Abstract
Many Mendelian randomization (MR) studies have been published recently, with inferences on the causal relationships between risk factors and diseases that have potential implications for clinical research. In nephrology, MR methods have been applied to investigate potential causal relationships of traditional risk factors, lifestyle factors, and biomarkers from omics technologies with kidney function or CKD. This primer summarizes the basic concepts of MR studies, highlighting methods used in recent applications, and emphasizes key elements in conducting and reporting of MR studies that are important for interpreting the results.
Keywords: Epidemiology and outcomes
CKD is a heterogeneous disorder with numerous risk factors and complications.1 Some risk factors may be causal, whereas others are risk markers that are not causal for the disease itself, such as elevated levels of C-reactive protein (CRP).2 Differentiating causal factors from risk markers can help to prioritize or refine potential treatment targets.
Mendelian randomization (MR) investigation is an analytic tool that uses genetic variants as proxies of an exposure to assess whether the effect of this exposure on an outcome may be causal using observational data.3,4 This kind of analysis is of particular interest in assessing risk associations for which the causal relationships are uncertain. For example, MR studies examining the relationship of HDL cholesterol with coronary heart disease (CHD)5 suggest low HDL levels are unlikely to have causal effects on CHD. Consequently, therapeutic elevation of HDL levels may not be a promising approach to reduce CHD risk. In contrast, genetic variants in genes affecting LDL cholesterol levels are associated with CHD, supporting a causal role of LDL, and the beneficial effect of lowering LDL levels on CHD risk.6
A recent increase in annual citations of seminal MR publications7 indicates growing interest in the tool. In nephrology, MR methods have been applied to investigate the potential causal relationships of traditional risk factors (blood pressure and adiposity),8–10 lifestyle and mental health factors (sleep duration, coffee consumption, and depressive symptoms),11–13 and biomarkers from omics technologies14,15 with kidney function or CKD (Table 1). To extend previous work that introduced the concepts of MR studies,16 this primer will summarize those concepts, highlight methods used in recent applications of MR investigations in nephrology,8,9,12,13,17 and emphasize key elements in conducting and reporting of MR studies that are important for their interpretation.
Table 1.
Opportunities for MR investigations in nephrology. The opportunities are presented in three broad categories, with citations from recent publications indicating the increasing interest in the application of MR methods
| Opportunity |
|---|
| Clinical, lifestyle, and mental health factors. Recent MR publications have investigated the bidirectional causal effect between kidney function (eGFR and albuminuria) with blood pressure,8,9 and compared the causal and observed effects of adiposity on CKD.10 In addition, the causal effects of lifestyle and mental health factors on kidney function and CKD have been evaluated, such as the potential effects of sleep duration, coffee consumption, and depressive symptoms.11–13 |
| Circulating and urinary biomarkers from high-throughput technologies. Omics technologies allow for the quantification of a large number of analytes from blood or urine samples, providing abundant data for biomarker discovery.53 MR analyses have been applied in evaluating the potentially causal role of circulating proteins and urinary metabolites on kidney function and CKD.14,15 |
| Tissue-specific biomarkers from high-throughput technologies. As a basis for the development of new treatments for CKD, mechanistic insights into molecular functions have long relied on animal models or specific cell types. As more tissue- or cell type-specific biomarkers from humans become available, MR analysis represents a complementary approach to evaluate potentially causal pathways for CKD. In an omics setting, the selection and validation of genetic instruments for novel biomarkers may add additional complexities, depending on our knowledge of the biomarkers and their proximity to the disease. Methods and applications in this area are emerging.54–56 |
Basic Concepts of MR
An MR study is an epidemiologic investigation on the basis of observational data that uses genetic variants as instrumental variables or proxies of an exposure to evaluate the potential causal effect of this exposure on an outcome.3 A key limitation of observational studies regarding causal inference is confounding, which refers to an observed association between an exposure and an outcome induced by an association with measured or unmeasured third factors, the confounders (Table 2).3 Even when measured confounders are controlled for, unmeasured confounding may still be present. For example, the measurement errors of eGFR are higher in the normal range, compared with those in the lower ranges.18 Other key concepts of epidemiologic studies, such as a relevant study population, and validity and reliability of the measurements of exposure and outcome, are also relevant to MR studies.19
Table 2.
Glossary
| Term | |
|---|---|
| Confounder | A factor that is associated with the exposure and not a consequence of the exposure. When this factor is also associated with the outcome, it can induce a spurious association between the exposure and the outcome (Figure 1).57 The counterfactual framework58 can also be used to define a confounder. |
| Consistent estimate | An estimate that will approach the true value as the sample size increases. |
| F statistic | A ratio of two chi-square statistics. In the context of MR, an F statistic can evaluate the strength of association between a genetic variant and the exposure.31 |
| Instrumental variable | A variable that is associated with the exposure, not related to confounders, and affects the outcome only through the exposure (Figure 1).21 |
| One- and two-sample MR study | A one-sample MR study uses individual-level data to estimate the causal effect of the exposure on the outcome.3 A two-sample MR study derives the causal estimate from the associations of the genetic instruments with the exposure and the outcome in two different study populations, such as two sets of GWAS summary statistics.25 |
| Pleiotropy | Horizontal pleiotropy refers to a genetic variant influencing an outcome through multiple parallel pathways. Balanced pleiotropy is present when the horizontal pleiotropic effects of multiple genetic variants average to zero. Vertical pleiotropy refers to a genetic variant influencing an outcome through the exposure as a mediator. Only horizontal pleiotropy is a threat to the causal inference of MR studies. |
| Population stratification | Systematic differences in genotype frequencies between subpopulations of a sample. Differences that correlate with the distributions of the exposure and the outcome can introduce a spurious association between the exposure and the outcome in the absence of causality.34 In other words, population stratification can act as a confounder.59,60 Methods to control for population stratification in the analysis include restriction to specific ancestry groups and controlling for population substructure.61 |
| Reverse causation | An association between an exposure and an outcome that is in fact induced by the effect of the outcome on the exposure. In other words, the primary phenotype is mis-specified. |
Randomized controlled trials have been considered the gold standard for evaluating causal relationships in medicine, because randomization can result in the independence of the treatment, which is considered an exposure, from both measured and unmeasured confounders.20 However, clinical trials may not be feasible for many exposures, including lifestyle factors such as smoking or some physiologic measures, such as kidney function. MR studies take advantage of the random distribution of genetic variants during meiosis, analogous to the random assignment of treatments in clinical trials. This random assignment enables an evaluation of causal relationships using data from observational studies. If a genetic variant satisfies the assumptions of an instrumental variable, it can serve as an instrument or a proxy for the lifetime levels of the exposure due to genetics, and can be used for estimating the causal effect of the exposure on the outcome.3,4
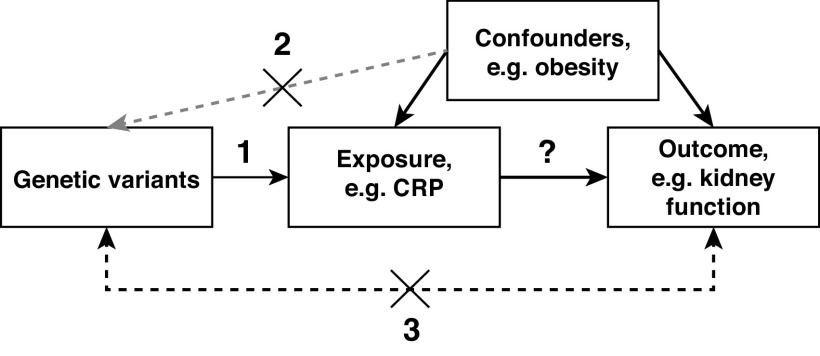
The assumptions for a genetic variant to be a valid instrument are that (1) the genetic variant is associated with the exposure, also known as the relevancy assumption; (2) it is independent of measured or unmeasured confounders, also known as the independence assumption; and (3) it can only influence the outcome through the exposure, also known as the exclusion restriction assumption (Figure 1).3,21 The third assumption excludes horizontal pleiotropy (Table 2), that is, a genetic variant that influences the outcome through an alternate pathway or pathways. This assumption also excludes reverse causation, namely, a genetic variant that affects the outcome, which in turn leads to changes in the exposure (Table 2),22 a situation that can also be considered a mis-specification of the primary phenotype. The inference of a causal relationship between an exposure and an outcome is valid only when these three underlying assumptions are met. Therefore, the assessment of these assumptions is among the key elements in conducting and reporting of MR studies (Table 3).23,24 Rigorous assessments of the three assumptions strengthen the credibility of the causal inference from an MR study.
Figure 1.
Conceptual diagram of the assumptions that must be fulfilled for genetic variants to represent valid instruments of the exposure. The numbers represent each of the assumptions: (1) the genetic variant is associated with the exposure, (2) the genetic variant is independent of measured or unmeasured confounders, and (3) the genetic variant can only influence the outcome through the exposure. For example, obesity could lead to higher levels of inflammatory biomarkers, such as CRP, and lower kidney function, but CRP itself is not the cause for lower kidney function. Therefore, obesity is a confounder. For assumption 2, the gray arrow indicates that confounders do not change germline genetic variants. Some MR publications show an arrow from the genetic variant to the confounder (e.g., Sekula et al.16; Bowden et al.39; Slob & Burgess38). This direction indicates that the use of a genetic variant that is associated with a confounder as an instrument also violates assumption 2, namely the independence of a genetic instrument from confounders.
Table 3.
Key elements to be included in a manuscript reporting an MR study
| Elements |
|---|
| 1. Rationale for the MR analysis |
| 2. Description and justification for the data used |
| 3. Description and justification of the genetic variants used |
| 4. Discussion of whether the genetic variants are likely to satisfy the three instrumental variable assumptions |
| 5. Graphical presentation of the data, such as scatter plots of the genetic associations when several instruments are evaluated |
| 6. Results of additional testing to assess the robustness of the main findings |
Adapted from reference 24.
Emerging Resources for Conducting One- and Two-sample MR studies
This section summarizes emerging resources for conducting MR studies. The next section discusses methods for assessing the three assumptions for valid genetic instruments.
A one-sample MR study directly uses individual-level data to estimate the causal effect of an exposure on an outcome. Data from emerging and existing large-scale biobank studies (Table 4), such as the UK Biobank, are powerful resources to perform one-sample MR studies. In two-sample MR studies, the associations of the genetic instruments with the exposure and with the outcome are estimated in two different study populations, respectively, such as from two sets of summary statistics of a genome-wide association study (GWAS). The many publicly available summary statistics from large-scale GWAS (Table 4) have improved the statistical power of two-sample MR studies and enabled investigation of many combinations of exposure-outcome pairs. To reduce biases, members of the two study populations should share the same ancestry and have similar characteristics.25 Overlap of the two study populations may lead to biased estimates.26
Table 4.
Examples of large-scale datasets that enable the conduct of MR studies
| Examples |
|---|
| Biobanks and studies for one-sample MR studies |
| The HUNT study enrolled 125,000 Norwegians in Nord-Trøndelag County, Norway, in three separate periods in the 1980s, 1990s, and 2000s.62 |
| The BioBank Japan enrolled 291,274 patients with any of 47 common diseases from 2003 to 2008. Participants were followed annually until 2013. Follow-up surveys were conducted for participants with any of 32 diseases.63 |
| The Million Veteran Program is a US study of veterans that examines the effects of genes, lifestyle, and military exposures on health and diseases. Since its launch in 2011, over 825,000 veterans have joined the program.64 |
| The Norwegian Mother and Child Cohort Study included 114,000 children, 95,000 mothers, and 75,000 fathers across Norway. Participants have been followed at regular intervals since 1999.65,66 |
| The UK Biobank is a prospective cohort study that enrolled approximately 500,000 individuals aged 40–69 across the United Kingdom with extensive genetic and phenotypic data.67 |
| The UK Millennium Cohort Study is a nationally representative birth cohort study of children born around the turn of the millennium, and their parents.68 |
| Large-scale genome-wide association summary statistics for two-sample MR studies |
| CKDGen Consortium website (https://ckdgen.imbi.uni-freiburg.de/)69: GWAS summary statistics for eGFR, urine albumin-creatinine ratio, blood urea nitrogen, serum urate, CKD, and eGFR decline. |
| GWAS catalog (https://www.ebi.ac.uk/gwas/)70: summary statistics of hundreds of published GWAS, including kidney outcomes and biomarkers. |
Methods for estimating causal effects differ for the one- and the two-sample settings. We refer the reader to the recent guidelines for MR investigation for details.24 Software tools have been developed to facilitate the use of multiple genetic instruments for an exposure to increase statistical power, and for the harmonization of genetic associations between the exposure and the outcome in the two-sample setting.27,28 Tools for conducting power analysis assuming all genetic variants are valid instruments are also available.29,30
Methods for Assessing the Instrumental Variable Assumptions of Genetic Variants
The first assumption, the relevancy assumption, can be evaluated by performing a statistical test on the association between the genetic variant and the exposure (Table 5). When a genetic variant has a weak association with the exposure, its association in one study may not be observed in another. As a result, the causal estimate generated from weak genetic instruments may have more biases.24,31,32 To avoid potentially biased estimates from weak instruments, the use of genetic variants with strong associations with the exposure has been proposed. A suggested statistical criterion is an F statistic (Table 2) >10 in the association between the genetic instruments and the exposure.31 Statistical significance in GWAS (P value <5E–08, corresponding to an F statistic of 30) has also been used.8,9,33
Table 5.
Summary of approaches for assessing the three assumptions on the validity of genetic instruments in MR studies
| Assumptions |
|---|
| Assumption 1: the genetic variant is associated with the exposure, the relevancy assumption. |
| Approaches to address this assumption |
| Test the association between the genetic variant and the exposure in a relevant population. For example, if an MR study aims to draw causal inference among women, the genetic variant needs to be associated with the exposure among women to be considered a valid instrument.24 |
| Test instrument strength. Genetic variants with weak associations with the exposure could lead to weak instrument bias.31,32 To reduce this potential bias, a suggested statistical criterion is requiring an F statistic (Table 2) >10 in the association between the genetic instruments and the exposure.31 Statistical significance in GWAS (P value <5E–08, corresponding to an F statistic of 30) has also been used.8,9,33 |
| Assumption 2: the genetic variant is independent of measured or unmeasured confounders, the independency assumption. This assumption is also known as the exchangeability assumption when confounding is defined using a counterfactual framework.58 |
| Approach to address this assumption |
| Evaluate the association of genetic variants with potential confounders and consider excluding the associated variants as instruments.25 Genetic association statistics for many exposures and outcomes are publicly available for addressing this assumption, such as the GWAS Catalog.70 |
| Assumption 3: the genetic variant can only influence the outcome through the exposure, the exclusion restriction assumption. |
| Approaches to address this assumption when multiple genetic instruments are available |
| Conduct an Egger intercept test, which can provide an overall assessment of horizontal pleiotropy.71 |
| Use MR methods that are robust to pleiotropy, such as mode- or median-based methods, which do not require all genetic instruments to be valid.39,40 Consistent results among multiple methods support the causal inference from an MR study. |
| Apply Steiger filtering to exclude genetic variants that have stronger associations with the outcome than with the exposure. These genetic variants may represent reverse causation and are not valid instrumental variables of the exposure.45 |
| When multiple exposures are correlated due to widespread pleiotropy, multivariable MR analysis can be used to combine the genetic instruments from multiple exposures.42–44 |
There are no fail-safe methods to fully assess the second and third assumptions, but there are methods to evaluate them. Assessing the association between the genetic instruments and potential confounders can evaluate the second, independence assumption, on the absence of confounding. A potential source of confounding in genetics is population stratification (Table 2). This refers to differences in genotype frequencies in subpopulations that can underlie a spurious association between the exposure and the outcome.34 Appropriate methods (Table 2) can control confounding due to population stratification.35 For example, conducting a study among participants of similar ancestry can reduce confounding by population stratification.
The third assumption, the exclusion restriction assumption, requires a genetic variant only influence the outcome through the exposure. Genetic variants that affect the outcome through alternate pathways, or those that affect the exposure through the outcome, are not valid instruments for the exposure. When the genetic instruments are valid, several statistical methods can be used to provide consistent estimates of the causal effect of the exposure on the outcome.33,36 These are two-stage least squares regression, the use of a genetic score, and inverse-variance weighted methods to combine instruments.
An example of the importance of selecting valid genetic instruments is the relationship between the inflammation marker CRP and eGFR. A comprehensive study used two sets of genetic variants strongly associated with CRP levels as instruments. The first set included CRP-associated genetic variants located in the CRP gene, making them more likely to directly affect CRP levels. The second set included all CRP-associated genetic variants across the genome, some of which were also associated with other inflammatory biomarkers, suggesting confounding or pleiotropy. Both sets of instruments were analyzed using an MR method that assumed all genetic instruments were valid. The first set did not provide support for a causal effect of CRP levels on eGFR, whereas the second set did.37 However, inferring true biologic relationships between genetic variants and an exposure is often difficult, including for the majority of genetic variants with replicated associations identified in recent GWAS. As discussed below, for these genetic variants, statistical methods are available to evaluate the potential presence of horizontal pleiotropy and reverse causation, threats to the third assumption of MR analysis (Table 5).
An extensive and detailed list of the MR methods that address horizontal pleiotropy is available in the recent guidelines for MR investigations.24 Below we briefly discuss the assumptions of some of these methods to illustrate that multiple methods may be necessary for conducting a comprehensive MR study when using multiple genetic instruments with largely unknown biologic function.
Inverse variance weighted methods, the most commonly used methods in two-sample MR studies, provide consistent estimates of causal effect when all genetic instruments are valid or the horizontal pleiotropic effects of all variants average to zero,38 also known as balanced pleiotropy (Table 2). Because the balanced pleiotropy assumption cannot be assessed, methods have been developed that are more robust to horizontal pleiotropy. The median-based methods can provide consistent estimates when <50% of the information comes from invalid instruments.39 The mode-based methods assume the effect sizes of valid instruments form the mode of the effect sizes of all instruments, such that it can be used to estimate the causal effect of the exposure on the outcome.40 The Egger regression method assumes the overall effect of horizontal pleiotropy, if it exists, is not correlated with the effects of the genetic instruments on the exposure, also known as the instrument strength independent of direct effect assumption.41 The instrument strength independent of direct effect assumption may not hold if a genetic instrument influences the outcome through alternate pathways that are correlated with exposure levels. When the pleiotropic effects of genetic variants are uncertain, the inference is more strongly supported if the results from multiple methods with different assumptions support a causal effect of an exposure on an outcome.
Many recent MR studies reported results from multiple methods, including two studies on the causal effects of albuminuria and kidney function on blood pressure.8,9 Finally, when multiple exposures are highly correlated due to shared pathways leading to horizontal pleiotropy, for example with lipids (high-density cholesterol, low-density cholesterol, and triglycerides), multivariable MR methods can combine their respective genetic instruments to estimate the causal effects of the exposures on an outcome.42–44
Reverse causation refers to a situation in which a genetic variant is associated with the exposure through the outcome, which makes it an unsuitable genetic instrument. These variants can be removed on the basis of the results from the Steiger test, which evaluates whether the effect of a genetic variant on the outcome is larger than its effect on the exposure.45 The two studies on the causal effects of albuminuria and kidney function on blood pressure reported results of the Steiger test.8,9
The second assumption on the lack of confounding and the third assumption on the absence of horizontal pleiotropy can only be assessed and not fully verified. Therefore, it is recommended that an MR study use a variety of robust methods with different assumptions to assess the reliability of causal inference from any given method, and exercise caution in inferring causal relationships.24,38
Potential for Further Development of Methods and Application of MR Analysis
Although many MR studies have been published, several areas could benefit from further development of methods and applications of the tool. For example, current MR methods have not been designed to evaluate causal feedback loops, which underlie much of physiologic homeostasis, such as the autoregulation of renal blood flow.46 In addition, commonly used MR methods assume a linear causal effect of the exposure on the outcome, ignoring the possibility of threshold effects. Methods for evaluating nonlinear causal relationships have been proposed for the one-sample setting.47
Large-scale biobanks with longitudinal design (Table 3), and omics data, provide opportunities to extend MR studies in several areas. First, biobanks enable well-powered MR studies using the one-sample design with more flexibility in modeling exposure-outcome relationships, including nonlinear ones; interactions between genetic variants and exposures; the study of disease progression; and the assessment of causal relationships across the lifespan.48–50 Many MR studies have used data from exposures and outcomes measured at a single time point. The interpretation of the causal estimates from such studies simply assumes the effect of the genetic instruments on the exposure is stable over time. However, although genetic variants are present since conception, epigenetic or environmental influences might affect the relationship between a genetic instrument and the exposure across the lifespan. Analyzing large-scale longitudinal data could identify and evaluate a critical period when the exposure can influence the outcome.
Second, a well-powered one-sample approach enables direct comparison of the exposure-outcome relationship as assessed using MR versus conventional epidemiologic analyses in the same study sample.10,35 Recent investigations of the relationship between adiposity and CKD were subject to a one-sample approach.10 Selection of genetic instruments and the genetic effects on the exposure can be derived from relevant external data to achieve more generalizable causal estimates.10,36
Third, large-scale family studies could enable within-family MR analyses that may provide more robust causal estimates through better control of subtle population substructure than studies among unrelated individuals.51,52 Finally, omics data provide a rich resource for MR investigation to identify and assess possible causal relationships between novel biomarkers and diseases (Table 1).
MR analysis is a valuable tool for evaluating the potential causal effect of an exposure on an outcome using data from observational studies. An example is the contribution of MR studies in inferring CRP as a biomarker of risk rather than a causal factor.2 There has been tremendous growth in the availability of data, methods, and tools for MR studies, and the emergence of guidelines and reporting standards.23,24 These new developments will improve the quality of MR studies and advance our knowledge of the causal relationships between epidemiologic risk factors and clinical outcomes in nephrology.
Disclosures
A. Kottgen reports receiving honoraria from Sanofi Genzyme; reports being a scientific advisor or member of American Journal of Kidney Diseases, American Kidney Fund, JASN, Kidney International, and Nature Reviews Nephrology. The remaining author has nothing to disclose.
Funding
The work of A. Köttgen has been funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 431984000 – SFB 1453.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group: KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Available at: https://kdigo.org/wp-content/uploads/2017/02/KDIGO_2012_CKD_GL.pdf. Accessed January 5, 2021 [Google Scholar]
- 2.Markozannes G, Koutsioumpa C, Cividini S, Monori G, Tsilidis KK, Kretsavos N, et al.: Global assessment of C-reactive protein and health-related outcomes: An umbrella review of evidence from observational studies and Mendelian randomization studies. Eur J Epidemiol 36: 11–36, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G: Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat Med 27: 1133–1163, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Smith GD, Ebrahim S: ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32: 1–22, 2003 [DOI] [PubMed] [Google Scholar]
- 5.Vitali C, Khetarpal SA, Rader DJ: HDL cholesterol metabolism and the risk of CHD: New insights from human genetics. Curr Cardiol Rep 19: 132, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Ference BA, Robinson JG, Brook RD, Catapano AL, Chapman MJ, Neff DR, et al.: Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med 375: 2144–2153, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Davey Smith G, Holmes MV, Davies NM, Ebrahim S: Mendel’s laws, Mendelian randomization and causal inference in observational data: substantive and nomenclatural issues. Eur J Epidemiol 35: 99–111, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas ME, Aragam KG, Emdin CA, Bick AG, Hemani G, Davey Smith G, et al.; International Consortium for Blood Pressure : Genetic Association of Albuminuria with Cardiometabolic Disease and Blood Pressure. Am J Hum Genet 103: 461–473, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Z, Coresh J, Qi G, Grams M, Boerwinkle E, Snieder H, et al.: A bidirectional Mendelian randomization study supports causal effects of kidney function on blood pressure. Kidney Int 98: 708–716, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu P, Herrington WG, Haynes R, Emberson J, Landray MJ, Sudlow CLM, et al.: Conventional and genetic evidence on the association between adiposity and CKD. J Am Soc Nephrol 32: 127–137, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park, S, Lee, S, Kim, Y, Lee, Y, Kang, MW, Kim, K, et al.: Causal effects of positive affect, life satisfaction, depressive symptoms, and neuroticism on kidney function: A Mendelian randomization study. JASN 32: 1484–1496, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S, Lee S, Kim Y, Lee Y, Kang MW, Kim K, et al.: Short or long sleep duration and CKD: A Mendelian randomization study. J Am Soc Nephrol 31: 2937–2947, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy OJ, Pirastu N, Poole R, Fallowfield JA, Hayes PC, Grzeszkowiak EJ, et al.: Coffee consumption and kidney function: A Mendelian randomization study. Am J Kidney Dis 75: 753–761, 2020 [DOI] [PubMed] [Google Scholar]
- 14.Cheng Y, Li Y, Benkowitz P, Lamina C, Köttgen A, Sekula P: The relationship between blood metabolites of the tryptophan pathway and kidney function: A bidirectional Mendelian randomization analysis. Sci Rep 10: 12675, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matias-Garcia PR, Wilson R, Guo Q, Zaghlool SB, Eales J, Xu X, et al.: Plasma proteomics of renal function: A trans-ethnic meta-analysis and Mendelian randomization study [published online ahead of print June 16, 2021] J Am Soc Nephrol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekula P, Del Greco M F, Pattaro C, Köttgen A: Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol 27: 3253–3265, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanktree MB, Thériault S, Walsh M, Paré G: HDL cholesterol, LDL cholesterol, and triglycerides as risk factors for CKD: A Mendelian randomization study. Am J Kidney Dis 71: 166–172, 2018 [DOI] [PubMed] [Google Scholar]
- 18.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, et al.; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coggon D, Rose G, Barker DJP: Epidemiology for the uninitiated. Available at: https://www.bmj.com/about-bmj/resources-readers/publications/epidemiology-uninitiated/. Accessed January 5, 2021
- 20.Stel VS, Jager KJ, Zoccali C, Wanner C, Dekker FW: The randomized clinical trial: An unbeatable standard in clinical research? Kidney Int 72: 539–542, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Davies NM, Holmes MV, Davey Smith G: Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 362: k601, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davey Smith G, Hemani G: Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23: R89–R98, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davey Smith G, Davies NM, Dimou N, Egger M, Gallo V, Golub R, et al.: STROBE-MR: Guidelines for strengthening the reporting of Mendelian randomization studies. PeerJ Preprints, 7, 2019 [Google Scholar]
- 24.Burgess S, Davey Smith G, Davies N, Dudbridge F, Gill D, Glymour M, et al.: Guidelines for performing Mendelian randomization investigations [version 2; peer review: 2 approved]. Wellcome Open Research, 4, 2020 [DOI] [PMC free article] [PubMed]
- 25.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG; EPIC- InterAct Consortium: Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur J Epidemiol 30: 543–552, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S, Davies NM, Thompson SG: Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 40: 597–608, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yavorska O, Staley J, Burgess S: Mendelian Randomization, CRAN, 2020 [Google Scholar]
- 28.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al.: The MR-Base platform supports systematic causal inference across the human phenome. eLife 7: 7, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freeman G, Cowling BJ, Schooling CM: Power and sample size calculations for Mendelian randomization studies using one genetic instrument. Int J Epidemiol 42: 1157–1163, 2013 [DOI] [PubMed] [Google Scholar]
- 30.Burgess S: Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol 43: 922–929, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgess S, Thompson SG; CRP CHD Genetics Collaboration: Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 40: 755–764, 2011 [DOI] [PubMed] [Google Scholar]
- 32.Pierce BL, Ahsan H, Vanderweele TJ: Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 40: 740–752, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess S, Butterworth A, Thompson SG: Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37: 658–665, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardon LR, Palmer LJ: Population stratification and spurious allelic association. Lancet 361: 598–604, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Smith GD, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S: Clustered environments and randomized genes: A fundamental distinction between conventional and genetic epidemiology. PLoS Med 4: e352, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgess S, Thompson SG: Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol 42: 1134–1144, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prins BP, Abbasi A, Wong A, Vaez A, Nolte I, Franceschini N, et al.; PAGE Consortium; International Stroke Genetics Consortium; Systemic Sclerosis consortium; Treat OA consortium; DIAGRAM Consortium; CARDIoGRAMplusC4D Consortium; ALS consortium; International Parkinson’s Disease Genomics Consortium; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; CKDGen consortium; GERAD1 Consortium; International Consortium for Blood Pressure; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Inflammation Working Group of the CHARGE Consortium : Investigating the causal relationship of C-reactive protein with 32 complex somatic and psychiatric outcomes: A large-scale cross-consortium Mendelian randomization study. PLoS Med 13: e1001976, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slob EAW, Burgess S: A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol 44: 313–329, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowden J, Davey Smith G, Haycock PC, Burgess S: Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40: 304–314, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartwig FP, Davey Smith G, Bowden J: Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 46: 1985–1998, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowden J, Davey Smith G, Burgess S: Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int J Epidemiol 44: 512–525, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, et al.: Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med 17: e1003062, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burgess S, Thompson SG: Multivariable Mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol 181: 251–260, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanderson E, Davey Smith G, Windmeijer F, Bowden J: An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol 48: 713–727, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hemani G, Tilling K, Davey Smith G: Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 13: e1007081, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inscho EW: Lewis K. Dahl Memorial Lecture. Mysteries of renal autoregulation. Hypertension 53: 299–306, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burgess S, Davies NM, Thompson SG; on behalf of, E-IC: Instrumental variable analysis with a nonlinear exposure–outcome relationship. Epidemiology 25: 877–885, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paternoster L, Tilling K, Davey Smith G: Genetic epidemiology and Mendelian randomization for informing disease therapeutics: Conceptual and methodological challenges. PLoS Genet 13: e1006944, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holmes MV, Ala-Korpela M, Smith GD: Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol 14: 577–590, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y-Q, Burgess S, Staley JR, Wood AM, Bell S, Kaptoge SK, et al.: Body mass index and all cause mortality in HUNT and UK Biobank studies: linear and non-linear mendelian randomisation analyses. BMJ 364: l1042, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brumpton B, Sanderson E, Heilbron K, Hartwig FP, Harrison S, Vie GÅ, et al.; Within-family Consortium; 23andMe Research Team : Avoiding dynastic, assortative mating, and population stratification biases in Mendelian randomization through within-family analyses. Nat Commun 11: 3519, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies NM, Howe LJ, Brumpton B, Havdahl A, Evans DM, Davey Smith G: Within family Mendelian randomization studies. Hum Mol Genet 28[R2]: R170–R179, 2019 [DOI] [PubMed] [Google Scholar]
- 53.Rhee EP: How omics data can be used in nephrology. Am J Kidney Dis 72: 129–135, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richardson TG, Hemani G, Gaunt TR, Relton CL, Davey Smith G: A transcriptome-wide Mendelian randomization study to uncover tissue-dependent regulatory mechanisms across the human phenome. Nat Commun 11: 185, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou D, Jiang Y, Zhong X, Cox NJ, Liu C, Gamazon ER: A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat Genet 52: 1239–1246, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eales JM, Jiang X, Xu X, Saluja S, Akbarov A, Cano-Gamez E, et al.: Uncovering genetic mechanisms of hypertension through multi-omic analysis of the kidney. Nat Genet 53: 630–637, 2021 [DOI] [PubMed] [Google Scholar]
- 57.Celentano DD, Moyses S: Gordis Epidemiology E-Book, 6th Ed., Elsevier, 2018, p 427 [Google Scholar]
- 58.Hernán M, Robins J: Causal Inference: What If, Boca Raton, Chapman & Hall/CRC, 2020 [Google Scholar]
- 59.Hellwege, JN, Keaton, JM, Giri, A, Gao, X, Velez Edwards, DR, Edwards, TL: Population Stratification in Genetic Association Studies. Curr Protocol Human Genetics 95: 1.22.21–21.22.23, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith GD, Ebrahim S: Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 33: 30–42, 2004 [DOI] [PubMed] [Google Scholar]
- 61.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D: Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909, 2006 [DOI] [PubMed] [Google Scholar]
- 62.Krokstad S, Langhammer A, Hveem K, Holmen TL, Midthjell K, Stene TR, et al.: Cohort Profile: the HUNT Study, Norway. Int J Epidemiol 42: 968–977, 2013 [DOI] [PubMed] [Google Scholar]
- 63.Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y, et al.; BioBank Japan Cooperative Hospital Group : Overview of the BioBank Japan Project: Study design and profile. J Epidemiol 27: S2–S8, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, et al.: Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70: 214–223, 2016 [DOI] [PubMed] [Google Scholar]
- 65.Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, et al.: Cohort Profile Update: The Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol 45: 382–388, 2016 [DOI] [PubMed] [Google Scholar]
- 66.Magnus P, Irgens LM, Haug K, Nystad W, Skjaerven R, Stoltenberg C; MoBa Study Group: Cohort profile: The Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol 35: 1146–1150, 2006 [DOI] [PubMed] [Google Scholar]
- 67.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al.: The UK Biobank resource with deep phenotyping and genomic data. Nature 562: 203–209, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fitzsimons E, Moulton V, Hughes DA, Neaves S, Ho K, Hemani G, et al.: Collection of DNA samples and genetic data at scale in the UK Millennium Cohort Study. CLS Working Paper 2020/7. London: UCL Centre for Longitudinal Studies. [DOI] [PubMed] [Google Scholar]
- 69.Köttgen A, Pattaro C: The CKDGen Consortium: The CKDGen Consortium: ten years of insights into the genetic basis of kidney function. Kidney Int 97: 236–242, 2020 [DOI] [PubMed] [Google Scholar]
- 70.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al.: The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47: D1005–D1012, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burgess S, Thompson SG: Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 32: 377–389, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bowden J, Del Greco M F, Minelli C, Davey Smith G, Sheehan N, Thompson J: A framework for the investigation of pleiotropy in two-sample summary data mendelian randomization. Stat Med 36: 1783–1802, 2017. 28114746 [DOI] [PMC free article] [PubMed] [Google Scholar]