ABSTRACT
Increased macroautophagy/autophagy and lysosomal activity promote tumor growth, survival and chemo-resistance. During acute starvation, autophagy is rapidly engaged by AMPK (AMP-activated protein kinase) activation and MTOR (mechanistic target of rapamycin kinase) complex 1 (MTORC1) inhibition to maintain energy homeostasis and cell survival. TFEB (transcription factor E3) and TFE3 (transcription factor binding to IGHM enhancer 3) are master transcriptional regulators of autophagy and lysosomal activity and their cytoplasm/nuclear shuttling is controlled by MTORC1-dependent multisite phosphorylation. However, it is not known whether and how the transcriptional activity of TFEB or TFE3 is regulated. We show that AMPK mediates phosphorylation of TFEB and TFE3 on three serine residues, leading to TFEB and TFE3 transcriptional activity upon nutrient starvation, FLCN (folliculin) depletion and pharmacological manipulation of MTORC1 or AMPK. Collectively, we show that MTORC1 specifically controls TFEB and TFE3 cytosolic retention, whereas AMPK is essential for TFEB and TFE3 transcriptional activity. This dual and opposing regulation of TFEB and TFE3 by MTORC1 and AMPK is reminiscent of the regulation of another critical regulator of autophagy, ULK1 (unc-51 like autophagy activating kinase 1). Surprisingly, we show that chemoresistance is mediated by AMPK-dependent activation of TFEB, which is abolished by pharmacological inhibition of AMPK or mutation of serine 466, 467 and 469 to alanine residues within TFEB. Altogether, we show that AMPK is a key regulator of TFEB and TFE3 transcriptional activity, and we validate AMPK as a promising target in cancer therapy to evade chemotherapeutic resistance.
Abbreviations: ACACA: acetyl-CoA carboxylase alpha; ACTB: actin beta; AICAR: 5-aminoimidazole-4-carboxamide ribonucleotide; AMPK: AMP-activated protein kinase; AMPKi: AMPK inhibitor, SBI-0206965; CA: constitutively active; CARM1: coactivator-associated arginine methyltransferase 1; CFP: cyan fluorescent protein; CLEAR: coordinated lysosomal expression and regulation; DKO: double knock-out; DMEM: Dulbecco’s modified Eagle’s medium; DMSO: dimethyl sulfoxide; DQ-BSA: self-quenched BODIPY® dye conjugates of bovine serum albumin; EBSS: Earle’s balanced salt solution; FLCN: folliculin; GFP: green fluorescent protein; GST: glutathione S-transferases; HD: Huntington disease; HTT: huntingtin; KO: knock-out; LAMP1: lysosomal associated membrane protein 1; MEF: mouse embryonic fibroblasts; MITF: melanocyte inducing transcription factor; MTORC1: MTOR complex 1; PolyQ: polyglutamine; RPS6: ribosomal protein S6; RT-qPCR: reverse transcription quantitative polymerase chain reaction; TCL: total cell lysates; TFE3: transcription factor binding to IGHM enhancer 3; TFEB: transcription factor EB; TKO: triple knock-out; ULK1: unc-51 like autophagy activating kinase 1.
KEYWORDS: AMP-activated protein kinase, autophagy, drug resistance, lysosomal biogenesis, mechanistic target of rapamycin kinase, phosphorylation, transcription factor EB, transcription factor E3
Introduction
Macroautophagy/autophagy, the process of cellular self-degradation, regulates many processes including energy supply during development and in response to acute nutrient stress [1,2]. Lysosomal biogenesis and autophagic activities are rapidly upregulated in response to starvation, oxidative stress, organelle damage and chemotherapy resistance [3]. Degradation of cargos inside the lysosome is a multistep process that is conserved in eukaryotes from yeast to multicellular organisms [4]. Fragments resulting from autophagic protein degradation are recycled to produce energy, allowing cells to survive under harsh circumstances. Deregulation of these pathways has been linked to various human disorders, including myopathy, neurodegeneration, cancer and metabolic syndrome [5,6].
The MiT/TFE (microphthalmia family of bHLH-LZ) transcription factor family, namely TFEB (transcription factor EB) and TFE3 (transcription factor binding to IGHM enhancer 3) [7] are master transcriptional regulators of autophagy and lysosomal biogenesis. TFEB and TFE3 homo- or hetero-dimerize and bind to gene promoter regions to activate a panel of genes involved in autophagy, lysosomal biogenesis, lipid metabolism, innate immune response and pathogen resistance [8–11]. They are redundant in some cellular contexts while indispensable in others [8,12,13].
TFEB and TFE3 have been linked with tumorigenesis. Both proteins have been implicated in chromosomal translocations resulting in the fusion of partial TFEB or TFE3 coding regions with strong promoters, such as MALAT1, PRCC, ASPSCR1, SFPQ, NONO and CLTC [6,14]. Such fusion events enhance TFEB and TFE3 expression and activity and is strongly associated with juvenile renal cell carcinoma and alveolar soft part sarcoma [14,15]. The oncogenic effect of these transcription factors is likely due to altered gene expression. Increased autophagy and lysosomal biogenesis may activate several pathways promoting cell survival, tumor growth and progression [2]. Typically, autophagy is induced during cancer therapy, which protects cancer cells and leads to drug resistance and refractory cancer [16]. Therefore, reducing TFEB and TFE3 activity might be important in cancer therapy settings.
Interestingly, TFEB and TFE3 also play important roles in non-dividing neurons to clear toxins and misfolded proteins through regulation of autophagic pathways [17]. Neurodegenerative diseases, including Alzheimer disease, Parkinson disease, and Huntington disease (HD) are characterized by the accumulation of intracellular aggregates in the brain [18]. Specifically in HD, expansion of a CAG trinucleotide repeat in the first exon of the HTT (huntingtin) gene generates a protein containing an expanded polyglutamine (polyQ) tract, leading to pathogenic misfolding [17]. Increased autophagic/lysosomal activity was shown to reduce aggregates, revert symptoms and to restore cognitive capabilities [19–22]. Understanding the pathways modulating TFEB and TFE3 activity is therefore crucial to identify new targets for treatment of autophagy and lysosomal biogenesis related diseases, such as Alzheimer disease, Parkinson disease and HD.
To date, the best characterized regulator of TFEB and TFE3 is MTOR complex 1 (MTORC1). MTORC1 is a mediator of cellular growth and proliferation that primarily integrates stress and growth signals by phosphorylating downstream targets to promote anabolic processes such as protein synthesis [23]. Under nutrient replete conditions, MTORC1 negatively regulates catabolic processes, including autophagy, by directly phosphorylating and inhibiting TFEB (serine residues 211 and 142) and TFE3 (serine residue 321). These phosphorylation events inhibit TFEB and TFE3 activation by promoting their cytoplasmic retention [24–27]. Conversely, under nutrient deficient conditions, these repressive phosphorylation events are removed, resulting in the nuclear translocation of TFEB and TFE3 and activation of autophagy and lysosomal biogenesis [8,11,28]. Similarly, MTORC1 also inhibits autophagy by direct phosphorylation of ULK1 (unc-51 like autophagy activating kinase 1) under nutrient replete conditions [29–31].
AMP-activated protein kinase (AMPK) activates TFEB by blocking the activity of MTORC1 [32] and by increasing the levels of CARM1 (coactivator associated arginine methyltransferase 1), an important cofactor for TFEB activity [33]. AMPK is an energy sensor and plays an essential role in the control of cellular bioenergetics [34] by responding to various stresses including those that induce changes in the cellular AMP:ATP ratio or modulation in intracellular calcium [35]. Upon its activation, AMPK maintains energy homeostasis by promoting energy production and limiting energy expenditure [35]. AMPK activation negatively regulates MTORC1 in two different ways. First, through direct phosphorylation and activation of TSC1 (TSC complex subunit 1)-TSC2, AMPK inactivates RHEB (Ras homolog, mTORC1 binding), leading to subsequent MTORC1 inhibition [36]. Second, through phosphorylation of RPTOR (regulatory associated protein of MTOR complex 1), AMPK promotes the binding of the chaperones YWHA/14-3-3 and MTORC1 inactivation [37]. Moreover, AMPK also regulates autophagosome maturation and autophagy by direct phosphorylation of ULK1 [29–31].
We and others have shown that AMPK can be chronically activated by the loss of FLCN (folliculin), FNIP1 (folliculin interacting protein 1) or FNIP2 [38–44]. FLCN was also identified as a negative regulator of TFEB and TFE3 nuclear localization and activity [45,46]. FLCN is the gene associated with the Birt-Hogg-Dubé neoplastic syndrome in humans, characterized by predisposition to renal cell carcinoma, skin fibrofolliculoma, lung cysts and increased risk for spontaneous pneumothorax [47]. FLCN is thought to activate MTORC1 through its GTPase activating protein activity (GAP) toward RRAGC (Ras related GTP binding C) and RRAGD upon nutrient replenishment [48,49]. RRAGC, RRAGD, and the other members RRAGA and RRAGB (ras related GTP binding A and B), must be in an active conformation to anchor MTORC1 at the lysosome and to allow sensing of amino acids [50]. Therefore, FLCN loss leads to inhibition of RRAGC/RRAGD and removal of MTORC1 from the lysosomal surface, reducing phosphorylation and cytoplasmic sequestration of TFEB and TFE3 [51–53].
In this study, we assessed the role of AMPK in the regulation of TFEB and TFE3 activity under several conditions of cellular stress. We showed that although the subcellular localization of TFEB and TFE3 was regulated by MTORC1, AMPK was required for increased TFEB and TFE3 transcriptional activity, since deletion of AMPK completely blocked TFEB and TFE3 activity upon nutrient starvation, inhibition of MTORC1 or deletion of FLCN. We identified AMPK-dependent phosphorylation sites within TFEB in a highly conserved serine cluster (S466, S467 and S469), which were dispensable for proper subcellular TFEB localization but required for its transcriptional activity. Serine to alanine substitution mutations were sufficient to inhibit TFEB transcriptional activity in the nucleus, even in the context of MTORC1 inhibition or in presence of a constitutively nuclear TFEB mutant (S142A/S211A), which cannot be phosphorylated by MTORC1. AMPK-dependent phosphorylation of TFEB was required for increased resistance to the chemotherapy drug doxorubicin. Our findings shed new light on the interplay of two major metabolic kinases, AMPK and MTORC1, which both regulate autophagy and lysosomal biogenesis via TFEB and TFE3.
Results
AMPK is required for TFEB and TFE3 activity
We first studied the differential TFEB and TFE3 subcellular localization status in wild-type (WT) mouse embryonic fibroblasts (MEFs) compared to prkaa1/AMPKα1 prkaa2/AMPKα2 double knock-out (prkaa DKO) MEFs in a nutrient starvation context, using Earle’s Balanced Salt Solution (EBSS). As a positive control, we used the allosteric AMPK activator AICAR. EBSS starvation for 2 h induced nuclear translocation of TFEB and TFE3 in both WT and prkaa DKO MEFs, while 2 h AICAR treatment induced nuclear translocation only in WT cells (Figure 1A, quantified in B). EBSS inhibited MTORC1 signaling, as shown by reduction in the phosphorylated forms of the known downstream RPS6 (ribosomal protein S6) in both WT and prkaa DKO cells (Figure 1C), suggesting that MTORC1 inhibition upon EBSS starvation was independent of AMPK. Indeed, MTORC1 can directly sense levels of amino acids independently of AMPK activity [50,54–56]. Nonetheless, AMPK was activated by EBSS starvation as shown by the increase in its phosphorylated form (p-AMPK) and phosphorylation of its known downstream target ACACA (acetyl-CoA carboxylase alpha) (Figure 1C). AICAR also activated AMPK and inhibited MTORC1 in WT cells, but not in prkaa DKO cells, pointing to a specific action of AMPK (Figure 1D). We did not observe a reduction in TFEB and TFE3 protein levels upon above mentioned treatments or in prkaa DKO cells when assessed by fluorescence intensity or western blot. Surprisingly, EBSS-dependent increases in DQ-BSA fluorescence intensity, which measures lysosomal protease activity and serves as a proxy for autophagosome/lysosome activity [32,57], was significantly abrogated in prkaa DKO cells (Figure 1E, quantified in F) despite evident TFEB and TFE3 nuclear localization upon EBSS starvation. DQ-BSA is a quenched fluorogenic substrate for intracellular lysosomal proteases. Cleavage of the fluorescent peptides within the lysosome relieves quenching, resulting in a fluorescent signal. As expected, AICAR-dependent increase in DQ-BSA fluorescence intensity was abrogated in prkaa DKO cells (Figure 1E, quantified in F). Treatment with EBSS and AICAR also increased the number of lysosomes in WT but not prkaa DKO cells, as assessed by counting the average number of LAMP1 (lysosomal associated membrane protein 1) puncta per cell (Figure 1G). This increase in number was not accompanied by a difference in lysosome size (Figure 1H). Moreover, we measured TFEB and TFE3 transcriptional activity using a luciferase reporter containing the TFEB and TFE3 consensus promoter region (Coordinated Lysosomal Expression and Regulation; CLEAR). Treatment of WT MEFs with EBSS and AICAR induced expression from the CLEAR luciferase reporter, which was not observed in prkaa DKO cells (Figure 1I,J). Furthermore, the induction of known TFEB and TFE3 target genes such as Atp6v1c1, Atp6v0d1, Ctsa, Ctsd, Gabarap and Sdha by EBSS starvation and AICAR treatment was strongly reduced in prkaa DKO cells as measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR) (Figure 1K,L). Taken together, these results suggest that although TFEB and TFE3 nuclear localization is not dependent on AMPK upon EBSS starvation, AMPK plays an essential role in regulating TFEB and TFE3 transcriptional activity.
Figure 1.
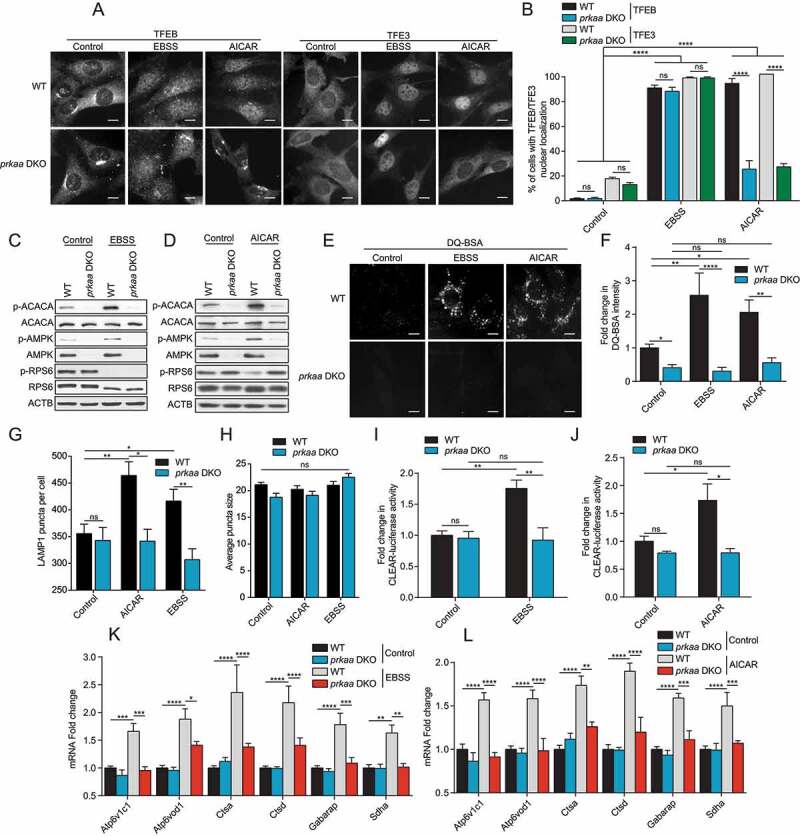
AMPK is required for complete TFEB and TFE3 activity upon EBSS starvation and AICAR treatment. (A) WT and prkaa DKO MEFs were incubated in complete media (Control), starved (EBSS) or in presence of AICAR (2 mM) for 2 h. Cells were fixed, permeabilized, and stained with an antibody against TFEB or TFE3. Scale bar: 20 μm. Images are representative of three independent experiments. (B) Quantification of the percentage of indicated MEF lines with nuclear TFEB or TFE3 upon treatments as indicated in (A) (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, ****P < 0.0001; n > 200 cells per condition). (C-D) Immunoblot of protein lysates of indicated MEF lines treated as indicated in (A). Data are representative of three independent experiments. (E) Representative images of indicated MEF lines after 1 h of incubation with DQ-BSA-Red followed by a 2 h chase in complete media (Control), starved (EBSS) or in presence of AICAR (2 mM) prior to fixation. Scale bar: 20 μm. Images are representative of four independent experiments. (F) Relative lysosomal activity, as determined by DQ-BSA assay, in indicated MEF lines upon treatment as indicated in (E) (mean ± SEM of four independent experiments, two-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01, ****P < 0.0001; n > 200 cells per condition). (G) Quantification of the number of LAMP1 puncta upon treatment as indicated in (A). (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01; n > 50 cells per condition). (H) Quantification of the average LAMP1 puncta size upon treatment as indicated in (A). (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, n > 50 cells per condition). (I-J) Relative Tfeb and Tfe3 transcriptional activity, as determined by CLEAR-luciferase promoter activity normalized against CMV-Renilla in indicated MEF lines upon treatment as indicated in (A) (mean ± SEM of the luminescence fold change from five independent experiments, two-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01). (K-L) Relative quantitative real‐time PCR analysis of Tfeb and Tfe3 target genes mRNA transcript levels in indicated MEF lines upon treatment as indicated in (A) (mean ± SEM of the RNA fold change of indicated mRNAs from five independent experiments, two-way ANOVA, *P < 0.05, ***P < 0.001, ****P < 0.0001)
MTORC1 inhibition requires AMPK for complete TFEB and TFE3 activity
We showed above that EBSS starvation of WT cells led to simultaneous AMPK activation and MTORC1 inhibition. We also observed MTORC1 inhibition upon EBSS starvation in prkaa DKO cells, which is probably due to the ability of MTORC1 to sense amino acid levels independently of AMPK. Since AMPK is a negative upstream regulator of MTORC1, it is conceivable that the observed inhibition of MTORC1, and not the activation of AMPK, led to the translocation of TFEB and TFE3 in both cases. To test this hypothesis, we examined whether MTORC1 inhibition using Torin1 would show a similar AMPK-dependent effect on TFEB and TFE3 transcriptional activity. To extend our observations beyond MEF cells, we generated human HEK293T prkaa DKO using CRISPR-Cas9. Gene knock-out was confirmed by western blot using an antibody recognizing both the α1 and α2 subunits (Figure 2A). Torin1 treatment for 2 h led to inhibition of MTORC1 signaling in both WT MEFs and HEK293T and prkaa DKO cell lines (Figure 2A,B), as shown by a decrease in levels of pRPS6, and led to nuclear localization of TFEB and TFE3 in both WT and prkaa DKO cells (Figure 2C, quantified in D and E). Since TFEB is not well expressed in HEK293T cells we focused only on TFE3 localization in these cells. In line with results in Figure 1, whereas Torin1 treatment led to an increase in lysosomal activity measured by DQ-BSA fluorescence in WT cells, this effect was abolished in prkaa DKO cells (Figure 2F, quantified in G). This was accompanied with an increase in lysosome number in WT cells compared to prkaa DKO when treated with Torin1 (Figure 2H), but no statistical difference in terms of lysosome size was observed (Figure 2I). In addition, TFEB and TFE3 transcriptional activity was induced upon treatment with Torin1, which was not observed in prkaa DKO cells by both CLEAR luciferase reporter (Figure 2J,K) and RT-qPCR (Figure 2L,M) assays in MEFS and HEK293T cells. These results suggest that TFEB and TFE3 transcriptional activity is dictated by an MTORC1-independent but AMPK-dependent mechanism, whereas nuclear translocation is only dependent on MTORC1.
Figure 2.
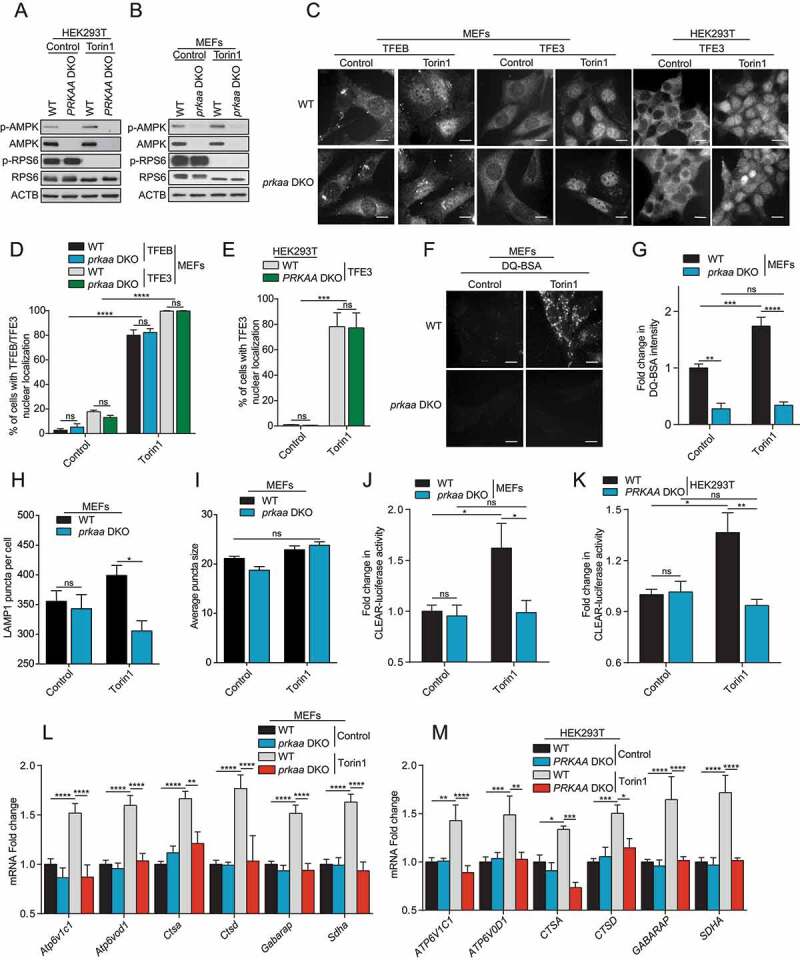
MTORC1 inhibition requires AMPK for complete TFEB and TFE3 activity. (A-B) Immunoblot of protein lysates of HEK293T (A) and MEFs (B) with indicated genotypes incubated in presence of DMSO (Control) or torin1 (1 μM) for 2 h. Data are representative of three independent experiments. (C) Representative images of cell lines treated as indicated in (A-B). Cells were fixed, permeabilized, and stained with an antibody against TFEB or TFE3. Scale bar: 20 μm. Images are representative of three independent experiments. (D-E) Quantification of the percentage of MEFs (D) and HEK293T (E) with nuclear TFEB or TFE3 upon treatments as indicated in (A-B) (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, ***P < 0.001, ****P < 0.0001; n > 200 cells per condition). (F) Representative images of MEFs after 1 h of incubation with DQ-BSA-Red followed by a 2-h chase in complete media in presence of DMSO (Control) or torin1 (1 μM) prior to fixation. Scale bar: 20 μm. Images are representative of four independent experiments. (G) Relative lysosomal activity, as determined by DQ-BSA assay, in MEFs upon treatment as indicated in (F) (mean ± SEM of four independent experiments, two-way ANOVA, ns = not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001; n > 200 cells per condition). (H) Quantification of the number of LAMP1 puncta in MEFs upon treatment as indicated in (A). (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, *P < 0.05; n > 50 cells per condition). (I) Quantification of the average LAMP1 puncta size in MEFs upon treatment as indicated in (A). (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, n > 50 cells per condition). (J-K) Relative Tfeb and Tfe3 transcriptional activity, as determined by CLEAR-luciferase promoter activity normalized against CMV-Renilla in MEFs (J) and HEK293T (K) upon treatment as indicated in (A) (mean ± SEM of the luminescence fold change from four independent experiments, two-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01). (L-M) Relative quantitative real‐time PCR analysis of Tfeb and Tfe3 target genes mRNA transcript levels in MEFs (L) and HEK293T (M) upon treatment as indicated in (A) (mean ± SEM of the RNA fold change of indicated mRNAs from four independent experiments, two-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
AMPK is required for FLCN-dependent activation of TFEB and TFE3
To further substantiate our observations that AMPK is required for TFEB and TFE3 activity, we tested conditions where TFEB and TFE3 are constitutively active and localized to the nucleus, such as in flcn KO cells [45,58]. To this end, we generated flcn KO + prkaa DKO MEFs (TKO; Figure 3A). Upon loss of FLCN, TFEB and TFE3 were localized permanently to the nucleus, which remained unchanged upon deletion of AMPK in TKO cells (Figure 3B, quantified in C). These results demonstrate that the nuclear localization of these transcription factors, in the context of FLCN deletion, is independent of AMPK as we observed above with EBSS and Torin1 treatment. However, the transcriptional activity of TFEB and TFE3 observed in flcn KO cells was abolished upon deletion of AMPK in TKO cells, as measured by DQ-BSA assay (Figure 3D, quantified in E), CLEAR luciferase reporter assay (Figure 3F) and RT-qPCR of known TFEB and TFE3 target genes (Figure 3G), suggesting that AMPK is required for FLCN-dependent transcriptional activation of TFEB and TFE3.
Figure 3.
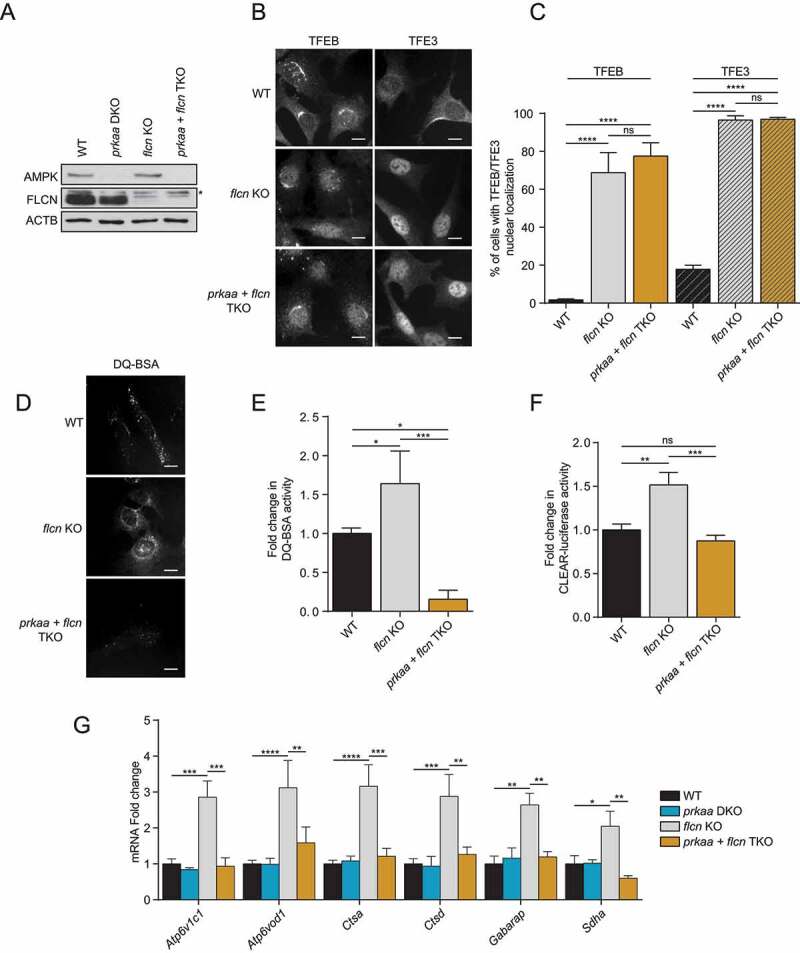
AMPK is required for FLCN-dependent activation of TFEB and TFE3. (A) Immunoblot of protein lysates of MEFs with indicated genotypes were incubated in complete media. Data are representative of three independent experiments. * represents nonspecific bands. (B) Representative images of indicated MEF lines incubated in complete media. Cells were fixed, permeabilized, and stained with an antibody against TFEB or TFE3. Scale bar: 20 μm. Images are representative of three independent experiments. (C) Quantification of the percentage of nuclear TFEB or TFE3 in MEFs incubated in complete media (mean ± SEM of three independent experiments, one‐way ANOVA, ns = not significant, ****P < 0.0001; n > 200 cells per condition). (D) Representative images of indicated MEF lines after 1 h of incubation with DQ-BSA-Red followed by a 2 h chase in complete media prior to fixation. Scale bar: 20 μm. Images are representative of three independent experiments. (E) Relative lysosomal activity, as determined by DQ-BSA assay in MEFs upon treatment as indicated in (D) (mean ± SEM of three independent experiments, one‐way ANOVA, *P < 0.05, ***P < 0.001; n > 200 cells per condition). (F) Relative Tfeb and Tfe3 transcriptional activity, as determined by CLEAR-luciferase promoter activity normalized against CMV-Renilla in MEFs incubated in complete media (mean ± SEM of the luminescence fold change from six independent experiments, one-way ANOVA, ns = not significant, **P < 0.01, ***P < 0.001) (G) Relative quantitative real‐time PCR analysis of Tfeb and Tfe3 target genes mRNA transcript levels in MEFs incubated in complete media (mean ± SEM of the RNA fold change of indicated mRNAs from four independent experiments, two-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
AMPK is required for clearance of polyQ protein aggregates in a model of Huntington disease
To substantiate the physiological relevance of our results we used a cellular model of Huntington disease (HD), which is dependent on stimulation of autophagy and lysosomal biogenesis. Several groups have reported that MTORC1 inhibition leads to clearance of intracellular protein aggregates [59–61]. To this end, we generated stable WT, prkaa DKO, flcn KO and prkaa flcn TKO MEFs expressing a doxycycline-inducible fusion protein consisting of the cyan fluorescent protein (CFP) merged to a chain of 94 glutamine residues. Following a three-day induction with doxycycline, the polyQ chains form fluorescent aggregates mimicking aggregates formed in HD neurons that require autophagy and lysosomal activity for their clearance [62]. Treatment with AICAR or torin1 led to a decrease in polyQ aggregates in WT cells but not in prkaa DKO cells (Fig. S1A, quantified in B). In addition, prkaa DKO cells displayed increased accumulation of polyQ aggregates upon treatment with vehicle consistent with impaired autophagic and lysosomal activity (Fig. S1A, quantified in B). Consistently with the constitutive TFEB and TFE3 nuclear localization and increased activity upon FLCN loss, flcn KO cells displayed a reduction in protein aggregates (Fig. S1C, quantified in D). Interestingly, the additional AMPK loss in flcn KO increased the number of cells with protein aggregates to a level similar to WT cells (Fig. S1C, quantified in D). Moreover, we observed a reduction of insoluble protein aggregates in WT cells but not in prkaa DKO as measured by fractionation of soluble versus insoluble protein (Fig. S1E, quantified in F). These results suggest that the presence and activity of AMPK is required for clearance of insoluble protein aggregates upon treatment with a MTORC1 inhibitor.
AMPK phosphorylates TFEB and TFE3 on highly conserved serine cluster 466, 467, 469
Since AMPK is required for TFEB and TFE3 transcriptional activity and is a serine/threonine kinase, we next assessed whether AMPK regulates TFEB and TFE3 activity via phosphorylation. Since TFEB and TFE3 appear to be regulated in a similar fashion, we focused our phosphorylation studies on TFEB for simplicity. We developed an in vitro phosphorylation assay using recombinant AMPK and TFEB. After incubation with [γ-32P] ATP, we observed AMPK-dependent TFEB phosphorylation, which was enhanced after the addition of AMP, an allosteric AMPK activator (Figure 4A). The level of phosphorylation on TFEB was similar to another well-characterized AMPK substrate, ULK1 (Figure 4B). It is noteworthy to mention that some phosphorylation on ULK1 could be due to autophosphorylation. Strikingly, the phosphorylation efficiency of AMPK toward TFEB was more significant than toward ACACA, suggesting that TFEB is a better substrate than this very well characterized substrate (Figure 4C). Interestingly, immunoprecipitation of endogenous AMPK revealed that AMPK and TFEB form a complex in HEK293T cells, which was slightly increased upon the addition of AICAR (Figure 4D). In order to reveal potential AMPK phosphorylation sites on TFEB, we purified recombinant fragments of GST-TFEB from bacteria, performed a kinase assay with purified AMPK, and analyzed GST-TFEB with mass spectrometry. This approach identified phosphorylation of the serine residues 467 and 469 (Fig. S2A and B). The C-terminal region is highly conserved throughout evolution and is maintained in other MiT/TFE family members (Figure 4E). Moreover, these two serine residues and the adjacent serine (S466) resemble an AMPK phosphorylation motif (Figure 4F). Although the phosphorylation motif is not a perfect fit, it has been previously shown that AMPK can potently phosphorylate its substrate without the canonical motif, such as ULK1 (Figure 4F) [63–65]. Importantly, while WT GST-TFEB is efficiently phosphorylated by AMPK in vitro, a truncation of the 36 C-terminal residues led to a dramatic reduction in phosphorylation (Figure 4G). Furthermore, although single point mutations of the three serine residues to alanine (S466, S467 or S469) did not reduce phosphorylation to a large extent, a triple S466A S467A S469A mutation led to a significant inhibition of AMPK-dependent TFEB phosphorylation in vitro (Figure 4H). Similarly, point mutations of serine residues at 567, 568, 570 in TFE3 (corresponding to S466, S467, S469 in TFEB) to alanine also led to a significant reduction of AMPK-dependent TFE3 phosphorylation in vitro (Figure 4I). Importantly, we generated a phospho-specific antibody raised against a triple-phosphorylated peptide p-S466 p-S467 p-S469 corresponding to the human C-terminal region of TFEB and p-S567 p-S568 p-S570 of TFE3 to measure potential phosphorylation by AMPK in cultured cells. This antibody recognized a phosphorylated TFEB fragment incubated in presence of purified AMPK, but not a fragment harboring serine to alanine mutations (Figure 4J). As the phosphorylated peptide used to immunize the rabbits is perfectly conserved in TFE3, the antibody recognized a phosphorylated TFE3 fragment incubated in presence of AMPK as well, but not the fragment S567A S568A S570A (Figure 4K). To assess potential phosphorylation by AMPK in cultured cells and to better discriminate between TFEB and TFE3, we immunoprecipitated both p-TFEB and p-TFE3 from HEK293T cells using this antibody and measured the relative abundance of each phospho-protein using specific antibodies against total TFEB and TFE3. Starvation in EBSS revealed a 2.9-fold increase in the abundance of phosphorylated TFEB and a 2.6-fold increase in phosphorylated TFE3, which was not apparent in prkaa DKO cells (Figure 4L, quantified in M). Similarly, treatment with AICAR increased the abundance of phosphorylated TFEB by 2.5-fold and TFE3 by 2-fold (Figure 4L, quantified in M). Therefore, we conclude that AMPK phosphorylates TFEB and TFE3 on this highly conserved C-terminal serine cluster in vitro and in cultured cells.
Figure 4.
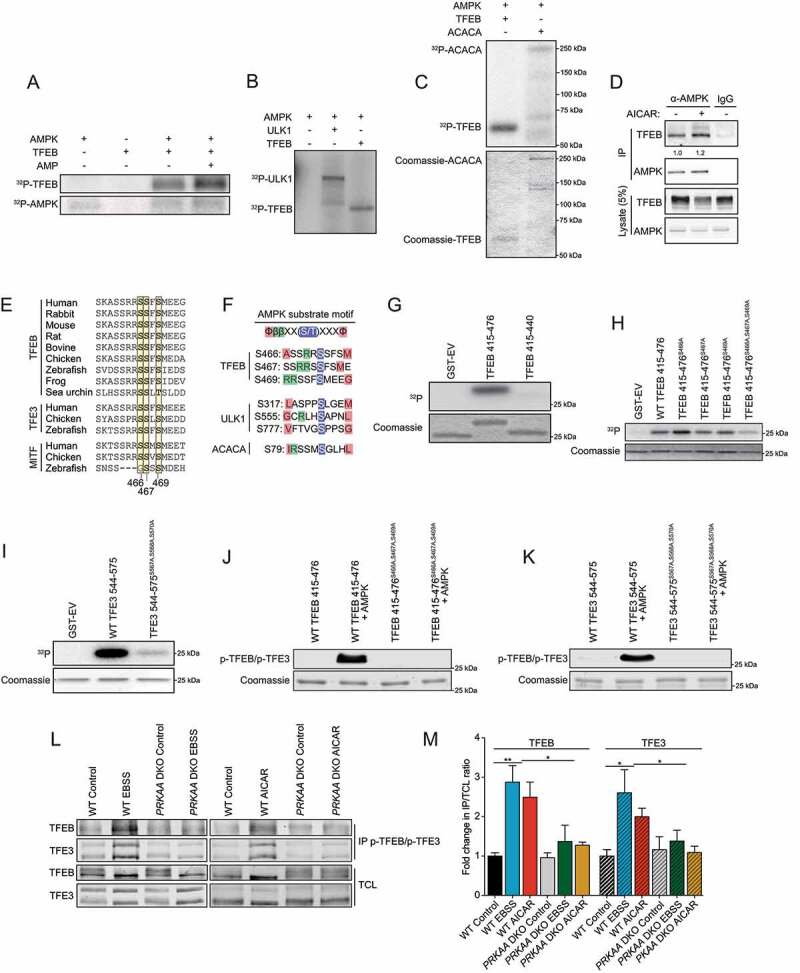
AMPK phosphorylates TFEB and TFE3 on highly conserved serine cluster 466, 467, 469. (A) In vitro kinase assays. Purified AMPK (0.1 μg) and recombinant TFEB (0.5 μg) were incubated with [γ-32P]ATP with or without AMP and analyzed by SDS-PAGE followed by autoradiography. Data are representative of three independent experiments. (B) In vitro kinase assays. Purified AMPK (0.1 μg), recombinant TFEB (0.5 μg), and recombinant ULK1 (0.5 μg) incubated as described in (A) (C) In vitro kinase assays. Purified AMPK (0.05 μg), recombinant TFEB (0.5 μg), and recombinant ACACA (0.5 μg) incubated as described in (A). (D) Endogenous protein complexes were immuno-purified from HEK293T cells treated with vehicle or 2 mM AICAR for 2 h with AMPKα1/2 antibody and analyzed by immunoblotting with indicated antibodies. Nonspecific IgG was used as a negative control. Data are representative of three independent experiments, quantification of TFEB protein levels normalized to AMPK protein level, Student’s t-test, p = 0.03. (E) Alignment of TFEB, TFE3, and MITF amino acid C-terminal regions from various species. (F) AMPK substrate motif alignment surrounding the serine residues 466, 467 and 469 in human TFEB, ULK1, and ACACA. (G) In vitro kinase assays using [γ-32P]ATP, Purified AMPK, and GST-TFEB fragments purified from bacteria as described and analyzed by SDS-PAGE followed by Coomassie staining and autoradiography. Data are representative of three independent experiments. (H) In vitro kinase assays using [γ-32P]ATP, Purified AMPK, and GST-TFEB fragments purified from bacteria as described and analyzed by SDS-PAGE followed by Coomassie staining and autoradiography. Data are representative of three independent experiments. (I) In vitro kinase assays using [γ-32P]ATP, Purified AMPK, and GST-TFE3 fragments purified from bacteria as described and analyzed by SDS-PAGE followed by Coomassie staining and autoradiography. Data are representative of three independent experiments. (J-K) In vitro kinase assay using cold ATP, Purified AMPK, and GST-TFEB or GST-TFE3 fragments purified from bacteria and analyzed by immunoblotting with the phospho-specific TFEB and TFE3 antibody. (L) Endogenous phosphorylated TFEB and TFE3 were immunoprecipitated with pTFEB/pTFE3 antibody followed by immunoblotting using either total TFEB or total TFE3 antibodies. WT HEK293T or prkaa DKO cells were incubated in complete media (Control), starved (EBSS), or in presence of AICAR (2 mM) for 2 h. Data are representative of three independent experiments. (M) Quantification of the fold change in pTFEB and pTFE3 purified compared to TFEB and TFE3 in total cell lysates (TCL) ratio upon treatment as described in (L) (mean ± SEM of three independent experiments, two-way anova, *P < 0.05; **P < 0.01)
Phosphorylation of the serine cluster (S466 S467 S469) is required for TFEB activity
To address the biological importance of the potential AMPK-mediated TFEB phosphorylation, we generated MEF cell lines stably expressing either full-length WT TFEB-GFP-3xFLAG or mutant TFEB-GFP-3xFLAGS466A,S467A,S469A. Expression levels of WT and mutant TFEB proteins were comparable (Figure 5A). Using a phospho-specific antibody for TFEB Ser211, the major MTORC1 phosphorylation site, we observed a reduction in p-S211-TFEB levels in both the WT and mutant upon AICAR and torin1 treatment demonstrating TFEB dephosphorylation, which should lead to nuclear translocation of WT and mutant TFEB (Figure 5A). As expected, treatment with both compounds led to decreased MTORC1 activity as measured by immunoblot (Figure 5A). Moreover, both AICAR and Torin1 treatment led to nuclear localization of WT and TFEBS466A,S467A,S469A (Figure 5B, quantified in C), suggesting that phosphorylation of this serine cluster 466–469 is not important for TFEB nuclear localization. Strikingly, despite its nuclear localization and similar expression levels (Figure 5A), we observed no increase in TFEB transcriptional activity with TFEBS466A,S467A,S469A, when measured by staining for induction of autophagy with DQ-BSA (Figure 5D, quantified in E), luciferase assay with a CLEAR reporter construct (Figure 5F), number of lysosomes (Figure 5G) or by RT-qPCR of known TFEB and TFE3 target genes, upon treatment with AICAR (Figure 5H) or torin1 (Figure 5I). To confirm that TFE3 is regulated similarly by AMPK, we generated MEFs stably expressing full-length WT TFE3-GFP-3xFLAG or mutant TFE3-GFP-3xFLAGS567A,S568A,S570A. Both proteins are expressed at similar levels (Fig. S3A). AICAR treatment induced nuclear translocation of both proteins (Fig. S3B). However, TFE3 transcriptional activity was not induced in TFE3S567A,S568A,S570A upon AICAR treatment, as measured by DQ-BSA assay (Fig. S3C), and CLEAR luciferase reporter assay (Fig. S3D). These data phenocopy the observations made in the context of AMPK loss (Figure 2), reinforcing an essential role for the presumed AMPK-mediated phosphorylation of TFEB on S466 S467 S469 and TFE3 on S567 S568 S570 to promote autophagy.
Figure 5.
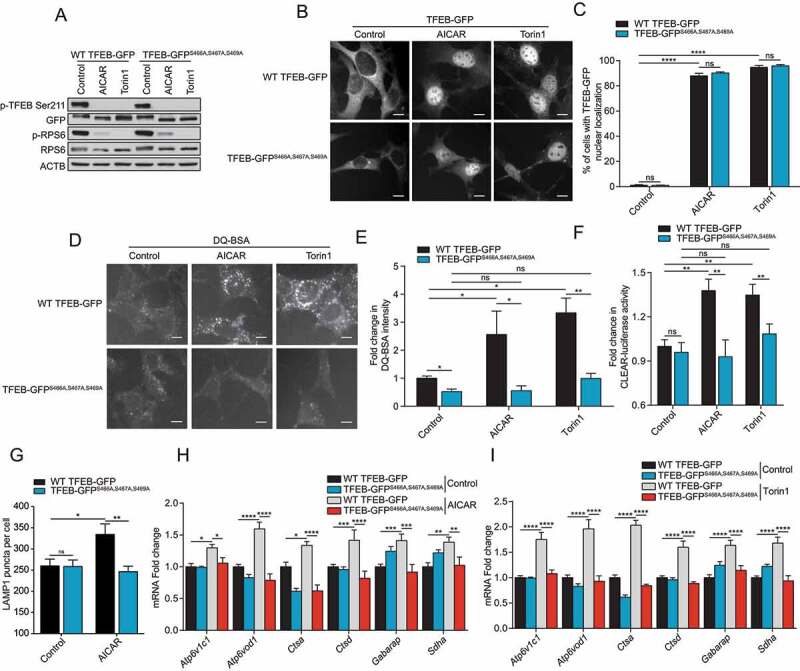
S466 S467 S469 phosphorylation is required for TFEB activity. (A) Immunoblot of protein lysates of MEFs overexpressing WT TFEB-GFP and TFEB-GFPS466A,S467A,S469A incubated in complete media (Control) or in presence of AICAR (2 mM) or Torin1 (1 μM) for 2 h. Data are representative of three independent experiments. (B) Representative images of MEFs overexpressing WT TFEB-GFP and TFEB-GFPS466A,S467A,S469A upon treatment as indicated in (A). Cells were fixed and the GFP fluorescence directly observed by microscopy. Scale bar: 20 μm. Images are representative of three independent experiments. (C) Quantification of the percentage of MEFs overexpressing WT TFEB-GFP and TFEB-GFPS466A,S467A,S469A with nuclear TFEB-GFP upon treatments as indicated in (A) (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, ****P < 0.0001; n > 200 cells per condition). (D) Representative images of MEFs expressing WT TFEB-GFP and TFEB-GFPS466A,S467A,S469A after 1 h of incubation with DQ-BSA-Red followed by a 2 h chase in complete media in presence of DMSO (Control), AICAR (2 mM) or Torin1 (1 μM) prior to fixation. Scale bar: 20 μm. Images are representative of four independent experiments. (E) Relative lysosomal activity, as determined by DQ-BSA assay, in MEFs overexpressing WT TFEB-GFP and TFEB-GFPS466A,S467A,S469A upon treatment as indicated in (D) (mean ± SEM of four independent experiments, two-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01; n > 200 cells per condition). (F) Relative Tfeb and Tfe3 transcriptional activity, as determined by CLEAR-luciferase promoter activity normalized against CMV-Renilla in MEFs expressing WT TFEB-GFP and TFEB-GFPS466A,S467A,S469A upon treatment as indicated in (A) (mean ± SEM of the luminescence fold change from four independent experiments, two-way ANOVA, ns = not significant, **P < 0.01). (G) Quantification of the number of LAMP1 puncta in MEFs overexpressing WT TFEB-GFP and TFEB-GFPS466A,S467A,S469A upon treatment as indicated in (A) (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, *P < 0.05; **P < 0.01; n > 50 cells per condition). (H-I) Relative quantitative real‐time PCR analysis of Tfeb and Tfe3 target genes mRNA transcript levels in MEFs expressing WT TFEB-GFP and TFEB-GFPS466A,S467A,S469A upon treatment as indicated in (A) (mean ± SEM of the RNA fold change of indicated mRNAs from four independent experiments, two-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
S466A S467A S469A blocks MTORC1-regulated TFEB activity
Next, we investigated whether the MTORC1 and the likely AMPK phosphorylation sites on TFEB affect its activity by using a constitutively active (CA) TFEB mutant. We generated stable MEF lines expressing TFEB-GFPS142A,S211A (TFEB-GFP[CA]) mutants (MTORC1 phosphorylation sites important for TFEB nuclear translocation) in combination with S466A S467A S469A mutants (presumed AMPK phosphorylation sites) (Figure 6A). Expression of the CA mutant resulted in a strong nuclear localization, as expected, which was not affected by the additional S466A S467A S469A mutations (Figure 6B, quantified in C). Strikingly, the increased transcriptional activity of the TFEB-GFP[CA] mutant was abolished upon mutation of the serine residues S466A S467A S469A, measured by induction of lysosomal activity with DQ-BSA (Figure 6D, quantified in E), number of LAMP1 puncta (Figure 6F), CLEAR-luciferase reporter assay (Figure 6G), and RT-qPCR of TFEB and TFE3 target genes (Figure 6H).
Figure 6.
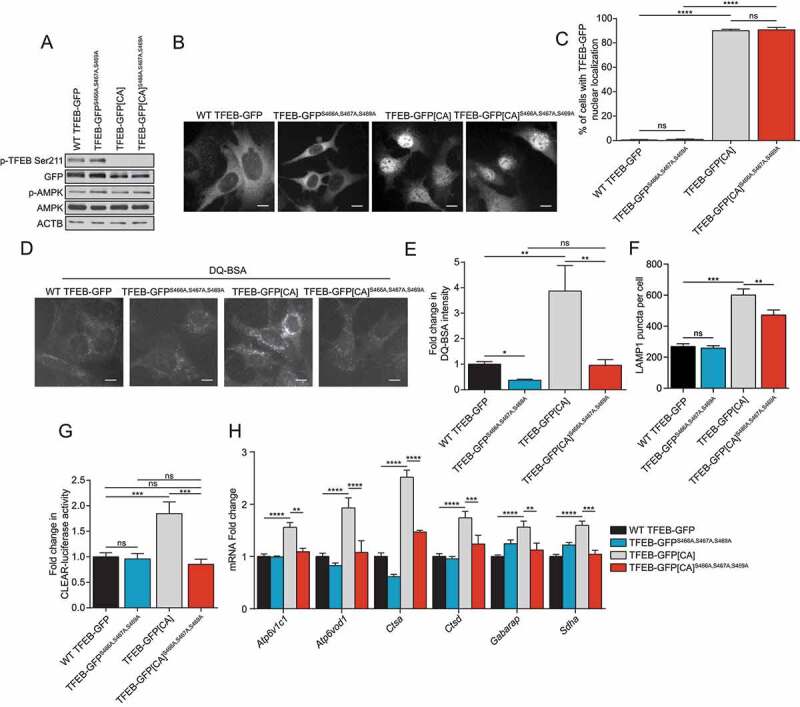
S466A S467A S469A blocks MTORC1-regulated TFEB activity. (A) Immunoblot of protein lysates of MEFs expressing WT TFEB-GFP, TFEB-GFPS466A,S467A,S469A, TFEB-GFPS142A,S211A (TFEB-GFP[CA]) or TFEB-GFP[CA]S466A,S467A,S469A incubated in complete media. Data are representative of three independent experiments. (B) Representative images of indicated MEF lines incubated in complete media. Cells were fixed and the GFP fluorescence directly observed by microscopy. Scale bar: 20 μm. Images are representative of three independent experiments. (C) Quantification of the percentage of MEFs with nuclear TFEB-GFP in complete media (mean ± SEM of three independent experiments, one-way ANOVA, ns = not significant, ****P < 0.0001; n > 200 cells per condition). (D) Representative images of MEFs after 1 h of incubation with DQ-BSA-Red followed by a 2 h chase in complete media prior to fixation. Scale bar: 20 μm. Images are representative of four independent experiments. (E) Relative lysosomal activity, as determined by DQ-BSA assay, in MEFs upon treatment as indicated in (D) (mean ± SEM of four independent experiments, one-way ANOVA, ns = not significant, **P < 0.01; n > 200 cells per condition). (F) Quantification of the number of LAMP1 puncta in MEFs overexpressing WT TFEB-GFP, TFEB-GFPS466A,S467A,S469A, TFEB-GFPS142A,S211A (TFEB-GFP[CA]) or TFEB-GFP[CA]S466A,S467A,S469A incubated in complete media (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, **P < 0.01; ***P < 0.001; n > 50 cells per condition). (G) Relative Tfeb and Tfe3 transcriptional activity, as determined by CLEAR-luciferase promoter activity normalized against CMV-Renilla in MEFs were incubated in complete media (mean ± SEM of the luminescence fold change from five independent experiments, one-way ANOVA, ns = not significant, ***P < 0.001). (H) Relative quantitative real‐time PCR analysis of TFEB and TFE3 target genes mRNA transcript levels in MEFs were incubated in complete media (mean ± SEM of the RNA fold change of indicated mRNAs from four independent experiments, two-way ANOVA, **P < 0.01, ***P < 0.001, ****P < 0.0001)
Inhibition of TFEB and AMPK sensitized cells to chemotherapy
To further substantiate the physiological relevance of our results with respect to cancer therapy, we measured cell survival following treatment with a chemotherapeutic drug (doxorubicin) in MEFs. First, we tested whether expression of the TFEB gain-of-function mutant (TFEB-GFP[CA]) results in resistance to chemotherapy. Overexpression of the TFEB-GFP[CA] mutant, but not WT TFEB, conferred increased colony forming ability and resistance to 0.5 nM doxorubicin for 7 days by 108%, which was abolished by mutation of the S466A S467A S469A sites (Figure 7A, quantified in B). These observations prompted us to target this drug resistance mechanism pharmacologically. Since no inhibitor of TFEB activity is available, we tested whether inhibition of AMPK, which in turn would block TFEB transcriptional activity, could abolish drug resistance. To this end, we used and validated the specific AMPK inhibitor SBI-0206965. SBI-0206965 was initially identified as an ULK1 inhibitor but was later shown to be more specific toward AMPK [66]. First, we observed that treatment with doxorubicin led to potent activation of AMPK and TFEB, which was reduced upon addition of SBI-0206965 (Figure 7C–F). Doxorubicin, at a concentration of 0.25 nM for 7 days, did not reduce viability of WT cells (Figure 7G, quantified in H). However, the combination with SBI-0206965 significantly sensitized the cells to doxorubicin and decreased their colony forming ability (Figure 7G, quantified in H). At 1.5 µM, SBI-0206965 reduced the percentage of viable cells by 92%. As this AMPK inhibitor is not a clinical grade inhibitor and did not completely block AMPK activation, further work would be required to identify more potent compounds for validation studies in animals and humans.
Figure 7.
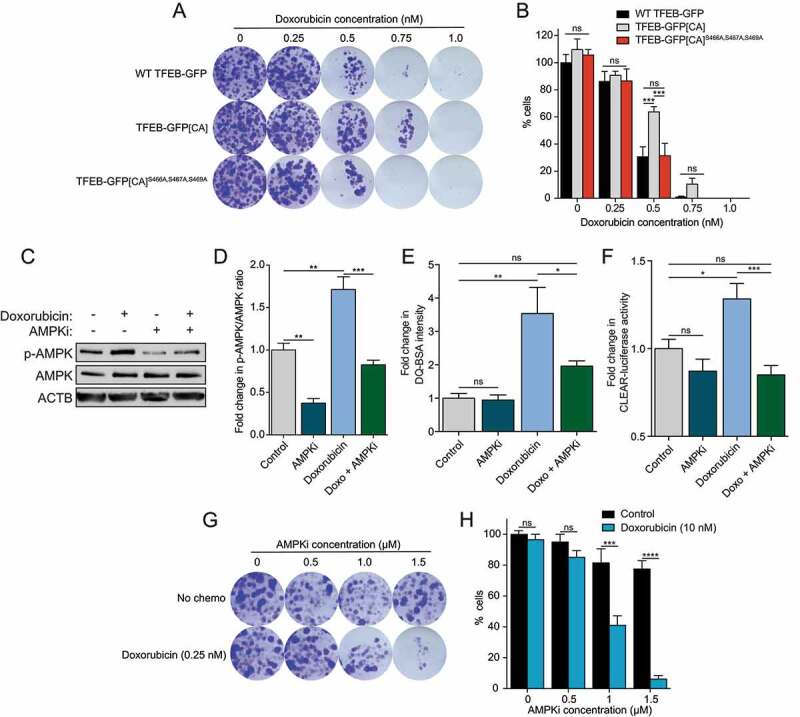
S466A S467A S469A and AMPK inhibition reduce doxorubicin resistance. (A) Representative images of MEFs incubated for 7 days in complete media in presence of indicated doxorubicin concentrations, fixed and stained with crystal violet. Images are representative of three independent experiments. (B) Quantification of the percentage of cells remaining upon treatments as indicated in (A) (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, ***P < 0.001). (C) Immunoblot of protein lysates of WT MEFs incubated in complete media in presence of doxorubicin (10 nM) and/or SBI-0206965 (AMPKi, 5 μM) for 24 h. Data are representative of three independent experiments. (D) Quantification of the p-AMPK:AMPK ratio upon treatments as indicated in (C) (mean ± SEM of three independent experiments, one-way ANOVA, **P < 0.01, ***P < 0.001). (E) Relative lysosomal activity, as determined by DQ-BSA assay, in MEFs upon treatment as indicated in (C) (mean ± SEM of four independent experiments, one-way ANOVA, ns = not significant, **P < 0.01; n > 200 cells per condition). (F) Relative Tfeb and Tfe3 transcriptional activity, as determined by CLEAR-luciferase promoter activity normalized against CMV-Renilla in MEFs upon treatment as indicated in (C) (mean ± SEM of the luminescence fold change from five independent experiments, one-way ANOVA, ns = not significant, ***P < 0.001). (G) Representative images of MEFs incubated for 7 days in complete medßia in presence of doxorubicin (0.25 nM) and indicated SBI-0206965 (AMPKi) concentrations, fixed and stained with crystal violet. Images are representative of three independent experiments. (H) Quantification of the percentage of cells remaining upon treatments as indicated in (G) (mean ± SEM of three independent experiments, two-way ANOVA, ns = not significant, ***P < 0.001, ****P < 0.0001)
Collectively, our data demonstrate that AMPK activation regulates TFEB and TFE3 via two separate mechanisms: a) inhibition of MTORC1 activity that subsequently promotes TFEB and TFE3 nuclear translocation and b) stimulates transcriptional activation. Importantly, we show that AMPK phosphorylates TFEB and TFE3 on specific serine residues and confirm that these residues are essential for their activity. Finally, we show that chemotherapy activates AMPK and TFEB leading to drug resistance, which can be blocked by inhibition of AMPK.
Discussion
We have previously shown that AMPK and FLCN act upstream of TFEB and TFE3 in regulation of the innate immune response in C. elegans and mammalian cells [67]. We reported that FLCN loss constitutively activates AMPK and induces TFEB and TFE3-dependent antimicrobial genes expression. Similar results were obtained upon expression of a constitutively active AMPK mutant in nematodes or pharmacological AMPK activation in mammalian cells. In addition, LPS treatment of macrophages led to acute energy stress, AMPK-mediated TFEB and TFE3 activation, which resulted in increased pro-inflammatory cytokine secretion and phagocytosis. Ablation of AMPK abolished TFEB and TFE3 translocation and activation [67]. These genetic data support the idea that AMPK lies upstream of TFEB and TFE3 and that AMPK regulation of TFEB and TFE3 is necessary for proper autophagy. Here, we report evidence supporting a novel mechanism of TFEB and TFE3 regulation through phosphorylation of TFEB and TFE3 by AMPK (Fig. S4).
In line with previous studies, we showed that severe nutrient starvation such as EBSS starvation (low glucose, no amino acids, no FBS) resulted in both AMPK activation and MTORC1 inhibition [68,69]. The energy drop resulting from the lack of nutrients is acutely sensed by AMPK, which quickly inhibits MTORC1 to stop anabolic processes such as protein synthesis and increase catabolic processes such as autophagy and lysosomal biogenesis. AMPK inhibits MTORC1 via direct phosphorylation of TSC1/TSC2 or by direct phosphorylation of RPTOR [36,37]. Our results showed that MTORC1 was inhibited upon 2 h EBSS starvation in both WT and prkaa DKO cells. This confirms that MTORC1 senses the lack of nutrients such as amino acids independently of AMPK when cells are chronically energy starved. MTORC1 senses the presence of amino acids through a multi-protein complex including the RRAGA, RRAGB, RRAGC, and RRAGD guanine nucleotide status (GTP or GDP loaded) [2,50,70,71]. Activated proteins are required to anchor MTORC1 at the lysosomes and phosphorylate TFEB and TFE3 [70]. RRAGA, RRAGB, RRAGC, and RRAGD activation status is regulated by several guanine-exchange factors (GEF) and GTP-ase activating proteins (GAP), particularly by the ragulator complex, which uses ATP6V (V-type ATPases) to detect the abundance of amino acids [50,54,55,72,73]. In this report, while inactivation of MTORC1 upon EBSS treatment was sufficient to promote TFEB and TFE3 localization to the nucleus independently of AMPK, TFEB and TFE3 activity was abrogated in prkaa DKO cells, suggesting that AMPK dictates the TFEB and TFE3 transcriptional activation status in the nucleus independently of MTORC1. Importantly, the MTORC1 inhibitor Torin1 promoted AMPK-independent nuclear translocation of TFEB and TFE3 but required the presence of AMPK for their transcriptional activation. Despite a slight increase in AMPK activation upon Torin1 treatment, our results suggested that the basal level of AMPK activity is sufficient to promote TFEB and TFE3 activity. However, the kinetics of AMPK phosphorylation remain unclear. Further work would be required to determine if the phosphorylation by AMPK happens at the lysosome or in the nucleus, and if the phosphorylation by MTORC1 precedes those of AMPK.
Consistent with our results, AMPK governs lineage specification by promoting autophagy through transcriptional mechanisms including TFEB [32]. Embryonic stem cells that were deficient for AMPK failed to properly differentiate and had highly deregulated expression of lysosomal genes. Active AMPK resulting from differentiation removed the inhibitory regulation on TFEB by inhibiting MTORC1 downstream of AMPK. Furthermore, AMPK is required to support non-small-cell lung cancer growth by regulating lysosomal gene expression via TFE3 [74]. Upon glucose starvation, AMPK activity is required to promote TFE3 nuclear localization and transcriptional activity. Our results are consistent with previous studies, but we add another layer of complexity in TFEB regulation. AMPK was required not only to inhibit MTORC1 to promote TFEB and TFE3 nuclear localization, but also to stimulate TFEB and TFE3 transcriptional activity upon acute energy stress.
The only reported TFEB transcriptional coactivator is CARM1, which is induced in an AMPK-dependent manner upon chronic glucose starvation for 12–24 h [33]. Chronic glucose starvation promoted AMPK-dependent FOXO3 (forkhead box O3) phosphorylation in the nucleus, which transcriptionally repressed SKP2 (S-phase kinase associated protein 2) leading to increased levels of CARM1. The kinetics of starvation and activation observed in this study were different from our study; increased levels of CARM1 protein and reduction of SKP2 required 12–24 h of glucose starvation. We observed an increase in TFEB and TFE3 activity after 15 minutes of EBSS starvation with a peak after 2 h. Moreover, in this study SKP2 silencing in prkaa DKO MEFs only partially restored autophagic activity, suggesting that AMPK also regulates TFEB and TFE3 activity via other pathways. This is consistent with a distinct response upon acute versus chronic starvation stress; AMPK quickly promotes autophagy via direct phosphorylation of ULK1, TFEB and TFE3, and inhibition of MTORC1, while long-term starvation might further promote autophagy epigenetically by transcriptionally modulating TFEB and TFE3 co-activators, such as CARM1.
TFEB subcellular localization and activity is regulated by phosphorylation through multiple kinases. We report that AMPK may activate TFEB via phosphorylation on a highly conserved cluster of serine residues near the C-terminus. Mutation of S466A S467A S469A led to abrogation of TFEB activation upon AMPK activation or MTORC1 inhibition. It is conceivable that a consequence of AMPK-mediated phosphorylation of S466 S467 S469 is a conformational change that renders TFEB transcriptionally active. Further work is required to delineate the mechanism through which mutation of this region reduces TFEB transcriptional activity. One possibility is that these AMPK serine phosphorylation sites control the interaction of TFEB and TFE3 with yet unknown transcriptional co-activators.
Interestingly, AKT1 (AKT serine/threonine kinase 1) is reported to phosphorylate TFEB at serine 467, but this phosphorylation led to repression of TFEB nuclear translocation independently of MTORC1 [75]. In this study, mutation of S467 to alanine led to increased TFEB nuclear localization. In our hands this mutant S467A had no effect on TFEB nuclear translocation. In agreement, the triple mutant S466A S467A S469A also did not affect nuclear localization. However, the triple-mutant resulted in reduced autophagy, lysosomal activity, CLEAR reporter activity and TFEB and TFE3 target gene expression. AMPK and AKT1 might both phosphorylate the C-terminal region in a different hierarchy which could result in different activity. Similar regulation by AMPK and AKT1 of a different transcription factor was recently described [76]. Consistent with our findings, it is reported that a TFEB mutant harboring a C-terminal truncated of 15 residues, which contains the S466 S467 S469 AMPK phosphorylation sites, lacked transcriptional activity [77]. Phosphorylation of S462, 463, 467, and 469 is shown to be important for the stability and activity of TFEB [77]. In our study, we showed that phosphorylation of the C-terminal region of TFEB by AMPK promoted TFEB transcriptional activity, but we did not observe an effect on protein levels upon mutation of S466A S467A S469A compared to WT. The C-terminal region of TFEB and TFE3 might be tightly regulated via phosphorylation through multiple kinases; the phosphorylation status of the many serine residues located at this region could either promote or inhibit TFEB and TFE3 activity depending on the phosphorylation pattern.
We were not able to determine how many of the three serine residues were phosphorylated in cultured cells from our mass spectrometry experiments. However, it was necessary to mutate all three serine residues for full abrogation of transcriptional activity in cells. Single or double combination mutations of the three serines led to partial responses. It is likely that AMPK phosphorylates the adjacent serine(s) if only one or two of the three serines are mutated to alanine. Moreover, we could not fully demonstrate that AMPK phosphorylates all three sites of TFEB in cells. However, we developed a phospho-specific antibody against a peptide containing all three phospho-serines of TFEB and TFE3 and were able to validate this antibody in vitro and in cultured cells. More work involving phospho-specific antibodies and in vivo metabolic labeling is required to determine the exact site(s) of phosphorylation.
Several chromosomal translocations of TFEB and TFE3 with strong promoters, such as MALAT1, PRCC, ASPSCR1, SFPQ, NONO and CLTC, have been strongly linked with increased TFEB and TFE3 activity, renal cell carcinoma an d alveolar soft part sarcoma [6,14,15]. Knock-down of the MiT/TFE family in pancreatic ductal adenocarcinoma cells reduced their growth in xenograft assays, while overexpression increased their survival in response to nutrient stress and promoted tumorigenesis in orthotopic injection [78,79]. In addition to increased proliferation, TFEB and TFE3 are linked to chemotherapy resistance through lysosomal sequestration. Several anticancer drugs, such as doxorubicin, have been shown to be trapped in lysosomes due to their hydrophobic weak base properties [80,81]. Therefore, stresses such as nutrient starvation and chemotherapy treatment, augment lysosomal biogenesis, increase the number and size of lysosomes per cell, enhance drug sequestration and thus, confer drug resistance [82,83]. In our work, we reported that AMPK-mediated phosphorylation of TFEB was required for chemotherapy resistance. Moreover, the AMPK inhibitor SBI-0206965 sensitized the cells to doxorubicin, suggesting that inhibiting AMPK might be a potential target to reduce chemotherapy resistance. Our study represents a proof of concept and would benefit from the development of more potent inhibitors of AMPK.
Finally, it was reported that AMPK also regulates autophagy via non-transcriptional regulation such as direct phosphorylation of the autophagy initiating kinase ULK1 [29,84]. Our data reveal strong similarities between the dual and opposing regulations of ULK1 and TFEB and TFE3 by AMPK and MTORC1. AMPK directly phosphorylates and activates ULK1 (on serines S317, S467, S555, S574, S637, and S777), while MTORC1 inhibits ULK1 by directly phosphorylating serine residue S757. Together, our present and previous results demonstrate that acute nutrient starvation leads to a) AMPK-mediated inhibition of MTORC1 that promotes ULK1 dephosphorylation at S757 and TFEB dephosphorylation at S142 and S211, which are necessary steps for activation, and b) AMPK-mediated phosphorylation of ULK1 and TFEB at multiple sites, which complete the activation of both proteins. Both, AMPK-mediated phosphorylation of ULK1 and TFEB and inhibition of MTORC1-dependent dephosphorylation of TFEB and ULK1 are required for activity of these autophagy regulatory proteins. Strikingly, AMPK and MTORC1 regulate both critical regulators of autophagy ULK1 and TFEB and TFE3 in a similar manner. In conclusion, these results suggest that AMPK activates autophagy via a dual “fail-safe” mechanism involving not only inactivation of MTORC1 but also phosphorylation of ULK1, TFEB and TFE3.
Material and methods
Cell lines culture and treatments
Primary MEFs were isolated from C57BL/6 E12.5 Flcn floxed mice (generously provided by Dr. L.S. Schmidt, NCI, Bethesda, MD, USA) or Flcn Prkaa1 Prkaa2 floxed mice and cultured as described previously [41]. flcn and flcn prkaa KO MEFs were generated after immortalization of primary Flox/Flox MEFs with SV40 large T and retroviral infection with a Cre recombinase, followed by puromycin selection (Bioshop Canada, PUR333.25). WT and prkaa DKO MEFs cells were generously provided by Dr. Benoit Viollet (Institut Cochin INSERM, Paris, France). HEK293T PRKAA DKO cells were generated by co-transfection of lentivirus containing CRISPR-Cas9 encoding plasmid and target gRNA of both AMPK subunits (PRKAA1; gRNA sequence GAAGATCGGCCACTACATTC, and PRKAA2; gRNA sequence AAGATCGGACACTACGTGCT) and single-cell cloned. HEK293T control cell lines were also single-cell cloned after transduction with non-coding gRNA and CRISPR-Cas9. Cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Wisent, 319–005CL) supplemented with 10% fetal bovine serum (FBS) (Wisent, 080–150), 100 U/ml penicillin+100 µg/ml streptomycin (Wisent, 450–201-EL), and 50 µg/mL gentamycin (Wisent, 450–135) in 5% CO2 at 37°C. MEFs stably overexpressing TFEB-GFP-3xFLAG were transduced with retrovirus, selected with zeocin and single-cell cloned. MEFs stably expressing inducible poly94Q-CFP chains were transduced with lentivirus and selected with hygromycin B (Thermo Fisher Scientific, 10687010). Poly-glutamine aggregates were induced by addition of doxycyclin (5 µg/mL; Sigma-Aldrich, D9891) for 3 days to culture media prior to drug treatments. For EBSS starvation experiments, cells were washed twice with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) and incubated in Earle’s Balanced Salt Solution (EBSS; Sigma-Aldrich, E2888) for 2 h. For drug treatment experiments, cells were incubated for 2 h in medium containing one of the following reagents: Dimethyl sulfoxide (DMSO, 0.1%; Bioshop Canada, DMS666), torin1 (1 μM; Tocris, 10-1-4247), and AICAR (2 mM; Enzo Life Sciences, 89158–090). For chemotherapy drug resistance experiments, cells were incubated for either 24 h or 7 days in medium containing one or a combination of the following reagents: DMSO (Bioshop Canada, DMS666), Doxorubicin (Sigma-Aldrich, D1515), and SBI-0206965 (APExBIO, A8715-5). The final DMSO concentration never exceeded 0.1% and this concentration was shown to have no detrimental effect on all the studied cells.
Antibodies
The FLCN rabbit polyclonal antibody was generated by the McGill animal resource center using recombinant GST-FLCN. The p-TFEB S211 and p-TFEB S466 S467 S469/p-TFE3 S567 S568 S570 antibodies were generated by Yenzym Antibodies, LLC. TFEB (Bethyl Laboratories, A303-673A), TFE3 (Cell Signaling Technology, 14779S), PRKAA/AMPKα (Cell Signaling Technology, 2532), p-PRKAA/AMPKα (Thr172) (Cell Signaling Technology, 2531), ACACA (Cell Signaling Technology, 3676), p-ACACA (S79) (Cell Signaling Technology, 3661), RPS6 (Cell Signaling Technology, 2217), p-RPS6 (Cell Signaling Technology, 4858), ACTB/ß-Actin (Santa Cruz Biotechnology, sc-47778), TUBA1A/tubulin (Sigma-Aldrich, T9026), Anti-Polyglutamine-Expansion (Millipore, MAB1574), anti-GFP (Clontech, 632460), LAMP1 (Abcam, ab24170) antibodies are commercially available.
Immunofluorescence
Cells were gently washed with PBS and fixed in petri dishes with 3.7% formaldehyde (Bioshop Canada, FOR201.500) at room temperature for 30 min. After fixation, cells were washed twice with PBS and then permeabilized with 0.3% Triton X-100 (Bioshop Canada, TRX506.100) in PBS at room temperature for 10 min. Cells were incubated in 5% BSA (Bioshop Canada, ALB001.500) in PBS for 1 h and then with TFEB or TFE3 primary antibodies in 5% BSA in PBS for 2.5 h at 37°C. Cells were washed three times with PBS and incubated with the corresponding secondary antibodies conjugated to Alexa Fluor 488 (Thermo Fisher Scientific, A-11008) and DAPI (2 μg/ml) (Molecular Probes, D-1306) in 5% BSA in PBS for 30 min at 37°C. PBS-washed dishes were covered with cover slips and observed with Axioskop microscope (Zeiss, Germany). Scoring of the percentage of cells displaying nuclear localization was done manually using at least three different frames containing at least 100 cells.
Lysosome number and size
Cells were grown in 6-wells on coverslips. After treatment as described above, cells were gently washed twice with PBS and fixed with 4% paraformaldehyde (Bioshop Canada, par070.500) at room temperature for 15 min. After fixation, cells were washed twice with PBS, permeabilized with 0.1% saponin (Alfa Aesar, J63209-AK) in PBS at room temperature for 10 min. Cells were incubated with LAMP1 primary antibody in 0.1% saponin, 1% FBS in PBS for 2 h in a humidity chamber at 37°C. Cells were washed three times with PBS and incubated with Alexa Fluor 594 anti-rabbit (Thermo Fisher, A32740), Alexa Fluor 647 Phalloidin (Thermo Fisher Scientific, A22287), and DAPI (2 μg/ml) in 0.1% saponin, 1% FBS in PBS for 1 h in a humidity chamber at 37°C. PBS-washed coverslips were mounted on microscope slides and observed on confocal microscope LSM-800 (Zeiss) with 64x oil-immersion objectives at a zoom of 0.8 and acquired using LSM software. Lysosome number and size were quantified using Metamorph (Molecular Devices, USA) from at least 50 individual cells per condition.
DQ-BSA assay
MEFs were incubated with 5 μg/mL DQ Red BSA (Thermo Fisher Scientific, D12051) for 1 h and washed twice with 37°C PBS. EBSS or DMEM containing AICAR, Torin1, or vehicle were added for an additional 2 h. Cells were then fixed and stained for TFEB or TFE3 and DAPI as described above. Grey pixels from pictures acquired on Axioskop microscope (Zeiss, Germany) were quantified using ImageJ (NIH, USA).
Protein extraction and immunoblotting
Cells were washed twice with PBS, lysed in AMPK lysis buffer (10 mM Tris-HCl, pH 8.0, 0.5 mM CHAPS [Bioshop Canada, CHA001], 1.5 mM MgCl2 1 mM EGTA, 10% glycerol, 5 mM NaF [Bioshop Canada, SFL001], 0.1 mM sodium orthovanadate [Na3VO4; Sigma-Aldrich, 450243], 1 mM benzamidine [Sigma-Aldrich, 434760], 5 mM sodium pyrophosphate [NaPPi; Sigma-Aldrich, 221368], supplemented with a complete protease inhibitor cocktail [Roche, 5056489001] and dithiothreitol [DTT; 1 mM]), sonicated 2 × 30 s at maximum power, and cell lysates were cleared by centrifugation at 13000 x g. Proteins were separated on SDS-PAGE gels and revealed by western blot as previously described [41] using the antibodies listed above, and quantified using ImageJ (NIH, USA).
HTT94Q aggregation assay
Cells stably expressing doxycycline-inducible HTT94Q-CFP fusion plasmids were plated and incubated with 5 μg/mL of doxycycline for 3 days prior to treatment with AICAR (2 mM), torin1 (1 μM), or vehicle for 2 h. Cells were fixed in petri dishes with 3.7% formaldehyde at room temperature for 30 min. pTreTight-Htt94Q-CFP was a gift from Nico Dantuma (Addgene, 23966). PBS-washed dishes were covered with cover slips and observed with Axioskop microscope (Zeiss, Germany). Scoring of the percentage of cells displaying CFP puncta was done manually using at least three different frames containing at least 200 cells.
For soluble/insoluble HTT94Q-CFP quantification, Cells were lysed in soluble lysis buffer (1% Triton X-100, phosphatase (Sigma-Aldrich, 4906837001) and protease inhibitors in PBS) on ice for 30 min, followed by centrifugation at 15,000 x g for 30 min at 4°C. Soluble fraction was collected and the pellets were washed 4 times in soluble lysis buffer. The remaining pellets were solubilized using insoluble lysis buffer (1% SDS, 1% Triton X-100, phosphatase/protease inhibitors in PBS) at 60°C for 1 h. After centrifugation at 15,000 x g for 30 min at 4°C, the insoluble fraction was collected. After revelation by western blot as described above, multiple exposures were used to quantify the relative amount of insoluble HTT94Q-CFP using ImageJ (NIH).
CLEAR-luciferase reporter assays
MEFs were seeded in 6-well plates and transfected for 6–8 h with 1 μg of plasmid 4XCLEAR-luciferase reporter (a gift from Albert La Spade, Addgene, 66800) and 0.1 ug of CMV-Renilla Luciferase (Promega, E2261) using 5 μL of polyethylenimine (PEI; Polysciences, 23966–1) at 1 mg/mL stock concentration. 24 h after transfection, cells were treated with AICAR, torin1, EBSS, or vehicle for 2 h. Proteins were extracted using 100 μL of Passive Lysis Buffer from Dual-Luciferase reporter assay systems (Promega, E1980) according to manufacturer’s instruction and assayed using FLUOstar Omega (BMG Labtech, France). Samples were normalized against non-transfected controls and CMV-Renilla values.
Quantitative real-time PCR
After treatments described above, cells were collected, and total RNA was isolated and purified using Total RNA Mini Kit (Geneaid, RB300) according to the manufacturer’s instructions. For quantitative real-time PCR analysis, 0.75 μg of total RNA was reverse-transcribed using the iScript Reverse Transcription Supermix for RT-qPCR (BioRad, 1708841). SYBR Green reactions using the SYBR Green qPCR supermix (BioRad, 1725125) and specific primers (Table 1) were performed using an AriaMX Real-time PCR system (Agilent Technologies, USA). Relative expression of mRNAs was determined after normalization against housekeeping genes Tbp, B2m, or RNA18S.
Co-immunoprecipitations
HEK293T cells were lysed in lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 1 mM DTT, 5% glycerol, 1 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich, P7626), 2 μg/ml pepstatin (Sigma-Aldrich, P5318), 2 μg/ml leupeptin (Sigma-Aldrich, L2884), and 2 μg/ml aprotinin Sigma-Aldrich, A1153)) in the presence of phosphatase inhibitors (after one wash with cold PBS. Cellular debris was removed by centrifugation at 13,000 x g for 10 min at 4°C and the lysate pre-cleared for 30 min with G-agarose beads. The cell lysates were immunoprecipitated for 2 h at 4°C, using and anti-AMPK antibodies (2 µg) or p-TFEB/p-TFE3 antibody (2 µg). 50% protein G-agarose beads (30 µl; Millipore, 16–266) were added to lysates after 1 h. Beads were washed once with lysis buffer for 5 min, followed by 2 washes with wash buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 1 mM phenylmethanesulfonyl fluoride, 2 μg/ml pepstatin, 2 μg/ml leupeptin, and 2 μg/ml aprotinin). SDS-PAGE buffer was added to the washed beads and half of the volume was separated on a 4–15% polyacrylamide, precast Mini-PROTEAN TGS gel (Bio-Rad, 4561094) followed by transfer on Trans-Blot Turbo Transfect Pack (0.2 μm) nitrocellulose (Bio-Rad, 1620115), and western blotting was performed as described with indicated antibodies.
Protein purification from cells
HEK293T were seeded in 150 mm plates and transfected for 8 h with 10 μg of a TFEB-GFP-3xFLAG plasmid using 100 mM PEI. After 15 min on ice, debris was cleared by centrifugation for 10 min at 13000 x g at 4°C. Lysates were pre-cleared on rotator using protein A + G mix (Millipore, 16–156 and 16–266) for 1 h at 4°C, followed by TFEB immunoprecipitation for 3 h using magnetic Flag beads (Sigma-Aldrich, M8823-5ML). Beads were washed 3 times with lysis buffer and TFEB or TFE3 eluted using 200 ng/μL 3X-flag-peptide (Sigma-Aldrich, F4799-4 MG).
Protein purification from bacteria
TFEB fragments were cloned in frame with GST in pGEX-2 T (GE Healthcare, 27–1542-01). Overnight bacterial culture (E. coli, DH5α or BL21) was diluted 1:100 and incubated with shaking at 37°C in LB + ampicillin (100 μg/ml) until the OD600 of 0.6. Protein expression was induced by adding 0.5 mM IPTG (Bioshop Canada, IPT002.1) and incubation for an extra 2.5 h at 37°C. Bacteria were pelleted 10 min at 7700 x g, re-suspended in ice-cold PBS and sonicated 3 × 10 sec at maximum power. Triton X-100 to a final concentration of 1% was added and mixed at 4°C for 30 min. Debris was pelleted for 10 min at 12000 x g and the supernatant incubated with PBS-washed glutathione-sepharose 4 FastFlow beads (GE Healthcare, 17075601) overnight at 4°C. The beads were washed in PBS and the proteins eluted by incubating for 15 min with 50 mM Tris-HCl, pH 8.0, 10 mM glutathione (Bioshop Canada, GTH001.5).
Kinase assay
Bacteria or cell purified proteins or recombinant TFEB (LD Biopharma, hTF-1724), ULK1 (Sigma-Aldrich, SRP5096-10UG), ACACA (Sigma-Aldrich, A6986-10UG) were incubated in presence of [γ-32P] ATP (Perkin-Elmer, BLU502 H250UC) on ice for 10 min in reaction buffer (25 mM HEPES, pH 7.4, 50 mM KCl, 10 mM MgCl2, 100 μM ATP (Bioshop Canada, ATP007), 150 μM AMP (Bioshop Canada, AMP005), 5 mM sodium pyrophosphate (NaPPi), complete protease inhibitor cocktail (Roche, 5056489001). Recombinant active AMPK (Millipore, 14–840) was added and incubated for 30 min at 30°C. Reaction was stopped by adding Laemmli sample buffer and incubation at 95°C for 5 min. Proteins were resolved on acrylamide gels, Coomassie Brilliant Blue-stained, dried, and radioactivity was revealed with Typhoon Trio (GE Healthcare, USA).
Mass spectrometric analysis of TFEB phosphorylation
TFEB fragments were purified, incubated in presence of AMPK in a kinase assay reaction buffer, and sent to PhenoSwitch Bioscience (Sherbrooke, Canada) for analysis. TFEB fragments were digested using 8 μl proteomics grade Trypsin in 50 mM ammonium bicarbonate, at a concentration of 12 ng/μl, overnight. The digested protein samples were acidified in 0.1% trifluoroacetic acid (TFA) in 50% acetonitrile (ACN) and subjected to phosphopeptide enrichment, using titanium dioxide beads, without competition. Briefly, peptide digested in 400 μl 50% ACN, 0.1% TFA were incubated for 30 min with 3–4 μl of TiO2 beads (Canadian Life Science, ACE-111-0503 T). The beads were pelleted by centrifugation for 30 sec in a tabletop centrifuge and washed once with 400 μl 50% ACN, 0.1% TFA. After pelleting the TiO2 beads a second time, bound phosphopeptides were eluted with 100 μl 5% ammonium hydroxide in 50% ACN. The eluted phosphopeptide samples were dried in a speed vac. The enriched phosphopeptide samples were dissolved in 25 μl of demineralized water, 0.1% FA. They were subjected to reverse phase separation, on a 15-cm column Acclaim PepMap HPLC (Thermo Fisher Scientific, 164567) using a nano UHPLC Easy nLC-1000 (Thermo Fisher Scientific, USA) running a flow rate of 350 nl per min and a water/ACN gradient spanning 3–35% ACN in 100 min. The eluted peptides were analyzed using a Q-Exactive HF orbitrap mass spectrometer (Thermo Fisher Scientific, USA), operating at 120,000 resolution for precursor scans, in the mass range from 350 to 1500 m/z, with a trap fill set to 5e6 and at 30,000 resolution for MSMS scans in the mass range from 100–2000 m/z, with a trap fill set to 5e4. Dynamic exclusion was set to 8 sec and the instrument was run at 25 MSMS analyses per duty cycle. The resulting .raw files were converted into Mascot Generic Files (mgf) using Peak Distiller (Matrix Sciences, USA) and searched against the Uniprot reviewed human proteome (UP000005640) using Mascot v. 2.5 (Matrix Sciences, USA). Precursor mass tolerance was set at 5 ppm and MSMS mass tolerance was set at 50 mmu. Variable modifications were allowed for oxidized methionine and for phosphorylations on serines, threonines and tyrosines. Data were thereafter re-searched, using X!Tandem and the data were validated and false discovery rates (FDR) were determined, using Scaffold 4 (Proteome Software, USA). Data were filtered, setting the peptide FDR cutoff to 2%.
Doxorubicin resistance assays
MEFs were incubated with the indicated concentrations of doxorubicin and/or SBI-0206965 (APExBIO, A8715-5) and/or vehicle (DMSO) for 7 days. Viability was assessed in 6-well plates. After 7 days, cells were fixed in petri dishes with 3.7% formaldehyde at room temperature for 60 min. After fixation, cells were incubated 60 min with 0.1% crystal violet solution, thoroughly washed in water and allowed to dry. Quantification was performed using ImageJ software.
Statistical analysis
Data are expressed as mean ±SEM. All experiments were performed at least three times as indicated. Statistical analyses for all data were performed using student’s t-test, one-way ANOVA or two-way ANOVA as indicated using GraphPad Prism 7 software. Statistical significance is indicated in figures (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Supplementary Material
Acknowledgments
M.P., L.E-H., were supported by FRQS/CIHR, and FRQS, respectively. This work was supported by grants to A.P. from the Kidney Foundation of Canada, Terry Fox foundation (TFF-166128), CIHR (PJT-165829) and the Cancer Research Society. We would like to thank Paula Coehlo, the McGill University Life Sciences Complex Advanced BioImaging Facility (ABIF), and McGill University Mass Spectrometry Facility for their support and advices.
Funding Statement
This work was supported by the Canadian Institutes of Health Research [PJT-165829]; Cancer Research Society; Kidney Foundation of Canada (KFOC) [KFOC100021]; Terry Fox Research Institute [TFF-166128].
Disclosure statement
The authors have no competing interest to report and have no potential or real conflicts of interest to declare.
Supplemental material
Supplemental data for this article can be accessed here.
References
- [1].Hale AN, Ledbetter DJ, Gawriluk TR, et al. Autophagy: regulation and role in development. Autophagy 2013;9:951–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Paquette M, El-Houjeiri L, Pause A.. MTOR pathways in cancer and autophagy. Cancers (Basel). 2018;18:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Raben N, Puertollano R.. TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu Rev Cell Dev Biol. 2016;32:255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Huber LA, Teis D. Lysosomal signaling in control of degradation pathways. Curr Opin Cell Biol. 2016;39:8–14. [DOI] [PubMed] [Google Scholar]
- [5].Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. [DOI] [PubMed] [Google Scholar]
- [6].Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016;129:2475–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sardiello M, Palmieri M, Di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. [DOI] [PubMed] [Google Scholar]
- [8].Martina JA, Diab HI, Lishu L, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Palmieri M, Impey S, Kang H, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011;20:3852–3866. [DOI] [PubMed] [Google Scholar]
- [10].Settembre C, De Cegli R, Mansueto G, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pastore N, Vainshtein A, Klisch TJ, et al. TFE3 regulates whole-body energy metabolism in cooperation with TFEB. EMBO Mol Med. 2017;9:605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pastore N, Brady OA, Diab HI, et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy. 2016;12:1240–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kauffman EC, Ricketts CJ, Rais-Bahrami S, et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol. 2014;11:465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Argani P. MiT family translocation renal cell carcinoma. Semin Diagn Pathol. 2015;32:103–113. [DOI] [PubMed] [Google Scholar]
- [16].Buchser WJ, Laskow TC, Pavlik PJ, et al. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Martini-Stoica H, Xu Y, Ballabio A, et al. The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci. 2016;39:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol Dis. 2011;43:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tsunemi T, Ashe TD, Morrison BE, et al. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Decressac M, Mattsson B, Weikop P, et al. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci USA. 2013;110:E1817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xiao Q, Yan P, Ma X, et al. Neuronal-Targeted TFEB accelerates lysosomal degradation of APP, reducing Aβ generation and amyloid plaque pathogenesis. J Neurosci. 2015;35:12137–12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xiao Q, Yan P, Ma X, et al. Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J Neurosci. 2014;34:9607–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. [DOI] [PubMed] [Google Scholar]
- [24].Martina JA, Chen Y, Gucek M, et al. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Roczniak-Ferguson A, Petit CS, Froehlich F, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J. 2012;31:1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vega-rubin-de-celis S, Peña-Llopis S, Konda M, et al. Multistep regulation of TFEB by MTORC1. Autophagy. 2017;13:464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol. 2013;200:475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bach M, Larance M, James DE, et al. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J. 2011;440:283–291. [DOI] [PubMed] [Google Scholar]
- [31].Egan D, Kim J, Shaw RJ, et al. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Young NP, Kamireddy A, Van Nostrand JL, et al. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes Dev. 2016;30:535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shin H-JR, Kim H, Oh S, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Villanueva-Paz M, Cotán D, Garrido-Maraver J, et al. AMPK regulation of cell growth, apoptosis, autophagy, and bioenergetics. Exp Suppl. 2016;107:45–71. [DOI] [PubMed] [Google Scholar]
- [35].Hardie DG, Schaffer BE, Brunet A. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 2016;26:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Inoki K, Ouyang H, Zhu T, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. [DOI] [PubMed] [Google Scholar]
- [37].Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hasumi H, Baba M, Hasumi Y, et al. Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc Natl Acad Sci USA. 2015;112:E1624–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baba M, Hong S-B, Sharma N, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci USA. 2006;103:15552–15557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yan M, Audet-Walsh É, Manteghi S, et al. Chronic AMPK activation via loss of FLCN induces functional beige adipose tissue through PGC-1α/ERRα. Genes Dev. 2016;30:1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Possik E, Jalali Z, Nouët Y, et al. Folliculin regulates ampk-dependent autophagy and metabolic stress survival. PLoS Genet. 2014;10:e1004273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hudon V, Sabourin S, Dydensborg AB, et al. Renal tumour suppressor function of the Birt-Hogg-Dubé syndrome gene product folliculin. J Med Genet. 2010;47:182–189. [DOI] [PubMed] [Google Scholar]
- [43].Possik E, Ajisebutu A, Manteghi S, et al. FLCN and AMPK confer resistance to hyperosmotic stress via remodeling of glycogen stores. PLoS Genet. 2015;11:e1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yan M, Gingras M-C, Dunlop EA, et al. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J Clin Invest. 2014;124:2640–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hong S-B, Oh H, Valera VA, et al. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One. 2010;5:e15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wada S, Neinast M, Jang C, et al. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 2016;30:2551–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tee AR, Pause A. Birt-Hogg-Dubé: tumour suppressor function and signalling dynamics central to folliculin. Fam Cancer. 2013;12:367–372. [DOI] [PubMed] [Google Scholar]
- [48].Tsun Z-Y, Bar-Peled L, Chantranupong L, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Péli-Gulli M-P, Sardu A, Panchaud N, et al. Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-activating protein complex for the rag family GTPase Gtr2. Cell Rep. 2015;13:1–7. [DOI] [PubMed] [Google Scholar]
- [50].Sancak Y, Bar-Peled L, Zoncu R, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010;141:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Meng J, Ferguson SM. GATOR1-dependent recruitment of FLCN-FNIP to lysosomes coordinates Rag GTPase heterodimer nucleotide status in response to amino acids. J Cell Biol. 2018;217:2765–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hosios AM, Manning BD. Lysosomal catch-and-release controls mTORC1. Nat Cell Biol. 2018;20:996–997. [DOI] [PubMed] [Google Scholar]
- [53].Shen K, Huang RK, Brignole EJ, et al. Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature. 2018;556:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bar-Peled L, Schweitzer LD, Zoncu R, et al. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Han JM, Jeong SJ, Park MC, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. [DOI] [PubMed] [Google Scholar]
- [57].Marwaha R, Sharma M. DQ-Red BSA trafficking assay in cultured cells to assess cargo delivery to Lysosomes. Biol Protoc. 2017;7:e2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Baba M, Toyama H, Sun L, et al. Loss of folliculin disrupts hematopoietic stem cell quiescence and homeostasis resulting in bone marrow failure. Stem Cells. 2016;34:1068–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. [DOI] [PubMed] [Google Scholar]
- [60].Renna M, Jimenez-Sanchez M, Sarkar S, et al. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem. 2010;285:11061–11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Berger Z, Ravikumar B, Menzies FM, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet. 2006;15:433–442. [DOI] [PubMed] [Google Scholar]
- [62].Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. [DOI] [PubMed] [Google Scholar]
- [63].Scott JW, Norman DG, Hawley SA, et al. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J Mol Biol. 2002;317:309–323. [DOI] [PubMed] [Google Scholar]
- [64].Ducommun S, Deak M, Sumpton D, et al. Motif affinity and mass spectrometry proteomic approach for the discovery of cellular AMPK targets: identification of mitochondrial fission factor as a new AMPK substrate. Cell Signal. 2015;27:978–988. [DOI] [PubMed] [Google Scholar]
- [65].Marin TL, Gongol B, Martin M, et al. Identification of AMP-activated protein kinase targets by a consensus sequence search of the proteome. BMC Syst Biol. 2015;9:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dite TA, Langendorf CG, Hoque A, et al. AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965. J Biol Chem. 2018;293:8874–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].El-Houjeiri L, Possik E, Vijayaraghavan T, et al. The Transcription Factors TFEB and TFE3 link the FLCN-AMPK signaling axis to innate immune response and pathogen resistance. Cell Rep. 2019;26:3613–3628.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Löffler AS, Alers S, Dieterle AM, et al. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy. 2011;7:696–706. [DOI] [PubMed] [Google Scholar]
- [69].Mack HID, Zheng B, Asara JM, et al. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sancak Y, Peterson TR, Shaul YD, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Sekiguchi T, Hirose E, Nakashima N, et al. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. [DOI] [PubMed] [Google Scholar]
- [72].Su M-Y, Morris KL, Kim DJ, et al. Hybrid structure of the RagA/C-Ragulator mTORC1 activation complex. Mol Cell. 2017;68:835–846.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Stransky LA, Forgac M. Amino acid availability modulates Vacuolar H+-ATPase assembly. J Biol Chem. 2015;290:27360–27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Eichner LJ, Brun SN, Herzig S, et al. Genetic analysis reveals AMPK is required to support tumor growth in murine kras-dependent LUNG CANCER MODELs. Cell Metab. 2019;29:285–302.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Palmieri M, Pal R, Nelvagal HR, et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Saline M, Badertscher L, Wolter M, et al. AMPK and AKT protein kinases hierarchically phosphorylate the N-terminus of the FOXO1 transcription factor, modulating interactions with 14-3-3 proteins. J Biol Chem. 2019;294:13106–13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ferron M, Settembre C, Shimazu J, et al. A RANKL-PKCβ-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Slade L, Pulinilkunnil T. The mitf/tfe family of transcription factors: master regulators of organelle signaling, metabolism, and stress adaptation. Mol Cancer Res. 2017;15:1637–1643. [DOI] [PubMed] [Google Scholar]
- [79].Perera RM, Stoykova S, Nicolay BN, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zhitomirsky B, Assaraf YG. Lysosomes as mediators of drug resistance in cancer. Drug Resist Updat. 2016;24:23–33. [DOI] [PubMed] [Google Scholar]
- [81].Herlevsen M, Oxford G, Owens CR, et al. Depletion of major vault protein increases doxorubicin sensitivity and nuclear accumulation and disrupts its sequestration in lysosomes. Mol Cancer Ther. 2007;6:1804–1813. [DOI] [PubMed] [Google Scholar]
- [82].Zhitomirsky B, Assaraf YG. Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosome-dependent cancer multidrug resistance. Oncotarget. 2015;6:1143–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gotink KJ, Rovithi M, De Haas RR, et al. Cross-resistance to clinically used tyrosine kinase inhibitors sunitinib, sorafenib and pazopanib. Cell Oncol. 2015;38:119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


