Abstract
Neuroinflammation is a common feature in neurodegenerative diseases, modulated by the Alzheimer’s disease risk factor, apolipoprotein E (APOE). In the brain, apoE protein is synthesized by astrocytes and microglia. We examined primary cultures of astrocytes and microglia from human APOE (E2, E3 and E4) targeted-replacement mice. Astrocytes secreted two species of apoE, whereas cellular apoE consisted of only one. Both forms of secreted astrocytic apoE were bound during glycoprotein isolation, and enzymatic removal of glycans produced a convergence of the two forms of apoE to a single form; thus, the two species of astrocyte-secreted apoE are differentially glycosylated. Microglia released only a single species of apoE, while cellular apoE consisted of two forms; the secreted apoE and one of the two forms of cellular apoE were glycosylated. We treated the primary glia with either endogenous (TNFα) or exogenous (LPS) pro-inflammatory stimuli. While LPS had no effect on astrocytic apoE, APOE2 and APOE3 microglia increased release of apoE; APOE4 microglia showed no effect. APOE4 microglia showed higher baseline secretion of TNFα compared to APOE2 and APOE3 microglia. TNFα treatment reduced the secretion and cellular expression of apoE only in APOE4 astrocytes. The patterns of apoE species produced by astrocytes and microglia were not affected by inflammation. No changes in APOE mRNA were observed in astrocytes after both treatments. Together, our data demonstrate that astrocytes and microglia differentially express and secrete glycosylated forms of apoE and that APOE4 astrocytes and microglia are deficient in immunomodulation compared to APOE2 and APOE3.
Keywords: apolipoprotein E, inflammation, glia cells, Alzheimer’s disease, post-translational modifications
TABLE OF CONTENT
1) Moification and secretion of apoE isoforms differ between astrocytes and microglia, based in part on the presence of O-linked glycans.
2) Inflammation affects apoE secretion and expression in an isoform- and cell-specific manner.
INTRODUCTION
Neuroinflammation plays a fundamental role in the progression and severity of Alzheimer’s disease (AD), by facilitating or exacerbating both amyloid β (Aβ) and neurofibrillary tangles formation (Barroeta-Espar et al., 2019; Heppner, Ransohoff, & Becher, 2015; Newcombe et al., 2018; B. Zhang et al., 2013). Neuroinflammation is characterized by activated glial cells and increased levels of cytokines and chemokines. Chronic inflammatory processes with activation of astrocytes and microglia can lead to overproduction of neurotoxic molecules and pro-inflammatory cytokines that lead to neuronal damage and death (Fernandez, Hamby, McReynolds, & Ray, 2019). Thus, regulating the activation of astrocytes and microglia is a target for overcoming inflammation and neurodegeneration in the brain.
An immunomodulatory protein important to AD pathogenesis is apolipoprotein E (apoE). ApoE is a 34-kDa protein that is involved in a variety of functions, including lipid transport, neurite outgrowth and neuronal plasticity, and regulation of Aβ structure and clearance (LaDu et al., 2001; Manelli, Stine, Van Eldik, & LaDu, 2004). ApoE also modulates the inflammatory response of microglia and astrocytes (Barger & Harmon, 1997; Laskowitz, Goel, Bennett, & Matthew, 1997; Laskowitz, Horsburgh, & Roses, 1998; Laskowitz, Matthew, et al., 1998; Laskowitz, Sheng, et al., 1997). Human apoE exists in three major isoforms, apoE2, apoE3 and apoE4, encoded by the ε2, ε3 and ε4 alleles, respectively. The APOE ε4 allele not only increases an individual's risk for AD (Di Battista, Heinsinger, & Rebeck, 2016; Fernandez et al., 2019; Hyman et al., 1996), but also for the outcome from neurological injuries with dramatic brain inflammation (e.g., intracranial hemorrhage, closed head injury, stroke) (Fernandez et al., 2019; Kockx, Traini, & Kritharides, 2018). Conversely, the APOE ε2 allele is protective in these conditions (Fernandez et al., 2019). Increasing evidence suggests that apoE reduces central nervous system (CNS) inflammation in an isoform-specific manner, with apoE4 displaying the least anti-inflammatory activity (Brown et al., 2002; Vitek, Brown, & Colton, 2009). Moreover, application of apoE or apoE-mimetic peptides modulates the inflammatory response in vitro and in vivo (Cheng et al., 2018; Laskowitz et al., 2017; J. Liu et al., 2018; Tu et al., 2017; Wei et al., 2013). For example, an apoE-mimetic peptide COG1410 reduced levels of TNFα, IL-1β, IL-6 and IL-12 in both APOE3 and APOE4 mice experiencing sepsis (H. Wang, Christensen, Vitek, Sullivan, & Laskowitz, 2009). Furthermore, mice that lack the APOE gene exhibit increased pro-inflammatory cytokines in the CNS, suggesting that the presence of apoE regulates immune function (Laskowitz, Lee, Schmechel, & Staats, 2000; Laskowitz et al., 2001; Y. Liu et al., 2015). Thus, there is a direct link between the effects of apoE on glial activation and the production of cytokines.
The apoE protein undergoes post-translational modifications. ApoE is modified by O-glycosylation at different sites and to a greater extent in cerebrospinal fluid (CSF) than in plasma (Flowers, Grant, Woods, & Rebeck, 2019; Hu, Meuret, Go, Yassine, & Nedelkov, 2020; Lee et al., 2010; Rebeck et al., 1998). In addition, C-terminal cleavage of CNS apoE occurs in an isoform- and cell-type dependent manner (Bien-Ly et al., 2011; Brecht et al., 2004), and might exacerbate the effects of inflammation and behavioral deficits in mice (Harris et al., 2003). ApoE modifications are associated with biochemical differences in the brain such that larger forms of apoE are solubilized in saline, whereas the smaller form is solubilized only in the presence of detergent (DiBattista, Dumanis, Newman, & Rebeck, 2016). Given this complexity, understanding CNS apoE functions requires a better discernment of the different forms of apoE, particularly during their response to inflammatory insult. Here we assessed the effects of inflammation on the various types of apoE in primary microglia and astrocytes of different APOE genotypes.
MATERIAL AND METHODS
Animals.
We used C57BL/6J male and female mice pups (P1-P2) expressing human APOE2, APOE3 or APOE4 under the control of the endogenous murine APOE promoter, that have been previously validated (Sullivan et al., 1997; P. T. Xu et al., 1996). All studies were carried out following the Guide for the Care and Use of Laboratory Animals as adopted by the U.S. National Institutes of Health and approved by Georgetown University Animal Care and Use Committee.
Tissue collection.
Mice were deeply anesthetized, blood was drawn from the tail, and then the mice were intracardially perfused with 0.1 M phosphate-buffered saline solution (PBS, pH 7.4). Brain cortex, heart, liver, adipose tissue, and lymph nodes were dissected. TBS and TBSX brain cortex fractions were obtained as previously described (DiBattista et al., 2016). Briefly, tissue was homogenized using a dounce homogenizer in Tris-buffered saline solution (TBS, pH 7.4) supplemented with protease and phosphatase inhibitors (Thermo Scientific™ Pierce™, Waltham, MA) at 4 °C. Homogenates were centrifuged at 47,000 rpm at 4 °C for 45 min, and the supernatants were collected (TBS fraction). The insoluble pellets were sonicated in TBSX buffer (TBS, 1% Triton X-100, pH 7.4) supplemented with protease and phosphatase inhibitors, and centrifuged at 47,000 rpm at 4 °C for 45 min. The supernatant solutions were collected (TBSX fraction). High speed centrifugation was performed in an Optima TLX centrifuge in a TLA 120.2 rotor (Beckman). Heart, liver, adipose tissue, and lymph nodes were homogenized using a dounce homogenizer in 1x RIPA buffer (100 mg of tissue per mL of buffer) supplemented with protease and phosphatase inhibitors. Samples were sonicated and centrifuged at 16,000 x g for 10 min at 4 °C. The supernatant was collected and considered the total protein samples. To collect plasma, whole blood was collected into commercially EDTA-treated tubes, and centrifuged for 15 min (2,000 x g at 4 °C). Protein concentrations were measured using the bicinchoninic acid (BCA) Protein Assay Kit (Pierce, Rockford, IL, USA).
Primary cultures of glial cells.
Glia were isolated from the cerebral cortex of 1–2 day-old APOE targeted-replacement mice according to an established protocol (Daniele, Edwards, & Maguire-Zeiss, 2014) with slight modifications. Briefly, cortices were collected and mechanically dissociated in ice-cold serum-free Opti-MEM media (Invitrogen™, Waltham, MA). After centrifugation and resuspension in microglia growth media (Opti-MEM supplemented with 2% fetal bovine serum (FBS), 3% fetal horse serum (FHS), 1 mM sodium pyruvate, 0.6% glucose, 1 mM L-glutamine, and antibiotic and antimycotic), cells were expanded for 21 days and the flasks were shaken at 200 rpm for 3 hrs. to isolate microglia. The supernatant was centrifuged and microglial pellet was resuspended in microglia growth media and seeded in 12-well plates (2×105 cells/well). Opti-MEM supplemented with 10% FBS, and antibiotic and antimycotic was added to the flask and allowed to shake overnight at 37 °C to remove oligodendrocytes and remaining microglia. The next day, the media was discarded and the adhered cells were then trypsinized, collected and centrifuged. These astrocytes were seeded at density of 1×105 cells/well. Glial cell purity was confirmed to be ~95% using immunofluorescence for Iba-1 (microglial marker, Cat. No. 019-19741, FUJIFILM Wako Pure Chemical Corp., Richmond, VA), GFAP (astrocytic marker, Cat. No. 3670, Cell Signaling Technologies, Beverly, MA) and ProLong™ Diamond Antifade Mountant, with DAPI (Invitrogen™, Waltham, MA) for nuclear counterstain. For biochemical experiments, primary mouse astrocytes and microglia cells were maintained overnight, and then the medium was replaced with serum-free Opti-MEM containing either PBS (control) or experimental agents. All reagents were prepared in sterile PBS and used at the following range of concentrations: Lipopolysaccharide, E. coli 0111:B4 (LPS, Cat. No. 437627), from 5 ng/mL to 500 ng/mL, was purchased from Millipore Sigma (St. Louis, MI). The tumor necrosis factor-alpha (TNFα, Cat. No. 410-MT), at 3 ng/mL and 30 ng/mL, was purchased from R&D Biosystems Inc. (Minneapolis, MN). Interleukin-10 (IL-10, Cat. No. ab222176) at 100 ng/mL, the JNK inhibitor SP600125 (Cat. No. ab120065) at 10 μM, the MEK1 inhibitor PD989059 (Cat. No. ab120234) at 50 μM, the p38 MAPK inhibitor, doramapimod (BIRD, Cat. No. ab142166) at 1 μM, the IkappaB kinase inhibitor IKK-16 (Cat. No. ab216471) at 2.5 μM were each purchased from Abcam (Cambridge, MA). Experiments were performed in serum-free pre-warmed Opti-MEM. Cells were allowed to adjust to the media for 1 hr. before applying the reagents and then maintained for 24 hrs. Conditioned media and cells were collected for western blot and ELISA experiments.
Western blot.
Conditioned media (25 μL) and cell lysates (2.5 or 5.0 μg) were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PDVF) membrane overnight at 4 °C. Membranes were blocked in 5% nonfat milk in PBS-0.5% Tween-20 (PBS-T), and were probed with polyclonal goat anti-apoE (1:2000, Cat. No. K74190G, Lot. No. 3F17515, Meridian Life Science, Inc., Memphis, TN) and α-tubulin (1:5000, Cat. No. ab4074, Abcam, Cambridge, MA) antibodies overnight at 4 °C with gentle agitation. α-tubulin was used for western blot normalization because its expression did not change with treatment. After three washing steps with PBS-T, immune complexes were detected with appropriate horseradish peroxidase-conjugated secondary antibodies (1:2000) and chemiluminescence detection using SuperSignal™ West Dura Extended Duration Substrate (ThermoFisher Scientific, Inc, Waltham, MA) and imaged via Amersham Imager 600 (GE Healthcare).
Enzyme-linked immunosorbent assay (ELISA).
TNFα levels were determined using the TNFα mouse ELISA kit (Cat. No. ab100747, Abcam, Cambridge, MA), according to manufacturer’s instructions. ApoE levels were determined using the Human Apolipoprotein E ELISA kit (Cat. No. ab108813, lot No. GR3218897-5, Abcam, Cambridge, MA), according to manufacturer’s instructions. 25 μL of cell lysate and 50 μL of conditioned media were loaded per well. Absorbance was measured at 450 nm using a SpectraMax iD3 microplate reader (Molecular Devices, San Jose, CA). The concentration of apoE was determined against a seven-point standard curve. The quantity of apoE was expressed as total amount of apoE protein (μg) per number of cells seeded. To determine the percentage of secreted and cellular apoE, the amount of apoE detected in a well was normalized to the total volume of conditioned media (500 μL for astrocytes, 250 μL for microglia) or cell lysate collected (200 μL for astrocytes, 125 μL for microglia), respectively, during the experiment. The total amount of apoE was considered to be the sum of secreted and cellular apoE measured, per number of cells plated in a well.
Glycoprotein isolation.
Conditioned media was concentrated at least ten-fold with polyethersulfone (PES) membrane microcentrifuge filters (Macrosep Advance Centrifugal Devices, Pall Laboratory, MWCO 10K) in a refrigerated Sorvell Legend X1R centrifuge at 2500 rpm before performing glycoprotein isolation. The Pierce™ Glycoprotein Isolation Kit, WGA (Cat. No. 89805, Thermo Scientific™ Pierce™, Waltham, MA) based on Wheat Germ Agglutinin binding was used according to manufacturer’s instructions with minor modifications. To elute protein from the resin, resin was incubated with elution buffer for 30 min.
Enzymatic deglycosylation reaction of glycoproteins.
The Protein Deglycosylation Mix II kit (Cat. No. P6044S, New England Biolabs Inc., Ipswich MA) was used to remove O-linked oligosaccharides from glycoproteins according to manufacturer’s instructions with minor modifications. Briefly, the glycoproteins (concentrated conditioned media or cell lysates) were denatured by heating the samples at 75 °C for 10 min. Samples were allowed to cool before initiating enzymatic deglycosylation, and incubated at room temperature for 30 min in the presence of the Protein Deglycosylation Mix with occasional shaking. Samples were transfer to a water bath at 37 °C, and incubated overnight. The enzymatic reaction was stopped with the addition of loading buffer.
Quantitative RT-PCR (qRT-PCR).
Total RNA was isolated from cultures using TRIzol™ Plus RNA Purification Kit (Cat. No. 12183555, Invitrogen™, Waltham, MA). cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (Cat. No. 4368814, Applied Biosystems, Foster City, CA). cDNA (1:2 dilution, 4 μl) was amplified by real-time PCR using Power SYBR™ Green PCR Master Mix (Cat. No. K9021, Applied Biosystems, Foster City, CA). Samples were standardized to GAPDH. Synthetic oligonucleotides were used for human apoE (forward: 5’-TTCCTGGCAGGATGCCAGGC-3’ and reverse: 5’-GGTCAGTTGTTCCTCCAGTTC-3’), mouse Lcn2 (forward: 5’-AATGACTCTCACGGGGATTG-3’ and reverse: 5’-AGTGGTGGGGATGACTTCAG-3’), mouse Serpina3n (forward: 5’-CCCTGAGGAAGTGGAAGAAT-3’ and reverse: 5’-CCTGATGCCCAGCTTTGAAA-3’), mouse TNFα (forward: 5’-GGTGCCTATGTCTCAGCCTCTT-3’ and reverse: 5’-GCCATAGAACTGATGAGAGGGAG-3’), and mouse GAPDH (forward: 5’-GTGTTTCC-TCGTCCCGTAGA-3’ and reverse: 5’-AATCCGTTCACACCGACCTT-3’). Each individual sample was analyzed in triplicate and RNA levels were reported as fold difference compared to control (vehicle or APOE3 genotype). Analysis of the qRT-PCR data was done on SDS 2.3 (Applied Biosystems, Foster City, CA) and relative expression was calculated using RQ Manager software (Applied Biosystems, Foster City, CA). The resulting data was analyzed using the double delta Ct method.
Hoechst 33342/propidium iodine (Hoechst/PI) staining.
Cell survival was measured using Hoechst/propidium iodide (PI) staining 24 hrs. after LPS treatment (5-5000 ng/mL) or 2 min after 10% methanol application (as a positive control for cell death). APOE4 astrocytes were washed with pre-warmed serum-free media and incubated simultaneously with Hoechst 33342 (staining healthy cells, Cat. No. 62249, ThermoFisher Scientific) and propidium iodide (staining apoptotic and necrotic cells, Cat. No. 40017, Biotium), 0.5uL each in serum-free media, for 5 min at 37 °C. Media was removed and cells were washed three times with pre-warmed serum-free media. Astrocytes were visualized with the inverted fluorescent microscope Olympus IX71. Hoechst/PI-positive cells were then counted using ImageJ and cell viability was calculated by subtracting Hoechst/PI-positive cells from the total number of astrocytes (Hoechst-positive cells) and expressed as a percentage of the total number of astrocytes.
Statistical analysis.
Western blot results were quantified using Image J software (National Institutes of Health, Bethesda, MA, USA). The density signal of secreted apoE was normalized to the total amount of protein found in a cell lysate by BCA protein assay (Thermo Scientific™ Pierce™). After this normalization step, the values were expressed as relative percent of the average of vehicle treatment (0 ng/mL drug treatment), which was set as 100%. The total amount of cellular apoE was normalized to α-tubulin when it was compared between different treatments. To calculate the percentage of the highest molecular weight band or “upper band”, densitometry of the lower band was subtracted from the densitometry of total apoE detection and expressed as percentage of total apoE. Data were expressed as mean ± standard error of the mean (SEM) from at least three independent experiments with each treatment in duplicate. For Figures 1 through 6, a Two-tailed Student T-test, a one-way ANOVA or a two-way ANOVA were used when appropriate. When statistical differences were achieved, a Tukey’s multiple comparison test was used. GraphPad Prism (GraphPad Software, San Diego, CA) was used for statistical analysis with p<0.05 considered as statistically significant.
Figure 1. Secreted and cellular apoE ratio and abundance are different between glia cell-types.
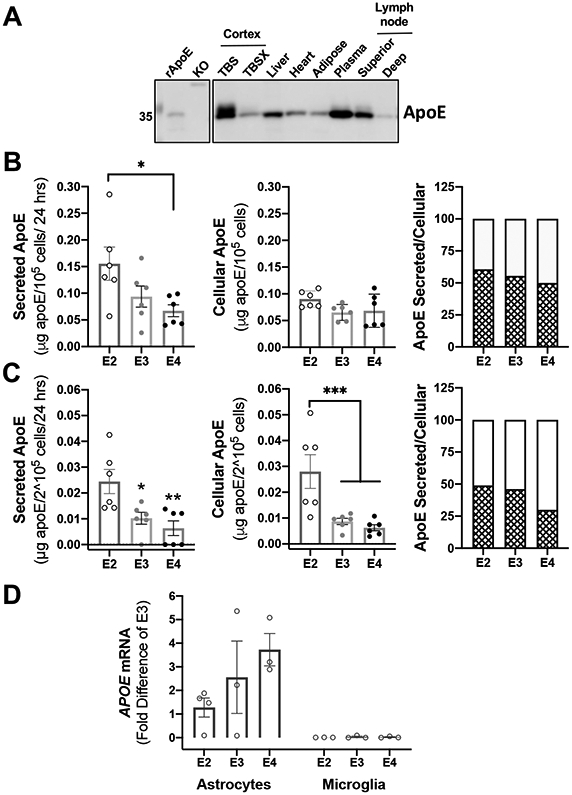
(A) Tissue from human APOE2 targeted-replacement mice was isolated and processed according to material and methods. 2.5-10 μg of protein lysates was analyzed by western blotting with antibodies against apoE. TBS:=soluble fraction; TBSX:=insoluble fraction; KO:=total homogenate of cortex from apoE knockout (KO) mice. TBS and TBSX fractions were isolated as described in DiBattista et al. (2016). Representative image of one of two experiments. (B-C) Astrocytes (B) and microglia (C) derived from human APOE2, APOE3 and APOE4 targeted-replacement mice were isolated and seeded in serum-free media. Conditioned media and cells were harvested after 24 hrs. of incubation. 50 μL of conditioned media and 25 μL of cell lysates were analyzed by ELISA (n=6 independent cell cultures). *p<0.05, **p<0.005, ***p<0.0005. A one-way ANOVA was used to assess outcome measures from genotype in fractions from secreted and cellular apoE in cell cultures (astrocytes and microglia). ApoE ratio was calculated as the mean percentage of secreted apoE (pattern bars) and cellular apoE (blank bars) to total apoE (see Material and Methods). (D) Basal levels of APOE mRNA expression are expressed as fold difference from APOE3 astrocytes (n=3-4 independent cell cultures).
Figure 6. Astrocytic apoE4 secretion reduction after TNFα activation is unaltered across several inflammatory signaling pathways.
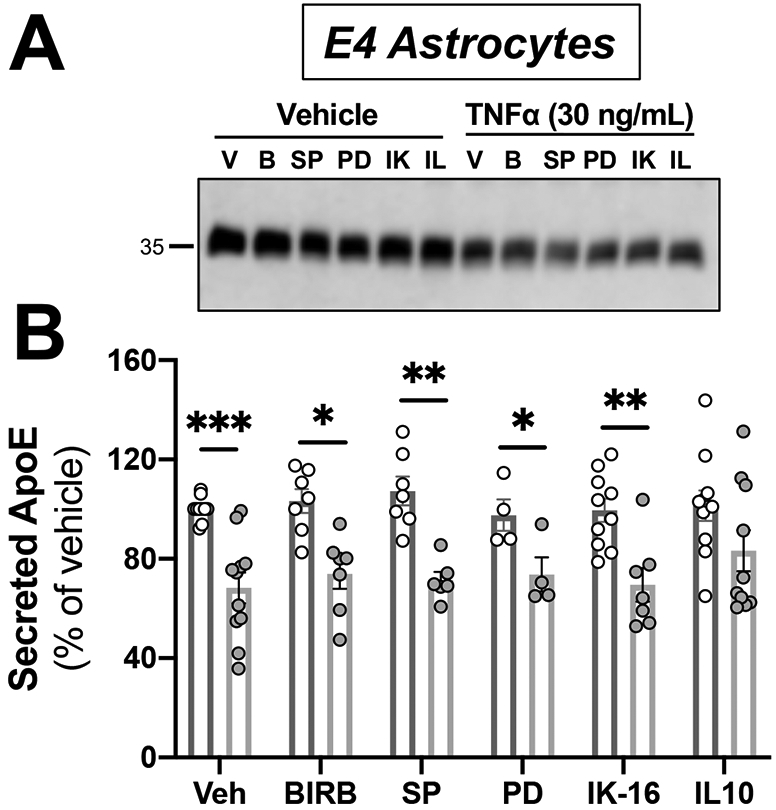
Astrocytes from human APOE4 targeted-replacement mice were treated with 30 ng/mL of TNFα and inhibitors. Conditioned media was collected at 24 hrs., and was analyzed by western blotting with antibodies against apoE. Veh = vehicle; BIRB = Doramapimod, p38 MAPK inhibitor (1 μM); SP = SP600125, JNK kinase inhibitor (10 μM); PD = PD98059, MEK inhibitor (50 μM); IK-16 = IKK-16, IkappaB inhibitor (2.5 μM); IL10 = interleukin 10 (100 ng/mL). (A) Representative immunoblots showing the changes in secreted apoE by APOE4 astrocytes. (B) Quantification of western blots. Bar graphs represent the mean ± SEM (n=5-8 individual experiments). A two-way ANOVA was used to assess outcome measures on interactions between treatment and pretreatment. *p<0.05, **p<0.005 compared to APOE4 treated with TNFα.
RESULTS
ApoE species differ between astrocytes and microglia.
ApoE is a 34-kDa glycosylated apolipoprotein expressed in several organs, including liver, brain, spleen, kidney and lung (Elshourbagy, Liao, Mahley, & Taylor, 1985). Under denaturing conditions, peripheral apoE (in liver, heart, adipose tissue, plasma and lymph nodes) migrated generally as a single species, whereas brain apoE migrated as several species of similar size (Figure 1A). Consistent with previous studies (DiBattista et al., 2016), TBS-soluble samples from mice cortex contained higher molecular weight apoE species compared to TBSX-soluble samples, which presented as a single species. In the uninjured brain, apoE is synthesized and secreted primarily by astrocytes, although expression also occurs in microglia (Q. Xu et al., 2006). We examined whether APOE genotype affected the levels of secreted and cellular apoE in astrocytes and microglia (Figure 1B and 1C). We used primary glia from human APOE targeted-replacement mice (Sullivan et al., 1997; P. T. Xu et al., 1996). APOE2, APOE3, and APOE4 astrocytes and microglia were grown for 24 hrs. in serum-deprived media; levels of total apoE were measured in conditioned media and cell lysates by ELISA. Astrocytes released at least six times more apoE than microglia under basal conditions (Figure 1B and 1C). APOE2 astrocytes secreted more apoE than APOE4 astrocytes (F(2,15)=4.151, p=0.037); there were no significant differences in cellular apoE levels across genotypes (F(2,15)=2.375, p=0.13). The proportion of secreted apoE relative to total apoE produced by astrocytes was similar among genotypes (at 50-60%) (Figure 1B). APOE mRNA levels did not differ significantly across APOE genotypes, although APOE4 astrocytes trended to more APOE mRNA compared to APOE2 astrocytes (p=0.057, Figure 1D). A higher level of APOE4 mRNA could reflect a compensatory mechanism for the reduced secretion of apoE4 protein (Chernick et al., 2018; Nuriel et al., 2017; Riddell et al., 2008).
In microglia, secreted apoE2 was present at significantly higher levels than apoE3 and apoE4 (F(2,15)=7.744, p=0.005). In contrast to astrocytes, microglia had more cellular apoE2 than apoE3 and apoE4 (F(2,15)=9.376, p=0.002). The proportion of secreted apoE relative to total apoE produced by microglia was higher in APOE2 microglia (49%) compared to APOE4 microglia (30%) (Figure 1C). Thus, a smaller percentage of apoE4 was secreted from microglia (30%) compared to astrocytes (50%). APOE mRNA in microglia under basal conditions was too low for quantification by our qRT-PCR methods (Figure 1E).
Since apoE undergoes post-translational modifications (O-glycosylation and C-terminal cleavage (Bien-Ly et al., 2011; Brecht et al., 2004; Flowers et al., 2019; Flowers & Rebeck, 2020)), we used SDS-PAGE to determine the migration pattern of apoE in cell lysates and conditioned media of APOE3 primary astrocytes and microglia (Figure 2A). ApoE3 secreted by astrocytes was characterized by a double band around 35-kDa, showing two distinct populations of apoE. In contrast, apoE3 secreted by microglia was characterized by only the single band above 35-kDa, showing one population of apoE. Cellular astrocytic apoE3 consisted primarily of the lower band at 35-kDa, whereas apoE3 in microglia displayed a double band of two forms of apoE. ApoE2 and apoE4 from glia showed similar band patterns compared to apoE3 (Figure 2B).
Figure 2. Secreted and cellular apoE migration patterns differ between glia cell types.
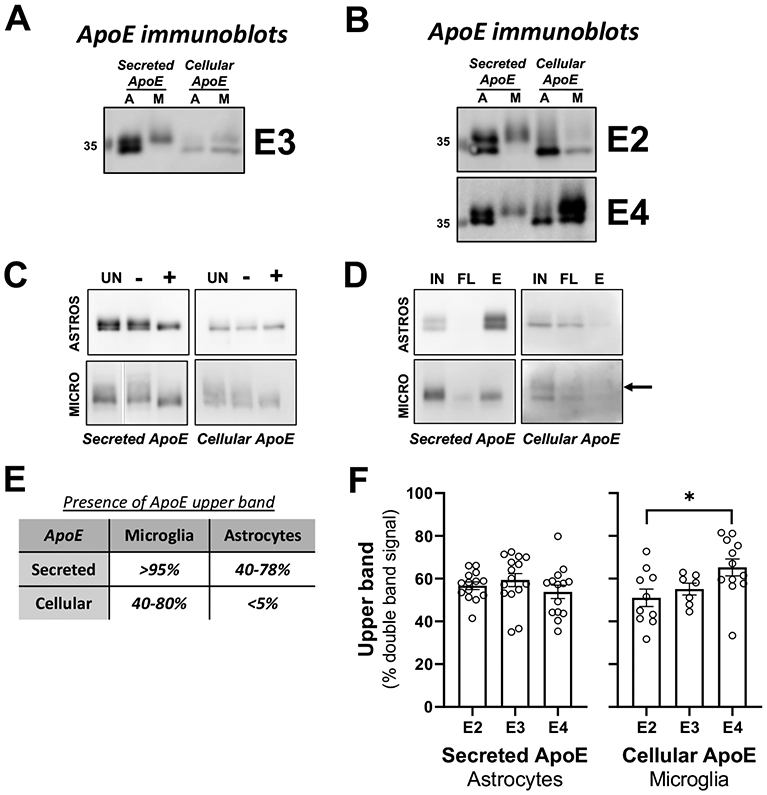
(A-B) Astrocytes (A) and microglia (M) derived from human APOE2, APOE3 and APOE4 targeted-replacement mice were isolated and seeded in serum-free media. Conditioned media and cells were harvested after 24 hrs. of incubation. 5-25 μL of conditioned media and 2.5-10 μg of cell lysates were analyzed by western blotting with antibodies against apoE. Representative image of 3 independently experiments. (A) apoE3; (B) apoE2 and apoE4. Concentrated conditioned media or cell lysate from APOE3 astrocytes and microglia underwent enzymatic deglycosylation (C) and glycoprotein isolation (D). (C) UN = untreated sample; ‘−’ = sample undergoing the deglycosylation procedure in the absence of the deglycosylation enzymes; ‘+’ = sample undergoing the deglycosylation procedure in the presence of the deglycosylation enzymes. (D) IN = input sample before resin binding; ‘FL’ = supernatant after resin binding and centrifugation; ‘E’ = sample eluted from resin. Arrow shows upper band in eluted sample (n=4 independent experiments). (E) Presence of apoE upper band in secreted and cellular samples. Range in percentage correspond to the three genotypes combined. (F) Pattern of expression is cell-type and isoform-specific. Double bands from conditioned media and total homogenate from astrocytes and microglia, respectively, were quantified and expressed as percentage of upper bands with respect to total apoE protein detected. A one-way ANOVA was used to assess outcome measures from percentage of upper band in different genotypes of secreted apoE in astrocytes (n=12-17) and cellular apoE in microglia (n=7-12). *p<0.05.
We performed two assays to determine whether the apparent difference in size of apoE species was due to a post-translational modification such as glycosylation or proteolysis. First, we isolated cell lysates and concentrated conditioned media from APOE3 astrocytes and microglia. The samples underwent enzymatic deglycosylation to strip the O-linked glycans from the apoE protein (Figure 2C). We reasoned that if the difference in apoE forms was due to glycosylation, then we would observe a convergence of the two species into one form. However, if a difference in apoE forms was due to apoE proteolysis, we could observe no shifts, or a shift of both species. Deglycosylation caused a shift in the migration of the larger apoE species in the SDS-PAGE gel such that there was single species of apoE. These data did not test whether the lower species of apoE might also be glycosylated to some extent. We conducted a glycoprotein isolation based on binding to Wheat Germ Agglutinin (Figure 2D): both the larger and smaller apoE species bound to this resin. Thus, our data suggest that both forms of apoE are O-glycosylsted, although the glycosylation differs between the forms.
Figure 2E illustrates the range values as a percentage of the presence of the “upper band” of apoE in relation to total apoE detection for all the genotypes combined. The chart illustrates that the presence of the upper band depends on the type of glial cell, and on whether apoE is secreted or intracellular. More than 95% of secreted apoE from microglia was of the higher molecular weight type, while the upper band detected in secreted apoE from astrocytes ranged from 40-78%. For cellular apoE, the higher molecular weight apoE form was prominent in microglia (40-80%), but nearly undetectable in astrocytes.
We tested whether there was an effect of APOE genotype on the relative proportion of each apoE species (Figure 2F). Because there was almost exclusively one form of apoE in conditioned media from microglia and in cell lysates from astrocytes, analysis was not conducted in those samples. In astrocyte conditioned media, there was no difference in the distribution of the higher molecular weight species among APOE genotypes. In microglia cell extracts, APOE4 cells contained significantly more (14%) of the higher molecular weight band than APOE2 cells (F(2,26)=3.961, p=0.032), indicating a difference in post-translational modifications among genotypes.
LPS does not affect apoE levels in astrocytes.
APOE genotype affects in vivo and in vitro responses to inflammatory conditions (Colton et al., 2002; Laskowitz, Fillit, Yeung, Toku, & Vitek, 2006; Laskowitz et al., 2010; Vitek et al., 2009); here we investigated whether inflammation affected the expression and secretion of apoE in astrocytes and microglia in an apoE isoform-dependent manner. Reactive gliosis is a common response to brain pathology in vivo, including injuries and neurodegeneration (Zamanian et al., 2012), and to culturing in vitro (Liddelow et al., 2017; Wu & Schwartz, 1998). To examine whether our culture conditions had inflammatory effects specific to one APOE genotype, we measured Serpina3n as a marker for reactive astrocytes and Lcn2 as a marker for both reactive astrocytes and microglia (Zamanian et al., 2012). Culturing astrocytes in the presence or absence of FBS had no effects on Serpina3n or Lcn2 mRNA levels in cells of any of the APOE genotypes (data not shown).
We used LPS to cause an inflammatory response, treating both microglia and astrocytes. APOE2, APOE3 and APOE4 astrocytes were exposed to a range of LPS concentrations (5-500 ng/mL) for 24 hrs, and apoE was analyzed by western blots (Figure 3A). There were no changes in secreted or cellular apoE from astrocytes in response to different doses of LPS (Figure 3C and 3E). These results are consistent with previous studies showing that LPS up to 100 ng/mL had no effects on apoE expression or release (Aleong, Blain, & Poirier, 2008). As in Figure 2, secreted astrocytic apoE consisted of two species by immunoblot; LPS treatment did not change the ratio of the larger apoE form to total apoE detected in conditioned media across APOE genotypes. We tested whether LPS caused astrocytic cell death, potentially affecting apoE levels, but found no evidence of cell death up to 5000 ng/mL LPS (Figure 4A). Furthermore, LPS treatment had no effect on APOE mRNA levels 24 hrs. after treatment (Figure 3G).
Figure 3. Activation of glial cells by LPS is cell-type and genotype specific.
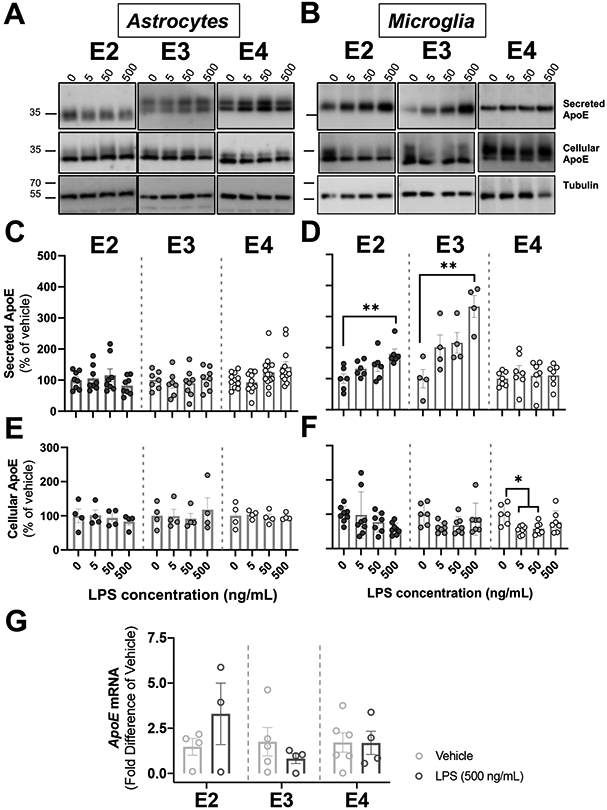
Glial cells derived from human APOE (E2, E3, E4) targeted-replacement mice were treated with several doses of LPS (5-500ng/mL). Conditioned media and cells were collected at 24 hrs., and were analyzed by western blotting with antibodies against apoE. Tubulin was used as a loading control in cell lysates. (A-B) Representative immunoblots showing the changes in secreted and cellular apoE by astrocytes (A) and microglia (B) in response to LPS. (C) ApoE secretion in astrocytes did not change after LPS application. Quantification of western blots. Bar graphs represent the mean ± SEM (n=6-8, 3-4 experiments, run in duplicate). (D) APOE2 and APOE3 microglia release more apoE at 24 hrs. than APOE4 microglia. Quantification of western blots. Bar graphs represent the mean ± SEM (n=4-6, 2-3 experiments, run in duplicate). **p<0.005 vs. untreated control. (E-F) Cellular apoE in astrocytes (E) and microglia (F) following LPS stimulation. Quantification of western blots. Bar graphs represent the mean ± SEM (Astrocytes: n=4, 2 experiments, run in duplicate; Microglia: n=6, 3 experiments, run in duplicate). (C-F) A one-way ANOVA was used to assess outcome measures from pharmacological manipulation, setting vehicles at 100%. (G) APOE mRNA expression was expressed as fold difference from vehicle. Bar graphs represent the mean ± SEM (n=3-6, 3 experiments, run in duplicate). A two-tailed Student T-test was used to assess outcome measures from pharmacological manipulation, setting vehicle treatment as control.
Figure 4. Basal TNFα protein levels are APOE genotype-dependent.
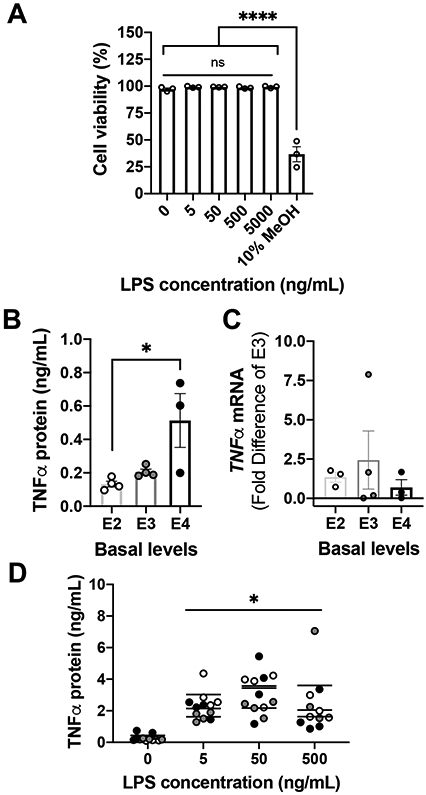
(A) Primary APOE4 astrocytes were exposed to LPS for 24 hrs. Cell loss was determined by Hoechst/PI staining. Data are presented as mean ± SEM, ****p < 0.0001 vs. control (0 LPS ng/mL), one-way ANOVA with Tukey’s post hoc test. Experiments were repeated with three independent cultures of astrocytes. (B-C) Microglia derived from human APOE (E2, E3 and E4) targeted-replacement mice were used. TNFα release was measured by ELISA (B) and TNFα mRNA expression was measured by qRT-PCR (C). (B) Untreated APOE4 microglia released more TNFα than untreated APOE2 microglia. Scatter dot plots represent the mean ± SEM (n=3-4 experiments). *p<0.05 vs. APOE2 microglia. One-way ANOVA with Tukey’s post hoc test. (C) APOE genotype did not affect TNFα mRNA expression levels. Samples were normalized to APOE3. Scatter dot plots represent the mean ± SEM (n=3-4 experiments). (D) TNFα release was measured by ELISA from APOE2, APOE3, and APOE4 microglia after the indicated doses of LPS. *p<0.0001 vs. untreated cultures. A two-way ANOVA was used to assess outcome measures from genotype and pharmacological manipulation in microglia.
LPS increases levels of secreted apoE2 and apoE3, but not apoE4, from microglia.
In contrast to astrocytes, LPS stimulation increased levels of secreted apoE in conditioned media of microglia (Figure 3B). More specifically, both APOE2 and APOE3 microglia had more secreted apoE at high doses of LPS compared to vehicle control: 80% and 230%, respectively [APOE2: (F(3,20)=5.015, p=0.009), APOE3: (F(3,12)=7.620, p=0.004)]. Secreted apoE from APOE4 microglia showed no effects of LPS (Figure 3D). There were no major effects of LPS on cellular levels of apoE in microglia across genotypes, although APOE4 microglia showed a decrease in cellular apoE, but only at low and moderate doses of LPS (F(3,23)=4.877, p=0.009) (Figure 3F). There were no effects of LPS on the relative abundance of the apoE isoforms visible on the western blots. APOE mRNA in microglia under basal and treated conditions were too low for quantification by our qRT-PCR methods (data not shown).
TNFα secretion by microglia after LPS stimulation.
Mixed glial cultures released TNFα 24 hrs. after LPS stimulation (Aleong et al., 2008; Welser-Alves & Milner, 2013), and this TNFα originated from microglia (Welser-Alves & Milner, 2013). Basal levels of TNFα were affected by APOE genotype in microglia, with APOE4 microglia demonstrating five times more TNFα than APOE2 microglia (F(2,8)=4.883, p=0.041) (Figure 4B); however, no differences in TNFα mRNA were detected across APOE genotypes (Figure 4C). LPS significantly increased release of TNFα by microglia in vitro to the same level across APOE genotypes (Figure 4D). On average, APOE2, APOE3 and APOE4 microglia secreted approximately 2.8 ± 0.3 ng/mL of TNFα after LPS, in agreement to levels detected ex vivo (Welser-Alves & Milner, 2013; Zhu et al., 2012).
TNFα affects apoE expression only in APOE4 astrocytes and microglia.
We tested the effects of TNFα on secreted and cellular apoE derived from astrocytes and microglia at 3 and 30 ng/mL of TNFα (Figure 5) (Liddelow & Barres, 2017; Liddelow et al., 2017). Conditioned media and cells were collected 24 hrs. after stimulation with TNFα. In astrocytes, TNFα had no effects on either apoE2 and apoE3 in conditioned media or cell lysates (Figure 5A, 5C, and 5E). In contrast, TNFα reduced the secreted levels of apoE in APOE4 astrocytes (F(2,32)=17.80, p<0.0001). We observed a reduction of apoE secretion of 26% after 3 ng/mL TNFα and 41% after 30 ng/mL TNFα (Figure 5C). The reduction of secreted apoE in APOE4 astrocytes was accompanied with a reduction in cellular apoE by 39% (F(2,22)=6.058, p=0.008) (Figure 5E), with no changes in the relative proportion of each apoE species (p=0.67, data not shown). To determine whether this change in apoE4 was coupled to a decrease in APOE4 transcripts, we performed qRT-PCR in astrocytes treated with 30 ng/mL TNFα compared to vehicle (Figure 5G). Although all APOE genotypes exhibit a similar trend towards a decrease after treatment, the change was not statistically significant (Figure 5G).
Figure 5. Activation of glial cells by TNFα reduced apoE secretion in APOE4 astrocytes, and increased apoE secretion in APOE4 microglia.
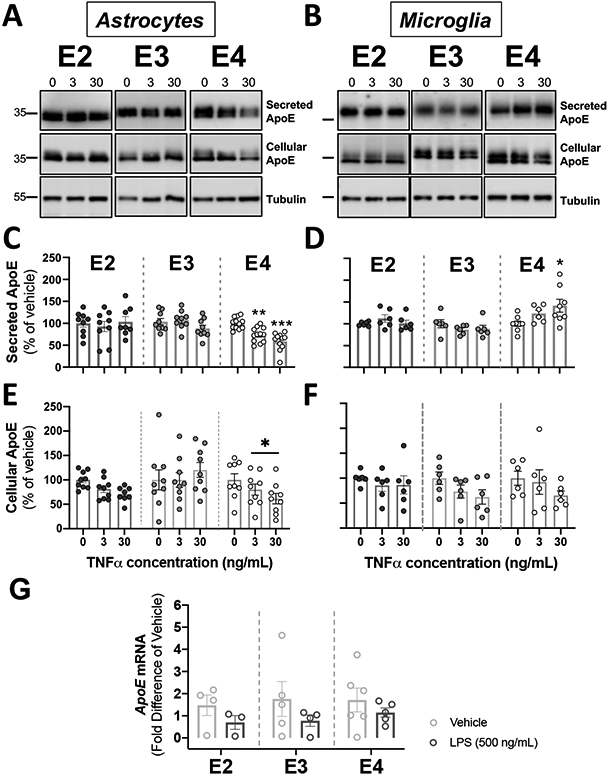
Glial cells derived from human APOE (E2, E3, E4) targeted-replacement mice were treated with 3-30 ng/mL of TNFα. Conditioned media and cells were collected at 24 hrs., and were analyzed by western blotting with antibodies against apoE. Tubulin was used as a loading control in cell lysates. (A-B) Representative immunoblots showing the changes in secreted and cellular apoE by astrocytes (A) and microglia (B) in response to TNFα. (C-D). Quantification of secreted apoE by western blots. (C) TNFα decreased apoE release in APOE4 astrocytes. Bar graphs represent the mean ± SEM (n=9-12, 3-4 experiments, run in triplicate). **p<0.01 ***p<0.005 compared to APOE4 untreated cultures. (D) TNFα increased apoE release in APOE4 microglia. Bar graphs represent the mean ± SEM (n=6-8, 3-4 experiments, run in duplicate). *p<0.05 compared to APOE4 untreated cultures. (E-F). Quantification of cellular apoE by western blots from cell lysates. (E) TNFα decreased cellular ApoE in APOE4 astrocytes. Bar graphs represent the mean ± SEM (n=7-9, 3 experiments, run in triplicate). *p<0.05 compared to APOE4 untreated cultures. (F) TNFα did not change cellular apoE in microglia. Bar graphs represent the mean ± SEM (n=6, 3 experiments, run in duplicate). (C-F) A one-way ANOVA was used to assess outcome measures from pharmacological manipulation, setting vehicles at 100%. (G) TNFα did not change APOE mRNA in astrocytes. APOE mRNA expression was expressed as fold difference from vehicle. Bar graphs represent the mean ± SEM (n=3-6, 3 experiments, run in duplicate). A two-tailed Student T-test was used to assess outcome measures from pharmacological manipulation, setting vehicle treatment as control.
In microglia, TNFα had no effects on apoE presence in conditioned media or cell lysates from APOE2 or APOE3 cells (Figure 5B, 5D, and 5F). APOE4 microglia had higher levels of secreted apoE after 30 ng/mL TNFα (F(2,16)=4.050, p=0.034) (Figure 5D) with no significant effects on cellular apoE (Figure 5F). Regardless of genotype, the relative proportion of the higher molecular mass species over total apoE remain unchanged throughout TNFα stimulation in microglia.
Secreted apoE levels was recovered by IL-10, but not by NF-κβ, ERK, p38 MAPK nor JNK pathways inhibition in APOE4 astrocytes.
Since the main effects of TNFα on cellular and secreted apoE were observed in APOE4 astrocytes, we conducted subsequent experiments with only those cells. We used 30 ng/mL of TNFα because this dose produced a robust effect on apoE secretion in astrocytes. TNFα signals through two transmembrane receptors, TNFR1 and TNFR2, resulting in activation of the NF-κβ, ERK, p38 MAPK, and JNK pathways (Parameswaran & Patial, 2010). To determine which pathway was responsible for the reduction in apoE4 secretion in astrocytes by TNFα, we used a panel of inhibitors (Figure 6): Doramapimod to inhibit the p38 MAPK inhibitor, SP600125 to inhibit JNK kinase, PD98059 to inhibit MEK, IKK-16 to inhibit the IκB kinase inhibitor in the NF-κβ pathway. IL-10 is a known inhibitor of TNFα activation and the NF-κβ pathway (Conti et al., 2003). After 24 hrs. in serum-deprived media, conditioned media was collected and secretion of apoE was analyzed by western blot. There was a main effect of TNFα stimulation reproducing our observed reduction in secreted apoE4 (F(1,82)=57.42, p<0.0001) (Figure 6). However, there was no effect of treatment (F(2,82)=0.6608, p=0.65), or interaction of TNFα stimulation x treatment (F(5,82)=0.5822, p= 0.71) across all inhibitors. Post-hoc analyses showed that the statistically significant reduction of secreted apoE4 levels by TNFα remained in the presence of all inhibitors, but not in the presence of IL-10.
DISCUSSION
The complexity of apoE in the CNS differs from apoE in the periphery, with unique post-translational modifications (Flowers et al., 2019; Kockx et al., 2018; Pitas, Boyles, Lee, Hui, & Weisgraber, 1987; Rebeck et al., 1998). In the CNS, apoE is expressed mainly from astrocytes and microglia/monocytes, but it is also expressed from choroid plexus ependymal cells and from injured neurons (Boyles, Pitas, Wilson, Mahley, & Taylor, 1985; Huang, Weisgraber, Mucke, & Mahley, 2004; Metzger et al., 1996; Polazzi et al., 2015; Q. Xu et al., 2006). We found that much of the apoE from astrocytes and microglia was post-translationally modified due to different levels of O-glycosylation. ApoE in CSF includes isoforms with various O-linked oligosaccharide chains containing zero, one, or two sialic acid residues (Brand, Mackman, & Curtiss, 1993; Flowers et al., 2019; Kockx et al., 2018; Lee et al., 2010). ApoE protein follows a classic secretory pathway, in which synthesis starts in the endoplasmic reticulum, followed by movement through the Golgi network, during which apoE is glycosylated and sialylated (Kockx et al., 2018). The effects of these modifications on apoE function or half-life are unknown.
Some of the complexity of CNS apoE is demonstrated in the forms of apoE secreted from astrocytes and microglia. Astrocytes have only one form of apoE in cells, but two forms of apoE in their conditioned media. Thus, the apoE modified by astrocytes in its synthesis must be immediately secreted. Microglial apoE is very similar to that observed in macrophage (Brand et al., 1993), namely two intracellular species and only one larger extracellular species. ApoE that is synthesized by human macrophage is extensively sialylated and glycosylated on the C-terminus, generating at least seven consistently observed glycoforms of secreted and intracellular apoE (Lee et al., 2010). A two-dimensional gel electrophoresis identified up to six sialylated apoE glycoforms in cells, while fewer glycoforms in plasma (Zannis, 1986; Zannis, vanderSpek, & Silverman, 1986). CSF apoE has abundant glycosylation at the C-terminal, affecting the structure of its lipid binding domain (Flowers & Rebeck, 2020). In mouse and human brains, the modified versions of apoE are soluble in the absence of detergent while the unmodified apoE requires detergent (DiBattista et al., 2016), demonstrating that this modification changes the biochemical properties of apoE.
ApoE secreted by astrocytes consisted of at least two species. We speculated that these forms could reflect differences in O-glycosylation or differences in apoE cleavage. ApoE4 is more susceptible than apoE3 to proteolytic cleavage by cathepsin D, thrombin, chymotrypsin-like serine protease and aspartic proteases in vitro and in vivo (Elliott et al., 2011; Harris et al., 2003; Huang et al., 2001; Marques, Owens, & Crutcher, 2004; Tamboli, Heo, & Rebeck, 2014). This proteolysis occurs in neurons but not in astrocytes (Brecht et al., 2004). We found that apoE secreted by astrocytes and microglia bound Wheat Germ Agglutinin, demonstrating that they are glycoproteins. Deglycosylation simplified the patterns of apoE in SDS-PAGE, demonstraing that O-glycosylation differences can account for the differences in the size of the observed apoE species.
APOE4 genotype was associated with lower levels of secreted apoE from astrocytes and microglia, as well as higher levels of the larger apoE species that remained inside microglia. In neurons, apoE3 and apoE4 have different intracellular trafficking profiles: apoE4 is retained in the endoplasmic reticulum (ER) and Golgi apparatus, and moves more slowly through the secretory pathways (Brodbeck et al., 2011). In astrocytes, apoE4 causes ER stress (Zhong, Ramaswamy, & Weisgraber, 2009). The amino acid difference of apoE4 promotes a misfolding based on an ionic interaction between Arg-61 in the N-terminal domain and Glu-255 in the C-terminal domain (Mahley, 2016; Mahley & Rall, 2000; Wilson et al., 1994; Wilson, Wardell, Weisgraber, Mahley, & Agard, 1991). ApoE4 in primary astrocytes showed enhanced degradation and reduced half-life compared to apoE3 (Riddell et al., 2008). These obervations led to the hypothesis that misfolded proteins accumulate in the ER and golgi apparatus, causing an activation of proteolytic processing pathways and a decrease in apoE abundance. Our observations of the effects of APOE genotype on modified forms of apoE from astrocytes and microglia may reflect a difference in intracellular trafficking between the major sources of CNS apoE (Nuriel et al., 2017; Riddell et al., 2008).
In our studies, APOE2 astrocytes and microglia secreted three times to five times more apoE than APOE4 cells, with APOE3 cells intermediate. This effect is consistent with previous studies showing that apoE levels in APOE2 targeted-replacement mice are higher than in APOE4 mice (Riddell et al., 2008), and that brain lysates from neonatal and old APOE3 mice present significantly higher levels of apoE protein compared to APOE4 mice (Vitek et al., 2009). Since mouse models and cell culture show anti-inflammatory properties of apoE in the CNS (Colton et al., 2002; Laskowitz et al., 2006; Laskowitz et al., 2010; Lynch et al., 2005; Pocivavsek, Burns, & Rebeck, 2009; Vitek et al., 2009), higher secreted basal levels of the protective apoE2 isoform might better regulate inflammatory signals in the brain.
We found that there are higher basal levels of TNFα in APOE4 microglia, supporting the hypothesis that APOE4 predisposes to greater inflammation in primary cells. In the periphery, apoE regulates macrophage inflammation in an isoform-specific manner, such that the APOE4 allele is associated with altered morphology and increased production of cytokines (Jofre-Monseny et al., 2007; H. Zhang, Wu, & Wu, 2011). Similarly, the impact of the apoE protein isoforms on immune cells in the brain can alter disease outcome in an additional manner by affecting the overall immune status of the organism. Recombinant apoE3 or apoE mimetic peptides both reduce LPS- or injury-mediated inflammation (Cheng et al., 2018; Dorey et al., 2017; James, Komisarow, Wang, & Laskowitz, 2020; L. Wang et al., 2019), and the presence of apoE4 is associated with overactive pro-inflammatory immune phenotypes such as elevated NO, TNFα, and IL-6 (Rieker et al., 2019; K. J. Zhang et al., 2011; Zhu et al., 2012). Moreover, expression of astrocytic apoE3 decreased the levels of IL-6, IL-1β, but not TNFα in a mouse model of AD where astrocytic APOE gene is inducible, whereas expression of astrocytic apoE4 increased IL-6, IL-1β and TNFα (C. C. Liu et al., 2017). Importantly, using this inducible mouse model, the authors showed that the expression of apoE4 during the initial seeding stage is sufficient to drive amyloid pathology and plaque-associated neuritic dystrophy, while the presence of apoE4 after the initial seeding stage has minimal impact of amyloid pathology, highlighting the importance of early alterations of apoE in AD (C. C. Liu et al., 2017).
In vivo, LPS injections of APOE3 and APOE4 mice did not change apoE protein expression (Vitek et al., 2009). There are fewer studies about how inflammatory stimuli affect the levels of apoE in glia in vitro (Aleong et al., 2008; Pocivavsek et al., 2009; Pocivavsek & Rebeck, 2009). We found that inflammation affected apoE in a genotype- and glial cell-type specific manner. LPS treatment caused increased levels of secreted apoE in APOE2 and APOE3 (but not APOE4) microglia, while no effects in astrocytes were observed. LPS, through activation of the toll-like receptors (TLR), induces the innate immune response. Astrocytes demonstrate little (Shen et al., 2016) or no (Y. Zhang et al., 2016) TLR4 in vitro. Acutely isolated astrocytes and microglia from murine cortex revealed ten times less expression of TLR4 mRNA in astrocytes compared to microglia, and 100 times more TLR2 mRNA in microglia than in astrocytes (http://www.brainrnaseq.org/). Finally, microglia contain over 40 times more TLR1 than astrocytes (http://www.brainrnaseq.org/) (Y. Zhang et al., 2016). Thus, the lack of the response of astrocytes in terms of apoE expression may be due to a lack or very low expression of TLR species. Inflammation induced by TNFα caused a reduction in levels of secreted and cellular apoE in APOE4 (but not APOE2 and APOE3) astrocytes. These data are consistent with findings in mixed glial cell culture, where there was a reduction of approximately 50% after TNFα application (Aleong et al., 2008). Overall, in our study, astrocytes and microglia responded differently to an exogenous inflammatory signal (LPS) compared to an endogenous inflammatory signal (TNFα).
Traumatic brain injury (TBI) produces neuroinflammation, thus elevating endogenous levels of inflammatory molecules, such as TNFα, IL-1β among others (Villapol, Loane, & Burns, 2017). TBI decreased soluble apoE after 24 hrs. of traumatic injury in both wild-type and human APOE-targeted replacement mice; the reduction was more persistent and pronounced in APOE4 mice than in APOE3 mice (Main et al., 2018), consistent with our findings on the effects of inflammation on apoE. In TBI, the reduction of apoE was correlated to an increased accumulation of Aβ40 (Washington & Burns, 2016). In humans, a 70% reduction in CSF apoE levels was observed following severe TBI (Kay et al., 2003). ApoE suppresses TNFα (Laskowitz, Goel, et al., 1997), and APOE4 mice might have a deficient negative feedback loop between apoE and TNFα resulting in an unregulated inflammatory response. The reduction of apoE 24 hrs. post-injury could be in part due to the elevation of endogenous TNFα.
We did not observe a recovery of apoE levels after inhibiting the activation of the TNFα signaling pathways over 24 hrs. TNFα signals through two transmembrane receptors resulting in activation of the NF-κβ, ERK, p38 MAPK and JNK pathways (Parameswaran & Patial, 2010), but pharmacological inhibition of these pathways did not affect apoE levels. Our results suggest that the reduction of apoE secretion and expression due to TNFα effects could be achieved by multiple inflammatory pathways, or through a less canonical signaling pathway activated by TNFα.
Primary cell cultures are useful tools to understand astrocytic and microglia functions, although there are concerns about how these cultures accurately mirror events in vivo due to the presence of FBS in the media (Foo et al., 2011; Prah et al., 2019; Y. Zhang et al., 2016). FBS contains growth factors, proteins and vitamins that can alter cell biology and phenotype (Aswad, Jalabert, & Rome, 2016), and mimic a more inflammatory profile. Astrocytes grown in serum-free media had a transcriptome more representative of in vivo astrocytes (Foo et al., 2011). In our experiments, cells grown for 24 hrs. in serum-free or serum-containing media showed no differences in APOE mRNA expression (data not shown). However, improving cell culture methods to resemble the environment that occurs in vivo will allow a more accurate understanding of astrocytic and microglia functions in both health and disease.
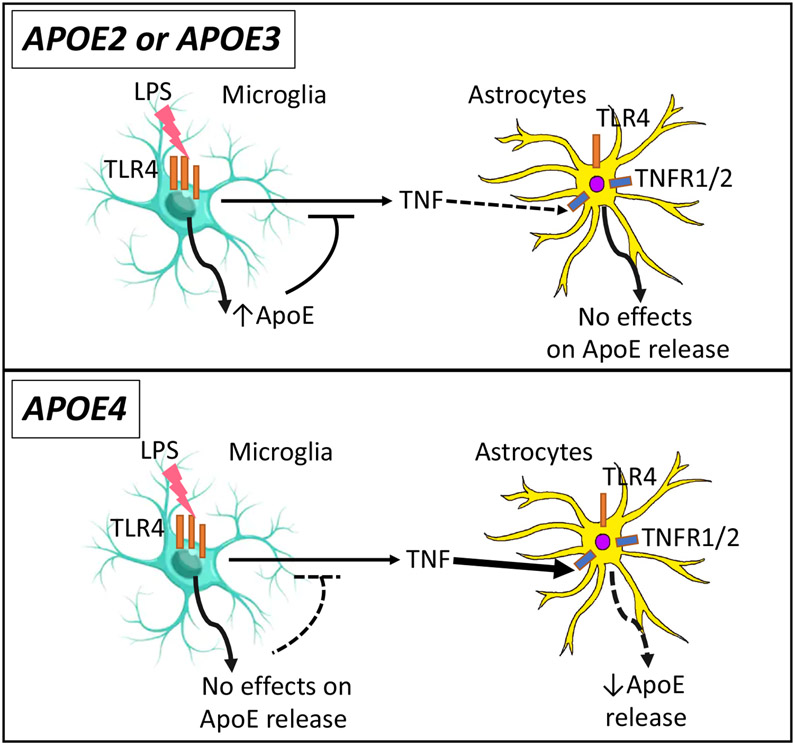
From these various findings, we propose the following model (Figure 7). The lower levels of apoE from APOE4 microglia and astrocytes sensitize an APOE4 brain to inflammatory insults (also evidenced by the higher resting levels of TNFα). ApoE interacts with microglia to regulate their response to inflammatory stimuli. LPS stimulation resulted in release of TNFα by microglia from all APOE genotypes, and increased apoE secretion in APOE2 and APOE3, but not in APOE4 microglia. In astrocytes, apoE2 and apoE3 levels are unchanged by inflammatory stimuli (TNFα), but apoE4 is reduced. The lower levels of apoE4 from both types of glia might not be sufficient to decrease inflammatory processes over time, such as through TNFα signaling. This model is also supported by studies editing APOE3 to APOE4 in astrocytes and microglia from iPSCs, which showed significantly altered expression of genes involved in lipid metabolism in astrocytes and immune responses in microglia-like cells (Lin et al., 2018). These pathways may affect the secretion of apoE and its effects in regulation of inflammation, affecting APOE-related pathogenesis.
Figure 7. Secretion of ApoE under endogenous inflammation.
ApoE acts as an anti-inflammatory molecule. The lower levels of apoE from APOE4 microglia predisposes to APOE4 brain reactivity to inflammatory insults (also evidenced by the higher resting protein levels of TNFα in APOE4 microglia). The higher resting levels of TNFα are associated with lower apoE levels in APOE4 microglia. Induction of inflammation by LPS increases microglial apoE secretion in APOE2 and APOE3, but not in APOE4 microglia, to eventually limit the inflammatory response. The observed elevation of apoE secretion in APOE4 microglia after TNFα might not be sufficient to inhibit TNFα signaling. While apoE2 and apoE3 levels are unchanged by inflammatory stimuli in astrocytes, apoE4 is reduced by TNFα. These opposing events create an imbalance in the inflammatory response of APOE4 microglia and astrocytes.
ACKNOWLEDGEMENTS
This work is supported by NIH R01 NS100714 and R01 AG067258. We would like to thank Hyunwook Nam and Nahdia Jones for technical assistance, and Christi Anne Ng for the illustration in Figure 7.
ABBREVIATIONS
- AD
Alzheimer’s disease
- Aβ
amyloid β
- ApoE
Apolipoprotein E
- APOE
Apolipoprotein E allele
- BCA
Bicinchoninic acid
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- ELISA
Enzyme-linked immunosorbent assay
- ER
Endoplasmic reticulum
- ERK
Extracellular signal-regulated kinase
- FBS
Fetal bovine serum
- FHS
Fetal horse serum
- IL-1β
Interleukin-1b
- IL-10
Interleukin-10
- IL-12
Interleukin-12
- IL-6
Interleukin-6
- IKK
IkappaB kinase
- LPS
Lipopolysaccharide
- MAPK
Mitogen-activated protein kinase
- MEK
Mitogen-activated protein kinase kinase
- NF-κβ
Nuclear factor kappa Beta
- PBS
Phosphate-buffered saline
- PDVF
Polyvinylidene difluoride
- PES
Polyethersulfone
- qRT-PCR
Quantitative real time-polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TBI
Traumatic brain injury
- TBS
Tris-buffered saline
- TLR
Toll-like receptor
- TNFα
Tumor necrosis factor-α
- WGA
Wheat germ agglutinin
Footnotes
CONFLICT OF INTERESTS
M.L., J.S. and G.K. declare that they have no conflict of interests with the content of this article. G.W.R. is a member of the APOE Scientific Advisory Board at Biogen.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Aleong R, Blain JF, & Poirier J (2008). Pro-inflammatory cytokines modulate glial apolipoprotein E secretion. Curr Alzheimer Res, 5(1), 33–37. doi: 10.2174/156720508783884666 [DOI] [PubMed] [Google Scholar]
- Aswad H, Jalabert A, & Rome S (2016). Depleting extracellular vesicles from fetal bovine serum alters proliferation and differentiation of skeletal muscle cells in vitro. BMC Biotechnol, 16, 32. doi: 10.1186/s12896-016-0262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger SW, & Harmon AD (1997). Microglial activation by Alzheimer amyloid precursor protein and modulation by apolipoprotein E. Nature, 388(6645), 878–881. doi: 10.1038/42257 [DOI] [PubMed] [Google Scholar]
- Barroeta-Espar I, Weinstock LD, Perez-Nievas BG, Meltzer AC, Siao Tick Chong M, Amaral AC, … Gomez-Isla T (2019). Distinct cytokine profiles in human brains resilient to Alzheimer's pathology. Neurobiol Dis, 121, 327–337. doi: 10.1016/j.nbd.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Wang C, & Huang Y (2011). C-terminal-truncated apolipoprotein (apo) E4 inefficiently clears amyloid-beta (Abeta) and acts in concert with Abeta to elicit neuronal and behavioral deficits in mice. Proc Natl Acad Sci U S A, 108(10), 4236–4241. doi: 10.1073/pnas.1018381108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles JK, Pitas RE, Wilson E, Mahley RW, & Taylor JM (1985). Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Invest, 76(4), 1501–1513. doi: 10.1172/JCI112130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand K, Mackman N, & Curtiss LK (1993). Interferon-gamma inhibits macrophage apolipoprotein E production by posttranslational mechanisms. J Clin Invest, 91(5), 2031–2039. doi: 10.1172/JCI116425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht WJ, Harris FM, Chang S, Tesseur I, Yu GQ, Xu Q, … Huang Y (2004). Neuron-specific apolipoprotein e4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J Neurosci, 24(10), 2527–2534. doi: 10.1523/JNEUROSCI.4315-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck J, McGuire J, Liu Z, Meyer-Franke A, Balestra ME, Jeong DE, … Huang Y (2011). Structure-dependent impairment of intracellular apolipoprotein E4 trafficking and its detrimental effects are rescued by small-molecule structure correctors. J Biol Chem, 286(19), 17217–17226. doi: 10.1074/jbc.M110.217380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CM, Wright E, Colton CA, Sullivan PM, Laskowitz DT, & Vitek MP (2002). Apolipoprotein E isoform mediated regulation of nitric oxide release. Free Radic Biol Med, 32(11), 1071–1075. doi: 10.1016/s0891-5849(02)00803-1 [DOI] [PubMed] [Google Scholar]
- Cheng X, Zheng Y, Bu P, Qi X, Fan C, Li F, … Cao Q (2018). Apolipoprotein E as a novel therapeutic neuroprotection target after traumatic spinal cord injury. Exp Neurol, 299(Pt A), 97–108. doi: 10.1016/j.expneurol.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernick D, Ortiz-Valle S, Jeong A, Swaminathan SK, Kandimalla KK, Rebeck GW, & Li L (2018). High-density lipoprotein mimetic peptide 4F mitigates amyloid-beta-induced inhibition of apolipoprotein E secretion and lipidation in primary astrocytes and microglia. J Neurochem, 147(5), 647–662. doi: 10.1111/jnc.14554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton CA, Brown CM, Cook D, Needham LK, Xu Q, Czapiga M, … Vitek MP (2002). APOE and the regulation of microglial nitric oxide production: a link between genetic risk and oxidative stress. Neurobiol Aging, 23(5), 777–785. doi: 10.1016/s0197-4580(02)00016-7 [DOI] [PubMed] [Google Scholar]
- Conti P, Kempuraj D, Kandere K, Di Gioacchino M, Barbacane RC, Castellani ML, … Theoharides TC (2003). IL-10, an inflammatory/inhibitory cytokine, but not always. Immunol Lett, 86(2), 123–129. doi: 10.1016/s0165-2478(03)00002-6 [DOI] [PubMed] [Google Scholar]
- Daniele SG, Edwards AA, & Maguire-Zeiss KA (2014). Isolation of cortical microglia with preserved immunophenotype and functionality from murine neonates. J Vis Exp(83), e51005. doi: 10.3791/51005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Battista AM, Heinsinger NM, & Rebeck GW (2016). Alzheimer's Disease Genetic Risk Factor APOE-epsilon4 Also Affects Normal Brain Function. Curr Alzheimer Res, 13(11), 1200–1207. doi: 10.2174/1567205013666160401115127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBattista AM, Dumanis SB, Newman J, & Rebeck GW (2016). Identification and modification of amyloid-independent phenotypes of APOE4 mice. Exp Neurol, 280, 97–105. doi: 10.1016/j.expneurol.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorey E, Bamji-Mirza M, Najem D, Li Y, Liu H, Callaghan D, … Zhang W (2017). Apolipoprotein E Isoforms Differentially Regulate Alzheimer's Disease and Amyloid-beta-Induced Inflammatory Response in vivo and in vitro. J Alzheimers Dis, 57(4), 1265–1279. doi: 10.3233/JAD-160133 [DOI] [PubMed] [Google Scholar]
- Elliott DA, Tsoi K, Holinkova S, Chan SL, Kim WS, Halliday GM, … Garner B (2011). Isoform-specific proteolysis of apolipoprotein-E in the brain. Neurobiol Aging, 32(2), 257–271. doi: 10.1016/j.neurobiolaging.2009.02.006 [DOI] [PubMed] [Google Scholar]
- Elshourbagy NA, Liao WS, Mahley RW, & Taylor JM (1985). Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc Natl Acad Sci U S A, 82(1), 203–207. doi: 10.1073/pnas.82.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CG, Hamby ME, McReynolds ML, & Ray WJ (2019). The Role of APOE4 in Disrupting the Homeostatic Functions of Astrocytes and Microglia in Aging and Alzheimer's Disease. Front Aging Neurosci, 11, 14. doi: 10.3389/fnagi.2019.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers SA, Grant OC, Woods RJ, & Rebeck GW (2019). O-glycosylation on cerebrospinal fluid and plasma apolipoprotein E differs in the lipid-binding domain. Glycobiology, 30(2), 74–85. doi: 10.1093/glycob/cwz084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers SA, & Rebeck GW (2020). APOE in the normal brain. Neurobiol Dis, 136, 104724. doi: 10.1016/j.nbd.2019.104724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC, Allen NJ, Bushong EA, Ventura PB, Chung WS, Zhou L, … Barres BA (2011). Development of a method for the purification and culture of rodent astrocytes. Neuron, 71(5), 799–811. doi: 10.1016/j.neuron.2011.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, … Huang Y (2003). Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer's disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc Natl Acad Sci U S A, 100(19), 10966–10971. doi: 10.1073/pnas.1434398100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner FL, Ransohoff RM, & Becher B (2015). Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci, 16(6), 358–372. doi: 10.1038/nrn3880 [DOI] [PubMed] [Google Scholar]
- Hu Y, Meuret C, Go S, Yassine HN, & Nedelkov D (2020). Simple and Fast Assay for Apolipoprotein E Phenotyping and Glycotyping: Discovering Isoform-Specific Glycosylation in Plasma and Cerebrospinal Fluid. J Alzheimers Dis. doi: 10.3233/JAD-200203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Liu XQ, Wyss-Coray T, Brecht WJ, Sanan DA, & Mahley RW (2001). Apolipoprotein E fragments present in Alzheimer's disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc Natl Acad Sci U S A, 98(15), 8838–8843. doi: 10.1073/pnas.151254698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Weisgraber KH, Mucke L, & Mahley RW (2004). Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer's disease. J Mol Neurosci, 23(3), 189–204. doi: 10.1385/JMN:23:3:189 [DOI] [PubMed] [Google Scholar]
- Hyman BT, Gomez-Isla T, West H, Briggs M, Chung H, Growdon JH, & Rebeck GW (1996). Clinical and neuropathological correlates of apolipoprotein E genotype in Alzheimer's disease. Window on molecular epidemiology. Ann N Y Acad Sci, 777, 158–165. doi: 10.1111/j.1749-6632.1996.tb34414.x [DOI] [PubMed] [Google Scholar]
- James ML, Komisarow JM, Wang H, & Laskowitz DT (2020). Therapeutic Development of Apolipoprotein E Mimetics for Acute Brain Injury: Augmenting Endogenous Responses to Reduce Secondary Injury. Neurotherapeutics. doi: 10.1007/s13311-020-00858-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofre-Monseny L, Loboda A, Wagner AE, Huebbe P, Boesch-Saadatmandi C, Jozkowicz A, … Rimbach G (2007). Effects of apoE genotype on macrophage inflammation and heme oxygenase-1 expression. Biochem Biophys Res Commun, 357(1), 319–324. doi: 10.1016/j.bbrc.2007.03.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay AD, Petzold A, Kerr M, Keir G, Thompson E, & Nicoll JA (2003). Alterations in cerebrospinal fluid apolipoprotein E and amyloid beta-protein after traumatic brain injury. J Neurotrauma, 20(10), 943–952. doi: 10.1089/089771503770195795 [DOI] [PubMed] [Google Scholar]
- Kockx M, Traini M, & Kritharides L (2018). Cell-specific production, secretion, and function of apolipoprotein E. J Mol Med (Berl; ), 96(5), 361–371. doi: 10.1007/s00109-018-1632-y [DOI] [PubMed] [Google Scholar]
- LaDu MJ, Shah JA, Reardon CA, Getz GS, Bu G, Hu J, … Van Eldik LJ (2001). Apolipoprotein E and apolipoprotein E receptors modulate A beta-induced glial neuroinflammatory responses. Neurochem Int, 39(5-6), 427–434. doi: 10.1016/s0197-0186(01)00050-x [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Fillit H, Yeung N, Toku K, & Vitek MP (2006). Apolipoprotein E-derived peptides reduce CNS inflammation: implications for therapy of neurological disease. Acta Neurol Scand Suppl, 185, 15–20. doi: 10.1111/j.1600-0404.2006.00680.x [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Goel S, Bennett ER, & Matthew WD (1997). Apolipoprotein E suppresses glial cell secretion of TNF alpha. J Neuroimmunol, 76(1-2), 70–74. doi: 10.1016/s0165-5728(97)00021-0 [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Horsburgh K, & Roses AD (1998). Apolipoprotein E and the CNS response to injury. J Cereb Blood Flow Metab, 18(5), 465–471. doi: 10.1097/00004647-199805000-00001 [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Lee DM, Schmechel D, & Staats HF (2000). Altered immune responses in apolipoprotein E-deficient mice. J Lipid Res, 41(4), 613–620. [PubMed] [Google Scholar]
- Laskowitz DT, Matthew WD, Bennett ER, Schmechel D, Herbstreith MH, Goel S, & McMillian MK (1998). Endogenous apolipoprotein E suppresses LPS-stimulated microglial nitric oxide production. Neuroreport, 9(4), 615–618. doi: 10.1097/00001756-199803090-00010 [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Sheng H, Bart RD, Joyner KA, Roses AD, & Warner DS (1997). Apolipoprotein E-deficient mice have increased susceptibility to focal cerebral ischemia. J Cereb Blood Flow Metab, 17(7), 753–758. doi: 10.1097/00004647-199707000-00005 [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Song P, Wang H, Mace B, Sullivan PM, Vitek MP, & Dawson HN (2010). Traumatic brain injury exacerbates neurodegenerative pathology: improvement with an apolipoprotein E-based therapeutic. J Neurotrauma, 27(11), 1983–1995. doi: 10.1089/neu.2010.1396 [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Thekdi AD, Thekdi SD, Han SK, Myers JK, Pizzo SV, & Bennett ER (2001). Downregulation of microglial activation by apolipoprotein E and apoE-mimetic peptides. Exp Neurol, 167(1), 74–85. doi: 10.1006/exnr.2001.7541 [DOI] [PubMed] [Google Scholar]
- Laskowitz DT, Wang H, Chen T, Lubkin DT, Cantillana V, Tu TM, … Dawson, H. N. (2017). Neuroprotective pentapeptide CN-105 is associated with reduced sterile inflammation and improved functional outcomes in a traumatic brain injury murine model. Sci Rep, 7, 46461. doi: 10.1038/srep46461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kockx M, Raftery MJ, Jessup W, Griffith R, & Kritharides L (2010). Glycosylation and sialylation of macrophage-derived human apolipoprotein E analyzed by SDS-PAGE and mass spectrometry: evidence for a novel site of glycosylation on Ser290. Mol Cell Proteomics, 9(9), 1968–1981. doi: 10.1074/mcp.M900430-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, & Barres BA (2017). Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity, 46(6), 957–967. doi: 10.1016/j.immuni.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, … Barres BA (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature, 541(7638), 481–487. doi: 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, … Tsai LH (2018). APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer's Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron, 98(6), 1141–1154 e1147. doi: 10.1016/j.neuron.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Zhao N, Fu Y, Wang N, Linares C, Tsai CW, & Bu G (2017). ApoE4 Accelerates Early Seeding of Amyloid Pathology. Neuron, 96(5), 1024–1032 e1023. doi: 10.1016/j.neuron.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhou G, Kolls BJ, Tan Y, Fang C, Wang H, & Laskowitz DT (2018). Apolipoprotein E mimetic peptide CN-105 improves outcome in a murine model of SAH. Stroke Vasc Neurol, 3(4), 222–230. doi: 10.1136/svn-2018-000152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu X, Dou H, Hua Y, Xu J, & Hui X (2015). Apolipoprotein E knockout induced inflammatory responses related to microglia in neonatal mice brain via astrocytes. Int J Clin Exp Med, 8(1), 737–743. [PMC free article] [PubMed] [Google Scholar]
- Lynch JR, Wang H, Mace B, Leinenweber S, Warner DS, Bennett ER, … Laskowitz DT (2005). A novel therapeutic derived from apolipoprotein E reduces brain inflammation and improves outcome after closed head injury. Exp Neurol, 192(1), 109–116. doi: 10.1016/j.expneurol.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Mahley RW (2016). Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J Mol Med (Berl), 94(7), 739–746. doi: 10.1007/s00109-016-1427-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, & Rall SC Jr. (2000). Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet, 1, 507–537. doi: 10.1146/annurev.genom.1.1.507 [DOI] [PubMed] [Google Scholar]
- Main BS, Villapol S, Sloley SS, Barton DJ, Parsadanian M, Agbaegbu C, … Burns MP (2018). Apolipoprotein E4 impairs spontaneous blood brain barrier repair following traumatic brain injury. Mol Neurodegener, 13(1), 17. doi: 10.1186/s13024-018-0249-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manelli AM, Stine WB, Van Eldik LJ, & LaDu MJ (2004). ApoE and Abeta1-42 interactions: effects of isoform and conformation on structure and function. J Mol Neurosci, 23(3), 235–246. doi: 10.1385/JMN:23:3:235 [DOI] [PubMed] [Google Scholar]
- Marques MA, Owens PA, & Crutcher KA (2004). Progress toward identification of protease activity involved in proteolysis of apolipoprotein e in human brain. J Mol Neurosci, 24(1), 73–80. doi: 10.1385/JMN:24:1:073 [DOI] [PubMed] [Google Scholar]
- Metzger RE, LaDu MJ, Pan JB, Getz GS, Frail DE, & Falduto MT (1996). Neurons of the human frontal cortex display apolipoprotein E immunoreactivity: implications for Alzheimer's disease. J Neuropathol Exp Neurol, 55(3), 372–380. doi: 10.1097/00005072-199603000-00013 [DOI] [PubMed] [Google Scholar]
- Newcombe EA, Camats-Perna J, Silva ML, Valmas N, Huat TJ, & Medeiros R (2018). Inflammation: the link between comorbidities, genetics, and Alzheimer's disease. J Neuroinflammation, 15(1), 276. doi: 10.1186/s12974-018-1313-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuriel T, Peng KY, Ashok A, Dillman AA, Figueroa HY, Apuzzo J, … Duff, K. E. (2017). The Endosomal-Lysosomal Pathway Is Dysregulated by APOE4 Expression in Vivo. Front Neurosci, 11, 702. doi: 10.3389/fnins.2017.00702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran N, & Patial S (2010). Tumor necrosis factor-alpha signaling in macrophages. Crit Rev Eukaryot Gene Expr, 20(2), 87–103. doi: 10.1615/critreveukargeneexpr.v20.i2.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitas RE, Boyles JK, Lee SH, Hui D, & Weisgraber KH (1987). Lipoproteins and their receptors in the central nervous system. Characterization of the lipoproteins in cerebrospinal fluid and identification of apolipoprotein B,E(LDL) receptors in the brain. J Biol Chem, 262(29), 14352–14360. [PubMed] [Google Scholar]
- Pocivavsek A, Burns MP, & Rebeck GW (2009). Low-density lipoprotein receptors regulate microglial inflammation through c-Jun N-terminal kinase. Glia, 57(4), 444–453. doi: 10.1002/glia.20772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocivavsek A, & Rebeck GW (2009). Inhibition of c-Jun N-terminal kinase increases apoE expression in vitro and in vivo. Biochem Biophys Res Commun, 387(3), 516–520. doi: 10.1016/j.bbrc.2009.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polazzi E, Mengoni I, Pena-Altamira E, Massenzio F, Virgili M, Petralla S, & Monti B (2015). Neuronal Regulation of Neuroprotective Microglial Apolipoprotein E Secretion in Rat In Vitro Models of Brain Pathophysiology. J Neuropathol Exp Neurol, 74(8), 818–834. doi: 10.1097/NEN.0000000000000222 [DOI] [PubMed] [Google Scholar]
- Prah J, Winters A, Chaudhari K, Hersh J, Liu R, & Yang SH (2019). A novel serum free primary astrocyte culture method that mimic quiescent astrocyte phenotype. J Neurosci Methods, 320, 50–63. doi: 10.1016/j.jneumeth.2019.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeck GW, Alonzo NC, Berezovska O, Harr SD, Knowles RB, Growdon JH, … Mendez, A. J. (1998). Structure and functions of human cerebrospinal fluid lipoproteins from individuals of different APOE genotypes. Exp Neurol, 149(1), 175–182. doi: 10.1006/exnr.1997.6710 [DOI] [PubMed] [Google Scholar]
- Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, … Reinhart, P. H. (2008). Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci, 28(45), 11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieker C, Migliavacca E, Vaucher A, Baud G, Marquis J, Charpagne A, … Pooler, A. M. (2019). Apolipoprotein E4 Expression Causes Gain of Toxic Function in Isogenic Human Induced Pluripotent Stem Cell-Derived Endothelial Cells. Arterioscler Thromb Vasc Biol, 39(9), e195–e207. doi: 10.1161/ATVBAHA.118.312261 [DOI] [PubMed] [Google Scholar]
- Shen Y, Qin H, Chen J, Mou L, He Y, Yan Y, … Zhou YD (2016). Postnatal activation of TLR4 in astrocytes promotes excitatory synaptogenesis in hippocampal neurons. J Cell Biol, 215(5), 719–734. doi: 10.1083/jcb.201605046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, … Maeda N (1997). Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem, 272(29), 17972–17980. doi: 10.1074/jbc.272.29.17972 [DOI] [PubMed] [Google Scholar]
- Tamboli IY, Heo D, & Rebeck GW (2014). Extracellular proteolysis of apolipoprotein E (apoE) by secreted serine neuronal protease. PLoS One, 9(3), e93120. doi: 10.1371/journal.pone.0093120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu TM, Kolls BJ, Soderblom EJ, Cantillana V, Ferrell PD, Moseley MA, … Laskowitz DT (2017). Apolipoprotein E mimetic peptide, CN-105, improves outcomes in ischemic stroke. Ann Clin Transl Neurol, 4(4), 246–265. doi: 10.1002/acn3.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villapol S, Loane DJ, & Burns MP (2017). Sexual dimorphism in the inflammatory response to traumatic brain injury. Glia, 65(9), 1423–1438. doi: 10.1002/glia.23171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek MP, Brown CM, & Colton CA (2009). APOE genotype-specific differences in the innate immune response. Neurobiol Aging, 30(9), 1350–1360. doi: 10.1016/j.neurobiolaging.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Christensen DJ, Vitek MP, Sullivan PM, & Laskowitz DT (2009). APOE genotype affects outcome in a murine model of sepsis: implications for a new treatment strategy. Anaesth Intensive Care, 37(1), 38–45. doi: 10.1177/0310057X0903700111 [DOI] [PubMed] [Google Scholar]
- Wang L, Hou H, Zi D, Habib A, Tan J, & Sawmiller D (2019). Novel apoE receptor mimetics reduce LPS-induced microglial inflammation. Am J Transl Res, 11(8), 5076–5085. [PMC free article] [PubMed] [Google Scholar]
- Washington PM, & Burns MP (2016). The Effect of the APOE4 Gene on Accumulation of Abeta40 After Brain Injury Cannot Be Reversed by Increasing apoE4 Protein. J Neuropathol Exp Neurol, 75(8), 770–778. doi: 10.1093/jnen/nlw049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Zheng M, Liang P, Wei Y, Yin X, Tang Y, & Xue Y (2013). Apolipoprotein E and its mimetic peptide suppress Th1 and Th17 responses in experimental autoimmune encephalomyelitis. Neurobiol Dis, 56, 59–63. doi: 10.1016/j.nbd.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Welser-Alves JV, & Milner R (2013). Microglia are the major source of TNF-alpha and TGF-beta1 in postnatal glial cultures; regulation by cytokines, lipopolysaccharide, and vitronectin. Neurochem Int, 65(1), 47–53. doi: 10.1016/j.neuint.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Mau T, Weisgraber KH, Wardell MR, Mahley RW, & Agard DA (1994). Salt bridge relay triggers defective LDL receptor binding by a mutant apolipoprotein. Structure, 2(8), 713–718. doi: 10.1016/s0969-2126(00)00072-1 [DOI] [PubMed] [Google Scholar]
- Wilson C, Wardell MR, Weisgraber KH, Mahley RW, & Agard DA (1991). Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science, 252(5014), 1817–1822. doi: 10.1126/science.2063194 [DOI] [PubMed] [Google Scholar]
- Wu VW, & Schwartz JP (1998). Cell culture models for reactive gliosis: new perspectives. J Neurosci Res, 51(6), 675–681. doi: [DOI] [PubMed] [Google Scholar]
- Xu PT, Schmechel D, Rothrock-Christian T, Burkhart DS, Qiu HL, Popko B, … Gilbert JR (1996). Human apolipoprotein E2, E3, and E4 isoform-specific transgenic mice: human-like pattern of glial and neuronal immunoreactivity in central nervous system not observed in wild-type mice. Neurobiol Dis, 3(3), 229–245. doi: 10.1006/nbdi.1996.0023 [DOI] [PubMed] [Google Scholar]
- Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, & Huang Y (2006). Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci, 26(19), 4985–4994. doi: 10.1523/JNEUROSCI.5476-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, & Barres BA (2012). Genomic analysis of reactive astrogliosis. J Neurosci, 32(18), 6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannis VI (1986). Genetic polymorphism in human apolipoprotein E. Methods Enzymol, 128, 823–851. doi: 10.1016/0076-6879(86)28109-4 [DOI] [PubMed] [Google Scholar]
- Zannis VI, vanderSpek J, & Silverman D (1986). Intracellular modifications of human apolipoprotein E. J Biol Chem, 261(29), 13415–13421. [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, … Emilsson V (2013). Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell, 153(3), 707–720. doi: 10.1016/j.cell.2013.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wu LM, & Wu J (2011). Cross-talk between apolipoprotein E and cytokines. Mediators Inflamm, 2011, 949072. doi: 10.1155/2011/949072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KJ, Zhang HL, Zhang XM, Zheng XY, Quezada HC, Zhang D, & Zhu J (2011). Apolipoprotein E isoform-specific effects on cytokine and nitric oxide production from mouse Schwann cells after inflammatory stimulation. Neurosci Lett, 499(3), 175–180. doi: 10.1016/j.neulet.2011.05.050 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, … Barres, B. A. (2016). Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron, 89(1), 37–53. doi: 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N, Ramaswamy G, & Weisgraber KH (2009). Apolipoprotein E4 domain interaction induces endoplasmic reticulum stress and impairs astrocyte function. J Biol Chem, 284(40), 27273–27280. doi: 10.1074/jbc.M109.014464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Nwabuisi-Heath E, Dumanis SB, Tai LM, Yu C, Rebeck GW, & LaDu MJ (2012). APOE genotype alters glial activation and loss of synaptic markers in mice. Glia, 60(4), 559–569. doi: 10.1002/glia.22289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.