Abstract
gutMGene (http://bio-annotation.cn/gutmgene), a manually curated database, aims at providing a comprehensive resource of target genes of gut microbes and microbial metabolites in humans and mice. Metagenomic sequencing of fecal samples has identified 3.3 × 106 non-redundant microbial genes from up to 1500 different species. One of the contributions of gut microbiota to host biology is the circulating pool of bacterially derived small-molecule metabolites. It has been estimated that 10% of metabolites found in mammalian blood are derived from the gut microbiota, where they can produce systemic effects on the host through activating or inhibiting gene expression. The current version of gutMGene documents 1331 curated relationships between 332 gut microbes, 207 microbial metabolites and 223 genes in humans, and 2349 curated relationships between 209 gut microbes, 149 microbial metabolites and 544 genes in mice. Each entry in the gutMGene contains detailed information on a relationship between gut microbe, microbial metabolite and target gene, a brief description of the relationship, experiment technology and platform, literature reference and so on. gutMGene provides a user-friendly interface to browse and retrieve each entry using gut microbes, disorders and intervention measures. It also offers the option to download all the entries and submit new experimentally validated associations.
INTRODUCTION
The human body is colonized by a large number of microbes, including up to 38 trillion bacteria, eukaryotes, viruses and archaea, most of which are present in the intestine (1,2). Much evidence indicates that alterations in the diversity and composition of gut microbes are closely related to the pathogenesis of many diseases such as cardiovascular diseases, metabolic diseases and nervous system diseases (3–5). Recently, researchers have come to explore the molecular mechanism of gut microbes, which can help to understand the associations between gut microbes and host. One of the most important ways microbes significantly impact host biology is through the production of small molecules, which accumulate in the intestine and circulate in host bodies (6,7).
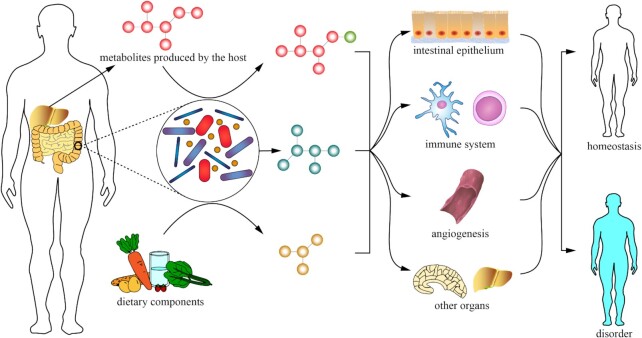
The relationships between gut microbes and host are highly mutually beneficial, as the host provides the microbes with nutrients and a shelter, while the microbes aid the host in acquiring energy from non-digestible food and synthesizing essential metabolites (8). Metabolites produced by microbes are mainly divided into three categories: produced by microbes from exogenously consumed compounds; produced by the host and biochemically modified by gut microbes; and synthesized de novo by gut microbes (9). Actually, it has been estimated that ∼10% of metabolites found in mammalian blood, where they can produce systemic effects on the immune system and host homeostasis through engaging specific host receptors and activating downstream signaling cascades, are derived from the gut microbiota, as shown in Figure 1 (10). For example, as the most thoroughly studied gut microbial metabolites, short-chain fatty acids (SCFAs) can be produced by a variety of microbes such as some members of Bacteroidetes and Firmicutes (11–14). Tolhurst et al. have proved that acetate and butyrate, the major types of SCFAs, promote the secretion of glucagon-like peptide 1 strongly linked to glucose homeostasis in L cells by coupling G protein-coupled receptor 43, which may help the treatment improvement of diabetes to some degree (15). Levy et al. have confirmed that taurine, biochemically modified by gut microbes, could enhance the activation of NLRP6 inflammasome and thereby increase the production of IL-18 in the intestinal epithelium, which supports epithelial barrier function and maintenance (16). Moreover, it has been substantiated that ATPs, synthesized de novo by gut microbes, have the ability to drive chemotaxis of and cytokine release by immune cells (17,18).
Figure 1.
Intestinal microbial metabolites affect host homeostasis.
To help researchers to better understand gut microbes, some important resources have been developed. For example, GMrepo (19) and gutMEGA (20) curated and annotated human gut metagenomes. gutMDisorder (21) collected experimentally validated associations between gut microbiota and diseases or intervention measures. Nevertheless, a public resource of high-quality curated target genes and metabolites of gut microbes remains unavailable. Thus, we developed a manually curated database entitled gutMGene to collect experimentally validated relationships between gut microbes, microbial metabolites and target genes from papers. This database is freely available at http://bio-annotation.cn/gutmgene.
DATA COLLECTION AND DATABASE CONTENT
To collect high-quality data, all the associations between gut microbes, microbial metabolites and genes were manually extracted from previously published studies. Here, we searched PubMed database with a list of keywords to acquire potentially relevant papers, such as ‘gut’, ‘intestinal’, ‘microbe’, ‘metabolite’, etc., and finally we selected >3000 papers. Then, we screened out papers documenting experimentally validated associations. Subsequently, we manually extracted microbe–metabolite, microbe–target and metabolite–target relationships in humans and mice from over 360 publications. As a result, 1331 curated relationships among 332 gut microbes, 207 microbial metabolites and 223 genes in humans, and 2349 curated relationships among 209 gut microbes, 149 microbial metabolites and 544 genes in mice were collected in the current version of gutMGene (Table 1).
Table 1.
The number of gut microbes, metabolites, genes and their relationships in humans and mice
| Species | No. of gut microbes | No. of metabolites | No. of genes | No. of relationships |
|---|---|---|---|---|
| Human | 332 | 207 | 222 | 1331 |
| Mouse | 209 | 149 | 544 | 2349 |
Each entry in the gutMGene contains two sections for documenting an association. The ‘Relationships between gut microbes, metabolites, and genes’ section documents intestinal microbe, sample type, substrate, metabolite, gene, brief description of the alteration in the gene under the intestinal microbe or metabolite, detailed description, species (human or mouse), experimental methods and measurement techniques. Especially, it contains a throughput description of gene expression experimental technologies, which include low-throughput [e.g. quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and western blot] and high-throughput experimental technologies (e.g. microarray or RNA-seq). The gut microbe, substrate, metabolite and gene are organized based on NCBI Taxonomy database (22), PubChem (23), HMDB (24), ChEBI (25) and NCBI gene database. The experimental method includes cell culture, in vitro bacterial culture, in vivo infection experiment, control experiment, supplementation of metabolite and so on. The measurement technique includes 16S rDNA sequence, high-performance liquid chromatography analysis, electrospray ionization mass spectrometry analysis, qRT-PCR, fluorescence microscopy, western blot assay, RNA-seq and so on. Meanwhile, the ‘Literature’ section documents a detailed description of the literature reference.
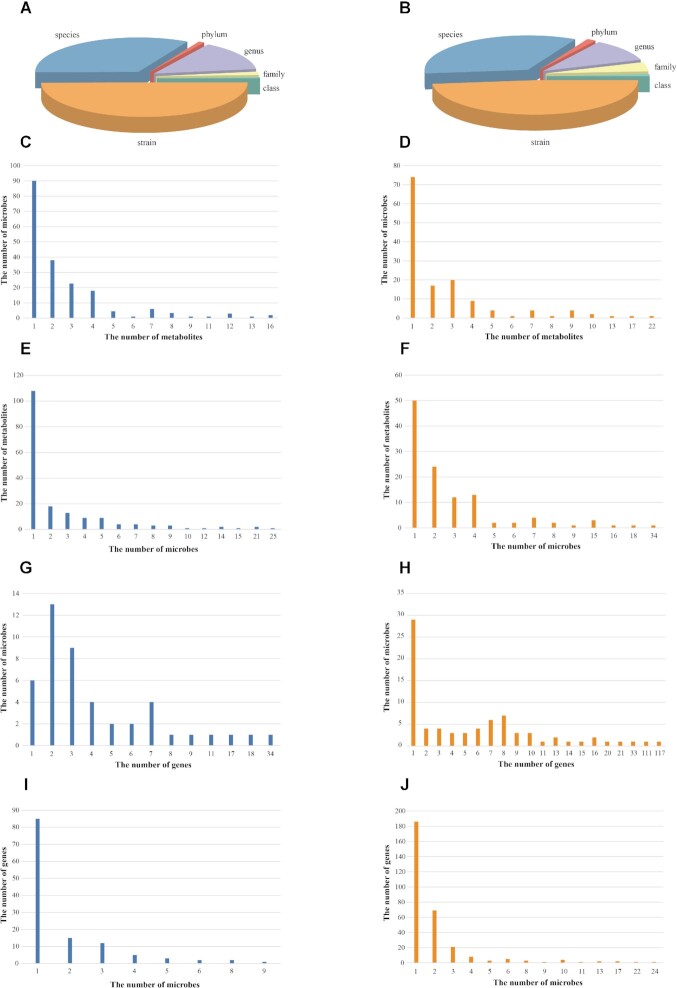
Figure 2A shows the distribution of gut microbes in humans among different classifications of NCBI Taxonomy database (22). Current version of gutMGene documents microbes according to phylum, class, family, genus, species and strain levels. One hundred sixty-nine (50.90%) microbes are at strain level, which make up almost half of our database. As in humans, a great many of the microbes (47.85%, 100/209) in mice are at strain level, which is shown in Figure 2B.
Figure 2.
The distribution of gut microbes, metabolites and genes in gutMGene. (A) The distribution of gut microbes in humans among different taxa. (B) The distribution of gut microbes in mice among different taxa. (C) Histogram of the number of microbes in humans associated with individual metabolite. (D) Histogram of the number of microbes in mice associated with individual metabolite. (E) Histogram of the number of metabolites associated with individual microbe in humans. (F) Histogram of the number of metabolites associated with individual microbe in mice. (G) Histogram of the number of microbes in humans associated with individual gene. (H) Histogram of the number of microbes in mice associated with individual gene. (I) Histogram of the number of genes associated with individual microbe in humans. (J) Histogram of the number of genes associated with individual microbe in mice.
Figure 2C and D shows the number of metabolites associated with individual microbe in humans and mice, respectively. 46.63% microbes from humans (90/193) and 53.24% from mice (74/139) are associated with only one metabolite, which could be a potential marker. Consistently, 60.00% metabolites in humans (111/185) and 43.10% in mice (50/116) are associated with only one microbe, which is shown in Figure 2E and F. Similarly, as shown in Figure 2G–J, microbes and genes have a consistent association pattern with gut microbes and metabolites except that the majority of human microbes are associated with two genes.
USER INTERFACE
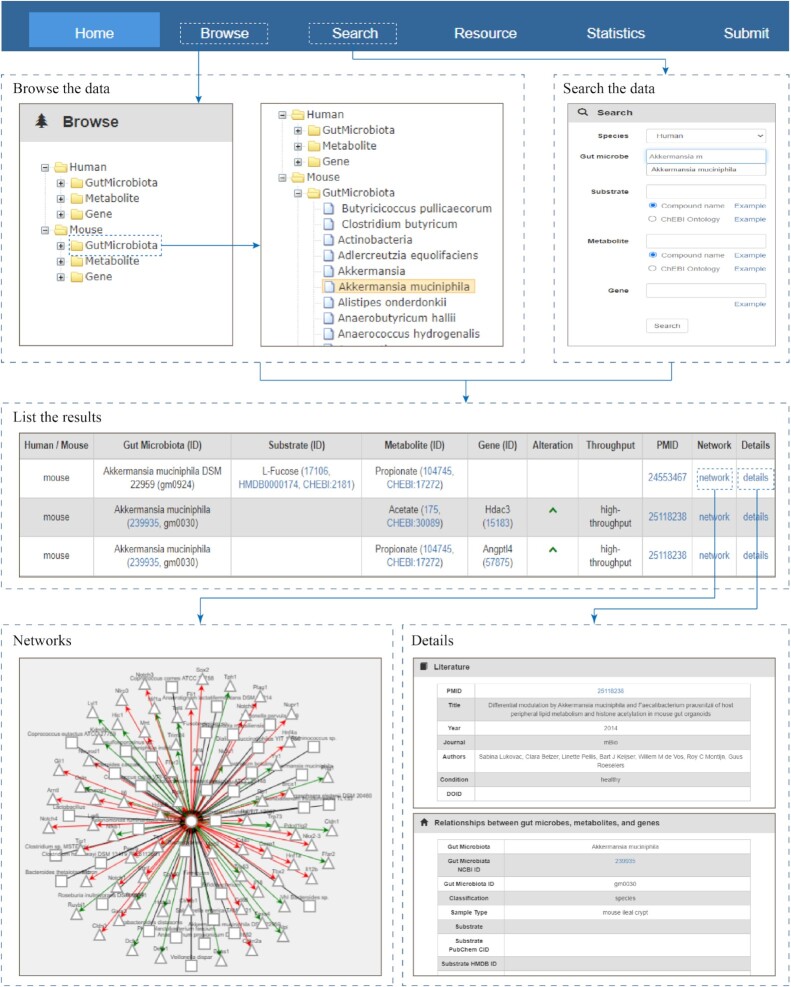
gutMGene provides a tree browser and a search engine to query detailed information about associations between gut microbe, metabolite and target gene. Figure 3 shows the schematic workflow.
Figure 3.
Schematic workflow of gutMGene.
The tree browser organizes the data according to species, using ‘Human’ and ‘Mouse’ as root categories. Each of the species involves three subcategories named ‘GutMicrobiota’, ‘Metabolite’ and ‘Gene’. By clicking ‘GutMicrobiota’, ‘Metabolite’ or ‘Gene’ category, all the names of gut microbes, metabolites or target genes belonging to the corresponding species would be listed as leaf nodes. Figure 3 shows a partial list of mouse gut microbes. After selecting a microbe ‘Akkermansia muciniphila’, all the associations between Akkermansia muciniphila and metabolites or target genes would be retrieved and shown in a table, where an association with a brief introduction is represented into one row. The identifier of microbe, substrate, metabolite, gene and PMID could be linked to NCBI Taxonomy database, PubChem, HMDB, ChEBI and PubMed for detailed description of these entities. In the result table, clicking the ‘detail’ link of a row would lead to the detailed information about an association. For an association between a microbe and a metabolite or a target gene in a row, clicking the ‘network’ link, microbes, metabolites and genes associated with these items would be shown in a network.
gutMGene provides a search engine to query associations by inputting a term of microbe, substrate, metabolite and gene. Additionally, ChEBI ontology (25) was imported for searching broad chemical classes of compounds by inputting a term of ChEBI ontology in the ‘Substrate’ or ‘Metabolite’ search box after selecting ‘ChEBI ontology’. For ease of use, these inputting terms could be autocompleted by selecting a species. After submitting the input items, entries that matched with these items exactly will be returned and shown in a table as above.
gutMGene provides a ‘Submit’ page for researchers to submit a traceable introduction about important associations that are not documented in the database. Once approved by the reviewer committee, the associations with detailed information will be included in the updated version. In addition, a ‘Resource’ page was offered for downloading all the data and associated networks between gut microbe and metabolite or gene in humans and mice.
FUTURE DEVELOPMENT
With the development of high-throughput technologies, the number of experiments discovering target genes of gut microbes and microbial metabolites increases exponentially. We will continue to manually curate newly validated relationships by prescreening papers in MEDLINE titles and abstracts using text-mining tools and reviewing the submission in web page. Meanwhile, with the emergence of more gut microbial resources, more links through gut microbes, metabolites and genes to other potential related resources would be provided to incorporate gutMGene in other resources for annotating the function of gut microbiota.
CONCLUSION
gutMGene is a comprehensive resource for documenting target genes of gut microbes and microbial metabolites, which provides an easy way to search, browse and download all the experiment-based associations in humans and mice. The current version of gutMGene documents 1331 curated relationships among 332 gut microbes, 207 microbial metabolites and 223 genes in humans, and 2349 curated relationships among 209 gut microbes, 149 microbial metabolites and 544 genes in mice. Recently, researchers come to explore the molecular mechanism of gut microbiota, which can help to understand the associations between gut microbiota and host. Since the influence of the gut microbiome is mainly achieved through metabolism, gutMGene provides a choice in exploring the role of gut microbes in the occurrence and development of diseases and predicting novel drugs using commensal bacteria as a medium.
DATA AVAILABILITY
This database is freely available at http://bio-annotation.cn/gutmgene.
Contributor Information
Liang Cheng, NHC and CAMS Key Laboratory of Molecular Probe and Targeted Theranostics, Harbin Medical University, Harbin 150028, Heilongjiang, China; College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Changlu Qi, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Haixiu Yang, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Minke Lu, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Yiting Cai, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Tongze Fu, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Jialiang Ren, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Qu Jin, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, Heilongjiang, China.
Xue Zhang, NHC and CAMS Key Laboratory of Molecular Probe and Targeted Theranostics, Harbin Medical University, Harbin 150028, Heilongjiang, China; McKusick–Zhang Center for Genetic Medicine, Peking Union Medical College, Beijing 100005, China.
FUNDING
Tou-Yan Innovation Team Program of the Heilongjiang Province [2019-15]; National Natural Science Foundation of China [61871160, 62172130]; Young Innovative Talents in Colleges and Universities of Heilongjiang Province [2018-69]; Heilongjiang Postdoctoral Fund [LBH-Q20030]. Funding for open access charge: National Natural Science Foundation of China [61871160].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sender R., Fuchs S., Milo R.. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016; 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thursby E., Juge N.. Introduction to the human gut microbiota. Biochem. J. 2017; 474:1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmad A.F., Dwivedi G., O’Gara F., Caparros-Martin J., Ward N.C.. The gut microbiome and cardiovascular disease: current knowledge and clinical potential. Am. J. Physiol. Heart Circ. Physiol. 2019; 317:H923–H938. [DOI] [PubMed] [Google Scholar]
- 4. Boulange C.L., Neves A.L., Chilloux J., Nicholson J.K., Dumas M.E.. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016; 8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dinan T.G., Cryan J.F.. The microbiome–gut–brain axis in health and disease. Gastroenterol. Clin. North Am. 2017; 46:77–89. [DOI] [PubMed] [Google Scholar]
- 6. Lin L., Zhang J.. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017; 18:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qi C., Wang P., Fu T., Lu M., Cai Y., Chen X., Cheng L.. A comprehensive review for gut microbes: technologies, interventions, metabolites and diseases. Brief. Funct. Genomics. 2021; 20:42–60. [DOI] [PubMed] [Google Scholar]
- 8. Neu J., Douglas-Escobar M., Lopez M.. Microbes and the developing gastrointestinal tract. Nutr. Clin. Pract. 2007; 22:174–182. [DOI] [PubMed] [Google Scholar]
- 9. Postler T.S., Ghosh S.. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017; 26:110–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., Siuzdak G.. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. U.S.A. 2009; 106:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wrzosek L., Miquel S., Noordine M.L., Bouet S., Joncquel Chevalier-Curt M., Robert V., Philippe C., Bridonneau C., Cherbuy C., Robbe-Masselot C.et al.. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013; 11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen D., Jin D., Huang S., Wu J., Xu M., Liu T., Dong W., Liu X., Wang S., Zhong W.et al.. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 2020; 469:456–467. [DOI] [PubMed] [Google Scholar]
- 13. Reichardt N., Duncan S.H., Young P., Belenguer A., McWilliam Leitch C., Scott K.P., Flint H.J., Louis P.. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014; 8:1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang F., Zou Q.. mAML: an automated machine learning pipeline with a microbiome repository for human disease classification. Database (Oxford). 2020; 2020:baaa050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tolhurst G., Heffron H., Lam Y.S., Parker H.E., Habib A.M., Diakogiannaki E., Cameron J., Grosse J., Reimann F., Gribble F.M.. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012; 61:364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levy M., Thaiss C.A., Zeevi D., Dohnalova L., Zilberman-Schapira G., Mahdi J.A., David E., Savidor A., Korem T., Herzig Y.et al.. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015; 163:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Faas M.M., Saez T., de Vos P.. Extracellular ATP and adenosine: the Yin and Yang in immune responses?. Mol. Aspects Med. 2017; 55:9–19. [DOI] [PubMed] [Google Scholar]
- 18. Yang F., Zou Q.. DisBalance: a platform to automatically build balance-based disease prediction models and discover microbial biomarkers from microbiome data. Brief. Bioinform. 2021; 10.1093/bib/bbab094. [DOI] [PubMed] [Google Scholar]
- 19. Wu S., Sun C., Li Y., Wang T., Jia L., Lai S., Yang Y., Luo P., Dai D., Yang Y.Q.et al.. GMrepo: a database of curated and consistently annotated human gut metagenomes. Nucleic Acids Res. 2020; 48:D545–D553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Q., Yu K., Li S., Zhang X., Zhao Q., Zhao X., Liu Z., Cheng H., Liu Z.X., Li X.. gutMEGA: a database of the human gut MEtaGenome Atlas. Brief. Bioinform. 2021; 22:bbaa082. [DOI] [PubMed] [Google Scholar]
- 21. Cheng L., Qi C., Zhuang H., Fu T., Zhang X.. gutMDisorder: a comprehensive database for dysbiosis of the gut microbiota in disorders and interventions. Nucleic Acids Res. 2020; 48:D554–D560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., McVeigh R., O’Neill K., Robbertse B.et al.. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vazquez-Fresno R., Sajed T., Johnson D., Li C., Karu N.et al.. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018; 46:D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C.. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016; 44:D1214–D1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This database is freely available at http://bio-annotation.cn/gutmgene.