Abstract
Auxin biology as a field has been at the forefront of advances in delineating the structures, dynamics, and control of plant growth networks. Advances have been enabled by combining the complementary fields of top-down, holistic systems biology and bottom-up, build-to-understand synthetic biology. Continued collaboration between these approaches will facilitate our understanding of and ability to engineer auxin's control of plant growth, development, and physiology. There is a need for the application of similar complementary approaches to improving equity and justice through analysis and redesign of the human systems in which this research is undertaken.
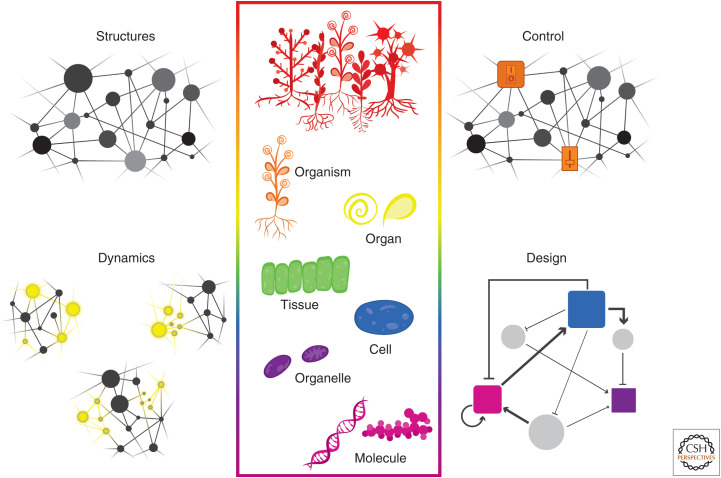
Nearly 20 years ago, Hiroaki Kitano published an article entitled “Systems Biology: A Brief Overview” (Kitano 2002). His conceptualization and articulation of the urgent need to “examine the structure and dynamics of cellular and organismal function” remains a major goal of molecular biology, biochemistry, development, and physiology. What would it mean to have “a continuous spectrum of knowledge” of plants from molecular/biochemical/biophysical details to signaling networks to intracellular communication to multicell phenomena to interorgan coordination to ecology? Kitano described four major domains of inquiry in pursuit of this awesome vision: (1) system structures, integrated maps that connect experimentally validated information like gene regulatory networks and biochemical pathways to the physical properties of individual cells and higher order assemblages; (2) system dynamics, comprehensive quantitative models of how system structures change over time in response to a variety of stimuli; (3) system control, a fully annotated dynamical model that is able to predict which interventions will be able to shift behavior of the system to meet distinct sets of specifications; and (4) system design, an automated process for constructing biological systems via well-articulated principles and simulation (Fig. 1). The intervening decades have seen remarkable achievements in the first two of these aims, largely under the umbrella of systems biology, but the second two have been more of a challenge. Synthetic biology, or engineering biology (Engineering Biology Research Consortium 2019), has in many ways emerged as a field to fill this gap. The decade ahead will require a framework that enables bridging between these two fields, a task that plant biologists are well positioned to pursue (Rhee et al. 2019; Henkhaus et al. 2020; Leydon et al. 2020).
Figure 1.
Kitano's four proposed domains of inquiry. The network maps shown represent the four paths of inquiry needed to build a continuous spectrum of knowledge from molecules to ecosystems. Structures represent experimentally validated connectivity maps of biological processes and pathways. Dynamics capture quantitatively how network behaviors change over time, often in response to stimuli. Control uses techniques like sensitivity analysis to identify which nodes in the network are best suited for engineering desired changes in network performance (phenotype). Design synthesizes knowledge arising from the other three domains to modify and construct novel biological systems with user-defined behaviors.
In this review, we will lay out our operating definitions of systems and synthetic biology. In doing so, we will also provide examples of how auxin biology has been at the forefront in advancing both fields. Finally, we will position researchers themselves as a part of a system that needs attention and study.
DEFINITIONS
One challenge to building bridges between the disciplines of systems and synthetic biology is philosophical. Systems biology can be rooted in any one or all of three distinct lenses: methodological, ontological, and epistemological (Mazzocchi 2012). The first, methodological, is really about how data are collected and modeled. The ontological and epistemological lenses refer respectively to which level(s) the researcher believes encode system function, and whether the direction of causality for functions is unidirectional across levels of organization. Fulvio Mazzocchi coined the term “pragmatic systems-biologist” as someone who can build and use tools that prioritize capturing the function of the intact system, while retaining membership in the ontological and epistemological reductionist camps that hold that the principles of chemistry and physics can explain the phenomena observed at higher orders (Mazzocchi 2012). In contrast, a systems-theoretic biologist believes that the arrow of causality points in both directions. In other words, systems properties like emergence and self-construction make biological systems essentially non-Turing computable (Mazzocchi 2012), which is a potentially serious obstacle for a synthetic biologist to surmount. Fortunately, physicists and computer scientists have a head start on building reasonable conceptual and pragmatic work-arounds for exactly this problem (Cooper 2012; Grozinger et al. 2019). Perhaps the most important takeaway from full-on systems theory is the clarion call to explicitly state the assumptions underlying data acquisition and their interpretation and acknowledge what is missing or unmeasurable. Although Mazzocchi might not consider it a desirable state, we would argue that the “pragmatic systems-biologist” is an ideal vantage point from which to blend systems and synthetic biology approaches (Table 1).
Table 1.
A useful but oversimplified guide to differences between systems and synthetic biology
| Biology type | Scope | Objective |
|---|---|---|
| Systems | Organismal, integration of multilevel and diverse types of networks, holistic | Behavior characterization, causal mechanisms, predictive modeling |
| Synthetic | Gene circuits, subnetworks, modules, reductionist | Parts characterization, engineered behavior, predictive modeling, model-guided design |
Synthetic biology can be defined as the application of engineering design principles to biological systems, particularly the model-based design-build-test-learn cycle (Pouvreau et al. 2018). Synthetic biology is often associated with the translation of biological knowledge for applications of human value. However, the bottom-up, build-to-understand approach has perhaps made its greatest impact in improving our understanding of biology (Elowitz and Lim 2010; Church et al. 2014). The undeniably, unabashedly artificial methodology of synthetic biology thus plays multiple useful roles as a companion with systems approaches. Echoing the strengths of systems theory, synthetic biology approaches also force the researcher to articulate assumptions in the design of the experiment, and frequently reveal gaps in knowledge. Confronted with these gaps, the synthetic biologist can continue to mine the system for additional parts and interactions, thereby testing both methodological and epistemological reductionism. Attempts to engineer the intensity, placement, and mode of feedback, for example, in synthetic and natural systems has produced critical insights into highly desirable qualities of engineered systems like stability and multicell coordination (Chen 2013).
USING SYSTEMS AND SYNTHETIC BIOLOGY TO IDENTIFY CONTROL AND LEARN TO DESIGN
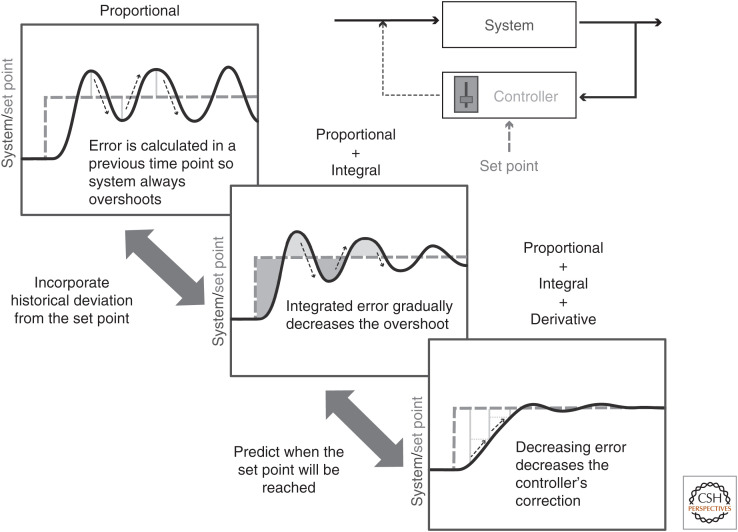
Control is critical to biological systems. Plants must remain at least moderately in control of the chaos of thermodynamic equilibrium to extract work, organize and build their fractal structures, and operate molecular machinery to ensure their survival. A control system, or controller, can be defined as a mathematical function that calculates an output that drives the system closer to the set point or goal value of the system, based on an input value that is typically the difference between the current state of the system and the desired set point (Baetica et al. 2019). For example, the simple temperature controller in an oven turns on a heating element whenever the actual temperature falls below the set point temperature and turns the element off when the temperature rises above the set point. In a biological system example, we can think about the expression level of a gene critical for maintenance of cellular identity as being the system state (Fig. 2). In many cases, a biological controller composed of intertwined metabolic and gene regulatory networks will detect whether this gene deviates from the average expression level required for maintenance of cellular identity and correct its expression. To be a good controller that maintains the set point without significant oscillations, rapidly yet smoothly returning to the set point after perturbation, the driving function must integrate the previous behavior of the system and be an accurate predictor of the future behavior or dynamics of a system (Chevalier et al. 2019; Whitby et al. 2021).
Figure 2.
A useful but oversimplified introduction to control in biological systems. The dynamics of a system (dark, solid lines) are regulated by a controller with the aim of maintaining a set point (lighter, dashed lines). The controller modulates the system by applying corrections to the system input (thin, dark, dashed lines), which are calculated based on various functions of the difference between the system and set point values (lightest lines and fill).
So how do we go about understanding control in biological systems? We do it largely in the same way we tune engineered control systems, by tinkering. In tuning a slightly more complex controller than in a typical oven—a controller that regulates the intensity of the heating element instead of simply turning it on or off, or the controller for the cruise control in a car—there are generally three “tuning knobs” or parameters engineers tinker with to optimize a controller's ability to swiftly and accurately return to the set point temperature or speed in our examples after perturbation (Fig. 2). The first of these tuning knobs drives the heating element or accelerator proportionally to the difference between the current and the set point values. However, proportional control alone, similar to on–off logic, only enacts control when there is a difference between the set point and current values. Proportional control alone therefore leads to some consistent error around the set point. The second tuning knob, integral control, solves this problem of consistent error by summing the error between the actual and set point values over a period of time and adjusting accordingly. So small deviations from the set point over time gradually affect stronger responses in integral control. In this way, integral control takes into account the history of the system. The third mode of control in built systems, derivative control, takes into account the future of the system by including the rate of change of the system's deviation from the set point. Derivative control therefore provides information about how fast a system is returning to its set point and prevents the system from overshooting this set point by adjusting the control proportionally to the rate of approach.
Unlike the temperature controller in an oven, which has a clear, human-engineered goal of maintaining the set point temperature by implementing a simple on–off logic, controllers in biological systems often operate at vastly different temporal and spatial scales from the organismal “goal” of fitness, or in the case of domesticated organisms, their value to humans. At many scales across biology, it is unclear what the goal is, or more appropriately, the set point of these biological controllers. Even the most complex engineered systems are built upon logic that is orders of magnitude less complex than biological systems. After all, “evolution is a tinkerer, not an engineer” (Jacob 1977; Oster and Rocklin 1979). However, this comparison is still useful in introducing the concepts of control in biological systems (more mathematically detailed comparison can be found in Del Vecchio et al. 2016, Chevalier et al. 2019, and Whitby et al. 2021).
For auxin, “the specificity in the system is not in the signal but in the cells that perceive it” (Leyser 2018). It may be tempting to consider auxin itself as the system being controlled, but auxin is most often the signal communicating the difference between the current state and set point of gene expression (Roosjen et al. 2018). In the context of feedback, the lines between the controller and controlled are blurred. Auxin's ability to regulate its own transport and metabolism is an integral part of the controller's function by generating negative feedback to reject noise and maintain signaling and cellular homeostasis as well as positive feedback to drive changes in the set point, signaling behavior, and cellular identity (Casanova-Sáez and Voß 2019; Semeradova et al. 2020). So, to understand auxin signaling as a control system we need to answer the question, what is the set point of auxin signaling? What does auxin signaling control? The answer to this question depends upon the scope of the system we wish to understand, that is, the specific cells, tissues, or organs of interest at particular times in development or during their response to stimuli. At the cellular level, auxin controls cell size, shape, growth axis, and identity in coordination with other signals and control systems. At the tissue and organ levels, these cellular behaviors are integrated into morphology and physiology. The nuclear auxin controller calculates transcriptional activity of a significant fraction of the genomes of all plants (Mutte et al. 2018). The set point for auxin signaling may then be whatever rate of transcription is needed to coordinate cell growth and division or higher-order outputs.
CONTROL AND DESIGN OF AUXIN SIGNALING
Beyond its centrality to plant biology, auxin signaling has the attractive feature of at once embodying elegant simplicity (few parts comprise a minimal pathway) and expansive complexity (each component belongs to large gene families and has multiple tuning knobs) (Leyser 2018). We know more about how cells recognize and respond to auxin than almost any other signaling process. The nuclear auxin signaling pathway is as stripped down as nuclear receptors are in animals; yet, like nuclear receptors, it incorporates several highly conserved eukaryotic functions such as ubiquitin-mediated degradation and transcriptional de-repression/activation. As described in great detail in other articles in this collection, auxin-induced transcription is mediated by dimerization of transcription factors called auxin response factors (ARFs). In low auxin conditions, ARFs are repressed by interaction with proteins called Aux/IAAs that form a bridge between the ARFs and TOPLESS (TPL), a member of the Groucho/Tup1 family of corepressors. When auxin accumulates, it facilitates a high-affinity interaction between the Aux/IAA substrates and an auxin receptor F-box protein. Interaction with the F-box brings the substrate to an SCF-type E3 ubiquitin ligase, leading to polyubiquitination and subsequent degradation by the proteasome.
This simple architecture produces complex behaviors through expression of different complements of signaling components. In Arabidopsis, for example, there are six auxin F-box receptors, 29 Aux/IAA substrates, and 23 ARF transcription factors, each with its own production/decay rate and relative affinity for other components. Whereas it had been long hypothesized that specificity in auxin responses were conferred by different component combinations, it has been impossible to rigorously test this model in plants because of coexpression of many family members in the same cell, and multilevel feedback on pathway components and auxin itself. The number of accessory components that are required make in vitro assays impractical. An early plant synthetic biology application was porting the few plant-specific nuclear auxin signaling components into yeast (Havens et al. 2012; Pierre-Jerome et al. 2014). By tagging different components with fluorescent reporters, time-lapse flow cytometry can be used to quantify the dynamics of each part in user-defined circuits. In addition, the inducibility of the system with a small molecule that is readily taken up by diverse eukaryotic cells with low toxicity makes auxin-induced degradation (AID) and gene activation a desirable technology for applications in nonplant cells. A similar “planti-fied” yeast approach has now also been applied to abscisic acid signaling (Ruschhaupt et al. 2019).
The synthetic dissection of the auxin pathway in yeast is in many ways paralleled by the use of “humanized” yeast to dissect the design principles of the animal cell nuclear factor κB (NF-κB) signaling pathway (Zhang et al. 2017). Similar to auxin, the NF-κB pathway is a rapidly activated signaling system that functions during growth and development, and in animals has a particularly key role in immune response. Extensive live-cell imaging, biochemical studies, and computational modeling have revealed NF-κB signaling to be an oscillator circuit with time-delayed negative feedback. The negative feedback loop consists of a transcription factor (NF-κB family) and an inhibitor (IκB family). In response to a signal, IκB is ubiquitinated and degraded, releasing inhibition on NF-κB, which can then up-regulate new IκBs to inhibit NF-κBs, and the cycle goes on. Recently, Zhang et al. (2017) reconstituted a minimal IκB/NF-κB feedback loop in yeast cells and rewired it into the yeast-mating pathway with the α factor pheromone as an input. They successfully recapitulated oscillatory NF-κB dynamics in single cells, with a periodicity close to that observed in mammalian cells but with sustained rather than damped oscillations. Further, the simplified nature of the system enabled quantitative exploration of the relationship between oscillation waveforms and parameters such as IκB degradation rate, which was found to tune the frequency of oscillatory dynamics. NF-κB researchers are also making headway in understanding how signaling dynamics are “read-out” to generate specific transcriptional responses by using an integrated platform that enables live-cell imaging of NF-κB dynamics coupled with single-cell RNA-seq to measure downstream transcriptional profiles associated with specific outputs (Lane et al. 2017). These approaches forge a path for future auxin synthetic biology research seeking to connect circuit function with context dependency and evolutionary innovation.
Recently, the power of a systems biology approach to identifying control parameters in auxin signaling was highlighted in a study of how cell division and growth are organized in the shoot apical meristem of Arabidopsis (Galvan-Ampudia et al. 2020). Synthetic reporters of auxin flux and signaling output were deployed to generate a cellular resolution time course of auxin signaling during floral organ initiation. This work revealed a mismatch between the measurements of input (auxin flux) and output (auxin signaling), demonstrating that how cells perceive auxin is changing during the transition from stem cell to floral organ progenitor. This behavior is well known and makes sense in light of the highly reticulated auxin signaling network. An auxin stimulus may lead to degradation of extant Aux/IAAs and expression of other Aux/IAAs and ARFs resulting in a different response upon a second exposure to auxin. Thus, auxin signaling and specifically the milieu of auxin signaling proteins—TIR1/AFB receptors, Aux/IAA substrate/repressors, and ARF transcription factors—act as a cellular memory, recording the previous states of auxin within the cell. This approach also helped define the responsibility of WUSCHEL in modulating auxin responses in the SAM stem cell niche (Ma et al. 2019). WUS restrains the start of an auxin signaling cascade that integrates geometry via spatiotemporal differences in auxin flux and perception contingent upon cellular histories to organize floral organ initiation. In light of our control example, WUS changes the set point of the auxin controller. These works clearly demonstrate a tenant of developmental signaling: current and future signaling dynamics are predicted by cell location and history. Perhaps not surprisingly, these results also usher in nearly the identical question as the synthetic biology studies described above: what duration of, or changes in, auxin flux patterning are perceived by the signaling pathway, and how is this signal converted into transcriptional activation of specific genes? Are auxin signals encoded in the frequency domain, the amplitude domain, or both? How many channels are there on auxin radio?
CASE STUDY: SYSTEMS AND SYNTHETIC BIOLOGY APPROACHES APPLIED TO THE Aux/IAAs
The story of understanding and now engineering the auxin response is a fair encapsulation of the history of technical innovations in modern plant biology (Abel and Theologis 2010). Starting with the first wave of molecular biology studies in the mid-1980s, much of the focus has been on the earliest auxin-induced transcripts (Walker and Key 1982; Hagen and Guilfoyle 1985; Theologis et al. 1985). The Aux/IAAs were among these genes, and subsequent work led to the identification of cis elements in their promoters that conferred inducibility in response to auxin or to treatment with inhibitors of translation (Ballas et al. 1993). This second quality led to a model in which auxin regulated a short-lived repressor protein (Koshiba et al. 1995), a premise that was eventually supported by cloning the genes affected in some of the first mutants isolated in the then new model plant Arabidopsis (Rouse et al. 1998), and eventually in elegant in vitro biochemical assays and crystallography that proved that the Aux/IAAs themselves were coreceptors for auxin (Dharmasiri et al. 2005; Kepinski and Leyser 2005; Tan et al. 2007). Luciferase fusions to Aux/IAAs were used to delimit the auxin degron, and subsequently revealed a range of AID rates among family members (Zenser et al. 2001; Dreher et al. 2006). Aux/IAAs were also used in an early yeast two-hybrid study that identified sequences in the carboxy terminus (now known as a PB1 domain; Korasick et al. 2014) as interaction surfaces allowing for Aux/IAA and ARF homo- and heterodimerization (Kim et al. 1997), an experiment that has recently been repeated on a larger scale (Vernoux et al. 2011). Synthetic assays of Aux/IAA functions in yeast combined with dynamic modeling demonstrated that degradation rate is among the most effective tuning knobs for altering downstream transcriptional dynamics (Pierre-Jerome et al. 2014). This Aux/IAA degradation tuning knob can be adjusted by varying sequences within the Aux/IAAs (Dreher et al. 2006; Guseman et al. 2015; Moss et al. 2015). In addition, natural or induced variation in the TIR1/AFB receptors can also impact degradation rate, and thus tune auxin sensitivity (Havens et al. 2012; Yu et al. 2013; Dezfulian et al. 2016; Wright et al. 2017; Ramos Báez et al. 2020).
These observations collectively suggest a model where the rate of auxin-induced Aux/IAA turnover acts as a checkpoint or timer for auxin-regulated developmental events. Evidence in support of this model came from an experiment analyzing transgenic plants expressing degradation rate variants of IAA14, a crucial determinant of lateral root initiation (Guseman et al. 2015). Progression through the well-established stages of lateral root development was strongly correlated with the engineered rates of IAA14 turnover. This straightforward result is unexpected given the multiple levels of feedback within the auxin pathway, which might be predicted to provide a strong buffering effect. This work led to a hypothesis that Aux/IAAs are auxin-initiated timers that synchronize developmental transitions. Additional support for this idea came from work connecting light perception to lateral root initiation, where the authors present a quantitative model whereby AID of Aux/IAAs allows for on-the-fly environmental resetting of developmental timing (Kircher and Schopfer 2018).
AID of Aux/IAAs has also fueled the development of a number of new technologies. In recent years, there has been widespread adoption of AID using different portions of Aux/IAA degrons as fusion tags to allow inducible degradation of proteins of interest in a wide array of nonplant organisms (Nishimura et al. 2009, 2020; Zhang et al. 2015; Bence et al. 2017). Similarly, Aux/IAA degrons have also been incorporated into cell–cell communication systems. In yeast, auxin-responsive synthetic transcription factors were expressed in one strain and mixed with a second strain that was engineered to synthesize auxin (Khakhar et al. 2016). Recently, this idea was expanded to act as a way to penalize cheaters in a quorum-sensing circuit engineered into mammalian cells (Ma et al. 2020). Synthetic transcription factors that incorporate the Aux/IAA degron have also been used to semirationally design plant development (Khakhar et al. 2018). Finally, the engineering of an orthogonal auxin-TIR1 pair that interfaces with the endogenous auxin response has been a major breakthrough in resolving controversial areas of auxin biology, notably very rapid auxin-induced growth responses (Fendrych et al. 2018; Uchida et al. 2018). These types of approaches alter the control logic of endogenous nuclear auxin control systems and add exogenous control points.
THE NEXT SET OF QUESTIONS WELL SUITED FOR SYNTHETIC AND/OR SYSTEMS APPROACHES
Remarkably given all of the progress made to date, there remain a host of unanswered questions in auxin biology, many of which will likely benefit from incorporating synthetic and systems approaches. From the perspective of the molecular signaling system itself, there remain key gaps in knowledge in regard to fundamental questions like what components are working in the nucleus, in the cytoplasm, or associated with various membranes. This question has been thrust to the fore with new findings about the activity of TIR1/AFB1 in triggering rapid growth responses (Prigge et al. 2020), as well as with revelations about auxin sequestration in the endoplasmic reticulum (Barbez and Kleine-Vehn 2013) and regulated nucleocytoplasmic partitioning of at least some ARFs (Powers et al. 2019). These same findings also bring out questions about coordination or competition between different signaling events, as well as presenting new potential mechanisms for partitioning auxin action within the cell body during anisotropic growth responses.
There are also areas where we have seen great progress, but still lack complete understanding. For example, work on mosses and liverworts, in addition to an ever-growing number of angiosperms, has been critical for connecting the diversification of auxin signaling components to evolutionary innovations (Matthes et al. 2019; Blázquez et al. 2020; Israeli et al. 2020), and this rich source of information has yet to be fully incorporated into control models or leveraged in engineering efforts. There is also much to learn about how promoter architecture and different classes of ARFs feed into these dynamics (Lavy et al. 2016; Kato et al. 2017; Flores-Sandoval et al. 2018; Galli et al. 2018; Freire-Rios et al. 2020; Lanctot et al. 2020). The challenges in this regard call to mind the work on one of the most extensively studied enhancers regulating eve2 in Drosophila embryogenesis. In their review article “The Appeasement of Doug: A Synthetic Approach to Enhancer Biology,” Vincent et al. (2016) document several failures at reconstituting the function of eve2 by minimizing, multimerizing, and even recoding spacer sequences between known transcription factor-binding sites. These authors argue, and we concur, that both successes and failures in enhancer reconstitution are informative and worthy of public report and advocate the use of iterative design-build-test-learn cycles to continue deciphering the design principles of regulatory logic in (auxin) biology.
From an engineering standpoint, we have yet to fully explore the natural features of transcription factors, such as modularity and cooperative binding. Bashor et al. (2019) recently demonstrated the ability to engineer cooperativity into synthetic transcription factors to generate nonlinear gene circuit behavior in yeast cells. They used a computational model-driven approach that enabled them to explore circuit configuration and behavior space for a large set of highly modular synthetic transcription factors. These highly tunable synthetic transcription factors enabled construction of circuits capable of such complex behaviors such as persistence filtering (activation only in response to an input of sufficiently long duration) and temporal decoding (bandpass filters that respond only to inputs of specified frequencies). How do the interactions between ARFs, and between ARFs and other transcription factors, impact native signaling dynamics? How could they be manipulated to change critical control parameters? The next generation of mathematical models, particularly if built with an eye to design, could be of great use in generating and evaluating experiments in this arena.
So how is information about development encoded in auxin signals? How can we learn to reprogram this controller to be better adapted to our changing climate to improve the lives of farmers and humanity more broadly? To answer these questions requires examination of a variety of tissues and how different auxin controllers are implemented in these systems. A beautiful example of such work examined auxin regulation of tomato leaf shape (Israeli et al. 2019). The Aux/IAA gene Entire regulates leaflet formation in tomatoes and is one of the rare examples of a clear phenotype associated with a loss of function in an Aux/IAA. Of course, auxin controllers are not operating in isolation, and many questions still remain as to how auxin and other signaling pathways interact. Other pathways modulate auxin responses and their effects on plant growth, development, and behavior. In addition to WUSCHEL and the shoot apical meristem cell network discussed above, interactions with the circadian clock are well known and have recently been dissected in circadian-regulated floral opening using systems approaches (Atamian and Harmer 2016; Ke et al. 2018). Hormone interactions in development are well founded, and our understanding of how auxin and cytokinin interact to control development and floral organ size have recently been advanced through a combination of synthetic signaling reporters and quantitative time-lapse microscopy (Zhu et al. 2020). Just as we have perturbed auxin controllers to study their behavior and logic, so too have other organisms sabotaged auxin signaling for their own gain (Zhang et al. 2020; Su et al. 2021). Over the past year, several auxin biosynthetic genes and gene circuits have been implicated in regulating plant–microbe interactions, pushing once again our system boundaries of auxin's realm of controls from organism to ecosystem (Kunkel and Harper 2018).
SCIENTISTS ARE THEMSELVES EMBEDDED IN A SYSTEM THAT NEEDS TO BE UNDERSTOOD AND INTENTIONALLY (SYNTHETICALLY) ENGINEERED FOR EQUITY AND JUSTICE
In 2016, the World Economic Forum brought together a group of young scientists from all over the world to discuss ethical conduct of research. The results of this workshop were published in 2018 as a code of conduct resting on seven principles: (1) engage with the public, (2) pursue the truth, (3) minimize harm, (4) engage with decision makers, (5) support diversity, (6) be a mentor, and (7) be accountable (World Economic Forum Young Scientists Community 2020). This expanded definition of responsible conduct of research is provocative and highlights the ways in which our research programs are themselves systems, embedded in a series of higher-order systems.
We are on the precipice of the next green revolution. With full knowledge of our past successes and failures, we must face this next transition with full attention to innovation in both the approaches we take and in which people participate in the work. In Kitano's domains, we might first focus on the individual stakeholders—what they can contribute, what they need, what success would look like for them—and then how they interact with one another. We can then use this information to build predictable models and guide rational design of our research systems (e.g., Byars-Winston et al. 2018). When we think of our mentees, this would necessitate enthusiastic adoption of best practices, especially those that make our groups truly welcoming and inclusive environments for all (Committee on Effective Mentoring in STEMM et al. 2019; Henkhaus et al. 2020). Professor Beronda Montgomery has cogently laid out a path for how we can use what we know from studying plants and microbes to inform how we approach our mentoring, our collaborative relationships, and our institutional structures (Montgomery 2018, 2020a,b). When we think of our students, we can develop course-based undergraduate research experiences (CUREs) that leverage the excitement around systems and synthetic approaches (Campbell et al. 2014; Murren et al. 2019; Govindan et al. 2020). This type of experience offers more equitable access to research and could substantially broaden the pool of future plant biologists (Lopatto et al. 2008; Bangera and Brownell 2014; Brownell et al. 2015).
Beyond the people directly engaged in the research, there are other stakeholders that historically have been marginalized, excluded, or disrespected. These include the public on whose behalf we are working, farmers working in an enormous range of socioeconomic and environmental conditions, indigenous communities with deep knowledge of specific plants and practices, agronomists, and decision makers in governmental and nongovernmental organizations, just to name a few. How would our science be different if we were regularly engaging with these audiences, not to “educate” them but to listen, to learn, and to build long-term relationships based on trust. There is a need for funding and leadership to foster scientific exchanges and collaborations between scientists in different parts of the world. Perhaps one of the most effective ways to minimize harm with technology is to transfer expertise to scientists who live in the communities where new varieties are needed, allowing for local conversations about values and priorities to guide the decision of which products are developed. We must reckon with how our choices as plant scientists can minimize harm and maximize accountability.
ACKNOWLEDGMENTS
We would like to thank Dr. Daria Chrobok of DC Sci-Art for collaborating with us to produce the figures. Research in the Wright Plant Synthetic Biology Laboratory is supported by the USDA National Institute of Food and Agriculture (Hatch Project 1021738). Research in the AuxSynBio Laboratory at Whitman College is supported by a National Science Foundation (IOS-1546873) award to Brit Moss. Systems and synthetic biology work in the Nemhauser Laboratory is supported by the National Institutes of Health (R01-GM107084), National Science Foundation (IOS-1546873), and the Howard Hughes Medical Institute Faculty Scholar Program.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Abel S, Theologis A. 2010. Odyssey of auxin. Cold Spring Harb Perspect Biol 2: a004572. 10.1101/cshperspect.a004572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamian HS, Harmer SL. 2016. Circadian regulation of hormone signaling and plant physiology. Plant Mol Biol 91: 691–702. 10.1007/s11103-016-0477-4 [DOI] [PubMed] [Google Scholar]
- Baetica AA, Westbrook A, El-Samad H. 2019. Control theoretical concepts for synthetic and systems biology. Curr Opin Syst Biol 14: 50–57. 10.1016/j.coisb.2019.02.010 [DOI] [Google Scholar]
- Ballas N, Wong LM, Theologis A. 1993. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of Pea (Pisum sativum). J Mol Biol 233: 580–596. 10.1006/jmbi.1993.1537 [DOI] [PubMed] [Google Scholar]
- Bangera G, Brownell SE. 2014. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Sci Educ 13: 602–606. 10.1187/cbe.14-06-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E, Kleine-Vehn J. 2013. Divide Et Impera—cellular auxin compartmentalization. Curr Opin Plant Biol 16: 78–84. 10.1016/j.pbi.2012.10.005 [DOI] [PubMed] [Google Scholar]
- Bashor CJ, Patel N, Choubey S, Beyzavi A, Kondev J, Collins JJ, Khalil AS. 2019. Complex signal processing in synthetic gene circuits using cooperative regulatory assemblies. Science 364: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence M, Jankovics F, Lukácsovich T, Erdélyi M. 2017. Combining the auxin-inducible degradation system with CRISPR/Cas9-based genome editing for the conditional depletion of endogenous Drosophila melanogaster proteins. FEBS J 284: 1056–1069. 10.1111/febs.14042 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Nelson DC, Weijers D. 2020. Evolution of plant hormone response pathways. Annu Rev Plant Biol 71: 327–353. 10.1146/annurev-arplant-050718-100309 [DOI] [PubMed] [Google Scholar]
- Brownell SE, Hekmat-Scafe DS, Singla V, Chandler Seawell P, Conklin Imam JF, Eddy SL, Stearns T, Cyert MS. 2015. A high-enrollment course-based undergraduate research experience improves student conceptions of scientific thinking and ability to interpret data. Life Sci Educ 14: ar21. 10.1187/cbe.14-05-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byars-Winston A, Womack VY, Butz AR, McGee R, Quinn SC, Utzerath E, Saetermoe CL, Thomas SB. 2018. Pilot study of an intervention to increase cultural awareness in research mentoring: implications for diversifying the scientific workforce. J Clin Transl Sci 2: 86–94. 10.1017/cts.2018.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AM, Eckdahl T, Cronk B, Andresen C, Frederick P, Huckuntod S, Shinneman C, Wacker A, Yuan J. 2014. Pclone: synthetic biology tool makes promoter research accessible to beginning biology students. Life Sci Educ 13: 285–296. 10.1187/cbe.13-09-0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Sáez R, Voß U. 2019. Auxin metabolism controls developmental decisions in land plants. Trends Plant Sci 24: 741–754. 10.1016/j.tplants.2019.05.006 [DOI] [PubMed] [Google Scholar]
- Chen S, Harrigan P, Heineike B, Stewart-Ornstein J, El-Samad H. 2013. Building robust functionality in synthetic circuits using engineered feedback regulation. Curr Opin Biotechnol 24: 790–796. 10.1016/j.copbio.2013.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier M, Gómez-Schiavon M, Ng AH, El-Samad H. 2019. Design and analysis of a proportional-integral-derivative controller with biological molecules. Cell Syst 9: 338–353.e10. 10.1016/j.cels.2019.08.010 [DOI] [PubMed] [Google Scholar]
- Church GM, Elowitz MB, Smolke CD, Voigt CA, Weiss R. 2014. Realizing the potential of synthetic biology. Nat Rev Mol Cell Biol 15: 289–294. 10.1038/nrm3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Effective Mentoring in STEMM, Board on Higher Education and Workforce, Policy and Global Affairs, National Academies of Sciences, Engineering, and Medicine. 2019. The science of effective mentorship in STEMM (ed. Byars-Winston A, Dahlberg ML). National Academies Press, Washington, DC. www.nap.edu/catalog/25568 [PubMed] [Google Scholar]
- Cooper B. 2012. The incomputable reality. Nature 482: 465. 10.1038/482465a [DOI] [PubMed] [Google Scholar]
- Del Vecchio D, Dy AJ, Qian Y. 2016. Control theory meets synthetic biology. J R Soc Interface 13: 20160380. 10.1098/rsif.2016.0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezfulian MH, Jalili E, Roberto DKA, Moss BL, Khoo K, Nemhauser JL, Crosby WL. 2016. Oligomerization of SCFTIR1 is essential for Aux/IAA degradation and auxin signaling in Arabidopsis. PLOS Genet 12: e1006301. 10.1371/journal.pgen.1006301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. 2006. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714. 10.1105/tpc.105.039172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M, Lim WA. 2010. Build life to understand it. Nature 468: 889–890. 10.1038/468889a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engineering Biology Research Consortium. 2019. Engineering biology: A research roadmap for the next-generation bioeconomy. roadmap.ebrc.org/resources/2019roadmap
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459. 10.1038/s41477-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Sandoval E, Eklund DM, Hong SF, Alvarez JP, Fisher TJ, Lampugnani ER, Golz JF, Vázquez-Lobo A, Dierschke T, Lin SS, et al. 2018. Class C ARFs evolved before the origin of land plants and antagonize differentiation and developmental transitions in Marchantia polymorpha. New Phytol 218: 1612–1630. 10.1111/nph.15090 [DOI] [PubMed] [Google Scholar]
- Freire-Rios A, Tanaka K, Crespo I, van der Wijk E, Sizentsova Y, Levitsky V, Lindhoud S, Fontana M, Hohlbein J, Boer DR, et al. 2020. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. Proc Natl Acad Sci 117: 24557–24566. 10.1073/pnas.2009554117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Khakhar A, Lu Z, Chen Z, Sen S, Joshi T, Nemhauser JL, Schmitz RJ, Gallavotti A. 2018. The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nat Commun 9: 4526. 10.1038/s41467-018-06977-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Cerutti G, Legrand J, Brunoud G, Martin-Arevalillo R, Azais R, Bayle V, Moussu S, Wenzl C, Jaillais Y, et al. 2020. Temporal integration of auxin information for the regulation of patterning. eLife 9: e55832. 10.7554/eLife.55832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan B, Pickett S, Riggs B. 2020. Fear of the CURE: a beginner's guide to overcoming barriers in creating a course-based undergraduate research experience. J Microbiol Biol Educ 21: 21.2.48. 10.1128/jmbe.v21i2.2109, www.asmscience.org/content/journal/jmbe/10.1128/jmbe.v21i2.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger L, Amos M, Gorochowski TE, Carbonell P, Oyarzún DA, Stoof R, Fellermann H, Zuliani P, Tas H, Goñi-Moreno A. 2019. Pathways to cellular supremacy in biocomputing. Nat Commun 10: 5250. 10.1038/s41467-019-13232-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman JM, Hellmuth A, Lanctot A, Feldman TP, Moss BL, Klavins E, Villalobos LIAC, Nemhauser JL. 2015. Auxin-induced degradation dynamics set the pace for lateral root development. Development 142: 905–909. 10.1242/dev.117234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ. 1985. Rapid induction of selective transcription by auxins. Mol Cell Biol 5: 1197–1203. 10.1128/MCB.5.6.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL. 2012. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol 160: 135–142. 10.1104/pp.112.202184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkhaus N, Bartlett M, Gang D, Grumet R, Jordon-Thaden I, Lorence A, Lyons E, Miller S, Murray S, Nelson A, et al. 2020. Plant science decadal vision 2020–2030: reimagining the potential of plants for a healthy and sustainable future. Plant Direct 4: e00252. 10.1002/pld3.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli A, Capua Y, Shwartz I, Tal L, Meir Z, Levy M, Bar M, Efroni I, Ori N. 2019. Multiple auxin-response regulators enable stability and variability in leaf development. Curr Biol 29: 1746–1759.e5. 10.1016/j.cub.2019.04.047 [DOI] [PubMed] [Google Scholar]
- Israeli A, Reed JW, Ori N. 2020. Genetic dissection of the auxin response network. Nat Plants 6: 1082–1090. 10.1038/s41477-020-0739-7 [DOI] [PubMed] [Google Scholar]
- Jacob F. 1977. Evolution and tinkering. Science 196: 1161–1166. 10.1126/science.860134 [DOI] [PubMed] [Google Scholar]
- Kato H, Kouno M, Takeda M, Suzuki H, Ishizaki K, Nishihama R, Kohchi T. 2017. The roles of the sole activator-type auxin response factor in pattern formation of Marchantia polymorpha. Plant Cell Physiol 58: 1642–1651. 10.1093/pcp/pcx095 [DOI] [PubMed] [Google Scholar]
- Ke M, Gao Z, Chen J, Qiu Y, Zhang L, Chen X. 2018. Auxin controls circadian flower opening and closure in the waterlily. BMC Plant Biol 18: 143. 10.1186/s12870-018-1357-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Khakhar A, Bolten NJ, Nemhauser J, Klavins E. 2016. Cell–cell communication in yeast using auxin biosynthesis and auxin responsive CRISPR transcription factors. ACS Synth Biol 5: 279–286. 10.1021/acssynbio.5b00064 [DOI] [PubMed] [Google Scholar]
- Khakhar A, Leydon AR, Lemmex AC, Klavins E, Nemhauser JL. 2018. Synthetic hormone-responsive transcription factors can monitor and re-program plant development. eLife 7: e34702. 10.7554/eLife.34702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. 1997. Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci 94: 11786–11791. 10.1073/pnas.94.22.11786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Schopfer P. 2018. The plant hormone auxin beats the time for oscillating light-regulated lateral root induction. Development 145: dev169839. 10.1242/dev.169839 [DOI] [PubMed] [Google Scholar]
- Kitano H. 2002. Systems biology: a brief overview. Science 295: 1662–1664. 10.1126/science.1069492 [DOI] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC. 2014. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci 111: 5427–5432. 10.1073/pnas.1400074111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Ballas N, Wong LM, Theologis A. 1995. Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum). J Mol Biol 253: 396–413. 10.1006/jmbi.1995.0562 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Harper CP. 2018. The roles of auxin during interactions between bacterial plant pathogens and their hosts. J Exp Bot 69: 245–254. 10.1093/jxb/erx447 [DOI] [PubMed] [Google Scholar]
- Lanctot A, Taylor-Teeples M, Oki EA, Nemhauser JL. 2020. Specificity in auxin responses is not explained by the promoter preferences of activator ARFs. Plant Physiol 182: 1533–1536. 10.1104/pp.19.01474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane K, Van Valen D, DeFelice MM, Macklin DN, Kudo T, Jaimovich A, Carr A, Meyer T, Pe'er D, Boutet SC, et al. 2017. Measuring signaling and RNA-Seq in the same cell links gene expression to dynamic patterns of NF-κB activation. Cell Syst 4: 458–469.e5. 10.1016/j.cels.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Prigge MJ, Tao S, Shain S, Kuo A, Kirchsteiger K, Estelle M. 2016. Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins. eLife 5: e13325. 10.7554/eLife.13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leydon AR, Gala HP, Guiziou S, Nemhauser JL. 2020. Engineering synthetic signaling in plants. Annu Rev Plant Biol 71: 767–788. 10.1146/annurev-arplant-081519-035852 [DOI] [PubMed] [Google Scholar]
- Leyser O. 2018. Auxin signaling. Plant Physiol 176: 465–479. 10.1104/pp.17.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopatto D, Alvarez C, Barnard D, Chandrasekaran C, Chung HM, Du C, Eckdahl T, Goodman AL, Hauser C, Jones CJ, et al. 2008. Undergraduate research: genomics education partnership. Science 322: 684–685. 10.1126/science.1165351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Miotk A, Šutiković Z, Ermakova O, Wenzl C, Medzihradszky A, Gaillochet C, Forner J, Utan G, Brackmann K, et al. 2019. WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat Commun 10: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Budde MW, Mayalu MN, Zhu J, Murray RM, Elowitz MB. 2020. Synthetic mammalian signaling circuits for robust cell population control. bioRxiv 10.1101/2020.09.02.278564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes MS, Best NB, Robil JM, Malcomber S, Gallavotti A, McSteen P. 2019. Auxin EvoDevo: conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Mol Plant 12: 298–320. 10.1016/j.molp.2018.12.012 [DOI] [PubMed] [Google Scholar]
- Mazzocchi F. 2012. Complexity and the reductionism–holism debate in systems biology. Wiley Interdiscip Rev Syst Biol Med 4: 413–427. 10.1002/wsbm.1181 [DOI] [PubMed] [Google Scholar]
- Montgomery B. 2018. From deficits to possibilities: mentoring lessons from plants on cultivating individual growth through environmental assessment and optimization. Public Philos J 1: 1– 12. osf.io/g83s9 [Accessed October 29, 2020]. [Google Scholar]
- Montgomery BL. 2020a. Lessons from microbes: what can we learn about equity from unculturable bacteria? mSphere 5: e01046. msphere.asm.org/content/5/5/e01046-20, 10.1128/mSphere.01046-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BL. 2020b. Planting equity: using what we know to cultivate growth as a plant biology community. Plant Cell 32: 3372–3375. 10.1105/tpc.20.00589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss BL, Mao H, Guseman JM, Hinds TR, Hellmuth A, Kovenock M, Noorassa A, Lanctot A, Villalobos LIAC, Zheng N, et al. 2015. Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol 169: 803–813. 10.1104/pp.15.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murren CJ, Wolyniak MJ, Rutter MT, Bisner AM, Callahan HS, Strand AE, Corwin LA. 2019. Undergraduates phenotyping Arabidopsis knockouts in a course-based undergraduate research experience: exploring plant fitness and vigor using quantitative phenotyping methods. J Microbiol Biol Educ 20: 1–9. 10.1128/jmbe.v20i2.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GKS, Weijers D. 2018. Origin and evolution of the nuclear auxin response system. eLife 7: e33399. 10.7554/eLife.33399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Yamada R, Hagihara S, Iwasaki R, Uchida N, Kamura T, Takahashi K, Torii KU, Fukagawa T. 2020. A super-sensitive auxin-inducible degron system with an engineered auxin-TIR1 pair. Nucleic Acids Res 48: e108–e108. 10.1093/nar/gkaa748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster G, Rocklin S. 1979. Optimization models in evolutionary biology. In Some mathematical questions in biology (ed. Levin S), Vol. X, pp. 21–88. American Mathematical Society, Providence, RI. [Google Scholar]
- Pierre-Jerome E, Jang SS, Havens KA, Nemhauser JL, Klavins E. 2014. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc Natl Acd Sci 111: 9407–9412. 10.1073/pnas.1324147111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau B, Vanhercke T, Singh S. 2018. From plant metabolic engineering to plant synthetic biology: the evolution of the design/build/test/learn cycle. Plant Sci 273: 3–12. 10.1016/j.plantsci.2018.03.035 [DOI] [PubMed] [Google Scholar]
- Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, et al. 2019. Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana. Mol Cell 76: 177–190.e5. 10.1016/j.molcel.2019.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, et al. 2020. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9: e54740. 10.7554/eLife.54740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos Báez R, Buckley Y, Yu H, Chen Z, Gallavotti A, Nemhauser JL, Moss BL. 2020. A synthetic approach allows rapid characterization of the maize nuclear auxin response circuit. Plant Physiol 182: 1713–1722. 10.1104/pp.19.01475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Birnbaum KD, Ehrhardt DW. 2019. Towards building a plant cell atlas. Trends Plant Sci 24: 303–310. 10.1016/j.tplants.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosjen M, Paque S, Weijers D. 2018. Auxin response factors: output control in auxin biology. J Exp Bot 69: 179–188. 10.1093/jxb/erx237 [DOI] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. 1998. Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373. 10.1126/science.279.5355.1371 [DOI] [PubMed] [Google Scholar]
- Ruschhaupt M, Mergner J, Mucha S, Papacek M, Doch I, Tischer SV, Hemmler D, Chiasson D, Edel KH, Kudla J, et al. 2019. Rebuilding core abscisic acid signaling pathways of Arabidopsis in yeast. EMBO J 38: e101859. 10.15252/embj.2019101859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeradova H, Montesinos JC, Benkova E. 2020. All roads lead to auxin: post-translational regulation of auxin transport by multiple hormonal pathways. Plant Commun 1: 100048. 10.1016/j.xplc.2020.100048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P, Zhao L, Li W, Zhao J, Yan J, Ma X, Li A, Wang H, Kong L. 2021. Integrated metabolo-transcriptomics and functional characterization reveals that the wheat auxin receptor TIR1 negatively regulates defense against Fusarium graminearum. J Integr Plant Biol 63: 340–352. 10.1111/jipb.12992 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Theologis A, Huynh TV, Davis RW. 1985. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol 183: 53–68. 10.1016/0022-2836(85)90280-3 [DOI] [PubMed] [Google Scholar]
- Uchida N, Takahashi K, Iwasaki R, Yamada R, Yoshimura M, Endo TA, Kimura S, Zhang H, Nomoto M, Tada Y, et al. 2018. Chemical hijacking of auxin signaling with an engineered auxin–TIR1 pair. Nat Chem Biol 14: 299–305. 10.1038/nchembio.2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. 10.1038/msb.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent BJ, Estrada J, DePace AH. 2016. The appeasement of Doug: a synthetic approach to enhancer biology. Integr Biol (Camb) 8: 475–484. [DOI] [PubMed] [Google Scholar]
- Walker JC, Key JL. 1982. Isolation of cloned cDNAs to auxin-responsive poly(A)+RNAs of elongating soybean hypocotyl. Proc Natl Acad Sci 79: 7185–7189. 10.1073/pnas.79.23.7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M, Cardelli L, Kwiatkowska M, Laurenti L, Tribastone M, Tschaikowski M. 2021. PID control of biochemical reaction networks. IEEE Trans Automat Contr 10.1109/TAC.2021.3062544 [DOI] [Google Scholar]
- World Economic Forum Young Scientists Community. 2020. World economic forum code of ethics for researchers. widgets.weforum.org/coe/#code
- Wright RC, Zahler ML, Gerben SR, Nemhauser JL. 2017. Insights into the evolution and function of auxin signaling F-Box proteins in Arabidopsis thaliana through synthetic analysis of natural variants. Genetics 207: 583–591. 10.1534/genetics.117.300092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Moss BL, Jang SS, Prigge M, Klavins E, Nemhauser JL, Estelle M. 2013. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol 162: 295–303. 10.1104/pp.113.215582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J. 2001. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci 98: 11795–11800. 10.1073/pnas.211312798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF. 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142: 4374–4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZB, Wang QY, Ke YX, Liu SY, Ju JQ, Lim WA, Tang C, Wei P. 2017. Design of tunable oscillatory dynamics in a synthetic NF-κB signaling circuit. Cell Syst 5: 460–470.e5. 10.1016/j.cels.2017.09.016 [DOI] [PubMed] [Google Scholar]
- Zhang H, Li L, He Y, Qin Q, Chen C, Wei Z, Tan X, Xie K, Zhang R, Hong G, et al. 2020. Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc Natl Acad Sci 117: 9112–9121. 10.1073/pnas.1918254117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen W, Mirabet V, Hong L, Bovio S, Strauss S, Schwarz EM, Tsugawa S, Wang Z, Smith RS, et al. 2020. Robust organ size requires robust timing of initiation orchestrated by focused auxin and cytokinin signalling. Nat Plants 6: 686–698. 10.1038/s41477-020-0666-7 [DOI] [PMC free article] [PubMed] [Google Scholar]