Abstract
Genome-wide association studies (GWAS) have identified thousands of non-coding single-nucleotide polymorphisms (SNPs) associated with human traits and diseases. However, functional interpretation of these SNPs remains a significant challenge. Our recent study established the concept of 3′ untranslated region (3′UTR) alternative polyadenylation (APA) quantitative trait loci (3′aQTLs), which can be used to interpret ∼16.1% of GWAS SNPs and are distinct from gene expression QTLs and splicing QTLs. Despite the growing interest in 3′aQTLs, there is no comprehensive database for users to search and visualize them across human normal tissues. In the 3′aQTL-atlas (https://wlcb.oit.uci.edu/3aQTLatlas), we provide a comprehensive list of 3′aQTLs containing ∼1.49 million SNPs associated with APA of target genes, based on 15,201 RNA-seq samples across 49 human Genotype-Tissue Expression (GTEx v8) tissues isolated from 838 individuals. The 3′aQTL-atlas provides a ∼2-fold increase in sample size compared with our published study. It also includes 3′aQTL searches by Gene/SNP across tissues, a 3′aQTL genome browser, 3′aQTL boxplots, and GWAS-3′aQTL colocalization event visualization. The 3′aQTL-atlas aims to establish APA as an emerging molecular phenotype to explain a large fraction of GWAS risk SNPs, leading to significant novel insights into the genetic basis of APA and APA-linked susceptibility genes in human traits and diseases.
INTRODUCTION
Genome-wide association studies (GWAS) have identified >100 000 single-nucleotide polymorphisms (SNPs) associated with complex traits and diseases in humans. However, the functional interpretation the effects of these SNPs is difficult, as the SNPs often lie in non-coding regions and do not directly affect protein-coding regions. To bridge the gap between GWAS non-coding variants and human phenotypes, a quantitative trait locus (QTL)-based analysis has been used to evaluate the effects on various molecular phenotypes. In particular, gene expression (eQTL) and splicing (sQTL) analyses have successfully explained the target genes and functional mechanisms of numerous GWAS risk loci (1–4). Despite massive efforts on eQTLs and sQTLs, a large fraction of GWAS risk loci remains unexplained (5,6).
Alternative polyadenylation (APA), which occurs in >70% of human genes, is a major mechanism of post-transcriptional regulation under diverse biological conditions, tissues and cell types (7–10). By changing the position of polyadenylation sites, APA can generate transcripts with either long or short 3′ untranslated regions (3′UTRs) that contain different cis-regulatory elements, such as binding sites of RNA-binding proteins or miRNAs. This leads to altered translation efficiency, cellular localization, and stability of transcripts (9,11) independent of gene expression or splicing. Disruption of the key APA regulators (e.g. PABPN1, CDK12 and NUDT21) leading to global APA changes has been linked to serious human diseases, including oculopharyngeal muscular dystrophy (12), neuropsychiatric disease (13), leukemia (14), and glioblastoma (15).
In addition, mounting evidence suggests that genetic variations that affect APA usage in individual genes can confer the risks of many diseases, including Parkinson's disease (16), systemic lupus erythematosus (17,18), multiple cancer types (19), and diabetes (20,21). For example, a common variant (rs356165) at the 3′UTR of SNCA (coding α-synuclein protein) can increase SNCA long 3′UTR usage, which enhances the accumulation of α-synuclein protein in mitochondria and further contributes to a high risk of Parkinson's disease (16). Similarly, rs10954213 at the IRF5 3′UTR locus can shorten the 3′UTR of IRF5, which alters mRNA stability and further results in high systemic lupus erythematosus susceptibility (17). Taking the advantage of large-scale transcriptome and genotype data from the Genotype-Tissue Expression (GTEx) project (version 7), our recent study established the concept of 3′UTR APA quantitative trait loci (3′aQTLs), which can be used to interpret ∼16.1% of GWAS SNPs and are largely distinct from eQTLs and sQTLs (22). 3′aQTLs provide consequential supplements to the functional interpretation of non-coding variants. Despite the growing interest in 3′aQTLs, there is no comprehensive database for users to search and visualize these 3′aQTLs across human normal tissues.
We developed a database for 3′aQTLs, termed 3′aQTL-atlas. 3′aQTL-atlas contains ∼1.49 million SNPs associated with the APA of target genes, based on 15,201 RNA-seq samples across 49 human GTEx (version 8) tissues isolated from 838 individuals. The 3′aQTL-atlas not only provides a ∼2-fold increase in sample size compared with our published study (22) but also includes 3′aQTL searches by Gene/SNP across tissues, a 3′aQTL genome browser, 3′aQTL boxplots, and GWAS-3′aQTL colocalization event visualization. The 3′aQTL-atlas aims to establish APA as an emerging and important molecular phenotype to explain a large fraction of GWAS risk SNPs, leading to significant novel biological insights into the genetic basis of APA and APA-linked susceptibility genes in a wide spectrum of human traits and diseases.
DATA COLLECTION AND PROCESSING
GTEx data collection and processing
We downloaded the RNA-seq BAM files of 17 382 human normal samples across 54 tissues in 948 individuals from the GTEx project (dbGaP, phs000424.v8.p2) (2). The original RNA-seq reads were aligned to the human genome (hg38/GRCh38) using STAR v2.5.3a (23), with the alignment parameters described in the GTEx study (2). We removed the BAM files that were either generated from diseased tissues and or tissue types with small sample sizes. We also removed RNA-seq BAM files from individuals without genotype data, which were not included in the GTEx analysis freeze. The remaining BAM files were then sorted and converted into bedGraph format using bedtools v2.17.0 (24). Genotype data from the GTEx v8 release were called from whole-genome sequencing (WGS) data (2). Briefly, WGS reads were aligned to the human genome (hg38/GRCh38) with BWA (25). Variants in the variant call format (VCF) file were called using GATK HaplotypeCaller v3.5 (26). After filtering low-quality samples by the GTEx Consortium, the final analysis freeze set contained variants called from 838 donors. The final variants were imputed and phased using SHAPEIT v2 (27). The associated sample description files were downloaded from the GTEx Portal (www.gtexportal.org).
Quantification of APA usage using DaPars2
Our previously developed DaPars2 (28) was used to calculate relative APA usage, which was measured by the percentage of distal polyA site usage index (PDUI), from standard RNA-seq data. DaPars2 applies a two-normal mixture model to allow for the joint quantification of APA usage for multiple samples (28). Using the University of California Santa Cruz (UCSC) Table Browser (29), we first downloaded the human genes annotation file (hg38/GRCh38). Then, the script ‘DaPars_Extract_Anno.py’ was used to extract a 3′UTR annotation for each transcript. Afterwards, we calculated the sequencing depth for each sample, which was used as an input of DaPars2 for normalizing the sequencing depth difference of samples, by SAMtools v1.9 (25). Finally, multiple RNA-seq samples were jointly analyzed to identify de novo APA sites and to calculate the APA usage of each transcript in each sample with DaPars2 (28).
3′aQTL mapping for each tissue
3′aQTL analysis was performed separately for each tissue, using the genotype and normalized PDUI values as previously described (Figure 1A) (22). Briefly, we split the VCF data for each tissue with BCFtools (25) and further transformed them into genotype matrix files using BioAlcidae v.2.27.1 (30). Only variants with a minor allele frequency ≥ 0.01 were included in further analyses. For each tissue type, we used linear regression by MatrixEQTL (31) to test the association between normalized PDUI values and SNPs within a 1-Mb interval of the 3′UTR region. Both known covariates (e.g. sex, RNA integrity number [RIN], platform, and top five genotype principal components) and unobserved covariates calculated by PEER (32) were included in the analysis. The number of PEER covariates for each tissue was selected based on suggestions from the GTEx Consortium: 15, 30 and 35 PEER factors were chosen for tissue sample sizes of <150, 150–250 and >250, respectively (1). By randomly sampling individual labels, 1000 rounds of permutation analysis were conducted to obtain empirical P-values for each APA gene. Then, these P-values were adjusted with the R package qvalue v.2.0.0 (33).
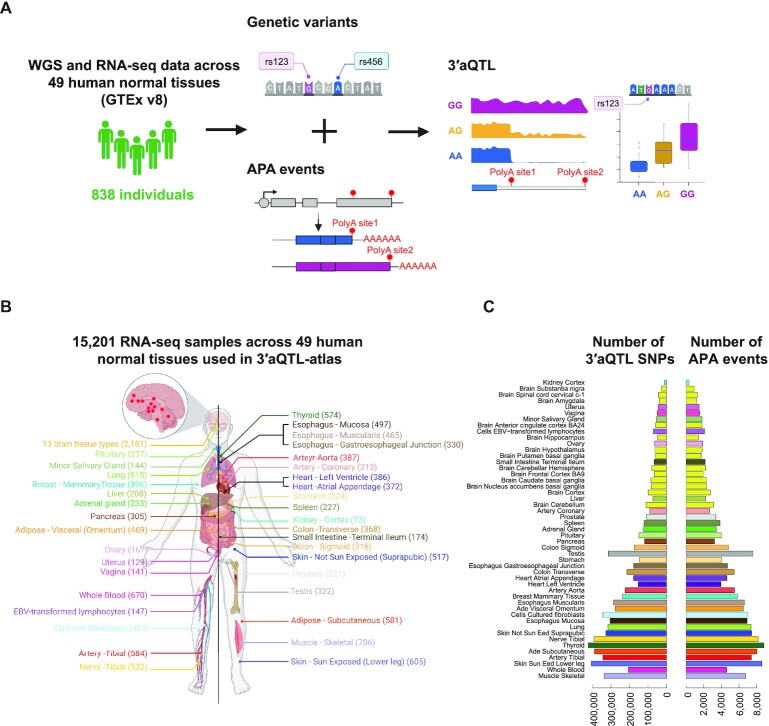
Figure 1.
Data processing and data statistics in 3′aQTL-atlas. (A) Schematic of overall data processing in the 3′aQTL-atlas. (B) Distribution of the number of RNA-seq samples for each tissue used in the 3′aQTL-atlas. (C) Distribution of the number of APA events and significant 3′aQTL SNPs (FDR ≤ 0.05) for each tissue, sorted by the tissue sample sizes. Each color code indicates a tissue of origin. APA, alternative polyadenylation; WGS, whole-genome sequencing; 3′aQTL, 3′ untranslated region APA quantitative trait loci.
Identification of GWAS-associated 3′aQTLs
To identify human diseases and traits associated with SNPs, we obtained GWAS risk SNPs (referred to as tag SNPs) from the National Human Genome Research Institute GWAS catalog (34) (accessed 1 June 2021). SNPs with no dbSNP accessions were removed. Previous studies have suggested that causal variants are often not the tag SNPs themselves, but the SNPs in linkage disequilibrium (LD) with tag SNPs (35,36). Thus, we extracted a list of SNPs that were in strong LD of European ancestry with the GWAS catalog tag SNP (referred to as LD SNPs). Using the LD cutoff of r2 ≥ 0.8, we finally identified 1 711 210 LD SNPs and tag SNPs, which we refer to as GWAS-associated SNPs. GWAS-associated 3′aQTLs were defined when the lead 3′aQTLs were overlapped with these GWAS-associated SNPs.
Construction of the 3′aQTL-atlas website
The 3′aQTL-atlas website was constructed on a Linux-based Apache Web server (http://www.apache.org). All processed and annotated data in the 3′aQTL-atlas were stored in MySQL. The R package LocusCompare (37) and in-house R scripts were used to perform data analyses and data plotting. The interactive web pages were implemented using HTML, CSS, JavaScript and PHP languages, with several JavaScript libraries (JQuery.js, DataTable.js, NG-Circos.js and IGV.js) and Bootstrap framework (a popular framework for developing interactive websites). The 3′aQTL-atlas is freely available and does not require registration or login for access.
RESULTS
Sample summary and 3′aQTL landscape of human tissues
In this version of the 3′aQTL-atlas, we analyzed 15,201 RNA-seq samples across 49 human normal tissues from GTEx version 8 (Figure 1A, B). The RNA-seq sample sizes for each tissue ranged from 73 in the kidney cortex to 706 in skeletal muscle, with a median of 310 (Figure 1B). With the FDR <0.05, a total of 1.49 million common genetic variants associated with 3′aQTLs were identified; the median was 30 427 variants per tissue type, with a minimum of 9938 in the kidney cortex and a maximum of 424 628 in thyroid (Figure 1C). The number of 3′aQTL APA events was highly correlated (Pearson correlation P-value < 2.2e−16, r = 0.91) with the sample size in each tissue (Figure 1C and Supplementary Figure S1). The strong correlation between 3′aQTL number and sample size suggests that more 3′aQTLs will continue to be identified as additional RNA-seq datasets become available.
Data searching, browsing, and visualizing by four modules
We developed a user-friendly website (3′aQTL-atlas; https://wlcb.oit.uci.edu/3aQTLatlas) for searching, browsing, and visualizing 1.49 million common genetic variants associated with 3′aQTLs across 49 human normal tissues. The 3′aQTL-atlas consists of four modules (Figure 2A): a 3′aQTL search by Gene/SNP (Figure 2B), 3′aQTL genome browser (Figure 2C), 3′aQTL boxplot (Figure 2D), and GWAS-3′aQTL colocalization event visualization (Figure 2E). In addition, a list of GWAS-associated 3′aQTLs are also provided for users to deeply investigate the mechanisms of 3′aQTLs in human traits and diseases.
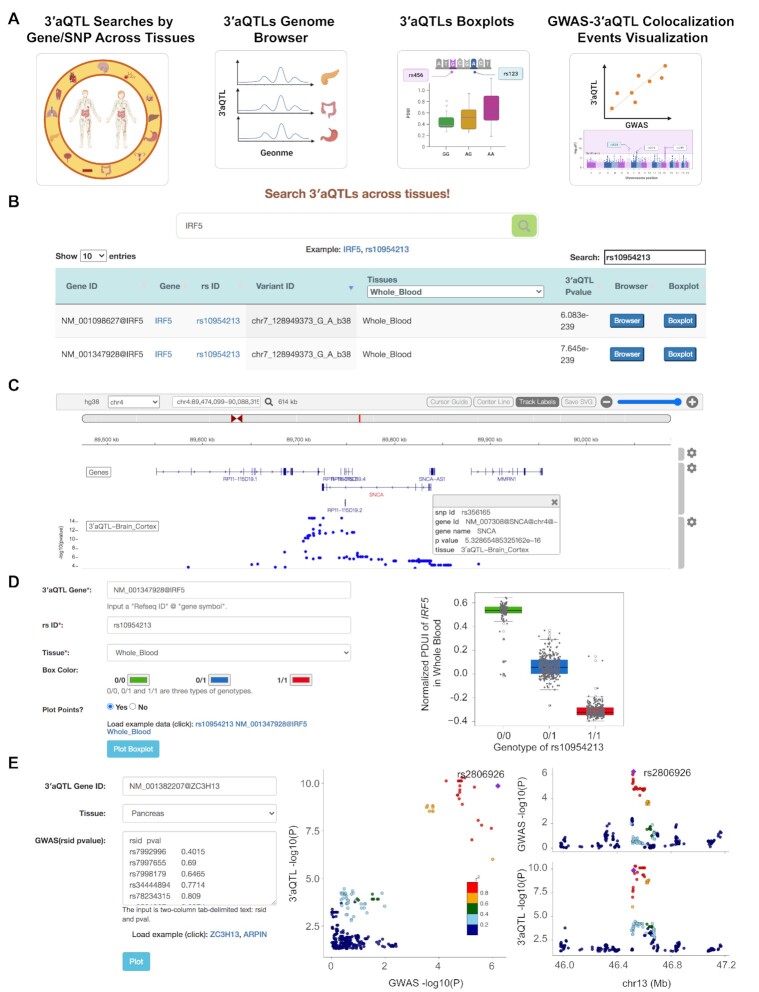
Figure 2.
Web interface of 3′aQTL-atlas. (A) 3′aQTL-atlas consists of four modules: 3′aQTL search by Gene/SNP, 3′aQTL genome browser, 3′aQTL boxplot, and GWAS-3′aQTL colocalization event visualization. (B) 3′aQTL query interface and sample results in the ‘3′aQTL search by Gene/SNP’ module. (C) An example of the genome browser view shows the 3′aQTLs of brain cortex tissue at the SNCA locus. (D) Interface of the ‘3′aQTL boxplot’ module and an example of the 3′aQTL boxplot for IRF5 and rs10954213 in whole blood. (E) Interface of the ‘GWAS-3′aQTL colocalization event visualization’ module and an example of the LocusCompare plot at the ZC3H13 locus with T2D GWAS P-values and 3′aQTL P-values in pancreas tissue. GWAS, genome-wide association studies; SNP, single-nucleotide polymorphism; WGS, whole-genome sequencing; 3′aQTL, 3′ untranslated region alternative polyadenylation quantitative trait loci.
In the ‘3′aQTL search by Gene/SNP’ module, users can search the 3′aQTLs across 49 human tissues using the gene name or SNP rs ID. It will return a table with the RefSeq transcript ID, gene symbol, SNP rs ID, chromosome position, tissue types, and P-value of each 3′aQTL item for the queried gene or SNP. For example, querying IRF5 will return 12 308 significant 3′aQTL items. Users can further filter these 3′aQTLs by selecting tissue names in the ‘Tissues’ column (e.g. Whole Blood) or inputting custom filter key words (e.g. rs10954213) into the ‘Search’ field at the top-right corner of the table (Figure 2B). We also provide the ‘Browser’ and ‘Boxplot’ button for each 3′aQTL item to allow users to visualize the 3′aQTL item in the genome browser and boxplot figures.
In the ‘3′aQTL genome browser’ module, users can explore the 3′aQTLs across human tissues in an interactive genome browser using the gene symbol (e.g., IRF5), SNP rs ID (e.g. rs10954213), or genome position (e.g. chr7:128687612–129200035). For example, by searching ‘SNCA’ in the genome browser, we can find all significant 3′aQTLs for SNCA (blue points) in Brain Cortex tissue, which confirms previous results that common variants can change the APA usage of SNCA in the brain (16) (Figure 2C). Clicking the dot of interest will show details of the SNP, including rs ID and P-value (Figure 2C). Only 3′aQTLs of the queried gene are labeled in blue in the genome browser, whereas 3′aQTLs of other genes are labeled in grey. The genome browser also provides gene structure annotation, GWAS catalog risk SNPs (34), and PolyA_DB3 polyA sites (38) tracks, which allow users to integrate these data with 3′aQTLs. In addition, users can download the figures of the browser tracks in SVG format by clicking on the ‘Save SVG’ button at the top-right corner of the genome browser.
In the ‘3′aQTL boxplot’ module, we designed an online tool that allows users to customize boxplots for each 3′aQTL. For example, users can draw the boxplot by inputting the gene ID (e.g. NM_001347928@IRF5), rs ID (e.g. rs10954213), and tissue name (e.g. Whole Blood) (Figure 2D). The color of the boxplot is user defined, and the whole plot can be downloaded as a publishable PDF document.
In the ‘GWAS-3′aQTL colocalization event visualization’ module, we provide an online server for the widely used R package LocusCompare (37), which allows users to visualize GWAS-3′aQTLs colocalization events using their own GWAS data. For example, by inputting the gene ID (e.g. NM_001382207@ZC3H13), tissue name (e.g. Pancreas), and a two-column text with the rs ID and corresponding P-value, users can visualize the GWAS-3′aQTLs colocalization event at the ZC3H13 locus (Figure 2E). The plots can also be downloaded in PDF format.
To link 3′aQTL variants to human genetic traits and diseases, we also provide a list of GWAS-associated 3′aQTLs, which were defined when the lead 3′aQTL variants are the GWAS catalog (34) tag SNPs or SNPs in strong LD with tag SNPs. This allows users to investigate the mechanisms of 3′aQTLs in human traits and diseases.
Downloading data and figures
We provide download functions for all four modules of the 3′aQTL-atlas. In the ‘3′aQTL search by Gene/SNP’ module, users can download a table file for all queried results. In the ‘3′aQTL genome browser’ module, the genome browser can be downloaded in SVG format by clicking on the ‘Save SVG’ button (e.g. Figure 2C). For the other two modules, users can download a customized boxplot for each 3′aQTL (e.g. Figure 2D), as well as LocusCompare plots (37) for the GWAS-3′aQTLs colocalization events, in PDF format (e.g. Figure 2E). We also provide a download page (https://wlcb.oit.uci.edu/3aQTLatlas/Download.php), where users can download all the 3′aQTLs across 49 human normal tissues and a table of all human trait- and disease- associated 3′aQTLs for further analysis.
SUMMARY AND FUTURE DIRECTIONS
Increasing evidence suggests that genetic variants impacting APA usage play crucial roles in human diseases and traits (22,39,40). Here, we comprehensively evaluated the effects of genetic variants on 3′UTR usage across 49 human normal tissue types from the GTEx project and provide a user-friendly database, 3′aQTL-atlas, for users to query, browse, visualize, and download the 3′aQTLs. To the best of our knowledge, 3′aQTL-atlas is the first database for users to explore the genetic effects on 3′UTR usage in large-scale human normal tissues. In recent years, similar QTL resources, such as eQTL and sQTL, utilizing calculations from GTEx human normal tissues have been wildly used for functional interpretations of GWAS risk loci. The 3′aQTL-atlas aims to establish APA as another emerging and important molecular phenotype to explain a large fraction of GWAS risk SNPs, leading to significant novel biological insights into the genetic basis of APA and APA-linked susceptibility genes in human traits and diseases. With the increasing number of RNA-seq datasets from the GTEx project and other consortium projects, such as the Trans-Omics for Precision Medicine program (41), we will continue to update the 3′aQTL-atlas to include 3′aQTLs from more individuals and tissue types. This would make the 3′aQTL-atlas an important resource for the genetic research community. In summary, the 3′aQTL-atlas provides significant supplements to interpret the function of non-coding GWAS risk variants and offers beneficial resources for exploring the genetic basis of APA in human phenotypic diversity and a wide spectrum of human diseases.
DATA AVAILABILITY
The data used for the analyses described in this manuscript were obtained from dbGaP accession number phs000424.v8.p2
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Li laboratory and Dr Jingyi Jessica Li for helpful discussions.
Contributor Information
Ya Cui, Division of Computational Biomedicine, Department of Biological Chemistry, School of Medicine, University of California, Irvine, Irvine, CA 92697, USA.
Fanglue Peng, Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030, USA.
Dan Wang, Department of Medicine, Division of Cardiology, University of California, Los Angeles, Los Angeles, CA, 90095, USA.
Yumei Li, Division of Computational Biomedicine, Department of Biological Chemistry, School of Medicine, University of California, Irvine, Irvine, CA 92697, USA.
Jason Sheng Li, Division of Computational Biomedicine, Department of Biological Chemistry, School of Medicine, University of California, Irvine, Irvine, CA 92697, USA.
Lei Li, Division of Computational Biomedicine, Department of Biological Chemistry, School of Medicine, University of California, Irvine, Irvine, CA 92697, USA.
Wei Li, Division of Computational Biomedicine, Department of Biological Chemistry, School of Medicine, University of California, Irvine, Irvine, CA 92697, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health; NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS; National Institutes of Health [R01CA193466 to W.L.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. GTEx Consortium Genetic effects on gene expression across human tissues. Nature. 2017; 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020; 369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martin D., Watt S., Yan Y., Kundu K., Ecker S.et al.. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016; 167:1398–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Franzen O., Ermel R., Cohain A., Akers N.K., Di Narzo A., Talukdar H.A., Foroughi-Asl H., Giambartolomei C., Fullard J.F., Sukhavasi K.et al.. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science. 2016; 353:827–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gamazon E.R., Segre A.V., van de Bunt M., Wen X., Xi H.S., Hormozdiari F., Ongen H., Konkashbaev A., Derks E.M., Aguet F.et al.. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 2018; 50:956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yao D.W., O’Connor L.J., Price A.L., Gusev A.. Quantifying genetic effects on disease mediated by assayed gene expression levels. Nat. Genet. 2020; 52:626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoque M., Ji Z., Zheng D., Luo W., Li W., You B., Park J.Y., Yehia G., Tian B.. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat. Methods. 2013; 10:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elkon R., Ugalde A.P., Agami R.. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 2013; 14:496–506. [DOI] [PubMed] [Google Scholar]
- 9. Tian B., Manley J.L.. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017; 18:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hong W., Ruan H., Zhang Z., Ye Y., Liu Y., Li S., Jing Y., Zhang H., Diao L., Liang H.et al.. APAatlas: decoding alternative polyadenylation across human tissues. Nucleic Acids Res. 2020; 48:D34–D39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taliaferro J.M., Vidaki M., Oliveira R., Olson S., Zhan L., Saxena T., Wang E.T., Graveley B.R., Gertler F.B., Swanson M.S.et al.. Distal alternative last exons localize mRNAs to neural projections. Mol. Cell. 2016; 61:821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jenal M., Elkon R., Loayza-Puch F., van Haaften G., Kuhn U., Menzies F.M., Oude Vrielink J.A., Bos A.J., Drost J., Rooijers K.et al.. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012; 149:538–553. [DOI] [PubMed] [Google Scholar]
- 13. Gennarino V.A., Alcott C.E., Chen C.A., Chaudhury A., Gillentine M.A., Rosenfeld J.A., Parikh S., Wheless J.W., Roeder E.R., Horovitz D.D.et al.. NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. Elife. 2015; 4:e10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee S.H., Singh I., Tisdale S., Abdel-Wahab O., Leslie C.S., Mayr C.. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature. 2018; 561:127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Masamha C.P., Xia Z., Yang J., Albrecht T.R., Li M., Shyu A.B., Li W., Wagner E.J.. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014; 510:412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rhinn H., Qiang L., Yamashita T., Rhee D., Zolin A., Vanti W., Abeliovich A.. Alternative alpha-synuclein transcript usage as a convergent mechanism in Parkinson's disease pathology. Nat. Commun. 2012; 3:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Graham R.R., Kyogoku C., Sigurdsson S., Vlasova I.A., Davies L.R., Baechler E.C., Plenge R.M., Koeuth T., Ortmann W.A., Hom G.et al.. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:6758–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hellquist A., Zucchelli M., Kivinen K., Saarialho-Kere U., Koskenmies S., Widen E., Julkunen H., Wong A., Karjalainen-Lindsberg M.L., Skoog T.et al.. The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. J. Med. Genet. 2007; 44:314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stacey S.N., Sulem P., Jonasdottir A., Masson G., Gudmundsson J., Gudbjartsson D.F., Magnusson O.T., Gudjonsson S.A., Sigurgeirsson B., Thorisdottir K.et al.. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat. Genet. 2011; 43:1098–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garin I., Edghill E.L., Akerman I., Rubio-Cabezas O., Rica I., Locke J.M., Maestro M.A., Alshaikh A., Bundak R., del Castillo G.et al.. Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Locke J.M., Da Silva Xavier G., Rutter G.A., Harries L.W.. An alternative polyadenylation signal in TCF7L2 generates isoforms that inhibit T cell factor/lymphoid-enhancer factor (TCF/LEF)-dependent target genes. Diabetologia. 2011; 54:3078–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L., Huang K.L., Gao Y., Cui Y., Wang G., Elrod N.D., Li Y., Chen Y.E., Ji P., Peng F.et al.. An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability. Nat. Genet. 2021; 53:994–1005. [DOI] [PubMed] [Google Scholar]
- 23. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poplin R., Ruano-Rubio V., DePristo M.A., Fennell T.J., Carneiro M.O., Van der Auwera G.A., Kling D.E., Gauthier L.D., Levy-Moonshine A., Roazen D.et al.. Scaling accurate genetic variant discovery to tens of thousands of samples. 2018; bioRxiv doi:24 July 2018, preprint: not peer reviewed 10.1101/201178. [DOI]
- 27. Delaneau O., Zagury J.F., Marchini J.. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013; 10:5–6. [DOI] [PubMed] [Google Scholar]
- 28. Feng X., Li L., Wagner E.J., Li W.. TC3A: the cancer 3′ UTR atlas. Nucleic Acids Res. 2018; 46:D1027–D1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindenbaum P., Redon R.. bioalcidae, samjs and vcffilterjs: object-oriented formatters and filters for bioinformatics files. Bioinformatics. 2018; 34:1224–1225. [DOI] [PubMed] [Google Scholar]
- 31. Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012; 28:1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stegle O., Parts L., Piipari M., Winn J., Durbin R.. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012; 7:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Storey J.D., Tibshirani R.. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J.et al.. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McClellan J., King M.C.. Genetic heterogeneity in human disease. Cell. 2010; 141:210–217. [DOI] [PubMed] [Google Scholar]
- 36. Cowper-Sal lari R., Zhang X., Wright J.B., Bailey S.D., Cole M.D., Eeckhoute J., Moore J.H., Lupien M.. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat. Genet. 2012; 44:1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu B., Gloudemans M.J., Rao A.S., Ingelsson E., Montgomery S.B.. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet. 2019; 51:768–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang R., Nambiar R., Zheng D., Tian B.. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res. 2018; 46:D315–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng Z., Huang D., Wang J., Zhao K., Zhou Y., Guo Z., Zhai S., Xu H., Cui H., Yao H.et al.. QTLbase: an integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res. 2020; 48:D983–D991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang Y., Zhang Q., Miao Y.R., Yang J., Yang W., Yu F., Wang D., Guo A.Y., Gong J.. SNP2APA: a database for evaluating effects of genetic variants on alternative polyadenylation in human cancers. Nucleic Acids Res. 2020; 48:D226–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M.et al.. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021; 590:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used for the analyses described in this manuscript were obtained from dbGaP accession number phs000424.v8.p2