Abstract
The Eukaryotic Pathogen, Vector and Host Informatics Resource (VEuPathDB, https://veupathdb.org) represents the 2019 merger of VectorBase with the EuPathDB projects. As a Bioinformatics Resource Center funded by the National Institutes of Health, with additional support from the Welllcome Trust, VEuPathDB supports >500 organisms comprising invertebrate vectors, eukaryotic pathogens (protists and fungi) and relevant free-living or non-pathogenic species or hosts. Designed to empower researchers with access to Omics data and bioinformatic analyses, VEuPathDB projects integrate >1700 pre-analysed datasets (and associated metadata) with advanced search capabilities, visualizations, and analysis tools in a graphic interface. Diverse data types are analysed with standardized workflows including an in-house OrthoMCL algorithm for predicting orthology. Comparisons are easily made across datasets, data types and organisms in this unique data mining platform. A new site-wide search facilitates access for both experienced and novice users. Upgraded infrastructure and workflows support numerous updates to the web interface, tools, searches and strategies, and Galaxy workspace where users can privately analyse their own data. Forthcoming upgrades include cloud-ready application architecture, expanded support for the Galaxy workspace, tools for interrogating host-pathogen interactions, and improved interactions with affiliated databases (ClinEpiDB, MicrobiomeDB) and other scientific resources, and increased interoperability with the Bacterial & Viral BRC.
INTRODUCTION
The Eukaryotic Pathogen, Vector and Host Informatics Resource (VEuPathDB.org) is a Bioinformatics Resource Center (https://www.niaid.nih.gov/research/bioinformatics-resource-centers) funded by the National Institute of Allergy and Infectious Diseases (https://www.niaid.nih.gov/) at the National Institutes of Health (https://www.nih.gov/) with additional support for specific projects provided by Wellcome (UK) (https://wellcome.org/). Funded as independent Bioinformatics Resource Centers since 2004, VectorBase (1) and EuPathDB (2) merged in 2019 to form VEuPathDB. This combined resource leverages the strengths of both entities, including sophisticated data mining, visualization, and analysis tools, as well as staff and outreach personnel, bringing improved service to the vector, parasite, and fungal research communities worldwide.
The challenge of combining mature projects into a single, cost effective, sustainable and scalable bioinformatics resource required careful changes on several fronts. The underlying architecture of VEuPathDB couples the Genomics Unified Schema (GUS) (3) and the highly flexible Strategies Web Development Kit (WDK) (4) from EuPathDB with VectorBase's efficient bioinformatic pre-processing workflows and pipelines processed through the European Bioinformatics Institute (EBI) (https://www.ebi.ac.uk/). An ontology-driven framework applied across all projects ensures consistent representation across diverse taxa and data sources. In addition, community expertise is leveraged to improve annotation and enhance the quality of the information available, providing a dynamic and ever improving community resource.
VEuPathDB provides a central resource for public access to computational platforms, analysis tools and data mining of genome-scale research data for over 500 organisms comprising invertebrate vectors, eukaryotic pathogens (protists and fungi) and relevant free-living or non-pathogenic species or hosts. While all fourteen VEuPathDB projects share the same web architecture, thirteen (Table 1) support data mining of parasite, vector, or host omics data, while OrthoMCL DB (https://orthomcl.org/orthomcl/) offers a platform for exploring orthology relationships across >500 organisms, including VEuPathDB organisms as well as species from archaea, bacteria and eukaryote that are not supported in VEuPathDB.
Table 1.
VEuPathDB resources and organisms supported
*Datasets can represent genome sequences or other omics-scale data e.g. an RNA-Seq developmental time series study, a proteomics analysis, or a phenotype screen. The sum of the project datasets is larger than the number of datasets contained in the parent project, VEuPathDB, because some common dataset, e.g. the taxonomy, are present in each project.
VEuPathDB projects integrate a wide range of data types including genome sequence and annotation, transcriptomics, proteomics, epigenomics, metabolomics, population resequencing, clinical data, surveillance data, host-pathogen interactions, and orthology profiles across all integrated organisms. Raw data are integrated from public repositories such as the NCBI (https://www.ncbi.nlm.nih.gov/) or GO Consortium (http://geneontology.org/) (5,6). Data are analysed according to standard workflows to ensure that data from different sources are comparable. These analysis results underly our unique Search Strategies system and enable in silico experiments that easily query across datasets, data types and organisms.
VEuPathDB also supports private analysis of Omics data via Galaxy (7), a menu driven analysis platform that does not require command line programming. With published workflows, pre-loaded annotated genome sequences for the organisms we support, and tools for privately porting analysis results to VEuPathDB’s public data mining tools, users can privately analyse their own data and compare their analyses with public data in VEuPathDB.
VEuPathDB’s unique infrastructure, Search Strategy system, and Galaxy interface support a comprehensive data mining experience accessible to research scientists without computational training. VEuPathDB facilitates the discovery of meaningful biological relationships from large volumes of data. Upgrades to the infrastructure and workflows made over the past 5 years support numerous updates to our web interface, tools, searches and strategies, and Galaxy workspace. Individual changes are chronicled in the News (Figure 1F) and this manuscript describes new content, features and tools that increase the data mining power of VEuPathDB.
Figure 1.
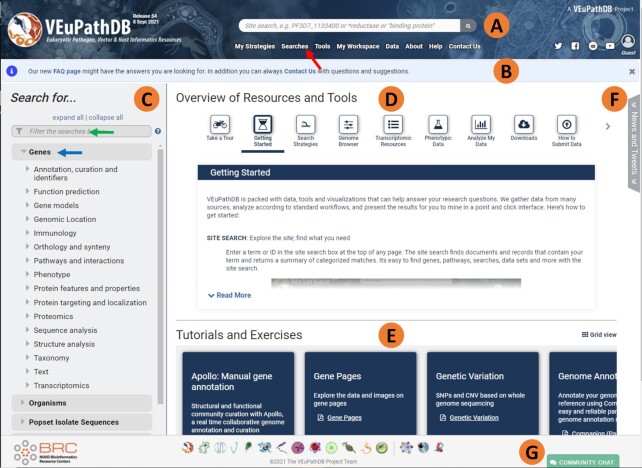
VEuPathDB redesigned homepage. (A) The banner section is present on all webpages and contains the site search box and access to all searches, data, and tools. (B) Just below the banner is a section reserved for general community announcements (C) The left-hand search panel contains categorized searches of data in the resource. (D) The Overview of resources and tools section is in the centre of the page, providing quick vignettes to help our users get started with a specific topic. (E) Links to more detailed step-by-step instructional exercises are in the centre of the page just above the footer. (F) The news and tweets section is an expandable tab (collapsed by default) offering users access to recent news releases and tweets. (G) The footer section includes hyperlinked logos to all VEuPathDB component and affiliated sites in addition to the Gitter chat button.
New in VEuPathDB
Although VEuPathDB is routinely updated with new data, features, and tools, the current resources also harbour significant improvements to the web interface, tools, infrastructure and analysis workflows.
Web interface improvements
The web interface was redesigned to reflect current trends in web architecture and to emphasize easy access to data, searches and help information.
Home page redesign
The new VEuPathDB home page (Figure 1) merges positive aspects of both VectorBase and EuPathDB to facilitate a user's ability to quickly learn and navigate the new resource. Shared by all sites (Table 1), the home page features a banner that appears above a main panel that has left and central portions. Present on all pages, the banner (Figure 1A), consists of a background of images (graciously provided by the community) of representative organisms supported in the databases. The left side of the banner includes the logo and name of the resource and information about release date and version. The centre of the banner includes a site search box that allows both gene identifier and free text searches (see site search section below). Menu items below the site search box include quick access to the ‘My Strategies’ section, all searches, tools, the ‘My Workspace’ section, data, about and help pages and the ‘Contact Us’ link. Social media, login, registration, and user profile links appear on the right. Announcements are made just underneath the banner (Figure 1B).
The left side of the main panel (Figure 1C) provides access to all available searches in the project. Users navigate to searches using either the expandable menu (blue arrow), or the filter above the menu that quickly refines the list of searches based on key words (green arrow). As an example, a user interested in finding any search that identifies genes with signal peptides may start to type the word ‘signal’ in the search filter to view only searches whose titles and descriptions contain the word signal.
The main panel's central portion, ‘Overview of Resources and Tools’, provides topical information useful for getting started with VEuPathDB resources (Figure 1D). A list of scrollable vignette buttons appears at the top which, when clicked, reveal helpful information below. Below the vignettes is a ‘Tutorials and Exercises’ section which provides access to detailed step-by-step tutorials downloadable in PDF format (Figure 1E). To the right of the vignettes is an expandable ‘News and Tweets’ section for quick exploration of website news and recent tweets (Figure 1F). The footer at the bottom of the homepage includes clickable icons for each of the VEuPathDB resources (Figure 1G). In addition, the footer includes a community chat button that enables users to chat with each other using Gitter (https://developer.gitter.im/docs/welcome), an open-source instant messaging and chat room system.
Strategies improvements
VEuPathDB’s My Strategies, a unique and powerful data mining system to find relationships across datasets, data types and organisms, received significant infrastructure and interface updates. While the workflows within My Strategies have not changed, infrastructure improvements (see Infrastructure and Workflows section) provide better programmatic access to data via the new REST API, and interface updates improve usability with clear language and interactive graphic panels.
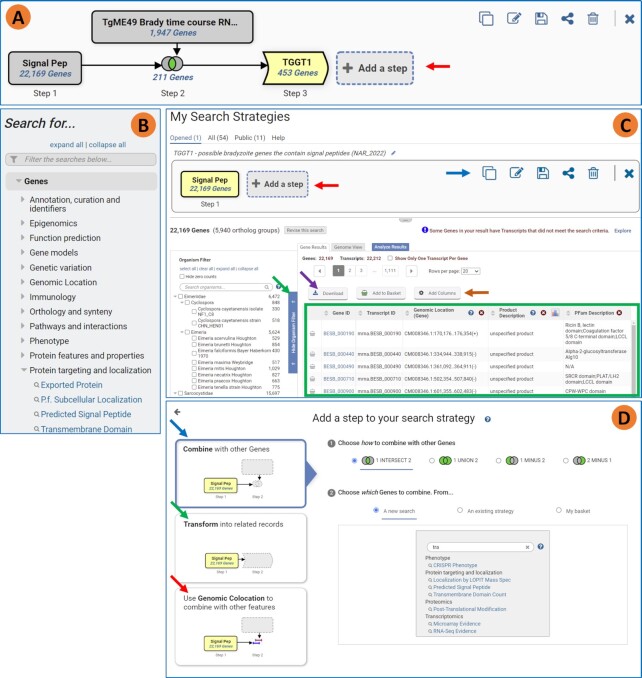
Multistep strategies (Figure 2A) are built one step at a time, choosing the first search from either the Search For… menu on the home page (Figure 2B) or the Searches menu in the banner (Figure1A, red arrow). Redesigned search pages include hoverable icons that reveal updated help information. Search results appear in a newly designed My Search Strategies page (Figure 2C). The top graphic panel shows the growing strategy and offers options to copy, edit description, save, share, delete and close the strategy, respectively (Figure 2C, blue arrow). Saving a strategy retains the order of steps and parameter values but does not retain the resulting ID lists since subsequent database versions may contain new data that alter the IDs returned. The result table, which includes the list of returned IDs and associated data (Figure 2C, green box), can be downloaded via the Download tool (Figure 2C, purple arrow). Columns of associated data can be added to the result table via the Add Columns tool (Figure 2C, orange arrow). The share strategy option generates a unique URL for the strategy which can be sent to colleagues. Once a strategy is saved it can also be made public to all website users. The collapsible organism filter (Figure 2C, green arrow), displays the distribution of results across the organisms searched and is categorized by taxonomy. Columns within search result tables can be rearranged by dragging to change positions and the column headers contain help information.
Figure 2.
Updated strategy system interface. (A) example strategy showing graphic interface for exploring relationships across data sets and organisms. Final strategy can be found here https://toxodb.org/toxo/app/workspace/strategies/import/037c32a7060e4c90. Upper right corner shows actions that can be performed on the strategy: copy, add/edit description, save, share, delete and close. (B) Home page Search for menu often used to choose the first search in a strategy. (C) Result of first step with the graphic of the growing strategy in the top panel, redesigned vertical organism filter on the left and the gene result table on the right. (D) Redesigned Add Step popup. The three choices when adding a step are aligned on the right and details appear in the right panel once a left panel option is chosen.
Strategies are extended (Figure 2D) by clicking ‘Add a step’ from the graphic panel (Figure 2A, C, red arrows). Options for extending a strategy include combine with similar records, transform to related records and genomic colocation (Figure 2D, blue, green, red arrows). Once an option is chosen, details for configuring that option appear on the right.
Tools
To increase the data mining power of VEuPathDB, the following tools have been updated or added.
Site search
The banner on all VEuPathDB pages now includes a tool that performs a site-wide text (Figure 1A) search and returns a categorized list of pages that contain the term or phrase of interest. Designed to reveal the full scope of data and information in VEuPathDB, site search results not only include genomic feature record pages (genes, pathways, etc.) but also other pages such as datasets, news items and tutorials. Category filters offer the opportunity to explore the results based on organism and ‘found in’ fields. The details panel appears to the right of the categorized results and links directly to each returned page. When site search results correspond to VEuPathDB records (genes, SNPs, metabolic pathways, etc), the results can be exported to the Strategies for download or further exploration in combination with searches against the data types.
Gene records
Gene records compile all available data for a gene in a single web page. Help information on record pages is updated for clarity and easily accessed by hovering over the help icon (blue question mark icon) to reveal additional information or definitions. Several new tools and options are available from gene pages and described here according to the data section in which they appear.
Orthology and synteny
A gene's sequence can be aligned with the sequences of its orthologs and paralogs using the Clustal Omega (8) alignment tool. Users are able to select: the set of genes from the ‘Orthologs and Paralogs within VEuPathDB’ table; the sequence type (protein, CDS, or genomic); and the output format (Mismatch highlighted, FASTA, PHYLIP, STOCKHOLM, VIENNA).
Transcriptomics
A new RNA-Seq Transcript Summary Graph summarizes a gene's expression values (x-axis, e.g. TPM) across the many RNA-Seq experiments (y-axis) integrated into VEuPathDB. The summary graph uses Plotly (https://plotly.com/), an interactive graphical interface that allows users to switch between linear and log2 scales, display the gene's expression in each sample, zoom and pan. This graph is especially useful for observing overall trends in expression of a gene, identifying outliers (samples) that may deserve further inquiry.
Protein features and properties
This section includes six new tools for submitting the gene's protein product for analysis with: BLAST-P (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) (9), big-PI Predictor (http://mendel.imp.ac.at/sat/gpi/gpi_server.html) (10), InterPro (http://www.ebi.ac.uk/interpro/) (11), MitoProt (12), STRING (https://string-db.org/cgi/about.pl) (13), and WoLF PSORT (https://wolfpsort.hgc.jp/) (14). Clicking a tool's Submit button, opens a new tab and initiates the respective query. Protein-protein BLAST-P is available against the following databases: Non-redundant protein sequences, Reference proteins, UniProtKB/Swiss-Prot (15), Model Organisms, Patented protein sequences, Protein Data Bank (16) proteins, Metagenomic proteins and Transcriptome Shotgun Assembly proteins. Glycosylphosphatidylinositol anchor, mitochondrial targeting, and subcellular localization site prediction data are retrieved with the GPI Modification Site Predictor (big-PI Predictor), MitoProt and WoLF PSORT options, respectively. The STRING tool accesses visualizations of known and predicted protein-protein interactions in the STRING database that includes information about direct and indirect protein associations.
Function prediction
A new GO Slim table appears above the GO Terms table for genes where GO associated data are available. The GO Slim ontology, developed by the GO consortium (5,6), provides a broad overview of the more granular ontology content presented in the GO Terms table.
Pathways and interactions
Metabolic Pathway Reaction tables now include reaction compound names, with ChEBI IDs (17) available on mouse-over of the Equation column entries, and a column indicating whether the Enzyme Commission (EC) number associated with a gene is an exact match to the EC number in the metabolic pathway.
JBrowse
To dynamically visualize annotations on genomes, VEuPathDB embeds JBrowse (18), an open-source, robust, scalable, configurable genome browser that offers improved browsing and zooming speed as well as the ability to save and share personalized views. Data tracks informing on transcriptomics, proteomics, comparative genomics, epigenomics, genetic variation and the results of in-house sequence analyses are easily accessible through the filterable Select Tracks menu. Synteny views that highlight synteny across multiple genomes are created with our custom JBrowse plugin.
Apollo
Community annotation offers a value-added perspective to genome curation. Apollo (19) is an open source software enabling users to inspect, refine and add gene models to the current genome annotations. VEuPathDB offers Apollo for the genomes that are under active curation by VEuPathDB or the scientific community. To contribute structural gene model changes, users inspect transcriptomic data as JBrowse tracks, create new or modified gene models, and save them in Apollo. One of the main advantages of Apollo is that it instantly presents the updates made by other researchers to the whole community in real-time. In some specific cases, for example when different users propose conflicting edits, VEuPathDB will analyse the evidence supporting the proposed changes, but time constraints prevent review and curation of every submission. VEuPathDB has a pipeline that reviews some aspects of the annotations (e.g. missing start or stop codon, or a stop codon in the open reading frame) and, when appropriate, integrates changes into the official genome annotation.
Pathway record
The Cytoscape JS (20) drawing interface is updated with semantic zooming. When zoomed out, pathway nodes appear as glyphs. Circles denote compounds. Yellow boxes designate enzymes where a gene matches the EC number in VEuPathDB, while clear boxes indicate that there is no matching EC number in the VEuPathDB records. This important distinction allows at-a-glance recognition of pathways that are clearly presents in an organism (mostly yellow nodes) or likely absent (mostly white nodes). Zoom reveals molecular structures for main compound nodes. At high zoom levels, enzymes and side nodes appear as molecular structures, terms or EC numbers. Clicking on a node reveals node details. This feature can be activated at any zoom level.
Searches
Several new searches against the pre-analysed data offer the ability to probe post-translational modifications, paralog counts, phenotype data, and antisense transcriptional regulation. A new parameter in RNA-Seq searches, the floor parameter, improves the accuracy of fold change search results when gene expression is very low.
Genes by post-translational modifications
This new search returns genes whose protein products are experimentally determined to contain post-translational modifications based on evidence from proteomics mass spectroscopy. Depending on data availability, modifications available to search include phosphorylation, acetylation, ubiquitination, and several species of methylated arginine. Flexible user-defined parameters also allow an inverse search for genes whose protein products do not contain any modified residues.
Genes by paralog count
Available in all VEuPathDB sites, this search uses data from VEuPathDB’s OrthoMCL (21) analysis (see OrthoMCL section below) to return genes with a user-specified number of paralogs. Flexible search parameters can be configured to return single-copy genes or to highlight multigene families in any organism.
Genes by phenotype evidence
Phenotype data in VEuPathDB ranges from curated phenotypes to genome-wide screens that use technologies such as RNAi or CRISPR that assign fitness scores to genes. The new ‘Genes by Phenotype Evidence’ search provides a landing page where the available datasets and searches are listed. Curated phenotype searches return genes based on their assigned phenotype which is chosen from a dropdown menu or filter parameter of terms used in the study. The phenotype text search allows users to specify their own query terms and use wildcards. For other types of phenotype data, custom searches have been created in collaboration with the data providers.
RNA-Seq Sense-Antisense search
Available from the ‘Genes by RNA-Seq Evidence’ landing page, this new search is only an option for RNA-Seq datasets derived from stranded libraries. The search returns genes that exhibit simultaneous changes in sense and antisense transcript abundance and can be configured to specify the direction and magnitude of change of both antisense and sense transcripts between two groups of samples. For example, the search could be used to find genes in which antisense transcript abundance is upregulated 2-fold while sense transcript abundance is downregulated 2-fold between two samples. This could be an indication that sense transcription is suppressed by antisense inhibitory RNAs.
RNA-Seq search floor parameter
VEuPathDB continues to offer simple fold-change searches that return genes with transcript abundance that varies between groups of samples. While not statistically robust, these searches improve access to datasets that lack sufficient replicates for formal differential expression analysis and allow flexible sample groupings during the fold-change calculation. However, these searches can return misleading results when very low expression values are used in the denominator of a fold-change calculation. The new floor parameter prevents misleading fold change values by applying a threshold to very low expression values (default is equivalent to 10 reads). Expression values that fall below this threshold are reset to the threshold value, preventing cases where genes with very low expression levels are reported as having high fold-change values. Users can reduce this parameter to 5 reads or raise it in pre-defined increments up to 100 reads.
Search results
VEuPathDB searches return a table of IDs with columns of associated data (Figure 2C, green box) for records that meet the search criteria. The result page of a search that returns a list of genes, the Add Columns button (Figure 2C, orange arrow) now allows the user to add normalised expression values, graphs, and other data associated with each gene. Images and graphs within data columns are displayed with increased resolution. The search results, including the record ID and user-specified columns, can be saved locally via the Download button (Figure 2C, purple arrow). The Download tool offers additional formats, such as FASTA files containing protein/DNA sequences or GFF files. The Results Table interface now retains custom column configurations and sorting after the search strategy has been saved or when a saved strategy is shared.
Enrichment analyses
Opportunities to explore functional enrichment in gene search results vary from simple word clouds to formal gene ontology (GO) and metabolic pathway enrichment analyses that use Fisher's Exact test in combination with multiple test corrections. For GO enrichment analysis, a parameter is now available to limit the analysis to GO terms in the slim subset, which reduces redundancy in the enrichment results, making them easier to interpret. A simple word cloud visualization of enrichment results is available for GO and metabolic pathway enrichments. GO enrichment data can also be exported to REVIGO (22), facilitating data visualisation through a range of interactive tools including treemaps and graphs. Lastly, it is now easier to find genes in your gene list that contribute to the enrichment of a particular term. Enrichment results now include a column listing the number of genes in your gene list annotated with the enriched term. This column also serves as a link which runs a pre-configured search returning these genes.
Downloads
New options are available for retrieving sequences in FASTA format. When downloading sequences from the gene page, the sequences are FASTA formatted with the description line (header) indicating the gene ID. When downloading sequences from a search result (FASTA-sequence retrieval, configurable) or from the sequence retrieval tool, the FASTA header can be configured to contain only the gene ID or a full FASTA header with sequence formatted as a single line or limited to 60 characters per a line (default). Processed genomes, their annotations, as well as other genome-scale data are available for download from the banner menu under Data, Download data files. For search results, IDs and associated data can be downloaded via the download tool accessed by clicking Download from the search result page (Figure 2C, purple arrow).
MapVEu
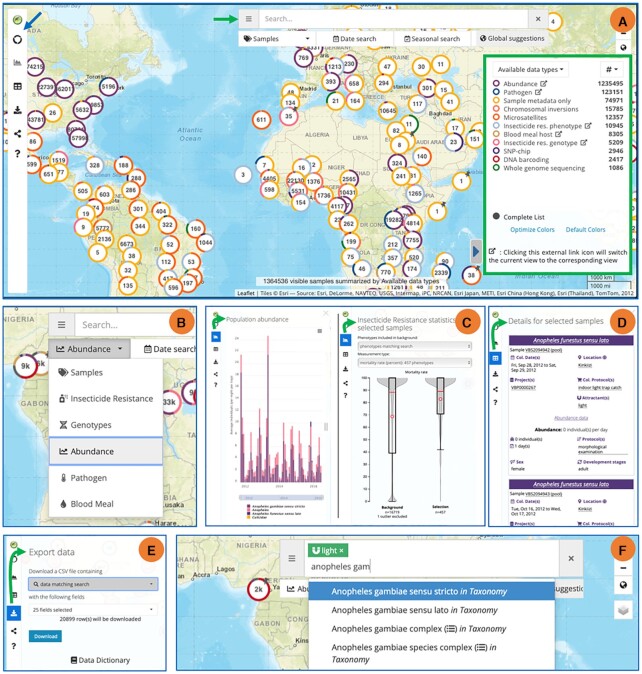
The VectorBase PopBio tool has been updated and renamed to MapVEu, VEuPathDB’s visualization and filtering tool for spatially resolved data (Figure 3). MapVEu integrates genomic, phenotypic and population data for traits such as insecticide resistance genotypes and phenotypes, genetic variation with microsatellites, chromosomal inversions and SNPs, population abundance, pathogen infection status and blood meal identification. To maximize reusability, data are standardized by our curators using a structured ontology (e.g. identical trap type names, species names, trap set-collect dates etc.). The map can be used both to discover which kinds of data are available in a geographic location (Figure 3A, B) and to view plots and charts summarising the data (Figure 3C). Specialized views (Figure 3B) presently exist for viewing population surveillance, insecticide resistance (phenotype and genotype), vector blood meal source analysis, and vector pathogen status. New features or views could be added at any time when sufficient data exists (e.g. microsatellites) and geospatial data from other VEuPathDB non-vector species will soon be added. Raw data can be downloaded as a .csv file (Figure 3E). Using the autocomplete search box, metadata search filters can be added (Figure 3F).
Figure 3.
MapVEu tool for visualizing and downloading spatially resolved data. (A) The MapVEu interface includes a left-side menu for accessing tools (blue arrow), a search and filtering bar at the top of the screen (green arrow), and a lower right-side legend (green box). Circular markers indicate the location of data collections. Clicking on these markers will zoom and disaggregate them into more spatially resolved points. (B) Tool for choosing data type of interest (enlarged search and filtering bar seen in A, green arrow). The map can display several data types including Samples, Insecticide Resistance, Genotypes, Abundance, Pathogens and Blood Meals. Users can select one of several views to see a specific data type on the map. (C) For many data types, specialized representations are available from the graph tab of the tool menu. Shown here are species abundance counts and insecticide resistance assays. (D) The tool menus’ data table tab highlights the depth of metadata recorded from each sample. (E) All data can be downloaded using the export data tab. F. Visible data can be filtered by adding search terms to the filter bar, which has auto-complete functionality.
Workspaces
VEuPathDB Workspaces offer users the ability to analyse their own data and make comparisons with data that are public in VEuPathDB. A new My Workspace menu in the header provides access to our updated Galaxy instance and a new My Data Sets page for tracking private data ported to VEuPathDB. The Galaxy workspace is updated with VEuPathDB-specific RNA-Seq, variant calling and OrthoMCL workflows as well as new bioinformatic tools that can be used in workflows and new custom tools for exporting workflow results to VEuPathDB.
Once at Galaxy, the user can upload their own data, which can be processed by the tools and workflows. Workflow and analysis results can be analysed in Galaxy, downloaded to the user's computer, or exported (using the new export tool) to a VEuPathDB project for visualization and further analysis. Data and workflows can be shared with selected colleagues to facilitate collaboration, but importantly, a user can control sharing to maintain data privacy. New export tools support the transfer of BigWig and RNA-Seq expression results to VEuPathDB projects, where user data can be viewed (in JBrowse and on gene pages) and queried (in RNA-Seq searches). In addition to RNA-Seq workflows, two additional workflow types deserve mention. First, ‘Workflow to map your proteins to OrthoMCL groups’ takes as input a user-uploaded proteome and places each protein into the appropriate ortholog group using our OrthoMCL algorithm; this analysis is useful for functional annotation of a newly-sequenced organism. Second, our ‘Variant calling’ workflows identify DNA sequence variants (and their effects, such as intergenic or non-synonymous) from user-uploaded DNA-Seq FASTQ files, employing VEuPathDB genomes as the reference. Finally, it should be re-iterated that the Galaxy platform contains other easy-to-use bioinformatic tools (e.g. statistics, text manipulation, ChIP peak calling, editing and filtering various sequence file types) that may aid the user in the analysis of their genomic data.
Data
VEuPathDB supports many datatypes including genomic sequence and annotation, transcriptomics, proteomics, epigenomics, metabolomics, comparative genomics data, host-response data, isolate and population level data, clinical data, and surveillance data. Our bimonthly releases add new data in these categories. Discussed below are major additions or improvements to data content in the last 5 years.
Annotation and curation
Selected genomes are manually curated by VEuPathDB expert curators (Plasmodium genomes were curated by the Parasite Genomics group at the Wellcome Sanger Institute until January 2021) and regularly updated in our bimonthly releases. Currently, VEuPathDB curates 11 Plasmodium, 10 Kinetoplastida and 17 Fungal genomes (listed here: https://tinyurl.com/VEuPathDBCuratedGenomes). Functional annotation attributes dominate the curatorial efforts and include gene names, synonyms, product descriptions, GO annotations, EC numbers, annotator notes, literature citations, and previous systematic identifiers. A summary of annotation changes made in FungiDB, PlasmoDB and TriTrypDB over the last four years can be found in Table 2. Functional annotations have also been integrated or updated from other external databases such as MIPS Functional Catalogue (23), AspGD (24), SGD (25), CGD (26), PomBase (27) and the Neurospora crassa e-Compendium (28) for fungal genomes in addition to providing link outs from other resources such as RefSeq (29), MEROPS (30) and CAZy (31) databases.
Table 2.
Summary of functional annotation updates made by VEuPathDB curators
| Annotation field | PlasmoDB | TriTrypDB | FungiDB | Total |
|---|---|---|---|---|
| Product descriptions | 14 408 | 9932 | 4062 | 28 402 |
| Gene names | 4171 | 2955 | 1958 | 9084 |
| Gene synonyms | 42 | 257 | 245 | 544 |
| Gene Ontology Terms | 23 480 | 70 748 | 7984 | 102 212 |
| Enzyme Commission numbers | 477 | 198 | 99 | 774 |
| PMIDs | 14 288 | 5127 | 9409 | 28 824 |
| Comments (notes from annotators) | 3 | 7622 | 21 461 | 29 086 |
| External database references added | 1 | 47 133 | 94 182 | 141 316 |
| Reviewed user comments per gene | 328 | 7400 | 1080 | 8808 |
| Genes with new functional annotations | 20 276 | 60 708 | 86 872 | 167 856 |
Curated phenotypes
Phenotypes, curated from the published literature using a uniform approach and ontologies from the OBO Foundry (32), are integrated into VEuPathDB at the bimonthly release. New curated phenotypes are available for several Aspergillus and Cryptococcus genomes and Fusarium graminearum PH-1. New phenotypic data from PHI-base (33), Aspergillus fumigatus transcription factors knockout collection (34) and several Neurospora crassa knockout projects are displayed in new custom tables on gene pages of FungiDB and are searchable from the ‘Identify Genes based on Phenotype Evidence’ gene search. VEuPathDB also works with OBO Foundry ontologies to develop new terms to better represent the biology of eukaryotic parasites and fungal pathogens.
GO slim
GO Slim ontologies, abbreviated versions of the full GO ontologies, are useful when fine-grained descriptions of a gene's function are encumbering. In addition to the full GO ontologies, GO Slim terms are now available on gene pages and searches or enrichment analyses that access GO ontologies offer an option to limit the results to GO Slim to avoid excessive detail.
Affiliated databases
The VEuPathDB infrastructure has been leveraged by other resources to enable mining and exploration of other datatypes. The ClinEpiDB (https://clinepidb.org/ce/app) (35) platform, funded by the Bill and Melinda Gates Foundation, is an epidemiology focused resource that allows users to explore data from multiple clinical projects including large scale population-based studies. The MicrobiomeDB (https://microbiomedb.org/mbio/app) (36) platform enables the exploration and analysis of data from various microbiome datasets.
Outreach
VEuPathDB earnestly aspires to meet the needs of the many research communities we serve. Our outreach team interacts with users and stake holders to discuss data integration, provide updates and instruction, receive feature suggestions, and understand issues and concerns. In addition to our email help desk, social media presence and YouTube channel, a webinar series begun in March 2020 serves as a point of contact for community meetings, in-depth presentations about specific topics, and demonstrations of new features or data at each bimonthly release (Table 3). As demand for virtual formats increases, we also offer online workshops and conference support. Virtual workshops still include hands on exercises that students complete on their own, but students and instructors assemble in a virtual room for screensharing demonstrations and discussions. Our traditional conference booth at scientific meetings can be accomplished with a virtual table for individual discussion and demonstrations.
Table 3.
VEuPathDB Community Resource access points
Infrastructure and data processing
Core infrastructure
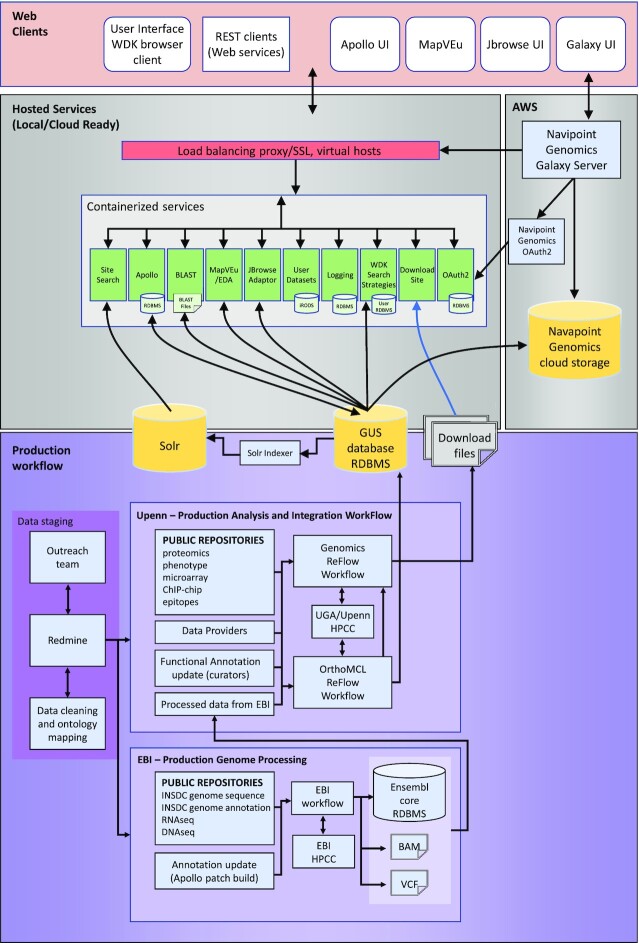
Significant updates to the VEuPathDB core infrastructure (Figure 4) accommodate the merger and improve scalability, flexibility, supportability, data flow and the user experience. Heavy data processing of genome sequences and their associated functional data (e.g. RNA-sequencing) is now performed at the EBI, leveraging their large compute cluster and mature data processing pipelines of the ENSEMBL (37) project. The EBI-processed data are then loaded into VEuPathDB GUS databases using very efficient workflows which are tailored to facilitate data integration and access by our searches. The data are provided to the user with a new RESTful (https://searchapparchitecture.techtarget.com/definition/RESTful-API) StrategiesWDK system that enables advanced searches, full Boolean operations for joining search results, and an operator (colocation tool) that co-locates record objects that are mapped on the genome sequence. The user interface is now driven by a React/Redux (https://react-redux.js.org/) client application that enables faster, more flexible tool development and facilitates a better user experience. Increasingly, tools are being containerized and provided to the application via micro-services to facilitate a more open, flexible, interoperable and cloud-ready infrastructure. Currently this includes the Solr (https://lucene.apache.org/solr/) search engine supporting our new site-wide search and plans to expand access and the functionality of MapVEu and Apollo.
Figure 4.
VEuPathDB data production workflow and architecture. The complete pathway from data acquisition to web presentation and utilization by users is detailed. Production activities and systems are represented in the bottom purple box and the services and presentation layers are represented in the pink and grey boxes. Data enter the system in the data staging box where they are identified and prioritized by Outreach and entered into the Redmine issue tracking system. Once data are cleaned and structured, datasets are available to the processing and integration workflows. Genome sequence, annotation, RNA Seq and DNA sequencing reads are processed at the EBI (bottom box) and passed back to Penn (top box) for data integration and subsequent processing including integration of functional data and ortholog assignment. Data are prepared by these workflows for presentation in the form of relational databases and indexed flat files. The web clients provide access to users via a set of services that communicate with the back-end data stores. The system also includes a user data analysis system (right side) enabling users to analyse their own data and, for some datatypes, import their results into VEuPathDB for analysis and integration with publicly available data.
Based on the new REST API, our web services facilitate much greater programmatic access to the resource and enhanced functionality. The results of complex strategies can now be returned via web services in different formats (json, fasta, gff, tab delimited, etc), and we provide you with the code needed for your own customized search (see https://tinyurl.com/veupath-web-services). To facilitate greater transparency and tool reuse, our codebase has been migrated to the GIT repository (https://github.com/VEuPathDB).
OrthoMCL 6
Significant upgrades to the OrthoMCL workflow result in an iterative build process that enables bi-monthly updates in sync with other VEuPathDB projects. The OrthoMCL algorithm (21) clusters proteins into ortholog groups based on BLAST similarity across many genomes that span the tree of life, documenting protein similarity across many species. Launched in April 2020, OrthoMCL 6 became the foundation for OrthoMCL.org (38) as well as the orthology relationships in the VEuPathDB sites. OrthoMCL 6 contains two changes that we believe improve orthology prediction and provide scalability. First, to improve prediction, we chose a set of 150 ‘core’ species that are evolutionarily separated across Eukaryota, Bacteria, and Archaea. This species diversity not only allows for grouping of a wide array of protein families, but also avoids problems that arise when organisms are too closely related (such as unnecessarily placing closely related genes into separate ortholog groups). Second, for scalability, we developed a strategy to map proteins from additional species, so-called ‘peripheral’ species, into the ortholog groups that were originally formed using proteins from the 150 core species. Any protein that fails to map to a core group (due to sequence divergence) is termed a ‘residual’ protein. Together, all residual proteins are subject to a second round of the OrthoMCL algorithm to form ‘residual’ groups. The current OrthoMCL version 6.7 contains 150 core and 505 peripheral species and, as new organisms are integrated into VEuPathDB sites, they will be processed for orthology as peripheral species at the OrthoMCL website.
FUTURE DIRECTIONS
Having successfully navigated the merger of two NIH funded Bioinformatic Resource Centers in a single year, VEuPathDB is emerging as a streamlined and efficient data mining resource that represents the best of each individual resource with substantial infrastructure improvements behind the scenes (Figure 4). While some bioinformatic analyses are universal and shared by the vector and eukaryotic pathogen communities, opportunities abound for cross-fertilization of ideas and tool development. Our new user community is larger, more diverse and adopting new or emerging technologies that VEuPathDB will endeavor to support.
Scalability
Our enhanced post-merger infrastructure and expanded web services were built with scalability, efficiency, and interoperability in mind. VEuPathDB must be nimble enough to respond to community needs which change as technology evolves. The challenges we anticipate include increasing amounts of data from a rapidly changing research landscape with increased emphasis on pathogen variation and host-pathogen interactions. A unified database that contains data from all supported organisms will enhance scalability and improve cross-species as well as host-pathogen interrogations. To improve the user experience of a unified database, we are also developing a configurable interface that allows users to customize all aspects of the site to their preferred subset of organisms, e.g. Ascomycota, or anaerobic microbes.
Host-Pathogen interactions
Support for this rich, multi-dimensional data is a key priority for VEuPathDB, including the development of tools for analysis and visualization of host-pathogen interactions and other systems-level studies. Host-pathogen interaction data are now available for many VEuPathDB targeted pathogen genomes, including cases with vector hosts. As research inquiries in these areas expand, more diverse data types are also available, including high-throughput metabolic, immune, and single cell profiling data that include rich metadata describing the experiments, samples, and clinical characteristics.
MapVEu
This powerful geoinformatics resource enables fast browsing and enhanced visualization of, currently, vector associated geospatial data. Further development efforts at VEuPathDB will concentrate on expanding geolocation mapping tools to the pathogen species hosted by VEuPathDB. Another priority is to develop an interface for visualizing multiple markers from a user-selected group of MapVEu selected samples and an enhanced functionality to compare different geo-selected samples.
Containerization
Software containers package code and software dependencies together, helping to solve the problem of inadequate software reliably between computing environments. As VectorBase and EuPathDB merged, we retained legacy services, while adding newly developed services into containerized environments. On the data production side, workflows continue to make use of reusable and disposable containerized environments. When appropriate, we will continue to implement new services as containers to make the code more easily deployable in different environments.
Cross-silo integration
Given the explosion in specialized data and bioinformatics tool resources, we can address the needs of our user communities by enabling easy data re-use and portability. To this end, we provide bulk data downloads and download of search results in a variety of standard formats that can be uploaded and analyzed with applications outside of VEuPathDB. We are working to make it even easier for users to directly take results and analyze them with other applications, such as GO enrichment and the Galaxy platform. Future plans include a shared Gateway website accessing both VEuPathDB and BV-BRC, the bacterial and viral resource, and tighter linkages with our affiliated databases MicrobiomeDB and ClinEpiDB. Additional planned efforts include tighter integration with CyVerse (39) and its vast suite of tools such as the CoGe (40) comparative genomics package. Our goal, together with other service providers is to facilitate an ecosystem of interoperability that maximizes utility and ease of use for our use communities wowrldwide.
Support for users analysing their own data
Increasingly users are generating datasets via routine or novel omics technologies and would like to analyse these data in the context of VEuPathDB without the need to submit the data for public release. As noted above, we have implemented user workspaces and a custom Galaxy instance where users can upload and analyse their own data against the updated reference genome sequences, annotation, expression and orthology data provided by VEuPathDB. With the addition of planned cross-silo integration, we will be enhancing users’ ability to take their data, partially processed in VEuPathDB or our Galaxy site, to additional sites that offer highly specialized services. Depending on the analyses performed elsewhere and the data format, users may bring back results, e.g. GFF files or lists of gene IDs to VEuPathDB for use in strategies or visualization in JBrowse. The goal is to make the movement and analysis of user data easier and to provide robust mechanisms for data portability and proven analysis tools against the same reference datasets to facilitate scientific discovery.
ACKNOWLEDGEMENTS
The authors wish to thank members of the VEuPathDB research communities for their willingness to share genomic-scale datasets, often prior to publication and for numerous comments and suggestions from our scientific advisors and the scientific community at large, which have helped to improve the functionality of VEuPathDB resources. We also thank past and present staff associated with the VEuPathDB BRC project, Nathan Dunn from the Apollo team and our research laboratory colleagues whose contributions have facilitated the creation and maintenance of this database resource.
Contributor Information
Beatrice Amos, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
Cristina Aurrecoechea, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
Matthieu Barba, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
Ana Barreto, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Genetics, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Evelina Y Basenko, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
Wojciech Bażant, Wellcome Centre for Integrative Parasitology, University of Glasgow, Glasgow G12 8TA, UK.
Robert Belnap, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
Ann S Blevins, Department of Pathology, School of Veterinary Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Ulrike Böhme, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
John Brestelli, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Genetics, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Brian P Brunk, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Mark Caddick, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
Danielle Callan, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Genetics, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Lahcen Campbell, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
Mikkel B Christensen, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
George K Christophides, Department of Life Sciences, Imperial College London, South Kensington Campus, London SW7 2AZ, UK.
Kathryn Crouch, Wellcome Centre for Integrative Parasitology, University of Glasgow, Glasgow G12 8TA, UK.
Kristina Davis, Center for Research Computing, University of Notre Dame, Notre Dame, IN 46556, USA.
Jeremy DeBarry, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
Ryan Doherty, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Yikun Duan, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Michael Dunn, Center for Research Computing, University of Notre Dame, Notre Dame, IN 46556, USA.
Dave Falke, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
Steve Fisher, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Genetics, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Paul Flicek, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
Brett Fox, Center for Research Computing, University of Notre Dame, Notre Dame, IN 46556, USA.
Bindu Gajria, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Gloria I Giraldo-Calderón, Department of Biological Sciences, Eck Institute for Global Health, University of Notre Dame, Notre Dame, IN 46556, USA; Departamento de Ciencias Biológicas y Departamento de Ciencias Básicas Médicas, Universidad Icesi, Calle 18 No. 122-135, Cali, Colombia.
Omar S Harb, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Elizabeth Harper, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Christiane Hertz-Fowler, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
Mark J Hickman, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Connor Howington, Center for Research Computing, University of Notre Dame, Notre Dame, IN 46556, USA.
Sufen Hu, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Jay Humphrey, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
John Iodice, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Genetics, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Andrew Jones, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
John Judkins, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Sarah A Kelly, Department of Life Sciences, Imperial College London, South Kensington Campus, London SW7 2AZ, UK.
Jessica C Kissinger, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA; Department of Genetics, University of Georgia, Athens, GA 30602, USA; Institute of Bioinformatics, University of Georgia, Athens, GA 30602, USA.
Dae Kun Kwon, Department of Civil & Environmental Engineering & Earth Sciences, University of Notre Dame, Notre Dame, IN 46556, USA.
Kristopher Lamoureux, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
Daniel Lawson, Department of Life Sciences, Imperial College London, South Kensington Campus, London SW7 2AZ, UK.
Wei Li, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Kallie Lies, Center for Research Computing, University of Notre Dame, Notre Dame, IN 46556, USA.
Disha Lodha, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
Jamie Long, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Robert M MacCallum, Department of Life Sciences, Imperial College London, South Kensington Campus, London SW7 2AZ, UK.
Gareth Maslen, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
Mary Ann McDowell, Department of Biological Sciences, Eck Institute for Global Health, University of Notre Dame, Notre Dame, IN 46556, USA.
Jaroslaw Nabrzyski, Center for Research Computing, University of Notre Dame, Notre Dame, IN 46556, USA.
David S Roos, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Samuel S C Rund, Department of Biological Sciences, Eck Institute for Global Health, University of Notre Dame, Notre Dame, IN 46556, USA.
Stephanie Wever Schulman, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Achchuthan Shanmugasundram, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
Vasily Sitnik, European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge CB10 1SD, UK.
Drew Spruill, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
David Starns, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
Christian J Stoeckert, Jr, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Genetics, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Sheena Shah Tomko, Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Haiming Wang, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
Susanne Warrenfeltz, Center for Tropical & Emerging Global Diseases, University of Georgia, Athens, GA 30602, USA.
Robert Wieck, Center for Research Computing, University of Notre Dame, Notre Dame, IN 46556, USA.
Paul A Wilkinson, Institute of Systems, Molecular & Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
Lin Xu, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Biology, University of Pennsylvania, Philadelphia, PA 19104, USA.
Jie Zheng, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Genetics, School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
DATA AVAILABILITY
VEuPathDB is a suite of projects that support many communities including: AmoebaDB (https://amoebadb.org); CryptoDB (https://cryptodb.org); FungiDB (https://fungidb.org); GiardiaDB (https://giardiadb.org); MicrosporidiaDB (https://microsporidiadb.org); OrthoMCL (https://orthomcl.org); PiroplasmaDB (https://piroplasmadb.org); PlasmoDB (https://plasmodb.org); ToxoDB (https://toxodb.org); TrichDB (https://trichdb.org); TriTrypDB (https://tritrypdb.org); VectorBase (https://www.vectorbase.org); and VEuPathDB (https://veupathdb.org). VEuPathDB makes most source code publicly available in a GitHub repository (https://github.com/VEuPathDB).
Release 54 (September 8, 2021) of VEuPathDB contains 1775 datasets. The release dates, versions and sources can be accessed at (https://veupathdb.org/veupathdb/app/search/dataset/AllDatasets/result) and links therein.
FUNDING
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. 75N93019C00077 and Wellcome Trust grants 218288/Z/19/Z and 212929/Z/18/Z. No NIAID contract funds were used to fund the planning, preparation, submission or publication of this manuscript. Funding for the open access charge was provided by the University of Pennsylvania.
Conflict of interest statement. None declared.
REFERENCES
- 1. Giraldo-Calderon G.I., Emrich S.J., MacCallum R.M., Maslen G., Dialynas E., Topalis P., Ho N., Gesing S., VectorBase C., Madey G. et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015; 43:D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aurrecoechea C., Barreto A., Basenko E.Y., Brestelli J., Brunk B.P., Cade S., Crouch K., Doherty R., Falke D., Fischer S. et al. EuPathDB: the eukaryotic pathogen genomics database resource. Nucleic Acids Res. 2017; 45:D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davidson S.B., Crabtree J., Brunk B., Schug J., Tannen V., Overton G.C., Stoeckert J., C. J. K2/Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Syst. J. 2001; 40:512–531. [Google Scholar]
- 4. Fischer S., Aurrecoechea C., Brunk B.P., Gao X., Harb O.S., Kraemer E.T., Pennington C., Treatman C., Kissinger J.C., Roos D.S. et al. The Strategies WDK: a graphical search interface and web development kit for functional genomics databases. Database (Oxford). 2011; 2011:bar027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The Gene Ontology, C. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019; 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jalili V., Afgan E., Gu Q., Clements D., Blankenberg D., Goecks J., Taylor J., Nekrutenko A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020; 48:8205–8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sievers F., Higgins D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018; 27:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 10. Eisenhaber B., Bork P., Eisenhaber F. Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 1999; 292:741–758. [DOI] [PubMed] [Google Scholar]
- 11. Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.Y., El-Gebali S., Fraser M.I. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019; 47:D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Claros M.G., Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996; 241:779–786. [DOI] [PubMed] [Google Scholar]
- 13. Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021; 49:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007; 35:W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. UniProt, C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L. et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunn N.A., Unni D.R., Diesh C., Munoz-Torres M., Harris N.L., Yao E., Rasche H., Holmes I.H., Elsik C.G., Lewis S.E. Apollo: democratizing genome annotation. PLoS Comput. Biol. 2019; 15:e1006790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016; 32:309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L., Stoeckert C.J. Jr, Roos D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003; 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011; 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mewes H.W., Ruepp A., Theis F., Rattei T., Walter M., Frishman D., Suhre K., Spannagl M., Mayer K.F., Stumpflen V. et al. MIPS: curated databases and comprehensive secondary data resources in 2010. Nucleic Acids Res. 2011; 39:D220–D224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cerqueira G.C., Arnaud M.B., Inglis D.O., Skrzypek M.S., Binkley G., Simison M., Miyasato S.R., Binkley J., Orvis J., Shah P. et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014; 42:D705–D710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R. et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012; 40:D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skrzypek M.S., Binkley J., Binkley G., Miyasato S.R., Simison M., Sherlock G. The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017; 45:D592–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lock A., Rutherford K., Harris M.A., Hayles J., Oliver S.G., Bahler J., Wood V. PomBase 2018: user-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic. Acids. Res. 2019; 47:D821–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perkins D., Radford A., Sachs M. The Neurospora Compendium: Chromosomal Loci. 2000; Academic Press. [Google Scholar]
- 29. O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., D. Ako-Adjei. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rawlings N.D., Barrett A.J., Finn R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016; 44:D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014; 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith B., Ashburner M., Rosse C., Bard J., Bug W., Ceusters W., Goldberg L.J., Eilbeck K., Ireland A., Mungall C.J. et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007; 25:1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Urban M., Cuzick A., Seager J., Wood V., Rutherford K., Venkatesh S.Y., De Silva N., Martinez M.C., Pedro H., Yates A.D. et al. PHI-base: the pathogen-host interactions database. Nucleic Acids Res. 2020; 48:D613–D620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Furukawa T., van Rhijn N., Fraczek M., Gsaller F., Davies E., Carr P., Gago S., Fortune-Grant R., Rahman S., Gilsenan J.M. et al. The negative cofactor 2 complex is a key regulator of drug resistance in Aspergillus fumigatus. Nat. Commun. 2020; 11:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruhamyankaka E., Brunk B.P., Dorsey G., Harb O.S., Helb D.A., Judkins J., Kissinger J.C., Lindsay B., Roos D.S., San E.J. et al. ClinEpiDB: an open-access clinical epidemiology database resource encouraging online exploration of complex studies. Gates Open Res. 2019; 3:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oliveira F.S., Brestelli J., Cade S., Zheng J., Iodice J., Fischer S., Aurrecoechea C., Kissinger J.C., Brunk B.P., Stoeckert C.J. Jr et al. MicrobiomeDB: a systems biology platform for integrating, mining and analyzing microbiome experiments. Nucleic Acids Res. 2018; 46:D684–D691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yates A.D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R. et al. Ensembl 2020. Nucleic Acids Res. 2020; 48:D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen F., Mackey A.J., Stoeckert C.J. Jr, Roos D.S. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic. Acids. Res. 2006; 34:D363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Merchant N., Lyons E., Goff S., Vaughn M., Ware D., Micklos D., Antin P. The iPlant collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 2016; 14:e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grover J.W., Bomhoff M., Davey S., Gregory B.D., Mosher R.A., Lyons E. CoGe LoadExp+: a web-based suite that integrates next-generation sequencing data analysis workflows and visualization. Plant Direct. 2017; 1: 10.1002/pld3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
VEuPathDB is a suite of projects that support many communities including: AmoebaDB (https://amoebadb.org); CryptoDB (https://cryptodb.org); FungiDB (https://fungidb.org); GiardiaDB (https://giardiadb.org); MicrosporidiaDB (https://microsporidiadb.org); OrthoMCL (https://orthomcl.org); PiroplasmaDB (https://piroplasmadb.org); PlasmoDB (https://plasmodb.org); ToxoDB (https://toxodb.org); TrichDB (https://trichdb.org); TriTrypDB (https://tritrypdb.org); VectorBase (https://www.vectorbase.org); and VEuPathDB (https://veupathdb.org). VEuPathDB makes most source code publicly available in a GitHub repository (https://github.com/VEuPathDB).
Release 54 (September 8, 2021) of VEuPathDB contains 1775 datasets. The release dates, versions and sources can be accessed at (https://veupathdb.org/veupathdb/app/search/dataset/AllDatasets/result) and links therein.