Abstract
Background
Despite significant advances in multiple myeloma (MM) therapy, disease relapse and treatment resistance remain major obstacles in clinical management. Herein, we have studied the clinical utility of miRNAs in improving patients’ risk-stratification and prognosis.
Methods
miRNA-seq was performed in CD138+ plasma cells of MM, smoldering multiple myeloma (sMM) and monoclonal gammopathy of undetermined significance (MGUS) patients. The screening MM cohort consisted of 138 patients. miRNA levels of CD138+ plasma cells were quantified by RT-qPCR following 3′-end RNA polyadenylation. Disease progression and patients’ death were used as clinical end-point events. Internal validation was conducted by bootstrap analysis. Clinical net benefit on disease prognosis was assessed by decision curve analysis. Kruykov et al. 2016 served as validation cohort (n = 151).
Results
miRNA-seq highlighted miR-181a to be upregulated in MM vs. sMM/MGUS, and R-ISS III vs. I patients. Screening and validation cohorts confirmed the significantly higher risk for short-term progression and worse survival of the patients overexpressing miR-181a. Multivariate models integrating miR-181a with disease established markers led to superior risk-stratification and clinical benefit for MM prognosis.
Conclusions
CD138+ overexpression of miR-181a was strongly correlated with inferior disease outcome and contributed to superior prediction of MM patients early progression, supporting personalised prognosis and treatment decisions.
Subject terms: Tumour biomarkers, Myeloma, Prognostic markers, miRNAs, Preclinical research
Background
Multiple myeloma (MM) is the second most frequently diagnosed hematologic malignancy following non-Hodgkin lymphoma, accounting for ~13% of all hematological neoplasms [1, 2]. The disease progresses from a pre-malignant precursor stage, monoclonal gammopathy of undetermined significance (MGUS), through an indolent intermediate condition known as smoldering multiple myeloma (sMM), to a fully symptomatic myeloma that requires treatment [3]. Within the progression from MGUS to MM, several complex genetic events emerge, including cytogenetic abnormalities, primary or secondary chromosomal translocations and activation of oncogenes [4].
Recently, considerable progress has been made regarding treatment strategies resulting in significantly prolonged patients’ survival [5, 6]. Nevertheless, despite the notable improvements in patient care, MM remains incurable due to frequent patients’ recurrence and emergence of drug resistance in its clinical course [3, 7]. Currently, MM prognosis relies on established clinical markers unified in the Revised MM International Staging System (R-ISS). Although R-ISS is a reliable staging system, patients of the same stage and with similar clinicopathological features could present greatly heterogeneous disease course, supporting the need for novel prognostic markers [8]. In this regard, the elucidation of disease molecular landscape could offer an alternative approach for the identification of modern molecular indicators of disease course to dissect heterogeneity and to support tailored patient’s management.
It is nowadays evident that the non-coding RNAs (ncRNAs) family, which constitutes >75% of the human genome, regulates essential human physiologic and pathologic processes. MicroRNAs (miRNAs) represent a principal class of functional small non-coding RNAs (~22 nucleotides) that play a pivotal role in the regulation of gene expression at the post-transcriptional level [9, 10]. miRNAs exert their regulatory function by binding to the 3′-untranslated region (3′-UTR) of mRNAs and recruiting the RNA-induced silencing complex (miRISC), thereby leading to translational repression and mRNA decay [9]. Their function is crucial for diverse cellular processes, including cell proliferation, differentiation, apoptosis and cellular homeostasis, while the deregulation of miRNAs expression has become a hallmark of the majority of human malignancies [11, 12]. Compelling findings have shown that numerous miRNAs are actively implicated in the pathobiology of MM, holding also a great clinical potential in modern molecular diagnostics and prognostics for MM management [6, 13].
Herein, using miRNA-seq we investigated miRNA profiles in association with MM and its precursor stages, aiming to reveal potential MM-related miRNAs of prognostic significance. Based on miRNA-seq data and the screening of the identified miRNA candidates, miR-181a clinical value in MM prognosis and treatment outcome was further evaluated, for the first time, in our screening (n = 138 patients) and an external validation (n = 151 patients) MM cohort. Our study highlighted that CD138+ plasma cells overexpression of miR-181a is associated with higher risk for short-term disease progression and worse survival outcome following treatment, independently of MM patients’ clinicopathological data. Ultimately, implementation of miR-181a levels with the established disease markers resulted in improved risk-stratification and superior positive prediction of patient’s short-term progression and poor treatment response, supporting miR-181a utility in modern management of MM patients.
Materials and methods
Screening cohort
A screening cohort of 193 consecutive patients diagnosed with MM (n = 138), sMM (n = 30) and MGUS (n = 25), with a median age of 69 years, was included in the study. Bone marrow (BM) samples were obtained at the time of diagnosis at the Department of Clinical Therapeutics, “Alexandra” Hospital, Athens, Greece. Diagnosis was based on the standard criteria of the International Myeloma Working Group (IMWG) [14]. All MM patients were newly diagnosed and non-previously treated.
At the time of diagnosis whole blood count, biochemical analysis, serum and urine M-protein levels assessment were performed. Trephine biopsy was also performed, and cytogenetic abnormalities were detected using conventional cytogenetic protocols and interphase fluorescence in situ hybridization (FISH) at BM aspirate. Whole body low dose computed tomography (WBLDCT) was used to assess bone disease. Focusing on MM cohort, 22 patients had initial diagnosis of MGUS or sMM before the development of symptomatic MM. According to the R-ISS staging system 24.7%, 47.8% and 20.3% of the patients were classified in stage I, II and III, respectively. The median B2M and LDH levels were 4.5 mg/l and 165.5 U/l, respectively, while 79 patients presented with ≥60% BM infiltration by plasma cells. Renal impairment and bone disease were detected in 5.8% and 65.2% of the patients at diagnosis, respectively.
Of the enrolled patients, 121 patients received bortezomib-based regimens, while 11 of them were treated with lenalidomide-based regimen. High-dose melphalan therapy with autologous stem cell transplantation (HDM/ASCT) was administrated to 37 patients. Response was assessed monthly with blood and urine tests, according to IMWG criteria [15]. Post-treatment follow-up was adequately completed for 133 patients (96.4%) whereas 5 patients (3.6%) were excluded from the survival analysis due to insufficient monitoring data. During a median follow-up time (reverse Kaplan–Meier method) of 22 months (95% CI: 19.97–24.02), disease relapse and death were detected in 32 (24.1%) and 29 (21.8%) MM patients, respectively. Focusing on patients’ clinical outcome, the mean event-free survival (EFS) and overall survival (OS) were 26.34 (95% CI: 24.63–28.05) and 27.31 (95% CI: 25.76–28.85) months, respectively, while patients displayed a mean progression-free survival (PFS) of 23.9 months (95% CI: 22.04–25.77). Detailed patients’ clinicopathological characteristics are summarised in Table 1.
Table 1.
Clinicopathological features of the screening cohort.
| Variable | No. of patients n = 193 |
|---|---|
| Disease | |
| MGUS | 25 (13.0%) |
| sMM | 30 (15.5%) |
| MM | 138 (71.5%) |
| Multiple myeloma patients (n = 138) | |
| R-ISS stage | |
| R-ISS I | 34 (24.6%) |
| R-ISS II | 66 (47.8%) |
| R-ISS III | 28 (20.3%) |
| Missing data | 10 |
| ISS stage | |
| ISS I | 39 (28.3%) |
| ISS II | 42 (30.4%) |
| ISS III | 54 (39.2%) |
| Missing data | 3 |
| Prior sMM/MGUS | |
| Yes | 22 (15.9%) |
| No | 114 (82.6%) |
| Missing data | 2 |
| Gender | |
| Male | 77 (55.8%) |
| Female | 61 (44.2%) |
| Therapy | |
| Bortezomib-based regimens | 121 (87.7%) |
| Lenalidomide-dexamethasone | 11 (8.00%) |
| Other | 2 |
| Missing data | 4 |
| Bone disease | |
| Yes | 90 (65.2%) |
| No | 36 (26.1%) |
| Missing data | 12 |
| HDM/ASCT | |
| Yes | 37 (26.8%) |
| No | 98 (71.0%) |
| Missing data | 3 |
| B2M (mg/L) | |
| Median (min–max) | 4.5 (1.5–36.9) |
| LDH (U/L) | |
| Median (min–max) | 165.5 (71.0–570.0) |
| Marrow plasma cells | |
| <60% | 57 (41.3%) |
| ≥60% | 79 (57.2%) |
| Missing data | 2 |
| Response to 1st line | |
| sCR | 26 (18.8%) |
| CR | 18 (13.1%) |
| VGPR | 46 (33.3%) |
| PR | 31 (22.5%) |
| SD | 8 (5.80%) |
| PD | 2 (1.50%) |
| Missing data | 7 |
| Disease monitoring | |
| Follow-up patients | 133 |
| Relapse | 32 (24.1%) |
| Death/alive | 29 (21.8%) |
| Progression/progression-free survival | 48 (36.0%)/85 (64.0%) |
| Excluded from follow-up | 5 |
HDM/ASCT high-dose melphalan therapy with autologous stem cell transplantation, sCR stringent complete response, CR complete response, VGPR very good partial response, PR partial response, SD stable disease, PD progressed disease.
Prior to sampling, the study was approved by the Ethics Committee of “Alexandra” Hospital, Athens, Greece, and all patients who participated were informed and signed an informed consent according to 1975 Declaration of Helsinki ethical standards, as revised in 2008.
Validation cohort
The Kryukov et al. 2016 (n = 151) [16] cohort was used as an independent validation MM cohort. Kryukov et al. 2016 performed transcription profiling by array using Affymetrix GeneChip Human Gene 1.0 ST Array in CD138+ plasma cells from 151 newly diagnosed untreated MM patients. The clinical and normalised expression publicly available data were downloaded by EMBL-EBI ArrayExpress (accession number ArrayExpress: E-MTAB-1038 and E-MTAB-4032).
Isolation of CD138+ plasma cells—extraction of total RNA
BM aspirate samples were collected in EDTA tubes. Mononuclear cells were isolated from BM aspirate using the Ficoll-Paque technique. CD138+ plasma cells were afterwards selected by positive magnetic cell sorting selection with immunomagnetic microbeads coated with an anti-CD138 monoclonal antibody (MACS CD138 microbeads, Miltenyi-Biotec GmbH, Bergisch Gladbach, Germany).
Total RNA was isolated from purified CD138+ plasma cells using TRI-Reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer’s protocol, dissolved in RNA Storage Solution (Ambion, Austin, TX, USA) and stored at −80 °C until analysis. RNA concentration was estimated using the Qubit RNA broad range assay in the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA).
miRNA-seq
Next-generation sequencing (NGS) libraries were constructed from 4 sMM patients, 4 MGUS patients, 4 R-ISS I, 8 R-ISS II and 8 R-ISS III MM patients. NGS libraries were constructed with the QIAseq miRNA Library Kit (Qiagen, Hilden, Germany), using 500 ng of total RNA as template per library. In brief, adapters were ligated sequentially to the 3′ and 5′ ends of miRNAs, UMI-based cDNA synthesis was carried out followed by cDNA cleanup and finally amplification was implemented with a universal forward primer and indexing reverse primers. The quality assessment of the created miRNA-seq libraries was performed with the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), using the Agilent High Sensitivity DNA Kit (Agilent). Finally, the subsequent miRNA-seq was performed using an Illumina NextSeq 550 (Illumina, San Diego, CA, USA), leading to the production of 75 bp single-sequencing reads for each library.
Bioinformatics analysis
The quality control of the sequenced libraries was performed with FastQC software, whereas adapter trimming was accomplished with TrimGalore algorithm. Sequencing reads with lengths <16 nt and >40 nt were filtered out of the raw datasets that were used for downstream analysis. In the next step, miRDeep2 was used for the elucidation of the miRNA expression profiling across the tested samples [17]. For each investigated sample, only miRNAs with >10 unnormalised read counts and positive miRDeep2 score were taken into consideration for further analysis.
Polyadenylation of total RNA and first-strand cDNA synthesis
Two hundred nanograms of total RNA were polyadenylated at 3′-end in a 10 μl reaction, containing 1 U of recombinant E. coli Poly(A) Polymerase (New England Biolabs Inc., Ipswich, MA) and 800 mM ATP, at 37 °C for 60 min. Subsequently, 10 min incubation at 65 °C was performed for polymerase inactivation.
Reverse transcription of the polyadenylated RNA was held in a 20 μl reaction mixture consisting of 200 U M-MLV Reverse Transcriptase (Invitrogen), 40 U RNaseOUT Recombinant Ribonuclease Inhibitor (Invitrogen), 10 mM dNTP Mix and 250 mM oligo-dT adapter 5′-GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN-3′ (V = G, A, C and N = G, A, T, C), at 37 °C for 60 min. Heat inactivation of M-MLV was performed at 70 °C for 15 min.
Quantitative real-time PCR (qPCR)
The quantification of miRNA levels was performed by a SYBR-Green fluorescent-based quantitative real-time PCR (qPCR) assay. Specific forward primers for miR-1–3p, miR-125b-5p, miR-181a-5p, miR-503-5p, miR-218-5p, miR-10a-5p and small nucleolar RNA, C/D box 48 (SNORD48), frequently annotated as RNU48, were synthesised according to their published sequences and in silico specificity analysis (Table S1).
The qPCR assays were carried out in the 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). The 10 μl reaction mixtures contained Kapa SYBR® Fast Universal 2× qPCR Master Mix (Kapa Biosystems, Inc., Woburn, MA), 200 nM of each PCR primer and 1 ng of cDNA template. The thermal protocol comprised of a polymerase activation step at 95 °C for 3 min, followed by 40 cycles of denaturation step at 95 °C for 15 sec and finally primer annealing and elongation step at 60 °C for 1 min. Following amplification, the reaction specificity was evaluated via melting curve analysis and agarose gel electrophoresis. All reactions were performed in duplicates in order to obtain reproducible results. The 2−ΔΔCT relative quantification (RQ) method was conducted for the analysis of miRNA expression levels, using RNU48 as endogenous reference control for normalisation purposes.
Gene Ontology (GO) enrichment analysis
miRDB target-prediction tool was used to identify the potential target genes of miR-181a [18]. To minimise the prediction error rates only targets with miRDB Target Score ≥80 were selected. Functional annotation of miR-181a target genes was performed with Enrichr database platform (https://maayanlab.cloud/Enrichr/) [19]. GO enrichment analysis (http://www.geneontology.org/) was conducted categorising target genes into groups according to three classification standards, Biological Processes (BPs), molecular functions (MFs) and cellular components (CCs). BPs, CCs and MFs with a p-value < 0.05, and combined enrichment score >20 were retained following analysis. The significantly associated terms were imported to REVIGO where they were clustered based on their relatedness and any redundancy was excluded [20].
Statistical analysis
Statistical analysis was performed by IBM SPSS Statistics 20 software (IBM Corp., Armonk, New York, USA). Sapiro–Wilk and Kolmogorov–Smirnov tests were applied to test the normal distribution of the data. The non-parametric Mann–Whitney U test and Kruskal–Wallis test were used appropriately to assess the correlation of miRNA levels with categorical clinicopathological features.
Kaplan–Meier survival curves using log-rank test, and Cox proportional regression analysis were implemented for patients’ survival analysis. The X-tile algorithm was applied for the optimal selection of the cut-off values of miRNA levels. Disease relapse and patients’ death were used as clinical end-point events for EFS, as well as OS and cancer-specific survival (CSS), respectively. PFS was evaluated by patients’ relapse and/or death (whichever came first). Internal validation was performed by bootstrap Cox proportional regression analysis based on 1000 bootstrap samples. Finally, decision curve analysis (DCA), in order to evaluate miR-181a clinical benefit in patients’ prognosis and treatment outcome, was performed according to Vickers et al. [21], by STATA 16 software (StataCorp LLC, College Station, TX, USA).
Results
miRNA expression profiling of CD138+ plasma cells by miRNA-seq
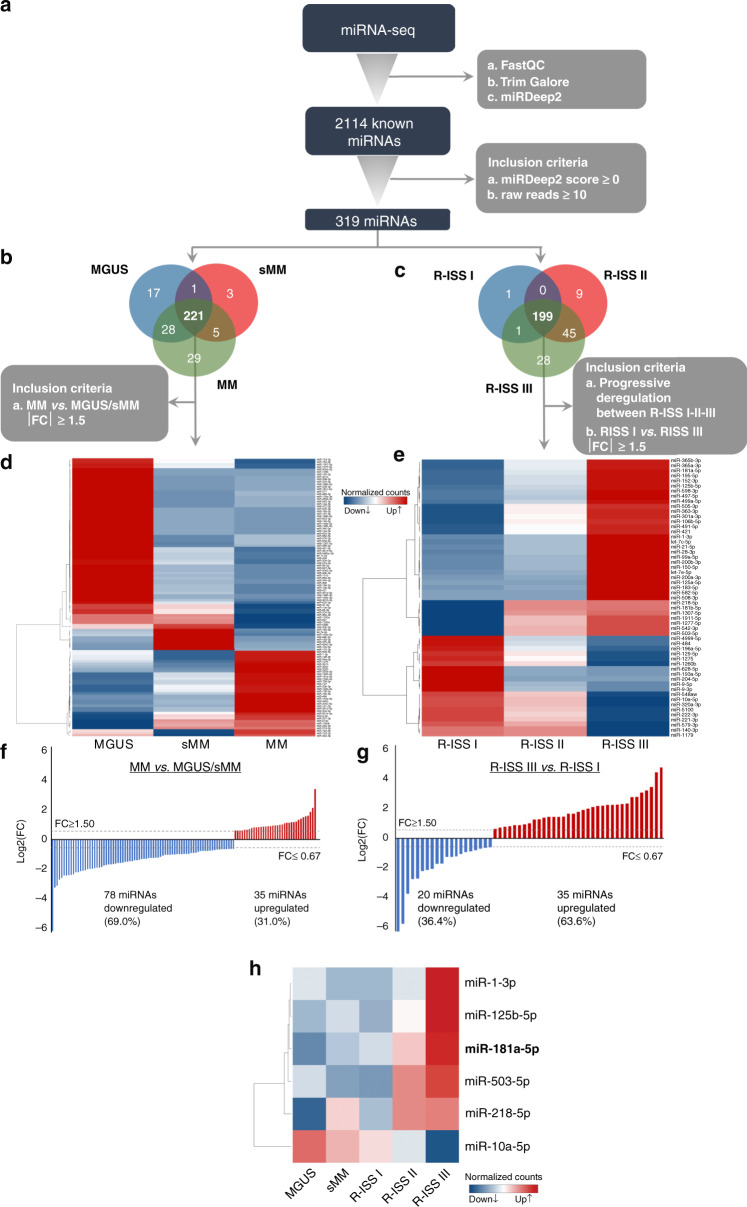
To investigate the alterations of miRNA expression in MM, miRNA˗seq was performed in CD138+ plasma cells from MM, sMM and MGUS patients using the Illumina NextSeq 550 system. The main steps of miRNA-seq workflow are schematically presented in Fig. 1. Following filtering out noise using FastQC, TrimGalore and miRDeep2 softwares, 2114 different known miRNAs were successfully mapped on the human genome sequence. To further filter our results, we excluded miRNAs with <10 raw read counts and negative miRDeep2 score, narrowing down to 319 miRNAs that were selected for downstream analysis (Fig. 1a). Our analysis was divided in two parts in order to explore the changes in the transcriptomic level between a. MM and sMM/MGUS and b. R-ISS I, II and III MM patients.
Fig. 1. miRNAs profile of CD138+ plasma cells.
Workflow diagram of miRNA-seq analysis of CD138+ plasma cells in MM, sMM and MGUS patients (a). Venn diagram representing shared miRNAs between MM, sMM and MGUS (b) and between R-ISS I-II-III (c) patients. Heatmap of the differently expressed miRNAs between MM, sMM and MGUS (d) and between R-ISS I-II-III (e) patients. Bar graph of miRNAs log2FC in MM vs. MGUS/sMM (f) and in R-ISS III vs. R-ISS I stages (g). Heatmap of the concurrently altered miRNAs between MGUS, sMM and R-ISS stages. Colour gram depicts high (red) and low (blue) expression levels of miRNAs (h). FC fold change.
Focusing on the differences between MM vs. sMM/MGUS, 221 of the 319 miRNAs were found to be concurrently expressed in MM, sMM and MGUS patients (Fig. 1b). Among them, 113 miRNAs were identified to be differentially expressed between MM and sMM/MGUS samples with fold change (FC) ≥ 1.5 or FC ≤ 0.67 (Fig. 1d; Table S2), of which, 35 (31.0%) miRNAs were found to be upregulated in MM compared to sMM/MGUS (Fig. 1f). Similarly, 199 of the 319 miRNAs were present in all 3 R-ISS stages (Fig. 1c). Applying: (a) progressive deregulation of miRNA expression levels between R-ISS I-II-III, and (b) FC ≥ 1.5 or FC ≤ 0.67 between R-ISS I and R-ISS III, 55 miRNAs met the abovementioned criteria (Fig. 1e; Table S3), of which, 35 miRNAs (63.6%) were found to be overexpressed in R-ISS III compared to R-ISS I stage (Fig. 1g).
Overall, six miRNAs (miR-1–3p, miR-125b-5p, miR-181a-5p, miR-503-5p, miR-218-5p and miR-10a-5p) were concurrently deregulated between both MM vs. sMM/MGUS and R-ISS III vs. R-ISS I patients (Fig. 1h) and selected for further expression analysis and clinical evaluation in a subset screening cohort of 45 MM patients (~33% of our successfully followed-up screening cohort). No statistically significant differences of clinicopathological and follow-up data observed between patients of the subset (n = 45) and the complete (n = 138) screening MM cohorts. Notably, the survival analysis highlighted miR-181a to be the only candidate displaying significant association (unfavourable) with both OS (Kaplan–Meier: p = 0.016; univariate Cox: HR = 8.455; p = 0.044) and PFS (Kaplan–Meier: p = 0.036; univariate Cox: HR = 2.880; p = 0.047) of MM patients (Fig. S1), while no statistically significant correlation was documented either for OS or PFS for the other candidates (miR-1–3p, miR-125b-5p, miR-503-5p, miR-218-5p and miR-10a-5p). Additionally, target-prediction and GO analysis of miR-181a (Fig. S2) resulted in 27 BPs, 9 CCs and 10 MFs to be significantly enriched, including B cell apoptotic process and response to glucocorticoids, in line with the documented role of miR-181a in B-lymphoid cells differentiation [22]. In this regard, the strong association of miR-181a overexpression with the worse survival outcome of the subset cohort prompted us to further evaluate its clinical significance in improving risk-stratification and treatment prognosis of MM patients.
CD138+ overexpression of miR-181a is associated with short-term post-treatment progression and worse survival outcome
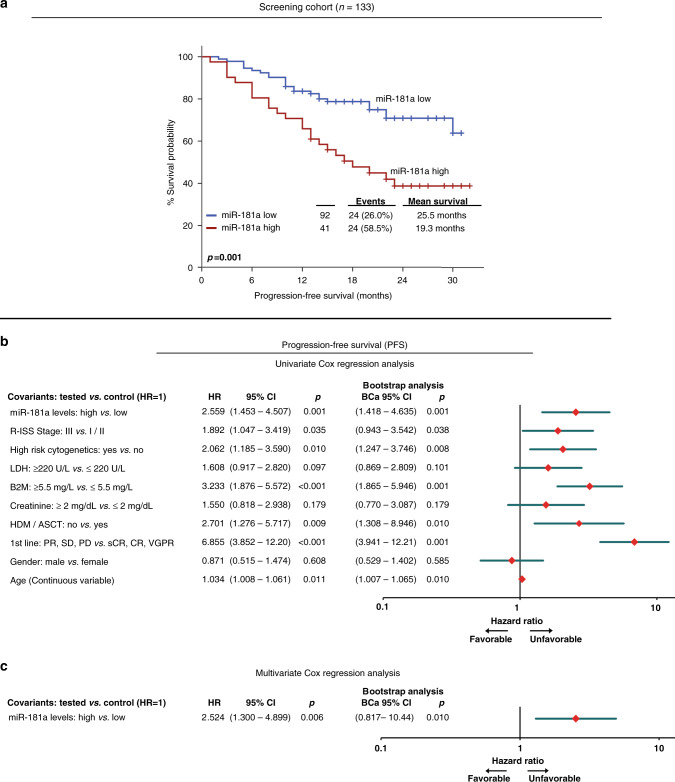
Kaplan–Meier and Cox regression analyses were performed to evaluate the clinical significance of miR-181a levels of CD138+ plasma cells for the post-treatment survival outcome of MM patients (Figs. 2 and 3; Table S4). According to the X-tile algorithm, the 70th percentile of miR-181a levels was adopted as the optimal cut-off value.
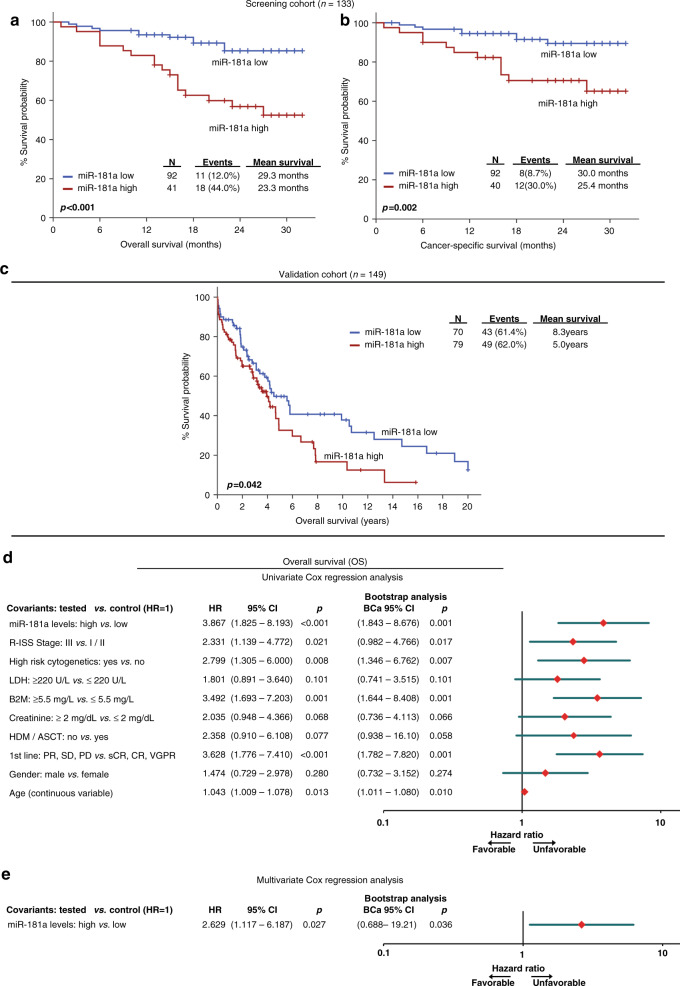
Fig. 2. CD138+ overexpression of miR-181a is strongly correlated with worse survival outcome following treatment.
a–c Kaplan–Meier survival curves for overall survival (OS; a) and cancer-specific survival (CSS; b) of the MM screening cohort, as well as OS (c) of the Kruykov et al. 2016 validation cohort, according to miR-181a expression. p-values calculated by log-rank test. d, e Forest plots of the univariate (d) and multivariate (e) Cox regression analysis for the OS of MM patients. Multivariate analysis adjusted for miR-181a, R-ISS stage, high-risk cytogenetics, B2M, LDH and creatinine levels, gender, age and response to 1st-line therapy. Internal validation was performed by bootstrap Cox proportional regression analysis based on 1000 bootstrap samples. HR Hazard Ratio, 95% CI 95% confidence interval of the estimated HR, BCa 95% CI bootstrap bias-corrected and accelerated 95% CI of the estimated HR based on 1000 bootstrap samples.
Fig. 3. Patients overexpressing miR-181a are at significantly higher risk for short-term disease progression.
a Kaplan–Meier survival curves for the progression-free survival (PFS; a) of the MM patients according to CD138+ expression of miR-181a. p-values calculated by log-rank test. b, c Forest plots of the univariate (b) and multivariate (c) Cox regression analysis for the MM patients’ PFS. Multivariate analysis adjusted for miR-181a, R-ISS stage, high-risk cytogenetics, B2M, LDH and creatinine levels, gender, age and response to 1st-line therapy. Internal validation was performed by bootstrap Cox proportional regression analysis based on 1000 bootstrap samples. HR Hazard Ratio, 95% CI 95% confidence interval of the estimated HR, BCa 95% CI bootstrap bias-corrected and accelerated 95% CI of the estimated HR based on 1000 bootstrap samples.
Kaplan–Meier curves illustrated the significantly shorter OS expectancy of MM patients with CD138+ overexpression of miR-181a (p < 0.001; Fig. 2a). Notably, the adverse prognostic utility of miR-181a overexpression was maintained regarding MM-specific survival (p = 0.002; Fig. 2b). To confirm miR-181a prognostic value in MM, the Kryukov et al. 2016 cohort (n = 151) was analyzed as external independent validation cohort. Supporting our findings, the evaluation of the validation cohort affirmed the unfavourable prognostic nature of CD138+ overexpression of miR-181a for the MM patient’s survival (p = 0.042; Fig. 2c). Finally, univariate Cox regression analysis strengthened the inferior OS of the MM patients with increased miR-181a (HR: 3.867; 95% CI: 1.825–8.193; p < 0.001; Fig. 2d). Focusing on MM post-treatment progression, Kaplan–Meier curves highlighted also that patients with CD138+ overexpression of miR-181a presented significantly shorter PFS (p = 0.001; Fig. 3a), which was also confirmed by univariate Cox analysis (HR: 2.559; 95% CI: 1.453–4.507; p = 0.001; Fig. 3b), compared to patients with lower miR-181a levels.
As MIR181A1 is located on 1q32.1, and gain of chromosome 1q (+1q) consists of one of the most common recurrent cytogenetic abnormalities in MM [23, 24], we sought to investigate the potential impact of +1q in miR-181a overexpression and clinical value in MM (Fig. S3). In this regard, +1q was not associated either with miR-181a levels (p = 0.423) or with patients’ OS (Kaplan–Meier: p = 0.400; univariate Cox: p = 0.405) and PFS (Kaplan–Meier: p = 0.591; univariate Cox: p = 0.595). Finally, multivariate Cox regression analysis clearly confirmed the ability of CD138+ overexpression of miR-181a to predict the adverse clinical outcome of MM patients independently of +1q.
Furthermore, to evaluate the independent prognostic value of miR-181a in MM, multivariate Cox regression models, adjusted to patients’ R-ISS stage, high-risk cytogenetics [+1q21, t(4;14), del(17p13), t(14;16), t(11;14), del(13q)], B2M, LDH and creatinine levels, gender, age, HDM/ASCT and response to 1st-line therapy, were performed. The analysis highlighted miR-181a elevated levels as independent predictor of MM patients’ worse survival (HR: 2.629; 95% CI: 1.117–6.187; p = 0.027; Fig. 2e) and higher risk for post-treatment progression (HR: 2.524; 95% CI: 1.300–4.899; p = 0.006; Fig. 3c). The survival analysis of patients’ EFS, although not proven to be statistically significant, pointed out the shorter EFS intervals of patients with upregulated miR-181a levels (p = 0.100; Fig. S4).
miR-181a overexpression significantly enhances the prognostic ability of the clinically established MM markers
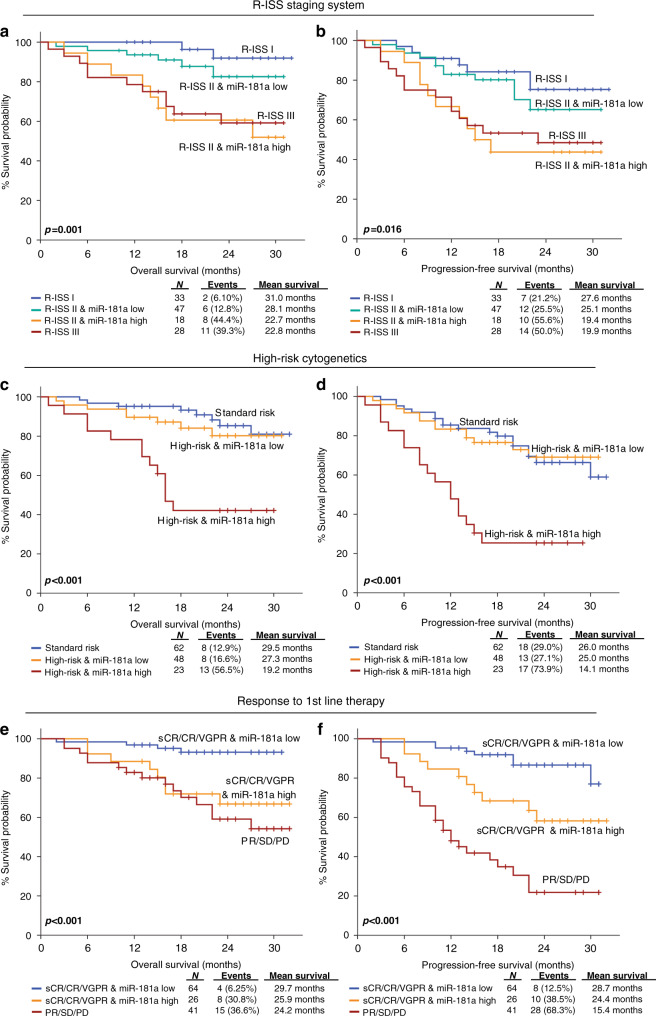
Prompted by the independent prognostic significance, we evaluated further the potential of miR-181a to improve patients’ prognosis by the established and clinically used markers of MM (Fig. 4). In this regard, R-ISS stage, high-risk cytogenetics [+1q21, t(4;14), del(17p13), t(14;16), t(11;14), del(13q)] and response to 1st-line therapy, represent the most widely used prognostic indicators of the disease.
Fig. 4. Evaluation of CD138+ miR-181a levels increases risk-stratification efficacy and results to superior clinical benefit in MM prognosis.
Kaplan–Meier survival curves for the overall survival (OS) and progression-free Survival (PFS) of MM patients according to CD138+ expression of miR-181a combined with R-ISS stage (a, b), high-risk cytogenetics (c, d) and response to 1st-line therapy (e, f). p-values calculated by log-rank test. sCR stringent complete response, CR complete response and VGPR very good partial response, PR partial response, SD stable disease, PD progressive disease.
Integration of miR-181a levels with these markers clearly improved patients’ prognostication. More precisely, Kaplan–Meier curves highlighted that the combination of R-ISS stage with miR-181a overexpression could provide a better stratification of MM patients’ OS (p = 0.001; Fig. 4a) and PFS (p = 0.016; Fig. 4b) expectancy, as R-ISS II patients overexpressing miR-181a displayed significantly shorter survival and analogous to R-ISS III patients, compared to R-ISS II patients with lower miR-181a levels, resembling R-ISS I. Similarly, the analysis of CD138+ miR-181a levels was able also to further stratify the risk for adverse disease progression between patients in high-risk cytogenetics group. In this regard, high-risk patients with elevated miR-181a levels showed significantly inferior disease outcome, in terms of OS (p < 0.001; Fig. 4c) and PFS (p < 0.001; Fig. 4d), compared to high-risk patients having lower miR-181a levels, resembling standard-risk group.
Finally, focusing on patients’ response to 1st-line therapy, patients’ stratification was significantly improved by the evaluation of miR-181a expression. More precisely, miR-181a overexpression could effectively predict patients with optimal treatment responses (Stringent Complete Response—sCR, Complete Response—CR and Very Good Partial Response—VGPR) at higher risk for disease progression (p < 0.001; Fig. 4e) and worse survival outcome (p < 0.001; Fig. 4f), compared to optimal treatment responders expressing lower miR-181a levels.
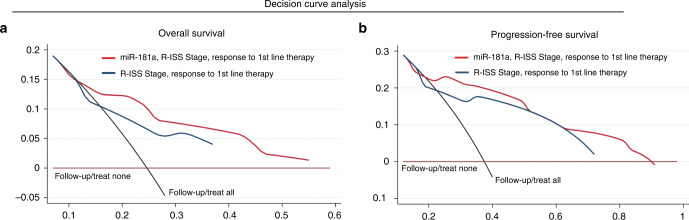
DCA revealed the advanced clinical benefit in ΜΜ prognosis by CD138+ miR-181a evaluation
DCA was performed, according to Vickers et al. [21], in order to study the clinical benefit of the multivariate prediction model incorporating miR-181a levels of CD138+ plasma cells along with the established clinically used MM markers (Fig. 5). In this regard, the analysis demonstrated the significantly augmented clinical net benefit of the model integrating miR-181a overexpression in predicting patients OS (Fig. 5a) and PFS (Fig. 5b) compared to R-ISS stage and response to 1st-line therapy model. The superiority of miR-181a-dependent prediction model, even at low threshold probabilities, is pivotal for the efficient risk-stratification and management of the MM patients.
Fig. 5. Decision curve analysis demonstrated the improved clinical net benefit of multivariate models integrating miR-181a with the established clinical markers.
a, b Decision curves of the multivariate prediction models for the overall survival (OS; a) and progression-free survival (PFS; b) of MM patients. Net benefit is plotted against various ranges of threshold probabilities.
Discussion
Despite significant advancements in MM therapy, the majority of the patients will become refractory and relapse [25, 26], while the highly heterogenous treatment response of MM patients hinders the reliable disease prognosis and the personalised patients’ management [27]. Thus, the identification of novel tools to improve patients’ risk-stratification and prediction of disease course is of first clinical priority. In addition to their role in normal homeostasis, miRNAs have been strongly implicated in tumorigenesis and progression of various human malignances, as well as in modern cancer diagnostics [28, 29].
Focusing on MM pathogenesis, miR-125b and miR-34a display a tumour suppressive function and are significantly downregulated in MM patients [30, 31]. In particular, ectopic expression of miR-125b has been recently found to impair MM cell proliferation by upregulating miR-34a, which in turn inhibits the IL-6R/STAT3 pathway [32] and represses CDK6, BCL2, and NOTCH1 expression [33]. On the other hand, miR-21 exerts a well-documented oncogenic role, the expression of which is induced by IL-6 in a STAT3-dependent manner [34]. Interestingly, a recent study highlighted that IL-17 producing CD4+ T cells (Th17) cells with elevated miR-21 levels induced tumour growth and osteoclast-dependent bone impairment, whereas suppression of miR-21 resulted in inhibiting Th17-mediated MM proliferation and osteoclasts activity [35]. Regarding drug resistance, miR-29b can influence the bortezomib sensitivity to MM cells by targeting Sp1 [36] and MCL1 [37], whereas overexpression of miR-221/222 promotes dexamethasone resistance through the inhibition of autophagy [38] and PUMA/BAK/BAX apoptotic pathway [39]. Lastly, aberrant levels of miR-137 [40] and miR-410 [41] in CD138+ plasma cells have been strongly correlated with early disease progression and poor treatment outcome of MM patients, indicating a strong clinical value in MM prognosis.
In the present study, by performing miRNA-seq in CD138+ plasma cells of patients with MM and its precursor conditions (sMM/MGUS), we investigated MM-related miRNA profiles and, for the first time, we have demonstrated the clinical utility of miR-181a in improving patients’ prognosis and prediction of disease outcome.
Our miRNA-seq analysis revealed six miRNA candidates (miR-1–3p, miR-125b-5p, miR-181a-5p, miR-503-5p, miR-218-5p and miR-10a-5p) to be concurrently deregulated in MM vs. sMM/MGUS, as well as in R-ISS III vs. R-ISS I patients. The levels of the six miRNAs were quantified in a subset MM cohort (n = 45) and, following survival analysis, miR-181a was found as the only candidate of prognostic significance both in OS and PFS of MM patients. The miR-181a is a member of the evolutionarily conserved miR-181 family (miR-181a/b/c/d), encoded by the MIR181A1 (1q32.1) and MIR181A2 (9q33.3) genes. Focusing on normal hematopoiesis, miR-181a was of the first miRNAs reported to be preferentially expressed in the BM, modulating B and T lymphocytes differentiation [22, 42]. Focusing on MM, Pichiorri et al. were the first to report a significant upregulation of miR-181a levels in BM plasma cells of MM/MGUS patients compared to healthy individuals [43]. Additional studies have also reported the upregulation of miR-181a in MM patients compared to healthy controls, as well as in advanced disease stages and patients with poor response to 1st-line therapy [44, 45], without, however, miR-181a clinical value in patients’ prognosis and post-treatment outcome to be evaluated in clinical setting. Supporting the central role of miR-181a in hematopoiesis, our target prediction and GO enrichment analysis highlighted the significant enrichment of target genes related to hematopoietic cell differentiation and apoptosis, as well as response to glucocorticoids.
The survival analysis of our MM screening cohort (n = 138) demonstrated the independent unfavourable prognostic value of miR-181a for MM outcome, as CD138+ plasma cells overexpression of miR-181a was associated with significantly higher risk for short-term relapse and worse survival expectancy of the patients. Moreover, multivariate Cox regression models highlighted the poor treatment outcome of the MM patients overexpressing miR-181a independently of the established disease markers and patients clinicopathological data, including response to 1st-line therapy, R-ISS stage, high-risk cytogenetics, HDM/ASCT, B2M, LDH and creatinine levels, age and gender. In line with our findings, the analysis of Kryukov et al. 2016 [16] external validation cohort (n = 151) clearly verified the worse survival outcome of the MM patients with CD138+ overexpression of miR-181a compared to those with lower levels.
Of the most common cytogenetic abnormalities in MM is +1q, which correlates with disease progression from MGUS/sMM to fully symptomatic MM and inferior patients’ outcome. More precisely, +1q has been associated with higher tumour burden, early relapse and drug resistance, while recent studies have highlighted that +1q can predict the worse survival of MM patients independently of other high-risk cytogenetic abnormalities, ISS stage and patients’ age [46, 47]. Several genes located in 1q, including cell cycle regulator CKS1B, BCL2 antiapoptotic family member MCL1 and RNA editing enzyme ADAR1, have been proposed as key regulators of pathogenesis in +1q myeloma, by altering MAPK/ERK and JAK/STAT3 signaling pathways and thus modulating cell proliferation and survival. Strikingly, overexpression of CKS1B, MCL1 and ADAR1 is highly associated with +1q patients and aggressive clinical course [23, 24]. Although MIR181A1 gene maps on 1q32.1, the analysis of our screening cohort did not highlight any correlation of +1q with either miR-181a levels, indicating no significant impact of +1q on miR-181a clinical value for the patients’ prognosis. In this regard, multivariate Cox analysis confirmed that CD138+ overexpression of miR-181a can effectively predict worse survival outcome independently of +1q.
Our findings are in agreement with previous studies regarding the expression regulation and oncogenic role of miR-181a in MM. More precisely, overexpression of miR-181a in RPMI8226 MM cells promoted cells proliferation, while miR-181a silencing resulted in significantly reduced survival and proliferation rates, and stimulated apoptosis [48, 49]. Moreover, miR-181a knockdown in vivo using xenograft mouse models of MM, strongly inhibited tumour growth [43, 49]. To explain this tumour-promoting role of miR-181a in MM, the direct targeting of HOXA11 and the MEG3/miR-181a/HOXA11 regulatory network has been demonstrated in vitro, in which the downregulation of MEG3 lncRNA, sponging miR-181a, results in HOXA11 targeting-related MM progression [50].
Beside MM, the tumorigenic and/or clinical role of miR-181a has been reported in numerous solid human malignancies, including ovarian [51], breast [52], colorectal [53], pancreatic [54] and gastric [55] cancers, as well as in childhood ALL and AML. Notably, the elevated miR-181a enhances G1/S transition and cell proliferation in pediatric AML, by regulating the tumour suppressor ATM [56]. Moreover, in childhood ALL, increased cell growth and proliferation was promoted by the miR-181a-mediated targeting of WIF-1 and the downstream stimulation of Wnt/β-catenin signalling [57], while miR-181a knockdown clearly inhibited NOTCH1-induced T-ALL development [58]. More recently also, elevated miR-181a exosomal levels detected in childhood ALL patients, while silencing of exosomal miR-181a prevented the exosomal-induced leukemic cell proliferation in vitro [59].
Taking advantage of the independent clinical value of miR-181a in MM patients’ outcome, we have further investigated miR-181a ability to improve the prognostic performance of the established and clinically used MM markers, including R-ISS stage, high-risk cytogenetics and response to 1st-line therapy. In this regard, the evaluation of miR-181a expression significantly improved the risk-stratification specificity of MM patients, resulting in the advanced positive prediction of patients’ poor treatment outcome and short-term progression within R-ISS II, high-risk cytogenetics and optimal treatment response patients. Consequently, as clinically evaluated by DCA, the integration of miR-181a expression along with the established disease markers lead to a superior MM prognosis prediction model.
Although previous studies have demonstrated the tumour-promoting role of miR-181a in MM, the lack of further in vitro validation of miR-181a regulatory role in CD138+ plasma cells is the main limitation of the study. Future studies, focusing on the in-depth characterisation of miR-181a role in MM will unveil the causal link between its function and the strong clinical value on patients’ risk-stratification and treatment prognosis. Moreover, different cut-off values were adopted in our screening and validation cohorts, which is mainly attributed to the different methodologic approaches applied in miR-181a quantification (specific RT-qPCR in screening vs. high-throughput expression profiling in validation cohort). Certainly, multi-institutional large-scale studies could identify the optimal cut-off value of CD138+ miR-181a levels to be utilised in clinical setting.
In conclusion, upregulated miR-181a levels of CD138+ plasma cells observed by miRNA-seq in MM compared to sMM/MGUS patients and in advanced R-ISS stages, while the analysis of the screening and validation cohorts highlighted the significantly higher risk for short-term progression and worse survival outcome of MM patients overexpressing miR-181a. Additionally, multivariate regression models verified CD138+ elevated miR-181a levels as independent predictor of poor treatment and survival outcome of MM patients. Finally, evaluation of miR-181a expression with the clinically used disease markers, including R-ISS stage, high-risk cytogenetics and response to 1st-line therapy, resulted in enhanced risk-stratification specificity, superior positive prediction of patients’ poor treatment outcome and higher clinical benefit compared to the established markers alone, supporting miR-181a utility in modern diagnostics and management of MM patients.
Supplementary information
Acknowledgements
We thank all patients and families for their participation. We thank IEMBITHEK (Greece) for partially funded the study. We thank Bodossaki Foundation (Athens, Greece) for partially supported the study by fellowship to PGA.
Author contributions
Conception and design: AS, ET, MAD, MA; Development of methodology: MAP, AMP, MA; Acquisition of data: MAP, AMP, PGA, KMP, CIL, PM, NMK, DP, EEP, MG, EK, MA; Analysis and interpretation of data: MAP, AMP, PGA, KMP, MA, MAD, ET, AS; Acquired and managed patients: AMP, MG, EK, MAD, ET; Drafting of the manuscript: MAP, AMP, PGA, MA; Critical revision of the manuscript: MAD, ET, AS; Study supervision: AS, ET, MAD, MA; Approval of the submitted and final version: All authors.
Data availability
All the data are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of “Alexandra” Hospital, Athens, Greece, and all patients who participated were informed and signed an informed consent according to 1975 Declaration of Helsinki ethical standards, as revised in 2008.
Consent for publication
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maria-Alexandra Papadimitriou, Aristea-Maria Papanota.
These authors jointly supervised this work: Evangelos Terpos, Andreas Scorilas.
Contributor Information
Evangelos Terpos, Email: eterpos@med.uoa.gr.
Andreas Scorilas, Email: ascorilas@biol.uoa.gr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01602-8.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed]
- 3.Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol. 2020;95:548–67. doi: 10.1002/ajh.25791. [DOI] [PubMed] [Google Scholar]
- 4.Boyle EM, Deshpande S, Tytarenko R, Ashby C, Wang Y, Bauer MA, et al. The molecular make up of smoldering myeloma highlights the evolutionary pathways leading to multiple myeloma. Nat Commun. 2021;12:293. doi: 10.1038/s41467-020-20524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimopoulos MA, Moreau P, Terpos E, Mateos MV, Zweegman S, Cook G, et al. Multiple myeloma: EHA-ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-up. Hemasphere. 2021;5:e528. doi: 10.1097/HS9.0000000000000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papanota AM, Karousi P, Kontos CK, Ntanasis-Stathopoulos I, Scorilas A, Terpos E. Multiple myeloma bone disease: implication of MicroRNAs in its molecular background. Int J Mol Sci. 2021;22:2375. [DOI] [PMC free article] [PubMed]
- 7.Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, et al. Multiple myeloma. Nat Rev Dis Prim. 2017;3:17046. doi: 10.1038/nrdp.2017.46. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Calle V, Slack A, Keane N, Luft S, Pearce KE, Ketterling RP, et al. Evaluation of revised international staging system (R-ISS) for transplant-eligible multiple myeloma patients. Ann Hematol. 2018;97:1453–62.. doi: 10.1007/s00277-018-3316-7. [DOI] [PubMed] [Google Scholar]
- 9.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 11.Diamantopoulos MA, Tsiakanikas P, Scorilas A. Non-coding RNAs: the riddle of the transcriptome and their perspectives in cancer. Ann Transl Med. 2018;6:241. doi: 10.21037/atm.2018.06.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D, Yang X, Liu M, Zhang Z, Xing E. Roles of miRNA dysregulation in the pathogenesis of multiple myeloma. Cancer Gene Ther. 2021. https://www.nature.com/articles/s41417-020-00291-4. [DOI] [PMC free article] [PubMed]
- 14.Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:e538–48. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P, et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17:e328–e46.. doi: 10.1016/S1470-2045(16)30206-6. [DOI] [PubMed] [Google Scholar]
- 16.Kryukov F, Nemec P, Radova L, Kryukova E, Okubote S, Minarik J, et al. Centrosome associated genes pattern for risk sub-stratification in multiple myeloma. J Transl Med. 2016;14:150. doi: 10.1186/s12967-016-0906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–52. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak. 2006;26:565–74. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt TM, Barwick BG, Joseph N, Heffner LT, Hofmeister CC, Bernal L, et al. Gain of Chromosome 1q is associated with early progression in multiple myeloma patients treated with lenalidomide, bortezomib, and dexamethasone. Blood Cancer J. 2019;9:94. doi: 10.1038/s41408-019-0254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt TM, Fonseca R, Usmani SZ. Chromosome 1q21 abnormalities in multiple myeloma. Blood Cancer J. 2021;11:83. doi: 10.1038/s41408-021-00474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landgren O, Rajkumar SV. New developments in diagnosis, prognosis, and assessment of response in multiple myeloma. Clin Cancer Res. 2016;22:5428–33.. doi: 10.1158/1078-0432.CCR-16-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimopoulos MA, Jakubowiak AJ, McCarthy PL, Orlowski RZ, Attal M, Blade J, et al. Developments in continuous therapy and maintenance treatment approaches for patients with newly diagnosed multiple myeloma. Blood Cancer J. 2020;10:17. doi: 10.1038/s41408-020-0273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasche L, Chavan SS, Stephens OW, Patel PH, Tytarenko R, Ashby C, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun. 2017;8:268. doi: 10.1038/s41467-017-00296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–22.. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 29.Avgeris M, Panoutsopoulou K, Papadimitriou MA, Scorilas A. Circulating exosomal miRNAs: clinical significance in human cancers. Expert Rev Mol Diagn. 2019;19:979–95.. doi: 10.1080/14737159.2019.1673732. [DOI] [PubMed] [Google Scholar]
- 30.Morelli E, Leone E, Cantafio ME, Di Martino MT, Amodio N, Biamonte L, et al. Selective targeting of IRF4 by synthetic microRNA-125b-5p mimics induces anti-multiple myeloma activity in vitro and in vivo. Leukemia. 2015;29:2173–83. doi: 10.1038/leu.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y, Yang W, Zhang L, Liu K, Luo Z. Long non-coding RNA ANRIL and its target microRNAs (microRNA-34a, microRNA-125a and microRNA-186) relate to risk stratification and prognosis in multiple myeloma. Hematology. 2021;26:160–9. doi: 10.1080/16078454.2021.1872275. [DOI] [PubMed] [Google Scholar]
- 32.Misso G, Zarone MR, Lombardi A, Grimaldi A, Cossu AM, Ferri C, et al. miR-125b upregulates miR-34a and sequentially activates stress adaption and cell death mechanisms in multiple myeloma. Mol Ther Nucleic Acids. 2019;16:391–406. doi: 10.1016/j.omtn.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18:6260–70.. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leone E, Morelli E, Di Martino MT, Amodio N, Foresta U, Gulla A, et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin Cancer Res. 2013;19:2096–106. doi: 10.1158/1078-0432.CCR-12-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossi M, Altomare E, Botta C, Gallo Cantafio ME, Sarvide S, Caracciolo D, et al. miR-21 antagonism abrogates Th17 tumor promoting functions in multiple myeloma. Leukemia. 2021;35:823–34.. doi: 10.1038/s41375-020-0947-1. [DOI] [PubMed] [Google Scholar]
- 36.Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012;3:e436. doi: 10.1038/cddis.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Y, Zhang Y, Liu W, Huang Y, Shen X, Jing R, et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019;10:106. [DOI] [PMC free article] [PubMed]
- 38.Xu J, Su Y, Xu A, Fan F, Mu S, Chen L, et al. miR-221/222-mediated inhibition of autophagy promotes dexamethasone resistance in multiple myeloma. Mol Ther. 2019;27:559–70.. doi: 10.1016/j.ymthe.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao JJ, Chu ZB, Hu Y, Lin J, Wang Z, Jiang M, et al. Targeting the miR-221-222/PUMA/BAK/BAX pathway abrogates dexamethasone resistance in multiple myeloma. Cancer Res. 2015;75:4384–97.. doi: 10.1158/0008-5472.CAN-15-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin Y, Zhang S, Deng S, An G, Qin X, Li F, et al. Epigenetic silencing of miR-137 induces drug resistance and chromosomal instability by targeting AURKA in multiple myeloma. Leukemia. 2017;31:1123–35.. doi: 10.1038/leu.2016.325. [DOI] [PubMed] [Google Scholar]
- 41.Yang N, Chen J, Zhang H, Wang X, Yao H, Peng Y, et al. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017;8:e2975. doi: 10.1038/cddis.2017.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emming S, Chirichella M, Monticelli S. MicroRNAs as modulators of T cell functions in cancer. Cancer Lett. 2018;430:172–8. doi: 10.1016/j.canlet.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105:12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–80. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li YJ, Li DP, Yan ZL, Qi KM, Chen LL, Zhang ZY, et al. Potential relationship and clinical significance of miRNAs and Th17 cytokines in patients with multiple myeloma. Leuk Res. 2014;38:1130–5. doi: 10.1016/j.leukres.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Giri S, Huntington SF, Wang R, Zeidan AM, Podoltsev N, Gore SD, et al. Chromosome 1 abnormalities and survival of patients with multiple myeloma in the era of novel agents. Blood Adv. 2020;4:2245–53.. doi: 10.1182/bloodadvances.2019001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdallah N, Greipp P, Kapoor P, Gertz MA, Dispenzieri A, Baughn LB, et al. Clinical characteristics and treatment outcomes of newly diagnosed multiple myeloma with chromosome 1q abnormalities. Blood Adv. 2020;4:3509–19.. doi: 10.1182/bloodadvances.2020002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng J, Thakur A, Zhang S, Dong YF, Wang XQ, Yuan RL, et al. Expressions of miR-181a and miR-20a in RPMI8226 cell line and their potential as biomarkers for multiple myeloma. Tumor Biol. 2015;36:8545–52. doi: 10.1007/s13277-015-3600-2. [DOI] [PubMed] [Google Scholar]
- 49.Liu N, Yang J, Yuan R, Peng J, Liu L, Guo X. Effects of miR181a on the biological function of multiple myeloma. Oncol Rep. 2019;42:291–300.. doi: 10.3892/or.2019.7160. [DOI] [PubMed] [Google Scholar]
- 50.Shen XX, Bai H, Zhu HY, Yan Q, Yang Y, Yu WJ, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate HOXA11 expression by sponging miR-181a in multiple myeloma. Cell Physiol Biochem. 2018;49:87–100. doi: 10.1159/000492846. [DOI] [PubMed] [Google Scholar]
- 51.Panoutsopoulou K, Avgeris M, Magkou P, Mavridis K, Dreyer T, Dorn J, et al. miR-181a overexpression predicts the poor treatment response and early-progression of serous ovarian cancer patients. Int J Cancer. 2020;147:3560–73.. doi: 10.1002/ijc.33182. [DOI] [PubMed] [Google Scholar]
- 52.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123:150–63. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishimura J, Handa R, Yamamoto H, Tanaka F, Shibata K, Mimori K, et al. microRNA-181a is associated with poor prognosis of colorectal cancer. Oncol Rep. 2012;28:2221–6. doi: 10.3892/or.2012.2059. [DOI] [PubMed] [Google Scholar]
- 54.Meijer LL, Garajova I, Caparello C, Le Large TYS, Frampton AE, Vasile E, et al. Plasma miR-181a-5p downregulation predicts response and improved survival after FOLFIRINOX in pancreatic ductal adenocarcinoma. Ann Surg. 2020;271:1137–47.. doi: 10.1097/SLA.0000000000003084. [DOI] [PubMed] [Google Scholar]
- 55.Mi Y, Zhang D, Jiang W, Weng J, Zhou C, Huang K, et al. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. 2017;389:11–22. doi: 10.1016/j.canlet.2016.12.033. [DOI] [PubMed] [Google Scholar]
- 56.Liu X, Liao W, Peng H, Luo X, Luo Z, Jiang H, et al. miR-181a promotes G1/S transition and cell proliferation in pediatric acute myeloid leukemia by targeting ATM. J Cancer Res Clin Oncol. 2016;142:77–87. doi: 10.1007/s00432-015-1995-1. [DOI] [PubMed] [Google Scholar]
- 57.Lyu X, Li J, Yun X, Huang R, Deng X, Wang Y, et al. miR-181a-5p, an inducer of Wnt-signaling, facilitates cell proliferation in acute lymphoblastic leukemia. Oncol Rep. 2017;37:1469–76.. doi: 10.3892/or.2017.5425. [DOI] [PubMed] [Google Scholar]
- 58.Fragoso R, Mao T, Wang S, Schaffert S, Gong X, Yue S, et al. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet. 2012;8:e1002855. doi: 10.1371/journal.pgen.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haque S, Vaiselbuh SR. Silencing of exosomal miR-181a reverses pediatric acute lymphocytic leukemia cell proliferation. Pharmaceuticals (Basel). 2020;13:241. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data are available from the corresponding authors on reasonable request.