Abstract
Two alleles of the Drosophila melanogaster Rfc4 (DmRfc4) gene, which encodes subunit 4 of the replication factor C (RFC) complex, cause striking defects in mitotic chromosome cohesion and condensation. These mutations produce larval phenotypes consistent with a role in DNA replication but also result in mitotic chromosomal defects appearing either as premature chromosome condensation-like or precocious sister chromatid separation figures. Though the DmRFC4 protein localizes to all replicating nuclei, it is dispersed from chromatin in mitosis. Thus the mitotic defects appear not to be the result of a direct role for RFC4 in chromosome structure. We also show that the mitotic defects in these two DmRfc4 alleles are the result of aberrant checkpoint control in response to DNA replication inhibition or damage to chromosomes. Not all surveillance function is compromised in these mutants, as the kinetochore attachment checkpoint is operative. Intriguingly, metaphase delay is frequently observed with the more severe of the two alleles, indicating that subsequent chromosome segregation may be inhibited. This is the first demonstration that subunit 4 of RFC functions in checkpoint control in any organism, and our findings additionally emphasize the conserved nature of RFC's involvement in checkpoint control in multicellular eukaryotes.
Control of cell cycle progression is critical for ensuring the precise duplication and distribution of the genome during cell division. One convenient way to envisage the cell cycle is as distinct phases, in which gap phases intervene before and between DNA replication and mitosis. However, a more accurate picture of the cell cycle emerges when the cell's ability to faithfully monitor crucial events is taken into account. Not only must the genome be completely and correctly replicated, but any remaining lesions in the sister chromatids must also be repaired prior to the entry into mitosis. Critically, the symmetric attachment of sister kinetochores to microtubules, achieving biorientation on the mitotic spindle, must occur prior to sister chromatid separation at the metaphase-to-anaphase transition. The concept of cell cycle checkpoints was initially proposed more than a decade ago with the characterization of the rad9 (for radiation sensitive) mutant in budding yeast (16). Since then, an explosion of studies of many diverse systems has served to illuminate three important checkpoints that normal cells progress through on the way to the completion of cell division: the DNA replication and DNA damage checkpoints (which may jointly be considered as DNA structure checkpoints) and the kinetochore attachment checkpoint. Precise molecular components have been identified in different model systems, primarily through genetic analysis of each of these checkpoints (6). Mutation of checkpoint components has been observed in many human cancers (9, 31).
Though much of the original identification and analysis of cell cycle checkpoints was achieved with budding and fission yeast, studies with Drosophila melanogaster and humans have highlighted the fact that the mechanisms observed in unicellular eukaryotes appear largely to be conserved in metazoans. In flies, genes important for checkpoint control have been discovered through the analysis of mutations affecting female meiosis or mutagen sensitivity (4, 14), embryogenesis (10, 37), and larval development (2). The genes studied in flies all appear to have orthologues in other species.
The life cycle of Drosophila not only requires mitosis at different times in development but also utilizes different types of cell cycles. Thus the extremely rapid, synchronized nuclear divisions of early embryogenesis (consisting of S and M phases only) are powered by maternally provided gene products. The cell cycle expands to include gap phases roughly concomitant with cellularization, and, from this point forward, most of the maternally provided gene products essential for cell division are depleted, and dependency on zygotic gene expression ensues. Though many of the larval tissues undergo endoreduplication cycles (repeated replication in the absence of cell division), resulting in highly polyploid cells, the imaginal discs (precursors of adult tissues) and neuroblasts remain diploid and mitotically active, with a conventional cell cycle punctuated by gap phases. A genetic dissection of the process of imaginal disc development was initiated by Shearn et al. when they screened for late-larval-phase lethal mutations in Drosophila (32, 33). These mutants die late in development because metamorphosis cannot be completed in the absence of imaginal discs. Gatti and Baker postulated that many of them might be dying because of cell cycle defects, as zygotic mutations disrupting the cell cycle specifically affect proliferating tissues but not the bulk of larval tissues, which are polyploid (11). With the goal of better understanding higher-order chromosome dynamics, we have been analyzing a number of mutations which have been selected from the original Shearn and Garen collection and which have been described as affecting mitotic chromosome structure by Gatti and Baker. This pool of mutations represents a powerful opportunity to examine the cell cycle in an organism amenable to genetic and cytological analysis.
One of the lines we have been examining, known as the l(3)e20 line, was initially characterized as being involved in cell proliferation and chromosome condensation (11, 42, 43). In results described below, we have found that a high degree of chromosome “pulverization” can be observed in mitosis in l(3)e20 mutants. In addition, we noticed the unusual phenotype due to chromosomes that appear normally condensed but that have precociously separated sister chromatids. We found that l(3)e20 is allelic to a mutant known as l(3)a18, which in turn was shown to be a mutant for one of the small subunits of the replication factor C (RFC) complex (15).
RFC was originally isolated from human cells as a factor essential for DNA replication using the in vitro simian virus 40 system (reviewed in references 19 and 46). It was later isolated from other systems such as yeast and calf thymus and shown to be important for the loading of proliferating cell nuclear antigen, the processivity factor for DNA polymerases δ and ɛ. RFC is present in cells as a heteropentameric protein complex, containing one large subunit with a molecular mass of ∼140 kDa and four small subunits (molecular masses, ∼40 kDa). Sequence similarities between the identified RFC subunits has allowed the recognition of eight conserved RFC “boxes,” seven of which are found in all five subunits (8). RFC also has other roles, as mutations of Rfc2 and Rfc5 in Saccharomyces cerevisiae and Rfc2 and Rfc3 in Schizosaccharomyces pombe produce mutants that are also defective in checkpoint control in response to DNA damage (12, 26, 28, 34, 40). An intriguing twist to the DNA damage checkpoint story was revealed when it was shown that the Rad24 protein of S. cerevisiae can complex with the four small RFC subunits (instead of RFC1), thus suggesting that this alternative complex may in fact be functioning as the sensor in the DNA structure-specific checkpoint pathway (13).
In this paper, we describe a detailed cytological analysis of mutations in an RFC subunit in Drosophila. Because of DmRfc4 mutations, a greatly decreased number of neuroblasts undergo DNA replication in larval brains and those that do replicate show a very high frequency of abnormal mitoses. The RFC4 protein is localized to replicating nuclei but is dispersed from chromatin in mitosis. Importantly, DmRfc4 mutants are defective in the checkpoint response to either blocked DNA replication or damaged DNA, although the spindle checkpoint is intact. These results importantly indicate that RFC4 is essential for checkpoint control (thus joining the other small subunits) and further reveal that the role of RFC in checkpoint control is conserved between yeast and metazoans.
MATERIALS AND METHODS
Fly stocks.
The wild-type strain used was Canton S. We received the l(3)e20 line from Maurizio Gatti (University of Rome); it was originally generated in an ethyl methane sulfonate screen by Allen Shearn (Johns Hopkins University). The cytological location was determined by testing for complementation with these deficiencies: Df(3L)TE1, Df(3L)HR298, Df(3L)C175, Df(3L)GN34, Df(3L)GN19, Df(3L)GN24, and Df(3L)GN50 (Bloomington Stock Center). Deficiencies removing the gene uncovered the lethality and the mitotic defects for transheterozygous animals. l(3)e20 flies were also crossed to 20 different lethal complementation groups that localize to 63F-64A. Each cross (5 heterozygous virgin female flies were crossed to 3 heterozygous male flies) was done in duplicate; 50 to 100 progeny were examined for each cross.
DAPI staining of larval neuroblasts.
DAPI (4′,6-diamidino-2-phenylindole; Sigma) staining of neuroblasts was performed as described previously (17). Larval brains were dissected from the rest of the larval tissues in Ephrussi-Beadle Ringers solution (EBR; 129 mM NaCl, 4.7 mM KCl, 1.9 mM CaCl2, 10 mM HEPES, pH 6.9) and then fixed for 30 s in a droplet of 45% acetic acid on a siliconized coverslip. A poly-l-lysine-coated slide was placed over the droplet of acetic acid and pressed to flatten the tissue into a monolayer. After the slide was frozen in liquid nitrogen, the coverslip was flicked from the slide with a razor blade. The slide was incubated for 5 min in phosphate-buffered saline (PBS; 150 mM NaCl, 10 mM NaHPO4, pH 7.2) to remove the acid. The slide was then incubated for 5 min in PBS–0.1% Triton X-100 (Tx) and then for 5 min in PBS–0.1% Tx–0.1 μg of DAPI/ml. Excess DAPI was removed by a 5-min wash in PBS–0.1% Tx. At times, hypotonic swelling of tissues for 10 min in 0.5% sodium citrate was performed prior to fixation. Coverslips were mounted onto the slides using Mowiol 4-88 (Calbiochem). The slides were viewed using either a Zeiss Axiophot or an Olympus Provis epifluorescence microscope, and images were photographed onto 35-mm 400 ASA slide film or digitally captured using a Photometrics Sensys cooled charge-coupled device camera and Vysis Quips SmartCapture software.
Salivary gland squash preparations.
Appropriately staged larvae were rinsed, and the salivary glands were dissected out in EBR. The salivary glands were fixed in 45% acetic acid for 30 s at room temperature and then transferred to a droplet of lactic acid-water-acetic acid in a ratio of 1:2:3 with 5% glycerol for 4 to 5 min on a poly-l-lysine-treated slide. The glands were then squashed between the slide and a siliconized coverslip, with gentle tapping to disrupt the nuclei. After the slide was frozen in liquid nitrogen, the coverslip was flicked from the slide with a razor blade. The squashed glands were then processed as for neuroblasts (above). The slides were examined on an Olympus Provis epifluorescence microscope, and images were digitally captured.
Isolation of larval genomic DNA.
Genomic DNA was isolated from third-instar larvae. Fifteen larvae with each genotype were collected and rinsed in EBR prior to being frozen at −20°C in 1.5-ml tubes. They were homogenized with a motorized pestle in 200 μl of cold grinding buffer (60 mM NaCl, 10 mM EDTA, 5% sucrose, 0.15 mM spermine, 0.15 mM spermidine, 5 mg of RNase/ml, 10 mM Tris, pH 7.5). Two hundred microliters of prewarmed (to 37°C) lysis buffer (0.1 M EDTA, 1.25% sodium dodecyl sulfate, 5% sucrose, 0.3 M Tris, pH 9.0) was immediately added. The larval extract was incubated at 65°C for 30 min. The extract was cooled to room temperature, and 8 M potassium acetate was added to a final concentration of 1 M. The solution was incubated on ice for 45 min and then centrifuged for 5 min at 4°C. The supernatant was removed and extracted twice with phenol-chloroform. The DNA was ethanol precipitated and resuspended in distilled water or 10 mM Tris–1 mM EDTA (pH 8.0) (2 μl per larva).
PCR amplification and cloning of DmRfc4 sequences.
Primers were designed from the wild-type DmRfc4 sequence to amplify the mutant alleles for sequencing. The 5′ primer (at −29 to −13 relative to the starting ATG; 5′-GGGTCGACTTGTGCGAATTTGTGAA-3′) included a SalI restriction site (underlined) for cloning purposes. The 3′ primer (at +1147 to +1164; 5′-GGGGATCCACTAACGGTGCCCAGTT-3′) included a BamHI restriction site (underlined). PCRs were performed in a Biometra Personal Cycler. The genomic DNA was first denatured for 3 min at 95°C. Twenty cycles were performed under the following conditions: denaturation at 95°C for 45 s, annealing at 55°C for 60 s, and elongation at 72°C for 90 s. The cycles were followed by 10 min of elongation at 72°C. In a 100-μl reaction mixture, 200 pmol of each primer, 1 μg of genomic DNA, 250 μM deoxynucleoside triphosphates, 0.25 mM MgCl2, and 5 U of Pwo polymerase (Boehringer Mannheim) were used. PCR products were electrophoresed on a 1.5% low-melting-point agarose (FMC; SeaPlaque) in TAE (40 mM Tris-acetate, 1 mM EDTA). The desired fragment was isolated from an excised gel slice using Wizard Minipreps DNA purification resin (Promega) according to the manufacturer's instructions. The isolated fragment was digested with BamHI and SalI (New England Biolabs) and gel purified a second time. The digested fragments were ligated into a similarly digested and gel-purified pBluescript II KS(+) vector (Stratagene) using T4 DNA ligase (Promega) overnight at 16°C. The ligated DNA was transformed into JM109 bacterial cells by electroporation with a Bio-Rad Gene Pulser II (200 Ω, 25 μF, 2.5 kV). DNA from three independent colonies (for each genotype) was prepared using a Qiagen MidiPrep plasmid isolation kit for sequencing and then was precipitated with polyethylene glycol (PEG) 8000 (400 mM NaCl, 1.3% PEG 8000) on ice for 10 min. The DNA was centrifuged at 10,000 × g for 15 min at 4°C. After an ethanol wash, the DNA was resuspended in distilled water, and 500 ng of DNA was used in a sequencing reaction (ABI Prism dye terminator cycle sequencing ready reaction kit; Perkin-Elmer). The T7 and T3 primers were used. An ABI Prism sequencer and software were used to read the sequence.
Immunofluorescence on whole-mount larval brains.
Brains were dissected from third-instar larvae in PBS (140 mM NaCl, 2.7 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 1.8 mM KH2PO4, 10 mM Na2HPO4, pH 7.4) and immediately fixed in PBS–4% paraformaldehyde (PFA; TAAB Laboratories) for 15 to 20 min. The dissected brains were then rinsed three times with PBS for 5 min each. The brains were blocked in PBS–10% normal goat serum (NGS; Sigma) for 1 h. The brains were washed three times for 15 min each in PBS–0.05% Tx (PBTx). The brains were incubated with a primary antibody (mouse monoclonal anti-DmRFC4 at 1:4) diluted in PBTx overnight at 4°C. After three 15-min washes in PBTx, the brains were blocked again with PBS–10% NGS for 30 min and then incubated with the secondary antibody (biotinylated horse anti-mouse antibody at 1:100; Vector Laboratories) in PBTx for 1 h. The PBTx washes were repeated. The brains were blocked again for 30 min, incubated with streptavidin-Texas red (Vector) at 1:1,000 for 1 h, and washed five times with PBS (DAPI was included at 0.1 μg/ml in the third wash). The brains were mounted in 90% glycerol and viewed as described above.
BrdU incorporation in larval brains.
Third-instar larvae were fed 50 mg of bromodeoxyuridine (BrdU; Boehringer Mannheim)/ml in Drosophila instant food (Sigma) for 2 h. Inclusion of food coloring allowed rapid determination of which larvae had ingested the drug. Visualization of the BrdU was as described above with the following modifications. Prior to the first NGS block, the brains were incubated in freshly prepared 2 N HCl for 1 h. The acid was removed with three PBS washes, which were followed by incubation with rat anti-BrdU antibody (1:2 dilution; Harlan SeraLab). After being washed, the brains were incubated in dichlorotriazinylamino-fluorescein-conjugated goat anti-rat antibody (1:100 dilution; Harlan SeraLab) for 1 h. Brains were then washed, stained, mounted, and viewed as described above. The relative fluorescence intensity of the incorporated BrdU was determined from digitally captured images using Adobe Photoshop, version 5.5.
Immunofluorescence of cultured cells.
Drosophila Dmel2 cells (Gibco) were cultured in Drosophila serum-free medium (Gibco) at 27°C. Cells were centrifuged at 1,000 × g for 5 min in a Hereaus Megafuge 1.0R and hypotonically swollen in 0.25× EBR at a concentration of 5 × 106/ml for 5 min at room temperature (RT). Cells were centrifuged again and resuspended at the same concentration in fresh 0.25× EBR. Aliquots of 5 × 105 cells and 1 × 106 cells were then centrifuged onto poly-l-lysine-coated slides (BDH) using a Hereaus cytospin column without the blotting pads for 10 min at 4,000 × g in the Megafuge at RT. The cells were immediately fixed for 3 min in 4% PFA in PBS at RT. After two 5-min washes in PBS, cells were permeabilized with three 10-min washes in PBTx and then blocked in PBS–3% bovine serum albumin (BSA; Sigma) for 1 h at RT. Cells were washed for 5 min in PBTx and incubated for 1.5 h at RT in a mouse monoclonal anti-RFC4 antibody (1:2 to 1:10 dilution) in PBS–0.3% BSA. After three 5-min washes in PBTx, the cells were incubated for 1.5 h at RT in Alexa 594 goat anti-mouse conjugate (1:500 dilution; Molecular Probes) in PBS–0.3% BSA and then washed again and stained with 50 ng of DAPI/ml. Coverslips were mounted onto the slides with Mowiol and viewed as described above.
BrdU incorporation in cultured Drosophila cells.
Dmel2 cells were grown on sterile poly-l-lysine-coated slides (BDH) from a starting concentration of 5 × 105/ml for 72 h to allow them to adhere and grow on the slides. Cells were incubated for 5 or 45 min in media containing 6 μg of BrdU/ml and then washed twice for 2 min in PBS and fixed for 3 min in 4% PFA in PBS at RT. After a 5-min wash in PBS, cells were permeabilized with three 10-min washes in PBTx and the blocking and anti-RFC4 immunofluorescence steps were performed as described above except for omitting the DAPI counterstain. After the anti-RFC4 immunofluorescence was complete, the cells were immediately fixed for 1 h at RT in 4% PFA in PBS, washed for 5 min in PBTx, and then incubated for 30 min in freshly prepared 2 N HCl. After a 5-min wash in PBTx, cells were blocked for 30 min in PBS–3% BSA and washed for 5 min in PBTx and then incubated with rat anti-BrdU antibody (1:2 dilution) in PBS–0.3% BSA for 16 h at 4°C. After three 10-min washes, the cells were incubated with Alexa 488 goat anti-rat conjugate (1:500 dilution; Molecular Probes) in PBS–0.3% BSA for 1.5 h at RT. The washes were repeated, and the cells were stained with 50 ng of DAPI/ml. Coverslips were mounted onto the slides with Mowiol and viewed as described above.
In vivo checkpoint studies.
Mature third-instar wild-type and DmRFC4 mutant larvae were placed on instant fly food containing food coloring and 1 mg of aphidicolin (Sigma), 50 mg of hydroxyurea (Sigma), 0.5 mg of etoposide (Calbiochem), 50 mg of caffeine (Calbiochem), 50 mg of colchicine (Sigma)/ml or no drug for 2 h. For UV irradiation, larvae were exposed to 200 mJ at 254 nm using a Stratalinker (Stratagene) and then placed on instant food to recover for 2 h. The larval brains were dissected and squashed as described above. Between 4 and 11 brains were examined for each condition. The mitotic index (MI) was calculated by counting the number of mitotic cells in three different fields at ×400 magnification, dividing by the total number of cells within the three fields, and then multiplying by 100. These drug treatments were also performed in an organ culture of freshly dissected brains on a total of 6 to 42 brains for each condition essentially as described previously (21).
RESULTS
l(3)e20 results in mitotic chromosomal abnormalities.
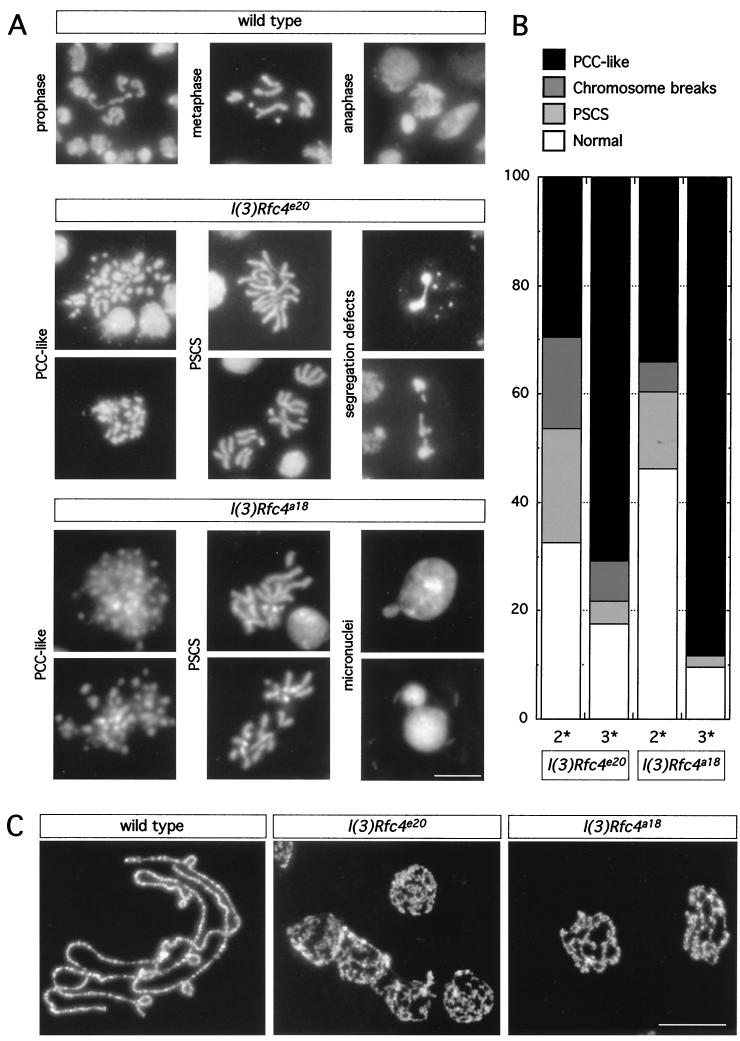
With aceto-orcein to stain larval neuroblasts, the mitotic phenotype for l(3)e20 was originally described as having the following characteristics: extreme reduction of imaginal disc tissue, a very low MI, irregular chromosome condensation, and extensive chromosome fragmentation (11). To characterize the mitotic phenotype due to the l(3)e20 mutation in greater detail, we examined brains from second- and third-instar homozygous l(3)e20 larvae stained with DAPI to visualize the chromatin (Fig. 1A). The major advantage of using DAPI is that only the chromatin is stained; thus defects in mitotic chromosome structure or behavior can be best resolved (17). We observed that the MI (percentage of mitotic cells/total number of cells) was decreased to 0.41% in homozygous mutants, in comparison to 0.94% in wild-type brains. More than 80% of the observed metaphase figures in third-instar l(3)e20 larval brains were abnormal, with the phenotypes falling into two types (Fig. 1A and B). The majority of the metaphase figures exhibited a pulverized appearance, with extreme effects on condensation. These metaphases are highly reminiscent of S-phase prematurely condensed chromosomes (PCCs) originally observed when interphase cells were fused with mitotic cells (27). At a lower percentage, we observed normally condensed mitotic figures containing chromosome breaks and the striking defect of precocious sister chromatid separation (PSCS), in which some or all of the sister chromatids within a mitotic cell had separated, without an apparent anaphase configuration (Fig. 1B). Of the anaphases we observed, greater than 50% showed defects in chromosome segregation exhibited as chromosomal bridges or lagging chromosomes. Not surprisingly, micronuclei, a consequence of unequal chromosome segregation, were also seen.
FIG. 1.
Chromosome phenotypes from wild-type and homozygous l(3)Rfc4e20 and l(3)Rfc4a18 third-instar larval brains and salivary glands. (A) Larval brains were dissected from third-instar larvae and hypotonically swollen before fixation. The neuroblasts were stained with DAPI to visualize the DNA. Figures representative of the major categories of phenotype, PCC and PSCS, and chromosome segregation defects are shown. Scale bar = 10 μm. (B) Quantitation of phenotypes in second- (2*) and third-instar (3*) larval brains from l(3)Rfc4e20 and l(3)Rfc4a18 larvae. The prevalence of PCC-like figures increases as the animals age and residual wild-type maternal RFC4 is depleted. (C) Larval salivary glands were dissected from wild-type and homozygous l(3)Rfc4e20 and l(3)Rfc4a18 third-instar larvae, fixed, and stained with DAPI. The nuclei of the cells in the salivary gland were disrupted to spread the polytene chromosomes. Compared to those from the wild type, the polytene chromosomes from homozygous l(3)Rfc4e20 and l(3)Rfc4a18 larvae appear underreplicated and the banding pattern is disrupted. Scale bar = 100 μm.
We also examined salivary gland polytene chromosome architecture in third-instar l(3)e20 homozygous larvae (Fig. 1C). In stark contrast to chromosomes from wild-type salivary glands, which are highly polyploid and reproducibly banded, polytene chromosomes from homozygous mutant salivary glands were substantially underreplicated and showed very little evidence of banding. Thus chromosomes from both diploid and polyploid tissues were abnormal, suggestive of a defect in a gene essential for DNA replication.
Mutations in DmRfc4 are responsible for the observed chromosomal defects.
Because of these intriguing phenotypes, we set out to identify the gene mutated in l(3)e20. We first mapped the lethal mutation by genetic crosses to Drosophila lines deficient for overlapping regions within the genomic interval 64A6 to 64A10 on the left arm of the third chromosome [the region to which l(3)e20 was originally mapped by recombination]. Since l(3)e20 was a chemically induced (ethyl methane sulfonate) mutant, we attempted to obtain a transposon-induced (P element) allele that would facilitate the cloning of the affected gene. Although inducing a nearby P element (from a different stock) to mobilize to new sites failed to yield new lethal alleles of l(3)e20, another chemical mutagenesis screen of the genomic region between 63E and 64A led us to a noncomplementing allele and thus to the ultimate identification of the gene. This screen identified 20 lethal complementation groups in this genomic interval (15). When an allele of each of these complementation groups was further screened, l(3)a18, an allele from the l(3)64Ai complementation group, was found to fail to complement l(3)e20. This complementation group had been identified as the Drosophila orthologue of human Rfc40, the gene encoding the Rfc4 subunit of human RFC (15). The proof that l(3)e20 was an allele of DmRfc40 is provided below. To simplify nomenclature, we propose to rename this gene DmRfc4, in accordance with the nomenclature of the yeast subunits (the human proteins are named after their molecular weights).
The chromosomal phenotypes for the l(3)Rfc4a18 allele were qualitatively similar to those for the l(3)Rfc4e20 allele but were more penetrant, suggesting that this allele was more severe (Fig. 1A and B). The MI was even lower, at 0.20%, than that for l(3)Rfc4e20, with ∼90% of the observed mitotic figures in l(3)Rfc4a18 appearing abnormal. The same two classes of mitotic defects were observed, with PCC-like figures more prevalent and chromosome breaks and PSCS figures less common than in l(3)Rfc4e20. Segregation defects were also observed. For both alleles, the frequencies of normal mitotic figures and PSCS were higher in neuroblasts from younger second-instar larvae, while PCC-like figures peaked in third-instar larvae and pupae (Fig. 1B). The younger larvae would be expected to contain a higher level of residual wild-type maternal product, resulting in a weaker phenotype. We conclude that PCC-like figures represent the most extreme manifestation of mitotic defect in DmRfc4 mutants.
Polytene chromosomes appeared similar in larvae homozygous for either of the two alleles (Fig. 1C). As RFC is a complex actively involved in DNA replication, this observation of poorly formed polytene chromosomes indicates that the endoreduplication cycles required to generate the polytene chromosomes are affected. However, this is in marked contrast to the normal appearance of polytene chromosomes from salivary glands from animals mutant in subunit 2 of the origin recognition complex (DmORC2) (21).
Both DmRfc4 alleles encode truncated proteins.
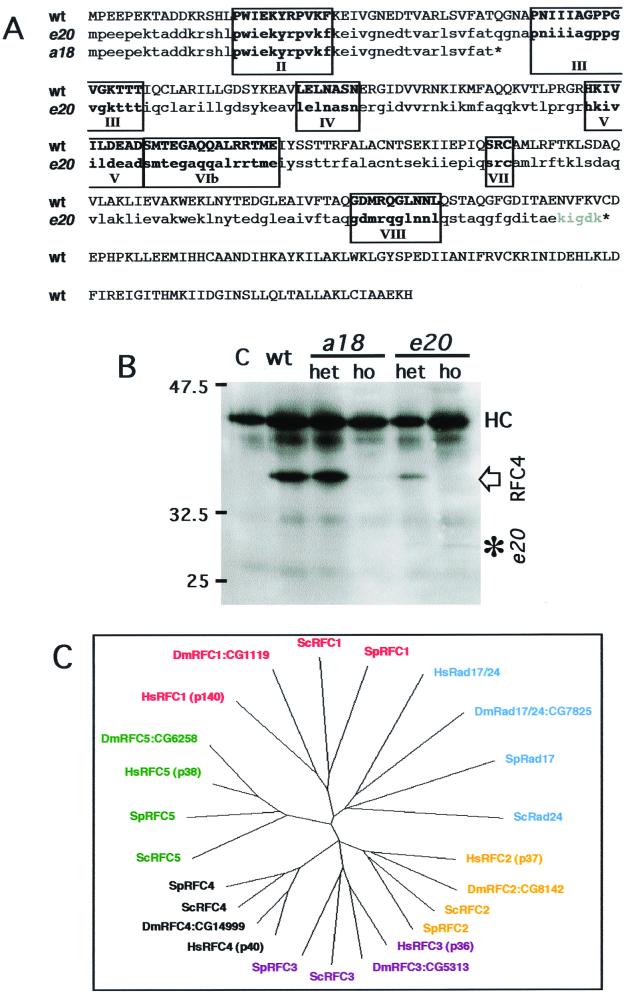
To further confirm that these two alleles both result from mutations in the DmRfc4 gene, genomic DNA from both l(3)Rfc4e20 and l(3)Rfc4a18 homozygous larvae was amplified by PCR using primers designed from the wild-type DmRfc4 sequence (15). The PCR-amplified DNA fragments were cloned and subsequently sequenced. l(3)Rfc4a18 carried a single base change (C136T), resulting in a premature stop codon (Fig. 2A). The putative RFC4a18 product is only 45 amino acids long and, if produced, would include only box II of the conserved RFC boxes (8). The lesion in l(3)Rfc4e20 is an in-frame deletion of bases 740 to 1090 (351 bp in total) including the wild-type stop codon; this deletion results in a C-terminal truncation of the protein. After incorporating five additional amino acids, translation is terminated at the next stop codon. The potential RFC4e20 product could include all of the conserved RFC boxes (Fig. 2A).
FIG. 2.
Identification of the mutations in l(3)Rfc4e20 and l(3)Rfc4a18 and phylogenetic relationships between RFC family members from D. melanogaster, Homo sapiens, S. pombe, and S. cerevisiae. (A) DNA from l(3)Rfc4e20 and l(3)Rfc4a18 homozygous larvae was amplified by PCR, cloned, and sequenced to determine the molecular lesions in l(3)Rfc4e20 and l(3)Rfc4a18. The lesion in l(3)a18 is a single base change (C136T), which resulted in a premature stop codon yielding a putative product of 45 amino acids. The lesion in l(3)e20 is a 351 bp in-frame deletion. The predicted protein products are shown beneath the wild-type (wt) gene open reading frame (ORF). The conserved RFC boxes are in boldface and numbered. (B) Immunoprecipitation of the DmRFC4 protein from wild-type, heterozygous, and homozygous larval extracts. RFC4 was present in wild-type and heterozygous larvae carrying both alleles at the expected size of 40 kDa. In extracts from both homozygotes, the wild-type protein was detected at very low levels, which may represent residual maternal product. In extracts from homozygous l(3)a18 larvae, the mutant protein could not be detected. In l(3)e20 larval extracts, a protein band at the predicted molecular mass for the mutant form of DmRFC4e20 (29 kDa) was observed, thus confirming the sequencing data. C, antibody-only control for immunoprecipitation (no larval extract); HC, immunoglobulin heavy chain of the anti-DmRFC4 antibody used for immunoprecipitation and detected by the secondary reagent during the immunoblotting of the immunoprecipitates. (C) Phylogenetic relationship between the five RFC subunits (and the related RFC1 products, SpRad17 and ScRad24) (obtained from ClustalX analysis and drawn with TreeViewPPC). The molecular weights of the human proteins (as this is the conventional nomenclature) and the Celera Gene nomenclature (CG number) for the Drosophila genes are given. For all proteins used in the phylogenetic analysis, the Drosophila ORF is more closely related to the human gene than to the yeast sequences.
A monoclonal antibody against the N-terminal 55 amino acids of DmRFC4 (15) was used to examine the wild-type and mutant proteins. The wild-type DmRFC4 protein could be immunoprecipitated from wild-type and heterozygous extracts of either allele and migrated at the expected size of 40 kDa (Fig. 2B). In addition, a protein of the predicted molecular size of 29 kDa could be immunoprecipitated from l(3)Rfc4e20 larval extracts, suggesting that this form may be present in heterozygotes and homozygotes, albeit at a much lower level. The truncated form is possibly not immunoprecipitated as efficiently as the full-length form or possibly not synthesized to the same level. It is also feasible that only the full-length form can complex with the other RFC subunits (the C terminus being required for complex formation), leaving the truncated form with an uncertain fate. A protein with the predicted size of 5 kDa was not detectable in l(3)Rfc4a18 extracts (it may be unstable or not produced, or electrophoresis conditions may not have been optimal). Thus, the immunoprecipitation independently confirmed the prediction made from sequencing the homozygous mutant DNA.
As this is the first detailed cytological description of mitotic defects in an Rfc4 mutant in any organism, we were interested to know whether all the RFC subunits could be identified in Drosophila as in budding and fission yeast and humans (only DmRfc1 and DmRfc4 had been previously reported) (1, 15). By analysis of the sequenced Drosophila genome, we were able to identify by homology all five subunits of the RFC complex and also the homologue of SpRad17 and ScRad24 (an Rfc1-related gene) (13, 24). A phylogenetic tree depicting the relationship of Drosophila protein sequences with their yeast and human counterparts is shown in Fig. 2C. For all subunits included in this analysis, the Drosophila protein was more similar to the human protein than to either of the yeast orthologues.
The pattern of DmRFC4 expression resembles that of actively replicating larval neuroblasts.
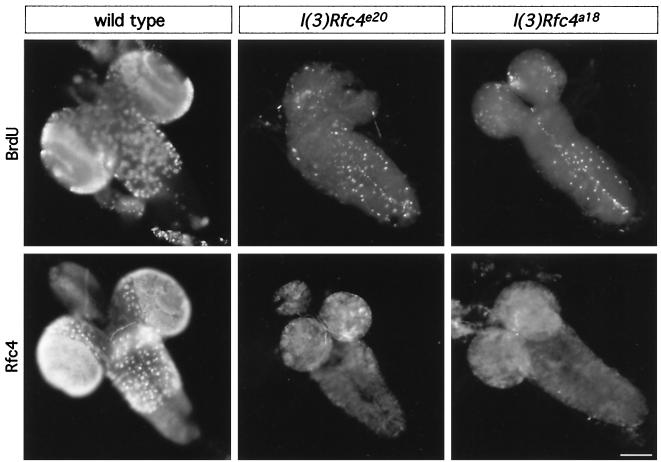
We next analyzed the mutant phenotype further in terms of DNA replication. We examined the expression of the RFC4 protein and also the level of replication in larval brains after incorporation with BrdU (Fig. 3). In wild-type brains, the pattern of DmRFC4 detectable by immunofluorescence closely resembled the BrdU pattern in zones of proliferation (most notable in the optic lobes of the larval brain). Thus DmRFC4 is likely present in actively replicating cells (with all wild-type brains exhibiting BrdU incorporation after feeding or in vitro incubation). In contrast, immunofluorescence of homozygous mutant brains carrying either DmRfc4 allele revealed very little BrdU incorporation (only 50% of the larval brains incorporated any BrdU at all) or expression of the RFC4 protein within the brain. Therefore, mutations in DmRfc4 clearly lead to a reduction in the number of cells actively undergoing DNA replication throughout the brain.
FIG. 3.
BrdU incorporation and RFC4 localization in intact larval brains. Larval brains were dissected from wild-type and homozygous l(3)Rfc4e20 and l(3)Rfc4a18 third-instar larvae after the larvae were fed BrdU for 2 h and processed for BrdU detection (top). Patterns characteristic of proliferation zones are observed in wild-type brains, while mutant brains show only low levels of BrdU incorporation. Another set of larval brains was processed for localization of DmRFC4 (bottom). Only background immunofluorescence was observed when visualizing DmRFC4 in l(3)Rfc4e20 and l(3)Rfc4a18 third-instar larval brains. Scale bar = 100 μm.
Although both the MI and the replicative activity (RA) of the mutant alleles were lower than in wild-type larvae, quantitation of these two parameters relative to one another indicated that the accumulation of cells in mitosis differed between wild-type and mutant cells. The MI/RA ratio was roughly fivefold higher in DmRFC4 mutants than in wild-type cells (Table 1), suggesting that it was not only progression into or through S phase that was disrupted but also subsequent progression through the cell cycle, leading ultimately to accumulation of mitotic cells with aberrant chromosome structure and behavior.
TABLE 1.
Quantitation of the ratio between MI and RA in wild-type and DmRfc4 larval neuroblasts
| Genotype | MI (%) | RA | MI/RA | Normalized MI/RA |
|---|---|---|---|---|
| Wild type | 0.94 | 57.48 | 0.0164 | 1 |
| l(3)Rfc4e20 | 0.41 | 4.43 | 0.0926 | 5.65 |
| l(3)Rfc4a18 | 0.20 | 2.74 | 0.073 | 4.45 |
DmRFC4 is absent from mitotic chromosomes.
One might expect that cells defective for a protein with a role only in DNA replication would arrest in S phase. That the homozygous l(3)Rfc4e20 and l(3)Rfc4a18 neuroblasts exhibit striking mitotic defects suggested to us a broader role for DmRFC4 in chromatin dynamics or cell cycle control. We thus set out to determine the subcellular localization of the DmRFC4 protein in a number of Drosophila cultured cell lines to see whether it was present on interphase and/or mitotic chromatin.
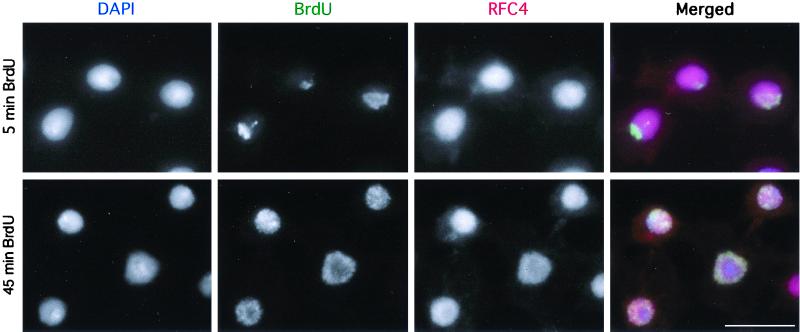
Identical results were obtained with clone 8 (derived from wing imaginal discs), 1182 and Schneider line 2 (both derived from Drosophila embryos), and Dmel2 (derived from S2) cells; however, only the data for Dmel2 cells are shown (Fig. 4 and 5). More than 95% of interphase cells contained DmRFC4 within nuclei (Fig. 4). Cells were incubated in BrdU prior to immunofluorescence to be able to detect actively replicating cells (Fig. 4, top [5-min incubation]). After a 45-min incubation with BrdU (Fig. 4, bottom), the nuclei show a greater level of BrdU incorporation and all are positive for RFC4. These cells are likely to be in S phase, though some have progressed into G2 phase. RFC4 appears to be distributed homogeneously throughout nuclei and is not restricted to the active sites of replication. We conclude that RFC4 is nuclear in interphase and that all replicating nuclei in these cell lines contain RFC4.
FIG. 4.
Detection of DmRFC4 by immunofluorescence in interphase Drosophila cultured cells. Dmel2 cells were incubated in BrdU for 5 or 45 min and processed for immunofluorescence detection of RFC4 and incorporated BrdU. All replicating cells show homogeneous nuclear staining for RFC4, which is not restricted to the sites of active replication (5-min BrdU incubation). Scale bar = 10 μm.
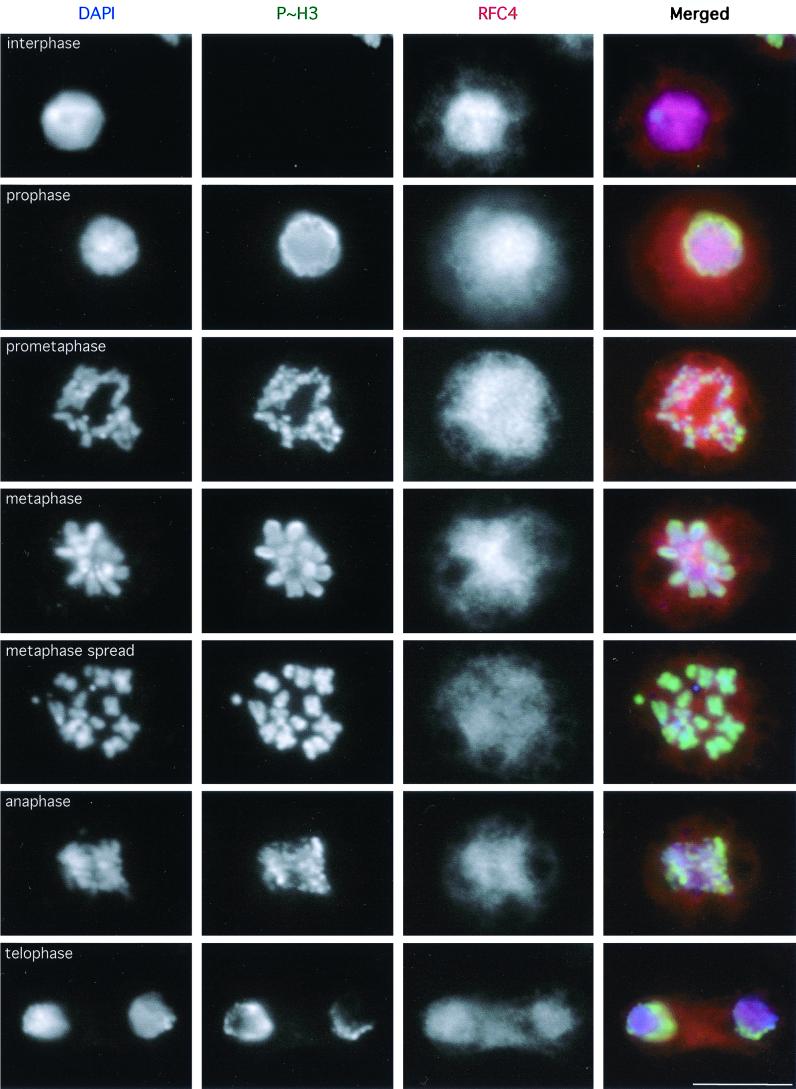
FIG. 5.
Detection of DmRFC4 by immunofluorescence in mitotic Drosophila cultured cells. Dmel2 cells were processed for immunofluorescence detection of RFC4. These cells were also stained for the mitosis-specific phosphorylated form of histone H3 to unambiguously distinguish mitotic cells (18). RFC4 appears to be homogeneously distributed throughout mitotic cells from prophase through telophase. A metaphase cell processed to spread the chromosomes is also shown. Scale bar = 10 μm.
Mitotic Dmel2 cells stained for RFC4 are presented in Fig. 5. Independent confirmation of mitotic state was given by colabeling for the mitosis-specific phosphorylated form of histone H3 (18). Only a homogeneous cellular staining of RFC4 was observed in mitotic cells, with no detectable concentration of the protein on mitotic chromosomes. Though it is possible that a small fraction of the cellular RFC4 remains associated with the chromatin during mitosis, the majority of the protein appears not to be associated with mitotic chromosomes (this was similarly observed when HeLa cells were stained with the DmRFC4 antibody; data not shown). We have also analyzed the localization of RFC4 in “mitotic spreads,” cells that have been hypotonically swollen prior to spinning onto coverslips (an example of which is shown in Fig. 5). Even when individual chromosomes are well spread, we have been unable to detect any localization of RFC4 on mitotic chromosomes. Therefore we believe that the mitotic defects observed in the l(3)Rfc4e20 and l(3)Rfc4a18 neuroblasts are unlikely to result from a direct role for DmRFC4 in mitotic chromosome structure.
While even telophase nuclei appear not to contain appreciable amounts of RFC4, by the time nuclei enter S phase RFC4 is nuclear. Thus RFC4 is likely imported into nuclei sometime during G1 phase. This is in contrast to the localization of subunit 2 of the origin recognition complex in Drosophila, which binds chromosomes already in anaphase (21). Therefore the cell cycle kinetics with which assorted replication components associate with chromatin appear to differ.
Cell cycle checkpoint defects in DmRfc4 mutants.
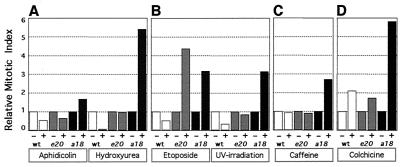
If the mitotic phenotype observed in the l(3)Rfc4e20 and l(3)Rfc4a18 neuroblasts is not due to a direct structural role of RFC4 in mitotic chromosomes, then perhaps the protein is important for ensuring proper progression through the cell cycle. The quantitation of the MI/RA ratio also suggests that progression through the cell cycle is abnormal in the DmRfc4 mutants, such that mitotic cells with chromosomal aberrations accumulate (Table 1). To test this hypothesis, we performed various treatments on l(3)Rfc4e20 and l(3)Rfc4a18 larvae to ascertain the integrity of cell cycle checkpoints when the DmRfc4 gene is mutated. As BrdU can be detected in wild-type neuroblast nuclei within 10 min after ingestion of food (unpublished observations), drugs fed in a similar manner should also be taken up by the neuroblasts with similar timing. For these experiments, therefore, wild-type and homozygous mutant third-instar larvae were placed for 2 h on instant Drosophila food containing a drug to perturb the cell cycle at various points. To determine subsequent cell cycle progression, the MIs for brains from treated larvae were normalized to that for brains from untreated controls to determine the response to various treatments within actively cycling neuroblasts (Fig. 6; numbers are given in Table 2). The DNA replication checkpoint was monitored by feeding larvae with hydroxyurea or aphidicolin, while the DNA damage checkpoint was activated by etoposide treatment or UV irradiation. The spindle assembly-kinetochore attachment checkpoint was examined following colchicine treatment. Caffeine was also used as a treatment, as it has been shown to inhibit two conserved phosphatidylinositol-related kinases: the ATM checkpoint kinase in human cells (3) and the Rad3 kinase in fission yeast (23). These treatments were also performed in organ culture of brains dissected from larvae, with similar results (data not shown).
FIG. 6.
In vivo checkpoint studies. Third-instar larvae were fed for 2 h on instant food with or without a drug (or irradiated and then left for 2 h). (A) Aphidicolin or hydroxyurea; (B) etoposide or UV irradiation; (C) caffeine; (D) colchicine. The larval brains were dissected, and the MI was determined after DAPI staining. The MIs were normalized to the MI for untreated brains in each case. Whereas wild-type neuroblasts respond as expected to perturbation of the cell cycle (arresting prior to mitosis in response to inhibition of DNA replication or DNA damage or in mitosis after spindle depolymerization), DmRfc4 mutant neuroblasts are unable to respond properly to inhibition of DNA replication or induction of DNA damage, although the kinetochore attachment checkpoint can be activated. While caffeine appears to have no effect on either wild type or l(3)Rfc4e20 neuroblasts, l(3)Rfc4a18 cells respond by arrest or delay in mitosis, as evidenced by an increased incidence of PCC-like figures.
TABLE 2.
Statistical analysis of the MI of wild-type and DmRfc4 larval neuroblasts following drug treatments
| Treatment | Genotypea | Drug | No. of animals | MI (%) | Normalized MI | t | P |
|---|---|---|---|---|---|---|---|
| Aphidicolin | wt | − | 9 | 0.945 | |||
| wt | + | 4 | 0.515 | 0.545 | 2.818 | 0.025–0.01 | |
| e20 | − | 10 | 0.410 | ||||
| e20 | + | 6 | 0.268 | 0.654 | 1.224 | 0.4–0.2 | |
| a18 | − | 11 | 0.198 | ||||
| a18 | + | 4 | 0.330 | 1.665 | 1.074 | 0.4–0.2 | |
| Hydroxyurea | wt | − | 9 | 0.945 | |||
| wt | + | 4 | 0.074 | 0.078 | 5.577 | <0.001 | |
| e20 | − | 10 | 0.410 | ||||
| e20 | + | 4 | 0.394 | 0.961 | 0.116 | >0.500 | |
| a18 | − | 11 | 0.198 | ||||
| a18 | + | 4 | 1.070 | 5.398 | 5.326 | <0.001 | |
| Etoposide | wt | − | 9 | 0.945 | |||
| wt | + | 4 | 0.497 | 0.527 | 2.904 | 0.025–0.01 | |
| e20 | − | 10 | 0.410 | ||||
| e20 | + | 3 | 1.790 | 4.365 | 8.973 | <0.001 | |
| a18 | − | 11 | 0.198 | ||||
| a18 | + | 4 | 0.627 | 3.163 | 2.295 | 0.05–0.025 | |
| UV irradiation | wt | − | 9 | 0.945 | |||
| wt | + | 4 | 0.317 | 0.336 | 4.028 | 0.005–0.001 | |
| e20 | − | 10 | 0.410 | ||||
| e20 | + | 3 | 0.349 | 0.850 | 0.396 | >0.500 | |
| a18 | − | 11 | 0.198 | ||||
| a18 | + | 4 | 0.622 | 3.139 | 3.496 | 0.005–0.001 | |
| Caffeine | wt | − | 9 | 0.945 | |||
| wt | + | 3 | 0.882 | 0.934 | 0.340 | >0.500 | |
| e20 | − | 10 | 0.410 | ||||
| e20 | + | 3 | 0.368 | 0.898 | 0.258 | >0.500 | |
| a18 | − | 11 | 0.198 | ||||
| a18 | + | 4 | 0.534 | 2.698 | 2.189 | 0.05–0.025 | |
| Colchicine | wt | − | 9 | 0.945 | |||
| wt | + | 3 | 1.983 | 2.099 | 2.099 | <0.001 | |
| e20 | − | 10 | 0.410 | ||||
| e20 | + | 6 | 0.705 | 1.719 | 1.719 | 0.10–0.05 | |
| a18 | − | 11 | 0.198 | ||||
| a18 | + | 5 | 1.152 | 5.813 | 5.813 | <0.001 |
wt, wild type; e20, l(3)Rfc4e20; a18, l(3)Rfc4a18.
Treatment of wild-type larvae with DNA replication inhibitors resulted in a decreased MI as predicted: cells arrest in response, and therefore a decreased number enter mitosis (Fig. 6A). l(3)Rfc4e20 and l(3)Rfc4a18 larvae did not halt progression through the cell cycle in response to aphidicolin and hydroxyurea, as neither showed a reduction in MI. l(3)Rfc4e20 neuroblasts appeared to progress as though untreated with an unchanged MI, while l(3)Rfc4a18 neuroblasts showed a higher MI, strikingly indicating mitotic arrest. Thus, DmRfc4 mutant larvae appeared deficient in the ability to arrest cell cycle progression prior to entry into mitosis when DNA replication was perturbed.
The response of the l(3)Rfc4e20 and l(3)Rfc4a18 homozygous larvae to DNA damage was investigated following etoposide treatment or UV irradiation. Both treatments caused wild-type neuroblasts to arrest progression through the cell cycle and resulted in a corresponding drop in MI (Fig. 6B). For the DmRfc4 alleles, there was no decrease in the MI, indicating that both alleles failed to arrest the cell cycle prior to mitosis in response to DNA damage. As seen above in the activation of DNA replication checkpoints, l(3)Rfc4a18 neuroblasts again showed an elevated MI in response to both treatments while l(3)Rfc4e20 neuroblasts did so only in response to etoposide. Therefore, neither DmRfc4 allele was able to prevent the onset of mitosis in the presence of damaged DNA. Strikingly, though caffeine has no effect on the MI in wild-type cells or l(3)Rfc4e20 neuroblasts, the MI is dramatically increased in l(3)Rfc4a18 neuroblasts, demonstrating their enhanced sensitivity to this treatment (Fig. 6C).
Not all checkpoint function is abrogated in DmRfc4 mutants however. Wild-type, l(3)Rfc4e20, and l(3)Rfc4a18 larvae all exhibited an intact spindle assembly-kinetochore attachment checkpoint, as cells arrested in mitosis in response to colchicine depolymerization of microtubules (Fig. 6D). Indeed, the increase in MI achieved by l(3)Rfc4a18 neuroblasts is quite surprising (and perhaps more likely to occur as it is the more severe allele) and suggests a metaphase arrest resulting not only from entry into mitosis in the presence of chromosomal defects but also from delay in exiting mitosis.
DISCUSSION
We have shown in this study that Drosophila l(3)e20 and l(3)a18, which cause severe defects in mitotic chromosome structure, are mutant alleles of the Drosophila Rfc4 gene. RFC is known to recognize the primer-template junction and play a role in loading proliferating cell nuclear antigen to allow processive replication. In light of this, it is not surprising that mutations in DmRFC4 cause replication defects, evidenced as underreplicated polytene chromosomes and greatly diminished BrdU incorporation in larval brains. However, the observation of distinct mitotic defects is striking and unexpected and has not been previously shown. We observed PCC-like figures and PSCS in both alleles, with the PCC-like figures being the more severe manifestation of mitotic defects, having a higher frequency later in development and in the more severe l(3)Rfc4a18 allele. The PCC-like figures likely arise from cells attempting to enter mitosis prematurely without the completion of DNA replication or repair of DNA damage (these figures are positive for mitosis-specific histone H3 phosphorylation).
The mutation in l(3)Rfc4a18 is a single base change resulting in a premature stop codon. The putative DmRFC4a18 product, if synthesized, is only 45 amino acids in length, and includes only box II of the conserved RFC boxes. A protein corresponding to this could not be detected. The l(3)Rfc4e20 mutation is a 351-bp in-frame deletion and generates a truncated protein of 29 kDa, which we were able to detect by immunoprecipitation from larval extracts. Since RFC boxes II to VIII are contained in the putative DmRfc4e20 product, this polypeptide may have some residual function. However, as the mutation is lethal, some or all of the 102 C-terminal amino acids must be essential for a fully functional DmRFC4 in vivo. In vitro studies have shown that the C termini of all five of the RFC subunits are important for formation of a stable RFC complex (44, 45). The C-terminal region is highly conserved; this region of DmRFC4 has 42% identity (69% similarity) to the S. cerevisiae homologue and 70% identity (81% similarity) to the human protein.
The DmRFC4 protein is localized to zones of proliferation in wild-type larval brains. In mutant larval brains, very little if any protein could be detected, and a correspondingly low number of actively replicating cells was detected. Examination of cultured cells confirmed that the DmRFC4 protein was present in replicating nuclei and that the protein was homogeneously distributed throughout mitotic cells. The diffuse distribution of DmRFC4 throughout mitosis suggested that the mitotic defects observed in the mutants were most likely due to an indirect role for RFC in chromosome structure and behavior, hence our decision to analyze a possible role for DmRFC4 in cell cycle checkpoint function. The analysis of the fate of condensin and condensin subunits in mutants such as RFC4 and ORC2, which affect mitotic chromosome architecture in addition to playing crucial roles in DNA replication, should help shed light on the precise defects observed in chromosome structure (21, 38, 47).
We have obtained evidence that DNA structure-specific checkpoints are defective in DmRfc4 mutants. Neither DmRfc4 mutant allele was capable of responding properly by arresting cell cycle progression when DNA replication was inhibited by either hydroxyurea or aphidicolin treatment or DNA damage was induced by etoposide or UV irradiation. In all cases, the treated cells continued to progress into mitosis under circumstances where wild-type cells arrested in interphase. This study presents not only the first demonstration of a role for RFC in checkpoint control in a higher eukaryote but also the first indication that RFC4 may play the same role as that shown for the other small RFC subunits in fission and budding yeast. Mutations in ScRfc2, ScRfc5, SpRfc2, and SpRfc3 all result in defective replication as well as entry into mitosis with chromosomal abnormalities, a phenotype strikingly similar to that produced by the DmRfc4 mutations described in this report (12, 26, 28, 34, 40). The Scrfc5-1 phenotypes can be suppressed by overexpression of Tel1, Rad53, and Rad 24 (35, 39). Tel1 is an S. cerevisiae family member of the ATM group of phosphatidylinositol-related kinases, which have a fundamental role in the signaling of unreplicated or damaged DNA to prevent premature progression into mitosis (29). Rad53 is a protein kinase that acts downstream of Tel1 in the signaling cascade in S. cerevisiae (30, 41). Rad24 has now been shown to associate with the four small RFC subunits in a complex that lacks RFC1 (13, 24). Taken together, these results suggest that the small RFC subunits play a critical role in the pathway signaling incomplete DNA replication or the presence of DNA damage and that the complex functioning in the damage checkpoint pathway contains Rad24 instead of RFC1. As a Drosophila orthologue of SpRad17 and ScRad24 also exists (albeit with no evident mutations), it will be important to compare the function of this protein and its association with other RFC subunits in flies with those of proteins in fission and budding yeasts. The genetic interaction of cutlet, a Drosophila orthologue of the S. cerevisiae CHL12/CTF18 gene (which has homology with genes for the small RFC subunits), with DmRFC4 has also recently been reported (20). However, cell cycle progression in the presence of irreparable DNA damage appears to be controlled by the Mec1 kinase (orthologues being Rad3p in S. pombe and MEI-41 in flies), which may play a primary role in the S-phase damage-sensing pathway (25).
We believe the detectable effects on DNA structure checkpoints to be specific, as not all checkpoint function is abolished in DmRfc4 mutants. DmRfc4 mutant neuroblasts are able to respond to the colchicine-induced depolymerization of microtubules by arresting in mitosis like wild-type cells, and therefore the kinetochore assembly checkpoint is intact. Intriguingly, in a number of experiments, neuroblasts defective in RFC4 showed a greatly elevated MI relative to that of untreated neuroblasts, suggesting that DmRfc4 mutants must accumulate in mitosis. It is feasible that the activation of the mitotic checkpoint contributes to the elevated MI observed when DmRfc4 mutant neuroblasts pass through the replication and damage checkpoints (perhaps as a result of centrosome inactivation [36, 48]). In this way, cells are protected from aberrant chromosome segregation when DNA structure-specific checkpoint control fails.
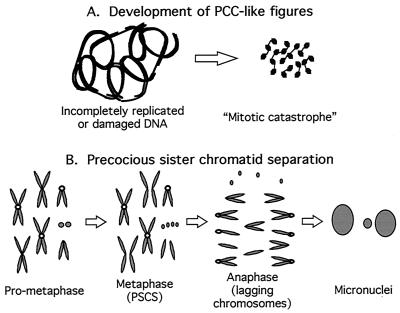
When Rfc4 is mutated in Drosophila, the transmission of signals from damaged DNA is abrogated and cells prematurely enter mitosis with either incompletely replicated or repaired DNA, evidenced as PCC-like figures or “mitotic catastrophe,” as depicted in Fig. 7A. Such figures are also observed when ATR is disrupted in mice (5); therefore it is a phenotype like those produced by disruptions of other genes important for checkpoint control. Whether such mitotic defects are evident in Drosophila mei-41 (ATM orthologue) or mus304 mutations is currently unknown. The other striking mitotic defect that we observe in DmRfc4 mutants is PSCS (diagrammed in Fig. 7B). The prematurely separated chromatids appeared to be fully replicated, as evidenced by a normal structure and the absence of gaps or decondensed regions. As this phenotype was more prevalent in younger animals carrying both alleles (which would be predicted to have more of the wild-type maternal product), this suggests that perhaps the latest-replicating regions are the first to be affected as the RFC4 protein is depleted. As a result, the structures necessary to form mitotic checkpoint machinery on kinetochores may not assemble properly, leading to inadequate kinetochore function and premature separation of sister chromatids. Intriguingly, checkpoint components have been shown to be necessary for the integrity and silencing of the late-replicating telomeres in yeast (7, 22). To further address this point, we are currently investigating the state of checkpoint components and proteins important for chromosome architecture in the DmRfc4 mutants in order to pinpoint the mechanisms that give rise to the mitotic phenotypes observed.
FIG. 7.
Model for DmRfc4 mutant phenotypes in mitosis. The mitotic phenotypes observed when DmRfc4 is mutated can be classified in two categories, that of PCC-like and that of PSCS figures. In wild-type cells, a block to replication or presence of DNA damage would result in cell cycle arrest, as the signal is passed through a phosphorylation cascade, resulting ultimately in inhibition of CDK1 activity and preventing entry into mitosis. In Rfc4 mutants, this arrest is not accomplished and cells proceed into mitosis. (A) Development of PCCs. PCC is a state of mitotic catastrophe resulting from the premature condensation of chromatin during S or G2 phase without the completion of DNA replication and/or the repair of damaged DNA. (B) Development of PSCS. This mitotic phenotype may result from abnormal progression through mitosis, possibly due to an aberrant structure at the kinetochore. A normal centromere (open circle) properly mediates spindle attachment and chromosome segregation. Improperly replicated centromeres may not accurately assemble centromeric proteins. Therefore, the lack of attachment between the sister chromatids and the spindle observed during metaphase results in lagging chromatids during anaphase. Micronuclei are formed from the inaccurately segregated chromosomes.
ACKNOWLEDGMENTS
We thank Maurizio Gatti for the l(3)e20 flies and for his encouragement in our endeavors to examine mutations affecting chromosome condensation. We are grateful to Bill Earnshaw and members of the Heck lab for lively discussions contributing to the understanding of chromosome structure. Neville Cobbe is thanked for his statistically significant skills. M.M.S.H. acknowledges Stuart MacNeill's insight into RFC phylogeny.
This work was supported by a Senior Research Fellowship in the Biomedical Sciences from the Wellcome Trust to M.M.S.H.
S. A. Krause and M.-L. Loupart contributed equally to this work.
REFERENCES
- 1.Allen B L, Uhlmann F, Gaur L K, Mulder B A, Posey K L, Jones L B, Hardin S H. DNA recognition properties of the N-terminal DNA binding domain within the large subunit of replication factor C. Nucleic Acids Res. 1998;26:3877–3882. doi: 10.1093/nar/26.17.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu J, Bousbaa H, Logarinho E, Li Z, Williams B C, Lopes C, Sunkel C E, Goldberg M L. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasina A, Price B D, Turenne G A, McGowan C H. Caffeine inhibits the checkpoint kinase ATM. Curr Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 4.Brodsky M H, Sekelsky J J, Tsang G, Hawley R S, Rubin G M. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 2000;14:666–678. [PMC free article] [PubMed] [Google Scholar]
- 5.Brown E J, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke D J, Gimenez-Abian J F. Checkpoints controlling mitosis. Bioessays. 2000;22:351–363. doi: 10.1002/(SICI)1521-1878(200004)22:4<351::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Craven R J, Petes T D. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:2378–2384. doi: 10.1128/mcb.20.7.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor-C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasika G K, Lin S-C J, Zhao S, Sung P, Tomkinson A, Lee E Y-H P. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- 10.Fogarty P, Campbell S D, Abu-Shumays R, Phalle B S, Yu K R, Uy G L, Goldberg M L, Sullivan W. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr Biol. 1997;7:418–426. doi: 10.1016/s0960-9822(06)00189-8. [DOI] [PubMed] [Google Scholar]
- 11.Gatti M, Baker B S. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- 12.Gray F C, MacNeill S A. The Schizosaccharomyces pombe rfc3+ gene encodes a homologue of the human hRFC36 and Saccharomyces cerevisiae Rfc3 subunits of replication factor C. Curr Genet. 2000;37:159–167. doi: 10.1007/s002940050514. [DOI] [PubMed] [Google Scholar]
- 13.Green C M, Erdjument-Bromage H, Tempst P, Lowndes N F. A novel Rad24 checkpoint complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 14.Hari K L, Santerre A, Sekelsky J J, McKim K S, Boyd J B, Hawley R S. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- 15.Harrison S D, Solomon N, Rubin G M. A genetic analysis of the 63E–64A genomic region of Drosophila melanogaster: identification of mutations in a replication factor C subunit. Genetics. 1995;139:1701–1709. doi: 10.1093/genetics/139.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 17.Heck M M S, Pereira A, Pesavento P, Yannoni Y, Spradling A C, Goldstein L S B. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendzel M J, Wei Y, Mancini M A, van Hooser A, Ranali T, Brinkley B R, Bazett-Jones D P, Allis C D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 19.Hubscher U, Maga G, Podust V N, editors. DNA replication accessory proteins. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 20.Jaffe A B, Jongens T A. Structure-specific abnormalities associated with mutations in a DNA replication accessory factor in Drosophila. Dev Biol. 2001;230:161–176. doi: 10.1006/dbio.2000.0117. [DOI] [PubMed] [Google Scholar]
- 21.Loupart M-L, Krause S A, Heck M M S. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol. 2000;10:1547–1556. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- 22.Matsuura A, Naito T, Ishikawa F. Genetic control of telomere integrity in Schizosaccharomyces pombe: rad3(+) and tel1(+) are parts of two regulatory networks independent of the downstream protein kinases chk1(+) and cds1(+) Genetics. 1999;152:1501–1512. doi: 10.1093/genetics/152.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moser B A, Brondello J M, Baber-Furnari B, Russell P. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol Cell Biol. 2000;20:4288–4294. doi: 10.1128/mcb.20.12.4288-4294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neecke H, Lucchini G, Longhese M P. Cell cycle progression in the presence of irreparable DNA damage is controlled by a Mec1- and Rad53-dependent checkpoint in budding yeast. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noskov V N, Araki H, Sugino A. The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4914–4923. doi: 10.1128/mcb.18.8.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao P N, Johnson R T. Mammalian cell fusion studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds N, Fantes P A, MacNeill S A. A key role for replication factor C in DNA replication checkpoint function in fission yeast. Nucleic Acids Res. 1999;27:462–469. doi: 10.1093/nar/27.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotman G, Shiloh Y. ATM: a mediator of multiple responses to genotoxic stress. Oncogene. 1999;18:6135–6144. doi: 10.1038/sj.onc.1203124. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 31.Schar P. Spontaneous DNA damage, genome instability, and cancer—when DNA replication escapes control. Cell. 2001;104:329–332. doi: 10.1016/s0092-8674(01)00220-3. [DOI] [PubMed] [Google Scholar]
- 32.Shearn A, Garen A. Genetic control of imaginal disc development in Drosophila. Proc Natl Acad Sci USA. 1974;71:1393–1397. doi: 10.1073/pnas.71.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shearn A, Rice T, Garen A, Gehring W. Imaginal disc abnormalities in lethal mutants of Drosophila. Proc Natl Acad Sci USA. 1971;68:2594–2598. doi: 10.1073/pnas.68.10.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai K K, Shimoda C, Nojima H. Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sibon O C, Kelkar A, Lemstra W, Theurkauf W E. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- 37.Sibon O C M, Stevenson V A, Theurkauf W E. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- 38.Steffensen S, Coelho P A, Cobbe N, Vass S, Costa M, Hassan B, Prokopenko S N, Bellen H, Heck M M S, Sunkel C E. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr Biol. 2001;11:295–307. doi: 10.1016/s0960-9822(01)00096-3. [DOI] [PubMed] [Google Scholar]
- 39.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugimoto K, Shimomura T, Hashimoto K, Araki H, Sugino A, Matsumoto K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc Natl Acad Sci USA. 1996;93:7048–7052. doi: 10.1073/pnas.93.14.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 42.Szabad J, Bryant P. The mode of action of “Discless” mutations in Drosophila melanogaster. Dev Biol. 1982;93:240–256. doi: 10.1016/0012-1606(82)90256-1. [DOI] [PubMed] [Google Scholar]
- 43.Taubert H, Szabad J. Genetic control of cell proliferation in female germ line cells of Drosophila: mosaic analysis of five discless mutations. Mol Gen Genet. 1987;209:545–551. doi: 10.1007/BF00331161. [DOI] [PubMed] [Google Scholar]
- 44.Uhlmann F, Cai J, Gibbs E, O'Donnell M, Hurwitz J. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J Biol Chem. 1997;272:10058–10064. doi: 10.1074/jbc.272.15.10058. [DOI] [PubMed] [Google Scholar]
- 45.Uhlmann F, Gibbs E, Cai J, O'Donnell M, Hurwitz J. Identification of regions within the four small subunits of human replication factor C required for complex formation and DNA replication. J Biol Chem. 1997;272:10065–10071. doi: 10.1074/jbc.272.15.10065. [DOI] [PubMed] [Google Scholar]
- 46.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 47.Warren W D, Steffensen S, Lin E, Coelho P, Loupart M-L, Cobbe N, Lee J, McKay M J, Orr-Weaver T, Heck M M S, Sunkel C E. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- 48.Yu K R, Saint R B, Sullivan W. The Grapes checkpoint coordinates nuclear envelope breakdown and chromosome condensation. Nat Cell Biol. 2000;2:609–615. doi: 10.1038/35023555. [DOI] [PubMed] [Google Scholar]