Abstract
EXO1 interacts with MSH2 and MLH1 and has been proposed to be a redundant exonuclease that functions in mismatch repair (MMR). To better understand the role of EXO1 in mismatch repair, a genetic screen was performed to identify mutations that increase the mutation rates caused by weak mutator mutations such as exo1Δ and pms1-A130V mutations. In a screen starting with an exo1 mutation, exo1-dependent mutator mutations were obtained in MLH1, PMS1, MSH2, MSH3, POL30 (PCNA), POL32, and RNR1, whereas starting with the weak pms1 allele pms1-A130V, pms1-dependent mutator mutations were identified in MLH1, MSH2, MSH3, MSH6, and EXO1. These mutations only cause weak MMR defects as single mutants but cause strong MMR defects when combined with each other. Most of the mutations obtained caused amino acid substitutions in MLH1 or PMS1, and these clustered in either the ATP-binding region or the MLH1-PMS1 interaction regions of these proteins. The mutations showed two other types of interactions: specific pairs of mutations showed unlinked noncomplementation in diploid strains, and the defect caused by pairs of mutations could be suppressed by high-copy-number expression of a third gene, an effect that showed allele and overexpressed gene specificity. These results support a model in which EXO1 plays a structural role in MMR and stabilizes multiprotein complexes containing a number of MMR proteins. A similar role is proposed for PCNA based on the data presented.
Postreplicative DNA mismatch repair (MMR) enhances the fidelity of DNA replication by repairing errors made by the replicative DNA polymerases. Studies of the Escherichia coli MutHLS MMR system have been instrumental in providing insights into the general mechanism of MMR (for reviews, see references 40 and 50). A central player in E. coli MMR is MutS, which is the mismatch recognition factor. After MutS binds a mismatch, the MutL protein binds to MutS and activates the MutH endonuclease, which nicks hemimethylated DNA at unmethylated GATC sites. Subsequently, the action of one of a number of redundant single-stranded DNA-specific exonucleases and a DNA helicase, UvrD, degrades the mismatch-containing DNA from the nick (27, 50, 78). The resulting gap is filled in by the replicative machinery, including DNA polymerase III. Eukaryotic MMR is related to bacterial MMR in that it utilizes MutS- and MutL-related proteins, but it appears to be more complex (for reviews, see references 40, 41, and 50). Instead of a single MutS protein, eukaryotic MMR utilizes three MutS-related proteins, MSH2, MSH3, and MSH6, that form two different heterodimeric complexes (1, 15, 21, 48, 53). The MSH2 and MSH6 proteins form a complex that is important for the recognition of single-base mispairs and small insertion-deletion loops, whereas the MSH2 and MSH3 proteins form a complex that recognizes insertion-deletion loops (21, 48, 62). Similarly, eukaryotic MMR uses three MutL-related proteins, MLH1, PMS1, and MLH3, that also form heterodimeric complexes (20, 45, 51, 57, 58). Although the exact function of these MutL homologues is unclear, it is known that MLH1 and PMS1 form a complex that interacts with the MSH complexes and is the major MutL-related complex required for MMR (25, 26, 45, 57, 58). A second MutL-related complex is composed of MLH1 and MLH3, and this complex seems to only play a minor role in MMR (20, 51). Several proteins that function in DNA replication, including DNA polymerase δ, PCNA, RFC, and RPA, have also been suggested to be involved in MMR (10, 13, 19, 33, 39, 46, 47, 76, 79). The importance of MMR in humans is evident from the fact that defects in MMR genes, predominantly MSH2, MSH6, and MLH1, underlie inherited cancer predisposition (32, 56) and can also underlie various sporadic cancers (6, 7, 16, 37, 59, 68).
In addition to the eukaryotic MMR proteins discussed above, a 5′-to-3′ double-stranded DNA exonuclease called EXO1 may be involved in the excision step of MMR. EXO1 belongs to the RAD2/XPG family of endo- and exonucleases, many of which are known to function in DNA repair (67, 70). EXO1 is known to function in mitotic and meiotic recombination (18, 38, 67, 75). In addition, several observations support the idea that EXO1 may be important in MSH2-dependent MMR. EXO1 was first suggested to be involved in MMR because exo1 mutations caused a hyper-rec phenotype when recombination between intragenic markers was examined (67). EXO1 has been demonstrated to physically interact with MSH2 in two-hybrid assays and in coimmunoprecipitation experiments (70) and EXO1 also interacts with MLH1 (59a, 74a). exo1 null mutants have increased mutation rates, which is consistent with a role for EXO1 in MMR, and EXO1 has an epistatic interaction with MSH2 (70, 72). Because the mutator phenotype of a exo1 null mutant is weak compared to a msh2 null mutant, the epistatic interaction of the two genes has been debated because it is not possible to distinguish whether the phenotype of the exo1 msh2 double mutant is equal to that of the msh2 single mutant or if it is the sum of the phenotypes of the exo1 and msh2 single mutants. Because of this, further investigation of the role of EXO1 in MMR is needed.
The weak mutator phenotype of exo1 single mutants has suggested that other exonucleases that function in eukaryotic MMR must be present in vivo, just as is the case in E. coli, where at least three exonucleases, including Exo1, ExoVII, and RecJ, are known to act in MMR (27, 40, 50, 78). Analysis of the Saccharomyces cerevisiae genome sequence has revealed putative exonucleases, but none of the obvious candidates from these searches appear to play a major role in MMR. For example, RAD27 has been demonstrated to function in DNA replication, recombination, and many DNA repair processes, but it may only play a minor role in MMR if at all (34, 71). DIN7 has recently been shown to be important for mitochondrial DNA repair, and a role for YEN1 is not yet apparent because a null mutation in this gene either alone or in combination with other mutations such as exo1 does not result in an increased mutator phenotype (17, 34, 70). It has also been suggested that the exonuclease activities associated with DNA polymerases δ and ɛ may function at the excision step of MMR along with EXO1 and RAD27 (72). However, it has been difficult to test this because mutations in these genes can cause a number of different phenotypes and are often lethal when combined with each other. These observations further underscore the need for additional studies designed to identify exonucleases that might function in MMR.
To further investigate the role of EXO1 in MMR, we carried out a genetic screen in the yeast S. cerevisiae to identify proteins that are functionally interacting and redundant with yeast EXO1. Mutagenesis of a strain carrying a deletion of the EXO1 gene revealed 19 mutator mutants that had a strong mutator phenotype that was dependent on the absence of the EXO1 gene. Identification of the exo1-dependent mutator (edm) genes revealed functional interactions of EXO1 with a majority of known MMR gene products, as well as with two proteins that function in DNA replication. The functional interactions of EXO1 with MMR proteins were confirmed in a screen for mutator mutants whose phenotype required the presence of a weak PMS1 allele. These studies illustrate that the functional importance of EXO1 in MMR is possibly due to its critical interactions with other MMR proteins.
MATERIALS AND METHODS
Strains and media.
Yeast cells were grown in YEPD (1% yeast extract, 2% Bacto Peptone, and 2% dextrose, with or without 2% Bacto Agar) or SD (0.67% yeast nitrogen base and 2% dextrose, with or without 2% Bacto Agar) medium (11, 61). SD medium was supplemented with the appropriate dropout mix of amino acids (Bio 101, Inc., Vista, Calif.); these plates are referred to as selective plates. Plates containing all amino acids are called synthetic complete (SC) plates. Canavanine plates contained SD medium with all amino acids except arginine, with 60 μg of canavanine (Sigma, St. Louis, Mo.) added per ml prior to pouring the plates. Sporulation medium consisted of 0.1% yeast extract, 1% potassium acetate, and 0.05% dextrose, with or without 2% Bacto Agar. Bacterial cells harboring plasmids were grown using Luria-Bertani medium (1% Bacto Tryptone, 0.5% yeast extract, and 0.5% NaCl, with or without 2% Bacto Agar) containing 100 μg of ampicillin per ml.
All strains used in this study were derived from an S288c strain background and are listed in Table 1. The strains were created by standard procedures involving crosses, tetrad dissection, gene disruption, and transformations. The lys2::InsE-A10 frameshift reversion substrate was introduced into strain RKY2700 (LYS2) using plasmid p93-10A (pRDK706; obtained from D. A. Gordenin, National Institute of Environmental Health Sciences) (73). The resulting strain RKY3590 (lys2::InsE-A10) was then used to create strain RKY4168 (exo1::URA3 lys2::InsE-A10) in which the chromosomal copy of the wild-type EXO1 gene had been replaced by the URA3 gene through transformation with a PCR product generated using pM53 (from R. Shiestl, Harvard School of Public Health) as template DNA and the primers 5′-ATGCTCTCATAGAATTATATTTGATATTGCTTTTTGGACCACATTAAAATAgcggataacaatttcacacagga-3′ and 5′-TTA ATTCTTGTCTTGAGGCATTTCGACGAGATTTTCATTTGAAAAATAT ACgccagggttttcccagtcacga-3′ (uppercase indicates homology to the disrupted region; lowercase indicates homology to the vector). Strain RKY4168 (exo1::URA3 lys2::InsE-A10) was used for the ethyl methanesulfonate (EMS) mutagenesis experiment described below and the strains obtained from this mutant screen that are exo1-dependent are RKY4170 to RKY4188. Strain RKY3590 (MATa lys2::InsE-A10) was crossed with RKY2704 (MATα), and upon sporulation of the resulting diploid followed by tetrad dissection, the strain RKY3686 (MATα lys2::InsE-A10) was obtained. Strain RKY3686 (MATα lys2::InsE-A10) was crossed with strain RKY4168 (MATa exo1::URA3 lys2::InsE-A10), and upon sporulation of the diploid followed by tetrad dissection, the strain RKY4169 (MATα exo1::URA3 lys2::InsE-A10) was obtained. A PCR product generated using pPS729 (from P. Silver, Harvard Medical School) as template DNA and the primers 5′-ATGGATCAAAAGGCGTCATATTTTATCAATGAGAAGCTCTTCACTGAGGTGgcctcctctagtacactc-3′ and 5′-TTATTTTGCCTTTCTTTT GAAAAAGCTTTCCAATGTTCCTTGCTTTTTTAGcgcgcctcgttcagaatg-3′ was used for disrupting POL32 with HIS3 in strains RKY3686 (lys2::InsE-A10) and RKY4169 (exo1::URA3 lys2::InsE-A10) to create strains RKY4206 (pol32::HIS3 lys2::InsE-A10) and RKY4207 (exo1::URA3 pol32::HIS3 lys2::InsE-A10), respectively.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotypea |
|---|---|
| RKY2700 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 |
| RKY2704 | MATα ura3-52 leu2Δ1 his3Δ200 hom3-10 |
| RKY3590 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 |
| RKY3591 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 msh2::hisG URA3 hisG |
| RKY3686 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 |
| RKY4168 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exol::URA3 |
| RKY4169 | MATα ura3-52 leu2Δ1 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 |
| RKY4170 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exol::URA3 mlh1-G19D |
| RKY4171 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-A28T |
| RKY4172 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-A41T |
| RKY4173 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-P157L |
| RKY4174 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-T227I |
| RKY4175 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-R265K |
| RKY4176 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-R547K |
| RKY4177 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-A130V |
| RKY4178 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-G160D |
| RKY4179 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-G305S |
| RKY4180 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-T749I |
| RKY4181 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-D774N |
| RKY4182 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-D901N |
| RKY4183 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 msh2-M541I |
| RKY4184 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 msh2-S762F |
| RKY4185 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 msh3-G824R |
| RKY4186 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 pol30-E143S |
| RKY4187 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 pol32-Q46STP |
| RKY4188 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 rnr1-G271S |
| RKY4189 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 mlh1-R547K |
| RKY4190 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 pms1-A130V |
| RKY4191 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 pol30-E143S |
| RKY4192 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-A130V |
| RKY4193 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-G19D |
| RKY4194 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-A41T |
| RKY4195 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-R265K |
| RKY4196 | MATα ura3-52 leu2Δ1 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-R547K |
| RKY4197 | MATα ura3-52 leu2Δ1 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-A130V |
| RKY4198 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 msh3::hisG |
| RKY4199 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 mlh1-R547K msh3::hisG |
| RKY4200 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 pol30-E143S msh3::hisG |
| RKY4201 | MATα ura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 rnr1-G271S msh3::hisG |
| RKY3684 | MATaura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 msh6::hisG |
| RKY4202 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 mlh1-R547K msh6::hisG |
| RKY4203 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 pol30-E143S msh6::hisG |
| RKY4204 | MATα ura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 rnr1-G271S msh6::hisG |
| RKY4205 | MATα ura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 rnr1-G271S |
| RKY4206 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 pol32::HIS3 |
| RKY4207 | MATα ura3-52 leu2Δ1 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 pol32::HIS3 |
| RKY4208 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2-Bgl ade2Δ1 ade8-104 msh3 pol32::HIS3 |
| RKY4209 | MATα ura3-52 leu2Δ1 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 pol30-E143S |
| RKY4210 | MATα ura3-52 leu2Δ1 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 rnr1-G271S |
| RKY4211 | MATα ura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 exo1::URA3 mlh1-T227I |
| RKY4212 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 pms1-T749I |
| RKY4213 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 pms1-A130V msh6::hisG |
| RKY4214 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 pms1-A130V msh3::hisG |
| RKY4252 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 hom3-10 lys2::InsE-A10 exo1::URA3 msh2-S762F |
The ade8-104 mutation is a deletion of nucleotides 104 and 105 of the ADE8 gene.
Mutant screens.
The mutants isolated in this study were obtained by random mutagenesis with the DNA alkylating agent EMS (49). Strain RKY4168 (exo1::URA3 lys2::InsE-A10) was grown in YEPD at 30°C to late log phase and then treated with 3% EMS (Sigma) in water for 30 min at 23°C. Appropriate dilutions of the cell suspension were then plated onto YEPD and were incubated at 30°C for 4 days. The resulting colonies were replica plated onto lysine-deficient, threonine-deficient, canavanine-containing plates and were incubated at 30°C for 3 days before screening for mutants that showed increased papilation on at least one of the indicator plates and therefore exhibited a mutator phenotype. The mutator strains obtained were retested by streaking for single colonies and carrying out patch tests as described below. To determine if any of the mutants had an edm phenotype, all of the mutant yeast strains (RKY4170 to RKY4188) and the control strains RKY3590 (EXO1) and RKY4168 (exo1::URA3) were transformed with either the CEN6/ARSH4 plasmid pRDK834 (EXO1, TRP1) or the 2μm plasmid pRDK480 (EXO1, LEU2) and the control plasmids pRDK838 (pRS314, TRP1) and YEP213 (LEU2) (12, 63). Plasmid pRDK834 contains full-length EXO1 and was obtained from a library of pRS200-derived yeast genomic DNA containing plasmids obtained from Phil Hieter (University of British Columbia) (pRS200 is a slight modification of pRS314). Plasmid pRDK480 (EXO1, LEU2) was also derived from a genomic DNA library made in the 2μm vector YEP213 and has been previously described (70). Mutants whose mutator phenotype as measured by patch tests (see below) was suppressed by the introduction of an EXO1 plasmid but not by the appropriate control plasmid were considered to be edm mutants. The resulting edm mutants were then backcrossed to RKY4169 (exo1Δ) both to determine if the mutator phenotype was a single gene trait and to obtain derivatives having the opposite mating type. A virtually identical screen was subsequently performed using strain RKY4190 (pms1-A130V lys2::InsE-A10) to identify pms1-A130V-dependent mutator (pdm) mutants. pRDK436 (PMS1) and pRS425 (control) plasmids were used to assess the pms1-A130V-dependent phenotype of the mutants essentially as described above.
Mutator patch tests.
Qualitative patch tests were used to assess the mutator phenotype of the different mutants in lys2::InsE-A10. Specifically, three or more colonies of a given strain were first patched onto YEPD plates or selective plates in the case of plasmid containing transformants. After 1 to 3 days of incubation at 30°C, the patches were replica plated onto lysine-deficient plates (selection was maintained if the strains contained plasmids). The mutator phenotype of the different strains was assessed after 2 to 3 days of incubation at 30°C. All experiments were carried out at least twice.
Fluctuation analysis.
Mutation rates were determined by fluctuation analysis (44, 48). Briefly, each strain was streaked out on a YEPD plate to obtain single colonies, and independent colonies were used to grow five overnight cultures in YEPD (5 ml) at 30°C. Appropriate dilutions of cells from each culture were then plated onto SC plates and onto SC plates lacking lysine. The number of colonies grown on each plate was scored after 3 days of incubation at 30°C. For each strain, the average mutation rate was calculated from four independent fluctuation experiments as described by Lea and Coulson (44).
Complementation analysis.
To identify the EDM genes, patch tests were used to determine if wild-type copies of candidate MMR genes or if plasmids from a yeast genomic DNA library could complement the mutator phenotype of individual mutants. Low-copy-number CEN/ARS plasmids (12, 63) were used for this purpose, and they include pRDK363 (MSH2, LEU2), pRDK444 (MSH3, LEU2), pRDK439 (MSH6, LEU2), pRDK835 (MLH1, TRP1), pRDK433 (PMS1, LEU2), pRDK837 (POL30, TRP1), pRDK842 (RAD27, TRP1), or the pRS200-derived TRP1 CEN6/ARSH4 yeast genomic DNA library obtained from Phil Hieter (made by digesting pRS200 with BamHI and BglII and inserting a Sau3A partial digest of yeast genomic DNA).
Mutation detection by DNA sequencing.
To identify mutations in the EDM genes, genomic DNA was prepared from the edmx exo1Δ double mutant strains (RKY4170 to RKY4188) using the glass bead method (35) and was used as template DNA in PCR reactions to amplify the chromosomal copy of MLH1, PMS1, MSH2, MSH3, POL30, POL32, and RNR1. The PCR products were then treated with shrimp alkaline phosphatase (USB Corp., Cleveland, Ohio) and exonuclease 1 (USB Corp.) and sequenced using an PE ABI 3700 DNA sequencer. The respective wild-type genes were also amplified from the unmutagenized parent strain RKY4168 (exo1::URA3) and sequenced as controls.
Unlinked noncomplementation analysis.
To detect an unlinked noncomplementation phenotype in diploids containing mutations in two different EDM genes, the MATα strains RKY4169 (exo1Δ), RKY4193 (mlh1-G19D exo1Δ), RKY4194 (mlh1-A41T exo1Δ), RKY4195 (mlh1-R265F exo1Δ), RKY4196 (mlh1-R547 exo1Δ), RKY4197 (pms1-A130V exo1Δ), and RKY4252 (msh2-S762F exo1Δ) were crossed with the MATa strains RKY3590 (wild type) and RKY4168 (exo1Δ) and the edmx exo1Δ strains RKY4170 to RKY4181 and RKY4183 to RKY4188 (see Table 1). The generation of diploid strains in each case was confirmed by using mating-type tests with the mating-type tester strains RKY1109 (MATa thr4) and RKY1110 (MATa thr4). Patch tests of the diploids were used to assess the presence of a mutator phenotype using the lys2::InsE-A10 assay. To determine if the unlinked noncomplementation observed in many of the diploids was exo1 dependent, the diploid strains RKY4240 (exo1Δ/exo1Δ MLH1/mlh1-A41T), RKY4241 (exo1Δ/exo1Δ PMS1/pms1-A130V), RKY4242 (exo1Δ/exo1Δ MLH/mlh1-R265K), RKY4243 (exo1Δ/exo1Δ MLH1/mlh1-G19D), RKY4244 (exo1Δ/exo1Δ MLH1/mlh1-A41T PMS1/pms1-A130V), RKY4245 (exo1Δ/exo1Δ pms1-A130V/pms1-A130V), RKY4246 (exo1Δ/exo1Δ PMS1/pms1-A130V MLH1/mlh1-R265K), and RKY4247 (exo1Δ/exo1Δ PMS1/pms1-A130V MLH1/mlh1-G19D) were transformed with pRDK838 (pRS314) or plasmid pRDK834 containing wild-type EXO1, and patch tests were used to detect mutator phenotypes using the lys2::InsE-A10 assay.
High- and low-copy-number suppression studies.
For the high- and low-copy-number suppression studies, the pairs of 2μm plasmids pRDK436 (PMS1) and pRS425 (control) or pRDK833 (POL30) and pRS424 (control) or the CEN6/ARSH4 plasmids pRDK835 (MLH1; a low-copy-number plasmid was used because overexpression of MLH1 from a 2μm plasmid causes a dominant mutator phenotype in wild-type cells [60]) and pRS314 (control) were transformed into RKY3590 (wild type), RKY4168 (exo1Δ), and the edmx exo1Δ strains RKY4170 to RKY4188 (12, 63). The transformants were then analyzed for a mutator phenotype in the lys2::InsE-A10 assay using patch tests.
RESULTS
Screening for exo1-dependent mutator mutations.
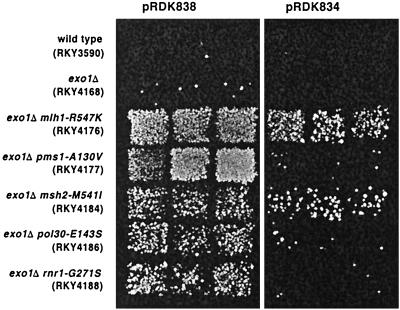
Biochemical and genetic studies have suggested that MMR involves the action of many proteins, possibly in the context of a multiprotein complex (40, 41, 50). The majority of studies have centered on understanding the MSH2-dependent mismatch recognition step, whereas the events occurring downstream of this step have yet to be elucidated in detail. In particular, EXO1 has been shown to physically interact with MSH2 (70) and, more recently, with MLH1 (59a, 74a), but it is not clear if EXO1 functionally interacts with MSH2 or another MMR protein(s). In addition, because the mutator phenotype of exo1Δ cells is weak compared to msh2Δ mutants, it is possible that other redundant or compensatory exonucleases function in MMR (34, 70, 72). To identify proteins that are functionally interacting and/or redundant with yeast EXO1, we screened for mutations (called edm mutations) that cause a strong mutator phenotype only in the absence of EXO1. The lys2::InsE-A10 frameshift assay, which primarily measures −1 frameshifts in a tract of 10 A's, was used in this screen because it is highly sensitive for detecting MMR defects with the additional advantage of having a low background mutation rate in wild-type cells (73). A total of 8,000 EMS-mutagenized survivors were screened, and 19 mutants were obtained that had an edm phenotype in the lys2::InsE-A10 assay. Examples of the phenotype of the edm mutants derived from the screen are shown in Fig. 1. In contrast to the weak mutator phenotype observed with the exo1Δ single mutant, the edm mutants show a significantly increased mutator phenotype when transformed with the control plasmid pRDK838 (pRS314). However, upon transformation of the edm strains with the wild-type EXO1-containing plasmid pRDK834, a dramatic suppression of the strong mutator phenotype was observed. In addition, 37 strong mutator mutations were identified that, as well as the null mutations in MSH2, MSH6, MLH1, and PMS1, were not enhanced by an exo1Δ mutation; none of these mutations or null mutations in MSH2, MSH6, MLH1, and PMS1 were suppressed by overexpression of EXO1 (70, 74a; data not shown).
FIG. 1.
Characterization of the mutator phenotype of the exo1-dependent mutator mutants using patch tests. The indicated strains were transformed with either the control vector or the vector containing the EXO1 gene. Then, three colonies each were patched onto a master plate and replica plated onto an SD-Lys plate to evaluate the lys2::InsE-A10 reversion properties of each strain as described in Materials and Methods.
Quantitation of the exo1-dependent mutator phenotype of selected mutants once again revealed a striking increase in the mutation rate of the edm mutants in the absence of EXO1 but a significantly reduced mutation rate in the presence of wild-type EXO1. The mutation rates of three different edm mutants were determined in the presence or absence of a wild-type copy of EXO1 using the lys2::InsE-A10 and hom3-10 frameshift reversion assays, as well as the canavanine resistance forward mutation assay. In comparison to the exo1 single mutant strain RKY4168, which has a mutation rate that is only 11-fold higher than that of the wild-type strain RKY3590 in the lys2::InsE-A10 assay, the absence of EXO1 in the D11 (RKY4192), J2 (RKY4176), and J10 (RKY4186) mutant strains caused a 350- to 7,500-fold increase in the mutation rate compared to the wild type (Table 2). In contrast, when wild-type EXO1 was introduced into the D11 (RKY4190), J2 (RKY4189), and J10 (RKY4191) mutant strains, a dramatic suppression of the mutator phenotype was observed (Table 2). Similar results were observed using the hom3-10 assay (data not shown). Increased mutation rates in the canavanine resistance forward mutation rate assay were also observed when EXO1 was absent compared to when a copy of EXO1 was present in the mutant strains; however, the rate differences were not as large as in the frameshift reversion assays (data not shown). This possibly reflects the fact that the canavanine resistance assay detects a wide variety of mutations, while the lys2::InsE-A10 and hom3-10 assays primarily detect frameshift mutations, which are characteristic of defects in MMR (48, 62, 73). Overall, these results indicate that the mutations identified cause a mutator phenotype that is dependent on the absence of the EXO1 gene product.
TABLE 2.
Mutation rate analysis of representative edm mutants using the lys2::InsE-A10 frameshift reversion assay
| Straina | Relevant genotype | lys2::InsE-A10 reversion rate (fold increase)b |
|---|---|---|
| RKY3590 | Wild type | 5.92 × 10−9 (1) |
| RKY3591 | msh2Δ | 1.12 × 10−4 (18,900) |
| RKY4168 | exo1Δ | 6.58 × 10−8 (11) |
| RKY4190 | pms1-A130V | 1.05 × 10−7 (18) |
| RKY4192 (D11) | exo1Δ pms1-A130V | 3.84 × 10−5 (6,486) |
| RKY4189 | mlh1-R547K | 5.129 × 10−6 (866) |
| RKY4176 (J2) | exoΔ mlh1-R547K | 4.48 × 10−5 (7,567) |
| RKY4191 | pol30-E143S | 9.634 × 10−8 (16) |
| RKY4186 (J10) | exo1Δ pol30-E143S | 2.067 × 10−6 (349) |
| RKY4198 | msh3Δ | 4.47 × 10−8 (8) |
| RKY3684 | msh6Δ | 4.21 × 10−7 (71) |
The original mutant identification number is indicated in parentheses where applicable.
The rate of accumulating frameshift mutations in each of the indicated strains was determined by fluctuation analysis using the lys2::InsE-A10 frameshift reversion assay as described in Materials and Methods. The values in parentheses after the reversion rates indicate the fold increases in reversion rates relative to the wild-type rate.
Identification of EDM genes.
The synergistic effect caused by combination of edm and exo1 mutations could occur because the wild-type counterparts of the two proteins identified by these mutations normally have overlapping functions or because they interact with one another. It is also possible that EXO1 and the EDM gene products act sequentially in the repair of DNA damage. To determine the nature of the exo1-dependent mutations, the EDM genes were identified. For this purpose, it was first necessary to determine if the edm mutations were dominant or recessive and if a single gene was affected in each edm mutant strain. The exo1Δ edmx double mutant strains were crossed with an exo1Δ single mutant strain (RKY4169), and the resulting diploids were analyzed for a dominant or recessive mutator phenotype (data not shown). None of the diploids revealed a mutator phenotype, suggesting that all of the edm mutant alleles were recessive. Sporulation and tetrad analysis of the diploids further revealed that a single EDM gene was affected in each of the edm mutant strains (data not shown).
The recessive nature of the edm mutations allowed identification of the edm genes by complementation analysis using plasmids containing a wild-type copy of known MMR and DNA replication genes and using a yeast genomic DNA library. In this manner the mutant genes in all 19 of the edm strains were identified and, in 17 of the 19 cases, the EDM genes were found to be known MMR genes. In particular, 13 of the edm mutations mapped to MLH1 or PMS1, two mapped to MSH2, one affected MSH3, and one affected the PCNA-encoding gene POL30. It is striking that almost all of the known MMR genes, with the exception of MSH6 and MLH3, were identified. This is not surprising in the case of MSH6 because it is predominantly important in the repair of base-base mispairs (48), whereas the lys2::InsE-A10 assay used in the screen is specific for detecting frameshift mutations (73); an msh6Δ mutation caused a small increase in the lys2::InsE-A10 assay that was 0.3% of that caused by an msh2Δ mutation (Table 2). Probably, mutations in the MLH3 gene were not identified because MLH3 has been shown to play a very minor role in MMR (20, 51). EXO1 has previously been shown to physically interact with MSH2 and, more recently, with MLH1, and the fact that mutations in MSH2 and MLH1 were obtained that showed an enhanced exo1-dependent mutator phenotype is consistent with a physical and functional interaction between EXO1 and these two proteins (59a, 70, 74a). A physical or functional interaction of yeast EXO1 with PMS1, MSH3, and POL30 has not been shown prior to this study.
In addition to the 17 edm MMR mutations, two edm mutations were obtained that affected other DNA synthesis genes (see below), namely, RNR1, which encodes the large subunit of ribonucleotide reductase, and POL32, which encodes one of the small subunits of the DNA polymerase δ holoenzyme (9, 22, 30). Because RNR1 is important in maintaining cellular deoxynucleoside triphosphate (dNTP) pools, it is conceivable that alterations in the activity of RNR1 in combination with a small defect in MMR, such as in an exo1Δ mutant, may result in an enhanced mutator phenotype. Thus, our remaining studies did not generally include the rnr1 exo1Δ double mutant. In contrast, that pol32 was identified in our screen as a mutator mutation in an exo1Δ background suggests that it may be directly important in MMR in vivo since it is known to be a component of the DNA polymerase δ holoenzyme; it also interacts with PCNA, DNA polymerase α, and WRN, and it has been shown to play a role in mutagenic bypass repair (9, 22, 28–30, 36).
Identification of mutations in the edm mutant genes.
To determine the nature of the defects in MLH1, PMS1, MSH2, MSH3, POL30, POL32, or RNR1 that cause the exo1-dependent mutator phenotype, the genomic copy of these genes from the respective edm mutant strains was sequenced. The sequencing results are summarized in Table 3. Single nucleotide mutations were observed in the respective edm genes in all 19 mutant strains. Each mutation caused a single amino acid change, except for the pol32 mutation, which was a nonsense mutation that changed glutamine 46 of the POL32 open reading frame to a stop codon. Because the nonsense mutation was present at the beginning of the 351-amino-acid open reading frame of POL32, this mutation was likely to cause a null phenotype. Consistent with this, complete deletion of the POL32 gene in an exo1Δ strain resulted in the same enhanced mutator phenotype seen with the exo1Δ pol32-Q46STP double mutant (data not shown). It is not clear whether the msh3-G824R mutation is a weak allele or a loss-of-function allele of MSH3 because even msh3 null mutations cause only weak mutator phenotypes in the assays used here. In contrast, the mlh1, pms1, msh2, pol30, and rnr1 mutant alleles appear to be hypomorphic because they confer weak mutator phenotypes in EXO1+ cells compared to the deletion alleles of these genes, which cause a strong mutator phenotype (MLH1, PMS1, or MSH2) or result in cell inviability (POL30 or RNR1). While most of the mutations observed were missense mutations, it is conceivable that, had more mutants been screened, it would have been possible to identify rare nonsense and frameshift mutations that were weak alleles like the other mutations identified here.
TABLE 3.
exo1-dependent mutator genes and mutations identifieda
| Mutant strainb | Mutator phenotype ratingc | EDM gene | Nucleotide change | Amino acid change |
|---|---|---|---|---|
| RKY4170 (H9) | 5 | MLH1 | G56A | G19D |
| RKY4171 (H3) | 4 | MLH1 | G82A | A28T |
| RKY4172 (C5) | 5 | MLH1 | G121A | A41T |
| RKY4173 (E15) | 4 | MLH1 | C470T | P157L |
| RKY4174 (C15) | 4 | MLH1 | C680T | T227I |
| RKY4175 (H2) | 5 | MLH1 | G794A | R265K |
| RKY4176 (J2) | 5 | MLH1 | G1640A | R547K |
| RKY4177 (D11) | 5 | PMS1 | C389T | A130V |
| RKY4178 (C14) | 4 | PMS1 | G479A | G160D |
| RKY4179 (F13) | 3 | PMS1 | G913A | G305S |
| RKY4180 (E13) | 3 | PMS1 | C2246T | T749I |
| RKY4181 (C11) | 4 | PMS1 | G2320A | D774N |
| RKY4182 (C2)d | 5 | PMS1 | G2701A | D901N |
| RKY4183 (B9) | 4 | MSH2 | C2285T | S762F |
| RKY4184 (J1) | 2 | MSH2 | G1623A | M541I |
| RKY4185 (D15) | 2 | MSH3 | G2470A | G824R |
| RKY4186 (J10) | 3 | POL30 | G427A | E143S |
| RKY4188 (J3) | 3 | RNR1 | G811T | G271S |
| RKY4187 (I1) | 3 | POL32 | C136T | Q46STP |
The EDM genes were identified by complementation analysis, and then the sequence of each relevant gene was determined. The base and amino acid changes given are numbered assuming that the A of the initiating ATG of each gene is numbered 1.
See Table 2, footnote a.
The numbers are a qualitative rating of the mutator phenotype of each exo1Δ edmx combination in patch tests like those shown in Fig. 1: 5, complete defect like that seen in an msh2Δ mutant; 4, 75% defect; 3, 50% defect; 2, 25% defect; 1, ∼1% defect like that seen in an exo1Δ mutant; −, wild type.
The phenotype of this mutant was partially suppressed only in the presence of a 2μm plasmid containing EXO1.
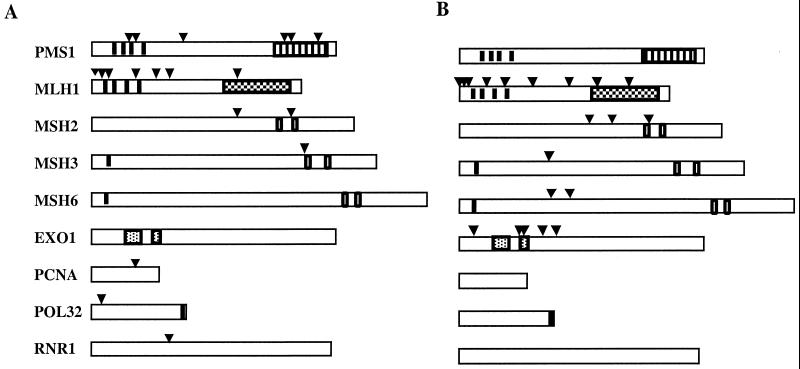
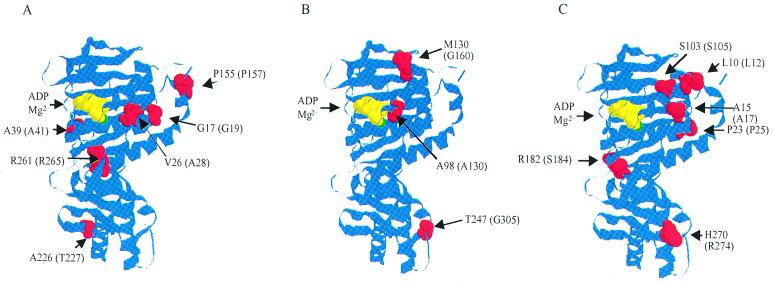
To identify the regions in MLH1, PMS1, MSH2, MSH3, and POL30 that are important for functional interaction with EXO1, the amino acid changes observed in these proteins were mapped onto the linear and in some cases the three-dimensional structures of these proteins. Mapping the amino acid changes observed in MLH1 onto the linear protein sequence revealed that six of the seven affected amino acids were in the N-terminal half of the protein (Fig. 2A). This region of the MutL family of proteins contains four ATP-binding motifs and other residues that are likely to be important for preserving an intact ATP-binding site (3–5), and mapping the amino acids described here onto the MutL structure shows that they all may be close to the ATP-binding site (Fig. 3A). In particular, the A28T and A41T amino acid changes occur within motif I of the ATP-binding site and the G19D change is adjacent to this motif (references 4 and 74 and references cited within). The P157L change affects a residue adjacent to motif IV (references 4 and 74 and references cited within) that is conserved among many MutL homologues (although not among human or rat MLH1), while the R265K change also affects a highly conserved sequence, common among the MutL proteins, which maps close to the ATP-binding site in the MutL proteins in a region thought to be important for ATP binding (3) (Fig. 3). Interestingly, missense mutations have been identified in human MLH1 in cases of hereditary nonpolyposis colorectal carcinoma (HNPCC) that alter the equivalent of the alanine 41 and arginine 265 residues (cited in reference 4). Besides the six-amino-acid changes observed in the N-terminal half of MLH1, a single-amino-acid change, R547K, was found in the C-terminal portion of the protein. This part of MLH1 contains the PMS1 interaction domain (residues 501 to 756 [24, 54]). Overall, the amino acid changes observed in or surrounding the ATPase motifs of MLH1 are likely to affect ATP binding or hydrolysis, and the single amino acid change observed in the C-terminal part of MLH1 could affect the interaction of MLH1 with PMS1.
FIG. 2.
Schematic representation of the position of the exo1-dependent and pms1-A130V-dependent mutator mutations. The relative positions of the exo1-dependent (A) and pms1-A130V-dependent (B) mutator mutations on the indicated genes are indicated by the arrows. The black vertical bars in PMS1 and MLH1 indicate the position of motifs important for ATP binding. The stippled boxes in PMS1 and MLH1 indicate the position of motifs important for PMS1-MLH1 interactions. The two open vertical boxes in MSH2, MSH3, and MSH6 indicate motifs important for ATP hydrolysis. The single vertical black bars in MSH3, MSH6, and POL32 indicate the motif that is important for interaction with PCNA. The two stippled boxes in EXO1 indicate motifs thought to be important for exonuclease activity.
FIG. 3.
Maps of the positions of the amino acid residues affected by the exo1-dependent and pms1-A130V-dependent mutator mutations onto the crystal structure of the ADP-bound form of the N-terminal fragment of MutL. The indicated structures were generated using the RasMol program from Roger Sayle (University of California at San Diego) with coordinates of the 40-kDa ATPase fragment of E. coli MutL complexed with ADP from the Protein Data Bank (ID1B62). The ribbon diagram of MutL is in blue. The ADP is indicated in yellow. The residues affected by the exo1-dependent mlh1 mutations (A), the exo1-dependent pms1 mutations (B), and the pms1-A130V-dependent mlh1 mutations are indicated in red. The relevant E. coli MutL amino acid residue numbers are indicated followed by the relevant yeast amino acid residue numbers in parentheses.
Mapping of the amino acid changes obtained in the second MutL homologue protein, PMS1, revealed a similar clustering of amino acids in either the N-terminal region that contains the ATP-binding site or the C-terminal region that contains the MLH1 interaction domain. Specifically, the A130V, G160D, and G305S amino acid changes observed in PMS1 lie within or around key residues in the ATP-binding region based on the three-dimensional structure of the N-terminal fragment of E. coli MutL (Fig. 2A and 3B). Of these, alanine 130 in yeast, which is alanine 98 in E. coli, is located in the ATP-binding motif III, which is highly conserved among the MutL family of proteins (74). Three amino acid changes that caused an exo1-dependent mutator phenotype were observed in the C-terminal region of PMS1, which is the MLH1 interaction domain (24, 54) (Fig. 2). Similar to the mutations identified in MLH1, there appear to be two classes of PMS1 mutations that cause an exo1-dependent mutator phenotype: the first class of PMS1 mutations is likely to perturb ATP binding or hydrolysis, while the second class is likely to affect the interaction with MLH1 and may indirectly affect the ATP binding or hydrolysis by the MLH1-PMS1 heterodimer.
Similar to that observed with PMS1 and MLH1, the amino acid changes in MSH2 and MSH3 that confer an exo1-dependent mutator phenotype are also located in or surrounding the region in these proteins that is important for ATP binding and hydrolysis (31, 43, 52, 66). The M541I amino acid change in MSH2 is significantly upstream of the phosphate-binding loop (p-loop) consensus sequence; however, the S762F amino acid change affects a highly conserved serine of the Walker B domain that is important for binding Mg2+ (Fig. 2A). The single-amino-acid change (G824R) observed in MSH3 was also found to reside in the highly conserved p-loop domain of MSH3 (Fig. 2A). It is possible that these msh2 and msh3 mutations affect ATP binding or hydrolysis or affect the structural transitions that occur in these proteins on ATP binding. Finally, the single-amino-acid change observed in PCNA mapped near the monomer-monomer boundary of the homotrimeric crystal structure of this protein (42). It is possible that this pol30 mutation could affect the stability of the PCNA trimer, as has been observed for other mutations that alter amino acids in this region of PCNA (2).
edm mutations show unlinked noncomplementation.
If the basis for the exo1-dependent mutator phenotype caused by the different edm alleles is that in the absence of EXO1 they destabilize a protein complex that is critical for MMR, then this could be tested using other genetic assays, such as unlinked noncomplementation. Unlinked noncomplementation refers to the observation of a mutant phenotype in a diploid cell that is heterozygous for recessive mutations in genes encoding different interacting proteins (65, 77). Noncomplementation of the mutant alleles occurs despite the wild-type copies of the proteins present in the diploid because only a small fraction of the heteromeric complexes are fully functional. To determine if unlinked noncomplementation could be observed between any of the exo1-dependent mutator mutations, seven different MATα strains, i.e., exo1Δ, mlh1-A41T exo1Δ, pms1-A130V exo1Δ, mlh1-R265K exo1Δ, mlh1-G19D exo1Δ, mlh1-R547K exo1Δ, and msh2-S762F exo1Δ strains, were crossed with the different MATa strains listed in Table 4 (pms1-D901N exo1Δ was not analyzed as this strain does not mate). The resulting diploid strains were subsequently analyzed for the presence of a mutator phenotype using the lys2::InsE-A10 patch assay (Table 4). As expected, a mutator phenotype was not observed when a wild-type strain was crossed with either exo1 or any of the edmx exo1 double mutant strains because of the recessive nature of the mutations. When the exo1 single mutant was crossed with each of the edmx exo1 double mutant strains, only a weak mutator phenotype was observed in the diploids because they were homozygous for the exo1 deletion mutation and contained one wild-type copy of each EDM gene. As expected, no allele was observed to complement itself. For example, in contrast to the weak mutator phenotype of the diploid that is homozygous for the exo1 mutation and heterozygous for the pms1-A130V allele, a strong mutator phenotype was observed in diploids that were homozygous for both exo1Δ and pms1-A130V mutations. Interestingly, in addition to these expected results, several cases of unlinked noncomplementation were observed in diploids that were heterozygous for mlh1 and pms1 mutations in an exo1-deficient background. For example, when the pms1-A130V exo1Δ double mutant was crossed with any of the mlh1 exo1 double mutants, a strong mutator phenotype resulted (Table 4).
TABLE 4.
Analysis of exo1-dependent mutator mutations for unlinked noncomplementationa
| Haploid strain mated | Mutator phenotype rating observed in the diploid strains constructed
|
||||||
|---|---|---|---|---|---|---|---|
| exo1Δ (RKY4169) | mlh1-A41T exo1Δ (RKY4194) | pms1-A130V exo1Δ (RKY4197) | mlh1-R265K exo1Δ (RKY4195) | mlh1-G19D exo1Δ (RKY4193) | mlh1-R547K exo1Δ (RKY4196) | msh2-S762F exo1Δ (RKY4252) | |
| Wild type (RKY3590) | − | − | − | − | − | − | − |
| exo1Δ (RKY4168) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| mlh1-G19D exo1Δ (RKY4170) | 1 | 5 | 3 | 5 | 5 | 5 | 1 |
| mlh1-A28T exo1Δ (RKY4171) | 1 | 5 | 3 | 4 | 5 | 5 | 1 |
| mlh1-A41T exo1Δ (RKY4172) | 1 | 5 | 3 | 5 | 5 | 5 | 1 |
| mlh1-P157L exo1Δ (RKY4173) | 1 | 5 | 3 | 4 | 5 | 5 | 1 |
| mlh1-T227I exo1Δ (RKY4174) | 1 | 5 | 2 | ND | ND | 4 | 1 |
| mlh1-R265K exo1Δ (RKY4175) | 1 | 5 | 3 | 5 | 5 | 5 | 1 |
| mlh1-R547K exo1Δ (RKY4176) | 1 | ND | 3 | 5 | 5 | 5 | 1 |
| pms1-A130V exo1Δ (RKY4177) | 1 | 3 | 5 | 3 | 3 | 3 | 1 |
| pms1-G160D exo1Δ (RKY4178) | 1 | 2 | 4 | 2 | 1 | 2 | 1 |
| pms1-G305S exo1Δ (RKY4179) | 1 | 2 | 4 | 1 | 1 | 1 | 1 |
| pms1-T749I exo1Δ (RKY4180) | 1 | 2 | 4 | 2 | 2 | 2 | 1 |
| pms1-D774N exo1Δ (RKY4181) | 1 | 3 | 4 | ND | 1 | 3 | 1 |
| msh2-M541I exo1Δ (RKY4183) | 1 | 1 | 1 | 1 | 1 | 1 | 4 |
| msh2-S762F exo1Δ (RKY4184) | 1 | 1 | 1 | 1 | ND | 1 | 2 |
| msh3-G824R exo1Δ (RKY4185) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| pol30-E143S exo1Δ (RKY4186) | 1 | 2 | 1 | 1 | 1 | 1 | 1 |
| rnrl-G271S exo1Δ (RKY4188) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| pol32-Q46STP exo1Δ (RKY4187) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
The haploid strain listed at the top of each column was mated to the haploid strain listed at the beginning of each row, and then the mutator phenotype of each resulting diploid strain was assessed in patch tests like those shown in Fig. 1. The numbers given are a qualitative rating of the mutator phenotype of each diploid: 5, complete defect like that seen in an msh2Δ mutant; 4, 75% defect; 3, 50% defect; 2, 25% defect; 1, ∼1% defect like that seen in an exo1Δ mutant; −, wild type; ND, not determined. After each genotype, the strain identification number is given in parentheses.
MLH1 and PMS1 are well known to interact with each other (24, 45, 54, 58). Because the mlh1 and pms1 alleles identified here cause strong mutator phenotypes in the absence of EXO1, it was of interest to determine if the unlinked noncomplementation observed between the mlh1 and pms1 alleles was exo1 dependent. In other words, if the mlh1 and pms1 mutations obtained cause destabilization of an MMR complex only in the absence of EXO1 in vivo, then the reintroduction of EXO1 into the diploids showing unlinked noncomplementation should overcome this defect. For this purpose, three different diploid strains that showed unlinked noncomplementation, RKY4244 (pms1-A130V × mlh1-A41T), RKY4246 (pms1-A130V × mlh1-R265K), and RKY4247 (pms1-A130V × mlh1-G19D)—as well as the control strains RKY4240 (exo1Δ × mlh1-A41T), RKY4241 (exo1Δ × pms1-A130V), RKY4242 (exo1Δ × mlh1-R265K), and RKY4243 (exo1Δ × mlh1-G19D)—were transformed with either an empty vector plasmid, pRDK838(pRS314) or a plasmid containing wild-type EXO1, pRDK834. The transformants were then analyzed for their mutator phenotype by patch tests using the lys2::InsE-A10 mutator assay. In contrast to the strains transformed with the empty vector pRDK838, which showed a mutator phenotype due to unlinked noncomplementation of the pms1 and mlh1 alleles in an exo1 background, suppression of this phenotype was observed when the same strains were transformed with pRDK834 which contains wild-type EXO1 (data not shown). These results support the idea that EXO1 plays an important role in the functional interaction between other proteins that function in MMR, especially MLH1 and PMS1.
Overexpression of specific MMR proteins alleviates the defect of edm mutations.
If the mutations identified here cause destabilization of a higher-order MMR complex in the absence of EXO1, then it is conceivable that increasing the level of other wild-type MMR proteins could restabilize the MMR complex. To investigate this possibility, we determined if increased expression of MLH1, PMS1, or PCNA could overcome the defect of one or more of the edm mutants. RKY3590 (wild type), RKY4168 (exo1Δ), and the edmx exo1Δ double mutant strains listed in Table 5 were transformed with the 2μm plasmid pRDK436 (PMS1) or pRDK833 (POL30) or the low-copy-number plasmid pRDK835 (MLH1; note that overexpression of MLH1 by a 2μm plasmid causes a dominant mutator phenotype in wild-type cells [this study and reference 60]). The transformants were then analyzed for their mutator phenotype in the lys2::InsE-A10 assay in comparison to the transformants containing the empty vector plasmid pRS425, pRS424, or pRS314.
TABLE 5.
Suppression of the edm phenotype by increased expression of MMR proteinsa
| Relevant genotype | Suppression rating with plasmid used to transform each strain:
|
||||
|---|---|---|---|---|---|
| EXO1 low copy number (pRDK834) | MLH1 low copy number (pRDK835) | PMS1 high copy number (pRDK436) | POL30 low copy number (pRDK837) | POL30 high copy number (pRDK833) | |
| Wild type (RKY3590) | − | − | − | − | − |
| exo1Δ (RKY4168) | +++ | − | − | − | +++ |
| mlh1-G19D exo1Δ (RKY4170) | ++ | ND | − | ND | ND |
| mlh1-A28T exo1Δ (RKY4171) | +++ | +++ | − | ND | +++ |
| mlh1-A41T exo1Δ (RKY4172) | ++ | +++ | − | ND | + |
| mlh1-P157L exo1Δ (RKY4173) | +++ | +++ | ++ | ND | +++ |
| mlh1-T227I exo1Δ (RKY4174) | +++ | +++ | ++ | − | +++ |
| mlh1-R265K exo1Δ (RKY4175) | ++ | +++ | − | ND | +++ |
| mlh1-R547K exo1Δ (RKY4176) | ++ | +++ | ++ | − | +++ |
| pms1-A130V exo1Δ (RKY4177) | +++ | − | +++ | − | +++ |
| pms1-G160D exo1Δ (RKY4178) | ++ | − | +++ | − | +++ |
| pms1-G305S exo1Δ (RKY4179) | +++ | + | +++ | + | +++ |
| pms1-T749I exo1Δ (RKY4180) | +++ | + | +++ | − | +++ |
| pms1-D774N exo1Δ (RKY4181) | +++ | − | +++ | − | +++ |
| pms1-D901N exo1Δ (RKY4182) | + | − | +++ | ND | +++ |
| msh2-M541I exo1Δ (RKY4183) | ++ | − | ++ | − | +++ |
| msh2-S762F exo1Δ (RKY4184) | ++ | − | ND | − | +++ |
| msh3-G824R exo1Δ (RKY4185) | ++ | − | ++ | ++ | +++ |
| pol30-E143S exo1Δ (RKY4186) | +++ | − | − | +++ | +++ |
| pol32-Q46STP exo1Δ (RKY4187) | +++ | ++ | − | − | +++ |
| rnr1-G271S exo1Δ (RKY4188) | +++ | ++ | − | ++ | +++ |
Each of the indicated edm strains was transformed with the indicated plasmid, and the mutator phenotype was assessed in patch tests using the lys2::InsE-A10 assay as described in Materials and Methods. The values reflect a qualitative rating of the suppression observed: +++, complete suppression to wild-type reversion levels; ++, two-thirds suppression; +, one-third suppression; −, no suppression; ND, not determined. After each genotype, the strain or plasmid identification number is indicated in parentheses.
Increased expression of MLH1 was found to suppress the phenotypes of a limited number of mutants (Table 5). First, the addition of a second copy of wild-type MLH1 in the pms1-G305S exo1 or pms1-T749I exo1 double mutant caused a mild suppression of the mutator phenotype. Suppression by low levels of wild-type MLH1 was allele specific since it was not observed in the other pms1 exo1 double mutants. Suppression of the phenotype of some pms1 exo1 double mutants by elevated levels of MLH1 was not surprising since MLH1 and PMS1 form a heterodimer. Second, and more surprisingly, the mutator phenotypes of the rnr1-G271S exo1 and the pol32-Q46STP exo1 mutants were significantly suppressed by adding a second copy of wild-type MLH1. This suggests a functional interaction between MLH1 and either RNR1 or POL32.
Allele-specific suppression of the mutator phenotype of some of the mlh1 exo1 double mutants by overexpression of PMS1 was also observed (Table 5). Not surprisingly, expression of PMS1 from a 2μm plasmid partially suppressed the mutator phenotype of the mlh1-R547K exo1 mutant since mlh1-R547K changes an amino acid in the PMS1 interaction region of MLH1. In addition, partial suppression of the mutator phenotype was observed for two other mlh1 mutations that cause amino acid changes in the N-terminal half of MLH1, where the ATP-binding pocket is located. This suggests that the N-terminal region of MLH1 is also important in the functional interaction with PMS1 and/or other proteins during MMR. Besides the suppression of the mutator phenotype of some of the mlh1 exo1 mutants by pRDK436 (PMS1), the msh2-M541I exo1 and msh3-G824R exo1 mutants were partially suppressed by the 2μm PMS1 plasmid. This observation is consistent with the idea that the defect in msh2-S762F exo1 and msh3-G824R exo1 mutants is also associated with instability of a multiprotein MMR complex.
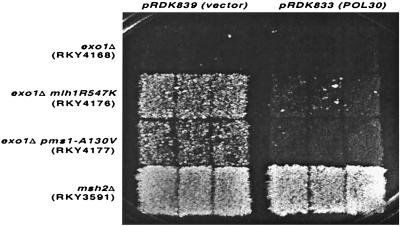
One of the interesting features of the proteins identified here is that most of them (i.e., MLH1-PMS1, MSH2-MSH3, MSH2-MSH6, and POL32) have been previously shown to physically interact with the DNA replication and repair factor, PCNA (9, 13, 19, 22, 23, 30, 33, 76). Thus, PCNA may play a crucial role in the functional interaction of MMR proteins in vivo. To test this idea, a control high-copy-number plasmid, pRDK839 (pRS424), or a high-copy-number plasmid containing the PCNA-encoding gene POL30, pRDK833, was introduced into RKY3590 (wild type), RKY4168 (exo1Δ), and all the edm strains. Compared to the transformants containing the control plasmid pRDK839, the POL30-containing plasmid pRDK833 suppressed the mutator phenotype of all the edm strains as well as the exo1Δ single mutant strain (Table 5 and Fig. 4). To confirm that the PCNA-dependent suppression was specific, the plasmids pRDK839 (control) and pRDK833 (POL30) were introduced into strains RKY3591 (msh2Δ), RKY2751 (mlh1Δ), and RKY2750 (pms1Δ), which contain complete deletion mutations in essential MMR genes. In contrast to the suppression of the mutator phenotypes of the exo1 and exo1 edm strains by the high-copy-number POL30 plasmid, pRDK833, there was no suppression of the mutator phenotypes of the msh2Δ, mlh1Δ, or pms1Δ strain by the POL30 plasmid (Fig. 4 and data not shown). These results suggest that PCNA alleviates the MMR defects observed in the exo1 and exo1 edm strains by stabilizing the functional interactions within the MMR complex in the absence of EXO1.
FIG. 4.
Suppression of representation exo1-dependent mutator mutations by increased expression of PCNA. The indicated strains were transformed with either the control vector or the vector containing the POL30 gene encoding PCNA. Then, three colonies each were patched onto a master plate and replica plated onto an SD-Lys plate to evaluate the lys2::InsE-A10 reversion properties of each strain as described in Materials and Methods.
Isolation of pms1-dependent mutator mutations.
If the genetic interactions discussed above reflect protein-protein interactions that are required for MMR, then it should be possible to obtain similar mutations in EXO1 and other known and unknown MMR repair genes in a similar screen starting with one of the edm single mutants. To test this possibility, the pms1-A130V single mutant strain RKY4190 was mutagenized with EMS as described for the exo1Δ strain RKY4168, and the survivors were screened for an increased mutator phenotype. Of 10,000 colonies that were screened for an enhanced mutator phenotype in the lys2::InsE-A10 assay, 43 mutants were obtained that had a pdm phenotype. Because the starting strain contained a missense mutation in the PMS1 gene (pms1-A130V), it was necessary to determine if the pdm mutants contained a second mutation in PMS1. Sequencing the PMS1 gene in all 43 pdm mutants revealed that 12 mutants contained a second mutation in the PMS1 gene. To identify the remaining 31 pdm mutations, the obvious candidate genes, such as MLH1, EXO1, MSH2, MSH3, MSH6, and POL30, were sequenced in the pdm strains. Eight of the mutants had a mutation in the MLH1 gene, five had a mutation in the EXO1 gene, two had a mutation in MSH2, two had a mutation in MSH6, and one had a mutation in MSH3 (Table 6). No mutations were identified in the POL30 gene. The remaining 13 pdm mutants had relatively weak mutator phenotypes and, because of this, they were not analyzed further.
TABLE 6.
pms1-A130V-dependent mutator genes and mutations identifieda
| Mutant no. | Mutator phenotype rating | PDM gene | Nucleotide change | Amino acid change |
|---|---|---|---|---|
| B5-1 | 4 | MLH1 | G34A | V12M |
| C35-1 | 4 | MLH1 | G49A | A17T |
| C38-4b | 4 | MLH1 | C74T | P25L |
| C40-1 | 3 | MLH1 | C314T | S105L |
| C22-2 | 3 | MLH1 | C551T | S184F |
| A13-1 | 4 | MLH1 | G281T | R274I |
| C16-3 | 2 | MLH1 | G1207A | D403N |
| B12-1 | 3 | MLH1 | C1531T | L511F |
| C37-6 | 4 | MLH1 | G1869A | M623I |
| C37-4 | 3 | MSH2 | G1410A | M470I |
| C5-3 | 3 | MSH2 | C1633T | R545K |
| B17-1 | 2 | MSH2 | G2062A | G688R |
| B18-1 | 3 | MSH3 | G994A | G332R |
| B10-1 | 3 | MSH6 | G1004A | G335D |
| C36-1 | 2 | MSH6 | G1235A | R412K |
| C3-1 | 4 | EXO1 | C169T | N57STP |
| A9-1 | 4 | EXO1 | G677A | C226Y |
| C43-1 | 3 | EXO1 | G707A | G236D |
| C14-2 | 4 | EXO1 | C1069T | N357STP |
| B16-1 | 4 | EXO1 | C1186T | N396STP |
The PDM genes were identified by complementation analysis, and then the sequence of each relevant gene was determined. The base and amino acid changes given are numbered assuming that the A of the initiating ATG of each gene is numbered “1.” The numbers in the mutator phenotype column are a qualitative rating of the mutator phenotype of each exo1Δ pdmx combination in patch tests like those shown in Fig. 1: 5, complete defect like that seen in an msh2Δ mutant; 4, 75% defect; 3, 50% defect; 2, 25% defect; 1, ∼1% defect like that seen in an exo1Δ mutant; −, wild type.
The phenotype of this mutant was only partially (∼2-fold) suppressed in the lys2::InsE-A10, hom3-10, and CAN assays.
Further analysis of the pdm mutations revealed that four of the eight mlh1 mutations caused amino acid changes located in the N terminus of MLH1 in the vicinity of the ATP-binding site (4, 5, 74), two of the MLH1 mutations caused amino acid substitutions located in the central region of the linear MLH1 amino acid sequence, and the remaining two mlh1 mutations caused amino acid substitutions in the PMS1 interaction domain of MLH1 (24, 54) (Fig. 2B and 3C). One of the MLH1 mutations, P251, has been described as causing a ∼50% MMR defect in lys2A14 and his7-2 frameshift reversion assays and a complete MMR defect in a CAN assay and was interpreted as causing a partial MMR defect (60). The observation here that this mutation was only partially suppressed by a PMS1 plasmid, which indicates that this allele causes a strong, but not complete, mutator phenotype on its own and is only partially dependent on pms1-A130V, is consistent with previously published results (60). The two MSH2 mutations caused amino acid substitutions in the C-terminal half of the protein, with the G688R amino acid change being located within the Walker A ATPase domain (14, 31, 43, 52, 66) (Fig. 2B). In contrast, the mutations identified in MSH3 and MSH6 caused amino acid substitutions in the N-terminal half of these proteins in regions of these proteins that are thought to interact with DNA (8, 14, 43, 52) (Fig. 2). Given that MSH3 and MSH6 are redundant with regard to the repair of frameshift mispairs, it was surprising to find pdm mutations in MSH3 or MSH6. It is possible that these mutations, in combination with pms1-A130V, are dominant (14). Three of the exo1 mutations identified were nonsense mutations in the N-terminal half of the EXO1 gene and are presumably null mutations. The two exo1 missense mutations identified caused amino acid changes within one of the conserved exonuclease domains of EXO1 and could be loss-of-function mutations, as well as significantly altering the structure or expression of EXO1 (64, 70). Overall, these results suggest that PMS1 has critical functional interactions with MLH1, as expected from previous studies (25, 26, 45, 57, 58). Additionally, PMS1 also appears to have important functional interactions with EXO1, MSH2, MSH6, and MSH3.
DISCUSSION
In the present study, we have described a genetic approach for the identification and characterization of mutations in MMR genes that cause little or no defect as single mutations but which cause strong defects when combined with other mutations that also cause little or no defect as single mutations. This approach is a generalized extension of previous studies that have examined interactions between individual MMR genes, including genes encoding DNA polymerases (20, 34, 38, 48, 73, 74). One experiment involved the isolation of mutations that increased the frameshift mutator phenotype of an exo1 mutation and resulted in the identification of mutations in the majority of known MMR genes, including MLH1, PMS1, MSH2, MSH3, and POL30, as well as POL32 and RNR1, which have not been previously implicated in MMR. A second experiment involved the isolation of mutations that increased the frameshift mutator phenotype of a pms1 mutation and resulted in the identification of mutations in many of the known MMR genes, including EXO1, MLH1, MSH2, MSH3, and MSH6. These mutations also showed two other types of genetic interactions: specific pairs of mutations were observed to show unlinked noncomplementation in doubly heterozygous diploid strains, and the defect caused by pairs of mutations could be suppressed by high-copy-number expression of a third gene, an effect that showed considerable mutant allele specificity and overexpressed gene specificity. Previous studies have shown that MSH2 exists as a stable complex with MSH3 or MSH6 (1, 15, 21, 48, 53), that MLH1 exists as a stable complex with PMS1 (24, 45, 54, 58), and that interactions have been detected between POL30 (PCNA) and MLH1, MSH3, and MSH6 (13, 19, 23, 33, 76) and between EXO1 and both MSH2 and MLH1 (59a, 70, 74a). Combined with these previous results, as expanded upon below, our results support the hypothesis that higher-order protein complexes are formed during MMR that may simultaneously contain MSH2-MSH6 (or alternately MSH2-MSH3), MLH1-PMS1, EXO1, POL30 (PCNA), and DNA polymerase δ (POL32 and other subunits). Furthermore, such higher-order complexes likely involve multiple protein-protein interactions and protein-protein interactions that may be dependent on interactions with a third protein. Particularly key interactions appear to be interactions with EXO1 and POL30 (PCNA). We are presently attempting to isolate some of the higher-order protein complexes predicted by the genetic results presented here and study the biochemical properties of these complexes.
An alternate hypothesis that might explain the edm mutations is if exo1Δ mutations cause a weak defect in the editing of mutations caused by an error-prone pathway. Given the polarity of EXO1, such a pathway is unlikely to edit DNA polymerase misincorporation errors but could be a pathway that edits the 5′ end of Okazaki fragments (71) or a function that acts during recombination (18, 38, 67, 75). If MMR normally repairs such errors, then combining an exo1Δ mutation with a weak MMR-defective mutation might result in the saturation of MMR with errors and hence an increase in mutation rates. In this circumstance, the observed mutator phenotypes, unlinked noncomplementation, and suppression of mutations by overexpression of other MMR proteins would still reflect the destabilization of MMR complexes by the edm and pdm mutations. However, the mutator phenotypes measured would reflect the combination of increased errors due to exo1 mutations and destabilization of MMR complexes due to the edm and pdm mutations. One point in contradiction to this hypothesis is the observation that exo1Δ mutations do not increase the mutation rate caused by null mutations in MSH2, MSH6, MLH1, or PMS1, whereas mutations in the editing exonuclease functions of DNA polymerases that increase misincorporation rates do increase the mutation rate caused by null mutations in MMR genes (69, 72 ,74a; also data not shown).
The genetic screen to identify proteins that are functionally interacting or redundant with EXO1 was performed to better understand the in vivo role of EXO1. The observation that a majority of exo1-interacting mutations was obtained in five known MMR genes (MSH2, MSH3, MLH1, PMS1, and POL30) is consistent with, but does not prove, that EXO1 plays a direct role in MMR. Because EXO1 has been proposed to be a redundant exonuclease that functions in MMR (34, 71, 73), it was surprising that no genes encoding potential exonucleases were identified. Some mutations like the pol2-04 editing exonuclease mutation should have been found (72), but if, like pol2-04, they were specific missense mutations in essential genes, they might not be identified unless much higher numbers of mutants were examined. It is possible that exo1 mutations may be lethal in combination with mutations in other genes encoding exonucleases that function in MMR and other aspects of DNA metabolism (71, 72). Alternatively, it is possible that there are many redundant exonucleases, as in E. coli, that function in MMR so that mutations in even two such genes will not yield a strong mutator phenotype (27, 40, 50, 78). It is also possible that EXO1 plays an important role in the formation of protein complexes that function in MMR and that the mutator phenotype caused by an exo1 mutation reflects this structural role of EXO1, possibly in addition to an enzymatic role in the degradation of DNA during MMR. A number of results support the view that EXO1 could stabilize a multiprotein complex. Many of the exo1-dependent mutator mutations affect MLH1 and MSH2, which are known to interact with EXO1. These could represent cases where EXO1 stabilizes MLH1 and MSH2 so that the mlh1 and msh2 mutations by themselves do not destabilize these proteins but do so in the absence of the EXO1 interaction. This interpretation is supported by the observation that overexpression of a third protein that interacts with the protein containing the amino acid substitution (e.g., POL30 or PMS1 with exo1 mlh1, POL30 with exo1 msh3, and PMS1 with exo1 mlh1) (Table 5) suppresses the MMR defect. This is because if the edm mutations were to weaken the interaction between the mutant protein and a normal partner protein and if this interaction was further weakened by the lack of the EXO1 interaction, overexpression of a different interacting protein would stabilize the weakened protein-protein interaction. Many of the exo1-dependent mutator mutations affect PMS1, MSH3, and POL30, which are not known to interact with EXO1. This is also true for the pms1-dependent exo1 mutations identified (Table 6). Thus, EXO1 interactions with MLH1 and MSH2 could be critical in maintaining a conformation important for the interactions between these latter proteins and PMS1, MSH3, and POL30 so that the mutation by itself does not destabilize these proteins but does so in the absence of the EXO1 interaction with MSH2 and MLH1. This interpretation is supported by the observation that the overexpression of a third protein that does not interact with either mutant protein but does interact with MSH2 or MLH1 (e.g., POL30 with exo1 pms1 and exo1 msh2) (Table 5) suppresses the MMR defect.
In light of the fact that EXO1 interacts directly with MLH1 but not with PMS1 (74a), a striking feature of the data presented here is the high proportion of exo1-dependent mutations that were in either MLH1 or PMS1 and the high proportion of pms1-dependent mutations that were in either MLH1 or EXO1. The mlh1 and pms1 mutations clustered in two homologous regions of each protein, the C-terminal MLH1-PMS1 interaction region (24, 54) and the region around the N-terminal ATP-binding site (4, 5), which have been suggested to participate in ATP-binding-regulated interactions between the N-terminal regions of MLH1 and PMS1 (74). This appears to reflect the importance of both the MLH1-PMS1 interaction and the interaction between EXO1 and MLH1-PMS1. Two aspects of the data further support the view that the N-terminal regions of MLH1 and PMS1 interact during MMR. First, overexpression of PMS1 partially suppresses the defect caused by two N-terminal mlh1 edm mutations (Table 5) and, second, unlinked noncomplementation was observed between N-terminal pms1 edm mutations and a C-terminal mlh1 edm mutation and between N-terminal mlh1 edm mutations and C-terminal pms1 edm mutations (Table 4). These effects seem unlikely if MLH1 and PMS1 interact only at their C termini. Finally, the fact that the expression of EXO1 suppresses a broad spectrum of mutations present in interaction regions of MLH1 and PMS1 supports the view that the EXO1-MLH1 interaction is important for the stability of the entire MLH1-PMS1 complex.
It was surprising to obtain mutations in the genes encoding POL30 (PCNA), POL32, and RNR1 because these proteins had not previously been shown to interact with either EXO1 or PMS1. RNR1 and POL32 had not previously been implicated in MMR, although DNA polymerase δ, of which POL32 is a subunit, is required for MMR (47). Combined with previous observations that PCNA (POL30) interacts with MLH1, MSH3, and MSH6 (19, 23, 30, 33, 76), the exo1-dependent pol30 mutation and the observation that overexpression of POL30 suppresses essentially all exo1- and pms1-dependent mutations support the view that interactions with PCNA are important in maintaining complexes of MMR proteins. It is possible that POL30 and EXO1 each interact with many MMR proteins and complexes, and hence a combination of mutations in both genes has a general effect on the stability of complexes that function in MMR. The observation of an exo1-dependent pol32 mutation may also reflect a critical role of PCNA because POL32 is a subunit of DNA polymerase δ that is not required for DNA polymerase activity but is involved in DNA polymerase δ PCNA interactions (9, 22, 30). Thus, this involvement of POL32 in MMR could reflect the coupling of polymerase δ to MMR proteins via PCNA interactions. It is not clear from our data if RNR1 plays a direct role in MMR. This is because the rnr1 mutation identified increases the mutation rate of msh2 and mlh1 null mutants (data not shown), a result that is more consistent with the rnr1 mutation altering dNTP pools in a way that leads to increased mutation rates.
Previous studies have shown that inherited mutations in two MMR genes, hMSH2 and hMLH1, underlie the majority of HNPCC cases, a common cancer susceptibility syndrome associated with high penetrance, early onset, and a diversity of tumor types (56). Inherited mutations in hMSH6 also appear to cause inherited cancer susceptibility, although associated with a lower penetrance and later onset than classical HNPCC (32). Finally, somatic inactivation of MMR genes, notably the silencing of hMLH1, is associated with a variety of sporadic cancers (6, 7, 16, 37, 59, 68). The results described here demonstrating that interactions between weak alleles of MMR genes can produce strong mutator phenotypes have interesting implications for the genetics of human cancer susceptibility. These results suggest that weak alleles could exist in humans without causing cancer susceptibility by themselves but could result in increased cancer susceptibility in individuals who had inherited two or more such alleles. Because the inheritance of two or more independent alleles would not show vertical, dominant transmission as seen in HNPCC, this view suggests that some sporadic cancer cases might have a polygenic basis. The observation of unlinked noncomplementation between multiple weak alleles is particularly interesting in this regard. Individuals carrying alleles showing unlinked noncomplementation would be expected to show some mutator phenotype in normal tissues, as has been observed in individuals carrying potentially dominant mutations in hMLH1 and hPMS2 (55). The types of experiments described here in which interacting weak alleles can be identified and tested provide a model on which to base tests for polygenic inheritance of MMR defects in humans.
ACKNOWLEDGMENTS
We thank the Kolodner lab for helpful discussions and comments on the manuscript, Mike Liskay and Rick Fishel for communicating unpublished results, and John Weger and Jill Green for DNA sequencing.
This work was supported by National Institutes of Health grant GM50006 to R.D.K.
REFERENCES
- 1.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayyagari R, Impellizzeri K J, Yoder B L, Gary S L, Burgers P M. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 4.Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 5.Bergerat A, de Massy B, Gadelle D, Varoutas P C, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 6.Bevilacqua R A, Simpson A J. Methylation of the hMLH1 promoter but no hMLH1 mutations in sporadic gastric carcinomas with high-level microsatellite instability. Int J Cancer. 2000;87:200–203. doi: 10.1002/1097-0215(20000715)87:2<200::aid-ijc7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 7.Borresen A-L, Lothe R A, Meling G I, Lystad S, Morrison P, Lipford J, Kane M F, Rognum T O, Kolodner R D. Somatic mutations in the hMSH2 gene in microsatellite unstable colorectal carcinomas. Hum Mol Genet. 1995;11:2065–2072. doi: 10.1093/hmg/4.11.2065. [DOI] [PubMed] [Google Scholar]
- 8.Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J Biol Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 9.Burgers P M, Gerik K J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Merrill B J, Lau P J, Holm C, Kolodner R D. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol Cell Biol. 1999;19:7801–7815. doi: 10.1128/mcb.19.11.7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Umezu K, Kolodner R D. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- 12.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 13.Clark A B, Valle F, Drotschmann K, Gary R K, Kunkel T A. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J Biol Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 14.Das Gupta R, Kolodner R D. Novel dominant mutations in Saccharomyces cerevisiae MSH6. Nat Genet. 2000;24:53–56. doi: 10.1038/71684. [DOI] [PubMed] [Google Scholar]
- 15.Drummond J T, Li G M, Longley M J, Modrich P. Isolation of an hMSH2–p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 16.Eshleman J R, Markowitz S D. Microsatellite instability in inherited and sporadic neoplasms. Curr Opin Oncol. 1995;7:83–89. [PubMed] [Google Scholar]
- 17.Fikus M U, Mieczkowski P A, Koprowski P, Rytka J, Sledziewska-Gojska E, Ciesla Z. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics. 2000;154:73–81. doi: 10.1093/genetics/154.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores-Rozas H, Clark D, Kolodner R D. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 20.Flores-Rozas H, Kolodner R D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genschel J, Littman S J, Drummond J T, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 22.Gerik K J, Li X, Pautz A, Burgers P M. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- 23.Gu L, Hong Y, McCulloch S, Watanabe H, Li G M. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerrette S, Acharya S, Fishel R. The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem. 1999;274:6336–6341. doi: 10.1074/jbc.274.10.6336. [DOI] [PubMed] [Google Scholar]
- 25.Habraken Y, Sung P, Prakash L, Prakash S. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2-MSH6 and MLH1-PMS1 protein complexes. J Biol Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 26.Habraken Y, Sung P, Prakash L, Prakash S. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 27.Harris R S, Ross K J, Lombardo M J, Rosenberg S M. Mismatch repair in Escherichia coli cells lacking single-strand exonucleases ExoI, ExoVII, and RecJ. J Bacteriol. 1998;180:989–993. doi: 10.1128/jb.180.4.989-993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang M E, de Calignon A, Nicolas A, Galibert F. POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr Genet. 2000;38:178–187. doi: 10.1007/s002940000149. [DOI] [PubMed] [Google Scholar]
- 29.Huang M E, Le Douarin B, Henry C, Galibert F. The Saccharomyces cerevisiae protein YJR043C (Pol32) interacts with the catalytic subunit of DNA polymerase alpha and is required for cell cycle progression in G2/M. Mol Gen Genet. 1999;260:541–550. doi: 10.1007/s004380050927. [DOI] [PubMed] [Google Scholar]
- 30.Hughes P, Tratner I, Ducoux M, Piard K, Baldacci G. Isolation and identification of the third subunit of mammalian DNA polymerase delta by PCNA-affinity chromatography of mouse FM3A cell extracts. Nucleic Acids Res. 1999;27:2108–2114. doi: 10.1093/nar/27.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiricny J, Nystrom-Lahti M. Mismatch repair defects in cancer. Curr Opin Genet Dev. 2000;10:157–161. doi: 10.1016/s0959-437x(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 33.Johnson R E, Kovvali G K, Guzder S N, Amin N S, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 34.Johnson R E, Kovvali G K, Prakash L, Prakash S. Role of yeast Rth1 nuclease and its homologs in mutation avoidance, DNA repair, and DNA replication. Curr Genet. 1998;34:21–29. doi: 10.1007/s002940050362. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser C, Michaelis S, Mitchell A. A ten-minute DNA prep from yeast. Methods Yeast Genet. 1994;1994:141–143. [Google Scholar]
- 36.Kamath-Loeb A S, Johansson E, Burgers P M, Loeb L A. Functional interaction between the Werner Syndrome protein and DNA polymerase delta. Proc Natl Acad Sci USA. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kane M F, Loda M, Gaida G M, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 38.Kirkpatrick D T, Ferguson J R, Petes T D, Symington L S. Decreased meiotic intergenic recombination and increased meiosis I nondisjunction in exo1 mutants of Saccharomyces cerevisiae. Genetics. 2000;156:1549–1557. doi: 10.1093/genetics/156.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kokoska R J, Stefanovic L, Buermeyer A B, Liskay R M, Petes T D. A mutation of the yeast gene encoding PCNA destabilizes both microsatellite and minisatellite DNA sequences. Genetics. 1999;151:511–519. doi: 10.1093/genetics/151.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 41.Kolodner R D, Marsischky G T. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 42.Krishna T S, Kong X P, Gary S, Burgers P M, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 43.Lamers M H, Perrais A, Enzlin J H, Winterwerp H H K, de Wind N, Sixma T K. The crystal structure of DNA mismatch repair protein MutS binding to a G:T mismatch. Nature. 2000;406:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 44.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1948;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 45.Li G M, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Y L, Shivji M K, Chen C, Kolodner R, Wood R D, Dutta A. The evolutionarily conserved zinc finger motif in the largest subunit of human replication protein A is required for DNA replication and mismatch repair but not nucleotide excision repair. J Biol Chem. 1998;273:1453–1461. doi: 10.1074/jbc.273.3.1453. [DOI] [PubMed] [Google Scholar]
- 47.Longley M J, Pierce A J, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 48.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 49.Merrill B J, Holm C. A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defects of mec1 mutants. Genetics. 1999;153:595–605. doi: 10.1093/genetics/153.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 51.Nakagawa T, Datta A, Kolodner R D. Multiple functions of MutS- and MutL-related heterocomplexes. Proc Natl Acad Sci USA. 1999;95:14186–14188. doi: 10.1073/pnas.96.25.14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structure of mismatch repair protein MutS and its complex with a DNA substrate. Nature. 2000;406:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 53.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 54.Pang Q, Prolla T A, Liskay R M. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol Cell Biol. 1997;17:4465–4473. doi: 10.1128/mcb.17.8.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons R, Li G M, Longley M, Modrich P, Liu B, Berk T, Hamilton S R, Kinzler K W, Vogelstein B. Mismatch repair deficiency in phenotypically normal human cells. Science. 1995;268:738–740. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- 56.Peltomaki P, Vasen H. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 57.Prolla T A, Christie D M, Liskay R M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prolla T A, Pang Q, Alani E, Kolodner R D, Liskay R M. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 59.Salvesen H B, MacDonald N, Ryan A, Iversen O E, Jacobs I J, Akslen L A, Das S. Methylation of hMLH1 in a population-based series of endometrial carcinomas. Clin Cancer Res. 2000;6:3607–3613. [PubMed] [Google Scholar]
- 59a.Schmutte, C., M. M. Sadoff, S. Guerrette, S. Acharya, and R. Fishel. Interactions of the human exonuclease I with DNA mismatch repair proteins hMSH2, hMSH3 and hMLH1. J. Biol. Chem., in press. [DOI] [PubMed]
- 60.Shcherbakova P V, Kunkel T A. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 62.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sokolsky T, Alani E. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics. 2000;155:589–599. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stearns T, Botstein D. Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. Genetics. 1988;119:249–260. doi: 10.1093/genetics/119.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Studamire B, Quach T, Alani E. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol Cell Biol. 1998;18:7590–7601. doi: 10.1128/mcb.18.12.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szankasi P, Smith G R. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 68.Thibodeau S N, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 69.Tishkoff D X, Amin N S, Viars C S, Arden K C, Kolodner R D. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- 70.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 72.Tran H T, Gordenin D A, Resnick M A. The 3′→5′ exonucleases of DNA polymerases δ and ɛ and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tran H T, Keen J D, Kricker M, Resnick M A, Gordenin D A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tran P T, Liskay R M. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLα. Mol Cell Biol. 2000;20:6390–6398. doi: 10.1128/mcb.20.17.6390-6398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74a.Tran, P. T., J. A. Simon, and R. M. Liskay. Interaction of EXO1 with components of MutLα in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 75.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 77.Vinh D B, Welch M D, Corsi A K, Wertman K F, Drubin D G. Genetic evidence for functional interactions between actin noncomplementing (Anc) gene products and actin cytoskeletal proteins in Saccharomyces cerevisiae. Genetics. 1993;135:275–286. doi: 10.1093/genetics/135.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Viswanathan M, Lovett S T. Single-strand DNA-specific exonucleases in Escherichia coli. Roles in repair and mutation avoidance. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xie Y, Counter C, Alani E. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics. 1999;151:499–509. doi: 10.1093/genetics/151.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]