Summary
Background
We aimed to understand the relationship between serum biomarker concentration and lesion type and volume found on computed tomography (CT) following all severities of TBI.
Methods
Concentrations of six serum biomarkers (GFAP, NFL, NSE, S100B, t-tau and UCH-L1) were measured in samples obtained <24 hours post-injury from 2869 patients with all severities of TBI, enrolled in the CENTER-TBI prospective cohort study (NCT02210221). Imaging phenotypes were defined as intraparenchymal haemorrhage (IPH), oedema, subdural haematoma (SDH), extradural haematoma (EDH), traumatic subarachnoid haemorrhage (tSAH), diffuse axonal injury (DAI), and intraventricular haemorrhage (IVH). Multivariable polynomial regression was performed to examine the association between biomarker levels and both distinct lesion types and lesion volumes. Hierarchical clustering was used to explore imaging phenotypes; and principal component analysis and k-means clustering of acute biomarker concentrations to explore patterns of biomarker clustering.
Findings
2869 patient were included, 68% (n=1946) male with a median age of 49 years (range 2-96). All severities of TBI (mild, moderate and severe) were included for analysis with majority (n=1946, 68%) having a mild injury (GCS 13-15). Patients with severe diffuse injury (Marshall III/IV) showed significantly higher levels of all measured biomarkers, with the exception of NFL, than patients with focal mass lesions (Marshall grades V/VI). Patients with either DAI+IVH or SDH+IPH+tSAH, had significantly higher biomarker concentrations than patients with EDH. Higher biomarker concentrations were associated with greater volume of IPH (GFAP, S100B, t-tau;adj r2 range:0·48-0·49; p<0·05), oedema (GFAP, NFL, NSE, t-tau, UCH-L1;adj r2 range:0·44-0·44; p<0·01), IVH (S100B;adj r2 range:0.48-0.49; p<0.05), Unsupervised k-means biomarker clustering revealed two clusters explaining 83·9% of variance, with phenotyping characteristics related to clinical injury severity.
Interpretation
Interpretation: Biomarker concentration within 24 hours of TBI is primarily related to severity of injury and intracranial disease burden, rather than pathoanatomical type of injury.
Funding
CENTER-TBI is funded by the European Union 7th Framework programme (EC grant 602150).
Keywords: Brain injury, Traumatic; Biomarkers; Neuroimaging; Computed tomography
Research in context.
Evidence before this study
We searched key terms relating to traumatic brain injury (TBI), biomarkers and neuroimaging on MEDLINE and EMBASE to review the literature assessing the relationships between serum biomarkers and acute neuroimaging findings published before June 1st 2021, with the terms: (“brain injuries, traumatic” or “traumatic brain injury” or (brain adj2 trauma*) or “TBI”) AND (“biological marker” or “biomarkers”) AND (“diagnostic imaging” or “neuroimaging”). We found previous approaches have been limited by focusing on presence or absence of lesions, small sample sizes, focus on a small number of biomarkers (often S100B and NSE); with methodological heterogeneity making comparisons difficult. S100B has been consistently found to correspond to volume of contusions. The largest study assessing lesion volumes (n = 115) demonstrated correlations between S100B, GFAP, total tau and NSE levels to volumes of contusion, intraventricular haemorrhage, subarachnoid haemorrhage and total volume of intracerebral haemorrhage (except NSE). In a study of 81 patients with severe TBI GFAP levels were demonstrated to be greater in focal mass lesion as compared to diffuse injury whilst UCH-L1 has been primarily raised in diffuse injury.
Added value of this study
To our knowledge this study represents the largest and most comprehensive study to date (2869 patients encompassing the entire injury spectrum) investigating a panel of six of the most frequently studied TBI blood biomarkers in relation to acute neuroimaging findings (GFAP, NFL, NSE, S100B, t-tau and UCH-L1). This is the first study to utilise an automated lesion segmentation method based on deep convolutional neural networks to allow for volumetric analysis of four categories of intracranial lesion. Further, the large sample size and sampling of six biomarkers allows for direct inter-biomarker comparisons in relation to specific lesion types and volumes, giving unique insights into relationships between pathological subtype and serum biomarker concentration. We demonstrate a positive association between acute biomarker levels and intracranial lesion burden with significant positive associations between serum biomarker concentrations and volumes of intraparenchymal haemorrhage and intracerebral oedema. Biomarker concentrations were higher with an increasing number of different lesions types and in severe diffuse injury as compared to focal mass lesion whilst unsupervised biomarker clustering revealed two natural clusters, with clear phenotyping characteristics that related to clinical injury severity. We further demonstrate differences between patients with parenchymal injuries versus extra-parenchymal injuries. However, there was substantial overlap in injury types, making it difficult to use individual biomarker signatures to identify individual pathoanatomical lesion types.
Implications of all available evidence
Blood biomarkers in TBI relate to the burden of cranial and intracranial disease and are elevated in the majority of lesion types with concentrations reflecting injury severity. While we found significant differences between intraparenchymal versus extraparenchymal injury patterns, biomarker concentrations were unable to distinguish lesions by pathoanatomical injury type. This is important for the interpretation of biomarker levels in both research and clinical contexts.
Alt-text: Unlabelled box
Introduction
Traumatic brain injury (TBI) has been described as the most complex disease in the most complex organ;1 and much of this complexity is secondary to the large heterogeneity of lesions that may occur. Computed Tomography (CT) remains the most commonly utilised radiological method for the diagnosis of TBI associated intracranial lesions and for determining the need for emergent management.2 The cost and radiation burden associated with neuroimaging, particularly in those with a mild injury, has led to increasing exploration of blood biomarkers to aid in diagnosis, detection of neuro-worsening and prognostication.3,4 Biomarkers also offer the potential to better characterise the heterogeneity of TBI. However, the promise of blood biomarkers in these contexts remains unproven, and their role in clinical care uncertain.
Prior literature has focused on the diagnostic ability of proteomic biomarkers for the detection of intracranial pathology.5, 6, 7 Yet the impact of intracranial injury pattern, lesion type and lesion burden on serum biomarker concentrations remains unclear. The varied cellular origin and pathophysiological drivers of proteomic biomarker release suggest that patterns of biomarker elevation may provide discrete and identifiable signatures of different types of intracranial injuries.8 Biomarker levels could also be expected to scale with both injury volume and severity.9, 10, 11 The relative impact of these two factors - lesion specificity and injury burden - in driving the levels of specific biomarkers remains unclear, and is responsible for uncertainty in the interpretation of biomarker levels following TBI.
This study examines six biomarkers that may be released from different cell types or cell components, after TBI: glial fibrillary acidic protein (GFAP), neurofilament light (NFL), neuron specific enolase (NSE); S100 calcium protein B (S100B), ubiquitin carboxy-terminal hydrolase-L1 (UCH-L1) and total tau (t-tau). NSE and UCH-L1 are both enriched in the neuronal stroma; NFL and tau protein are structural axonal proteins associated with axonal injury; whilst S100B and GFAP are proteins secreted from astrocytes and microglia following TBI (Fig. S1).8
The Collaborative European NeuroTrauma Effectiveness Research in TBI (CENTER-TBI) core study collected data across the entire spectrum of TBI and lesion type including this panel of biomarkers within 24 hours.12,13 We aimed to assess the impact of injury detected using CT radiological parameters, including traumatic intracranial lesion type and lesion volume, on serum biomarker concentration in patients following all severities of TBI.
Methods
Study design, Ethics and Participants
The CENTER-TBI core study is a prospective observational study of patients with all severities of TBI, conducted in 65 clinical sites from 18 countries between December 19th 2014 and December 17th 2017 (https://clinicaltrials.gov/ct2/show/NCT02210221).12,13 The CENTER-TBI inclusion criteria were a clinical diagnosis of TBI, and presentation within 24 hrs of injury with a clinical indication for CT scanning. Patient data were accessed using Neurobot platform (RRID/SCR_017004, Core data version 2·1).
The CENTER-TBI study (EC grant 602150) has been conducted in accordance with all relevant laws of the EU if directly applicable or of direct effect and all relevant laws of the country where the Recruiting sites were located, including but not limited to, the relevant privacy and data protection laws and regulations (the “Privacy Law”), the relevant laws and regulations on the use of human materials, and all relevant guidance relating to clinical studies from time to time in force including, but not limited to, the ICH Harmonised Tripartite Guideline for Good Clinical Practice (CPMP/ICH/135/95) (“ICH GCP”) and the World Medical Association Declaration of Helsinki entitled “Ethical Principles for Medical Research Involving Human Subjects”. Informed Consent by the patients and/or the legal representative/next of kin was obtained, accordingly to the local legislations, for all patients recruited in the Core Dataset of CENTER-TBI and documented in the electronic case report form. A full list of recruitment sites, Ethical committees, approval numbers and approval dates can be found in the supplemental material (Table S1), and online.14
All CENTER-TBI participants with an interpretable computed tomography (CT) scan performed on admission and complete proteomic biomarker panel measured within 24 hours of injury were included in the analysis. The study is reported in accordance with the STROBE recommendations
Neuroimaging
Central review of CT scans was performed in accordance to the Common Data Element (CDE) scheme for TBI (https://commondataelements.ninds.nih.gov/).15,16 The presence or absence of the following pathoanatomical lesions were reported on the first available CT scan: skull fracture, acute subdural haematoma (aSDH), mixed density SDH (mSDH), extradural haematoma (EDH), traumatic subarachnoid haemorrhage (tSAH), intraventricular haemorrhage (IVH), diffuse axonal injury (DAI) and intraparenchymal haemorrhage (IPH) by a central review panel consisting of three protocol-trained reviewers. All readers were blinded to clinical information except for gender, age and care path stratum. Information was entered directly into digital custom-made multi-tiered structured templates based on the National Institute of Neurological Disorders and Stroke (NINDS) TBI CDEs.15 Good inter-rater reliability was reported between all reviewers.16
In addition, the Marshall CT classification, a CT derived metric of image classification commonly used in TBI research, was reported.17
Patients were separated into nine groups depending on the centrally reported CT findings: No acute abnormality, mixed lesion (patients with any two or more lesion types), and seven categories of isolated pathology types (skull fracture, aSDH, mSDH, EDH, tSAH (which included patients with IVH due to only 11 patients having isolated IVH), DAI, IPH). Given the high association of skull fractures with EDH due to mechanism of injury these were classified as one lesion (EDH). If a patient in the isolated skull fracture group had a Marshall CT classification II or above, demonstrating signs of diffuse intracranial injury, the patient was categorized into the mixed lesion group owing to likely undefined underlying brain injury.
To quantify lesion burden, a convolutional neural network (Brain Lesion Analysis and Segmentation Tool for CT (BLAST-CT), www.github/biomedia-mira/blast-ct/) developed and validated for use in TBI,18 was used to produce automated voxel based volumetric assessment of IPH, total extra-axial haemorrhage (EAH; SDH, EDH and tSAH),IVH and intracerebral oedema. CT images with outlying lesion volumes were visually inspected to confirm the plausibility of the lesion volumes provided from the CNN.
Biomarker sampling
Blood samples for biomarker analysis were collected within 24 hours of injury. S100B and NSE were measured with a clinical-use automated system, using an electrochemiluminecesence immunoassay kit (Elecsys S100 and NSE assays on the Cobas 8000 modular analyzer, Roche Diagnostics, Mannheim, Germany). GFAP, UCH-L1, t-tau, and NFL were analysed using Single Molecule Arrays (SiMoA) based assay on the SR-X benchtop assay platform (Quanterix Corp., Lexington, MA). For biomarkers measured using the SiMoA based assay, the limit of detection (LoD) was as follows: GFAP 1.32 pg/ml, NFL 0.0971 pg/ml, t-tau 0.0236 pg/ml and UCH-L1 1.34 pg/ml. All samples for GFAP, NFL and t-tau were over the LoD, whilst <1% (n=19) of UCH-L1 samples were below the LoD. Complete details of biomarker sampling and assays can be found at a previous open access publication.7
Statistics
Unless otherwise specified Wilcoxon signed rank test was used for comparisons of continuous data and χ2 statistics for categorical variables. The serum biomarker values were skewed, therefore the natural log of biomarker values was used for analyses following initial data inspection.
Baseline characteristics were summarized using standard descriptive statistics. Categorical variables are presented as frequencies and percentages, with continuous variables presented as median (interquartile range). We compared the subsets of patients recruited to the CENTER-TBI Core Study who were included in this analysis with those who were excluded because of non-availability of 24 hour biomarkers, an early CT, or both.
Due to only nine patients having an isolated mixed density SDH (which may indicate ongoing bleeding or subacute pathology) they were included in basic descriptive analysis, but excluded from further analysis. The Dunn Kruskal-Wallis test with Benjamini-Hochberg correction for multiple comparisons was used for group-wise comparison between biomarker levels in different Marshall CT categories and CT pathology groups.19 In order to improve comparability between biomarkers, we calculated Z-scores for each biomarker in each lesion group, against a reference population of patients with no acute imaging findings.
Hierarchical clustering was used to determine acute imaging phenotypes based upon the qualitative CT reporting. Optimal number of clusters was determined via the elbow method. Biomarker concentrations for patients with the distinct combinations of pathology highlighted on cluster analysis were compared via the Dunn Kruskal-Wallis test with Benjamini-Hochberg correction for multiple comparisons.
The area under the receiver operating characteristic (ROC) curve (AUC) was used to determine the discriminative ability of the six blood biomarkers for the detection of any traumatic abnormality, and individual subgroups of isolated pathoanatomical lesion types. A case was considered the presence of the pathological group of interest, and a non-case being patients with normal CT scans. Confidence intervals were derived via the DeLong method.
Polynomial regression was used to assess the relationship between biomarker levels and volume of the different intracranial lesions as derived from BLAST-CT. The overall sample was separated into patients with the presence of either of the four derived lesion classes with regression performed on these subsets. To assess the classical pathoanatomical lesion categories contained within the total EAH group, total EAH was subdivided into three further classes dependent on the presiding EAH lesion type as recorded on qualitative CT reporting; aSDH, EDH and tSAH. Four models were used, with the covariates of final model examined for linearity and inclusion of the appropriate polynomial term of variables that were demonstrated to have a non-linear relationship to the dependent variable on univariate analysis. The inclusion of the polynomial terms led to improvement in model fit, with visual inspection of the variance of residuals demonstrating a satisfactory distribution and were therefore included in the final model. Covariates were selected based on factors known to influence lesion volume or biomarker concentration. Model 1 – Univariate, Model 2 - Adjustment for age and sex, Model 3 – Model 2 additionally adjusted for time to biomarker, time to scan, GCS and extracranial abbreviated injury score, Model 4 - Model 3 additionally adjusted for–volume of IVH, volume of IPH, volume of post traumatic oedema, total brain volume, and polynomial terms for all variables found to have a non-linear association with biomarker concentration.
Total case numbers are reported throughout with missing patient demographic data presented.
To understand if biomarker concentrations related to radiological variables beyond the above groups an unsupervised approach using principal component analysis (PCA) and subsequent K-means clustering was performed including all patients. Scaled variables of the complete biomarker panel were inputted into the PCA algorithm, with creation of a loading plot to explore biomarker covariance. K-means clustering was used to ascertain natural clustering of biomarker inputs. Optimal number of clusters was selected via the silhouette method with subsequent descriptive comparison of the demographic, biochemical and radiological features of these clusters.
Data analysis was conducted via R (version 3·6·2, https://www.R-project.org/) in RStudio (version 1·2·5033, http://www.rstudio.com).
Role of funding source
The funders had no role in the study design, collection, analysis and interpretation of data, nor in the writing of the report or in publication decisions. All authors had full access to the study data and had final responsibility for the decision to submit for publication.
Results
Patient cohort
Of 4509 patients in the CENTER-TBI core study 2869 (63·6%) had CT and biomarker data available. Further demographic information is available in the supplemental material (Fig. S2, Table S2, Table S3). 2682 (93·5%) scans were accessible for the volumetric analysis. The analysis population was 68% male with a median age of 49 years (range 2-96). The median time elapsed between injury to biomarker sample was 13 hours (IQR [6·2-19]) with median time between injury and CT imaging of 2 hours (IQR [1·4-3·2]).
The majority of patients (n=1800, 63%) had traumatic lesions present on CT imaging (Table S4). Of these 1313 (73%) had two or more different lesion types. The most frequent pattern of injury observed was a combination of aSDH, IPH, tSAH and skull fracture (n=181). In relation to patients with only a single injury classification only on central reporting, 184 had tSAH, 89 aSDH, 86 isolated skull fractures, 47 EDH, 41 IPH, 31 DAI and 9 had a mixed density SDH.
How do conventional pathoanatomical groups relate to biomarker levels and signatures?
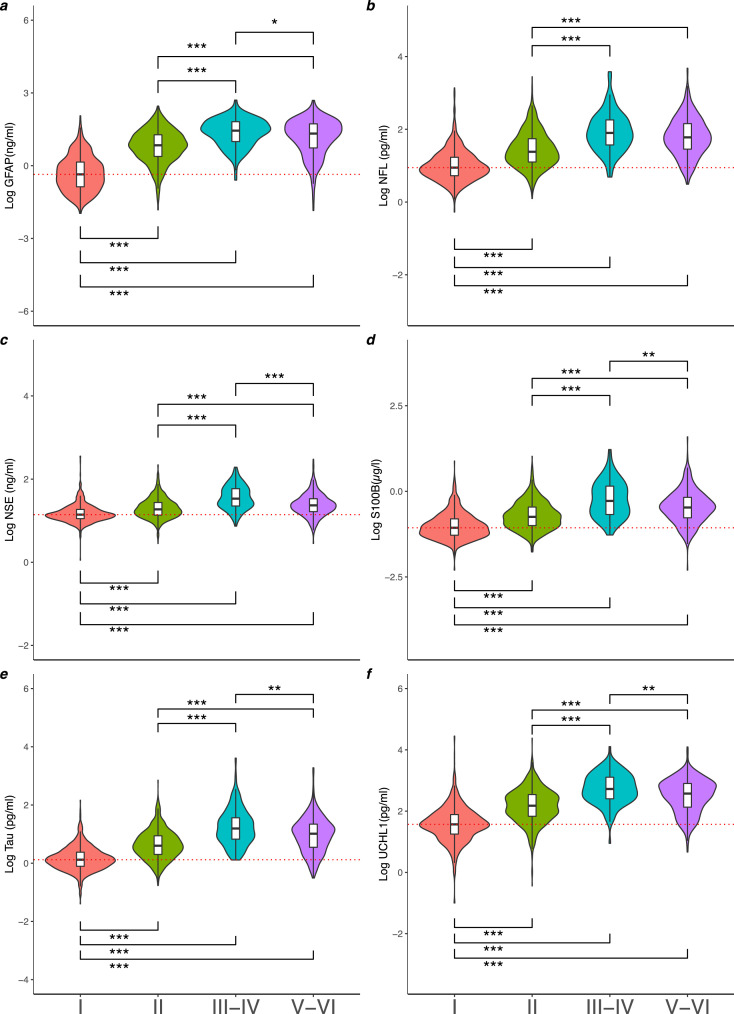
The levels of all biomarkers varied with Marshall CT grade (Figure 1, Table S5), with higher levels seen in severe diffuse injury (Marshall grade III-IV), when compared to Marshall grade I and Marshall grade II (Dunn Kruskal-Wallis test with Benjamini-Hochberg correction, all p<0·005). Biomarker concentrations were significantly higher in severe diffuse injury compared to focal mass lesion in all biomarkers (Median (IQR), p values derived from Dunn Kruskal-Wallis test with Benjamini-Hochberg correction: GFAP ng/ml [27·81 (9·87-64·88) vs 21·06 (5·41-53·24) p= 0·03], NSE ng/ml [33·91 (22·54-58·87) vs 23·34 (16·50-33·73) p=0·000], S100B µg/L [0·52 (0·21-1·42) vs 0·34 (0·17-0·67) p=0·004], t-tau pg/ml [15·6 (6·7-36·44) vs 10·34 (3·52-21·93) p=0·002], UCH-L1 [527·86 (253·25-1283·25) vs 373·59 (134·74-798·24) p=0·002]), aside from NFL (NFL pg/ml [79·7 (37·19-181·27) vs 60·16 (28·51-143·23) p=0·085]).
Figure 1.
The Log of serum GFAP (a), NFL (b), NSE (c), S100B (d), total-tau (e) and UCH-L1 (f) concentration by Marshall CT score.Violin plots and boxplots provide median, range and 25-75th percentile of the log10 biomarker concentration per Marshall CT score grouping I (n=1154), II (n=1120), II-IV (n=120) and V-VI (n=475). P values determined by the Dunn Kruskal-Wallis test with Benjamini-Hochberg correction for multiple comparisons was used for group wise comparison across different CT findings. Significance levels are displayed for statistically significant group wise comparisons, * = p< 0·05, ** = p< 0·01, *** = p< 0·001. Red dotted line indicates the median of Marshall Score I group.
Do biomarker levels or signatures vary across isolated lesion types and/or pre-specified lesion groupings?
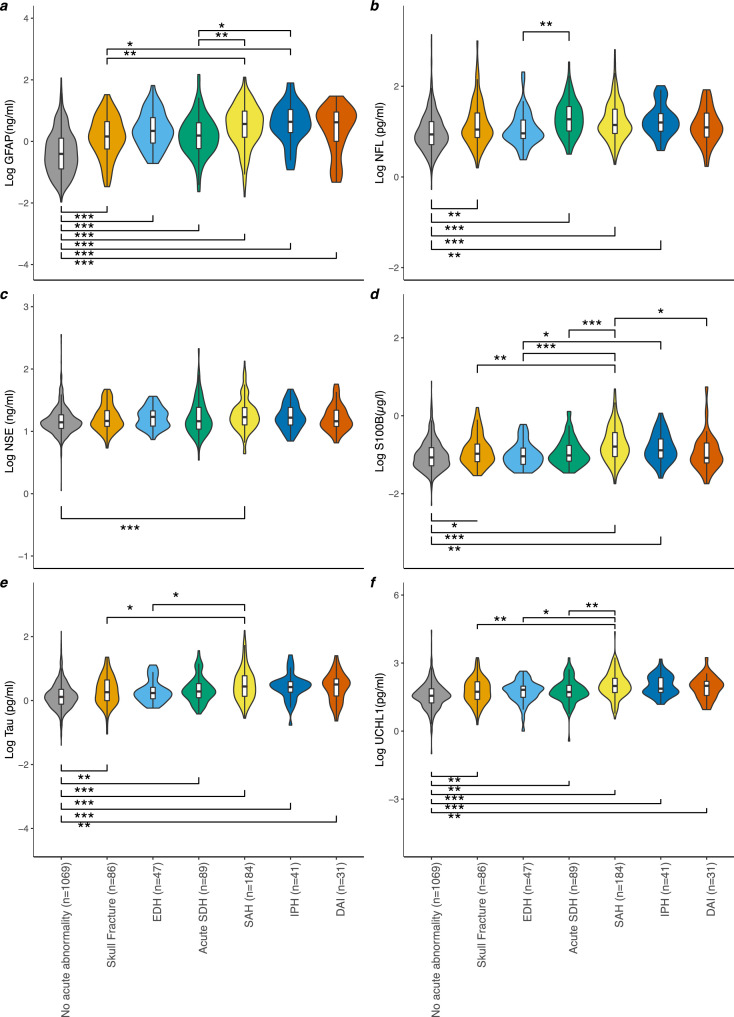
The levels of biomarkers by lesion type are seen in Figure 2 and Table S6. Significant differences between GFAP levels were found for all isolated pathology groupings when compared to patients with no visible lesions, and it was the only biomarker to be significantly raised in the EDH group. Conversely, for NSE, significant differences were only seen in the tSAH cohort. In general, higher biomarker levels were observed in the tSAH and IPH groups. This was significant in a proportion of comparisons on inter-group testing (Figure 2).
Figure 2.
The Log of serum GFAP (a), NFL (b), NSE (c), S100B (d), total-tau (e) and UCH-L1 (f) concentration by CT pathology. Violin plots and boxplots provide median, range and 25-75th percentile of the log10 biomarker concentration per pathoanatomical grouping/isolated lesion type: (No acute abnormality (n=1069), Skull fracture (n=86), EDH (n=47), Acute SDH (n=89), SAH (n=184), IPH (n=41), DAI (n=31), Mixed Lesion (n=1313). P values determined by the Dunn Kruskal-Wallis test with Benjamini-Hochberg correction for multiple comparisons was used for group wise comparison across different CT findings. Significance levels are displayed for statistically significant group wise comparisons, * = p< 0·05, ** = p< 0·01, *** = p< 0·001. Patients with mixed lesion on CT (defined as 2 or more lesions types) (n=1362) and those with isolated mixed density SDH (n=9) are not included as a boxplot.
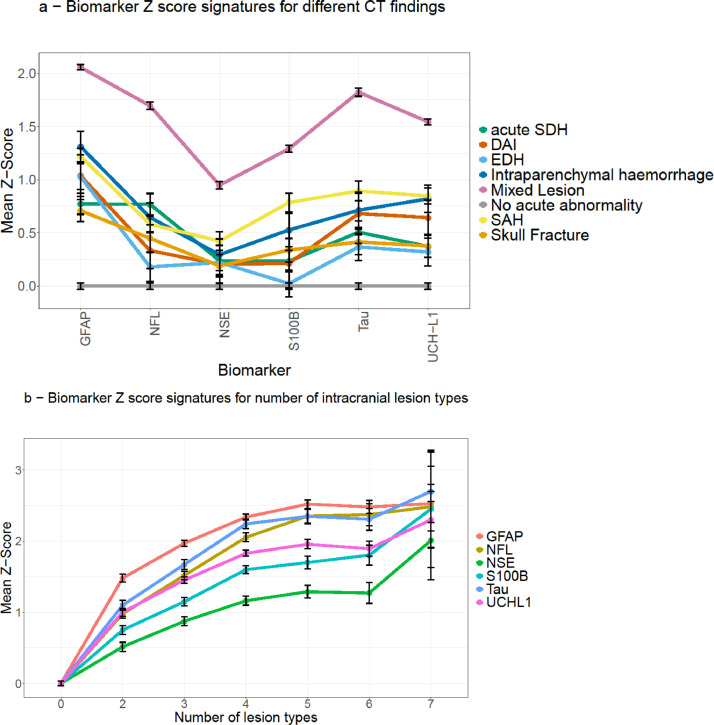
The mean Z-scores of all biomarkers were elevated in all isolated lesion categories, and further elevated in the mixed lesion group (Figure 3, Fig. S3). GFAP showed the highest Z-score in all pathology groups, while conversely, NSE showed the lowest Z-scores for most traumatic CT findings (Figure 3). The cohort with an isolated skull fracture had a positive mean Z-score for all biomarkers. The Z-score increased as the number of different lesion types increased; this increase was not related to the addition of any specific lesion type (Figure 3).
Figure 3.
Z-score signatures across different pathoanatomical groups. The Biomarker concentrations presented as means and standard errors of Z scores with the reference group being patients with a normal CT scan following TBI. Panel a shows mixed lesion (n=1313) and isolated pathology groups (Skull fracture (n=86), EDH (n=47), Acute SDH (n=89), SAH (n=184), IPH (n=41), DAI (n=31)) in relation to biomarker expression with patients with normal CT (n=1069) as the reference group. Panel b shows the number of different intracranial lesion types reported on each CT image for patients with mixed lesions, in relation to biomarker expression.
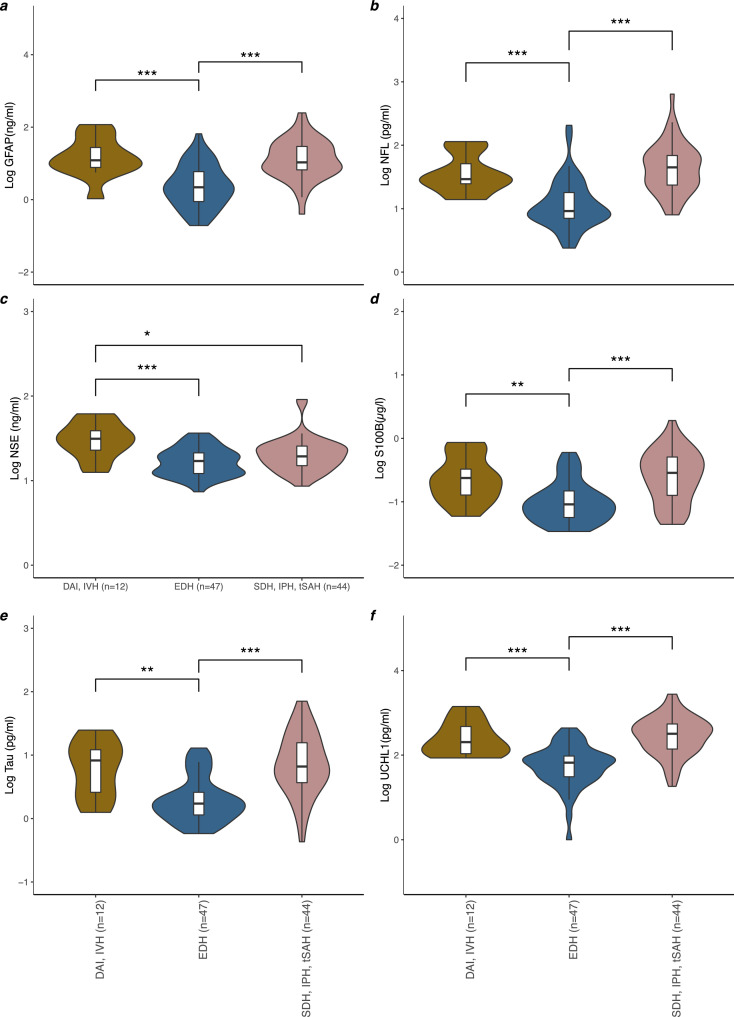
Hierarchical clustering found 70·6% of the variance was explained by three dimensions, representing three distinct phenotypes; DAI+IVH, SDH+IPH+tSAH, and isolated EDH (Fig. S4), with lowest levels of all biomarkers in the EDH group compared to the other two (Figure 4). Except for NSE, there was no significant difference found between the DAI+IVH, or SDH+IPH+tSAH groups.
Figure 4.
The Log of serum GFAP (a), NFL (b), NSE (c), S100B (d), total-tau (e) and UCH-L1 (f) concentration between patients with DAI and IVH (n = 12), EDH alone (n = 47) or SDH, IPH and tSAH (n = 44). Violin plots and boxplots provide median, range and 25-75th percentile of the log10 biomarker concentration per pathological phenotypes as determined by hierarchical clustering of qualitative CT reports: DAI and IVH (n = 12), EDH alone (n = 47) or SDH, IPH and tSAH (n = 44). P values determined by the Dunn Kruskal-Wallis test with Benjamini-Hochberg correction for multiple comparisons was used for group wise comparison across different CT findings. Significance levels are displayed for statistically significant group wise comparisons, * = p< 0·05, ** = p< 0·01, *** = p< 0·001.
Ability for detection of isolated lesions, compared to the normal population
All biomarkers had fair (AUC >0·7) discrimination for detection of any traumatic abnormality on CT, with a range of AUCs [95% CI] between (NSE) 0.70 (0.68-0.72) to (GFAP) 0·88 [0·87-0·90]). Of all biomarkers, GFAP showed the best discrimination for the detection of isolated lesion types in all lesions aside from acute SDH, where it was NFL. For the detection of isolated lesion types AUC [95% CI] ranged from: skull fracture (0·56 [0·49,0·63] to 0·70 [0·65,0·76]), EDH (0·51 [0·43,0·59] to 0·79 [0·73,0·84]), SDH (0·54 [0·47,0·61] to 0·73 [0·67,0·78], SAH (0·62 [0·58,0·67] to 0·81 [0·78,0·85]), IPH (0·60 [0·51,0·69] to 0·83 [0·77,0·89]) and DAI (0·46 [0·35,0·56] to 0·76 [0·66,0·87]). Plotted ROC curves, AUC and optimal cut offs of all biomarkers and pathoanatomical lesion types are available in the supplemental material (Fig. S5, Fig. S6, Table S8, Table S9).
Relation between lesion volume and biomarker levels
Table 1 and Tables S10-S13 show data on comparisons between biomarker levels and lesion volumes, for univariate linear regression and multivariable polynomial regression respectively. On univariate analysis all biomarkers showed a significant positive association with volume of total EAH, IPH and traumatic cerebral oedema, with all biomarkers aside from NSE and NFL demonstrating significant association with IVH volume. On multivariable analysis there was a significant positive association between volume of IPH and the serum concentration of GFAP, S100B, Tau, but not NFL, NSE or UCH-L1. Significant positive association was shown between volume of traumatic intracerebral oedema and the serum concentration of GFAP, NFL, NSE, Tau and UCH-L1, but not S100B. No significant association was also shown between the volume of EAH and any of the biomarkers. Of all the biomarkers, only S100B demonstrated a significant positive association with volume of IVH.
Table 1.
Multivariable polynomial regression of CNN derived lesion volumes to biomarker levels.
| Lesion Type (n) | Biomarker | Coefficient (SE) | 95% Confidence Interval | P-value |
|---|---|---|---|---|
| Total EAH (n=1294) | GFAP | -3.65 (2.68) | -8.91, 1.6 | 0.172 |
| NFL | 0.1 (0.06) | -0.02, 0.22 | 0.094 | |
| NSE | 0.13 (0.12) | -0.1, 0.36 | 0.279 | |
| S100B | 0.12 (0.08) | -0.04, 0.27 | 0.135 | |
| t-tau | 0.1 (0.06) | -0.01, 0.21 | 0.090 | |
| UCH-L1 | 0.49 (2.79) | -4.99, 5.97 | 0.861 | |
| Oedema (n=934) | GFAP | 8.66 (2.28) | 4.19, 13.12 | 0.000*** |
| NFL | 0.22 (0.06) | 0.11, 0.33 | 0.000*** | |
| NSE | 0.28 (0.12) | 0.05, 0.52 | 0.017* | |
| S100B | 1.94 (2.52) | -3, 6.88 | 0.440 | |
| t-tau | 0.17 (0.06) | 0.06, 0.28 | 0.003** | |
| UCH-L1 | 0.16 (0.06) | 0.04, 0.27 | 0.008** | |
| IVH (n=212) | GFAP | 0.16 (0.1) | -0.04, 0.37 | 0.117 |
| NFL | 0.03 (0.12) | -0.21, 0.27 | 0.809 | |
| NSE | -0.08 (0.25) | -0.56, 0.41 | 0.760 | |
| S100B | 0.32 (0.16) | 0.01, 0.64 | 0.042* | |
| t-tau | 0.12 (0.11) | -0.1, 0.34 | 0.293 | |
| UCH-L1 | 0.22 (0.13) | -0.04, 0.48 | 0.090 | |
| IPH (n=659) | GFAP | 5.55 (1.95) | 1.73, 9.38 | 0.004** |
| NFL | 0.1 (0.06) | -0.02, 0.22 | 0.114 | |
| NSE | 0.02 (0.13) | -0.23, 0.28 | 0.853 | |
| S100B | 0.21 (0.09) | 0.04, 0.38 | 0.013* | |
| t-tau | 0.13 (0.06) | 0, 0.25 | 0.044* | |
| UCH-L1 | 0.12 (0.07) | -0.02, 0.25 | 0.082 |
Results of multivariable polynomial regression of the natural log of lesion volume to biomarker level. Adjustment for time to biomarker, time to scan, age, sex, Glasgow Coma Score (GCS), extracranial abbreviated injury score, volume of other lesion types and calculated total brain volume with polynomial terms of non-linear variables. *** p<0·001, ** p<0·01, * p<0·05.
The estimated regression coefficient, confidence intervals and p-values are presented for univariate linear (Tables S14-S16) and multivariable polynomial regressions (Table 2) of the EAH subgroup analysis. Analysis of the subgroups within the EAH cohort showed that volume of tSAH was significantly positively correlated with t-tau, but not with GFAP, NFL, NSE, S100B or UCH-L1 levels. No biomarkers showed a significant association with the volume of acute SDH. S100B had a significant negative association with EDH volume, with no significant associations demonstrated with GFAP, NFL, NSE, t-tau or UCH-L1.
Table 2.
Subset analysis of extra-axial haemorrhage group. Multivariable polynomial regression of CNN derived lesion volumes to biomarker levels.
| Lesion Type (n) | Biomarker | Coefficient (SE) | 95% Confidence Interval | P-value |
|---|---|---|---|---|
| Acute SDH (n=132) | GFAP | -0.15 (0.13) | -0.42, 0.11 | 0.253 |
| NFL | -0.27 (0.22) | -0.71, 0.16 | 0.214 | |
| NSE | -0.68 (0.37) | -1.41, 0.05 | 0.067 | |
| S100B | -1.87 (3.27) | -8.34, 4.6 | 0.569 | |
| t-tau | -2.85 (2.87) | -8.53, 2.82 | 0.322 | |
| UCH-L1 | -0.16 (0.19) | -0.52, 0.21 | 0.400 | |
| tSAH (n=406) | GFAP | 0.12 (2.39) | -4.57, 4.81 | 0.960 |
| NFL | 0.14 (0.1) | -0.05, 0.33 | 0.144 | |
| NSE | 0.19 (0.19) | -0.18, 0.55 | 0.319 | |
| S100B | 0.19 (0.13) | -0.06, 0.44 | 0.133 | |
| t-tau | 0.21 (0.09) | 0.03, 0.38 | 0.019* | |
| UCH-L1 | 2.46 (2.46) | -2.37, 7.29 | 0.317 | |
| EDH (n=54) | GFAP | -2.01 (2.34) | -6.74, 2.72 | 0.395 |
| NFL | -0.47 (2.33) | -5.18, 4.24 | 0.841 | |
| NSE | -0.01 (0.75) | -1.52, 1.51 | 0.991 | |
| S100B | -5.78 (2.5) | -10.83, -0.72 | 0.026* | |
| t-tau | -0.34 (0.4) | -1.14, 0.47 | 0.402 | |
| UCH-L1 | -1.07 (2.19) | -5.49, 3.35 | 0.626 |
Results of multivariable polynomial regression of the natural log of extra-axial lesion, as separated by isolated extra-axial lesion type, volume to biomarker level. Adjustment for time to biomarker, time to scan, age, sex, Glasgow Coma Score (GCS), extracranial abbreviated injury score, volume of IVH, volume of IPH, volume of post traumatic oedema and total brain volume with polynomial terms of non-linear variables. *** p<0·001, ** p<0·01, * p<0·05.
Biomarker PCA and Cluster analysis
PCA analysis of the biomarker panel concentrations demonstrated 83·9% of the variance was explained via the first two components (Fig. S7, Tables S17 and S18). GFAP, NFL, S100B, t-tau and UCH-L1 all showed significant covariance, with NSE showing the least covariance with other members of the biomarker panel. NSE explained the majority of the variance for dimension 2 (Fig. S8). K-means clustering identified two clusters which demonstrated distinct phenotypes based on clinical and radiological findings (Table S19). Raised levels of all biomarkers were observed in the cluster two with higher severity of injury demonstrated by larger numbers and volumes of lesions, lower GCS, total injury severity score, and a greater percentage of patients admitted to ICU compared to cluster one.
Discussion
To our knowledge, this is the largest and most comprehensive study to date examining the relationship between serum concentration of a panel of proteomic biomarkers on one hand; and neuroimaging characterization of injury class (based on the Marshall CT Classification), traumatic lesion type, and intracranial lesion volume, on the other. We demonstrated that a primary driver of acute biomarker concentrations in the acute phase after TBI was lesion burden, with parenchymal injury burden having higher biomarker concentrations than extra-parenchymal injury.
Most biomarker levels scaled directly with intracranial lesion burden, both overall, and in relation to the volume of individual lesion types. We found the greater the volume of intracerebral oedema the greater the serum concentration of all biomarkers studied aside from S100B, with greater volumes of IPH associated with GFAP, S100B and t-tau concentration. We, however, found no significant association between volume of extra-axial hemorrhage and biomarker concentration. This demonstrates a clear difference between intraparenchymal injury (IPH, oedema) and extraparenchymal injury (extra-axial haemorrhage) in their respective influence on biomarker concentration. The positive association on volumetric analysis echo the observed increase in biomarker concentration with both the greater degree of diffuse injury on Marshall CT classification and the increased number of lesion types in the mixed lesion category. These results indicate that it is the burden of intra-axial injury, and therefore extent of brain parenchymal injury, has the largest impact on serum biomarker concentration.
A strength of this analysis is the large sample size providing sufficient numbers of isolated intracranial injuries for direct comparison of multiple biomarkers across the different isolated lesion types. Further, the use of the BLAST-CT algorithm allowed for automated voxel based volumetric assessment giving volumetric measurements in an unprecedentedly large cohort allowing for novel analysis of the impact of the volume of intracranial pathologies on a panel of multiple different TBI biomarkers. By using a multivariable approach with adjustment for factors known to influence biomarker concentrations including; basic demographics, imaging and biomarker timings, and coexisting traumatic pathology, we provide a more detailed and comprehensive analysis of volumetric CT findings than previously reported. We demonstrate a direct relationship between contusion volume on CT and serum levels of S100B, GFAP, and t-tau with a poor association with SDH and EDH volume, building upon previous smaller analyses.9, 10, 11 NSE association has previously shown inconsistent relationship with intraparenchymal lesion volume,9, 10, 11,20 and we confirm a lack of correlation in our much larger cohort. Only S100B had a significant positive association with volume of IVH, consistent with its presence in ependymal cells and choroid epithelium or following diffusion across a damaged choroidal barrier from the cerebrospinal fluid.21
We performed a cluster analysis of lesions reported which extends the findings, across the entire disease spectrum, of two recently reported clusters of lesion types concentration that had focused on mild injury only.22 Lower biomarker levels were observed in patients with EDH when compared to DAI+IVH and SDH+IPH+tSAH echoing the results from volumetric analysis and indicating a difference in biomarkers signatures between parenchymal and extraparenchymal injury.
It has previously been suggested that UCH-L1 is primarily raised in diffuse injury, whilst GFAP is raised in focal mass lesions.23 We were unable to replicate this finding; serum concentrations of all biomarkers were higher in severe diffuse injury when compared to focal mass lesion, with no differences between GFAP and UCH-L1. We also demonstrated a lack of statistically significant variation in biomarker profiles between different isolated lesion types. There exists a substantial overlap between pathoanatomical lesions types, with lesions rarely presenting in isolation and the majority of patients with intracranial pathology having a mixed lesion injury.24 Most injuries will extend across multiple cell types or components, and this heterogeneity will make it difficult to utilize biomarker signatures to correctly identify individual lesion classes.
Of note, this study demonstrated significantly raised levels of GFAP, S100B, NFL, t-tau and UCH-L1 in the isolated skull fracture group with no demonstrable underlying parenchymal injury on CT imaging, in comparison to those with normal CT scans. Within TBI research, skull fractures are often not considered to represent a significant CT finding, and are often classified as showing no acute abnormality. Previous publications have reported elevation in some TBI biomarker levels in skull and facial fractures, but these usually fail to reach significance.25, 26, 27 Our current analysis, based on a substantially larger sample volume, shows that these elevations are significant at a group level. We have also previously shown that some biomarkers may be elevated in patients who are clinically characterized as non-TBI orthopaedic trauma.28 Combined, these findings suggest that injuries that result in skull fracture or major extracranial injury can cause sufficient insult to the brain, or disruption to the blood brain barriers, to cause elevation of brain injury biomarkers in blood; even if such insult is undetectable clinically and by CT. This is consistent with previous studies demonstrating an increase in biomarkers in CT negative patients, who demonstrate abnormalities on magnetic resonance imaging (MRI) investigation.6,7 Such isolated biomarker elevation may also provide the diagnostic substrate of occult neurological injury in patients who experience persistent post-concussion syndrome despite the absence of intracranial findings on acute neuroimaging.29
The ability for acute biomarkers to detect the presence of isolated DAI would be of particular clinical importance due to the relatively poor sensitivity of CT in comparison to MRI.30 If biomarkers could identify those patients at higher risk of underlying DAI then it may help to inform which patients may benefit most from MRI investigation. Tau, and the neurofilament proteins NFL and neurofilament heavy, hold most promise as potential biomarkers of DAI, owing in part to their proposed axonal and dendritic origins, respectively, and prior studies suggest an increased concentration of these axonal markers in the presence of DAI. However, the evidence is inconclusive with small sample sizes, and significant confounders in analyses.31, 32, 33, 34 We found that all biomarkers studied demonstrated poor discrimination for patients with DAI, including in a selected population of patients with isolated DAI detected on CT imaging. Although it is entirely possible that the poor sensitivity of CT compared to MRI for assessing underlying DAI is likely to have led to misclassification bias. Future studies utilising the improved diagnostic ability of structural and quantitative MRI imaging techniques for DAI may allow for greater clarity to this question of whether biomarkers are able to detect occult DAI.
The majority of biomarker concentration variance on PCA was explained by two dimensions, with covariance observed in all biomarkers aside from NSE. These findings are in-keeping with previous PCA analysis of the same biomarker panel in a critical care cohort,35 but here are extended across the severity spectrum. K-means clustering found two natural clusters, which separated out more severe and mild intracranial injuries, with the former characterized by worse clinical features and CT evidence of intracranial pathology. The lack of clustering of specific pathoanatomical lesion types and different signatures of biomarker elevation are consistent with our conclusion that their levels are primarily driven by injury severity rather than injury type.
Caveats and limitations
There are several limitations to our study. 2869/4509 (64%) of patients in CENTER-TBI met our inclusion criteria, with those excluded having a slightly greater injury severity, with lower baseline GCS, less patients in the ED stratum, more patients in the Admission/ICU stratum and a greater injury severity score (ISS) (Table S2, Table S3). The patient population of CENTER-TBI, and of this analysis, is largely white and European, and this may limit generalizability of findings in non-white or non-European populations. The requirement for a clinical indication for a CT for recruitment in CENTER-TBI may also miss patients with mild injuries who did not meet the criteria for a CT scan, though still presented with a TBI. Substantial variety exists in the kinetic profile of different biomarkers,36 and the use of a 24 hour time frame for biomarker sampling in this analysis may have an impact on the diagnostic ability of biomarkers with shorter half-lives. Where possible multivariable adjustment has been made to account for time to biomarker sampling. In addition, in Fig. S9 we present the mean Z-score change of each biomarker as separated by time of collection in each of the pathology groups to demonstrate the temporal profile of each biomarker as dependent on pathology present. The use of the acute time point in this study means that the findings are most applicable to those presenting to hospital acutely following injury, and not to those with delayed presentations. Future studies assessing how the temporal profile of biomarkers corresponds to the injury and lesion progression will be key to understand if biomarkers and level changes can either predict neuroworsening and/or are reflective of ongoing injury. The CT negative population was used as a reference for the derivation of Z-score changes in biomarker concentrations. As demonstrated previously,6,7,30 the absence of CT pathology does not exclude underlying brain injury and there may therefore be an element of dampening of results with the use of CT-negative patients as the reference group. ROC curve analysis was not externally validated in a separate cohort, therefore limiting the reproducibility and generalizability of these results. Finally, although this study has an unprecedentedly large sample size in relation to TBI volumetric CT analysis, small sample sizes in the EAH subgroup analysis limit the transferability of these results.
Conclusion
In conclusion, lesion burden, as demonstrated by intracranial lesion volume, lesion number, degree of diffuse injury, and clustering in regard to injury severity, was shown to be positively associated with acute biomarker concentrations following TBI. The complex and heterogenous nature of TBI creates substantial overlap between pathoanatomical lesions, making it difficult for biomarkers to identify individual lesion types. Future studies investigating the temporal signatures of biomarker and lesion progression will be important to further understand how to interpret and use biomarkers in research and clinical practice.
Contributors
All persons who meet authorship criteria are listed as authors, and all authors certify that they have participated in the concept, design, analysis, writing, or revision of the manuscript. All authors participated in the reported analyses and interpretation of results relevant to their domain of interest. DPW, DKM and VN prepared the draft manuscript and coordinated its finalisation. DPW, FV, DKM and VN performed data extraction, data curation, statistical analyses and drafted the figures and tables. ES, ZYe and AIRM provided study design and statistical advice. Data was verified by DPW, EC and VN. MM, KK and BG performed the CNN analysis. EC, AB, KA, KKWW, HX, and ZYang conducted the biomarker assay analysis, data extraction and curation. ThvdV, JV, TD, ENK and VFJN performed the structured reporting and analyses of neuro-images. All authors have read and approved the final manuscript.
Declaration of interests
DKM reports: grants from the European Union (EU), the National Institute for Health Research UK supporting the submitted work; grants from GlaxoSmithKline Ltd and Lantmannen AB, consulting fees from Calico LLC, GlaxoSmithKline Ltd, Lantmannen AB and NeuroTrauma Sciences LLC, and personal fees from Integra Neurosciences outside the submitted work. BG has received grants from European Commission and UK Research and Innovation Engineering and Physical Sciences Research Council, during the conduct of this study; and is Scientific Advisor for Kheiron Medical Technologies, Advisor and Scientific Lead of the HeartFlow-Imperial Research Team, outside the submitted work. MM reports a ERC grant agreement and consultancy fees from Triradiate Industries, outside the submitted work. ES reports a FP7 grant from the EU supporting the submitted work, and royalties for the book “Clinical Prediction Models” published by Springer. AIRM reports a FP7 grant from the EU supporting the submitted work, and grants from NeuroTrauma Sciences, Hannelore Kohl Foundation, and IntegraLife Sciences, personal fees from PresSura Neuro as DSMB chairman outside of the submitted work. KKWW reports a FP7 grant from the EU supporting the submitted work, and as a shareholder of Gryphon Bio, Inc. VFJN reports an Academy of Medical Sciences/The Health Foundation Clinician Scientist Fellowship, during the conduct of this study; a grant from Roche Pharmaceuticals, and honorarium for talks from Neurodiem, outside the submitted work. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
The CENTER-TBI study was supported by the European Union 7th Framework Programme (EC grant 602150), with additional project support from OneMind, Hannelore Kohl Foundation, NeuroTrauma Sciences and Integra Neurosciences. Development of the BLAST-CT algorithm was supported by the European Research Council Horizon 2020 (EC grant 757173). Individual sources of funding were the Engineering and Physical Sciences Research Council (EP/R511547/1, to BG, KK), the National Institute for Health Research UK (DKM), and the Academy of Medical Sciences/The Health Foundation (VFJN).
Data sharing
Data access is conditional to an approved study proposal; there are no end dates to the availability. The CENTER-TBI data used in this study is available to researchers who provide a methodologically sound study proposal that is approved by the CENTER-TBI management committee to achieve the aims in the approved proposal. Proposals may be submitted online to CENTER-TBI (https://www.center-tbi.eu/data). A data access agreement is required, and all access must comply with regulatory restrictions imposed on the original study. No patient-identifiable information is made available, and all data have been anonymised. Study protocols and additional information for CENTER-TBI about data collection, recruitment, and participating centres is available online.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103777.
Appendix. Supplementary materials
References
- 1.Maas AIR. Prefactory comments: Promise and enigma of biomarkers for brain injury [Internet]. Vol. 3 DEC. Frontiers in Neurology. Frontiers. 2012 doi: 10.3389/fneur.2012.00173. www.frontiersin.org [cited 2021 May 17]. p. 173. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin HS, Diaz-Arrastia RR. Diagnosis, prognosis, and clinical management of mild traumatic brain injury. Vol. 14. The Lancet Neurology. Lancet Publishing Group. 2015:506–517. doi: 10.1016/S1474-4422(15)00002-2. [DOI] [PubMed] [Google Scholar]
- 3.Hall EJ, Brenner DJ. Cancer risks from diagnostic radiology: The impact of new epidemiological data. Vol. 85. British Journal of Radiology. 2012 doi: 10.1259/bjr/13739950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stiell IG, Clement CM, Rowe BH, Schull MJ, Brison R, Cass D, et al. Comparison of the Canadian CT Head Rule and the New Orleans Criteria in patients with minor head injury. JAMA. 2019;294(12) doi: 10.1001/jama.294.12.1511. http://www.ncbi.nlm.nih.gov/pubmed/16189364 [Internet]. 2005 Sep 28 [citedAug 26]1511–8Available from. [DOI] [PubMed] [Google Scholar]
- 5.Bazarian JJ, Biberthaler P, Welch RD, Lewis LM, Barzo P, Bogner-Flatz V, et al. Serum GFAP and UCH-L1 for prediction of absence of intracranial injuries on head CT (ALERT-TBI): a multicentre observational study. Lancet Neurol [Internet] 2018 Sep 1;17(9):782–789. doi: 10.1016/S1474-4422(18)30231-X. [cited 2020 Jul 7]Available from: 10.1016/ [DOI] [PubMed] [Google Scholar]
- 6.Yue JK, Yuh EL, Korley FK, Winkler EA, Sun X, Puffer RC, et al. Association between plasma GFAP concentrations and MRI abnormalities in patients with CT-negative traumatic brain injury in the TRACK-TBI cohort: a prospective multicentre study. Lancet Neurol. 2019 Oct 1;18(10):953–961. doi: 10.1016/S1474-4422(19)30282-0. [DOI] [PubMed] [Google Scholar]
- 7.Czeiter E, Amrein K, Gravesteijn BY, Lecky F, Menon DK, Mondello S, et al. Blood biomarkers on admission in acute traumatic brain injury: Relations to severity, CT findings and care path in the CENTER-TBI study. EBioMedicine [Internet]. 2020 Jun 1;56 doi: 10.1016/j.ebiom.2020.102785. http://creativecommons.org/licenses/by-nc-nd/4.0/ [cited 2020 Jul 7]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zetterberg H, Blennow K. Vol. 12. Nature Publishing Group; 2016. Fluid biomarkers for mild traumatic brain injury and related conditions; pp. 563–574. (Nature Reviews Neurology). [DOI] [PubMed] [Google Scholar]
- 9.Herrmann M, Jost S, Kutz S, Ebert AD, Kratz T, Wunderlich MT, et al. Temporal profile of release of neurobiochemical markers of brain damage after traumatic brain injury is associated with intracranial pathology as demonstrated in cranial computerized tomography. J Neurotrauma [Internet] 2000;17(2):113–122. doi: 10.1089/neu.2000.17.113. http://www.ncbi.nlm.nih.gov/pubmed/10709869 Feb [cited 2020 Feb 25]Available from. [DOI] [PubMed] [Google Scholar]
- 10.Raabe A, Grolms C, Keller M, Döhnert J, Sorge O, Seifert V. Correlation of computed tomography findings and serum brain damage markers following severe head injury. Acta Neurochir (Wien) [Internet] 1998;140(8):787–792. doi: 10.1007/s007010050180. http://www.ncbi.nlm.nih.gov/pubmed/9810445 [cited 2020 Feb 25]Available from. [DOI] [PubMed] [Google Scholar]
- 11.Carabias CS, Gomez PA, Panero I, Eiriz C, Castaño-León AM, Egea J, et al. Chitinase-3-Like Protein 1, Serum Amyloid A1, C-Reactive Protein, and Procalcitonin Are Promising Biomarkers for Intracranial Severity Assessment of Traumatic Brain Injury: Relationship with Glasgow Coma Scale and Computed Tomography Volumetry. World Neurosurg [Internet] 2020 Feb 1;134 doi: 10.1016/j.wneu.2019.09.143. https://pubmed.ncbi.nlm.nih.gov/31606503/ [cited 2020 Aug 4]e120–43Available from. [DOI] [PubMed] [Google Scholar]
- 12.Steyerberg EW, Wiegers E, Sewalt C, Buki A, Citerio G, De Keyser V, et al. Case-mix, care pathways, and outcomes in patients with traumatic brain injury in CENTER-TBI: a European prospective, multicentre, longitudinal, cohort study. Lancet Neurol. 2019 Oct 1;18(10):923–934. doi: 10.1016/S1474-4422(19)30232-7. [DOI] [PubMed] [Google Scholar]
- 13.Maas AIR, Menon DK, Steyerberg EW, Citerio G, Lecky F, Manley GT, et al. Collaborative European neurotrauma effectiveness research in traumatic brain injury (CENTER-TBI): A prospective longitudinal observational study. Neurosurgery [Internet] 2015;76(1):67–80. doi: 10.1227/NEU.0000000000000575. http://www.ncbi.nlm.nih.gov/pubmed/25525693 Jan [cited 2019 Aug 20]Available from. [DOI] [PubMed] [Google Scholar]
- 14.Ethical Approval //CENTER-TBI.eu [Internet]. [cited 2020 Jul 7]. Available from: https://www.center-tbi.eu/project/ethical-approval
- 15.Haacke EM, Duhaime AC, Gean AD, Riedy G, Wintermark M, Mukherjee P, et al. Common data elements in radiologic imaging of traumatic brain injury. J Magn Reson Imaging [Internet] 2010 Sep 1;32(3):516–543. doi: 10.1002/jmri.22259. https://onlinelibrary.wiley.com/doi/full/10.1002/jmri.22259 [cited 2021 Sep 1]Available from. [DOI] [PubMed] [Google Scholar]
- 16.Vande Vyvere T, Wilms G, Claes L, Martin Leon F, Nieboer D, Verheyden J, et al. Central versus Local Radiological Reading of Acute Computed Tomography Characteristics in Multi-Center Traumatic Brain Injury Research. J Neurotrauma [Internet] 2019 Apr 1;36(7):1080–1092. doi: 10.1089/neu.2018.6061. [cited 2020 Jul 7]Available from: https://www.liebertpub.com/doi/10.1089/neu.2018.6061. [DOI] [PubMed] [Google Scholar]
- 17.Marshall LF, Marshall SB, Klauber MR, Van Berkum Clark M, Eisenberg H, Jane JA, et al. The diagnosis of head injury requires a classification based on computed axial tomography. J Neurotrauma. 1992;9 (SUPPL. 1) [PubMed] [Google Scholar]
- 18.Monteiro M, Newcombe VFJ, Mathieu F, Adatia K, Kamnitsas K, Ferrante E, et al. Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head CT using deep learning: an algorithm development and multicentre validation study. Lancet Digit Heal [Internet] 2020 Jun 1;2(6):e314–e322. doi: 10.1016/S2589-7500(20)30085-6. t [cited 2020 Jul 7]Available fromhttps://www.thelancet.com/journals/landig/article/PIIS2589-7500 (20)30085-6/fulltex. [DOI] [PubMed] [Google Scholar]
- 19.Dunn OJ. Multiple Comparisons Using Rank Sums. Technometrics. 1964;6(3):241–252. [Google Scholar]
- 20.Skogseid IM, Nordby HK, Urdal P, Paus E, Lilleaas F. Increased serum creatine kinase BB and neuron specific enolase following head injury indicates brain damage. Acta Neurochir (Wien) [Internet] 1992;115(3–4):106–111. doi: 10.1007/BF01406367. https://pubmed.ncbi.nlm.nih.gov/1605077/ [cited 2020 Aug 4]Available from. [DOI] [PubMed] [Google Scholar]
- 21.Steiner J, Bernstein HG, Bielau H, Berndt A, Brisch R, Mawrin C, et al. Evidence for a wide extra-astrocytic distribution of S100B in human brain. BMC Neurosci [Internet] 2007;8 doi: 10.1186/1471-2202-8-2. [cited 2021 May 13]2Available from: /pmc/articles/PMC1769505/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuh E, Jain S, Piscia D, Al E. Intracranial Hemorrhage Subtypes on Computed Tomography and Adverse Outcome After Mild Traumatic Brain Injury: A TRACK-TBI Study with External Validation in CENTER-TBI. JAMA Neurol. 2021 doi: 10.1001/jamaneurol.2021.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mondello S, Papa L, Buki A, Bullock MR, Czeiter E, Tortella FC, et al. Neuronal and glial markers are differently associated with computed tomography findings and outcome in patients with severe traumatic brain injury: A case control study. Crit Care [Internet] 2011 Jul 24;15(3) doi: 10.1186/cc10286. http://www.ncbi.nlm.nih.gov/pubmed/21702960 [cited 2020 Feb 25]R156. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vande Vyvere T, De La, Rosa E, Wilms G, Nieboer D, Steyerberg E, Maas AIR, et al. Prognostic Validation of the NINDS Common Data Elements for the Radiologic Reporting of Acute Traumatic Brain Injuries: A CENTER-TBI Study. J Neurotrauma [Internet] 2020 Jun 1;37(11):1269–1282. doi: 10.1089/neu.2019.6710. https://www.liebertpub.com/doi/10.1089/neu.2019.6710 [cited 2021 Apr 19]Available from. [DOI] [PubMed] [Google Scholar]
- 25.Pandey S, Singh K, Sharma V, Pandey D, Jha RP, Rai SK, et al. A prospective pilot study on serum cleaved tau protein as a neurological marker in severe traumatic brain injury. Br J Neurosurg [Internet] 2017 May 4;31(3):356–363. doi: 10.1080/02688697.2017.1297378. http://www.ncbi.nlm.nih.gov/pubmed/28293977 [cited 2020 Feb 25]Available from. [DOI] [PubMed] [Google Scholar]
- 26.Papa L, Silvestri S, Brophy GM, Giordano P, Falk JL, Braga CF, et al. GFAP out-performs S100β in detecting traumatic intracranial lesions on computed tomography in trauma patients with mild traumatic brain injury and those with extracranial lesions. J Neurotrauma [Internet] 2014 Nov 15;31(22):1815–1822. doi: 10.1089/neu.2013.3245. http://www.ncbi.nlm.nih.gov/pubmed/24903744 [cited 2020 Feb 25]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papa L, Mittal MK, Ramirez J, Silvestri S, Giordano P, Braga CF, et al. Neuronal Biomarker Ubiquitin C-Terminal Hydrolase Detects Traumatic Intracranial Lesions on Computed Tomography in Children and Youth with Mild Traumatic Brain Injury. J Neurotrauma [Internet] 2017 Jul 1;34(13):2132–2140. doi: 10.1089/neu.2016.4806. http://www.ncbi.nlm.nih.gov/pubmed/28158951 [cited 2020 Feb 25]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posti JP, Hossain I, Takala RSK, Liedes H, Newcombe V, Outtrim J, et al. Glial Fibrillary Acidic Protein and Ubiquitin C-Terminal Hydrolase-L1 Are Not Specific Biomarkers for Mild CT-Negative Traumatic Brain Injury. J Neurotrauma [Internet] 2017 Apr 1;34(7):1427–1438. doi: 10.1089/neu.2016.4442. https://pubmed.ncbi.nlm.nih.gov/27841729/ [cited 2021 Mar 19]Available from. [DOI] [PubMed] [Google Scholar]
- 29.Polinder S, Cnossen MC, Real RGL, Covic A, Gorbunova A, Voormolen DC, et al. A Multidimensional Approach to Post-concussion Symptoms in Mild Traumatic Brain Injury [Internet] Frontiers in Neurology. Frontiers Media S.A. 2018;Vol. 9 doi: 10.3389/fneur.2018.01113. www.frontiersin.org [cited 2020 Jul 21]. p1113. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metting Z, Rödiger LA, De Keyser J, van der Naalt J. Structural and functional neuroimaging in mild-to-moderate head injury. Lancet Neurology. 2007;Vol. 6:699–710. doi: 10.1016/S1474-4422(07)70191-6. [DOI] [PubMed] [Google Scholar]
- 31.Tomita K, aki Nakada T, Oshima T, Motoshima T, Kawaguchi R, Oda S. Tau protein as a diagnostic marker for diffuse axonal injury. PLoS One [Internet] 2019 Mar 1;14(3) doi: 10.1371/journal.pone.0214381. http://www.ncbi.nlm.nih.gov/pubmed/30901365 [cited 2020 Feb 25]e0214381Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ljungqvist J, Zetterberg H, Mitsis M, Blennow K, Skoglund T. Serum Neurofilament Light Protein as a Marker for Diffuse Axonal Injury: Results from a Case Series Study. J Neurotrauma [Internet] 2017 Mar 1;34(5):1124–1127. doi: 10.1089/neu.2016.4496. http://www.ncbi.nlm.nih.gov/pubmed/27539721 [cited 2020 Feb 25]Available from. [DOI] [PubMed] [Google Scholar]
- 33.Skandsen T, Clarke G., Einarsen C., Vik A, Feyling C, Zetterberg H, et al. Levels of blood biomarkers in patients with mtbi were related to injury severity, but not to the post-concussive symptoms. J Neurotrauma. 2018;35(16):A209. [Google Scholar]
- 34.Žurek J, Bartlová L, Fedora M. Hyperphosphorylated neurofilament NF-H as a predictor of mortality after brain injury in children. Brain Inj [Internet] 2011;25(2):221–226. doi: 10.3109/02699052.2010.541895. http://www.ncbi.nlm.nih.gov/pubmed/21219092 [cited 2020 Feb 25]Available from. [DOI] [PubMed] [Google Scholar]
- 35.Thelin E, Al Nimer F, Frostell A, Zetterberg H, Blennow K, Nyström H, et al. A Serum protein biomarker panel improves outcome prediction in human traumatic brain injury. J Neurotrauma [Internet] 2019 Oct 15;36(20):2850–2862. doi: 10.1089/neu.2019.6375. https://www.liebertpub.com/doi/10.1089/neu.2019.6375 [cited 2020 Jul 29]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dadas A, Washington J, Diaz-Arrastia R, Janigro D. Vol. 14. Dove Medical Press Ltd.; 2018. Biomarkers in traumatic brain injury (TBI): A review [Internet] pp. 2989–3000.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6231511/ (Neuropsychiatric Disease and Treatment). [cited 2020 Jul 9]. pAvailable from. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.