Abstract
Cell fusion induced by human immunodeficiency virus type 1 (HIV-1) is usually assessed by counting multinucleated giant cells (syncytia) visualized by light microscopy. Currently used methods do not allow quantification of syncytia, nor do they estimate the number of cells involved in cell fusion. We describe two fluorescence-based methods for the detection and quantification of HIV-1-induced in vitro syncytium formation. The lymphoblastoid cell lines MT-2 and SupT1 were infected with syncytium-inducing (SI) HIV-1 isolates. Syncytia were detected by DNA staining with propidium iodide using flow cytometry to determine cell size or by two-color cytoplasmic staining of infected cell populations by using fluorescence microscopy. Both methods were able to detect and quantify HIV-induced syncytia. The methods could distinguish between SI and non-SI HIV isolates and could be used with at least two separate types of CD4+ T-cell lines. Small syncytia can be readily identified by the two-color cytoplasmic staining method. Both methods were also shown to be useful for evaluating antiretroviral compounds, as demonstrated by the accurate assessment of HIV inhibition by azidothymidine (zidovudine), dideoxycytidine (zalcytibine), and hydroxyurea. These fluorescence-based assays allow a rapid and practical method for measuring HIV replication and anti-HIV activity of potential inhibitory compounds.
The rate of human immunodeficiency virus type 1 (HIV-1) disease progression shows considerable variation between patients (23). The underlying pathogenic mechanisms that determine the progression rate of HIV infection in vivo remain largely unknown. HIV-1 isolates from the peripheral blood of infected individuals have been shown to differ in biological properties such as replication rate, cell tropism, and syncytium-inducing (SI) capacity (26, 32). Early in the course of infection, viral isolates grown in vitro are predominantly slowly replicating, macrophage tropic, and non-SI (NSI or C5 variants) (1, 8). Over time, an increasing percentage of HIV-infected individuals harbor rapid replicating, predominantly T-cell-tropic viruses (X4 variants), and these virus isolates induce syncytia in T-cell lines (12–15). Among HIV-infected individuals with fewer than 50 CD4+ T cells/mm3, about half harbor the SI (X4) phenotype in vitro (20). The appearance of the SI phenotype in infected people is correlated with an accelerated rate of decline in CD4+ T cells and a more rapid progression to AIDS (3, 7, 11, 19, 21, 30, 31), suggesting that SI variants might be causally involved in CD4+ T-cell depletion.
Standard methods to determine the SI phenotype in vitro are performed by adding plasma or peripheral blood mononuclear cells (PBMCs) from infected patients to highly permissive CD4+ T-cell lines, followed by light microscopic visualization of large “balloon” cells (16, 19). Problems associated with this method include the fact that the visual interpretation of syncytia is subjective and there is considerable variability in the size of syncytia, depending upon the HIV-1 isolate tested, the multiplicity of infection (MOI), and the T-cell line used. Furthermore, this method does not determine the number of cell equivalents fused in a single syncytium, and therefore underestimates the extent of syncytia formation (4, 5, 25, 29). Consequently, use of syncytia formation as an end-point marker of infection for the evaluation of antiviral drugs or antibodies is not quantitative. Because of this, the study of HIV-1 inhibitors generally requires the determination of either culture supernatant p24 antigen or RT (reverse transcriptase) activity (9, 18, 22). p24 antigen testing is expensive, and measurement of RT activity generally requires concentration of virus from cell culture supernatants.
The use of flow cytometry (measured by a fluorescence-activated cell sorter [FACS]) to quantify HIV-induced syncytia has previously been described; however, published methods did not measure syncytia populations directly, but estimated fusion indirectly by measuring the disappearance of cocultured cells (27, 28, 31). To determine whether a direct method of quantification of HIV-induced syncytia using a FACS was feasible, we evaluated propidium iodide (PI) DNA staining and size measurements with a FACS in order to detect and quantify syncytia. Furthermore, we developed a color fusion assay for the assessment of syncytia formation between differentially stained HIV-infected MT-2 cells. The potential application of these assays was demonstrated by using known anti-HIV drugs, including 3′-azido-3′-deoxythimidine (zidovudine [ZDV]), 2′-3′-dideoxycytidine (zalcytibine [ddC]), anti-CD4 monoclonal antibody, and the ribonucleotide reductase inhibitor hydroxyurea (HU).
MATERIAL AND METHODS
HIV viral isolates.
HIV isolates used in these studies were obtained from the AIDS Research and Reference Reagent Program. A clinical HIV-1 isolate (catalog no. 1073,) and human T-cell leukemia virus type IIIb (HTLV-IIIb) (catalog no. 398) were used in these studies. Both isolates are genotype B and SI (X4) phenotype. An NSI HIV isolate (92UG031) of genotype A served as a control virus in some experiments (catalog no. 1741). HIV-1 stocks were derived from tissue culture supernatant fluids, with the infectious titer determined by end-point dilution in MT-2 cells (for the clinical isolate and HTLV-IIIb) or PBMCs (for the NSI isolate) as previously described (10). HIV-1 p24 antigen concentration was determined by using a commercial immunoassay (Organon-Teknika Corp., Durham, N.C.) (10).
Cell culture.
The lymphoblastoid cell lines MT-2 and SupT1 used in these studies were obtained from the AIDS Research and Reference Reagent Program (catalog no. 237 and 100, respectively). MT-2 and SupT1 were grown in RPMI 1640 medium supplemented with 2 mM l-glutamine, 10% fetal calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin sulfate per ml. The titer of the HIV clinical isolate was determined in MT-2 cells, and the titer of HTLV-IIIb was determined in SupT1 cells. For experimental purposes, MT-2 cells were pelleted and 106 cells were infected with HIV-1 (MOI = 1). Parallel mock infections were performed by using supernatant of uninfected MT-2 cells in place of the virus pool. Cells were incubated for 2 h, and supplemented RPMI 1640 medium was added. Cultures were maintained for up to 72 h at 37°C at 5% CO2 atmosphere. The NSI isolate was propagated in PBMC cultures as previously described (10) except where noted. For experiments measuring p24 antigen production in the culture supernatant, cells were washed 16 h postinfection. Antiviral compounds HU, AZT, and ddC were obtained from Sigma Chemical Co. (St. Louis, Mo.).
DNA staining.
Cells were washed in phosphate-buffered saline (PBS) and were resuspended in 100 μl of PBS followed by the addition of 1 ml of freshly prepared 3.7% paraformaldehyde solution. Cells were fixed for 15 min at room temperature followed by one wash with PBS and one wash in PBST (0.02% Tween 20, 0.5% bovine serum albumin in PBS). Fixed cells were incubated with 0.5 ml of RNase A in PBST (1 mg/ml) and were washed prior to staining with PI solution (50 μg/ml in PBST).
Color fusion assay.
A color fluorescence cell-labeling system, which was previously used to detect electrofusion products by Jaroszeski et al. (17), was used in these studies. Two vital cytoplasmic dyes (Molecular Probes, Eugene, Oreg.) were used to directly measure and quantitate HIV-induced syncytia. MT-2 cells (106) were pelleted, and half were stained with 3 μM 5- (and 6)-{[(4-chloromethyl)benzoyl]amino}tetramethylrhodamine (CMTMR) dye (red) and the other half with 5-chloromethylfluorescein diacetate (CMFDA) dye (green) for 30 min at 37°C in 5% CO2. Cells were washed and incubated for 45 min in fresh media prior to infection with HIV-1. Two hours postinfection, the red-stained cell population was mixed with the green-stained cells and incubated in a total volume of 5 ml in supplemented RPMI 1640 medium for 48 h. Cells were fixed in 3.7% paraformaldehyde and analyzed by confocal microscopy for double staining, indicating HIV-induced fusion. Quantitation was accomplished by measuring the number and size of dually stained yellow fusion products per field and by calculating cell volume equivalents. Briefly, the mean diameter of uninfected cells was determined, and this value was used to represent a single-cell diameter. The diameter of HIV-1-infected yellow fusion products was also measured, and this value was used to calculate the number of cell-equivalents present in syncytia based on the relative increase in diameter compared to a single cell.
Flow cytometry.
Flow cytometric analysis was performed by using a fluorescence-activated cell scanner (Becton-Dickinson FACScan). The PI emission signals, using the standard fluorescein isothiocyanate-PI filter set, as well as forward light scatter and side light scatter signals, were determined. For quantitative analysis, data were collected within a set time frame of 100 s for each sample. Analysis was performed using the WinMDI software (Windows Multiple Document Interface, Flow Cytometry Application).
Blocking of HIV attachment with anti-CD4 monoclonal antibody.
MT-2 cells were pelleted and incubated with 10 μg of anti-CD4 monoclonal antibody (Ancell, Bayport, Minn.) per ml for 45 min at 4°C. Cells incubated with a nonreactive murine isotype-specific antibody at equal concentration served as the negative control. Cells were washed once in media and were infected with HIV-1 (MOI = 1) for 2 h at 4°C. Supplemented RPMI 1640 medium was added, and cells were incubated for 72 h at 37°C in a humidified 5% CO2 atmosphere. Cells were fixed, stained for DNA, and analyzed by flow cytometry.
Inhibition of viral replication with antiviral drugs.
MT-2 cells were pelleted and infected with HIV-1 (MOI = 1) for 2 h at 37°C. Supplemented RPMI 1640 medium containing either AZT (0.5 μM), ddC (20 μM), or HU (25 to 400 μM) was added to a final volume of 5 ml. Cells were incubated for 48 to 72 h at 37°C in a humidified 5% CO2 atmosphere.
RESULTS
Detection and quantitation of HIV-infected MT-2 cells by flow cytometry.
To determine the utility of flow cytometry for detecting HIV syncytia, mock-infected and HIV-infected MT-2 cells were fixed 72 h postinfection, stained with PI, and analyzed in a set time frame of 100 s by flow cytometry. The results are shown in Fig. 1A to D. Analysis of the forward scatter versus the DNA content identified two well-defined cell populations (R1 and R2) in uninfected control cultures (Fig. 1A), representing cells in different stages of the cell cycle (Fig. 1B) (2). The majority of cells (52%) were in the G1 phase of the cell cycle, whereas cells of the R2 region (43%) represent the S, G2, or M phases of the cell cycle. Analysis of HIV-infected MT-2 cells (Fig. 1C) revealed a more heterogenous population containing cells with a broad spectrum of size and DNA content (Fig. 1D). The percentage of cells in the G1 phase was significantly reduced (27%).
FIG. 1.
Detection and quantitation of HIV-infected MT-2 cells by flow cytometry. HIV-infected MT-2 cells were fixed 72 h postinfection, stained with PI, and analyzed in a set time frame of 100 s by flow cytometry. The analysis of the forward scatter (FSC) versus the DNA content (PI-H) in mock-infected (A) and HIV-infected (C) cultures was compared to the DNA histograms (B and D).
The total number of cells counted in 100 s was 48,930 events for the mock-infected control versus 17,670 events for HIV-infected cells. Despite a reduced number of cells per volume unit, HIV-infected MT-2 cells contained subpopulations of increased size and cells with a DNA content greater than 2 N (R3), representing multinucleated, fused cells (syncytia). The number of cell equivalents present in these multinucleated cells (R3) was estimated by subtracting the number of cells of HIV R1 and R2 from the total number of cells counted in mock-infected R1 and R2 as follows: (mock-infected R1 + R2 − HIV-infected R1 + R2 = HIV R3.
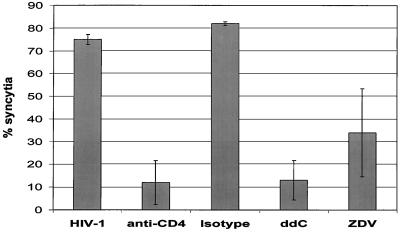
The increase in syncytia formation determined by FACS correlated with an increase of HIV-1 p24 antigen in the culture supernatant (Fig. 2), indicating that the syncytia formation measured by the FACS fusion method was directly related to virus replication. Syncytia formation was inhibited by blocking viral entry with an anti-CD4 monoclonal antibody, but not with a nonreactive isotype control antibody (Fig. 3). The antiviral drugs ZDV and ddC were evaluated to determine if the FACS assays could be used to test new anti-HIV drugs. In the flow cytometric assay, the nucleoside analogs inhibited syncytium formation by 40 and 60%, respectively (Fig. 3). To determine if this FACS method is applicable for the detection and quantitation of syncytia formation by using different cell lines and virus strains, we infected SupT1 cells with the HIV clinical isolate, HTLV-IIIb, and an NSI control isolate. SupT1 cells infected with HTLV-IIIb or the clinical isolate demonstrated 46 and 38% syncytia, respectively, whereas the NSI isolate did not induce any syncytia in the SupT1 cells (data not shown). The FACS method was able to detect syncytia induction in both MT-2 and SupT1 cells and using different HIV-1 isolates.
FIG. 2.
Syncytia formation correlates with viral replication. Syncytia formation was evaluated by FACS, and HIV-1 replication was determined by measuring p24 antigen concentration in cell culture supernatants.
FIG. 3.
Inhibition of syncytia formation by anti-CD4 antibody and antiretroviral drugs. MT-2 cells infected with HIV-1 demonstrated 74% HIV-1 syncytia in this experiment. Prior to infection, CD4 receptors on MT-2 cells were blocked with anti-CD4 antibodies or nonspecific antibodies of the same isotype (anti-CD4 and Isotype, respectively). HIV-1-infected MT-2 cells were also cultured in the presence or absence of the antiviral compounds ddC (20 μM) or ZDV (0.5 μM).
Syncytia formation is the major cause of cell loss in HIV-1-infected MT-2 cell cultures.
Although lysis of syncytia occurs in HIV-1 infection in vitro, significant lysis does not appear to occur prior to 72 h postinfection (29). To demonstrate that the reduced cell number measured in HIV-infected cultures is predominantly due to formation of multinucleated giant cells and not to the cell lysis frequently observed in HIV-1-infected cells, we developed a two-color fusion assay, allowing microscopic detection of double-stained fusion cells. HIV-infected MT-2 cells were stained with a vital red or green fluorescent dye. Equal numbers of red- and green-stained cells were mixed and incubated for 48 h. HIV-1-induced fusion between red fluorescent (Fig. 4E) and green fluorescent (Fig. 4F) cells resulted in yellow-stained syncytia, indicating colocalization of both dyes (Fig. 4G), that could be detected by fluorescence microscopy. Mock-infected cultures contained only red (Fig. 4A) or green (Fig. 4B) fluorescent cells and did not contain dually stained yellow fusion products (Fig. 4C). When uninfected MT-2 cells stained green were mixed with HIV-infected cells stained red, dually stained fusion products were also identified in the HIV-infected cells but not in the mock-infected controls (data not shown).
FIG. 4.
Mock-infected (panels A to D) or HIV-infected (panels E to H) MT-2 cells were stained with a vital red or green fluorescent dye. Equal numbers of red- and green-stained cells were mixed and incubated for 48 h. HIV-1-induced fusion between red fluorescent (E) and green fluorescent (F) cells can be detected as yellow-stained syncytia (G). Panels D and H show phase contrast microscopy of stained cells.
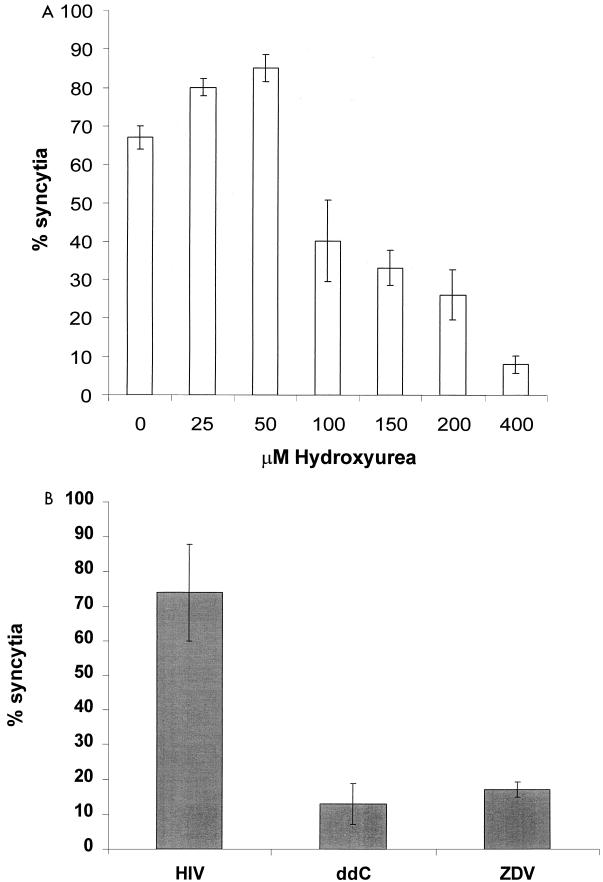
The number of cell equivalents present in syncytia was estimated by determining the increase in cell volume compared to that of the average size of a single nucleated cell in the mock-infected control. Estimated cell volume was calculated by measuring the cell diameter of dually stained cells, using the smallest transverse diameter for irregular-shaped cells. The number of cell equivalents present in syncytia corresponded closely to the total cell number of the mock-infected control (Fig. 5). The combination of finding yellow cell-cell fusion products (Fig. 4) and the fact that the corrected number of cells (based on cell measurements in Fig. 5) in the HIV-infected MT-2 culture argues against significant cell lysis in our system. The anti-HIV activity of various known antiretroviral drugs was also analyzed by using this fusion assay. ZDV and ddC and the ribonucleotide reductase inhibitor HU were shown to inhibit HIV-1 growth in vitro (Fig. 6).
FIG. 5.
Syncytia formation is the major cause of cell loss in infected MT-2 cell cultures. Cell numbers were determined in HIV-infected MT-2 cells and uninfected controls. By using the fluorescent dyes CMFDA and CMTMR, the number of cells present in the syncytia was estimated based on diameter of dually fluorescent cells. The estimated number of cells present in HIV-infected cultures (HIV-Corrected) was determined by adding the number of single fluorescent cells to the estimated number of cell equivalents present in dually fluorescent syncytia. By using a 72-h infection time period, the number of cell equivalents (HIV-Corrected) in the syncytia was approximately equal to the total number of cells present in the mock-infected control.
FIG. 6.
Inhibition of syncytium formation by antiviral drugs. Infected MT-2 cells were cultured in the presence or absence of the antiviral compounds ZDV (0.5 μM), ddC (20 μM) (B), or an increasing concentration of HU (A). Syncytium formation was quantitated by confocal microscopy.
DISCUSSION
Two distinct biological phenotypes of HIV have been described on the basis of their ability or inability to produce cytopathic effects in MT-2 cells: the SI and NSI phenotypes (6). Previous reports demonstrate that progression to AIDS is associated with increasing viral burden, deterioration of immunological status, and emergence of drug-resistant strains with more cytopathic viral phenotypes (3, 11, 21, 24). In vitro methods to determine SI phenotypes in HIV-1 clinical isolates are of prognostic value and provide an important tool to study HIV pathogenesis (7). In this report, we describe two new methods to determine the HIV-1 SI phenotype.
Due to the use of light microscopy in the standard syncytium assay (16), quantitation of syncytium induction is not possible. In addition, small syncytia may be easily overlooked, since it requires the fusion of eight cells before the syncytia diameter will double. Therefore, these fluorescence-based methods offer several advantages over the standard syncytium assay. The assays described above do not require measurement of p24 antigen or concentration of cell culture supernatants to detect RT activity. The yellow fusion products are easy to differentiate from single unfused cells, and, thus, quantitation of syncytia is greatly facilitated. Calculation of the cell volume, based on measurement of the cell diameters of fused cells, allows quantitation of the number of single-cell equivalents present in syncytia.
Using this method, we were able to show that decreased cell numbers in HIV-infected MT-2 cell cultures are predominantly due to syncytium formation, rather than to virus-induced cell death, confirming the observation of Sylwester et al. (29). Using the flow cytometric assay, we were able to show a correlation between virus growth and syncytia formation (Fig. 2). Thus, assays for syncytia quantitation, as described here, can provide an inexpensive alternative to p24 enzyme-linked immunosorbent assay testing of HIV-1 SI isolates. Furthermore, these methods can be applied to the measurement of HIV-1 inhibitors.
ACKNOWLEDGMENTS
We thank Donna Klinzman and Jim McCoy for assistance with cell cultures and virus stocks and Robert Cook and David R. Soll for helpful discussions.
This work was supported by a Veterans Administration Merit Review grant (to J.T.S.) and by National Institutes of Health grants R21AA0906 (to J.T.S.), 1RO1AA12671 (to J.T.S.), and 1RO1A140040 (to D.R.S.). In addition, The University of Iowa Flow Cytometry Core Program was utilized for these studies.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. Immunoflourescence and cell sorting: use of flow cytometry for DNA analysis. In: Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Current protocols in immunology. New York, N. Y: John Wiley and Sons; 1991. pp. 5.7–5.7.2. [Google Scholar]
- 3.Åsjö B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, Fenyö E M. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet. 1986;ii:660–662. [PubMed] [Google Scholar]
- 4.Barbeau B, Fortin J-F, Genois N, Tremblay M J. Modulation of human immunodeficiency virus type-1-induced syncytium formation by the conformational state of LFA-1 determined by a new luciferase-based syncytium quantitative assay. J Virol. 1998;72:7125–7136. doi: 10.1128/jvi.72.9.7125-7136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benyoucef S, Hober D, Shen L, Ajana F, Gérard Y, Bocket-Mouton L, Mouton Y, Wattré P. A microassay for determination of the cytopathogenicity of human immunodeficiency virus type-1 isolates. Microbiol Immunol. 1996;40:381–388. doi: 10.1111/j.1348-0421.1996.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 6.Berger E A, Doms R W, Fenyö E-M. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Alexander S, Bare C, Linquist-Stepps V, Bogan M, Moreno M E, Gibson L, Wieder E D, Kosek J, Stoddart C A, McCune J M. CCR5- and CXCR4-utilizing strains of human immunodeficiency virus type 1 exhibit differential tropism and pathogenesis in vivo. J Virol. 1998;72:10108–10117. doi: 10.1128/jvi.72.12.10108-10117.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Björndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 10.Cook R T, Stapleton J T, Klinzman D, Ballas Z K. Effect of a single ethanol ingestion on lymphocyte subsets and in vitro HIV replication. J Investig Med. 1997;45:265–271. [PubMed] [Google Scholar]
- 11.Coombs R W, Welles S L, Hooper C, Reichelderfer P S, D'Aquila R T, Japour A J, Johnson V A, Kuritzkes D R, Richman D D, Kwok S, Todd J, Jackson J B, DeGruttola V, Crumpacker C S, Kahn J. Association of plasma human immunodeficiency virus type 1 RNA level with risk of clinical progression in patients with advanced infection. J Infect Dis. 1996;174:704–712. doi: 10.1093/infdis/174.4.704. [DOI] [PubMed] [Google Scholar]
- 12.D'Souza M P, Harden V A. Chemokines and HIV-1 second receptors. Nat Med. 1996;2:1293–1300. doi: 10.1038/nm1296-1293. [DOI] [PubMed] [Google Scholar]
- 13.Fauci A S. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 15.Hori T, Sakaida H, Sato A, Nakajima T, Shida H, Yoshie O, Uchiyama T. Detection and delineation of CXCR-4 (fusin) as an entry and fusion cofactor for T cell-tropic HIV-1 by three different monoclonal antibodies. J Immunol. 1998;160:180–188. [PubMed] [Google Scholar]
- 16.Japour A J, Fiscus S A, Arduino J-M, Mayers D L, Reichelderfer P S, Kuritzkes D R. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J Clin Microbiol. 1994;32:2291–2294. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaroszeski M J, Gilbert R, Heller R. Detection and quantitation of cell-cell electrofusion products by flow cytometry. Anal Biochem. 1994;216:271–275. doi: 10.1006/abio.1994.1041. [DOI] [PubMed] [Google Scholar]
- 18.Judice J K, Tom J Y K, Huang W, Wrin T, Vennari J, Petropoulos C J, McDowell R S. Inhibition of HIV type 1 infectivity by constrained α-helical peptides: implications for the viral fusion mechanism. Proc Natl Acad Sci USA. 1997;94:13426–13430. doi: 10.1073/pnas.94.25.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koot M, Vos A H V, Keet R P M, de Goede R E Y, Dercksen M W, Terpstra F G, Coutinho R A, Miedema F, Tersmette M. HIV-1 biological phenotype in long-term infected individuals evaluated with an MT-2 cocultivation assay. AIDS. 1992;6:49–54. doi: 10.1097/00002030-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 20.Kozal M J, Ramachandran R V, Shafer R W. Prevalence of HIV-1 syncytium-inducing phenotype. Ann Intern Med. 1994;120:811. doi: 10.7326/0003-4819-120-9-199405010-00019. [DOI] [PubMed] [Google Scholar]
- 21.Kozal M J, Shafer R W, Winters M A, Katzenstein D A, Aguiniga E, Halpern J, Merigan T C. HIV-1 syncytium-inducing phenotype, virus burden, codon 215 reverse transcriptase mutation and CD4 cell decline in zidovudine-treated patients. J Acquir Immune Defic Syndr. 1994;7:832–838. [PubMed] [Google Scholar]
- 22.Lee M H, Sano K, Morales F E, Imagawa D T. Sensitive reverse transcriptase assay to detect and quantitate human immunodeficiency virus. J Clin Microbiol. 1987;25:1717–1721. doi: 10.1128/jcm.25.9.1717-1721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy J A. HIV pathogenesis and long-term survival. AIDS. 1993;7:1401–1410. doi: 10.1097/00002030-199311000-00001. [DOI] [PubMed] [Google Scholar]
- 24.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 25.Moir S, Poulin L. Expression of HIV env gene in a human T cell line for a rapid and quantifiable cell fusion assay. AIDS Res Hum Retrovir. 1996;12:811–820. doi: 10.1089/aid.1996.12.811. [DOI] [PubMed] [Google Scholar]
- 26.Saha K, Bentsman G, Chess L, Volsky D J. Endogenous production of β-chemokines by CD4+, but not CD8+, T-cell clones correlates with the clinical state of human immunodeficiency virus type 1 (HIV-1)-infected individuals and may be responsible for blocking infection with non-syncytium-inducing HIV-1 in vitro. J Virol. 1998;72:876–881. doi: 10.1128/jvi.72.1.876-881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schols D, Pauwels R, Baba M, Desmyter J, De Clercq E. Syncytium formation and destruction of bystander CD4+ cells cocultured with T cells persistently infected with human immunodeficiency virus as demonstrated by flow cytometry. J Gen Virol. 1989;70:2397–2408. doi: 10.1099/0022-1317-70-9-2397. [DOI] [PubMed] [Google Scholar]
- 28.Schols D, Pauwels R, Desmyter J, De Clercq E. Flow cytometric method to monitor the destruction of CD4+ cells following their fusion with HIV-infected cells. Cytometry. 1990;11:736–743. doi: 10.1002/cyto.990110611. [DOI] [PubMed] [Google Scholar]
- 29.Sylvester A, Shutt D, Murphy S, Soll D R. HIV-induced syncytia are self-perpetuating and the primary cause of T cell death in culture. J Immunol. 1997;158:3996–4007. [PubMed] [Google Scholar]
- 30.van 't Wout A B, Blaak H, Ran L J, Brouwer M, Kuiken C, Schuitemaker H. Evolution of syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J Virol. 1998;72:5099–5107. doi: 10.1128/jvi.72.6.5099-5107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witvrouw M, De Clercq E. Sulfated polysaccharides extracted from sea algae as potential antiviral drugs. Gen Pharmacol. 1997;29:497–511. doi: 10.1016/s0306-3623(96)00563-0. [DOI] [PubMed] [Google Scholar]
- 32.Wong M T, Dolan M J, Kozlow E, Doe R, Melcher G P, Burke D S, Boswell R N, Vahey M. Patterns of virus burden and T cell phenotype are established early and are correlated with the rate of disease progression in human immunodeficiency virus type 1-infected persons. J Infect Dis. 1996;173:877–887. doi: 10.1093/infdis/173.4.877. [DOI] [PubMed] [Google Scholar]