Abstract
Chromosomal replicators in budding yeast contain an autonomously replicating sequence (ARS) that functions in a plasmid, but certain ARSs are silent as replication origins in their natural chromosomal context. In chromosome III, the HML ARS cluster (ARS302-ARS303-ARS320) and ARS301 flank the transcriptionally silent mating-type locus HML, and all of these ARSs are silent as replication origins. ARS301 and ARS302 function in transcriptional silencing mediated by the origin recognition complex (ORC) and a heterochromatin structure, while the functions of ARS303 and ARS320 are not known. In this work, we discovered replication fork pause sites at the HML ARS cluster and ARS301 by analyzing DNA replication intermediates from the chromosome via two-dimensional gel electrophoresis. The replication fork pause at the HML ARS cluster was independent of cis- and trans-acting mutations that abrogate transcriptional silencing at HML. Deletion of the HML ARS cluster led to loss of the pause site. Insertion of a single, heterologous ARS (ARS305) in place of the HML ARS cluster reconstituted the pause site, as did multiple copies of DNA elements (A and B1) that bind ORC. The orc2-1 mutation, known to alter replication timing at origins, did not detectably affect the pause but activated the silent origin at the HML ARS cluster in a minority of cells. Delaying the time of fork arrival at HML led to the elimination of the pause sites at the HML ARS cluster and at the copy of ARS305 inserted in place of the cluster. Loss of the pause sites was accompanied by activation of the silent origins in the majority of cells. Thus, replication fork movement near HML pauses at a silent origin which is competent for replication initiation but kept silent through Orc2p, a component of the replication initiator. Possible functions for replication fork pause sites in checkpoints, S-phase regulation, mating-type switching, and transcriptionally silent heterochromatin are discussed.
In the yeast Saccharomyces cerevisiae, chromosomal replicators can be isolated as autonomously replicating sequences (ARSs). ARS elements allow high-frequency transformation of yeast cells (32, 62) and serve as replication origins in plasmids (9, 30). ARS function in yeast requires important modular sequences. A highly conserved region, called the ARS consensus sequences (ACS), is the core of a larger functional sequence called the A element (29, 41). The A element is essential, and point mutations in the ACS can abolish ARS activity (28, 44, 66). The B domain, located 3′ to the T-rich strand of the ACS, is also essential for ARS function and contains multiple functionally important modules called B elements (28, 36, 41, 49, 64). One of these, B2, contains a DNA-unwinding element (DUE) that is thought to facilitate entry of the replication machinery into the double helix at the origin (5, 36). The origin recognition complex (ORC) binds the A and B1 elements and interacts with other replication proteins to render an origin competent for initiation (3, 50, 56; reviewed in reference 31).
While all chromosomal replicators identified in yeast coincide with an ARS element, not all ARS elements serve as active chromosomal origins (16, 46, 67). ARS elements which are not associated with an active chromosomal origin are called silent origins. Some of the silent origins function as transcriptional silencers (7), although active origins can also serve as transcriptional silencers (52, 53). ORC binds to ARS elements that are silent origins and contributes to transcriptional silencing (2, 21, 22). The function of silent origins which are not transcriptional silencers is not known, and the reason why these silent origins are present in single-cell organisms such as budding yeast is not clear. In multicellular organisms such as Drosophila melanogaster, silencing of a large number of active origins on chromosomes is postulated to be one of the mechanisms to prolong the S phase and reduce the cell growth rate during development and differentiation (6, 60). Recently, evidence was reported that silent origins in yeast can be activated at their native chromosomal locations under special circumstances (59, 67).
Duplication of DNA in eukaryotic chromosomes initiates at multiple origins with different origins activated at different times in S phase. In general, replication forks derived from chromosomal origins move bidirectionally away from the origins, and replication terminates when forks from different origins collide with each other and when forks reach both ends of the chromosomes. At certain loci, replication forks are arrested or temporarily stalled prior to termination (10, 14, 15, 23, 39). Temporary slowdown or arrest of a replication fork in the chromosome results in the accumulation of replication intermediates at a particular chromosomal location and can be detected as a heavy hybridization signal along the arc of replication fork intermediates after two-dimensional (2D) gel electrophoresis (10). In yeast chromosomes, replication fork barriers have been identified in the ribosomal gene cluster, and replication fork pause sites have been reported at the centromeres and at tRNA genes (10, 14, 23). Protein-DNA interactions are thought to be important in mediating some replication fork pauses (23, 39). Also, active transcription has been shown to pause replication fork movement (14). One function of pause sites and fork barriers is to block the replication forks from entering into actively transcribed genes (10, 14). Replication fork pause sites occur in chromosomes from a variety of eukaryotes and prokaryotes (reviewed in reference 54). However, aside from the role of fork pause sites at actively transcribed genes, little is known about their biological functions in eukaryotic chromosomes. In addition, more remains to be learned about the nature of the DNA elements and trans-acting factors that determine replication fork pausing.
Here we report that replication fork pause sites are present near HML, a transcriptionally silent mating-type locus in budding yeast. Fork pause sites map to silent origins of replication including the HML ARS cluster (ARS302-ARS303-ARS320) and ARS301. Although two of theses ARS elements are transcriptional silencers, we found that fork pausing is independent of mutations that abrogate transcriptional silencing at HML. Replication fork pausing is tightly linked to origin silencing. HML ARS elements at silent origins are required for fork pausing. A heterologous ARS element inserted in place of the HML ARS cluster becomes a silent origin and reconstitutes the replication fork pause site. Multiple copies of DNA elements that bind ORC also reconstitute the pause site. The orc2-1 mutation, known to affect replication timing, did not detectably affect the pause signal but activated the silent origin at the HML ARS cluster in a subpopulation of cells. Delaying the fork arrival at HML resulted in the disappearance of the pause and the activation of associated silent origins in the majority of cells. Our results show that replication fork movement near HML pauses at a silent origin which is competent for replication initiation but kept silent through Orc2p, a component of the replication initiator. Possible functions of replication fork pause sites in checkpoint signaling, S-phase regulation, and mating-type switching and in transcriptionally silent heterochromatin are discussed.
MATERIALS AND METHODS
Reagents.
Restriction enzymes, T4 DNA ligase, Klenow polymerase, and T4 DNA polymerase were obtained from New England Biolabs, Inc. [α-32P]dATP was purchased from Amersham International. BND-cellulose for replication intermediate enrichment was obtained from Sigma Chemical Company. 5-Fluoro-orotic acid was from Toronto Research Chemicals Inc. Media reagents were from Difco Laboratories and American Biorganics, Inc.
Bacteria, plasmids, and yeast strains.
Escherichia coli strain DH5α was used for plasmid transformation and propagation. The plasmids used in yeast integration are all YIP5 derivatives that carry an ampicillin resistance gene and a bacterial replication origin for propagation and selection in E. coli. They also contain a URA3 gene used for selection and counterselection during the two-step method for the integration of mutations into yeast chromosome III.
Yeast strains used in this study were YPH98 (MATa ade2-101 lys2-801 ura3-52 trp1-1 leu2-1) (obtained from Philip Hieter, Johns Hopkins University) and its derivatives YWY1 (YPH98 with a 1,054-bp PvuII/XhoI fragment containing the HML ARS302-ARS303-ARS320), YWY2 (YWY1 with a 549-bp NruI/ClaI fragment containing ARS305 in place of the HML ARS302-ARS303-ARS320), YWY3 (YWY2 with ORI305 and ORI306 chromosomal origins deleted), YWY6 (YPH98 with a 1,074-bp PvuII/BlpI deletion of the part of the HML cassette), YWY8 (YWY1 with a 299-bp mutated ARS305 derivative [AB1GCΔ {36}, termed mt305] in place of the HML ARS302-ARS303-ARS320), YWY12 (YWY1 with five copies of a 299-bp mutated ARS305 derivative [mt305] in place of the HML ARS302-ARS303-ARS320); DMY1 (HMLα MATa HMRa ura3-52 leu2-3,112 ade2-1 lys1-1 his5-2 can1-100); DMY2 (DMY1 sir3::LEU2); DMY94 (DMY1 E+::URA3 1 D242); DMY94 (DMY1 E− D79-113::URA3 I D242) (DMY1 and its derivatives were obtained from James Broach, Princeton University) (40); JRY4556 (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 orc5-1,3) (obtained from Jasper Rine, University of California, Berkeley) (22); and W303-1A (MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3 can1-100), and its derivative W303-1A orc2-1 (obtained from Bruce Stillman, Cold Spring Harbor Laboratory (35).
For creation of YPH98 derivatives, a two-step mutagenesis method was used as previously described (28). Briefly, YIP5-based integration vectors harboring the desired mutations were first linearized with a unique restriction enzyme and transformed into the competent yeast YPH98 cells by lithium acetate transformation as described elsewhere (28). Positive chromosomal integrants were selected after plating on synthetic dextrose minimal medium without uracil. The transformants were picked and inoculated in a small volume of YPD liquid medium (nonselective) and grown overnight. The cell concentration was determined by optical density at 600 nm. A total of 104 cells were plated on 5′-fluoro-orotic acid plates for selection against the URA3+ transformants. Genomic DNA from the resulting colonies was screened for the desired mutation by the Southern blot hybridization.
Genomic DNA isolation for 2D gels.
Most of the yeast strains were grown at 30°C. In the case of the orc2-1 strain and the isogenic W303-1A strain, cultures were grown overnight at 23°C and then shifted to either 30 or 37°C for 2 h. Cells were grown to 1.3 × 107 1.5 × 107 cells/ml, and genomic DNA was isolated by CsCl gradient centrifugation followed by restriction endonuclease digestion. Completely digested DNA samples were precipitated, resuspended in Tris-EDTA (pH 7.5) and combined with BND-cellulose to enrich for replication intermediates. Bound DNA was eluted with 1.8% caffeine (30). The caffeine wash samples were precipitated and resuspended in small volume of Tris-EDTA and were analyzed by 2D gel electrophoresis as described previously (9), with minor modifications. First-dimension electrophoresis was carried out in 0.4% agarose gel in 1× Tris-acetate-EDTA buffer containing 0.1 μg of ethidium bromide per ml for 18 to 20 h at 1 V/cm. First-dimension sample lanes were cut from the gel and embedded into 1% agarose gel, and the second dimension was carried out at 5 V/cm for 8 to 10 h 4°C in 1× Tris-borate-EDTA buffer containing 0.5 μg of ethidium bromide per ml. The second-dimension gels were Southern blotted to a nylon membrane (Gene Screen Plus; DuPont) using a pressure blotter (Stratagene) and hybridized to 32P-labeled DNA probes (28). The radioactive signals were detected and analyzed with a PhosphorImager (Molecular Dynamics STORM).
RESULTS
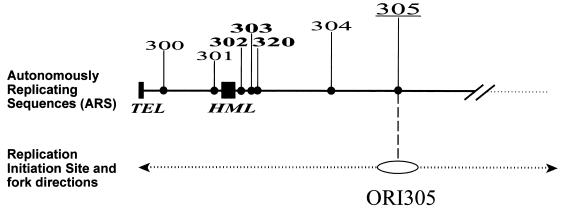
The silent mating-type locus, HML, is located near the left end of chromosome III and is flanked by a number of ARS elements which are silent as chromosomal replication origins (Fig. 1). Only one of the ARS elements, ARS305, is associated with an active replication origin, ORI305, in the left 40 kb of the chromosome. Since there are no other active origins in that region, a replication fork that originates at ORI305 and moves leftward is responsible for duplicating the entire left 40 kb of chromosome III, including the silent origins at the HML ARS cluster (ARS302-ARS303-ARS320) and at ARS301 (Fig. 1).
FIG. 1.
Left arm of S. cerevisiae chromosome III. ARS elements within the 40-kb region of chromosome III are marked. The ARS305 replicator (305), which is associated with an active origin, ORI305, is underlined. ARS elements in the HML ARS cluster (302, 303, 320) are marked in bold. ORI305 is the only active origin within this part of the chromosome. The HML locus is normally replicated by a fork derived from ORI305, as indicated by the arrow.
Replication forks pause near the HML locus.
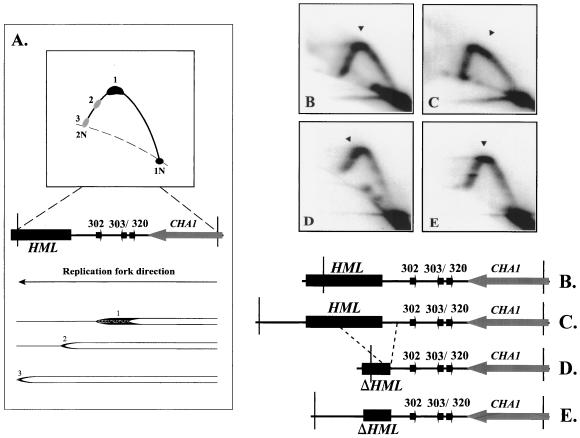
When replication intermediates at the HML ARS cluster are analyzed by 2D gel electrophoresis, a simple Y arc indicative of passive replication is detected (Fig. 2A and 2B). No bubble arc is detected, consistent with a normally silent origin at the HML ARS cluster. In addition to the simple Y arc, a heavy hybridization signal is obvious at the peak of the Y arc (Fig. 2B). The heavy hybridization signal indicates the accumulation of replication forks, i.e., a fork pause site. The major fork pause signal corresponds to replication intermediates that accumulated at the HML ARS cluster region, since the peak of the Y arc represents the center of the analyzed EcoRI/FspI restriction fragment and the HML ARS cluster is centrally located as shown in the map (Fig. 2B). In addition to the major pause site at the peak, two minor fork pause sites were also detected on the late Y arc, which map within the HML cassette of the analyzed EcoRI/FspI restriction fragment (Fig. 2B). The three-spot pattern has been detected in several different yeast strains at the HML ARS cluster region (see below). Since the HML ARS cluster appears to correspond to the major fork accumulation site seen during 2D gel analysis, it was of interest to determine whether the major pause is mediated by those ARS elements that are silent as chromosomal replication origins.
FIG. 2.
Replication fork movement pauses at the HML ARS cluster in chromosome III. (A) Schematic illustration of replication fork pause signals (spots 1, 2, and 3) detected on the Y arc after 2D gel electrophoresis of a DNA fragment containing the HML ARS cluster. A 3.9-kb EcoRI/FspI genomic fragment containing the centrally located HML ARS cluster (ARS302-ARS303-ARS320 [302 303/ 320]) is diagrammed. ARS elements are indicated as small arrows. The filled box indicates part of the HML cassette, and the shaded arrow indicates part of the CHA1 gene. The direction of fork migration is indicated. Major (1) and minor (2 and 3) pause sites are depicted as accumulated replication forks. (B to D) Two-dimensional gel electrophoresis of replication intermediates from particular strains probed for specific restriction fragments shown in the maps. Arrowheads in the 2D gel patterns denote the center of the pause signal. (B) A 3.9-kb EcoRI/FspI genomic fragment from the parental strain YPH98; (C) a 5.4-kb EcoRI genomic fragment from the YPH98 strain; (D) a 2.8-kb EcoRI/FspI genomic fragment from a mutant strain with a 1.07-kb deletion of part of the HML cassette (YWY6). (E) A 3.35-kb EcoRI genomic fragment from the YWY6 strain.
The major replication fork pause site colocalizes with the HML ARS cluster.
To determine whether the major pause site observed on the 2D gel is associated with the HML ARS cluster, we first changed the position of the HML ARS cluster relative to the ends of the analyzed genomic fragments. If the major pause site is associated with the HML ARS cluster, then changing the position of the HML ARS cluster would shift the location of the major pause site on the Y arc correspondingly.
The EcoRI/FspI genomic fragment that centers the position of the HML ARS cluster results in detection of the major pause site at the peak of the Y arc (Fig. 2B). Digestion by EcoRI alone yields a genomic fragment that shifts the HML ARS cluster off center to the right. Analysis of the EcoRI-digested HML genomic fragment on the 2D gel shows that the major pause site is indeed shifted toward the right side of the Y arc within the early Y-arc portion. Additionally, the extent of the major pause signal shift is consistent with the off-center location of the HML ARS cluster on the fragment (Fig. 2C). An EcoRI/FspI genomic fragment from a mutant yeast strain with a portion of the HML locus deleted was also analyzed. The deletion results in the shift of the HML ARS cluster off center to the left of the EcoRI/FspI fragment. Two-dimensional gel analysis of the fragment in the deleted strain revealed that the major pause was shifted off center to the left, within late Y-arc region (Fig. 2D). This pause signal in the late Y-arc region returned to the peak of the Y arc when the same strain was analyzed by EcoRI digestion, which places the HML ARS cluster back to the center of the genomic fragment (Fig. 2E). Thus, the positions of the HML ARS cluster on the analyzed restriction fragments map with the positions of the major pause site on the Y arc. These results suggest that the major replication fork pause site colocalizes with the HML ARS cluster region.
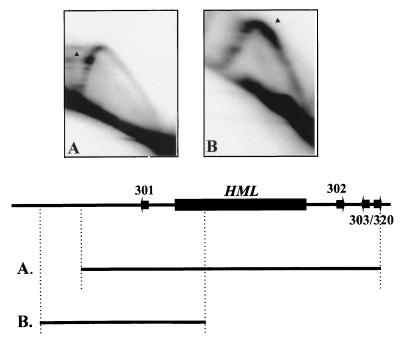
ARS301 near the HML cassette is associated with a replication fork pause site.
In addition to the HML ARS cluster, another ARS element, ARS301, located within the E silencer near HML, is also inactive as a chromosomal origin. Despite the absence of detectable origin inactivity, ARS301 forms a prereplication complex and is competent for replication initiation in the chromosome under normal growth conditions (58, 67). When an XhoI genomic fragment containing ARS301 is analyzed on a 2D gel, a strong replication fork pause signal is detected. The position of ARS301 on the restriction fragment is consistent with the off-center location of the pause signal on the 2D gel (Fig. 3A). Moreover, when we examined the BamHI/FspI genomic fragment that shifted ARS301 off center to the right, we found the fork pause site to be shifted accordingly to the right of the peak and into the early Y-arc region, suggesting that the replication fork pause is associated with the ARS301 region (Fig. 3B). The results for ARS301, along with those for the HML ARS cluster, suggest that in general, replication forks pause at regions containing ARS elements that are silent origins near the HML locus of chromosome III.
FIG. 3.
Mapping of a replication fork pause site associated with ARS301 on the left end of chromosome III. Arrowheads denote the center of the pause signal. (A) A 5.4-kb XhoI genomic fragment from the YPH98 strain. ARS301 is located off center to the left of the XhoI fragment. (B) A 3.05-kb BamHI/FspI genomic fragment from the YPH98 strain. ARS301 is located off center to the right of the BamHI/FspI fragment. In both maps, a fork that moves unidirectionally from the centromeric side (right) is responsible for the replication of ARS301 region.
Deletion of the HML ARS cluster eliminates the major replication fork pause site.
Restriction fragment shift experiments suggested that the major pause site colocalizes with the HML ARS cluster region. To determine if the ARS elements are important for the pause, we examined whether deletion of those ARSs could eliminate the major pause signal. To this end, we constructed a yeast strain in which all three ARSs in the HML ARS cluster were deleted. Analysis of the EcoRI/FspI fragment of the mutant strain by 2D gel electrophoresis shows that the major pause signal seen in the wild-type strain at the peak of the Y arc is absent. The mutant strain (Fig. 4B) lacks the heavy hybridization signal seen at the peak of the Y arc in the parental strain (Fig. 4A). Analysis of a BamHI/FspI fragment of the HML locus shows that the heavy accumulation of replication intermediates at the peak of the Y arc in the parental strain (Fig. 4C) is again absent in the mutant (Fig. 4D). Our deletion results indicate that the region encompassing the HML ARS cluster is required for mediating the replication fork pause near the HML locus.
FIG. 4.
Deletion of the HML ARS cluster leads to the loss of the major replication fork pause site, as determined by 2D gel electrophoresis of replication intermediates from the indicated strains probed for specific restriction fragments diagrammed in the maps. Arrowheads denote a pause signal. (A) A 3.9-kb EcoRI/FspI genomic fragment from the parental YPH98 strain; (B) a 2.85-kb EcoRI/FspI genomic fragment from a mutant strain (YWY1) that has the HML ARS cluster deleted; (C) a 3.54-kb BamHI/FspI genomic fragment from the parental YPH98 strain; (D) a 2.48-kb BamHI/FspI genomic fragment from a mutant strain (YWY1) that has the HML ARS cluster deleted.
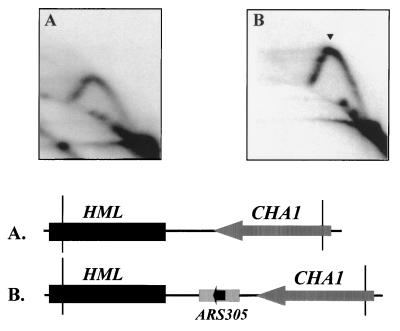
Replacement of the HML ARS cluster with a heterologous ARS element recapitulates the major pause site.
The mapping and deletion experiments described above suggested that ARS elements near the HML locus could be responsible for the replication fork pause. Different ARS elements share structural similarities. The ACS, which is essential for ORC binding, is present in all ARSs isolated. The less conserved B1 element, which contributes to ORC binding, is also present in several ARS elements analyzed in detail (29, 36, 41, 49, 64). If the ARS elements themselves are important for the observed replication fork pause near the HML locus, a heterologous ARS element might be able to regenerate the fork pause. To test this possibility, we replaced the HML ARS cluster with ARS305. There is no sequence similarity between the deleted fragment containing HML ARS cluster and the heterologous ARS305 fragment other than the ACS. ARS305 was derived from a chromosomal origin, ORI305. The fragment of ARS305 used to replace the HML ARS cluster is able to function as chromosomal origin when inserted at a different location in chromosome III (R. Y. Huang, M. J. Eddy, and D. Kowalski, unpublished data). However, when inserted at the HML region, ARS305 is a silent origin, as indicated by a complete Y arc and the absence of a bubble arc on the 2D gel (Fig. 5B). Moreover, the major pause site which is absent in the mutant strain with the HML ARS cluster deleted (Fig. 5A) was reconstituted in the strain with ARS305 inserted (Fig. 5B). Two minor pause sites difficult to detect in the strain deleted for the HML ARS cluster are seen on the late Y arc in the strain with ARS305 inserted. The above experiment indicates that a heterologous ARS element from an active chromosomal replication origin is silenced when inserted near the HML locus and can recapitulate the major pause site. Additionally, the deleted sequence encompassing the HML ARS cluster is not essential for origin silencing or fork pausing at the heterologous ARS element inserted near HML.
FIG. 5.
ARS305 can functionally substitute for the HML ARS cluster in generating a replication fork pause, as determined by 2D gel electrophoresis of replication intermediates from the indicated strains probed for specific restriction fragments diagrammed in the maps. The arrowhead denotes a pause signal. (A) A 2.85-kb EcoRI/FspI genomic fragment from the parental YPH98 strain; (B) a 3.45-kb EcoRI/FspI genomic fragment from a mutant yeast strain (YWY2) that has the HML ARS cluster replaced by a copy of ARS305. The 549-bp copy of ARS305 was inserted into the PstI site near the HML locus with the HML ARS cluster deleted.
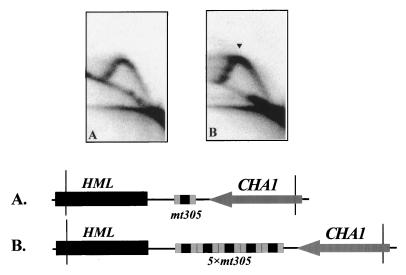
A and B1 elements which bind ORC contribute to the generation of the pause signal.
Sequences important for ARS function have been demonstrated to be necessary for full replication origin activity in the chromosome. In the case of ARS305, the A element is essential for plasmid ARS activity and for chromosomal origin function, and the B1 element contributes to replication efficiency (28, 29). These elements from ARS305 are functionally exchangeable with the corresponding elements in ARS1 (36). The A element is the primary binding site for ORC and the B1 element also contributes to ORC binding (3, 50, 56). Since the copy of the ARS305 that replaced the HML ARS cluster remained silent at HML and reconstituted the pause, we asked whether sequences essential for ARS305 origin activity are important for the generation of replication fork pause. To test this possibility, we replaced the HML ARS cluster with a mutated ARS305 derivative. The derivative contains the wild-type A and B1 elements of ARS305 with the 3′ sequences, including the B4 module and DUE, replaced by a randomly generated sequence that greatly weakens ARS function (36). The HML locus of the resulting mutant strain was analyzed for a replication fork pause by 2D gel electrophoresis. As shown in Fig. 6A, replacement of the HML ARS cluster with the mutated ARS305 derivative is unable to reconstitute the major pause site. However, when five copies of the mutated ARS305 derivative are concatenated and inserted in place of the HML ARS cluster, the major pause site is recapitulated. When the major pause site is restored, the minor pause signals on the late Y arc also become clearer (Fig. 6B). Therefore, five concatenated copies of the mutated ARS305 derivative containing five copies of the A and B1 elements are able to reconstitute the major pause signal. The capability of multiple copies of the A and B1 elements to stall the fork movement suggests that these ORC-binding elements contribute to the generation of replication fork pause at the HML locus.
FIG. 6.
Multiple copies of the A and B1 elements can functionally substitute for the HML ARS cluster in generating a replication fork pause, as determined by 2D gel electrophoresis of replication intermediates from the indicated strains probed for the specific restriction fragment diagrammed in the maps. The HML ARS cluster was substituted by a mutated ARS305 derivative (mt305) which contains the A and B1 elements and mutated sequences in place of the B4 element and the easily unwound sequences. (A) One copy of mt305 replacing the natural HML ARS cluster was analyzed on a 3.2-kb EcoRI/FspI genomic fragment. (B) Five copies of mt305 replacing the HML ARS cluster were analyzed on a 4.4-kb EcoRI/FspI genomic fragment. The arrowhead denotes a pause signal.
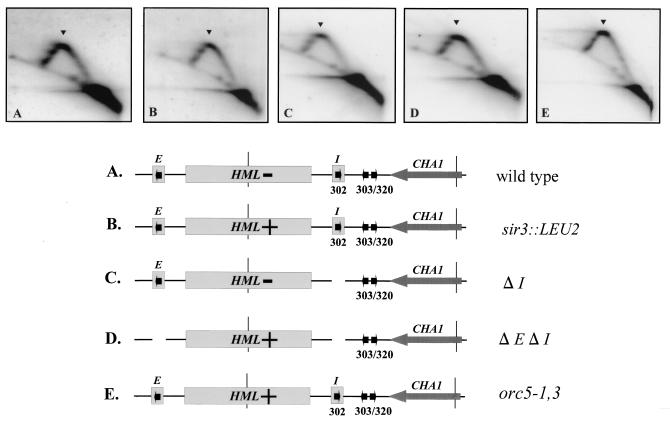
Replication fork pause site at the HML ARS cluster is independent of mutations that abrogate transcriptional silencing at HML.
Detection of the replication fork pause sites at the transcriptionally silent HML locus raises the possibility that regulation of transcriptional silencing may be involved in stalling the fork progression. Fork pauses at HML are associated with ARS elements required for transcriptional silencing. ARS301 and ARS302 comprise the E and I transcriptional silencers, respectively. Furthermore, trans-acting factors such as ORC have dual roles in both replication and silencing and bind to these ARS elements. The more compact heterochromatin-like structure at HML that is important for transcriptional silencing may also contribute to the slowdown of fork migration. Despite the existence of common functional elements between transcriptional silencers and DNA replication origins such as ARSs and ORC, silencing of replication origin activity at HML has been shown to be independent of transcriptional silencing (67). However, whether generation of a replication fork pause at HML is linked to the transcriptional silencing is not known.
To test the involvement of transcriptional silencing in regulating the fork pauses at HML, we examined certain cis- and trans-acting mutations that abrogate transcriptional silencing for their effects on the replication fork pause at the HML ARS cluster. Deletions that removed the cis-acting silencer elements E and I were tested. These elements bind several proteins that contribute to transcriptional silencing, including ORC, Rap1p, and Abf1p, and also contribute to establishing a heterochromatin-like structure (21, 37, 45). Deletion of both E and I elements abrogates transcriptional silencing at HML (40). We examined an ORC mutant, orc5-1,3, which is defective in transcriptional silencing but competent for initiation of DNA replication (22). We also looked at the disruption of the SIR3 gene, sir3::LEU2, that abrogates silencing of HML transcription (40). As seen in Fig. 7, the major fork pause at the HML ARS cluster is apparent in all cases, and the three-spot pattern (one major and two minor) similar to those in YPH98 derivatives is present in all of the above strains. The results indicate that replication fork pausing near the HML ARS cluster is independent of cis- and trans-acting mutations that abrogate transcriptional silencing at HML.
FIG. 7.
Detection of replication fork pause sites at the HML ARS cluster is independent of mutations that abrogate transcriptional silencing at HML, as determined by 2D gel electrophoresis of replication intermediates from the indicated strains probed for the 3.9-kb EcoRI/FspI fragment diagrammed in the maps. The maps in all panels show positions of the HML locus, the E and I silencer elements, and the ARS elements and the transcriptional status of HML (−, silent; +, active). A HindIII/BamHI fragment from was used as a probe in the hybridization. Arrowheads denote a replication pause signal. (A) DMY1, the parental strain; (B) DMY2, a strain with a mutation disrupting the SIR3 gene (shown as sir3::LEU2); (C) DMY94, a strain with the I silencer deleted (ΔI); (D) DMY95, a strain with E and I silencers deleted (ΔE ΔI); (E) JRY4556, a strain with a mutation in ORC5 (orc5-1,3) which is defective in transcriptional silencing but competent in DNA replication.
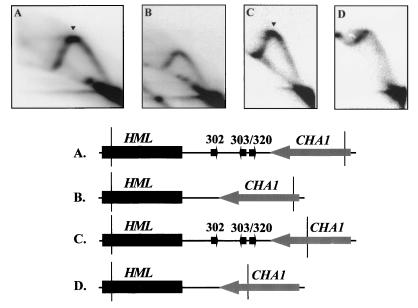
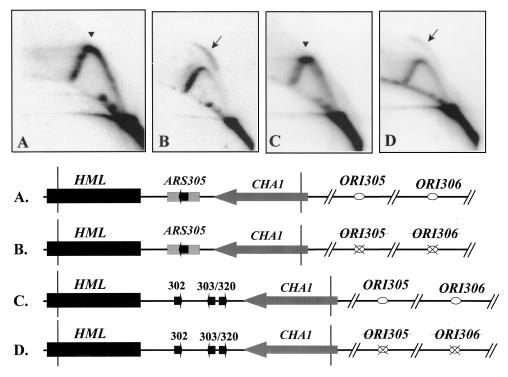
Deletion of ORI305 and ORI306 leads to loss of the replication fork pause signal and activation of silent origins at HML.
Forks that pause at HML ARS305 are derived from active origins present only on one side of the HML locus. The closest active origins, ORI305 and ORI306, are located 25 and 59 kb away from HML (13, 28, 72). These are two of the earliest firing origins in chromosome III (51). ORI305 and ORI306 are presumed to supply most, if not all, of the forks that pause at HML. If this is so, deletion of ORI305 and ORI306 would move the source of the replication forks to ORI307, which is 93 kb away (13), and significantly delay the fork arrival at the HML locus. If this hypothesis is true, deletion of ORI305 and ORI306 would be expected to diminish fork pause signal at the HML locus.
To test this hypothesis, we examined fork pausing at ARS305 inserted in place of the HML ARS cluster after the deletion of active distal origins, ORI305 and ORI306. The results showed that the major fork pause, previously identified by a heavy spot at the peak of the Y arc at ARS305 (Fig. 8A), is not present after deletion of ORI305 and ORI306 (Fig. 8B). Instead, the copy of ARS305 near the HML region was no longer silenced and showed origin activity at a high level. Origin activation is indicated by the appearance of a distinct high-rising arc, termed a bubble arc. A high level of origin activity is reflected by the high ratio of bubble arc signal to the early Y-arc signal. Activation of ARS305 inserted near HML is about 75% as efficient as in its native ORI305 location (29), as estimated from the bubble arc/early Y-arc ratio. Thus, loss of the pause signal is accompanied by activation of the silent origin in most of cells. Origin activation was also seen at the natural silent origin in the HML ARS cluster when ORI305 and ORI306 were deleted (67). Activation of the silent origin is accompanied by the loss of the major fork pause (compare Fig. 8C and D). ARS elements in the HML ARS cluster function inefficiently in plasmids and in the chromosome (67). Thus, origin activation does not occur in every cell, and a low ratio of bubble arc to early Y-arc signals is seen (Fig. 8D). Although a complete Y arc indicative of passive replication is seen in some cells, a pause signal at the peak of the Y arc is not apparent (Fig. 8D). Our results indicate that deletion of the adjacent early-firing origins from the chromosome reduces the pause signal, suggesting that fork arrival at HML from the adjacent early-firing origins was delayed. Furthermore, loss of the pause signal at the silent origins is accompanied by the activation of the silent origins in the chromosome.
FIG. 8.
Deletion of the adjacent active origins from the chromosome leads to the activation of silent origins near HML and to the disappearance of the replication fork pause sites, as determined by 2D gel electrophoresis of replication intermediates from the indicated strains probed for specific restriction fragments diagrammed in the maps. Arrowheads denote a pause signal; arrows point to a replication bubble arc. (A) A 3.45-kb EcoRI/FspI genomic fragment from a mutant yeast strain (YWY2) that has the HML ARS cluster replaced by a copy of ARS305; (B) a 3.45-kb EcoRI/FspI genomic fragment from a mutant yeast strain (YWY4) that has the HML ARS cluster replaced by a copy of ARS305 and that has ORI305 and ORI306 deleted; (C) a 3.9-kb EcoRI/FspI genomic fragment from YPH98; (D) a 3.9-kb EcoRI/FspI genomic fragment from a mutant strain that has ORI305 and ORI306 deleted.
ORC2 maintains origin silencing at a replication fork pause site.
All ARS elements and chromosomal origins tested require ORC for replication function. The observations that the heterologous ARS305 origin is silenced at HML and can recapitulate the pause, and that DNA elements A-B1 which bind ORC contribute to the pause, suggest that ORC may play a role in fork pausing. Orc2p, one of the six subunits of ORC, is known to regulate both origin activity and the time at which origins fire in S phase (21, 35, 48, 61). The orc2-1 mutation can have either a positive or a negative effect on activity, depending upon the specific origin, and can advance the activation time of origins that fire late in S phase (61). The silent origin at the HML ARS cluster fires late in S phase when activated by deletion of adjacent origins (M. Vujcic, M. J. Eddy, and D. Kowalski, unpublished data). ORC is known to bind to ARS elements that are silent origins in the chromosome (2). We examined the possible role of ORC in replication fork pausing at the silent origin in the HML ARS cluster using the temperature-sensitive orc2-1 mutant.
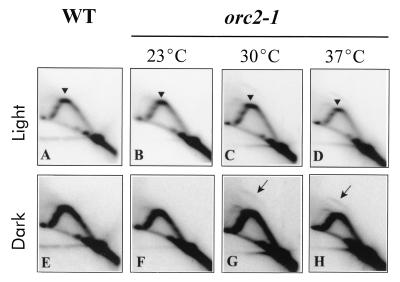
We performed 2D gel electrophoresis of replication intermediates on genomic DNA isolated from cultures of orc2-1 and an isogenic wild-type strain grown at 23, 30, and 37°C. Replication intermediates were probed for the silent origin at the HML ARS cluster. The orc2-1 mutation had no detectable effect on fork pausing at the silent origin at all temperatures tested (Fig. 9B to D). Additionally, dark exposures of the radioactive signals reveal that the silent origin at the HML ARS cluster remains silent in the wild-type strain (Fig. 9E) and in the orc2-1 mutant at 23°C (Fig. 9F). The elevated temperatures had no effect on the silent origins in the wild-type strain (data not shown). However, the silent origins were activated in the temperature-sensitive orc2-1 mutant at 30 and 37°C, as indicated by the appearance of a bubble arc (Fig. 9G and H). The low ratio of the bubble arc to the early Y arc indicates that the silent origin becomes active in a subpopulation of cells. A corresponding low-level reduction in the pause signal contributed by the majority of cells would be difficult to detect. The results indicate that ORC2 plays a role in maintaining origin silencing at the replication fork pause site.
FIG. 9.
Effect of orc2-1 mutation on fork pausing and silent origin activation at the HML ARS cluster. DNA replication intermediates were isolated from cultures of wild-type (WT) cells and the orc2-1 mutant grown at the permissive temperature (23°C) overnight and then shifted to an elevated temperature (30 or 37°C) for 2 h prior to DNA isolation. Southern blots of 2D gels were probed for the HML ARS cluster. (A to D) Light exposure for analysis of pause sites; (E to H) dark exposure for analysis of the activation of the silent origin. Arrowheads denote a pause signal; arrows point to a replication bubble arc.
DISCUSSION
Yeast ARS elements are known to function as replication origins, and in exceptional cases, they function as transcriptional silencers. We have discovered that ARS elements can also serve as replication fork pause sites in an S. cerevisiae chromosome. Natural fork pause sites were detected at ARS elements on both sides of a transcriptionally silent mating locus, HML. The pause sites occur at the HML ARS cluster on the centromeric side of HML and at ARS301 on the telomeric side. Since the HML locus is transcriptionally silent, fork pauses at this locus cannot be caused by head-on collisions of transcription and replication machinery seen at other loci in yeast chromosomes (14). Fork pause sites are also known to occur at yeast centromeres, and a fork barrier occurs in the ribosomal DNA locus outside the replication origin (10, 23). Given our findings that deletion of the HML ARS cluster eliminates the pause signal and that substitution of the HML ARS cluster with the heterologous ARS305 regenerates the pause, we conclude that ARS elements are responsible for pausing replication fork movement in the chromosome.
ARS elements that pause replication fork movement near HML are silent as chromosomal replication origins. Both the HML ARS cluster and ARS301 are known to be silent origins (16, 67). As shown here, ARS305 derived from the active chromosomal origin, ORI305, is silenced when inserted at the HML locus. Instead of functioning as a replication origin, the ARS305 insert functions as a fork pause site at HML. Fork pausing at the ARS305 insert is independent of the HML ARS cluster, which was deleted in this strain. Deletion of two distal active origins, ORI305 and ORI306, results in the loss of the fork pause and the activation of the ARS305 insert as a chromosomal origin at HML. Activation of silent origins at the native HML ARS cluster and at the native ARS301 location was also seen when the distal active origins were deleted (67). Here we found that activation of the silent origin at the HML ARS cluster results in loss of the replication fork pause. The establishment of both origin silencing and fork pausing at a copy of ARS305 inserted near HML, together with the loss of fork pausing seen when the silent origins are activated, indicates that origin silencing is tightly linked to replication fork pausing. The capacity of the silent origins at the pause sites to become active replication origins demonstrates that the silent origins are competent for replication initiation in the chromosome.
ARS elements are known to bind ORC, which is required for replication origin activity in budding yeast (3, 50, 56). ORC also functions in transcriptional silencing at HM loci, and at HML, ORC binds the silencer elements ARS301 and ARS302 (2, 21, 40). A cell cycle-dependent alternation between an ORC-dependent postreplicative state and a Cdc6p-dependent prereplicative state occurs at ARS301 (58). These observations, together with the direct demonstration of origin activation at ARS301 after deletion of adjacent origins (67), suggest that ORC and associated replication proteins are responsible for the initiation competency of silent origins. As shown here, origin silencing and fork pausing near HML are tightly linked. Thus, the fork pause may consist of a complex of ARS DNA with proteins, such as ORC and associated replication proteins, that confer initiation competence to replication origins.
The A and B1 elements of ARS305 interact with ORC (34). The A element is essential for initiation competence in the chromosome and the B1 element is important (28, 29). Our finding that multiple copies of A and B1 elements recapitulate the major fork pause site indicates that DNA elements known to interact with ORC contribute to the replication fork pause. One copy of the A and B1 elements is not sufficient to generate the pause signal at the HML locus, while one copy of ARS305 is sufficient. In addition to A and B1 elements, ARS305 contains a DUE and a B4 element which are functionally important (36). Besides ORC binding, initiation competence at an origin requires Cdc6p and the MCM2-7 complex, a DNA helicase (26, 73). The ARS1 association with Mcm2p, but not with ORC, is diminished by disruption of the B2 element (73), which contains a DUE (36). Mutations of the DUE and the B4 element in the ARS305 derivative tested likely interfere with the unwinding events important for initiation. This mutation, like the B2/DUE mutation in ARS1 (73), may weaken the stable association of the MCM2-7 in the complex with ARS. This could explain why one copy of A-B1 alone is insufficient to generate a pause signal, while one copy of A-B1 within the context of a functional ARS element, ARS305, is sufficient. Interestingly, a mutation in another initiation protein, Mcm10p (26), can have the opposite effect at an active replication origin. The mcm10 mutation appears to stabilize the association of initiation proteins with the ARS and leads to origin inactivation and replication fork pausing (42).
ORC2 plays a regulatory role in initiation and may have either positive or negative effects on active replication origins, depending upon the specific origin (61). The observation that the orc2-1 mutation activates the silent origin at the HML ARS cluster indicates a role for wild-type ORC2 in the maintenance of origin silencing at the replication fork pause site. ORC2 is known to regulate replication timing at origins. The orc2-1 mutation advances the time that certain late-activated origins fire in S phase (61). Interestingly, the silent origins initiate replication late in S phase when activated by deleting adjacent early-firing origins (Vujcic et al., unpublished). The late activation time likely accounts for their passive replication by a fork from an adjacent early-firing origin (ORI305 and ORI306). A sufficient advance in the origin activation time at the HML ARS cluster in a subpopulation of cells could account for the activation of the silent origin seen in the orc2-1 mutant. The orc2-1 mutant could also affect origin silencing by delaying replication timing at adjacent active origins or reducing their activity. Replication timing experiments on the orc2-1 mutant will be required to fully understand the mechanism by which ORC2 affects origin silencing at the fork pause site. The simplest interpretation of all of our results is that a late-programmed initiation complex at the silent origin is responsible for pausing a replication fork that arrives relatively early in S phase. Consistent with this, the silent origin is activated and the natural pause signal is lost when fork arrival is delayed until later in S phase by deleting adjacent, early-firing origins. Also consistent is the observation that an active origin is silenced in the late-replicating context of HML and a pause site is created.
RAD53 and MEC1, which encode protein kinases that function in S-phase checkpoints, also function to regulate replication timing at origins. In the presence of DNA damage or replication stress, these checkpoint kinases suppress the firing of certain late-activated origins (57, 61). The RAD53 pathway attenuates the activity of the Cdc7p-Dbf4p kinase and, at late-activated origins, inhibits recruitment of the single-strand binding protein RPA into the initiation complex (63, 69). The silent origins at the pause sites are programmed to activate late in S phase (Vujcic et al., unpublished). Recently we found that a rad53 mutation activates silent origins at the HML ARS cluster and at ARS301 in the presence of DNA damage, while a mec1 mutation activates only ARS301 (68). Thus, these checkpoint kinase mutations activate silent origins at the two major pause sites, the HML ARS cluster and ARS301. The available evidence suggests that the wild-type checkpoint kinases suppress activation of a late-programmed initiation complex at silent origins after DNA damage.
The rad53 and mec1 mutations show no detectable activation at the silent origins in the absence of DNA damage, unlike the orc2-1 mutation studied here. In the presence of DNA damage, the natural pause sites at the silent origins near HML become stronger. Additionally, a novel pause site is created at an active replication origin, and origin activity is inhibited (68). Thus, fork pause sites at replication origins appear to be inducible and may function to arrest replication fork progression and inhibit origin activation during the DNA damage response. In this way, replication fork pause sites at origins could function to coordinate initiation and elongation of replication in the chromosome during the S-phase checkpoint induced by DNA damage.
Another possible function of natural pause sites is in signaling during cell cycle regulation in the absence of DNA damage or replication stress such as nucleotide deprivation. In bacteria, replication fork progression through DNA is kept under surveillance to avoid inactivation of stalled forks and subsequent mutations (54). A similar process may exist in yeast (12). The encounter of a replication fork with a natural pause site may trigger a checkpoint signal in an unperturbed S phase. The signal could convey to cell cycle regulators that replication of the chromosome is incomplete and thereby delay the exit from S phase. Thus, natural pause sites in the chromosome could serve to monitor replication fork progression and link it to S-phase progression in the cell cycle.
We found that the occurrence of the replication fork pause in the HML ARS cluster is independent of cis- and trans-acting mutations that abrogate transcriptional silencing. Also, the occurrence of the pause site in the HML ARS cluster does not require ARS302, the I transcriptional silencer, which was deleted in two strains examined. The I silencer establishes a boundary between active and inactive chromatin (4), and so this boundary alone is not responsible for the fork pause. Remaining in the HML ARS cluster in these deletion mutants are ARS303 and ARS320, silent origins which are not essential for transcriptional silencing in a plasmid but may contribute to the potency of transcriptional silencing in the chromosome (40). Our results suggest that replication fork pausing could be a primary function of ARS303 and ARS320. The replication fork from ORI305 encounters these ARS elements prior to progressing into HML (Fig. 1). HML is known to be duplicated late in S phase, and the average rate of replication fork movement in the HML region (1.3 kb/min) is much lower than at earlier replicated regions of the chromosome (3.6 kb/min) (51). The molecular mechanism for the rate reduction is unknown. One hypothesis is that fork movement is generally slowed by the heterochromatin structure at HML, although there is no direct evidence for this possibility. Alternatively, fork movement could be slowed only at specific pause sites. The major pause sites that we discovered at the HML ARS cluster and at ARS301 likely contribute to, or possibly determine, the reduced rate of replication fork movement in the HML region.
Another possible function of pause sites at initiation-competent silent origins is to facilitate inheritance of a late-replicating heterochromatin structure and transcriptional silencing. Transcriptional silencing at HM loci requires passage through S phase and ORC function (21, 22, 43). The late-replicating heterochromatin structure must be faithfully duplicated to maintain transcriptional silencing. At HMLα, the heterochromatin structure includes ORC, associated silent information regulator and other specific proteins, a protected HO endonuclease cleavage site, an exposed transcription promoter region for α1 and α2 coding regions, phased nucleosomes, and specifically hypoacetylated histones (4, 8, 70; reviewed in reference 24). Replication fork proteins, such as PCNA, components of the replication initiation complex in addition to ORC, such as Cdc7p, and chromatin assembly factors also contribute to heterochromatin-mediated transcriptional silencing (18, 20, 71). HML transcriptional silencers are substantially more potent in their native context than in other chromosome locations or in a plasmid (24, 40). In the chromosome, the replication fork from the early-firing ORI305 arrives at the HML ARS cluster relatively early in S phase (51). The pause site at the HML ARS cluster could function to delay the duplication of the HML locus until later in S phase when transcription factors that bind to active, early-replicated genes are less abundant. Alternatively, the delay in duplication could be linked to a time in late S phase when factors required for heterochromatin and transcriptional repression are available or modified. The replication fork pause site at the HML ARS cluster could also function to recruit or localize such factors to HML. The silent origin at the fork pause site is programmed to activate replication late in S phase (Vujcic et al., unpublished). It is interesting in this regard that a histone deacetylase which contributes to formation of a transcriptionally repressive chromatin structure is targeted to replication foci only during late S phase in human cells (55).
Recently, a role for a replication fork pause site in imprinting and cell type switching in Schizosaccaromyces pombe was identified. The pause site occurs at the imprinting site at the mat1 locus and involves swi1p and swi3p (11). The replication fork pause is thought to facilitate placement of an RNA primer at a specific position by lagging-strand DNA replication. Leading-strand replication in a cell that inherits the imprint is thought to result in a DNA break that initiates mat1 switching. In S. cerevisiae, a different mechanism for switching mating type is used (24). The HO endonuclease induces a DNA break at the MAT locus in the G1 phase of the cell cycle, and the mating-type switch occurs before MAT replicates. If the pause sites flanking HML function in mating-type switching in S. cerevisiae, they must employ a mechanism distinct from that used in S. pombe.
Strong parallels exist between budding yeast and D. melanogaster in terms of the involvement of ORC and other replication proteins in late-replicating heterochromatin structures and gene silencing. Sequence-specific DNA binding by D. melanogaster ORC (DmORC) regulates initiation of replication, and a large number of early embryonic replication origins are silent in differentiated cells (1, 6, 60). DmOrc2p is enriched in heterochromatin and mutations in the gene affect replication timing, as is the case in budding yeast (39, 47, 61). DmORC localizes the non-DNA binding protein HP-1 into heterochromatin likely through DmOrc1p, similar to the role of yeast Orc1p in recruiting Sir1p (27, 65; reviewed in reference 19). DmORC2 can complement the transcriptional silencing defect of orc2-1 in budding yeast (17). Mutations in DmORC2 and in the gene encoding the replication fork protein DmPCNA suppress heterochromatin-mediated gene silencing in Drosophila (25, 47), as do mutations in homologous genes in budding yeast. Finally, the specific pattern of histone hypoacetylation is identical in heterochromatin in Drosophila and budding yeast (8). Fork pause sites have been proposed to occur late in S phase in heterochromatic satellite sequences in diploid cells and to be responsible for DNA truncation in the shortened S phase of polytene cells (33). The remarkable similarities described above suggest that replication fork pause sites at initiation-competent silent origins in budding yeast may also occur in late-replicating heterochromatin in Drosophila as well as in other metazoans.
Our discovery of replication fork pause sites at initiation-competent silent origins in budding yeast opens the possibility of identifying them in other species and determining their function(s) within chromosomes. Further studies of replication pause sites in yeast will be necessary to test their possible roles in checkpoints, S-phase regulation, and mating-type switching and in transcriptionally silent heterochromatin.
ACKNOWLEDGMENTS
We thank James Broach, Phil Hieter, Carol Newlon, Jasper Rine, and Bruce Stillman for yeast strains and plasmids. Thanks go to Martha Eddy for technical assistance and to Ruea Huang and John Yates for helpful comments on the manuscript.
This work was supported in part by shared resources of Roswell Park Cancer Center Support Grant (P30 CA16056) and by grant GM-30614 from the National Institutes of Health.
REFERENCES
- 1.Austin R J, Orr-Weaver T L, Bell S P. Drosophila ORC specifically binds to ACE3, and origin of DNA replication control element. Genes Dev. 1999;13:2639–2649. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell S P, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 4.Bi X, Braunstein M, Shei G J, Broach J R. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc Natl Acad Sci USA. 1999;96:11934–11939. doi: 10.1073/pnas.96.21.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielinsky A K, Gerbi S A. Chromosomal ARS1 has a single leading strand start site. Mol Cell. 1999;3:477–486. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal A B, Kriegstein H J, Hogness D S. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Brand A H, Micklem G, Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- 8.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 10.Brewer B J, Fangman W L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 11.Dalgaard J Z, Klar A J. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 12.Desany B A, Alcasabas A A, Bachant J B, Elledge S J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshpande A M, Newlon C S. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4305–4313. doi: 10.1128/mcb.12.10.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshpande A M, Newlon C S. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 15.Dhar V, Schildkraut C L. Role of EBNA-1 in arresting replication forks at the Epstein-Barr virus oriP family of tandem repeats. Mol Cell Biol. 1991;11:6268–6278. doi: 10.1128/mcb.11.12.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubey D D, Davis L R, Greenfeder S A, Ong L Y, Zhu J G, Broach J R, Newlon C S, Huberman J A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrenhofer-Murray A E, Gossen M, Pak D T, Botchan M R, Rine J. Separation of origin recognition complex functions by cross-species complementation. Science. 1995;270:1671–1674. doi: 10.1126/science.270.5242.1671. [DOI] [PubMed] [Google Scholar]
- 18.Ehrenhofer-Murray A E, Kamakaka R T, Rine J. A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics. 1999;153:1171–1182. doi: 10.1093/genetics/153.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eissenberg J C, Elgin S C. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 20.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foss M, McNally F J, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 22.Fox C A, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- 23.Greenfeder S A, Newlon C S. Replication forks pause at yeast centromeres. Mol Cell Biol. 1992;12:4056–4066. doi: 10.1128/mcb.12.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haber J E. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 25.Henderson D S, Banga S S, Grigliatti T A, Boyd J B. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J. 1994;13:1450–1459. doi: 10.1002/j.1460-2075.1994.tb06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye B K. Mcm10 and the MCM2–7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000;14:913–926. [PMC free article] [PubMed] [Google Scholar]
- 27.Huang D W, Fanti L, Pak D T, Botchan M R, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang R Y, Kowalski D. A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J. 1993;12:4521–4531. doi: 10.1002/j.1460-2075.1993.tb06141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang R Y, Kowalski D. Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res. 1996;24:816–823. doi: 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huberman J A, Spotila L D, Nawotka K A, el-Assouli S M, Davis L R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 31.Kelly T J, Brown G W. Regulation of chromosome replication. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 32.Kingsman A J, Clarke L, Mortimer R K, Carbon J. Replication in Saccharomyces cerevisiae of plasmid pBR313 carrying DNA from the yeast trp1 region. Gene. 1979;7:141–152. doi: 10.1016/0378-1119(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 33.Leach T J, Chotkowski H L, Wotring M G, Dilwith R L, Glaser R L. Replication of heterochromatin and structure of polytene chromosomes. Mol Cell Biol. 2000;20:6308–6316. doi: 10.1128/mcb.20.17.6308-6316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee D G, Bell S P. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 36.Lin S, Kowalski D. Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol Cell Biol. 1997;17:5473–5484. doi: 10.1128/mcb.17.9.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loo S, Fox C A, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loupart M, Krause S, Heck M S. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol. 2000;10:1547–1556. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- 39.MacAlpine D M, Zhang Z, Kapler G M. Type I elements mediate replication fork pausing at conserved upstream sites in the Tetrahymena thermophila ribosomal DNA minichromosome. Mol Cell Biol. 1997;17:4517–4525. doi: 10.1128/mcb.17.8.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahoney D J, Broach J R. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol Cell Biol. 1989;9:4621–4630. doi: 10.1128/mcb.9.11.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 42.Merchant A M, Kawasaki Y, Chen Y, Lei M, Tye B K. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:3261–3271. doi: 10.1128/mcb.17.6.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller A M, Nasmyth K A. Role of DNA replication in the repression of silent mating type loci in yeast. Nature. 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- 44.Miller C A, Kowalski D. cis-acting components in the replication origin from ribosomal DNA of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5360–5369. doi: 10.1128/mcb.13.9.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 46.Newlon C S, Collins I, Dershowitz A, Deshpande A M, Greenfeder S A, Ong L Y, Theis J F. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- 47.Pak D T, Pflumm M, Chesnokov I, Huang D W, Kellum R, Marr J, Romanowski P, Botchan M R. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 48.Pasero P, Braguglia D, Gasser S M. ORC-dependent and origin-specific initiation of DNA replication at defined foci in isolated yeast nuclei. Genes Dev. 1997;11:1504–1518. doi: 10.1101/gad.11.12.1504. [DOI] [PubMed] [Google Scholar]
- 49.Rao H, Marahrens Y, Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Mol Cell Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds A E, McCarroll R M, Newlon C S, Fangman W L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–94. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rivier D H, Ekena J L, Rine J. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics. 1999;151:521–529. doi: 10.1093/genetics/151.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivier D H, Rine J. An origin of DNA replication and a transcription silencer require a common element. Science. 1992;256:659–663. doi: 10.1126/science.1585179. [DOI] [PubMed] [Google Scholar]
- 54.Rothstein R, Michel B, Gangloff S. Replication fork pausing and recombination or “gimme a break”. Genes Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- 55.Rountree M R, Bachman K E, Baylin S B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 56.Rowley A, Cocker J H, Harwood J, Diffley J F. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santocanale C, Diffley J F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 58.Santocanale C, Diffley J F. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- 59.Santocanale C, Sharma K, Diffley J F. Activation of dormant origins of DNA replication in budding yeast. Genes Dev. 1999;13:2360–2364. doi: 10.1101/gad.13.18.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol Cell Biol. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 62.Struhl K, Stinchcomb D T, Scherer S, Davis R W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Theis J F, Newlon C S. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 66.Van Houten J V, Newlon C S. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol Cell Biol. 1990;10:3917–25. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vujcic M, Miller C A, Kowalski D. Activation of silent replication origins at autonomously replicating sequence elements near the HML locus in budding yeast. Mol Cell Biol. 1999;19:6098–6109. doi: 10.1128/mcb.19.9.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, Beerman T A, Kowalski D. Antitumor drug adozelesin differentially affects active and silent origins of DNA replication in yeast checkpoint kinase mutants. Cancer Res. 2001;61:3787–3794. [PubMed] [Google Scholar]
- 69.Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss K, Simpson R T. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα Mol. Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 72.Zhu J, Newlon C S, Huberman J A. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]