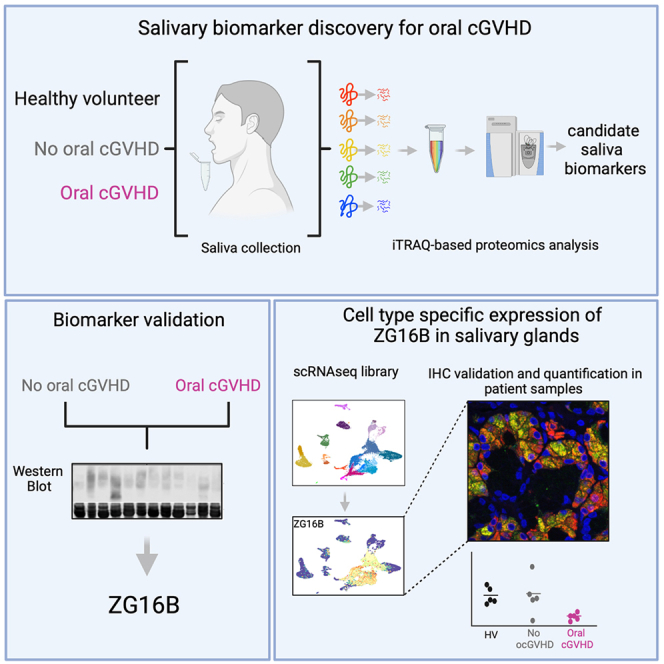
Summary
Chronic graft-versus-host disease (cGVHD) targets include the oral mucosa and salivary glands after allogeneic hematopoietic stem cell transplant (HSCT). Without incisional biopsy, no diagnostic test exists to confirm oral cGVHD. Consequently, therapy is often withheld until severe manifestations develop. This proteomic study examined saliva and human salivary gland for a biomarker profile at first onset of oral cGVHD prior to initiation of topical steroid therapy. Whole saliva collected at onset of biopsy-proven oral GVHD was assessed using liquid chromatography–coupled tandem mass spectrometry with identification of 569 proteins, of which 77 significantly changed in abundance. ZG16B, a secretory lectin protein, was reduced 2-fold in oral cGVHD saliva (p <0.05), and significantly decreased in salivary gland secretory cells affected by cGVHD. Single-cell RNA-seq analysis of healthy MSG localized ZG16B expression to two discrete acinar cell populations. Reduced ZG16B expression may indicate specific cGVHD activity and possibly general salivary gland dysfunction.
Subject areas: Pathophysiology, Proteomics, Transcriptomics
Graphical abstract
Highlights
-
•
Salivary glands are targets of chronic graft-versus-host disease (cGVHD)
-
•
Saliva and salivary glands at onset of oral cGVHD have reduced ZG16B protein
-
•
ZG16b gene expression localizes to two types of salivary gland excretory cells
-
•
ZG16B expression loss may indicate cGVHD activity or general salivary gland damage
Pathophysiology; Proteomics; Transcriptomics
Trial Registration: Clinicaltrials.gov NCT00331968, NCT00520130, and NCT01851382
Introduction
Chronic graft-versus-host disease (cGVHD) is a major cause of non-relapse morbidity and mortality that occurs after allogeneic hematopoietic stem cell transplantation (HSCT) (Copelan, 2006). This complex multi-organ disorder is the result of a break in immune tolerance marked by donor T cell recognition of a genetically different host after allogeneic HSCT (Zeiser and Blazar, 2017). Major target organs include skin, oral mucosa, liver, eyes, gut, lungs, and exocrine tissues including the salivary glands (Flowers et al., 2002). The oral mucosa and/or salivary glands are impacted in 30% to 70% of cGVHD patients (Bassim et al., 2015; Flowers et al., 2002; Martin et al., 2009). Damage to exocrine tissues such as the salivary glands is insidious, can be permanent and results in reduced production of saliva caused by progressive and silent damage to salivary glands (Mays et al., 2013). These manifestations have a severe negative impact on quality of life, nutrition, and oral health in post-transplant patients (Bassim et al., 2014; Castellarin et al., 2012; Elad et al., 2015; Meier et al., 2011).
A major challenge in clinical HSCT is the clear early diagnosis of cGVHD. Biomarkers in conjunction with clinical exam could aid this process and are being actively pursued for diagnosis of cGVHD and other post-transplant complications (Juric et al., 2016; Kariminia et al., 2016; Sarantopoulos et al., 2007; Weissinger et al., 2017; Yu et al., 2016). These studies have focused on peripheral blood, although several pilot studies have looked at organ-specific biofluids including saliva, urine, and tear fluid (Bassim et al., 2012; Cocho et al., 2016; Devic et al., 2014; Gerber-Hollbach et al., 2018; Imanguli et al., 2007; Riemens et al., 2012; Tibrewal et al., 2013; Weissinger et al., 2017). Changes in peripheral blood biomarkers are not necessarily related to organ-specific changes, including changes in the mouth where the local oral biofluid, saliva, is likely a more accurate reflection of the local condition. Saliva, produced by acinar cells and released into excretory ducts which deliver it to the mouth, is a complex oral fluid consisting of water with electrolytes, minerals, nucleic acids, mucus, and proteins including cytokines, mucins, and other glycoproteins (Wang et al., 2015). Saliva can be collected noninvasively, and its large array of protein content has already been informative for detection of oral (Hu et al., 2007, 2008; Nomura et al., 2012) and systemic diseases (Zhang et al., 2010a, 2010b). Consequently, it offers an alternative and accessible resource for biomarker discovery. To date, the few published proteomic studies screening saliva for oral cGVHD biomarkers have used pooled patient samples (Bassim et al., 2012; Chiusolo et al., 2013; Devic et al., 2014; Souza et al., 2017) with variable methods and outcomes.
Currently, diagnosis of salivary gland GVHD per the 2014 NIH consensus criteria requires incisional biopsy for histologic confirmation (Jagasia et al., 2015; Shulman et al., 2015). Consequently, this is not often done, hindering diagnosis of salivary gland GVHD and delaying assignment of proper therapy for transplant recipients until irreversible manifestations, such as glandular fibrosis, develop. Accurate, more advanced, and less invasive testing methods are necessary.
In this study, we used whole saliva to search for salivary biomarkers of oral cGVHD at disease onset and prior to initiation of topical steroid therapy. We employed isobaric tags for relative and absolute quantitation (iTRAQ) labeling strategy to allow multiplexing of peptide pools to reduce variability and liquid chromatography–coupled tandem mass spectrometry for protein identification followed by western blot (WB) verification. This identified a clear reduction in a poorly characterized salivary protein, zymogen granule 16 homolog B (ZG16B), at onset of oral cGVHD, whose gene expression localizes to two types of salivary gland excretory cells. Single-cell gene expression analysis of human labial minor salivary gland maps ZG16B, which is co-expressed with MUC7, to serous and seromucous acinar cells. Our findings point to a potentially useful salivary marker of oral cGVHD that may have larger implications as a universal indicator of salivary gland damage or dysfunction.
Results
Proteomic analysis of new-onset oral cGVHD detects potential biomarkers
To characterize global changes in the salivary proteome at onset of oral cGVHD, we analyzed individual whole unstimulated saliva samples using iTRAQ-labeled shotgun proteomics. Five-minute, unstimulated whole saliva samples were collected immediately prior to a labial minor salivary gland (MSG) biopsy in the post-transplant patients. The clinical pathology report from the MSG was used to determine salivary gland cGVHD status per 2006 NIH consensus criteria (Shulman et al., 2006) for the discovery cohort. The specific criteria for diagnosis of cGVHD in the minor salivary gland per this 2006 criteria includes periductal inflammation, damaged intralobular ducts, fibroplasia in periductal stroma, and inflammation with destruction of acinar tissue, though not all features need to be present for cGVHD diagnosis. Samples from 3 post-transplant patients with normal MSG (no oral cGVHD), from 3 patients with cGVHD in the MSG, and from 2 age- and sex-matched healthy volunteers were included in the discovery cohort. For the discovery cohort and the subsequent validation cohort, all oral cGVHD patients also met the 2006 NIH consensus criteria for clinical diagnosis of mouth cGVHD including the presence of diagnostic features of oral lichen planus-like changes, or distinctive features (oral dryness, mucoceles, ulcers, pseudomembranes, or mucosal atrophy) with alternative causes clearly ruled out (Filipovich et al., 2005). Patients were enrolled in clinical trials that included oral exams at pre-specified calendar intervals post-transplant as well as problem-focused exams as needed. Demographic details related to these and all patient samples are provided in Table S1.
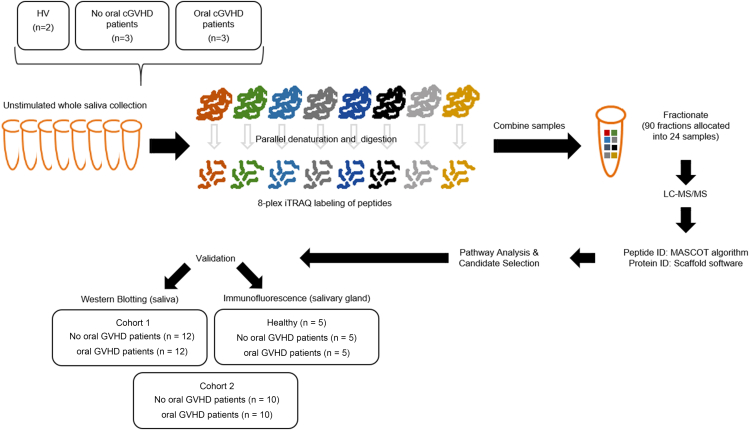
The individual sample peptide sets were labeled with one of 8 iTRAQ tags, pooled, fractionated, and then subjected to liquid chromatography–coupled tandem mass spectrometry (Figure 1).
Figure 1.
Shotgun LC-MS/MS workflow and analysis
Proteins extracted from eight saliva samples from two HVs, three post-transplanted patients without oral GVHD, and three post-transplanted patients with oral GVHD were proteolytically digested and labeled with isobaric iTRAQ tags. Peptides are then pooled at equal concentrations, fractioned using HPLC, and analyzed by mass spectrometer. Database search and bioinformatics procedures were used for protein identification, quantification, and selection of putative candidate biomarkers. Validation was performed by either WB analysis or immunofluorescence of two different cohorts of patients and five additional HVs.
The discovery dataset contained 569 confidently identified proteins. Quantitative testing between post-transplant +/-oral cGVHD samples was done using a Kruskal–Wallis test with Bonferroni correction for multiple comparisons on the log2 fold change values for iTRAQ ratios in Scaffold, resulting in the identification of 77 significantly differentially expressed proteins (DEP). Four additional filters were applied to the DEP list to select a panel of 7 candidate proteins for further validation: (1) Ingenuity Pathway Analysis (IPA, Ingenuity Systems) of list of significantly altered proteins, (2) consideration of biologic function with particular review of salivary gland and immune response proteins, (3) scrutiny of level of each protein in individual samples and the associated inter-sample variation, and (4) availability of reagents for downstream analysis. This identified calmodulin, ezrin, MMP9, PIP, alpha-1-antichymotripsin (a1ACT), and ZG16B as candidate markers for further downstream validation (Table 1).
Table 1.
Validation panel details
| Entrya | Protein namea | Gene symbolsa | p valueb |
|---|---|---|---|
| P01011 | Alpha-1-antichymotrypsin | a1ACT | 0.0001 |
| P0DP23 | Calmodulin | CALM1 | 0.0046 |
| P15311 | Ezrin | EZR | 0.0001 |
| P14780 | Matrix metalloproteinase 9 | MMP 9 | 0.0028 |
| P12273 | Prolactin-inducible protein | PIP | 0.0001 |
| Q96DA0 | Zymogen granule protein 16 homolog B | ZG16B | 0.0001 |
Entry, gene symbols, and protein names correspond to Uniprot Knowledgebase (http://www.uniprot.org/).
The p values were calculated by a Bonferroni-corrected Mann–Whitney test for non-GVHD versus GVHD saliva iTRAQ data.
MMP9, PIP, a1ACT, and Ezrin protein decrease in saliva from oral cGVHD patients
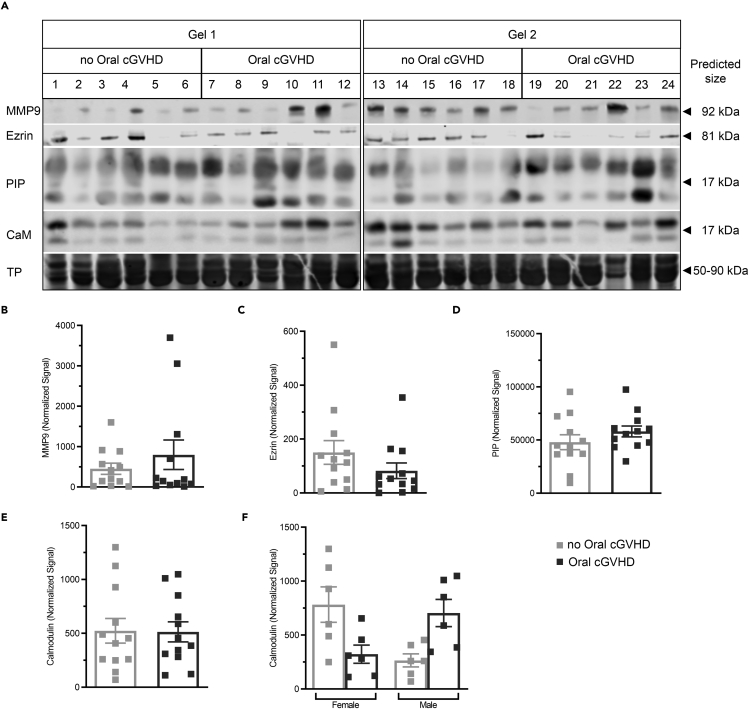
To test the discovery findings, the protein levels of calmodulin, ezrin, MMP9, PIP, alpha-1-antichymotripsin (a1ACT), and ZG16B were measured in saliva from individual post-transplant patients without and with oral cGVHD. Clinical characteristics for all cohorts are detailed in Table S1. Immunoblots for each protein measured in cohort 1 are shown in Figures 2A and 3A and for cohort 2 in Figure S1. Densiometric analysis to compare protein quantity between samples/patients was done for each of specific immunoreactive band and normalized to total protein (TP) (Figures 2B–2D). Mean values ±SEM were compared (post-HSCT no oral cGVHD vs. post-HSCT oral cGVHD). MMP9, PIP (Figures 2A and 2C, respectively), and a1ACT (Figures S1A and S1D) trended non-significantly toward elevation. Salivary ezrin was similarly decreased non-significantly in patients with oral cGVHD. Calmodulin protein levels did not change between post-HSCT patients with and without oral cGVHD (Figure 2E), though subset analysis on this cohort found a non-significant trend of inverse directionality of protein levels for sex and disease (Figure 2F, Kruskal–Wallis test with Dunn's correction for multiple comparisons); however, a larger cohort would be required for adequate power to interrogate these relationships.
Figure 2.
MMP9, PIP, a1ACT, and Ezrin protein decreases in saliva from oral cGVHD patients
(A–F) (A) Quantitative WB analysis of MMP9 (predicted MW of ∼92 kDa), ezrin (predicted MW of ∼81 kDa), PIP (predicted MW of ∼17 kDa), and calmodulin (predicted MW of ∼17 kDa) in the saliva samples from post-HSCT patients with (n = 12) or without (n = 12) oral cGVHD of cohort 1 was undertaken in 2 blots. Equal protein loading (10 μg/lane) and consistent electrotransfer of samples for the WB were confirmed by staining the entire nitrocellulose membrane using Revert Total Protein (TP) Stain immediately prior to blotting as shown by the representative proteins (range 50–90 kDa) visualized by the membrane stain in the bottom panel of (A). Full image for Revert staining of all membranes is shown in Figure S4. Densitometric analysis of (B) MMP9, (C) ezrin, and both glycosylated and nonglycosylated forms of (D) PIP and (E) calmodulin included the whole window shown and indicated no statistically significant differences between groups (p >0.05) calculated by Mann–Whitney test (unpaired, 2-tailed). Subgroup analysis of calmodulin expression (F) shows sex- and GVHD-related differences in expression that were not statistically significant (Kruskal–Wallis test with Dunn's correction for multiple comparisons). All values were normalized by total protein by lane and are plotted as mean ± SEM. Each datapoint represents one patient. See also Figures S1 and S5.
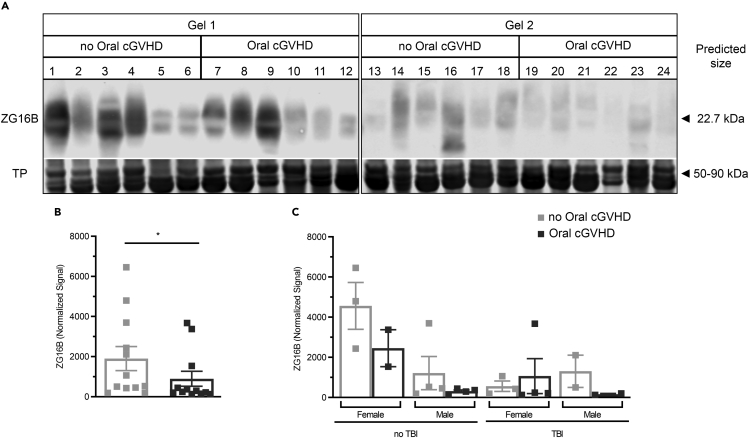
Figure 3.
Decreased ZG16B expression detected in the saliva of post-HSCT patients with oral cGVHD compared with non-affected post-HSCT patients
(A) WB for ZG16B (predicted MW of ∼22.7 kDa) in saliva samples from post-HSCT patients with (n = 12) or without (n = 12) oral cGVHD in 2 blots (top panel). TP staining (Revert, LI-COR) was used as normalization control as shown by the representative proteins (range 50–90 kDa) visualized in the bottom panel. Image for Revert staining of all membranes is shown in Figure S4
(B) Quantification of salivary ZG16B protein was performed in the whole area depicted in the blot. Values are plotted as mean ± SEM. Group-wise differences were calculated using Mann–Whitney test, unpaired, 2-tailed significance set at p≤0.05.
(C) Quantitative data comparing level of ZG16B between male and female and patients receiving or not TBI in the conditioning regimen. Each square represents 1 patient. Bars indicate mean ± SD. There were no statistical differences between groups tested at a 95% confidence level. See also Figures S1 and S5.
These 4 proteins, with the exception of calmodulin, displayed the same general tendency on WB as in the iTRAQ screen trending toward decreased secretion of specific proteins though the decrement was not statistically significant on WB (Figure 2B).
ZG16B in saliva is decreased, but post-translational modification is not affected at onset of oral cGVHD
In contrast to the other proteins tested, the relative intensity of ZG16B protein detected by WB was markedly decreased (2-fold) in post-HSCT patients with oral cGVHD (898.1 ± 371.8) compared with post-HSCT patients without oral cGVHD (1905± 598.6 p = 0.05, Mann–Whitney test, unpaired, 2-tailed: Figures 3A and 3B). Similar results were measured in both cohorts (Figure S1). Subgroup analysis split by total body irradiation (TBI) experience and sex suggested a trend toward overall reduction of ZG16B expression in saliva from patients with TBI experience; however, by ANOVA mixed-effects model analysis, this was not statistically significant. Analysis of the overall reduction by patient sex was not significant (Figure 3C).
Previous reports indicate that ZG16B contains an N-linked glycosylation site (Kanagawa et al., 2011; Kim et al., 2009). In the present study, the molecular size of the salivary ZG16B protein detected by WB was ∼30 kDa, which is higher than the predicted molecular weight (MW) of ∼22,7 kDa (Table S2 and Figure 3). To verify if the discrepancy in mobility was a result of differences in post-translational glycosylation, saliva samples from cohort 2 (Figure S2) were treated with the glycosidase PNGase F to remove N-linked oligosaccharides. The enzymatic deglycosylation produced ZG16B-specific bands with lower MW in both cohorts, irrespective of GVHD status (Figure S2). This substantiates that the N-linked carbohydrates were likely responsible for the increase in the mass observed on SDS–PAGE gel. No evident qualitative differences were observed between the N-glycosylation profile in the saliva of patients with or without oral cGVHD (Figure S2).
Taken together, the changes in ZG16B in saliva could not be attributed to differences in post-translational modification, indicating that the decrease detected is due to protein expression changes in salivary glands potentially due to cell-specific pathologies.
ZG16B in serous and seromucous acinar cells of MSG
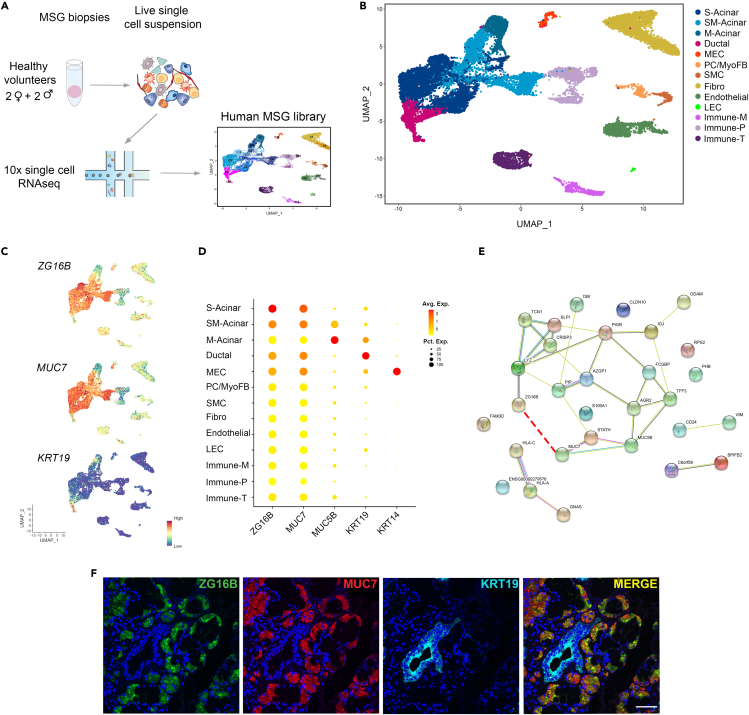
ZG16B was recently identified among the top transcribed genes expressed in the human sublingual and submandibular major salivary glands (Saitou et al., 2020). Still, the expression by specific cell populations was not investigated. To address the cell-specific source and distribution of ZG16B within the salivary gland, we performed single-cell RNA sequencing (scRNAseq) on labial minor salivary gland (MSG) specimens using the 10x Genomics platform. We analyzed the transcriptome profile of 21,402 cells derived from the combination of 4 healthy volunteer MSG, after quality control filtering (see STAR Methods, Table S2 and Figure 4A). To identify transcriptionally distinct cell populations, we analyzed the integrated data using a typical pipeline using Seurat software, including normalization, integration, dimensionality reduction, and subsequent unsupervised cell clustering (Satija et al., 2015). This analysis resulted in the identification of 20 distinct cell clusters (C0–C20) as visualized by Uniform Manifold Approximation and Projection (UMAP, Figure S3A). All clusters identified contained cells from each replicate, supporting the high reproducibility of the approach (Figure S3B). To define the identity of each cell cluster, we focused on both top DEGs and cell-specific signature genes according to published data (Figures S3C and S3D and Table S3). The gene expression patterns of these clusters identified them as acinar, ductal, myoepithelial (MECs), smooth muscle cells (SMCs), fibroblasts, endothelial, lymphatic endothelial (LECs), and immune cells (Figure S3A and Table S3).
Figure 4.
Single-cell RNA-seq and Immunofluorescence analysis of HV's MSG confirms RNA and protein expression of ZG16B in serous (S-) and seromucous (SM-) acinar cells
(A) Left, Experimental workflow of 10x single-cell RNAseq analysis of normal MSG. After dissociation, cells were captured in microfluidic oil droplets, lysed, sequenced, and analyzed.
(B) Right, UMAP embedding of 16,289 high-quality cells that were clustered into 14 populations using the Seurat algorithm.
(C) UMAP plots showing the expression of ZG16B (top), MUC7 (middle), and KRT19 across clusters.
(D) Dot plots illustrate the expression of ZG16B and known epithelial markers. The color of each plot reflects the average expression level from low (yellow) to high (red), and the size of each dot reflects the percentage of positive cells for each gene.
(E) STRING network analysis of genes shared between S-Acinar and SM-Acinar clusters shown in (C) The light blue lines represent database evidence; the purple lines represent experimental evidence; the yellow lines represent text mining evidence; and the black lines represent co-expression evidence. The red dotted line adds a new association between ZG16B and MUC7.
(F) Representative immunofluorescence staining pattern of co-expression of ZG16B (green) and MUC7 (red) in S- and SM-acinar cells. DAPI (blue) indicates nucleated cells and KRT19 (Cyan) labels ductal cells. Magnification 40x, scale bar = 50μm. See also Tables S2 and S3 and Figure S3.
Our analysis revealed heterogeneity within most cell types identified. Acinar cells were identified based on several distinct markers and revealed 6 clusters. They were grouped into three main categories; mucous acini (M-Acinar; C2), characterized by high expression of MUC5B, TFF3, and BPIFB2; serous acini comprised 4 clusters (S-Acinar; C3, C4, C8, and C13), expressing MUC7, LYZ, and PIP; or seromucous acini (SM-Acinar; C0), containing a mixed expression of both serous and mucous acini markers (Figure S3 and Table S3). C6 and C17 were characterized by enriched expression of ductal markers (KRT19, KRT7, and WFDC2) and basal ductal makers (KRT14 and KRT5), respectively. C16 displayed high expression of canonical myoepithelial cells (MECs) markers, such as TAGLN, ACTA2, and KRT14. Stromal populations were either defined as pericytes (PC; C14), expressing RGS5, NOTCH3, and PDGFRB; SMCs (C15), as identified by expression of ACTA2, MYH1, and MYL9; and fibroblasts (C1 and C10), expressing DCN, COL1A1, and FBLN1. The presence of PECAM, CDH5, and AQP1 in C5 and C19 identified these clusters as endothelial cells. An additional population which not only expresses PECAM1, but also LYVE1 and PROX1 was identified as LEC (C20). Finally, multiple immune cell populations expressing PTPRC were observed including myeloid cells (C11), T cells (C7), and plasma cells (C9, C12, and C18). Based on this, we generated curated clusters for further analysis of ZG16B in our MSG library (Figure 4B).
As observed in Figures 4C and 4D, the ZG16B gene was enriched in serous and seromucous acinar cells. Interestingly, both the expression level and distribution pattern of the ZG16B gene are similar to MUC7, a known serous acinar marker (Alos et al., 2005; Veerman et al., 2003). In accordance with the scRNAseq analysis, ZG16B protein exhibited strong co-expression with MUC7 in serous acinar cells in MSG sections of HVs. In contrast, no expression was observed in ductal cells, marked by KRT19 staining (Figure 4F).
Although ZG16B is highly expressed in saliva and salivary glands, little is known about its specific function. To gain further functional insight into the potential physical and functional interactions of ZG16B with other proteins, we used the STRING database (Szklarczyk et al., 2017) to perform a protein–protein interaction (PPI) network analysis of the 20 DEGs from S-Acinar and SM-acinar clusters in which ZG16B is highly expressed (Table S3). As expected, enriched biological processes and pathways related to these genes are mainly involved in salivary secretion and the antimicrobial humoral response (Table S3). Interestingly, the interaction of ZG16B and MUC7 is not predicted by the STRING database (Figure 4G), suggesting that this report marks the first reported co-expression of these two molecules in acinar cells at the level of both RNA and protein.
Presence of oral cGVHD is associated with loss of acinar-specific ZG16B in minor salivary glands
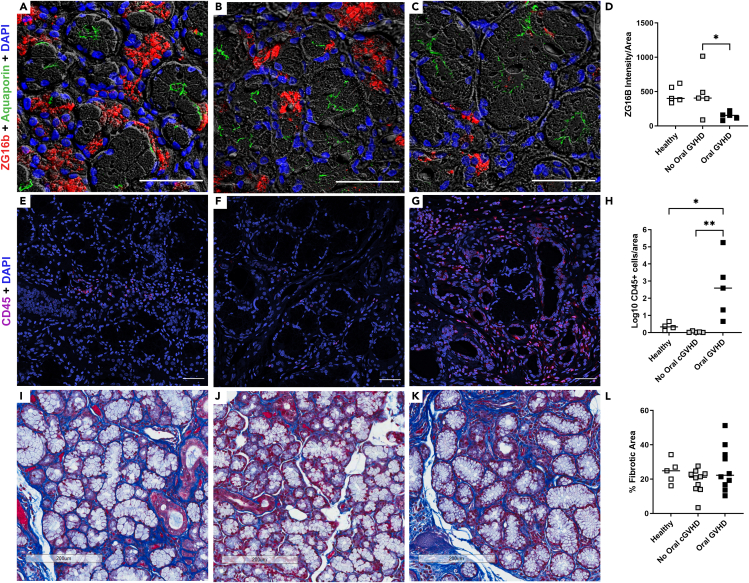
Immunohistochemistry on patient tissue and scRNAseq analysis demonstrated that ZG16B was localized either to the cytoplasm of acinar cells or to an extracellular location, suggesting likely production and release of ZG16B by acinar cells (Figure 5). To further investigate, we assessed ZG16B protein expression in FFPE MSG sections from post-HSCT patients with oral cGVHD or without oral cGVHD by IHC to confirm if the decrease in salivary levels of ZG16B is related to changes within acinar structures. Quantitative IHC identified significantly reduced ZG16B expression levels in MSG tissues of patients with oral cGVHD that are 3-fold lower than those in normal tissues or in post-transplant patients without oral cGVHD (Figures 5A–5D). A one-way ANOVA with Tukey's correction for multiple comparisons indicates significant (∗p ≤0.01) differences between the groups and specific differences between no oral cGVHD and oral cGVHD (p<05) and healthy and oral cGVHD. A representative image from each group for the figure was selected based on one that corresponded to average intensity values for that group obtained during quantification of ZG16B staining. Wider field views of these images are shown in Figure S4.
Figure 5.
Onset of oral cGVHD is associated with loss of acinar-specific ZG16B and lymphocyte infiltration in minor salivary glands
(A–C) IHC staining of ZG16B (red) in labial MSG of (A) healthy volunteers (n = 5), (B) post-HSCT patients without oral cGVHD (n = 5), and (C) post-HSCT patients with oral cGVHD (n = 5). DAPI (blue) indicates nucleated cells and aquaporin 5 (green) labels the apical membrane of acinar cells. Magnification 400x, scale bars = 50 μm.
(D) Quantification of ZG16B staining was performed as described in STAR Methods. The mean and individual values are shown for n = 5 per group. A one-way ANOVA with Tukey's correction for multiple comparisons indicates significant (∗p ≤0.01) differences between the groups and specific differences between no oral cGVHD and oral cGVHD (p<05) and healthy and oral cGVHD. (E–G) IHC staining of CD45 to mark lymphocytes (red) and nuclei (blue) in labial MSG of (E) healthy volunteers (n = 5), (F) post-HSCT patients without oral cGVHD (n = 5), and (G) post-HSCT patients with oral cGVHD (n = 5). Magnification 400x, scale bars = 100 μm.
(H) Quantification of CD45+ cells was performed as described in STAR Methods. The mean and individual values are shown for n = 4 to 5 per group. A one-way ANOVA with Tukey's correction for multiple comparisons indicates significant (∗p ≤0.01) differences between the groups and specific differences between no oral cGVHD and oral cGVHD (p<01) and healthy and oral cGVHD (p<0.05). (I–K) Representative images of Masson's trichrome-stained histological sections of (I) healthy volunteers (n = 5), (J) post-HSCT patients without oral cGVHD (n = 10), and (K) post-HSCT patients with oral cGVHD (n = 10) labial MSG from cohort 2 are shown. Blue staining marks collagen deposition.
(L) Quantification of MSG fibrosis area adjusted by total area from Masson trichrome-stained sections shown as individual values with mean. A one-way ANOVA with Tukey's correction for multiple comparisons indicate no significant (∗p ≤0.05) differences between the groups Magnification 20x, scale bars = 200 μm. See also Figure S4.
Salivary gland damage and subsequent dysfunction in cGVHD is the presumed product of an immune response direct to host tissues. Lymphocytes were quantitated using CD45 as a marker in subsequent sections of the biopsy specimens used for ZG16B IHC. Significantly, more CD45+cells were identified in cGVHD specimens versus normal tissues or post-transplant patients without oral cGVHD (Figures 5E–5H; P<0.05, one-way ANOVA with Tukey's correction for multiple comparisons). Within 14 matched cases, area-corrected ZG16B expression level negatively correlated with area-corrected CD45+ cells (Pearson correlation, r = −.50, 2-tailed p value =0.07).
Decrease of acinar markers in patients with cGVHD could be associated with loss of normal parenchyma due to fibrosis. To assess whether loss of ZG16B was related to salivary gland fibrosis, histological sections were stained with Masson's trichrome. No significant differences in fibrotic tissue replacement between healthy volunteers (24.26% ± 3.1), non-affected post-transplant patients (18.9% ± 2.21), and post-transplant patients with oral cGVHD (26.20% ± 4.07) were observed (Figures 5I–5L, one-way ANOVA with Tukey's correction for multiple comparisons). However, structural changes including disruption of acinar structures and widening of inter-acinar stromal tissues are generally observed in these cGVHD MSG (Figure 5K).
Taken together, these data support ZG16B as a protein secreted primarily from serous and seromucous acinar cells, whose reduction could be used as a potential molecular marker for evaluating not only the presence of oral cGVHD but potentially also salivary gland damage and dysfunction.
Discussion
In the present study, we sought to identify salivary markers of cGVHD onset in post-transplant patients and to characterize the source of a protein that was significantly diminished in saliva with onset of oral cGVHD. Here, we use an unbiased approach in individual patient samples (not pooled) to identify a sparsely characterized protein, zymogen granule 16b, that is reduced in the saliva of patients with biopsy-proven salivary gland cGVHD and trace its production to a specific population of serous acinar cells within the salivary gland, suggesting that a reduction in the salivary level of this protein reflects damage to salivary acinar units, which are responsible for production of saliva.
Defining organ-specific biomarkers for onset of cGVHD, among other disease milestones, is a significant challenge in the clinical care of transplant patients. The identification and use of organ-specific cGVHD biomarkers can guide the initiation of topical or systemic therapy. Clinical signs of salivary gland cGVHD include patient-reported xerostomia, or the perception of dry mouth, which can be caused by changes in the actual amount of saliva being produced or by alterations in the physical characteristics of saliva. Saliva production is affected by several classes of medications, including antidepressants and dehydration (Guggenheimer and Moore, 2003). Given the unclear etiology, xerostomia or reduced salivary flow rates alone are insufficient to diagnose oral or salivary gland cGVHD. Unstimulated whole saliva, which is a mixture of salivary gland secretions, crevicular fluid, oral epithelial cells, neutrophils, bronchoalveolar and nasal secretions along with food debris, and components of the microbiome can be passively collected in any clinical setting (Ní Ríordáin et al., 2015). Collection of gland-specific saliva is a possible method for a cleaner sampling of salivary gland secretions; however, this procedure is limited to specialty clinics and research centers and would not be practical for widespread use in the setting of clinical post-transplant care and monitoring.
In the case of post-transplant xerostomia, a labial MSG biopsy can be used to clarify the etiology. If a salivary biomarker test could replace the biopsy to clarify identification of true salivary gland damage, this would be a benefit to the patient. An ideal such test would (1) identify oral and salivary gland cGVHD before irreversible end-stage damage occurs in the organ and (2) would trigger the initiation of treatment including topical and systemic steroids and other targeted agents.
Clinical proteomic studies to identify biomarkers for cGVHD have focused on peripheral blood products and systemic disease. Although excellent studies in large cohorts have been reported, it remains challenging to identify clear peripheral biomarkers within the heterogeneous cGVHD population (Paczesny, 2018; Wolff et al., 2018). Site-specific investigations focused on the oral cavity and saliva have also been reported (Presland, 2017). Two proteomic studies have focused on whole human saliva to find biomarkers for oral cGVHD (Bassim et al., 2012; Devic et al., 2014). Both studies pooled samples in the discovery phase which tends to extinguish the variability between individuals and may explain some of the lack of reproducibility of the findings. In the first study, our group identified 102 differentially expressed salivary proteins in oral cGVHD patients using LC-MS/MS and flagged a reduction of salivary lactoperoxidase, lactotransferrin, and several members of the cysteine proteinase inhibitor family in oral cGVHD patients, suggesting impaired oral antimicrobial host immunity in these patients (Bassim et al., 2012). Devic and colleagues used iTRAQ combined with high-performance liquid chromatography/electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) to identify 82 proteins significantly changed as a result of oral cGHVD, and focused on expression of IL-1 receptor antagonist and cystatin B in pooled oral cGVHD patient saliva (Devic et al., 2014). There is little overlap in the findings between these two studies, highlighting the need for further work with well-matched patient cohorts and consistent methods. In the current study, iTRAQ labeling of individual patient samples was combined with HPLC-LC-MS/MS of carefully matched healthy volunteers and patients with and without oral cGVHD to quantitatively identify 569 salivary proteins, of which 77 were differently expressed in oral cGVHD. When 6 proteins were selected for validation using western blotting of saliva in two discreet patient cohorts, one protein, ZG16B, was present in consistently lower amounts in cGVHD saliva and was further investigated.
ZG16B, also known as pancreatic adenocarcinoma upregulated factor (PAUF), is a secretory lectin protein that is a highly expressed gene in human salivary gland tissue (Mullins et al., 2006; Saitou et al., 2020; Sasahira et al., 2017). ZG16B shares sequence homology with its paralog, ZG16p, containing a NH2-terminal signal sequence, putative related lectin domains, and an N-linked glycosylation site (Kanagawa et al., 2011; Kim et al., 2009). ZG16p is expressed in pancreatic acinar cells and digestive tract and is involved in granule content secretion during exocytosis (Cronshagen et al., 1994). Earlier studies have implicated ZG16B as an exocytosis regulator in secretory granules of lacrimal gland acinar cells (Perumal et al., 2015, 2016). Similar to data from pancreas and lacrimal gland, our study demonstrates ZG16B expression in normal acinar cells, both at the mRNA and protein levels. Specifically, ZG16B was expressed predominantly in serous and seromucous acinar cells co-expressing MUC7, suggesting that it could be involved in establishing and maintaining exocytosis of serous secretions.
Serous acinar cells produce a watery secretion composed of high levels of proteins, including amylase and antimicrobial peptides, ions, and water (de Paula et al., 2017). In addition to saliva secretion, our functional analysis pointed to a role of ZG16B in antimicrobial activity, which is in agreement with previous published data (Ambatipudi et al., 2010). Additionally, other studies report high ZB16B expression in reflex tears (Perumal et al., 2015) and decreased levels of ZG16B protein in the tear fluid of patients with multiple sclerosis (Salvisberg et al., 2014) and dry eye syndrome (Perumal et al., 2016).
In the present study, lower ZG16B protein expression tracks with salivary gland damage and lymphocyte infiltration, as the lowest amount of ZG16B quantified on IHC was present in the salivary glands with majorly disrupted acinar units. This suggests that reduction in ZG16B expression may mark general salivary gland damage, something that should be comparatively evaluated in other diseases, including radiation-induced sicca and Sjogren's syndrome, that cause structural damage within the exocrine salivary gland.
Much work remains to define what normal ranges are for ZB16B in saliva including normal variation in its expression. Proposing use of a marker that decreases, rather than elevates, is a challenge. It is unclear if this level alone will be a clinically useful salivary test for onset of cGVHD, and work in larger cohorts should be done to test both normal ranges of ZG16b and those at onset of oral cGVHD to determine the receiver operating characteristic curve for this biomarker. It could be used as part of a constellation of other markers, including reduced saliva production, altered appearance of the oral mucosa, foamy saliva, patient-reported xerostomia, and/or oral mucosal sensitivity to identify early signs of oral damage by GVHD. The present study analyzed unpooled samples from individual patients to provide better insight into biological variations, which by nature of the iTRAQ technology, limited our cohort in the discovery phase to eight, which is small and could explain some inconsistencies observed between discovery proteomics and validation in our data. However, rigorous validation was built into this study, and included WB of saliva, immunohistochemistry on tissue to identify the protein source and single-cell RNAseq to further track the cellular source of ZG16B in the human salivary gland. Further biochemical studies are needed to clarify the relationship between ZG16B, salivary gland physiology and cGVHD. Proteomic profiling of saliva in real time may be a clinically useful tool in the future to identify oral cGVHD onset. Studies in larger cohorts enrolling more affected patients need to be carried out in future.
Limitations of the study
This study demonstrated that lower ZG16B protein expression tracks with salivary gland damage in a well-defined small cohort of post-transplant patients, suggesting a relationship with oral cGVHD and general salivary gland damage. However, better understanding of the role of ZG16B in normal physiology is essential to defining its role in disease. Proposing use of a disease biomarker that decreases, rather than elevates, is a challenge and may not lead to a clinically useful salivary test for onset of cGVHD. Work in larger cohorts should be done to test both normal ranges of ZG16B and those at onset of oral cGVHD to determine the receiver operating characteristic curve for this biomarker.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Polyclonal anti-AQP5 (G19) | Santa Cruz | Cat# sc-9890, RRID:AB_2059877 |
| Rabbit Monoclonal Anti-Calmodulin (EP799Y) | Abcam | Cat# ab45689, RRID:AB_725815 |
| Rabbit Polyclonal anti-Ezrin | Cell Signaling | Cat# 3145S, RRID:AB_2100309 |
| Rat Monoclonal anti-KRT19 (TROMA-3) | Sigma-Aldrich | Cat# MABT913, RRID:AB_2892523 |
| Rabbit Monoclonal anti-MMP9 (D6O3H) | Cell Signaling | Cat# 13667, RRID:AB_2798289 |
| Rabbit Monoclonal Anti-GCDFP 15 (PIP, EP1582Y) | Abcam | Cat# ab62363, RRID:AB_940649 |
| Rabbit Polyclonal anti-MUC7 | Atlas Antibodies | Cat# HPA006411, RRID:AB_1854204 |
| Mouse Monoclonal anti-A1ACT (a1ACT; 71A1) | Thermo Fisher Scientific | Cat# LF-MA0166, RRID:AB_1954828 |
| Rabbit Polyclonal anti-CD45 | Abcam | Cat# ab10558, RRID:AB_442810 |
| Rabbit Polyclonal anti-ZG16B | Sigma-Aldrich | Cat# HPA041125-100UL; RRID:AB_10794264 |
| Alexa Fluor 488-AffiniPure Bovine Anti-Goat IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 805-545-180, RRID:AB_2340883 |
| Alexa Fluor® 594 AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 711-585-152, RRID:AB_2340621 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson ImmunoResearch Labs | Cat# 712-605-153, RRID:AB_2340694 |
| IRDye 800CW Donkey anti-Rabbit IgG antibody | Li-COR Biosciences | Cat# 926-32213, RRID:AB_621848 |
| IRDye 680RD Donkey anti-Mouse IgG antibody | Li-COR Biosciences | Cat# 926-68072, RRID:AB_10953628 |
| Biological samples | ||
| Patient samples (saliva and MSG biopsies) | NIH Clinical Center | NIH IRB-approved, Clinicaltrials.gov registered protocols: NCT01851382, NCT00092235, and NCT03602599 |
| Chemicals, peptides, and recombinant proteins | ||
| Trypsin, TPCK treated, 10-Pack | AB Sciex | Cat# 4352157 |
| Xylene Substitute | Sigma-Aldrich | Cat# A5597-1GAL |
| Normal donkey serum | Jackson ImmunoResearch Labs | Cat# 017-000-121 |
| Antibody diluent | DAKO | Cat# S0809 |
| Fluoromount-G | Thermo Fisher Scientific | Cat# 00-4958-02 |
| DAPI | Thermo Fisher Scientific | Cat# 62248 |
| Collagenase, type 2 | Worthington | Cat# LS004177 |
| Deoxyribonuclease I from bovine pancreas | Sigma-Aldrich | Cat# DN25-1G |
| PBS 1X | Quality Biological, Inc | |
| Odyssey Blocking Buffer (TBS) | Li-COR Biosciences | Cat# 927-50000 |
| 10X TBS pH 7.4 | Quality Biological | Cat# 351-086-101 |
| 4X Protein Sample Loading Buffer | Li-COR Biosciences | Cat# 928-40004 |
| Heat Inactivated Fetal Bovine Serum (FBS) | Quality Biological, Inc | Cat# 110-001-101HI |
| RPMI 1640 | GIBCO | Cat# 61870127 |
| Penicillin-Streptomycin | GIBCO | Cat# 15140122 |
| Tween 20 | Quality Biological, Inc | Cat# A611-M147-13 |
| Critical commercial assays | ||
| Pierce BCA protein assay kit | Thermo Fisher Scientific | Cat# 23227 |
| iTRAQ Reagent-8Plex Multiplex Kit | AB Sciex | Cat# 4390812 |
| iTRAQ Reagent- Multiplex Buffer Kit | AB Sciex | Cat# 4381664 |
| PNGase F | New England Biolabs | Cat# P0704L |
| Masson’s Trichrome 2000Stain Kit | StatLab | Cat# KTMTR2 EA |
| Zenon Alexa Fluor 594 Rabbit IgG labeling kit | Thermo Fisher Scientific | Cat# Z25307 |
| Deposited data | ||
| scRNAseq of healhy volunteer MSG | This paper | GEO record GSE180544 |
| Saliva Proteomics | This paper | Center for Open Science OSF project “Salivary proteomics in chronic graft-versus-host disease” |
| Salivary ZG16B decreased in oral cGVHD_raw LiCor Western blot scans | This paper | Center for Open Science OSF project “Salivary proteomics in chronic graft-versus-host disease” are available from Mendeley Data at doi:10.17632/7g5cftcbpv.1 |
| Software and algorithms | ||
| ImageJ 1.52a | (Schindelin et al., 2012; Schneider et al., 2012) | https://imagej.nih.gov/ij; RRID:SCR_003070 |
| Biorender | Biorender | https://biorender.com/; RRID:SCR_018361 |
| GraphPad Prism 8.4.1 | GraphPad | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| Proteome Discoverer (v. 1.4) | Thermo Fisher Scientific | Proteome Discoverer, RRID:SCR_014477 |
| Scaffold Q+ (v. 4.3.0) | Proteome Software | N/A |
| Protein Prophet | (Nesvizhskii et al., 2003) | N/A |
| i-Tracker (v.1.1) | (Shadforth et al., 2005) | N/A |
| Ingenuity Pathway Analysis | QIAGEN | RRID:SCR_008653 |
| Image Studio Lite (v.5.2.5) | Li-COR Biosciences | https://www.licor.com/bio/image-studio-lite/; RRID:SCR_013715 |
| Volocity 6.3 | PerkinElmer | https://www.perkinelmer.com/lab-solutions/resources/docs/BRO_VolocityBrochure_PerkinElmer.pdf; RRID:SCR_002668 |
| Adobe Photoshop CC | Adobe Systems | https://www.adobe.com/products/photoshop.html; RRID:SCR_014199 |
| Cell Ranger Software Suite (v.3.0.1) | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation; RRID:SCR_016957 |
| Seurat R Package (v.3) | (Stuart et al., 2019) | https://satijalab.org/seurat/; RRID:SCR_016341 |
| Clustree R package | (Zappia and Oshlack, 2018) | https://cran.r-project.org/web/packages/clustree/index.html ; RRID:SCR_016293 |
| Enrichr | (Chen et al., 2013) | https://maayanlab.cloud/Enrichr/; RRID: SCR_001575 |
| STRING (v.10) | (Szklarczyk et al., 2017) | https://string-db.org/; RRID:SCR_005223 |
| SoupX | (Young and Behjati, 2020) | https://github.com/constantAmateur/SoupX; RRID:SCR_019193 |
| Other | ||
| SwissProt | UniProtKB/Swiss-Prot | https://www.expasy.org/proteomics/; RID:SCR_004426 |
| Mini Gel Tank and Blot Module Set | Thermo Fisher Scientific | Cat# NW2000 |
| Revert™ 700 Total Protein Stain | Li-COR Biosciences | Cat# 926-11011 |
| NuPAGE™ 4 to 12%, Bis-Tris | Thermo Fisher Scientific | Cat# NP0323BOX |
| MACS SmartStrainers (70 μm) | Miltenyi Biotec | Cat# 130-110-916 |
| GentleMACS C tubes | Miltenyi Biotec | Cat#130-093-237 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jacqueline W. Mays (jacqueline.mays@nih.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Patients
Samples (saliva and labial MSG tissue) were obtained from adult patients at the NIH Clinical Center enrolled on IRB-approved protocols that allowed for saliva and tissue analysis (NCT01851382, NCT00092235, and NCT03602599) in accordance with the Declaration of Helsinki. Whole unstimulated 5-min saliva samples obtained at or near the time of oral cGVHD diagnosis for new-onset patients used for the described studies. Full demographic details, including age and sex and sample size, for all patient cohorts, are detailed in Table S1.
Method details
Saliva collection and preparation
Whole unstimulated saliva samples were collected per a standardized protocol in which the patient expectorated into a sterile tube on ice for 5 min at the time of oral evaluation (Bassim et al., 2012). Saliva was centrifuged at 8,600 × g for 5 min at 4°C, aliquoted and stored at −80°C. Prior to mass spectrometry or WB analysis, saliva samples were thawed and centrifuged at 2,600 × g for 15 min at 4°C to isolate the supernatant. Subsequently, protein concentration was determined by bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific), and subaliquots were prepared and stored at −80°C.
Sample preparation
Equal amounts of protein (50 μg each) from each saliva sample were precipitated with 6 volumes of chilled acetone at −20°C overnight, followed by centrifugation for 10 min at 5000 RCF. Subsequently, proteins were denatured, reduced, digested and labelled following the protocol recommended by AB Sciex. Briefly, the pellet was resuspended in 20 μL of dissolution buffer containing 1 μL of denaturant followed by 2 μL of reducing agent. Samples were incubated for 1 h at 60°C. After centrifugation, 1 μL of cysteine blocking reagent was added and samples were incubated for additional 10 min at room temperature (RT). Proteins were digested with trypsin (AB Sciex) and incubated overnight at 37°C. The peptides resulting from trypsin digestion of salivary proteins from 8 different samples were each labeled with a different isobaric tag (113–121) from an iTRAQ 8-plex kit (AB Sciex) and combined.
HPLC-LC-MS/MS
HPLC-LC-MS/MS was performed at the NHLBI Proteomics Core as previously described (Li et al., 2012). The peptides were fractionated using Basic Reverse Phase chromatography on an offline Agilent HPLC 1200. Concatenation (pooling equal interval fraction) of HPLC fractions was done prior to LC-MS/MS analysis. The peptide fractions were separated on an Eksigent nanoLC Ultra HPLC and analyzed on a Thermo Orbitrap Elite mass spectrometer.
iTRAQanalysis
The LC-MS/MS data were searched using the MASCOT algorithm within Proteome Discoverer (Thermo Electron Corp) against the human Swissprot protein database to obtain peptide and protein identifications. Scaffold Q+ (Proteome Software Inc., Portland, OR) was used to quantitate Label Based Quantitation (iTRAQ) peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 80.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 90.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. Channels were corrected by the matrix [0.000,0.000,0.929,0.0689,0.00220];[0.000,0.00940,0.930,0.0590,0.00160];[0.000,0.0188,0.931,0.0490,0.001000];[0.000,0.0282,0.932,0.0390,0.000700];[0.000600,0.0377,0.933,0.0288,0.000];[0.000900,0.0471,0.933,0.0188,0.000]; [0.00140,0.0566,0.933,0.00870,0.000];[0.000,0.000,0.000,0.000,0.000];[0.00270,0.0744,0.921,0.00180,0.000] in all samples according to the algorithm described in i-Tracker (Shadforth et al., 2005). Normalization was performed iteratively (across samples and spectra) on intensities, as described in Statistical Analysis of Relative Labeled Mass Spectrometry Data from Complex Samples Using ANOVA (Oberg et al., 2008). Medians were used for averaging. Spectra data were log-transformed, pruned of those matched to multiple proteins, and weighted by an adaptive intensity weighting algorithm. Of 27,719 spectra in the experiment at the given thresholds, 22,698 (82%) were included in quantitation. Differentially expressed proteins between groups were determined by applying Mann-Whitney Test with unadjusted significance level p <0.05 with Bonferroni correction for multiple comparisons to the Log2 Fold Change values in Scaffold.
Pathway analysis and candidate selection
The iTRAQ ratios were used to make group-wise comparisons between (1) the normal volunteer saliva and all post-transplant patients and (2) between the post-transplant patients with and without oral cGVHD. Quantitative testing between groups was done using a Kruskal-Wallis test with Bonferroni correction for multiple comparisons on the Log2 Fold Change values in Scaffold. The list of significantly altered proteins (p<0.05) between post-transplant patients was considered for validation. The list of significantly altered proteins for each of the 2 comparisons was explored with Ingenuity Pathway Analysis (IPA, Ingenuity Systems, content version 18841524, build version 313398M). Proteins with overall low or negligible expression were excluded as were proteins with heterogeneity in increased and decreased expression among samples within the same group (oral cGVHD versus no oral cGVHD). Keratin family proteins were excluded as these were considered contamination from shed oral epithelium. Four filters were then applied to the data to select a panel of 7 candidate proteins for further validation: (1) Ingenuity Pathway Analysis (IPA, Ingenuity Systems) of list of significantly altered proteins, (2) consideration of biologic function with particular review of salivary gland and immune response proteins, with the most weight given to proteins with known immunologic or salivary function (3) scrutiny of level of each protein in individual samples and the associated inter-sample variation, and (4) availability of reagents for downstream analysis including Western blotting.
Quantitative LI-COR Western blotting
Saliva samples for WB analyses were precipitated with 90% ice-cold ethanol. Total protein concentration was quantified using Pierce BCA protein assay. Equal amounts of protein (5–10μg per lane, depending on the experiment) were mixed 2-mercaptoethanol (Sigma-Aldrich) and 4x protein loading buffer (LI-COR) and heated at 70°C for 10 min. Samples were separated on NuPAGE 4 to 12% Bis-Tris gradient gels (Thermo Fisher Scientific) and then transferred to nitrocellulose membranes using the Mini Gel Tank and Blot Module Set (Thermo Fisher Scientific). Membranes were blocked in 5% nonfat dry milk in 0.1% Tween Tris-buffered saline (TBST) or Odyssey TBS Blocking Buffer (Li-COR) for 60 min at RT. Primary antibodies, MMP9 (1:500, Cell Signaling), Ezrin (1:1000, Cell Signaling), a1ACT (1:100, Thermo Fisher Scientific), PIP (1:2500; Abcam), CaM (1:500, Abcam) and ZG16B (1:1500, Sigma-Aldrich), were incubated overnight at 4°C in 0.2% Tween LI-COR blocking buffer. Membranes were washed in TBST then probed with donkey anti-mouse or donkey anti-rabbit secondary antibodies labelled with IR-Dye 680 or 800cw (LI-COR) in 0.2% Tween LI-COR blocking buffer for 90 min at RT. Following a final wash in Phosphate Buffered Saline (PBS, pH 7.4, Corning Life Sciences) for 60 min, membranes were visualized on a LI-COR Odyssey CLx Imaging System (LI-COR). Equal protein loading and consistent electrotransfer of samples for the Western blots were confirmed by staining the entire nitrocellulose membrane with Revert Total Protein Stain (TP; LI-COR), a reversible total protein stain, immediately prior to blocking the membranes. The antigen-specific bands were quantified using the Image Studio Lite Version 5.2.5 (LI-COR). Bar graphs were created in Prism from densitometric analysis data values that were normalized by total protein by lane and are plotted as mean ± SEM.
Deglycosylation with PNGase F
Ten μg of ethanol-precipitated saliva was denatured with Glycoprotein Denaturing Buffer (New England Biolabs, NEB) at 95°C for 5 min. Denatured samples cooled to 37°C, after which Glyco Buffer2, 10% NP-40 (1% final concentration) and 500 Units of PNGase F (all from NEB) were added to the sample and the reaction was incubated at 37°C for 1 h. After treatment, samples were subjected to immunoblot analysis as described above.
Immunohistochemistry
Labial MSG biopsies were obtained from post-HSCT patients and healthy volunteer controls. Immunohistochemical staining was performed on 5 μM formalin-fixed paraffin-embedded (FFPE) tissue sections. Slides were dewaxed in xylene substitute (Sigma-Aldrich), rehydrated in graded ethanol; antigen retrieval was performed using EDTA buffer (0.01M, pH 8.0) with pressurized heating for 10 min and blocked in 5% (vol/vol) donkey serum. Primary antibodies were incubated overnight at 4°C in antibody diluent (Dako) - ZG16B (1:150; Sigma-Aldrich), AQP5 (1:100; Santa Cruz), KRT19 (1:50, Sigma-Aldrich), and CD45 (1:200, Abcam) followed by incubation with appropriate secondary antibodies (1:100 in PBS, Jackson ImmunoResearch Labs). MUC7 (1:50; Atlas Antibodies) was conjugated with Alexa Fluor 594 using a Zenon labeling Kit (Thermo Fisher Scientific) and incubated for 2 h at RT. The nuclei were counterstained with DAPI (1:2000; Thermo Fisher Scientific) for 5 min at RT and mounted using Fluoromount-G mounting media (Thermo Scientific). Tissue sections were scanned using a Nikon A1R confocal microscope (fitted with a Plan Fluor 40x/1.30 oil objective) using the NIS Elements imaging software (Nikon Instruments Inc., Melville, NY). Images acquired from Nikon were visualized and processed using Fiji Is Just ImageJ ImageJ 1.52a (National Institutes of Health). Image stacks were z-projected using "Sum Slices" projection for 40x images and "Max Intensity" projection for full tissue tile-scan images in ImageJ software. CD45 and total area were quantified using ImageJ tools while ZG16B staining and total gland area were measured using Volocity 6.3 (PerkinElmer) software.
Masson's trichrome staining
Staining was done on MSG FFPE sections per manufacturer instructions with a Masson's Trichrome 2000 Stain Kit (Fisher Scientific). Slides were scanned on an Aperio CS2 Scan Scope (Leica Biosystems), and digital images were quantitated per Dahab et al (2004), with some modifications (Dahab et al., 2004). Briefly, Photoshop was used to count fibrosis-positive blue pixels in the tissue. The lasso tool was used to restrict the area of interest prior to fibrosis measurement, which allowed negation of parts of the image that were not salivary gland. The area was taken by the “record measurement” function, which contains an area subcomponent, and fibrotic area was calculated by the ratio of fibrotic area divided by the total area of the sample.
Tissue dissociation and single cell isolation
MSG tissue biopsies from four healthy volunteers were mechanically dissociated using the m_lung_01.01 program on the gentleMACS dissociator (Miltenyi Biotec). Tissues were then enzymatically digested with 610 U/mL Collagenase type 2 (Worthington) and 0.04 mg/mL DNase I (Sigma) for 40 min at 37°C with agitation. Cells were further dissociated using m_lung_02.01 program on the gentleMACS dissociator. The digested tissue was filtered through a 70 μm cell strainer (Miltenyi Biotec), washed and resuspended in RPMI 1640 (supplemented with 2% FBS, 50U/mL penicillin and 50 μg/mL streptomycin).
Single-cell RNA-seq
Single-cell RNA-seq library preparation was conducted at the NIDCR Genomics and Computational Biology Core using a Chromium Single Cell v3 method (10X Genomics) following the manufacturer's protocol. The libraries were then pooled and sequenced on four lanes on a NextSeq500 sequencer (Illumina). Read processing was performed using the 10x Genomics workflow. Briefly, the Cell Ranger Single-Cell Software Suite (v3.0.1) was used for demultiplexing, barcode assignment, and unique molecular identifier (UMI) quantification (http://software.10xgenomics.com/single-cell/overview/welcome). The reads were aligned to the GRCh38 reference genome using a pre-built annotation package obtained from the 10X Genomics website (support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/advanced/references" title="https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/advanced/references">https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/advanced/references). All four sequencing lanes per sample were merged using the ‘cellranger mkfastq’ function and UMI counts processed using the ‘cellranger count’ function.
In total, we sequenced 25,809 cells from four samples to an average depth of 20,170 reads per cell. The median of genes and UMIs detected per cells were 835 and 2,160, respectively. Secondary analysis and filtering were performed using SoupX and Seurat v3 R packages (Satija et al., 2015; Young and Behjati, 2020). Ambient RNA contamination was removed from scRNA-seq data using SoupX. Raw and filtered feature matrices were used as an input for SoupX. The contamination rate was calculated using autoEstCont() and the counts were corrected by adjustCounts() command. The following metrics were used to flag poor-quality cells using Seurat: number of genes detected, total number of UMIs, and percentage of molecules mapped to mitochondrial genes. Data for specific cells were not included in subsequent analyses when fewer than 200 genes and more than 5000 genes were detected. Cells with more than 20% of UMIs mapping to mitochondrial genes were defined as non-viable or apoptotic and were also excluded from the analyses. In addition, genes expressed in fewer than 3 cells were not included.
After filtering, Seurat v3 R package was used to perform SCTransform normalization and integration of the datasets as well as dimensionality reduction, clustering, plot and differential expression. To assign epithelial and mesenchymal cells to distinct clusters based on differentially expressed transcripts, significant dimensions were first defined by principal component analysis (PCA). Statistically significant 10 PCs were applied to graph-based clustering using Seurat's ‘FindNeighbors’ and ‘FindClusters’ function, and its resolution was 0.1 to 1.2, resulting in 12 to 33 clusters. A resolution of 0.5 was selected following clustering tree analysis using Clustree R package (Zappia and Oshlack, 2018). Cluster representations were performed by non-liner dimensional reduction using the Uniform Manifold Approximation and Projection (UMAP) algorithm. Marker identification was performed with default settings using the ‘FindAllMarkers’ function.
Pathway and gene functional analysis
Based on the DEGs from curated acinar clusters identified in our scRNAseq data, pathway analysis was performed with using the Enrichr platform (Chen et al., 2013; Table S3). STRING (v.10) (Szklarczyk et al., 2017) from the top 20 DEGs was used to construct the protein-protein interaction network.
Quantification and statistical analysis
Statistical tests as described in the Results and figure legends were carried out using GraphPad Prism version 8.4.1 software. Data were generally non-normally distributed, therefore non-parametric statistics were used for most tests to address non-normal sample distribution. The n in all tests indicates the number of unique patients or the single sample derived from an individual patient (i.e. the number of salivary glands when each is from an individual unique patient). Alpha was set at 0.05 for all tests except where correction for multiple comparisons was applied, when a smaller alpha was used, which is marked in the description of the test. Differences were considered significant at p<0.05.
Additional resources
The Collection of Saliva and/or Peripheral Blood From Healthy Volunteers for Research Trial has been registered on ClinicalTrials.gov (Identifier: NCT01851382, URL: https://clinicaltrials.gov/ct2/show/NCT01851382).
The Natural History Study of Clinical and Biological Factors Determining Outcomes in Chronic Graft-Versus-Host Disease Trial has been registered on ClinicalTrials.gov (Identifier: NCT00092235, URL: https://clinicaltrials.gov/ct2/show/NCT00092235).
The Chronic Graft-versus-Host Disease in the Oral Cavity of Patients Following Allogeneic Hematopoietic Stem Cell Transplant and Including Healthy Controls Trial has been registered on ClinicalTrials.gov (Identifier: NCT03602599, URL: https://clinicaltrials.gov/ct2/show/NCT03602599).
Acknowledgments
The authors extend gratitude to the patients and their families for their critical participation in this research. This work would not be possible without the teamwork of the NIH multidisciplinary cGVHD team and the support of the NIDCR Dental Clinic and Licia Masuch and funding from the NIDCR intramural program (ZIA DE000747). Invaluable technical expertise was provided by James Melvin (NIDCR), the NIDCR Combined Technical Research Core (ZIC DE000729), the NIDCR/NIDCD Genomics and Computational Biology Core (ZIC DC000086), the NIDCR Imaging Core (ZIC DE00075) and Steve Swatkoski and Marjan Gucek from the NHLBI Proteomics Core.
Intramural programs of the NIDCR and NCI, National Institutes of Health.
Author contributions
Conceptualization, J.W.M and A.C.C.S.; Methodology, J.W.M., A.C.C.S., and J.T.M.; Clinical, J.W.M., C.W.B., and S.Z.P.; Investigation, A.C.C.S., J.T.D., M.H.A., S.D., M.C., J.T.R., K.A. F.T.H., and J.W.M.; Data Analysis and Coding, A.C.C.S., M.H.A., and D.M.; Writing, A.C.C.S., M.H.A., and J.W.M
Declaration of interests
No competing interests from the authors.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103592.
Supplemental information
Data and code availability
Single-cell RNA-seq and Proteomics data have been deposited at GEO (GSE180544) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original Western blot images and shotgun proteomic datasets (are available from Mendeley Data at doi:10.17632/7g5cftcbpv.1) have been deposited at the Center for Open Science OSF project file for "Salivary proteomics in chronic graft-versus-host disease" and are publicly available as of the date of publication. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code. All codes were used in this study in alignment with recommendations made by authors of R packages in their respective user guide, which can be accessed at the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alos L., Lujan B., Castillo M., Nadal A., Carreras M., Caballero M., de Bolos C., Cardesa A. Expression of membrane-bound mucins (MUC1 and MUC4) and secreted mucins (MUC2, MUC5AC, MUC5B, MUC6 and MUC7) in mucoepidermoid carcinomas of salivary glands. Am. J. Surg. Pathol. 2005;29:806–813. doi: 10.1097/01.pas.0000155856.84553.c9. [DOI] [PubMed] [Google Scholar]
- Ambatipudi K.S., Hagen F.K., Delahunty C.M., Han X., Shafi R., Hryhorenko J., Gregoire S., Marquis R.E., Melvin J.E., Koo H., et al. Human common salivary protein 1 (CSP-1) promotes binding of Streptococcus mutans to experimental salivary pellicle and glucans formed on hydroxyapatite surface. J. Proteome Res. 2010;9:6605–6614. doi: 10.1021/pr100786y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassim C.W., Ambatipudi K.S., Mays J.W., Edwards D.A., Swatkoski S., Fassil H., Baird K., Gucek M., Melvin J.E., Pavletic S.Z. Quantitative salivary proteomic differences in oral chronic graft-versus-host disease. J. Clin.Immunol. 2012;32:1390–1399. doi: 10.1007/s10875-012-9738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassim C.W., Fassil H., Dobbin M., Steinberg S.M., Baird K., Cole K., Joe G., Comis L.E., Mitchell S.A., Grkovic L., et al. Malnutrition in patients with chronic GVHD. Bone Marrow Transpl. 2014;49:1300–1306. doi: 10.1038/bmt.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassim C.W., Fassil H., Mays J.W., Edwards D., Baird K., Steinberg S.M., Cowen E.W., Naik H., Datiles M., Stratton P., et al. Oral disease profiles in chronic graft versus host disease. J. Dent Res. 2015;94:547–554. doi: 10.1177/0022034515570942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin P., Stevenson K., Biasotto M., Yuan A., Woo S.B., Treister N.S. Extensive dental caries in patients with oral chronic graft-versus-host disease. Biol. Blood Marrow Transpl. 2012;18:1573–1579. doi: 10.1016/j.bbmt.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiusolo P., Giammarco S., Fanali C., Bellesi S., Metafuni E., Sica S., Iavarone F., Cabras T., Messana I., Leone G., et al. Salivary proteomic analysis and acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Biol. Blood Marrow Transpl. 2013;19:888–892. doi: 10.1016/j.bbmt.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Cocho L., Fernandez I., Calonge M., Martinez V., Gonzalez-Garcia M.J., Caballero D., Lopez-Corral L., Garcia-Vazquez C., Vazquez L., Stern M.E., et al. Biomarkers in ocular chronic graft versus host disease: Tear cytokine- and chemokine-based predictive model. Invest Ophthalmol. Vis. Sci. 2016;57:746–758. doi: 10.1167/iovs.15-18615. [DOI] [PubMed] [Google Scholar]
- Copelan E.A. Hematopoietic stem-cell transplantation. N. Engl. J. Med. 2006;354:1813–1826. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- Cronshagen U., Voland P., Kern H.F. cDNA cloning and characterization of a novel 16 kDa protein located in zymogen granules of rat pancreas and goblet cells of the gut. Eur. J.Cell Biol. 1994;65:366–377. [PubMed] [Google Scholar]
- Dahab G.M., Kheriza M.M., El-Beltagi H.M., Fouda A.M., El-Din O.A. Digital quantification of fibrosis in liver biopsy sections: Description of a new method by Photoshop software. J. Gastroenterol.Hepatol. 2004;19:78–85. doi: 10.1111/j.1440-1746.2004.03183.x. [DOI] [PubMed] [Google Scholar]
- de Paula F., Teshima T.H.N., Hsieh R., Souza M.M., Nico M.M.S., Lourenco S.V. Overview of human salivary glands: Highlights of morphology and developing processes. Anat. Rec. (Hoboken) 2017;300:1180–1188. doi: 10.1002/ar.23569. [DOI] [PubMed] [Google Scholar]
- Devic I., Shi M., Schubert M.M., Lloid M., Izutsu K.T., Pan C., Missaghi M., Morton T.H., Mancl L.A., Zhang J., et al. Proteomic analysis of saliva from patients with oral chronic graft-versus-host disease. Biol. Blood Marrow Transpl. 2014;20:1048–1055. doi: 10.1016/j.bbmt.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elad S., Raber-Durlacher J.E., Brennan M.T., Saunders D.P., Mank A.P., Zadik Y., Quinn B., Epstein J.B., Blijlevens N.M., Waltimo T., et al. Basic oral care for hematology-oncology patients and hematopoietic stem cell transplantation recipients: A position paper from the joint task force of the Multinational Association of Supportive Care in Cancer/International Society of Oral Oncology (MASCC/ISOO) and the European Society for Blood and Marrow Transplantation (EBMT) Support Care Cancer. 2015;23:223–236. doi: 10.1007/s00520-014-2378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovich A.H., Weisdorf D., Plavletic S., Socie G., Wingard R.R., Lee S.J., Martin P., Chien J., Prezepiorka D., Couriel D., et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol. Blood Marrow Transplant. 2005;11:945–955. doi: 10.1016/j.bbmt.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Flowers M.E., Parker P.M., Johnston L.J., Matos A.V., Storer B., Bensinger W.I., Storb R., Appelbaum F.R., Forman S.J., Blume K.G., et al. Comparison of chronic graft-versus-host disease after transplantation of peripheral blood stem cells versus bone marrow in allogeneic recipients: Long-term follow-up of a randomized trial. Blood. 2002;100:415–419. doi: 10.1182/blood-2002-01-0011. [DOI] [PubMed] [Google Scholar]
- Gerber-Hollbach N., Plattner K., O'Leary O.E., Jenoe P., Moes S., Drexler B., Schoetzau A., Halter J.P., Goldblum D. Tear film proteomics reveal important differences between patients with and without ocular GvHD after allogeneic hematopoietic cell transplantation. Invest Ophthalmol.Vis. Sci. 2018;59:3521–3530. doi: 10.1167/iovs.18-24433. [DOI] [PubMed] [Google Scholar]
- Guggenheimer J., Moore P.A. Xerostomia: Etiology, recognition and treatment. J. Am. Dent Assoc. 2003;134:61–69. doi: 10.14219/jada.archive.2003.0018. [DOI] [PubMed] [Google Scholar]
- Hu S., Arellano M., Boontheung P., Wang J., Zhou H., Jiang J., Elashoff D., Wei R., Loo J.A., Wong D.T. Salivary proteomics for oral cancer biomarker discovery. Clin.Cancer Res. 2008;14:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Wang J., Meijer J., Ieong S., Xie Y., Yu T., Zhou H., Henry S., Vissink A., Pijpe J., et al. Salivary proteomic and genomic biomarkers for primary Sjogren's syndrome. Arthritis Rheum. 2007;56:3588–3600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanguli M.M., Atkinson J.C., Harvey K.E., Hoehn G.T., Ryu O.H., Wu T., Kingman A., Barrett A.J., Bishop M.R., Childs R.W., et al. Changes in salivary proteome following allogeneic hematopoietic stem cell transplantation. Exp. Hematol. 2007;35:184–192. doi: 10.1016/j.exphem.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagasia M.H., Greinix H.T., Arora M., Williams K.M., Wolff D., Cowen E.W., Palmer J., Weisdorf D., Treister N.S., Cheng G.S., et al. National institutes of health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. The 2014 diagnosis and staging working group report. Biol. Blood Marrow Transpl. 2015;21:389–401 e381. doi: 10.1016/j.bbmt.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric M.K., Shevtsov M., Mozes P., Ogonek J., Crossland R.E., Dickinson A.M., Greinix H.T., Holler E., Weissinger E.M., Multhoff G. B-cell-based and soluble biomarkers in body liquids for predicting acute/chronic graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Front Immunol. 2016;7:660. doi: 10.3389/fimmu.2016.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagawa M., Satoh T., Ikeda A., Nakano Y., Yagi H., Kato K., Kojima-Aikawa K., Yamaguchi Y. Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related beta-prism fold. Biochem.Biophys. Res. Commun. 2011;404:201–205. doi: 10.1016/j.bbrc.2010.11.093. [DOI] [PubMed] [Google Scholar]
- Kariminia A., Holtan S.G., Ivison S., Rozmus J., Hebert M.J., Martin P.J., Lee S.J., Wolff D., Subrt P., Abdossamadi S., et al. Heterogeneity of chronic graft-versus-host disease biomarkers: Association with CXCL10 and CXCR3+ NK cells. Blood. 2016;127:3082–3091. doi: 10.1182/blood-2015-09-668251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.A., Lee Y., Jung D.E., Park K.H., Park J.Y., Gang J., Jeon S.B., Park E.C., Kim Y.G., Lee B., et al. Pancreatic adenocarcinoma up-regulated factor (PAUF), a novel up-regulated secretory protein in pancreatic ductal adenocarcinoma. Cancer Sci. 2009;100:828–836. doi: 10.1111/j.1349-7006.2009.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ferraris J.D., Yu D., Singh T., Izumi Y., Wang G., Gucek M., Burg M.B. Proteomic analysis of high NaCl-induced changes in abundance of nuclear proteins. Physiol. Genomics. 2012;44:1063–1071. doi: 10.1152/physiolgenomics.00068.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P.J., Storer B.E., Rowley S.D., Flowers M.E., Lee S.J., Carpenter P.A., Wingard J.R., Shaughnessy P.J., DeVetten M.P., Jagasia M., et al. Evaluation of mycophenolate mofetil for initial treatment of chronic graft-versus-host disease. Blood. 2009;113:5074–5082. doi: 10.1182/blood-2009-02-202937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays J.W., Fassil H., Edwards D.A., Pavletic S.Z., Bassim C.W. Oral chronic graft-versus-host disease: Current pathogenesis, therapy, and research. Oral Dis. 2013;19:327–346. doi: 10.1111/odi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J.K., Wolff D., Pavletic S., Greinix H., Gosau M., Bertz H., Lee S.J., Lawitschka A., Elad S. Oral chronic graft-versus-host disease: Report from the International Consensus Conference on clinical practice in cGVHD. Clin.Oral Investig. 2011;15:127–139. doi: 10.1007/s00784-010-0450-6. [DOI] [PubMed] [Google Scholar]
- Mullins J.J., Mullins L.J., Dunbar D.R., Brammar W.J., Gross K.W., Morley S.D. Identification of a human ortholog of the mouse Dcpp gene locus, encoding a novel member of the CSP-1/Dcpp salivary protein family. Physiol. Genomics. 2006;28:129–140. doi: 10.1152/physiolgenomics.00153.2006. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Ní Ríordáin R., Shirlaw P., Alajbeg I., Al Zamel G.Y., Fung P.L., Yuan A.D., McCreary C., Stoopler E.T., De Rossi S.S., Lodi G., et al. World workshop on oral medicine VI: Patient-reported outcome measures and oral mucosal disease: current status and future direction. Oral Surg. Oral Med. Oral Pathol.Oral Radiol. 2015;120:152–160.e111. doi: 10.1016/j.oooo.2015.01.023. [DOI] [PubMed] [Google Scholar]
- Nomura Y., Shimada Y., Hanada N., Numabe Y., Kamoi K., Sato T., Gomi K., Arai T., Inagaki K., Fukuda M., et al. Salivary biomarkers for predicting the progression of chronic periodontitis. Arch. Oral Biol. 2012;57:413–420. doi: 10.1016/j.archoralbio.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Oberg A.L., Mahoney D.W., Eckel-Passow J.E., Malone C.J., Wolfinger R.D., Hill E.G., Cooper L.T., Onuma O.K., Spiro C., Therneau T.M., et al. Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J. Proteome Res. 2008;7:225–233. doi: 10.1021/pr700734f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczesny S. Biomarkers for posttransplantation outcomes. Blood. 2018;131:2193–2204. doi: 10.1182/blood-2018-02-791509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal N., Funke S., Pfeiffer N., Grus F.H. Proteomics analysis of human tears from aqueous-deficient and evaporative dry eye patients. Sci. Rep. 2016;6:29629. doi: 10.1038/srep29629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal N., Funke S., Wolters D., Pfeiffer N., Grus F.H. Characterization of human reflex tear proteome reveals high expression of lacrimal proline-rich protein 4 (PRR4) Proteomics. 2015;15:3370–3381. doi: 10.1002/pmic.201400239. [DOI] [PubMed] [Google Scholar]
- Presland R.B. Application of proteomics to graft-versus-host disease: From biomarker discovery to potential clinical applications. Expert Rev. Proteomics. 2017;14:997–1006. doi: 10.1080/14789450.2017.1388166. [DOI] [PubMed] [Google Scholar]
- Riemens A., Stoyanova E., Rothova A., Kuiper J. Cytokines in tear fluid of patients with ocular graft-versus-host disease after allogeneic stem cell transplantation. Mol. Vis. 2012;18:797–802. [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Gaylord E.A., Xu E., May A.J., Neznanova L., Nathan S., Grawe A., Chang J., Ryan W., Ruhl S., et al. Functional specialization of human salivary glands and origins of proteins Intrinsic to human saliva. Cell Rep. 2020;33:108402. doi: 10.1016/j.celrep.2020.108402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvisberg C., Tajouri N., Hainard A., Burkhard P.R., Lalive P.H., Turck N. Exploring the human tear fluid: Discovery of new biomarkers in multiple sclerosis. Proteomics Clin. Appl. 2014;8:185–194. doi: 10.1002/prca.201300053. [DOI] [PubMed] [Google Scholar]
- Sarantopoulos S., Stevenson K.E., Kim H.T., Bhuiya N.S., Cutler C.S., Soiffer R.J., Antin J.H., Ritz J. High levels of B-cell activating factor in patients with active chronic graft-versus-host disease. Clin.Cancer Res. 2007;13:6107–6114. doi: 10.1158/1078-0432.CCR-07-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahira T., Kurihara M., Nishiguchi Y., Nakashima C., Kirita T., Kuniyasu H. Pancreatic adenocarcinoma up-regulated factor has oncogenic functions in oral squamous cell carcinoma. Histopathology. 2017;70:539–548. doi: 10.1111/his.13097. [DOI] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadforth I.P., Dunkley T.P., Lilley K.S., Bessant C. i-Tracker: For quantitative proteomics using iTRAQ. BMC Genomics. 2005;6:145. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman H.M., Cardona D.M., Greenson J.K., Hingorani S., Horn T., Huber E., Kreft A., Longerich T., Morton T., Myerson D., et al. NIH Consensus development project on criteria for clinical trials in chronic graft-versus-host disease: II. The 2014 pathology working group report. Biol. Blood Marrow Transpl. 2015;21:589–603. doi: 10.1016/j.bbmt.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman H.M., Kleiner D., Lee S.J., Morton T., Pavletic S.Z., Farmer E., Moresi J.M., Greenson J., Janin A., Martin P.J., et al. Histopathologic diagnosis of chronic graft-versus-host disease: National institutes of health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: II. Pathology working group report. Biol. Blood Marrow Transpl. 2006;12:31–47. doi: 10.1016/j.bbmt.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Souza M.M., de Paula F.M., Hsieh R., Macedo M.C., Corral M.A., Nunes T.B., De Paula F., Lourenco S.V. Could mucin 16 and colony-stimulating factor 2-receptor beta possible graft versus host disease biomarkers? Medical hypotheses. Med. Hypotheses. 2017;100:89–93. doi: 10.1016/j.mehy.2017.01.017. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibrewal S., Sarkar J., Jassim S.H., Gandhi S., Sonawane S., Chaudhary S., Byun Y.S., Ivanir Y., Hallak J., Horner J.H., et al. Tear fluid extracellular DNA: Diagnostic and therapeutic implications in dry eye disease. Invest Ophthalmol.Vis. Sci. 2013;54:8051–8061. doi: 10.1167/iovs.13-12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerman E.C., van den Keijbus P.A., Nazmi K., Vos W., van der Wal J.E., Bloemena E., Bolscher J.G., Amerongen A.V. Distinct localization of MUC5B glycoforms in the human salivary glands. Glycobiology. 2003;13:363–366. doi: 10.1093/glycob/cwg037. [DOI] [PubMed] [Google Scholar]
- Wang Q., Yu Q., Lin Q., Duan Y. Emerging salivary biomarkers by mass spectrometry. Clin.Chim. Acta. 2015;438:214–221. doi: 10.1016/j.cca.2014.08.037. [DOI] [PubMed] [Google Scholar]
- Weissinger E.M., Human C., Metzger J., Hambach L., Wolf D., Greinix H.T., Dickinson A.M., Mullen W., Jonigk D., Kuzmina Z., et al. The proteome pattern cGvHD_MS14 allows early and accurate prediction of chronic GvHD after allogeneic stem cell transplantation. Leukemia. 2017;31:654–662. doi: 10.1038/leu.2016.259. [DOI] [PubMed] [Google Scholar]
- Wolff D., Greinix H., Lee S.J., Gooley T., Paczesny S., Pavletic S., Hakim F., Malard F., Jagasia M., Lawitschka A., et al. Biomarkers in chronic graft-versus-host disease: Quo vadis? Bone Marrow Transpl. 2018;53:832–837. doi: 10.1038/s41409-018-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience. 2020;9:giaa151. doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Storer B.E., Kushekhar K., Abu Zaid M., Zhang Q., Gafken P.R., Ogata Y., Martin P.J., Flowers M.E., Hansen J.A., et al. Biomarker panel for chronic graft-versus-host disease. J. Clin.Oncol. 2016;34:2583–2590. doi: 10.1200/JCO.2015.65.9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia L., Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience. 2018;7:giy083. doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser R., Blazar B.R. Pathophysiology of chronic graft-versus-host disease and therapeutic targets. N. Engl. J. Med. 2017;377:2565–2579. doi: 10.1056/NEJMra1703472. [DOI] [PubMed] [Google Scholar]
- Zhang L., Farrell J.J., Zhou H., Elashoff D., Akin D., Park N.H., Chia D., Wong D.T. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology. 2010;138:949–957. doi: 10.1053/j.gastro.2009.11.010. e941-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xiao H., Karlan S., Zhou H., Gross J., Elashoff D., Akin D., Yan X., Chia D., Karlan B., et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One. 2010;5:e15573. doi: 10.1371/journal.pone.0015573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq and Proteomics data have been deposited at GEO (GSE180544) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original Western blot images and shotgun proteomic datasets (are available from Mendeley Data at doi:10.17632/7g5cftcbpv.1) have been deposited at the Center for Open Science OSF project file for "Salivary proteomics in chronic graft-versus-host disease" and are publicly available as of the date of publication. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code. All codes were used in this study in alignment with recommendations made by authors of R packages in their respective user guide, which can be accessed at the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.