Abstract
The transcriptional regulatory network in prokaryotes controls global gene expression mostly through transcription factors (TFs), which are DNA-binding proteins. Chromatin immunoprecipitation (ChIP) with DNA sequencing methods can identify TF binding sites across the genome, providing a bottom-up, mechanistic understanding of how gene expression is regulated. ChIP provides indispensable evidence toward the goal of acquiring a comprehensive understanding of cellular adaptation and regulation, including condition-specificity. ChIP-derived data's importance and labor-intensiveness motivate its broad dissemination and reuse, which is currently an unmet need in the prokaryotic domain. To fill this gap, we present proChIPdb (prochipdb.org), an information-rich, interactive web database. This website collects public ChIP-seq/-exo data across several prokaryotes and presents them in dashboards that include curated binding sites, nucleotide-resolution genome viewers, and summary plots such as motif enrichment sequence logos. Users can search for TFs of interest or their target genes, download all data, dashboards, and visuals, and follow external links to understand regulons through biological databases and the literature. This initial release of proChIPdb covers diverse organisms, including most major TFs of Escherichia coli, and can be expanded to support regulon discovery across the prokaryotic domain.
INTRODUCTION
The transcriptional regulatory network (TRN) enables global regulation of gene expression in response to environmental stimuli. A precise understanding of its constituents, mechanisms, and condition-specific activity is of fundamental interest in biology. To that end, chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) or combined with lambda exonuclease digestion (ChIP-exo) have been developed to map DNA-binding proteins to their binding sites in a genome-wide manner at near single base-pair resolution (1,2). The process is initiated by cross-linking DNA and DNA-bound transcription factors (TFs). Subsequently, chromatin is fragmented with sonication. A monoclonal antibody against a specific TF, or other DNA-binding proteins, is then used to immunoprecipitate specific DNA-protein complexes, from which precipitated DNA fragments are isolated and sequenced. The sequence of the DNA fragments is mapped back onto the reference genome for determination of the binding sites, enabling detailed localization of DNA-TF interactions (3), which play a major role in our understanding of the TRN.
The generation and analysis of ChIP data is time consuming, laborious, and expensive, which motivates broad dissemination of detailed results when possible. While several databases of ChIP results exist (4–8), they largely focus on eukaryotic organisms. To our knowledge, an online resource with easy accessibility to ChIP data broadly across the prokaryotic domain is a previously unmet need.
Driven by the rapidly declining price of next-generation sequencing (NGS), the life sciences are undergoing a big data revolution. As DNA sequences and RNA transcriptomes rapidly fill online databases, and tools to analyze and explain them become increasingly complex, it becomes increasingly important for mechanistic, experimental data types such as ChIP to become widely accessible. For example, transcription factor binding sites (TFBSs) in the genome are often mutated across related strains (9) or in evolution studies (10); the reasons for and effects of these mutations may become clear from an analysis of ChIP data for the relevant TFs. In addition, differential gene expression methods and more advanced transcriptomic analysis (11–14) have yielded gene groupings that may be the result of co-regulation, and ChIP can provide very valuable evidence for the direct mechanisms underlying those observations. In addition, TFBSs identified by ChIP serve as the basis to learn the chromatin network (15), and provide the information to reconstruct the regulons and TRNs (16).
The analysis of ChIP-derived data involves the characterization of individual binding peaks as well as global comparisons of peaks between different genomes (17). For each peak, the location, strength and shape are important for determining the structure and local function of the TF. Each peak can also be associated with its downstream genes and operons, which generates a set of putatively regulated genes for comparisons against regulons defined from other data sources (18). These comparisons can help elucidate the ‘core’ regulon by discerning between direct and indirect effects, and reveal condition-specific binding behavior (19,20). Also, sequence motif enrichment can be performed to identify common nucleotide motifs in TFBSs (21), which may indicate the specific DNA features that influence TF binding. The sequence motifs can be both compared to literature consensus motifs and used in hypothesis generation of novel TFBSs based on similarity to the motif (22).
Here, we present proChIPdb, the chromatin immunoprecipitation database and interactive web tool for prokaryotic organisms (available at prochipdb.org). This site provides users with ChIP-derived data, curated binding peaks, and additional plots presented as interactive web pages (termed dashboards) for a variety of TFs across the prokaryotic domain, with a particular focus on Escherichia coli for the initial release. We have included publicly-available ChIP-derived data and supplemented it with additional high-quality data from our research group that is also available on the Gene Expression Omnibus (GEO) (23). This data may be combined with other knowledge types which are currently out of the scope of proChIPdb, such as RNA-seq data, to provide a complete understanding of bacterial regulons. Once users begin by selecting a particular organism, strain and TF, they will then have access to an interactive dashboard featuring a whole-genome view of the binding intensity for all available samples and conditions, as well as curated tables of peak locations, heights, and downstream genes. Up to four additional plots characterize the overall binding width, location relative to downstream genes, enriched motif(s), and concordance of target genes with those from other data sources. A metadata panel provides links to the data source and external databases, such as RegulonDB (24) and iModulonDB (14). proChIPdb serves as a valuable addition to the online database ecosystem by providing a bottom-up, mechanistic understanding of the elements of prokaryotic TRNs via clear and interactive presentations of this important data type.
MATERIALS AND METHODS
Data sources and acquisition
All data was generated by using chromatin immunoprecipitation methods, as described in their respective publications (Supplementary Table S1). All raw data is available via the National Center for Biotechnology Information (NCBI) GEO accessions (https://www.ncbi.nlm.nih.gov/geo/), the Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra), or the European Bioinformatics Institute (EBI) ArrayExpress (https://www.ebi.ac.uk/arrayexpress/experiments/) unless otherwise indicated from their respective publications (23,25,26). All raw data is accessible through journal publications or public databases.
Data processing and peak calling
For data retrieved from publications, processing was performed as described therein, with public file types entering our processing pipeline wherever appropriate (i.e., public .fastq read files were processed with our pipeline, but public aligned .bw files or lists of peaks were included as published). Sequence read .fastq files generated from ChIP-seq/-exo were mapped onto individual reference genomes using Bowtie with default options to generate SAM output files (27). After mapping, each nucleotide of the genome will be associated with a number of mapped reads; these are stored as bigWig files using deeptools (28). When binding peak sites were not available in the TF’s related publication, the MACE program (https://code.google.com/p/chip-exo/) was used to define peak candidates from biological duplicates for each experimental condition with sequence depth normalization (29). MACE was chosen as the best algorithm for this purpose because it was designed specifically for the chip-exo data which makes up the vast majority of proChIPdb data. As noise levels vary across samples, the signal to noise ratio is a valuable measure for peak height. The noise level was set to the top 5% of signals at genomic positions (30–32). To reduce false-positive peaks, peaks with a signal-to-noise (S/N) ratio <1.5 were removed, along with peaks without expected bimodal shape (33).
Each peak was assigned to its nearest gene and associated operons, according to genomic position. The nearest gene was included regardless of distance, but target operons must fall within a distance cutoff of 500 bp and be transcribed downstream of the peak (Supplementary Figure S1). Operon annotations were obtained from RegulonDB (34). Although detailed algorithms for the prediction of target genes in eukaryotes exist (35), the simple operon structure of prokaryotes allows us to predict target genes by location alone.
Motif analysis from ChIP-seq/exo peaks
Apart from previously created binding motifs directly captured from cited literature sources, binding motif analysis was completed using the MEME-ChIP tool from the MEME software suite (36,37). Sequences of each binding peak were extracted from individual reference genomes, and the sequence of each binding was extended by 20 bp at each end to allow for adjacent sequences to be included in the analysis. The default MEME-ChIP settings were used, except for changes in the parameters to broaden the search results (minimum width reduced to -meme-minw = 5 bp, maximum width increased to -meme-maxw = 45 bp, and number of motifs increased to -meme-nmotifs = 4) and to filter the results to the top significant hit with E-value < 1e–3 (-filter-thresh = 0.001) If the motifs were retrieved from a publication, the motif search was analyzed by their respective methods.
Web design and implementation
The proChIPdb site was implemented using a simple web stack. The server side is hosted by GitHub Pages (pages.github.com) with an HTTPS protocol. Local computations to generate necessary files were performed in Python 3.8 with Jupyter notebooks (jupyter.org). The client side makes use of Bootstrap 4.5 (getbootstrap.com) to manage page layout, including support for mobile users. Button symbols are provided by Google Material Icons (fonts.google.com/icons). Tables were made using Tabulator (tabulator.info). The genome viewer is an igv.js element (github.com/igvteam/igv.js/) (38). Additional plots utilize HighCharts with a non-commercial license (highcharts.com). Other javascript packages used include: jQuery (jquery.com), Popper.js (popper.js.org), URLSearchParams (developer.mozilla.org), PapaParse (papaparse.com), and Intro.js (introjs.com).
RESULTS
A web-based platform for browsing prokaryotic ChIP data
We developed proChIPdb (prochipdb.org), a chromatin immunoprecipitation database for prokaryotic organisms, to vastly improve the accessibility of TFBS information for a range of TFs across diverse species. Users, such as micro- and systems biologists, are given the ability to easily search or browse through available TFs and target genes. Detailed dashboards with clickable and hoverable features provide genome-wide overviews of TF activity as well as high-resolution access to every binding peak.
To generate this database, we searched for ChIP-based literature on NCBI PubMed, collected all available processed data from the selected publications, downloaded sequence files from databases, and processed the data with the streamlined pipeline summarized in Figure 1. We curated metadata for each dataset based on published details, and captured any reported TFBSs and motifs to include in the site. We also computed our own motifs for each TF and condition, and report our results along with the published ones, where applicable. For our in-house data, we computed these features as described in Materials and Methods above. Details about the database size as of its initial release are presented in Table 1.
Figure 1.
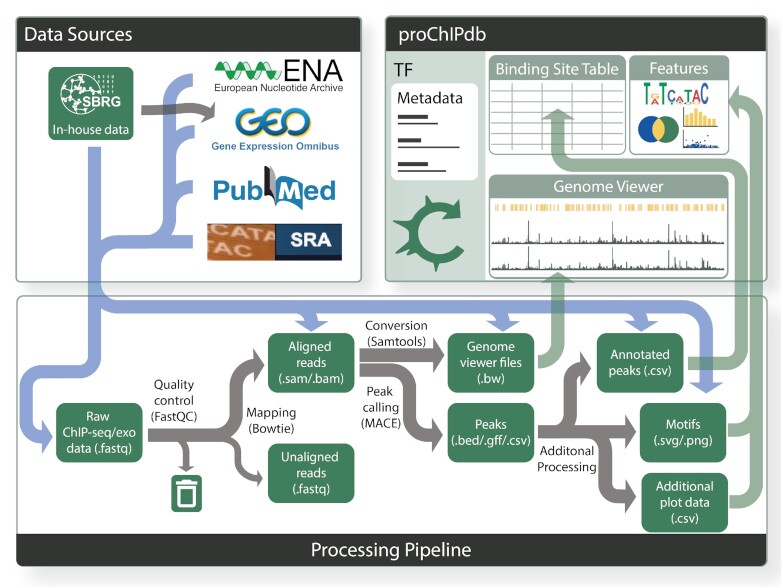
Overview of proChIPdb's sources, pipeline, and content. Gray arrows in this figure present the flow of data. Data sources (upper left) include the European Nucleotide Archive, Gene Expression Omnibus (GEO), Pubmed, the Sequence Read Archive, and in-house data (which has been posted to GEO). This data enters the processing pipeline (lower) through the various file types indicated by the blue flow arrows. Processing through the pipeline occurs from left to right via the gray arrows, with green rectangles indicating data types and files and black text indicating processing steps and tools. Towards the right of the pipeline, data feeds through the green arrows into the proChIPdb site (upper right), which consists of a binding site table, genome viewer, and feature visualization panel.
Table 1.
Key statistics for the datasets underlying proChIPdb. Individual columns to match the dataset pages for ‘Escherichia coli’ and ‘All Other Organisms’ are delineated as well as total counts across proChIPdb
| Dataset description | ChIP-seq and ChIP-exo data for E. coli | ChIP-seq and ChIP-exo data for all other organisms on proChIPdb | Total proChIP data | |
|---|---|---|---|---|
| Number of organisms | 1 | 13 | 14 | |
| Number of unique strains | 4 | 14 | 18 | |
| Number of TF pages | 65 | 35 | 100 | |
| Number of samples | ChIP-exo samples | 184 (92.9%) | 28 (38.4%) | 212 (78.2%) |
| ChIP-seq samples | 14 (7.1%) | 45 (61.6%) | 59 (21.8%) | |
| Total count | 198 | 73 | 271 | |
| Percent of TFs with curated TFBSs available | 92.3% (60 of 65) | 85.7% (30 of 35) | 90.0% (90 of 100) | |
proChIPdb can be consulted to obtain evidence of transcriptional regulation that is of broad interest to the scientific community. Its major strengths include (i) providing detailed ChIP data in an interactive and user-friendly online interface, (ii) encouraging TFBS exploration from other perspectives via external links to knowledge bases and (iii) enabling search by TFs and target genes to easily consolidate information for researchers with specific questions. A useful ‘about’ page (prochipdb.org/about.html) is provided, and all data is available for download and custom analysis. We include an email address (be-chip-pro@eng.ucsd.edu) to encourage feedback and collaboration.
Information-dense, interactive TF dashboards
At the center of proChIPdb are the TF dashboards, which consolidate information across all available samples and conditions for a single TF in a reference strain, such as the E. coli K-12 MG1655 Fur dashboard (https://prochipdb.org/tf_dashboard.html?organism=e_coli&tf=Fur&genome=NC_000913.3&i=3). The elements of the dashboard are summarized in Figure 2 and include the metadata panel for basic information and external links, the binding site table for enumerating TFBSs, the genome viewer for directly browsing ChIP data, and the feature visualization panel for summary plots, motifs, and comparisons with public regulons. The information icon, denoted with an ‘?’, adjacent to each panel header can be hovered over to view details about the content of the panel. The following sections detail the individual parts of these dashboards.
Figure 2.
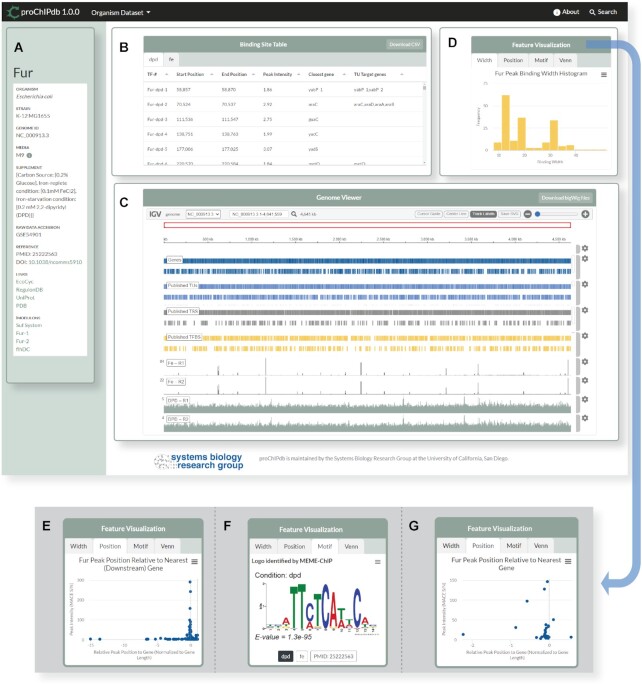
TF dashboard webpage for E. coli K-12 MG1655 Fur regulator. (A) Metadata and relevant links. (B) Binding site table with curated list of binding peaks. Each tab contains the TFBSs for a unique condition (DPD versus Fe supplementation in this example). (C) Embedded Integrative Genomics Viewer (igv.js) component with annotation tracks and genome-wide peaks from raw data. (D–G) Feature visuals, which contains tabs for the various additional plots. (D) The active tab shows a histogram of binding peak widths. (E) A scatterplot of peak positions relative to their closest downstream gene. (F) The consensus binding sequence motifs.( G) Venn diagram comparing proChIPdb identified target genes versus literature regulon genes.
Regulator metadata panel and synergy with external databases
In the left-side metadata panel, metadata and relevant links are provided (Figure 2A). These include the organism name, strain, reference genome, media used and supplement(s) added, if applicable. Information buttons adjacent to the media and supplement details provide additional details about the condition used. In the Fur example, two separate conditions were used: iron supplementation and 2,2-dipyridyl (DPD) supplementation. These conditions were selected because the former suppresses Fur binding and the latter activates it (30). By providing several conditions, this dashboard is particularly informative about how various relevant conditions can affect binding. There is also a raw data accession number and DOI to the relevant publication in this panel.
The metadata panel also provides relevant links to external databases, including: EcoCyc (39), RegulonDB (24,40) Uniprot (41), the Protein Data Bank (42), Pseudomonas Genome DB (43) and AureoWiki (44). This enables users to easily acquire up to date knowledge about the TF, including any available structural information.
The final section of the metadata panel contains links to iModulonDB (imodulondb.org) (14), which can provide a different perspective on the regulon. iModulons are machine learning derived groups of independently modulated genes which appear as coherent signals in transcriptomic datasets (45–47). Often, iModulons will have a 1:1 mapping with regulons. In other cases (such as Fur), they may capture the effects of combinations of regulators or separate non-linear responses into multiple iModulons. If the regulator's signal is not particularly strong for the transcriptomic dataset's particular environmental conditions, then it may not appear as an iModulon at all. Understanding how regulatory molecules such as those explored on proChIPdb generate transcriptomic signals is an active area of research, especially since regulator binding alone has been unable to predict expression data (48). For this reason, it may be quite valuable to compare the target genes enumerated by proChIPdb with those of the iModulons in the provided links. For instance, the case may be that the genes with the strongest binding are also co-regulated in an iModulon. Alternatively, the target genes which share regulation with another TF may end up more strongly regulated by the latter as evidenced by iModulon membership.
TFBS enumeration and interactive genome viewer
The first main dashboard element is the binding site table viewer (Figure 2B). Tabs across the top of the panel allow users to select from the available conditions (iron (Fe) and DPD in the case of Fur). The tables list each of the observed binding peak locations, which are individually identified using a ‘TF-condition-#’ identifier such as Fur-dpd-1. Identification numbers are assigned by genome location. The genomic location, peak intensity in S/N ratio, closest gene, and all target genes potentially regulated operons are each included as rows. Elements of the table are filled in from existing literature where reported, or computed and curated as described in Materials and Methods. Each of the columns can be selected to allow for sorting by that value; for example, it may be useful to sort the table by binding peak intensity to see the strongest binding events or by gene names if a particular gene is of interest. In most cases, the transcriptional unit is a hyperlink to the corresponding operon page on RegulonDB (34). This allows users to find promoter diagrams and relevant citations for each set of target genes.
Below the table, an IGV.js (38) genome viewer provides a whole genome view of the ChIP data at the core of proChIPdb (Figure 2C). A top toolbar enables genome navigation, various utilities such as a cursor guide, and the ability to save the current view as an SVG file. The first four tracks present various genomic annotations: genes, published transcriptional units (TUs), published transcription start sites (TSSs), and published TFBSs, which are sourced from the Bitome's database of genomic features (49) that are expanded from RegulonDB (34). By default, the viewer shows the entirety of the prokaryote's genome and allows the user to quickly see which sites may be of interest.
Users can manually zoom in by interacting with the viewer controls or by clicking on a row in the binding site table to jump to the binding peak in the viewer. At this zoomed in view, users can quickly acquire valuable information about which genes are near the TFBS, which known TFs may be interacting, and how binding may change depending on the various conditions present. Users can also see the specific nucleotide sequence of the binding site when the viewer is sufficiently zoomed in. It is important to compare the peak to the surrounding noise and pay special attention to the y-axis labels of the read tracks, as high relative baselines and low overall read numbers may indicate fairly weak binding.
Additional feature visualization
The top right corner of the dashboard contains a panel with four tabs, each of which present insights across all the observed peaks (Figure 2D). The plots can be downloaded in various file formats by clicking the menu button in the upper right. The first tab is a histogram of binding widths, which provides important evidence for the length of the TF’s recognized DNA sequences. Each bar in the plot can be hovered over to learn its exact height and bin bounds.
The second tab shows the peak position relative to the nearest target gene as a scatter plot (Figure 2E). Its x-axis is centered around each target gene's start point and normalized to the length of the gene, such that 0 would indicate the first nucleotide of the coding sequence and 1 would indicate the last nucleotide (with respect to the coding direction). The y-axis is the S/N ratio of the peak. Each peak is placed according to its maximum binding strength nucleotide. Hovering over the points will reveal the corresponding peak, gene and values. This plot allows users to determine the distance of the TFBS from the gene, giving various clues about function: whether it is located within a promoter, inside a coding sequence itself, or some distance before the gene.
The third tab presents the enriched motif(s) as sequence logo images, if applicable (Figure 2F). Sub-tabs across the bottom of the panel correspond to each of the conditions represented in the dashboard; they can be clicked on to view the motif we computed for the condition. If available, the motif computed by the source publication is also presented under an additional tab labeled by the source publication's PMID. The menu button in the upper right corner enables download of both the sequence logo image and the underlying position weight matrix (PWM). The provided motifs may be extremely valuable for predicting TFBSs in new sequences and can also be compared to previously published consensus motifs. For additional tools enabling motif analysis, see MEME and JASPAR (8,37).
Finally, the Venn diagram tab presents a comparison between proChIPdb's target genes and the corresponding published regulon (such as those available on EcoCyc (39) (Figure 2G). Hovering over a section of the Venn diagram will list the specific genes that fall into each subset. Because evidence of regulation exists both on proChIPdb and in published regulons, the genes in the overlapping portion have very strong evidence of regulation. The genes in literature alone may reflect binding under different conditions, while genes in proChIPdb alone are opportunities to expand the target genes as evidence is added from other data sources.
Search by TF and/or target gene
At the upper left corner of every page on proChIPdb, there is a link to our search page. There, users can enter a TF name, gene name, gene locus, PMID, or accession number to receive a list of all TF dashboards that may be of interest to them. If the search term matches any TFs, those will be listed first along with the organism and strain. Below that, any gene whose name or locus matches the search term and is also the target of a TF will be listed. If a synonym of one of our genes is searched, the results will show the proChIPdb preferred name as well as the synonym which matched the search term. Gene results include additional information, such as which TF binds nearby and the S/N intensity of the peak. Clicking an element will link users to the dashboard for the potentially regulating TF, allowing them to quickly access the data for the peak.
By enabling gene search, proChIPdb provides a powerful workflow for exploring ChIP data. For any gene, users can quickly learn not only which regulators it interacts with, but also directly explore the data underlying those interactions. This more nuanced view of the relative strength and condition-specificity of regulatory interactions will empower more detailed and predictive TRN elucidation.
FUTURE DIRECTIONS
proChIPdb will continue to grow as ChIP datasets are generated. The initial release presented here is a valuable step forward for the dissemination of this valuable data type, but additional features should be added to increase the ease of analysis. These include the implementation of a robust back-end to process submitted user data with respect to our database (for instance, CentriMo functionality (50)), search by DNA or protein sequence, and motif comparison tools (51,52). Though RNA-seq data is currently out of the scope of proChIPdb, evidence of significant expression changes in target genes upon TF knockout would be a valuable inclusion. We also plan to add a ‘Frequently Asked Questions’ section to our about page after receiving user questions through our provided email (be-chip-pro@eng.ucsd.edu).
CONCLUSIONS
As ChIP data is highly informative, but unfortunately laborious and expensive to generate, it is paramount that it be disseminated broadly in a manner that is easy to search, browse, and download. proChIPdb is an information-rich, interactive database of prokaryotic ChIP data that meets this need and allows researchers to define regulons from the bottom up, both through a nuanced understanding of TFBSs across whole genomes and through synergy with other databases. The ability to search the proChIPdb database for target genes to quickly acquire evidence of TF binding will empower anyone with a particular interest in a gene's promoter region. The nucleotide resolution of our genome viewer and pre-called binding peaks make the workflow for perusing ChIP data simple. The additional plots put summary information such as binding width and motif analysis directly into users’ hands. proChIPdb should be regularly updated and expanded with newly available prokaryotic ChIP data to ultimately achieve a comprehensive database of protein–DNA interactions. The existence of this information-rich and easily browsable database should motivate advances in ChIP technology development and empower a deep understanding of TRNs that will advance the life sciences.
DATA AVAILABILITY
proChIPdb is freely available at https://prochipdb.org and can be accessed with a JavaScript-enabled browser. The download links in the toolbars throughout the website enable download of all data and facilitate custom analysis. In addition, all database files can be downloaded as a tarball from Zenodo (https://zenodo.org/record/5168081) or accessed from the public GitHub repository (https://github.com/SBRG/ChIPdb).
ACCESSION NUMBERS
All accession numbers are provided in Supplementary Table S1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Patrick Phaneuf, Dr Anand Sastry, Dr Jongoh Shin, Dr Donghui Choe, and Ina Bang for informative discussions and Marc Abrams for reviewing and editing the manuscript.
Author contributions: K.T.D. led database development. Y.G., D.K. and B.K.C. generated and acquired data. K.R. drafted the paper. K.T.D., K.R., S.M.C. and T.A. wrote software to process and visualize data. B.O.P. provided mentorship, conception, resources and guidance in planning and implementation. All authors participated in writing the paper.
Contributor Information
Katherine T Decker, Department of Bioengineering, University of California, San Diego, La Jolla, CA, 92093, USA.
Ye Gao, Department of Bioengineering, University of California, San Diego, La Jolla, CA, 92093, USA.
Kevin Rychel, Department of Bioengineering, University of California, San Diego, La Jolla, CA, 92093, USA.
Tahani Al Bulushi, Department of Bioengineering, University of California, San Diego, La Jolla, CA, 92093, USA.
Siddharth M Chauhan, Department of Bioengineering, University of California, San Diego, La Jolla, CA, 92093, USA.
Donghyuk Kim, School of Energy and Chemical Engineering, Ulsan National Institute of Science and Technology, Ulsan 44919, Korea.
Byung-Kwan Cho, Department of Biological Sciences and KI for the BioCentury, Korea Advanced Institute of Science and Technology, Daejeon, 34141, Republic of Korea.
Bernhard O Palsson, Department of Bioengineering, University of California, San Diego, La Jolla, CA, 92093, USA; Department of Pediatrics, University of California, San Diego, La Jolla, CA, 92093, USA; Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Building 220, Kemitorvet, 2800 Kgs. Lyngby, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novo Nordisk Foundation [NNF10CC1016517]. Funding for open access charge: Novo Nordisk Foundation [NNF10CC1016517].
Conflict of interest statement. None declared.
REFERENCES
- 1. Raha D., Hong M., Snyder M.. ChIP-Seq: a method for global identification of regulatory elements in the genome. Curr. Protoc. Mol. Biol. 2010; Chapter 21:Unit 21.19.1–Unit 21.19.14. [DOI] [PubMed] [Google Scholar]
- 2. Rhee H.S., Pugh B.F.. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr. Protoc. Mol. Biol. 2012; Chapter 21:Unit 21.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gao Y., Yurkovich J.T., Seo S.W., Kabimoldayev I., Dräger A., Chen K., Sastry A.V., Fang X., Mih N., Yang L.et al.. Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2018; 46:10682–10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eckweiler D., Dudek C.-A., Hartlich J., Brötje D., Jahn D.. PRODORIC2: the bacterial gene regulation database in 2018. Nucleic Acids Res. 2018; 46:D320–D326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pachkov M., Balwierz P.J., Arnold P., Ozonov E., van Nimwegen E.. SwissRegulon, a database of genome-wide annotations of regulatory sites: recent updates. Nucleic Acids Res. 2013; 41:D214–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yevshin I., Sharipov R., Valeev T., Kel A., Kolpakov F.. GTRD: a database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 2017; 45:D61–D67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Czipa E., Schiller M., Nagy T., Kontra L., Steiner L., Koller J., Pálné-Szén O., Barta E.. ChIPSummitDB: a ChIP-seq-based database of human transcription factor binding sites and the topological arrangements of the proteins bound to them. Database. 2020; 2020:baz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranašić D.et al.. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020; 48:D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spivakov M., Akhtar J., Kheradpour P., Beal K., Girardot C., Koscielny G., Herrero J., Kellis M., Furlong E.E.M., Birney E.. Analysis of variation at transcription factor binding sites in Drosophila and humans. Genome Biol. 2012; 13:R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ballester B., Medina-Rivera A., Schmidt D., Gonzàlez-Porta M., Carlucci M., Chen X., Chessman K., Faure A.J., Funnell A.P.W., Goncalves A.et al.. Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife. 2014; 3:e02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grimes T., Potter S.S., Datta S.. Integrating gene regulatory pathways into differential network analysis of gene expression data. Sci. Rep. 2019; 9:5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J., Liu J., Lee D., Lou S., Chen Z., Gürsoy G., Gerstein M.. DiNeR: a Differential graphical model for analysis of co-regulation network rewiring. BMC Bioinformatics. 2020; 21:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fahrenbach J.P., Andrade J., McNally E.M.. The CO-Regulation Database (CORD): a tool to identify coordinately expressed genes. PLoS One. 2014; 9:e90408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rychel K., Decker K., Sastry A.V., Phaneuf P.V., Poudel S., Palsson B.O.. iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning. Nucleic Acids Res. 2021; 49:D112–D120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lundberg S.M., Tu W.B., Raught B., Penn L.Z., Hoffman M.M., Lee S.-I.. ChromNet: learning the human chromatin network from all ENCODE ChIP-seq data. Genome Biol. 2016; 17:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao Y., Lim H.G., Verkler H., Szubin R., Quach D., Rodionova I., Chen K., Yurkovich J.T., Cho B.K., Palsson B.O.. Unraveling the functions of uncharacterized transcription factors in Escherichia coli using ChIP-exo. Nucleic Acids Res. 2021; 49:9696–9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao Y., Poudel S., Seif Y., Shen Z.Y., Palsson B.O.. Elucidating the CodY regulon in Staphylococcus aureus USA300 substrains. 2021; bioRxiv doi:09 January 2021, preprint: not peer reviewed 10.1101/2021.01.08.426013. [DOI] [PMC free article] [PubMed]
- 18. Myers K.S., Park D.M., Beauchene N.A., Kiley P.J.. Defining bacterial regulons using ChIP-seq. Methods. 2015; 86:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peano C., Wolf J., Demol J., Rossi E., Petiti L., De Bellis G., Geiselmann J., Egli T., Lacour S., Landini P.. Characterization of the Escherichia coli σ(S) core regulon by Chromatin Immunoprecipitation-sequencing (ChIP-seq) analysis. Sci. Rep. 2015; 5:10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choudhary K.S., Kleinmanns J.A., Decker K., Sastry A.V., Gao Y., Szubin R., Seif Y., Palsson B.O.. Elucidation of regulatory modes for five two-component systems in escherichia coli reveals novel relationships. mSystems. 2020; 5:e00980-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hashim F.A., Mabrouk M.S., Al-Atabany W.. Review of different sequence motif finding algorithms. Avicenna J. Med. Biotechnol. 2019; 11:130–148. [PMC free article] [PubMed] [Google Scholar]
- 22. MacIsaac K.D., Fraenkel E.. Sequence analysis of chromatin immunoprecipitation data for transcription factors. Methods Mol. Biol. 2010; 674:179–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muñiz-Rascado L., García-Sotelo J.S., Alquicira-Hernández K., Martínez-Flores I., Pannier L., Castro-Mondragón J.A.et al.. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016; 44:D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Athar A., Füllgrabe A., George N., Iqbal H., Huerta L., Ali A., Snow C., Fonseca N.A., Petryszak R., Papatheodorou I.et al.. ArrayExpress update - from bulk to single-cell expression data. Nucleic Acids Res. 2019; 47:D711–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leinonen R., Sugawara H., Shumway M.International Nucleotide Sequence Database Collaboration . The sequence read archive. Nucleic. Acids. Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang L., Chen J., Wang C., Uusküla-Reimand L., Chen K., Medina-Rivera A., Young E.J., Zimmermann M.T., Yan H., Sun Z.et al.. MACE: model based analysis of ChIP-exo. Nucleic Acids Res. 2014; 42:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo S.W., Kim D., Latif H., O’Brien E.J., Szubin R., Palsson B.O.. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 2014; 5:4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seo S.W., Kim D., Szubin R., Palsson B.O.. Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in escherichia coli K-12 MG1655. Cell Rep. 2015; 12:1289–1299. [DOI] [PubMed] [Google Scholar]
- 32. Ogasawara H., Ohe S., Ishihama A.. Role of transcription factor NimR (YeaM) in sensitivity control of Escherichia coli to 2-nitroimidazole. FEMS Microbiol. Lett. 2015; 362:1–8. [DOI] [PubMed] [Google Scholar]
- 33. Kharchenko P.V., Tolstorukov M.Y., Park P.J.. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008; 26:1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Santos-Zavaleta A., Salgado H., Gama-Castro S., Sánchez-Pérez M., Gómez-Romero L., Ledezma-Tejeida D., García-Sotelo J.S., Alquicira-Hernández K., Muñiz-Rascado L.J., Peña-Loredo P.et al.. RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 2019; 47:D212–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O’Connor T., Grant C.E., Bodén M., Bailey T.L.. T-Gene: improved target gene prediction. Bioinformatics. 2020; 36:3902–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic. Acids. Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robinson J.T., Thorvaldsdóttir H., Turner D., Mesirov J.P.. igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). 2020; bioRxiv doi:05 May 2020, preprint: not peer reviewed 10.1101/2020.05.03.075499. [DOI] [PMC free article] [PubMed]
- 39. Keseler I.M., Mackie A., Santos-Zavaleta A., Billington R., Bonavides-Martínez C., Caspi R., Fulcher C., Gama-Castro S., Kothari A., Krummenacker M.et al.. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017; 45:D543–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santos-Zavaleta A., Sánchez-Pérez M., Salgado H., Velázquez-Ramírez D.A., Gama-Castro S., Tierrafría V.H., Busby S.J.W., Aquino P., Fang X., Palsson B.O.et al.. A unified resource for transcriptional regulation in Escherichia coli K-12 incorporating high-throughput-generated binding data into RegulonDB version 10.0. BMC Biol. 2018; 16:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The protein data bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S.L.. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016; 44:D646–D653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fuchs S., Mehlan H., Bernhardt J., Hennig A., Michalik S., Surmann K., Pané-Farré J., Giese A., Weiss S., Backert L.et al.. AureoWiki ? The repository of the Staphylococcus aureus research and annotation community. Int. J. Med. Microbiol. 2018; 308:558–568. [DOI] [PubMed] [Google Scholar]
- 45. Sastry A.V., Gao Y., Szubin R., Hefner Y., Xu S., Kim D., Choudhary K.S., Yang L., King Z.A., Palsson B.O.. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat. Commun. 2019; 10:5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rychel K., Sastry A.V., Palsson B.O.. Machine learning uncovers independently regulated modules in the Bacillus subtilis transcriptome. Nat. Commun. 2020; 11:6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sastry A.V., Poudel S., Rychel K., Yoo R., Lamoureux C.R., Chauhan S., Haiman Z.B., Al Bulushi T., Seif Y., Palsson B.O.. Mining all publicly available expression data to compute dynamic microbial transcriptional regulatory networks. 2021; bioRxiv doi:02 July 2021, preprint: not peer reviewed 10.1101/2021.07.01.450581. [DOI]
- 48. Fang X., Sastry A., Mih N., Kim D., Tan J., Yurkovich J.T., Lloyd C.J., Gao Y., Yang L., Palsson B.O.. Global transcriptional regulatory network for Escherichia coli robustly connects gene expression to transcription factor activities. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:10286–10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lamoureux C.R., Choudhary K.S., King Z.A., Sandberg T.E., Gao Y., Sastry A.V., Phaneuf P.V., Choe D., Cho B.-K., Palsson B.O.. The Bitome: digitized genomic features reveal fundamental genome organization. Nucleic Acids Res. 2020; 48:10157–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bailey T.L., Machanick P.. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012; 40:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S.. Quantifying similarity between motifs. Genome Biol. 2007; 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mahony S., Benos P.V.. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007; 35:W253–W258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
proChIPdb is freely available at https://prochipdb.org and can be accessed with a JavaScript-enabled browser. The download links in the toolbars throughout the website enable download of all data and facilitate custom analysis. In addition, all database files can be downloaded as a tarball from Zenodo (https://zenodo.org/record/5168081) or accessed from the public GitHub repository (https://github.com/SBRG/ChIPdb).