Graphical abstract
Keywords: Epigenetics, DNA methylation, H3K9me3, Heterochromatin
Highlights
-
•
Exposure to Pb significantly lowered meCpG and H3K9me3 levels in HEK293T cells.
-
•
The observed epigenetic changes can be partially attributed to transcriptional changes.
-
•
A compensatory increase of meCpG and H3K9me3 levels occurs after Pb exposure is ceased.
-
•
Pb-induced changes do not immediately recover suggesting long-term damages.
Abstract
Lead (Pb) is a heavy metal contaminant commonly found in air, soil, and drinking water due to legacy uses. Excretion of ingested Pb can result in extensive kidney damages due to elevated oxidative stress. Epigenetic alterations induced by exposure to Pb have also been implied but remain poorly understood. In this work, we assessed changes in repressive epigenetic marks, namely DNA methylation (meCpG) and histone 3 lysine 9 tri-methylation (H3K9me3) after exposure to Pb. Live cell epigenetic probes coupled to bimolecular fluorescence complementation (BiFC) were used to monitor changes in the selected epigenetic marks. Exposure to Pb significantly lowered meCpG and H3K9me3 levels in HEK293T cells suggesting global changes in constitutive heterochromatin. A heterodimeric pair of probes that tags chromatin regions enriched in both meCpG and H3K9me3 further confirmed our findings. The observed epigenetic changes can be partially attributed to aberrant transcriptional changes induced by Pb, such as overexpression of TET1 after Pb exposure. Lastly, we monitored changes in selected heterochromatin marks after removal of Pb and found that changes in these markers do not immediately recover to their original level suggesting potential long-term damages to chromatin structure.
1. Introduction
Extensive epidemiological studies have shown that exposure to environmental chemicals, especially heavy metals, at different stages of life is associated with various diseases. For instance, developmental exposure to lead (Pb), including prenatal or childhood exposure, can result in disrupted neuropsychiatric function and reduced cognitive ability (Senut et al., 2012, Mitra et al., 2017). Exposure to Pb in human is commonly assessed via blood lead level. The accumulating Pb in body can be either excreted by the kidneys via glomerular filtration and tubular secretion; or deposited in bones (Mitra et al., 2017). Given that kidney is one of the major excretion routes of Pb from body, exposure to Pb can induce kidney damages. These damages can occur via oxidative stress and lipid peroxidation (Rana et al., 2018) by decoupling the respiratory chains in mitochondria (Reyes et al., 2013). Studies using Wistar rats chronically exposed to Pb revealed that Pb can accumulate in kidney causing oxidative damages to cells; lead to severe morphological modifications of the tubule epithelial cells; and result in unbalanced biochemical functions (Navarro-Moreno et al., 2009). Interestingly, small inclusion bodies (∼5 µm) were also found inside the nuclei of kidney cells with size significantly larger than those found in cells with subacute exposure to Pb (∼1 μm) (Navarro-Moreno et al., 2009).
Exposure to Pb has been linked to aberrant epigenome changes and alterations in epigenetic enzyme activities. Specifically, Pb can modulate DNA methyltransferase activity as a non-competitive inhibitor (Sanchez et al., 2017) and result in global DNA hypomethylation (Hou et al., 2012, Nye et al., 2015) as shown in both in vitro and ex vivo assays. Similar changes in DNA methylation as well as alterations in H3K9ac, H3K4me2 and H3K27me3 levels were reported in brain tissues of mice with early life Pb exposure (Eid et al., 2016). Interestingly, Pb induced alterations in epigenome are potentially sex-dependent. For example, female mice exposed to Pb during a prenatal stage demonstrate lower levels of H3K9me3 (Schneider et al., 2016) and DNA methylation in hippocampus (Sánchez-Martín et al., 2015), while no significant changes were observed in the hippocampus of males (Schneider et al., 2016).
Changes in epigenome content have also been observed in acute or chronic kidney injuries (reviewed in 12). For instance, C57BL/6 mice injured via renal ischemia–reperfusion exhibited global DNA demethylation (Zhao et al., 2017). Acute kidney injury is often associated with alterations in H3 acetylation levels (Marumo et al., 2008, Zager et al., 2011); upregulation of H3K4me3 (Naito et al., 2009, Munshi et al., 2011), and H3K27me3 (Zhou et al., 2018). These epigenomic alterations are also observed in kidney cells exposed to Pb. Specifically, human embryonic kidney (HEK) 293 cells exposed for 24 h and 48 h to Pb (100 and 300 µM) demonstrated hypomethylation in LINE-1 promoter (Li et al., 2013), and in a murine model, prenatal exposure to Pb led to sex-dependent hypomethylation of intracisternal A particle (IAP) retrotransposons in female kidneys compared to the male counterpart (Montrose et al., 2017). Although Pb exposure has been shown to alter both DNA methylation and H3K9me3, which are key constituents of constitutive heterochromatin, there are no reports that systematically evaluate both epigenetic marks in kidney cells. The lack of information warrants further studies on how Pb exposure can affect heterochromatin conformation.
Epigenetic modifications, including DNA methylation and histone posttranslational modifications (PTMs), are dynamic, varying in time and across different cell lineages (Aguilar and Craighead, 2013). DNA methylation varies during the life span of individuals due to intrinsic and extrinsic factors, like aging and exposure to environmental chemicals (Almén et al., 2014, Feil and Fraga, 2012); and among tissues as determined by their underlying biological functionality (Rakyan et al., 2008). Histone PTMs are also highly dynamic and can be altered by micro-environment (Prompsy et al., 2020, Garcia-Gomez et al., 2018). Instead of performing static measurements at selected time points, it is thus highly desirable to track epigenetic changes in live cells. Few technologies exist that enable in situ tracking of epigenetic features, including FRET (Fluorescence Resonance Energy Transfer) and BiFC (Bimolecular Fluorescence Complementation) probes. Specifically, a FRET-based sensor using a reader domain (i.e., heterochromatin protein 1a (HP1a)) (Lin et al., 2004) was developed to monitor intracellular H3K9me3 levels. This approach has also been used to quantify histone acetylation levels (Sasaki et al., 2009, Nakaoka et al., 2016). Recently, a series of live-cell compatible epigenetic probes have been developed using engineered epigenetic reader domains including works from our group. These probes have enabled us to trace intracellular levels of histone PTMs, such as acetylation (Sanchez et al., 2017) and methylation (Sánchez et al., 2019), as well as methylated DNA (Kim et al., 2014, Sánchez et al., 2020). Most recently monitoring changes in DNA methylation or H3K9me3 at specific gene regions were also achieved via BiFC probes (Lungu et al., 2017).
Utilizing novel epigenetic probes developed in our laboratory, we monitored dynamic changes in selected epigenetic hallmarks of constitutive heterochromatin, namely methylated DNA and H3K9me3, via a human embryonic kidney cell line (HEK293T) model responding to Pb exposure. Engineered BiFC probes were used to monitor individual changes in selected epigenetic marks. Furthermore, given the ability to mix-and-match different epigenetic probes, for example combining the different split probes or hetero-dimer probes, we can track dual epigenetic marks co-localized. The dynamic and persistent nature of the acquired epigenetic changes were also assessed via an “on” and “off” Pb exposure experiment.
2. Materials and methods
2.1. Live-cell protein sensors to track epigenetic changes in situ
Epigenetic “readers” with high affinity and selectivity towards targeted epigenetic marks were selected based on literature reports and our previous experimental validation work. Briefly, detection of meCpG was conducted via a probe constituted by the methyl binding domain (MBD) of MBD1 protein (Kim et al., 2014, Jørgensen et al., 2006, Mendonca et al., 2021), and H3K9me3 was detected via a probe constituted by the chromodomain of the chromodomain Y chromosome (CDY) (Mendonca et al., 2021, Fischle et al., 2008). The detailed amino acid sequence of the construct was summarized in Table S1 (Supporting Information). BiFC was used to enhance the signal-to-noise ratio as previously demonstrated in literature (Kodama and Hu, 2012, Alongkronrusmee et al., 2018). All constructs contained an SV40 nuclear localization signal (SV40 NLS) in the N-terminus followed by the recognition domain and split half of mEGFP in the C-terminus of the construct as illustrated in Fig. S1A (Supporting Information). A negative control pair using the same expression vector without the epigenetic “reader” domain was also constructed, as we described in our previous work (Sánchez et al., 2020).
The detection scheme is illustrated in Fig. S1B (Supporting Information). Specifically, a homo-dimer probe consists of the same epigenetic recognition motif. It is thus used to track a single type of epigenetic modification as illustrated in Fig. S1B top (Supporting Information). A hetero-dimer pair consists of two probes fused to two different reader domains suitable for simultaneous monitoring of two different epigenetic marks in a proximity as illustrated in Fig. S1B bottom (Supporting Information). BiFC signal is typically generated when the two halves of a fluorescent protein is within a distance of ∼ 7 nm (Fan et al., 2008). We thus expect that a homo- and hetero-dimer set will give strong fluorescence signal when the modification(s) of interest are abundant within one nucleosomal distance. Unbound probes will not fluoresce and thus provide low background signals and potentially provide more accurate tagging of functionally distinct heterochromatin regions.
2.2. Mammalian cell culture and transfection
Human Embryonic Kidney 293 T (HEK293T) cells were used as a model system to study Pb toxicity on kidneys. HEK293T cells were cultured as described previously (Sanchez et al., 2017). Briefly, cells were cultured in Dulbecco modified Eagle medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) of a Penicillin-Streptomycin solution (Gibco®, CA, US), and incubated at 37 °C with 5% CO2. Engineered sensors were transiently transfected into HEK293T conducted using Lipofectamine® 3000 (Life Technologies, MD, US) following the manufacture’s protocol.
2.3. Chemical treatment of HEK293T cells with lead (Pb)
We selected to work with Pb concentrations of 0, 100, and 500 ppb based on current and historical EPA regulation standard (15 ppb since 1991, 50 ppb since 1975) (Dignam, T.; Kaufmann, R. B.; LeStourgeon, L.; Brown, M. J., Control of Lead Sources in the United States, 2019), as well as, their abundance in the environment. For example, Pb of concentration up to 101 ppb was detected in drinking water in the recent Flint water crisis (Hanna-Attisha et al., 2016). Food can be another source of exposure including rice (Ankar-Brewoo et al., 2020), poultry (Trampel et al., 2003), and fruits (Parveen et al., 2020, Türkdoğan et al., 2003). Exposure to Pb can cause high blood Pb level. For instance, workers with occupational exposure to Pb for 36–44 days can have blood Pb levels of 49.1 ± 14.1 µg dL-1 (equivalent to ∼ 491 ± 141 ppb) (Dobrakowski et al., 2016). The Pb bioavailability in kidneys is also estimated using urinal Pb concentrations, which is found to be ∼ 22.28 µg / (g of creatinine) (Gil et al., 2011). Typical Pb levels in human blood and urine are below 10 µg dL-1 (equivalent to ∼ 100 ppb) (HEALTH, C. O. E.; Lanphear, B. P.; Lowry, J. A.; Ahdoot, S.; Baum, C. R.; Bernstein, A. S.; Bole, A.; Brumberg, H. L.; Campbell, C. C.; Lanphear, B. P.; Pacheco, S. E.; Spanier, A. J.; Trasande, L., Prevention of Childhood Lead Toxicity. Pediatrics, 2016). Our selected Pb concentration (500 ppb) thus reflects potentially the highest occupational Pb level found in blood and urine samples of exposed population. In a single-dose pharmacokinetic study of Pb using a rat model, the maximum concentration of Pb reaches a plateau value after ∼ 12 h of treatments in various tissues including bone, kidney, and liver; and this concentration had a significant reduction after 40 h of administration (Dalley et al., 1990). We thus chose an exposure time of 24 h to mimic the Pb effects on selected epigenetic marks in a short-term acute exposure model.
A Pb-relaxation experiment was conducted to assess the short-term effects of Pb on the persistence of H3K9me3 and meCpG changes. HEK293T cells transfected with the homo-dimer pair were exposed to the selected Pb doses for 24 h. After that, culture medium was aspirated. Cells were washed twice with PBS before fresh culture medium was added. Cells were continuously cultured for another 24 h and imaged every 8 h during the experimental course.
2.4. Immuno-fluorescence staining
Immunofluorescence staining was performed similarly as we described previously (Lin et al., 2021). Briefly, cells treated with Pb were fixed and permeabilized using cold methanol for 10 mins; and blocked with 1% BSA in PBS for 1 h followed by incubation with primary antibodies for 1 h at room temperature. Primary antibodies, including anti-5mC (#61480, Active motif, CA) and anti-H3K9me3 (ab8898, Abcam, MA), and secondary antibodies, including goat-anti-rabbit Alexa Fluor 568 (ab175471, Abcam, MA) and donkey-anti-mouse Alexa Fluor 647 (a31571, Invitrogen, CA), were used. For 5mC antibody staining, an additional denaturating and neuralization step were included prior to adding primary antibodies.
2.5. Fluorescent microscopy
Cells transfected with the selected BiFC protein sensors were imaged using a Nikon Eclipse Ti-2 inverted microscope equipped with a 488 nm laser line using a 63×/1.49NA oil objective. A FITC (excitation 460–500 nm, emission 510–560 nm) filter set was applied to collect BiFC mEGFP. Z-stack images of cells were collected using Nikon EZ-C1 software. Collected images were analyzed using Cellprofiler (Carpenter et al., 2006) to get the Integrated Intensity per Nuclei (IIN) as we detailed in our previous work (Sanchez et al., 2017, Sánchez et al., 2019, Sánchez et al., 2020).
2.6. Flow cytometry
Flow cytometry data were collected using a BD FACS AriaTM (Becton Dickson, San Jose, CA). A 488 nm excitation laser line and a 530/30 nm FITC filter were used for capturing the GFP fluorescent intensity. All cytometry data were analyzed using FCS Express software (De Novo Software, Glendale, CA). Reported data comes from independent assays run in triplicate.
2.7. Quantitative polymerase chain reaction (qPCR)
HEK293T cells treated with different concentrations of Pb were harvested. Total RNA was extracted using an RNA purification kit (PureLink, Thermo Fisher Scientific, U.S.) following the manufacturer’s protocol. RT-qPCR was carried out using QuantStudio 3 (Thermo Fisher Scientific, U.S.) as we described in our previous work (Lin et al., 2021). Primers used in qPCR are summarized in Table S2 (Supporting Information). β-Actin (ACTB) was used as the reference gene. qPCR was performed following the MIQE requirements (Bustin et al., 2019).
2.8. Statistical analysis
All statistical analysis and error propagation calculations were performed in OriginPro 2018 (Origin Lab Corp, North Hampton, MA) statistical software. An analysis of variance (ANOVA) followed by Tukey’s HSD post-hoc test was used with p = 0.05. Graphpad Prism was also used for analysis. All comparisons are with the control group unless otherwise annotated in the figures or texts.
3. Results
3.1. Exposure to low doses of Pb does not elicit immediate cytotoxic effects or phenotypic changes in the human cell line HEK293T
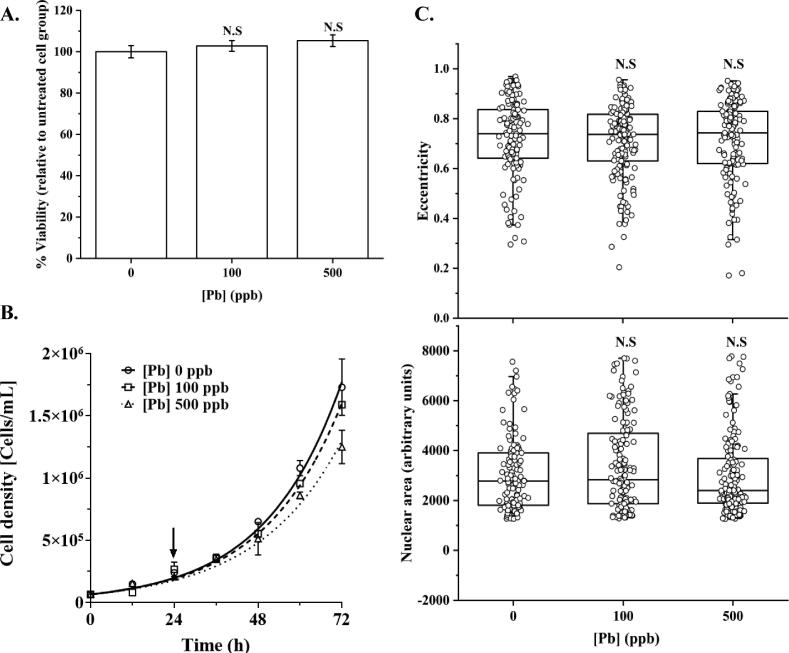
We started by examining the cytotoxicity of selected Pb concentrations on HEK293T cells. No significant changes were observed in the percentage of viability relative to the untreated control via MTT assay (Fig. 1A). These results suggest that the selected Pb doses do not have a significant cytotoxic effect. Fig. 1B summarizes the effect of Pb on the growth of HEK293T. Increasing Pb concentrations slightly slow down cell growth, but the changes were found not to be statistically significant (p > 0.05, one-way ANOVA followed by Tukey’s HSD post-hoc test). We then analyzed changes in the nuclear cell morphology by assessing the nuclear size and eccentricity of exposed cells to Pb (Fig. 1C). Exposure to Pb does not significantly alter nuclear size or eccentricity. Collectively, HEK293T cells exposed to a low dose of Pb (≤500 ppb) did not induce significant changes in either viability, cell growth rates, or morphologies.
Fig. 1.
No significant toxicity effects are induced in HEK293T cells by exposure to Pb. A. Cell survival was assessed by MTT assay. The results represent the means of 3 biological experiments with six technical repeats. No significant (p > 0.05, one-way ANOVA followed by Tukey’s HSD post-hoc test) changes were observed at selected Pb doses, 100 and 500 ppb. B. Growth curves of HEK293T after treating with Pb, 100 and 500 ppb. Pb treatment slow down cell growth, but no significant (p > 0.05, one-way ANOVA followed by Tukey’s HSD post-hoc test) effect was found in the doubling time (15.2 h, 15.7 h and 16.7 h for 0, 100, and 500 ppb of Pb, respectively). The arrow indicates the time when Pb was added to the cell culture. Data is presented as the mean of 3 biological repeats. C. Exposure to Pb revealed no significant (p > 0.05, one-way ANOVA followed by Tukey’s HSD post-hoc test) modification in the nuclear size or eccentricity of HEK293T cells. Data presented correspond to at least 25 cells analyzed per biological repeat, N = 3.
3.2. Exposure to Pb downregulates H3K9me3 and alters its distribution in HEK293T cells
To monitor changes in H3K9me3 level upon Pb exposure, we transfected HEK293T cells with our H3K9me3 in situ probe sets. Our probes exhibit distinctive fluorescent patterns with large size foci enriched primarily inside the cell nucleus and small puncta near the nuclear periphery. This pattern is consistent with what was commonly observed for H3K9me3 patterns inside cells (Politz et al., 2013). To ensure that the observed features are due to the presence of specific epigenetic “readers” domains, we transfected cells with the negative control, i.e., BiFC-mEGFP lacking the epigenetic domain, as shown in Fig. S2 (Supporting Information). As expected, cells transfected with the BiFC-mEGFP pair did not generate any distinctive features. Cells transfected with only half of the BiFC-pair, such as m-EGFP-N- or mEGFP-C-terminus did not fluoresce.
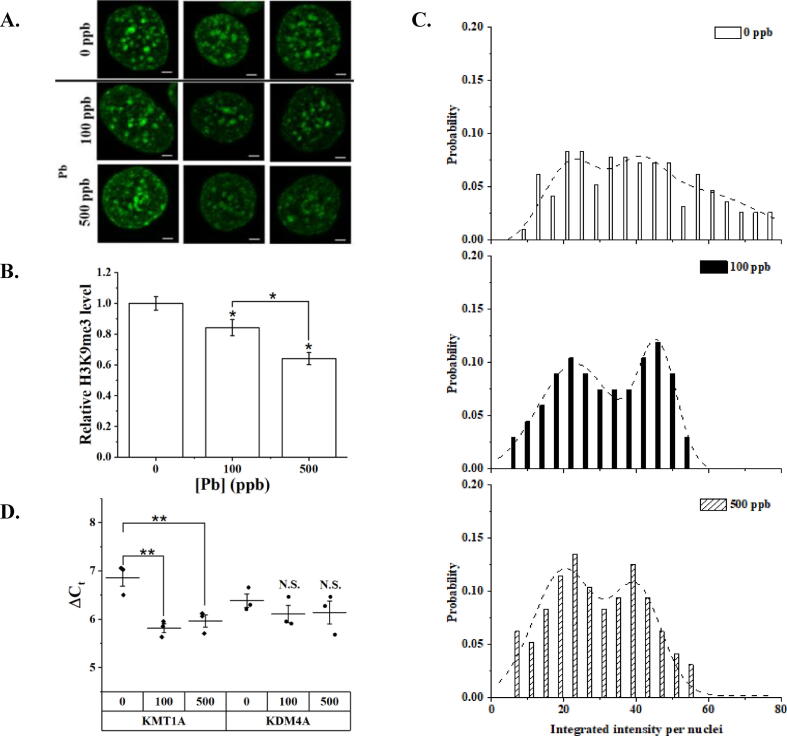
Projected 2D images of transfected cells under varying Pb concentrations are summarized in Fig. 2A. The bright puncta near the nuclear periphery are expected to be perinuclear heterochromatin as reported in the literature (Politz et al., 2013), while bright foci inside the cell nucleus were expected to be repressive heterochromatin not actively transcribing (Mutlu et al., 2018, Bonnet-Garnier et al., 2018). After Pb treatment, we noticed significant reductions in both the number of bright foci and decrease in foci intensity, suggesting the potential onset of chromatin instability. We quantified changes in H3K9me3 by measuring the Integrated Intensity per Nuclei (IIN). We and others have previously demonstrated that IIN correlates with global epigenetic modification level per cell (Sanchez et al., 2017, Sánchez et al., 2019, Sánchez et al., 2020, Lungu et al., 2017, Hayashi-Takanaka et al., 2011). IIN analysis between the control and Pb-treated groups shows that Pb exposure significantly (p < 0.05, one-way ANOVA followed by Tukey’s HSD post-hoc test) lowers the global level of H3K9me3, by 16% and 36% for Pb concentrations of 100 and 500 ppb, respectively (see Fig. 2B).
Fig. 2.
H3K9me3 levels in HEK293T cells exposed to Pb. A. Representative 2D projected images of HEK293T cells after 24 h of transfection with H3K9me3 probes and pre-exposed for 24 h to Pb, 100 or 500 ppb. Scale bar = 2 µm. B. Relative H3K9me3 levels to the control, or 0 ppb of Pb, using the integrated intensity per nuclei (IIN) obtained from image analysis. Data is presented as the mean ± S.D., N = 3. C. Probability density distribution of the IIN obtained from single cell image analysis, n ≥ 80. Presented data corresponds to at least 25 cells per biological repeat, N = 3. D. Expression of mRNAs coding for KMT1A and KDM4A. Data is expressed as cycle threshold (Ct) difference to β-actin presented as dot plots with indicated mean value. Data is presented as the mean ± S.D., N = 3. N.S.: stands for no significant difference. * represents p < 0.05 and ** represents p < 0.01 from a one-way ANOVA followed by Tukey’s HSD post-hoc test.
Since we carried out a single-cell-based analysis, we were able to plot the probability distribution histograms of IIN, reflecting inter-cellular heterogeneity distribution in H3K9me3 as in Fig. 2C. The histograms exhibit a positively skewed distribution that can be well-fitted using a bimodal distribution modeled by a mixture Gaussian model with R2 = 0.86, 0.98, 0.94 for cells treated with 0, 100, and 500 ppb of Pb, respectively. Randomly distributed residuals were obtained for all fittings, suggesting good fitting quality. Two subpopulations of cells are identified with fitting parameters summarized in Table S3A (Supporting Information). Pb treatment has resulted in a noticeable narrowing of the distribution of high-H3K9me3 subpopulation and an increment in the probability density of low-H3K9me3 subpopulation. This observation is supported by changes in width (w) and area (A) parameters of modeled distributions (Table S3A (Supporting Information). Compared to the untreated group the peak that represents high-H3K9me3 subpopulation shows a reduction in the peak width and area upon Pb treatment. Conversely, the peak representing the low-H3K9me3 subpopulation exhibited higher width and area than the control group. Suggesting an increase in the low-H3K9me3 subpopulation owed to Pb treatment. However, the center value (xc) parameter, which represents the maximum IIN of each subpopulation, is similar across Pb treatments.
To verify our findings, we carried out a parallel experiment by immuno-fluorescence staining. Typical images of HEK293T cells exposed to Pb for 24 h and stained with anti-H3K9me3 antibody are shown in Fig. S3A (Supporting Information). Consistent with our observations using BiFC probes, we observed ∼ 14.3 and 34% reductions in H3K9me3 levels for cells exposed to 100 and 500 ppb of Pb, respectively (see Fig. S3B (Supporting Information)). Single-cell analysis revealed an increment in the population with low H3K9me3 levels as the Pb concentration increases (Fig. S3C (Supporting Information).
To further verify the observed heterogeneous changes in HEK293 after Pb treatments, we analyzed similarly treated cell populations via FACS. Our results are shown in Fig. S4A and S4B (Supporting Information). A similar bimodal distribution with a low- and a high-fluorescent-intensity species were obtained agreeing with our microscopy-based image analysis (see Fig. 2C). Pb treatments significantly reduced the cell subpopulation with a high level of H3K9me3 as well as increased the cell subpopulation with a low level of H3K9me3. The drop in the median fluorescence intensity, which is expected to correlate with a drop in H3K9me3 levels, of Pb treated samples were of 12% and 21% for 100 ppb and 500 ppb of Pb (Fig. S4B (Supporting Information)), respectively, which is reasonably close to the obtained results via image analysis.
RT-qPCR was then performed to reveal alterations in epigenetic enzymes. We specifically determined changes in the transcriptional levels of KMT1A and KDM4A (Fig. 2D and Table S4 (Supporting Information)), which is a histone methyltransferase and demethylase, respectively. The selected epigenetic enzymes are highly specific for H3K9me3 (Hyun et al., 2017). Although statistical difference among Pb treatments was observed for KMT1A (p < 0.01, one-way ANOVA followed by Tukey’s HSD post hoc test), unveiling an overexpression of mRNA for the histone methyltransferase upon Pb exposure, the observed change does not explain the reduction in H3K9me3 levels due to Pb exposure. Furthermore, no statistical difference among treatments was observed in the mRNA level of KDM4A (p > 0.05, one-way ANOVA followed by Tukey’s HSD post hoc test).
3.3. Exposure to low-dose of Pb affects meCpG distribution and abundance in HEK293T cells
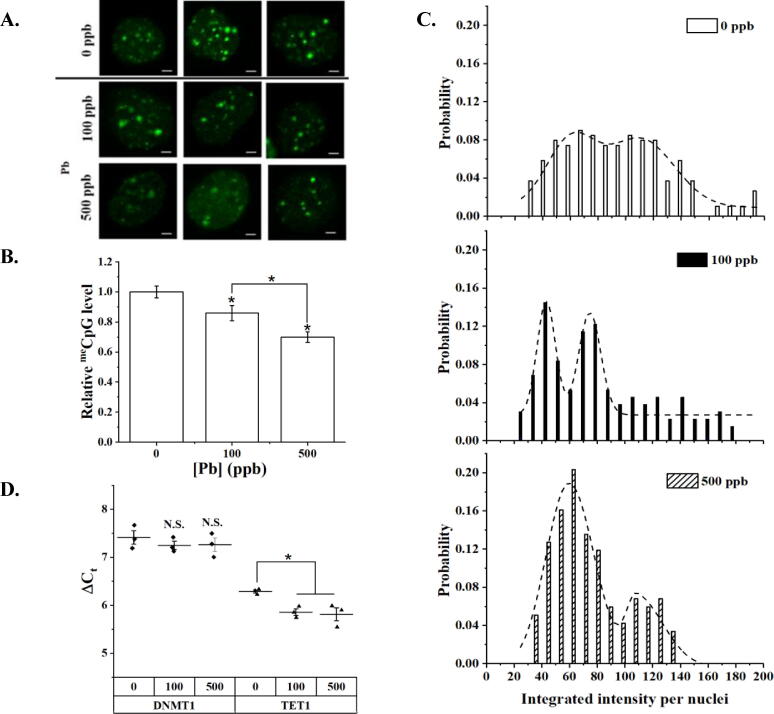
Like described in the previous section, cells were transfected with a set of meCpG BIFC probes and treated with Pb of varying doses. Untreated cells exhibited clustered meCpG islands as bright foci located mainly within the cell nucleus (Fig. 3A) representing mainly repressive chromatin regions (Seki et al., 2005, Latham et al., 2008). Bright meCpG clusters can be found close to the periphery but not on the periphery as previously observed for H3K9me3. The observed meCpG pattern is consistent with what was reported in the literature (Lungu et al., 2017, Jørgensen et al., 2006). Pb treatment has led to reductions in both the number and brightness of meCpG foci. IIN analysis showed that Pb-treatment significantly (p < 0.05, one-way ANOVA followed by Tukey’s HSD post hoc test) reduces the global level of meCpG when compared to untreated cells by 14% and 30% for 100 and 500 ppb of Pb, respectively (Fig. 3B).
Fig. 3.
meCpG levels in HEK293T cells exposed to Pb. A. Representative 2D projected images of HEK293T cells after 24 h of transfection with meCpG probes and pre-exposed for 24 h to Pb, 100 or 500 ppb. Scale bar = 2 µm. B. Relative meCpG levels to the control or 0 ppb of Pb using the integrated intensity per nuclei and integrated foci intensity per nuclei obtained from image analysis. Data is presented as the mean ± S.D., N = 3. C. Probability density distribution of the integrated intensity per nuclei and integrated foci intensity per nuclei obtained from single cell image analysis, n ≥ 80. Presented data corresponds to at least 25 cells analyzed per biological repeat, N = 3. D. Expression of mRNAs coding for DNMT1 and TET1. Data is expressed as cycle threshold (Ct) difference to β-actin presented as dot plots with indicated mean value. Data is presented as the mean ± S.D., N = 3. N.S.: stands for no significant difference. * represents p < 0.05 from a one-way ANOVA followed by Tukey’s HSD post hoc test.
The probability distribution histograms were constructed; fitted similarly as described previously; and illustrated in Fig. 3C. The histograms exhibit a positively skewed distribution shape which was better fitted by a bimodal distribution with R2 = 0.89, 0.90, 0.97 for 0, 100, and 500 ppb of Pb, respectively. The fitting parameters were summarized in Table S3B (Supporting Information). Pb treatment has resulted in a significant narrowing of the high-meCpG subpopulation and an increase in the abundance of the low-meCpG subpopulation. This trend was further verified using FACS as shown in Fig. S4C and S4D (Supporting Information). The drop in the median fluorescence intensity of FACS measurements was found to be 12% and 21% for cells treated with 100 ppb and 500 ppb of Pb, respectively (Fig. S4D (Supporting Information)), respectively.
Validation experiments were carried out via immuno-fluorescence with typical cell images shown in Fig. S5A (Supporting Information). We found ∼ 15 and 40% drop in meCpG levels for cells treated with 100 and 500 ppb Pb, respectively (Fig. S5B (Supporting Information). Single-cell analysis suggested an increment in the population with low meCpG levels for cells treated with higher concentrations of Pb as shown in Fig. S5C (Supporting Information) consistent with our findings using in situ probes.
RT-qPCR analysis was performed to determine changes in the transcriptional levels of DNMT1 and TET1 (Fig. 3D and Table S4 (Supporting Information)). DNMT1 is a maintenance methyltransferase responsible for the maintaining of meCpG, while TET1 is responsible for the removal of meCpG (De Carvalho et al., 2010). Statistical difference (p < 0.05, one-way ANOVA followed by Tukey’s HSD post hoc test) between Pb-treated and the control group was observed for TET1, unveiling an overexpression of mRNA for the DNA demethylase upon Pb exposure. No statistical difference was observed in the mRNA level of DNMT1. Collectively, changes in the transcription of epigenetic enzymes governing the level of mRNA can partially explain the observed drop in meCpG due to Pb exposure.
3.4. Detection of heterochromatin regions enriched in meCpGs and H3K9me3
One of the advantages of our BiFC protein sensors lies in their ability to monitor different epigenetic marks based on the complementation of the conjugated split mEGFP. Taken advantage of this, we have paired BIFC probes targeting different epigenetic modifications, namely a pair of meCpG-N-mEGFP and H3K9me3-C-mEGFP (labeled as meCpG-N-H3K9me3-C pair) and another pair of H3K9me3-N-mEGFP and meCpG-C-mEGFP (labeled as H3K9me3-N-meCpG-C pair, see also Fig. S1 (Supporting Information)). Different from homo-pairs used in the previous two sections, these hetero-pairs uniquely enable us to explore changes in chromatin marked with a high density of meCpG and H3K9me3 simultaneously. Such chromatin regions are commonly known as constitutively suppressed chromatin and are enriched in genes that are permanently silenced (Nicetto and Zaret, 2019).
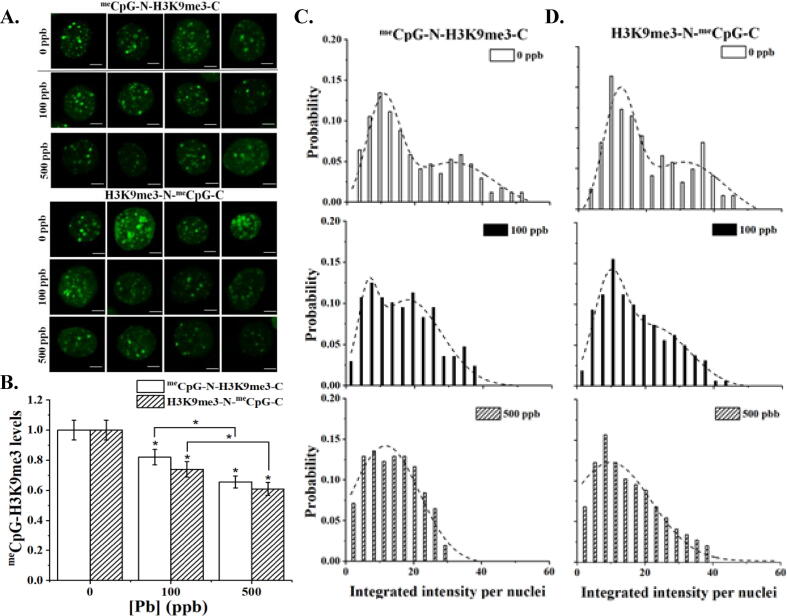
The two hetero-pairs were transfected into HEK293 cells with typical fluorescent patterns shown in Fig. 4A. Untreated cells exhibit bright foci primarily inside the cell nucleus similar to those observed previously in cells transfected with meCpG probe pair. The size of foci, however, are smaller in cells transfected with the hetero-pair. No significant difference was observed between cells transfected with different hetero-pair of probes (p > 0.05, one-way ANOVA followed by Tukey’s HSD post hoc test). Fig. 4A further illustrates the effects of Pb treatments. Exposure to Pb resulted in significant reductions in both foci number and intensity. Signifcantly, the brighter foci within the nucleus seem to completely diminish after Pb treatments. Intensity measurements confirmed our visual observations as shown in Fig. 4B. Although the percentage of reductions are slightly different between two hetero-pairs (meCpG-N-H3K9me3-C pair: 18 ± 5.1% and 34 ± 4.0% for 100 and 500 ppb of Pb; and H3K9me3-N-meCpG-C pair: 26 ± 5.3% and 39 ± 4.4% for 100 and 500 ppb of Pb, respectively), the difference was not found statistically significant (p > 0.05, one-way ANOVA followed by Tukey’s HSD post hoc test).
Fig. 4.
Cross detection of meCpG and H3K9me3 levels in HEK293T cells exposed to Pb capture by meCpG-N-H3K9me3-C pair and H3K9me3-N-meCpG-C pair. A. Representative 2D projected cells from Z-stacks transfected with either set of hetero-dimer sensors. B. Changes in the relative cross detected meCpG and H3K9me3 levels induced by Pb exposure. Data is presented as the mean ± S.D., N = 3. * represents p < 0.05 from a one-way ANOVA followed by Tukey’s HSD post hoc test. C. Probability distribution histograms of IIN from HEK293T cells exposed to various Pb concentrations and transfected with either set of hetero-dimer sensors. Presented data corresponds to at least 25 cells analyzed per biological repeat, N = 3.
We then analyzed the intensity histogram obtained using the hetero-pair as shown in Fig. 4C. Similar to meCpG and H3K9me3 probes, untreated cells exhibited a bimodal distribution (R2 0.90 – 0.94) with fitting parameters summarized in Table S3C-D (Supporting Information). Pb treatments, however, seem to promote the diminishing of the high fluorescent subpopulation. As a result, the intensity histogram of cells treated with 500 ppb of Pb is better fitted using a unimodal distribution (with R2 = 0.94 – 0.97). Cells treated with 100 ppb of Pb are more like a transition state in between with R2 = 0.96 and 0.86 for bi- and uni-modal distribution, respectively. Essentially, the cell sub-population with IIN values>40 from the control group vanishes upon Pb treatment (Fig. 4C). These results suggest that the heterodimer set of probes can be more sensitive to capture changes related to the pair of epigenetic marks that are sensing.
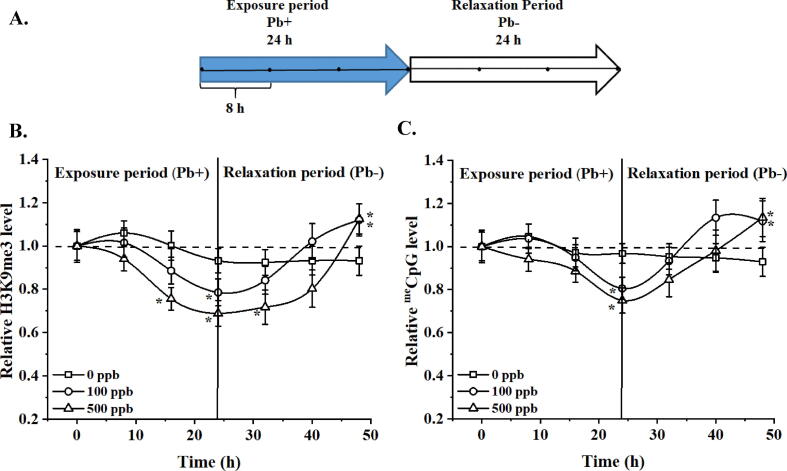
3.5. Short-term effect on Pb-induced changes of H3K9me3 and meCpGs
All epigenetic modifications are dynamic, we thus evaluated the dynamic alterations during and after Pb treatments. All cells were exposed to Pb for 24 h followed by 24 h of relaxation as shown in Fig. 5A. Imaging was performed at an interval of 8 h during the 24 h of exposure and 24 h of relaxation time-period.
Fig. 5.
Overcompensation in H3K9me3 and meCpG levels in HEK293T cells is observed after a Pb relaxation period. A. Schematic time course of HEK293T cells exposed to Pb and relaxation cycle. Imaging was conducted at 8 h intervals, denoted by the dark dots inside the arrows. B-C. Relative H3K9me3 and meCpG levels after HEK293T “ON and OFF” exposure to Pb. One-way ANOVA was conducted at each time point to establish statistical difference with regard to the control group (0 ppb of Pb). Presented data corresponds to at least 25 cells analyzed per biological repeat, N = 3. *, denotes statistical difference (p < 0.05, one-way ANOVA followed by Tukey’s HSD post-hoc test) at the specific time point.
All fluorescent intensity was normalized to the registered IIN at 0 h. Untreated cells were used to account for endogenous changes of selected epigenetic marks. Relative changes in H3K9me3 and meCpG were recorded as a function of time as shown in Fig. 5B and 5C, respectively. Cells treated with 500 ppb of Pb seem to have a faster decreasing rate and an early on-set time for both H3K9me3 and meCpG. By the end of 24 h, cells in the control group showed a reduction of ∼ 5% in H3K9me3 while cells treated with 100 and 500 ppb showed a reduction of ∼ 22% and 32% in H3K9me3, respectively. The reductions were found to be significant compared to the untreated control (p < 0.05, one-way ANOVA followed by Tukey’s HSD post hoc test). Pb was removed from culture medium at the end of 24 h followed by three times washing in Pb-free culture medium. The recovery rate and path observed during the relaxation period were found to be Pb-dose-dependent. For the control group, the H3K9me3 level remained more or less constant (±5%) till the end of the relaxation period (48 hrs). Cells exposed to 100 and 500 ppb of Pb slowly start to recover with cells exposed to 100 ppb recover at a faster rate compared to cells exposed to 500 ppb of Pb. Towards the end of relaxation, Pb-treated groups exhibit a ∼ 20% increase in H3K9me3 for 100 and 500 ppb treated samples, (p < 0.05, one-way ANOVA followed by Tukey’s HSD post-hoc test), suggesting an overcompensation effect. A similar trend was observed for meCpG with cells treated with 500 ppb of Pb exhibit faster reduction and slower recovery rate compared to sampled exposed to 100 ppb of Pb as shown in Fig. 5C.
As a benchmark comparison, we assessed the relative changes of H3K9me3 and meCpG towards the end of the relaxation stage via immunostaining (Fig. S6 (Supporting Information)). Compared to the untreated group (0 ppb), cells exposed to 100 and 500 ppb Pb for 24 h followed by 24 h relaxation showed an increment of ∼ 27 and ∼ 21% in H3K9me3 levels, respectively (Fig. S6A (Supporting Information)); while for meCpG this increment was of ∼ 12% and ∼ 15%, respectively (Fig. S6B (Supporting Information)) consistent with our observations via in situ probes.
4. Discussion
We focused on Pb concentrations of 100 and 500 ppb in this work, because of the recent incidences of Pb contamination in city water supplies (Hanna-Attisha et al., 2016) as well as its potential exposure to Pb via contaminated food sources (Trampel et al., 2003). Although the action level of Pb was recently lowered to 15 ppb by EPA, high Pb concentrations (>100 ppb) have been found in drinking water in the U.S. and other regions worldwide (Hanna-Attisha et al., 2016, Chen et al., 2012, Bryant, 2004). Furthermore, similar or higher Pb concentration were reported in blood or urine samples of worker with occupational exposure to Pb (Dobrakowski et al., 2016, Gil et al., 2011). Typical Pb levels in the blood and urine of human are lower than 100 ppb (HEALTH, C. O. E.; Lanphear, B. P.; Lowry, J. A.; Ahdoot, S.; Baum, C. R.; Bernstein, A. S.; Bole, A.; Brumberg, H. L.; Campbell, C. C.; Lanphear, B. P.; Pacheco, S. E.; Spanier, A. J.; Trasande, L., Prevention of Childhood Lead Toxicity. Pediatrics, 2016). The higher concentration of Pb being studied here, namely 500 ppb, thus reflect the highest Pb level that can be found in exposed population potentially due to substantial exposure to occupational hazards. The selected Pb concentrations do not illicit acute toxicity in human kidney cells and have minimal effects on cell morphology. These findings are consistent with literature reports in which phenotypical changes are only observed in animals or cell cultures at a concentration 100 – 1000 times higher than what we used in this work (Martinez et al., 2004, Akinyemi et al., 2019).
Exposure to Pb can result in a significant reduction in both meCpG and H3K9me3 in a dose-dependent manner. A similar trend was observed in previous literature. Specifically, zebrafish embryos exposed to 100 and 500 ppb of Pb for 72 h post-fertilization exhibited a global meCpG reduction of ∼ 13 and 29%, respectively (Sanchez et al., 2017). Similarly, SH-SY5Y exposed to even lower Pb concentrations, such as 15 and 50 ppb, for 96 h showed DNA hypomethylation (Lin et al., 2021). DNA hypomethylation has also been observed in vivo studies. For instance, male mice exposed to Pb via consuming drinking water at ∼ ppm level developed DNA hypomethylation in the brain cortex (Dou et al., 2019). While the changes in meCpG can be partially explained by the increase in TET1 transcription, Pb can also inhibit the activity of DNMT1 as suggested in literature (Sanchez et al., 2017). Collectively, they can lead to the observed reduction in meCpG.
Alterations in H3K9me3, however, cannot be fully accounted for by changes in the transcription of its governing epigenetic enzymes, such as H3K9me3 writer (KMT1A) and eraser (KDM4A) selected in this work. There are, however, other epigenetic enzymes that may modulate H3K9me3 levels in an independent or synergistic manner that are not accounted for in the current experimental design and may contribute to the observed trend here. SH-SY5Y cells exposed to Pb exhibit similar changes in H3K9me3 as we observed in this work (Lin et al., 2021). We expect that these changes are a result of cross-talking between meCpG and H3K9me3. DNA methylation can facilitate the recruitment of lysine methyltransferase for H3K9me3 and subsequently induce higher levels of H3K9me3 in DNA methylated-rich regions (Becker et al., 2016).
The application of heterodimeric pairs enabled tracking of heterochromatin that are commonly enriched in both meCpG and H3K9me3. The observed reductions in heterochromatin content are consistent with changes observed in either H3K9me3 or meCpG. Before treatment, we observed two subpopulations in HEK293 cells, one with high intensity and the other with low intensity. After Pb treatments, the subpopulation with a high intensity almost completely diminishes. Our results thus unequivocally suggest that Pb exposure can perturb the stability of heterochromatin regions which may lead to an abnormal gene transcriptional profile. Heterochromatin disruption is common to different cancer types (Fischer et al., 2010) and is associated with abnormal gene expression and phenotypical behavior (Carone and Lawrence, 2013). Loss of constitutive heterochromatin has also been associated with aging (Tsurumi and Li, 2012). Our results thus suggest that Pb treatment can lead to a reduction in both meCpG and H3K9me3 levels, cause disruptions in heterochromatin stability, and induce an abnormal phenotype that closely resembles cells in a pre-senescent state characteristic by redistribution of the constitutive heterochromatin (Chandra et al., 2015) and an imbalance between repressive and active histone marks (Chandra and Kirschner, 2016).
The probability distribution of H3K9me3 or meCpG in Pb treated cells shows a bimodal distribution, suggesting two sub-populations with distinctive levels of epigenetic modifications. Pb treatment caused a reduction of cells of high epigenetic modifications. A similar trend was observed with the hetero-dimer pair. Our results aligns with previous reports, where this bimodal distribution is a common feature reported in single-cell analysis of epigenetics, given the heterogeneous epigenetic content within a cell population (Grosselin et al., 2019, Castillo-Fernandez et al., 2020).
Given the dynamic nature of epigenetic marks, we tracked changes in H3K9me3 and meCpG responding to Pb treatments. As expected, high doses of Pb are associated with a faster reduction in both H3K9me3 and meCpG. Recovery was observed after cells were exchanged into a Pb-free culture medium, the recovery occurs faster for cells treated with a lower dose of Pb. Pb-treated cells end up with higher levels of meCpG and H3K9me3 after recovering for 24 h, suggesting an overcompensation effect. Overcompensation effect has been commonly reported in the literature as part of the hormetic dose–response to toxins (Calabrese et al., 2009, Vaiserman, 2011). The altered epigenetic profiles can be potentially recovered by the intrinsic epigenome repairing systems that utilize epigenetic enzymes (Flatau et al., 1984). The partial recovery and overcompensation as observed in this work have also been demonstrated in other studies (Lin et al., 2021, Xie et al., 2021, Wang et al., 2018). For instance, mouse fibroblasts (C3H 10 T1/2 C18) exhibit dynamic changes in DNA methylation after treating with a DNA methyltransferase inhibitor (5-aza-2′deoxycytidine) followed by a recovery period of 24 h. The treated cells ended up with an overall increase of 2.5% in DNA methylation after treatment (Flatau et al., 1984), suggesting an overcompensation effect consistent with our observations in this work. Similar observations were made in SUM159 cells after treating with A-196 a specific inhibitor of the histone-lysine methyltransferase SUV4-20 (Wang et al., 2018).
Another interesting aspect of Pb exposure is sex-dependence. There is growing evidence in literature suggsing that epigenetic changes induced by Pb is sex-sepcific (Schneider et al., 2016, Sánchez-Martín et al., 2015, Montrose et al., 2017). The selected human cell line HEK293T here was originated from a female human fetus (Libertini et al., 2015). Unsurprisingly, the trend we observed in this work is similar to a previous study using SH-SY5Y cells, a neuroblastoma cell line with a female origin (Mendell et al., 2018). Future work focusing on cell lines with male origins can provide further insights into the sex-dependent effects of Pb exposure on epigenome.
5. Conclusions
Exposure to Pb can cause deterioration of multiple organs in animals. Here we assessed the effects of Pb on selected epigenetic marks, namely meCpG and H3K9me3. Using HEK293T as a human cell line model, we found that exposure to Pb significantly downregulates meCpG and H3K9me3, which are characteristic epigenetic marks of constitutive heterochromatin, while minimal perturbations were observed in cellular viability, morphology, and proliferative properties. Constitutive heterochromatin regions enriched in both meCpG and H3K9me3 exhibit significant reductions in volume suggesting the onset of potential genome instability. The epigenetic changes induced in HEK293T by Pb exposure were found to be dynamic. An overcompensation was observed after the cessation of Pb exposure suggesting a dynamic regulation mechanism facilitating the restoration of homeostasis with potential lingering long-term effects.
CRediT authorship contribution statement
Oscar F. Sanchez: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Visualization, Supervision, Project administration. Li F. Lin: Methodology, Validation, Formal analysis, Investigation, Data curation. Junkai Xie: Methodology, Validation, Formal analysis, Investigation, Data curation, Visualization. Jennifer L. Freeman: Conceptualization. Chongli Yuan: Conceptualization, Methodology, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Science Foundation [CBET-1512285, CBET-1705560 & EF-1935226]. Support from the Purdue Center for Cancer Research Pilot Grant Program is gratefully acknowledged. Funding: OS was supported by CBET-1512285, LL and JX was supported by CBET-1705560 & EF-1935226, CY was supported by CBET-1512285, CBET-1705560 & EF-1935226.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crtox.2021.12.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Senut M.-C., Cingolani P., Sen A., Kruger A., Shaik A., Hirsch H., Suhr S.T., Ruden D. Epigenetics of early-life lead exposure and effects on brain development. Epigenomics. 2012;4(6):665–674. doi: 10.2217/epi.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P., Sharma S., Purohit P., Sharma P. Clinical and molecular aspects of lead toxicity: An update. Crit. Rev. Clin. Lab. Sci. 2017;54(7–8):506–528. doi: 10.1080/10408363.2017.1408562. [DOI] [PubMed] [Google Scholar]
- Rana M.N., Tangpong J., Rahman M.M. Toxicodynamics of Lead, Cadmium, Mercury and Arsenic- induced kidney toxicity and treatment strategy: A mini review. Toxicol. Rep. 2018;5:704–713. doi: 10.1016/j.toxrep.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, J.L., Molina-Jijón, E., Rodríguez-Muñoz, R., Bautista-García, P., Debray-García, Y., Namorado, M.D.C., Tight junction proteins and oxidative stress in heavy metals-induced nephrotoxicity. BioMed research international2013,2013. [DOI] [PMC free article] [PubMed]
- Navarro-Moreno L.G., Quintanar-Escorza M.A., González S., Mondragón R., Cerbón-Solorzáno J., Valdés J., Calderón-Salinas J.V. Effects of lead intoxication on intercellular junctions and biochemical alterations of the renal proximal tubule cells. Toxicol. In Vitro. 2009;23(7):1298–1304. doi: 10.1016/j.tiv.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Sanchez O.F., Lee J., Hing N.Y.K., Kim S.-E., Freeman J.L., Yuan C. Lead (Pb) exposure reduces global DNA methylation level by non-competitive inhibition and alteration of dnmt expression. Metallomics. 2017;9(2):149–160. doi: 10.1039/c6mt00198j. [DOI] [PubMed] [Google Scholar]
- Hou L., Zhang X., Wang D., Baccarelli A. Environmental chemical exposures and human epigenetics. Int. J. Epidemiol. 2012;41(1):79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye M.D., Hoyo C., Murphy S.K. In vitro lead exposure changes DNA methylation and expression of IGF2 and PEG1/MEST. Toxicol. In Vitro. 2015;29(3):544–550. doi: 10.1016/j.tiv.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid A., Bihaqi S.W., Renehan W.E., Zawia N.H. Developmental lead exposure and lifespan alterations in epigenetic regulators and their correspondence to biomarkers of Alzheimer's disease. Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring. 2016;2:123–131. doi: 10.1016/j.dadm.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J.S., Anderson D.W., Kidd S.K., Sobolewski M., Cory-Slechta D.A. Sex-dependent effects of lead and prenatal stress on post-translational histone modifications in frontal cortex and hippocampus in the early postnatal brain. Neurotoxicology. 2016;54:65–71. doi: 10.1016/j.neuro.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Martín F.J., Lindquist D.M., Landero-Figueroa J., Zhang X., Chen J., Cecil K.M., Medvedovic M., Puga A. Sex-and tissue-specific methylome changes in brains of mice perinatally exposed to lead. Neurotoxicology. 2015;46:92–100. doi: 10.1016/j.neuro.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ding C., Xue W., Ding X., Zheng J., Gao Y., Xia X., Li S., Liu J., Han F. Genome-wide DNA methylation analysis in renal ischemia reperfusion injury. Gene. 2017;610:32–43. doi: 10.1016/j.gene.2017.02.005. [DOI] [PubMed] [Google Scholar]
- Marumo T., Hishikawa K., Yoshikawa M., Fujita T. Epigenetic regulation of BMP7 in the regenerative response to ischemia. J. Am. Soc. Nephrol. 2008;19(7):1311–1320. doi: 10.1681/ASN.2007091040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zager R.A., Johnson A.C., Becker K. Acute unilateral ischemic renal injury induces progressive renal inflammation, lipid accumulation, histone modification, and “end-stage” kidney disease. Am. J. Physiol.-Renal Physiol. 2011;301(6):F1334–F1345. doi: 10.1152/ajprenal.00431.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M., Bomsztyk K., Zager R.A. Renal ischemia-induced cholesterol loading: transcription factor recruitment and chromatin remodeling along the HMG CoA reductase gene. Am. J. Pathol. 2009;174(1):54–62. doi: 10.2353/ajpath.2009.080602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi R., Johnson A., Siew E.D., Ikizler T.A., Ware L.B., Wurfel M.M., Himmelfarb J., Zager R.A. MCP-1 gene activation marks acute kidney injury. J. Am. Soc. Nephrol. 2011;22(1):165–175. doi: 10.1681/ASN.2010060641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Xiong C., Tolbert E., Zhao T.C., Bayliss G., Zhuang S. Targeting histone methyltransferase enhancer of zeste homolog-2 inhibits renal epithelial-mesenchymal transition and attenuates renal fibrosis. FASEB J. 2018;32(11):5976–5989. doi: 10.1096/fj.201800237R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Yang X., Xu M., Zhang J., Sun N. Epigenetic marker (LINE-1 promoter) methylation level was associated with occupational lead exposure. Clin. Toxicol. 2013;51(4):225–229. doi: 10.3109/15563650.2013.782410. [DOI] [PubMed] [Google Scholar]
- Montrose L., Faulk C., Francis J., Dolinoy D. Perinatal lead (Pb) exposure results in sex and tissue-dependent adult DNA methylation alterations in murine IAP transposons. Environ. Mol. Mutagen. 2017;58(8):540–550. doi: 10.1002/em.22119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar C.A., Craighead H.G. Micro- and nanoscale devices for the investigation of epigenetics and chromatin dynamics. Na.t Nano. 2013;8(10):709–718. doi: 10.1038/nnano.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almén M.S., Nilsson E.K., Jacobsson J.A., Kalnina I., Klovins J., Fredriksson R., Schiöth H.B. Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. Gene. 2014;548(1):61–67. doi: 10.1016/j.gene.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Feil R., Fraga M.F. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2012;13(2):97. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Rakyan V.K., Down T.A., Thorne N.P., Flicek P., Kulesha E., Gräf S., Tomazou E.M., Bäckdahl L., Johnson N., Herberth M., Howe K.L., Jackson D.K., Miretti M.M., Fiegler H., Marioni J.C., Birney E., Hubbard T.J.P., Carter N.P., Tavaré S., Beck S. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18(9):1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompsy P., Kirchmeier P., Marsolier J., Deloger M., Servant N., Vallot C. Interactive analysis of single-cell epigenomic landscapes with ChromSCape. Nat. Commun. 2020;11(1):5702. doi: 10.1038/s41467-020-19542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gomez A., Rodríguez-Ubreva J., Ballestar E. Epigenetic interplay between immune, stromal and cancer cells in the tumor microenvironment. Clin. Immunol. 2018;196:64–71. doi: 10.1016/j.clim.2018.02.013. [DOI] [PubMed] [Google Scholar]
- Lin C.-W., Jao C.Y., Ting A.Y. Genetically encoded fluorescent reporters of histone methylation in living cells. J. Am. Chem. Soc. 2004;126(19):5982–5983. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Ito T., Nishino N., Khochbin S., Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc. Natl. Acad. Sci. 2009;106(38):16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka S., Sasaki K., Ito A., Nakao Y., Yoshida M. A Genetically Encoded FRET Probe to Detect Intranucleosomal Histone H3K9 or H3K14 Acetylation Using BRD4, a BET Family Member. ACS Chem. Biol. 2016;11(3):729–733. doi: 10.1021/cb501046t. [DOI] [PubMed] [Google Scholar]
- Sanchez O.F., Mendonca A., Carneiro A.D., Yuan C. Engineering Recombinant Protein Sensors for Quantifying Histone Acetylation. ACS Sensors. 2017 doi: 10.1021/acssensors.7b00026. [DOI] [PubMed] [Google Scholar]
- Sánchez O.F., Mendonca A., Min A., Liu J., Yuan C. Monitoring Histone Methylation (H3K9me3) Changes in Live Cells. ACS Omega. 2019;4(8):13250–13259. doi: 10.1021/acsomega.9b01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-E., Chang M., Yuan C. One-pot approach for examining the DNA methylation patterns using an engineered methyl-probe. Biosens. Bioelectron. 2014;58:333–337. doi: 10.1016/j.bios.2014.02.064. [DOI] [PubMed] [Google Scholar]
- Sánchez O.F., Lin L., Bryan C.J., Xie J., Freeman J.L., Yuan C. Profiling epigenetic changes in human cell line induced by atrazine exposure. Environ. Pollut. 2020;258 doi: 10.1016/j.envpol.2019.113712. [DOI] [PubMed] [Google Scholar]
- Lungu C., Pinter S., Broche J., Rathert P., Jeltsch A. Modular fluorescence complementation sensors for live cell detection of epigenetic signals at endogenous genomic sites. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-00457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen, H.F., Adie, K., Chaubert, P., Bird, A.P. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Research2006,34 (13), e96-e96. [DOI] [PMC free article] [PubMed]
- Mendonca A., Sánchez O.F., Xie J., Carneiro A., Lin L., Yuan C. Identifying distinct heterochromatin regions using combinatorial epigenetic probes in live cells. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2021;194725 doi: 10.1016/j.bbagrm.2021.194725. [DOI] [PubMed] [Google Scholar]
- Fischle W., Franz H., Jacobs S.A., Allis C.D., Khorasanizadeh S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK (S/T) motifs. J. Biol. Chem. 2008;283(28):19626–19635. doi: 10.1074/jbc.M802655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y., Hu C.-D. Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. Biotechniques. 2012;53(5):285–298. doi: 10.2144/000113943. [DOI] [PubMed] [Google Scholar]
- Alongkronrusmee, D., Watts, V.J., van Rijn, R.M., Bimolecular Fluorescence Complementation Methodology to Study G Protein-Coupled Receptor Dimerization in Living Cells. In Receptor-Receptor Interactions in the Central Nervous System, Springer: 2018; pp 205-221.
- Fan J.-Y., Cui Z.-Q., Wei H.-P., Zhang Z.-P., Zhou Y.-F., Wang Y.-P., Zhang X.-E. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein–protein interactions in living cells. Biochem. Biophys. Res. Commun. 2008;367(1):47–53. doi: 10.1016/j.bbrc.2007.12.101. [DOI] [PubMed] [Google Scholar]
- Dignam, T.; Kaufmann, R. B.; LeStourgeon, L.; Brown, M. J., Control of Lead Sources in the United States, 1970-2017: Public Health Progress and Current Challenges to Eliminating Lead Exposure. J Public Health Manag Pract2019,25 Suppl 1, Lead Poisoning Prevention (Suppl 1 LEAD POISONING PREVENTION), S13-S22. [DOI] [PMC free article] [PubMed]
- Hanna-Attisha M., LaChance J., Sadler R.C., Champney Schnepp A. Elevated Blood Lead Levels in Children Associated With the Flint Drinking Water Crisis: A Spatial Analysis of Risk and Public Health Response. Am. J. Public Health. 2016;106(2):283–290. doi: 10.2105/AJPH.2015.303003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankar-Brewoo G.M., Darko G., Abaidoo R.C., Dalsgaard A., Johnson P.-N., Ellis W.O., Brimer L. Health risks of toxic metals (Al, Fe and Pb) in two common street vended foods, fufu and fried-rice, in Kumasi. Ghana. Scientific African. 2020;7 [Google Scholar]
- Trampel D.W., Imerman P.M., Carson T.L., Kinker J.A., Ensley S.M. Lead contamination of chicken eggs and tissues from a small farm flock. J. Vet. Diagn. Invest. 2003;15(5):418–422. doi: 10.1177/104063870301500503. [DOI] [PubMed] [Google Scholar]
- Parveen R., Abbasi A.M., Shaheen N., Shah M.H. Accumulation of selected metals in the fruits of medicinal plants grown in urban environment of Islamabad, Pakistan. Arab. J. Chem. 2020;13(1):308–317. [Google Scholar]
- Türkdoğan M.K., Kilicel F., Kara K., Tuncer I., Uygan I. Heavy metals in soil, vegetables and fruits in the endemic upper gastrointestinal cancer region of Turkey. Environ. Toxicol. Pharmacol. 2003;13(3):175–179. doi: 10.1016/S1382-6689(02)00156-4. [DOI] [PubMed] [Google Scholar]
- Dobrakowski M., Boroń M., Czuba Z.P., Birkner E., Chwalba A., Hudziec E., Kasperczyk S. Blood morphology and the levels of selected cytokines related to hematopoiesis in occupational short-term exposure to lead. Toxicol. Appl. Pharmacol. 2016;305:111–117. doi: 10.1016/j.taap.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Gil F., Hernández A.F., Márquez C., Femia P., Olmedo P., López-Guarnido O., Pla A. Biomonitorization of cadmium, chromium, manganese, nickel and lead in whole blood, urine, axillary hair and saliva in an occupationally exposed population. Sci. Total Environ. 2011;409(6):1172–1180. doi: 10.1016/j.scitotenv.2010.11.033. [DOI] [PubMed] [Google Scholar]
- Health, C.O.E., Lanphear, B.P., Lowry, J.A., Ahdoot, S., Baum, C.R., Bernstein, A. S., Bole, A., Brumberg, H.L., Campbell, C.C., Lanphear, B.P., Pacheco, S.E., Spanier, A.J., Trasande, L. Prevention of Childhood Lead Toxicity. Pediatrics2016,138 (1). [DOI] [PubMed]
- Dalley J., Gupta P., Hung C. A physiological pharmacokinetic model describing the disposition of lead in the absence and presence of L-ascorbic acid in rats. Toxicol. Lett. 1990;50(2–3):337–348. doi: 10.1016/0378-4274(90)90027-j. [DOI] [PubMed] [Google Scholar]
- Lin L.F., Xie J., Sánchez O.F., Bryan C., Freeman J.L., Yuan C. Low dose lead exposure induces alterations on heterochromatin hallmarks persisting through SH-SY5Y cell differentiation. Chemosphere. 2021;264 doi: 10.1016/j.chemosphere.2020.128486. [DOI] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., Golland P., Sabatini D.M. Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2019;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Politz J.C.R., Scalzo D., Groudine M. Something Silent This Way Forms: The Functional Organization of the Repressive Nuclear Compartment. Annu. Rev. Cell Dev. Biol. 2013;29(1):241–270. doi: 10.1146/annurev-cellbio-101512-122317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu B., Chen H.-M., Moresco J.J., Orelo B.D., Yang B., Gaspar J.M., Keppler-Ross S., Yates J.R., Hall D.H., Maine E.M., Mango S.E. Regulated nuclear accumulation of a histone methyltransferase times the onset of heterochromatin formation in <em>C. elegans</em> embryos. Science. Advances. 2018;4(8):eaat6224. doi: 10.1126/sciadv.aat6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet-Garnier A., Kiêu K., Aguirre-Lavin T., Tar K., Flores P., Liu Z., Peynot N., Chebrout M., Dinnyés A., Duranthon V., Beaujean N. Three-dimensional analysis of nuclear heterochromatin distribution during early development in the rabbit. Chromosoma. 2018;127(3):387–403. doi: 10.1007/s00412-018-0671-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takanaka Y., Yamagata K., Wakayama T., Stasevich T.J., Kainuma T., Tsurimoto T., Tachibana M., Shinkai Y., Kurumizaka H., Nozaki N., Kimura H. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun, K., Jeon, J., Park, K., Kim, J., Writing, erasing and reading histone lysine methylations. Experimental & Molecular Medicine2017,49 (4), e324-e324. [DOI] [PMC free article] [PubMed]
- Seki Y., Hayashi K., Itoh K., Mizugaki M., Saitou M., Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 2005;278(2):440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Latham T., Gilbert N., Ramsahoye B. DNA methylation in mouse embryonic stem cells and development. Cell Tissue Res. 2008;331(1):31–55. doi: 10.1007/s00441-007-0537-9. [DOI] [PubMed] [Google Scholar]
- De Carvalho D.D., You J.S., Jones P.A. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20(10):609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicetto D., Zaret K.S. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 2019;55:1–10. doi: 10.1016/j.gde.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xu Z., Liu M., Huang Y., Fan R., Su Y., Hu G., Peng X., Peng X. Lead exposure assessment from study near a lead-acid battery factory in China. Sci. Total Environ. 2012;429:191–198. doi: 10.1016/j.scitotenv.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Bryant S. Lead-contaminated drinking waters in the public schools of Philadelphia. J. Toxicol. Clin. Toxicol. 2004;42(3):287–294. doi: 10.1081/clt-120037429. [DOI] [PubMed] [Google Scholar]
- Martinez C., Nagae M., Zaia C., Zaia D. Acute morphological and physiological effects of lead in the neotropical fish Prochilodus lineatus. Braz. J. Biol. 2004;64(4):797–807. doi: 10.1590/s1519-69842004000500009. [DOI] [PubMed] [Google Scholar]
- Akinyemi A.J., Miah M.R., Ijomone O.M., Tsatsakis A., Soares F.A.A., Tinkov A.A., Skalny A.V., Venkataramani V., Aschner M. Lead (Pb) exposure induces dopaminergic neurotoxicity in Caenorhabditis elegans: Involvement of the dopamine transporter. Toxicol. Rep. 2019;6:833–840. doi: 10.1016/j.toxrep.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J.F., Farooqui Z., Faulk C.D., Barks A.K., Jones T., Dolinoy D.C., Bakulski K.M. Perinatal lead (Pb) exposure and cortical neuron-specific DNA methylation in male mice. Genes (Basel) 2019;10(4):274. doi: 10.3390/genes10040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J.S., Nicetto D., Zaret K.S. H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet. 2016;32(1):29–41. doi: 10.1016/j.tig.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A.H., Zhao C., Li Q.K., Gustafson K.S., Eltoum I.E., Tambouret R., Benstein B., Savaloja L.C., Kulesza P. The cytologic criteria of malignancy. J. Cell. Biochem. 2010;110(4):795–811. doi: 10.1002/jcb.22585. [DOI] [PubMed] [Google Scholar]
- Carone D.M., Lawrence J.B. Heterochromatin instability in cancer: From the Barr body to satellites and the nuclear periphery. Semin. Cancer Biol. 2013;23(2):99–108. doi: 10.1016/j.semcancer.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi A., Li W. Global heterochromatin loss: a unifying theory of aging? Epigenetics. 2012;7(7):680–688. doi: 10.4161/epi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T., Ewels P.A., Schoenfelder S., Furlan-Magaril M., Wingett S.W., Kirschner K., Thuret J.-Y., Andrews S., Fraser P., Reik W. Global reorganization of the nuclear landscape in senescent cells. Cell reports. 2015;10(4):471–483. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T., Kirschner K. Chromosome organisation during ageing and senescence. Curr. Opin. Cell Biol. 2016;40:161–167. doi: 10.1016/j.ceb.2016.03.020. [DOI] [PubMed] [Google Scholar]
- Grosselin K., Durand A., Marsolier J., Poitou A., Marangoni E., Nemati F., Dahmani A., Lameiras S., Reyal F., Frenoy O., Pousse Y., Reichen M., Woolfe A., Brenan C., Griffiths A.D., Vallot C., Gérard A. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet. 2019;51(6):1060–1066. doi: 10.1038/s41588-019-0424-9. [DOI] [PubMed] [Google Scholar]
- Castillo-Fernandez J., Herrera-Puerta E., Demond H., Clark S.J., Hanna C.W., Hemberger M., Kelsey G. Increased transcriptome variation and localised DNA methylation changes in oocytes from aged mice revealed by parallel single-cell analysis. Aging Cell. 2020;19(12) doi: 10.1111/acel.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese V., Cornelius C., Dinkova-Kostova A.T., Calabrese E.J. Vitagenes, cellular stress response, and acetylcarnitine: relevance to hormesis. BioFactors. 2009;35(2):146–160. doi: 10.1002/biof.22. [DOI] [PubMed] [Google Scholar]
- Vaiserman A.M. Hormesis and epigenetics: is there a link? Ageing Res. Rev. 2011;10(4):413–421. doi: 10.1016/j.arr.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Flatau E., Gonzales F., Michalowsky L., Jones P. DNA methylation in 5-aza-2'-deoxycytidine-resistant variants of C3H 10T1/2 C18 cells. Mol. Cell. Biol. 1984;4(10):2098–2102. doi: 10.1128/mcb.4.10.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Lin L., Sánchez O.F., Bryan C., Freeman J.L., Yuan C. Pre-differentiation exposure to low-dose of atrazine results in persistent phenotypic changes in human neuronal cell lines. Environ. Pollut. 2021;271 doi: 10.1016/j.envpol.2020.116379. [DOI] [PubMed] [Google Scholar]
- Wang T., Holt M.V., Young N.L. The histone H4 proteoform dynamics in response to SUV4-20 inhibition reveals single molecule mechanisms of inhibitor resistance. Epigenetics Chromatin. 2018;11(1):29. doi: 10.1186/s13072-018-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libertini E., Lebreton A., Lakisic G., Dillies M.-A., Beck S., Coppée J.-Y., Cossart P., Bierne H. Overexpression of the heterochromatinization factor BAHD1 in HEK293 cells differentially reshapes the DNA methylome on autosomes and X chromosome. Front. Genet. 2015;6:339. doi: 10.3389/fgene.2015.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell A.L., Chung B.Y., Creighton C.E., Kalisch B.E., Bailey C.D., MacLusky N.J. Neurosteroid metabolites of testosterone and progesterone differentially inhibit ERK phosphorylation induced by amyloid β in SH-SY5Y cells and primary cortical neurons. Brain Res. 2018;1686:83–93. doi: 10.1016/j.brainres.2018.02.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.