Abstract
Secreted and nuclear forms of fibroblast growth factor 3 (FGF3) have opposing effects on cells. The secreted form stimulates cell growth and transformation, while the nuclear form inhibits DNA synthesis and cell proliferation. By using the yeast two-hybrid system we have identified a nucleolar FGF3 binding protein (NoBP) which coimmunoprecipitated and colocalized with FGF3 in transfected COS-1 cells. Characterization of the NoBP binding domain of FGF3 exactly matched the sequence requirements of FGF3 for its translocation into the nucleoli, suggesting that NoBP might be the nucleolar binding partner of FGF3 essential for its nucleolus localization. Carboxyl-terminal domains of NoBP contain linear nuclear and nucleolar targeting motifs which are capable of directing a heterologous protein β-galactosidase to the nucleus and the nucleoli. While NoBP expression was detected in all analyzed proliferating established cell lines, NoBP transcription was rapidly downregulated in the promyelocytic leukemia cell line HL60 when induced to differentiate. Analysis on the expression pattern of NoBP mRNA throughout the cell cycle in HeLa cells synchronized by lovastatin demonstrated a substantial upregulation during the late G1/early S phase. NoBP overexpression conferred a proliferating effect onto NIH 3T3 cells and can counteract the inhibitory effect of nuclear FGF3, suggesting a role of NoBP in controlling proliferation in cells. We propose that NoBP is the functional target of nuclear FGF3 action.
In mammals, the fibroblast growth factor (FGF) family is currently comprised of 20 genes encoding structurally related proteins with molecular masses in the range of 20 to 40 kDa. In vitro, the FGFs demonstrate the ability to regulate cell proliferation, differentiation, cell motility, extension of neurites, and cell survival, depending on the context. In vivo, many members of this family of intercellular signaling molecules have been shown to be crucial for normal development, while their inappropriate activity has been implicated in a wide range of pathological conditions, including skeletal dysplasias, tumorigenesis, and metastasis (3, 4, 10, 23, 25, 28, 34).
FGFs have been shown to bind three different types of transmembrane receptor. A cystein-rich receptor which binds FGFs and transforming growth factor β (TGFβ) with high affinity. This receptor resides in the secretory pathway as well as on the cell surface. Its function is nuclear, although there is evidence to suggest that it influences the intracellular trafficking of FGFs (7, 26, 30, 33, 39). Intercellular signaling by FGFs is mediated by high-affiniy cell surface receptors (FGFR) with intrinsic tyrosine kinase activity (12, 15). However, there is also a requirement for a lower-affinity heparan sulfate-containing proteoglycan receptor which forms part of the multimeric signaling complex (23). There are four different genes encoding high-affinity FGFRs, although receptor complexity is expanded by alternative splicing that gives rise to receptor isoforms with different ligand binding specifities (29, 36, 37). However, there is good evidence that several FGFs, including FGF2 and FGF3, can signal by directly entering the nucleus, thereby providing a cell with the potential to respond directly to intracrine signals, in addition to autocrine or paracrine signals, via cell surface receptors (9, 16, 18, 27).
FGF3 was identified as a proto-oncogene in virally induced mouse mammary tumors. However, subsequent analyses revealed that it is not normally expressed in the mammary gland but rather is primarily restricted to prenatal mouse development. In situ hybridization revealed a dynamic pattern of expression from gastrulation to birth, suggesting potential roles in mouse development (14, 32, 38). The biosynthesis of FGF3 is unusual in that a single CUG initiation codon is the major translation start site which gives rise to a protein that is directed in similar proportions to the cell nucleus and the secretion pathway. The dual fate of FGF3 is achieved by finely balanced opposing signals near the amino terminus: an internal signal peptide for vectorial translation across the endoplasmic reticulum and a bipartite nuclear localization signal (NLS). The import of FGF3 into the nucleus is mediated by karyopherin α1 (NPI-1), the NLS binding subunit of a heterodimeric receptor of the nuclear import machinery. The N-terminal targeting signals of FGF3 are weak signals since substitution with stronger signals changes the balance between the secretory pathway and nuclear uptake. These weak signals are mechanistically important to allow competition between the intracellular trafficking pathways. To overcome the disadvantage of a weak bipartite NLS, an additional NLS is located in the body of the protein, which also interacts with karyopherin α1 to enhance nuclear uptake without disturbing the balance of the competing N-terminal targeting motifs (2). A C-terminal motif was found to be necessary for efficient nucleolar association but was dispensable for the nuclear import of FGF3. Cells expressing low levels of an FGF3 mutant, lacks the signal peptide and therefore is exclusively nuclear, proliferate very poorly. The growth-inhibitory effect depends on the nucleolar localization of FGF3. Cells transfected with cDNAs in which the encoded FGF3 lacked the C-terminal motif essential for nucleolar accumulation exhibited growth rates similar to those of the nontransfected cells (2, 18, 20).
Nuclear localization of FGFs is not an exclusive property of FGF3, since FGF1 and FGF2 have been shown to localize to the nucleus by two apparently independent processes (34). Moreover, FGF2 shows an intracellular route to the nucleus, as well as an extracellular uptake and transport to the nucleolus. Extracellular uptake of FGF2 to the nucleolus occurs during late G1 phase of the cell cycle in growing aortic endothelial cells and is correlated with increase rRNA transcription (6). Intracellular nuclear transport occurs primarily with amino-terminally extended isoforms of FGF2 that are initiated at CUG codons. In contrast to FGF3, the nuclear localization of FGF2 appears not to involve a classical NLS but rather requires methylated glycine-arginine-rich sequences within the amino-terminal extension (5, 24).
To gain some insight into the signaling pathway used by nuclear FGF3, we used a yeast two-hybrid screen to identify genes encoding possible FGF3 interacting proteins. We identified a human gene of unknown function whose 305-amino-acid product interacts with FGF3 in vitro and in vivo. The protein contains a nuclear and a nucleolar targeting signal and accumulates in the nucleoli. We therefore named the protein NoBP, for nucleolar binding protein. NoBP transcription is regulated in a cell cycle-dependent fashion, and overexpression of NoBP in NIH 3T3 cells resulted in proliferation under serum-reduced conditions. Significantly, overexpression of NoBP in nuclear FGF3-expressing mouse mammary epithelial cells can abrogate the inhibitory effect of nuclear FGF3.
MATERIALS AND METHODS
Cell culture.
COS-1, HeLa, and NIH 3T3 cells were routinely maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS). HC11 mouse mammary epithelial cells were maintained in RPMI 1640 medium supplemented with 8% FCS, 10 ng of epidermal growth factor (EGF) per ml, and 5 μg of insulin per ml as described previously (20). HL60 cells were maintained in RPMI 1640 medium supplemented with 10% FCS. To induce granulocyte differentiation, HL60 cells were resuspended at a concentration of 2 × 105 cells/ml in the growth medium supplemented with 1.2% dimethyl sulfoxide (DMSO), and for the induction of monocyte-macrophage differentiation the growth medium was supplemented with 3.3 × 10−8 M tetradecanoyl phorbol acetate (TPA). Differentiation was monitored by examining the morphological appearance and by the ability to reduce nitroblue tetrazolium. For growth rate comparisons, equal numbers of each cell type were seeded in DMEM with the indicated concentration of FCS. At the times indicated, triplicate samples were harvested with trypsin, and four independent dilutions were counted in a hemocytometer. For transient DNA transfections, 20 μg of purified plasmid DNA was introduced into 5 × 105 COS-1 cells by electroporation (450 V/250 μF) using a Bio-Rad Gene-Pulser. At between 48 and 72 h after transfection, the cells were harvested for immunoblot analysis or processed for immunofluorescence. For stable DNA transfections, purified plasmid DNAs were introduced by lipofection using Transfast (Promega) as recommended by the manufacturer.
RNA isolation and Northern blot hybridization.
Total cellular RNA was extracted from cultured cell lines and mouse tissues by guanidium isothiocyanate and cesium trifluoroacetate gradient purification. For Northern blot analysis, 20 μg of total RNA was fractionated in denaturing Glyoxal gels, transferred to Hybond N (Amersham), and hybridized with 32P-labeled probes under stringent conditions (1).
Plasmid constructions.
pNoBP1.1 was constructed by inserting the anti-RGS(His)6 epitope upstream and in frame of the coding region of human NoBP cDNA. The modified NoBP cDNA was then inserted into the expression vector pKC4 under control of the early simian virus 40 (SV40) promoter. To obtain the plasmids pNoBP1.2, pNoBP1.3, and pNoBP1.4, PCR was used to delete the carboxyl-terminal 44, 86, and 139 amino acids of NoBP, respectively. The vector pKC4.16, which expresses a mutant FGF3 lacking the signal peptide, has been described previously (18). pNoBP1.5 was produced by deleting the 162 N-terminal amino acids and inserting 5′ the sequence encoding the anti-RGS(His)6 epitope. The β-galactosidase–NoBP fusion proteins were based on the expression plasmids pGAL1.0 and pGAL1.1, which have been previously described. Using PCR, partial sequences of NoBP, including the His-tag, were amplified with 3′ primers that introduced an XhoI site and 5′ primers that introduced an XbaI site. The resulting PCR fragments were inserted into the single XhoI site and the XbaI site of pGAL1.1, replacing the fgf3 sequences. The expression plasmid pDobs4.16 was described previously (20). A retrovirus vector based on Moloney murine leukemia virus was used to construct pBabe NoBP1.1 by inserting the NoBP cDNA from pKCNoBP1.1 as a blunted HindIII/EcoRI fragment into the SnaBI and EcoRI sites of the pBabe neo vector.
Cell cycle analysis.
Medium was removed from growing HeLa cells and replaced with fresh medium containing 60 μM lovastatin for 33 h. At time zero, cells were stimulated by replacing the medium with fresh medium plus 6 mM mevalonic acid. At the indicated times, the level of DNA synthesis was estimated by labeling the cells with 5 μCi of 3H-radiolabeled thymidine per ml for 1 h at 37°C in medium lacking thymidine and hypoxanthine (17). DNA synthesis was then assayed as described previously (21).
Immunofluorescence.
COS-1 cells grown on glass coverslips were transfected with the appropriate plasmids, and 48 h later they were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min. The cells were then permeabilized with 0.2% Triton X-100 in PBS and processed as previously described (22).
Immunoblot analysis and in vitro translation.
The procedures used for preparing cell lysates have been described in detail elsewhere (18). Samples from equivalent numbers of cells were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12.5 or 15% polyacrylamide gels, transferred to nitrocellulose membranes (Schleicher & Schuell), and then probed with rabbit polyclonal antibody to FGF3 or a mouse monoclonal antibody against the RGS-His tag (Qiagen). The immunoreactive proteins were detected by enhanced chemiluminescence using horseradish peroxidase-coupled anti-rabbit immunoglobulin antibodies as described by the manufacturer (Amersham International). Mouse FGF3 and NoBP cDNAs in pGem4Z were used in an in vitro translation system (Promega) to generate 35S-labeled products for use in binding assays as described in the text.
GST fusion proteins.
To obtain the glutathione S-transferase (GST)–NoBP constructs, DNA sequences encoding the appropriate amino acids were amplified by PCR using Pfu polymerase (Stratagene) and oligonucleotides with SmaI and EcoRI recognition sequences. The PCR products were cloned into SmaI-EcoRI-digested pGEX (Pharmacia). All expression constructs were verified by DNA sequence analysis and transformed into Escherichia coli DH5α for the expression of the fusion proteins.
GST-NoBP fusion protein affinity chromatography.
An overnight 30-ml E. coli culture containing pGEX-NoBP or a control GST plasmid was diluted 10-fold into Luria-Bertani or ampicillin medium and grown at 37°C to an optical density at 600 nm of 1.0 before induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Bacteria were lysed by pulse sonication in lysis buffer (1% Triton X-100, 1.5% N-laurylsarcosine, 25 mM triethanolamine, and 1 mM EDTA in PBS). Then, 200 μl of a 50% slurry of glutathione (GSH)-agarose beads (Molecular Probes) was added, and the mixture was incubated at 4°C overnight. After six washes in PBS–1% Triton X-100 (PBS-TX), agarose beads containing bound proteins were analyzed by SDS-PAGE, followed by Coomassie blue staining and immunoblot analysis. Interactions between GST-NoBP and 35S-labeled FGF3 or cell-associated proteins were analyzed using 0.5-ml aliquots of fgf3-transfected COS-1 cells or 2 μl of 35S-labeled FGF3 to which 50 μl of the prepared GST- or GST-NoBP-glutathione-agarose beads was added. The binding reactions were incubated at 4°C overnight. The beads were washed, resuspended in PBS-TX buffer, and poured into a column. The columns were extensively washed with an excess of PBS-TX, and the retained proteins were analyzed by SDS-PAGE. FGF3 was detected by immunoblotting or fluororadiography.
Immunoprecipitation of His-NoBP from transfected COS-1 cells.
COS-1 cells transfected with vectors containing His-NoBP cDNA or pKC4.16 (18) were washed twice with PBS and lysed in ice-cold lysis buffer (50 mM Tris, pH 8.0; 150 mM NaCl; 1% Nonidet P-40; 0,5% sodium deoxycholate; 0.1% SDS; 0.02% sodium azide; 1 mM phenylmethylsulfonyl fluoride; 10 μg of aprotinin per ml). The lysates were incubated at 4°C overnight with anti-PentaHis (Quiagen) preadsorbed onto 40 μl of protein G-Sepharose (Pharmacia). The precipitates were washed four times with sodium phosphate wash buffer, eluted in Laemmli loading buffer, and subjected to SDS-PAGE and Western blot analysis with anti-His monoclonal antibody or anti-FGF3 serum.
Yeast two-hybrid screening.
The bait contained fgf3 cDNA encoding the entire protein (4.12 [18]) fused in frame to the DNA-binding domain of Gal4. The cDNA insert from pKC4.12 was inserted at the NcoI/SalI restriction sites of vector pAS2 (Clontech). The bait was used to screen a human B-cell lymphoma cDNA library (a generous gift of Steve Elledge [8]) cloned in pACT. After cotransfection of the pAS2-construct and the pACT library into Y190 yeast cells, positive clones were selected on triple-minus plates (Leu− Trp− His−) containing 25 mM 3-aminotriazol and assayed for β-galactosidase activity. Positive cDNA clones were cycloheximide selected and tested by cotransfection and by mating with control bait vectors and with the original pAS2 vector to confirm the interaction. Mutations in the NoBP constructs were made by PCR, and the sequence was confirmed by DNA sequencing before the mutations were analyzed in the yeast two-hybrid assay. The N-terminal and C-terminal deletion mutants of FGF3 were previously described (20), and the inserts were transfered into the NcoI/SalI restriction sites of the pAS2 vector.
RESULTS
Identification of an FGF3 NoBP.
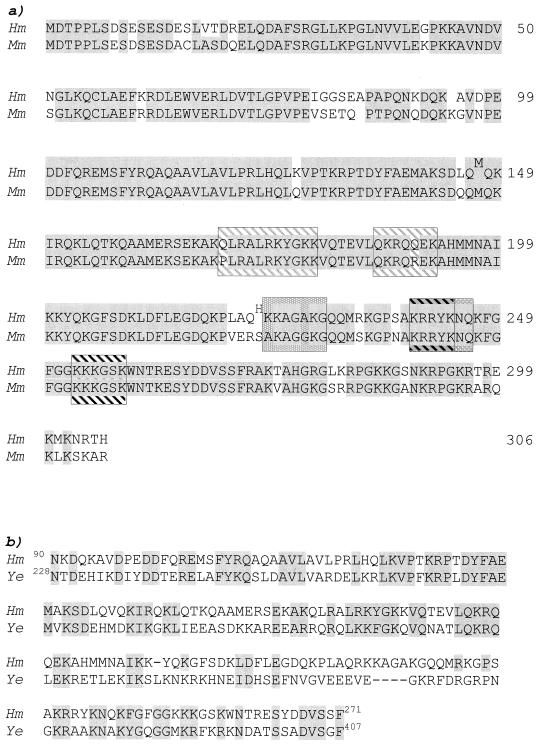
A yeast two-hybrid screen was used to search for FGF3 binding proteins. A cDNA library prepared from human B-cell lymphoma cells and containing in-frame fusions with the activator domain of GAL4 in the vector pACT was screened with a bait containing a full-length FGF3 cDNA fused to the DNA binding domain of GAL4 in the vector pAS2. From a total of 2.5 × 107 transformants, 29 clones were isolated. Inserts from 19 independent clones that gave strong signals with the FGF3 bait were isolated, sequenced, and compared to sequences in GenBank using the BLAST search program. Four identical clones (F2, F3, F17, and F31) were found to be very similar to a partial human cDNA designated p40, which was submitted as a sequence encoding a nucleolear protein of unknown function (D. Henning et al., GenBank NM_006824). The NoBP is 306 amino acids long, with a calculated molecular weight of 39,000. The amino acid derived sequence of the clone F2 was found to begin at amino acid 53 of the published cDNA sequence. Features of NoBP include a consensus sequence for casein kinase II phosphorylation at the N terminus, two overlapping basic domains which may function as NLSs, and a GR- and GK-rich region at its C terminus. A search of the mouse EST database revealed overlapping cDNA sequences which would encode a protein which shares 82% amino acid sequence identity with the human NoBP protein (Fig. 1), and a BLAST search of the GenBank database with the entire NoBP coding cDNA sequence revealed 57% identity with a hypothetical Caenorhabditis elegans protein of 40 kDa, and a 32% identity with a hypothetical protein of 49 kDa encoded by the open reading frame (ORF) YKL172W in Saccharomyces cerevisiae. (EMB CAA82014.1). The predicted human NoBP product aligned with the sequences of C. elegans and S. cerevisiae proteins (Fig. 1b) demonstrates the highest homology acids over the central region of 100 amino acids (residues 103 to 203) with 62% identity and 55% similarity, respectively, allowing for substitution of chemically similar amino acids.
FIG. 1.
(a) Amino acid sequence of human NoBP (Hm) and comparison to mouse NoBP (Mm). Alignment of the deduced amino acid sequence of human and mouse cDNAs using the DNAsis program. Identical amino acids are indicated by gray boxes. Two amino acid differences were found in the human NoBP compared to the sequence present in the GenBank database (accession no. NM_006824) and are indicated as superscripts. The basic amino acid-rich regions involved in targeting are boxed. (b) Amino acid sequence alignment of the middle region of human NoBP and the yeast ORF YKL172w. Identical and chemically similar amino acid residues are boxed in gray.
To determine whether the p49 from S. cerevisiae could interact with mouse FGF3, the hypothetical ORF encoding p49 (YKL172W) was amplified by PCR and cloned into the pACT vector to create an in-frame fusion with the activator domain of GAL4. The generated plasmid, pACTY49, was cotransformed into yeast with the FGF3 bait vector. A good positive signal was obtained, indicating conservation of the FGF3 interacting region in the NoBP homologs (data not shown).
NoBP associates with FGF3 in vitro and in vivo.
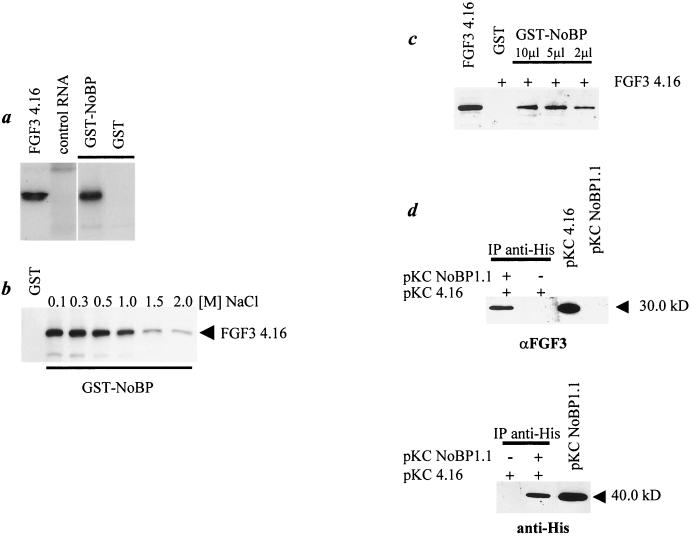
To investigate the interaction between FGF3 and NoBP, we used recombinant NoBP generated as a GST fusion protein immobilized on GSH-Sepharose beads to bind to in vitro-translated 35S-FGF3. The result was a strong retention of 35S-FGF3 by GST-NoBP, demonstrating that the two proteins can interact and that posttranslational modifications of the proteins are unlikely to be needed for the interaction (Fig. 2a). Moreover, binding under conditions of increasing NaCl concentration demonstrated that the complex can form in 1 M NaCl, which is indicative of a strong interaction (Fig. 2b). Similarly, an extract of COS-1 cells expressing a mutant of FGF3 (pKC4.16) that resides exclusively in the nucleus and/or nucleolus, was mixed with GST-NoBP bound to GSH-Sepharose beads, washed, subjected to SDS-PAGE, and immunoblotted for FGF3. The results confirmed the ability of FGF3 to bind NoBP in vitro. To show that FGF3 and NoBP may form an intracellular association, COS-1 cells were cotransfected with vectors expressing NoBP (pKCNoBP1.1) and FGF3 (pKC4.16), and cell extracts were immunoprecipitated with anti-Penta His monoclonal antibody, which detects the histidine tag at the N terminus of NoBP encoded by pKCNoBP1.1. After separation by SDS-PAGE, FGF3-related proteins were detected by immunoblotting using a polyclonal antibody against FGF3 (Fig. 2d, upper panel). To detect the expression of NoBP, a monoclonal anti-RGS(His) antibody was used in similar immunoblots (Fig. 2d, lower panel). The results demonstrate that detection of FGF3 was dependent on the presence of NoBP.
FIG. 2.
Interaction between NoBP and FGF3. (a) In vitro-translated 35S-labeled FGF3 (FGF3 4.16) was incubated with GST beads containing the NoBP fusion protein or GST beads as indicated. After an extensive washing, the retained proteins were analyzed as described in the text. (b) To assess the affinity of FGF3 binding to NoBP bound to GST beads, the beads were loaded with 35S-labeled FGF3 at increasing concentrations of NaCl as indicated. (c) Cell extracts from Fgf-3-transfected COS-1 cells were affinity precipitated with GST-NoBP-loaded beads. Bound proteins were eluted, subjected to SDS–12.5% PAGE, and examined for the presence of FGF3 by immunoblotting with a rabbit anti-FGF3 peptide antibody. (d) COS-1 cells were transfected with RGS(His)6-tagged full-length NoBP cDNA (pKCNoBP1.0) or with the control vector alone, together with pKC4.16. The cell lysates were immunoprecipitated with the anti-PentaHis monoclonal antibody (Qiagen) and immunoblotted with the anti-FGF3 peptide antibody or with an anti-RGS(His) monoclonal antibody (Qiagen).
Localization of the sequences necessary for the FGF3 and NoBP association.
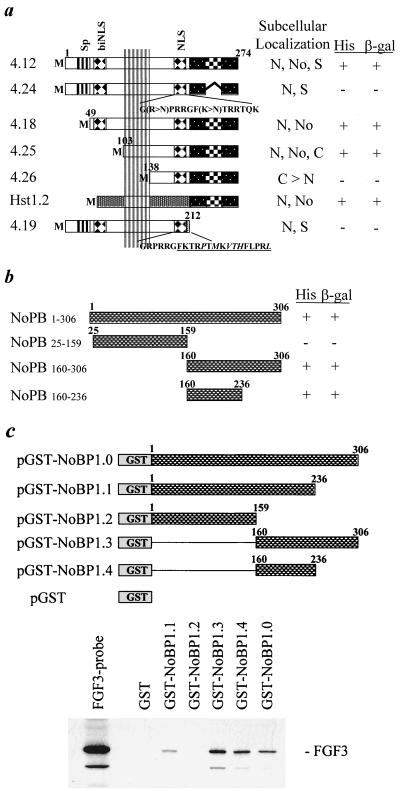
Previous mutation analyses have implicated different basic domains in the carboxy-terminal region of FGF3 as important for nuclear uptake and nucleolar association (20). To determine whether the nucleolar retention sequences are necessary for NoBP binding, several deletion and point mutants of FGF3 previously described (Fig. 3a) were analyzed for NoBP binding in a yeast two-hybrid screen. The results are summerized in Fig. 3a and show a perfect correlation between nucleolar localization in COS-1 cells and colony formation in the yeast screen. Mutant FGF3 analysis and the results obtained with the FGF chimera demonstrated that the sequences necessary for nucleolar localization have to be presented in a certain structural context, suggesting a nucleolar retention via binding to interacting proteins rather than a linear nucleolar targeting motif (19). The FGF3 mutant 4.26 lacking the sequences encoded by the second exon which is highly conserved in the FGF family and thought to be essential for their structural integrity is no longer located in the nucleoli and no longer interacted with NoBP in the yeast two-hybrid binding assay. The deleted N-terminal FGF3 sequences could be functionally replaced by the corresponding sequences of the exclusively secreted FGF family member FGF4 (Hst1.2). The mutant 4.19, in which the FGF4 C terminus is substituted for that of FGF3, was not detected in the nucleoli and did not bind to NoBP. These findings support the idea that, in fact, binding of FGF3 to NoBP is essential for its nucleolar accumulation.
FIG. 3.
Deletion analysis of the FGF3 and NoBP binding domains in the yeast two-hybrid system. (a) fgf3 cDNAs encoding wild-type or mutant FGF3 proteins with previously characterized subcellular localizations (18, 20) were cloned into the pAS2 vector and examined for NoBP binding in the two-hybrid system. Interactions between FGF3 and NoBP were scored based on growth on His− plates and β-galactosidase (β-gal) activity. The fgf3 cDNA encodes a signal peptide (Sp), a bipartite NLS (biNLS), a second NLS, and a C-terminal motif essential for nucleolar accumulation (checkered pattern). The part of the FGF proteins encoded by the second exon is marked by vertical stripes. The subcellular localization of the mutants are summarized to the right of each depicted cDNA as follows: N, nuclear; No, nucleolar; S, secreted; and C, cytoplasmic. (b) Truncation mutations of NoBP were generated by cleavage with appropriate restriction enzymes and then cloned into pACT2. Interactions between the NoBP mutations and wild-type FGF3 were scored as in panel a. (c) The binding of FGF3 to mutations of NoBP was assessed by affinity precipitation. Deletion mutations of NoBP fused to GST are indicated. The fusion proteins were expressed in E. coli, and extracts were bound to GSH-Sepharose beads as described in the text. The washed beads were incubated with 35S-labeled FGF3, and the retained protein was analyzed as described in Fig. 2b.
To locate the domain in NoBP that interacts with FGF3, truncation mutants were generated and assayed for FGF3 binding in the yeast two-hybrid screen (Fig. 3b). The results indicate that a fragment composed of amino acids 160 to 236 retained the ability to bind FGF3. This result was substantiated using a similarly truncated NoBP as GST-fusion protein in an in vitro binding assay with 35S-labeled in vitro-translated FGF3 (Fig. 3c). The results show that the region of NoBP between amino acids 160 and 236 is sufficient for FGF3 binding.
Nuclear FGF3 and NoBP colocalize in cells.
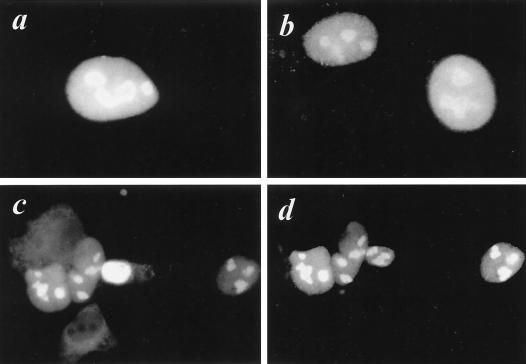
The association between FGF3 and NoBP was further investigated by immunofluorescence microscopy. FGF3 and His-tagged NoBP protein were coexpressed by transfecting COS-1 cells with pKC4.16 (FGF3) and pKCNoBP1.1 (NoBP) (Fig. 4). The nuclear and nucleolar distribution of both proteins appears to be virtually identical. For staining of the cotransfected cells, a rabbit polyclonal antibody against FGF3 and a monoclonal anti-His antibody was used with species-specific secondary antibodies conjugated to different fluorescence dyes (Texas red and fluorescein). Extensive colocalization of nuclear FGF3 and NoBP was observed. In experiments with single-staining antibodies, no crossover was observed between the channels for Texas red and fluorescein.
FIG. 4.
Colocalization of nuclear FGF3 and NoBP in COS-1 cells by immunofluorescence microscopy. COS-1 cells transiently transfected with pKC4.16 encoding nuclear FGF3 (a) or with RGS(His)-tagged NoBP (pKC-NoBP1.1) (b) or cotransfected with pKC4.16 and pKC-NoBP1.0 (c and d) were grown on coverslips for 48 h, fixed in 4% paraformaldehyde, and permeabilized with Triton X-100 as described in the text. The coverslips were then stained with a rabbit polyclonal peptide antibody against FGF3 or with anti-RGS(His) monoclonal antibody (Qiagen) against the His epitope. The NoBP His-tagged protein complexes were visualized with goat anti-mouse immunoglobulin tagged with fluorescein and the anti-FGF3 immunocomplexes were detected with goat anti-rabbit immunoglobulin tagged with Texas red.
Localization of elements in NoBP necessary for nuclear uptake and nucleolar association.
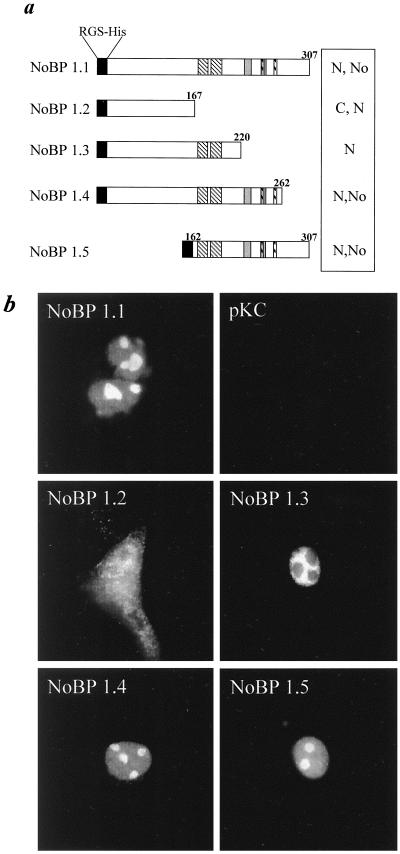
The NoBP sequence contains several clusters of basic residues located in the C-terminal half of the protein as depicted schematically in Fig. 5. To test the importance of these motifs on nuclear uptake, a series of deletion mutations were introduced into pKCNoBP1.1 which removed combinations of these potential targeting motifs. Deletion of the C terminus from amino acid 167, which encompasses all of the basic elements, leads to an even distribution of the protein between the nucleus and cytoplasm, but without nucleolar association. Deletion of the carboxyl terminus from amino acid residue 220, which conserves a lysine-, arginine-, and glutamine-rich domain resulted in exclusively a nuclear location of the product, but with exclusion from the nucleoli. In contrast, deletion of the carboxy-terminal 45-amino-acid residues did not affect the nuclear or nucleolar distribution. Moreover, deletion of the N-terminal half of NoBP (amino acids 1 to 166), did not change the subcellular location of the truncated protein. These results indicate that the domain between amino acids 167 and 220 contains sequences that are important for nuclear localization, while the sequences between amino acids 220 and 262 are necessary for nucleolar association (Fig. 5).
FIG. 5.
Deletion mutations affecting nuclear and nucleolar localization of NoBP. (a) Deletion mutations of pKC-NoBP1.1. Domains rich in basic amino acids are depicted schematically by the various stripped boxes (see Fig. 1). (b) Subcellular distribution of NoBP-related products of the deletion mutations depicted in panel a were analyzed by immunofluorescence using the anti-RGS(His) monoclonal antibody. Examples of the staining patterns are shown and are summarized alongside the depiction of each mutation shown in panel a. N, nuclear; No, nucleolar; C, cytoplasmic.
Motifs in the carboxyl terminus of NoBP can confer nuclear and nucleolar localization to a heterologous protein.
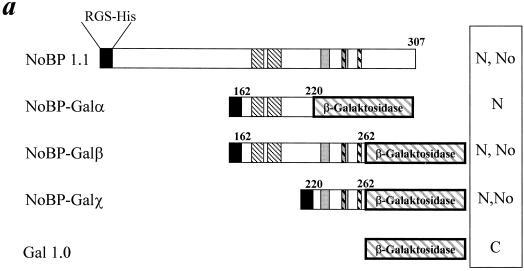
A more stringent test of nuclear and nucleolar targeting function is whether the candidate C-terminal motifs are sufficient to direct a heterologous cytoplasmic protein into the nucleus and associate it with the nucleolus. Segments of the carboxy terminus of NoBP were fused with the bacterial gene β-galactosidase as depicted in Fig. 6. After transfection of the various constructs into COS-1 cells, their intracellular localization was determined by immunofluorescence by using antibodies to β-galactosidase. The parental vector, Gal1.0 was located in the cell cytoplasm while, in contrast, constructs with segments of the carboxyl terminus of NoBP fused at the N terminus of the β-galacosidase accumulated in the nucleus. The fusion protein containing NoBP sequences between amino acids 162 and 262 and amino acids 220 and 262 both showed nuclear and nucleolar staining indistinguishable from that of NoBP. However, the segments at 162 and 220 only showed a nuclear staining pattern with exclusion from the nucleoli, a result again consistent with the deletion analysis of NoBP (Fig. 5). Taken together, the deletion analysis and signal transfer experiments suggest that there is a nucleolar targeting motif contained in amino acids 220 to 262 which is necessary and sufficient to direct a heterologous cytoplasmic protein into the nucleus and associate it with the nucleoli.
FIG. 6.
Localization of NoBP nuclear and nucleolar localization signals. (a) Segments of the carboxy terminus of NoBP, encompassing amino acids 162 to 262, were fused to the coding domain of β-galactosidase. The shading used to mark the different basic domains in the C-terminal region of NoBP are the same as in Fig. 5. (b) The location of the NoBP–β-galactosidase fusion proteins was determined by staining with a monoclonal antibody against β-galactosidase, followed by the addition of a secondary antibody conjugated to fluorescein.
NoBP is widely expressed in mouse tissue and cell lines.
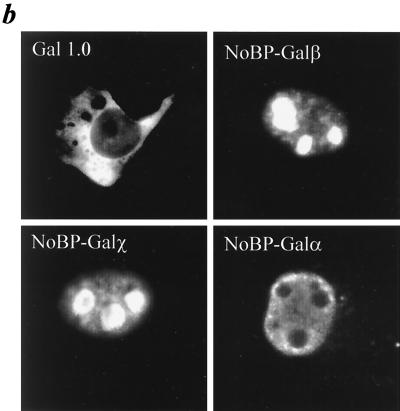
The expression of NoBP in adult mouse tissues and several cell lines of mouse and human origin was assessed by Northern blot analysis. A single transcript of 1.5 kb was detected in all mouse tissues and cell lines examined. The highest levels of expression were found in the lung, heart, spleen, and kidney. The broad expression of NoBP in mouse tissue, which does not express FGF3, suggests that NoBP has a common function in all cells and presumably interacts with partners other than FGF3 (Fig. 7).
FIG. 7.
Expression of NoBP transcripts in human and mouse cell lines and adult mouse tissues. Northern blots containing 10 μg of total RNA from different established human cell lines (a) or mouse cell lines (b) were hybridized with 32P-labeled human or mouse NoBP cDNA probe. (c) Northern blot of 10 μg of total RNA from different adult mouse tissues was done with a 32P-labeled mouse NoBP cDNA probe. The size of the NoBP-specific transcripts are indicated.
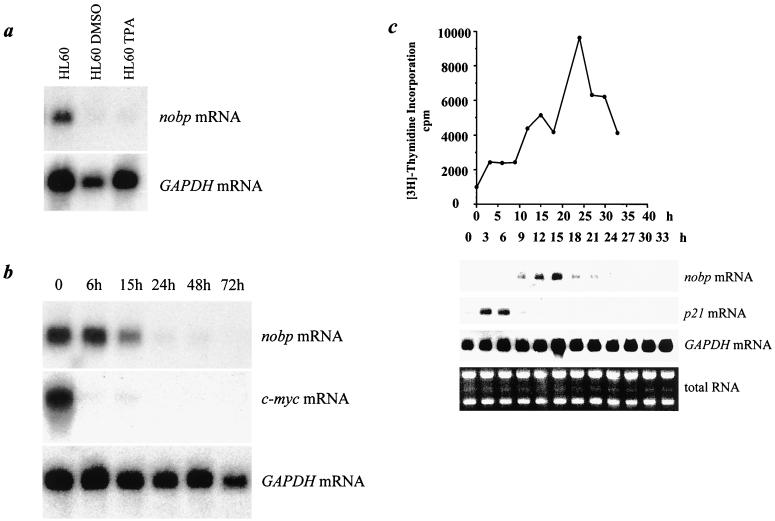
Modulation of NoBP transcripts in differentiating HL60 cells and cell cycle synchronized HeLa cells.
To determine whether NoBP is regulated during the cell cycle, we examined NoBP RNA expression in two different systems: differentiating HL60 cells and HeLa cells reentering the cell cycle from a lovastatin block. HL60 promyelocytic leukemia cells can be induced to differentiate into monocytes by treatment with a phorbol ester such as TPA or into granulocytes by treatment with DMSO (19). Differentiation is preceded by a withdrawal from the cell cycle. To examine expression of NoBP in differentiated cells, total RNA was prepared from HL60 cells exposed to DMSO or TPA for 3 days and then analyzed by Northern blotting. No NoBP-specific transcription was detectable in the differentiated HL60 cell populations. To investigate the kinetics NoBP transcript disappearance, HL60 cells were monitored for 3 days as they ceased to proliferate and differentiate in response to TPA treatment. Under these conditions, NoBP transcripts diminished in the absence of proliferation, but not as rapidly as c-myc transcripts (Fig. 8a). The kinetics of disappearance will be a function of transcriptional repression and messenger half-life which were not separable in this experiment; nevertheless, downregulation of NoBP appears to parallel exit from the cell cycle.
FIG. 8.
NoBP transcripts are downregulated in HL60 cells following differentiation. (a) Northern blot analysis of RNA samples were taken after 72 h of treatment of HL60 cells with TPA or DMSO. As a control for RNA loading, the blot was reprobed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (b) Kinetics of NoBP RNA (nobp) downregulation following treatment with TPA to induce monocyte-macrophage differentiation. (c) Transcriptional regulation of nobp expression in lovastatin-synchronized HeLa cells. HeLa cells were cultured in 60 μM lovastatin for 33 h, at which time lovastatin was replaced with medium containing mevalonate as described in the text. Cell samples were harvested at the indicated times after lovastatin removal, and the extracted RNA was analyzed by Northern blotting. The membranes were probed with 32P-labeled cDNA to nobp, p21, and GAPDH. RNA loading was also monitored by ethidium bromide staining of the RNA gel. The DNA synthesis rate was monitored by pulse labeling the DNA with [3H]thymidine as described in the text.
To further investigate the association of NoBP transcription with the cell cycle, lovastatin, an inhibitor of the cholesterol biosynthetic pathway, was used to synchronize HeLa cells in early G1 (17). After 33 h in lovastatin, the cultures were refed with medium supplemented with mevalonate to stimulate their reentry into the cell cycle. After release from the lovastatin block, there was a delay in DNA synthesis for several hours before an increase in incorporation of [3H]thymidine was observed (Fig. 8c). Immediately after release there was a peak of p21 (CIP1/WAF1) RNA expression (11), followed by a peak of NoBP transcription in late G1 and early S phases. However, NoBP expression decreased before the peak of DNA synthesis. These results demonstrate a strong correlation between proliferation and the expression NoBP mRNA.
NoBP expression confers a growth-stimulating effect on NIH 3T3 cells.
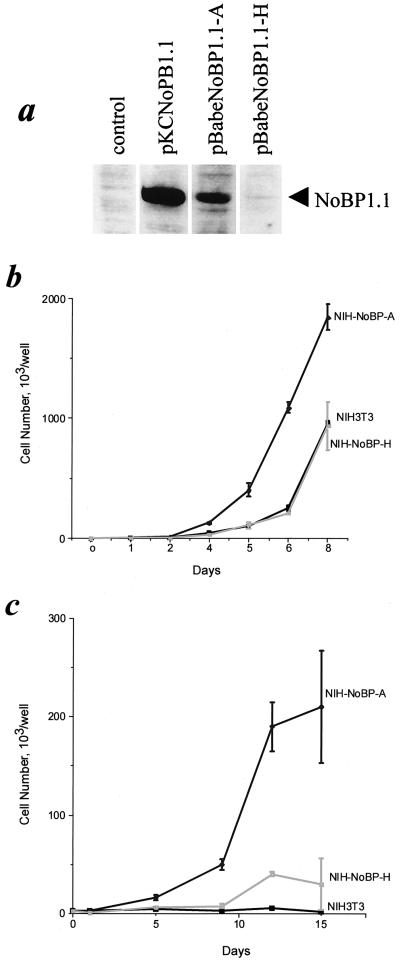
To determine whether the level of NoBP expression affected the proliferation rate, we tested the effect of overexpressing NoBP. NIH 3T3 cells were transfected with pBABE-NoBP1.1, and stably expressing clones were selected. Clones were selected on the basis of high or moderate levels of NoBP protein expression and analyzed for their proliferation potential (Fig. 9a). Expression of NoBP in NIH 3T3 did not lead to a transformed morphology and, with a full serum supply, a growth stimulatory effect was only detected in the cell line expressing high levels of NoBP (Fig. 9b). Still even in culture medium supplemented with 10% serum, NoBP is able to induce a twofold increase in the growth rate of NIH 3T3 cells. However, under serum-reduced conditions, the cell line expressing high and moderate levels of NoBP grew significantly better than the parental cells. The cell line with the low level of NoBP expression still grew at a 10-fold-higher rate than the parental NIH 3T3 cell line and the NoBP-overexpressing cell line (NIH-NoBP-A) induced a 100-fold-higher growth rate compared to the control cells. These findings suggest that higher expression levels of NoBP can confer a clear growth-promoting effect on NIH 3T3 cells without changing the cell morphology (Fig. 9c).
FIG. 9.
NoBP is growth stimulatory for NIH 3T3 cells. Two stable clones of NIH 3T3 cells expressing human NoBP (designated pBabeNoBP1.1-A and pBabeNoBP1.1H) were selected and tested for growth in complete medium or in low-serum conditions. (a) Immunoblot analysis of His-tagged NoBP expression in the two transfected NIH cell lines are compared to control COS-1 cells transiently transfected with pKCNoBP1.1. Protein samples corresponding to the same number of cells were analyzed. Cells were seeded at 2.5 × 103 cells per well in 24-well dishes and grown for 8 days in DMEM containing 10% FCS (b) or 2% FCS (c) for 15 days. Cells were harvested and counted, and the cell numbers were plotted as the means of triplicate determinations, with bars indicating the standard error of the mean.
Expression of NoBP can antagonize the inhibitory effect of nucleolar FGF3 on HC11 cells.
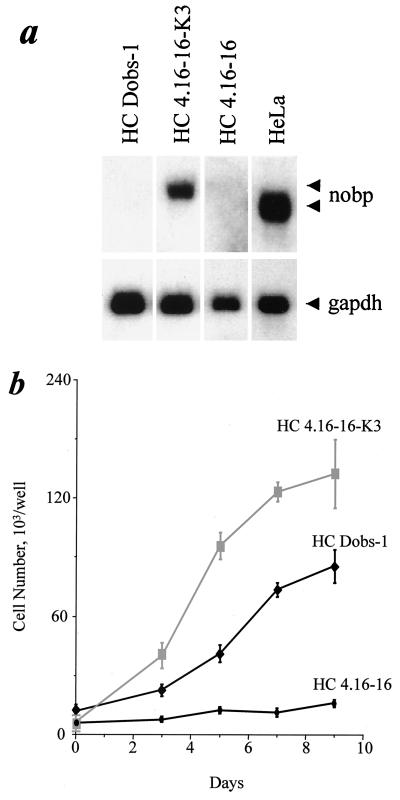
Expression of the nucleolar isoform of FGF3 in mammary epithelial HC11 cells inhibits cell proliferation (20). To determine whether overexpression of NoBP could reverse the block, a cell line (HC4.16-16 [20]) expressing nucleolar FGF3 was cotransfected with an SV40-based vector expressing the human NoBP cDNA and selected with a vector carrying the puromycin-resistant gene. Transfected cells were selected by resistance to puromycin, and several colonies were chosen and passaged in complete medium supplemented with FCS, EGF, and insulin. A cell line expressing a high level of NoBP mRNA was selected (Fig. 10, upper panel). The growth rate of this cell line (HC4.16-16-K3) was compared with those of the parental cell line HC4.16-16 under low-serum conditions. A cell line of HC11 cells transfected with the empty vector DNA was used as control (HC Dobs-1). Cells were seeded at a low density in medium supplemented with 2.5% serum, EGF, and insulin. Under these conditions, there was a significant correlation of high growth rate with high NoBP expression (Fig. 10b). The HC11 cells transfected with the 4.16 cDNA which encodes the nucleolus-associated form of FGF3 grew at a much lower growth rate than the control cells. However, when these cells were transfected with the NoBP cDNA and were expressing similar levels of NoBP transcripts as the HeLa cells, the growth-inhibitory effect of FGF3 was antagonized and NoBP induced a 10-fold increase in the growth rate compared to the parental cell line (HC4.16-16). These findings clearly demonstrate that expression of NoBP can counter the inhibitory effect of nuclear FGF3 and suggest that the growth-inhibitory effect of nuclear FGF3 may be due to an interaction of nucleolar FGF3 with NoBP, slowing entry into the S phase of the cell division cycle.
FIG. 10.
NoBP expression can reverse the inhibitory effect of nuclear FGF3 in HC11 cells. (a) An HC11 cell line expressing the nuclear form of FGF3 (HC4.16-16) was transfected with a vector encoding human His-tagged NoBP protein and a high-expression cell clone was selected (HC4.16-16-K3), followed by Northern blot analysis. (b) The nuclear FGF3 expressing cell line HC4.16-16, a control vector transfected cell clone (HC Dobs-1), and the NoBP and nuclear FGF3 expressing cell clone (HC4.16-16-K3) were seeded at 2.5 × 103 cells per well in 24-well dishes and grown for 9 days in DMEM containing 2% FCS, EGF, and insulin. Cells were harvested and counted, and the cell counts are plotted as the means of triplicate determinations, with bars indicating the standard error of the mean.
DISCUSSION
Many FGF family members have important intercellular signaling functions during mammalian development (34). For autocrine and paracrine signaling, the FGFs are secreted and activate receptors on the same or adjacent cells. However, several members of the FGF gene family encode isoforms that have a nuclear location (4). These include FGF1, FGF2, and FGF3, as well as the more recently described FGF homology factors (FGF11 to FGF14) (31). The latter encode proteins that do not contain signal sequences for secretion and may primarily function as nuclear proteins. In contrast, FGF3 encodes isoforms that are either exclusively secreted or have a dual localization. Thus, the amino-terminally extended isoform either enters the secretion pathway or is translocated to the nucleus or nucleolus. This allows the same protein to potentially have two signaling pathways: one autocrine or paracrine through secretion and receptor activation and a second directly acting on some nuclear targets. In an effort to establish possible components of a nuclear FGF3 signaling pathway, we used a yeast two-hybrid assay to identify an FGF3 binding protein that normally resides in the nucleolus. The affinity of FGF3 for NoBP is sufficiently strong to account for its nucleolar localization. Moreover, a mutation analysis of FGF3 shows a correlation between nucleolar localization and binding to NoBP. A search of the databases showed that NoBP was previously identified as a nucleolar protein of no known function. Here we demonstrate that it not only binds to FGF3 but is regulated in a cell cycle-dependent manner. Unlike FGF3, NoBP is widely distributed in different tissues and cell lines, and there appear to be homologues in flys and yeast, suggesting that it may have a function common to most, if not all, eukaryotic cells.
A mutation analysis of NoBP shows that FGF3 binding and its ability to translocate to the nucleus and associate with the nucleolus reside in the carboxy-terminal region of the protein (Fig. 3 and 5). A preliminary deletion analysis suggests that sequences between amino acids 162 and 220 encode the nuclear localization motif. This region is rich in basic amino acids (Fig. 1). Inclusion of amino acid sequences that are more carboxy terminal confer nucleolar association. Moreover, amino acid sequences between 220 and 262 are sufficient to confer nuclear localization and nucleolar association on β-galactosidase that normally resides in the cytoplasm. Interestingly, amino acids 162 to 220 will translocate β-galactosidase to the nucleus, indicating some redundancy of NLSs. The carboxy-terminal domain of NoBP is quite complex, encoding nuclear localization sequences, the nucleolar association domain, as well as a region that binds FGF3. How proteins which have no obvious RNA-binding domain elements accumulate in the nucleoli is not well understood.
Expression of human NoBP in NIH 3T3 cells resulted in a reduced requirement for serum and could keep them in a proliferating state under conditions in which they would normally quiesce. Furthermore, expression of NoBP RNA is regulated during the cell cycle, peaking during the late G1 and early S phases, supporting the idea that NoBP may be involved in controlling progress through the cell division cycle. The high degree of sequence homology between the mammalian NoBP and the putative S. cerevisiae gene product p49 suggests that the function of these proteins may be conserved during evolution. Homozygous deletion of p49 is lethal, establishing an essential function (data not shown). Recently, two groups demonstrated that the yeast homologue of NoBP is an essential nucleolar protein required for pre-rRNA processing (13, 35). The sequences critical for the essential activity of the yeast homologue comprise the most conserved region between the yeast and human protein and two putative NLSs (13). The FGF3 binding domain of human NoBP corresponds to the C-terminal part of the conserved region. Since the synthesis of ribosomes is essential for growing cells, mechanisms involved in the control of ribosome synthesis are therefore expected to determine the coordination between cell growth and cell division. Also, candidate c-myc target genes implicated in cell growth control regulate rRNA transcription and processing. In the light of these new findings, it is interesting to notice that immunogold electron microscopy locates FGF3 within the dense fibrillar components, which are regarded as the site of active transcription of rRNA genes and processing of pre-rRNA (20). Taken together, these findings suggest NoBP may be essential for integrating growth-regulatory signals with gene transcription and RNA processing.
Dual subcellular localization of FGF3 appears to reflect two opposing biological effects. Hence, secreted FGF3 induces cell proliferation through cell surface tyrosine kinase receptors, while in the same HC11 cells a nuclear-targeted FGF3 suppresses proliferation in the G1 phase of the cell cycle. This dual arrangement of signaling might hypothetically cause an FGF3-producing cell to send a paracrine signal inducing a mitogenic response, while its nuclear form blocks autocrine proliferation by an intracrine signal. We would suggest that NoBP serves as a target for the inhibition of cell proliferation by nuclear FGF3.
ACKNOWLEDGMENTS
We thank A. Eldredge for the kind gift of the human B-lymphoma cDNA library.
This work was supported by a grant from the DFG to P.K.
K.R. and M.A. contributed equally to this work.
REFERENCES
- 1.Antoine M, Kiefer P. Isolation, characterization and expression of the Xenopus laevis ribosomal protein S6 gene. Gene. 1999;231:127–135. doi: 10.1016/s0378-1119(99)00100-6. [DOI] [PubMed] [Google Scholar]
- 2.Antoine M, Reimers K, Dickson C, Kiefer P. Fibroblast growth factor 3, a protein with dual subcellular localization, is targeted to the nucleus and nucleolus by the concerted action of two nuclear localization signals and a nucleolar retention signal. J Biol Chem. 1997;272:29475–29481. doi: 10.1074/jbc.272.47.29475. [DOI] [PubMed] [Google Scholar]
- 3.Baird A, Klagsbrun M. The fibroblast growth factor family. Cancer Cells. 1991;3:239–243. [PubMed] [Google Scholar]
- 4.Basilico C, Moscatelli D. The FGF family of growth factors and oncogenes. Adv Cancer Res. 1992;59:115–165. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- 5.Bikfalvi A, Klein S, Pintucci G, Rifkin D B. Biological roles of fibroblast growth factor-2. Endocrinol Rev. 1997;18:26–45. doi: 10.1210/edrv.18.1.0292. [DOI] [PubMed] [Google Scholar]
- 6.Bouche G, Gas N, Prats H, Baldin V, Tauber J, Teissie J, Amalric F. Basic fibroblast growth factor enters the nucleolus and stimulates the transcription of ribosomal genes in ABAE cells undergoing G0→G1 transition. Proc Natl Acad Sci USA. 1987;84:6770–6774. doi: 10.1073/pnas.84.19.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrus L W, Zuber M E, Lueddecke B A, Olwin B B. Identification of a cysteine-rich receptor for fibroblast growth factors. Mol Cell Biol. 1992;12:5600–5609. doi: 10.1128/mcb.12.12.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durfee T, Mancini M A, Jones D, Elledge S J, Lee W H. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that co-localizes to centers for RNA processing. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesel R E, Maciag T. Molecular mechanisms of angiogenesis: fibroblast growth factor signal transduction. FASEB J. 1995;9:919–925. doi: 10.1096/fasebj.9.10.7542215. [DOI] [PubMed] [Google Scholar]
- 10.Goldfarb M. The fibroblast growth factor family. Cell Growth Differ. 1990;1:439–445. [PubMed] [Google Scholar]
- 11.Gray-Bablin J, Rao S, Keyomarsi K. Lovastatin induction of cyclin-dependent kinase inhibitors in human breast cells occurs in a cell cycle-independent fashion. Cancer Res. 1997;57:604–609. [PubMed] [Google Scholar]
- 12.Green P J, Walsh F S, Doherty P. Promiscuity of fibroblast growth factor receptors. Bioessays. 1996;18:639–646. doi: 10.1002/bies.950180807. [DOI] [PubMed] [Google Scholar]
- 13.Huber M D, Dworet J H, Shire K, Frappier L, McAlear M A. The budding yeast homolog of the human EBNA1-binding protein 2 (Ebp2p) is an essential nucleolar protein required for pre-rRNA processing. J Biol Chem. 2000;275:28764–28773. doi: 10.1074/jbc.M000594200. [DOI] [PubMed] [Google Scholar]
- 14.Jakobovits A, Shackleford G, Varmus H, Martin G. Two proto-oncogenes implicated in mammary carcinogenesis, int-1 and int-2, are independently regulated during mouse development. Proc Natl Acad Sci USA. 1986;83:7806–7810. doi: 10.1073/pnas.83.20.7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson D, Williams L. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 16.Joy A, Moffett J, Neary K, Mordechai E, Stachowiak E K, Coons S, Rankin-Shapiro J, Florkiewicz R Z, Stachowiak M K. Nuclear accumulation of FGF-2 is associated with proliferation of human astrocytes and glioma cells. Oncogene. 1997;14:171–183. doi: 10.1038/sj.onc.1200823. [DOI] [PubMed] [Google Scholar]
- 17.Keyomarsi K, Sandoval L, Band V, Pardee A B. Synchronization of tumor and normal cells from G1 to multiple cell cycles by lovastatin. Cancer Res. 1991;51:3602–3609. [PubMed] [Google Scholar]
- 18.Kiefer P, Acland P, Pappin D, Peters G, Dickson C. Competition between nuclear localization and secretory signals determines the subcellular fate of a single CUG-initiated form of FGF3. EMBO J. 1994;13:4126–4136. doi: 10.1002/j.1460-2075.1994.tb06730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiefer P, Bacher M, Pfluger K H. Relationship of calcitonin mRNA expression to the differentiation state of HL 60 cells. Leukoc Lymphoma. 1994;13:501–507. doi: 10.3109/10428199409049641. [DOI] [PubMed] [Google Scholar]
- 20.Kiefer P, Dickson C. Nucleolar association of fibroblast growth factor 3 via specific sequence motifs has inhibitory effects on cell growth. Mol Cell Biol. 1995;15:4364–4374. doi: 10.1128/mcb.15.8.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiefer P, Mathieu M, Close M J, Peters G, Dickson C. FGF3 from Xenopus laevis. EMBO J. 1993;12:4159–4168. doi: 10.1002/j.1460-2075.1993.tb06100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiefer P, Peters G, Dickson C. Retention of fibroblast growth factor 3 in the Golgi complex may regulate its export from cells. Mol Cell Biol. 1993;13:5781–5793. doi: 10.1128/mcb.13.9.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klagsbrun M, Baird A. A dual receptor system is required for basic fibroblast growth factor activity. Cell. 1991;67:229–231. doi: 10.1016/0092-8674(91)90173-v. [DOI] [PubMed] [Google Scholar]
- 24.Klein S, Carroll J A, Chen Y, Henry M F, Henry P A, Ortonowski I E, Pintucci G, Beavis R C, Burgess W H, Rifkin D B. Biochemical analysis of the arginine methylation of high molecular weight fibroblast growth factor-2. J Biol Chem. 2000;275:3150–3157. doi: 10.1074/jbc.275.5.3150. [DOI] [PubMed] [Google Scholar]
- 25.Klein S, Roghani M, Rifkin D. Fibroblast growth factors as angiogenesis factors: new insights into their mechanism of action. EXS. 1997;79:159–192. doi: 10.1007/978-3-0348-9006-9_7. [DOI] [PubMed] [Google Scholar]
- 26.Köhl R, Antoine M, Olwin B B, Dickson C, Kiefer P. Cysteine-rich fibroblast growth factor receptor alters secretion and intracellular routing of fibroblast growth factor 3. J Biol Chem. 2000;275:15741–15748. doi: 10.1074/jbc.M903271199. [DOI] [PubMed] [Google Scholar]
- 27.Maciag T, Friesel R E. Molecular mechanisms of fibroblast growth factor-1 traffick, signaling and release. Thromb Haemost. 1995;74:411–414. [PubMed] [Google Scholar]
- 28.McKeehan W, Wang F, Kan M. The heparan sulfate-fibroblast growth factor family: diversity of structure and function. Prog Nucleic Acid Res Mol Biol. 1998;59:135–176. doi: 10.1016/s0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- 29.Miki T, Bottaro D, Fleming T, Smith C, Burgess W, Chan A, Aaronson S. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mourelatos Z, Gonatas J O, Cinato E, Gonatas N K. Cloning and sequence analysis of the human MG160, a fibroblast growth factor and E-selectin binding membrane sialoglycoprotein of the Golgi apparatus. DNA Cell Biol. 1996;15:1121–1128. doi: 10.1089/dna.1996.15.1121. [DOI] [PubMed] [Google Scholar]
- 31.Smallwood P M, Munoz-Sanjuan I, Tong P, Macke J P, Hendry S H, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci USA. 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamp G, Fantl V, Poulsom R, Jamieson S, Smith R, Peters G, Dickson C. Nonuniform expression of a mouse mammary tumor virus-driven int-2/Fgf-3 transgene in pregnancy-responsive breast tumors. Cell Growth Differ. 1992;3:929–938. [PubMed] [Google Scholar]
- 33.Steegmaier M, Levinovitz A, Isenmann S, Borges E, Lenter M, Kocher H P, Kleuser B, Vestweber D. The E-selectin-ligand ESL-1 is a variant of a receptor for fibroblast growth factor. Nature. 1995;373:615–20. doi: 10.1038/373615a0. [DOI] [PubMed] [Google Scholar]
- 34.Szebenyi G, Fallon J. Fibroblast growth factors as multifunctional signaling factors. Int Rev Cytol. 1999;185:45–106. doi: 10.1016/s0074-7696(08)60149-7. [DOI] [PubMed] [Google Scholar]
- 35.Tsujii R, Miyoshi K, Tsuno A, Matsui Y, Toh-e A, Miyakawa T, Mizuta K. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus nuclear antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells. 2000;5:543–553. doi: 10.1046/j.1365-2443.2000.00346.x. [DOI] [PubMed] [Google Scholar]
- 36.Vainikka S, Partanen J, Bellosta P, Coulier F, Birnbaum D, Basilico C, Jaye M, Alitalo K. Fibroblast growth factor receptor-4 shows novel features in genomic structure, ligand binding and signal transduction. EMBO J. 1992;11:4273–4280. doi: 10.1002/j.1460-2075.1992.tb05526.x. . (Erratum, 12:810, 1993.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner S, Duan D, de Vries C, Peters K, Johnson D, Williams L. Differential splicing in the extracellular region of fibroblast growth factor receptor 1 generates receptor variants with different ligand-binding specificities. Mol Cell Biol. 1992;12:82–88. doi: 10.1128/mcb.12.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson D, Bhatt S, McMahon A. Expression pattern of the FGF-related proto-oncogene int-2 suggests multiple roles in fetal development. Development. 1989;105:131–136. doi: 10.1242/dev.105.1.131. [DOI] [PubMed] [Google Scholar]
- 39.Zuber M E, Zhou Z, Burrus L W, Olwin B B. Cysteine-rich FGF receptor regulates intracellular FGF-1 and FGF-2 levels. J Cell Physiol. 1997;170:217–227. doi: 10.1002/(SICI)1097-4652(199703)170:3<217::AID-JCP1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]