Abstract
The Drosophila mod(mdg4) gene products counteract heterochromatin-mediated silencing of the white gene and help activate genes of the bithorax complex. They also regulate the insulator activity of the gypsy transposon when gypsy inserts between an enhancer and promoter. The Su(Hw) protein is required for gypsy-mediated insulation, and the Mod(mdg4)-67.2 protein binds to Su(Hw). The aim of this study was to determine whether Mod(mdg4)-67.2 is a coinsulator that helps Su(Hw) block enhancers or a facilitator of activation that is inhibited by Su(Hw). Here we provide evidence that Mod(mdg4)-67.2 acts as a coinsulator by showing that some loss-of-function mod(mdg4) mutations decrease enhancer blocking by a gypsy insert in the cut gene. We find that the C terminus of Mod(mdg4)-67.2 binds in vitro to a region of Su(Hw) that is required for insulation, while the N terminus mediates self-association. The N terminus of Mod(mdg4)-67.2 also interacts with the Chip protein, which facilitates activation of cut. Mod(mdg4)-67.2 truncated in the C terminus interferes in a dominant-negative fashion with insulation in cut but does not significantly affect heterochromatin-mediated silencing of white. We infer that multiple contacts between Su(Hw) and a Mod(mdg4)-67.2 multimer are required for insulation. We theorize that Mod(mdg4)-67.2 usually aids gene activation but can also act as a coinsulator by helping Su(Hw) trap facilitators of activation, such as the Chip protein.
The mechanisms by which enhancers transcriptionally activate promoters located several kilobases away are unknown. Some of these mechanisms are inhibited by insulator sequences. Insulators block activation of a promoter by an enhancer only when positioned between the enhancer and promoter (for reviews, see references 1, 12, 22, 43, and 56). Insulators block activation without inhibiting enhancers or repressing promoters. Thus, an enhancer prevented by an insulator from activating one promoter can still activate another promoter that is not separated by the insulator (4, 51). Similarly, a promoter insulated from some enhancers can still be activated by other enhancers that are not separated by the insulator. Insulators, therefore, interfere specifically with communication between enhancers and promoters.
Insulators help control gene expression. For example, multiple insulators organize enhancer-promoter interactions in the bithorax HOX gene complex of Drosophila (reviewed in references 39 and 56). Two insulators that bind BEAF-32 or the Zw5 protein flank a Drosophila hsp70 heat shock gene cluster and may prevent activation of neighboring genes by heat shock factor (16, 33, 34, 58). Insulators flanking the chicken β-globin gene complex that bind the CCTC-binding factor (CTCF) protein are at the boundaries of the transcriptionally active domain (7, 50). An insulator that binds CTCF in a methylation-sensitive manner helps mediate imprinting of the mouse Igf2 gene (2, 26, 53, 54).
Several models have been proposed to explain how insulators block activation. These include the suggestion that insulators regulate chromatin structure to form boundaries between domains of closed and open chromatin (7, 21, 33) and the idea that insulators are decoys that mimic a promoter and trap enhancers in futile interactions (22). An alternative idea is that insulators thwart proteins that act between enhancers and promoters to facilitate communication (11, 12, 41, 42). Such facilitator proteins could, for example, modify chromatin structure to bring an enhancer and promoter closer together or transmit an activation signal down the chromosome from an enhancer to a promoter.
The Drosophila proteins Chip and Nipped-B are putative facilitators of enhancer-promoter communication (41, 42, 48). Both proteins potentiate activation by diverse enhancers and have close mammalian homologues. They were originally identified by screening for mutations that increase insulation by gypsy transposon insertions in the cut and Ultrabithorax genes (41, 48).
The Su(Hw) protein is required for the gypsy transposon insulator to block enhancer-promoter interactions. Mutations in the suppressor of Hairy-wing [su(Hw)] gene decrease (suppress) the severity of the mutant phenotypes caused by gypsy insertions in many different genes (40, 49), and a sequence that binds Su(Hw) protein is the only part of gypsy required for insulation (23, 28). Su(Hw) interacts with the Chip protein in vitro, suggesting that Chip is a direct molecular target of the gypsy insulator (55).
The modifier of mdg4 [mod(mdg4)] gene also determines how gypsy insertions alter gene expression. Unlike su(Hw), however, the effects of mod(mdg4) differ from gene to gene. The mod(mdg4)u1 mutation was identified on the basis that it increases the severity of the mutant phenotype caused by a gypsy insertion at the yellow locus (18). The y2 gypsy insertion is positioned such that it blocks only enhancers that activate yellow in wing and body cuticle (24). In mod(mdg4)u1 mutants, yellow expression is also reduced in bristles and larval mouthparts, and this repressive effect requires the Su(Hw) protein (18, 21). In this and other cases, the mod(mdg4)u1 mutation appears to convert Su(Hw) from an insulator to a repressor protein (5, 19, 21). In contrast to the effects on yellow, mod(mdg4)u1 has effects similar to those of su(Hw) mutations and suppresses the mutant phenotypes caused by gypsy insertions at other loci, such as cut (18). In these cases, the mod(mdg4)u1 mutation appears to reduce insulation by gypsy without causing repression.
The mod(mdg4) gene encodes several proteins (3, 9, 21, 27). Mod(mdg4)-67.2 is the major product and is present at the majority of the few hundred sites on polytene chromosomes that bind mod(mdg4) proteins (4) [Mod(mdg4)-67.2 is encoded by the mRNA designated mod2.2 in reference 21]. Mod(mdg4)-67.2 interacts with Su(Hw) in vitro, and Su(Hw) is present at almost half the sites on polytene chromosomes that bind mod(mdg4) proteins (20, 21). The mod(mdg4)u1 mutation is a Stalker transposon insertion into the C-terminal exon unique to the Mod(mdg4)-67.2 mRNA (3, 21). It is likely, therefore, that a change in the activity of Mod(mdg4)-67.2 is responsible for the various effects that the mod(mdg4)u1 mutation has on the gypsy insulator.
Most mod(mdg4) mutations are recessive lethal, but mod(mdg4)u1 is homozygous viable, indicating that it is not a null allele. It has not been determined whether mod(mdg4)u1 simply reduces the activity of Mod(mdg4)-67.2. This lack of knowledge about the effects of mod(mdg4)u1 on Mod(mdg4)-67.2 activity has made it difficult to understand the precise role of Mod(mdg4)-67.2 in Su(Hw) insulator activity. Until now, the effect of the mod(mdg4) gene on the gypsy insulator has been deduced from flies homozygous for mod(mdg4)u1 or heterozygous for mod(mdg4)u1 and another mutation in mod(mdg4). Thus, Mod(mdg4)-67.2 may be a coinsulator that helps Su(Hw) block enhancers, or alternatively, Mod(mdg4)-67.2 may be a facilitator of enhancer-promoter communication that is targeted by Su(Hw). In the latter scenario, mod(mdg4)u1 would produce a mutant Mod(mdg4)-67.2 protein that is immune to Su(Hw).
Indeed, there is evidence that some mod(mdg4) products support gene activation. The first lethal mod(mdg4) alleles [originally named E(var)3-93D mutations] were isolated as dominant mutations that increase silencing of the white gene caused by heterochromatic position effect (10). This suggests that mod(mdg4) products promote white expression. mod(mdg4) mutations also increase the severity of homeotic transformations caused by mutations in trithorax group (trxG) genes, suggesting that mod(mdg4) gene products are themselves trxG proteins and promote expression of genes in the bithorax complex (9, 20). Consistent with this idea, expression of the Antennapedia complex is also reduced in mod(mdg4) mutants (20).
Here we report the results of experiments designed to clarify the role of Mod(mdg4)-67.2 in Su(Hw)-mediated insulation at the cut locus. Our results indicate that a multimer of wild-type Mod(mdg4)-67.2 interacts with Su(Hw) and contributes to insulation. We find that mod(mdg4)u1 is antimorphic and interferes with wild-type mod(mdg4) activity in insulation but is virtually wild type with regard to counteracting heterochromatin-mediated variegation of white gene expression. We also present evidence that Chip may be a molecular target of the gypsy insulator.
MATERIALS AND METHODS
Genetic crosses.
All crosses were conducted at 25°C unless indicated otherwise. Various mod(mdg4) mutant stocks and deficiencies were provided by Pavel Georgiev (Institute of Gene Biology, Moscow, Russia), Haini Cai (University of California, Berkeley), Manfred Frasch (Mt. Sinai School of Medicine, New York, N.Y.), Rainer Dorn (Martin Luther University, Halle, Germany), Mark Brennan (University of Louisville, Louisville, Ky.), Valerie Budnik (University of Massachusetts, Amherst), and the Indiana University (Bloomington) stock center (Table 1).
TABLE 1.
Dominant effects of mutant mod(mdg4) alleles on the ctK and In(1)wm4 mutant phenotypesa
| Allele | Mutagen | Viability | Effect of allele on phenotype ofb:
|
Source or reference | |
|---|---|---|---|---|---|
| ctK | In(1)wm4 | ||||
| mod(mdg4)ul | Stalker | Viable | S | O | 20 |
| mod(mdg4)T6 | EMS | Viable | S | O | 23 |
| mod(mdg4)neo129 | P element | Semilethal | sc | E | 12 |
| mod(mdg4)bpd1 | P element | Viable | O | NT | 27 |
| mod(mdg4)bpdEX57 | P excision | Lethal | s | O | 27 |
| mod(mdg4)20 | EMS | Lethal | O | NT | M. Frasch |
| mod(mdg4)117 | EMS | Lethal | O | NT | M. Frasch |
| mod(mdg4)269 | EMS | Lethal | O | NT | M. Frasch |
| mod(mdg4)324 | EMS | Lethal | O | NT | M. Frasch |
| mod(mdg4)334 | EMS | Lethal | O | NT | M. Frasch |
| mod(mdg4)340 | EMS | Lethal | sd | NT | M. Frasch |
| Df(3R)e-BS2 | X ray | Lethal | s | E | 3 |
EMS, ethyl methanesulfonate; S, strong suppression; s, weak suppression; O, no significant effect; E, strongly enhanced; NT, not tested.
Letters in the ctK column indicate the dominant effect on the wing-nicking phenotype displayed by ctK males. Selected wings representing the median effects of mod(mdg4)u1, mod(mdg4)T6, mod(mdg4)bpdEX57, and Df(3R)e-BS2 are shown in Fig. 1. Letters in the In(1)wm4 column indicate the dominant effect on the variegated eye color phenotype caused by heterochromatin-mediated silencing of white displayed by In(1)wm4 males. In most cases the effects were determined when the indicated allele was heterozygous with mod(mdg4)u1, but the mod(mdg4)neo129 allele has previously been reported to enhance variegation when heterozygous with wild-type mod(mdg4) (12). mod(mdg4)u1 had no significant effect when homozygous or when heterozygous with the wild type. Selected eyes representing the median phenotypes for the alleles tested are shown in Fig. 2.
At 29° some mod(mdg4)neo129 homozygotes were recovered, and these demonstrated strong but incomplete suppression of the ctK mutant phenotype.
With mod(mdg)340/+ only a small fraction of the wings showed weak suppression.
To observe the dominant effects of various mod(mdg4) mutations or deletions on the partial cut wing phenotype displayed by ctK, virgin cm ctK females were crossed to mod(mdg4) mutant males and progeny ctK males with the mod(mdg4) mutation were scored for the cut wing phenotype. Similarly, to observe the dominant effects of mod(mdg4) mutations on the ct53d cut wing phenotype, y w67c23 ct53d females were crossed to mod(mdg4) mutant males. Wings were mounted and photographed as described previously (11). The representative wings displayed in Fig. 1 were selected by arranging several wings in order of the severity of their cut wing phenotype and choosing the median wings to photograph. To quantify the phenotypes displayed by some genotypes, 10 randomly selected wings for each genotype were mounted and photographed using a 4× objective and a digital camera (Qimaging Microimager II) mounted on a Nikon microscope. The percentage of margin lacking bristles was determined for each wing using the curve measuring tool in Northern Eclipse 6.0 software (Empix Imaging). Statistical analysis was performed using Statview 5.0 software (SAS Institute).
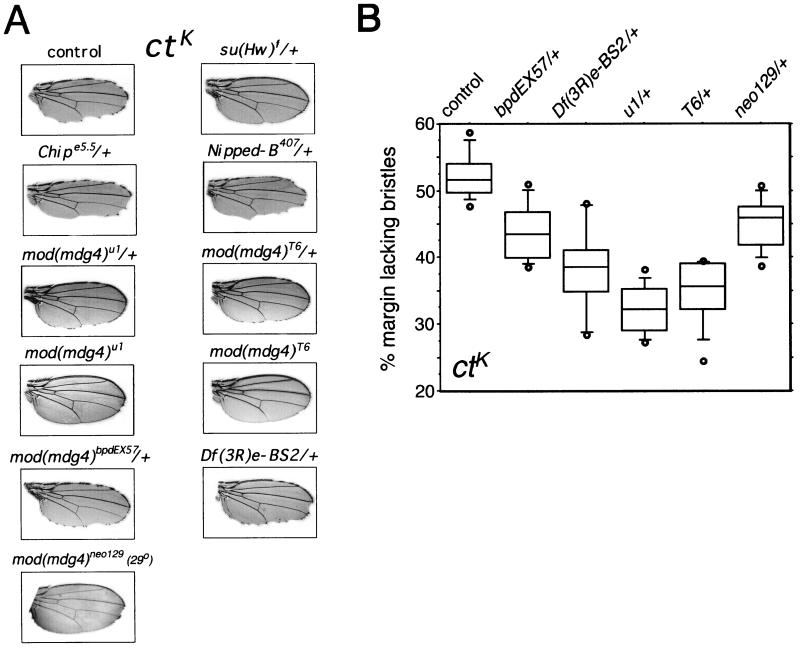
FIG. 1.
Dominant effects of mod(mdg4) mutations on the ctK cut wing phenotype. (A) Representative wings from the indicated genotypes. The mod(mdg4)u1 and mod(mdg4)T6 mutations strongly suppress the cut wing phenotype, and the mod(mdg4)bpdEX57 mutation and the Df(3R)e-BS2 deficiency that deletes the mod(mdg4) gene weakly suppress it. Rare mod(mdg4)neo129 homozygotes generated at 29°C show strong but incomplete suppression. The wings shown represent the median phenotypes for the indicated genotypes. (B) Box plot of the suppression of the ctK mutant phenotype by selected mod(mdg4) alleles. The percentage of the wing margin lacking bristles was measured from 10 randomly chosen wings for each genotype, and a box plot was generated. The five horizontal lines from top to bottom for each genotype are the 90th, 75th, 50th, 25th, and 10th percentiles. The circles are the minimum and maximum values observed for each genotype.
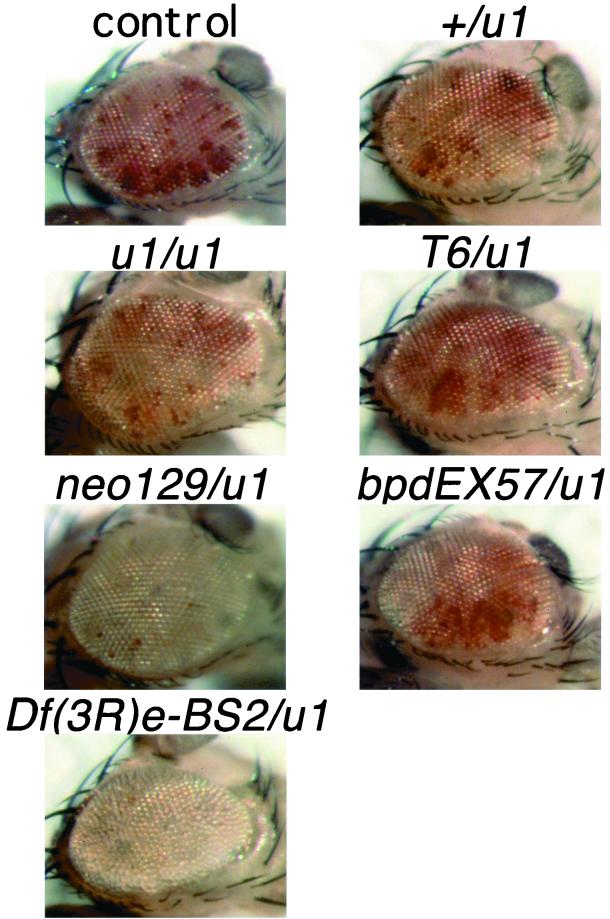
To examine the effects of various mod(mdg4) mutations on the position-effect variegation of white expression displayed by In(1)wm4, In(1)wm4 ctL-32 females were crossed to various mod(mdg4) mutant males and the male progeny were examined for the variegation of white expression in the adult eye. For each mod(mdg4) mutation, 20 or more flies were examined and compared side-by-side with controls. Initial comparisons were conducted by one person, and the flies were then coded and scored again in a blind fashion by a second person. In all cases the original scoring and rescoring were in agreement. The representative eyes shown in Fig. 2 were chosen by photographing several eyes for each genotype, arranging them in order of phenotypic severity, and selecting one with the median phenotype.
FIG. 2.
Effects of mod(mdg4) mutations on the In(1)wm4 variegation phenotype. The panels show representative eyes from In(1)wm4 males with the indicated combinations of mod(mdg4) alleles. mod(mdg4)neo129 and Df(3R)e-BS2 strongly enhance the variegation when heterozygous with mod(mdg4)u1. mod(mdg4)neo129 and deficiencies also strongly enhance the variegation when they are heterozygous with wild-type mod(mdg4) (10). The eyes shown represent the median phenotype displayed by the indicated genotype.
Structural analysis of mod(mdg4) mutations.
The mod(mdg4)bpd1 and mod(mdg4)bpdEX57 mutations were balanced over In(3LR)TM6,Tb ubi-GFP (provided by Kathryn Anderson, Sloan-Kettering Institute, New York, N.Y.), and homozygous mutant embryos and larvae lacking green fluorescent protein were collected using a fluorescence dissection microscope. Genomic DNA was prepared as described elsewhere (37). Genomic DNA samples were analyzed for the presence or absence of specific sequences by PCR using several different primers. These include a primer against the inverted repeat of the P element (5′-GAC GGG ACC ACC TTA TGT TA-3′), a primer against the first exon in the mod(mdg4)-67.2 mRNA (5′-GAC GCG TTC TGC GGG TCG-3′), and a primer against coding sequences present in the second exon (5′-CCA GCA CAA GCT GAA TTG CTC-3′).
Northern blot hybridization.
Total RNA from wild-type flies and mod(mdg4) mutants was isolated from different stages of development using the Trizma reagent (Sigma). The 2.3-kb mod(mdg4)-67.2 transcripts were detected by Northern blot hybridization performed as described elsewhere (13). Radioactively labeled single-stranded antisense RNA probes were prepared from a pGEM vector (Pharmacia) containing the EcoRI-BstEII fragment from the mod(mdg4)-67.2 cDNA (provided by Elizabeth Blackwood, University of California, San Diego). This probe hybridizes only to sequences deleted from the terminal exon in the mod(mdg4)u1 mutant. As an internal standard, ethidium bromide-stained rRNA was measured by densitometric scanning of a photographic negative.
Affinity chromatography.
Affinity chromatography experiments were conducted as described elsewhere (55) using various glutathione S-transferase (GST) fusion proteins (see below) expressed in Escherichia coli and bound to glutathione beads and various 35S-labeled proteins prepared by translation in vitro. Briefly, binding reactions were conducted at 4°C for 1 h in phosphate-buffered saline containing 1% Triton X-100, 2% nonfat milk, protease inhibitors, and 1 mM dithiothreitol. Each reaction mixture contained ∼10−13 pmol of 35S-labeled proteins and beads with ∼10 to 20 μg of GST fusion protein in a total volume of 200 μl. After extensive washing, the beads were boiled in 40 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and the amount of labeled protein that was retained was quantified with a phosphorimager after SDS-PAGE.
GST-fusion constructs.
To make the GST-Mod(mdg4)-67.2 fusion, the open reading frame encoding amino acids 1 to 610 from the Mod(mdg4)-67.2 cDNA clone was amplified by PCR (primers 5′-ATT GGA TCC AAG ATG GCG GAC GAC GAG CAA-3′ and 5′-ATA TGA TAT CGG CAT ATT CCG TGT CAC TTC T-3′) and cloned into the BamHI and SmaI sites of pGEX-2T (Pharmacia).
The GST-Su(Hw)-CTD (C-terminal domain) fusion was made by cloning the EcoRI fragment from pGEM-3Z-SUHW* (35) into the EcoRI site of pGEX-1ZT (Patricia Cortes, Rockefeller University, personal communication), which was constructed by insertion of a polylinker into the EcoRI site of pGEX-1λ 84 (Pharmacia). GST-Su(Hw)-CTD fusions with various deletions in the Su(Hw)-CTD were made by cloning EcoRI-SmaI fragments from the previously described deletion constructs in the pCasper vector (35) into the EcoRI-blunted HindIII sites of pGEX-1ZT.
In vitro translation constructs.
The full-length Su(Hw) construct and the Su(Hw) construct lacking the CTD were described elsewhere (55). A full-length Mod(mdg4)-67.2 in vitro translation construct was made by cloning the BamHI-EcoRV fragment of the PCR product described above into the BglII-EcoRV sites of the pING14.1 vector (S. Ingles and I. Brierly [55]). To make in vitro translation vectors for Mod(mdg4)-67.2u1 and the Mod(mdg4)-67.2-CTD (CTD residues 307 to 610), products generated by PCR [5′-ATT AGG ATC CAA GAT GGC GGA CGA C-3′ and 5′-ATT AGG ATC CGC CAT GGA ATT GCC CAC GAA A-3′ primers for Mod(mdg4)-67.2u1; 5′-ATT AGA TAT CCT ACA CAT TGA AGA TTA GCT TGA-3′ and 5′-ATA TGA TAT CGG CAT ATT CCG TGT CAC TTC T-3′ primers for Mod(mdg4)-67.2-CTD] were cloned into the BglII-EcoRV sites of pING14.1. To generate Mod(mdg4)-67.2T6, we used primers 5′-ATT AGG ATC CAA GAT GGC GGA CGA C-3′ and 5′-TTC AAA TCT CAA ACT CCT CGA A-3′, and the product was ligated to the pCRII-TOPO vector (Invitrogen). To produce the Mod(mdg4)-67.2 N-terminal domain (residues 1 to 308), the NcoI-EcoRI fragment was deleted from pGEX-2T- Mod(mdg4)-67.2 by digestion and blunt-end ligation, and the BamHI-blunted EcoRI fragment of the resulting plasmid was cloned into the BglII-EcoRV sites of pING14.1. All clones produced by PCR were sequenced at the 5′ end of the coding region, and the Mod(mdg4)-67.2u1 clone was sequenced at both the 5′ and 3′ ends.
RESULTS
Some mod(mdg4) mutations decrease insulation by the ctKgypsy insertion.
If Mod(mdg4)-67.2 is required for insulation by Su(Hw) and gypsy, loss-of-function mutations in mod(mdg4) should reduce insulation by a gypsy insertion. In contrast, if Mod(mdg4)-67.2 facilitates enhancer-promoter communication, then loss-of-function mutations in mod(mdg4) either would have no effect on insulation or might increase insulation. The results described below indicate that some loss-of-function alleles of mod(mdg4) can reduce insulation by gypsy at cut, but more weakly than does mod(mdg4)u1.
We used the ctK gypsy insertion in cut to compare the abilities of several mutant alleles of mod(mdg4) to alter insulation. Gypsy insertions in cut block activation of the cut promoter by a wing margin-specific enhancer located some 85 kb upstream of the promoter (11, 31). The ctK gypsy only partially insulates the wing margin enhancer of cut, resulting in an intermediate cut wing phenotype, presumably because it contains fewer Su(Hw)-binding sites than most gypsy elements (Fig. 1A) (29). The ctK mutant phenotype is more sensitive to the levels of Su(Hw) activity than most gypsy insertions and is almost completely suppressed by heterozygous su(Hw) mutations (Fig. 1A) (49). ctK is also highly sensitive to the activities of the Chip and Nipped-B proteins, which facilitate activation by the wing margin enhancer, and is made more severe by heterozygous Chipe5.5 and Nipped-B407 loss-of-function mutations (Fig. 1A). We reasoned therefore that ctK might also be very sensitive to Mod(mdg4)-67.2 activity and would allow us to test whether both viable and lethal alleles of mod(mdg4) have dominant effects on insulation.
As illustrated in Fig. 1 and summarized in Table 1, heterozygous mod(mdg4)u1 and mod(mdg4)T6 mutations suppressed the cut wing phenotype of ctK. Similar to mod(mdg4)u1, the mod(mdg4)T6 allele is homozygous viable and produces a Mod(mdg4)-67.2 protein truncated near the C terminus (M. Brennan, personal communication). The suppression by heterozygous mod(mdg4)u1 and mod(mdg4)T6 was not as strong as that observed with a heterozygous su(Hw) mutation (Fig. 1A). Homozygous mod(mdg4)u1 and mod(mdg4)T6 mutations suppressed more strongly than did the heterozygous mutations, although the suppression was still incomplete (Fig. 1A).
In contrast to the suppression by mod(mdg4)u1 and mod(mdg4)T6, most lethal mod(mdg4) mutations tested had little or no dominant effect on the ctK mutant phenotype (Table 1). However, the mod(mdg4)bpdEX57 allele and the Df(3R)e-BS2 deficiency, which deletes mod(mdg4), both suppressed ctK, but more weakly than did mod(mdg4)u1 and mod(mdg4)T6 (Table 1; Fig. 1). Although weak, the suppression was reproducible in repeated experiments.
Because most lethal mod(mdg4) alleles had no effect on the ctK phenotype, we were concerned that the weaker suppression observed with mod(mdg4)bpdEX57 and Df(3R)e-BS2 might be caused by mutations in other genes besides mod(mdg4). The mod(mdg4)bpdEX57 mutation was generated by excision of a P element inserted 27 bp upstream of the putative transcription start site for the mod(mdg4)-67.2 transcript in the mod(mdg4)bpd1 mutation (25). The mod(mdg4)bpdEX57 mutation also fails to complement mutations in tinman, the gene just upstream of mod(mdg4). We performed PCR analysis on genomic DNA isolated from mod(mdg4)bpdEX57 and mod(mdg4)bpd1 homozygous mutant larvae and confirmed that the P element is absent in mod(mdg4)bpdEX57 (see Materials and Methods). Northern blot hybridization with a probe that hybridizes only to exon sequences downstream of the Stalker insertion in mod(mdg4)u1 revealed that mod(mdg4)bpdEX57/mod(mdg4)u1 adults contain less than 5% wild-type levels of the 2.3-kb mod(mdg4)-67.2 mRNA (not shown). These results confirm that mod(mdg4)bpdEX57 is a loss-of-function mutation. Although the tinman gene is mutant or absent in the mod(mdg4)bpdEX57 and Df(3R)e-BS2 chromosomes, the tinEC40 null allele in the absence of mod(mdg4) mutations had no dominant effect on the ctK mutant phenotype (not shown).
If the effects of the mod(mdg4)bpdEX57 and Df(3R)e-BS2 chromosomes on the ctK phenotype are the result of defects in other genes that regulate cut, it is likely that they would not be specific for gypsy insertions but would also modify the phenotypes of other sensitive mutations in cut. The ct53d lesion is a small deletion in the wing margin enhancer that partially reduces enhancer strength, and the ct53d phenotype is dominantly enhanced by mutations in several genes that regulate cut, including Chip, scalloped, vestigial, mastermind, Nipped-A, and l(2)41Af, but is not affected by su(Hw) mutations (30, 41, 48). None of the several heterozygous mod(mdg4) mutations tested, including mod(mdg4)u1 and mod(mdg4)bpdEX57, suppressed or enhanced the partial cut wing phenotype displayed by ct53d males (not shown).
The inability of most lethal mod(mdg4) mutations to suppress the ctK mutant phenotype suggests that they may affect essential Mod(mdg4) proteins that are not involved in insulation or that they may not reduce Mod(mdg4) protein activity enough to suppress the mutant phenotype. Experiments with the mod(mdg4)neo129 cold-sensitive allele are consistent with the notion that the level of mod(mdg4) activity can be critical. At 25°C, mod(mdg4)neo129 is homozygous lethal, but at 29°C, it is semilethal and a few mod(mdg4)neo129 homozygotes survive to adulthood (9). At 25 and 29°C, we found that mod(mdg4)neo129 had only a very slight dominant effect on the ctK mutant phenotype, but the few mod(mdg4)neo129 homozygotes produced at 29°C displayed strong but incomplete suppression (Fig. 1A). Based on this and the weak dominant suppression of the ctK mutant phenotype by mod(mdg4)bpdEX57 and Df(3R)e-BS2 loss-of-function alleles, we conclude that at least some Mod(mdg4) proteins contribute to insulation by the ctK gypsy insertion.
The results portrayed in Fig. 1 also indicate that the mod(mdg4)u1 and mod(mdg4)T6 mutations are not simple loss-of-function (hypomorphic) mutations. They suppressed ctK more strongly than did the loss-of-function alleles, suggesting that their products actively interfere with the activities of the remaining dose of wild-type Mod(mdg4) proteins. We considered the possibility that the mod(mdg4)u1 and mod(mdg4)T6 chromosomes might contain other mutations that increase their effects on ctK. It is unlikely, however, that both chromosomes, which were isolated independently and are homozygous viable, would contain similar additional mutations. It is much more likely that they have similar effects because they both produce similarly truncated Mod(mdg4)-67.2 proteins. We conclude, therefore, that the mod(mdg4)u1 and mod(mdg4)T6 mutations are antimorphic and inhibit the function of wild-type mod(mdg4) in insulation by gypsy in a dominant-negative manner.
The mod(mdg4)u1 and mod(mdg)T6 mutations do not substantially alter position-effect variegation of In(1)wm4.
Some alleles of mod(mdg4) were isolated as dominant mutations that increase heterochromatin-mediated silencing [position effect variegation (PEV)] of the white gene in the In(1)wm4 chromosomal rearrangement (10). The dominant-negative effects of mod(mdg4)u1 and mod(mdg4)T6 on insulation by gypsy raised the question of whether these mutations are also antimorphic with regard to enhancement of PEV. Unexpectedly, mod(mdg4)u1/+, mod(mdg4)T6/mod(mdg4)u1, and mod(mdg4)bpdEX7/mod(mdg4)u1 did not detectably enhance the PEV displayed by In(1)wm4 (Fig. 2). In contrast, mod(mdg4)neo129/mod(mdg4)u1 and Df(3R)e-BS2/mod(mdg4)u1 both strongly enhanced variegation (Fig. 2). mod(mdg4)u1/mod(mdg4)u1 flies displayed a weak enhancement of the PEV, but this is likely to be a background effect, because mod(mdg4)T6/mod(mdg4)u1 had the opposite effect and mildly suppressed the PEV. Previously it had been observed that mod(mdg4)neo129 and deficiencies that remove mod(mdg4) also dominantly enhanced the PEV displayed by In(1)wm4 when heterozygous with wild-type mod(mdg4) (10). The enhancement of PEV that was observed when mod(mdg4)neo129 and Df(3R)e-BS2 were heterozygous with mod(mdg4)u1 indicates that the mod(mdg4)u1 chromosome does not contain a counteracting dominant suppressor-of-variegation mutation that would mask any effects of mod(mdg4)u1 on PEV. We conclude, therefore, that mod(mdg4)u1 and mod(mdg4)T6 are essentially wild type with regard to their effects on variegation of In(1)wm4.
The Mod(mdg4)-67.2 protein interacts with the insulation domain of Su(Hw).
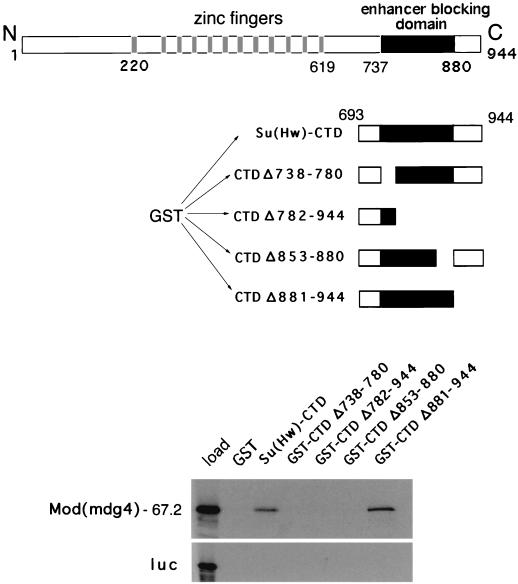
It has previously been reported that Mod(mdg4) proteins interact with full-length Su(Hw) in vitro (21). Our laboratory has previously shown that a region of some 140 amino acids (amino acids 737 to 880) in Su(Hw) contains the residues required for strong enhancer-blocking activity in vivo (35). To test whether this insulation region interacts with the Mod(mdg4)-67.2 protein, we expressed a fusion protein in which GST was fused to the CTD of Su(Hw) [Su(Hw)-CTD, residues 673 to 943] in E. coli and bound the fusion protein to glutathione beads to make an affinity matrix. We also constructed a series of similar affinity matrices with various deletion mutants of Su(Hw)-CTD (Fig. 3). Full-length 35S-Mod(mdg4)-67.2 protein was prepared by translation in vitro and incubated with the wild-type and mutant GST-Su(Hw)-CTD beads. Consistent with previous reports that Mod(mdg4) proteins interact with full-length Su(Hw) (23), Mod(mdg4)-67.2 bound to the wild-type GST-Su(Hw)-CTD beads (Fig. 3, lane 3) but not to GST control beads (Fig. 3, lane 2). In contrast, Mod(mdg4)-67.2 did not bind to any of the mutant forms of GST-Su(Hw)-CTD in which portions of the insulation domain (residues 737 to 880) were deleted (Fig. 3, lanes 4, 5, and 6).
FIG. 3.
The Mod(mdg4)-67.2 protein interacts with a region of Su(Hw) that is required for insulation. The Su(Hw) protein contains 12 zinc fingers (gray boxes) and an enhancer-blocking region within residues 737 to 880 (black box). The Su(Hw)-CTD and the indicated mutant forms fused to GST were expressed in E. coli and bound to glutathione beads. The autoradiograms of SDS-PAGE gels show the amount of 35S-Mod(mdg4)-67.2 and 35S-luciferase control protein (luc) prepared by translation in vitro that bound to the various GST-Su(Hw)-CTD beads and to GST-only control beads. The left-hand lane (load, lane 1) shows the amount of input labeled proteins. Mod(mdg4)-67.2 binds to the GST-Su(Hw)-CTD beads (lane 3), but deletions that affect the enhancer-blocking region of Su(Hw) prevent binding of Mod(mdg4)-67.2 (lanes 4, 5 and 6). All the results were reproduced in multiple independent experiments.
To test for other Su(Hw) domains that may interact with Mod(mdg4)-67.2, GST was fused to full-length Mod(mdg4)-67.2 and the fusion protein bound to beads. Full-length Su(Hw) prepared by translation in vitro bound to the GST-Mod(mdg4)-67.2 beads, but Su(Hw) with the C-terminal residues (673 to 943) deleted did not (data not shown). These results indicate that the CTD is the only portion of Su(Hw) that interacts with Mod(mdg4)-67.2.
Mutant Mod(mdg4)-67.2 proteins produced by mod(mdg4)u1 and mod(mdg4)T6 do not interact with Su(Hw).
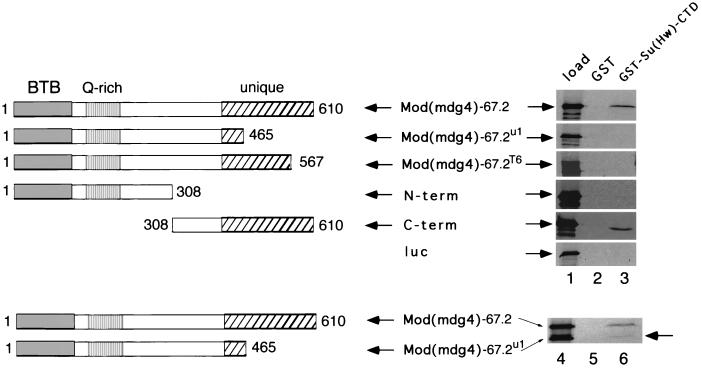
As shown above, the viable mod(mdg4)u1 and mod(mdg4)T6 mutations more strongly reduce insulation by ctK gypsy than do lethal and null alleles of mod(mdg4). The experiments of Fig. 4 show that the residues of Mod(mdg4)-67.2 truncated by the mod(mdg4)u1 and mod(mdg4)T6 mutations are required to interact with Su(Hw).
FIG. 4.
The truncated Mod(mdg4)-67.2u1 and Mod(mdg4)T6 proteins do not interact with Su(Hw). The Mod(mdg4)-67.2 protein contains a BTB/POZ motif (gray box) at the N terminus common to most if not all Mod(mdg4) proteins, a glutamine-rich (Q-rich) region, and a unique C terminus (cross-hatched box) (3). 35S-labeled fragments of Mod(mdg4)-67.2, including fragments that mimic the predicted mutant proteins produced by the mod(mdg4)u1 and mod(mdg4)T6 alleles, were prepared by translation in vitro and tested for their ability to bind GST-Su(Hw)-CTD beads and GST-only control beads. The autoradiograms of SDS-PAGE gels show the amount of input labeled protein (load, lane 1) and the amount that bound to the beads indicated at the tops of lanes 2 and 3. The full-length Mod(mdg4)-67.2 and the C-terminal fragment of Mod(mdg4)-7.2 both bound to the GST-Su(Hw)-CTD beads (lane 3), while the truncated Mod(mdg4)-67.2u1, Mod(mdg4)-67.2T6, and N-terminal fragment of Mod(mdg4)-67.2 did not (lane 3). As expected, the luciferase (luc) control protein also did not bind to any of the beads. In the lowest panel, nearly equal amounts of labeled full-length Mod(mdg4)-67.2 and Mod(mdg4)-67.2u1 were prepared by cotranslation in vitro. In this case, a small but significant amount of Mod(mdg4)-67.2u1 was retained (lane 6, arrow). All the results shown were reproducible in independent experiments.
The mod(mdg4)u1 mutation is a Stalker transposon insertion (18, 21). The residues affected are unique to the Mod(mdg4)-67.2 protein and are not present in other mod(mdg4) products. The Stalker insertion results in replacement of several C-terminal residues of Mod(mdg4)-67.2 by a single residue encoded by Stalker sequences (M. Brennan, personal communication). Thus, the Mod(mdg4)-67.2u1 protein contains wild-type residues 1 to 465 out of 610 (Fig. 4). The mod(mdg4)T6 mutation was induced by chemical mutagenesis and, similar to mod(mdg4)u1, is also homozygous viable (21; Mark Brennan, personal communication). The mod(mdg4)T6 mutation also results in truncation of Mod(mdg4)-67.2, resulting in a protein that contains wild-type residues 1 to 567 (Fig. 4; M. Brennan, personal communication). In contrast to the full-length Mod(mdg4)-67.2 protein, the Mod(mdg4)-67.2u1 and Mod(mdg4)-67.2T6 mutant proteins prepared by in vitro translation did not bind to GST-Su(Hw)-CTD beads (Fig. 4, lane 3).
The Mod(mdg4)-67.2u1 mutant protein interacts with wild-type Mod(mdg4)-67.2.
The genetic data described above suggest that the mod(mdg4)u1 mutant products interfere in a dominant-negative (antimorphic) fashion with the remaining dose of wild-type mod(mdg4) products. The predicted mod(mdg4) proteins contain a BTB/POZ domain at the N terminus (3). BTB/POZ domains of other proteins have been shown to self-interact (8, 32, 38), leading us to postulate that the Mod(mdg4)-67.2u1 protein might form inactive multimers with wild-type Mod(mdg4)-67.2.
An experiment depicted in Fig. 4 provides evidence that the Mod(mdg4)-67.2u1 protein interacts with wild-type Mod(mdg4)-67.2. When wild-type Mod(mdg4)-67.2 and Mod(mdg4)-67.2u1 were translated together and incubated with GST-Su(Hw)-CTD beads, a small but significant amount of Mod(mdg4)-67.2u1 protein was retained (Fig. 4, lane 6). Because the truncated Mod(mdg4)-67.2 protein bound to the Su(Hw) C-terminal region only in the presence of full-length Mod(mdg4)-67.2 protein, we infer that the two Mod(mdg4)-67.2 proteins interact with each other, presumably through the BTB/POZ domain or other N-terminal residues.
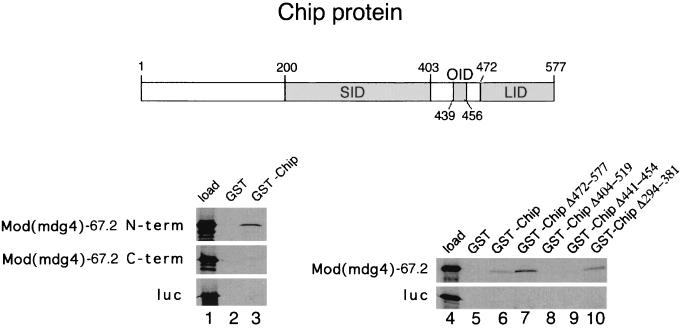
Mod(mdg4)-67.2 interacts with the Chip facilitator protein.
The putative facilitator protein Chip interacts with the zinc finger region of Su(Hw) in vitro, supporting the notion that the in vivo antagonism between Chip and Su(Hw) insulator activity is direct (41, 55). The experiments portrayed in Fig. 5 further support this idea by indicating that Chip also interacts with Mod(mdg4)-67.2. An N-terminal fragment of Mod(mdg4)-67.2 (residues 1 to 308) containing the BTB/POZ domain bound to GST-Chip beads, but a C-terminal fragment (residues 307 to 610) did not (Fig. 5, lane 3).
FIG. 5.
Mod(mdg4)-67.2 interacts with the Chip facilitator protein. The Chip protein contains multiple protein interaction regions (gray boxes) including the SID, the LID, and the OID that interacts with several homeodomain proteins and the Su(Hw) zinc finger region (55). 35S-labeled N-terminal and C-terminal fragments of Mod(mdg4)-67.2 (see Fig. 4) and a luciferase control protein (luc) were prepared by translation in vitro and tested for their ability to bind to GST control beads and GST-Chip fusion protein beads (55). The autoradiograms of SDS-PAGE gels on the left (lanes 1, 2, and 3) show the amount of input protein (load, lane 1) and the amount that was retained by the indicated beads. The N-terminal fragment bound to the GST-Chip beads, but the C-terminal fragment and the luciferase control did not (lane 3). The autoradiogram of an SDS-PAGE gel on the right (lanes 4 through 10) shows the binding of full-length 35S-labeled Mod(mdg4)-67.2 prepared by in vitro translation to beads containing fusions of GST to wild-type Chip and various deletion mutants of Chip (55). The load lane (lane 4) contains a tenth of the input protein. Deletions affecting only the SID (lane 10) or only the LID (lane 7) did not affect binding of Mod(mdg4)-67.2, while deletions affecting the OID (lanes 8 and 9) displayed substantially reduced binding. All the results shown were reproduced in independent experiments.
Experiments depicted in Fig. 5 also indicate that Mod(mdg4)-67.2 interacts with the same region of Chip that interacts with Su(Hw). The Chip self-interaction domain (SID) mediates Chip-Chip interactions, the LIM interaction domain (LID) interacts with LIM domains, and the other interaction domain (OID) supports self-interactions, interactions with several homeodomain proteins, and interactions with the zinc finger region of Su(Hw) (55). Full-length Mod(mdg4)-67.2 prepared by translation in vitro bound to beads containing mutant GST-Chip fusion proteins that lack the LID or SID (Fig. 5, lanes 7 and 10) but substantially less to beads containing GST-Chip mutants that lack the OID (Fig. 5, lanes 8 and 9). As shown previously, luciferase prepared by translation in vitro did not bind to any of the GST-Chip beads (55) (Fig. 5).
DISCUSSION
Mod(mdg4) proteins contribute to the insulator activity of Su(Hw).
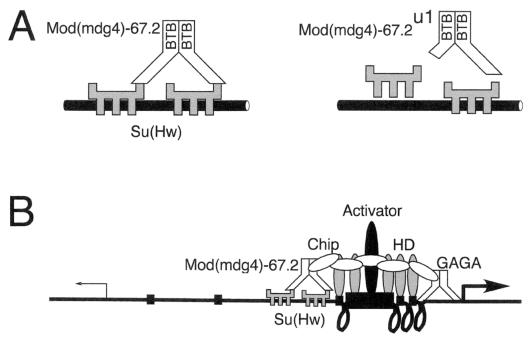
We found that certain loss-of-function alleles of mod(mdg4) reduce insulation by the Su(Hw) protein in the cut gene. This is evidence that mod(mdg4) products are not simply targets of Su(Hw) insulator activity but contribute to the insulator activity of Su(Hw). We also found that wild-type Mod(mdg4)-67.2, the major protein product of mod(mdg4), interacted with a region of Su(Hw) that our laboratory has previously shown is required for insulation in vivo, but the truncated versions of the Mod(mdg4)-67.2 proteins produced by the viable mod(mdg4)u1 and mod(mdg4)T6 alleles did not. This is consistent with the previous report that binding of Mod(mdg4) proteins to Su(Hw) binding sites on salivary gland polytene chromosomes is greatly reduced in mod(mdg4)u1 mutants (20). We also found that mod(mdg4)u1 and mod(mdg4)T6 more strongly reduce insulator activity than do null alleles of mod(mdg4) and that this antimorphic nature of mod(mdg4)u1 may stem from the ability of the mutant protein to interact with wild-type Mod(mdg4)-67.2 protein. To explain these observations, we propose a model in which a multimer of Mod(mdg4)-67.2 interacts with more than one Su(Hw) molecule to form the active insulator complex, and the truncated Mod(mdg4)-67.2 proteins produced by mod(mdg4)u1 and mod(mdg4)T6 destabilize this complex (Fig. 6A).
FIG. 6.
Proposed model for the role of Mod(mdg4)-67.2 protein in insulation. (A) To explain why truncated Mod(mdg4)-67.2 proteins have dominant-negative (antimorphic) effects on insulator activity, we propose, as shown on the left, that a multimer of Mod(mdg4)-67.2 interacts with more than one DNA-bound molecule of Su(Hw) to form a stable insulator complex. As shown on the right, truncated Mod(mdg4)-67.2u1 or Mod(mdg4)-67.2T6 protein would destabilize the complex, leading to reduced interaction between Mod(mdg4)-67.2 and Su(Hw) and/or reduced binding of Su(Hw) to DNA. (B) To explain how the Mod(mdg4)-67.2 protein contributes to the insulator activity of Su(Hw), we propose, as depicted on the left, that it traps facilitators such as Chip and blocks the previously postulated Chip-assisted spread of homeodomain protein (HD) binding between the enhancer and promoter (12, 55). As depicted on the right, this spread could create a series of loops that brings the enhancer and promoter closer together or could aid the binding of a surrogate activator near the promoter. Like the Mod(mdg4) proteins, GAGA factor is a group of BTB/POZ-containing trxG proteins that can also both aid activation and help insulate (44, 45). GAGA factor binds proximal to several promoters and potentiates activation of the engrailed promoter by a distal enhancer (46). We speculate, as shown with the promoter on the right, that when BTB/POZ proteins such as GAGA bind just upstream of a promoter, they anchor activators close to the promoter and thereby aid activation.
An alternative explanation for why mod(mdg4)u1 has a stronger effect on insulation than does deletion of mod(mdg4) is that Mod(mdg4)-67.2 may support both activation of cut and insulation by Su(Hw). In this case, mod(mdg4)u1 would suppress the ctK mutant phenotype more strongly than a null allele because the truncated Mod(mdg4)-67.2u1 protein can still activate cut but does not contribute to insulator activity, while a null allele would simultaneously reduce both insulation and activation. Our data suggest, however, that mod(mdg4) does not normally regulate cut in the absence of a gypsy insertion. Mutations in most genes known to regulate cut dominantly enhance the weak cut wing phenotype of ct53d mutants (41, 48), but none of the several mutant mod(mdg4) alleles tested had an effect on ct53d.
Interactions between Mod(mdg4)-67.2 and Su(Hw) are not required for the critical functions of Mod(mdg4)-67.2 and Su(Hw).
The available evidence supports the notion that the insulation region of Su(Hw) interacts with other proteins besides Mod(mdg4)-67.2. Although viable, females homozygous for most su(Hw) mutations are sterile because of a block in oogenesis (36). Our laboratory has previously reported that the insulation region of Su(Hw) shown here to interact with Mod(mdg4)-67.2 is required for female fertility (35). Females homozygous for mod(mdg4)u1 are fertile, indicating that interactions between the insulation region of Su(Hw) and Mod(mdg4)-67.2 are not necessary to support oogenesis. This raises the possibility that the insulation region of Su(Hw) interacts with other proteins besides Mod(mdg4)-67.2. Indeed, the viability of both su(Hw) and mod(mdg4)u1 mutants indicates that the interactions between Su(Hw) and Mod(mdg4)-67.2 are also not required for any of the essential functions of mod(mdg4).
Is Chip a target of the Su(Hw)-Mod(mdg4)-67.2 insulator?
In this study we observed a direct in vitro interaction between Mod(mdg4)-67.2 and the Chip protein. Our laboratory has previously shown by genetic experiments that Chip and the Su(Hw) insulator protein are specifically antagonistic to each other in vivo (41). It was suggested, therefore, that Chip may act between enhancers and promoters to aid communication. Consistent with this idea, it was found that Chip is widely expressed and potentiates activation by diverse enhancers in several genes (41, 42).
Our laboratory has proposed a model in which Chip aids enhancer-promoter communication by assisting the binding of homeodomain proteins to several sites between enhancers and promoters (Fig. 6B) (12, 55). This model was based in part on in vitro interactions between Chip and diverse homeodomain proteins and the positive effects that Chip has on Bicoid homeodomain protein activity in vivo (55). It was also motivated by the in vivo binding of several homeodomain proteins to many sites between enhancers and promoters (6). The domain of Chip that interacts with homeodomain proteins also interacts with the zinc finger region of Su(Hw), consistent with the notion that the Chip-homeodomain interaction is a direct target of Su(Hw) (55). The in vitro interactions between Chip and the Mod(mdg4)-67.2 protein reported here suggest that Chip is a direct target of both proteins in the Su(Hw)-Mod(mdg4)-67.2 insulator complex (Fig. 6B).
In our experiments the N-terminal region of Mod(mdg4)-67.2 interacted with Chip, while the C-terminal portion interacted with Su(Hw). Both Chip and Mod(mdg4)-67.2 appear to form multimers in vitro. These observations suggested that it might be possible to form higher-order in vitro complexes containing Chip, Mod(mdg4)-57.2, and Su(Hw). Our preliminary attempts, however, to form such complexes by simultaneously binding Mod(mdg4)-67.2 and Su(Hw) to GST-Chip beads were unsuccessful. It appeared that in the presence of both Su(Hw) and Mod(mdg)-67.2, Chip preferred to interact with Su(Hw). Because Su(Hw) and Mod(mdg4)-67.2 both interact with the same region of Chip, it is possible that they compete with each other. It is also feasible that interactions between Chip and Mod(mdg4)-67.2 are transitory in the context of the complete insulator protein complex.
The Mod(mdg4)-67.2 and GAGA BTB/POZ proteins have both insulator and activator activities.
The evidence that Mod(mdg4)-67.2 is an active component of the gypsy insulator that blocks gene activation appears at first glance to be contradictory to the evidence indicating that the mod(mdg4) gene is a member of the trxG of genes that activate genes in the bithorax complex (9, 20). Another trxG protein, however, also appears to have insulator activity. The GAGA factor encoded by the Trithorax-like (Trl) gene is similar to Mod(mdg4)-67.2 in that it contains a BTB/POZ motif at the N terminus, self-interacts, and supports activation of the bithorax complex (14, 15, 32, 47, 52). GAGA factor is also required for enhancer blocking by the insulator associated with the even-skipped (eve) promoter (45). This insulator activity requires GAGA binding sites just proximal to the transcription start site and is diminished by Trl mutations. Potential GAGA binding sites are found just proximal to many promoters in Drosophila, including sequences associated with insulator activity in the α1 tubulin gene promoter (44).
The GAGA-dependent insulator just proximal to the eve promoter does not prevent activation of the eve promoter by upstream enhancers even though it is positioned between them. Indeed, GAGA binding sites just proximal to the engrailed gene promoter potentiate activation by an upstream enhancer (46). To resolve the paradoxical insulator and activator activities of the GAGA and Mod(mdg4)-67.2 BTB/POZ proteins, therefore, we theorize that the function of promoter-proximal insulators is to aid activation of the promoters that contain them by helping to capture and anchor distal activator or facilitator proteins near the promoter (Fig. 6B). If so, it is feasible that the Mod(mdg4)-67.2 protein has a promoter-anchoring function in the bithorax complex, but when bound to Su(Hw), it anchors activator or facilitator proteins far from the promoter, thereby preventing activation.
ACKNOWLEDGMENTS
We thank Beth Blackwood for providing the mod(mdg4)-67.2 cDNA clone and sharing unpublished data, Mark Brennan for providing fly stocks and sharing unpublished data, and Pavel Georgiev, Rainer Dorn, Manfred Frasch, Haini Cai, Valerie Budnik, Kathryn Anderson, and the Indiana University stock center for providing fly stocks. We are also grateful to Joel Eissenberg for thoughtful comments on the manuscript.
This work was supported by NIH grant no. RO1 GM55683 to D.D.
REFERENCES
- 1.Bell A C, Felsenfeld G. Stopped at the border: boundaries and insulators. Curr Opin Genet Dev. 1999;9:191–198. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- 2.Bell A C, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 3.Büchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, Dorn R. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics. 2000;155:141–157. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 5.Cai H N, Levine M. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 1997;16:1732–1741. doi: 10.1093/emboj/16.7.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr A, Biggin M D. A comparison of in vivo and in vitro DNA binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. EMBO J. 1999;18:1598–1608. doi: 10.1093/emboj/18.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J H, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 8.Dhordain P, Albagli O, Ansieau S, Koken M H, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert J P, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- 9.Dorn R, Krauss V, Reuter G, Saumweber H. The enhancer of position-effect variegation of Drosophila, E(var)3–93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proc Natl Acad Sci USA. 1993;90:11376–11380. doi: 10.1073/pnas.90.23.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn R, Szidonya J, Korge G, Sehnert M, Taubert H, Archoukieh E, Tschiersch B, Morawietz H, Wustmann G, Hoffmann G, Reuter G. P transposon-induced dominant enhancer mutations of position-effect variegation in Drosophila melanogaster. Genetics. 1993;133:279–290. doi: 10.1093/genetics/133.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorsett D. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics. 1993;134:1135–1144. doi: 10.1093/genetics/134.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsett D. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr Opin Genet Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 13.Dorsett D, Viglianti G A, Rutledge B J, Meselson M. Alteration of hsp82 gene expression by the gypsy transposon and suppressor genes in Drosophila melanogaster. Genes Dev. 1989;3:454–468. doi: 10.1101/gad.3.4.454. [DOI] [PubMed] [Google Scholar]
- 14.Espinas M L, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, Azorin F. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J Biol Chem. 1999;274:16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 15.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 16.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gdula D A, Gerasimova T I, Corces V G. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgiev P, Gerasimova T. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol Gen Genet. 1989;220:121–126. doi: 10.1007/BF00260865. [DOI] [PubMed] [Google Scholar]
- 19.Georgiev P, Kozycina M. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics. 1996;142:425–436. doi: 10.1093/genetics/142.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerasimova T, Corces V G. Polycomb and Trithorax group proteins mediate the function of the chromatin insulator. Cell. 1998;92:511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 21.Gerasimova T, Gdula D, Gerasimov D, Simonova O, Corces V G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 22.Geyer P K. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- 23.Geyer P K, Corces V G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 24.Geyer P K, Green M M, Corces V G. Tissue-specific transcriptional enhancers may act in trans on the gene located on the homologous chromosome: the molecular basis of transvection in Drosophila. EMBO J. 1990;9:2247–2256. doi: 10.1002/j.1460-2075.1990.tb07395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorczyca M, Popova E, Jia X X, Budnik V. The gene mod(mdg4) affects synapse specificity and structure in Drosophila. J Neurobiol. 1999;39:447–460. doi: 10.1002/(sici)1097-4695(19990605)39:3<447::aid-neu10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 26.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 27.Harvey A J, Bidwai A P, Miller L K. Doom, a product of the Drosophila mod(mdg4) gene, induces apoptosis and binds to baculovirus inhibitor-of-apoptosis proteins. Mol Cell Biol. 1997;17:2835–2843. doi: 10.1128/mcb.17.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holdridge C, Dorsett D. Repression of hsp70 heat shock gene transcription by the suppressor of Hairy-wing protein of Drosophila melanogaster. Mol Cell Biol. 1991;11:1894–1900. doi: 10.1128/mcb.11.4.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoover K K, Gerasimova T I, Chien A J, Corces V G. Dominant effects of suppressor of Hairy-wing mutations on gypsy-induced alleles of forked and cut in Drosophila melanogaster. Genetics. 1992;132:691–697. doi: 10.1093/genetics/132.3.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jack J. Molecular organization of the cut locus of Drosophila melanogaster. Cell. 1985;42:869–876. doi: 10.1016/0092-8674(85)90283-1. [DOI] [PubMed] [Google Scholar]
- 31.Jack J, Dorsett D, DeLotto Y, Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 1991;113:735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- 32.Katsani K R, Hajibagheri M A, Verrijzer C P. Cooperative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 34.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Shen B, Rosen C, Dorsett D. The DNA-binding and enhancer-blocking domains of the Drosophila suppressor of Hairy-wing protein. Mol Cell Biol. 1996;16:3381–3392. doi: 10.1128/mcb.16.7.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klug W S, Bodenstein D, King R C. Oogenesis in the suppressor of Hairy-wing mutant of Drosophila melanogaster. I. Phenotypic characterization and transplantation experiments. J Exp Zool. 1968;167:151–156. doi: 10.1002/jez.1401670203. [DOI] [PubMed] [Google Scholar]
- 37.Levis R, Bingham P, Rubin G M. Physical map of the white locus of Drosophila melanogaster. Proc Natl Acad Sci USA. 1982;79:564–568. doi: 10.1073/pnas.79.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Lopez-Guisa J M, Ninan N, Weiner E J, Rauscher III F J, Marmorstein R. Overexpression, purification, characterization, and crystallization of the BTB/POZ domain from the PLZF oncoprotein. J Biol Chem. 1997;272:27324–27329. doi: 10.1074/jbc.272.43.27324. [DOI] [PubMed] [Google Scholar]
- 39.Mihaly J, Hogga I, Barges S, Galloni M, Mishra R K, Hagstrom K, Muller M, Schedl P, Sipos L, Gausz J, Gyurkovics H, Karch F. Chromatin domain boundaries in the bithorax complex. Cell Mol Life Sci. 1998;54:60–70. doi: 10.1007/s000180050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modolell J, Bender W, Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci USA. 1983;80:1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morcillo P, Rosen C, Dorsett D. Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics. 1996;144:1143–1154. doi: 10.1093/genetics/144.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morcillo P, Rosen C, Baylies M K, Dorsett D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Müller J. The benefits of selective insulation. Curr Biol. 2000;10:R241–R244. doi: 10.1016/s0960-9822(00)00374-2. [DOI] [PubMed] [Google Scholar]
- 44.O'Donnell K H, Chen C T, Wensink P C. Insulating DNA directs ubiquitous transcription of the Drosophila melanogaster α1-tubulin gene. Mol Cell Biol. 1994;14:6398–6408. doi: 10.1128/mcb.14.9.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orihara M, Hosono C, Kojima T, Saigo K. Identification of engrailed promoter elements essential for interactions with a stripe enhancer in Drosophila embryos. Genes Cells. 1999;4:205–218. doi: 10.1046/j.1365-2443.1999.00254.x. [DOI] [PubMed] [Google Scholar]
- 47.Read D, Butte M J, Dernburg A F, Frasch M, Kornberg T B. Functional studies of the BTB domain in the Drosophila GAGA and Mod(mdg4) proteins. Nucleic Acids Res. 2000;28:3864–3870. doi: 10.1093/nar/28.20.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rollins R A, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rutledge B J, Mortin M A, Schwartz E, Thierry-Mieg D, Meselson M. Genetic interactions of modifier genes and modifiable alleles in Drosophila melanogaster. Genetics. 1988;119:391–397. doi: 10.1093/genetics/119.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saitoh N, Bell A C, Recillas-Targa F, West A G, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken α-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott K S, Geyer P K. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soeller W C, Oh C E, Kornberg T B. Isolation of cDNAs encoding the Drosophila GAGA transcription factor. Mol Cell Biol. 1993;13:7961–7970. doi: 10.1128/mcb.13.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava M, Hsieh S, Grinberg A, Williams-Simons L, Huang S P, Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 2000;14:1186–1181. [PMC free article] [PubMed] [Google Scholar]
- 54.Szabo P, Tang S H, Rentsendorj A, Pfeifer G P, Mann J R. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 55.Torigoi E, Bennani-Baiti I M, Rosen C, Gonzalez K, Morcillo P, Ptashne M, Dorsett D. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc Natl Acad Sci USA. 2000;97:2686–2691. doi: 10.1073/pnas.050586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Udvardy A. Dividing the empire: boundary chromatin elements delimit the territory of enhancers. EMBO J. 1999;18:1–8. doi: 10.1093/emboj/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao K, Hart C M, Laemmli U K. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]