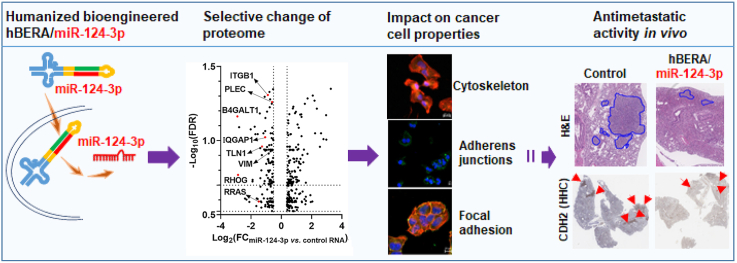
Abstract
With the understanding of microRNA (miRNA or miR) functions in tumor initiation, progression, and metastasis, efforts are underway to develop new miRNA-based therapies. Very recently, we demonstrated effectiveness of a novel humanized bioengineered miR-124-3p prodrug in controlling spontaneous lung metastasis in mouse models. This study was to investigate the molecular and cellular mechanisms by which miR-124-3p controls tumor metastasis. Proteomics study identified a set of proteins selectively and significantly downregulated by bioengineered miR-124-3p in A549 cells, which were assembled into multiple cellular components critical for metastatic potential. Among them, plectin (PLEC) was verified as a new direct target for miR-124-3p that links cytoskeleton components and junctions. In miR-124-3p-treated lung cancer and osteosarcoma cells, protein levels of vimentin, talin 1 (TLN1), integrin beta-1 (ITGB1), IQ motif containing GTPase activating protein 1 (IQGAP1), cadherin 2 or N-cadherin (CDH2), and junctional adhesion molecule A (F11R or JAMA or JAM1) decreased, causing remodeling of cytoskeletons and disruption of cell–cell junctions. Furthermore, miR-124-3p sharply suppressed the formation of focal adhesion plaques, leading to reduced cell adhesion capacity. Additionally, efficacy and safety of biologic miR-124-3p therapy was established in an aggressive experimental metastasis mouse model in vivo. These results connect miR-124-3p−PLEC signaling to other elements in the control of cytoskeleton, cell junctions, and adhesion essential for cancer cell invasion and extravasation towards metastasis, and support the promise of miR-124 therapy.
KEY WORDS: RNA therapy, MiR-124-3p, Metastasis, Proteomics, Junctions, Cell adhesion, Bioengineer, Imaging
Graphical abstract
The antimetastatic activity of bioengineered miR-124-3p is attributable to selective downregulation of proteins underlying multiple cellular components, including cytoskeleton, cell junctions, and focal adhesion, critical for invasion and extravasation.
1. Introduction
Cancer remains a leading cause of death worldwide1, and systemic metastasis of primary tumor accounts for about 90% of cancer deaths2, 3, 4. By the time an advanced tumor such as osteosarcoma is diagnosed, around 15%–20% of patients likely have macrometastases; the most common of which are pulmonary metastases (85%–90%), and the rest of patients are believed to have micrometastases which are undetectable by current diagnostic means5. In addition, 11%–36% of patients with non-small cell lung cancer (NSCLC), the most common type of lung cancer6,7, exhibit distant metastasis, and up to 93% of patients show malignant dissemination when post-mortem analyses were conducted8. Since there are limited treatment options for metastatic diseases9, large efforts are underway to improve the understanding of metastatic processes, identify potential targets, develop new therapeutics, and define pharmacological actions of effective agents.
While small molecules and antibodies remain the most common forms of drugs in clinical use and under investigations, RNAs or oligoribonucleotides have emerged as a new class of therapeutics that hold great promise to expand the druggable targets from proteins to the genome and transcripts which far outnumber proteins10, 11, 12, 13. Since 2018, three siRNA drugs, patisiran, givosiran, and lumasiran, have been already approved by the United States Food and Drug Administration for clinical use. Among them, patisiran and lumasiran follow the mechanism of actions of genome-derived microRNAs (miRNAs or miRs), through base pairing with the 3′-untranslated regions (3′ UTR) of targeted transcripts14,15. Indeed, miRNAs are a superfamily of small noncoding RNAs (ncRNAs) governing posttranscriptional gene regulation behind essentially all cellular processes, and some miRNAs have been revealed to play important roles in the pathogenesis of various human diseases including cancer14, 15, 16, 17.
With an improved understanding of the important roles of miRNAs (e.g., miR-34 and miR-124) in tumorigenesis and metastasis, there is growing interest in developing miRNA-based therapies10,12,16, 17, 18. Some miRNA-based therapies such as miR-16 and miR-34 have entered into clinical trials19,20. However, current miRNA research and development rely mainly on the miRNA mimics made in vitro by chemical synthesis, which are different from natural RNAs produced and folded in vivo12,21. Our efforts have led to the development of a novel ncRNA bioengineering technology to achieve high-yield, large-scale, and cost-effective in vivo fermentation production of bioengineered RNAi agents (BERAs) by using tRNA-fused pre-miRNA as a carrier to accommodate payload miRNAs or other small RNAs22, 23, 24. Acting as a “prodrug”, BERAs are precisely processed to target miRNAs in human cells to selectively regulate target gene expression22, 23, 24, 25, 26, 27, 28, 29, 30. Co-administration of biologic miRNAs with chemotherapeutic drugs can synergistically inhibit the viability of human carcinoma cells through co-targeting of pharmacokinetic and pharmacodynamic factors31,32. We have further established the effectiveness of several BERAs in the control of tumor growth in different xenograft mouse models, including let-7c-5p against advanced hepatocellular carcinoma27, miR-1291-5p against pancreatic cancer26, and miR-34a-5p against NSCLC22. Very recently, we have produced a small set of humanized BERAs (hBERAs) and identified hBERA/miR-124-3p as a potent inhibitor against human osteosarcoma cell viability and invasiveness in vitro24. Moreover, hBERA/miR-124-3p significantly reduces tumor growth and pulmonary metastasis in the orthotopic osteosarcoma xenograft and spontaneous metastases mouse models in vivo24.
In this study, we aimed at delineating the molecular and cellular mechanisms by which hBERA/miR-124-3p controls metastasis. We first conducted a liquid chromatography tandem mass spectrometry (LC−MS/MS)-based proteomics study that allowed us to identify a set of proteins selectively and significantly suppressed by biologic miR-124-3p prodrug that are assembled into multiple cellular components critical for cell adhesion and metastasis. After verifying several downregulated proteins in both NSCLC A549 and osteosarcoma 143B and MG-63 cells, we identified and validated plectin (PLEC) as a direct target for miR-124-3p. Further, we employed confocal imaging technology to demonstrate the actions of hBERA/miRNA-124-3p in remodeling cell cytoskeleton, disrupting adherens junctions, and reducing focal adhesion plaques, leading to reduced cell adhesion capacity. In addition, the effectiveness and safety of miR-124-3p therapy in the control of lung metastasis was established in experimental metastasis mouse models in vivo. These findings shall provide insights into biologic miRNA-124-3p prodrug mechanisms of actions in the control of metastasis and development of miRNA therapeutics.
2. Materials and methods
2.1. Materials
RPMI 1640 medium, trypsin, phosphate-buffered saline (PBS), fetal bovine serum, opti-MEM, RIPA buffer, bicinchoninic acid (BCA) Protein Assay Kit, and Lipofectamine 3000 Transfection reagents were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Protease inhibitor cocktail and Trizol reagent were bought from Sigma–Aldrich (St. Louis, MO, USA). Clarity Western Enhanced Chemiluminescence Substrates, blotting-grade blocker, and polyvinylidene difluoride (PVDF) membranes were purchased from Bio-Rad (Hercules, CA, USA). In vivo-jetPEI was purchased from Polyplus-transfection (New York, NY, USA). All other chemicals were purchased from Sigma–Aldrich or Thermo Fisher Scientific Inc.
2.2. Human cell culture
The human lung carcinoma A549 (CCL-185), osteosarcoma 143B (CRL-8303), MG-63 (CRL-1427), and embryonic kidney HEK293 (CRL-1573) cells were purchased from American Type Culture Collection (Manassas, VA, USA). Carcinoma cells and HEK293 cells were maintained in RPMI 1640 and DMEM media, respectively, containing 10% fetal bovine serum, at 37 °C in a humidified atmosphere with 5% carbon dioxide.
2.3. Expression and purification of hBERA/miR-124-3p and control RNA
Humanized biologic hBERA/miR-124-3p and control RNA, namely human leucyl tRNA with a sephadex aptamer (LSA), were produced through in vivo fermentation production, as described very recently24. The hBERA/miR-124-3p prodrug is assembled by using the human leucyl tRNA, and the corresponding LSA was revealed as a valid control RNA to determine miR-124-3p function24, which was chosen according to our previous findings on BERAs22,23,26,30. Briefly, HST08 E. coli competent cells (Clontech Laboratories, Mountain View, CA, USA) were transformed with hBERA/miR-124-3p or LSA expression plasmids and incubated at 37 °C for 15–16 h. Total RNAs were isolated by using Tris-HCl saturated phenol method, recombinant RNAs were further purified with an ENrich™ Q 10 × 100 column on an NGC Quest 10 Plus Chromatography fast protein liquid chromatography (FPLC) system (Bio-Rad), as reported22,24. FPLC-purified RNAs were quantified with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), and purities were determined by using a high-performance liquid chromatography (HPLC) method, as described25. Endotoxin levels were measured by using the Pyrogent-5000 kinetic LAL assay (Lonza, Walkersville, MD), following manufacturer's instructions. The hBERA/miR-124-3p and LSA with high purity (>98%) and low endotoxin activity (<5 EU/μg RNA) were used in the following studies.
2.4. Proteomics study
A549 cells were plated at 300,000 cells/well in a 6-well plate, grown overnight, and transfected with 15 nmol/L of RNAs or empty Lipofectamine 3000 (vehicle). Three replicates were conducted for each group, and cell pellets collected at 72 h post-transfection were submitted to the UC Davis Proteomics Core for digestion and LC−MS/MS analyses. Briefly, proteins were extracted in 2% sodium lauryl sulfate in 50 mmol/L triethyl ammonium bicarbonate (TEAB). Protein concentration was determined by BCA assay, and 150 μg/sample was used for S-Trap based digestion (Protifi, Farmingdale, NY, USA) by following the manufacturer's instructions33,34. The digested peptides were released from the S-trap column, vacuum centrifuged to dryness and resuspended in 2% acetonitrile containing 0.1% trifluoroacetic acid. Peptide concentrations were determined by using Pierce Quantitative Fluorometric Peptide Assay (Thermo Fisher Scientific).
LC−MS/MS analysis was performed using an Ultimate 3000 nano-LC system coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Peptides were separated on an Easy-spray 100 μm × 25 cm C18 column over a 90-min gradient elution with solvent A (0.1% formic acid) and solvent B (100% acetonitrile with 0.1% formic acid), from 2% solution B to 50% for 60 min followed by a 50%–99% solution B in 6 min and then held for 3 min before changed back to 2% in 2 min. Thermo Scientific Fusion Lumos mass spectrometer running in Data Independent Acquisition mode. Six gas phase fractionated (GPF) chromatogram library injections were made using staggered 4 Da isolation widows, GPF1 400–500 m/z, GPF2 500–600 m/z, GPF3 600–700 m/z, GPF4 700–800 m/z, GPF5 800–900 m/z, and GPF6 900–1000 m/z. Data were acquired using a collision energy of 35, resolution of 30,000, maximum inject time of 54 ms and a AGC target of 50,000. Each sample was run with staggered isolation windows of 12 Da in the range 400–1000 m/z.
MS raw data were analyzed using Scaffold DIA (2.0.0) (Proteome Software, Portland, OR, USA). Raw data files were converted to mzML format using ProteoWizard (3.0.11748). The Reference Spectral Library was created by Encyclopedia (0.9.2). Reference samples were individually searched against uniprot-proteome_UP000005640.fasta with a peptide mass tolerance of 10.0 ppm and a fragment mass tolerance of 10.0 ppm. Peptides identified in each search were filtered by Percolator (3.01.nightly-13-655e4c7-dirty) to achieve a maximum false discovery rate (FDR) of 0.0135. Individual search results were combined and peptides were again filtered to an FDR threshold of 0.01 for inclusion in the reference library. Analytic samples were aligned based on retention times and individually searched against the reference Spectral library created from the six GPF runs described above with a peptide mass tolerance of 10.0 ppm and a fragment mass tolerance of 10.0 ppm. Peptides identified in each sample were filtered by Percolator to achieve a maximum FDR of 0.01. Individual search results were combined and peptide identifications were assigned posterior error probabilities and filtered to an FDR threshold of 0.01 by Percolator. Peptide quantification was performed by EncyclopeDIA (0.9.2). For each peptide, the 5 highest quality fragment ions were selected for quantitation. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis were grouped to satisfy the principles of parsimony. Proteins with a minimum of 2 identified peptides were thresholded to achieve a protein FDR threshold of 0.0135.
2.5. Pathway and miRNA enrichment analyses
Differentially expressed proteins in cells following miR-124-3p treatment were identified stringently (fold change≥30%, FDR<0.02, and intensity>200,000) by using EdgeR package. Gene Ontology (GO) networks, functions, and pathways were generated from those downregulated proteins by using STRING (https://string-db.org/)36. In addition, miRNA target enrichment analysis was conducted by using MIENTURNET (http://userver.bio.uniroma1.it/apps/mienturnet/)37.
2.6. Plasmid construction and luciferase reporter assays
The PLEC 3′ UTR segment (0–1024 nucleotides from stop codon), consisting of two putative miRNA response elements (MREs) for miR-124-3p identified by using TargetScan (http://www.targetscan.org/) was cloned downstream of the firefly luciferase gene within dual-luciferase pEZX-MT06 vector (GeneCopoeia, Rockville, MD, USA). To validate the interactions between miR-124-3p and MREs, three mutations were designed and created. The four plasmids were constructed by GeneCopoeia and validated by DNA sequencing. Dual luciferase reporter assays were conducted as previously described26,32. Briefly, HEK293 cells were co-transfected with 50 ng PLEC 3′ UTR-luciferase reporter plasmids plus hBERA/miR-124-3p or LSA (5 nmol/L) using Lipofectamine 3000, or vehicle alone. After 48 h, luciferase activity was determined by using the dual-luciferase reporter assay system (Promega, Madison, WI, USA) following the manufacturer's instructions, on a SpectraMax® M3 microplate reader (Molecular Devices, Sunnyvale, CA, USA). The firefly luciferase activity was normalized to corresponding Renilla luciferase activity and then normalized to vehicle control group.
2.7. Reverse transcription quantitative real-time PCR (qPCR)
Cells (2.0 × 105 cells/well) were grown in 24-well plate and transfected with hBERA/miR-124-3p or LSA (15 nmol/L for A549, and 10 nmol/L for 143B and MG-63 cells) using Lipofectamine 3000. Total RNAs were extracted at 48 h post-transfection using a Direct-zol RNA MiniPrep Kit (Zymo Research, Irvine, CA, USA). Reverse-transcription was conducted with NxGen M-MuLV reverse transcriptase (Lucigen, Middleton, WI, USA) and random hexamers (for U6) or stem-loop primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCATT-3′ (for miR-124-3p). Real-time PCR analysis was carried out on a CFX96 Touch real-time PCR system (Bio-Rad) using specific primers: forward 5′-GCGCTAAGGCACGCGGTG-3′ and reverse 5′-GTGCAGGGTCCGAGGT-3′ for miR-124-3p, and forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′ for U6. The relative expression levels of mature miR-124-3p were calculated by the comparative threshold cycle (Ct) method using formula 2−ΔΔCt.
2.8. Immunoblot analyses
Cells (3.0 × 105 cells/well) were seeded in 6-well plates and transfected with hBERA/miR-124-3p, LSA (15 nmol/L for A549, and 10 nmol/L for 143B and MG-63 cells), or vehicle. Cells were harvested at 48 h post-treatment, and total proteins were isolated with RIPA lysis buffer supplemented with complete protease inhibitors. Protein concentrations were determined with BCA kit. Thirty μg of proteins were resolved with 6%, 10%, or 12% polyacrylamide gel electrophoresis gel and transferred on to a PVDF membrane. Following a 2-h blocking with 5% milk, PVDF membranes were incubated with selective antibodies against PLEC (sc-33649, Santa Cruz, USA; 1:200), integrin beta-1 (ITGB1; sc-374429, Santa Cruz; 1:100), IQ motif containing GTPase activating protein 1 (IQGAP1; 20648S, Cell signaling Technology, USA; 1:1000), vimentin (VIM; 5741S, Cell signaling Technology; 1:1000), N-cadherin or cadherin 2 (CDH2; 13116s, Cell signaling Technology; 1:1000), talin-1 (TLN1; PA5-82162, Thermo Fisher Scientific; 1:200), junctional adhesion molecule A (JAMA or JAM1 or F11R; ab269948, Abcam, USA; 1:1000 dilution), or β-actin (A5441, Sigma–Aldrich; 1:3000), and then incubated with horseradish peroxidase-conjugated secondary antibodies (anti-rabbit, 1:10,000, Jackson ImmunoResearch, West Grove, PA, USA; anti-mouse, 1:3000, Cell Signaling Technology). Individual protein bands were visualized with ChemiDoc MP Imaging System (Bio-Rad) after incubation with Clarity Western enhanced chemiluminescent blotting substrates, and band densities were determined by Image Lab software (Bio-Rad) and normalized to corresponding β-actin levels.
2.9. Confocal imaging studies
A549 (2.0 × 104 cells/well), 143B (1.0 × 104 cells/well), and MG-63 (1.0 × 104 cells/well) cells were plated on 8-well chamber slides overnight for attachment. Subsequently, cells were treated with vehicle, LSA, or hBERA/miR-124-3p (15 nmol/L for A549, 10 nmol/L for 143B and MG-63). At 48-h post-transfection, the medium was removed and cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min. Cells were then permeabilized with 0.25% Triton X-100 for 10 min, washed 3 times, blocked with 1% BSA in PBS for 30 min, and stained with selective anti-PLEC (sc-33649, Santa Cruz; 1:50), anti-CDH2 (13116s, Cell signaling Technology; 1:200), and anti-vinculin antibody (V9131, Sigma–Aldrich; 1:400) in blocking buffer overnight at 4 °C, followed by the incubation with anti-mouse or anti-rabbit Alexa Fluor® 488-conjugated secondary antibody and then 4ʹ,6-diamidino-2-phenylindole (DAPI; #4083, Cell signaling Technology) for staining nuclei or Alexa Fluor® 594 Phalloidin (#12877, Cell signaling Technology) for staining F-actin. Images were acquired by using a Zeiss Axio Observer.z1 Microscope coupled to a Zeiss LSM 710 Scanning Device (Zeiss, Oberkochen, Germany).
2.10. Cell adhesion assay
Adhesion assays were performed by using the Vybrant™ Cell Adhesion Assay Kit (V-13181, Thermo Fisher Scientific) according to the instructions. Briefly, cells (3.5 × 105 cells/well) were seeded into 6-well plates and treated with hBERA/miR-124-3p or LSA (15 nmol/L for A549, and 10 nmol/L for 143B and MG-63 cells), or vehicle. After 48 h, cells were detached with Cell Dissociation Buffer, washed with PBS, resuspended in RPMI medium containing Calcein AM (5 μmol/L), and then incubated at 37 °C for 30 min. Cells (1.0 × 105 cells/100 μL) were added to 96-well plates precoated with Collagen type I for attachment. To monitor cell adhesion capacity over time, cells were allowed to adhere for 1, 2, and 4 h at 37 °C, after which the experimental wells were aspirated and washed four times to remove non-adherent cells. The fluorescence intensity was measured using a SpectraMax® M3 microplate reader at an excitation of 494 nm and emission of 517 nm. The percentage of adhesion was determined by dividing the corrected fluorescence of adherent cells by the total corrected fluorescence of cells added to each well.
2.11. Experimental lung metastasis mouse models and miR-124 therapy studies
All animal procedures were approved by the Institutional Animal Care and Use Committee of University of California, Davis (UC Davis, USA). Five-to six-week-old female non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA) and adaptively fed at least 1 week before experiments. Experimental lung metastasis mouse models were established through intravenous (i.v.) injection of 143B cells38. Briefly, each mouse was inoculated with 143B-GFP-Luc cells (3.0 × 106) suspended in 250 μL saline through tail vein injection. Tumor growth was monitored by bioluminescence imaging after the injection of d-luciferin potassium salt solution (150 mg/kg, intraperitoneally) at 5 min post-anesthesia, by using a Spectral Lago X Imaging System (Spectral Instruments Imaging, Tucson, AZ, USA), and normalized to the same exposure time. Mice showing similar lung bioluminescence signals were divided randomly into two groups and subjected to in vivo-jetPEI-formulated hBERA/miR-124-3p or control LSA treatments (30 μg, i.v., 3 times per week for 3 weeks). At the end of the study, all mice were euthanized and lungs were dissected. Following ex vivo imaging, lung tissues were fixed with 10% formalin and subjected to hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) examination of proliferation and apoptosis biomarkers, as well as miR-124-3p target CDH2, at UC Davis Center for Genomic Pathology. Slides were scanned by using an Aperio AT2 ScanScope (Leica Biosystems, Danvers, MA, USA). Additionally, blood sample was collected from each mouse, and serum was isolated and subjected to blood chemistry analyses by the Comparative Pathology Laboratory at UC Davis.
2.12. Statistical analysis
Values are presented as mean ± standard deviation (SD). Depending on the numbers of groups and variances, data were analyzed by using Student's t-test, one-way or two-way ANOVA with Bonferroni post-tests (Prism; GraphPad Software, San Diego, CA, USA). Difference was considered as statistically significant when the probability was less than 0.05 (P < 0.05).
3. Results
3.1. Bioengineered miR-124-3p selectively controls human NSCLC A549 cell proteome underlying multiple cellular components critical to the metastatic potential of cells
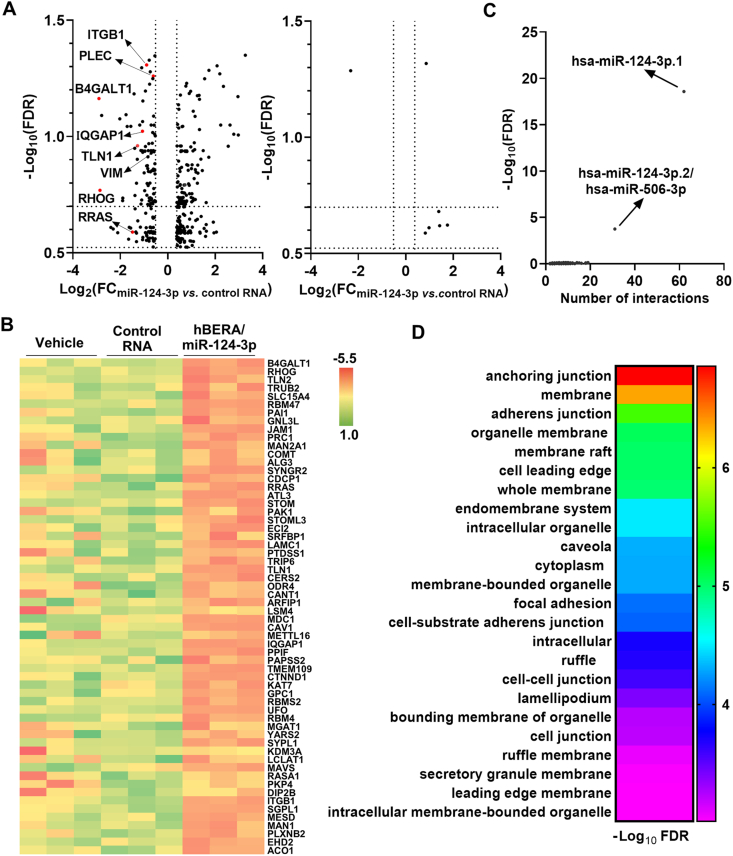
To understand the molecular and cellular mechanisms underlying the antitumoral and antimetastatic activities of biologic miR-124-3p prodrugs24, we first carried out an unbiased proteomics study to compare global protein expression profiles in A549 cells treated with hBERA/miR-124-3p, control LSA, or vehicle. Our data showed that the levels of 147 proteins were downregulated and 229 proteins were upregulated over 30% by hBERA/miR-124-3p (FDR<0.3), compared with the control LSA treatment (Fig. 1A and Supporting Information Table S1). Among these downregulated proteins, many [e.g., ITGB1, IQGAP1, Rho-related GTP-binding protein (RHOG), and RAS-related protein (RRAS)] are reported targets of miR-124-3p39, 40, 41, 42 while some (e.g., PLEC) have not been verified yet. By contrast, LSA had nominal effects on the proteome, as compared with vehicle treatment (Fig. 1A), supporting the use of LSA as the control RNA. Therefore, identification of the selective effects of miR-124-3p on targeted protein expression may be achieved by comparing miR-124-3p to either LSA or vehicle controls. This is obvious by inspecting the top 60 most dramatically suppressed proteins (Fig. 1B).
Figure 1.
Bioengineered miR-124-3p selectively controls the levels of many proteins involved in cellular components critical to the metastatic potential of cells. (A) Volcano plots of significantly-altered proteins (FC>30% and FDR<0.3) in A549 cells treated with hBERA/miR-124-3p versus control LSA, as well as LSA versus vehicle. 147 proteins were downregulated by hBERA/miR-124-3p, among which some targets are reported and many are not verified yet. Further, control RNA showed minimal impact on cell proteome, compared with vehicle treatment. n = 3/group. (B) Heatmap of the top 60 most downregulated proteins by hBERA/miR-124-3p, compared to LSA or vehicle treatments. (C) Specificity of hBERA/miR-124-3p in the regulation of miR-124-3p targeted genes is supported by miRNA enrichment analyses, which readily identified hsa-miR-124-3p underlying genes encoding the proteins significantly downregulated by hBERA/miR-124-3p. (D) Major biological processes were revealed by Gene Ontology (GO) enrichment analysis of downregulated proteins.
To further verify the specificity of bioengineered miRNA-124-3p in the regulation of miR-124-3p targeted proteins, computational miRNA enrichment analysis was performed. The results indicated that only miR-124-3p or miR-506-3p, within the same miR-124/506 family, was significantly (FDR<0.01) enriched when inputting the hBERA/miRNA-124-3p-downregulated proteins (Fig. 1C), demonstrating the specificity of bioengineered hBERA/miR-124-3p in the control of miR-124-3p target gene expression.
In addition, functional pathway analysis was conducted for a set of 88 downregulated proteins identified from the above proteins (Fig. 1A) by using more stringent criteria (FDR<0.2). The results showed that miR-124-3p-suppressed proteins (e.g., ITGB1, VIM, PLEC, IQGAP1, TLN1, CDH2, JAM1) formed a network of interactions in the control of many cellular elements, including anchoring junction, adherens junction, and focal adhesion (Fig. 1D), which are critical factors for carcinoma cell invasiveness and metastasis43,44. Collectively, these findings indicate that biologic miR-124-3p selectively modulates the expression of targeted proteins governing a network of important cellular components critical for the metastatic potential of cancer cells.
3.2. Verification of the impact of humanized miRNA-124-3p prodrug on multiple targeted proteins important in cytoskeleton, cell adhesion, and junctions
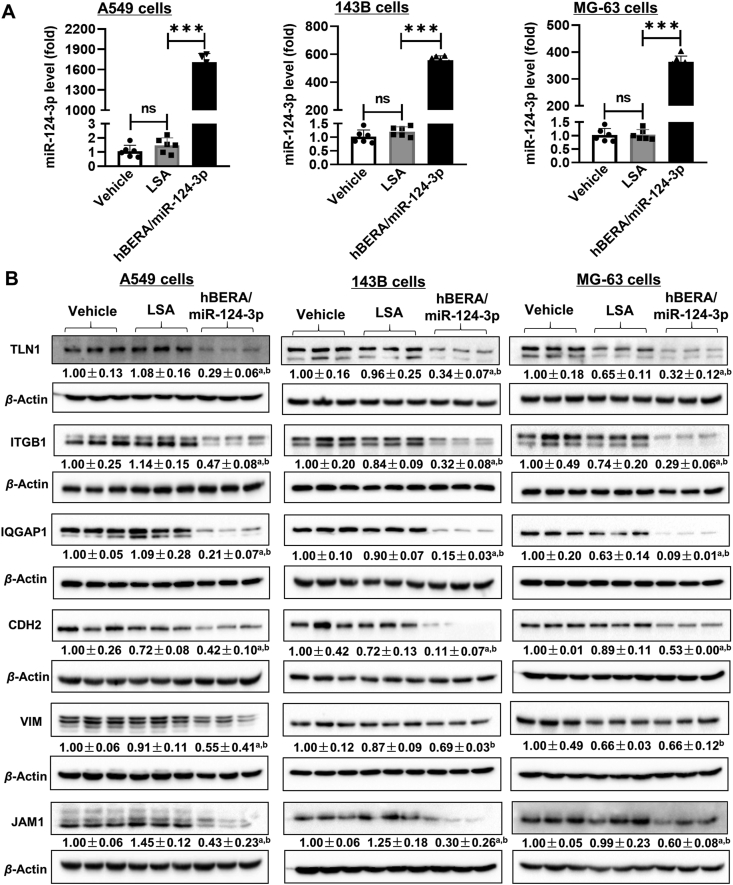
A number of validated miR-124-targeted proteins, such as TLN1, ITGB1, IQGAP1, CDH2, VIM, and JAM1, assembled into multiple cellular components critical for cancer metastasis (Fig. 1) were thus chosen for targeted analyses towards verification of proteomic findings. As shown in Fig. 2A, mature miR-124-3p was specifically released from biologic hBERA/miR-124-3p agents in cells, as manifested by over 1600-, 460-, and 350-fold higher levels in A549, 143B, and MG-63 cells, respectively, than LSA or vehicle treatments. Immunoblot analyses were conducted with selective antibodies and the data demonstrated that proteins levels of TLN1, ITGB1, IQGAP1, CDH2, and JAM1, which are important elements in cytoskeleton, cell adhesions, and junctions43, 44, 45, were all reduced remarkably by miR-124-3p in both NSCLC A549 and osteosarcoma 143B and MG-63 cells (Fig. 2B). Interestingly, while hBERA/miR-124-3p significantly suppressed the protein levels of VIM in A549 cells, the effects were absent in 143B and MG-63 cells when compared with LSA treatment (Fig. 2B). Together, these results verified the reduction of some targeted proteins in human NSCLC and osteosarcoma cells by humanized biologic miRNA-124-3p prodrug.
Figure 2.
Bioengineered miR-124-3p modulates the expression of multiple cytoskeleton, adhesion, and junction proteins. (A) hBERA/miR-124-3p was precisely processed to target miR-124-3p in human NSCLC A549, osteosarcoma 143B, and MG-63 cells, as quantitated by selective stem-loop reverse transcription qPCR assay. (B) The levels of many proteins involving cytoskeletal architecture, junctions, and adhesion were downregulated consequently, as determined by immunoblot analyses. Values are mean ± SD. ∗∗∗P < 0.001; ns, not significant (P ≥ 0.05); aP < 0.05, as compared to LSA control; bP < 0.05, compared to vehicle treatment (one-way ANOVA with Bonferroni post hoc tests).
3.3. PLEC is identified and verified as a direct target for miR-124-3p
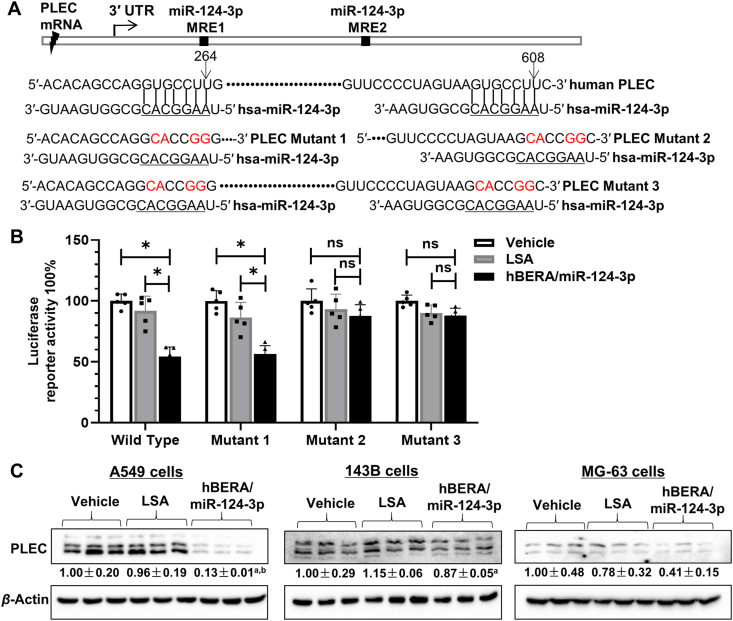
Among those downregulated proteins assembled into important cellular components (Fig. 1), PLEC is a giant protein critical for cytoskeleton organization and dynamics, as well as linkage between cytoskeleton and junctions of plasma membrane46,47. Interestingly, PLEC was revealed as a direct target for miR-124-3p that has not been validated yet, consisting of two well-conserved MREs for miR-124-3p (Fig. 3A). To investigate the interactions between PLEC and miR-124-3p, PLEC 3′ UTR-luciferase reporter plasmids including wild type and three mutants were constructed, and dual luciferase reporter assays were performed. The results showed that wild-type PLEC 3′ UTR-luciferase activities were inhibited more than 40% by miR-124-3p, compared with control LSA or vehicle treatments (Fig. 3B). The impact of miR-124-3p on PLEC 3′ UTR reporter activities retained when the proximal MRE1 was disrupted alone (mutant 1), whereas the effects disappeared when the distal MRE2 was mutated (mutants 2 and 3) (Fig. 3B), suggesting the importance of distal MRE within PLEC 3′ UTR for the interactions with miRNA-124-3p.
Figure 3.
PLEC is verified as a new target for miR-124-3p. (A) Two conserved MRE sites were identified for miR-124-3p within the 3′ UTR of PLEC. The proximal MRE1 and distal MRE2 were altered alone or together to generate specific mutants 1–3. (B) Luciferase reporter assay demonstrated the interactions of miR-124-3p with the MRE sites. When both MRE sites were disrupted (mutant 3), PLEC 3ʹ UTR-luciferase activities were not changed by miR-124-3p. ∗P < 0.05; ns, not significant (two-way ANOVA with Bonferroni post hoc tests). (C) Immunoblot analyses revealed the impact of miR-124-3p on PLEC protein levels in human NSCLC A549 as well as osteosarcoma 143B and MG-63 cells. Values are mean ± SD. aP < 0.05, as compared to LSA control; bP < 0.05, compared to vehicle treatment (one-way ANOVA with Bonferroni post hoc tests).
We further carried out immunoblot and immunofluorescence studies to determine the impact of miR-124-3p on PLEC protein outcomes in NSCLC and osteosarcoma cells. Consistent with proteomic findings (Fig. 1), immunoblot studies showed that PLEC protein levels were reduced approximately 80% by bioengineered miR-124-3p in A549 cells (Fig. 3C). Rather, the effects of miR-124-3p on PLEC protein levels were smaller or statistically insignificant in 143B and MG-63 cells. Immunofluorescent studies revealed the same results, as indicated by the intensity of staining for PLEC (Supporting Information Fig. S1), with a dramatic reduction of the numbers of A549, 143B, and MG-63 cells by biologic miR-124-3p. Together, these results demonstrated that miR-124-3p directly acts on PLEC 3′ UTR and suppresses the levels of PLEC protein.
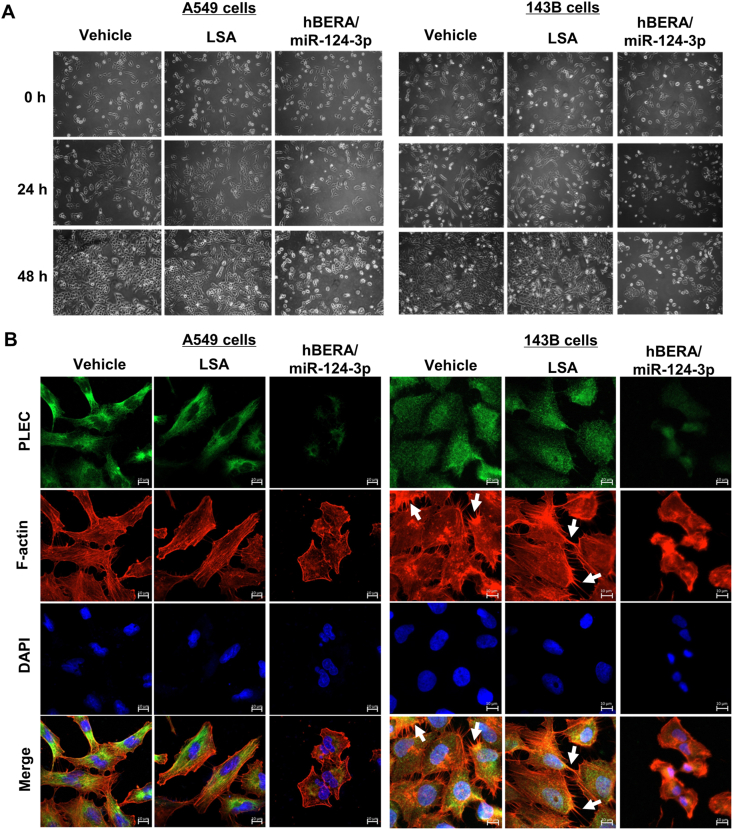
3.4. Biologic miRNA-124-3p alters the morphology of human carcinoma cells by inducing cytoskeletal network remodeling
An obvious change of cell morphology was immediately noted after the treatment with miRNA-124-3p. The NSCLC A549 (Fig. 4A) and osteosarcoma MG-63 (Supporting Information Fig. S2A) cells transfected with hBERA/miR-124-3p underwent apparent changes from spindle to oval shapes and became less clustered, aligned with a lower number of cells. On the other hand, 143B cells seemed largely compressed after treated with miR-124-3p, besides the gradual decrease in number of cells that were less associated (Fig. 4A).
Figure 4.
Alteration of the morphology and redistribution of cytoskeleton of human NSCLC and osteosarcoma cells by biologic miR-124-3p. (A) Phase contrast images (10× magnification) of cells taken with a digital camera under an Olympus CK×31 inverted microscope at different time points following various treatments. A549 and 143B cells gradually changed from spindle to oval shapes or largely compressed by miR-124-3p, with the decrease of cell numbers and clusters. (B) Confocal imaging studies revealed a redistribution of cytoskeletal networks by miR-124-3p, as manifested by F-actin (red) and PLEC (green) staining. In addition, the microvilli or lamellipodia- and filopodia-like structures (white arrowheads) within the 143B cells were sharply impaired by miR-124-3p. Nuclei stained with DAPI are labeled in blue. Scale bar, 10 μm.
The alteration of cell morphology mirrors the change of cytoskeleton that was further investigated by confocal imaging of F-actin and PLEC staining (Fig. 4B and Fig. S2B). The stress fibers or F-actin bundles were distinctly observed in the cytoplasm of cells subjected to control RNA or vehicle treatments. In contrast, only punctuated or short rod-like F-actin was present in cells treated with biologic miRNA-124-3p (Fig. 4B). Moreover, the F-actin staining revealed an impairment of the microvilli, or lamellipodia- and filopodia-like structures, within the highly metastatic 143B cells by miR-124-3p, compared with LSA and vehicle treatments. In addition, beside a lower intensity, the distribution of cytoskeletal PLEC was changed from the periphery towards center of miR-124-3p-treated cells, in line with the re-organization of F-actin (Fig. 4B and Fig. S2B). These results demonstrated an important role for miRNA-124-3p in the control of cytoskeletal and cellular morphology, attributed to the downregulation of multiple targeted proteins (Figure 1, Figure 2), including the cytoskeletal linker PLEC (Fig. 3).
3.5. MiRNA-124-3p prodrug impairs human carcinoma cell–cell adherens junctions
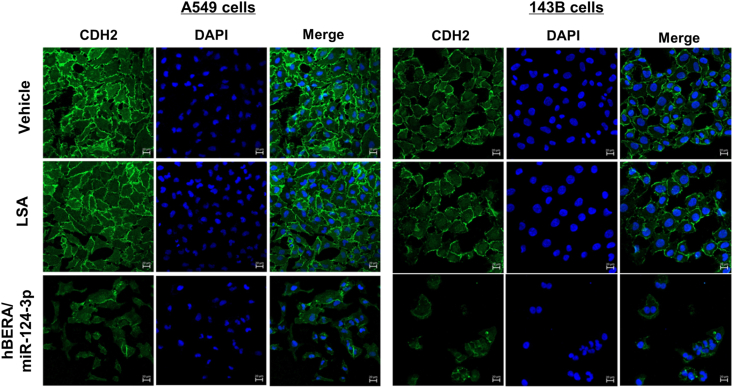
Because proteomics studies revealed many downregulated proteins assembled into cell adherens and anchoring junctions (Fig. 1) essential for the integration of individual cells into sphere and metastasis, among which some (e.g., IQGAP1, JAM1, CDH2, and PLEC) were verified by immunoblot studies (Figure 2, Figure 3), we employed confocal microscopy to investigate the effects of bioengineered miR-124-3p on cell−cell adherens junctions. CDH2, a calcium-dependent adhesion protein that preferentially mediates the adherens junctions of adjacent cells, was chosen for immunofluorescence imaging studies. Our results showed that treatment with hBERA/miR-124-3p not only dramatically reduced the intensity of CDH2 signals but also interrupted its distribution along the cytoplasm membranes of A549 and 143B cells (Fig. 5), as well as MG-63 cells (Supporting Information Fig. S3), implicated to the impairment of cell−cell adherens junctions by biologic miRNA-124-3p agent.
Figure 5.
Adherens junctions are disrupted by miR-124-3p in human NSCLC A549 and osteosarcoma 143B cells. Compared with control LSA and vehicle treatments, hBERA/miRNA-124-3p largely decreased the CDH2 immunostaining signals (green), indicating an impairment of cell−cell adherens junctions. DAPI-stained nuclei are in blue. Scale bar, 20 μm.
3.6. Bioengineered miRNA-124-3p largely disrupts the focal adhesion complexes in human carcinoma cells
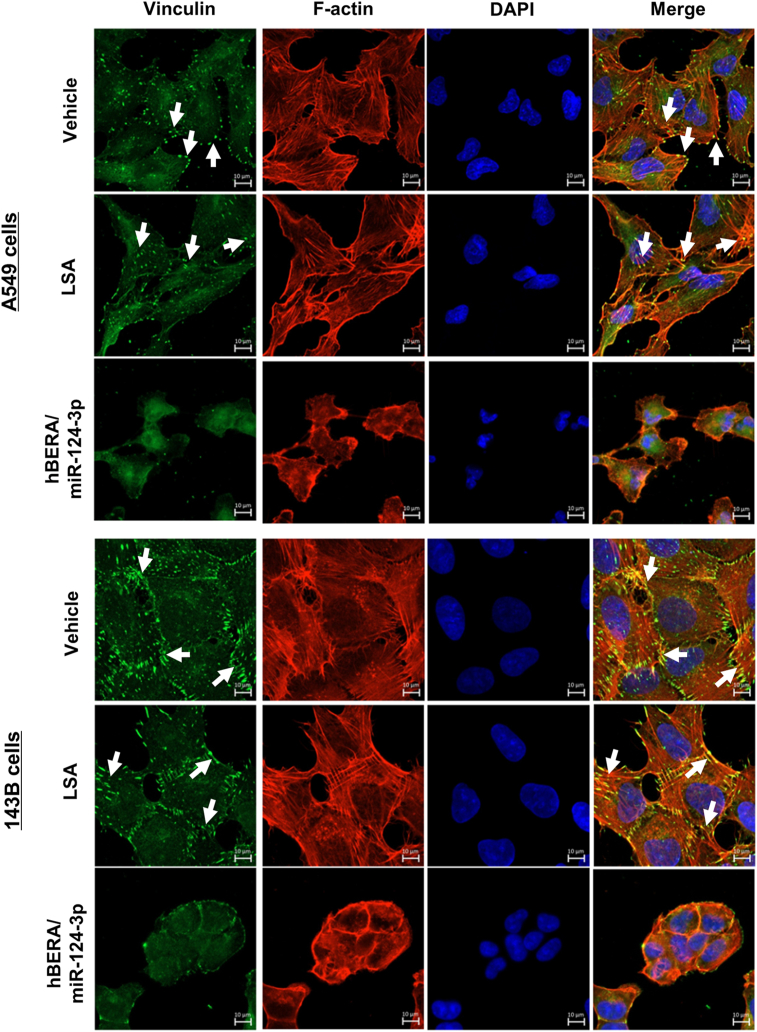
We further sought to delineate the impact of biologic miRNA-124-3p on focal adhesions, which anchor the cytoskeleton to the membrane and extracellular matrix (ECM) and are critical for cancer metastasis. As proteomic (Fig. 1) and immunoblot studies (Figure 2, Figure 3) demonstrated significant reduction of multiple proteins (e.g., ITGB1, TLN1, and PLEC) that are elements of focal adhesion complexes, we chose to use confocal microcopy to examine vinculin plaques, a binder partner for talin, integrin proteins, and actin cytoskeleton48. Our data showed that vinculin-containing focal adhesion complexes were aligned densely along the extension of actin cytoskeleton and enriched at the plasma membrane regions of the control RNA- or vehicle-treated A549 and 143B cells (Fig. 6), as well as MG-63 cells (Supporting Information Fig. S4). Following the treatment with miR-124-3p, the shortened or distorted actin filaments were accompanied with sparse and fewer focal adhesion plaques that seemed unable to aggregate along plasma membranes in cells. These results demonstrated the effectiveness of bioengineered miR-124-3p in suppressing mature focal adhesion complexes attached to the end of the stress fibers and anchoring cells to ECMs.
Figure 6.
MiR-124-3p controls focal adhesion plaques in human NSCLC A549 and osteosarcoma 143B cells. Confocal images showed that there were much sparser and less vinculin-containing focal adhesion plaques (green, white arrowheads) distributed along the distorted or compressed actin filaments (red) or enriched at the plasma membrane regions in cells treated with miR-124-3p, as compared with control LSA or vehicle treatments. Nuclei are labeled in blue. Scale bar, 10 μm.
3.7. Biologic miRNA-124-3p molecule significantly reduces carcinoma cell adhesion capacity
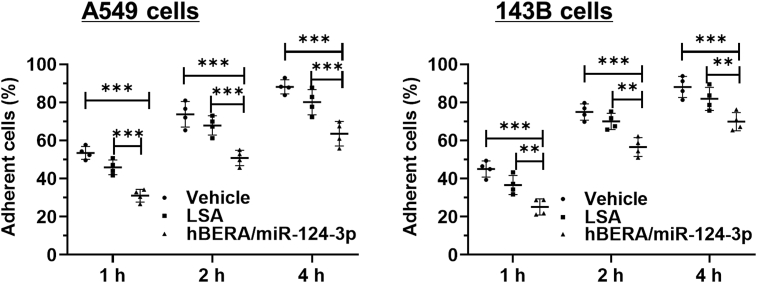
We thus directly measured the effects of hBERA/miRNA-124-3p on cell–matrix adhesion capacity that is essential for carcinoma cell invasion and metastasis. As shown in Fig. 7 and Supporting Information Fig. S5, both NSCLC A549 and osteosarcoma 143B and MG-63 cell adhesion capacities were inhibited by biologic miR-124-3p prodrug. For instance, 45%–52%, 68%–72%, and 78%–83% of control LSA- or vehicle-treated A549 cells achieved adherence at 1, 2, and 4 h, respectively, after seeding. By contrast, around 30%, 50%, and 62% of miR-124-3p-treated A549 cells were adherent to the surface at 1, 2, and 4 h, respectively, after seeding, which is equivalent to a 20%–45% suppression of cell adhesion. These data indicated the efficacy of bioengineered miR-124-3p in the control of cell adhesion, attributed to the suppression of adhesion proteins (Figure 1, Figure 2), and consequently disrupt cell–cell junctions and focal adhesion complexes (Figure 5, Figure 6).
Figure 7.
MiR-124-3p significantly inhibits the adhesion capacity of human NSCLC A549 and osteosarcoma 143B cells. Cells were treated with hBERA/miR-124-3p, control LSA, or vehicle for 48 h, adherent cells were determined over time with the Vybrant™ Cell Adhesion Assay Kit. Values are mean ± SD. ∗∗P < 0.01, ∗∗∗P < 0.001 (two-way ANOVA with Bonferroni post hoc tests).
3.8. Bioengineered miRNA-124-3p prodrug effectively inhibits lung metastasis in an experimental metastasis mouse model in vivo and does not cause any liver or kidney toxicity
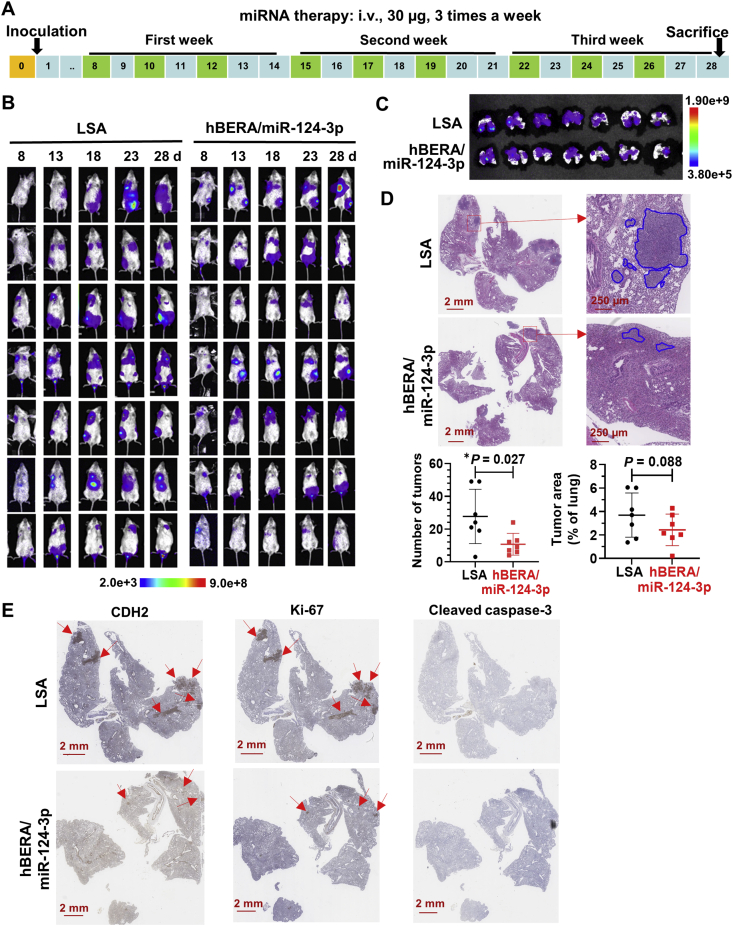
Lastly, we established an aggressive experimental lung metastasis mouse model for the assessment of efficacy and safety of fully-humanized biologic miRNA-124-3p (Fig. 8A). The formation and progression of lung metastasis was monitored by bioluminescence imaging of live animals, and the results showed that miR-124-3p treatment group exhibited relatively weaker bioluminescent signals (Fig. 8B). This was also indicated by imaging ex vivo whole lung tissues excised from individual mice at the end of therapy study (Fig. 8C). Further histopathological studies verified the presence of pulmonary metastasis in all samples, and mice treated with hBERA/miR-124-3p had significantly less lung tumor nodules than control LSA (Fig. 8D). Metastatic tumor areas were also lower in the miRNA therapy group, but it is not statistically significant. Moreover, IHC staining for miR-124-3p target (CDH2) as well as proliferation (Ki-67) and apoptosis (cleaved caspase-3) biomarkers demonstrated miR-124 on-target effects as well as the association of reduction of cell proliferation with the suppression of lung metastasis (Fig. 8E). In addition, fully-humanized miR-124 prodrug was well tolerated in mice. Changes in body weights were not different between the treatment groups despite that all mice showed 3%–7% gain of body weights at the end of the study (Supporting Information Fig. S6A). Except that the total bilirubin concentrations were slightly elevated in three mice (one in LSA group and two in miR-124-3p treatment group) (Fig. S6I), all other blood biomarkers for hepatic and renal functions were within the normal ranges derived from BALB/c strain mice and did not differ between miR-124-3p and LSA treatment groups (Fig. S6B–S6H). Together, these results demonstrate the effectiveness and safety of biologic miR-124-3p therapy in the control of lung metastasis in vivo.
Figure 8.
MiR-124-3p effectively reduces lung metastasis in vivo. (A) Schematic illustration of miRNA therapy in an aggressive experimental metastasis mouse model. (B) Live animal bioluminescent imaging of lung metastasis in mice subjected to miR-124-3p and control LSA treatments. Images were taken on Days 8, 13, 18, 23, and 28 post-inoculation of 143B cells. (C) Bioluminescent imaging of ex vivo lung tissues at the end of the study. (D) Representative H&E stains showed the reduction of the number and extent of metastatic tumors in lung tissues by miR-124-3p treatment. Values are mean ± SD. ∗P < 0.05 (unpaired Student's t-test). (E) Representative IHC staining for CDH2, Ki-67, and cleaved caspase-3. While the apoptosis marker cleaved caspase-3 was hardly detectable, CDH2 and Ki-67 levels were much lower in therapy group than the control, indicating the on-target effects and reduction of proliferation for miR-124 therapy.
4. Discussion
Genome-derived miR-124-3p has been established as a tumor suppressor through targeting of many (proto)oncogenes involved in cancer cell proliferation, apoptosis, stemness, and invasion, and is ubiquitously dysregulated in various types of human malignancies including lung, breast, prostate, brain, liver, and osteosarcoma49, 50, 51, 52, 53, 54. Restoration of miR-124 expression and function, as well as other oncolytic miRNAs that are lost or downregulated in carcinoma cells, represents a new therapeutic approach for the treatment of cancer16,17. This miRNA replacement therapy reintroduces endogenous miRNAs into cancerous cells and is expected to be more tolerable than other approaches that employ antisense oligonucleotides to inhibit oncogenic miRNAs overexpressed in tumors18. Previous miRNA replacement therapies are mainly restricted to the use of chemo-engineered miRNA mimics prepared in vitro12. After establishing a novel approach for in vivo fermentation production of true biologic RNA molecules, we found that chimeric tRNA/pre-miRNA agents exhibited a rather surprisingly favorable stability within human carcinoma cells25. Moreover, commercial in vivo-jetPEI improves the stability of bioengineered RNAs in serum and offers efficient delivery to lung, liver, and xenograft tissues in mouse models25,27,55. The effectiveness of in vivo-jetPEI-formulated, bioengineered miR-124-3p prodrug for the control of spontaneous metastasis was further demonstrated with mouse models24 while optimal delivery systems such as lipid nanoparticles and exosomes10,13,56 are to be explored. Using unparalleled hBERA/miR-124-3p agents combined with unbiased proteomics and confocal imaging of NSCLC and osteosarcoma cells, the present study established the selective effects of miR-124-3p in the control of proteome underlying many basic cellular components such as the cytoskeleton, cell junctions, and adhesion. In addition, we demonstrated the efficacy and safety of miR-124 therapy for the control of metastasis in an aggressive experimental metastasis mouse model. These findings not only provide new insights into understanding miR-124-3p actions in the inhibition of tumor metastasis at the molecular and cellular levels but also support the development of miRNA therapies.
One miRNA may control disease initiation and progression through the regulation of multiple targets. While transcriptomics analyses are able to determine the miRISC-bound messages or global transcripts altered by a miRNA towards the understanding of important targets57, proteomics studies directly identify the influence of miRNA on the proteome outcome58,59, either through mRNA degradation or translation inhibition mechanisms. Using quantitative mass spectrometry-based method following stable isotope labelling with amino acids (SILAC), Baek et al.58 analyzed the change of nuclear-localized proteins in human cervical carcinoma HeLa cells by chemo-engineered miR-124 mimics, and found that the most downregulated proteins matched the miR-124-3p seed sequence. In the present study, S-Trap and LC−MS/MS-based shotgun proteomics technology was employed to reveal global protein change in human NSCLC A549 cells by bioengineered miR-124 agent, and computational miRNA enrichment analyses identified miR-124 underlying the most significantly suppressed proteins. Indeed, a number of miR-124 targets including TLN1, PLOD3, STOM, VIM, and PLEC were identified in both studies. However, many well-known miR-124 targets, such as B4GALT1, CDH2, IQGAP1, ITGB1, RRAS, and JAM1, were only identified in the current study but not by previous proteomics study58. In addition, many of the keratin (KRT) family proteins such as KRT1, 2, 5, 9, 10, 18, and 79 were largely upregulated by miR-124 mimics in HeLa cells58, but not by biologic miR-124 in A549 cells in this study. On the other hand, the human leukocyte antigens (HLA) including HLA-A, HLA-B, and HLA-C were greatly increased in A549 cells by miR-124 in this study, whereas HLA-A and HLA-B remained unchanged and HLA-C was not identified in HeLa cells following miR-124 treatment58. This discrepancy is likely due to the focus on different types of carcinoma cells (cervical versus pulmonary) and cellular proteins (nuclear versus whole cell) between previous and present studies, besides the use of different miR-124 agents and proteomics techniques. These findings are also indicative of the presence of common and cell-specific miRNA−transcript interactions or factors important for particular cell structures and functions. Indeed, the reduction of PLEC protein outcome by miR-124-3p in A549 cells was revealed by the proteomic study and confirmed by immunoblot analyses, whereas the effects were minimal in 143B and MG-63 cells. Therefore, different types of human carcinoma cells may exhibit variable sensitivities or behaviors to the same miRNA, highlighting the importance of experimental determination and verification.
Interference with cytoskeleton elements, actin microfilaments, microtubules, or intermediate filaments (IFs) disrupts the capacity of tumor cells to invade, migrate, and metastasize to distal organs60. Vimentin, a type III multifunctional IF protein in the control of cell morphogenesis and migration as well as a biomarker of epithelial–to–mesenchymal transition (EMT)61, has been shown to be required for lung adenocarcinoma metastasis by maintaining the interactions between heterotypic tumor cells and cancer associated fibroblasts during collective invasion62. Agreeing with these findings, the present study identified a selective reduction of vimentin by novel bioengineered miR-124-3p agent, associated with the redistribution of cytoskeleton in both NSCLC and osteosarcoma cells, as well as an alteration of overall cell morphology. Our study is also among the first to verify PLEC as a new direct target for miR-124-3p and define the effects in NSCLC cells, which was reported very recently by another group in neuroblastoma cells63. PLEC, a cytolinker protein essential for maintaining the assembly and function of IFs and the integrity of the cytoskeleton64,65, is commonly upregulated in tumor tissues and carcinoma cells66,67. Previous studies have showed that PLEC deficiency or knockdown reduces the formation of actin protrusions and compromises cell migration and invasion47,66, and vimentin is recruited to the mitotic cortex in a PLEC-dependent manner during cell division68. Indeed, PLEC directly interacts with vimentin IFs in invasive carcinoma cells and the disruption of the PLEC−vimentin IF interaction sharply suppresses invadopodia formation and reduces transendothelial migration and metastasis69. The current study revealed the disruption of PLEC and F-actin networks as well as the microvilli or lamellipodia-like structures in miR-124-3p-treated 143B cells, which supports the roles of PLEC-vimentin interactions in cell invasiveness and offers insights into the antimetastatic activity of bioengineered miR-124-3p24.
Other skeletal regulatory proteins, such as IQGAP1, contain distinct domains that directly bind to actin and promote cross-linking of actin filaments45. TLN1, a member of the adaptor protein family, plays a critical role in structurally linking integrins to the actin cytoskeleton70. One study has demonstrated that TLN-1 deficiency leads to the suppression of cell protrusions, actin dynamics, and actin barbed end formation, which ultimately impairs the ability of cells to invade71. Therefore, the suppression of IQGAP1 and TLN1 by miR-124-3p revealed in the present study may also contribute to the cytoskeleton remodeling underlying cell invasion and metastasis24.
Adherens junctions initiate cell−cell interactions and mediate the maturation and maintenance of their adhesions. Cadherins, such as CDH2, are a major group of transmembrane proteins involved in adherens junctions and also bind to many cytoplasmic proteins, such as catenins72. Cadherin-mediated cell−cell adhesion plays a key role in the morphogenetic processes during development, as well as EMT processes in tumor metastasis4,73. Our finding that CDH2 is downregulated by miR-124-3p, associated with distinct morphology, is in agreement with the diminished cell−cell adhesion of miR-124-3p-treated cancer cells. This was accompanied by the reduction of cell viability by miR-124-3p, as demonstrated in the present and previous studies24. Indeed, carcinoma cells integrate with each other to promote collective survival and metastatic potential, and circulating tumor cells clusters have greater metastatic ability than single cells74. The suppression of CDH2 was also confirmed in metastatic lung tissues following miR-124 therapy, demonstrating the on-target actions of bioengineered miR-124-3p prodrug and its contribution to the antimetastatic activity of miR-124-3p. Furthermore, the tight junctions that function as a fence in the prevention of mixing membrane lipids between the apical and basolateral membranes and regulate the transportation of molecules between cells, are important components in tumor progression and metastasis43. JAM1 (or JAMA or F11R), a multifunctional transmembrane immunoglobulin preferentially concentrated at tight junctions and supporting tight junction formations75,76, has been considered as a potential target for the treatment of cancer metastasis77. Consistent with a very recent finding JAM1 as a direct target for miR-124-3p in the inhibition of stem-like properties of nasopharyngeal carcinoma cells78, the present study disclosed the suppression of JAM1 protein levels in both NSCLC and osteosarcoma cells following the restoration of miR-124-3p expression. These findings suggest an important role for miR-124-3p in the regulation of cadherin and JAM proteins governing cell junctions for cell−cell adhesion and cancer metastasis.
Anchoring junctions attach cells to the ECM and overexpression of anchoring junction proteins and signaling pathways are commonly associated with greater degrees of cancer cell invasion and metastatic potential4,44. ITGB1 is a member of the transmembrane integrin proteins connecting actin cytoskeleton with ECM79 and participates in focal adhesion dynamics essential for cancer cell invasive behavior80. TLN1 directly binds to ITGB1, links integrins to the actin cytoskeleton, and regulates integrin activation81. Silencing TLN1 significantly reduces focal adhesion signaling and tumor metastasis82. In the present study, both ITGB1 and TLN1 were sharply suppressed by bioengineered miR-124-3p in NSCLC and osteosarcoma cells, providing a molecular explanation for the reduced vinculin-containing focal adhesion complexes distributed along the altered actin filaments. As a result, miR-124-3p-treated carcinoma cells exhibited a remarkably lower level of cell−ECM adhesion capacity, demonstrating the actions of bioengineered miR-124-3p prodrug in the control of proteins and signaling pathways underlying cell junctions, adhesion, and invasiveness.
5. Conclusions
The present study has established the selective actions of bioengineered miR-124-3p in the modulation of global proteomic profiles in human NSCLC cells. The downregulated proteins are assembled into multiple cellular components critical for metastatic potential. Among them, the cytolinker protein PLEC is regulated by miR-124-3p through direct targeting. Furthermore, the effects of miR-124-3p on cytoskeleton remodeling, adherens junctions, focal adhesion, and cell−cell and cell−surface adhesion capacity have been demonstrated in both NSCLC and osteosarcoma cells. Additionally, the efficacy and safety of miR-124-3p therapy in the management of lung metastasis was established in an experimental metastasis mouse model. These results suggest a central role for miR-124-3p in the control of carcinoma cell invasion and metastasis through the regulation of a collection of cytoskeleton, junction and adhesion proteins, supporting the development of miR-124 therapy for the treatment of cancer and metastasis.
Acknowledgments
This study is supported by the National Cancer Institute (No. R01CA225958 to Ai-Ming Yu, USA), National Institutes of Health. Ai-Xi Yu is supported by Hubei Province Scientific and Technological Innovation Key Project (No. 2019ACA136, China) and Hubei Province Medical Leader Talent Project (No. LJ20200405, China). Linglong Deng was supported by a fellowship from the Chinese Scholarship Council (No. 201906270202, China). The authors also appreciate the access to the Mouse Biology and Molecular Pharmacology Shared Resources funded by the UC Davis Comprehensive Cancer Center Support Grant awarded by the National Cancer Institute (No. P30CA093373, USA), National Institutes of Health.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.07.027.
Contributor Information
Ai-Xi Yu, Email: yuaixi@whu.edu.cn.
Ai-Ming Yu, Email: aimyu@ucdavis.edu.
Author contributions
Ai-Ming Yu, Ai-Xi Yu, and all other authors participated in research design. Linglong Deng, Hannah Petrek, Mei-Juan Tu, and Neelu Batra conducted the experiments and data analyses. All authors contributed to data interpretation and writing of the manuscript. All authors have read and agreed to the final version of the manuscript.
Conflicts of interest
The authors report no conflicts of interest. The authors are responsible for the content and writing of this article.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 3.Steeg P.S. Targeting metastasis. Nat Rev Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5:402–418. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meazza C., Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev Anticancer Ther. 2016;16:543–556. doi: 10.1586/14737140.2016.1168697. [DOI] [PubMed] [Google Scholar]
- 6.Travis W.D., Brambilla E., Burke A.P., Marx A., Nicholson A.G. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol. 2015;10:1240–1242. doi: 10.1097/JTO.0000000000000663. [DOI] [PubMed] [Google Scholar]
- 7.Osmani L., Askin F., Gabrielson E., Li Q.K. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109. doi: 10.1016/j.semcancer.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood S.L., Pernemalm M., Crosbie P.A., Whetton A.D. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40:558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Pitroda S.P., Chmura S.J., Weichselbaum R.R. Integration of radiotherapy and immunotherapy for treatment of oligometastases. Lancet Oncol. 2019;20:e434–e442. doi: 10.1016/S1470-2045(19)30157-3. [DOI] [PubMed] [Google Scholar]
- 10.Yu A.M., Choi Y.H., Tu M.J. RNA drugs and RNA targets for small molecules: principles, progress, and challenges. Pharmacol Rev. 2020;72:862–898. doi: 10.1124/pr.120.019554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu A.M., Jian C., Yu A.H., Tu M.J. RNA therapy: are we using the right molecules?. Pharmacol Ther. 2019;196:91–104. doi: 10.1016/j.pharmthera.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charbe N.B., Amnerkar N.D., Ramesh B., Tambuwala M.M., Bakshi H.A., Aljabali A.A.A., et al. Small interfering RNA for cancer treatment: overcoming hurdles in delivery. Acta Pharm Sin B. 2020;10:2075–2109. doi: 10.1016/j.apsb.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 17.Petrek H., Yu A.M. MicroRNAs in non-small cell lung cancer: gene regulation, impact on cancer cellular processes, and therapeutic potential. Pharmacol Res Perspect. 2019;7 doi: 10.1002/prp2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bader A.G., Brown D., Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Zandwijk N., Pavlakis N., Kao S.C., Linton A., Boyer M.J., Clarke S., et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 20.Hong D.S., Kang Y.K., Borad M., Sachdev J., Ejadi S., Lim H.Y., et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer. 2020;122:1630–1637. doi: 10.1038/s41416-020-0802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho P.Y., Yu A.M. Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA. 2016;7:186–197. doi: 10.1002/wrna.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho P.Y., Duan Z., Batra N., Jilek J.L., Tu M.J., Qiu J.X., et al. Bioengineered noncoding RNAs selectively change cellular miRNome profiles for cancer therapy. J Pharmacol Exp Therapeut. 2018;365:494–506. doi: 10.1124/jpet.118.247775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Q.X., Wang W.P., Zeng S., Urayama S., Yu A.M. A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Res. 2015;43:3857–3869. doi: 10.1093/nar/gkv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P.C., Tu M.J., Ho P.Y., Batra N., Tran M.M.L., Qiu J.X., et al. In vivo fermentation production of humanized noncoding RNAs for functional and experimental therapeutic studies. Theranostics. 2021;11:4858–4871. doi: 10.7150/thno.56596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W.P., Ho P.Y., Chen Q.X., Addepalli B., Limbach P.A., Li M.M., et al. Bioengineering novel chimeric microRNA-34a for prodrug cancer therapy: high-yield expression and purification, and structural and functional characterization. J Pharmacol Exp Therapeut. 2015;354:131–141. doi: 10.1124/jpet.115.225631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu M.J., Ho P.Y., Zhang Q.Y., Jian C., Qiu J.X., Kim E.J., et al. Bioengineered miRNA-1291 prodrug therapy in pancreatic cancer cells and patient-derived xenograft mouse models. Cancer Lett. 2019;442:82–90. doi: 10.1016/j.canlet.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jilek J.L., Zhang Q.Y., Tu M.J., Ho P.Y., Duan Z., Qiu J.X., et al. Bioengineered Let-7c inhibits orthotopic hepatocellular carcinoma and improves overall survival with minimal immunogenicity. Mol Ther Nucleic Acids. 2019;14:498–508. doi: 10.1016/j.omtn.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrek H., Yan Ho P., Batra N., Tu M.J., Zhang Q., Qiu J.X., et al. Single bioengineered ncRNA molecule for dual-targeting toward the control of non-small cell lung cancer patient-derived xenograft tumor growth. Biochem Pharmacol. 2021;189:114392. doi: 10.1016/j.bcp.2020.114392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X., Tian Y., Tu M.J., Ho P.Y., Batra N., Yu A.M. Bioengineered miR-27b-3p and miR-328-3p modulate drug metabolism and disposition via the regulation of target ADME gene expression. Acta Pharm Sin B. 2019;9:639–647. doi: 10.1016/j.apsb.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umeh-Garcia M., Simion C., Ho P.Y., Batra N., Berg A.L., Carraway K.L., et al. A novel bioengineered miR-127 prodrug suppresses the growth and metastatic potential of triple-negative breast cancer cells. Cancer Res. 2020;80:418–429. doi: 10.1158/0008-5472.CAN-19-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jilek J.L., Tu M.J., Zhang C., Yu A.M. Pharmacokinetic and pharmacodynamic factors contribute to synergism between Let-7c-5p and 5-fluorouracil in inhibiting hepatocellular carcinoma cell viability. Drug Metab Dispos. 2020;48:1257–1263. doi: 10.1124/dmd.120.000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi W., Tu M.J., Liu Z., Zhang C., Batra N., Yu A.X., et al. Bioengineered miR-328-3p modulates GLUT1-mediated glucose uptake and metabolism to exert synergistic antiproliferative effects with chemotherapeutics. Acta Pharm Sin B. 2020;10:159–170. doi: 10.1016/j.apsb.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.HaileMariam M., Eguez R.V., Singh H., Bekele S., Ameni G., Pieper R., et al. S-Trap, an ultrafast sample-preparation approach for shotgun proteomics. J Proteome Res. 2018;17:2917–2924. doi: 10.1021/acs.jproteome.8b00505. [DOI] [PubMed] [Google Scholar]
- 34.Ludwig K.R., Schroll M.M., Hummon A.B. Comparison of in-solution, FASP, and S-Trap based digestion methods for bottom-up proteomic studies. J Proteome Res. 2018;17:2480–2490. doi: 10.1021/acs.jproteome.8b00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia J., Claude-Taupin A., Gu Y., Choi S.W., Peters R., Bissa B., et al. Galectin-3 coordinates a cellular system for lysosomal repair and removal. Dev Cell. 2020;52:69–87.e68. doi: 10.1016/j.devcel.2019.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licursi V., Conte F., Fiscon G., Paci P. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinf. 2019;20:545. doi: 10.1186/s12859-019-3105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jian C., Tu M.J., Ho P.Y., Duan Z., Zhang Q., Qiu J.X., et al. Co-targeting of DNA, RNA, and protein molecules provides optimal outcomes for treating osteosarcoma and pulmonary metastasis in spontaneous and experimental metastasis mouse models. Oncotarget. 2017;8:30742–30755. doi: 10.18632/oncotarget.16372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunt S., Jones A.V., Hinsley E.E., Whawell S.A., Lambert D.W. MicroRNA-124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS Lett. 2011;585:187–192. doi: 10.1016/j.febslet.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 40.Furuta M., Kozaki K.I., Tanaka S., Arii S., Imoto I., Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 41.Franke K., Otto W., Johannes S., Baumgart J., Nitsch R., Schumacher S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2012;31:2908–2921. doi: 10.1038/emboj.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Z., Chen Q., Li C., Wang L., Qian X., Jiang C., et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol. 2014;16:1341–1353. doi: 10.1093/neuonc/nou084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin T.A. The role of tight junctions in cancer metastasis. Semin Cell Dev Biol. 2014;36:224–231. doi: 10.1016/j.semcdb.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Bhat A.A., Uppada S., Achkar I.W., Hashem S., Yadav S.K., Shanmugakonar M., et al. Tight junction proteins and signaling pathways in cancer and inflammation: a functional crosstalk. Front Physiol. 2018;9:1942. doi: 10.3389/fphys.2018.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White C.D., Erdemir H.H., Sacks D.B. IQGAP1 and its binding proteins control diverse biological functions. Cell Signal. 2012;24:826–834. doi: 10.1016/j.cellsig.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiche G. Role of plectin in cytoskeleton organization and dynamics. J Cell Sci. 1998;111:2477–2486. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- 47.Andrä K., Nikolic B., Stöcher M., Drenckhahn D., Wiche G. Not just scaffolding: plectin regulates actin dynamics in cultured cells. Genes Dev. 1998;12:3442–3451. doi: 10.1101/gad.12.21.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziegler W.H., Gingras A.R., Critchley D.R., Emsley J. Integrin connections to the cytoskeleton through talin and vinculin. Biochem Soc Trans. 2008;36:235–239. doi: 10.1042/BST0360235. [DOI] [PubMed] [Google Scholar]
- 49.Hatziapostolou M., Polytarchou C., Aggelidou E., Drakaki A., Poultsides G.A., Jaeger S.A., et al. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zu L., Xue Y., Wang J., Fu Y., Wang X., Xiao G., et al. The feedback loop between miR-124 and TGF-β pathway plays a significant role in non-small cell lung cancer metastasis. Carcinogenesis. 2016;37:333–343. doi: 10.1093/carcin/bgw011. [DOI] [PubMed] [Google Scholar]
- 51.Cong C., Wang W., Tian J., Gao T., Zheng W., Zhou C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer Biomarkers. 2018;21:449–454. doi: 10.3233/CBM-170672. [DOI] [PubMed] [Google Scholar]
- 52.Li L., Luo J., Wang B., Wang D., Xie X., Yuan L., et al. Microrna-124 targets flotillin-1 to regulate proliferation and migration in breast cancer. Mol Cancer. 2013;12:163. doi: 10.1186/1476-4598-12-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi X.B., Xue L., Ma A.H., Tepper C.G., Gandour-Edwards R., Kung H.J., et al. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 2013;32:4130–4138. doi: 10.1038/onc.2012.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silber J., Lim D.A., Petritsch C., Persson A.I., Maunakea A.K., Yu M., et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Q.Y., Ho P.Y., Tu M.J., Jilek J.L., Chen Q.X., Zeng S., et al. Lipidation of polyethylenimine-based polyplex increases serum stability of bioengineered RNAi agents and offers more consistent tumoral gene knockdown in vivo. Int J Pharm. 2018;547:537–544. doi: 10.1016/j.ijpharm.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ha D., Yang N., Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karginov F.V., Conaco C., Xuan Z., Schmidt B.H., Parker J.S., Mandel G., et al. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 60.Etienne-Manneville S. Microtubules in cell migration. Annu Rev Cell Dev Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- 61.Battaglia R.A., Delic S., Herrmann H., Snider N.T. Vimentin on the move: new developments in cell migration. F1000Res. 2018;7:F1000. doi: 10.12688/f1000research.15967.1. Faculty Rev-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richardson A.M., Havel L.S., Koyen A.E., Konen J.M., Shupe J., WGt Wiles, et al. Vimentin Is Required for Lung Adenocarcinoma metastasis via heterotypic tumor cell-cancer-associated fibroblast interactions during collective invasion. Clin Cancer Res. 2018;24:420–432. doi: 10.1158/1078-0432.CCR-17-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nolan J.C., Salvucci M., Carberry S., Barat A., Segura M.F., Fenn J., et al. A context-dependent role for miR-124-3p on cell phenotype, viability and chemosensitivity in neuroblastoma in vitro. Front Cell Dev Biol. 2020;8:559553. doi: 10.3389/fcell.2020.559553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gregor M., Osmanagic-Myers S., Burgstaller G., Wolfram M., Fischer I., Walko G., et al. Mechanosensing through focal adhesion-anchored intermediate filaments. Faseb J. 2014;28:715–729. doi: 10.1096/fj.13-231829. [DOI] [PubMed] [Google Scholar]
- 65.Jiu Y., Lehtimaki J., Tojkander S., Cheng F., Jaalinoja H., Liu X., et al. Bidirectional interplay between vimentin intermediate filaments and contractile actin stress fibers. Cell Rep. 2015;11:1511–1518. doi: 10.1016/j.celrep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 66.Katada K., Tomonaga T., Satoh M., Matsushita K., Tonoike Y., Kodera Y., et al. Plectin promotes migration and invasion of cancer cells and is a novel prognostic marker for head and neck squamous cell carcinoma. J Proteomics. 2012;75:1803–1815. doi: 10.1016/j.jprot.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 67.Bausch D., Thomas S., Mino-Kenudson M., Fernandez-del C.C., Bauer T.W., Williams M., et al. Plectin-1 as a novel biomarker for pancreatic cancer. Clin Cancer Res. 2011;17:302–309. doi: 10.1158/1078-0432.CCR-10-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serres M.P., Samwer M., Truong Quang B.A., Lavoie G., Perera U., Gorlich D., et al. F-actin interactome reveals vimentin as a key regulator of actin organization and cell mechanics in mitosis. Dev Cell. 2020;52:210–222.e217. doi: 10.1016/j.devcel.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sutoh Yoneyama M., Hatakeyama S., Habuchi T., Inoue T., Nakamura T., Funyu T., et al. Vimentin intermediate filament and plectin provide a scaffold for invadopodia, facilitating cancer cell invasion and extravasation for metastasis. Eur J Cell Biol. 2014;93:157–169. doi: 10.1016/j.ejcb.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Ben-Yosef T., Francomano C.A. Characterization of the human talin (TLN) gene: genomic structure, chromosomal localization, and expression pattern. Genomics. 1999;62:316–319. doi: 10.1006/geno.1999.6019. [DOI] [PubMed] [Google Scholar]
- 71.Beaty B.T., Wang Y., Bravo-Cordero J.J., Sharma V.P., Miskolci V., Hodgson L., et al. Talin regulates moesin-NHE-1 recruitment to invadopodia and promotes mammary tumor metastasis. J Cell Biol. 2014;205:737–751. doi: 10.1083/jcb.201312046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marie P.J., Hay E., Modrowski D., Revollo L., Mbalaviele G., Civitelli R. Cadherin-mediated cell–cell adhesion and signaling in the skeleton. Calcif Tissue Int. 2014;94:46–54. doi: 10.1007/s00223-013-9733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y., Lv Z., Zhang S., Wang Z., He L., Tang M., et al. Genetic fate mapping of transient cell fate reveals N-cadherin activity and function in tumor metastasis. Dev Cell. 2020;54:593–607.e595. doi: 10.1016/j.devcel.2020.06.021. [DOI] [PubMed] [Google Scholar]
- 74.Aceto N., Bardia A., Miyamoto D.T., Donaldson M.C., Wittner B.S., Spencer J.A., et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ebnet K., Suzuki A., Ohno S., Vestweber D. Junctional adhesion molecules (JAMs): more molecules with dual functions? J Cell Sci. 2004;117:19–29. doi: 10.1242/jcs.00930. [DOI] [PubMed] [Google Scholar]
- 76.Liu Y., Nusrat A., Schnell F.J., Reaves T.A., Walsh S., Pochet M., et al. Human junction adhesion molecule regulates tight junction resealing in epithelia. J Cell Sci. 2000;113:2363–2374. doi: 10.1242/jcs.113.13.2363. [DOI] [PubMed] [Google Scholar]
- 77.Bednarek R., Selmi A., Wojkowska D., Karolczak K., Popielarski M., Stasiak M., et al. Functional inhibition of F11 receptor (F11R/junctional adhesion molecule-A/JAM-A) activity by a F11R-derived peptide in breast cancer and its microenvironment. Breast Cancer Res Treat. 2020;179:325–335. doi: 10.1007/s10549-019-05471-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian Y., Tian Y., Tu Y., Zhang G., Zeng X., Lin J., et al. microRNA-124 inhibits stem-like properties and enhances radiosensitivity in nasopharyngeal carcinoma cells via direct repression of expression of JAMA. J Cell Mol Med. 2020;24:9533–9544. doi: 10.1111/jcmm.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwartz M.A., Schaller M.D., Ginsberg M.H. Integrins: emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- 80.Brockbank E.C., Bridges J., Marshall C.J., Sahai E. Integrin β1 is required for the invasive behaviour but not proliferation of squamous cell carcinoma cells in vivo. Br J Cancer. 2005;92:102–112. doi: 10.1038/sj.bjc.6602255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Calderwood D.A., Zent R., Grant R., Rees D.J., Hynes R.O., Ginsberg M.H. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J Biol Chem. 1999;274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 82.Sakamoto S., McCann R.O., Dhir R., Kyprianou N. Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 2010;70:1885–1895. doi: 10.1158/0008-5472.CAN-09-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.