Abstract
Background
The aim of this study was to evaluate whether histone deacetylase 4 S246/467/632A mutant (m-HDAC4) has enhanced function at histone deacetylase 4 (HDAC4) to attenuate cartilage degeneration in a rat model of osteoarthritis (OA).
Methods
Chondrocytes were infected with Ad-m-HDAC4-GFP or Ad-HDAC4-GFP for 24 h, incubated with interleukin-1β (IL-1β 10 ng/mL) for 24 h, and then measured by RT-qPCR. Male Sprague-Dawley rats (n = 48) were randomly divided into four groups and transduced with different vectors: ACLT/Ad-GFP, ACLT/Ad-HDAC4-GFP, ACLT/Ad-m-HDAC4-GFP, and sham/Ad-GFP. All rats received intra-articular injections 48 h after the operation and every 3 weeks thereafter. Cartilage damage was assessed using radiography and Safranin O staining and quantified using the OARSI score. The hypertrophic and anabolic molecules were detected by immunohistochemistry and RT-qPCR.
Results
M-HDAC4 decreased the expression levels of Runx-2, Mmp-13, and Col 10a1, but increased the levels of Col 2a1 and ACAN more effectively than HDAC4 in the IL-1β-induced chondrocyte OA model; upregulation of HDAC4 and m-HDAC4 in the rat OA model suppressed Runx-2 and MMP-13 production, and enhanced Col 2a1 and ACAN synthesis. Stronger Safranin O staining was detected in rats treated with m-HDAC4 than in those treated with HDAC4. The resulting OARSI scores were lower in the Ad-m-HDAC4 group (5.80 ± 0.45) than in the Ad-HDAC4 group (9.67 ± 1.83, P = 0.045). The OARSI scores were highest in rat knees that underwent ACLT treated with Ad-GFP control adenovirus vector (14.93 ± 2.14, P = 0.019 compared with Ad-HDAC4 group; P = 0.003 compared with Ad-m-HDAC4 group). Lower Runx-2 and MMP-13 production, and stronger Col 2a1 and ACAN synthesis were detected in rats treated with m-HDAC4 than in those treated with HDAC4.
Conclusions
M-HDAC4 repressed chondrocyte hypertrophy and induced chondrocyte anabolism in the nucleus. M-HDAC4 was more effective in attenuating articular cartilage damage than HDAC4.
Keywords: Histone deacetylase 4 S246/467/632A mutant, Chondrocyte hypertrophy, Nucleus translocation, Osteoarthritis
Background
Osteoarthritis (OA) is a major cause of chronic pain and joint dysfunction in the elderly [1]. Currently, there are no effective treatments to delay the progression of OA. The pathological changes in OA include progressive articular cartilage disruption and osteophyte formation [2]. Most OA studies focus on articular cartilage because cartilage damage is the major pathologic feature of OA.
Studies have demonstrated that chondrocyte hypertrophy plays a significant role in the cartilage degeneration of OA. Hypertrophic chondrocytes lose the synthesis ability of type II collagen (Col 2a1) and aggrecan (ACAN) and then synthetize cartilage matrix degrading enzymes. Matrix metalloproteinase-13 (MMP-13) is the main matrix degrading enzyme, contributing to cartilage matrix degradation and cartilage damage [3, 4].
Runx-2, a key regulator of chondrocyte hypertrophy, was increased in OA cartilage. Histone deacetylase 4 (HDAC4) suppresses chondrocyte hypertrophy by repressing the transcriptional activation of Runx-2, while HDAC4 is decreased in OA cartilage [5]. A previous study indicated that overexpression of HDAC4 in IL-1β-stimulated chondrocytes decreased the expression levels of Runx-2, Mmp-13, and upregulation of HDAC4 in articular cartilage attenuated OA progression in a rat OA model [6]. HDAC4 regulates gene expression by locating in the nucleus. However, the transduction of HDAC4 is mainly localized in the cytoplasm [7]. A significant feature of HDAC4 is translocating between the nucleus and cytoplasm of cells. Normally, HDAC4 is located in the cytoplasm, binding to 14-3-3 proteins [8, 9]. When the three serine residues (S246/467/632A) of HDAC4 are dephosphorylated, they detach from the 14-3-3 proteins and are translocated in the nucleus [10]. Data from our laboratory demonstrated that the HDAC4 S246/467/632A mutant (m-HDAC4) loses the ability to bind to 14-3-3 proteins and enters the nucleus to regulate gene expression in chondrocytes [7]. Thus, m-HDAC4, which is mainly located in the nucleus, may have a superior function to wild-type HDAC4 to repress chondrocyte hypertrophy during OA.
In the present study, we established in vitro and in vivo OA models, which were induced by IL-1β stimulation and rat anterior cruciate ligament transection (ACLT), respectively, to investigate the chondroprotective effect of m-HDAC4.
Methods
This study was approved by the Institutional Animal Welfare Committee of Shanxi Medical University. All methods were carried out in accordance with ARRIVE guidelines.
Construction and purification of adenoviral vectors
An adenoviral vector encoding m-HDAC4-GFP, HDAC4-GFP (Ad-m-HDAC4-GFP, Ad-HDAC4-GFP) were constructed and purified by Genechem company (GCPA0154819, Shanghai, China). Ad-GFP is a negative control.
Culture of rat costal chondrocyte and gene delivery in vitro
Costal chondrocytes were obtained from thorax of newborn Sprague-Dawley (SD) rats as as previously described [11]. Brifely, the ribs were dissected from the rat thorax and predigested with 0.2% collagenase II for 1 h,and further digested with 0.05% collagenase II for 3 h. The chondrocytes were cultured in DEM/F-12 medium (Hyclone, South Logan, UT, USA) containing 10% fetal bovine serum (Gibco, NSW, Australia). At passage 2, chondrocytes were divided into three group and treated with 200 multiplicities of infection (MOI) of Ad-m-HDAC4-GFP, Ad-HDAC4-GFP, or Ad-GFP (1 × 109 plaque-forming units [PFUs]/mL) for 24 h. The subcellular localization of HDAC4 and m-HDAC4 was determined by fluorescence microscopy (Leica DM i8, Wetzlar, Germany). The chondrocytes were then treated with 10 ng/mL IL-1β for 24 h (in vitro OA model). Total mRNA was isolated using real-time qPCR.
Rat ACLT OA model and ad-m-HDAC4, ad-HDAC4 intra-articular injection
Two-month-old male SD rats (n = 48) with healthy appearance, appetite and activity were purchased from the Shanxi Medical University Experimental Animal Department, each rat weighing about 230 g. All the experimental procedures were performed in the Shanxi Medical University Experimental Animal Department. During the experiment, all rats were housed in a temperature- and humidity-controlled environment with a 12 h light/12 h dark cycle. The indoor temperature was controlled at 22 °C-26°Cand the humidity was kept at 50-60%. The rats were conventional feeding with standard rat chow. All rats were adapted for 1 week prior to the operation. During operation, the rats were anesthetized by an intraperitoneal injection of 0.3% pentobarbital sodium (1 ml/100 g). ACLT and sham operations were performed on the right rat knees, as described previously [12]. The rats were randomly divided into four groups by random number table (n = 12 per group): (1) ACLT+Ad-GFP; (2) ACLT+Ad-HDAC4-GFP;(3) ACLT+Ad-m-HDAC4-GFP;and (4) sham+Ad-GFP. We calculated the sample size based on the range of degrees of freedom (DF). The acceptable range of degrees of freedom (DF) for the error term in an analysis of variance (ANOVA) is between 10 to 20. n = DF/k + 1,where k = number of groups, n = number of subjects per group. Therefore, the number of animals in each group ranges from 2 to 4. However, according to our experimental protocol and preliminary experiment, four femoral condyle cartilages were pooled together to achieve the minimum sample size required for RT-qPCR.In order to meet the statistical power, and make the experimental results more reliable, we prepared 3 pooled samples per group, so there were 12 rats in each group. Ad-HDAC4-GFP, Ad-m-HDAC4-GFP, and Ad-GFP were intra-articularly injected 48 h after operation, and every 3 weeks thereafter(1 × 109 PFUs per knee). Every random four rats in each cage were kept and drank freely. All rats were euthanized by intraperitoneal injection of overdose of pentobarbital sodium 2 months after operation. Throughout the study period, the researchers responsible for animal raising were blind to all subsequent operations involving animals, and the researchers conducting the experiment did not know the grouping.
Radiography
Rat knee anteroposterior and lateral radiographs were taken using a small-animal X-ray radiography system (UltraFocus, Faxitron, Tucson, AZ, USA) to evaluate OA changes 2 months after operation. The images were taken using “Automatic Exposure Control”.
Histology
The rat right tibial plateaus were harvested and fixed in 10% formalin for 48 h. Samples were immersed in a 10% EDTA solution for decalcification for 6 weeks, the EDTA solution was renewed once a week. Then the tibial plateaus were cut into two approximately equal halves along the frontal plane, and each half was embedded in a paraffin block. Six-μm frontal sections were cut at 0, 200, 400 μm intervals, and Safranin O/Fast Green staining was performed for three sections from each interval. Cartilage lesions were evaluated by two observers following the Osteoarthritis Research Society International (OARSI) grading system, and the scores were averaged for each rat.
Immunohistochemistry (IHC)
IHC was performed to detect the distribution of Runx-2, MMP-13, and type II collagen in cartilage sections. The knee joint slides were deparaffinized with xylene, rehydrated with ethanol of different concentrations. Endogenous peroxidase was blocked with 3% hydrogen peroxide for 10 min, and then digested with 0.1% trypsin for 30 min at 37 °C. 5% BSA blocking buffer was used to block nonspecific protein binding, and the sections were then incubated with a primary antibody against rat Runx-2 (bs-1134R; Bisso, Beijing, China), MMP-13 (ab39012; Abcam, Cambridge, UK), or type II collagen (ab34712; Abcam, Cambridge, UK) at 4 °C overnight. Thereafter, the sections were treated with horseradish peroxidase (HRP)-conjugated secondary antibody for 30 min at 37 °C and developed using a DAB chromogen. Images were taken with an automatic digital slide scanner (Pannoramic MIDI, 3DHISTECH, Budapest, Hungary).
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated from rat femoral condyle cartilage using the TRIzol™ Reagent. Four femoral condyle cartilages were dissected and pooled together, with three pooled cartilages per group. Total RNA was reverse transcribed to complementary DNA (cDNA) using PrimeScript™ RT Master Mix kit (TAKARA, Shiga, Japan), and RT-qPCR was performed using TB Green™ PCR Kit (TAKARA) with a two-step Real-Time PCR System (Applied Biosystems™ QuantStudio™ 6 Flex, Carlsbad, CA, USA). Relative transcript levels were calculated using the 2-ΔΔCt method, as previously described [13]. Primer sequences are listed in Table 1.
Table 1.
Sequences of primers
| Gene | Sequence(5′-3′) |
|---|---|
| rat Col2a1 |
F: GAGGGCAACAGCAGGTTCAC R: TGTGATCGGTACTCGATGATGG |
| rat ACAN |
F: CTGATCCACTGTCCAAGCACCATG R: ATCCACGCCAGGCTCCACTC |
| rat Col10a1 |
F: GGATGCCTCTTGTCAGTGCTAACC R: TCATAGTGCTGCTGCCTGTTGTAC |
| rat MMP-13 |
F: AACCAGATGTGGAGTGCCTGATG R:CACATCAGACCAGACCTTGAAGGC |
| rat Runx-2 |
F: AACAGCAGCAGCAGCAGCAG R: GCACGGAGCACAGGAAGTTGG |
| 18S rRNA |
F: CGGCTACCACATCCAAGGAA R: GCTGGAATTACCGCGGCT |
Statistical analysis
One-way analysis of variance (ANOVA) was used to analyze the differences in OARSI scores and gene expression levels of Runx-2, MMP-13, Col 10a1, aggrecan (ACAN), and Col 2a1. The least significant difference (LSD) multiple comparison test was used for pairwise comparisons following ANOVA. Differences were considered statistically significant at P values < 0.05. Statistical analyses were performed using SPSS version 19.0.
Results
Adenovirus-mediated transduction of m-HDAC4 located in nucleus and had an enhanced function to inhibit chondrocyte hypertrophy compared with HDAC4
We used fluorescence microscopy to investigate the subcellular localization of m-HDAC4 and HDAC4. We found that m-HDAC4 was located in the chondrocyte nuclei, while HDAC4 was located in the cytoplasm of chondrocytes, as indicated by green fluorescence at 24 h after adenovirus infection of the cells (Fig. 1a). RT-qPCR was used to detected the mRNA level of hypertrophic and anabolic molecules in chondrocytes. RT-qPCR results showed that, compared with the Ad-GFP treated cells (Runx-2:0.95 ± 0.04, Mmp-13:0.93 ± 0.07,Col 10a1: 0.85 ± 0.13, Col 2a1:0.83 ± 0.15,ACAN:0.94 ± 0.06), HDAC4 and m-HDAC4 decreased the expression levels of Runx-2(P = 0.001 Ad-HDAC4 compared with Ad-GFP; P < 0.001 Ad-m-HDAC4 compared with Ad-GFP. F = 47.347, df = 8), Mmp-13(P < 0.001 Ad-HDAC4 and Ad-m-HDAC4 compared with Ad-GFP. F = 112.643, df = 8), and Col 10a1(P < 0.001 Ad-HDAC4 and Ad-m-HDAC4 compared with Ad-GFP. F = 74.167, df = 8), and they increased the expression levels of Col 2a1(P = 0.012 Ad-HDAC4 compared with Ad-GFP; P < 0.001 Ad-m-HDAC4 compared with Ad-GFP. F = 99.810, df = 8) and ACAN(P = 0.001 Ad-HDAC4 compared with Ad-GFP; P < 0.001 Ad-m-HDAC4 compared with Ad-GFP. F = 30.561, df = 8). Relative to HDAC4, the mRNA levels of Runx-2(0.35 ± 0.12), Mmp-13(0.40 ± 0.02), and Col 10a1(0.09 ± 0.01) in the Ad-m-HDAC4 treated cells were lower than those in the Ad-HDAC4 treated cells (Runx-2: 0.56 ± 0.04, P = 0.018; Mmp-13: 0.51 ± 0.04, P = 0.025; Col 10a1: 0.32 ± 0.04,P = 0.011), while the levels of Col 2a1(2.46 ± 0.18) were higher than those in the Ad-HDAC4 treated cells(1.26 ± 0.10, P < 0.001) (Fig. 1b).
Fig. 1.
HDAC4 and m-HDAC4 inhibit chondrocyte hypertrophy in vitro OA model. a The green fluorescence of m-HDAC4-GFP was located in the nuclei of chondrocytes, while HDAC4-GFP was located in the cytoplasm of cells after infection with adenoviral vectors (MOI:200). Scale bars: 100 μm. b RT-qPCR showed that upregulation of m-HDAC4 decreased the mRNA expression of Runx-2, MMP-13, and Col 10a1 and increased the mRNA expression of Col 2a1 and ACAN in rat chondrocytes more effectively than HDAC4.* indicates P < 0.05
Radiology indicated that m-HDAC4 and HDAC4 reduced osteophyte formation in OA
We examined the OA changes of the rat knees by taking X-ray images at 2 months after operation (Fig. 2). The results showed less osteophyte formation along knee joint margins in the Ad-HDAC4 and Ad-m-HDAC4 treated rats, and the joint degeneration degree was no obvious difference between the two groups. Osteophyte formation was more evident in the Ad-GFP control group.
Fig. 2.
Radiographs of osteophyte formation in the rat knee 2 months after ACLT. The images show slight osteophyte formation along the knee joint margins in the Ad-HDAC4 and Ad-m-HDAC4 treated rats; however, the rats than underwent ACLT and treated with Ad-GFP demonstrated the most severe change (black arrow heads)
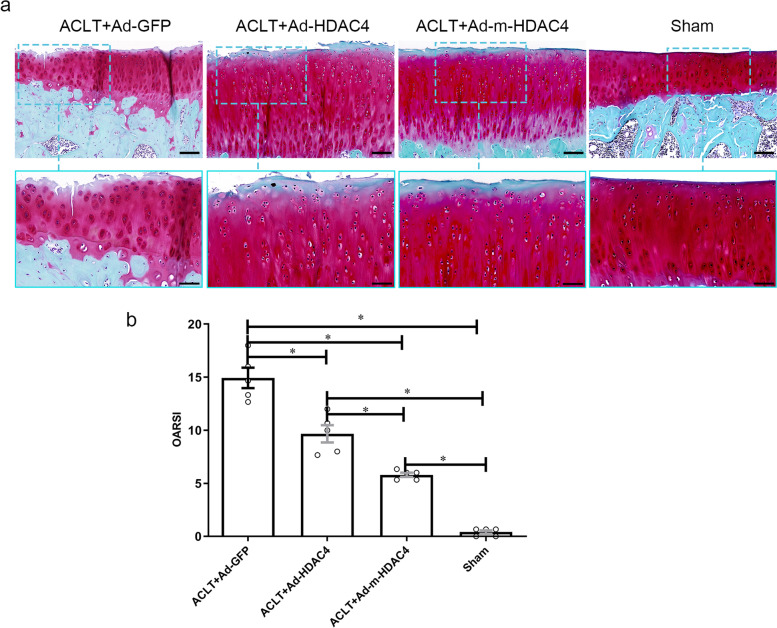
Safranin O staining indicated that m-HDAC4 has a stronger chondroprotective effect to attenuate cartilage degeneration, compared with HDAC4 in OA
We stained the tibia plateau cartilage with Safranin O/Fast green, which indicated proteoglycan loss and cartilage surface erosion (Fig. 3a). The results showed strong Safranin O staining and intact articular cartilage surface in the Ad-HDAC4 and Ad-m-HDAC4 treated rats. The Ad-m-HDAC4 treated rats showed stronger staining and a less-damaged cartilage surface than Ad-HDAC4 treated rats. The resulting OARSI scores were lower in the Ad-m-HDAC4 group (5.80 ± 0.45) than in the Ad-HDAC4 group (9.67 ± 1.83, P = 0.045). The OARSI scores were highest in rat knees that underwent ACLT treated with Ad-GFP control adenovirus vector (14.93 ± 2.14, P = 0.019 compared with Ad-HDAC4 group; P = 0.003 compared with Ad-m-HDAC4 group. F = 90.748, df = 19) (Fig. 3b).
Fig. 3.
HDAC4 and m-HDAC4 attenuate articular cartilage damage. a The Ad-m-HDAC4 treated rats had stronger Safranin O staining and less-damaged cartilage surface than the Ad-HDAC4 treated rats. The most severe Safranin O staining lost and cartilage surface damage were detected in rats that underwent ACLT operation and treated with Ad-GFP. Top panel scale bars: 100 μm; bottom panel scale bars: 50 μm. b OARSI scores were lower in in the Ad-m-HDAC4 treated rats than in the Ad-HDAC4 treated rats. The OARSI scores was highest in the ACLT rats and treated with Ad-GFP and sham-operated rats had the lowest scores. * indicates P < 0.05.
IHC showed that m-HDAC4 represses hypertrophy and enhances anabolism of cartilage more effectively than HDAC4
We compared the expression of Runx-2, MMP-13, and Col 2a1 in the four groups using IHC staining of cartilage (Fig. 4). The staining of Runx-2 and MMP-13 was lower in the Ad-HDAC4 and Ad-m-HDAC4 treated rats than in the rat that underwent ACLT treated with Ad-GFP. Moreover, Runx-2, MMP-13 staining were lower in the Ad-m-HDAC4 treated rats than in the Ad-HDAC4 treated rats. In contrast, Col 2a1 expression was higher in the Ad-HDAC4 and Ad-m-HDAC4 treated rats than in rats that underwent ACLT operation treated with Ad-GFP, and, compared to the Ad-HDAC4 group, Col 2a1 expression was higher in the Ad-m-HDAC4 group.
Fig. 4.
Runx-2 (a) and MMP-13 (b) IHC staining was increased in rats that underwent ACLT and treated with Ad-GFP, but it was lower in the Ad-m-HDAC4, Ad-HDAC4 and sham-operated groups. The staining of Runx-2 and MMP-13 was lower in the Ad-m-HDAC4 treated rats than in the Ad-HDAC4 treated rats. In contrast, Col 2a1 expression was higher in the Ad-m-HDAC4 and Ad-HDAC4 and sham-operated groups than in the Ad-GFP treated group. Compared to Ad-HDAC4 treated rats, Col 2a1 staining was greater in the Ad-m-HDAC4 treated rats (c)
RT-qPCR indicated that m-HDAC4 represses hypertrophy and enhances anabolism of cartilage more effectively than HDAC4
RT-qPCR results showed that the mRNA levels of Runx-2 and Mmp-13 were lower in the Ad-HDAC4(Runx-2: 1.16 ± 0.11, Mmp-13: 1.85 ± 0.21) and Ad-m-HDAC4(Runx-2: 0.90 ± 0.06, Mmp-13: 1.14 ± 0.10) treated rats than in the rat that underwent ACLT treated with Ad-GFP (Runx-2: 2.34 ± 0.11, P < 0.001 Ad-HDAC4 and Ad-m-HDAC4 compared with Ad-GFP, F = 104.477, df = 11; Mmp-13: 2.84 ± 0.11, P < 0.001 Ad-HDAC4 and Ad-m-HDAC4 compared with Ad-GFP, F = 105.524, df = 11), while the mRNA levels of Runx-2 and Mmp-13 were lower in the Ad-m-HDAC4 treated rats than in the Ad-HDAC4 treated rats (Runx-2: P = 0.029; Mmp-13: P < 0.001). In contrast, the mRNA levels of Col 2a1 and ACAN followed the opposite pattern. The mRNA levels of Col 2a1 and ACAN were higher in the Ad-HDAC4(Col 2a1: 0.21 ± 0.02, ACAN: 0.45 ± 0.08) and Ad-m-HDAC4(Col 2a1: 0.41 ± 0.11, ACAN: 0.66 ± 0.07) treated rats than in the rat that underwent ACLT treated with Ad-GFP (Col 2a1: 0.06 ± 0.01, P < 0.001 Ad-HDAC4 and Ad-m-HDAC4 compared with Ad-GFP, F = 124.582, df = 11; ACAN: 0.19 ± 0.06, P = 0.012 Ad-HDAC4 compared with Ad-GFP; P < 0.001 Ad-m-HDAC4 compared with Ad-GFP, F = 27.123, df = 11), while the mRNA levels of Col 2a1 and ACAN were higher in the Ad-m-HDAC4 treated rats than in the Ad-HDAC4 treated rats (Col 2a1: P = 0.003; ACAN: P = 0.027).(Fig. 5).
Fig. 5.
M-HDAC4 repress cartilage hypertrophy and enhance anabolism in rat OA model. a, b Levels of mRNA for Runx-2 and MMP-13 were lower in rats that were treated with Ad-m-HDAC4 or Ad-HDAC4 compared to the rats that underwent ACLT and treated with Ad-GFP. In addition, these 2 genes were expressed at a lower level in the Ad-m-HDAC4 treated rats than in the Ad-HDAC4 treated rats. In contrast, the mRNA levels of Col 2a1 (c) and ACAN (d) were elevated in rats that were treated with Ad-m-HDAC4 or Ad-HDAC4 compared to the rats that underwent ACLT and treated with Ad-GFP, and the mRNA expression of Col 2a1 and ACAN were greater in the Ad-m-HDAC4 treated rats than in the Ad-HDAC4 treated rats.* indicates P < 0.05
Discussion
HDAC4 subcellular translocation plays a vital role in neuronal death [14], myocyte differentiation [15], and chondrocyte differentiation [9]. The primary premise of HDAC4 activation is the nucleus. There are two forms of HDAC4 in cells: one is in the nucleus, which is the activate form, and the other is in the cytoplasm, which has no enzyme activity [16].
HDAC4 is highly important in repressing chondrocyte hypertrophy and regulating growth plate chondrocyte differentiation through relocation of proliferating chondrocytes in the nucleus to prehypertrophic chondrocytes in the cytoplasm. HDAC4 nuclear relocation decreased the gene expression of Runx2, MMP-13, and Col 10a1 and increased the expression of ACAN and Col 2a1 [17]. Our previous study indicated that upregulation of HDAC4 repressed the effect of IL-1β on the expression of catabolic factors in OA chondrocytes and attenuated cartilage degeneration by repressing Runx-2,MMP-13, and Col 10a1 in a rat ACLT OA model [6]. However, the transduction of HDAC4 in chondrocytes is mainly localized in the cytoplasm.
The 14-3-3 protein can promote cytoplasmic localization by binding to HDAC4. The 14-3-3 protein binds to three serine residues (S246, S467, and S632) at the N-terminal of HDAC4 [18], where binding with S246 masks the nuclear localization signal (NLS) and interferes with the binding of importin α and NLS. The 14-3-3 protein also contains a nuclear export signal (NES) when it binds to S467 in the form of a dimer, which is equivalent to providing a NES for HDAC4. Therefore, the 14-3-3 protein can keep HDAC4 in the cytoplasm by inhibiting entry and promoting exit from the nucleus [19–22]. HDAC4 has three serine residues that need to be phosphorylated to bind to the 14-3-3 protein, which requires multiple enzymes [23]. CaMKI phosphorylates S246 and S467, and CaMK II phosphorylates S467 and S632, exposing the 14-3-3 protein anchoring region of HDAC4, and binding to 14-3-3 protein promotes HDAC4 nucleation [24]. In contrast, protein phosphatase 2A (PP2A) can dephosphorylate these HDAC4 binding sites, thus leading HDAC4 into the nucleus, which led to our hypothesis that m-HDAC4, which is mainly located in the nucleus, may induce slower OA progression, compared to HDAC4.
In this study, we constructed an adenoviral vector encoding m-HDAC4 and GFP to infect the rat chondrocytes. The localization of m-HDAC4 was observed by fluorescent microscopy. We observed green fluorescence of m-HDAC4 mainly located in the chondrocyte nuclei, while HDAC4 green fluorescence was mainly located in the cytoplasm. This is consistent with a previous study [7]. We used an in vitro OA model to investigate the protective functions of m-HDAC4 and HDAC4 on IL-1β stimulated chondrocytes, and our in vitro results showed that the upregulation of m-HDAC4 and HDAC4 ameliorated IL-1β-induced chondrocyte hypertrophy; however, compared with HDAC4, m-HDAC4 had a stronger inhibitory effect on IL-1β-induced chondrocyte catabolism and promoting anabolism.
We used an ACLT OA rat model to investigate the chondroprotective effect of m-HDAC4 on OA in vivo, and our previous study indicated that adenoviral vectors intraarticular injection efficiently transduce HDAC4 gene in rat articular cartilage [6].
X-ray examination was used to detect osteophyte formation in rat knees. Normally, osteophyte formation is a secondary change in OA, and more osteophytes indicate more severe cartilage damage. Our results showed that the Ad-m-HDAC4 and Ad-HDAC4 treated rats had fewer osteophytes in periarticular bone than rats underwent ACLT treated with Ad-GFP group. Our radiological results demonstrate that m-HDAC4 upregulation attenuated joint damage degree.
We futher investigated the therapeutic function of m-HDAC4 by Safranin O staining. The articular cartilage has stronger staining and a smoother and more-intact cartilage surface in the Ad-m-HDAC4 group than in the Ad-HDAC4 group. Then, we used the OARSI score to evaluate rat articular cartilage damage. The OARSI score is a semi-quantitative grading method based on Safranin O staining. The score is positively correlated with articular cartilage damage [25, 26]. We found that the OARSI score is lower in the Ad-m-HDAC4 group than in the Ad-HDAC4 group. Thus, m-HDAC4 transduction had a stronger chondroprotective effect that decreased cartilage aggrecan loss and cartilage damage, compared to HDAC4.
HDAC4 represses the expression of Runx-2 and MMP-13, the key factors of chondrocyte hypertrophy and cartilage matrix degradation, to decrease cartilage damage. Thus, in our in vivo experiment, we measured Runx-2 and MMP-13 using IHC staining and RT-qPCR. The IHC results showed that there were fewer Runx-2 and MMP-13-positive cells in the Ad-m-HDAC4 treated rats than in the Ad-HDAC4 treated rats. The RT-qPCR results were consistent with the IHC results, and this inhibitory effect of HDAC4 depends on nucleus relocation [27, 17]. Compared to the cytoplasmic localization of HDAC4, the m-HDAC4 is mainly located in the nucleus, and these results indicate that m-HDAC4 attenuates OA cartilage damage by repressing the expression of Runx-2 and MMP-13 more effectively than HDAC4.
In addition, the expression of ACAN and Col 2a1 were upregulated in the Ad-m-HDAC4 and Ad-HDAC4 treated rats. Thus, HDAC4 and m-HDAC4 upregulation also increased chondrocyte anabolism. This suggests that HDAC4 and its triple mutant may promote chondrocyte proliferation. However, the mechanism of this function is remain unclear and should be investigated in future studies.
The limitations of the study should be mentioned. First, we used rat costal chondrocytes to construct an in vitro OA model to study the effects of M-HDAC4 on chondrocyte anabolism and catabolism. Although studies have shown that costal chondrocytes and articular chondrocytes have the same origin [28] and almost the same biological characteristics and functions [29], knee articular chondrocytes were more appropriate in the study of knee osteoarthritis. Second, this study only evaluated the chondroprotective effect of M-HDAC4, and did not detect the changes of subchondral bone. The changes of subchondral bone were equally important for the evaluation of the treatment effect of osteoarthritis. Third, we use an in vivo OA model of rats, and thus, our findings might not translate directly to humans, as anatomical differences between rats and humans may affect the chondroprotective effect of transgene.
Conclusions
In conclusion, our study demonstrated that m-HDAC4 and HDAC4 attenuated articular cartilage degeneration by repressing Runx-2 and MMP-13, and by inducing Col 2a1 and ACAN. The m-HDAC4 is more protective than HDAC4.
Acknowledgements
Not Applicable.
Abbreviations
- OA
Osteoarthritis
- HDAC4
Histone deacetylase 4
- m-HDAC4
Histone deacetylase 4 S246/467/632A mutant
- Runx-2
Runt-related transcription factor 2
- MMP-13
Matrix metalloproteinase 13
- Col 2a1
Type II collagen
- ACAN
Aggrecan
- ACLT
Anterior cruciate ligament transection
Authors’ contributions
PCL contributed to the conception of the study; XDG and FL performed the experiment, analysed the data and wrote the manuscript; YYG and XDC helped perform the data analyses. All authors discussed the results and revised the manuscript. The author(s) read and approved the final manuscript.
Funding
This research was funded by National Science Foundation for Young Scientists of China (Grant No. 81601949) and the Osteoarthritis Biological Sample Resource Sharing Service Platform Construction Project of Shanxi Province (Grant No. 201705D121010). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by the Institutional Animal Welfare Committee of Shanxi Medical University. The approval No.: 2016LL083.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Whittaker JL, Truong LK, Dhiman K, Beck C. Osteoarthritis year in review 2020: rehabilitation and outcomes. Osteoarthritis Cartilage. 2021;29(2):190–207. doi: 10.1016/j.joca.2020.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Bijlsma JW, Berenbaum F, Lafeber FP. Osteoarthritis: an update with relevance for clinical practice. Lancet. 2011;377(9783):2115–2126. doi: 10.1016/S0140-6736(11)60243-2. [DOI] [PubMed] [Google Scholar]
- 3.Orfanidou T, Iliopoulos D, Malizos KN, Tsezou A. Involvement of SOX-9 and FGF-23 in RUNX-2 regulation in osteoarthritic chondrocytes. J Cell Mol Med. 2009;13(9B):3186–3194. doi: 10.1111/j.1582-4934.2009.00678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei F, Zhou J, Wei X, Zhang J, Fleming BC, Terek R, Pei M, Chen Q, Liu T, Wei L. Activation of Indian hedgehog promotes chondrocyte hypertrophy and upregulation of MMP-13 in human osteoarthritic cartilage. Osteoarthr Cartil. 2012;20(7):755–763. doi: 10.1016/j.joca.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, Karsenty G, Olson EN. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119(4):555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 6.Gu XD, Wei L, Li PC, Che XD, Zhao RP, Han PF, Lu JG, Wei XC. Adenovirus-mediated transduction with histone Deacetylase 4 ameliorates disease progression in an osteoarthritis rat model. Int Immunopharmacol. 2019;75:105752. doi: 10.1016/j.intimp.2019.105752. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Wei X, Wang S, Jiao Q, Zhang Y, Du G, Wang X, Wei F, Zhang J, Wei L. Compression regulates gene expression of chondrocytes through HDAC4 nuclear relocation via PP2A-dependent HDAC4 dephosphorylation. Biochim Biophys Acta. 2016;1863(7 Pt A):1633–1642. doi: 10.1016/j.bbamcr.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Qin G, Zhao TC. HDAC4: mechanism of regulation and biological functions. Epigenomics. 2014;6(1):139–150. doi: 10.2217/epi.13.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan Y, Chen Q, Yang X, Haines P, Pei M, Terek R, Wei X, Zhao T, Wei L. Subcellular relocation of histone deacetylase 4 regulates growth plate chondrocyte differentiation through Ca2+/calmodulin-dependent kinase IV. Am J Physiol Cell Physiol. 2012;303(1):C33–C40. doi: 10.1152/ajpcell.00348.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI, Bazett-Jones DP, Yang XJ. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol. 2000;20(18):6904–6912. doi: 10.1128/MCB.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Ren X, Li S, Liang J, Zhao X, Wang T, Wang Z. Morphological, Immunocytochemical, and biochemical studies of rat costal chondrocytes exposed to IL-1β and TGF-β1. J Healthc Eng. 2017;2017:9747264. doi: 10.1155/2017/9747264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsaid KA, Zhang L, Waller K, Tofte J, Teeple E, Fleming BC, Jay GD. The impact of forced joint exercise on lubricin biosynthesis from articular cartilage following ACL transection and intra-articular lubricin's effect in exercised joints following ACL transection. Osteoarthr Cartil. 2012;20(8):940–948. doi: 10.1016/j.joca.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Du G, Zhan H, Ding D, Wang S, Wei X, Wei F, Zhang J, Bilgen B, Reginato AM, Fleming BC, Deng J, Wei L. Abnormal mechanical loading induces cartilage degeneration by accelerating Meniscus hypertrophy and mineralization after ACL injuries in vivo. Am J Sports Med. 2016;44(3):652–663. doi: 10.1177/0363546515621285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J Neurosci. 2005;25(41):9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miska EA, Langley E, Wolf D, Karlsson C, Pines J, Kouzarides T. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 2001;29(16):3439–3447. doi: 10.1093/nar/29.16.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9(1):45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Wei X, Lv Z, Sun X, Wang S, Zhang Y, Jiao Q, Wang X, Li Y, Wei L. Cyclic Equibiaxial tensile strain alters gene expression of chondrocytes via histone Deacetylase 4 shuttling. PLoS One. 2016;11(5):e0154951. doi: 10.1371/journal.pone.0154951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97(14):7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397(6715):172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 21.Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, Gamblin SJ, Yaffe MB. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4(2):153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang AH, Yang XJ. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol Cell Biol. 2001;21(17):5992–6005. doi: 10.1128/MCB.21.17.5992-6005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9(3):206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Backs J, Song K, Bezprozvannaya S, Chang S, Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J Clin Invest. 2006;116(7):1853–1864. doi: 10.1172/JCI27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pritzker KP, Gay S, Jimenez SA, Ostergaard K, Pelletier JP, Revell PA, Salter D, van den Berg WB. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthr Cartil. 2006;14(1):13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Gerwin N, Bendele AM, Glasson S, Carlson CS. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the rat. Osteoarthr Cartil. 2010;18(Suppl 3):S24–S34. doi: 10.1016/j.joca.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 27.Nishimori S, Lai F, Shiraishi M, Kobayashi T, Kozhemyakina E, Yao TP, Lassar AB, Kronenberg HM. PTHrP targets HDAC4 and HDAC5 to repress chondrocyte hypertrophy. JCI Insight. 2019;4(5):e97903. doi: 10.1172/jci.insight.97903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Sayed K, Haisch A, John T, Marzahn U, Lohan A, Müller RD, Kohl B, Ertel W, Stoelzel K, Schulze-Tanzil G. Heterotopic autologous chondrocyte transplantation--a realistic approach to support articular cartilage repair? Tissue Eng Part B Rev. 2010;16(6):603–616. doi: 10.1089/ten.TEB.2010.0167. [DOI] [PubMed] [Google Scholar]
- 29.Kitaoka E, Satomura K, Hayashi E, Yamanouchi K, Tobiume S, Kume K, Obinata M, Nagayama M. Establishment and characterization of chondrocyte cell lines from the costal cartilage of SV40 large T antigen transgenic mice. J Cell Biochem. 2001;81(4):571–582. doi: 10.1002/jcb.1075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.