Abstract
Infraspecific variation of the ITS rDNA region of some brown Parmeliae occurring in Poland is studied and compared with non-molecular characters. Haplotype networks are used to illustrate the variability within the species. Both newly-produced sequences from Central Europe and from all over the world, downloaded from the GenBank, are used.
The number of haplotypes found for each taxon ranged from five in Melaneliastygia to 12 in Melaneliahepatizon and Montaneliadisjuncta; however, their numbers correlate with the number of specimens tested. New haplotypes for Melaneliaagnata, M.hepatizon and Cetrariacommixta are found. Based on our 169-sample dataset, we could not infer any geographical correlation, either locally or world-wide. Many of the analysed haplotypes were widely distributed and the same haplotype was often shared between temperate and polar populations. A comparison of molecular, morphological, anatomical and chemical characters also shows no correlation.
Keywords: Cryptic species, haplotype, lichenised fungi, Parmeliaceae, phylogeny, taxonomy
Introduction
The brown Parmeliae (Esslinger 1977) have been an object of numerous studies (Guzow-Krzemińska and Węgrzyn 2003; Blanco et al. 2005; Crespo et al. 2010, 2011; Nelsen et al. 2011; Divakar et al. 2012; Thell et al. 2012; Leavitt et al. 2014, 2015) and, due to this exceptional attention, they are one of the best-studied assemblages in the family Parmeliaceae. These lichens are a polyphyletic group possessing foliose, a dark to medium brown thallus and usually lacking atranorin or usnic acid in the cortex (Esslinger 1977; Blanco et al. 2004).
For many years, one of the largest genera within this group was Melanelia Essl., segregated from Parmelia Ach. by Esslinger (1978) to accommodate species with brown, foliose thalli and an N– cortex layer. However, during the following years, it has been demonstrated that the genus Melanelia s. lat. was polyphyletic and several new genera were distinguished within it, such as Melanelixia O. Blanco et al., Melanohalea O. Blanco et al. (Blanco et al. 2004) and Montanelia Divakar et al. (Divakar et al. 2012). In traditional terms, brown Parmeliae includes other genera, such as Allantoparmelia (Vain.) Essl., Pleurosticta Petr. and some species of Xanthoparmelia (Vain.) Hale. Moreover, due to the historical taxonomic approach (Thell 1995; Rico et al. 2005) and the similarity in the morphological and anatomical features of thalli, Cetrariacommixta is also referred to this group.
Our studies have focused on the saxicolous species of Melanelia and Montanelia genera. According to Otte et al. (2005), species of these genera are arctic-alpine, circumpolar and occur on silicate rocks in the mountain areas of the Northern Hemisphere, including Arctic Regions (Divakar et al. 2012). Nowadays, Melanelia s. str. is restricted to a small clade of saxicolous, cetrarioid lichens and includes four species: M.agnata (Nyl.) A. Thell, M.hepatizon (Ach.) A. Thell, M.pseudoglabra (Essl.) Essl. and M.stygia (L.) Essl. According to Thell (1995), these species are characterised by broadly clavate asci with a small tholus and a broad axial body, a thick, paraplectenchymatous cortex and dumb-bell-shaped pycnoconidia. Montanelia, representing the parmelioid clade, includes eight species: M.disjuncta (Erichsen) Divakar, A. Crespo, Wedin & Essl., M.occultipanniformis S.D. Leav., Essl., Divakar, A. Crespo & Lumbsch, M.panniformis (Nyl.) Divakar, A. Crespo, Wedin & Essl., M.predisjuncta (Essl.) Divakar, A. Crespo, Wedin & Essl., M.saximontana (R.A. Anderson & W.A. Weber) S.D. Leav., Essl., Divakar, A. Crespo & Lumbsch, M.secwepemc S.D. Leav., Essl., Divakar, A. Crespo & Lumbsch, M.sorediata (Ach.) Divakar, A. Crespo, Wedin & Essl. and M.tominii (Oxner) Divakar, A. Crespo, Wedin & Essl. (Divakar et al. 2012; Leavitt et al. 2015; Leavitt et al. 2016). The characteristic features of the Montanelia genus are short and narrow lobes, with flat to convex lobe margins, a non-pored epicortex, cylindrical to fusiform conidia, a medulla containing orcinol depsides and flat, effigurate pseudocyphellae (absent only in M.sorediata; Divakar et al. 2012). Three of these species (M.disjuncta, M.panniformis and M.sorediata) have broad, intercontinental distributions, with no evidence of phylogeographic substructure (Leavitt et al. 2015).
The genera Melanelia and Montanelia have been the subject of a critical revision in Poland and data concerning their distribution, ecology and morphological, anatomical and chemical features are presented in previous papers (Szczepańska et al. 2015; Szczepańska and Kossowska 2017). However, recent molecular studies imply that both genera may include previously unrecognised species-level diversity (Divakar et al. 2012; Leavitt et al. 2014), especially within Icelandic populations of M.stygia (Xu et al. 2017).
One of the goals of this study was to assess the intraspecific internal transcribed spacer (ITS) rDNA variability in brown Parmeliae species. Investigations of genetic variation in lichen-forming symbionts have advanced considerably in recent years and resulted in interesting conclusions (Palice and Printzen 2004; Lindblom and Ekman 2006; Domaschke et al. 2012; Starosta and Svoboda 2020). Although brown Parmeliae appear to be well studied in taxonomic terms, there are insufficient molecular data to estimate their genetic variation. Most of the available data concern samples collected in a few regions of the world, such as Europe and North America. The North American species of this group were studied in Greenland and Canada (Leavitt et al. 2014; Leavitt et al. 2015), while samples from Europe originated mainly from the north – Iceland, Finland, Norway and Sweden (Blanco et al. 2004; Divakar et al. 2012; Xu et al. 2017). Therefore, we decided to fill in the gap in sampling and focused our study on samples collected in Central Europe. We have used phylogenetic trees and haplotype networks to investigate the extent of molecular differences between newly-generated sequences from samples collected in Central Europe (Austria, Czech Republic, Germany, Poland and Slovakia) and others originating from different geographical regions. Due to additional samplings from previously unexplored areas, it was possible to evaluate and compare the genetic variability of the studied specimens in Central Europe with samples from other regions of the world and to identify areas with the greatest haplotype diversity. In addition, we analysed morphological, anatomical and chemical characters of collected specimens to find a potential correlation between phenotypic characters and genetic variation of the studied taxa. By analysing genetic diversity and geographical distribution of identified haplotypes, as well as phenotypic characters of collected samples, we tried to better define and designate the species boundaries within analysed taxa. Special emphasis was placed on analysis of European, Greenlandic and Icelandic samples of M.agnata and M.stygia to revise the hypothesis assuming a semi-cryptic or cryptic nature of their potential species-level diversity.
Materials and methods
Taxon sampling
The study is based on collections from the AMNH, C and WRSL Herbaria, as well as the private material of Dr Maria Kossowska (hb. Kossowska). Our sampling focused on saxicolous representatives of the Parmeliaceae family occurring in Poland, with brown, foliose thalli, such as Cetrariacommixta, Melaneliaagnata, M.hepatizon, M.stygia, Montaneliadisjuncta and M.sorediata. We also included the holotype of Melaneliaagnata (Platysmaagnatum; Austria, Tirol, Gerölle unter dem Gneissfelsen zum wilden see. Auf dem Kraxentrag, Tirol, Brenner 225, Aug 1871, H-NYL 36086), borrowed from Herbarium of W. Nylander in Helsinki in our analyses.
Specimens for molecular study were selected after detailed morphological and chemical analyses. Due to DNA degradation, it was not possible to use samples collected more than three years prior to the DNA extraction procedure in most cases. As the Melaneliaagnata and M.stygia specimens from Greenland and Iceland were collected more than 10 years ago, we had to limit our phylogenetic analyses to ITS rDNA markers and used the sequences stored in GenBank. Before phylogenetic analysis, newly-obtained ITS rDNA sequences were subjected to a BLAST search (Altschul et al. 1997). The final ITS dataset used in this study includes 52 sequences newly generated and 117 sequences downloaded from GenBank (Table 1).
Table 1.
. The species and specimens used in the phylogenetic analyses and/or haplotype network analyses, sequences newly generated for this study are in bold.
| Species | Year of collection | Isolate | Locality | Collector (-s) | Voucher specimens (herbarium) | GenBank no. (ITS) |
|---|---|---|---|---|---|---|
| Cetrariellacommixta | 2007 | 36 | Poland, Sudety Mts | Kossowska, M. | Kossowska 107 (personal herbarium) | MZ029708 |
| Cetrariellacommixta | 2008 | 37 | Poland, Sudety Mts | Kossowska, M. | Kossowska 231 (personal herbarium) | MZ029709 |
| Cetrariellacommixta | 2016 | 97 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1137 (WRSL) | MZ029733 |
| Cetrariellacommixta | 2016 | 124 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1184 (WRSL) | MZ029753 |
| Cetrariellacommixta | 2018 | 129 | Germany, Bayerischer Wald | Szczepańska, K. | Szczepańska 1267 (WRSL) | MZ029758 |
| Cetrariellacommixta | Finland | Haikonen, V. | Haikonen 19093 (H) | AF451796 | ||
| Cetrariellacommixta | 1996 | Canada, British Columbia | Miao, V. & Taylor, T. | AF451797 | ||
| Cetrariellacommixta | Sweden | Wedin, M. | Wedin 8143 (UPS) | GU994554 | ||
| Cetrariellacommixta | Spain, Segovia | Rico, V. J. | 15555 (MAF) | GU994555 | ||
| Cetrariellacommixta | 2004 | CCO 01 | Sweden, Lule Lappmark | 1273926 (LD) | KC990132 | |
| Cetrariellacommixta | 6543 | Greenland, SEm, Tasilaq | Hansen, E. S. | Hansen ESH-10B.139 (C) | KF257934 | |
| Cetrariellacommixta | 6547 | Greenland, SWm, Qeqertaq | Hansen, E. S. | Hansen ESH-09.087 (C) | KF257935 | |
| Cetrariellacommixta | 6567 | Greenland, S, Igaliku | Hansen, E. S. | Hansen ESH-08.173 (C) | KF257936 | |
| Cetrariellacommixta | 6570 | Greenland, SWm, Midgard | Hansen, E. S. | Hansen ES-09.030 (C) | KF257937 | |
| Cetrariellacommixta | 6572 | Greenland, S, Aappilattoq | Hansen, E. S. | Hansen ES-04.070 (C) | KF257938 | |
| Cetrariellacommixta | 6573 | Greenland, SWm, Qeqertaq | Hansen, E. S. | Hansen ES-09.064 (C) | KF257939 | |
| Cetrariellacommixta | 2014 | Norway, Finnmark | Westberg, M. | O-L-195926 | KY266843 | |
| Melaneliaagnata | 2016 | 102 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1151 (WRSL) | MZ029737 |
| Melaneliaagnata | 2016 | 103 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1150 (WRSL) | MZ029738 |
| Melaneliaagnata | 2009 | 6549 | Greenland, SW m, Jensens Nunatakker | Hansen, E. S. | Hansen ESH-09.478 (C) | KF257940 |
| Melaneliaagnata | 2009 | 6553 | Greenland, SW m, Jensens Nunatakker | Hansen, E. S. | Hansen ESH-09.435 (C) | KF257941 |
| Melaneliaagnata | 2007 | 6563 | Greenland, N, Constable Bugt | Hansen, E. S. | Hansen ESH-07.464 (C) | KF257942 |
| Melaneliaagnata | 2002 | MX_MS2 | Iceland, Imi | Heiðmarsson, S. | LA29683 (AMHN) | KY508672 |
| Melaneliaagnata | 2005 | MX_MS3 | Iceland, Ino | Kristinsson, H. | LA27562 (AMHN) | KY963373 |
| Melaneliaagnata | 2008 | MX_MS4 | Iceland, Isu | Hjaltadóttir, A. | LA30974 (AMHN) | KY508673 |
| Melaneliaagnata | 2012 | MX_MS5 | Iceland, Ino | Heiðmarsson, S. | LA31859 (AMHN) | KY963374 |
| Melaneliaagnata | 2014 | Norway, Sor-Trondelag | Timdal, E. | O-L-196376 | MK812394 | |
| Melaneliaculbersonii | USA | Lendemer, J. | Lendemer 13821 (NY) | KR995286 | ||
| Melaneliahepatizon | 2016 | 83 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1051 (WRSL) | MZ029723 |
| Melaneliahepatizon | 2016 | 91 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1120 (WRSL) | MZ029717 |
| Melaneliahepatizon | 2016 | 95 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1136A (WRSL) | MZ029731 |
| Melaneliahepatizon | 2016 | 96 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1136B (WRSL) | MZ029732 |
| Melaneliahepatizon | 2016 | 98 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1138 (WRSL) | MZ029734 |
| Melaneliahepatizon | 2016 | 109 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1153 (WRSL) | MZ029741 |
| Melaneliahepatizon | 2016 | 110 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1154A (WRSL) | MZ029730 |
| Melaneliahepatizon | 2016 | 111 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1154B (WRSL) | MZ029743 |
| Melaneliahepatizon | 2016 | 113 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1144 (WRSL) | MZ029745 |
| Melaneliahepatizon | 2016 | 116 | Slovakia, Karpaty Mts | Szczepańska, K. | Szczepańska 1146 (WRSL) | MZ029746 |
| Melaneliahepatizon | 2016 | 117 | Slovakia, Karpaty Mts | Szczepańska, K. | Szczepańska 1147 (WRSL) | MZ029747 |
| Melaneliahepatizon | 2016 | 119 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1180 (WRSL) | MZ029748 |
| Melaneliahepatizon | 2016 | 122 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1182 (WRSL) | MZ029751 |
| Melaneliahepatizon | 2018 | 128 | Germany, Bayerischer Wald | Szczepańska, K. | Szczepańska 1269 (WRSL) | MZ029757 |
| Melaneliahepatizon | 1996 | Canada, British Columbia | Thell & Veer BC-9677 (LD) | AF141369 | ||
| Melaneliahepatizon | 2001 | DNA-AT934 | Italy, Trentino-Alto Adige (south Tirolia) | Feuerer T. & Thell A. s. n. | LD, HBG | AF451776 |
| Melaneliahepatizon | Sweden | Wedin, M. | Wedin 6812 (UPS) | DQ980016 | ||
| Melaneliahepatizon | Greenland, NWn, Siorapuluk | Hansen, E. S. | Hansen ESH-09B.164 (C) | KF257943 | ||
| Melaneliahepatizon | Greenland, NWn, Qaanaaq | Hansen, E. S. | Hansen ESH-09B.026 (C) | KF257944 | ||
| Melaneliahepatizon | Greenland, SEm, Tasilaq | Hansen, E. S. | Hansen ESH-10B.014 (C) | KF257945 | ||
| Melaneliahepatizon | Greenland, SWm, Nuuq | Hansen, E. S. | Hansen ESH-10A.019 (C) | KF257946 | ||
| Melaneliahepatizon | Greenland, S, Qaqortoq | Hansen, E. S. | Hansen ESH-08.036 (C) | KF257947 | ||
| Melaneliahepatizon | Greenland, S, Igaliku | Hansen, E. S. | Hansen ESH-08.170 (C) | KF257948 | ||
| Melaneliahepatizon | Greenland, S, Narssarsuag | Hansen, E. S. | Hansen ESH-08.263 (C) | KF257949 | ||
| Melaneliahepatizon | Greenland, S, Igaliku | Hansen, E. S. | Hansen ESH-08.215 (C) | KF257950 | ||
| Melaneliahepatizon | Greenland, SWm, Midgard | Hansen, E. S. | Hansen ESH-09.386 (C) | KF257951 | ||
| Melaneliahepatizon | Greenland, SWm, Frederikshab Isblink | Hansen, E. S. | Hansen ESH-09.324 (C) | KF257952 | ||
| Melaneliahepatizon | Greenland, S, Igaliku | Hansen, E. S. | Hansen ESH-08.477 (C) | KF257953 | ||
| Melaneliahepatizon | 2014 | Norway, Finnmark | Westberg, M. | O-L-195864 | KY266879 | |
| Melaneliahepatizon | 2003 | MH1 | Iceland, IAu | LA30501 (AMHN) | KY508674 | |
| Melaneliahepatizon | 2007 | MH3 | Iceland, IVe | LA30676 (AMHN) | KY508675 | |
| Melaneliahepatizon | 2007 | MH4 | Iceland, IVe | LA30674 (AMHN) | KY508676 | |
| Melaneliahepatizon | 2007 | MH5 | Iceland, IVe | LA30675 (AMHN) | KY508677 | |
| Melaneliahepatizon | 2007 | MH6 | Iceland, IVe | LA30673 (AMHN) | KY508678 | |
| Melaneliahepatizon | 2014 | MH9 | Iceland, INo | LA20781 (AMHN) | KY508679 | |
| Melaneliahepatizon | 2013 | MH10 | Iceland, INv | LA30117 (AMHN) | KY508680 | |
| Melaneliahepatizon | 2012 | MH11 | Iceland, Inv | LA31861 (AMHN) | KY963376 | |
| Melaneliahepatizon | 2014 | Norway, Hordaland | Timdal, E. | O-L-195807 | MK812512 | |
| Melaneliahepatizon | 2015 | Norway, Nord-Trondelag | Bendiksby, M. et al. | O-L-201254 | MK812070 | |
| Melaneliahepatizon | 2013 | Norway, Buskerud | Rui, S. & Timdal, E. | O-L-184723 | MK812188 | |
| Melaneliastygia | 2007 | 40 | Poland, Sudety Mts | Kossowska, M. | Kossowska 123 (personal herbarium) | MZ029710 |
| Melaneliastygia | 2009 | 42 | Austria, Tyrol | Szczepańska, K. | Szczepańska 737 (WRSL) | MZ029712 |
| Melaneliastygia | 2016 | 94 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1134 (WRSL) | MZ029719 |
| Melaneliastygia | 2016 | 104 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1152 (WRSL) | MZ029739 |
| Melaneliastygia | 2016 | 108 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1149 (WRSL) | MZ029740 |
| Melaneliastygia | 2016 | 112 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1160 (WRSL) | MZ029744 |
| Melaneliastygia | 2018 | 127 | Czech Republic, Šumava | Szczepańska, K. | Szczepańska 1265 (WRSL) | MZ029756 |
| Melaneliastygia | Finland, Nyland | Kuusinen, M. | FIN-9714 (LD) | AF115763 | ||
| Melaneliastygia | Italy | Feurerer, T & Thell, A. | DNA-AT922 (LD) | AF451775 | ||
| Melaneliastygia | Finland, Enonkoski | Haikonen, V. | Haikonen 20365 | AY611097 | ||
| Melaneliastygia | Austria, Steiermark | Hafellner, J. | Hafellner 51658 | AY611121 | ||
| Melaneliastygia | 2008 | 6551 | Greenland, S, Qaqortoq | Hansen, E. S. | Hansen ESH-08.036 (C) | KF257954 |
| Melaneliastygia | 2008 | 6569 | Greenland, S, Igaliku | Hansen, E. S. | Hansen ESH-08.478 (C) | KF257955 |
| Melaneliastygia | 1998 | MX_MS1 | Iceland, IAu | Kristinsson, H. | LA19972 (AMHN) | KY508681 |
| Melaneliastygia | 2014 | MX_MS3 | Iceland, IAu | Kristinsson, H. | LA20775 (AMHN) | KY508682 |
| Melaneliastygia | 2013 | MX_MS4 | Iceland, IAu | Kristinsson, H. | LA16894 (AMHN) | KY508683 |
| Melaneliastygia | 2000 | MX_MS2 | Iceland, IAu | Kristinsson, H. | LA28243 (AMHN) | KY963375 |
| Melaneliastygia | 2013 | Norway, Buskerud | Rui, S. & Timdal, E. | O-L-184736 | MK812608 | |
| Melaneliastygia | 2014 | Norway, Sor-Trondelag | Timdal, E. | O-L-196377 | MK812312 | |
| Montaneliadisjuncta | 2013 | 50 | Poland, Sudsty Forelands | Szczepańska, K. | Szczepańska 969 (WRSL) | MZ029713 |
| Montaneliadisjuncta | 2014 | 51 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 989 (WRSL) | MZ029714 |
| Montaneliadisjuncta | 2015 | 57 | Poland, Sudety Foothills | Szczepańska, K. | Szczepańska 1023 (WRSL) | MZ029715 |
| Montaneliadisjuncta | 2015 | 78 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1034 (WRSL) | MZ029716 |
| Montaneliadisjuncta | 2015 | 79 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1038 (WRSL) | MZ029711 |
| Montaneliadisjuncta | 2015 | 80 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1039 (WRSL) | MZ029720 |
| Montaneliadisjuncta | 2016 | 81 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1047 (WRSL) | MZ029721 |
| Montaneliadisjuncta | 2016 | 82 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1048 (WRSL) | MZ029722 |
| Montaneliadisjuncta | 2016 | 85 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1054 (WRSL) | MZ029724 |
| Montaneliadisjuncta | 2016 | 86 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1081 (WRSL) | MZ029725 |
| Montaneliadisjuncta | 2016 | 87 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1082 (WRSL) | MZ029726 |
| Montaneliadisjuncta | 2016 | 88 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1110 (WRSL) | MZ029727 |
| Montaneliadisjuncta | 2016 | 89 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1111 (WRSL) | MZ029728 |
| Montaneliadisjuncta | 2016 | 90 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1119 (WRSL) | MZ029729 |
| Montaneliadisjuncta | 2016 | 92 | Pland, Sudety Foothils | Szczepańska, K. | Szczepańska 1127 (WRSL) | MZ029755 |
| Montaneliadisjuncta | 2016 | 93 | Pland, Sudety Foothils | Szczepańska, K. | Szczepańska 1128 (WRSL) | MZ029718 |
| Montaneliadisjuncta | 2016 | 120 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1181A (WRSL) | MZ029749 |
| Montaneliadisjuncta | 2016 | 121 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1181B (WRSL) | MZ029750 |
| Montaneliadisjuncta | 2016 | 123 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1183 (WRSL) | MZ029752 |
| Montaneliadisjuncta | 2016 | 125 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1185 (WRSL) | MZ029754 |
| Montaneliadisjuncta | 2016 | 126 | Poland, Sudety Mts | Szczepańska, K. | Szczepańska 1230 (WRSL) | MZ029742 |
| Montaneliadisjuncta | 2018 | 130 | Czech Republic, Šumava | Szczepańska, K. | Szczepańska 1271 (WRSL) | MZ029759 |
| Montaneliadisjuncta | Austria, Steiermark | Mayrhofer 13743 | AY611077 | |||
| Montaneliadisjuncta | India | MAF-Lich 15512 | GU994556 | |||
| Montaneliadisjuncta | United Kingdom | Coppins 637 | JX974654 | |||
| Montaneliadisjuncta | Greenland, NWn, Siorapaluk | Hansen, E. S. | Hansen ESH-09B.363 (C) | KF257957 | ||
| Montaneliadisjuncta | 3921 | Canada, Yukon Territory | Spribille, T. | Spribille s.n. | KP771824 | |
| Montaneliadisjuncta | 3963 | Greenland, Northwest | Hansen, E. S. | Hansen ESH-09B.051 (C) | KP771827 | |
| Montaneliadisjuncta | 3995 | USA, Maine | Harris, R. | Harris 52938 (NY) | KP771828 | |
| Montaneliadisjuncta | 4503 | Norway, Tromso | Bjerke, J.W. | Bjerke WP286-2 (TLE) | KP771829 | |
| Montaneliadisjuncta | 4851 | Canada, Yukon Territory | Esslinger, T. L. | Esslinger BP94-3 (TLE) | KP771830 | |
| Montaneliadisjuncta | 5970 | USA, Alaska | Esslinger, T. L. | Esslinger 19403 (TLE) | KP771831 | |
| Montaneliadisjuncta | 6575 | Greenland, Northwest, Siorapaluk | Hansen, E. S. | Hansen ESH-09B.323 (C) | KP771833 | |
| Montaneliadisjuncta | MDISJUNCT | Sweden, Lycksele Lappmark | Wedin, M. | Wedin 7143 (UPS) | KP771834 | |
| Montaneliadisjuncta | MEDI637 | United Kingdom, Scotland | Coppins, B. | Coppins s.n (MAF) | KP771835 | |
| Montaneliadisjuncta | MESO773 | India, Uttaranchal | Divakar, P. K. | MAF-Lich 15512 | KP771837 | |
| Montaneliadisjuncta | 2014 | Norway, Finnmark, Vadso | Haugan, R. | O-L-198675 | KY266910 | |
| Montaneliadisjuncta | 2007 | MD8 | Iceland, INo | LA30657 (AMHN) | KY508686 | |
| Montaneliadisjuncta | Sweden | Wedin, M. | Wedin 7143 (UPS) | DQ980015 | ||
| Montaneliadisjuncta | USA | Lumbsch, H. T. | Lumbsch 2010/M7 (F) | JX126181 | ||
| Montaneliadisjuncta | USA, Maine | Harris 55589 (NY) | KF257960 | |||
| Montaneliadisjuncta | USA, Alaska | Esslinger 19403 (TLE) | KF257968 | |||
| Montaneliadisjuncta | Canada | Goward 08 | JX974658 | |||
| Montaneliadisjuncta | Canada, Yukon | Spribille s.n. (GZU) | KF257956 | |||
| Montaneliadisjuncta | Canada, Alberta | Holzinger 1061 (UBC) | KF257962 | |||
| Montaneliadisjuncta | Canada, British Columbia | Esslinger BP109-1 (TLE) | KF257964 | |||
| Montaneliadisjuncta | Canada, British Columbia | Esslinger BP97-01 (TLE) | KF257965 | |||
| Montaneliadisjuncta | Canada, Yukon | Esslinger BP94-2 (TLE) | KF257966 | |||
| Montaneliadisjuncta | Canada, Yukon | Esslinger BP94-3 (TLE) | KF257967 | |||
| Montaneliadisjuncta | Canada, New Brunswick | McMullin 7483 (TLE) | KF257969 | |||
| Montaneliadisjuncta | Canada, British Columbia | Goward 2008 (MAF) | KP771836 | |||
| Montaneliadisjuncta | Greenland, S, Igaliku | Hansen, E. S. | Hansen ESH-08.304 (C) | KF257958 | ||
| Montaneliadisjuncta | Greenland, NWn, Qaanaaq | Hansen, E. S. | Hansen ESH-09B.051 (C) | KF257959 | ||
| Montaneliadisjuncta | Greenland, S, Igaliku | Hansen, E. S. | Hansen ESH-08.216 (C) | KF257970 | ||
| Montaneliadisjuncta | Greenland, NWn, Siorapuluk | Hansen, E. S. | Hansen ESH-09B.323 (C) | KF257971 | ||
| Montaneliadisjuncta | 3956 | Greenland, Northwest | Hansen, E. S. | Hansen ESH-09B.363 (C) | KP771825 | |
| Montaneliadisjuncta | 3957 | Greenland, South | Hansen, E. S. | Hansen ESH-08.304 (C) | KP771826 | |
| Montaneliadisjuncta | 6574 | Greenland, South, Igaliku | Hansen, E. S. | Hansen ESH-08.216 (C) | KP771832 | |
| Montaneliadisjuncta | Norway, Tromso | Bjerke WP286-2 (TLE) | KF257961 | |||
| Montaneliadisjuncta | India, Uttar Pradesh | Divakar 15512 (MAF-Lich) | KF257972 | |||
| Montaneliadisjuncta | 2000 | MD2 | Iceland, Iau | LA28245 (AMHN) | KY963377 | |
| Montaneliadisjuncta | 2009 | MD5 | Iceland, Ino | LA31552 (AMHN) | KY963378 | |
| Montaneliadisjuncta | 2007 | MD3 | Iceland, Ino | LA30617 (AMHN) | KY508684 | |
| Montaneliadisjuncta | Canada, British Columbia | Goward 10-19 (UBC) | KF257963 | |||
| Montaneliadisjuncta | 2014 | Norway, Sor-Trondelag | Timdal, E. | O-L-196357 | MK811711 | |
| Montaneliadisjuncta | 2014 | Norway, Finnmark | Timdal, E. | O-L-195590 | MK811852 | |
| Montaneliadisjuncta | 2006 | MD4 | Iceland, Ino | LA27588 | KY508685 | |
| Montaneliasorediata | 2016 | 100 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1156 (WRSL) | MZ029735 |
| Montaneliasorediata | 2016 | 101 | Poland, Karpaty Mts | Szczepańska, K. | Szczepańska 1155 (WRSL) | MZ029736 |
| Montaneliasorediata | 4001 | USA, Pennsylvania | Lendemer, J. | Lendemer 13329 (NY) | KF257978 | |
| Montaneliasorediata | 4824 | Canada, British Columbia | Esslinger, T.L. | Esslinger BP111-1 (TLE) | KF257979 | |
| Montaneliasorediata | 4884 | USA, Alaska | Esslinger, T.L. | Esslinger BP73-6 (TLE) | KF257980 | |
| Montaneliasorediata | 5981 | Russia, Khabarovskiy Krai | Spribille, T. | Spribille 31972 (GZU) | KF257981 | |
| Montaneliasorediata | 6380 | Canada, Ontario | McMullin, T. | McMullin 8139 (TLE) | KF257982 | |
| Montaneliasorediata | B_8600 | Japan, Mt. Ohyama | Ohmura, Y. | Ohmura 9666 (TNS) | KM386101 | |
| Montaneliasorediata | MESO778 | Sweden, Vasterbotten | Wedin, M. | Wedin 6862 (UPS) | KP771845 | |
| Montaneliasorediata | 4001 | USA, Pennsylvania | Lendemer, J. | Lendemer 13329 (NY) | KP771846 | |
| Montaneliasorediata | 5981 | Russia, Khabarovskiy Krai | Spribille, T. | Spribille 31972 (GZU) | KP771847 | |
| Montaneliasorediata | 2014 | Norway, Telemark | Timdal, E. | O-L-195791 | MK811963 | |
| Montaneliasorediata | 2014 | Norway, Troms | Timdal, E. | O-L-195658 | MK811965 | |
| Montaneliasorediata | 2016 | Norway, Buskerud | Dahl, M. S., Kistenich, S. D., Timdal, E., Toreskaas, A. K. | O-L-204941 | MK811977 | |
| Montaneliasorediata | C_4670 | Canada, British Columbia | Bjork, C. | Bjork 15153 (UBC) | KM386102 |
Morphology and chemistry
The morphology and anatomy of the specimens were studied in detail with dissecting and light microscopes, following routine techniques. All specimens were examined for the assessment of the morphological characters, such as lobe width and morphology (flat/convex), the appearance of the upper surface (dull/glossy), the appearance of the lower surface (light/dark), apothecia morphology (sessile/constricted), appearance and position of pycnidia (marginal/laminal), appearance and position of the pseudocyphellae (marginal/laminal), size and shape of conidia (bacilliform/bifusiform), as well as ascospore size. For light microscopy, vertical sections of apothecia were cut by hand using a razor blade and mounted in water. Hymenium and conidia measurements were made in water and ascospore measurements were made in 10% potassium hydroxide (KOH). At least ten measurements of morphological variables and measurements of 20 spores and conidia were made for each sample and their minimum and maximum values were calculated.
The TLC analyses were undertaken in A and C solvent systems using the standardised method of Culberson (1972) and following Orange et al. (2001).
DNA extraction, PCR amplification and DNA sequencing
Genomic DNA was extracted from specimens after cell disruption in a Mixer Mill MM400 (Retsch, Haan, Germany) using a CTAB method according to the standard protocol of isolation (Doyle and Doyle 1987). The quality of the isolated DNA was determined using 1% TBE agarose electrophoresis. PCR reactions were performed in 20 μl reaction tubes that contained a Dream Taq reaction buffer containing MgCl2, a 0.2 mM dNTP mix, 1u DreamTaq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA), 0.5 mM each ITS1 and ITS4 primers and 0.8 μl of total genomic DNA. The adequate annealing temperature was determined using the gradient method. The PCR programme consisted of an initial denaturation at 95 °C for 6 min, according to a previous study (Szczepańska et al. 2020), followed by 30 cycles at 95 °C for 30 sec, 51.2 °C for 45 sec, 72 °C for 45 sec, with a final extension at 72 °C for 10 min. While performing PCR, the Veriti Thermal Cycler (Life Technologies, Carlsbad, CA, USA) was used. Amplification products were separated in 1% agarose gel, photographed and compared with the DNA mass ruler (Thermo Fisher Scientific Waltham, MA, USA). Bands corresponding to the ITS region were excised from the agarose gel and then purified by ethanol precipitation. Cleaned samples were sent to a sequencing service (Genomed, Warszawa, Poland). All laboratory analyses were performed at the Department of Botany and Plant Ecology at the Wrocław University of Environmental and Life Sciences.
Sequence alignment and phylogenetic analysis
The newly-generated sequences and selected representatives of brown saxicolous Parmeliaceae were aligned using the Guidance 2 server (Landan and Graur 2008; Penn et al. 2010; Sela et al. 2015) employing the MAFFT algorithm (Katoh et al. 2002) followed by elimination of terminal ends. The final alignment consisted of 117 sequences of 535 sites. Further, we used Partition Finder 2 (Lanfear et al. 2016) implemented at the CIPRES Science Gateway (Miller et al. 2010). Two different models were found for partitions: GTR+G for ITS1 and ITS2 and K80+G for the 18S and 5.8S regions.
Moreover, phylogenetic analysis of all Melanelia sequences was also performed. Newly-generated sequences and these downloaded from GenBank, together with representatives of Cetrariacommixta, which were further used as an outgroup, were aligned using the Guidance 2 server (Landan and Graur 2008; Penn et al. 2010; Sela et al. 2015) employing the MAFFT algorithm (Katoh et al. 2002) followed by elimination of unreliable columns. The final alignment consisted of 76 sequences of 803 sites. Further, we used jModeltest 2.1 (Darriba et al. 2012) implemented at the CIPRES Science Gateway (Miller et al. 2010) and the K80+G model was selected.
Bayesian analysis was carried out using a Markov Chain Monte Carlo (MCMC) method, in MrBayes v. 3.2.6 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) on the CIPRES Web Portal (Miller et al. 2010) using best models. Two parallel MCMC runs were performed, each using four independent chains and four million generations, sampling every 1000th tree. Posterior probabilities (PP) were determined by calculating a majority-rule consensus tree after discarding the initial 25% trees of each chain as the burn-in.
A Maximum Likelihood (ML) analysis was performed using RAxML-HPC2 v.8.2.10 (Stamatakis 2014) with 1000 ML bootstrap iterations (BS) and the GTRGAMMAI model for both analyses. Phylogenetic trees were visualised using FigTree v. 1.4.2 (Rambaut 2012) and modified in Inkscape (https://inkscape.org/).
Haplotype networks
Newly-generated sequences of the ITS rDNA marker, together with sequences downloaded from GenBank from specimens of Cetrariacommixta, Melaneliaagnata, M.hepatizon, M.stygia, Montaneliadisjuncta and M.sorediata, were aligned separately for each species using Seaview software (Galtier et al. 1996; Gouy et al. 2010). TCS networks (Clement et al. 2002) were created as implemented in PopART software (http://popart.otago.ac.nz). Nucleotide diversity per site was calculated using DnaSP v.6 software (Rozas et al. 2017).
Results
Phylogeny and haplotype networks
A total of 169 sequences were analysed in this study.
The RAxML tree did not contradict the Bayesian trees topologies for the strongly-supported branches and only the latter is shown with posterior probabilities. The bootstrap support values BS ≥ 70 and PP ≥ 0.95 were considered to be significant and are shown near the branches. In Fig. S1, three main, highly supported lineages representing Melanelia spp. (i.e. M.agnata, M.hepatizon and M.stygia), Montanelia spp. (i.e. M.disjuncta and M.sorediata) and Cetrariacommixta were distinguished. The newly-sequenced specimens clustered together with other representatives of the species downloaded from GenBank. Amongst them, Melaneliastygia is not monophyletic, but forms two separate well-supported clades.
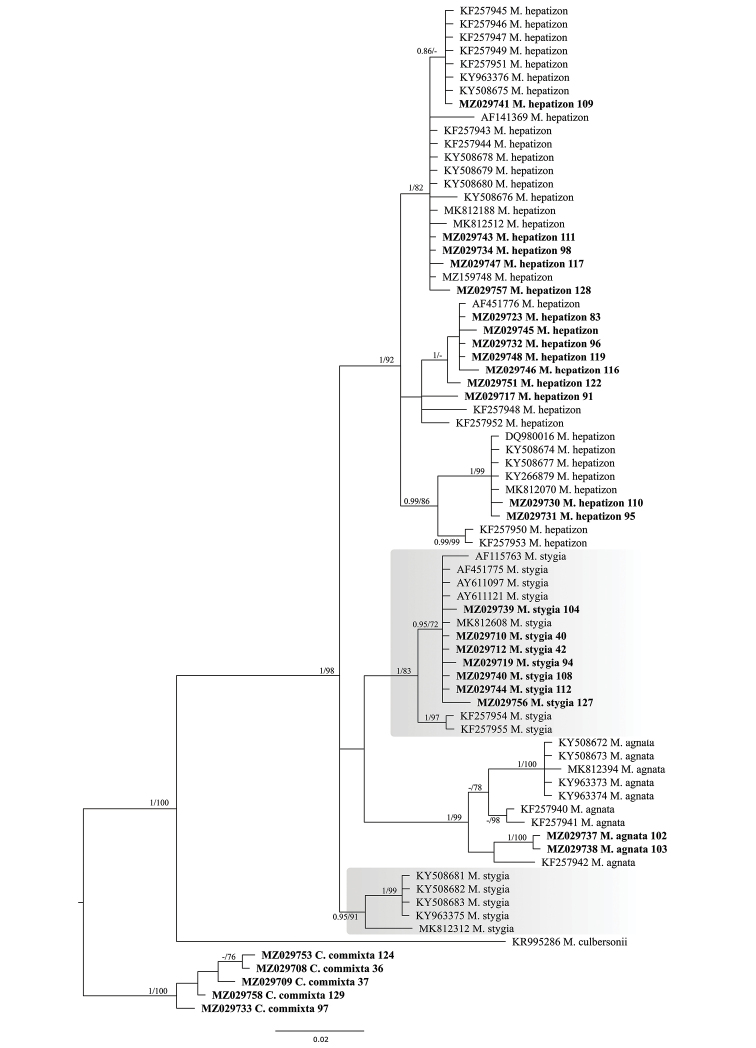
Moreover, to better understand phylogenetic relationships in the Melanelia, we performed additional analysis for all available ITS rDNA sequences from representatives of this genus. The Bayesian tree is presented in Fig. 1 with posterior probabilities and the bootstrap support values presented near the branches and with Cetrariacommixta as an outgroup. In this tree, Melaneliastygia also forms two separate, highly-supported clades.
Figure 1.
Phylogenetic relationships of Melanelia spp., based on Bayesian analysis of the ITS rDNA dataset. Posterior probabilities and Maximum Likelihood bootstrap values are shown near the internal branches. Newly-generated sequences are additionally described with isolate numbers following the species names and are marked in bold. GenBank accession numbers of sequences downloaded from GenBank are listed on the tree with species names.
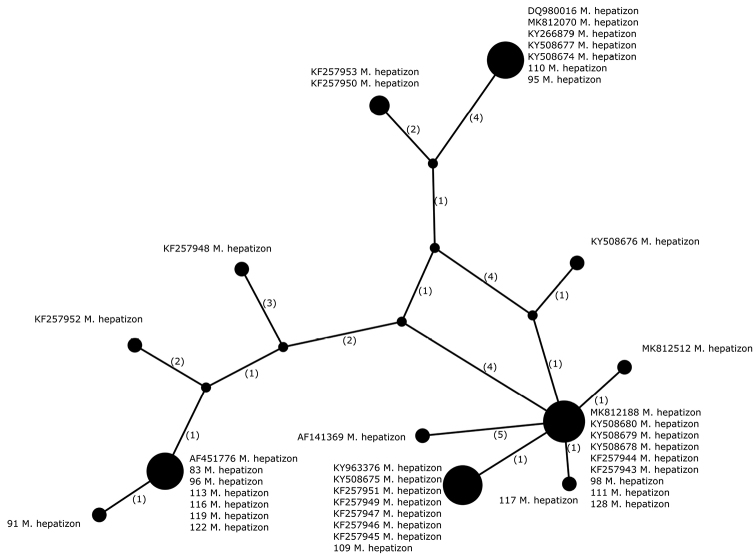
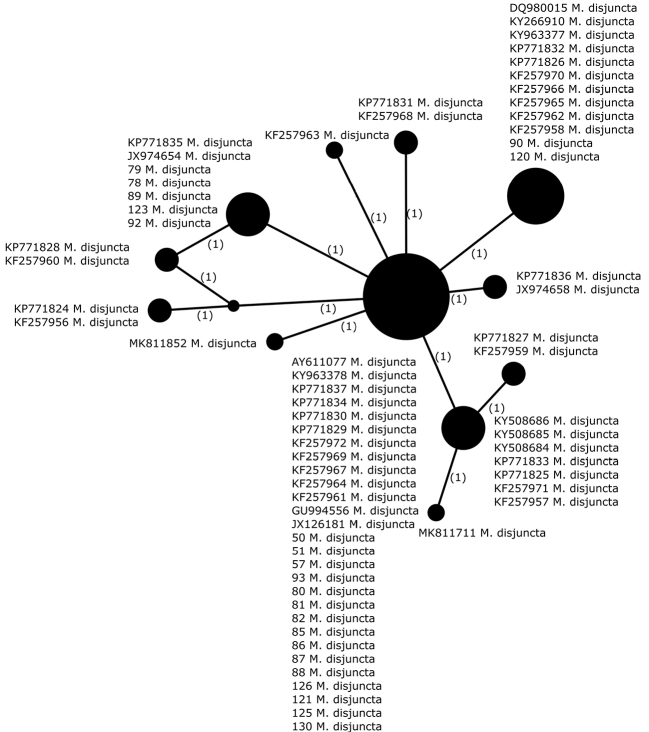
We constructed haplotype networks (Figs 2–7) to assess genetic variability within ITS rDNA marker for each species, including newly-collected specimens and data were downloaded from GenBank. The number of haplotypes found for each taxon ranged from five (in Melaneliastygia) to 12 (in Melaneliahepatizon and Montaneliadisjuncta); however, their numbers seem to be correlated with the abundance of specimens tested, which ranged from 10 (in Melaneliaagnata) to 67 (in Montaneliadisjuncta). Moreover, we also calculated nucleotide diversity for each dataset and found lower values for Montaneliadisjuncta and Cetrariacommixta (0.00380 and 0.00405, respectively) and higher values for Melaneliaagnata, M.hepatizon and M.stygia (0.01552, 0.01421 and 0.01418, respectively) (Table 2).
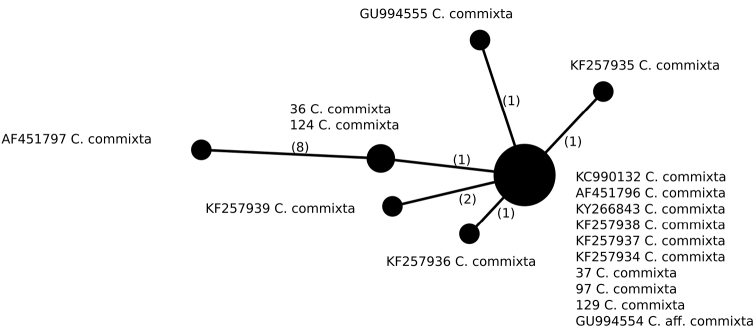
Figure 2.
Haplotype network, based on ITS rDNA sequences from specimens of Cetrariacommixta. Newly-generated sequences are described with isolate numbers preceding the species names. Sequences downloaded from GenBank are described with their accession numbers. Mutational changes are presented as numbers in brackets near lines between haplotypes.
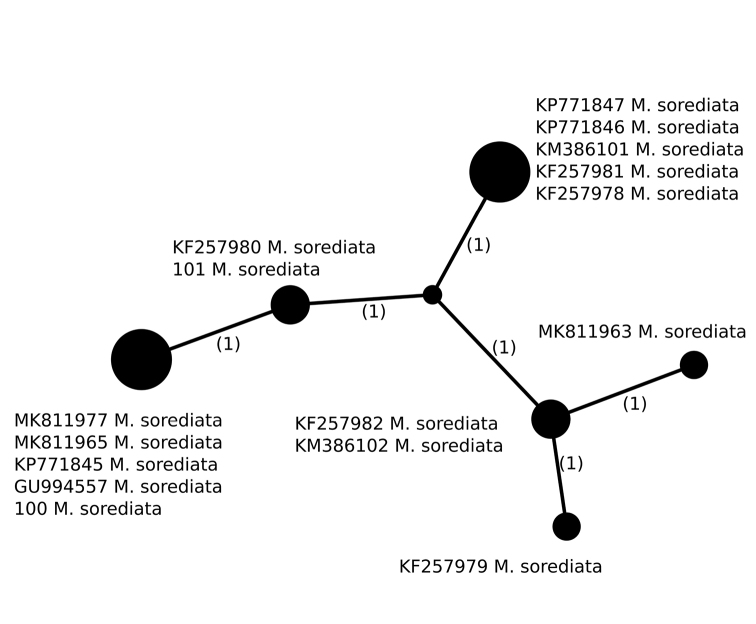
Figure 7.
Haplotype network, based on ITS rDNA sequences from specimens of Montaneliasorediata. Newly-generated sequences are described with isolate numbers preceding the species names. Sequences downloaded from GenBank are described with their accession numbers. Mutational changes are presented as numbers in brackets near lines between haplotypes.
Table 2.
List of haplotypes identified in this study and their geographical distribution. Nucleotide diversity for each species is also presented, and the newly generated sequences are in bold.
| Haplotypes number | North America | North Europe | Central Europe | West Europe | Asia | Nucleotide diversity |
|---|---|---|---|---|---|---|
| Cetrariacommixta | ||||||
| 1 | KF257934 Greenland | AF451796 Finland | 37 Poland | |||
| KF257937 Greenland | KY266843 Norway | 97 Poland | 0.00405 | |||
| KF257938 Greenland | KC990132 Sweden | 129 Germany | ||||
| GU994554 Sweden | ||||||
| 2 | 36 Poland | |||||
| 124 Poland | ||||||
| 3 | AF451797 Canada | |||||
| 4 | KF257939 Greenland | |||||
| 5 | KF257936 Greenland | |||||
| 6 | KF257935 Greenland | |||||
| 7 | GU994555 Spain | |||||
| Melaneliaagnata | ||||||
| 1 | KY508672 Iceland | |||||
| KY508673 Iceland | 0.01552 | |||||
| KY963373 Iceland | ||||||
| KY963374 Iceland | ||||||
| 2 | 102 Poland | |||||
| 103 Poland | ||||||
| 3 | KF257940 Greenland | |||||
| 4 | KF257941 Greenland | |||||
| 5 | KF257942 Greenland | |||||
| 6 | MK257942 Norway | |||||
| Melaneliahepatizon | ||||||
| 1 | KF257943 Greenland | KY508678 Iceland | 98 Poland | |||
| KF257944 Greenland | KY508680 Iceland | 111 Poland | 0.01421 | |||
| KY508679 Norway | 128 Germany | |||||
| MK812188 Norway | ||||||
| 2 | KF257945 Greenland | KY508675 Iceland | 109 Poland | |||
| KF257946 Greenland | KY508676 Iceland | |||||
| KF257947 Greenland | ||||||
| KF257949 Greenland | ||||||
| KF257951 Greenland | ||||||
| 3 | KY508674 Iceland | 95 Poland | ||||
| KY508677 Iceland | 110 Poland | |||||
| KY266879 Iceland | ||||||
| KY266879 Norway | ||||||
| DQ980016 Sweden | ||||||
| 4 | 83 Poland | AF451776 Italy | ||||
| 96 Poland | ||||||
| 113 Poland | ||||||
| 116 Slovakia | ||||||
| 119 Poland | ||||||
| 122 Poland | ||||||
| 5 | KF257950 Greenland | |||||
| KF257953 Greenland | ||||||
| 6 | KF257952 Greenland | |||||
| 7 | KF257948 Greenland | |||||
| 8 | AF141369 Canada | |||||
| 9 | KY963376 Iceland | |||||
| 10 | MK812512 Norway | |||||
| 11 | 91 Poland | |||||
| 12 | 117 Slovakia | |||||
| Melaneliastygia | ||||||
| 1 | AY611097 Finland | AY611121 Austria | AF451775 Italy | |||
| MK812608 Norway | 40 Poland | 0.01418 | ||||
| 42 Austria | ||||||
| 94 Poland | ||||||
| 104 Poland | ||||||
| 108 Poland | ||||||
| 112 Poland | ||||||
| 127 Czech Republic | ||||||
| 2 | KY508681 Island | |||||
| KY508682 Island | ||||||
| KY508683 Island | ||||||
| KY963375 Island | ||||||
| 3 | KF257954 Greenland | |||||
| KF257955 Greenland | ||||||
| 4 | AF115763 Finland | |||||
| 5 | MK812312 Norway | |||||
| Montaneliadisjuncta | ||||||
| 1 | KF257964 Canada | KY963378 Iceland | AY611077 Austria | GU994556 India | ||
| KF257967 Canada | KF257961 Norway | 50 Poland | KF257972 India | 0.00380 | ||
| KF257969 Canada | KP771829 Norway | 51 Poland | KP771837 India | |||
| KP771830 Canada | KP771834 Sweden | 57 Poland | ||||
| JX126181 USA | 80 Poland | |||||
| 81 Poland | ||||||
| 82 Poland | ||||||
| 85 Poland | ||||||
| 86 Poland | ||||||
| 87 Poland | ||||||
| 88 Poland | ||||||
| 93 Poland | ||||||
| 121 Poland | ||||||
| 125 Poland | ||||||
| 126 Poland | ||||||
| 130 Czech Republic | ||||||
| 2 | KF257962 Canada | KY963377 Iceland | 90 Poland | |||
| KF257965 Canada | KY266910 Norway | 120 Poland | ||||
| KF257966 Canada | DQ980015 Sweden | |||||
| KP771832 Greenland | ||||||
| KF257958 Greenland | ||||||
| KF257970 Greenland | ||||||
| KP771826 Greenland | ||||||
| 3 | KF257957 Greenland | KY508684 Iceland | ||||
| KF257971 Greenland | KY508685 Iceland | |||||
| KP771825 Greenland | KY508686 Iceland | |||||
| KP771833 Greenland | ||||||
| 4 | - | 78 Poland | ||||
| JX974654 United Kingdom | 79 Poland | |||||
| KP771835 United Kingdom | 89 Poland | |||||
| 92 Poland | ||||||
| 123 Poland | ||||||
| 5 | KF257956 Canada | |||||
| KP771824 Canada | ||||||
| 6 | JX974658 Canada | |||||
| KP771836 Canada | ||||||
| 7 | KF257963 Canada | |||||
| 8 | KF257959 Greenland | |||||
| KP771827 Greenland | ||||||
| 9 | KF257968 USA | |||||
| KP771831 USA | ||||||
| 10 | KF257960 USA | |||||
| KP771828 USA | ||||||
| 11 | MK811852 Norway | |||||
| 12 | MK811711 Norway | |||||
| Montaneliasorediata | ||||||
| 1 | MK811977 Norway | 100 Poland | ||||
| MK811965 Norway | 0.00830 | |||||
| GU994557 Sweden | ||||||
| KP771845 Sweden | ||||||
| 2 | KF257978 USA | KF257981 Russia | ||||
| KP771846 USA | KP771847 Russia | |||||
| KM386101 Japan | ||||||
| 3 | KF257980 USA | 101 Poland | ||||
| 4 | KM386102 Canada | |||||
| KF257982 Canada | ||||||
| 5 | KF257979 Canada | |||||
| 6 | MK811963 Norway | |||||
Characteristics of the studied species
. Cetraria commixta
(Nyl.) Th. Fr.
Lichenographia Scandinavica 1:109 (1871) ≡ Platysmacommixtum Nyl., Synopsis methodica lichenum 1:310 (1860) ≡ Melaneliacommixta (Nyl.) A. Thell, Nova Hedwigia 60:417 (1995) ≡ Cetrariellacommixta (Nyl.) A. Thell & Kärnefelt, Mycological Progress 3:309 (2004).
Description.
C.commixta is a foliose species with elongated, smooth and flat lobes, 0.25–2.5 mm broad, which are thick on the margins and rounded at the ends (Szczepańska and Kossowska 2017). Its upper surface is glossy, olive-brown to dark brown or almost black. The lower surface is pale brown, but darker in the centre, with single, dark rhizines. C.commixta possess rounded or slightly elongated pseudocyphellae, which are present only on the margins and edges of lobes and cylindrical, marginal pycnidia, producing hyaline, citriform conidia (3–4 × 1–1.5 µm). Apothecia are marginal, constricted at base, 0.2–7 mm diam., with hyaline, ellipsoid to oblong-ellipsoid ascospores (6–8 × 4–6 μm).
Chemistry.
α-collatolic acid (chemotype I) or no substances (chemotype III).
Distribution.
C.commixta is a circumpolar and arctic-alpine species (Otte et al. 2005), growing mainly in mountain sites, in open places with high precipitation, on natural acid, siliceous rocks in North America and Europe. Available molecular data concern samples collected in North America (Canada, Greenland), as well as North (Finland, Norway, Sweden) and West (Spain) Europe.
Haplotypes differentiation.
We identified seven different haplotypes (Fig. 2, Table 2) within C.commixta (n = 17) that differ from each other in one or two positions, except for a single Canadian sample that differs in at least eight positions. The most common haplotype was found in ten specimens occurring in Greenland and North and Central Europe, amongst them being three newly-sequenced specimens (samples 37 and 97 from Poland and sample 129 from Germany). Moreover, two Polish specimens (samples 36 and 124 from the Sudety Mountains) represent a unique haplotype that differs from the most common one in a single position. Five haplotypes identified in our dataset were represented by single specimens originating from Greenland (3 haplotypes), Canada or Spain.
. Melanelia agnata
(Nyl.) A. Thell
Nova Hedwigia 60:416 (1995) ≡ Platysmaagnatum Nyl., Flora, Jena 60:562 (1877) ≡ Cetrariaagnata (Nyl.) Kristinsson, Lichenologist 6:144 (1974).
Description.
M.agnata has foliose thallus with flat, smooth, 0.25–2 mm broad lobes which are thicker on the margins and rounded at the ends (Szczepańska and Kossowska 2017). The upper surface is glossy, olive-brown to dark brown. The lower surface is pale brown to dark brown in the centre, with single, dark rhizines. M.agnata possess pseudocyphellae which are larger on the lobe margins and smaller, punctiform on the upper surface of the lobes. Pycnidia are mainly marginal to laminal, partially immersed and globose with hyaline bacilliform conidia (4.5–5.5 × 1 µm). Apothecia are not seen in examined material.
Chemistry.
No secondary metabolites were detected by TLC.
Distribution.
M.agnata is a rare taxon occurring in arctic and boreal regions in North America and Europe, growing in open stands on siliceous and basalt rocks (Otte et al. 2005). Available molecular data concern samples collected only in North America (Greenland) and North Europe (Iceland, Norway).
Haplotypes differentiation.
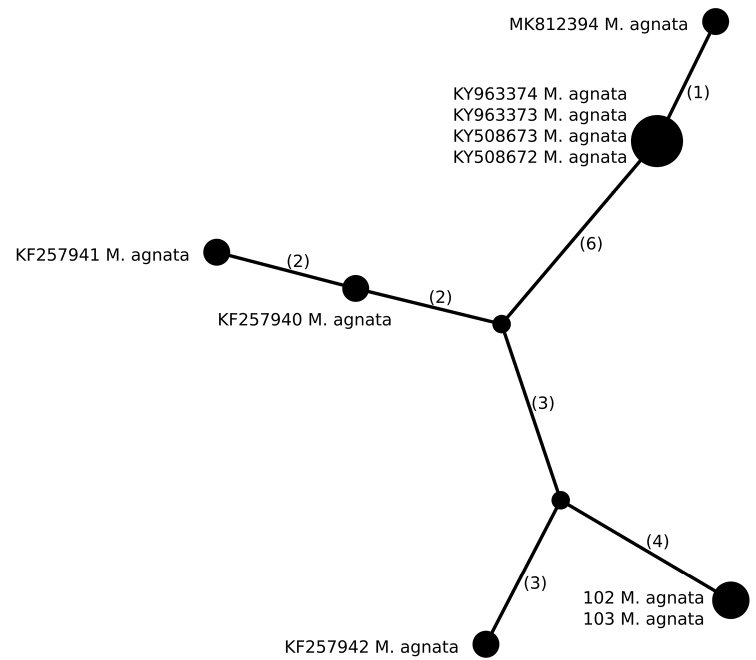
Six different haplotypes were identified in M.agnata (n = 10), of which two Polish specimens, collected in the Karpaty Mountains, have the same, not previously known, haplotype (Fig. 3, Table 2). It differs from other haplotypes in at least seven positions. However, the remaining specimens originate from Greenland, Iceland or Norway and no other samples from Central Europe have been sequenced until now. Four Icelandic specimens have the same haplotype, which is similar to the haplotype from Norwegian specimens. In contrast, Icelandic haplotypes differ from Greenlandic haplotypes in at least eight positions. Whether their genetic diversity supports conclusions from previous papers suggesting potentially unrecognised species lineages in the M.agnata genus (Leavitt et al. 2014; Xu et al. 2017) remains unresolved and should be further studied.
Figure 3.
Haplotype network, based on ITS rDNA sequences from specimens of Melaneliaagnata. Newly-generated sequences are described with isolate numbers preceding the species names. Sequences downloaded from GenBank are described with their accession numbers. Mutational changes are presented as numbers in brackets near lines between haplotypes.
. Melanelia hepatizon
(Ach.) A. Thell
Nova Hedwigia 60:419 (1995) ≡ Lichenhepatizon Ach., Lichenographiae Sueciae Prodromus 110 (1798) ≡ Cetrariahepatizon (Ach.) Vain., Termeszetrajzi Füzetek 22:278 (1899).
Description.
M.hepatizon is foliose species with flat lobes that are 0.25–2.5 mm broad and thick at the margins (Szczepańska and Kossowska 2017). Its upper surface is glossy, brown to almost black. The lower surface is dark brown to black, paler near the margins, with single, dark rhizines. Pseudocyphellae are mainly present on the margins and edges of lobes. Pycnidia are marginal, but sometimes also laminal, sessile, globose to stalked, slightly elongated or cylindrical with hyaline, bacilliform conidia (3–5 × 1 µm). Apothecia are marginal to laminal, sessile, with hyaline, ellipsoid to oblong-ellipsoid ascospores (6–8 × 4–6 μm).
Chemistry.
Stictic and norstictic acids.
Distribution.
M.hepatizon is a circumpolar and arctic-alpine species occurring from oceanic to continental sites on siliceous rocks in North America and Europe (Otte et al. 2005). Available molecular data concern samples collected in North America (Canada, Greenland) as well as North (Iceland, Norway, Sweden) and West (Italy) Europe.
Haplotypes differentiation.
A higher number of haplotypes was detected in M.hepatizon (n = 40), in which we identified 12 haplotypes (Fig. 4, Table 2). Amongst newly-sequenced specimens, we identified six haplotypes. Some are more common and were previously found in Greenland, Iceland, Italy, Norway or Sweden. In contrast, others were only found in newly-sequenced specimens, such as sample 91 from the Sudety Mountains in Poland and sample 117 from the Karpaty Mountains in Slovakia. However, no geographic pattern was found in the dataset.
Figure 4.
Haplotype network, based on ITS rDNA sequences from specimens of Melaneliahepatizon. Newly-generated sequences are described with isolate numbers preceding the species names. Sequences downloaded from GenBank are described with their accession numbers. Mutational changes are presented as numbers in brackets near lines between haplotypes.
. Melanelia stygia
(L.) Essl.
Mycotaxon 7:47 (1978) ≡ Lichenstygius L., Species Plantarum 2:1143 (1753).
Description.
M.stygia has foliose thallus, composed of 0.25–1.5 mm broad, smooth and usually distinctly convex lobes (Szczepańska and Kossowska 2017). The upper surface is glossy, dark brown to almost black. The lower surface is dark brown to black, paler near the margins, with single, dark rhizines. Pseudocyphellae in this species are numerous, rounded or slightly elongated and laminal – clearly visible on the upper surface of the lobes. Pycnidia are also common, globose, laminal and immersed with hyaline, bacilliform conidia (3.5–5 × 1 µm). Apothecia are laminal, constricted at the base and 0.5–6 mm in diameter. Ascospores are hyaline, ellipsoid to oblong-ellipsoid, 6–8 × 4–6 μm in size.
Chemistry.
Protocetraric and fumarprotocetraric acids (Race 1) or no substances detected (Race 6).
Distribution.
M.stygia is a circumpolar and arctic-alpine species occurring mainly on siliceous rocks in upper mountain areas in North America and Europe (Otte et al. 2005). Available molecular data concern only a few samples collected in North America (Greenland) and North (Iceland, Finland, Norway) and West (Italy) Europe.
Haplotypes differentiation.
Amongst five identified haplotypes in Melaneliastygia (n = 19), all newly-sequenced specimens (five from Poland, one from Austria and one from the Czech Republic) have the same haplotype, previously reported from Austria, Finland, Italy and Norway (Fig. 5, Table 2). It differs from the haplotype identified in another Finnish specimen in two positions. Two Greenlandic specimens have the same haplotype that differs from the most common one in five positions. Four Icelandic samples have an identical haplotype that differs from the Norwegian sample in five positions; however, these samples differ in at least 13 positions from other haplotypes of M.stygia. Moreover, these Icelandic and one Norwegian samples form a separate clade shown in Fig. 1, in contrast to the remaining specimens of M.stygia. These molecular data suggest that these lineages may represent phenotypically indistinguishable cryptic species.
Figure 5.
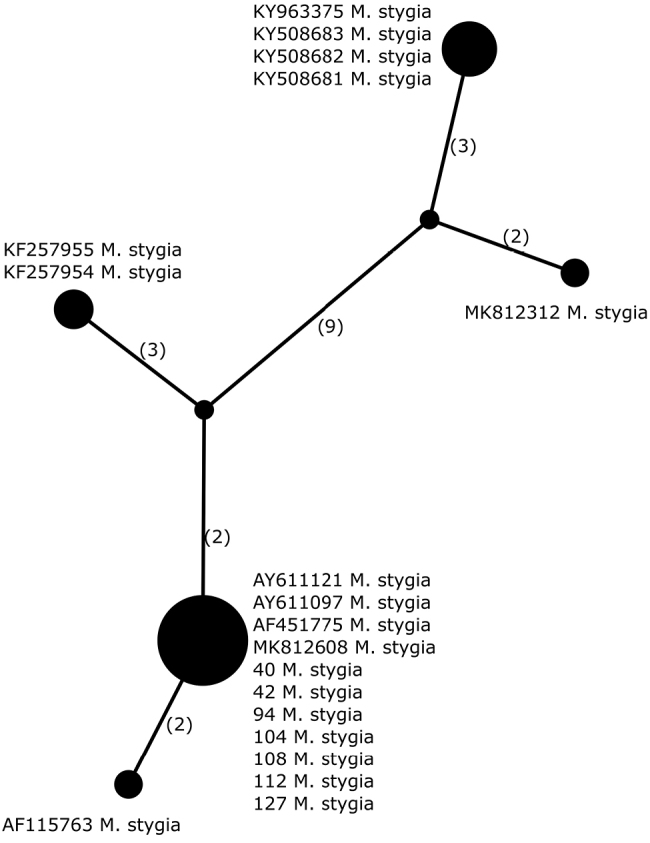
Haplotype network, based on ITS rDNA sequences from specimens of Melaneliastygia. Newly-generated sequences are described with isolate numbers preceding the species names. Sequences downloaded from GenBank are described with their accession numbers. Mutational changes are presented as numbers in brackets near lines between haplotypes.
. Montanelia disjuncta
(Erichsen) Divakar, A. Crespo, Wedin & Essl.
American Journal of Botany 99:2022 (2012) ≡ Parmeliadisjuncta Erichsen, Annales Mycologici 37:78 (1939) ≡ Melaneliadisjuncta (Erichsen) Essl., Mycotaxon 7:46 (1978).
Description.
M.disjuncta possess foliose thallus composed of 0.6–1.2 mm broad, flat to slightly convex and glossy lobes (Szczepańska et al. 2015). Its upper surface is smooth, olive-brown to dark brown. Pseudocyphellae are small, rather indistinct and submarginal. Its characteristic feature is the presence of the soralia (0.2–0.5 mm in diameter), which are punctiform, irregular, usually capitate and arise on the surface or at the margins of the lobes. Soredia are granular to isidioid, dark, but appearing white when abraded. Pycnidia are rare, conidia are 6–7 × 1 μm. Apothecia are not seen in the examined material.
Chemistry.
Perlatolic and stenosporic acids.
Distribution.
M.disjuncta is a circumpolar species growing mainly on siliceous rocks. The geographical range of this species consists of both continental and oceanic areas of Europe and North America (Esslinger 1977; Otte et al. 2005; Hansen 2013). Available molecular data concern samples collected in North America (Canada, Greenland, USA), North (Iceland, Norway, Sweden, United Kingdom) and Central (Austria) Europe, as well as Asia (India).
Haplotypes differentiation.
Twelve different haplotypes were identified in M.disjuncta (n = 67), of which the most common haplotype occurs in Europe, North America and Asia (Fig. 6, Table 2). The highest diversity was observed in North America (Canada, Greenland, USA), for which a total of nine different haplotypes were found, including six that were exclusive for this region. We identified three different haplotypes amongst the newly-collected samples (n = 22). The most common one also occurs in other European countries, Asia and North America. The second most common also occurs in Northern Europe and North America, while the third haplotype was previously identified in specimens collected in the United Kingdom. Moreover, four different haplotypes were identified amongst specimens collected in Norway, while five haplotypes were identified in Canadian samples, of which three are unique to Canada. Three haplotypes were identified in samples from both Iceland and Greenland, two of which are common for these areas and one haplotype is unique to Greenland. Some haplotypes are represented by more than one sample originating from particular areas, such as Alaska and Maine (USA), the Yukon Territory (Canada) or Greenland. The haplotypes identified in our dataset originated from different geographical areas and two of the most common haplotypes are widely distributed in the Northern Hemisphere. Based on the presented sampling, we could not indicate any geographical pattern, neither locally nor worldwide.
Figure 6.
Haplotype network, based on ITS rDNA sequences from specimens of Montaneliadisjuncta. Newly-generated sequences are described with isolate numbers preceding the species names. Sequences downloaded from GenBank are described with their accession numbers. Mutational changes are presented as numbers in brackets near lines between haplotypes.
. Montanelia sorediata
(Ach.) Divakar, A. Crespo, Wedin & Essl.
American Journal of Botany 99:2023 (2012) ≡ Parmeliastygiavar.sorediata Ach., Lichenographia Universalis 471 (1810) ≡ Melaneliasorediosa (Almb) Essl., Mycotaxon 7:47 (1978) ≡ Melaneliasorediata (Ach.) Goward & Ahti, Mycotaxon 28:94 (1987).
Description.
M.sorediata is a foliose species. Its lobes are flat to slightly convex, 0.2–0.6 mm broad, distinctly rugged and pitted at the ends (Szczepańska et al. 2017). The upper surface is smooth, dull, olive brown to dark brown. Characteristic soralia arise on the ends of the main lobes or on the smaller, erect side lobes. They are usually distinctly convex and capitate with granular to isidioid, dark soredia. Pseudocyphellae and pycnidia are absent. Apothecia are not seen in the examined material.
Chemistry.
Perlatolic and stenosporic acids.
Distribution.
M.sorediata is a probably circumpolar species that prefers siliceous substrates, usually in open and well-lit places. The species is mentioned as occurring in North America and Europe (Esslinger 1977; Otte et al. 2005). Available molecular data concern only a few samples collected in North America (Canada, USA), North Europe (Norway, Sweden) and Asia (India).
Haplotypes differentiation.
Six different haplotypes were identified in M.sorediata (n = 16), of which two Polish specimens, collected in the Karpaty Mountains, have two different haplotypes that differ in a single position (Fig. 7, Table 2). Interestingly, sample 101 has the same haplotype as the specimen collected in Alaska (KF257980), while sample 100 has the same haplotype as four Scandinavian specimens collected in Norway and Sweden. Another of the most common haplotypes is represented by specimens collected in Japan, Russia and the USA. Therefore, no specific geographic pattern was observed in the dataset.
Discussion
Although several studies focused on the phylogeny of brown Parmeliae, in the analysed datasets, there was an evident lack of molecular data concerning this group from Central Europe. The available data included only North America (mainly Greenland), Northern Europe (Scandinavian countries) and single sequences from specimens collected in Western Europe (Spain, Italy) and Asia (India, Russia). Having the opportunity to collect data from Poland, we focused on taxa occurring in this country, such as Cetrariacommixta, Melaneliaagnata, M.hepatizon, M.stygia, Montaneliadisjuncta and M.sorediata. Additionally, in analyses, we also included newly-generated sequences from samples collected in Austria, Czech Republic, Germany and Slovakia. By supplementing the dataset with new sequences from a previously-unexplored area, we wanted to study the intraspecific internal transcribed spacer (ITS) rDNA variability of mentioned species and analyse distribution patterns of individual haplotypes. Previously, Leavitt et al. (2014) reported mean genetic distance (given as the number of nucleotide substitutions per site) in brown Parmeliae and found higher values in the case of Melaneliaagnata and M.hepatizon (0.013) in contrast to Cetrariacommixta and M.stygia (0.002 and 0.007, respectively). In this study, we found the highest nucleotide diversity in Melaneliaagnata and M.hepatizon (0.01552 and 0.01421, respectively), but also in M.stygia (0.01418) as a result of additional sampling.
In our study, the haplotype networks illustrated that single-locus haplotypes and clades have no geographic clustering and cannot be useful in defining the species boundaries within brown Parmeliae. Haplotypes are dispersed amongst the sites and clades do not show apparent association with spatial location, as reported in literature data (Werth and Sork 2008; Starosta and Svoboda 2020). In addition, many of the analysed haplotypes of brown Parmeliae are widely distributed and, in many cases, the same haplotypes are shared between temperate and polar populations. What is more, all taxa, except Melaneliastygia, seem to be monophyletic and newly-sequenced specimens cluster together with other representatives of the species downloaded from GenBank. The extremely wide geographical distribution of mycobiont haplotypes has been observed in some other species, such as Cavernulariahultenii (Printzen et al. 2003), Cetrariaaculeata (Fernández-Mendoza et al. 2011) and Cladoniasubcervicornis (Printzen and Ekman 2003). In the first two cases, this phenomenon is assigned to lichens characterised by vegetative propagation and interpreted as evidence for ancestral polymorphisms and slow genetic drift (Printzen et al. 2003). This finding conforms well with the results of our study on Parmeliaceae, which are usually sterile species, reproducing by soredia (Montanelia) and conidia (Cetraria, Melanelia).
Although representatives of brown Parmeliae are known from both Hemispheres (Otte et al. 2005), all species studied in this paper represent circumpolar distribution and occur only on northern continents. The specimens used for the analyses originated mainly from mountain areas of Poland, both the Carpathians and the Sudetes; however, the range of sampling seems to be representative for this part of Europe. The number of analysed haplotypes representing different geographical regions was comparable for each taxon; nevertheless, the number of Melaneliaagnata and Montaneliasorediata samples remain very small. Due to the newly-generated molecular data covering Central Europe, we were able to compare the haplotype distribution in this area with other parts of the world. Unfortunately, the data available for discussed lichens taxa include, almost exclusively, specimens from North America and Northern Europe; the data concerning Asia and Southern Europe are not sufficient to make a reliable comparison possible. In almost all analysed taxa, stronger genetic differentiation was found amongst North American populations, with a few haplotypes unique for this part of the world, especially for Greenland. Specimens occurring in Central Europe have lower haplotype diversification and many of these haplotypes have wide geographical distribution (Table 2). Nevertheless, it seems that the number of analysed sequences is still insufficient to indicate high diversity areas (hotspots), species speciation centres or glacial refugia. Although the numbers of haplotypes correlated with the number of specimens tested, two species occurring in Poland (Melaneliaagnata and M.stygia) clearly indicate a very low level of genetic diversity. Both species are rare in Poland and their distribution is limited to the high mountain regions (Szczepańska and Kossowska 2017). Low genetic diversity and limited occurrence suggest considering both taxa as critically endangered in Poland.
In recent years, it has been proved that cryptic species-level lineages are very common amongst lichen-forming fungi (Crespo and Pérez-Ortega 2009; Crespo and Lumbsch 2010; Lumbsch and Leavitt 2011). At the same time, it has been shown that phenotypic variation is not always ‘sensitive’ enough for delimitation and description of new taxa. Modern methods of genetic analysis are recommended as an additional tool for this purpose (Molina et al. 2011; de Paz et al. 2012; Leavitt et al. 2013; Renner 2016). At the same time, it is necessary to include other evidence, such as chemistry, ecology, geography and morphology, for the proper delimitation of lichenised fungi species (Hawksworth 1976; Dayrat 2005; Crespo and Pérez-Ortega 2009). Such careful and versatile analysis of distinct phylogenetic lineages may lead to recognising some previously-overlooked characteristics (Kroken and Taylor 2001; del Prado et al. 2007; Frolov et al. 2016; Leavitt et al. 2016; Szczepańska et al. 2020). In the recent review paper, Lücking et al. (2021) proposed a detailed protocol for consistent taxonomy of lichen-forming fungi. The integrative taxonomy employing phylogeny, reproductive biology and phenotype should be used to delimit species (Lücking et al. 2020). Aime et al. (2021) recommended circumscription of new taxa, based on an appropriate sampling of multiple representatives from different collections for which multi-loci analyses should be performed. They also noted that description of a new species, based on single-locus phylogenetic analyses, could only be done in exceptional cases. The errors caused by contaminant sequences, laboratory mix-ups and chimeric sequences should be avoided for proper establishment of novel taxa, based on molecular data only (Lücking et al. 2021). Therefore, it is crucial to employ unlinked loci from different parts of the genome, even though the ITS rDNA marker is widely used in DNA barcoding of fungal taxa.
We analysed phenotypic diversity of samples representing individual haplotypes in our studies. However, in morphological, anatomical and chemical analyses, we observed that phenotypic characters of individuals representing different haplotypes are homogeneous and no visible distinctive features for samples with different geographic distribution were recognised. Recent molecular studies of one of the analysed genus – Melanelia, suggested previously unrecognised species-level diversity within this taxon (Divakar et al. 2012; Leavitt et al. 2014; Xu et al. 2017). However, the authors based their assumptions primarily on phylogenetic analyses without considering phenotypic features. Therefore, we have decided to analyse differences in morphology, anatomy and chemistry of M.stygia and M.agnata specimens originating from different geographic regions (Greenland, Iceland and Central Europe).
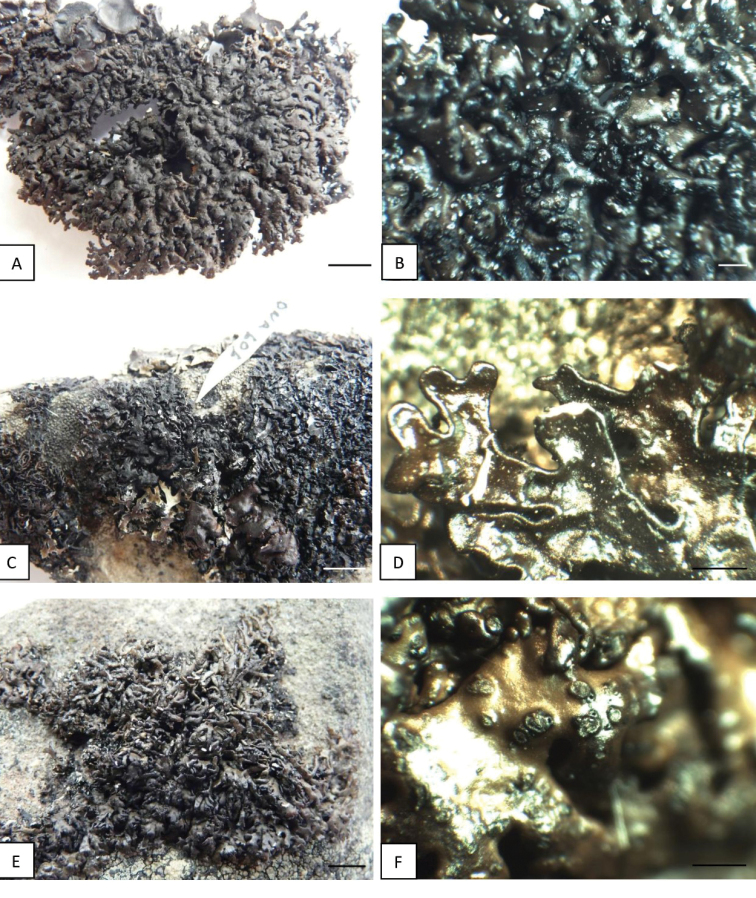
Melaneliaagnata is a rare lichen recorded in North America and some European countries, such as Austria, Iceland, Norway, Poland, Russia, Sweden, Switzerland and Slovakia (Westberg et al. 2004; Hawksworth et al. 2008; Szczepańska and Kossowska 2017). The analysed holotype of Melaneliaagnata is characterised by small (ca. 3.0 cm in diam.), foliose, olive-brown to dark-brown thallus, composed of flat, shiny, 0.25–2 mm broad, smooth lobes with thicker margins (Fig. 8A). Its lower surface is pale brown with single, dark rhizines. Polish (Figs 8G and H) and Greenlandic (Fig. 8E and F) specimens comply with the type. However, Icelandic material differs in a larger thallus size (up to 10 cm in diam.) and the appearance of the lobes, which are more convex than flat, 1–5 mm broad and distinctly wrinkled (Fig. 8C). Thell (1995) made an interesting taxonomic description of M.agnata, in which he noted that its thallus could reach up to 10 cm diam. However, in his research, Thell (1995) analysed only a few specimens, including one from Iceland (Kristinsson 14781, GZU, LD) and treated them all as a single taxon. A similar situation applies to conidia, reaching 5–7.5 µm in M.agnata, according to Thell (1995). Pycnidia observed in Icelandic specimens are usually marginal (Fig. 8D), very often double and produce bifusiform conidia, 4.5–6 × 1 µm, in contrast to the type specimen, which pycnidia are simple, marginal to laminal (Fig. 8B) with smaller conidia, at 3.5–5 × 1 µm. Pseudocyphellae are always whitish, rounded or irregular, marginal and laminal in all analysed material; they are much more abundant in specimens from Iceland (Fig. 8D). None of the Icelandic specimens had apothecia, so their anatomical analysis was impossible. All material was chemically homogeneous and no secondary metabolites were detected by thin-layer chromatography (TLC), which is consistent with other descriptions (Thell 1995; Xu et al. 2017).
Figure 8.
Melaneliaagnata specimens treated AMelaneliaagnata H-NYL 36086 (holotype) BMelaneliaagnata, H-NYL 36086 (holotype) CM.agnata, AMNH 27562 (Iceland) DM.agnata, AMNH 30974 (Iceland) EM.agnata, C 19019 (Greenland) FM.agnata, C 19019 (Greenland) GM.agnata, Szczepańska 1050, WRSL (Poland) HM.agnata, Szczepańska 1050, WRSL (Poland). Scale bars: 0.5 cm (A, C, E, G); 0.5 mm (B, D, F); 1 mm (H).
Melaneliastygia is a much more common species than M.agnata. In Europe, it was recorded in the upper mountain areas of Austria, the Czech Republic, Germany, Great Britain, Poland, Romania, Russia, Slovakia, Switzerland and Ukraine (Hawksworth et al. 2008).
After phenotypic studies, we have concluded that all material is homogeneous and none of the analysed morphological and anatomical features coincides with geographically-distinct M.stygia populations (Fig. 9A–F). However, some differences may be observed in the secondary chemistry. In his paper, Esslinger (1977) recognised six chemical races within M.stygia. He stated that some of them are broadly distributed and others are more frequent in particular regions. All the currently-examined samples originating from Greenland and Central Europe belong to Race 1, containing fumaroprotocetraric and protocetraric acids. Specimens from Iceland represent Race 6, without secondary metabolites. Both races are known to occur in Japan, North America and Europe; however, there is a possibility that Race 6 is the only chemical Race occurring in Iceland. Production of some secondary metabolites may be induced by environmental factors (Culberson 1986; Leavitt et al. 2011) and does not always correspond with molecular data. Moreover, chemical differences can be observed within some recognised haplotype groups and even in the same haplotype (Matteucci et al. 2017). At the same time, chemical characters may be successfully used to support delimitation of lichen taxa, but in any case, they cannot be treated as an exclusive diagnostic trait (Elix et al. 2009; Spribille et al. 2011; Leavitt et al. 2013; Onut-Brännström et al. 2018; Mark et al. 2019,).
Figure 9.
Melaneliastygia specimens treated AM.stygia, AMNH 28243 (Iceland) BM.stygia, AMNH 16894 (Iceland) CM.stygia, C 19893 (Greenland) DM.stygia, C 19893 (Greenland) EM.stygia, Szczepańska 1160, WRSL (Poland) FM.stygia, Szczepańska 737, WRSL (Austria). Scale bars: 0.5 cm (A, C, E);1 mm (B, D); 0.5 mm (F).
In conclusion, we can state that all of the potential species lineages within Melaneliaagnata and M.stygia are cryptic, with very slight morphological, anatomical and chemical variation. We were unable to distinguish any distinctive feature that could be considered diagnostic and useful for the delimitation of new species, except molecular variation. The phenotypic differences mentioned above may reflect environmental or climate conditions, such as temperature, light, humidity or substrate and may not be connected with genetic differences. However, this study was limited to a small number of samples and one genetic marker, ITS; therefore, we refrain from describing new species because further study is pending. We suggest that an extended phylogeographic study is necessary and an increase in the number of herbarium specimens would probably give additional information. Even though our analyses complement the knowledge on lichens in Central Europe, many areas remain insufficiently explored. Additional sampling from Asia and Southern Europe may bring new data on the phylogenetic and phenotypic diversity of species from the brown Parmeliae group.
Supplementary Material
Acknowledgements
The curators of AMNH, C and H, are gratefully acknowledged for the loan of specimens. The authors are also very grateful to the reviewers for their valuable comments and improvements to the manuscript. The publication is financed under the Leading Research Groups support project from the subsidy increased for the period 2020–2025 in the amount of 2% of the subsidy referred to Art. 387 (3) of the Law of 20 July 2018 on Higher Education and Science, obtained in 2019.
Citation
Szczepańska K, Guzow-Krzemińska B, Urbaniak J (2021) Infraspecific variation of some brown Parmeliae (in Poland) – a comparison of ITS rDNA and non-molecular characters. MycoKeys 85: 127–160. https://doi.org/10.3897/mycokeys.85.70552
Supplementary materials
Figure S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Katarzyna Szczepańska, Beata Guzow-Krzemińska, Jacek Urbaniak
Data type
Images.
Explanation note
Phylogenetic relationships of brown Parmeliae, based on Bayesian analysis of the ITS rDNA dataset. Posterior probabilities and Maximum Likelihood bootstrap values are shown near the internal branches. Newly-generated sequences are described with isolate numbers preceding the species names and are marked in bold. GenBank accession numbers of sequences downloaded from GenBank are listed on the tree with species names.
References
- Aime MC, Miller AN, Aoki T, Bensch K, Cai L, Crous PW, Hawksworth DL, Hyde KD, Kirk PM, Lücking R, May TW, Malosso E, Redhead SA, Rossman AY, Stadler M, Thines M, Yurkov AM, Zhang N, Schoch CL. (2021) How to publish a new fungal species, or name, version 3.0. IMA Fungus 12: е11. 10.1186/s43008-021-00063-1 [DOI] [PMC free article] [PubMed]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco O, Crespo A, Divakar PK, Esslinger TL, Hawksworth DL, Lumbsch H. (2004) Melanelixia and Melanohalea, two new genera segregated from Melanelia (Parmeliaceae) based on molecular and morphological data. Mycological Research 108: 873–884. 10.1017/S0953756204000723 [DOI] [PubMed] [Google Scholar]
- Blanco O, Crespo A, Divakar PK, Elix JA, Lumbsch HT. (2005) Molecular phylogeny of parmotremoid lichens (Ascomycota, Parmeliaceae). Mycologia 97: 150–159. 10.1080/15572536.2006.11832848 [DOI] [PubMed] [Google Scholar]
- Clement M, Snell Q, Walker P, Posada D, Crandall K. (2002) TCS: Estimating gene genealogies. Parallel and Distributed Processing Symposium, International Proceedings 2: е184. 10.1109/IPDPS.2002.1016585 [DOI]
- Crespo A, Pérez-Ortega S. (2009) Cryptic species and species pairs in lichens: a discussion on the relationship between molecular phylogenies and morphological characters. Anales del Jardín Botánico de Madrid 66: 71–81. 10.3989/ajbm.2225. [DOI] [Google Scholar]
- Crespo A, Lumbsch HT. (2010) Cryptic species in lichen-forming fungi. IMA fungus 1: 167–170. 10.5598/imafungus.2010.01.02.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo A, Divakar PK, Hawksworth DL. (2011) Generic concepts in parmelioid lichens, and the phylogenetic value of characters used in their circumscription. Lichenologist (London, England) 43: 511–535. 10.1017/S0024282911000570 [DOI] [Google Scholar]
- Crespo A, Kauff F, Divakar PK, del Prado R, Pérez-Ortega S, Amo de Paz G, et al. (2010) Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon 59: 1735–1753. 10.1002/tax.596008 [DOI] [Google Scholar]
- Culberson CF. (1972) Improved conditions and new data for identification of lichen products by standardized thin-layer chromatographic method. Journal of Chromatography A 72: 113–125. 10.1016/0021-9673(72)80013-X. [DOI] [PubMed] [Google Scholar]
- Culberson WL. (1986) Chemistry and sibling speciation in the lichen-forming fungi: ecological and biological considerations. Bryologist 89: 123–131. 10.2307/3242752 [DOI] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: е772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed]
- Dayrat B. (2005) Towards integrative taxonomy. Biological Journal of the Linnean Society 85: 407–417. 10.1111/j.1095-8312.2005.00503.x [DOI] [Google Scholar]
- de Paz GA, Cubas P, Crespo A, Elix JA, Lumbsch HT. (2012) Transoceanic dispersal and subsequent diversification on separate continents shaped diversity of the Xanthoparmeliapulla group (Ascomycota). PLоS ONE 7(6): e39683. 10.1371/journal.pone.0039683 [DOI] [PMC free article] [PubMed]
- del Prado R, Ferencová Z, Armas-Crespo V, de Paz GA, Cubas P, Crespo A. (2007) The arachiform vacuolar body: an overlooked shared character in the ascospores of a large monophyletic group within Parmeliaceae (Xanthoparmelia clade, Lecanorales). Mycological Research 111: 685–692. 10.1016/j.mycres.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Divakar PK, Del-Prado R, Lumbsch HT, Wedin M, Esslinger TL, Leavitt SD, Crespo A. (2012) Diversification of the newly recognized lichen-forming fungal lineage Montanelia (Parmeliaceae, Ascomycota) and its relation to key geological and climatic. American Journal of Botany 99: 2014–2026. 10.3732/ajb.1200258 [DOI] [PubMed] [Google Scholar]
- Domaschke S, Fernandez-Mendoza FA, García M, Martín M, Printzen C. (2012) Low genetic diversity in Antarctic populations of the lichen-forming ascomycete Cetrariaaculeata and its photobiont. Polar Research 31(1): е17353. 10.3402/polar.v31i0.17353 [DOI]
- Doyle JJ, Doyle JL. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Elix JA, Corush J, Lumbsch HT. (2009) Triterpene chemosyndromes and subtle morphological characters characterise lineages in the Physciaaipolia group in Australia (Ascomycota). Systematics and Biodiversity 7: 479–487 10.1017/S1477200009990223. [DOI] [Google Scholar]
- Esslinger TL. (1977) A chemosystematic revision of the brown Parmeliae. Journal of the Hattori Botanical Laboratory 42: 1–211. [Google Scholar]
- Esslinger TL. (1978) A new status for the brown Parmeliae. Mycotaxon 7: 45–54. [Google Scholar]
- Fernández-Mendoza F, Domaschke S, García MA, Jordan P, Martín MP, Printzen C. (2011) Population structure of mycobionts and photobionts of the widespread lichen Cetrariaaculeata. Molecular Ecology 20: 1208–1232. 10.1111/j.1365-294X.2010.04993.x [DOI] [PubMed] [Google Scholar]
- Frolov I, Vondrá J, Fernández-Mendoza F, Wilk K, Khodosovtsev A, Halıcı MG. (2016) Three new, seemingly-cryptic species in the lichen genus Caloplaca (Teloschistaceae) distinguished in two-phase phenotype evaluation. In Annales Botanici Fennici 53: 243–262. 10.5735/085.053.0413 [DOI] [Google Scholar]
- Galtier N, Gouy M, Gautier C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Computational Applied Biosciences 12: 543–548. 10.1093/bioinformatics/12.6.543 [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Guzow-Krzemińska B, Węgrzyn G. (2003) A preliminary study on the phylogeny of the genus Melanelia using nuclear large subunit ribosomal DNA sequences. Lichenologist 35: 83–86. 10.1006/lich.2002.0429 [DOI] [Google Scholar]
- Hansen ES. (2013) Lichens from three localities in Central West Greenland with notes on their climatic preferences. Botanica Lithuanica 19: 28–36. 10.2478/botlit-2013-0004 [DOI] [Google Scholar]
- Hawksworth DL. (1976) Lichen chemotaxonomy. In: Brown DH, Hawksworth DL, Bailey RH. (Eds) Lichenology: progress and problems.Academic Press, London, 139–184.
- Hawksworth DL, Blanco O, Divakar PK, Ahti T, Crespo A. (2008) A first checklist of parmelioid and similar lichens in Europe and some adjacent territories, adopting revised generic circumscriptions and with indications of species distributions. Lichenologist 40: 1–21. 10.1017/S0024282908007329 [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroken S, Taylor JW. (2001) A gene genealogical approach to recognize phylogenetic species boundaries in the lichenized fungus Letharia. Mycologia 93: 38–53. 10.1080/00275514.2001.12061278 [DOI] [Google Scholar]
- Landan G, Graur D. (2008) Local reliability measures from sets of co-optimal multiple sequence alignments. Pacific Symposium on Biocomputing 13: 15–24. [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. (2016) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Leavitt SD, Divakar PK, Ohmura Y, Wang L-S, Esslinger TL, Lumbsch HT. (2015) Who’s getting around? Assessing species diversity and phylogeography in the widely distributed lichen-forming fungal genus Montanelia (Parmeliaceae, Ascomycota). Molecular Phylogenetics and Evolution 90: 85–96. 10.1016/j.ympev.2015.04.029 [DOI] [PubMed] [Google Scholar]
- Leavitt SD, Fankhauser JD, Leavitt DH, Porter LD, Johnson LA, Clair LLS. (2011) Complex patterns of speciation in cosmopolitan “rock posy” lichens-Discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplacamelanophthalma species-complex (Lecanoraceae, Ascomycota). Molecular Phylogenetics and Evolution 59: 587–602. 10.1016/j.ympev.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Leavitt S, Fernández-Mendoza F, Pérez-Ortega S, Sohrabi M, Divakar P, Lumbsch T, Clair LLS. (2013) DNA barcode identification of lichen-forming fungal species in the Rhizoplacamelanophthalma species-complex (Lecanorales, Lecanoraceae), including five new species. MycoKeys 7: 1–22. 10.3897/mycokeys.7.4508. [DOI] [Google Scholar]
- Leavitt SD, Esslinger TL, Hansen ES, Divakar PK, Crespo A, Loomis BF, Lumbsch HT. (2014) DNA barcoding of brown Parmeliae (Parmeliaceae) species: a molecular approach for accurate specimen identification, emphasizing species in Greenland. Organisms Diversity & Evolution 14: 11–20. 10.1007/s13127-013-0147-1 [DOI] [Google Scholar]
- Leavitt SD, Esslinger TL, Divakar PK, Crespo A, Lumbsch HT. (2016) Hidden diversity before our eyes: Delimiting and describing cryptic lichen-forming fungal species in camouflage lichens (Parmeliaceae, Ascomycota). Fungal Biology 120: 1374–1391. 10.1016/j.funbio.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Lindblom L, Ekman S. (2006) Genetic variation and population differentiation in the lichen-forming ascomycete Xanthoriaparietina on the island Storfosna, central Norway. Molecular Ecology 15: 1545–1559. 10.1111/j.1365-294X.2006.02880.x [DOI] [PubMed] [Google Scholar]
- Lumbsch HT, Leavitt SD. (2011) Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Diversity 50: 59–72. 10.1007/s13225-011-0123-z [DOI] [Google Scholar]
- Lücking R, Aime MC, Robberts B, Miller AN, Ariyawansa HA, Aoki T, Cardinali G, Crous PW, Druzhinina IS, Geiser DM, Hawksworth DL, Hyde KD, Irinyi L, Jeewon R, Johnston PR, Kirk PM, Malosso E, May TW, Meyer W, Öpik M, Robert V, Stadler M, Thines M, Vu D, Yurkov AM, Zhang N, Schoch CL. (2020) Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal barcoding? IMA Fungus 11: е14. 10.1186/s43008-020-00033-z [DOI] [PMC free article] [PubMed]
- Lücking R, Leavitt SD, Hawksworth DL. (2021) Species in lichen-forming fungi: balancing between conceptual and practical considerations, and between phenotype and phylogenomics. Fungal Diversity. 10.1007/s13225-021-00477-7 [DOI]
- Mark K, Randlane T, Thor G, Hur JS, Obermayer W, Saag A. (2019) Lichen chemistry is concordant with multilocus gene genealogy in the genus Cetrelia (Parmeliaceae, Ascomycota). Fungal Biology 123: 125–139. 10.1016/j.funbio.2018.11.013 [DOI] [PubMed] [Google Scholar]
- Matteucci E, Occhipinti A, Piervittori R, Maffei ME, Favero-Longo SE. (2017) Morphological, secondary metabolite and ITS (rDNA) variability within usnic acid-containing lichen thalli of Xanthoparmelia explored at the local scale of rock outcrop in W-Alps. Chemistry & Biodiversity 14: e1600483. 10.1002/cbdv.201600483 [DOI] [PubMed]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Molina M, Del-Prado R, Divakar PK, Sánchez-Mata D, Crespo A. (2011) Another example of cryptic diversity in lichen-forming fungi: the new species Parmeliamayi (Ascomycota: Parmeliaceae). Organisms Diversity & Evolution 11: 331–342. 10.1007/s13127-011-0060-4 [DOI] [Google Scholar]
- Nelsen MP, Chavez N, Sackett-Hermann E, Thell A, Randlane T, Divakar PK, Rico VJ, Lumbsch HT. (2011) The cetrarioid core group revisited (Lecanorales: Parmeliaceae). Lichenologist 43: 537–551. 10.1017/S0024282911000508 [DOI] [Google Scholar]
- Onut-Brännström I, Johannesson H, Tibell L. (2018) Thamnoliatundrae sp. nov., a cryptic species and putative glacial relict. Lichenologist 50: 59–75 10.1017/S0024282917000615. [DOI] [Google Scholar]
- Orange A, James PW, White FJ. (2001) Microchemical methods for the identification of lichens. London: British Lichen Society.
- Otte V, Esslinger TL, Litterski B. (2005) Global distribution of the European species of the lichen genus Melanelia Essl. Journal of Biogeography 32: 1221–1241. 10.1111/j.1365-2699.2005.01268.x [DOI] [Google Scholar]
- Palice Z, Printzen C. (2004) Genetic variability in tropical and temperate populations of Trapeliopsisglaucolepidea: evidence against long range dispersal in a lichen with disjunct distribution. Mycotaxon 90: 43–54. [Google Scholar]
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. (2010) GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Research 38(Web Server issue): W23–W28. 10.1093/nar/gkq443 [DOI] [PMC free article] [PubMed]
- Printzen C, Ekman S. (2003) Local population subdivision in the lichen Cladoniasubcervicornis as revealed by mitochondrial cytochrome oxidase subunit 1 intron sequences. Mycologia 95: 399–406. 10.1080/15572536.2004.11833084 [DOI] [PubMed] [Google Scholar]
- Printzen C, Ekman S, Tønsberg T. (2003) Phylogeography of Cavernulariahultenii: evidence for slow genetic drift in a widely disjunct lichen. Molecular Ecology 12: 1473–1486. 10.1046/j.1365-294X.2003.01812.x [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2012) FigTree v.1.4.2. http://tree.bio.ed.ac.uk/software/figtree/
- Renner SS. (2016) A return to Linnaeus’s focus on diagnosis, not description: The use of DNA characters in the formal naming of species. Systematic Biology 65(6): 1085–1095. 10.1093/sysbio/syw032 [DOI] [PubMed] [Google Scholar]
- Rico VJ, van den Boom PP, Barrasa JM. (2005) Morphology, chemistry and distribution of Melaneliasorediella (Parmeliaceae) and similar species in the Iberian Peninsula. Lichenologist 37: 199–215. 10.1017/S0024282905014830 [DOI] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. (2017) DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Molecular Biology and Evolution 34: 3299–3302. 10.1093/molbev/msx248 [DOI] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. (2015) GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Research 43(Web Server issue): W7–W14. 10.1093/nar/gkq443 [DOI] [PMC free article] [PubMed]
- Spribille T, Klug B, Mayrhofer H. (2011) A phylogenetic analysis of the boreal lichen Mycoblastussanguinarius (Mycoblastaceae, lichenized Ascomycota) reveals cryptic clades correlated with fatty acid profiles. Molecular Phylogenetics and Evolution 59: 603–614. 10.1016/j.ympev.2011.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta J, Svoboda D. (2020) Genetic variability in the Physconiamuscigena group (Physciaceae, Ascomycota) in the Northern Hemisphere. Lichenologist 52: 305–317. 10.1017/S0024282920000134 [DOI] [Google Scholar]
- Szczepańska K, Kossowska M. (2017) Cetrariellacommixta and the genus Melanelia (Parmeliaceae, Ascomycota) in Poland. Herzogia 30: 272–288. 10.13158/heia.30.1.2017.272 [DOI] [Google Scholar]
- Szczepańska K, Pruchniewicz D, Sołtysiak J, Kossowska M. (2015) Lichen-forming fungi of the genus Montanelia in Poland and their potential distribution in Central Europe. Herzogia 28: 697–712. 10.13158/heia.28.2.2015.697 [DOI] [Google Scholar]
- Szczepańska K, Urbaniak J, Śliwa L. (2020) Taxonomic recognition of some species-level lineages circumscribed in nominal Rhizoplacasubdiscrepans s. lat. (Lecanoraceae, Ascomycota). PeerJ 8: e9555. [DOI] [PMC free article] [PubMed]
- Thell A. (1995) A new position of the Cetrariacommixta group in Melanelia (Ascomycotina, Parmeliaceae). Nova Hedwigia 60: 407–422. [Google Scholar]
- Thell A, Crespo A, Divakar PK, Kärnefelt I, Leavitt SD, Lumbsch HT, Seaward MRD. (2012) A review of the lichen family Parmeliaceae – history, phylogeny and current taxonomy. Nordic Journal of Botany 30: 641–664. 10.1111/j.1756-1051.2012.00008.x [DOI] [Google Scholar]
- Werth S, Sork VL. (2008) Local genetic structure in a North American epiphytic lichen, Ramalinamenziesii (Ramalinaceae). American Journal of Botany 95: 568–576. 10.3732/ajb.2007024 [DOI] [PubMed] [Google Scholar]
- Westberg M, Kärnefelt I, Thell A. (2004) Melaneliaagnata, an overlooked species, new to Sweden. Graphis Scripta 16(1): 23–27. [Google Scholar]
- Xu M, Heidmarsson S, Thorsteinsdottir M, Eiriksson FF, Omarsdottir S, Olafsdottir ES. (2017) DNA barcoding and LC-MS metabolite profiling of the lichen-forming genus Melanelia: Specimen identification and discrimination focusing on Icelandic taxa. PLоS ONE 12(5): e0178012. 10.1371/journal.pone.0178012 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Katarzyna Szczepańska, Beata Guzow-Krzemińska, Jacek Urbaniak
Data type
Images.
Explanation note
Phylogenetic relationships of brown Parmeliae, based on Bayesian analysis of the ITS rDNA dataset. Posterior probabilities and Maximum Likelihood bootstrap values are shown near the internal branches. Newly-generated sequences are described with isolate numbers preceding the species names and are marked in bold. GenBank accession numbers of sequences downloaded from GenBank are listed on the tree with species names.