ABSTRACT
Lung organogenesis requires precise timing and coordination to effect spatial organization and function of the parenchymal cells. To provide a systematic broad-based view of the mechanisms governing the dynamic alterations in parenchymal cells over crucial periods of development, we performed a single-cell RNA-sequencing time-series yielding 102,571 epithelial, endothelial and mesenchymal cells across nine time points from embryonic day 12 to postnatal day 14 in mice. Combining computational fate-likelihood prediction with RNA in situ hybridization and immunofluorescence, we explore lineage relationships during the saccular to alveolar stage transition. The utility of this publicly searchable atlas resource (www.sucrelab.org/lungcells) is exemplified by discoveries of the complexity of type 1 pneumocyte function and characterization of mesenchymal Wnt expression patterns during the saccular and alveolar stages – wherein major expansion of the gas-exchange surface occurs. We provide an integrated view of cellular dynamics in epithelial, endothelial and mesenchymal cell populations during lung organogenesis.
KEY WORDS: Lung development, Single-cell transcriptomics, Progenitor cells, RNA velocity, Type 1 pneumocyte, Mouse
Summary: Time-series single-cell transcriptomic analysis identifies previously unrecognized transitional states, population dynamics, fate trajectories and gene-expression evolution in the developing mouse lung.
INTRODUCTION
Organogenesis of the lung requires precisely timed and coordinated signaling between multiple cell types, as well as the adaptation of cellular functions during the transition from intrauterine to extrauterine life (Morrisey and Hogan, 2010). The later aspects of lung development are especially dynamic, with dramatic shifts in both spatial organization and function of epithelial, endothelial and mesenchymal cells required to coordinate sacculation and alveolarization (Herriges and Morrisey, 2014; Warburton et al., 2010). Although isolated single-cell genomic and lineage-tracing studies have provided important insights, there remains a need for an integrated understanding of the identity, localization and fate of parenchymal cells during lung development.
During the late terminal saccular stage [24 weeks to later fetal period in humans; embryonic day (E)17.5 to postnatal day (P)5 in mice; Warburton et al., 2010], the lung is especially susceptible to irreversible injury. The developmental program of the lung in the preterm infant can be permanently altered by exposure to hyperoxia and inflammation, resulting in persistent structural damage and lifelong respiratory impairment (Benjamin et al., 2020; Goss, 2018). What makes this stage so sensitive to injury and the molecular mechanisms that regulate normal developmental trajectories through this susceptible period remain incompletely understood. There is a crucial need for an unbiased, rigorous and systematic analysis of the complex cell states and interactions between them to provide context for understanding the functional mechanisms regulating lung development, homeostasis, injury and pathology.
To establish a comprehensive model of the cellular populations and their transcriptional changes during development, we performed single-cell RNA-sequencing (scRNA-seq) of 102,571 cells from lung tissue across nine time points from E12 to P14, and we created an easily searchable, deeply curated and temporally resolved single-cell atlas of the developing mouse lung. Applying transcriptional velocity-based fate-likelihood prediction (Lange et al., 2020 preprint) and RNA in situ hybridization (ISH), we characterize transcriptomic profiles of epithelial, endothelial and mesenchymal cells. We identify the emergence of a transitional population of epithelial cells [between early epithelial cells and alveolar type 1 (AT1) cells] beginning at E15 (suggesting early AT1 cell-fate commitment), and illustrate the developmental trajectory of the recently described Car4+ alveolar capillary (aCap) endothelial cells. We determine the transitions in Wnt ligand expression that accompany fundamental population shifts in the pulmonary mesenchyme during alveologenesis. We interrogate this cellular atlas to describe a previously unrecognized role of AT1 pneumocytes in the expression of genes associated with the vital process of elastin assembly and the supporting alveolar extracellular matrix. Finally, we provide an integrated view of the dynamics of the later stages of lung development, enhancing our understanding of the cell-specific trajectories during this vulnerable developmental period.
RESULTS
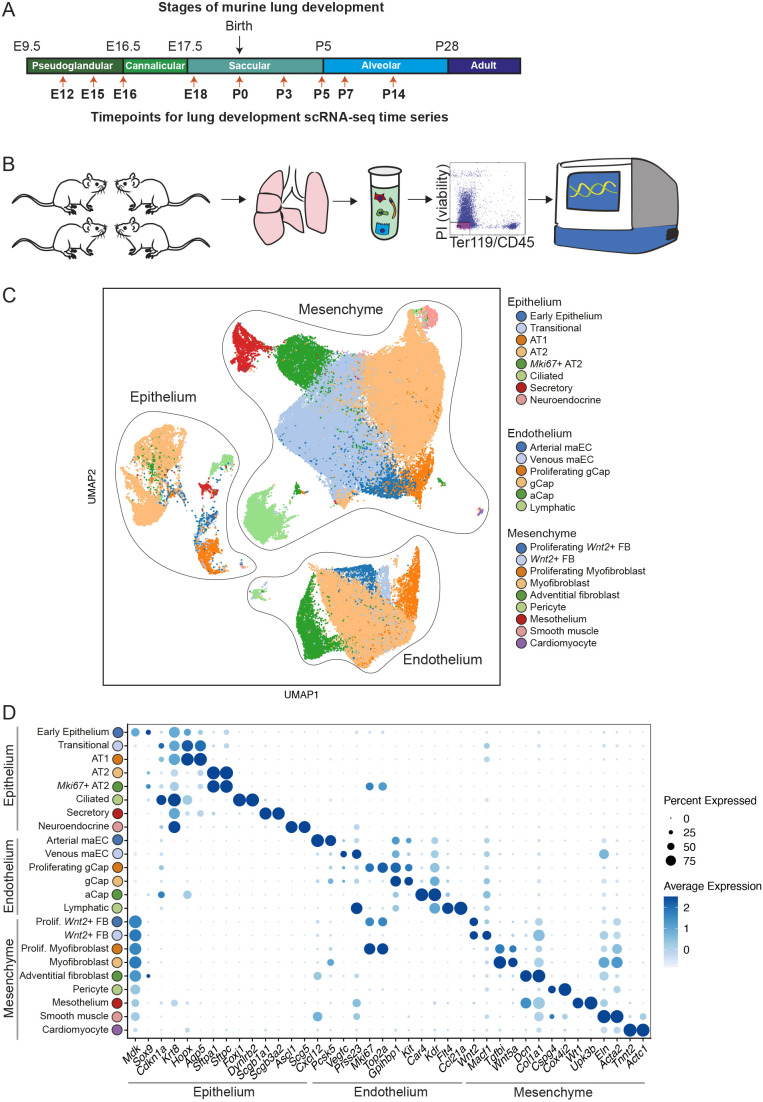
To assess the cellular landscape of the developing mouse lung (between E12 and P14), single-cell suspensions were generated from dissociated whole lung tissue (at least four mice per time point were pooled), parenchymal [CD45−/Ter119−] cells were sorted by fluorescence-activated cell sorting (FACS) and scRNA-seq was performed using the 10x Genomics Chromium platform (Fig. 1A,B). Depletion of cells expressing CD45 and Ter119 was carried out to remove immune and red blood cells, respectively.
Fig. 1.
Capturing the process of alveolarization at a single-cell resolution. (A) Schematic of time points sampled, overlaid onto histologically identified stages of murine lung development. (B) Experimental workflow: at least four mice were pooled per time point; lungs were harvested and single-cell suspensions were generated by enzymatic digest; viable, CD45− Ter119− cells were sorted by FACS for scRNA-seq library preparation. (C) UMAP embedding of all 102,571 cells that clustered, annotated by cell type. (D) Hallmark identifier genes for each cluster are plotted where the size of the dot indicates the proportion of cells within a cluster expressing that gene, and level of blue saturation indicates relative expression level.
After quality-control filtering, normalization and scaling (see Materials and Methods), 102,571 total single-cell transcriptomes were recovered, with a range of 3958 to 25,429 cells per time point, a median of 4509 (interquartile range 4293-4810) unique molecular identifiers (UMIs) per cell, and a median of 1799 genes (interquartile range 1494-2096) per cell (Fig. 1C,D; Fig. S1A-C). Unbiased clustering and cell-type annotation across all time points from E12 to P14 identified eight epithelial clusters, six endothelial clusters and nine mesenchymal clusters (Fig. 1C; Fig. S1A). The epithelial, endothelial and mesenchymal cells were independently extracted and separately analyzed to characterize cell-type-specific developmental changes over time.
Temporal evolution of population dynamics during lung development
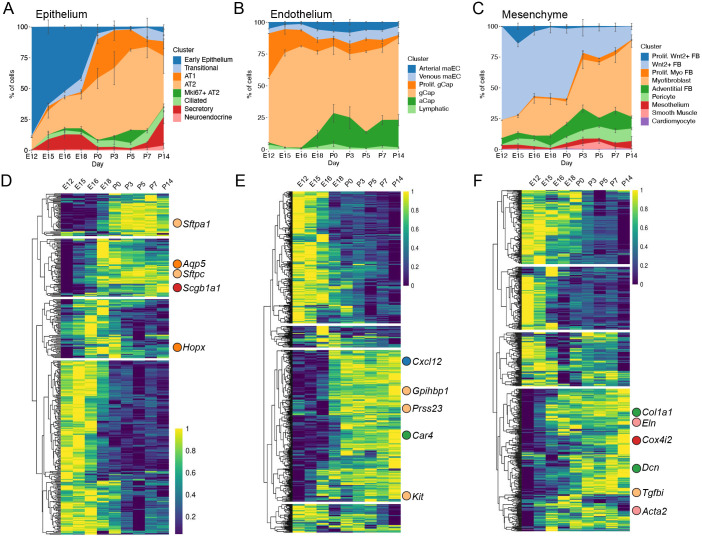
Analysis of the population structure of epithelial cells (n=10,918) from E12 to P14 identified eight distinct clusters (Fig. 1C,D) with notable changes over time in the proportional representation of the subpopulations (Fig. 2A). At E12, the predominant epithelial cell population is an early epithelial cluster identified by Mdk and Sox9 (cluster 1; Fig. 1D; Table S1), for which the relative number of cells in this population decreases significantly by time of birth (P0). By E18, all of the cell clusters expressing markers of mature lung alveolar epithelium appear, including clusters of transitional epithelial cells marked by Cdkn1a and Krt8 expression (cluster 2); alveolar type I epithelial (AT1) cells marked by Hopx and Aqp5 expression (cluster 3); a cluster of alveolar type II epithelial (AT2) cells marked by Sftpc and Sftpa1 expression (cluster 4); proliferating AT2 cells additionally expressing Mki67 and Top2a (cluster 5); secretory cells marked by Scgb1a1 and Scgb3a2 expression (cluster 6); ciliated cells marked by Foxj1 and Dynlrb2 expression (cluster 7); a small number of neuroendocrine cells marked by Ascl1 expression (cluster 8; Fig. 1D; Table S1). Our use of antibody labeling of each mouse lung sample before pooling single-cell suspensions and sequencing provided additional power to understand the relative abundance of each cell population at each time point. The relative proportion of subpopulations of epithelial, endothelial and mesenchymal cells recovered by scRNA-seq correlated highly with relative proportions of selected cell types measured by quantitative RNA ISH (Table S2) providing confidence that there was not a preferential loss of specific classes of cells during tissue dissociation or scRNA-seq analysis.
Fig. 2.
Cell populations in the lung change markedly over time. (A-C) Relative proportion of cell types as a function of time, as detected by scRNA-seq, in epithelium (A), endothelium (B) and mesenchyme (C). Error bars indicate standard error of the mean from at least four mice from each time point after E12. (D-F) Genes in epithelial cells (D) with statistically significant expression changes as a function of time (q<0.05) by Monocle3 analysis are plotted in a heatmap. Gene expression was log transformed and normalized from 0 to 1 and arranged using hierarchical clustering before plotting. Saturation shading of yellow to blue: high to low expression-level spectrum. Expression patterns were separated into four groups (k=4). When hallmark genes (Fig. 1D) were included in the heatmap, their location is indicated, with matching color to stacked area plots and UMAP above. Genes without statistically significant expression changes over time are not indicated. Similar analysis was performed for the endothelial cells (E) and the mesenchymal cells (F).
Compared with epithelial cells, endothelial cells (n=24,618) exhibited a more stable subpopulation structure over early stages of lung development (Fig. 2B). As early as E12, there were five distinct endothelial clusters: (1) macrovascular arterial endothelium (arterial maEC, expressing Vwf, Cxcl12 and Pcsk5); (2) macrovascular venous endothelium (venous maEC, expressing Vwf, Vegfc and Prss23); (3) general capillary (gCap, expressing Gpihbp1 and Kit), (4) proliferating gCap (expressing Mki67 and Gpihbp1); (5) lymphatic endothelium (expressing Flt4 and Ccl21a) (Figs 1C and 2B). At E18, a population of (6) endothelial cells emerged that is distinguished by expression of Car4 (aCap, alveolar capillary; Fig. 2B) (Gillich et al., 2020; Niethamer et al., 2020; Vila Ellis et al., 2020).
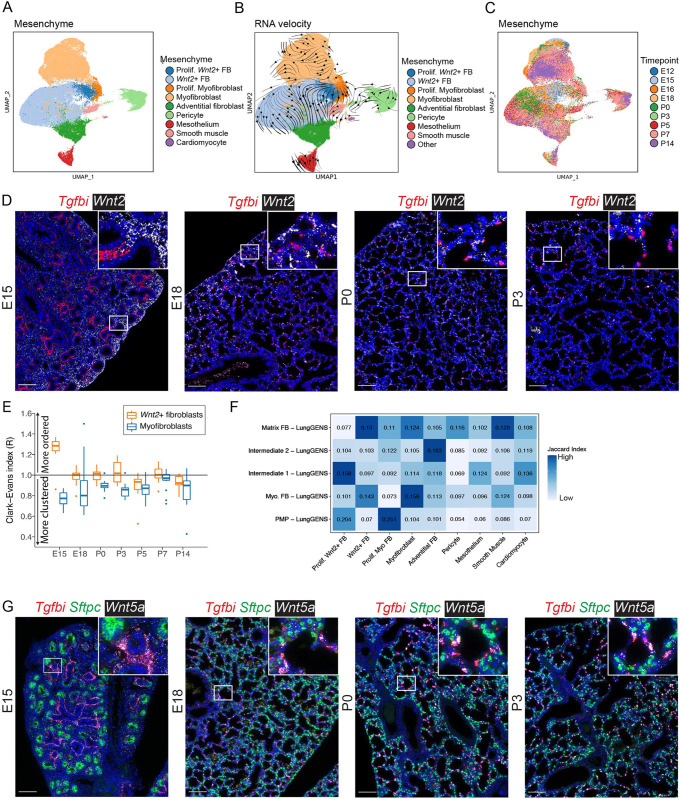
The majority of cells isolated and sequenced were pulmonary mesenchymal cells (n=67,035). Cluster analysis indicated nine distinct populations: (1) proliferating Wnt2+ fibroblasts (expressing Mki67, Top2a, Wnt2 and Macf1); (2) Wnt2+ fibroblasts (expressing Wnt2 and Macf1); (3) proliferating myofibroblasts (expressing Mki67, Top2a, Tgfbi and Wnt5a); (4) myofibroblasts (expressing Tgfbi and Wnt5a); (5) adventitial fibroblasts (expressing Dcn and Col1a1); (6) pericytes (expressing Cspg4 and Cox4i2); (7) mesothelium (expressing Wt1 and Upk3b); (8) smooth muscle (expressing Eln and Acta2); (9) cardiomyocytes (expressing Tnnt2 and Actc1) (Figs 1C and 2C). Although it is possible to further subdivide these clusters, parameters were chosen for clustering cell populations that were largely present during the entire developmental window in this study. The proliferating clusters of myofibroblasts and Wnt2+ fibroblasts were predominantly observed prenatally and distinguished by their expression of positive-acting cell-cycle genes (e.g. Mcm5, Pcna, Cdk1, Mki67). Both the proliferating and non-proliferating myofibroblast clusters expressed reference marker genes Acta2 and Tgfbi. Similarly, both the Wnt2+ fibroblast clusters expressed Wnt2 and Gyg (Table S1).
In contrast to the timing of changes in the epithelium and endothelium, there was a marked shift in the relative subpopulations of mesenchymal cells between P0 and P3 (Fig. 2B). The total population of all Wnt2+ fibroblasts decreased from 57.0±7.5% (mean±s.d.) of the total mesenchymal population to 25.1±6.4%, findings validated by RNA ISH (Table S2). During this time, the myofibroblasts increased from 17.9±5.2% to 39.8±16.5% of the total mesenchymal cell population.
Evaluation of time-dependent gene expression in epithelial, endothelial and mesenchymal cell populations was conducted using Monocle3 (Fig. 2D-F; Table S3). Genes that denote mature cell types (canonical marker genes) are associated with later expression programs. Specifically, it was observed that expression of markers of AT1 (Hopx, Aqp5), AT2 (Sftpc, Sftpa1), secretory (Scgb1a1), arterial (Cxcl12), gCap (Gpihbp1, Prss23, Kit), aCap (Car4), myofibroblast (Tgfbi), adventitial (Col1a1, Dcn) and smooth muscle (Eln, Acta2) cells became expressed at higher levels later in development.
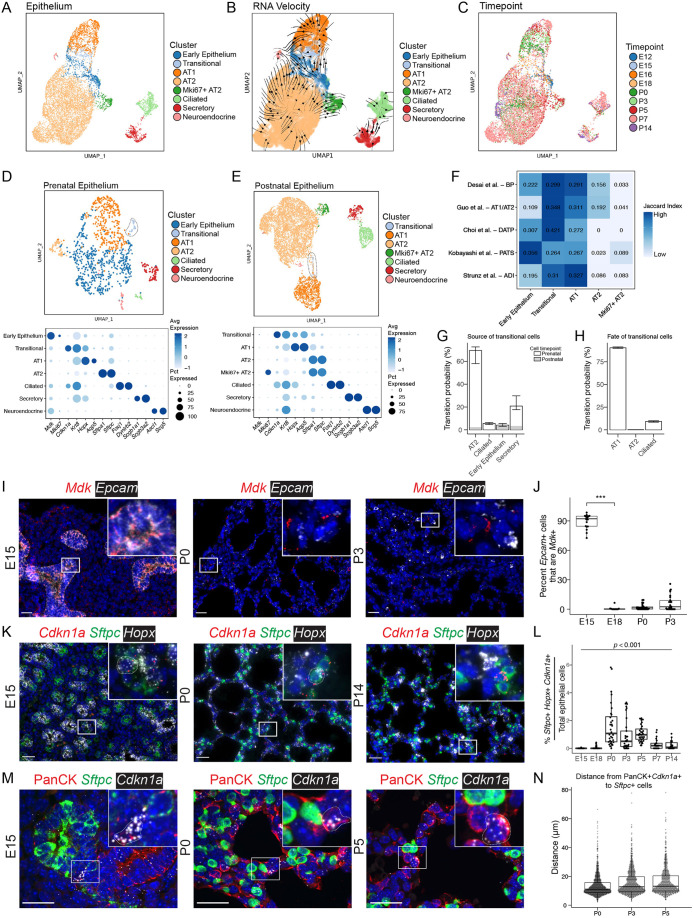
Trajectory analysis identifies a transitional state during alveolar epithelial differentiation
The pattern of epithelial population shifting and RNA velocity predicts differentiation from both early epithelial cells (present at E12 and E15) and AT2 cells to AT1 cells (Fig. 3A-C; Fig. S2A,B) in the developing lung. At E15, an epithelial subpopulation emerged that expresses markers of both AT2 cells (e.g. Sftpc) and AT1 cells (e.g. Hopx) at low levels. The expression of several genes, including Cdkn1a (also known as p21Cip1, a regulator of cell-cycle progression at G1) alongside higher Krt8 expression distinguishes these seemingly transitional cells. Although other epithelial cells outside this cluster expressed Cdkn1a, including ciliated cells, the combination of markers for both AT1 (Aqp5, Hopx) and AT2 (Sftpc, Sftpa1), as well as Cdkn1a and Krt8, defines this cluster (Fig. S2C). Separately analyzing prenatal (E12, E15, E16 and E18) and postnatal (P0, P3, P5, P7 and P14) epithelial cells further highlighted the distinct features of this transitional cluster of cells (Fig. 3D,E; Fig. S2D). These cells are marked by expression of Cdkn1a and enriched in Krt8 expression compared with other epithelial cells.
Fig. 3.
Distinct epithelial cell identities are established by E18. (A) UMAP embedding of lung epithelial cells (n=10,918) colored by cell type. Although epithelial cells from early lungs (E12 and E15) do occupy distinct UMAP space (‘early epithelium’), their broad spread is consistent with substantial heterogeneity and priming towards several nascent lineages. (B) RNA-velocity vectors were calculated using CellRank and overlaid on the UMAP embedding, with line thickness indicating velocity magnitude. (C) UMAP embedding colored by time point. (D) Epithelial cells from prenatal time points (E12, E15, E16 and E18) are projected on a UMAP and colored by cell type. The transitional epithelial cell cluster is circled in a dotted line. Marker gene expression by cluster in the prenatal epithelium is displayed in a dot plot (below), in which higher expression is represented as a darker color. The size of the dot indicates the proportion of cells expressing that marker. (E) Epithelial cells from postnatal time points (P0, P3, P5, P7 and P14) are projected on a UMAP and colored by cell type. The transitional epithelial cell cluster is encircled in a dotted line. Marker gene expression by cluster in the postnatal epithelium is displayed as a dot plot (below). (F) A row-normalized Jaccard index was calculated between clusters identified in the current analysis and transitional epithelial clusters identified in other studies. The intensity of color indicates relative similarity, with darker colors indicating higher similarity. (G) Cell-trajectory inference was calculated with CellRank, and probabilities of becoming a ‘transitional’ cell plotted. (H) Terminal state probabilities of the transitional cells. The bars are colored based on fraction of cells in each cluster present prenatally (gray) or postnatally (white). Central bars represent the median and range bars represent the interquartile range. (I) RNA ISH of Mdk (red) and Epcam (white, epithelial marker) at selected time points. For additional images see Fig. S2H. (J) Quantification of ISH using HALO for percent of Mdk+ epithelial cells (Epcam+Mdk+/Epcam+) cells over total epithelial cells. A non-parametric Kruskal–Wallis test was performed (***P<0.001). (K) RNA ISH of Cdkn1a (red, transitional cell marker), Sftpc (green, type II cell marker) and Hopx (white, type I cell marker) at selected time points. Cells that express Cdkn1a, Sftpc and Hopx are outlined in a dotted line. For channel-separated images see Fig. S3. (L) Quantification of ISH for percentage of transitional cells (Sftpc+Hopx+Cdkn1a+) cells over total epithelial cells. (M) Confocal micrographs of immunofluorescence and RNA ISH of pan-cytokeratin (PanCK, red), Sftpc (green) and Cdkn1a (white) at selected time points. Cells that express Cdkn1a and PanCK are outlined in a dotted line. For additional time points see Fig. S2I. Insets show magnification of boxed areas. (N) Distance from each transitional cell (PanCK+Cdkn1a+) to the nearest AT2 cell (Sftpc+) was calculated and plotted for each cell. Box and whisker plots show median values (middle bars) and first to third quartiles (boxes); whiskers indicate 1.5× the interquartile range, and dots are points outside the interquartile range. Scale bars: 25 µm.
Comparisons with cellular clusters identified in other studies using the Jaccard index (Kim et al., 2019; Mereu et al., 2020) (Fig. 3F) indicated that this cluster of transitional cells has similar expression patterns to bipotent progenitor (BP), AT1/AT2, damage-associated transient progenitors (DATP) and alveolar differentiation intermediate (ADI) cells that are associated with epithelial development and regeneration after injury (Choi et al., 2020; Desai et al., 2014; Guo et al., 2019; Strunz et al., 2020). There is some, but decreased, similarity between transitional cells and pre-alveolar transitional state (PATS) cells (Kobayashi et al., 2020). Notably, the PATS cells had a higher Jaccard index when compared with the early epithelium, but the latter also specifically expresses Mdk (Fig. 3D,I,J; Fig. S2H) unlike PATS cells. Cell-trajectory mapping based on mRNA-splicing (RNA velocity) analysis indicates that these transitional cells most frequently arise from AT2 cells postnatally, with contributions from the early epithelium prenatally (Fig. 3G). Trajectory mapping predicts that transitional cells have a 90.6% median probability of becoming AT1 cells (interquartile range 89.8-91.4%) (Fig. 3H). These transitional cells emerge at E15 and remain present through P5, with a decline in the relative abundance by P14 (7.1±4.8%) by sequencing. This relative decrease is supported in lung tissue by quantitative RNA ISH (Fig. 3K,L; Fig. S3). Although expression of Cdkn1a is a marker of the transitional cell population, this cluster does not have a high cell-cycle score, a multi-gene metric that identifies replicating cells (Fig. S2B). Immunodetection of pan-cytokeratin (PanCK) in addition to Sftpc and Cdkn1a (Fig. 3M; Fig. S2I) indicates that these transitional cells (PanCK+Cdkn1a+) appear cuboidal and closely associated (most within 10 µm) with Sftpc+ AT2 cells, suggesting a lineal relationship (Fig. 3N). Consistent with previous findings by others, our trajectory mapping also predicts that AT2 cells arise from the early epithelium or other AT2 cells (Barkauskas et al., 2013) and that, postnatally, AT2 cells can also give rise to AT1 cells (Fig. S2E,F).
Early specification is characteristic of the developing lung endothelium
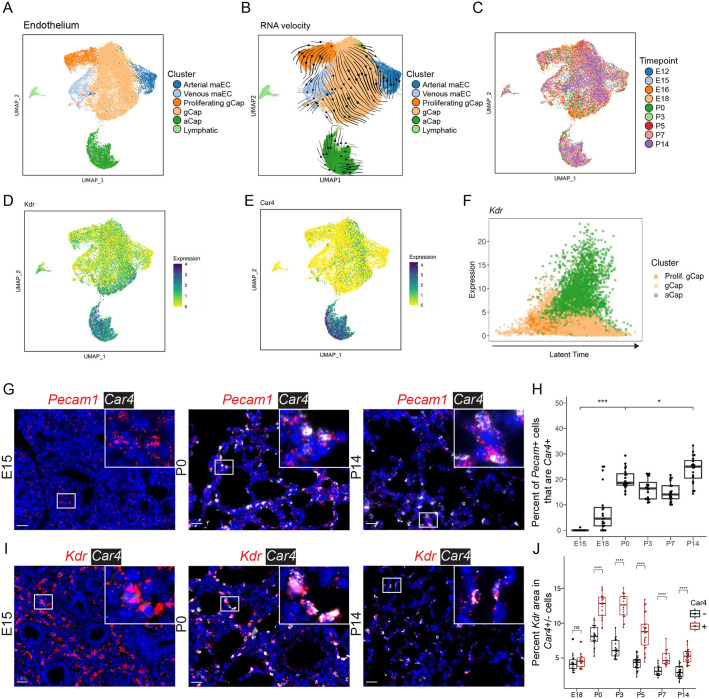
Although the lung endothelium demonstrated an overall stable population structure from E12 to P14, the population of Car4+ aCap cells emerged at E18 (Figs 2B and 4A-C; Fig. S4A). Velocity analysis of the endothelium suggests that gCap cells are capable of self-renewal (Fig. S4B,C), consistent with previous descriptions (Kawasaki et al., 2015), data that are congruent with the finding that there is a cluster of gCap cells with a high cell-cycle score, indicative of proliferation (Fig. S4B). Furthermore, using trajectory mapping, we defined a small subset of gCap cells with an elevated aCap fate probability (Fig. S4C,D). These cells began to express genes that are markers of the aCap population, including Igfbp7 and Kitl, in addition to Car4 expression (Table S4). Spatial examination of Car4 expression in lung tissue confirmed that, beginning at E18, aCap cells can be found in the alveoli by quantitative RNA ISH (Fig. 4G,H; Fig. S4F). As measured by a Jaccard index, the identified cellular clusters correspond well with previously described endothelial clusters (Gillich et al., 2020; Niethamer et al., 2020; Vila Ellis et al., 2020) (Fig. S5A).
Fig. 4.
Endothelial cells undergo specification by E15, with subsequent emergence of Car4+ aCap cells by E18. (A) UMAP embedding of lung endothelial cells (n=22,237) colored by cell type. (B) RNA-velocity vectors of endothelial cells were calculated with CellRank and overlaid on the UMAP embedding. (C) UMAP embedding colored by time point. (D) UMAP embedding of endothelial cells colored by Kdr expression: darker colors indicate greater expression. (E) UMAP of Car4 expression. (F) Latent time analysis: latent time increases along the x-axis, shows expression of Kdr in aCap cells. (G) RNA ISH of Pecam1 (red; endothelial cell marker) and Car4 (white; aCap cell marker). A complete series is presented in Fig. S4F. (H) Quantification of ISH was performed using HALO; fraction of Pecam+ cells that are Car4+ was plotted. Boxplots represent pooled data, with each dot representing the percentage of Pecam+Car4+/Pecam+ cells per image (n=20 images per time point, from n=3 individual mice). Increases were observed from E15 to P0 and from P0 to P14. (I) RNA ISH of Kdr (red), and Car4 (white). Additional images are presented in Fig. S5. Insets show magnification of boxed areas. (J) Quantification of Kdr expression in Car4− (black) and Car4+ (red) cells. *P<0.05, ***P<0.001, ****P<0.0001: Mann–Whitney U-test. Box and whisker plots show median values (middle bars) and first to third quartiles (boxes); whiskers indicate 1.5× the interquartile range, and dots are points outside the interquartile range. Scale bars: 25 µm.
To understand cellular maturation within each time point, we used mRNA-splicing-based quantification of transcriptome dynamics (scVelo; Bergen et al., 2020) to estimate cellular maturity by calculating ‘latent time’ for each cell. A different measure from developmental time (Fig. S6), latent time represents an inference of how long a particular cell has been in the tissue, independent of the age of the animal at each time point. As the aCap cells mature along the predicted trajectory defined by latent time, the expression of Kdr, a VEGF receptor, increases (Fig. 4F). Indeed, expression of Kdr is enriched in aCap cells compared with Car4− cells (Fig. 4D,E,I,J; Fig. S5B), consistent with a role of Vegf signaling in the regulation of capillary specialization (Vila Ellis et al., 2020).
Mesenchymal cells retain distinct identities but alter expression patterns and abundance during development
Compared with epithelial and endothelial cells, mesenchymal cells undergo more substantial population shifts after birth, suggesting both postnatal cellular replication and transitions (Figs 2C and 5A-C; Fig. S7A,B). Quantitative ISH was used to identify the myofibroblasts (marked by Tgfbi) and Wnt2+ fibroblasts in the developing lung (Fig. 5D; Fig. S7C). Examination of the spatial organization of these cells within the E15 lung found that most cells are Wnt2+ fibroblasts. There appears to be a redistribution of the Tgfbi-marked myofibroblasts from clusters of cells at E15 to more dispersed individual cells at E18, a redistribution that continues through to P0. Spatial analysis of at least 3200 cells per time point using the Clark and Evans aggregation index provided strong objective support for the greatest organizational changes occurring between E15 and P0, with Wnt2+ fibroblasts and myofibroblasts becoming both less aggregated over time and more dispersed around alveoli (Fig. 5E). Comparing these mesenchymal clusters to clusters presented in the LungGENS resource (Du et al., 2015; Guo et al., 2019) showed that the previously described proliferating mesenchymal progenitor (PMP) cells (Xie et al., 2018) can be specifically found in our study, but that PMP-like cells are separately located within the Wnt2+ and myofibroblast populations (Fig. 5F). These proliferating Wnt2+ fibroblasts and proliferating myofibroblasts also had a high cell-cycle score, indicative of replicative status (Fig. S7B). It was also noted that although Wnt2+ fibroblasts all broadly expressed Wnt2, the myofibroblasts (starting at E18) expressed Wnt5a (Fig. S7D,E; Table S1). Quantitative ISH supports this finding, as >90% (median) of Tgfbi+ myofibroblasts co-expressed Wnt5a (Fig. 5G; Fig. S8A,B). Using the measurement of latent time to investigate the expression patterns of Plin2, a lipofibroblast marker, demonstrated that Plin2 expression positively correlated with maturity by estimated latent time (Fig. S8C,D), and that these Plin2+ cells are likely a subset of the Wnt2+ fibroblasts.
Fig. 5.
The mesenchymal cell population undergoes dynamic population shifts between P0 and P3. (A) UMAP embedding of lung mesenchymal cells (n=67,035) colored by cell type. (B) RNA-velocity vectors overlaid on the UMAP depict inferred differentiation/maturation trajectories. (C) UMAP embedding colored by time point. (D) RNA ISH of Tgfbi (red; myofibroblast marker) and Wnt2 (white; Wnt2+ fibroblast marker) at E15, E18, P0 and P3. Images are stitched scans. A complete time series of representative images is presented in Fig. S7C. (E) HALO analysis of the Tgfbi and Wnt2 ISH and assignment of cell identities based on marker expression. A Clark-Evans clustering index was calculated from individual cells and aggregated by image. Values >1.0 indicate that cells are more regularly spaced apart (ordered), whereas values <1.0 indicate progressively increased clustering. Box and whisker plots show median values (middle bars) and first to third quartiles (boxes); whiskers indicate 1.5× the interquartile range, and dots are points outside the interquartile range. (F) A row-normalized Jaccard index was calculated between clusters identified in this study and those from other studies. Increased saturation of blue box shading indicates higher similarity. (G) RNA ISH of Tgfbi (red; myofibroblast marker), Sftpc (green; type II cell marker) and Wnt5a (white) at E15, E18, P0 and P3. Images are stitched scans. A complete time series of representative images is presented in Fig. S8A. Insets show magnification of boxed areas. Scale bars: 100 µm.
AT1 cell role in elastin assembly and extracellular matrix organization
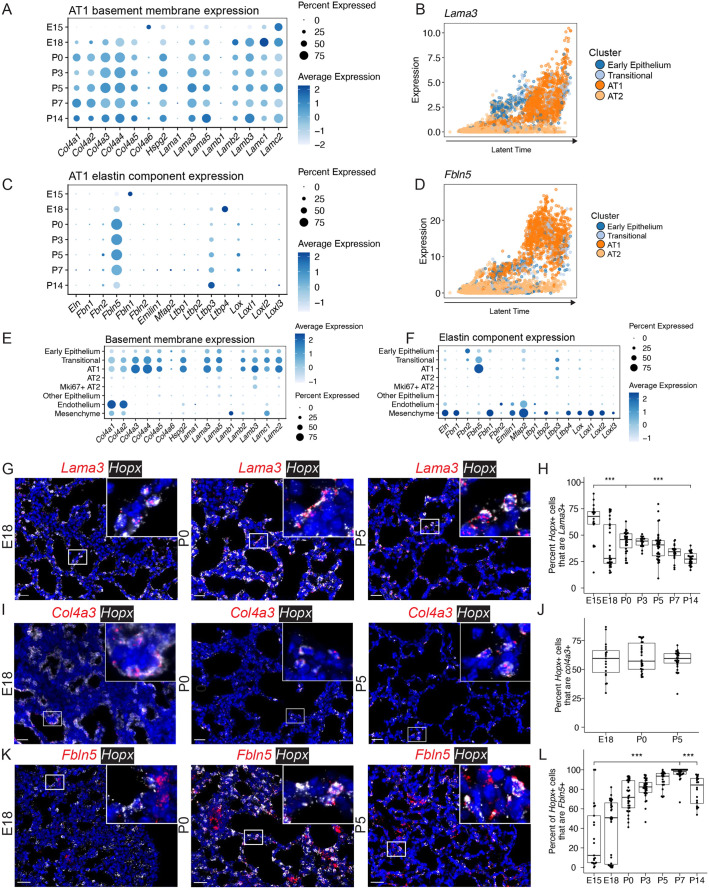
As an application of this single-cell atlas, we explored the extracellular matrix (ECM) expression profile of AT1 cells during later lung development. Previous work has suggested that proper elastin assembly and generation of basement membrane and ECM are crucial steps during the saccular and alveolar stages of lung development (Bourbon et al., 2005). Failure to establish this elastin network during development is a factor underlying long-term sequelae of bronchopulmonary dysplasia (Benjamin et al., 2020). We observed that AT1 cells expressed components of the basement membrane, including specific expression of Col4a3 and Col4a4 (Fig. 6A,B; Fig. S9A,B) and expression of the three genes that comprise laminin-332 (Lama3, Lamb3, Lamc2). Laminins are trimeric ECM proteins, composed of an α, β and γ subunit, located in the basement membrane, but the granular timing and cellular source of laminin gene expression are poorly defined. Our analysis found that expression of laminin-332, a principal laminin isoform of the mature alveolar basement membrane, is high at E18 in AT1 cells and continues throughout later development. Further, we observed that AT1 cells highly express Fbln5 (fibulin 5) (Fig. 6C,D,F; Figs S9B and S10A), which contributes to elastin assembly by binding tropoelastin and fibrillin 1, cross-linking enzymes required for this process (Yanagisawa et al., 2002). RNA ISH confirmed colocalization of AT1 marker Hopx expression with Lama3, Col4a3 and Fbln5 across development (Fig. 6G,I,K). Quantification of RNA ISH by automated image analysis (HALO, Indica Labs) demonstrated declining expression of Lama3, relatively consistent expression of Col4a3 and increasing expression of Flbn5 by Hopx+ cells across developmental time (Fig. 6H-L). Lama3 and Fbln5 expression were found to be positively correlated with latent time in AT1 cells (Fig. 6B,D). The expression profile of components of elastin assembly and expression of components of basement membrane and ECM in AT1 cells are unique among epithelial cells during development. Notably, AT2 cells have little to no detectable expression of these genes (Fig. 6E,F). Indeed, expression of Col4a3 and Col4a4 appears to be somewhat specific to AT1 cells when compared across all lung cell types (Fig. 6E; Figs S9B and S10B-D).
Fig. 6.
Epithelial alveolar type I (AT1) cells express components of elastin and the basement membrane. (A) Basement membrane gene expression in AT1 cells over time was plotted, with dot size indicating proportion of cells within a cluster expressing that gene and color saturation indicating relative expression level. (B) Latent time analysis shows expression of Lama3 in AT1 cells. (C) Expression of elastin components in AT1 cells over time is plotted. (D) Latent time analysis shows expression of Fbln5 in AT1 cells. (E) Basement membrane gene expression was plotted in alveolar epithelial cells and other cell types grouped into ‘Other Epithelium’, ‘Endothelium’, and ‘Mesenchyme’. (F) Elastin component gene expression in indicated cell types was plotted. (G) RNA ISH of Lama3 (red) and Hopx (white, AT1 marker). A complete series is presented in Fig. S9A. (H) ISH quantification using HALO. Box plots represent pooled data, each dot representing the percentage of Lama3+Hopx+/Hopx+ cells per image. (I) RNA ISH of Col4a3 (red) and Hopx (white). For additional images see Fig. S10B. (J) ISH quantification representing percentage of Col4a3+Hopx+/Hopx+ cells per image. (K) RNA ISH of Fbln5 (red) and Hopx (white). For additional images see Fig. S10A. (L) ISH quantification representing percentage of Fbln5+Hopx+/Hopx+ cells per image (***P<0.001 by Mann–Whitney U-test). Box and whisker plots show median values (middle bars) and first to third quartiles (boxes); whiskers indicate 1.5× the interquartile range, and dots are points outside the interquartile range. Scale bars: 25 µm.
DISCUSSION
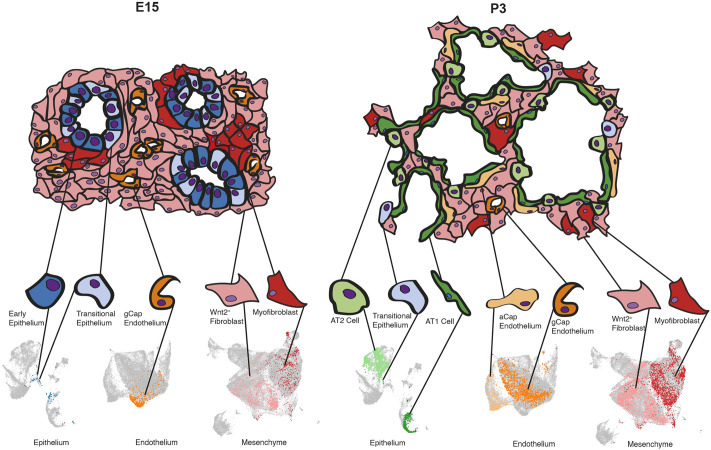
Using time-series single-cell transcriptomics, we generated an integrated cellular atlas of epithelial, mesenchymal and endothelial cells across lung development. We identified new temporal relationships between these cell types and defined the relative populations of major subtypes of these cells (Fig. 7). We applied emerging analysis paradigms to explore the trajectory and fate prediction of these cell types, contextualizing previous findings and discovering new features of developmental lung biology. These include a population of transitional epithelial cells with characteristics of intermediate progenitors and substantial similarity with regenerating epithelium after injury, a new role for AT1 cells in expressing ECM and Fbln5 suggesting that – in addition to expansion for gas-exchange – AT1 cells direct the construction of their own structural niche. These data now provide a developmental roadmap that identifies the stages of cell-type commitment in the lung, and a resource (https://sucrelab.org/lungcells) to define the cellular sources and kinetics of crucial developmental signaling and structural molecules.
Fig. 7.
Functional development of the lung is a result of spatial and temporal organization of multiple cell types. An abbreviated summary model of the developing lung compared between two representative time points: mid-embryogenesis (E15) and postnatal (P3). At E15, the lung epithelium is composed of indistinct, uncommitted cell types (as defined by marker-gene analysis) that are very rare or absent in adulthood. The prenatal endothelium in the lung parenchyma is composed of general capillary (gCap) endothelial cells, whereas the majority of cells in the remaining tissue are Wnt2+ fibroblasts, with interspersed clusters of myofibroblasts. By P3, many cell types observed in mature lungs are already present, including AT1, AT2 and transitional epithelial cells. The gCap endothelium has differentiated into Car4+ alveolar capillary (aCap) cells, alongside the gCap endothelium, and mesenchymal cells have undergone extensive spatial reorganization. Representative cell clusters are highlighted in the inset UMAPs derived from Fig. 1C.
Transitional cells in context of development and injury
Although the lung epithelium at E12 and E15 is primarily composed of unspecified early epithelial cells, a few AT1 and AT2 cells can be observed as early as E15, suggesting a relatively early fate commitment for at least a subset of pioneer alveolar epithelial cells, consistent with some previous findings (Frank et al., 2019; Gonzalez et al., 2019). Ongoing AT1 differentiation from AT2 cells occurs during postnatal alveologenesis, suggesting that multiple differentiation trajectories contribute to the maturing AT1 population (Desai et al., 2014). Indeed, the transitional cell cluster observed here has similarity to previously described bipotent progenitors (Desai et al., 2014) as well as AT1/AT2 cells (Guo et al., 2019). These transitional epithelial cells, which we identify with the marker gene Cdkn1a, have expression patterns similar to epithelial cells described in the adult lung arising from AT2 cells during recovery from injury – the previously described DATP (Choi et al., 2020) and ADI (Strunz et al., 2020) – with our early epithelial cells having more similarity with PATS cells (Kobayashi et al., 2020). Similar to the analysis of DATPs after injury, our trajectory analysis predicts that these Cdkn1a+ transitional cells arise from AT2 cells postnatally, and that >90% of these neonatal transitional cells ultimately become AT1 cells (Fig. 3I,J). In the adult mouse, DATPs appear to be essential for regeneration of the lung epithelium after injury (Choi et al., 2020). These findings about transitional cells rely on transcriptomic analysis and tissue localization in fixed tissue, methods that are not without limitations and inference error. Future mechanistic work is needed to elucidate the role of both early epithelium and transitional cell populations in the setting of neonatal injury and their governing mechanisms. Although Wnt responsiveness has been shown to be relevant to regeneration and repair in the adult lung (Zacharias et al., 2018), in this developmental dataset both early and transitional cell populations lack detectable expression of Wnt target gene Axin2. This suggests that active Wnt signaling is not a significant feature of cells in these states during normal development, though Wnt may still play a role up- or downstream of these cell states, or in the setting of injury.
Timing of endothelial specialization and source of aCap cells
This time-series demonstrates that the recently identified Car4+ aCap cells (Gillich et al., 2020; Niethamer et al., 2020; Vila Ellis et al., 2020) emerge at E18, persisting in relatively stable numbers through to P14. Trajectory analysis predicts that aCap cells arise from the gCap population, and the probability of gCap cells differentiating into aCap cells appears to be correlated with the expression of Igfbp7, Kitl and Tbx2. These specialized endothelial cells are closely related to AT1 epithelial cells spatially and temporally. We observed that the aCap cells appear at E18, at the same time as the neighboring AT1 cells emerge. With time, aCap endothelial cells begin to express higher levels of VEGF-receptor Kdr, as Vegfa levels concurrently increase in AT1 cells (Fig. S2G). This is consistent with the previous finding that VEGF is an essential factor for the formation of aCap cells that are found in association with alveolar structures (Vila Ellis et al., 2020).
Major subdivisions in pulmonary mesenchymal cells
The greatest diversity of cell populations and dynamics during the later stages of lung development can be found in the pulmonary mesenchyme. Prenatally, the predominant mesenchymal population expresses Wnt2. Later in development this population expresses other markers associated with lipofibroblasts (e.g. Plin2, Tcf21, Gyg) (Endale et al., 2017). Immediately before birth, the myofibroblast population begins to expand and express a different Wnt ligand, Wnt5a. The spatial and temporal shifts of these two fibroblast populations are highly dynamic between E15 and P5. Our previous work demonstrates a pathologic role for excessive Wnt5a expression by mesenchymal cells during neonatal hyperoxia injury (Sucre et al., 2020), and we speculate that increased Wnt5a expression by the myofibroblast population or expansion of this population after injury is a key driver of abnormal development observed after neonatal hyperoxia exposure.
The Wnt2+ populations we observed pre- and postnatally differ significantly from previously described Wnt2+ fibroblasts in adult lungs (Zepp et al., 2017). In the adult population, it has been reported that >90% of Wnt2-lineage-labeled fibroblasts express Pdgfra. In our developmental dataset, the opposite appears to be true, with Pdgfra expressed in higher levels in the Tgfbi+ myofibroblast population. However, our trajectory analysis found that some of the proliferating myofibroblasts have the predicted potential to become Wnt2+ fibroblasts. This suggests that, although there may be early overlap in these populations, the gene-expression profiles after commitment to a specific cell type are distinct. In addition to the age of the mice studied, there are technical reasons why our observations may differ from previous work. Our scRNA-seq covered all CD45− viable cells in the lung, without the use of fluorescent reporters to enrich for a specific target population. Although this allowed for a global, unbiased analysis of the structural cells in the lung, the ability to detect rare populations of cells is decreased. We observed expression of previously described Lgr6+ cells in the myofibroblast population beginning at E12, but rarer populations, such as the Lgr5+ mesenchymal cells, reported to be 1-2% of pulmonary mesenchymal cells in the adult lung (Lee et al., 2017), are not present in sufficient quantity to be detected by our analysis, which may reflect methodologic differences. In addition to the tissue validation we have carried out with RNA ISH, future work with reporter lines could be used to confirm our findings by lineage tracing, though these methods are not without their limitations related to fidelity and may overestimate or underestimate the number of cells in a particular population (Hsu, 2015).
Trajectory modeling over time
Profiling the developing lung over multiple time points has allowed greater insight into specific cellular transitions. Examining a single, jointly analyzed dataset comprising immature cells, mature cells and cells in transition provides a unique opportunity to make inferences as to the trajectory of individual cells. It is important to acknowledge, however, that such fate modeling is inherently predictive, and currently available techniques do not facilitate tracking large complex populations in a tissue over time. There are other notable limitations to this study, including the exclusion of immune cells. The resident and circulating immune cells of the lung have been shown to contribute significantly to both lung development and injury response (Stouch et al., 2016). In this study, we focused on the structural cells of the lung; future studies could similarly characterize the recruitment and polarization of immune cells during lung development. We are aware of the biases inherent in scRNA-seq, including that the single-cell dissociation may bias toward recovery and analysis of certain populations. Although the fractional representation of cell subpopulations in living tissue may be different, we anticipate that the relative changes over time are preserved. Indeed, this latter supposition is supported by the correlation between the relative proportion of cells recovered by scRNA-seq and selected cell types by RNA ISH (Table S2). In addition, although trajectory inferences about RNA velocity may be reversed if transcript abundance is regulated by RNA degradation, the correct prediction of already established lineage relationships in the epithelium and endothelium minimized this concern as a major confounder.
Prospective role for AT1 cells as central organizers of lung architecture
We have applied the findings of this developmental cell atlas to study the ECM gene expression profile of AT1 cells. Previous foundational work has demonstrated that proper elastin assembly is crucial for alveologenesis (Benjamin et al., 2016), and here we have identified a new role for AT1 cells in establishing the structural scaffold of the developing lung, as they express high levels of Fbln5, which is essential for elastin crosslinking and assembly. In addition, AT1 cells also appear to play a crucial role in establishing the basement membrane, expressing multiple genes that make up laminin (Nguyen et al., 2005; Schuger et al., 1990), which is required for branching morphogenesis. Others have previously reported that laminin isoforms expressed by the fetal lung differ from those in the adult alveolus; with laminin-111 and -511 expressed early and laminin-332 and -511 in the adult alveolar basement membrane (Miner et al., 1995, 1997; Nguyen and Senior, 2006). Our data precisely define the timing of expression of the three genes comprising laminin-332 beginning at E18 and identify AT1 cells as a cellular source of this laminin. AT1 cells also express significant amounts of Col4a3 and Col4a4, additional essential basement-membrane constituents. Notably, the aCap endothelial cells express Col4a1 and Col4a2 (Figs S9B and S10D,E), suggesting that the closely apposed AT1 and aCap cells play complementary and plausibly interactive roles in forming the alveolar basement membrane. To validate these surprising findings in AT1 cells, we integrated our epithelial data with a recently published single-cell study representing a similar developmental window (Zepp et al., 2021). Clustering of the joint dataset yielded highly similar clusters, with all cell types in this study (including the transitional cells described above) being present in the Zepp dataset (Fig. S11). Assessing this dataset, we validated that AT1 cells express Fbln5, Col4a3, Col4a4 and the three genes that comprise laminin-332 (Lama3, Lamb3, Lamc2) during the saccular-to-alveolar transition. Our study adds to existing work by pointing to a previously underappreciated role of AT1 cells, including their singular capacity among epithelial cells regarding expression of ECM and elastin assembly-associated genes. Although AT1 cells have been previously characterized as providing a functional surface for gas exchange and for regulating permeability and tight junctions of the alveolus (Warburton et al., 2010), taken together, our time series suggests that AT1 cells are a cellular source of essential ECM components as the tissue transitions through the saccular and alveolar stages. Future work with in vivo models is needed to characterize this prospective function of AT1 cells.
Asynchronous patterning and differentiation in lung development
The developmental stages of the lung were largely founded on histologically descriptive features (Warburton et al., 2010). This categorization often results in debate regarding the function and identity of cell types within the boundaries of each stage. In contrast to a histologic strategy of staging, we approached development from the perspective of cellular function as defined by the transcriptome. In aggregate, we found that different cell types commit to change asynchronously during development, suggesting that the timing of the saccular-to-alveolar transition is fluid and highly cell-type specific. Combined, the dynamic expansions and contractions of cell populations, including epithelial specification at around E15, the appearance of aCap endothelial cells at around E18 and an expansion of myofibroblasts at around P3, indicate tremendous coordination and interaction between epithelial, mesenchymal and endothelial cells at every time point. Indeed, it appears that many cell types in the lung possess the ability to signal to each other during key transitional periods (Fig. S12). This granular single-cell atlas of the developing lung provides insight into the relevant populations present between E12 and P14 and provides a foundation for future mechanistic studies into pathways governing the coordinated cell-cell communication across this specialized developmental stage.
MATERIALS AND METHODS
Animal sample collection and care
C57BL/6J mice were used for all experiments. Timed matings were performed as previously described (Plosa et al., 2020) and mice sacrificed at P0, P3, P5, P7 or P14 for scRNA-seq or lung block fixation/embedding. E12, E15, E16 and E18 lungs were isolated by removing pups from the uterus and isolating lung tissue. E12, E15, E16, E18, P0, P3 and P5 lungs were fixed in formalin. P7 and P14 mouse lungs were inflation-fixed by gravity filling with 10% buffered formalin and paraffin embedded. All animal work was approved by the Institutional Animal Care and Use Committee of Vanderbilt University (Nashville, TN, USA) and was in compliance with the Public Health Services policy on humane care and use of laboratory animals.
Single-cell data collection
Sample collection and single-cell sequencing was performed as previously described (Schuler et al., 2020). Briefly, lung lobes were harvested, minced and incubated for 30 min at 37°C in dissociation medium (RPMI-1640 with 0.7 mg/ml collagenase XI and 30 mg/ml type IV bovine pancreatic DNase). After incubation, tissue was disassociated into single-cell suspension by passage through a wide-bore pipet tip and filtration through a 40 µm filter. Single-cell lung suspension was then counted, aliquoted and blocked with CD-32 Fc block (BD Biosciences, 553142) for 20 min on ice. After a 2% fetal bovine serum staining buffer wash, cells were incubated with the conjugated primary antibodies anti-CD45 (BD Biosciences, 559864, 1:200) and anti-Ter119 (BioLegend, 116211, 1:200). For some of the P7 libraries (six of the twelve sequenced mice), epithelial enrichment was performed by sorting cells stained with an Epcam antibody (BD Biosciences, 563477, 1:200). In these samples, 5000 cells that were Epcam-high were added to the 10,000 cells collected during the CD45−/Ter119− sort. Across all time points, the relative proportions of cell sub-populations by RNA ISH was consistent with the sequencing data, indicating that the Epcam+ enrichment for some of the P7 mice did not impact cellular proportions after the in silico sort into epithelial, endothelial and mesenchymal cells.
In the same manner, fluorescence minus-one controls were blocked and stained with the appropriate antibody controls. Cells from individual mice were incubated with identifiable hashtags, resuspended in staining buffer, and treated with propidium iodide (PI) viability dye. CD45− Ter119− viable cells were collected by FACS using a 70 µm nozzle on a four-laser BD FACSAria III Cell Sorter. For epithelially enriched samples, Epcam+ cells were sorted. Single-color controls were used for compensation and fluorescence-minus-one controls were used for gating. Before flow sorting, hashing antibodies with unique oligos were added to samples from each individual mouse.
scRNA-seq library preparation and next-generation sequencing
scRNA-seq libraries were generated using the 10x Chromium platform 5′ library preparation kits (10x Genomics) following the manufacturer's recommendations and targeting 10,000-20,000 cells per sample. Next-generation sequencing was performed using an Illumina Novaseq 6000. Reads with read quality less than 30 were filtered out and CellRanger Count v3.1 (10x Genomics) was used to align reads onto the mm10 reference genome.
Analysis of single-cell sequencing data
Ambient background RNAs were cleaned from the scRNA-seq data using ‘SoupX’ (Young and Behjati, 2020) as described (Schuler et al., 2020), using the following genes to estimate the non-expressing cells, calculate the contamination fraction and adjust the gene-expression counts: Dcn, Bgn, Aspn, Ecm2, Fos, Hbb-bs, Hbb-bt, Hba-a1, Hba-a2, Lyz1, Lyz2, Mgp, Postn, Scgb1a1. Quality filtering removed cells with >10% mitochondrial mRNA, <0.5% mitochondrial mRNA, and cells with <700 detected genes.
Preliminary data analysis, dimensionality reduction, clustering, visualization and grouping of cells into broad categories (epithelial, endothelial, mesenchymal) was performed using Seurat v4.0.2 and SCTransform (Hafemeister and Satija, 2019; Stuart et al., 2019), with manual inspection of expression patterns of marker genes: Hbb-bs, Epcam, Foxj1, Scgb1a1, Scgb3a2, Abca3, Hopx, Col1a1, Dcn, Lum, Acta2, Wnt2, Wnt5a, Lgr6, Pdgfra, Pdgfrb, Cspg4, Wt1, Pecam1, Ccl21a, Vwf, Nrg1, Plvap, Car4, Mki67, Tnnt2, Mpz and Ptprc. Doublet removal was performed by removing all ‘cells’ labeled with more than one hashing antibody, indicating doublets from two individual mice. Further, cell clusters expressing non-physiologic marker combination Epcam+/Pecam1+ were also dropped at this stage. SCTransform was run with each sequencing reaction (corresponding to an individual time point) as a batch variable, and with the percentage of mitochondrial RNA as a regression variable.
Further data cleaning was carried out to remove gene counts for Gm42418 (also known as Rn18s-rs5), a likely rRNA (Kimmel et al., 2019). After removal of Gm42418, genes with expression in less than ten cells across the dataset were removed from further analysis. SCTransform (version 0.3.2.9008) (Hafemeister and Satija, 2019) was used with glmGamPoi (version 1.2.0) (Ahlmann-Eltze and Huber, 2020) to normalize data before PCA, UMAP embedding (McInnes et al., 2018) and cell clustering individually for each broad cell category (epithelial, endothelial, mesenchymal). Cell clusters for each broad category were identified using marker genes outlined in Table S1 in addition to marker gene detection using MAST (Finak et al., 2015). After annotation of all cell clusters, the broad tissue categories were merged and dimensionality reduction, clustering and visualization were conducted. Monocle3 (version 1.0.0) (Cao et al., 2019) was used to identify genes that exhibit changes associated with time. Gene-expression changes were evaluated individually in epithelial, endothelial and mesenchymal cells. NicheNet (version 1.0.0) (Browaeys et al., 2020) was used to evaluate potential receptor-ligand interactions associated with saccular (P0 and P3) to alveolar (P7 and P14) cell types. Data integration with another single-cell dataset (Zepp et al., 2021) was performed by downloading the reads from E12.5, E15.5, E17.5, P3 and P7 mice from the NCBI GEO record GSE149563 and processing with CellRanger 3.1. Epithelial cells (Epcam+ by in silico sort) were combined with the epithelial dataset generated in this study using the ‘IntegrateData’ function of Seurat following the SCTransform workflow. All charts and heatmaps as part of the scRNA-seq analysis were generated with ggplot2, and all parts of the analysis were run in R 4.0.2.
Predictions of RNA velocity were made using a velocyto–scVelo–CellRank pipeline. Spliced and unspliced mRNA counts were determined with velocyto (version 0.17.17) (La Manno et al., 2018) and analyzed using scVelo (version 0.2.3) with the dynamical model (Bergen et al., 2020). CellRank (version 1.4.0) (Lange et al., 2020 preprint) was then used to infer the initial and terminal cell states. Cell cycle scores were calculated using scVelo. All packages were run in Python 3.7.11. Specific parameter settings, a complete collection of all package versions, and code for all steps of the analysis is available at https://github.com/SucreLab/LungDevelopment.
RNA in situ hybridization
RNAScope technology (ACDBio) was used to perform all RNA ISH experiments according to the manufacturer's instructions. RNAScope probes to the following mouse genes were used: Sftpc, Hopx, Cdkn1a, Wnt2, Tgfbi, Pecam1, Car4, Flbn5, Mdk, Epcam, Kdr, Lama3, Wnt5a, Col4a1, Col4a3. Positive control probe (PPIB) and negative control probe (DapB) were purchased from the company and performed with each experimental replicate, with representative data shown (Fig. S7C).
Image acquisition and analysis
Fluorescent images were acquired using a Keyence BZ-X710 or BZ-X810 with BZ-X Viewer software and 40× objective. Wavelengths used for excitation included 405, 488, 561 and 647 nm. Automated image analysis was performed with HALO software (Indica Labs); an example of cellular segmentation and determination of co-localization is demonstrated in Fig. S7D. After HALO analysis the rate of false colocalization of non-physiological expression combinations (Sftpc+Wnt5a+ or Wnt2+Tgfbi+) was low, with a mean of <1% (Fig. S13). High-resolution images were acquired using a Nikon TiE inverted spinning-disk confocal microscope outfitted with a Yokogawa ×1 spinning disk head, a Photometrics Prime 95B camera and a Plan Apo 60×1.40 numerical aperture objective (Nikon Instruments). Lasers used for excitation included 405, 488, 561 and 647 nm, and emission filters were 455/50, 525/36, 641/75 and 700/74 (peak/bandwidth), respectively.
Supplementary Material
Acknowledgements
Flow cytometry experiments were performed in the VMC Flow Cytometry Shared Resource which is supported by the Vanderbilt-Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). We also thank Dr Christopher V. E. Wright for thoughtful discussions.
Footnotes
Competing interests
J.A.K. has received advisory board fees from Boehringer Ingelheim and Janssen Therapeutics, grants from Boehringer Ingelheim and Bristol-Myers-Squibb and research contracts with Genentech. T.S.B. has received advisory board fees from Boehringer Ingelheim, Orinove, GRI Bio, Morphic and Novelstar, and has research contracts with Genentech and Celgene.
Author contributions
Conceptualization: N.M.N., E.J.P., J.T.B., J.A.K., J.M.S.S.; Methodology: N.M.N., E.J.P., B.A.S., J.A.K., J.M.S.S.; Software: N.M.N., B.A.S., A.C.H., J.A.K.; Validation: N.M.N., E.J.P., J.T.B., C.B., M.R., A.N.H., J.A.K., J.M.S.S.; Formal analysis: N.M.N., B.A.S., A.C.H., C.B., N.E.B., J.A.K., J.M.S.S.; Investigation: N.M.N., E.J.P., J.T.B., B.A.S., A.C.H., C.S.J., P.G., C.J.T., D.N., B.K.M., J.A.K., J.M.S.S.; Data curation: N.M.N., J.A.K., J.M.S.S.; Writing - original draft: N.M.N., E.J.P., J.T.B., J.A.K., J.M.S.S.; Writing - review & editing: E.J.P., J.T.B., B.A.S., C.B., M.R., A.N.H., S.H.G., T.S.B., N.E.B., J.A.K., J.M.S.S.; Visualization: N.M.N., B.A.S., J.A.K., J.M.S.S.; Supervision: J.A.K., J.M.S.S.; Project administration: C.S.J., P.G.; Funding acquisition: J.A.K., J.M.S.S.
Funding
This work was supported by National Institutes of Health grants K08HL143051 (J.M.S.S.), K08HL130595 (J.A.K.), R01HL153246 (J.A.K.), R01HL145372 (J.A.K./N.E.B.), P01HL092470 (T.S.B.), K08HL127102 (E.J.P.), R03HL154287 (E.J.P.), K08HL133484 (J.T.B.), R01HL157373 (J.T.B.), and The Francis Family Foundation (J.A.K. and J.M.S.S.). Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199512.
References
- Ahlmann-Eltze, C. and Huber, W. (2020). glmGamPoi: fitting gamma-poisson generalized linear models on single cell count data. Bioinformatics 36, 5701-5702. 10.1093/bioinformatics/btaa1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas, C. E., Cronce, M. J., Rackley, C. R., Bowie, E. J., Keene, D. R., Stripp, B. R., Randell, S. H., Noble, P. W. and Hogan, B. L. M. (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025-3036. 10.1172/JCI68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin, J. T., van der Meer, R., Im, A. M., Plosa, E. J., Zaynagetdinov, R., Burman, A., Havrilla, M. E., Gleaves, L. A., Polosukhin, V. V., Deutsch, G. H.et al. (2016). Epithelial-derived inflammation disrupts elastin assembly and alters saccular stage lung development. Am. J. Pathol. 186, 1786-1800. 10.1016/j.ajpath.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin, J. T., Plosa, E., Sucre, J. M. S., van der Meer, R., Dave, S., Gutor, S. S., Nichols, D., Gulleman, P., Jetter, C. S., Han, W.et al. (2020). Neutrophilic inflammation during lung development disrupts elastin assembly and predisposes adult mice to COPD. J. Clin. Invest. 131, e139481 10.1172/JCI139481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen, V., Lange, M., Peidli, S., Wolf, F. A. and Theis, F. J. (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408-1414. 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- Bourbon, J., Boucherat, O., Chailley-Heu, B. and Delacourt, C. (2005). Control mechanisms of lung alveolar development and their disorders in bronchopulmonary dysplasia. Pediatr. Res. 57, 38R-46R. 10.1203/01.PDR.0000159630.35883.BE [DOI] [PubMed] [Google Scholar]
- Browaeys, R., Saelens, W. and Saeys, Y. (2020). NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159-162. 10.1038/s41592-019-0667-5 [DOI] [PubMed] [Google Scholar]
- Cao, J., Spielmann, M., Qiu, X., Huang, X., Ibrahim, D. M., Hill, A. J., Zhang, F., Mundlos, S., Christiansen, L., Steemers, F. J.et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496-502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J., Park, J.-E., Tsagkogeorga, G., Yanagita, M., Koo, B.-K., Han, N. and Lee, J.-H. (2020). Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366-382.e367. 10.1016/j.stem.2020.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, T. J., Brownfield, D. G. and Krasnow, M. A. (2014). Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190-194. 10.1038/nature12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y., Guo, M., Whitsett, J. A. and Xu, Y. (2015). ‘LungGENS’: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax 70, 1092-1094. 10.1136/thoraxjnl-2015-207035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endale, M., Ahlfeld, S., Bao, E., Chen, X., Green, J., Bess, Z., Weirauch, M. T., Xu, Y. and Perl, A. K. (2017). Temporal, spatial, and phenotypical changes of PDGFRα expressing fibroblasts during late lung development. Dev. Biol. 425, 161-175. 10.1016/j.ydbio.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak, G., McDavid, A., Yajima, M., Deng, J., Gersuk, V., Shalek, A. K., Slichter, C. K., Miller, H. W., McElrath, M. J., Prlic, M.et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, D. B., Penkala, I. J., Zepp, J. A., Sivakumar, A., Linares-Saldana, R., Zacharias, W. J., Stolz, K. G., Pankin, J., Lu, M. Q., Wang, Q.et al. (2019). Early lineage specification defines alveolar epithelial ontogeny in the murine lung. Proc. Natl. Acad. Sci. USA 116, 4362-4371. 10.1073/pnas.1813952116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillich, A., Zhang, F., Farmer, C. G., Travaglini, K. J., Tan, S. Y., Gu, M., Zhou, B., Feinstein, J. A., Krasnow, M. A. and Metzger, R. J. (2020). Capillary cell-type specialization in the alveolus. Nature 586, 785-789. 10.1038/s41586-020-2822-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, R., Leaffer, D., Chapin, C., Gillespie, A. M., Eckalbar, W. and Dobbs, L. (2019). Cell fate analysis in fetal mouse lung reveals distinct pathways for TI and TII cell development. Am. J. Physiol. Lung Cell. Mol. Physiol. 317, L653-L666. 10.1152/ajplung.00503.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss, K. (2018). Long-term pulmonary vascular consequences of perinatal insults. J. Physiol. 597, 1175-1184. 10.1113/JP275859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M., Du, Y., Gokey, J. J., Ray, S., Bell, S. M., Adam, M., Sudha, P., Perl, A. K., Deshmukh, H., Potter, S. S.et al. (2019). Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nat. Commun. 10, 37. 10.1038/s41467-018-07770-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister, C. and Satija, R. (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges, M. and Morrisey, E. E. (2014). Lung development: orchestrating the generation and regeneration of a complex organ. Development 141, 502-513. 10.1242/dev.098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, Y.-C. (2015). Theory and practice of lineage tracing. Stem Cells 33, 3197-3204. 10.1002/stem.2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T., Nishiwaki, T., Sekine, A., Nishimura, R., Suda, R., Urushibara, T., Suzuki, T., Takayanagi, S., Terada, J., Sakao, S.et al. (2015). Vascular repair by tissue-resident endothelial progenitor cells in endotoxin-induced lung injury. Am. J. Respir. Cell Mol. Biol. 53, 500-512. 10.1165/rcmb.2014-0185OC [DOI] [PubMed] [Google Scholar]
- Kim, T., Chen, I. R., Lin, Y., Wang, A. Y.-Y., Yang, J. Y. H. and Yang, P. (2019). Impact of similarity metrics on single-cell RNA-seq data clustering. Brief. Bioinform. 20, 2316-2326. 10.1093/bib/bby076 [DOI] [PubMed] [Google Scholar]
- Kimmel, J. C., Penland, L., Rubinstein, N. D., Hendrickson, D. G., Kelley, D. R. and Rosenthal, A. Z. (2019). Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res. 29, 2088-2103. 10.1101/gr.253880.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y., Tata, A., Konkimalla, A., Katsura, H., Lee, R. F., Ou, J., Banovich, N. E., Kropski, J. A. and Tata, P. R. (2020). Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934-946. 10.1038/s41556-020-0542-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno, G., Soldatov, R., Zeisel, A., Braun, E., Hochgerner, H., Petukhov, V., Lidschreiber, K., Kastriti, M. E., Lönnerberg, P., Furlan, A.et al. (2018). RNA velocity of single cells. Nature 560, 494-498. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, M., Bergen, V., Klein, M., Setty, M., Reuter, B., Bakhti, M., Lickert, H., Ansari, M., Schniering, J., Schiller, H. B.et al. (2020). CellRank for directed single-cell fate mapping. bioRxiv. 2020.10.19.345983. 10.21203/rs.3.rs-94819/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.-H., Tammela, T., Hofree, M., Choi, J., Marjanovic, N. D., Han, S., Canner, D., Wu, K., Paschini, M., Bhang, D. H.et al. (2017). Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170, 1149-1163.e1112. 10.1016/j.cell.2017.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes, L., Healy, J., Saul, N. and Großberger, L. (2018). UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3, 861. 10.21105/joss.00861 [DOI] [Google Scholar]
- Mereu, E., Lafzi, A., Moutinho, C., Ziegenhain, C., McCarthy, D. J., Álvarez-Varela, A., Batlle, E., Sagar, E., Grün, D., Lau, J. K.et al. (2020). Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 38, 747-755. 10.1038/s41587-020-0469-4 [DOI] [PubMed] [Google Scholar]
- Miner, J. H., Lewis, R. M. and Sanes, J. R. (1995). Molecular cloning of a novel laminin chain, α5, and widespread expression in adult mouse tissues. J. Biol. Chem. 270, 28523-28526. 10.1074/jbc.270.48.28523 [DOI] [PubMed] [Google Scholar]
- Miner, J. H., Patton, B. L., Lentz, S. I., Gilbert, D. J., Snider, W. D., Jenkins, N. A., Copeland, N. G. and Sanes, J. R. (1997). The laminin α chains: expression, developmental transitions, and chromosomal locations of α1-5, identification of heterotrimeric laminins 8-11, and cloning of a novel α3 isoform. J. Cell Biol. 137, 685-701. 10.1083/jcb.137.3.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey, E. E. and Hogan, B. L. M. (2010). Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell 18, 8-23. 10.1016/j.devcel.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, N. M. and Senior, R. M. (2006). Laminin isoforms and lung development: all isoforms are not equal. Dev. Biol. 294, 271-279. 10.1016/j.ydbio.2006.03.032 [DOI] [PubMed] [Google Scholar]
- Nguyen, N. M., Kelley, D. G., Schlueter, J. A., Meyer, M. J., Senior, R. M. and Miner, J. H. (2005). Epithelial laminin α5 is necessary for distal epithelial cell maturation, VEGF production, and alveolization in the developing murine lung. Dev. Biol. 282, 111-125. 10.1016/j.ydbio.2005.02.031 [DOI] [PubMed] [Google Scholar]
- Niethamer, T. K., Stabler, C. T., Leach, J. P., Zepp, J. A., Morley, M. P., Babu, A., Zhou, S. and Morrisey, E. E. (2020). Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. eLife 9, e53072. 10.7554/eLife.53072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plosa, E. J., Benjamin, J. T., Sucre, J. M., Gulleman, P. M., Gleaves, L. A., Han, W., Kook, S., Polosukhin, V. V., Haake, S. M., Guttentag, S. H.et al. (2020). beta1 Integrin regulates adult lung alveolar epithelial cell inflammation. JCI Insight 5, e129259. 10.1172/jci.insight.129259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuger, L., O'Shea, S., Rheinheimer, J. and Varani, J. (1990). Laminin in lung development: effects of anti-laminin antibody in murine lung morphogenesis. Dev. Biol. 137, 26-32. 10.1016/0012-1606(90)90004-3 [DOI] [PubMed] [Google Scholar]
- Schuler, B. A., Habermann, A. C., Plosa, E. J., Taylor, C. J., Jetter, C., Negretti, N. M., Kapp, M. E., Benjamin, J. T., Gulleman, P., Nichols, D. S.et al. (2020). Age-determined expression of priming protease TMPRSS2 and localization of SARS-CoV-2 in lung epithelium. J. Clin. Invest. 131, e140766. 10.1172/JCI140766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouch, A. N., McCoy, A. M., Greer, R. M., Lakhdari, O., Yull, F. E., Blackwell, T. S., Hoffman, H. M. and Prince, L. S. (2016). IL-1beta and inflammasome activity link inflammation to abnormal fetal airway development. J. Immunol. 196, 3411-3420. 10.4049/jimmunol.1500906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunz, M., Simon, L. M., Ansari, M., Kathiriya, J. J., Angelidis, I., Mayr, C. H., Tsidiridis, G., Lange, M., Mattner, L. F., Yee, M.et al. (2020). Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559. 10.1038/s41467-020-17358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W. M., III, Hao, Y., Stoeckius, M., Smibert, P. and Satija, R. (2019). Comprehensive integration of single-cell data. Cell 177, 1888-1902.e1821. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucre, J. M. S., Vickers, K. C., Benjamin, J. T., Plosa, E. J., Jetter, C. S., Cutrone, A., Ransom, M., Anderson, Z., Sheng, Q., Fensterheim, B. A.et al. (2020). Hyperoxia injury in the developing lung is mediated by mesenchymal expression of Wnt5A. Am. J. Respir. Crit. Care. Med. 201, 1249-1262. 10.1164/rccm.201908-1513OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila Ellis, L., Cain, M. P., Hutchison, V., Flodby, P., Crandall, E. D., Borok, Z., Zhou, B., Ostrin, E. J., Wythe, J. D. and Chen, J. (2020). Epithelial Vegfa specifies a distinct endothelial population in the mouse lung. Dev. Cell 52, 617-630.e616. 10.1016/j.devcel.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton, D., El-Hashash, A., Carraro, G., Tiozzo, C., Sala, F., Rogers, O., De Langhe, S., Kemp, P. J., Riccardi, D., Torday, J.et al. (2010). Lung organogenesis. Curr. Top. Dev. Biol. 90, 73-158. 10.1016/S0070-2153(10)90003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, T., Wang, Y., Deng, N., Huang, G., Taghavifar, F., Geng, Y., Liu, N., Kulur, V., Yao, C., Chen, P.et al. (2018). Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep. 22, 3625-3640. 10.1016/j.celrep.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa, H., Davis, E. C., Starcher, B. C., Ouchi, T., Yanagisawa, M., Richardson, J. A. and Olson, E. N. (2002). Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 415, 168-171. 10.1038/415168a [DOI] [PubMed] [Google Scholar]
- Young, M. D. and Behjati, S. (2020). SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9, giaa151. 10.1093/gigascience/giaa151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, W. J., Frank, D. B., Zepp, J. A., Morley, M. P., Alkhaleel, F. A., Kong, J., Zhou, S., Cantu, E. and Morrisey, E. E. (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251-255. 10.1038/nature25786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp, J. A., Zacharias, W. J., Frank, D. B., Cavanaugh, C. A., Zhou, S., Morley, M. P. and Morrisey, E. E. (2017). Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 170, 1134-1148.e1110. 10.1016/j.cell.2017.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp, J. A., Morley, M. P., Loebel, C., Kremp, M. M., Chaudhry, F. N., Basil, M. C., Leach, J. P., Liberti, D. C., Niethamer, T. K., Ying, Y.et al. (2021). Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science 371, eabc3172. 10.1126/science.abc3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.