 15248–56. 10.1021/ja055251s. [DOI] [PubMed] [Google Scholar]
- Tousignant A.; Pelletier J. N. Protein motions promote catalysis. Chem. Biol. 2004, 11 (8), 1037–42. 10.1016/j.chembiol.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Cornelissen N. V.; Michailidou F.; Muttach F.; Rau K.; Rentmeister A. Nucleoside-modified AdoMet analogues for differential methyltransferase targeting. Chem. Commun. (Cambridge, U. K.) 2020, 56 (14), 2115–2118. 10.1039/C9CC07807J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean I. J. W.; Sadler J. C.; Cuetos A.; Frese A.; Humphreys L. D.; Grogan G.; Hoskisson P. A.; Burley G. A. S-Adenosyl Methionine Cofactor Modifications Enhance the Biocatalytic Repertoire of Small Molecule C-Alkylation. Angew. Chem., Int. Ed. 2019, 58 (49), 17583–17588. 10.1002/anie.201908681. [DOI] [PubMed] [Google Scholar]
- Singh S.; Zhang J.; Huber T. D.; Sunkara M.; Hurley K.; Goff R. D.; Wang G.; Zhang W.; Liu C.; Rohr J.; Van Lanen S. G.; Morris A. J.; Thorson J. S. Facile chemoenzymatic strategies for the synthesis and utilization of S-adenosyl-(L)-methionine analogues. Angew. Chem., Int. Ed. 2014, 53 (15), 3965–9. 10.1002/anie.201308272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips T. M.; McPhillips S. E.; Chiu H. J.; Cohen A. E.; Deacon A. M.; Ellis P. J.; Garman E.; Gonzalez A.; Sauter N. K.; Phizackerley R. P.; Soltis S. M.; Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 2002, 9 (6), 401–6. 10.1107/S0909049502015170. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66 (2), 125–32. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. R.; Murshudov G. N. How good are my data and what is the resolution?. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69 (7), 1204–14. 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40 (4), 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66 (4), 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P. V.; Grosse-Kunstleve R. W.; Echols N.; Headd J. J.; Moriarty N. W.; Mustyakimov M.; Terwilliger T. C.; Urzhumtsev A.; Zwart P. H.; Adams P. D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68 (4), 352–67. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Huang J.; Rauscher S.; Nawrocki G.; Ran T.; Feig M.; de Groot B. L.; Grubmuller H.; MacKerell A. D. Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14 (1), 71–73. 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J. C.; Fraaije J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18 (12), 1463–1472. . [DOI] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics 1996, 14 (1), 33–8. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Parks L. W.; Schlenk F. The stability and hydrolysis of S-adenosylmethionine; isolation of S-ribosylmethionine. J. Biol. Chem. 1958, 230 (1), 295–305. 10.1016/S0021-9258(18)70564-4. [DOI] [PubMed] [Google Scholar]
- Iwig D. F.; Booker S. J. Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-L-methionine. Biochemistry 2004, 43 (42), 13496–509. 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.