Abstract
Several reports have linked activating protein 2α (AP-2α) to apoptosis, leading us to hypothesize that AP-2α is a substrate for caspases. We tested this hypothesis by examining the effects of tumor necrosis factor alpha (TNF-α) on the expression of AP-2 in breast cancer cells. Here, we provide evidence that TNF-α downregulates AP-2α and AP-2γ expression posttranscriptionally during TNF-α-induced apoptosis. Both a general caspase antagonist (zVADfmk) and a caspase 6-preferred antagonist (zVEIDfmk) inhibited TNF-α-induced apoptosis and AP-2α downregulation. In vivo tests showed that AP-2α was cleaved by caspases ahead of the DNA fragmentation phase of apoptosis. Recombinant caspase 6 cleaved AP-2α preferentially, although caspases 1 and 3 also cleaved it, albeit at 50-fold or higher concentrations. Activated caspase 6 was detected in TNF-α-treated cells, thus confirming its involvement in AP-2α cleavage. All three caspases cleaved AP-2α at asp19 of the sequence asp-arg-his-asp (DRHD19). Mutating D19 to A19 abrogated AP-2α cleavage by all three caspases. TNF-α-induced cleavage of AP-2α in vivo led to AP-2α degradation and loss of DNA-binding activity, both of which were prevented by pretreatment with zVEIDfmk. AP-2α degradation but not cleavage was inhibited in vivo by PS-431 (a proteasome antagonist), suggesting that AP-2α is degraded subsequent to cleavage by caspase 6 or caspase 6-like enzymes. Cells transfected with green fluorescent protein-tagged mutant AP-2α are resistant to TNF-α-induced apoptosis, further demonstrating the link between caspase-mediated cleavage of AP-2α and apoptosis. This is the first report to demonstrate that degradation of AP-2α is a critical event in TNF-α-induced apoptosis. Since the DRHD sequence in vertebrate AP-2 is widely conserved, its cleavage by caspases may represent an important mechanism for regulating cell survival, proliferation, differentiation, and apoptosis.
Activating protein 2 (AP-2) transcription factor, first cloned by Williams et al. (71), regulates many aspects of cell proliferation, differentiation, and death. AP-2 consists of a family of three isoforms encoded by different genes designated AP-2α, AP-2β, and AP-2γ (47, 51, 71), located on chromosomes 6p24, 6p12, and 20q13.3, respectively (24, 73). In situ hybridization showed that mouse embryos express AP-2α specifically in ectoderm-derived tissues, including craniofacial, gonad, kidney, and skin tissues. AP-2α expression in the adult is restricted to a few tissues, such as skin and kidney. AP-2α knockout mice die perinatally with severe multiple congenital defects involving the face, skull, and sensory organs (63, 77), while AP-2β knockout mice die postnatally because of polycystic kidney disease (48).
AP-2α targets many gene products, including insulin-like growth factor binding protein 5 (19), MMP-2 (55), c-erbB-2/HER2/neu (7, 29, 67), p21WAF/CIP1 (76), and keratins 5 and 14 (10). Additionally, the promoters of many genes contain AP-2 or AP-2-like consensus DNA-binding sequences, suggesting that they are also regulated by AP-2. Furthermore, overexpression of AP-2α in N-ras-transformed teratocarcinoma cells correlates with resistance to retinoic acid-induced cell differentiation (39). Similarly, high expression of AP-2α in breast cancer cells (7, 29, 67), medulloblastomas (26), and glioblastomas (6) correlates with poor prognosis. In contrast, we and others have recently demonstrated that loss of or reduction in AP-2α expression in melanoma cells correlates with increased tumorigenic and metastatic potential (1, 2, 35, 40). In addition, immunohistochemical examination of invasive breast cancer tissues indicated that AP-2α behaves like a tumor suppressor gene (25). In colorectal carcinoma, AP-2α in conjunction with p21 expression was found to correlate with recurrence-free survival (60). These observations indicate that AP-2α plays multiple roles in both normal development and tumor progression.
Based on the above observations, it is pertinent to question how AP-2 can play these roles of promoting cell death as well as cell survival. While we do not yet know the complete answer to this question, increasing evidence indicates that it does so by several mechanisms. First, in addition to the three AP-2 isoforms (α, β, and γ), there are many splice variants in mouse embryos (43, 50) and in humans. An alternatively spliced human protein, AP-2B, that differs in its C terminus and acts as a dominant negative to AP-2α has been cloned (8). Xenopus also appears to have multiple AP-2 splice variants (74).
Second, the ability of AP-2α to play multiple regulatory roles can also be seen in its response to different signal-transducing agents. For example, cyclic AMP (cAMP) (32), phorbol ester (42), retinoic acid (8), UV light, and singlet oxygen (27, 30) all stimulate AP-2α expression.
More importantly, the combinatorial power of AP-2α to regulate different genes lies in its interaction with multiple proteins. Evidence shows that the list of AP-2α interacting proteins is growing. It includes the retinoblastoma tumor suppressor protein (5), the oncoproteins c-Myc (4, 23) and simian virus 40 large T antigen (31, 45); human T-cell leukemia virus type 1 tax (46); the transcriptional coactivators PC4 (37) and poly-ADP-ribose polymerase (PARP) (38); the glucocorticoid receptor (75); and the zinc finger proteins KLF9 and KLF12 (33, 61). Simian virus 40 large T antigen inhibits AP-2α expression, while transcription factor egr-1, the glucocorticoid receptor, and AP-2α interact synergistically to activate the phenylethanolamine N-methyltransferase gene (75). The proteins c-Myc1 and c-Myc2 individually repress AP-2α trans-activation (4). Furthermore, posttranslational modifications such as cAMP-induced phosphorylation (22, 53) provide additional mechanisms by which AP-2α can increase its ability to regulate different genes.
In the past few years, several reports have linked AP-2 to programmed cell death in AP-2α (63, 77) and AP-2β (48) knockout mice. A similar link was reported for retinoic acid-induced downregulation of AP-2α in chicken embryos (64). Furthermore, AP-2α is a regulator of p21WAF-1/CIP-1 (76) and Bcl-2 (2, 48). We found such a link intriguing, and we hypothesized that AP-2α could be a regulator of programmed cell death. To address this question we studied the effect of tumor necrosis factor alpha (TNF-α), a well-known inducer of apoptosis (57), on the expression of the AP-2α gene. As we reported previously, our preliminary results showed that TNF-α downregulated the expression of AP-2α in breast cancer cells by an unknown mechanism (O. Nyormoi and M. Bar-Eli, Abstr. Proc. Am. Assoc. Cancer Res. 90th Annu. Meet., abstr. 2210, 1999). From this observation, we further hypothesized that AP-2α might be a substrate for caspases.
In this paper we provide the first evidence that (i) AP-2α is cleaved by caspase 6 (with high efficiency) and caspases 1 and 3 (with low efficiency) before the execution phase of TNF-α-induced apoptosis, (ii) all three caspases cleaved AP-2α at D19, which, when mutated to A19, abrogated cleavage of AP-2α and rendered cells resistant to TNF-α-induced apoptosis, (iii) caspase 6 cleavage of AP-2α induced loss of DNA-binding activity, and (iv) loss of DNA-binding activity and TNF-α-induced apoptosis were prevented by a caspase 6-preferred inhibitor (zVEIDfmk). Our results reveal a novel mechanism of AP-2α regulation and add weight to the hypothesis that AP-2α plays a pivotal regulatory role in determining the fate of cancer cells and that of normal cells during morphogenesis.
MATERIALS AND METHODS
Cell culture and TNF-α treatment of cells.
Sublines 9B2T and 9D3S used in this study were derived from the mammary epithelial cell line ZR-75-1 by limited dilution. The 9B2T and 9D3S sublines are morphologically distinct. The former appears epithelium like, whereas the latter is lymphoblast like. However, they have chromosome and biochemical markers in common with ZR-75-1, indicating that they arose from ZR-75-1. The parental line ZR-75-1 was obtained from Michael Tainsky (Wayne State University). Janet E. Price (University of Texas M. D. Anderson Cancer Center) provided MCF-7 and MDA-MB-435lung2 (435L2).
Cells were maintained in 100-mm plastic dishes in RPMI 1640 medium that was supplemented with 10% fetal bovine serum (HyClone, Logan, Utah), 100 U of penicillin, 100 μg of streptomycin/ml, and a 1× solution of sodium pyruvate (GIBCO-BRL Life Technologies, Grand Island, N.Y.) in a humidified chamber with 5% CO2 atmosphere. Cultures were replenished with fresh medium every 3 to 4 days and were split 1:4 whenever they reached about 90% confluence. Adherent cells were harvested by trypsinization in calcium-free phosphate-buffered saline (PBS). Nonattached and loosely attached 9D3S cells were harvested by repeatedly pipetting the medium up and down or by scraping them off the dish gently with a cell scraper.
To determine the effect of TNF-α (Alexis Corporation, San Diego, Calif.) on the expression of AP-2α and AP-2γ, breast cancer cells were seeded at 2 × 106 cells per 100-mm plastic dish overnight. TNF-α was added to a final concentration of 0.1, 1.0, 5, 10, and 20 ng/ml for 48 h.
The time course of the effect of TNF-α on AP-2α expression was determined in two stages. First, 9D3S cells were seeded at 2 × 106 cells per 100-mm plate overnight and then treated chronically with 20 ng of TNF-α/ml. Samples were taken after 0, 15, and 30 min and 1, 2, 3, 6, 12, 24, 48, and 72 h of treatment. Second, to refine the time course a similar experiment was performed with samples taken every 4 h between 24 and 48 h after treatment initiation. Two sets of samples for each time point, control cells and cells treated with TNF-α, were prepared for analysis for DNA fragmentation by flow cytometry (fluorescence-activated cell sorter [FACS]) and for Western blotting.
To inhibit the effect of TNF-α, cells were preincubated with cell-permeable caspase inhibitors (Enzyme Systems Products, Livermore, Calif.) (20 μM final concentration) for 2 h before adding TNF-α. Since caspase inhibitors are dissolved in dimethyl sulfoxide, all samples were treated with dimethyl sulfoxide, with the final concentration not exceeding 0.5%. We used these inhibitors while being fully aware that they have only relative specificity.
Protein extraction.
Subcellular fractions were prepared according to a modified method of Dignam et al. (17) in which 2 μM instead of 0.2 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma Chemicals, St. Louis, Mo.) together with other protease inhibitors was used. Briefly, cells were grown to 70 to 80% confluence, scraped off the plates, washed once with PBS and once with hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 2 μM PMSF, and 0.5 mM dithiothreitol), and chilled in hypotonic buffer in ice for 10 min. Cells were lysed in a 2-ml glass homogenizer with a loose-fitting pestle (type B). Lysates were separated into nuclei (pellet) and other components (supernatant) by centrifugation at 230 × g in a Tommy desktop centrifuge. The nuclear pellet was resuspended in a 0.2× volume of the original cell pellet of 0.02 M KCl followed by an equal volume of 1.2 M KCl and extracted by gentle agitation for 30 min at 4°C. The supernatant (containing cytoplasmic proteins and membrane fragments) and the nuclear extract were clarified at 20,000 × g at 4°C for 45 min. The protein concentration of each extract was determined using Bradford reagent according to the manufacturer's instructions (Bio-Rad, Hercules, Calif.). Extracts were divided into 50-μl aliquots and stored at −80°C until analyzed. All extraction buffers contained a cocktail of protease inhibitors including 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 10 μM iodoacetamide, 1 μM benzamidine, and 2 μM PMSF as previously described (49).
Western blot analysis.
AP-2α and AP-2γ were detected in cell extracts by Western blotting as previously described (49). Briefly, 10 to 25 μg of each sample was resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) in a Bio-Rad Mini Protein III gel apparatus, transferred to Immobilon-P membrane (Millipore Corp., Bedford, Mass.), and blocked with 3% nonfat milk in Tris-buffered saline consisting of 20 mM Tris base, 137 mM sodium chloride, and 0.05% Tween 20, pH 7.6 (TBST), for 1 h at room temperature or overnight at 4°C. This was followed by incubation with a 1:3,000 dilution of the C terminus-specific C-18 rabbit anti-AP-2 immunoglobulin G (IgG) or a 1:100 dilution of AP-2γ-specific mouse monoclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) in TBST containing 1% milk for 1 h at room temperature. Excess antibody was washed off two times for 15 min each time with 20 ml of TBST, and the membrane was incubated with a 1:5,000 dilution of horseradish peroxidase-conjugated donkey anti-rabbit or goat anti-mouse IgG (Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Membranes were washed as described above. The enhanced chemiluminescence reagents that detect bands of reactive proteins were used as described by the manufacturer (Amersham Pharmacia Biotech, Arlington Heights, Ill.). Chemiluminescent signals were captured on Kodak Bio-MAX MR X-ray films, which were obtained from Sigma.
To show whether TNF-α affects protein selectivity, we carried out Western blotting for nuclear proteins (NF-1 and PARP), cytoplasmic proteins (extracellular signal-regulated kinases 1 and 2 [ERK1 and ERK2]), and a membrane-bound protein (HER2/neu) with the relevant antibodies. Rabbit antibodies specific for NF-1, ERK1 and ERK2, and HER2/neu were purchased from Santa Cruz Biotechnology, Inc. The ERK-specific antibody recognizes both isoforms 1 and 2. PARP-specific mouse monoclonal antibody was purchased from PharMingen (San Diego, Calif.). All antibodies were used at 1:10,000 dilutions. The rest of the Western blotting procedure was as described above for AP-2.
RT-PCR.
Total RNA was extracted by the TRIZOL method according to the instructions of the manufacturer (GIBCO-BRL Life Technologies). Aliquots of 1 μg of total RNA were amplified by a semiquantitative reverse transcription-PCR (RT-PCR) procedure as described by the manufacturer (Clontech, Palo Alto, Calif.). To assess the level of AP-2α mRNA, a 1,098-bp fragment was amplified using a pair of human AP-2α-specific primers with the sequences 5′-CCT ACA GCC TGA ACC CCC TGC ACG C-3′ and 5′-TCA CTT TCT GTG CTT CTC CTC TTT G-3′, corresponding to nucleotides 278 to 303 and 1376 to 1351, respectively. To show equal loading, a 437-bp product was amplified using human glyceraldehyde-3-phosphate dehydrogenate (GAPDH)-specific primers consisting of the sequences 5′-GAG CCA CAT CGC TCA GAC-3′ and 5′-CTT CTC ATG GTT CAC ACC C-3′, corresponding to nucleotides 40 to 58 and 477 to 458, respectively. The PCR amplification program consisted of an initial denaturation at 94°C for 5 min followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s and then a final extension at 72°C for 5 min and an indefinite 4°C soak using GeneAmp PCR Systems 9700 (PE Applied Biosystems, Foster City, Calif.). The optimum amounts of primer pairs were 45 pmol for AP-2α and 5.6 pmol for GAPDH. Preliminary experiments of varying the amount of cDNA template and cycle number were performed to ensure that the analysis of the PCR products was performed during the exponential phase of amplification. PCR products were analyzed on a 1% agarose gel in 1× Tris borate EDTA (TBE) buffer consisting of 0.09 M Tris base, 0.09 M boric acid, and 0.002 M EDTA.
In vitro synthesis, cleavage, and inhibition of cleavage of AP-2α.
Radiolabeled AP-2α was synthesized using 1 μg (each) of wild-type AP-2α and mutant AP-2 M1-3 cloned into the pCDNA3.1 expression vector (Invitrogen, Carlsbad, Calif.) downstream of the T7 promoter. l-[35S]methionine (Amersham Pharmacia Biotech)-labeled AP-2 was synthesized at 30°C for 90 min using the TNT coupled rabbit reticulocyte lysate system according to the manufacturer's manual (Promega Corp., Madison, Wis.). The labeled products were stored at −80°C in 20-μl aliquots until utilized.
The radiolabeled in vitro-translated (IVT) AP-2α was incubated with recombinant human caspases as described by Chen et al. (14). The caspase reaction buffer consisted of 10 mM HEPES, 100 mM NaCl, 10 mM dithiothreitol, 1 mM EDTA, and 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), pH 7.2. The total reaction volume was 25 μl instead of 50 μl, and each cleavage reaction mixture consisted of 2 μl of radiolabeled IVT AP-2α or AP-2 M1-3 in the reaction buffer. The mixture was incubated at 37°C for 2 h. To inhibit caspase activity, 20 μM solutions of the caspase inhibitor (Enzyme Systems Products, La Jolla, Calif.) were preincubated with the tested units of recombinant caspase enzyme for 1 h before radiolabeled IVT AP-2α or AP-2 M1-3 was added. One unit is the amount of enzyme that will cleave 1 nmol of a synthetic substrate per h at 37°C. The cleavage reaction was stopped by adding 10 μl of SDS-PAGE sample buffer and incubating it at room temperature for 15 min. Samples were resolved by SDS–10% PAGE, transferred to Immobilon-P membranes, and visualized by autoradiography.
Detection of apoptosis by flow cytometry of propidium iodide-stained cells.
To demonstrate TNF-α-induced DNA fragmentation, cells were prepared as previously described (36). Briefly, cells were chronically treated with 20 ng of TNF-α/ml with or without preincubation with 20 μM caspase inhibitor. At least 106 cells were scraped off the plate, pelleted at 900 × g in a swing bucket centrifuge, washed once with PBS, and resuspended in 0.75 ml of a solution containing 50 μg of propidium iodide/ml, 3 mM sodium citrate, and 0.1% Triton X-100. Cell suspensions were incubated at 4°C for about 12 h before being analyzed for DNA fragmentation and cell cycle by an Epics Profile flow cytometer (Coulter Corp., Miami, Fla.).
Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining of apoptotic cells.
TNF-α-induced apoptosis was also assayed using the APO-BRDU kit according to the manufacturer's instructions (Phoenix Flow Systems, Inc., San Diego, Calif.). Briefly, cells were washed with PBS, fixed in formalin, permeablized in 70% ethanol, and labeled with bromodeoxyuridine triphosphate using terminal deoxynucleotidyltransferase. The labeled DNA was stained with fluorescein isothiocyanate-conjugated bromodeoxyuridine antibody. For sub-G0/G1 analysis, the same cells were stained with propidium iodide. The dually labeled cells were then analyzed by flow cytometry.
Analysis of activated caspase 6 by flow cytometry.
To detect TNF-α-induced activated caspase 6, cells were treated with 20 ng of TNF-α/ml for 48 h and then processed using an activated caspase 6 detection kit according to the manufacturer's instructions (Santa Cruz Biotechnology Inc.). Briefly, 2 × 106 cells were washed once with PBS, and 1/10 of the cells were incubated with caspase 6-preferred synthetic substrate VEID-7-amino-4-trifluoromethyl coumarin (VEID-AFC). Cells containing activated caspase 6 were detected by flow cytometry by measuring a shift in the light emitted by free AFC when released by caspase 6 cleavage of the VEID-AFC substrate.
TNF-α-induced caspase cleavage and proteasome degradation of AP-2α in vivo.
To test whether cytosolic extracts of TNF-α-treated cells could cleave and degrade IVT AP-2α, 9D3S cells were chronically treated with 20 ng of TNF-α/ml for 48 h and cytosolic extracts were made in the absence of iodoacetamide. Control cytoplasmic extract was tested at 20 μg/ml, whereas TNF-α-treated cytoplasmic extract was tested at 0.5, 1, 5, 10, and 20 μg/ml by incubation with 2 μl of l-[35S]methionine-labeled IVT AP-2α as described above. A couple of the samples were pretreated with 20 μM zVEIDfmk or zVADfmk. After 2 h of incubation at 37°C, the reaction mixtures were resolved by SDS-PAGE, transferred onto Immobilon-P membranes, and visualized by autoradiography.
To test whether AP-2α cleaved in vivo is degraded by proteasome, 9D3S cells were pretreated with 10 and 100 nM PS-341, a proteasome inhibitor (52), for 30 min. This was followed by treatment with 20 ng of TNF-α/ml for 24 h. Since the combined treatment induced more rapid cell death than treatment with each one alone, treatment had to be terminated after 24 h. Nuclear extracts were prepared and analyzed for AP-2α cleavage and degradation by Western blotting.
Mapping of caspase 6 cleavage site of AP-2α by site-directed mutagenesis.
A point mutation was introduced in the triplet codon of the aspartate residue that was the most likely to be the cleavage site for caspase 6 using a site-directed mutagenesis kit (second version) according to the manufacturer's protocol (Clontech). The selection primer was based on a unique AflII restriction site (CTTAAG) at position 998 of the pCDNA3.1 vector (Invitrogen) that is absent in the AP-2α insert. The AflII site was converted to a new NdeI site so that we could confirm the mutation at the selection primer-binding site by restriction enzyme analysis. The wild-type sequence 5′-CCG AGC TCG GTA CCA AGC TTA AGT TTA AAC CGC TGA TCA GCC-3′ was changed to the mutant sequence 5′-CCG AGC TCG GTA CCA AGC ATA TGT TTA AAC CGC TGA TCA GCC-3′, substituting A for T and T for A, respectively, as underlined. The sequence of the mutagenic primer was selected on the basis of the most likely caspase 6 cleavage site. The predicted site was at asp19, in which the wild-type sequence 5′-GAC CGT CAC GAC GGC ACC AGC-3′ was changed to 5′-GAC CGT CAC GCC GGC ACC AGC-3′. The change in the GAC codon to GCC substitutes alanine for aspartate. To confirm the mutation introduced by the mutagenic primers, the selected plasmids were sequenced using the sequencing primer 5′-GTG CTG GAT ATC TGC AGA ATT CCG GC-3′ located in the pCDNA3.1 vector upstream of the mutagenic primer binding sites in the AP-2α insert. The resulting plasmid was subsequently used as a template for in vitro synthesis of mutant AP-2α as described above.
To determine if IVT wild-type AP-2α or the mutant AP-2 M1-3 can be cleaved by caspases, 2 μl of [35S]methionine-labeled AP-2α was incubated with various units of activity of recombinant caspases 1, 3, 6, 7, 8, and 10 for 2 h at 37°C. Duplicate samples were preincubated with 20 μM zVEIDfmk followed by incubation with caspases. The reaction was stopped by the addition of 10 μl of SDS-PAGE sample buffer. Reaction products were resolved by SDS-PAGE, transferred to Immobilon membranes, air-dried, and autoradiographed.
EMSA.
To determine the DNA-binding activity of AP-2α, electrophoretic mobility shift assay (EMSA) was performed using a double-stranded oligonucleotide probe consisting of commercially prepared oligonucleotides comprising the sequence 5′-GAT CGA ACT GAC CGC CCG CCG CCC GT-3′, which corresponds to the AP-2α binding site as described by the manufacturer (Stratagene, San Diego, Calif.). Nuclear extracts from control, TNF-α-treated, and zVEIDfmk plus TNF-α-treated cells were prepared as described above. The DNA-binding reaction was initiated by incubating 5 μg of nuclear extracts, 1 μg of poly(dI-dC), and 3,000 cpm of oligonucleotide probe end labeled with [γ-32P]ATP (Amersham Pharmacia Biotech). The reaction mixture was incubated on ice for 30 min. To show that the shifted bands contained AP-2α, we incubated the reaction mixture with 1 μl of anti-AP-2α IgG or a nonspecific antibody, anti-NF-1 IgG (Santa Cruz Biotechnology, Inc.), for an additional 1 h. Furthermore, the specificity of the radiolabeled AP-2α-binding oligonucleotides was determined by competition experiments using a 100-fold excess of cold AP-2α or NF-1-binding oligonucleotides. Protein-DNA complexes were resolved on a 6% nondenaturing polyacryamide gel in TBE buffer, dried, and exposed to X-ray films overnight.
Transfection of cells with GFP-tagged wild-type and mutant AP-2α.
To test the hypothesis that caspase-mediated cleavage of AP-2 at the DRHD19 motif plays a critical role in TNF-α-induced apoptosis, we first tagged wild-type and mutant AP-2α with green fluorescent protein (GFP) at the N terminus using GFP Fusion TOPO cloning kit, version E (Invitrogen). The GFP tag was used to distinguish transfected from endogenous AP-2α. The expression plasmid constructs were transfected into 9D3S cells using lipofectamine 2000 (Promega). Stable transfectants were first selected in G418 followed by two rounds of sorting for GFP expression by FACS. The expression of the fusion protein was verified by Western blotting. The effects of TNF-α on AP-2α expression and apoptosis were analyzed by Western blotting and FACS analysis as described before.
RESULTS
TNF-α downregulates AP-2α expression in breast cancer cells.
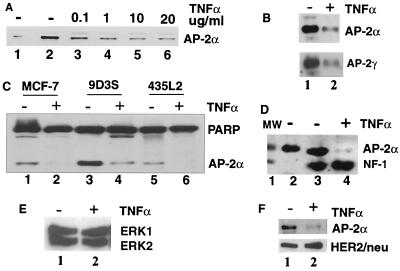
To determine whether AP-2α plays a role in cell death signaling, we investigated the effect of chronic 48-h TNF-α treatment on the expression of AP-2α in breast cancer cells. TNF-α treatment significantly reduced the expression of AP-2α in a concentration-dependent manner in 9D3S breast cancer cells (Fig. 1A, lanes 3 to 6) compared to that in the control (lane 2). Whereas 10 to 20 ng of TNF-α/ml is commonly used to induce biological effects in cells, 0.1 ng/ml was sufficient to induce a detectable reduction in the expression of AP-2α in the 9D3S cells (Fig. 1A, lane 3).
FIG. 1.
Western blot analysis of the effect of TNF-α on gene expression in breast cancer cells. (A) Treatment of 9D3S cells with increasing concentrations (0.1 to 20 ng/ml) of TNF-α for 48 h downregulated the expression of AP-2α in a concentration-dependent manner (lanes 3 through 6). Recombinant AP-2α for positive control and untreated control cells are shown in lanes 1 and 2, respectively. (B) Downregulation of AP-2α (upper panel, lane 2) and AP-2γ (lower panel, lane 2) in 9D3S cells. Corresponding controls are shown in lane 1 of each panel. (C) The expression of AP-2α and PARP in different cell lines after 48 h of treatment with TNF-α. AP-2α expression in control MCF-7, 9D3S, and 435L2 cells are shown in lanes 1, 3, and 5, respectively. In contrast, TNF-α treatment selectively downregulated AP-2α but not PARP expression (lanes 2, 4, and 6, respectively). Furthermore, NF-1 (panel D, lane 4), ERK1 and ERK2 (panel E, lane 2), and HER2/neu (panel F, lane 2) were not affected by TNF-α treatment. Lane 1 of panel D is a molecular weight (MW) marker.
Since there are three AP-2 isoforms (α, β, and γ) and since AP-2γ also has been shown to be upregulated in breast cancer specimens and cell lines, we wanted to determine whether they are all affected by TNF-α treatment. Two identical transfer membranes were blotted with rabbit anti-AP-2α and mouse monoclonal anti-AP-2γ, respectively, to avoid the possibility of cross-reaction, since the molecular weights of the two AP-2 isoforms are very close. Results show that both AP-2α (Fig. 1B, upper panel, lane 2) and AP-2γ (lower panel, lane 2) were downregulated by TNF-α treatment compared to the untreated control cells (lane 1 in both upper and lower panels). We did not have a good AP-2β-specific antibody to determine if its expression is also affected by TNF-α. The studies described herein will concentrate on the fate of AP-2α during TNF-α-induced apoptosis.
To determine whether the effect of TNF-α on AP-2α expression was not just limited to a particular cell line, we screened additional breast cancer cell lines, including MCF-7, 9D3S, and 435L2, all of which expressed variable levels of AP-2α (Fig. 1C, lanes 1, 3, and 5). TNF-α treatment for 48 h reduced AP-2α expression in all of them (Fig. 1C, lanes 2, 4, and 6), indicating that downregulation of AP-2α expression is not confined to one breast cancer cell line.
To determine whether TNF-α downregulates AP-2α selectively, we analyzed the expression of additional proteins, including nuclear, cytoplasmic, and membrane-bound proteins. Western blot analysis shows that the expressions of PARP (Fig. 1C, lanes 2, 4, and 6), NF-1 (Fig. 1D, lane 4), ERK1 and ERK2 (Fig. 1E, lane 2), and HER2/neu (Fig. 1F, lane 2) were not affected by TNF-α treatment. Taken together, these observations indicate that TNF-α treatment selectively downregulated the expression of AP-2α and AP-2γ during apoptosis.
TNF-α treatment did not affect AP-2α transcription.
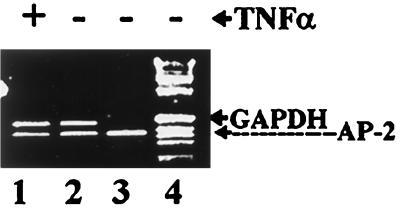
To determine whether TNF-α downregulates AP-2α expression at the transcriptional level, we compared the amount of AP-2α mRNA in control and TNF-α-treated 9D3S cells by semiquantitative RT-PCR. GAPDH was used for equal loading (Fig. 2, lanes 1 and 2). Thirty cycles were determined to be in the exponential phase of the PCR amplification. Results show that there was no significant difference between AP-2α transcripts in control (Fig. 2, lane 2) and treated (Fig. 2, lane 1) cells. Our analysis of AP-2α transcripts in cytoplasmic and nuclear preparations also demonstrated that TNF-α treatment did not affect their subcellular distribution (data not shown). Collectively, these results indicate that TNF-α does not downregulate AP-2α expression at the level of transcription, translation, or subcellular distribution of its transcripts.
FIG. 2.
RT-PCR analysis of AP-2α transcripts in total RNA preparations of control and TNF-α-treated 9D3S cells. Lane 4, 1-kb ladder markers; lane 3, AP-2α transcripts of plasmid DNA for positive control; lane 2, control without TNF-α treatment; lane 1, TNF-α-treated cells. GAPDH in lanes 1 and 2 was used to ensure equal loading.
TNF-α-induced downregulation of AP-2α correlates with TNF-α-induced apoptosis.
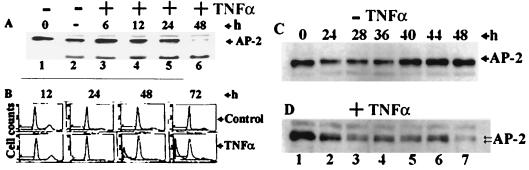
We next performed time course experiments for periods of 15 min to 72 h after treatment initiation to determine how early TNF-α begins to downregulate the expression of AP-2α and whether AP-2α downregulation correlates with cell death. Short-term treatment for 15 min to 2 h (data not shown) or 6 to 24 h had no significant effect on AP-2α expression (Fig. 3A, lanes 3 to 5). However, long-term treatment for 48 h reduced AP-2α expression to almost undetectable levels (Fig. 3A, lane 6). The lower nonspecific band in Fig. 3A was used to show equal loading and to demonstrate that not all nuclear proteins are downregulated following TNF-α treatment (see also Fig. 1C through F). Cell death rose to about 33% by 48 h and reached 40% at 72 h (Fig. 3B). Since TNF-α-induced apoptosis was noted at 48 h, we wanted to determine whether AP-2α degradation precedes apoptosis. To that end we analyzed the effect of TNF-α treatment at the crucial time points between 24 and 48 h following treatment. AP-2α expression in control, untreated cells during this period remained unchanged (Fig. 3C). In contrast, a twofold reduction in AP-2α expression was noted at 28 h after treatment and peaked at 48 h in a 20-fold reduction (Fig. 3D, lanes 3 and 7; compare panel C with panel D for each corresponding time point). Apoptosis was detected in 16.4, 33.6, and 40% of the cells treated with TNF-α for 24, 48, and 72 h, respectively (Fig. 3B). TNF-α-induced apoptosis at 48 h of treatment was also confirmed by the TUNEL assay (Fig. 7). These observations demonstrated that TNF-α induced AP-2α downregulation and apoptosis, with the former being an early event.
FIG. 3.
Time course of the effect of TNF-α treatment on AP-2α expression in 9D3S cells. (A) Western blot analysis of cells treated with 20 ng of TNF-α/ml for 6 to 48 h. Lane 1, recombinant AP-2α for positive control; lane 2, control cells without treatment; lanes 3 through 6, cells analyzed for AP-2α after 6, 12, 24, and 48 h of TNF-α treatment. Bands of unknown cross-reacting proteins above and below the AP-2α band show equal loading. (B) FACS analysis of DNA fragmentation in control cells after 12 to 72 h of incubation (upper panel) and cells treated with 20 ng of TNF-α/ml for 12 to 72 h (lower panel). Apoptosis was found in 16.4, 33.6, and 40% of cells after TNF-α treatment for 24, 48, and 72 h, respectively. (C) AP-2α expression in control 9D3S cells monitored every 4 h between 0 and 48 h. No significant difference in AP-2α expression was observed. (D) AP-2α expression in 9D3S cells treated with 20 ng of TNF-α/ml and monitored in the same way as the control cells in panel C. AP-2α expression was reduced by about twofold at times between 28 and 44 h of treatment. At 48 h the level of AP-2α was reduced by about 20-fold.
FIG. 7.
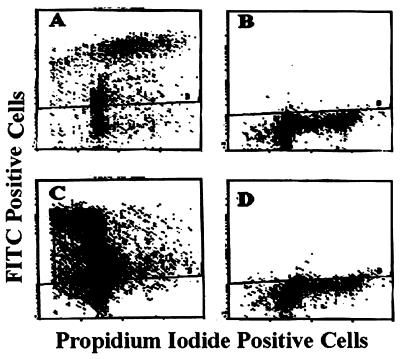
Detection of apoptosis by the TUNEL assay. Cells (9D3S) were treated with TNF-α for 48 h and assayed for nicked DNA by the APO-BRDU assay. In this FACS analysis, cells positive for nicked DNA are above the cross line, whereas negative cells are below it. (A) Positive control cells supplied by the manufacturer; (B) untreated control 9D3S cells; (C) TNF-α-treated cells; (D) cells preincubated with 20 μM zVEIDfmk for 2 h followed by TNF-α (20 ng/ml) treatment.
Caspase inhibitors prevent the effect of TNF-α on AP-2α expression and cell death.
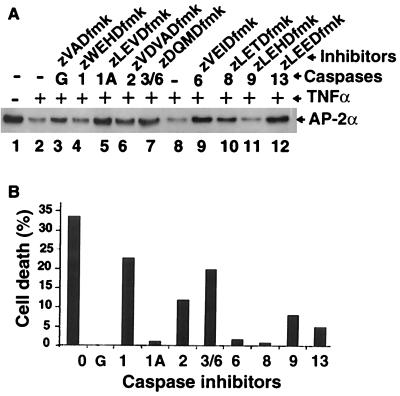
The correlation between cell death and AP-2α downregulation suggested that caspases might be involved, particularly since TNF-α is well known for activating caspases during apoptosis (12, 15, 20). We investigated this possibility by treating cells with several caspase inhibitors, being cognizant of the limitation of these reagents due to some degree of nonspecificity and promiscuity of caspases (44, 65). Antagonists of caspases 1 (zLEVDfmk), 6 (zVEIDfmk), and 13 (zLEEDfmk) inhibited both TNF-α downregulation of AP-2α expression (Fig. 4A, lanes 5, 9, and 12, respectively) and TNF-α-induced cell death (Fig. 4B, columns 1A, 6, and 13, respectively). Another group, including the general caspase inhibitor (zVADfmk) and inhibitors of caspases 2 (zVDVADfmk), 3 and 6 (zVQMDfmk), and 8 (zLETDfmk), moderately prevented the downregulation of AP-2α expression (Fig. 4A, lanes 3, 6, 7, and 10). However, the same inhibitors had different effects on TNF-α-induced cell death. The general caspase inhibitor and the inhibitor of caspase 8 prevented cell death almost completely (Fig. 4B, columns G and 8), whereas the inhibitor of caspase 2 and caspases 3 and 6 had only a moderate effect on apoptosis (Fig. 4B, columns 2 and 3/6). Finally, a third group consisting of a different caspase 1 inhibitor (zWEHDfmk) and caspase 9 inhibitor (zLEHDfmk) had no significant effect on the downregulation of AP-2α expression (Fig. 4A, lanes 4 and 11, respectively). The former had minimal effect, whereas the latter had some effect on cell death (Fig. 4B, columns 1 and 9, respectively). Although some degree of the nonspecificity of caspases and their inhibitors was involved, these results collectively show that some caspase antagonists inhibited TNF-α-induced AP-2α downregulation and cell death, suggesting that TNF-α-induced AP-2α downregulation and apoptosis are associated with degradation of AP-2α initiated possibly by caspases 1, 6, 8, and 13.
FIG. 4.
Inhibition of TNF-α-induced AP-2α downregulation and cell death by caspase inhibitors. (A) Western blot analysis of nuclear extracts. Lane 1, untreated control 9D3S cells; lane 2, TNF-α-treated 9D3S cells without caspase inhibitors. Inhibitors of caspases 1 (zLEVDfmk) (lane 5), 6 (lane 9), and 13 (lane 12) effectively prevented the downregulation of AP-2α expression by TNF-α. Inhibitors of caspases 2 (lane 6), 3 and 6 (lane 7), 8 (lane 10), and the general caspase inhibitor zVADfmk (lane 3) moderately prevented the effect of TNF-α on AP-2α expression. In contrast, the zWEHDfmk inhibitor of caspase 1 (lane 4) and the inhibitor of caspase 9 (lane 11) did not prevent the effect of TNF-α on AP-2α expression. Caspases 1 and 1A refer to the same caspase pretreated with different caspase 1 inhibitors. (B) FACS analysis of apoptosis by DNA fragmentation. TNF-α-treated 9D3S cells without inhibitors (column 0), cells pretreated with the general caspase inhibitor (column G), and inhibitors of caspases 1 (zLEVDfmk) (column 1A), 6 (column 6), and 8 (column 8) effectively blocked TNF-α-induced apoptosis. Inhibitors of caspases 13 (column 13), 9 (column 9), 2 (column 2), 3 and 6 (column 3/6), and a different caspase 1 inhibitor (zWEHDfmk) (column 1) were less effective in inhibiting TNF-α-induced cell death.
In vitro cleavage of AP-2α by recombinant caspases.
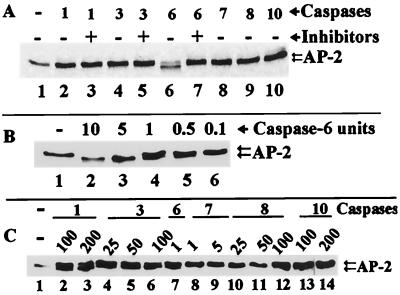
To determine more specifically which caspases can cleave AP-2α directly, we utilized IVT AP-2α as a substrate for recombinant caspases 1, 3, 6, 7, 8, and 10. Of the tested caspases, only caspase 6 cleaved AP-2α at 1 U per reaction, yielding a truncated fragment (Fig. 5A, lane 6), and this cleavage was inhibited by zVEIDfmk (Fig. 5A, lane 7). In contrast, caspase 7 did not cleave AP-2α at 1 U per reaction (Fig. 5A, lane 8). Caspases 1, 3, 8, and 10 did not cleave AP-2α even at a higher enzyme concentration of 12.5 U per reaction (Fig. 5A, lanes 2, 4, 9, and 10).
FIG. 5.
In vitro cleavage of IVT AP-2α by recombinant caspases. (A) Caspases 1, 3, 7, 8, and 10 (lanes 2, 4, 8, 9, and 10) did not cleave AP-2α. Inhibitors also had no effect on caspases 1 and 3 (lanes 3 and 5). Caspase 6 cleaved AP-2α (lane 6), and this cleavage was inhibited by zVEIDfmk (lane 7). Lane 1 represents control IVT AP-2α. (B) Titration of caspase 6 for AP-2α cleavage activity. Lane 1, IVT AP-2α control; lanes 2 through 6, decreasing concentrations of caspase 6 show complete cleavage (lane 2) and undetectable cleavage (lane 6). (C) Titration of caspases 1, 3, 7, 8, and 10. Lane 1, IVT AP-2α control; lane 7, caspase 6 cleavage at 1 U as a positive control; lanes 2 and 3, caspase 1 cleavage of AP-2α at 100 and 200 U; lanes 4 through 6, caspase 3 cleavage of AP-2α at 25, 50, and 100 U. Caspases 7 at 1 and 5 U (lanes 8 and 9), caspase 8 at 25, 50, and 100 U (lanes 10 through 12), and caspase 10 at 100 and 200 U (lanes 13 and 14) did not cleave AP-2α.
Furthermore, caspase 6 was titrated to determine the lowest amount of enzyme that can cleave AP-2α. We found that 0.5 U of caspase 6 per reaction was sufficient to cleave AP-2α (Fig. 5B, lane 5), and the cleavage occurred in a dose-dependent manner (Fig. 5B, lanes 2 to 6). Titration of caspases 1, 3, 7, 8, and 10 showed that more caspase 1 (100 to 200 U) and caspase 3 (25 to 100 U) were required per reaction to cleave AP-2α (Fig. 5C, lanes 2 and 3 and 4 to 6, respectively). Caspase 7 at 1 to 5 U (Fig. 5C, lanes 8 and 9), caspase 8 at 25 to 100 U (lanes 10 to 12), and caspase 10 at 100 to 200 U (lanes 13 and 14) per reaction did not cleave AP-2α. Recombinant caspase 13 was not available for testing to determine if it could cleave AP-2α directly. Collectively, the recombinant caspase studies suggest that AP-2α is preferentially cleaved by caspase 6.
TNF-α induction of apoptosis correlates with caspase 6 activation.
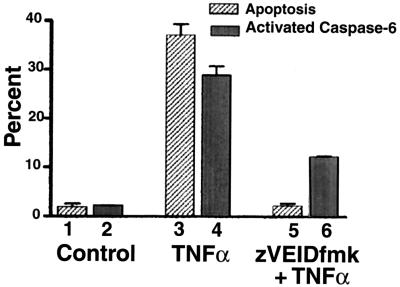
We next analyzed whether caspase 6 is indeed activated during TNF-α-induced apoptosis. Programmed cell death was detected by propidium iodide staining, and caspase 6 activation was detected by fluorescent light emitted by coumarin released from a synthetic caspase 6 substrate (see Materials and Methods). Results of flow cytometric analysis shown in Fig. 6 indicate that after 48 h, control cells had background levels of dead cells (Fig. 6, column 1) and activated caspase 6 (Fig. 6, column 2). In contrast, the percentages of dead cells and activated caspase 6-positive cells were elevated with TNF-α treatment alone (Fig. 6, columns 3 and 4, respectively), and both were inhibited by zVEIDfmk (Fig. 6, columns 5 and 6, respectively). A nonparametric paired t test analysis shows that columns 5 and 6 are significantly different from columns 3 and 4 (P < 0.05).
FIG. 6.
TNF-α induction of apoptosis correlates with caspase 6 activation. Breast cancer cells (9D3S) were treated with 20 ng of TNF-α/ml for 48 h. To block the effect of TNF-α, cells were preincubated for 2 h with zVEIDfmk. The averages of two experiments are shown. The percentages of apoptotic cells analyzed by FACS are shown in stripes, and the percentages of activated caspase 6-positive cells are shown in gray.
The correlation between TNF-α-induced apoptosis and caspase 6 activation was further confirmed by TUNEL and zVEIDfmk inhibition assays. TUNEL-positive control cells supplied by the manufacturer are shown in Fig. 7A. Untreated 9D3S cells were only 0.7% TUNEL positive (Fig. 7B). In contrast, TNF-α-treated cells were 79.8% TUNEL positive (Fig. 7C), and pretreatment with zVEIDfmk reduced the level of TUNEL-positive cells to 19% (Fig. 7D). We conclude from these observations that TNF-α-induced apoptosis is correlated with caspase 6 activation and that both events can be prevented by a caspase 6-preferred inhibitor, further suggesting that activation of caspase 6 might be a necessary but insufficient step in TNF-α-induced apoptosis (see below).
In vitro degradation of AP-2α by in vivo-activated caspase 6-like enzymes.
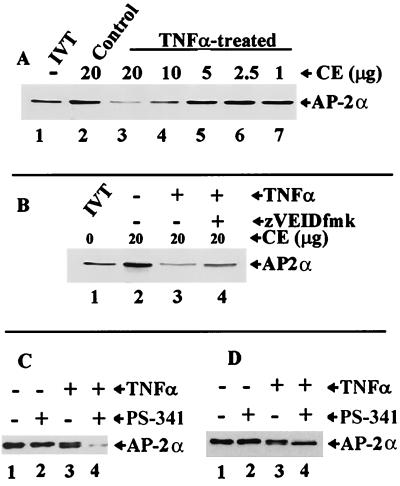
In the next set of experiments we wanted to confirm the involvement of caspase 6 in AP-2α degradation in vivo. To that end, cytoplasmic extracts from TNF-α-treated cells were used to determine if they contain activated caspases that could cleave AP-2α in vitro. As shown in Fig. 8A, 20 μg of cytoplasmic extracts from control cells did not cleave or degrade IVT AP-2α (Fig. 8A, lane 2), whereas extracts from TNF-α-treated cells did, and they did so in a concentration-dependent manner (Fig. 8A, lanes 3 to 7). As noted before, unlike AP-2α cleavage in vitro by recombinant caspase 1 (Fig. 5C, lanes 2 and 3), caspase 3 (Fig. 5C, lanes 5 and 6), and caspase 6 (Fig. 5A, lane 6), which yielded a truncated fragment, in vivo cleavage appears to render AP-2α susceptible to degradation by other proteases, possibly through the ubiquitin-proteasome pathway. AP-2α degradation by in vivo-activated enzymes (Fig. 8B, lane 3) can be partially inhibited by zVEIDfmk (Fig. 8B, lane 4). In addition, zVADfmk can block in vitro cleavage of AP-2α by cytoplasmic extracts of TNF-α-treated cells (data not shown). From these observations, we conclude that caspase 6 or a caspase 6-like enzyme is involved in the initiation of AP-2 degradation (possibly by proteasome) during apoptosis.
FIG. 8.
In vitro degradation of AP-2α by in vivo-activated caspase 6-like enzymes. (A) Caspase 6-like enzyme activity in cytoplasmic extracts from TNF-α-treated cells. Lane 1, undigested IVT AP-2α with only half the amounts of AP-2α that are loaded in lanes 2 through 7. Lane 2, AP-2α treated with control cytoplasmic extract (CE). Lanes 3 through 7, decreasing amounts of CE of TNF-α-treated cells show concentration-dependent digestion of AP-2α. (B) Inhibition of IVT AP-2α cleavage by a caspase 6 antagonist. Lane 1, control IVT AP-2α; lane 2, IVT AP-2α-treated cells with control CE; lane 3, AP-2α incubated with CE of TNF-α-treated cells showing significant digestion of AP-2α; lane 4, IVT AP-2α incubated with CE from cells preincubated with 20 μM zVEIDfmk followed by 20 ng of TNF-α/ml. (C) Effect of 10 nM proteasome inhibitor (PS-341) on AP-2α cleavage and degradation. Lane 1, Western blot analysis of AP-2α in nuclear extracts of control cells; lane 2, cells treated with 10 nM PS-341 alone; lane 3, cells treated with 20 ng of TNF-α/ml alone; lane 4, cells treated with a combination of 10 nM PS-341 and 20 ng of TNF-α/ml. (D) Effect of 100 nM PS-341 on AP-2α degradation. Lane 1, Western blot analysis of AP-2α in nuclear extracts of control cells; lane 2, cells treated with 100 nM PS-341 alone; lane 3, cells treated with 20 ng of TNF-α/ml alone; lane 4, cells treated with a combination of 100 nM PS-341 and 20 ng of TNF-α/ml.
The hypothesis that cleavage of AP-2α by caspase 6 or caspase 6-like enzymes renders it susceptible to subsequent degradation by proteasome was tested using a proteasome inhibitor, PS-341. We observed that 10 nM PS-341 alone had no effect on AP-2α (Fig. 8C, lane 2). At 20 ng/ml, TNF-α alone induced partial cleavage of AP-2α, seen as doublet bands in Fig. 8C (lane 3). In contrast, combination treatment of cells with 10 nM PS-341 followed by 20 ng of TNF-α/ml induced almost complete cleavage and degradation of AP-2α (Fig. 8C, lane 4). The increased effect of combined TNF-α plus PS-341 treatment on AP-2α cleavage and degradation (Fig. 8C, lane 4) suggests a synergistic mode of action. When 100 nM PS-341 was used alone, there was, again, no significant effect on AP-2α (Fig. 8D, lane 2). However, in combination with 20 ng of TNF-α/ml, PS-341 completely inhibited degradation of AP-2α without affecting cleavage, as only the truncated fragment was observed (Fig. 8D, lane 4). From these observations we conclude that caspase-mediated AP-2α cleavage in vivo led to degradation of AP-2α by PS-341-inhibitable proteasome.
Mapping of the cleavage site of caspase 6 in AP-2α.
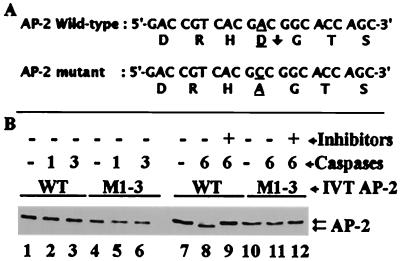
On the basis of the size of the cleavage product, detection by a C terminus-specific antibody, and caspase 6-preferred residues at the cleavage site (66), we predicted that the most likely cleavage site was at the N terminus at D19 of AP-2α (Fig. 9A, arrow). To ascertain if our prediction is correct, we changed the predicted cleavage sequence by site-directed mutagenesis. The mutation was confirmed by sequencing the DNA fragment containing the mutation. In vitro cleavage data presented in Fig. 9 show that 12.5 U of caspases 1 and 3 per reaction did not cleave wild-type or mutant AP-2α (Fig. 9B, lanes 2 and 3 and lanes 5 and 6, respectively). In another experiment, 100 U of caspase 1 and 50 U of caspase 3 did not cleave mutant AP-2α (data not shown) but did cleave wild-type AP-2α (Fig. 5C). In contrast, 1 U of caspase 6 cleaved wild-type AP-2α (Fig. 9B, lane 8) but not mutant AP-2 M1-3 (Fig. 9B, lane 11). Moreover, zVEIDfmk blocked the cleavage of wild-type AP-2α (Fig. 9B, lane 9). Taken together, the data show that in human breast cancer cells, caspase 6 (high efficiency) and caspases 1 and 3 (low efficiency) cleave AP-2α at D19 of the sequence DRHD located in the first region of exon 2, 7 amino acids downstream from the junction of exons 1 and 2.
FIG. 9.
Mapping of caspase 6 cleavage site by site-directed mutagenesis. (A) Nucleotide and amino acid sequences of wild-type (WT) AP-2α and mutant AP-2 M1-3. A downward arrow after D19 shows the caspase 6 cleavage site of WT AP-2α. A single base substitution of C for A in the GAC codon (underlined) mutated D19 to A19. (B) Control WT IVT AP-2α (lanes 1 and 7) and control mutant IVT AP-2 M1-3 (lanes 3 and 10). Using 12.5 U per reaction, caspases 1 and 3 did not cleave WT IVT AP-2α (lanes 2 and 3) or mutant AP-2 M1-3 (lanes 5 and 6). Caspase 6 cleaved only WT AP-2α at 1 U per reaction (lane 8), and a caspase 6-preferred inhibitor abrogated this cleavage (lane 9). In contrast, caspase 6 did not cleave mutant AP-2 M1-3 (lane 11), indicating that D19 is the cleavage site for caspase 6.
TNF-α-mediated loss of AP-2α DNA-binding activity.
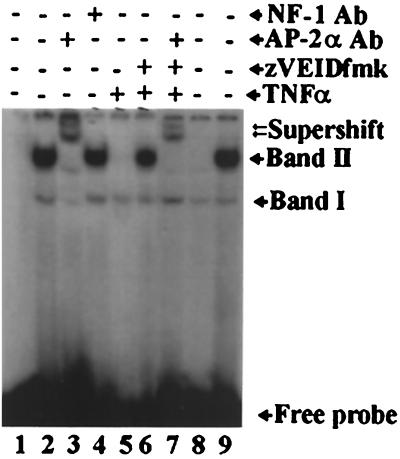
To determine whether TNF-α-induced caspase-mediated cleavage of AP-2α resulted in the loss of DNA-binding activity, nuclear extracts from control, TNF-α-treated, and zVEIDfmk-inhibited cells were examined on an EMSA gel. Nuclear extract from control cells formed two shifted bands, designated I and II, with an AP-2 consensus DNA-binding sequence (Fig. 10, lane 2). Band I appeared in all lanes (lanes 2 to 9) with approximately the same intensity, suggesting that it is nonspecific. This band was used to show equal loading, since it remains constant even after TNF-α treatment, and to demonstrate that not all nuclear proteins in TNF-α-treated cells are degraded (see also Fig. 1C through F and 3A). Band II was formed with nuclear extract from control cells (lane 2) and was super shifted by AP-2α-specific antibody (lane 3) but not by an NF-1-specific antibody (lane 4). Additionally, band II was out-competed by a 100-fold-excess of cold AP-2α probe (lane 8) but not by NF-1 probe (lane 9), demonstrating its AP-2α specificity. TNF-α treatment of cells abrogated the DNA-binding activity of nuclear extract (lane 5). Preincubation of cells with zVEIDfmk, however, preserved the DNA-binding activity of AP-2α (lane 6) and the ability to be super shifted by AP-2α antibody (lane 7). Taken together, these observations show that TNF-α-induced AP-2α downregulation leads to loss of AP-2α DNA-binding activity.
FIG. 10.
EMSA analysis of TNF-α-mediated loss of AP-2α DNA-binding activity. Lane 1, free probe. EMSA analysis detected two primary complexes, designated bands I and II. All nuclear extracts formed band I (lanes 2 through 9), suggesting that it is nonspecific. Band II is the AP-2α-specific complex, since it was formed with a control nuclear extract (lane 2) and was supershifted with AP-2α-specific antibody (AP-2α Ab) (lane 3) but not by a nonspecific NF-1 antibody (NF-1 Ab) (lane 4). Furthermore, band II was out-competed by a 100-fold excess of cold AP-2 consensus binding sequence (lane 8) but not by a nonspecific sequence (NF-1 consensus binding site) (lane 9). TNF-α treatment abrogated the DNA-binding activity of AP-2α (lane 5). However, the ability of cells to maintain their AP-2α DNA-binding activity was preserved by preincubation of cells with zVEIDfmk (lane 6, band II), which was super shifted with an AP-2α-specific antibody (lane 7).
Resistance to TNF-α-induced apoptosis by cells stably transfected with mutant AP-2α.
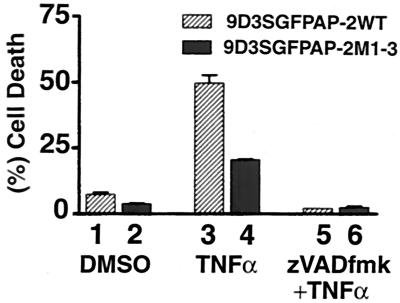
To determine if caspase-mediated cleavage of AP-2α plays a significant role in TNF-α-induced apoptosis, we compared cells stably transfected with wild-type and mutant GFP-AP-2α expression plasmids. Results of FACS analysis of TNF-α-treated cells are presented in Fig. 11. We verified the expression of wild-type and mutant AP-2α by FACS and Western blot analyses (see Materials and Methods). Control cells exhibited only background levels of cell death (columns 1 and 2). TNF-α-treated cells have significantly higher levels of apoptosis in the wild-type AP-2α transfectants (column 3) than in the mutant AP-2 M1-3 transfectants (column 4) in analysis by a nonparametric paired t test (P < 0.05). The general caspase inhibitor (zVADfmk) inhibited apoptosis in both wild-type and mutant AP-2α transfectants (columns 5 and 6). Replacement of wild-type with mutated AP-2α that can no longer be cleaved by caspases resulted in an inhibition of apoptosis by 50%. We conclude that cleavage of AP-2α by caspase 6 is a critical event in TNF-α-induced apoptosis.
FIG. 11.
Resistance to TNF-α-induced apoptosis by 9D3S cells stably transfected with mutant AP-2α. FACS analysis of propidium iodide-stained control cells (columns 1 and 2), TNF-α-treated cells (column 3 and 4), and cells pretreated with zVADfmk followed by TNF-α (column 5 and 6). DMSO, dimethyl sulfoxide.
DISCUSSION
This is the first study to demonstrate cleavage of AP-2α by caspases during TNF-α-induced apoptosis, and it adds AP-2α to a short but growing list of transcription factors that are cleaved by caspases (Table 1). This list includes STAT1 (41), NF-κB (58), SP1 (54, 59), and GATA-1 (16). Interestingly, in all these cases caspases cleave transcription factors and inactivate their transcriptional activities. Our observation that AP-2α is cleaved by caspases and is inactivated by subsequent degradation by proteasome suggests that this is an important mechanism of transcriptional regulation.
TABLE 1.
Transcription factors cleaved by caspases
| Factor | Cleavage site | Cleavage caspase(s) | Consequence of cleavage | Reference or source |
|---|---|---|---|---|
| STAT1 | MELD | 3 | Reduced transcription | 41 |
| NFκB | Unknown | 3 | Reduced transcription | 58 |
| SP1 | Unknown | 1 | Unknown | 54 |
| GATA-1 | Unknown | 3, 7, 8, 9, 10 | Arrest of erythropoiesis | 16 |
| AP-2α | DRHD | 6, 3, 1a | Loss of DNA-binding activity | This paper |
AP-2α is cleaved by caspase 6 with the highest efficiency, by caspase 3 with less efficiency, and by caspase 1 with the least efficiency.
The biological significance of this transcriptional regulation is indicated by several reports that link AP-2α to programmed cell death during embryonic morphogenesis. For example, increased apoptotic cell death was reported to be associated with the lack of AP-2 in AP-2α null mouse embryos (63, 77) and in AP-2β null mice (48). Similarly, retinoic acid-induced downregulation of AP-2α in chicken embryos was associated with programmed cell death (64). Here we have demonstrated that AP-2α is cleaved and degraded during TNF-α-induced apoptosis. We have also shown that AP-2 M1-3 that lacks the caspase 6 cleavage site had a 50% reduction in TNF-α-induced apoptosis in 9D3S cells. These studies provide a functional link between AP-2α and TNF-α-induced apoptosis. The level of resistance in mutant AP-2α-transfected cells is not 100%, because we used pooled clones that may express variable levels of the mutant AP-2α together with the endogenous wild-type AP-2α. Besides, the pooled clones do not comprise 100% of GFP–AP-2α-expressing cells, nor is it known if the fusion protein is expressed throughout the cell cycle. In any case, these observations collectively show that caspase 6-preferred cleavage of AP-2α plays a critical role in the regulation of programmed cell death during embryogenesis and cancer cell survival. In breast cancer cells, AP-2α may act as a survival factor by downregulating proapoptotic genes or by upregulating antiapoptotic genes.
Caspase-induced transcriptional regulation by AP-2 might be of great importance in tumor biology. For example, this mechanism may determine whether AP-2α overexpression promotes tumor progression, as was reported for some breast cancers (67), medulloblastomas (26), and glioblastomas (6), or suppresses tumor progression, as was reported for melanoma (35, 40), colorectal carcinoma (60), and some invasive breast cancer tissues (25). Indeed, metastatic melanoma cells do not express AP-2α, while nonmetastatic melanoma cells do (1, 2, 35, 40). Moreover, enforced expression of AP-2α in AP-2-negative A375SM metastatic melanoma cells rendered them susceptible to apoptosis (35). Of particular interest is the role that the two genes of the AP-2 family (AP-2α and AP-2γ) play in the biology of breast cancer. Both genes are known to be upregulated in breast cancer specimens and also in the cell lines used in our study. We found that TNF-α treatment of breast cancer cells downregulated AP-2α and AP-2γ. Since the two genes share sequence homology at the DRHD motif, it is conceivable that cleavage of AP-2 during TNF-α-induced apoptosis is not confined solely to AP-2α, suggesting that both proteins act as survival factors for breast cancer cells.
We want to point out that it is not at all unusual or unique that AP-2α plays multiple roles in different tumors. We predict that the specific protein with which AP-2 interacts at the transcriptional and posttranscriptional levels determines the specificity of each role. For example, KLF9 promotes AP-2α expression (35), whereas KLF12 represses AP-2α expression (61). There is also evidence that AP-2α can be modified posttranslationally by phosphorylation (22, 53). Furthermore, overexpression of AP-2α may lead to self interference or squelching (37, 39).
If a caspase-dependent mechanism of transcriptional regulation plays a critical role in embryogenesis and the biology of tumors, then it is crucial to locate where in the apoptotic pathway AP-2α is cleaved and degraded. To this end, our observation that caspase 6-mediated in vivo degradation of AP-2α precedes TNF-α-induced DNA fragmentation suggests that it is an early event. Similarly, it has been shown that nuclear death domain protein p84N5-induced apoptosis was associated with activation of caspase 6 prior to DNA fragmentation (18). The observation that caspase 6 cleaves AP-2α in MCF-7 cells, which lack caspase 3 (34), suggests that caspase 3 is not essential in this pathway. This is also supported by the observation that a caspase 3-preferred inhibitor did not significantly inhibit TNF-α-induced cell death (Fig. 4B, column 3/6), nor did recombinant caspase 3 cleave AP-2α at a low concentration (Fig. 5A, lane 4) compared to that of caspase 6 (Fig. 5B, lanes 2 to 5). Furthermore, the failure of the caspase 9 inhibitor to abrogate in vivo degradation of AP-2α (Fig. 4A, lane 11) suggests that the mitochondria are not involved in the cleavage of AP-2α. Finally, we believe that caspase 1 is not a primary participant in the TNF-α-induced AP-2α cleavage and apoptosis for two reasons. First, caspase 1 functions primarily in the proinflammatory response (69). Second, caspase 1 knockout mouse cells have no significant defects in development and apoptosis (69). This means that the observed cleavage of AP-2α by high concentrations of recombinant caspase 1 in vitro (Fig. 5C, lanes 2 and 3) and the inhibition of caspase-initiated degradation of AP-2α by a caspase 1-preferred antagonist (zLEVDfmk) in vivo (Fig. 4A, lane 5) may be due to the promiscuity of caspases and to the lack of absolute specificity of caspase inhibitors. Taken together, these observations suggest that caspase 6-preferred cleavage of AP-2α may belong to a novel TNF-α-induced apoptotic pathway that does not involve caspase 1, 3, or 9.
Our study also sheds light on the biochemistry of AP-2α. The observation that AP-2α is a caspase substrate has revealed a novel functional domain of this important transcription factor. The N terminus of AP-2α consists of exons 1 and 2. Although the trans-activating domain is located in exon 2, only part of this exon (covering residues 51 to 150) is required for trans-activation (72). As far as we are aware, the functions of exon 1 and the first 50 amino acids of exon 2 are not well known. In transient transfection studies, a deletion of this region produced a small inhibition of AP-2α trans-activation activity (72). Our data show that in vivo cleavage of AP-2α renders it susceptible to further degradation by proteasome, leading to a loss of DNA-binding activity (Fig. 10, lane 5). This is a novel observation that suggests that the function of the peptide consisting of the first 19 amino acids is to mask an internal destabilizing signal similar to one found in the yeast Cup9p protein (9). AP-2α could be cleaved and degraded in the cytoplasm before being translocated into the nuclei. Alternatively, it could be cleaved in the nuclei and then translocated into the cytoplasm, where it is degraded by proteasome.
We should point out that at low concentrations, PS-341 appeared to act synergistically with TNF-α to induce more degradation of AP-2α. It is conceivable that although 10 nM PS-341 is insufficient to inhibit AP-2α degradation, it may well be enough to inhibit the degradation of other key proteins, such as IκBα, which in turn might enhance the degradation of AP-2α and induction of apoptosis. PS-341 alone was reported by other investigators to induce apoptosis (J. J. Elliot, D. D. Lazarus, C. S. Pien, V. J. Pallombela, and J. Adams, Abstr. Proc. Am. Assoc. Cancer Res. 90th Annu. Meet., abstr. 836, 1999). Whatever the case may be, we propose that caspase 6 cleavage of AP-2α and its subsequent degradation by proteasome is a mechanism by which cells exercise temporal and spatial restrictions on the expression of AP-2α and possibly AP-2γ and their target genes.
In terms of specificity, the caspase 6-preferred cleavage site in AP-2α differs in sequence from the consensus VXXD (X represents any amino acid) site identified through peptide library screening (66) and is present in several of the identified caspase 6 substrates (Table 2). This observation is not unique to caspase 6 cleavage of AP-2α, because MDM2, FAK, IκB, topoisomerase, and TGEV nucleoprotein are also cleaved at noncanonical sites (Table 2), suggesting that recognition of the cleavage site is determined by more than the linear peptide sequence alone (62). While the DRHD sequence conforms to the DXXD consensus substrate sequence for caspase 3, it is not identical to the known caspase 3 or caspase 1 substrate. This is not surprising, because caspase 3, the most prolific enzyme to cleave proteins during apoptosis, does not cleave all of its substrates at a unique site. The DEVD sequence that is considered to be the consensus cleavage site of caspase 3 is found in only 5 of over 45 proteins that are cleaved by caspase 3 (12, 15, 20). Our data provide additional evidence that caspases do not have unique cleavage sites as restriction endonucleases do. Indeed, they are more promiscuous in their cleavage sites than was first thought.
TABLE 2.
Caspase 6 substrates
| Protein | Cleavage site | Consequence of cleavage | Reference or source |
|---|---|---|---|
| Cytokeratin 18 | VEVD | Keratin disruption | 11 |
| Lamin A | VEID | Nuclear lamin disassembly | 56 |
| NuMA | Unknown | Changed nuclear shape | 28 |
| MDM2 | DVPD | Inhibits trans-activation | 13 |
| FAK | VSWD | Loss of paxillin binding activity | 70 |
| IκB | DRHD | NF-κB inhibition | 3 |
| Topoisomerase I | PEDD/EEED | Unknown | 62 |
| β-Catenin | Unknown | Disrupts actin | 68 |
| TGEV | VVPD | Unknown | 21 |
| AP-2α | DRHD | Loss of DNA-binding activity | This paper |
In summary, we provide the first evidence that (i) AP-2α was cleaved preferentially by caspase 6 or caspase 6-like enzymes before DNA fragmentation during TNF-α-induced apoptosis, (ii) caspases 6, 1, and 3 cleaved AP-2α at asp19, the last two occurring at 50-fold or higher enzyme concentrations, (iii) caspase 6 or caspase 6-like enzyme cleavage of AP-2α in vivo leads to loss of AP-2α protein and its DNA-binding activity, (iv) loss of DNA-binding activity and TNF-α-induced apoptosis were prevented by zVEIDfmk, and (v) mutating D19 to A19 abolished cleavage of AP-2α by caspase 6 and rendered cells resistant to apoptosis. Our results revealed a novel functional domain of AP-2α that appears to provide a mechanism for temporal and spatial regulation of AP-2α expression and functions. Our observation also adds weight to the hypothesis that AP-2α plays a pivotal regulatory role in determining the fate of cancer cells as well as that of normal cells during embryogenesis.
ACKNOWLEDGMENTS
We thank Scott H. Kaufmann for critically reading the manuscript and Karen Ramirez for her excellent assistance with the FACS analysis.
This work was supported by NIH grants CA77055 to O.N. and CA76098 to M.B.-E.
REFERENCES
- 1.Bar-Eli M. Molecular mechanisms of melanoma metastasis. J Cell Physiol. 1997;173:275–278. doi: 10.1002/(SICI)1097-4652(199711)173:2<275::AID-JCP35>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Eli M. Role of AP-2 in tumor growth and metastasis of human melanoma. Cancer Metastasis Rev. 1999;18:377–385. doi: 10.1023/a:1006377309524. [DOI] [PubMed] [Google Scholar]
- 3.Barkett M, Xue D, Horvitz H R, Gilmore T D. Phosphorylation of IκB-α inhibits its cleavage by caspase CPP32 in vitro. J Biol Chem. 1997;272:29419–29422. doi: 10.1074/jbc.272.47.29419. [DOI] [PubMed] [Google Scholar]
- 4.Batsche E, Cremisi C. Opposite transcriptional activity between the wild type c-myc gene coding for c-Myc1 and c-Myc2 proteins and c-Myc1 and c-Myc2 separately. Oncogene. 1999;18:5662–5671. doi: 10.1038/sj.onc.1202927. [DOI] [PubMed] [Google Scholar]
- 5.Batsche E, Muchardt C, Behrens J, Hurst H C, Cremisi C. RB and c-Myc activate expression of the E-cadherin gene in epithelial cells through interaction with transcription factor AP-2. Mol Cell Biol. 1998;18:3647–3658. doi: 10.1128/mcb.18.7.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson L Q, Coon M R, Krueger L M, Han G C, Sarnaik A A, Wechsler D S. Expression of MXI1, a Myc antagonist, is regulated by Sp1 and AP2. J Biol Chem. 1999;274:28794–28802. doi: 10.1074/jbc.274.40.28794. [DOI] [PubMed] [Google Scholar]
- 7.Bosher J M, Williams T, Hurst H C. The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proc Natl Acad Sci USA. 1995;92:744–747. doi: 10.1073/pnas.92.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buettner R, Kannan P, Imhof A, Bauer R, Yim S O, Glockshuber R, Van Dyke M W, Tainsky M A. An alternatively spliced mRNA from the AP-2 gene encodes a negative regulator of transcriptional activation by AP-2. Mol Cell Biol. 1993;13:4174–4185. doi: 10.1128/mcb.13.7.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byrd C, Turner G C, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- 11.Caulin C, Salvesen G S, Oshima R G. Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J Cell Biol. 1997;138:1379–1394. doi: 10.1083/jcb.138.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan S L, Mattson M P. Caspase and calpain substrates: roles in synaptic plasticity and cell death. J Neurosci Res. 1999;58:167–190. [PubMed] [Google Scholar]
- 13.Chen L, Marechal V, Moreau J, Levine A J, Chen J. Proteolytic cleavage of the mdm2 oncoprotein during apoptosis. J Biol Chem. 1997;272:22966–22973. doi: 10.1074/jbc.272.36.22966. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y R, Meyer C F, Ahmed B, Yao B, Tan T H. Caspase-mediated cleavage and functional changes of hematopoietic progenitor kinase 1 (HPK1) Oncogene. 1999;18:7370–7377. doi: 10.1038/sj.onc.1203116. [DOI] [PubMed] [Google Scholar]
- 15.Cohen G M. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Maria R, Zeuner A, Eramo A, Domenichelli C, Bonci D, Grignani F, Srinivasula S M, Alnemri E S, Testa U, Peschle C. Negative regulation of erythropoiesis by caspase-mediated cleavage of GATA-1. Nature. 1999;401:489–493. doi: 10.1038/46809. [DOI] [PubMed] [Google Scholar]
- 17.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doostzadeh-Cizeron J, Yin S, Goodrich D W. Apoptosis induced by the nuclear death domain protein p84N5 is associated with caspase-6 and NF-[kappa]B activation. J Biol Chem. 2000;275:25336–25341. doi: 10.1074/jbc.M000793200. [DOI] [PubMed] [Google Scholar]
- 19.Duan C, Clemmons D R. Transcription factor AP-2 regulates human insulin-like growth factor binding protein-5 gene expression. J Biol Chem. 1995;270:24844–24851. doi: 10.1074/jbc.270.42.24844. [DOI] [PubMed] [Google Scholar]
- 20.Earnshaw W C, Martins L M, Kaufmann S H. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 21.Eleouet J F, Slee E A, Saurini F, Castagne N, Poncet D, Garrido C, Solary E, Martin S J. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J Virol. 2000;74:3975–3983. doi: 10.1128/jvi.74.9.3975-3983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia A, Campillos M, Marina A, Valdivieso F, Vazquez J. Transcription factor AP-2 activity is modulated by protein kinase A-mediated phosphorylation. FEBS Lett. 1999;444:27–31. doi: 10.1016/s0014-5793(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 23.Gaubatz S, Imhof A, Dosch R, Werner O, Mitchell P, Buettner R, Eilers M. Transcriptional activation by Myc is under negative control by the transcription factor AP-2. EMBO J. 1995;14:1508–1519. doi: 10.1002/j.1460-2075.1995.tb07137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaynor R B, Muchardt C, Xia Y R, Klisak I, Mohandas T, Sparkes R S, Lusis A J. Localization of the gene for the DNA-binding protein AP-2 to human chromosome 6p22.3-pter. Genomics. 1991;10:1100–1102. doi: 10.1016/0888-7543(91)90209-w. [DOI] [PubMed] [Google Scholar]
- 25.Gee J M, Robertson J F, Ellis I O, Nicholson R I, Hurst H C. Immunohistochemical analysis reveals a tumour suppressor-like role for the transcription factor AP-2 in invasive breast cancer. J Pathol. 1999;189:514–520. doi: 10.1002/(SICI)1096-9896(199912)189:4<514::AID-PATH463>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 26.Gilbertson R J, Perry R H, Kelly P J, Pearson A D, Lunec J. Prognostic significance of HER2 and HER4 co-expression in childhood medulloblastoma. Cancer Res. 1997;57:3272–3280. [PubMed] [Google Scholar]
- 27.Grether-Beck S, Olaizola-Horn S, Schmitt H, Grewe M, Jahnke A, Johnson J P, Briviba K, Sies H, Krutmann J. Activation of transcription factor AP-2 mediates UVA radiation- and singlet oxygen-induced expression of the human intercellular adhesion molecule 1 gene. Proc Natl Acad Sci USA. 1996;93:14586–14591. doi: 10.1073/pnas.93.25.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirata H, Takahashi A, Kobayashi S, Yonehara S, Sawai S H, Okazaki T, Yamamoto K, Sasada M. Caspases are activated in a branched protease cascade and control distinct downstream processes in Fas-induced apoptosis. J Exp Med. 1998;187:587–600. doi: 10.1084/jem.187.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollywood D P, Hurst H C. A novel transcription factor, OB2-1, is required for overexpression of the protooncogene c-erbB-2 in mammary tumor lines. EMBO J. 1993;12:2369–2375. doi: 10.1002/j.1460-2075.1993.tb05891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Domann F E. Redox modulation of AP-2 DNA binding activity in vitro. Biochem Biophys Res Commun. 1998;249:307–312. doi: 10.1006/bbrc.1998.9139. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y, Domann F E. Transcription factor AP-2 mRNA and DNA binding activity are constitutively expressed in SV40-immortalized but not normal human lung fibroblasts. Arch Biochem Biophys. 1999;364:241–246. doi: 10.1006/abbi.1999.1142. [DOI] [PubMed] [Google Scholar]
- 32.Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 33.Imhof A, Schuierer M, Werner O, Moser M, Roth C, Bauer R, Buettner R. Transcriptional regulation of the AP-2α promoter by BTEB-1 and AP-2rep, a novel wt-1/egr-related zinc finger repressor. Mol Cell Biol. 1999;19:194–204. doi: 10.1128/mcb.19.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janicke R U, Sprengart M L, Wati M R, Porter A G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- 35.Jean D, Gershenwald J E, Huang S, Luca M, Hudson M J, Tainsky M A, Bar-Eli M. Loss of AP-2 results in up-regulation of MCAM/MUC18 and an increase in tumor growth and metastasis of human melanoma cells. J Biol Chem. 1998;273:16501–16508. doi: 10.1074/jbc.273.26.16501. [DOI] [PubMed] [Google Scholar]
- 36.Jean D, Harbison M, McConkey D J, Ronai Z, Bar-Eli M. CREB and its associated proteins act as survival factors for human melanoma cells. J Biol Chem. 1998;273:24884–24890. doi: 10.1074/jbc.273.38.24884. [DOI] [PubMed] [Google Scholar]
- 37.Kannan P, Tainsky M A. Coactivator PC4 mediates AP-2 transcriptional activity and suppresses ras-induced transformation dependent on AP-2 transcriptional interference. Mol Cell Biol. 1999;19:899–908. doi: 10.1128/mcb.19.1.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kannan P, Yu Y, Wankhade S, Tainsky M A. PolyADP-ribose polymerase is a coactivator for AP-2-mediated transcriptional activation. Nucleic Acids Res. 1999;27:866–874. doi: 10.1093/nar/27.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kannan P, Buettner R, Chiao P J, Yim S O, Sarkiss M, Tainsky M A. N-ras oncogene causes AP-2 transcriptional self-interference, which leads to transformation. Genes Dev. 1994;8:1258–1269. doi: 10.1101/gad.8.11.1258. [DOI] [PubMed] [Google Scholar]
- 40.Karjalainen J M, Kellokoski J K, Eskelinen M J, Alhava E M, Kosma V M. Downregulation of transcription factor AP-2 predicts poor survival in stage I cutaneous malignant melanoma. J Clin Oncol. 1998;16:3584–3591. doi: 10.1200/JCO.1998.16.11.3584. [DOI] [PubMed] [Google Scholar]
- 41.King P, Goodbourn S. STAT1 is inactivated by a caspase. J Biol Chem. 1998;273:8699–8704. doi: 10.1074/jbc.273.15.8699. [DOI] [PubMed] [Google Scholar]
- 42.Luscher B, Mitchell P J, Williams T, Tjian R. Regulation of transcription factor AP-2 by the morphogen retinoic acid and by second messengers. Genes Dev. 1989;3:1507–1517. doi: 10.1101/gad.3.10.1507. [DOI] [PubMed] [Google Scholar]
- 43.Meier P, Koedood M, Phillipp J, Fontana A, Mitchell P J. Alternative mRNA encode multiple isoforms of transcription factor AP-2 during murine embryogenesis. Dev Biol. 1995;169:1–14. doi: 10.1006/dbio.1995.1121. [DOI] [PubMed] [Google Scholar]
- 44.Meiser P, Mesner W, Jr, Bible K C, Martins L M, Kottke T J, Srinivasula S M, Svingen P A, Chilcote T J, Basi G S, Tung J S, Krajewski S, Reed J C, Alnemri E S, Earnshaw W C, Kaufmann S H. Characterization of caspase processing and activation in HL-60 cell cytosol under cell-free conditions. Nucleotide requirement and inhibitor profile. J Biol Chem. 1999;274:22635–22645. doi: 10.1074/jbc.274.32.22635. [DOI] [PubMed] [Google Scholar]
- 45.Mitchell P J, Wang C, Tijian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 46.Mori N, Prager D. High levels of AP-2-binding activity in cell lines infected with human T-cell leukemia virus type I: possible enhancement of AP-2 binding by human T-cell leukemia virus type I tax. Cancer Res. 1996;56:779–782. [PubMed] [Google Scholar]
- 47.Moser M, Imhof A, Pscherer A, Bauer R, Amselgruber W, Sinowatz F, Hofstadter F, Schule R, Buettner R. Cloning and characterization of a second AP-2 transcription factor: AP-2β. Development. 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- 48.Moser M, Pscherer A, Roth C, Becker J, Mucher G, Zerres K, Dixkens C, Weis J, Guay-Woodford L, Buettner R, Fassler R. Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2β. Genes Dev. 1997;11:1938–1948. doi: 10.1101/gad.11.15.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nyormoi O. Proteolytic activity in amyotrophic lateral sclerosis IgG preparations. Ann Neurol. 1996;40:701–706. doi: 10.1002/ana.410400505. [DOI] [PubMed] [Google Scholar]
- 50.Ohtaka-Maruyama C, Hanaoka F, Chepelinsky A B. A novel alternative spliced variant of the transcription factor AP-2α is expressed in the murine ocular lens. Dev Biol. 1998;202:125–135. doi: 10.1006/dbio.1998.8997. [DOI] [PubMed] [Google Scholar]
- 51.Oulad-Abdelghani M, Bouillet P, Chazaud C, Dollé P, Chambon P. AP-2.2: a novel AP-2-related transcription factor induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Exp Cell Res. 1996;225:338–347. doi: 10.1006/excr.1996.0184. [DOI] [PubMed] [Google Scholar]
- 52.Palombella J V, Conner E M, Fuseler J W, Destree A, Davis J M, Laroux F S, Wolf R E, Huang J, Brand S, Elliott P J, Lazarus D, McCormack T, Parent L, Stein R, Adams J, Grisham M B. Role of the proteasome and NF-κB in streptococcal cell wall-induced polyarthritis. Proc Natl Acad Sci USA. 1998;95:15671–15676. doi: 10.1073/pnas.95.26.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park K, Kim K H. The site of cAMP action in the insulin induction of gene expression of acetyl-CoA carboxylase is AP-2. J Biol Chem. 1993;268:17811–17819. [PubMed] [Google Scholar]
- 54.Piedrafita F J, Pfahl M. Retinoid-induced apoptosis and Sp1 cleavage occur independently of transcription and require caspase activation. Mol Cell Biol. 1997;17:6348–6358. doi: 10.1128/mcb.17.11.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin H, Sun Y, Benveniste E N. The transcription factors Sp1, Sp3, and AP-2 are required for constitutive matrix metalloproteinase-2 gene expression in astroglioma cells. J Biol Chem. 1999;274:29130–29137. doi: 10.1074/jbc.274.41.29130. [DOI] [PubMed] [Google Scholar]
- 56.Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rath P C, Aggarwal B B. TNF-induced signaling in apoptosis. J Clin Immunol. 1999;19:350–364. doi: 10.1023/a:1020546615229. [DOI] [PubMed] [Google Scholar]
- 58.Ravi R, Bedi A, Fuchs E J, Bedi A. CD95 (Fas)-induced caspase-mediated proteolysis of NF-κB. Cancer Res. 1998;58:882–886. [PubMed] [Google Scholar]
- 59.Rickers A, Peters N, Badock V, Beyaert R, Vandenabeele P, Dorken B, Bommert K. Cleavage of transcription factor SP1 by caspases during anti-IgM-induced B-cell apoptosis. Eur J Biochem. 1999;261:269–274. doi: 10.1046/j.1432-1327.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- 60.Ropponen K M, Kellokoski J K, Lipponen P K, Pietilainen T, Eskelinen M J, Alhava E M, Kosma V M. p21WAF-1 expression in human colorectal carcinoma: association with p53, transcription factor AP-2 and prognosis. Br J Cancer. 1999;81:133–140. doi: 10.1038/sj.bjc.6690662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roth C, Schuierer M, Gunther K, Buettner R. Genomic structure and DNA binding properties of the human zinc finger transcriptional repressor AP-2rep. Genomics. 2000;63:384–390. doi: 10.1006/geno.1999.6084. [DOI] [PubMed] [Google Scholar]
- 62.Samejima K, Svingen P A, Basi G S, Kottke T, Mesner P W, Jr, Stewart L, Durrieu F, Poirier G G, Alnemri E S, Champoux J J, Kaufmann S H, Earnshaw W C. Caspase-mediated cleavage of DNA topoisomerase I at unconventional sites during apoptosis. J Biol Chem. 1999;274:4335–4340. doi: 10.1074/jbc.274.7.4335. [DOI] [PubMed] [Google Scholar]
- 63.Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell P J. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 64.Shen H, Wilke T, Ashique A M, Narvey M, Zerucha T, Savino E, Williams T, Richman J M. Chicken transcription factor AP-2: cloning, expression and its role in outgrowth of facial prominences and limb buds. Dev Biol. 1997;188:248–266. doi: 10.1006/dbio.1997.8617. [DOI] [PubMed] [Google Scholar]
- 65.Talanian R V, Quinlan C, Trautz S, Hackett M C, Mankovich J A, Banach D, Ghayur T, Brady K D, Wong W W. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 66.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstrom P A, Roy S, Vaillancourt J P, Chapman K T, Nicholson D W. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 67.Turner B C, Zhang J, Gumbs A A, Maher M G, Kaplan L, Carter D, Glazer P M, Hurst H C, Haffty B G, Williams T. Expression of AP-2 transcription factors in human breast cancer correlates with the regulation of multiple growth factor signalling pathways. Cancer Res. 1998;58:5466–5472. [PubMed] [Google Scholar]
- 68.Van de Craen M, Berx G, Van den Brande I, Fiers W, Declercq W, Vandenabeele P. Proteolytic cleavage of beta-catenin by caspases: an in vitro analysis. FEBS Lett. 1999;458:167–170. doi: 10.1016/s0014-5793(99)01153-9. [DOI] [PubMed] [Google Scholar]
- 69.Wang J, Lenardo M J. Roles of caspases in apoptosis, development, and cytokine maturation revealed by homologous gene deficiencies. J Cell Sci. 2000;113:753–757. doi: 10.1242/jcs.113.5.753. [DOI] [PubMed] [Google Scholar]
- 70.Wen L P, Fahrni J A, Troie S, Guan J L, Orth K, Rosen G D. Cleavage of focal adhesion kinase by caspases during apoptosis. J Biol Chem. 1997;272:26056–26061. doi: 10.1074/jbc.272.41.26056. [DOI] [PubMed] [Google Scholar]
- 71.Williams T, Admon A, Luscher B, Tijian R. Cloning and expression of AP-2, a cell-type specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- 72.Williams T, Tijian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 73.Williamson J A, Bosher J M, Skinner A, Sheer D, Williams T, Hurst H C. Chromosomal mapping of the human and mouse homologues of two new members of the AP-2 family of transcription factors. Genomics. 1996;35:262–264. doi: 10.1006/geno.1996.0351. [DOI] [PubMed] [Google Scholar]
- 74.Winning R S, Shea L J, Marcus S J, Sargent T D. Developmental regulation of transcription factor AP-2 during Xenopus laevis embryogenesis. Nucleic Acids Res. 1991;19:3709–3714. doi: 10.1093/nar/19.13.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong D L, Siddall B J, Ebert S N, Bell R A, Her S. Phenylethanolamine N-methyltransferase gene expression: synergistic activation by Egr-1, AP-2 and the glucocorticoid receptor. Brain Res Mol Brain Res. 1998;61:154–161. doi: 10.1016/s0169-328x(98)00225-3. [DOI] [PubMed] [Google Scholar]
- 76.Zeng Y-X, Soumasundaram K, El-Deiry W. AP-2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat Genet. 1997;15:78–81. doi: 10.1038/ng0197-78. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon A P, Flavell R A, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–341. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]