Abstract
Rett syndrome (RTT) is an X-linked neurodevelopmental disorder that is predominantly caused by alterations of the methyl-CpG-binding protein 2 (MECP2) gene. Disease severity and the presence of comorbidities such as gastrointestinal distress vary widely across affected individuals. The gut microbiome has been implicated in neurodevelopmental disorders such as Autism Spectrum Disorder (ASD) as a regulator of disease severity and gastrointestinal comorbidities. Although the gut microbiome has been previously characterized in humans with RTT compared to healthy controls, the impact of MECP2 mutation on the composition of the gut microbiome in animal models where the host and diet can be experimentally controlled remains to be elucidated. By evaluating the microbial community across postnatal development as behavioral symptoms appear and progress, we have identified microbial taxa that are differentially abundant across developmental timepoints in a zinc-finger nuclease rat model of RTT compared to WT. We have additionally identified p105 as a key translational timepoint. Lastly, we have demonstrated that fecal SCFA levels are not altered in RTT rats compared to WT rats across development. Overall, these results represent an important step in translational RTT research.
1. Introduction
RTT is an X-linked progressive neurodevelopmental disorder that affects up to 1 in 10,000 girls annually in the United States (Percy and Lane, 2005). RTT is characterized by seemingly typical development until 6–18 months of age followed by sometimes rapid neurodevelopmental regression. Greater than 95% of all cases of RTT harbor alterations in the methyl-CpG-binding protein 2 (MECP2) gene (Amir et al., 1999). However, severity of RTT symptoms can vary wildly across individuals due to differences in X-chromosome inactivation and specific gene mutation (Amir et al., 1999; Cuddapah et al., 2014). While the neurologic sequelae, loss of motor, cognitive, and social skills, are defining features in all Rett individuals, chronic issues related to other organ systems also characterize disease progression. A 2012 survey of nearly 1000 families of individuals with RTT revealed that comorbid gastrointestinal (GI) symptoms are present in a large majority (92%) of people with RTT (Motil et al., 2012) and represent a significant quality of life issue for long-term care. Symptoms such as constipation and oropharyngeal dysmotility predominate, however gastroparesis, gastroesophageal reflux, and biliary dysmotility may also occur. Importantly, severe sequelae of abnormal nutritional status such as low bone mineral density and fractures increase with age, suggesting an insidious disease course with respect to GI symptoms (Motil et al., 2012). Inadequate oral mechanical function surely affects nutritional intake and contributes to these sequelae. However, given that MeCP2 is expressed throughout the enteric nervous system in all gut regions (Wahba et al., 2015), it is not unreasonable to posit that changes in gut MeCP2 expression may directly contribute to GI symptoms. In fact, mechanisms whereby this may occur have recently been proposed (Wahba et al., 2015).
The trillions of bacteria that inhabit the gut are termed the gut microbiota and have long been known to impact GI disorders such as Crohn’s disease and irritable bowel syndrome (IBD) (Manichanh et al., 2006; Balsari et al., 1982). More recently, gut microbiota have been shown to be altered in a variety of neurological disorders, including Autism Spectrum Disorder (ASD) (de Theije et al., 2014), Parkinson’s Disease (Scheperjans et al., 2015), and epilepsy (Olson et al., 2018). The contribution of the gut microbiome not only to comorbid GI distress in these conditions but also disease pathogenesis itself are now being heavily investigated. For instance, a variety of animal and human studies of ASD and neuropsychiatric disorders describe a spectrum of alterations in gut microbiota as well as improvements in behavior phenotypes following treatments targeting the gut microbiome (Vuong et al., 2017). The microbiome has also been shown to be critical for appropriate brain development with respect to microglial cells (Erny et al., 2015), which function as central nervous system (CNS) immune mediators and appear to be aberrantly distributed and activated in the brains of individuals with ASD (Takano, 2015). Thus, we hypothesize that mechanisms of microbiome-mediated changes in gut/brain axis may be observed more broadly across other neurological disorders. In RTT, MECP2 mutation may directly alter GI motility, precipitating changes in the gut environment that favor certain bacterial taxonomies. Changes in microbiota ultimately affect nutrient metabolism and absorption, immune mediators, and neuroendocrine signaling (Sampson and Mazmanian, 2015), which may act in a feed-forward fashion to contribute to overall disease pathogenesis.
Previous studies have reported changes in the microbiome in patients with RTT compared to typically developing females (Strati et al., 2016) as well as changes in production of microbial metabolites such as short chain fatty acids (SCFAs) (Strati et al., 2016; Borghi et al., 2017), which are known to affect blood-brain barrier (BBB) integrity (Braniste et al., 2014) and have been implicated in ASD (MacFabe, 2015). However, the relationship between MeCP2-deficiency and the gut microbiome has remained unexplored in animal models of RTT. Given the emerging importance of the gut microbiome in neurologic disease, characterizing and understanding changes in the microbiome of animal models relative to humans with RTT is an appropriate next step to explore the role of the gut microbiome in disease pathogenesis.
RTT has historically been modeled in mice through a variety of genetic manipulations (Lombardi et al., 2015; Li and Pozzo-Miller, 2012). However, studying cognitive and social deficits such as those in RTT can represent a significant challenge in many animal models. Rats have been shown to participate in complex social interactions (Thor and Holloway, 1984) and have the ability to complete higher level cognitive tasks (Urcelay and Miller, 2010) relative to mice, offering another approach to this challenge. Recently, a novel zinc-finger nuclease model of RTT in rats was developed by Sage laboratories. In this model, the MeCP2 protein is absent due to a 71 base pair deletion in exon 4 of Mecp2. The motor, behavioral, and social deficits across development in this model have been previously described (Patterson et al., 2016; Veeraragavan et al., 2016). Importantly, development of abnormalities in MeCP2 Mecp2ZFN/+ heterozygous females begins as early as the third to fourth postnatal week, comparable to the developmental age of symptom onset in females with RTT. Severe weight, motor, and behavioral symptoms progress most profoundly in Mecp2ZFN/+ heterozygous female rats between 4 and 12 months of age, at which comparable developmental stage symptoms have generally also progressed and begun to stabilize in girls with RTT. As the microbiome changes rapidly throughout development (Voreades et al., 2014), an animal model that recapitulates developmental disease timepoints and can be easily studied at early phases of disease course is essential to characterize the impact of the microbiome on pathogenesis. The aim of the current study was to characterize the gut microbiome in a MeCP2 rat model of Rett syndrome across development.
2. Methods
2.1. Animals
All experiments were conducted in accordance with NIH guidelines and were carried out with approval from the Animal Care and Use Committee of the University of Alabama at Birmingham. All animals in the present study were bred as previously described (Patterson et al., 2016). Briefly, Sprague Dawley females lacking one copy of MeCP2 (Mecp2ZFN/+) were crossed to wildtype (WT) S100b eGFP (enhanced green fluorescent protein) males obtained from the National Bioresource Project Rat (Japan). Originally Wistar, WT S100b eGFP males were back crossed over 10 generations onto a Sprague Dawley background prior to crossing with Mecp2ZFN/+. For the current study only Mecp2ZFN/+ female rats were used as experimental animals. Mecp2ZFN/+ rats were weaned according to genotype at postnatal day 21 (PND 21). WT and Mecp2ZFN/+ experimental animals were not co-housed for this study, however, care was taken to balance number of cages across groups to prevent cage-effects. Animals were provided with food and water ad libitum, and kept under standard 12-h light-dark cycles.
2.2. Fecal Collection
Animals were individually removed from the home cage and placed into a clean, open, plastic container. Fresh fecal pellets were collected using clean tweezers and stored at −80C at time points indicated in the text.
2.3. DNA isolation
Rat fecal DNA was isolated using the DNA Fecal/Soil Microbe Miniprep Kit (Zymo Research) according to manufacturer’s instructions. For rats fecal isolation, one fecal pellet was used for each isolation. DNA was quantified and samples stored at −80C until sequenced.
2.4. Sequencing
Isolated DNA was quantitated prior to PCR and barcoded PCR amplification of the V4 region of the 16S rRNA gene (Woese and Gutell, 1989) was accomplished using degenerate primers originally taken from Caporaso et al., 2011 (Caporaso et al., 2011). We used primers as described by Kumar et al., 2014 (Kumar et al., 2014) for use on the Illumina MiSeq sequencer. PCR was carried out under conditions described by Kumar et al. PCR products were resolved on agarose gels; DNA isolated and purified using Qiagen kits; and then quantitated. The products were sequenced on the MiSeq platform, a single flowcell, single lane instrument that can generate approximately 9Gb of sequence data from our paired end 250 bp run. 2.5. 16S analysis Sequencing data was analyzed with QIIME (Caporaso et al., 2011). Forward and reverse reads were joined, and data was quality filtered and trimmed using BBMAP. Operational Taxonomic Units (OTUs) were picked using closed reference OTU picking with 97% sequence similarity. OTUs were assigned to taxonomies using a 97% similarity threshold with the greengenes database. Observed OTUs alpha diversity metrics, Bray Curtis beta diversity metrics, and individual taxonomy differences were assessed using QIIME. Rarefaction curves were generated for alpha diversity metrics (Fig. S1). OTU tables from QIIME were utilized to predict abundances of KEGG orthologs (KOs) and collapse KOs into KEGG pathways for functional analysis. KEGG pathways were analyzed and graphed using STAMP (Parks et al., 2014). Linear Discriminant effect size analysis (LEfSe) was performed with default parameters (Segata et al., 2011). OTU tables generated in QIIME were assigned LDA scores and graphed utilizing the Galaxy web application.
2.5. 16S analysis
Sequencing data was analyzed with QIIME (Caporaso et al., 2011). Forward and reverse reads were joined, and data was quality filtered and trimmed using BBMAP. Operational Taxonomic Units (OTUs) were picked using closed reference OTU picking with 97% sequence similarity. OTUs were assigned to taxonomies using a 97% similarity threshold with the greengenes database. Observed OTUs alpha diversity metrics, Bray Curtis beta diversity metrics, and individual taxonomy differences were assessed using QIIME. Rarefaction curves were generated for alpha diversity metrics (Fig. S1). OTU tables from QIIME were utilized to predict abundances of KEGG orthologs (KOs) and collapse KOs into KEGG pathways for functional analysis. KEGG pathways were analyzed and graphed using STAMP (Parks et al., 2014). Linear Discriminant effect size analysis (LEfSe) was performed with default parameters (Segata et al., 2011). OTU tables generated in QIIME were assigned LDA scores and graphed utilizing the Galaxy web application.
2.6. UPLC-MS/MS Analysis of Short Chain Fatty Acids
Standards and samples were prepared using the SOP for SCFA analysis developed in the DPMSR laboratory based on the published method of Han et al., 2015 (Han et al., 2015). A 12-point calibration curve from 10 mM to 0.975 uM for acetic acid, and 1 mM to 0.098 uM for the C3-C8 short chain fatty acids was prepared for quantitation of short chain fatty acids in study samples. The rat fecal samples were homogenized at 10 uL/mg volume to weight ratio in 50/50 v/v EtOH/water. The sample extracts were then derivatized using 1:1200 mM 3-nitrophenyl hydrazine (3-NPH) in 50% ethanol with 6% pyridine: 120 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC⋅HCl) in 50% ethanol in a 1:1 ratio of sample: derivatization reagent. At the end of the incubation, the reaction was quenched with the addition of 20× excess cold 10% ethanol in water with 1% formic acid. A 25 ul aliquot of the reaction solution from each sample was transferred and 25 ul aliquot of the stable isotope standards (SIS) solution was added. 5 uL injections were used for UPLC separation of the SCFAs, performed using a Exion AD liquid chromatograph (Sciex, Framingham, MA) with a Waters Acquity 2.1 mm × 50 mm 1.7 um BEH C18 column fitted with a Waters Acquity C18 1.7 um Vanguard guard column. Analytes were separated using a gradient from 15% solvent B to 100% solvent B in 9.5 min. Solvents A & B were 0.1% formic acid in water and acetonitrile, respectively. The total UPLC analysis time was approximately 13.5 min. The method uses electrospray ionization in negative mode introduced into a 6500+ QTrap mass spectrometer (Sciex) operating in the Multiple Reaction Monitoring (MRM) mode. MRM transitions (compound-specific precursor to product ion transitions) for each analyte and internal standard were collected over the appropriate retention time window and the data were analyzed using Skyline v19.1 (https://skyline.ms).
2.7. Statistics
Alpha diversity values were calculated with observed OUT, Chao1, Shannon, and Simpson metrics. Beta diversity analyses were calculated with bray-curtis distance. LDA score for LEfSe analysis was performed on differential abundance tables generated with QIIME. Differences between RTT and controls were assessed using Mann-Whitney U tests. All p-values for taxonomy differences and KEGG pathways are FDR adjusted. SCFA log2 values were analyzed using Mann-Whitney U tests.
3. Results
3.1. MeCP2-deficient rats have altered microbiota across development in parallel with behavioral symptoms of RTT
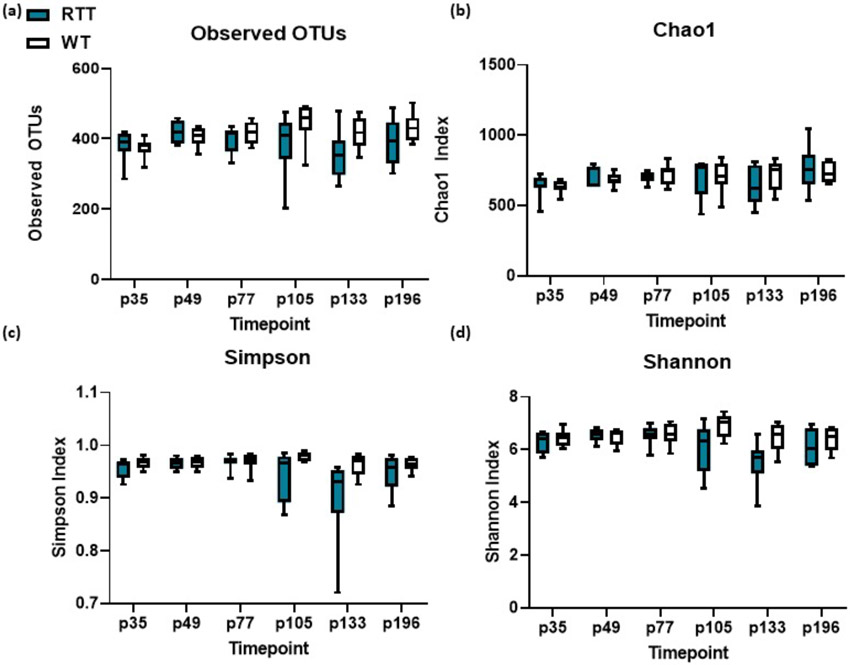
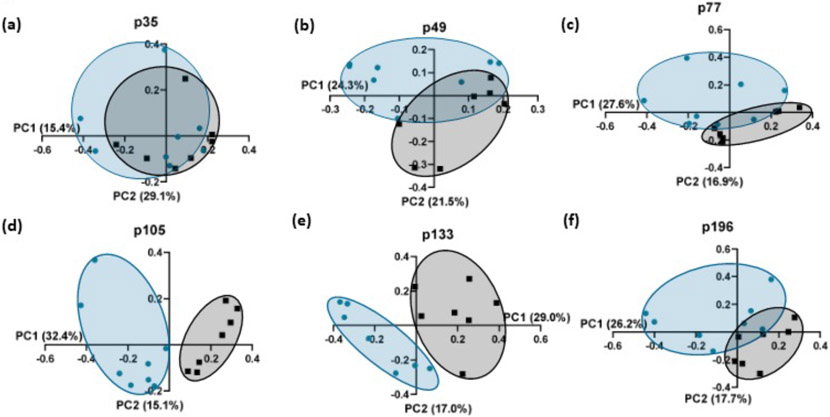
We first sought to characterize changes in microbiota at different developmental time points selected to parallel previously identified behavioral symptoms present in the female rat model, which range in appearance from p35 to p196. Alpha diversity measured by observed operational taxonomic units (OTUs) was not significantly different between the MeCP2-deficient rats at any of the selected timepoints (Fig. 1 a-d). Beta diversity measured by bray-curtis difference was also similar across groups at p35 (Fig. 2 a). However, Mecp2ZFN/+ rats begin to diverge from WT rats in beta diversity at p49, and continue to be significantly distinct through p196 (Fig. 2 b-f). Although beta diversity values separate at these timepoints, Firmicutes/Bacteroides ratio does not differ between RTT and WT rodents at any of the selected timepoints (Supp. Fig. 1). These clustering changes in beta diversity despite lack of changes in alpha diversity indicate that although the Mecp2ZFN/+ and WT rats have similar numbers of bacterial taxonomies in their guts throughout development, the makeup of bacterial taxonomies is altered across development at multiple timepoints associated with the development of behavioral symptoms in Mecp2ZFN/+ rats.
Fig. 1.
(a-d) Alpha diversity of HET and WT rats as measured by observed OTUs, Chao1, Simpson, and Shannon indices. n.s. using Mann-Whitney U tests.
Fig. 2.
(a) Beta diversity of p35 RTT (n = 8) and WT (n = 7) rats. n.s. (b) Beta diversity of p49 RTT (n = 8) and WT (n = 7) rats. p = 0.007. (c) Beta diversity of p77 RTT (n = 8) and WT (n = 7) rats. p = 0.019. (d) Beta diversity of p105 HET and WT rats. p = 0.001. (e) Beta diversity of p133 HET and WT rats. p = 0.001. (f) Beta diversity of p196 HET and WT rats. p = 0.003. All beta diversity graphs depict bray curtis distance, p values calculated using Kruskal Wallis metrics.
3.2. MeCP2-deficient rats show developmental shifts in the microbiome
In the current study, we also sought to characterize specific microbial shifts of RTT rats compared to WT rats across development as shown in Table 1. (For full taxonomies see Table S1). At p35, the gut microbiota of both Mecp2ZFN/+ and WT rats is characterized by dominance of Bacteroidetes and Firmicutes phyla, as is also apparent in the aforementioned human studies. Although behavior and metabolic symptoms begin to develop following p35, there are no specific shifts in microbial taxonomies at p35, p49, or p77 timepoints, suggesting a stable microbiome across early development.
Table 1.
Bold taxonomies were identified via literature search, and indicate taxonomies altered in the same direction in published studies in RTT patients. Table 1 depicts OTUs significantly altered in RTT rats compared to WT across developmental timepoints (Bootstrap Mann-Whitney U test, FDR adjusted). Bold taxonomies depict bacteria that were also found to be altered in the same direction in published work on the microbiome in human RTT patients.
| Age | OTU | RTT Mean |
WT Mean |
FDR p-value |
Human RTT studies |
|---|---|---|---|---|---|
| p21 | n.s | n.s | n.s | n.s | |
| p35 | n.s. | n.s. | n.s. | n.s. | |
| p49 | n.s. | n.s. | n.s. | n.s. | |
| n.s. | n.s. | n.s. | n.s. | ||
| p77 | n.s. | n.s. | n.s. | n.s. | |
| p105 | g_Turicibacter | 0.000001 | 0.000128 | 0.034 | |
| B. ovatus | 0.000506 | 0.000068 | 0.034 | genus in Borghi et al., 2017 | |
| L. ruminis | 0.000578 | 0.000024 | 0.034 | genus in Strati et al., 2016 | |
| A. municiphila | 0.000282 | 0.000676 | 0.034 | ||
| f_Mogibacteriaceae | 0.000587 | 0.003411 | 0.034 | ||
| B. uniformis | 0.002976 | 0.000874 | 0.034 | genus in Borghi et al., 2017 | |
| g_Desulfovibrio | 0.001522 | 0.004543 | 0.034 | ||
| g_Blautia | 0.068587 | 0.000649 | 0.034 | ||
| g_Oscillospira | 0.028362 | 0.094498 | 0.034 | ||
| g_Pediococcus | 0.000628 | 0.000047 | 0.044 | ||
| g_Odoribacter | 0.000311 | 0.001384 | 0.044 | ||
| g_Bacteroides | 0.066382 | 0.005045 | 0.044 | Borghi et al., 2017 | |
| f_Ruminococcaceae | 0.042262 | 0.096028 | 0.044 | Borghi et al., 2017 | |
| o_Clostridiales | 0.123888 | 0.292810 | 0.044 | ||
| 133 | n.s. | n.s. | n.s. | n.s. | |
| 196 | n.s. | n.s. | n.s. | n.s. | |
| 548 | n.s. | n.s. | n.s. | n.s. |
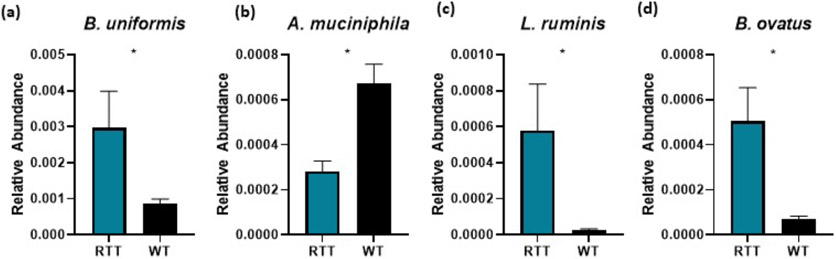
Of note, the gut microbiomes of RTT and WT rats significantly diverge at p105 across multiple diverse taxonomies. Importantly, we note shifts in Bacteroides spp., Lactobacillus spp., and family Ruminococcacaceae that mirror similar microbiome alterations in patients with RTT (Strati et al., 2016; Borghi et al., 2017) (Fig. 3 b-d). Additionally, A. muciniphila are lower in abundance in Mecp2ZFN/+ rats compared to WT (Fig. 3 a). Notably, A. muciniphila has been described as protective against epilepsy in rodents (Olson et al., 2018), and its reduction in abundance has also been reported in children with autism (Wang et al., 2011). Additionally, bacteria in genus Turicibacter were reduced in p105 RTT rats. Interestingly, reduction in Turicibacter is also reported in patients with ASD (Liu et al., 2019), and reduction in Turicibacter correlates with reduced microbiome stability in ASD patients (Berding and Donovan, 2020). Other taxonomies reduced in p105 RTT rodents included Desulfovibrio, Oscillospira, Odoribacter, and Clostridiales. Alternatively, Mogibacteriaceae, Blautia, and Pediococcus taxonomies were increased in p105 RTT rodents compared to WT.
Fig. 3.
Relative abundance of selected L7 OTUs (a) Relative abundance of B. uniformis. p = 0.034 (b) A. muciniphila. p = 0.034 (c) L. ruminis. p = 0.034. (d) B. ovatus. p = 0.034. All p values represent FDR-adjusted bootsrapped Mann-Whitney U tests of L7 relative abundances.
By p133, although beta diversity values still show separation, the microbiomes of WT and RTT rats do not significantly diverge at any level of taxonomy. This indicates that the microbial changes in RTT exist in a specific developmental window independent of age or weight related microbial shifts previously documented in rats (Flemer et al., 2017). This lack of specific taxonomy alterations persists at all of the later timepoints examined in this study. Taken together, these findings highlight p105 as an important translational timepoint as microbial community shifts at this timepoint display the most similarity to published studies in RTT patients.
3.3. Linear Discriminant Analysis reveals additional developmental shifts in the microbiome of MeCP2-deficient rats
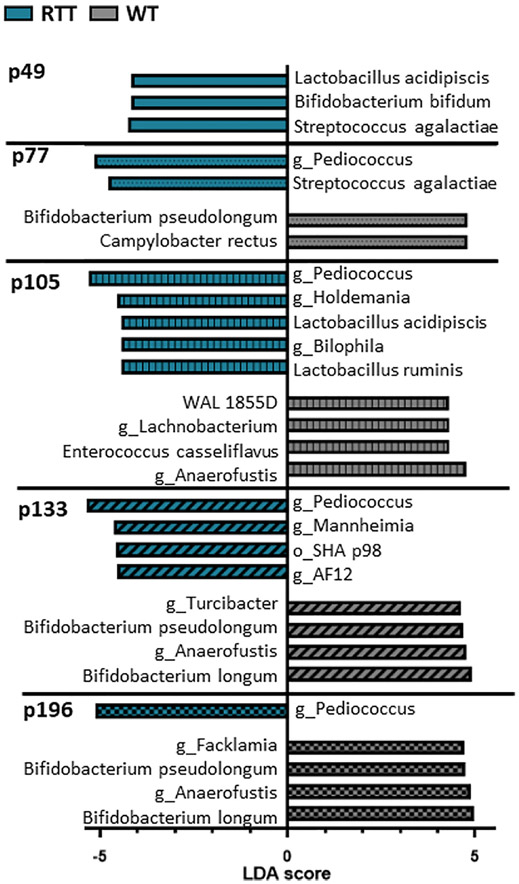
Previous studies have additionally utilized LEfSe scores to identify statistically significant differences in microbiota classifications (Segata et al., 2011) (Fig. 4, Table 2). We performed LEfSe analysis on RTT and WT rats to supplement our abundance data and further compare our findings to data on RTT patients. We found no significant differences between genotypes at the p35 timepoint, confirming that microbiota alterations due to genotype do not occur immediately following weaning. In accordance with significant alterations in beta diversity, RTT rats at p49 display enrichment in Streptococcus agalactiae, Bifidobacterium bifidum, and Lactobacillus acidipiscis. By p77 however, RTT samples begin to display reductions in Campylobacter rectus and Bifidobacterium pseudolongum compared to WT, while maintaining an enrichment in Streptococcus agalactiae and introducing an enrichment in genus Pediococcus. These enrichments are maintained through p105, and enrichments in Lactobacillus ruminis, genus Bilophila, and genus Holdemania appear at this timepoint. Importantly, these enrichments in Lactobacillus species mirror enrichments found in LefSe analyses in human RTT patients (Strati et al., 2016). Additionally, WT samples display significant enrichment in Enterococcus casseliflavus, understudied WAL 1855D, genus Anaerofustis, as well as genus Lachnobacterium. At p133, WT enrichment in Anaerofustis is maintained and WT samples are additionally enriched in Bifidobacterium longum, Bifidobacterium pseudolongum, and genus Turcibacter. RTT samples at p133 maintain enrichment in Pediococcus, but additionally are enriched in understudied bacteria SHA_98 and AF12, as well as genus Mannheimia. At p196 RTT samples are only enriched in Pediococcus, but WT samples are still enriched in Bifidobacterium longum, Bifidobacterium pseudolongum, genus Anaerofustis, and genus Facklamia.
Fig. 4.
Functional characterization of OTUs represented in the gut microbiota of RTT rats and WT rats across development. Significant OTUs have been identified by linear discriminant analysis in addition to effect size (LEfSe). Significance is represented by LDA >4. The p35 timepoint showed no significant OTUS. Teal histograms: OTUs enriched in RTT rats. Gray histograms: OTUs enriched in WT rats.
Table 2.
Bold taxonomies were identified via literature search, and indicate taxonomies altered in the same direction in published studies in RTT patients. Table 2 depicts OTUs significantly altered in RTT rats compared to WT across developmental timepoints (Lefse analysis, LDA cutoff of 4.0). Bold taxonomies depict bacteria that were also found to be altered in the same direction in LEfSe analyses in published work on the microbiome in human RTT patients.
| Age | OTU | RTT enriched or WT enriched |
LDA score |
Human RTT Studies |
|---|---|---|---|---|
| p35 | n.s. | n.s | n.s | |
| p49 | Streptococcus agalactiae | RTT | 4.26 | |
| Bifidobacterium bifidum | RTT | 4.17 | genus in Strati et al., 2016 | |
| Lactobacillus acidipiscis | RTT | 4.17 | genus in Strati et al., 2016 | |
| p77 | Campylobacter rectus | WT | 4.81 | |
| Bifidobacterium pseudolongum | WT | 4.79 | ||
| Streptococcus agalactiae | RTT | 4.77 | ||
| g_Pediococcus | RTT | 5.17 | ||
| p105 | g_Anaerofustis | WT | 4.77 | |
| Enterococcus casseliflavus | WT | 4.32 | ||
| g_Lachnobacterium | WT | 4.32 | ||
| WAL 1855D | WT | 4.30 | ||
| Lactobacillus ruminis | RTT | 4.42 | genus in Strati et al., 2016 | |
| g_Bilophila | RTT | 4.43 | ||
| Lactobacillus acidipiscis | RTT | 4.42 | genus in Strati et al., 2016 | |
| g_Holdemania | RTT | 4.56 | ||
| g_Pediococcus | RTT | 5.31 | ||
| p133 | Bifidobacterium longum | WT | 4.92 | |
| g_Anaerofustis | WT | 4.76 | ||
| Bifidobacterium pseudolongum | WT | 4.68 | ||
| g_Turicibacter | WT | 4.62 | ||
| g_AF12 | RTT | 4.54 | ||
| o_SHA_98 | RTT | 4.58 | ||
| g_Mannheimia | RTT | 4.62 | ||
| g_Pediococcus | RTT | 5.37 | ||
| p196 | Bifidobacterium longum | WT | 4.97 | |
| Anaerofustis | WT | 4.88 | ||
| Bifidobacterium pseudolongum | WT | 4.73 | ||
| Facklamia | WT | 4.71 | ||
| Pediococcus | RTT | 5.12 |
In summary, beginning at p77 RTT samples display sustained enrichment in Pediococcus as well as sustained deficiency in Bifidobacterium species. These results suggest that RTT rats may benefit from Bifidobacterium probiotic therapy in adulthood. Additionally, these results again highlight p105 as a translational timepoint at which the RTT rat microbiome most closely resembles changes that occur in the microbiomes of humans with RTT.
3.4. Impact of MecP2 mutation on fecal SCFA levels across development
As previous studies in humans with RTT showed alterations in fecal SCFA content (Strati et al., 2016; Borghi et al., 2017) and our sequencing results indicate alterations at p105 in SCFA-related microbes, we next examined the content of 12 SCFAs across development in fecal samples from p35, p105, and p196 RTT and WT rats. There were no measurable levels of 3-methyl valeric acid or octanoic acid in our samples. As expected, we found no differences in fecal SCFA levels in p35 compared to WT rodents (Table 3). However, although we report marked decreases in bacteria that produce SCFAs, such as lactic acid bacteria and Ruminococcacaceae at both p105 and p196, we additionally found no difference in any of the measured SCFAs in our WT vs our RTT samples at p105 and at p196 (Table 3). As SCFAs exhibit distinct biological gradients throughout the digestive tract (reviewed in Morrison and Preston, 2016), it is possible that fecal SCFA measurements may not be precise enough to catch site-specific alterations in SCFA content. Additionally, some SCFAs such as acetate are produced by a variety of microbiota (Miller and Wolin, 1996), indicating that the loss of specific microbes is not enough to alter overall SCFA production.
Table 3.
depicts fecal levels of acetic acid, propionic acid, isobutyric acid, butyric acid, 2-methyl butyric acid, isovaleric acid, valeric acid, isocaproic acid, and caproic acid in RTT and WT rats at p35, p105, and p196. There were no significant differences in RTT and WT samples at any developmental timepoint measured. Values depicted are log2 transformed data values.
| SCFA | Age | RTT Average pmol/mg |
RTT STE pmol/mg |
WT Average pmol/mg |
WT STE pmol/mg |
p-value |
|---|---|---|---|---|---|---|
| acetic acid | p35 | 14.75 | 0.27 | 14.68 | 0.02 | n.s. |
| p105 | 14.95 | 0.20 | 14.41 | 0.21 | n.s. | |
| p196 | 14.40 | 0.28 | 14.88 | 0.18 | n.s. | |
| propionic acid | p35 | 12.55 | 0.22 | 12.49 | 0.07 | n.s. |
| p105 | 12.78 | 0.19 | 12.13 | 0.32 | n.s. | |
| p196 | 12.12 | 0.23 | 12.66 | 0.15 | n.s. | |
| iso-butyric acid | p35 | 7.54 | 0.43 | 7.96 | 0.33 | n.s. |
| p105 | 8.15 | 0.40 | 7.50 | 0.54 | n.s. | |
| p196 | 7.55 | 0.40 | 8.38 | 0.23 | n.s. | |
| butyric acid | p35 | 11.30 | 0.49 | 11.73 | 0.13 | n.s. |
| p105 | 11.92 | 0.46 | 11.02 | 0.39 | n.s. | |
| p196 | 11.72 | 0.39 | 12.24 | 0.29 | n.s. | |
| 2-methyl butyric acid | p35 | 6.10 | 0.62 | 6.70 | 0.48 | n.s. |
| p105 | 6.65 | 0.56 | 6.31 | 0.59 | n.s. | |
| p196 | 6.24 | 0.53 | 7.29 | 0.23 | n.s. | |
| iso-valeric acid | p35 | 6.46 | 0.47 | 7.04 | 0.40 | n.s. |
| p105 | 7.09 | 0.50 | 6.60 | 0.58 | n.s. | |
| p196 | 6.39 | 0.40 | 7.42 | 0.24 | n.s. | |
| valeric acid | p35 | 7.77 | 0.66 | 8.67 | 0.19 | n.s. |
| p105 | 8.71 | 0.48 | 7.79 | 0.70 | n.s. | |
| p196 | 8.19 | 0.37 | 9.18 | 0.25 | n.s. | |
| iso-caproic acid | p35 | 2.59 | 0.38 | 1.68 | 0.22 | n.s. |
| p105 | 2.42 | 0.51 | 2.03 | 0.31 | n.s. | |
| p196 | 2.29 | 0.40 | 1.91 | 0.36 | n.s. | |
| caproic acid | p35 | 5.59 | 1.37 | 5.47 | 1.09 | n.s. |
| p105 | 5.28 | 1.42 | 3.29 | 0.70 | n.s. | |
| p196 | 3.00 | 0.53 | 3.74 | 0.70 | n.s. |
3.5. Timeline of microbial, behavioral, and motor shifts in female RTT rats
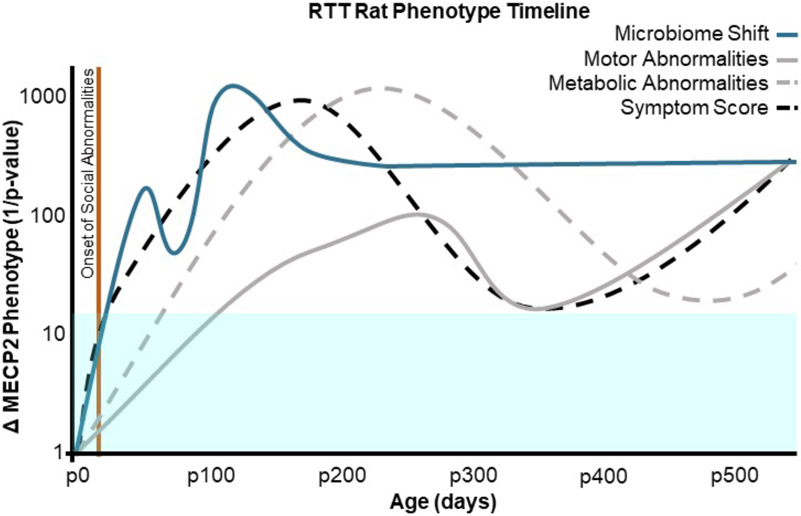
Rat models of RTT have previously been utilized to assess the longitudinal effects of MECP2 mutations on neural, motor, and metabolic symptoms. Fig. 5 (Supplementary table 2) depicts the timeline of gut microbiome shifts in female RTT rats in the current study compared to previously published longitudinal data on other RTT behavioral characteristics. Published work from our lab shows changes in brain weight in female RTT rats beginning at p21 and persisting through p540 (Patterson et al., 2016). Previous studies have additionally reported social abnormalities in the three chamber test in female RTT rat models as early as p35 (Wu et al., 2016). Although microbiome alteration has been reported to drive behavioral symptoms in rodent models of ASD (Sharon et al., 2019), and a healthy gut microbiome is essential for typical social development in rodents (Arentsen et al., 2015; Desbonnet et al., 2014), it is clear that neural correlates of RTT appear before significant microbiome alterations in female rat models of RTT.
Fig. 5.
Timeline of microbiome shifts in the current study compared to previously published longitudinal data on neural, motor, and metabolic abnormalities in female RTT rats. Microbiome shifts here are defined by Bray-Curtis beta diversity. Metabolic abnormalities describe severity of weight difference from WT and were taken from Patterson et al., 2016. Motor abnormalities here depict an average score on all catwalk categories measured in Patterson et al., 2016. Symptom score describes overall wellness described by Patterson et al., 2016. Onset of social abnormalities describes the age in Wu et al., 2016 performed the three chamber task on RTT and WT females rats and found significant differences in socialization in RTT rats. All values are inverse p-value scores indicating statistical significance. The blue box indicates a significance cutoff value of p = 0.05, all lines within this box indicate no statistically significant differences from WT rats.
Changes in body weight are known to affect the gut microbiome in both humans and rodents (Palleja et al., 2016; Ussar et al., 2015). Previous work from our lab and others shows weight gain in female RTT rats compared to WT beginning at p77 and persisting through late adulthood, although WT rodents do also experience considerable weight gain by p360 (Patterson et al., 2016). It is likely that weight gain drives later timepoint microbiome shifts, as body weight difference from WT is most robust at p196, and microbial changes also fluctuate at p196. However, it is possible that microbiome alterations at earlier timepoints actually drive some of these metabolic abnormalities in RTT rodents, as studies in germ free rodents indicate that the gut microbiome regulates weight gain (Bäckhed et al., 2004).
Difficulty walking is another common symptom of RTT in humans. Previous studies have reported rotor rod task deficiencies in female RTT et al., 2016). As rotor rod tasks do not account for complex gait changes, Patterson et al. also reported CatwalkTM gait analysis. Fig. 5 depicts an averaged score of female Catwalk gait analysis in comparison to microbial shifts identified in the current study. Most gait changes in female RTT rats were observed to onset at p180 and persist into late adulthood. These motor abnormalities largely appear after changes in the gut microbiome, suggesting a possible role for microbial community changes in affecting severity of motor symptoms in RTT rats.
4. Discussion
The human RTT microbiome has previously been characterized by two groups (Strati et al., 2016; Borghi et al., 2017). Previous studies indicated that the microbiomes of humans with RTT differ significantly in alpha and/or beta diversity compared to those of healthy controls (Strati et al., 2016; Borghi et al., 2017). In the current study, we found that Mecp2ZFN/+ differed from WT rats only in beta diversity measures. This indicates that Mecp2ZFN/+ rats experience changes in the diversity, but not number, of the taxonomies present compared to WT rats, suggesting an obvious potential effect on the gut/brain axis through changes in the microenvironment. In a large cohort of humans with RTT, Strati et al. found decreases in the relative abundance of Bacteroidetes in RTT compared to healthy controls, as well as increased Firmicutes/ Bacteroidetes ratio in RTT. Strati et al. also characterized RTT through genus level changes in the relative abundance of Actinomyces, Bifidobacterium, Clostridium XIVa Eggerthella, Enterococcus, Erysipelotrichaceae incertae sedis, Escherichia/Shigella, and Megasphaera. Borghi et al. found that although a cohort of humans with RTT had similar phylum level microbial communities to controls, RTT participants differed in family Bacteroidaceae as well as Clostridium and Suterella species, and that differences in Bacteroidaceae correlate with disease severity.
In the current study, we observed broad taxonomy shifts in the microbiomes of Mecp2ZFN/+ rats compared to WT rats beginning at p49, persisting through p105, and re-emerging at p196. Specifically, at p105, we observed significant changes in the abundance of B. ovatus, B. uniformis, L. ruminis, and A. muciniphila in Mecp2ZFN/+ rats compared to WT. Excitingly, these findings mirror alterations in similar bacterial taxonomies found in published studies of humans with RTT, humans with ASD, and rodent models of epilepsy. LefSe analysis also revealed significant differences in species in the genus Bifidobacterium as well as species in the genus Lactobacillus, further mirroring published findings in humans (Borghi et al., 2017). In contrast, we show decreases in Desulfovibrio, Odoribacter, Clostridiales, and Oscillospira, and increases in Blautia in p105 RTT rats compared to WT, while studies in humans have indicated that these species are altered in the opposite direction in patients with ASD (Tomova et al., 2015; Zhai et al., 2019; Coretti et al., 2017; Luna et al., 2017). This may point to differential microbiome profiles in RTT compared to ASD. Specifically, some of these microbiome differences may be due to sex-specific effects, as the current study examines a female model of RTT while many published studies on ASD describe male patients, and Oscillospira has been noted to change in abundance in male versus female ASD rodent models (Coretti et al., 2017).
LEfSe analysis revealed additional taxonomic differences in RTT rodents compared to WT. Specifically, RTT p49 samples begin to show an enrichment in L. acidipiscis, B. bifidum, and S. agalactiae. Interestingly, Streptococcus agalactiae is classified as a group B streptococcus (GBS) pathogen, and GBS exposure during gestation can lead to ASD-like behavioral symptoms in male rats (Allard et al., 2017) although it is unclear if adulthood GBS infection has effects on behavior. Over-abundance of S. agalactiae continues through p77, and a persistent overabundance of Pediococcus appears at this timepoints. Additionally, a reduction in Bifidobacterium and Campylobacter appear at p77, highlighting the possible benefits on Bifidobacterium-based probiotics in RTT rats beginning at this timepoint. P105 still represents the most severe microbiome alterations in the RTT rats, highlighted by additional alterations in Lactobacillus species which mirror alterations in humans with RTT (Strati et al., 2016). Additionally, a reduction in Anaerofustis begins at p105 and persists through all measured timepoints.
SCFAs are important metabolic products of carbohydrate and protein breakdown. Based on LDA effect size (LEfSe) in this study, several SCFA-producing bacterial classifications are reduced in RTT rats. Strati et al. and Borghi et al. also identified similar changes in LefSe score and abundance in Lactobacillus. Although increases in stool SCFA concentration in humans with RTT has been previously noted by Strati et al. and Borghi et al., and more broadly been implicated in ASD (MacFabe, 2015) we found no alterations in fecal SCFA levels across development in female RTT rats. SCFAs exhibit distinct biological gradients across various mucosal surfaces (reviewed in Morrison and Preston 2016), bringing to light the possibility that fecal SCFA measurements may not be precise enough to catch site-specific alterations in SCFA content. Additionally, genes encoding SCFA production are spread out across a wide variety of bacteria (Miller and Wolin, 1996), indicating that the loss of specific microbes may not be enough to alter fecal SCFA levels. Lastly, diet has been shown to have a large impact on fecal SCFA concentration (Flint et al., 2015), and our rodents were not fed a humanized diet, which could explain differences in human and rodent fecal SCFA levels. Whether or not differences might be identified at other time points corresponding to behavioral changes in the rat model or in rodents on humanized diets remains to be elucidated.
Finally, we compared the timeline of disease progression in previously published work on female RTT rats to our findings on developmental microbiome shifts. Social abnormalities in female RTT rodents compared to WT appear before the onset on microbiome changes (Miller and Wolin, 1996). The current study did not examine pre-weaning developmental timepoints, nor did we examine the maternal microbiome, so there is a possibility that microbial changes occur outside of the measured timepoints. The majority of microbiome alterations occur before the peaks of metabolic and motor symptoms, indicating the possible role of host-microbiota interaction on disease progression in RTT rats. Experimental manipulation of the gut microbiota at the key timepoints described in the current study is necessary to further elucidate the role of the gut microbiome in the severity and progression of RTT.
Supplementary Material
Acknowledgements
The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: Comprehensive Cancer Center (P30AR050948), Center for Clinical Translational Science (UL1TR001417), University Wide Institutional Core, Heflin Center for Genomic Sciences and Microbiome Center. Fecal short chain fatty acids (SCFA) were measured in the Duke Proteomics and Metabolomics Shared Resource.
Abbreviations:
- RTT
Rett syndrome
- MECP2
methyl-CpG-binding protein 2
- ASD
Autism Spectrum Disorder
- SCFA
short chain fatty acid
- IBD
Irritable bowel syndrome
- GI
gastrointestinal
- CNS
Central Nervous System
- BBB
blood brain barrier
- WT
wild type
- OUT
operational taxonomic unit
- QIIME
Quantitative Insights Into Microbial Ecology
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- LefSe
Linear discriminant analysis effect size
Footnotes
Ethical Statement
All experiments were conducted in accordance with NIH guidelines, the U.K. Animals (Scientific Procedures) Act 1986, and were carried out with approval from the Animal Care and Use Committee of the University of Alabama at Birmingham.
References
- Allard M-J, Bergeron JD, Baharnoori M, Srivastava LK, Fortier L-C, Poyart C, et al. , 2017. A sexually dichotomous, autistic-like phenotype is induced by group B streptococcus maternofetal immune activation. Autism Res 10 (2), 233–245. [DOI] [PubMed] [Google Scholar]
- Amir RE, den Veyver IBV, Wan M, Tran CQ, Francke U, Zoghbi HY, 1999. October. Rett syndrome is caused by mutations in X-linked MECP2 , encoding methyl-CpG-binding protein 2. Nat. Genet 23 (2), 185–188. [DOI] [PubMed] [Google Scholar]
- Arentsen T, Raith H, Qian Y, Forssberg H, Diaz Heijtz R., 2015. Host microbiota modulates development of social preference in mice. Microb. Ecol. Health Dis 26, 29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. , 2004. November 2. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A 101 (44), 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsari A, Ceccarelli A, Dubini F, Fesce E, Poli G, 1982. July. The fecal microbial population in the irritable bowel syndrome. Microbiologica 5 (3), 185–194. [PubMed] [Google Scholar]
- Berding K, Donovan SM, 2020. January. Dietary patterns impact temporal dynamics of fecal microbiota composition in children with Autism Spectrum Disorder. Frontiers in nutrition 6, 193, 10 [cited 2020 Aug 14];6. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6968728/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacherjee A, Winter MK, Eggimann LS, Mu Y, Gunewardena S, Liao Z, et al. , 2017. December 29. Motor, somatosensory, Viscerosensory and Metabolic impairments in a heterozygous female rat model of Rett Syndrome. Int. J. Mol. Sci 19 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi E, Borgo F, Severgnini M, Savini MN, Casiraghi MC, Vignoli A, 2017. February 7. Rett Syndrome: A Focus on Gut Microbiota. Int J Mol Sci [Internet] 18 (2) [cited 2019 Aug 9]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5343879/.10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY. The Gut Microbiota Mediates the Anti-Seizure Effects of the Ketogenic Diet. Cell. 2018. 14;173(7):1728–1741.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M, et al. , 2014. November 19. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med 6 (263), 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. , 2011. March 15. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108 (Supplement 1), 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coretti L, Cristiano C, Florio E, Scala G, Lama A, Keller S, et al. , 2017. March 28. Sex-related alterations of gut microbiota composition in the BTBR mouse model of autism spectrum disorder. Scientific reports 7, 45356 [cited 2020 Aug 14];7. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5368984/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah VA, Pillai RB, Shekar KV, Lane JB, Motil KJ, Skinner SA, et al. , 2014. March. Methyl-CpG-binding protein 2 (MEPC2) mutation type is associated with disease severity in Rett syndrome. J. Med. Genet 51 (3), 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF, 2014. February. Microbiota is essential for social development in the mouse. Mol. Psychiatry 19 (2), 146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. , 2015. July. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci 18 (7), 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemer B, Gaci N, Borrel G, Sanderson IR, Chaudhary PP, Tottey W, et al. , 2017. June 6. Fecal microbiota variation across the lifespan of the healthy laboratory rat. Gut Microbes 8 (5), 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP, Louis P, 2015. February. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc 74 (1), 13–22.40. 13. [DOI] [PubMed] [Google Scholar]; Takano T Role of Microglia in Autism: Recent Advances. Dev Neurosci. 2015;37(3):195–202. [DOI] [PubMed] [Google Scholar]
- Han Jun, Lin Karen, Sequeira Carita, Borchers Christoph H., January 7, 2015. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 854, 86–94. 10.1016/j.aca.2014.11.015. [DOI] [PubMed] [Google Scholar]
- Kumar A, Grover S, Batish VK, 2014. September 1. A multiplex PCR assay based on 16S rRNA and hly for rapid detection of L. monocytogenes in Milk. Food Measure 8 (3), 155–163. [Google Scholar]
- Li W, Pozzo-Miller L, 2012. Beyond widespread Mecp2 deletions to model Rett syndrome: conditional Spatio-temporal knockout, single-point mutations and transgenic rescue mice. Autism-Open Access. 2012 (Suppl. 1), 005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Li J, Wu F, Zheng H, Peng Q, Zhou H, 2019. January 29. Altered composition and function of intestinal microbiota in autism spectrum disorders: a systematic review. Transl Psychiatry [Internet] 9 (1), 43 [cited 2020 Aug 14];9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6351640/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi LM, Baker SA, Zoghbi HY, 2015. August 3. MECP2 disorders: from the clinic to mice and back. J. Clin. Invest 125 (8), 2914–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RA, Oezguen N, Balderas M, Venkatachalam A, Runge JK, Versalovic J, et al. , 2017. March. Distinct microbiome-Neuroimmune signatures correlate with functional abdominal pain in children with autism Spectrum disorder. Cell Mol Gastroenterol Hepatol. 3 (2), 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFabe DF, 2015. Enteric short-chain fatty acids: microbial messengers of metabolism, mitochondria, and mind: implications in autism spectrum disorders. Microb. Ecol. Health Dis 26, 28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. , 2006. February. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55 (2), 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TL, Wolin MJ, 1996. May. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl. Environ. Microbiol 62 (5), 1589–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DJ, Preston T, 2016. May 3. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7 (3), 189–200. 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motil KJ, Caeg E, Barrish JO, Geerts S, Lane JB, Percy AK, et al. , 2012. September. Gastrointestinal and nutritional problems occur frequently throughout life in girls and women with Rett syndrome. J. Pediatr. Gastroenterol. Nutr 55 (3), 292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY, 2018. The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 14 (173 (7)), 1728–1741.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleja A, Kashani A, Allin KH, Nielsen T, Zhang C, Li Y, et al. , 2016. June 15. Roux-en-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Medicine. 8 (1), 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG, 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson KC, Hawkins VE, Arps KM, Mulkey DK, Olsen ML, 2016. August 1. MeCP2 deficiency results in robust Rett-like behavioural and motor deficits in male and female rats. Hum. Mol. Genet 25 (15), 3303–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy AK, Lane JB, 2005. September 1. Rett syndrome: model of neurodevelopmental disorders. J. Child Neurol 20 (9), 718–721. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Mazmanian SK, 2015. May 13. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 17 (5), 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheperjans F, Aho V, Pereira PAB, Koskinen K, Paulin L, Pekkonen E, et al. , 2015. March. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord Off J Mov Disord Soc. 30 (3), 350–358. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. , 2011. June 24. Metagenomic biomarker discovery and explanation. Genome Biol. 12 (6), R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Cruz NJ, Kang D-W, Gandal MJ, Wang B, Kim Y-M, et al. , 2019. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 30 (177(6)), 1600–1618.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, et al. , 2016. July 30. Altered gut microbiota in Rett syndrome. Microbiome 4 (1), 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, 2015. Role of microglia in autism: recent advances. Dev. Neurosci 37 (3), 195–202. [DOI] [PubMed] [Google Scholar]
- de Theije CGM, Wopereis H, Ramadan M, van Eijndthoven T, Lambert J, Knol J, et al. , 2014. March. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav. Immun 37, 197–206. [DOI] [PubMed] [Google Scholar]
- Thor DH, Holloway WR, 1984. Social play in juvenile rats: a decade of methodological and experimental research. Neurosci. Biobehav. Rev 8 (4), 455–464. [DOI] [PubMed] [Google Scholar]
- Tomova A, Husarova V, Lakatosova S, Bakos J, Vlkova B, Babinska K, et al. , 2015. January. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav 138, 179–187. [DOI] [PubMed] [Google Scholar]
- Urcelay GP, Miller RR, 2010. January. On the generality and limits of abstraction in rats and humans. Anim. Cogn 13 (1), 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, et al. , 2015. September 1. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 22 (3), 516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeraragavan S, Wan Y-W, Connolly DR, Hamilton SM, Ward CS, Soriano S, et al. , 2016. Loss of MeCP2 in the rat models regression, impaired sociability and transcriptional deficits of Rett syndrome. Hum. Mol. Genet 01 (25(15)), 3284–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voreades N, Kozil A, Weir TL, 2014. Diet and the development of the human intestinal microbiome. Front Microbiol [Internet] 146 (6), 1564–1572 [cited 2019 Aug 9];5. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2014.00494/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong HE, Yano JM, Fung TC, Hsiao EY, 2017. The microbiome and host behavior. Annu. Rev. Neurosci 40 (1), 21–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba G, Schock SC, Claridge E, Bettolli M, Grynspan D, Humphreys P, et al. , 2015. MeCP2 in the enteric nervous system. Neurogastroenterol. Motil 27 (8), 1156–1161. [DOI] [PubMed] [Google Scholar]
- Wang LW, Tancredi DJ, Thomas DW, 2011. June. The prevalence of gastrointestinal problems in children across the United States with autism spectrum disorders from families with multiple affected members. J Dev Behav Pediatr JDBP. 32 (5), 351–360. [DOI] [PubMed] [Google Scholar]
- Woese CR, Gutell RR, 1989. May. Evidence for several higher order structural elements in ribosomal RNA. Proc. Natl. Acad. Sci. U. S. A 86 (9), 3119–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhong W, Cui N, Johnson CM, Xing H, Zhang S, et al. , 2016. Characterization of Rett syndrome-like phenotypes in Mecp2-knockout rats. J. Neurodev. Disord 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Cen S, Jiang J, Zhao J, Zhang H, Chen W, 2019. April 1. Disturbance of trace element and gut microbiota profiles as indicators of autism spectrum disorder: a pilot study of Chinese children. Environ. Res 171, 501–509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.