Abstract
The toxic metalloid inorganic arsenic (iAs) is widely distributed in the environment. Chronic exposure to iAs from environmental sources has been linked to a variety of human diseases. Methylation of iAs is the primary pathway for metabolism of iAs. In humans, methylation of iAs is catalyzed by arsenic (+ 3 oxidation state) methyltransferase (AS3MT). Conversion of iAs to mono- and di-methylated species (MAs and DMAs) detoxifies iAs by increasing the rate of whole body clearance of arsenic. Interindividual differences in iAs metabolism play key roles in pathogenesis of and susceptibility to a range of disease outcomes associated with iAs exposure. These adverse health effects are in part associated with the production of methylated trivalent arsenic species, methylarsonous acid (MAsIII) and dimethylarsinous acid (DMAsIII), during AS3MT-catalyzed methylation of iAs. The formation of these metabolites activates iAs to unique forms that cause disease initiation and progression. Taken together, the current evidence suggests that methylation of iAs is a pathway for detoxification and for activation of the metalloid. Beyond this general understanding of the consequences of iAs methylation, many questions remain unanswered. Our knowledge of metabolic targets for MAsIII and DMAsIII in human cells and mechanisms for interactions between these arsenicals and targets is incomplete. Development of novel analytical methods for quantitation of MAsIII and DMAsIII in biological samples promises to address some of these gaps. Here, we summarize current knowledge of the enzymatic basis of MAsIII and DMAsIII formation, the toxic actions of these metabolites, and methods available for their detection and quantification in biomatrices. Major knowledge gaps and future research directions are also discussed.
Keywords: Arsenic, Methylarsonous acid, Dimethylarsinous acid, Metabolism, Toxicity, Analysis
Introduction
As organisms evolved in the iAs-rich environment of the primordial sea, enzymatically catalyzed methylation of inorganic arsenic (iAs) developed as a mechanism to cope with the presence of this toxic metalloid (Zhu et al. 2014). Although other mechanisms mediate iAs detoxification in other branches of the tree of life, iAs methylation plays a dominant role in detoxification of iAs in complex organisms, including mammals. Methylation of iAs produces monomethylated (MAs), dimethylated (DMAs) and trimethylated (TMAs) arsenicals that contain either pentavalent arsenic (AsV) or trivalent arsenic (AsIII). Thus, in humans and many other mammals, exposure to iAs is associated with excretion of iAs and methylated arsenicals in urine (Thomas et al. 2001, 2007). Based on studies in microorganisms, a pathway for production of pentavalent and trivalent methylated As species during iAs methylation was first postulated by Challenger (Challenger 1947). Since then, the presence of MAsIII, MAsV, DMAsIII, DMAsV, TMAsIII, and trinethylarsine oxide (TMAsVO) has been confirmed in in vitro and animal models used in laboratory studies, as well as in urine and tissue samples collected in human cohorts.
The classification of iAs methylation as a process for detoxification of iAs is based on the fact that the methylated metabolites of iAs are rapidly excreted from the body and that deficiencies in mechanisms responsible for the formation of these metabolites are associated with the accumulation of iAs in tissues and with enhanced toxicity (Chen et al. 2011; Douillet et al. 2017; Drobna et al. 2009; Huang et al. 2018a, 2018b; Thomas et al. 2001; Vahter and Concha 2001; Yokohira et al. 2010, 2011). In addition, the methylated pentavalent arsenicals (MAsV, DMAsV, and TMAsVO) are less acutely toxic than iAs species that are typically associated with environmental exposures, trivalent arsenite (iAsIII) and pentavalent arsenate (iAsV) (Sarkar and Paul 2016; Vahter 2002; Vahter and Concha 2001). However, many laboratory studies show that methylated metabolites of iAs containing AsIII, specifically MAsIII and DMAsIII, are significantly more toxic than their pentavalent counterparts and are also more toxic than either iAsV or iAsIII. The first side-by-side comparison of the toxicities of trivalent and pentavalent iAs and methylated arsenicals in primary human and animal cells and cell lines derived from tissues that are targets for iAs exposure found that MAsIII to be the most toxic arsenical in all cell types (Styblo et al. 2000). Furthermore, DMAsIII is as toxic as iAsIII in some of these cell types. Additional work from this and other laboratories has identified MAsIII and DMAsIII as the most toxic and reactive metabolites in the pathway for iAs methylation, suggesting that, although methylation promotes clearance of As from the body, the formation of these metabolites also contributes to the adverse effects associated with iAs exposure.
Laboratory research of the metabolism and biological effects of the methylated trivalent arsenicals has been historically hampered by limited availability of pure and stable MAsIII and DMAsIII. Until recently, these compounds were chemically synthesized only in a few specialized academic laboratories. Among these, the laboratories of Dr. William Cullen (University of British Columbia) and Dr. Chris Le (University of Alberta) have been the main sources of MAsIII and DMAsIII for research laboratories worldwide. These laboratories have played a key role in supporting the toxicological research of MAsIII and DMAsIII. The supply of pure methylated trivalent arsenicals from these laboratories also supported development of novel methods for chemical analysis of MAsIII and DMAsIII. These methods allow for differential, oxidation state specific analysis of iAs and its methylated metabolites in many biological matrices. However, the rapid oxidation of MAsIII and DMAsIII to MAsV and DMAsV in oxic conditions remains a major challenge in determination of arsenic valency in samples collected in laboratory or field studies.
The goals of this review are to summarize information on mechanisms and functions of the enzymatic methylation of iAs, to examine evidence on formation and toxic effects of MAsIII and DMAsIII in biological systems, and to evaluate methods for the analysis of these arsenicals in biological matrices. Knowledge gaps and future research directions are also discussed.
Origins and mechanisms of enzymatically catalyzed methylation of iAs
Enzymatically catalyzed methylation of iAs is one of several adaptations that allows cells to thrive in environments that contain iAs. The role of iAs methylation is best understood in the context of all processes involved in arsenic metabolism at the cellular level. Figure 1 is a conceptual model for cellular metabolism of arsenic that includes pathways for entry and exit of this metalloid from the cell and pathways for chemical transformations of arsenic atoms that occur during their residence in the cell (Thomas 2007). Over the past several decades, research has characterized the molecular machinery involved in these processes and identified the genes that encode relevant transporters and enzymes. Thus, it is now possible to relate steps in cellular arsenic metabolism to functions of specific gene products and to explore phylogenetic relations among these genes as they appear in organism in all branches of the tree of life. As part of our consideration of the roles for arsenic methyltransferases, we begin with an overview of the biochemical processes that contribute to the cellular metabolism of arsenic.
Fig. 1.
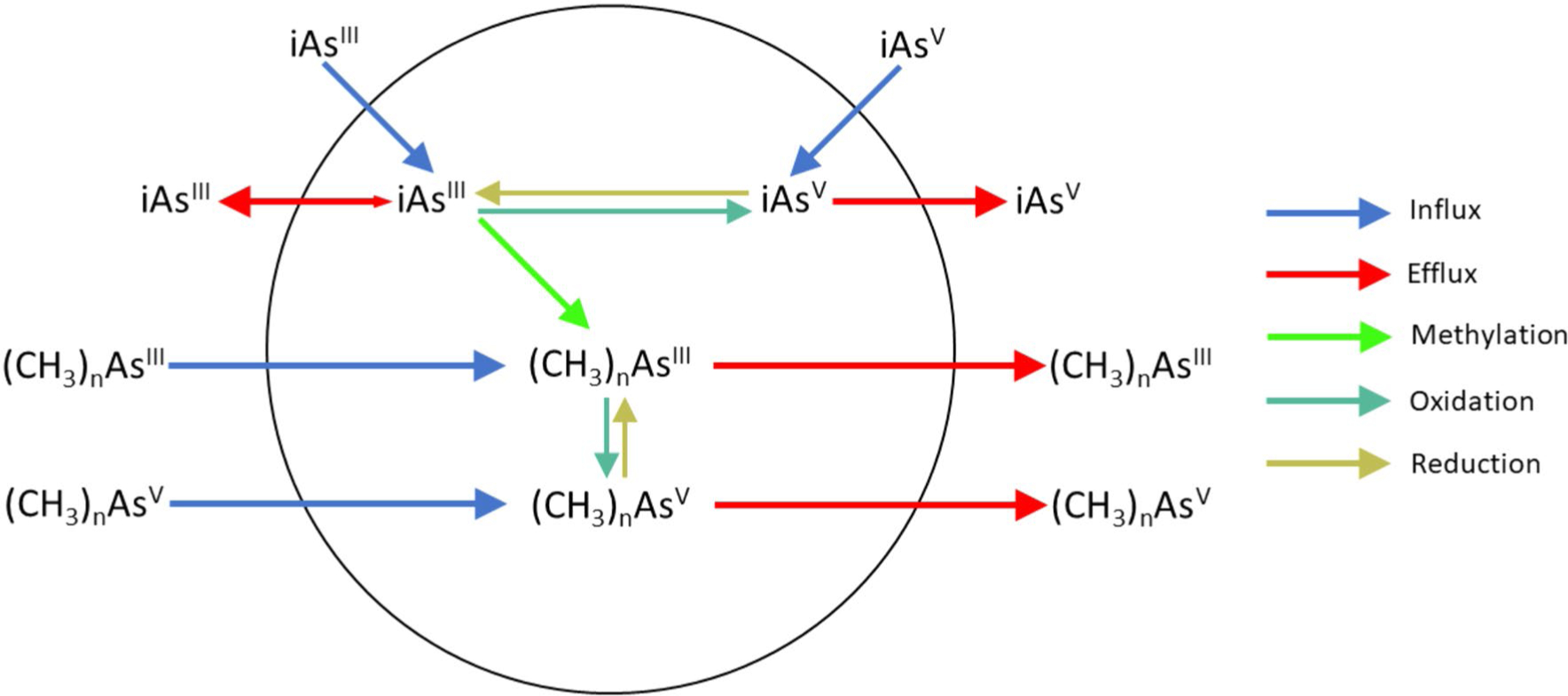
A conceptual model for the cellular metabolism of arsenic, including formation of the methylated trivalent and pentavalent arsenic metabolites. This model posits methylation without a change in oxidation state of arsenic and the ready interconversion of arsenic oxidation state in the cellular environment
Origins of iAs methylation
Nearly four billion years ago, primitive organisms in the primordial ocean adapted to the presence of iAs by acquisition of an array of genes that encode: (a) transmembrane transporters that facilitated arsenic fluxes, (b) oxidases and reductases that catalyzed reactions that altered the valence state of arsenic, and (c) methyltransferases that catalyzed reactions that coupled methyl groups to an arsenic atom (Zhu et al. 2014). High water temperature, low pH, and anoxic conditions produced higher levels of iAsIII in the primordial ocean. Primitive cells limited accumulation of iAs through development of transmembrane proteins that facilitated iAsIII efflux. Rising atmospheric oxygen levels during the Great Oxidation Event starting about 2.4 billion years ago oxidized iAsIII in the primordial ocean and increased levels of iAsV. Because transmembrane transporters were selective for iAsIII efflux, cells acquired iAsV reductases that catalyzed reduction of iAsV to iAsIII. The integration of existing pathways for detoxification of trivalent arsenicals with newly developed processes such as arsenate reduction provided additional protection of organisms against arsenic under a wider array of environmental conditions (Chen et al. 2020). Similarly, the transfer of genes encoding methyltransferases that catalyzed addition of methyl groups to an arsenic atom has been interpreted as a critical step in the adaptation of organisms to the presence of arsenic in the environment that facilitated the spread of organisms into arsenic-rich environments during the late Archean Eon and Proterozoic Eon (Chen et al. 2017a, b). In a broader context, repeated independent horizontal gene transfers among bacteria and from bacteria to eukaryotes provided organisms with the molecular machinery needed for tolerance to iAs present in the environment (Palmgren et al. 2017).
A global survey of soil microbiomes found a high frequency of occurrence of genes encoding an arsenic methyltransferase, suggesting an important role for arsenic methylation in the biogeochemical cycle for arsenic (Dunivin et al. 2019). Methylation of iAs by soil microbes likely fulfills multiple roles in the adaptation of life to environments contaminated with this toxic metalloid. For some microorganisms, production of monomethylarsonic acid (MAsV) may act as an antibiotic and confer a selective advantage against competing microbial species (Li et al. 2016; Chen et al. 2019; Garbinski et al. 2020). In other species, formation of trimethylarsine (TMAsIII), a volatile species that is released into the environment, not only reduces the celluar burden of arsenic but also contributes to global cycling of arsenic (Huang et al. 2016; Chen et al. 2014). The widespread occurrence of the capacity to form methylated metabolites of iAs in higher organisms, including humans, makes understanding the molecular basis of arsenic methylation central to understanding the risk associated with long-term exposure to iAs.
Enzymatic basis of iAs methylation
Studies in microorganisms in the nineteenth century suggested that methylated arsenicals were products of iAs metabolism (Cullen 2008). Continuing work on iAs methylation in microorganisms by Frederick Challenger and his associates lead to a postulated reaction scheme in which reduction of pentavalent arsenic to trivalency is followed by an oxidative methylation reaction that produces a methylated product containing pentavalent arsenic atom (Challenger 1947, 1951). As shown in Fig. 2, the Challenger scheme is a sequence of reactions in which reduction of arsenic from pentavalency to trivalency alternates with oxidation methylation of an arsenic atom.
Fig. 2.

The Challenger scheme for the methylation of arsenic
The Challenger scheme for iAs methylation significantly influenced the study of biomethylation processes because it made specific predictions about the formation of intermediates that contained either AsV or AsIII and focused attention on development of analytical methods that used the oxidation state of arsenic as a discriminating factor in the separation and quantitation of arsenicals in environmental and biological materials. The postulated existence of methylated intermediates containing arsenic in either valence state also lead to extensive studies of the effect of arsenic’s valency in methylated arsenicals on the toxic potency of these compounds. Finally, the Challenger scheme also served as a framework for interpretation of data on putative mechanisms by which arsenic methyltransferase catalyzes reactions that methylate arsenic atoms. Evaluating putative mechanisms for enzymatic catalysis in the context of the Challenger scheme also helped to identify chemically plausible pathways for methylation (Cullen 2014). As discussed below, recent evidence suggests that a pathway for methylation that does not involve obligatory oxidation of arsenic may be sufficient to describe the sequence of reactions catalyzed by arsenic methyltransferases.
Isolation and characterization of an arsenic methyltransferase from a mammalian source originates in early work that demonstrated that methylation of iAs in rat liver cytosol in an in vitro system is supported by an addition of glutathione (GSH) and S-adenosyl-L-methionine (AdoMet) (Buchet and Lauwerys 1985; Hirata et al. 1989). Later studies of iAs methylation in rat liver cytosol in an in vitro system characterized concentration dependencies for GSH and AdoMet in arsenic methylation reactions and showed that inhibitors of other non-DNA methyltransferases did not affect iAs methylation in this in vitro system, suggesting the reaction was catalyzed by a novel methyltransferase (Styblo et al. 1996). Because a simple in vitro system containing rat liver cytosol rapidly converts iAs to methylated species, purification of arsenic methyltransferase used rat liver as the starting material (Lin et al. 2002). This effort resulted in the identification of a ~43 kDa protein that catalyzed AdoMet-dependent methylation of substrates containing trivalent arsenic (iAsIII or MAsIII). This methyltransferase was initially identified as the product of the previously annotated cyt19 gene. Following its cloning, the gene was annotated as Arsenic (+ 3 oxidation state) MethylTransferase (AS3MT) (Walton et al. 2003; https://www.genenames.org). Characterization of rat liver As3mt led to identification of genes encoding closely related proteins in genomes of higher organisms ranging from sea squirts to humans (Thomas et al. 2007). Notably, As3mt orthologs in many higher organisms share a suite of conserved cysteine residues and contain sequence motifs that are common in methyltransferases that catalyze reactions that methylate proteins or small molecules. Early work showed that ArsM genes in microorganisms are orthologs of As3mt gene found in higher organisms and that expression of ArsM genes in these organisms is associated with capacity to form methylated products from iAs and confers resistance to iAs-induced cytotoxicity (Qin et al. 2006).
Common features of arsenic methyltransferases
Our understanding of molecular aspects of enzyme-catalyzed iAs methylation derives from studies with products of both the mammalian As3mt and ArsM genes that encode a large family of iAsIII methyltransferases in archaea, bacteria, fungi, algae, and plants. From studies with these proteins, it has been possible to identify residues critical for catalysis, to elucidate the mechanisms used in methylation, and to explore the role of oxidation–reduction reactions in arsenic methylation.
Function of conserved cysteinyl residues
The predicted amino acid sequences of arsenic methyltransferases from As3mt and ArsM gene families contain motifs commonly found in non-DNA methyltransferase (Thomas and Rosen 2013). In particular, locations of cysteinyl residues in these protein sequences are strongly conserved. The role of the conserved cysteinyl residues in catalytic function has been evaluated by their replacement with other amino acids. In recombinant rat As3mt, replacement of the conserved cysteinyl at position 156 (C156) with serine (C156S) eliminates its capacity to catalyze iAsIII methylation (Li et al. 2005). Systematic replacement of four conserved cysteinyl residues in human AS3MT (C150, C206, C226, C250) with serine affects structure and function of the modified proteins (Song et al. 2009). The secondary structures of C156S, C206S, and C250S mutant proteins differ from that of wild-type protein and are associated with a loss of capacity to catalyze iAsIII methylation. In contrast, the secondary structure and catalytic capacity of the C226S mutant do not differ from those of the wild-type protein. Functions of other cysteinyl residues in human AS3MT have been examined (Song et al. 2011). The C72S mutant protein display structural changes and do not catalyze iAsIII methylation. Structural changes in C334S and C360S mutant proteins lead slower turnover rates, suggesting that structural modifications affect efficiency of methylation reactions. Other cysteine to serine mutants (C32S and C61S) that do not catalyze iAsIII methylation could methylate MAsIII (Li et al. 2013a, b). By comparison, replacement of C156 or C206 lead to a complete loss of catalytic activity. Altered catalysis by these mutants provides evidence on the location and number of cysteinyl residues that bind iAsIII and MAsIII acid to the enzyme. Formation of critical disulfide bonds in human AS3MT affects its catalytic function (Wang et al. 2014). Reduction of a C250–C32 disulfide bond before iAsIII methylation alters the enzyme’s secondary structure. Thus, formation and cleavage of disulfide bonds between critical cysteinyl residues affects the binding of arsenicals to the active site and changes the secondary structure around the active site.
Additional insights into the role of cysteinyl residues in the structure and function of arsenic methyltransferases come from studies with ArsM. In particular, determination of the 3D structure of CmArsM from the thermophilic eukaryotic alga Cyanidioschyzon merolae sp. 5508 (CmArsM) informs our understanding of the relation between enzyme structure and function for both ArsM and As3mt arsenic methyltransferases (Ajees et al. 2012). Replacement of any of the conserved cysteinyl residues (C72, C174, and C224) in CrArsM abolishes capacity to methylate iAsIII; however, the replacement of C72 yields an enzyme that can methylate MAsIII (Marapakala et al. 2012). Studies of the binding of aromatic organic arsenicals phenylarsenite and reduced roxarsone that contain trivalent arsenic to CrArsM show that both bound to thiol moieties in C174 and C224, suggesting that MAsIII would also be held in a bidentate structure with these residues. The structure of the enzyme-organic arsenical complex includes a C44 as a conserved cysteinyl residue that forms a disulfide bond with C72 (Marapakala et al. 2015). As discussed below, formation of the C44–C72 disulfide bond may play a crucial role in binding of substrates to the enzyme.
The relation between conservation of cysteinyl residues and substrate specificity in arsenic methyltransferases has been examined using ArsM from the fungus Aspergillus fumigatus (Chen et al. 2017a, b). Although ArsM genes from most organisms encode four conserved cysteinyl residues into an enzyme that accepts either iAsIII or MAsIII as a substrate, the four arsM genes in the Aspergillus fumigatus genome encode ArsMs that conserves only the second, third and fourth cysteinyl residues. This loss affects substrate specificity of the gene products. For example, MAsIII but not iAsIII is a substrate for methylation by the fungal ArsM1. Existence of arsenic methyltransferases that discriminates among substrates could reflect different selective pressures that fine-tune substrate specificity and the central role of cysteinyl residues in control of enzyme function. The search for the minimal number of cysteinyl residues required for its function as an arsenic methyltransferase has been extended using BlArsM from Bacillus sp. CX-1, an arsenic methyltransferase with low sequence similarities (≤ 39%) to other ArsMs (Huang et al. 2018). Only two cysteinyl residues in this enzyme must be conserved for catalytic function; replacement of four of the six cysteinyl residues produces an enzyme that uses either iAsIII or MAsIII as substrates. The availability of these structurally diverse enzymes with similar catalytic functions should aid in understanding the role of cysteinyl residues in arsenic methyltransferases.
Mechanisms for arsenic methylation
Evaluating the role of cysteinyl residues in the catalytic function of arsenic methyltransferases provides insights into molecular processes involved in binding of reactants to the enzyme and transfer of a methyl group from AdoMet to an arsenic atom. Early work showed that endogenous reductants (e.g., thioredoxin/thioredoxin reductase/NADPH) support the activity of recombinant rat As3mt and that addition of GSH to reaction mixtures increases the rate of iAsIII methylation (Waters et al. 2004). The same effect of GSH on the rate of iAsIII methylation is observed in studies with recombinant human AS3MT and the common polymorph AS3MT(M287T) (Ding et al. 2012). Additional support for the role of GSH in the AS3MT-catalyzed methylation of iAsIII comes from a report that a complex of iAsIII with GSH (i.e., iAsIII-triglutathione) is the preferred substrate for this enzyme (Hayakawa et al. 2005). Notably, studies of protein fluorescence found that rates of binding of iAsIII or MAsIII to the active site of CmArsM increases in the presence of GSH (Marapakala et al. 2012). A role for arsenic-GSH complexes as substrates for methylation reactions is consistent with the presence of higher levels of GSH in cellular environments and the facile in vivo formation of arsenic-GSH complexes (Delnomdedieu et al. 1995).
Analysis of the order of binding of iAsIII and AdoMet to arsenic methyltransferases found that AdoMet is the substrate first bound to human AS3MT; this binding is associated with conformational changes (Wang et al. 2012). In CmArsM, AdoMet binding also changes protein conformation and positions this methyl donor for transfer to arsenic (Ajees et al. 2012). Mutational analysis of residues in human AS3MT demonstrates that a hydrogen bond network around the AdoMet binding site is required for catalytic activity (Li et al. 2013a).
Enzymatic catalysis and oxidation–reduction cycling
Information about the role of critical residues in the binding of substrates, changes in protein conformation, and structural data can be integrated to develop a model for enzyme-catalyzed methyl group transfer from AdoMet to arsenic. The current model builds on a scheme proposed less than a decade ago (Dheeman et al. 2014) that has been extended by studies with enzymes from diverse sources. Analysis of CmArsM identifies a structure in which iAsIII is 3-coordinately bound to conserved cysteinyl residues and the AdoMet binding site is occupied (Packianathan et al. 2018a). Here, AdoMet-induced conformational changes bring C44 close to C174 and C224 and permits 3-coordinate binding of iAsIII to cysteinyl residues. In this state, iAsIII is bound in proximity to AdoMet’s methyl group; this conformation facilitates transfer of the methyl group to the trivalent arsenic atom. Notably, the bound product of the first methylation step is MAsIII which can be the substrate for addition of another methyl group to the trivalent arsenic atom. Kinetic analysis shows that the relatively slower rate of the second methylation step reflects the slow reorientation of the AdoMet binding site to its initial position (Packianathan et al. 2018b). Upon reorientation, binding of AdoMet to this site starts the sequence steps that convert MAsIII to DMAsIII.
The alternate scheme for enzymatically catalyzed methylation of arsenic shown in Fig. 3 differs materially from the Challenger scheme. It lacks an obligatory cycle of oxidation and reduction of arsenic atoms during addition of methyl groups. Current kinetic and structural studies with arsenic methyltransferases provide evidence that these enzymes avidly bind AsIII and that the addition of a methyl group to bound arsenic does not result in its oxidation. Rather, use of a disulfide bond cascade using conserved cysteinyl residues functions to maintain bound arsenic as a trivalent species. In this model, the appearance of methylated arsenicals containing AsV is a secondary process, requiring the oxidation of the AsIII in methylated species that are the intermediates in enzymatically catalyzed methylation. Thus, the current model is sufficient to account for products containing AsIII or AsV without redox cycling of arsenic required by the Challenger scheme. The lack of redox cycling of arsenic during the methylation process eliminates the need for an additional reduction step in the pathway. The reduction of AsV had been posited to depend on endogenous reductants (e.g., GSH) or unique reductases that catalyze this step (Delnomdedieu et al. 1995; Zakharyan and Aposhian 1999; Zakharyan et al. 2001). If catalysis by arsenic methyltransferases does not generate intermediates that contain AsV, it is likely that the presence of these species is a result of spontaneous oxidation of the trivalent intermediates formed during the enzymatic reaction. Post synthesis oxidation of trivalent arsenic present in methylation products could account for the occurrence of methylated arsenicals containing either trivalent or pentavalent arsenic in tissues and excreta (Currier et al. 2011a, b).
Fig. 3.

Alternate scheme for methylation of arsenic
Roles of enzymatic methylation in metabolism and toxicity of iAs
There is a dichotomy at the center at any discussion of the significance of iAs methylation. The dichotomy is supported by the well-documented facts that: (a) conversion of iAs into methylated metabolites accelerates the rate of whole body clearance of arsenic, and (b) the methylated trivalent metabolites, specifically MAsIII and DMAsIII, are more reactive and toxic than inorganic arsenic. Thus, promotion of arsenic excretion through formation of methylated metabolites can be termed a detoxification process. Conversely, formation of more toxic species, MAsIII and DMAsIII, through methylation can be termed an activation or toxication process. In the following paragraphs, the dichotomous roles of iAs methylation are reviewed.
iAs methylation as a detoxification process
Studies in humans and other species illustrate the role of iAs methylation in the clearance of arsenic. Early studies in volunteers and workers who ingested or inhaled iAs found that mono- and di-methylated arsenical were quickly excreted in urine (Crecelius 1977; Smith et al. 1977). In volunteers who ingested 500 μg of arsenic as iAsIII, MAsV, or DMAsV, the urinary excretion rate for arsenic and the cumulative amount of arsenic excreted in urine is affected by the ingested form (MAs > DMAs > iAs) (Buchet et al. 1981). The form of arsenic ingested also affects the extent of metabolism. About 75% of arsenic ingested as iAsIII is excreted in urine as MAs or DMAs; about 13% of ingested MAsV is excreted as DMAs. In contrast, ingested DMAsV is excreted in urine without additional methylation.
Further insights into the role of arsenic methylation in the clearance of arsenic come from studies that examined patterns for urinary excretion of arsenic-containing species after oral administration of mono- and di-methylated species containing either trivalent or pentavalent arsenic to female B6C3F1 mice. Orally administered MAsV is rapidly cleared in urine with less than 10% of the administered arsenic converted to a dimethylated metabolite before excretion (Hughes et al. 2005). In contrast, MAsIII is highly retained in tissues; more than 90% of arsenic excreted in urine is a dimethylated metabolite. Related studies in female B6C3F1 mice that received single oral doses of DMAsV or DMAsIII found that the oxidation state of arsenic affects patterns of metabolism and rates of urinary clearance (Hughes et al. 2008). After DMAsV treatment, a rapid rise and fall in arsenic concentrations in tissues accompanies appearance of arsenic in urine. Compared to DMAsV, treatment with DMAsIII produces higher levels of arsenic in tissues. Notably, treatment with either dimethylated arsenical results in the presence of TMAsVO in urine, suggesting that either compound was a substrate for addition of a third methyl group. These studies demonstrated that the oxidation state of arsenic in methylated arsenicals affects the rate of whole body clearance with faster whole body clearance of arsenic after administration of MAsV or DMAsV. In addition, the oxidation state of arsenic in a methylated arsenical affects the likelihood that it will serve as a substrate for additional methylation reactions. For example, in mice MAsIII is readily converted to dimethylated product while MAsV is not extensively metabolized. Because the increased rate of clearance of methylated arsenical species, especially those containing AsV, has a salutary effect by promoting the excretion of arsenic, methylation of iAs can be regarded as a detoxification process. Additional evidence for the detoxification function of iAs methylation has been obtained in studies using As3mt knockout mice. The metabolism of iAs in these mice is characterized by slow whole body clearance and accumulation of iAs in tissues (Drobna et al. 2009; Hughes et al. 2010). The As3mt−/− mice are also more susceptible to As-induced toxicity than mice wild-type mice (Yokohira et al. 2011; Dodmane et al. 2013).
iAs methylation as an activation process
The Challenger scheme for iAs methylation that posits formation of methylated trivalent and pentavalent arsenicals has inspired many laboratory studies that compared effects of these intermediates on a wide range of biological end-points, using both in vitro and in vivo models. Attempts have also been made to assess roles of the methylated arsenicals containing AsIII or AsV as potential mediators of adverse effects associated with iAs exposure in human populations. The following sections summarizes reports of these studies.
Cytotoxicity of methylated trivalent arsenicals
Studies of the comparative cytotoxicity of methylated arsenicals are based in part on studies of the lethality of inorganic and methylated arsenicals in rodents. In acute lethality assays (LD50), both MAsV and DMAsV are less toxic than iAsV and iAsIII (Yamauchi and Fowler 1994). Cytotoxicity studies affirm that pentavalent methylated arsenicals, MAsV and DMAsV, are much less toxic for cultured cells than inorganic arsenicals. In addition, early work showed that oxidation state of arsenic affects toxicity of inorganic arsenicals; iAsIII was more cytotoxic than iAsV (Fischer et al. 1985, 1989; Bertolero et al. 1987). One of the first side-by-side comparisons of the cytotoxicity of arsenicals containing AsIII or AsV used primary and transformed cells derived from liver, a major site for iAs methylation, and from skin, lung, and urinary bladder that are targets for arsenic-induced tumors (Styblo et al 2000). This study found that the methylated trivalent arsenicals (MAsIII and DMAsIII) are more cytotoxic than their pentavalent counterparts and that MAsIII is more cytotoxic than iAsIII or iAsV. In addition, cytotoxicity of DMAsIII approaches or exceeds those of iAsIII and iAsV. Other studies with these cell types also found MAsIII to be more cytotoxic than iAsIII (Dodmane et al. 2013; Hirano et al. 2004; Petrick et al. 2000; Vega et al. 2001).
The cytotoxicity of MAsIII has been examined in other cell types, including leukemia NB4, Jurkat, and Namalwa cells (Chen et al. 2003), differentiated 3T3-L1 adipocytes (Walton et al. 2004), rat insulinoma (INS-1) cells (Dover et al. 2018), isolated murine pancreatic islets (Douillet et al. 2013), cultured human neurons, astrocytes, microglia, and brain microvascular endothelial cells (Yoshinaga-Sakurai et al. 2020), JEG-3 placental cells (Meakin et al. 2020), human monoblastoid (U937), osteosarcoma (HOS) and neuroblastoma (SK-N-SH) cells, Chinese hamster lung (V79–4) cells (Styblo et al. 2002). Without exception, these studies found MAsIII to be more cytotoxic than iAsIII. Notably, some of these studies found DMAsIII to be more cytotoxic than iAsIII.
Genotoxicity of methylated trivalent arsenicals
The genotoxic potential of trivalent methylated arsenicals has been assessed using in vitro systems containing naked viral (Phi 174 RF1) DNA and human peripheral lymphocytes (Mass et al. 2001). These investigators found that incubation with relatively high concentrations of MAsIII and DMAsIII caused nicks in viral DNA. In contrast, DNA nicking does not occur after incubation with iAsIII, iAsV or the methylated pentavalent arsenicals. Because DNA nicking does not require addition of chemical or enzymatic activation system, MAsIII and DMAsIII likely interacts directly with DNA. Both MAsIII and DMAsIII are tenfold more potent than iAsIII in causing DNA damage in cultured human lymphocytes. Damage to DNA by MAsIII and/or DMAsIII has also been reported in other studies. Although micromolar concentrations of MAsIII or DMAsIII induce oxidative DNA damage in cultured HeLa cells, DMAsIII alone causes DNA breaks in supercoiled and circular PM2 DNA (Schwerdtle et al. 2003). MAsIII also significantly increases micronuclei frequency in human peripheral lymphocytes, suggesting aneuploidogenic properties (Colognato et al. 2007).
The potential of methylated trivalent arsenicals to induce chromosomal aberrations, mutagenicity, sister chromatid exchange (SCE), and prophage inductions has also been examined (Kligerman et al. 2003). Both MAsIII and DMAsIII induce chromosomal aberrations and are potent clastogens in human lymphocytes and mutagens in mouse lymphoma cells. Neither MAsIII nor DMAsIII are potent SCE or prophage inducer. These findings suggest that DMAsIII and MAsIII act primarily as clastogens and are candidates for the ultimate genotoxic form of arsenic. In Chinese Hamster Ovary (CHO-9) cells, DMAsIII and MAsIII are also more genotoxic than DMAsV, MAsV, or iAs (Dopp et al. 2004). In this study, the high genotoxic potency of DMAsIII and MAsIII has been linked to a higher uptake and retention of DMAsIII and MAsIII compared to the methylated pentavalent and iAs species.
Cell transformation and carcinogenesis
The potential of methylated arsenicals to induce cell transformation and carcinogenesis has also been evaluated. UROtsa cells, a non-tumorigenic cell line derived from human urothelial cells, acquire the hallmarks of cancer cells when exposed to a low level of MAsIII (0.05 μM) for up to 52 weeks (Bredfeldt et al. 2006). MAsIII-induced changes in DNA methylation patterns may contribute to development of the tumorigenic phenotype (Wnek et al. 2010). In SV-HUC-1 cells, an immortalized human uroepithelial cell line, treatment with iAsIII or MAsIII produces changes in gene expression patterns that may be linked cellular transformation (Su et al. 2006). Notably, the concentration for iAsIII that induces these changes in gene expression are several-fold higher than the concentration of MAsIII that alter gene expression.
Only a few studies have evaluated the carcinogenicity of methylated trivalent arsenicals in animals. The transplacental carcinogenicity of MAsIII has been demonstrated in pregnant CD-1 mice that drank water-containing MAsIII at 0 (control), 12.5, or 25 parts per million (ppm) from gestational days 8 to 18 (Tokar et al. 2012). Mice so exposed in utero were evaluated for tumor occurrence up to two years of age. In female offspring, dose-related increases in total epithelial uterine tumors, oviduct hyperplasia, adrenal cortical adenoma, and total epithelial ovarian tumors were found. Male offspring showed dose-related increases in hepatocellular carcinoma, adrenal adenoma, and lung adenocarcinoma. Notably, patterns of proliferative lesions in mice exposed to MAsIII during gestation resemble those found in mice exposed to iAsIII during gestation (Waalkes et al. 2007). Notably, 2-year feeding studies with a MAsIII-amended diet identified intestinal tract alterations as a common response in male and female Fischer F344 rats and B6C3F1 mice (Arnold et al. 2003). However, at the dosage levels tested, neither species show evidence of treatment-related neoplasia. The low toxic and carcinogenic potency of MAsIII in mice may be related to its rapid whole body clearance.
Additional, albeit indirect, evidence on the role of the valence state of arsenic on the toxicity of methylated arsenicals comes from studies of toxic and carcinogenic effects of DMAsV in the urinary bladder of the rat. High dose levels studies in rats show that DMAsV treatment produces a spectrum of changes in the uroepithelium that culminates in carcinogenic transformation in this organ (Arnold et al. 2006). Notably, analysis of urine from rats treated with dimethylthioarsinic acid identified DMAsIII and TMAsVO as metabolites (Cohen et al. 2002; Adair et al. 2007). These findings suggested production of DMAsIII, a toxic and reactive species, caused requisite tissue injury needed for cellular transformation.
Enzyme inhibition
Early work showed that MAsIII or DMAsIII were more potent inhibitors of the purified yeast GSH reductase than iAsIII (Styblo et al. 1997). Similarly, MAsIII acid is a more potent inhibitor than iAsIII of the DTNB reductase activity of purified thioredoxin reductase or in cultured rat hepatocytes (Lin et al. 1999, 2001). Notably, after 3-h incubation, MAsIII is methylated by rat hepatocytes to DMAs and thioredoxin reductase activity is almost completely recovered, suggesting that the dimethylated product does not inhibit this enzyme. iAsIII and MAsIII also inhibit activities of purified bovine glutathione peroxidase and equine glutathione S-transferase (Chouchane and Snow 2001). MAsIII is a more potent inhibitor of the activity of purified porcine pyruvate dehydrogenase than iAsIII; parenterally administered MAsIII also inhibits pyruvate dehydrogenase activity in hamster kidneys (Petrick et al. 2001). When the co-factor is reduced, activities of pyruvate dehydrogenase and α-ketoglutarate dehydrogenase are inhibited by trivalent arsenicals (Bergquist et al. 2009). Here, the order of potency of these compounds is MAsIII > iAsIII > > DMAsIII. iAsIII inhibits complexes I and II in the inner membrane of isolated mitochondria; this inhibits electron transport chain, decreases mitochondrial ATP content, and increases ROS formation (Hosseini et al. 2013). A recent study found that MAsIII is more potent than iAsIII as an inhibitor of oxygen consumption rate and maximal respiration in INS-1 832/13 β-cell, suggesting that MAsIII also inhibits mitochondrial electron transport (Dover et al. 2018).
Disruption of cellular processes and signaling pathways
There is evidence for roles of MAsIII and DMAsIII in disruption of mechanisms involved in regulation of cellular processes, including inhibition of key signaling pathways. In a human urothelial cells line, trivalent arsenicals have been shown to affect activation of AP-1, one of the major transcription factors in mammalian cells (Drobna et al. 2003). MAsIII and DMAsIII are more potent than iAsIII in activating AP-1 and inducing its binding to DNA. MAsIII is also more potent than iAsIII in inducing the AP-1-dependent gene transcription in cells transiently transfected with an AP-1-dependent promoter-reporter construct. Because both MAsIII and DMAsIII induce phosphorylation of c-Jun, a component of the AP-1-DNA binding complex, and of extracellular signal-regulated kinase (ERK), the authors hypothesize that activation of an ERK-dependent signal transduction pathway is responsible for AP-1 activation in these cells.
The methylated arsenicals containing trivalent arsenic also disrupt pathways that control insulin release and action. A 48-h exposure to low concentrations of either MAsIII or DMAsIII (IC50 ≤ 100 nM) inhibits glucose-stimulated insulin release by isolated mouse pancreatic islets (Douillet et al. 2013). MAsIII also inhibits insulin secretion by cultured rat insulinoma INS-1 832/13 β cells that express human proinsulin (Dover et al. 2018). In both cell types, iAsIII is less potent than either MAsIII or DMAsIII as an inhibitor of insulin release. ATP production in mitochondria plays a key role in stimulation of glucose-dependent insulin secretion from β cells (Fu et al 2013). Studies examining mitochondrial metabolism in cultured INS-1 832/13 β cells exposed to iAsIII, MAsIII or DMAsIII found that oxygen consumption rate, a surrogate for mitochondrial function, is inhibited by exposure to 2 μM iAsIII or 0.375–0.5 μM MAsIII. These results suggest that these arsenicals inhibit insulin secretion by disrupting mitochondrial metabolism in β cells. Other studies using isolated pancreatic islets show that trivalent arsenicals also inhibit glucose stimulated Ca2+ influx, a process that triggers assembly and exocytosis of insulin-containing vesicles (Huang et al 2019). Not surprisingly, MAsIII or DMAsIII are more potent inhibitors of Ca2+ influx than iAsIII.
In related work, the effects of iAsIII and MAsIII acid on mitochondrial function have been examined in cultured rat aortic smooth muscle cells (Pace et al. 2016). In these cells, MAsIII acid is more cytotoxic than iAsIII. Treatment of cells with MAsIII decreases basal and maximal oxygen consumption rates and increases compensatory extracellular acidification rates, a proxy for glycolysis.
The methylated trivalent arsenicals have also been shown to disrupt insulin signaling and insulin-dependent processes in cultured cells from tissues that play essential roles in glucose metabolism. In cultured mouse hepatocytes stimulated with insulin, short-term exposure to iAsIII (0.5 μM) or MAsIII (0.2 μM) decreases cellular glycogen concentrations by inducing glycogen phosphorylase activity and inhibits activation of glycogen synthase (Zhang et al. 2017). Actions of both arsenicals are mediated through inhibition of insulin-dependent phosphorylation of protein kinase B/Akt, a factor involved in insulin-dependent regulation of glycogen phosphorylase and glycogen synthase activities. An earlier study found inhibition of insulin-dependent phosphorylation of Akt by iAsIII and MAsIII in cultured differentiated 3T3-L1 adipocytes (Paul et al. 2007a). Here, inhibition of insulin signaling is associated with reduction of the insulin-stimulated glucose uptake. DMAsIII also inhibits insulin-dependent glucose uptake in adipocytes but does not affect Akt phosphorylation or any other upstream step in the insulin-activated signaling cascade.
In sum, these studies suggest that methylated arsenicals containing trivalent arsenic are potent disruptors of cellular signaling and secretion processes. Disruption of energy metabolism through effects on mitochondrial metabolism may be central to these effects of arsenic exposure.
Immunotoxicity
Effects of methylated trivalent arsenicals on components of the immune system have been reported in several published studies. Exposure to low concentrations of iAsIII, MAsIII or DMAsIII stimulates secretion of the growth-promoting and proinflammatory cytokines in cultures of normal human keratinocytes (Vega et al 2001). Similarly, in cultured human bronchial epithelial cells, secretion of cytokines (e.g., IL-6, IL-8) induced by bacterial challenge is increased by concurrent exposure to MAsIII (Notch et al. 2015). MAsIII has also been shown to affect the pre-lymphoid development of cells in bone marrow (Ezeh et al. 2016). Addition of 50 nM MAsIII to medium of cultured mouse bone marrow pre-B cells inhibits phosphorylation of the transcription factor Signal Transducer and Activator of Transcription 5 and thereby blocks activation of the IL-7 signaling pathway required for pre-B cell maturation. Notably, a tenfold higher concentration of iAsIII in medium is needed to attain equivalent inhibition of cell maturation.
The effects of iAsIII, MAsIII, and DMAsIII exposures on inflammatory responses in Caco-2 cells, a human intestinal epithelial cell line, have also been examined (Calatayud et al. 2014). Here, levels of biomarkers of inflammation are measured in cells and media after exposure for 2, 4, 6, and 24 h to trivalent arsenicals in presence or absence of lipopolysaccharide (LPS). Exposure to each trivalent arsenical alone decreases TNF-α mRNA levels and TNF-α levels in media and increases IL-8 mRNA levels and IL-8 levels in media. Here the potency of DMAsIII exceeds iAsIII and MAsIII. Co-exposure to LPS further enhances effects induced by trivalent arsenicals. A follow-up study has examined effects of trivalent arsenicals in co-cultures of Caco-2 cells and peripheral blood mononuclear cells with or without LPS treatment (Calatayud et al. 2015). In this co-culture, trivalent arsenicals increase IL-6, IL-8, and TNF-α secretion. LPS co-exposure potentiates the proinflammatory response compared to that produced by arsenicals alone. Co-exposure to DMAsIII or iAsIII and LPS also increases the permeability of intestinal monolayer.
Dysregulation of gene transcription
A large body of evidence generated by laboratory and population studies suggests that iAs exposure is associated with a widespread dysregulation of gene transcription. The affected genes are linked to a variety of adverse mechanisms and diseases, e.g., oxidative stress (Lantz and Hays 2006), cancer (Bailey et al. 2012; Huang et al. 2004; Valko et al. 2006; Yang and Frenkel 2002), metabolic disease (Martin et al. 2017; Venkatratnam et al. 2021), or inflammation (States et al. 2012). The effects of iAs exposure on gene expression have been linked, in part, to dysregulation of epigenetic mechanisms that regulate gene expression, e.g., DNA methylation (Bailey et al. 2013; Bailey and Fry 2014; Smeester et al. 2011), histone modification (Barr et al. 2009; Costa 2019; Eckstein et al. 2017), or changes in chromatin structure (Riedmann et al. 2015). However, the extent to which the methylated trivalent arsenicals contribute to these outcomes remains unclear. Design of most of the published studies carried out in in vitro or in vivo systems in which iAs is methylated, including human studies, cannot differentiate between effects of the parent compound and effects of its methylated metabolites.
Only few studies have been able to link dysregulation of gene transcription to specific arsenic species. Distinctive gene expression profiles have been reported in immortalized human uroepithelial cells SV-HUC-1 cells exposed to iAsIII or the methylated trivalent arsenicals (Su et al 2006). Gene expression profiles in iAsIII or MAsIII-exposed cells differ significantly from those in DMAsIII-exposed cells. Notably, profiles in iAsIII or MAsIII-exposed cells resemble gene expression profiles in a tumorigenic uroepithelial cell line. These arsenicals appear to have distinctive effects on different epigenetic mechanisms. MAsIII has been found to be more potent than iAsIII in suppressing gene expression and protein level of Xeroderma pigmentosum complementation group C, a DNA damage recognition protein, in normal human skin fibroblast (Nollen et al. 2009). Suppression of phosphatase and tensin homolog protein (PTEN) in MAsIII-exposed normal human urothelial (UROtsa) cells may be a potential mechanism for malignant transformation of cells by this arsenical (Oliva-González et al. 2017). MAsIII also downregulates expression and activity of cytochrome P450 1A1 in human hepatocellular carcinoma (HepG2) cells (Elshenawy et al. 2017) which methylate arsenic (Drobna et al. 2006). Finally, exposure of K6/ODC mice to MAsIII in drinking water alters expression of multiple genes in the mouse skin; notably, an increased transcript abundance of Fosl1, Myc, and Rac1 oncogenes was detected (Delker et al. 2009). However, it is unclear if methylation of MAsIII contributes to gene transcription dysregulation in this mouse model.
Expression of microRNAs, short, non-coding RNAs that negatively regulate gene expression at the post-transcriptional level, can be altered by exposure to iAs (Beck et al. 2017, 2018; Cardoso et al. 2018; Ferragut Cardoso et al. 2020; Sollome et al. 2016). Inhibition of glucose-stimulated insulin secretion in iAsIII-exposed INS-1 832/13 cells alters expression of several microRNAs, including microRNA-146a, which is involved in regulation of beta-cell function (Beck et al. 2019). Interestingly, MAsIII which is more potent than iAsIII as inhibitor of insulin secretion by INS-1 832/13 cells (Dover et al. 2018), does not alter microRNA-146a expression.
Methylated trivalent arsenicals as indicators of disease risk in humans cohorts
The efficiency of iAs methylation is one of the key factors that affect susceptibility to adverse effects of iAs exposure (Gamboa-Loira et al. 2017; Kuo et al. 2017; Tseng 2007). In humans exposed to iAs, risks of developing iAs-associated diseases have been linked to proportions of iAs and its methylated metabolites in urine, which are thought to reflect the efficiency of iAs methylation. The evaluation of the roles of MAsIII and DMAsIII in disease initiation and progression is difficult. These species can be easily oxidized to MAsV and DMAsV (Gong et al. 2001). Thus, it is unclear whether measurement of stable arsenicals in urine (iAsIII, iAsV, MAsV and DMAsV) provides germane information about levels of MAsIII and DMAsIII in tissues or excreta. As a consequence, most population-based studies report total MAs and DMAs levels in urine and do not attempt to determine the oxidation state of As in methylated metabolites.
MAsIII and DMAsIII levels have been determined in some studies that analyzed freshly collected urines. Results from these studies has made it possible to examine associations between levels of these arsenicals in urine and disease phenotypes. In 2005, Valenzuela and associates reported presence of MAsIII and DMAsIII in urine of residents of Zimapan, Hidalgo state, Mexico, who were chronically exposed to iAs (Valenzuela et al. 2005). In this cohort, AS3MT (Met287Thr) variant was associated with a higher risk of premalignant skin lesions, a common indicator of chronic iAs toxicity (Valenzuela et al. 2009). Notably, the fraction of urinary As present as MAs is higher in individuals carrying this polymorphism than in other participants. In affected individuals, MAsIII accounts for a significant portion of urinary MAs, suggesting that this trivalent arsenical contributes to the cancer risk associated with iAs exposure. Later studies in Zimapan and Lagunera link urinary DMAsIII concentration to prevalence of diabetes mellitus (Drobna et al. 2013), another disease associated with chronic exposure to iAs (Kuo et al. 2017; Maull et al. 2012). In this cohort, carriers of AS3MT (Met287Thr) variant had higher concentrations of DMAsIII in urine and a higher likelihood of a diagnosis of diabetes (Drobna et al. 2013). Presence of MAsIII and/or DMAsIII in human urine has also been reported in several other studies (Le et al. 2000a, b; Aposhian et al. 2000). To date, only one population-based study has examined associations between a disease risk and concentrations of tri- and pentavalent arsenicals in cells originated from a tissue targeted by iAs exposure, specifically urinary bladder. This study found positive associations between risk of diabetes and concentrations of trivalent, but not pentavalent, arsenicals in exfoliated urothelial cells isolated from urine of the individuals exposed to iAs in drinking water (Currier et al. 2014).
Oxidation state-specific analysis of arsenic species in biological matrices
Understanding dose–response relations for toxic actions of iAs and its methylated metabolites depends upon the ability to accurately measure the levels of these compounds in complex biological matrices. Methods used for analysis of arsenic species in biological matrices have been reviewed (Francesconi and Kuehnelt 2004; Gong et al. 2002a; Hsu et al. 2011; Reid et al. 2020). This section focuses on oxidation-state specific methods designed to differentiate among inorganic and methylated arsenic species that contain AsIII or AsV. In broad terms, these methods involve two steps: (a) separation of As species based on arsenic oxidation-state and methylation status and (b) quantification of elemental arsenic in the separated fractions using established optical spectroscopic techniques or mass spectrometry. Differences in the approaches used to separate arsenic species influence the sensitivity and specificity of these methods and the requirement for sample processing and/or extraction before analysis. Two widely used analytical approaches are: (a) liquid chromatographic (LC) separation of arsenic species on the basis of charge or size of analytes and (b) hydride generation (HG) in which arsenicals are reduced to volatile arsines, cryogenically trapped, and separated by boiling points. For both approaches, separated arsenicals can be detected by various mass-dependent or spectrophotometric methods. The following paragraphs describe advantages and limitations associated with LC- and HG-based techniques in context of studies in which these techniques were used.
HG-based techniques
Analysis of arsenic species using HG is carried out in several steps. In the first step, samples are treated with a strong reductant, typically sodium borohydride. This treatment converts iAs and mono-, di- and tri-methylated arsenic species to their respective volatile arsines which are characterized by different boiling points (b.p.): arsine (AsH3, b.p. − 55 °C), methylarsine (CH3AsH2, b.p. 2 °C), dimethylarsine [(CH3)2AsH, b.p. 36 °C] and trimethylarsine [(CH3)3As, b.p. 52 °C]. The HG reaction mixture is passed through a gas–liquid separator and the gaseous phase containing arsines is carried in a stream of an inert gas (e.g., helium) into a cryogenic trap (CT), typically a U-tube filled with a chromatographic sorbent, that is submerged in liquid nitrogen. Cold-trapped arsines are sequentially volatized from the U-tube by controlled heating. Released arsines are then carried as separate fractions to an appropriate detector (Crecelius 1977). As summarized in Table 1, both optical and mass spectrometers have been used to detect and quantify elemental arsenic present in the arsines produced by HG.
Table 1.
Examples of studies in which HG-CT-based techniques were used for detection and quantification of MAsIII and DMAsIII in biological samples
| Technique | Sample pre-treatment | pHa | Sample | As species | References |
|---|---|---|---|---|---|
|
| |||||
| HG-CT-AAS | None | pH ≤ 2 and pH 6 | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | García-Montalvo et al. (2011) |
| None | pH ≤ 2 and pH 6 | Urine, hepatoma (HepG2) cells | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Del Razo et al. (2001) | |
| None | pH ≤ 2 and pH 6 | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Valenzuela et al. (2005) | |
| None | pH 1 and pH 6 | In vitro methylation system, primary hepatocytes, urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII, TMAsO | Devesa et al. (2004) | |
| ± 2% Cysteine | pH 6 | In vitro methylation system, cell culture | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Hernández-Zavala et al. (2008) | |
| ± 2% Cysteine | pH 6 | In vitro methylation system, cell culture | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Currier et al. (2013) | |
| ± 2% Cysteine | pH 6 | Homogenates from multiple tissues, plasma, blood cells | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII, TMAsO | Currier et al. (2016) | |
| ± 2% Cysteine | pH 6 | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Matoušek et al. (2008) | |
| ± 2% Cysteine | pH 6 | Liver homogenate | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Currier et al. (2011a) | |
| ± 2% Cysteine | pH 6 | UROtsa/F35, liver homogenate | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Currier et al. (2011b) | |
| ± 2% Cysteine | pH 6 | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Del Razo et al. (2011) | |
| HG-CT-ICP-MS | ± 2% Cysteine | pH 6 | River water (CRM SLRS-4), exfoliated bladder epithelial cells | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Matoušek et al. (2013) |
| ± 2% Cysteine | pH 6 | Exfoliated urothelial cells | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Currier et al. (2014) | |
Reports that described development of techniques for analysis of MAsIII and DMAsIII using mixtures if standards in water or a solvent but did not validate these methods in biological matrices are not included
pH of the hydride generation mixture
The original method for As speciation by HG used a strongly acidic solution (pH ~ 1) to generate arsines from both tri- and pentavalent iAs, MAs, and DMAs (Ng et al. 1998). Two methods are currently used for oxidation-state specific analysis. Both methods require two aliquots of a single sample for complete analysis. In the first method, arsines from both tri- and pentavalent arsenicals are generated from one sample aliquot at pH 1. The second aliquot undergoes HG at pH 6. Under these conditions, arsines are generated only from trivalent arsenicals. Concentrations of pentavalent arsenicals are then determined as the difference between the first and second measurement (Del Razo et al. 2001; Devesa et al. 2004). In the second method, trivalent arsenicals are measured in one sample aliquot after HG at pH 6. The second aliquot is treated with 2% cysteine prior to HG at pH 6. Cysteine reduces pentavalent arsenicals to trivalency. The concentrations of pentavalent arsenicals are again calculated as the differences between the concentrations measured in the cysteine-treated and in the untreated samples (Matoušek et al. 2008). The latter method is more convenient because the calibration is carried out using one HG condition (pH 6) and because the HG efficiencies for iAsIII, MAsIII and DMAsIII are almost identical, allowing calibration with a single arsenic standard, typically iAsIII (Matoušek et al. 2008). Although TMAsVO can be measured by this method, lower efficiency for hydride generation for this species may require a separate calibration using an authentic standard.
Advantages of the HG-based techniques
HG-CT methods were developed specifically for detection of iAs and the methylated arsenicals formed during enzymatically catalyzed iAs methylation. Although published methods are typically designed for analysis of samples with volumes between 0.1 and 0.5 ml, the HG procedure can easily be scaled-up to use much larger sample volumes. In addition, the HG-CT step can be repeated before release of arsines to a detector, which allows quantification of arsenicals present at very low concentrations. In particular, very low detection limits have been achieved by methods that coupled the HG-CT steps with arsenic detection by inductively coupled plasma-mass spectrometry (ICP-MS) (Matoušek et al. 2013). Notably, the HG-CT-based techniques can be used for quantitative analysis of AsIII and AsV-containing species in complex biological matrices without sample extraction. Elimination of sample processing before analysis minimizes the chance of conversion of AsIII to AsV during extraction. HG-CT techniques, specifically HG-CT-atomic absorption spectrometry (AAS) or HG-CT-atomic fluorescence spectrometry (AFS), provide nearly complete recoveries of As when used for analysis of complex matrices (tissue homogenates or cell lysates) (Currier et al. 2011a, b, 2013, 2014, 2016; Matoušek et al. 2013; Musil et al. 2014; Paul et al. 2007b, 2008). Finally, the relatively low cost of instrumentation needed for As speciation analysis by HG-CT as well as very low detection limits that can be attained makes this technique an attractive alternative to more costly LC-based techniques for As speciation.
Limitations of the HG-based techniques
A major limitation is the lack of commercial automated HG-CT systems that directly interface with existing detectors. Thus, a typical setup requires assembly of an online system consisting of individual, often custom-made, parts that support HG reaction, separation of gas phase from the reaction mixture, delivery of the gas phase into a cryotrap, heating the cryotrap, and delivery of the volatile arsine fractions into a detector. The HG and CT steps, including controlled heating of the U-tube, are typically carried out on-line and are fully automated. The flow of the reagents and timing of each step are controlled by a programable flow injection system. Although automated release of arsines from the cryotrap has been attempted (Matoušek et al. 2013), application of this method typically requires manual operations, including sample injection and removing the U-tube from liquid nitrogen prior to heating. Because all critical steps cannot be easily automated, autosamplers cannot be used and presence of an operator is required during the entire procedure. Analysis of a sample takes approximately 6 min, using a well-tuned-up system.
The HG-CT techniques can be used only for analysis or arsenic species that form hydrides. Some arsenicals that are commonly found in food and biological systems cannot be detected by these techniques. Published studies have shown that arsenobetaine and arsenocholine, two arsenicals found in urine of individuals-consuming seafood, do not generate arsines under conditions typically used for HG-CT-based analyses of arsenic species (Currier et al. 2013). Similarly, generation of dimethyl arsine from dimethylthioarsinic acid, a product of arsenic metabolism by microbiota, is much less efficient than from DMAsV or DMAsIII (Hernández-Zavala et al. 2008). Generation of hydrides from arsenosugars, arsenic species found in seaweed and some marine organisms, have been achieved only in a modified, strongly acidic HG reaction mixture (Marschner et al. 2018). Although viewed as a major limitation, the inability of HG-CT-based methods to detect arsenicals that do not form hydrides represents an advantage for analyses focusing on iAs and products of its methylation by AS3MT. It limits the possibility of false-positive signals caused by other arsenic species present in analyzed samples.
LC-based techniques
The LC techniques have been widely used for separation of arsenic species in aqueous and biological matrices. The most common methods involved separation of arsenic species by high-performance liquid chromatography (HPLC) coupled with an arsenic detector, typically ICP-MS. Several types of HPLC techniques have been used for separation of arsenicals, including anion exchange, cation exchange, reversed-phase, ion pair, or size exclusion chromatography (Francesconi and Kuehnelt 2004; Reid et al. 2020). In earlier studies focusing on iAs methylation, HPLC techniques were applied for analysis of iAsIII, iAsV, and the pentavalent methylated arsenicals. After recent optimizations, these techniques have also been utilized for analysis of MAsIII and DMAsIII (Gong et al. 2002a) (Table 2). These methods are discussed in the following sections.
Table 2.
Examples of studies in which HPLC-based techniques were used for detection and quantification of MAsIII and DMAsIII (including MAsIII and DMAsIII complexes with glutathione) in biological samples
| Separation | Column | Mobile phase | Detector | Sample | As species | References |
|---|---|---|---|---|---|---|
|
| ||||||
| Reversed phase/ion-pair | Phenomenex ODS(3) | 5 mM TBAH, 3 mM malonic acid, 5% methanol, pH 5.85 | HG-AFS | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV | Le et al. (2000a), Aposhian et al. (2000) |
| Phenomenex ODS(3) | 5 mM TBAH, 3 mM malonic acid, 5% methanol, pH 5.85 | HG-AFS | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Le et al. (2000b) | |
| Phenomenex ODS(3) | 4.7 mM TBAH, 2 mM malonic acid, 4% methanol, pH 5.85 or 5.9 | HG-AFS | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMPS-MAsIII | Gong et al. (2001) | |
| Phenomenex ODS(3) | 5 mM TBAH, 3 mM malonic acid, 5% methanol, pH 5.85 | HG-AFS | Red blood cells | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Lu et al. (2007) | |
| Phenomenex ODS(3) | 5 mM TBAH, 3 mM malonic acid, 5% methanol, pH 6.0 | ICP-MS | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Wang et al. (2004) | |
| Zorbax XDB | 10 mM TMAH, pH 9.0 | ICP-MS | SHIME | MAsIII, DMAsIII | Alava et al. (2012, 2013) | |
| Phenomenex ODS(3) | 4.7 mM TBAH, 2 mM malonic acid, 4% methanol, pH 5.85 | ICP-MS | In vitro methylation mixture | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII, DMMTA | Currier et al. (2013) | |
| Phenomenex ODS(3) | 5 mM TBAH, 3 mM malonic acid, 5% methanol, pH 5.65 | ICP-MS | Hair, fingernails, toenails | iAsIII, iAsV, MAsV, MAsIII, DMAsV, MMMTA, DMMTA | Chen et al. (2018) | |
| Phenomenex ODS(3) | 5 mM TBAH, 3 mM malonic acid, 5% methanol, pH 5.65 | ICP-MS | Saliva | iAsIII, iAsV, MAsV, MAsIII, DMAsV, MMMTA | Chen et al. (2013) | |
| Phenomenex Jupiter CIS 300A | 5 mM TBAH, 3 mM malonic acid, 5% methanol, pH 5.95 | ICP-MS | Cyano-bacterium Nostoc sp. PCC 7120 | iAsIII, iAsV, MAsV, MAsIII, DMAsV | Yan et al. (2015, 2017) | |
| Phenomenex Jupiter CIS 300A | 5 mM TBAH, 3 mM malonic acid, 5% methanol, pH 5.95 | ICP-MS | SHIME | iAsIII, iAsV, MAsV, MAsIII, DMAsV, MMMTA, | Chi et al. (2020) | |
| Phenomenex Luna CIS | 6 mM TBAH, 2 mM malonic acid, 5% methanol, pH 6.0 | ICP-MS | Hepatoma (HepG2) cells | MAsV, MAsIII, DMAsV, | Hippler et al. (2011) | |
| Phenomenex Aqua CIS | 0.1% formic acid (pH 2.6), acetonitrile gradient (0–40%) | ICP-MS | Urine | iAsIII(SG)3, MAsIII(SG)2, DMAs(GS) | Kala et al. (2004) | |
| Phenomenex Prodigy ODS-3 | 4.7 mM TBAH, 2 mM malonic acid, 4% methanol, pH 5.95 (adjusted with nitric acid) | ICP-MS | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, AsB | Rabieh et al. (2008) | |
| Waters Spherisorb C8 (at 10 °C) | 0.05% trifluoroacetate, acetonitrile gradient | ICP-MS | 8226/S Multiple myeloma cells | DMMTA, DMMTA(GS), DMAIII, DMAsIII(GS) | Stice et al. (2016) | |
| Excelpak ICS-A13 column | 3 mM NaH2PO4 at pH 6.0 with 1.0 mol l−1 NaOH | ICP-MS | Urine | MAsIII, DMAsIII, TMAsV after extraction to DDDC in CC14 and back-extraction to NaOH) | Okina et al. (2004) | |
| Ion exchange | Phenomenex Pheno-Sphere | 20 mM phosphate, 5% methanol, pH 7.0 | HG-AFS following on-line decomposition with 0.1 M NaOH | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMPS-MAsIII | Gong et al. (2002b) |
| Phenomenex Pheno-sphere SAX 80R | 20 mM Phosphate, 5% methanol (pH 5.8) (pretreated with DMPS) | HG-AFS | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII | Lu et al. (2003) | |
| Shodex Asahipak ES-502N 7C anion exchange | 15 mM citric acid, 10% HNO3, pH 2.0 | ICP-MS | Urine | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII, AsB, AsC | Mandal et al. (2001) | |
| Shodex Asahipak ES-502N 7C anion exchange | 15 mM citric acid, 10% HNO3, pH 2.0 | ICP-MS | Hair, fingernails | iAsIII, iAsV, MAsV, DMAsV, DMAsIII | Mandal et al. (2003) | |
| Shodex Asahipak ES-502N 7C anion exchange | 10–40 mM citric acid, pH 2.0–3.5 | ICP-MS | Urine, plasma, red blood cells, hair, fingernails | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII, AsB | Mandal et al. (2004) | |
| Shodex Asahipak ES-502N 7C anion exchange | 15 mM citric acid, 10% HNO3, pH 2.0 | ICP-MS | Urine, plasma, red blood cells | iAsIII, iAsV, MAsV, MAsIII, DMAsV, DMAsIII, AsB | Mandal et al. (2007) | |
| Shodex Asahipak ES-502N 7C anion exchange | 15 mM citric acid buffer, pH 2.0 | ICP-MS | Bile | iAsIII, MAsIII, iAsIII(SG)3, MAsIII(SG)2 | Bu et al. (2011) | |
| Shodex Rspak NN-614 cation exchange | 36 mM formic acid, 2 mM ammonium formate, pH 2.81 | ICP-MS | Red blood cells | DMAsV, DMAsIII | Shiobara et al. (2001) | |
| Shodex Rspak NN-614 cation exchange | 5 mM HNO3, 6 mM NH4NO3 | ICP-MS | Urine | MAsIII, DMAsIII (after extraction to DDDC in CC14 and back-extraction to NaOH) | Okina et al. (2004) | |
| Shodex Asahipak ES-502N 7C anion exchange | 15 mM citric acid (pH 2.0 adjusted with nitric acid) | ICP-MS | Bile | iAsIII, MAsV, iAsIII(SG)3, MAsIII(SG)2 | Suzuki et al. (2001) | |
| Hamilton PRP-X100 anion exchange | 20 mM phosphate-ammonia buffer, pH 6.0 | ICPQQQ-MS | Elaphomyces spp. (deer truffles) | iAsV, MAsV, MAsIII, DMAsV | Braeuer et al. (2018) | |
| Hamilton PRP-X100 anion exchange | 98% 10 mM ammonium phosphate (pH 8.25) and 2% methanol | ICP-MS | Blood, multiple tissues | DMAsV, DMAsIII | Twaddle et al. (2018a) | |
| Hamilton PRP-X100 anion exchange | 98% 10 mM ammonium phosphate (pH 8.25) and 2% methanol | ICP-MS | Blood, multiple tissues | iAsIII, iAsV, MAsV, DMAsV, MAsIII + DMAsIII | Twaddle et al. (2018b) | |
| Hamilton PRP-X100 anion exchange | 98% 10 mM ammonium phosphate (pH 8.25) and 2% methanol | ICP-MS | Blood, multiple tissues | DMAsV, DMAsIII | Twaddle et al. (2019) | |
| Hamilton PRP-X100 anion exchange | 10 mM ammonium dihydrogen phosphate, 10 mM ammonium nitrate, pH6.2 | ICP-MS | E. Coli | iAsIII, iAsV, MAsV, MAsIII, DMAsV, TMAsO | Yan et al. (2017) | |
| Hamilton PRP-X100 anion exchange | 10 mM ammonium dihydrogen phosphate, 10 mM ammonium nitrate, pH6.2 | ICP-MS | SHIME | iAsIII, iAsV, MAsV, MAsIII, DMAsV, MMMTA | Chi et al. (2020) | |
| Dionex AS11 and AG11 anion exchange | Water with 50 mM NaOH gradient (0–80%) | ICP-MS | Urine | iAsIII, iAsV, MAsV, DMAsV, MAsIII + DMAsIII + AsB | Xie et al. (2006) | |
| Dionex PCX-500 cation exchange | 70 mM nitric acid | ICP-MS | Urine | iAsIII, + iAsV + MAsV + DMAsV, MAsIII, DMAsIII, AsB | Xie et al. (2006) | |
| Size exclusion (Gel filtration) | Shodex Asahipak GS-220 HQ | 50 mM ammonium acetate buffer, pH 6.5, or 15 mM citric acid buffer, pH 2.0 | ICP-MS | Liver | MAsV, DMAsV, DMAsIII, DMTAV, DMTAIII, AsB | Suzuki et al. (2004) |
Reports that described development of techniques for analysis of MAsIII and DMAsIII using mixtures of standards in water or a solvent but did not validate these methods in biological matrices are not included
DDDC diethylammonium diethyldithiocarbamate, TBAH tetrabutylammoniumhydroxide, TMAH trimethylammoniumhydroxide, SHIME simulator of the human intestinal microbial ecosystem
Advantages of the LC-based techniques
The instruments required for LC-based speciation analysis of arsenic, including HPLC instruments and chromatographic columns, as well as spectrometers, are commercially available and can easily be configured for a high-throughput analysis. Software for operating these instruments and for data analysis is also available. For optimized HPLC techniques, all tri- and pentavalent arsenic species formed in the AS3MT-catalyzed reactions can be quickly separated on a single column (Reid et al. 2020; Currier et al. 2013). Separation of other biologically relevant arsenicals can also be achieved on a single column or by combining several LC techniques (Reid et al. 2020). In principle, negatively charged arsenic compounds can be separated using anion-exchange chromatography while positively charged arsenicals are best separated using cation-exchange chromatography (Francesconi and Kuehnelt 2004; Reid et al. 2020). In addition, using ICP-MS in tandem with an electrospray ionization mass spectrometer (ESI–MS) can provide information about molecular structures of arsenic species in chromatographic fractions (Reid et al. 2020). This is particularly important during analysis of biological samples in which arsenicals may be present in complexes with endogenous compounds, e.g., GSH (Raab et al. 2004). ICP-MS in tandem with ESI–MS can be also used to identify multiple arsenic species that are eluted with the same retention times (Reid et al. 2020). Thus, the LC-based techniques are suitable for routine analysis of known arsenic species, as well as for discovery of previously unidentified arsenic species.
Limitations of the LC-based techniques
Minimizing oxidation of unstable MAsIII and DMAsIII during sample preparation and chromatographic separation is the major challenge for LC-based techniques (Feldman et al. 1999; Gong et al. 2001). For example, bulk filtration of urine samples to remove particulates before sample injection to a chromatographic column or use of an in-line guard column between injector and the analytical column may promote the oxidation of MAsIII or DMAsIII. Analysis of more complex biological matrices requires liquid-phase extraction of arsenic species. A variety of solvents have been used with varied extraction efficiencies (Francesconi and Kuehnelt 2004; Luvonga et al. 2020). Sample extraction can lead to analyte dilution and adversely affect detection or quantification after LC separation. More importantly, MAsIII and/or DMAsIII may oxidize during extraction if an oxidizing solvent is used or if prolonged exposure to oxygen occurs. Notably, effects of the extraction by different solvents on oxidation states of arsenic in biological matrices have never been systematically studied.
Another challenge is associated with a potential loss of MAsIII and DMAsIII due to interaction with a mobile phase or a stationary phase of the column. A loss of DMAsIII on a Phenomenex Prodigy column with a tetrabutylammonium hydroxide-malonic acid–methanol mobile phase has been documented (Currier et al. 2013). However, no systematic research of the interaction between methylated trivalent arsenical and sorbents or solvents commonly used in LC techniques has been published. Thus, the oxidation state-specific analysis of arsenic species using LC techniques requires careful consideration and testing of chromatographic columns and mobile phases to minimize the oxidation of MAsIII and DMAsIII and to ensure a complete recovery of arsenic species during chromatography.
Knowledge gaps and future directions
Transport of arsenic species
The formation of methylated metabolites that contain either AsIII or AsV in the course of iAs metabolism is supported by results of the oxidation-specific analyses of arsenic species in a variety of biological systems. Overwhelming evidence also suggests that AS3MT is the primary catalyst for arsenic methylation in cells. Recent studies on the protein structure of AS3MT and the molecular processes involved in AS3MT-catalyzed reactions support the production of MAsIII and DMAsIII at the catalytical center of this enzyme (Marapakala et al. 2012). DMAsV and MAsV are likely formed by a rapid oxidation of the unstable trivalent species in oxygen-rich environment. The efficiency of iAs metabolism has been linked to susceptibility to iAs toxicity in human population studies using the proportions of arsenic metabolites in urine as indicators of the capacity to convert iAs to MAs and MAs to DMAs. However, it is reasonable to assume that the capacity to methylate iAs is not the only factor that determines the profiles of arsenic species in urine and the overall efficiency of iAs metabolism. For example, arsenic transporters that carry arsenic species from tissues to blood, bile or urine likely play a role. Several transporters have been proposed to participate in these processes, e.g., aquaglyceroporins, glucose (GLUT) transporters, organic anion transporters, or ATP-cassette transporters (Drobna et al. 2010; Garbinski et al. 2019; Leslie 2012; Liu 2010; Maciaszczyk-Dziubinska et al. 2012; Roggenbeck et al. 2016; Rosen and Liu 2009; Zangi and Filella 2012). However, a little is known about the specificity and affinity of these transporters for methylated arsenic species, specifically MAsIII and DMAsIII, and about contributions of these transporters to the whole-body metabolism and detoxification of iAs.
Protein binding of arsenicals
Another understudied issue is the binding of iAs and its metabolites to proteins in various cellular compartments. Binding to protein targets affects intracellular retention and distribution of arsenicals and the availability of these arsenicals for AS3MT-catalyzed reactions and for membrane transport. Although several studies have described subcellular distribution of arsenicals in mammalian cells (Dopp et al. 2008, 2010) and examined interactions between common protein motifs and arsenic (Kitchin and Wallace 2008; Lu et al. 2004; Shen et al. 2013; Yan et al. 2009, 2016; Zhang et al. 2015), these data are not sufficient to develop a comprehensive model for the role of protein–As interactions.
Data on binding of arsenicals, specifically MAsIII and DMAsIII, to cellular components would provide insights into the mechanistic basis of adverse effects associated with iAs exposure. Although reactions between iAs and protein and peptide thiol groups have been well-documented, our understanding of thiol interactions with MAsIII and DMAsIII are less complete.
Interactions between MAsIII and Zn finger motifs in the DNA-binding domain of the glucocorticoid receptor suggest a basis for some adverse effects of exposure to this arsenical (Spuches and Wilcox 2008). Similarly, identification of cysteinyl residues in alpha chain of rat hemoglobin as a high-affinity and high-capacity target for DMAsIII binding provides a molecular basis for the anomalous retention of DMAsIII in rat erythrocytes (Lu et al. 2007). Some studies have provided an indirect evidence for binding of methylated trivalent arsenicals to specific proteins. For example, MAsIII-dependent inhibition of insulin-stimulated phosphorylation of Akt by phosphoinositide-dependent protein kinase-1 (PDK-1) (Paul et al. 2007a; b) may involve an interaction with two closely spaced cysteines (Cys21 and Cys23) in the N-terminus of PDPK1 (Alessi et al. 1997; Dong et al. 1999). The availability of MAsIII and DMAsIII should permit additional studies to identify proteins that bind these arsenicals and to elucidate the functions of these protein-arsenical complexes.
Analysis of MAsIII and DMAsIII in samples collected in human studies
The ultimate evidence linking the formation of MAsIII and DMAsIII to adverse health effects associated with iAs exposure will likely come from human population-based or clinical studies. A major obstacle to obtaining such evidence is associated with the low stability of MAsIII and DMAsIII, as well as with limitations associated with non-invasive collection of biological samples from human cohorts. In addition, many of the current analytical methods are not optimized or validated for the oxidation-state specific analysis of arsenic species in biological matrices. Attempts have been made to overcome these obstacles. One study measured tri- and pentavalent arsenicals in human urine within 1 to 6 h after collection; these urine samples were frozen in dry ice immediately after collection (Del Razo et al. 2011). Using this approach, only small fraction of DMAsIII spiked into the urine samples was oxidized to DMAsV. Another approach to stabilize trivalent arsenicals in urine involves treatment of the donor with an arsenic chelator, 2,3-dimercapto-1-propane sulfonate (DMPS) (Gong et al. 2002b). Individuals in Romania who were exposed to high levels of iAs in their water supply received single oral doses of DMPS and urine was collected before and for up to 8 h post DMPS treatment. The presence of a MAsIII-DMPS complex in urines collected after DMPS treatment suggests that it is possible to use chelation to stabilize the oxidation state of As. However, it is probably not feasible to use this approach in population-based studies. In addition, treatment with a chelator is likely to modify the pattern of iAs metabolism, thus confounding evaluation of the associations between iAs metabolism and disease risk. Direct addition of an AsIII-complexing agent to freshly collected urine may be an alternative approach for stabilization of arsenic oxidation state before analysis. However, the capacity of some dithiols to reduce pentavalent arsenicals to trivalency may be an important caveat for this approach (Delnomdedieu et al. 1993, 1994).
Analysis of arsenic species in samples other than urine represents a conceptually different and challenging problem. Concentrations of tri- and pentavalent arsenicals in exfoliated urothelial cells isolated from urine of individuals exposed to iAs in drinking water have been measured (Currier et al. 2014). In this study, concentrations of iAsIII, MAsIII and DMAsIII in exfoliated cells positively correlated with indicators of the diabetogenic effects of iAs exposure. A similar approach could be used to analyze tri- and pentavalent arsenic species in other types of samples that can be collected using non-invasive procedures, e.g., buccal cells or nasal epithelial cells. In addition to providing information about the exposure to iAs and the disposition of arsenic species in tissues, this type of analyses may also help link the presence of individual arsenicals in tissues to adverse events related to iAs exposure.
Summary/conclusion
Although research has greatly expanded our knowledge of arsenic metabolism at the molecular, cellular, and organismic level, there are still lacunae to be explored. In particular, our understanding of linkages among genotypic variation in AS3MT, interindividual variation in capacity for arsenic methylation, and interindividual variation in susceptibility to arsenic-induced diseases is incomplete. As discussed above, current evidence suggests that polymorphisms in the human AS3MT gene affects the pattern of methylated arsenicals in urine and that these changes in absolute and relative contributions of MAs or DMAs to the yield of arsenic metabolites are linked to increased incidences of a variety of diseases associated with exposure to iAs. However, it is unclear how relatively modest changes in the excretion of MAs and DMAs related to AS3MT genotype can account for differences in disease incidence. It is possible that these differences reflect genotype-dependent changes in relative proportions of MAsIII and MAsV produced during AS3MT-catalyzed methylation. Notably, other factors unrelated to AS3MT polymorphism, e.g., sex or diet have also been shown to modulate urinary profiles of iAs and its methylated metabolites with possible links to disease risks. The roles of these factors in formation of the methylated trivalent metabolites remains unclear. Development and application of oxidation state-selective analytical methods may permit detailed study of formation of tri- and pentavalent MAs and DMAs in cellular systems and provide a fuller understanding of the role of MAsIII and DMAsIII in disease initiation and progression.
Acknowledgements
This work was funded by the National Institutes of Health (NIH) grant R01ES028721 to M.S. and R.C.F. and the UNC Superfund Research Program grant P42ES031007. Additional support was provided by the UNC Institute for Environmental Health Solutions. The authors would like to thank Madison Miller and Tara Brennan for their help with literature searches.
Funding
The authors declare they have no actual or potential competing financial interests.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest. This document was reviewed by the U.S. EPA Center for Computational Toxicology and Exposure for publication. Approval does not signify that the contents reflect the views of the Agency nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
References
- Adair BM, Moore T, Conklin SD, Creed JT, Wolf DC, Thomas DJ (2007) Tissue distribution and urinary excretion of dimethylated arsenic and its metabolites in dimethylarsinic acid- or arsenate-treated rats. Toxicol Appl Pharmacol 222:235–242. 10.1016/j.taap.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Ajees AA, Marapakala K, Packianathan C, Sankaran B, Rosen BP (2012) Structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation. Biochemistry 51:5476–5485. 10.1021/bi3004632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alava P, Tack F, Laing GD, de Wiele TV (2012) HPLC-ICP-MS method development to monitor arsenic speciation changes by human gut microbiota. Biomed Chromatogr 26(4):524–533. 10.1002/bmc.1700 [DOI] [PubMed] [Google Scholar]
- Alava P, Tack F, Laing GD, Van de Wiele T (2013) Arsenic undergoes significant speciation changes upon incubation of contaminated rice with human colon micro biota. J Hazard Mater 262:1237–1244. 10.1016/j.jhazmat.2012.05.042 [DOI] [PubMed] [Google Scholar]
- Alessi DR, Deak M, Casamayor A, Caudwell FB, Morrice N, Norman DG, Gaffney P, Reese CB, MacDougall CN, Harbison D, Ashworth A, Bownes M (1997) 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr Biol 7(10):776–789. 10.1016/s0960-9822(06)00336-8 [DOI] [PubMed] [Google Scholar]
- Aposhian HV, Gurzau ES, Le XC, Gurzau A, Healy SM, Lu X, Ma M, Yip L, Zakharyan RA, Maiorino RM, Dart RC, Tircus MG, Gonzalez-Ramirez D, Morgan DL, Avram D, Aposhian MM (2000) Occurrence of monomethylarsonous acid in urine of humans exposed to inorganic arsenic. Chem Res Toxicol 13(8):693–697. 10.1021/tx000114o [DOI] [PubMed] [Google Scholar]
- Arnold LL, Eldan M, van Gemert M, Capen CC, Cohen SM (2003) Chronic studies evaluating the carcinogenicity of monomethylarsonic acid in rats and mice. Toxicology 90:197–219. 10.1016/s0300-483x(03)00165-3 [DOI] [PubMed] [Google Scholar]
- Arnold LL, Eldan M, Nyska A, van Gemert M, Cohen SM (2006) Dimethylarsinic acid: results of chronic toxicity/oncogenicity studies in F344 rats and in B6C3F1 mice. Toxicology 223:82–100. 10.1016/j.tox.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Bailey KA, Fry RC (2014) Arsenic-associated changes to the epigenome: what are the functional consequences? Curr Environ Health Rep 1(1):22–34. 10.1007/s40572-013-0002-8 (eCollection 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Wallace K, Smeester L, Thai SF, Wolf DC, Edwards SW, Fry RC (2012) Transcriptional modulation of the ERK1/2 MAPK and NF-κB pathways in human urothelial cells after trivalent arsenical exposure: implications for urinary bladder cancer. J Can Res Updates 21(1):57–68 [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, García-Vargas G, Del Razo LM, Drobná Z, Stýblo M, Fry RC (2013) Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. J Biochem Mol Toxicol 27(2):106–115. 10.1002/jbt.21462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FD, Krohmer LJ, Hamilton JW, Sheldon LA (2009) Disruption of histone modification and CARM1 recruitment by arsenic represses transcription at glucocorticoid receptor-regulated promoters. PLoS ONE 4(8):e6766. 10.1371/journal.pone.0006766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Styblo M, Sethupathy P (2017) Arsenic exposure and type 2 diabetes: microRNAs as mechanistic links? Curr Diab Rep 17(3):18. 10.1007/s11892-017-0845-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Bommarito P, Douillet C, Kanke M, Del Razo LM, García-Vargas G, Fry RC, Sethupathy P, Stýblo M (2018) Circulating miRNAs Associated with Arsenic Exposure. Environ Sci Technol 52(24):14487–14495. 10.1021/acs.est.8b06457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Chandi M, Kanke M, Stýblo M, Sethupathy P (2019) Arsenic is more potent than cadmium or manganese in disrupting the INS-1 beta cell microRNA landscape. Arch Toxicol 93(11):3099–3109. 10.1007/s00204-019-02574-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergquist ER, Fischer RJ, Sugden KD, Martin BD (2009) Inhibition by methylated organo-arsenicals of the respiratory 2-oxo-acid dehydrogenases. J Organomet Chem 694(6):973–980. 10.1016/j.jorganchem.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolero F, Pozzi G, Sabbioni E, Saffiotti U (1987) Cellular uptake and metabolic reduction of pentavalent to trivalent arsenic as determinants of cytotoxicity and morphological transformation. Carcinogenesis 8:803–808. 10.1093/carcin/8.6.803 [DOI] [PubMed] [Google Scholar]
- Braeuer S, Borovička J, Goessler W (2018) A unique arsenic speciation profile in Elaphomyces spp. (“deer truffles”)-trimethylarsine oxide and methylarsonous acid as significant arsenic compounds. Anal Bioanal Chem 410(9):2283–2290. 10.1007/s00216-018-0903-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredfeldt TG, Jagadish B, Eblin KE, Mash EA, Gandolfi AJ (2006) Monomethylarsonous acid induces transformation of human bladder cells. Toxicol Appl Pharmacol 216:69–79. 10.1016/j.taap.2006.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu N, Wang HY, Hao WH, Liu X, Xu S, Wu B, Anan Y, Ogra Y, Lou YJ, Naranmandura H (2011) Generation of thioarsenicals is dependent on the enterohepatic circulation in rats. Metallomics 3(10):1064–1073. 10.1039/c1mt00036e [DOI] [PubMed] [Google Scholar]
- Buchet JP, Lauwerys R (1985) Study of inorganic arsenic methylation by rat liver in vitro: relevance for the interpretation of observations in man. Arch Toxicol 57:125–129. 10.1007/BF00343122 [DOI] [PubMed] [Google Scholar]
- Buchet JP, Lauwerys R, Roels H (1981) Comparison of the urinary excretion of arsenic metabolites after a single oral dose of sodium arsenite, monomethylarsonate, or dimethylarsinate in man. Int Arch Occup Environ Health 48:71–79. 10.1007/BF00405933 [DOI] [PubMed] [Google Scholar]
- Calatayud M, Gimeno-Alcaniz JV, Velez D, Devesa V (2014) Trivalent arsenic species induce changes in expression and levels of proinflammatory cytokines in intestinal epithelial cells. Toxicol Lett 224(1):40–46. 10.1016/j.toxlet.2013.09.016 [DOI] [PubMed] [Google Scholar]
- Calatayud M, Gimeno-Alcaniz JV, Devesa V, Velez D (2015) Proinflammatory effect of trivalent arsenical species in a co-culture of Caco-2 cells and peripheral blood mononuclear cells. Arch Toxicol 89(4):555–564. 10.1007/s00204-014-1271-1 [DOI] [PubMed] [Google Scholar]
- Cardoso APF, Al-Eryani L, States JC (2018) Arsenic-induced carcinogenesis: the impact of miRNA dysregulation. Toxicol Sci 165(2):284–290. 10.1093/toxsci/kfy128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challenger F (1947) Biological methylation. Sci Prog 35:396–416 [PubMed] [Google Scholar]
- Challenger F (1951) Biological methylation. Adv Enzymol Relat Subj Biochem 12:429–491. 10.1002/9780470122570.ch8.1951 [DOI] [PubMed] [Google Scholar]
- Chen GQ, Zhou L, Styblo M, Walton F, Jing Y, Weinberg R, Chen Z, Waxman S (2003) Methylated metabolites of arsenic trioxide are more potent than arsenic trioxide as apoptotic but not differentiation inducers in leukemia and lymphoma cells. Cancer Res 63(8):1853–1859 [PubMed] [Google Scholar]
- Chen B, Arnold LL, Cohen SM, Thomas DJ, Le XC (2011) Mouse arsenic (+3 oxidation state) methyltransferase genotype affects metabolism and tissue dosimetry of arsenicals after arsenite administration in drinking water. Toxicol Sci 124:320–326. 10.1093/toxsci/kfr246 [DOI] [PubMed] [Google Scholar]
- Chen B, Cao F, Yuan C, Lu X, Shen S, Zhou J, Le XC (2013) Arsenic speciation in saliva of acute promyelocytic leukemia patients undergoing arsenic trioxide treatment.Anal Bioanal Chem 405(6):1903–1911. 10.1007/s00216-012-6700-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun GX, Wang XX, Lorenzo Vd, Rosen BP, Zhu YG (2014) Volatilization of arsenic from polluted soil by Pseudomonas putida engineered for expression of the arsM Arsenic(III) S-adenosine methyltransferase gene. Environ Sci Technol 48:10337–10344. 10.1021/es502230b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li J, Jiang X, Rosen BP (2017a) Conserved cysteine residues determine substrate specificity in a novel As(III) S-adenosylmethionine methyltransferase from Aspergillus fumigatus. Mol Microbiol 104:250–259. 10.1111/mmi.13628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Sun GX, Rosen BP, Zhang SY, Deng Y, Zhu BK, Rensing C, Zhu YG (2017b) Recurrent horizontal transfer of arsenite methyltransferase genes facilitated adaptation of life to arsenic. Sci Rep 7(1):7741. 10.1038/s41598-017-08313-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Cao F, Lu X, Shen S, Zhou J, Le XC (2018) Arsenic speciation in hair and nails of acute promyelocytic leukemia (APL) patients undergoing arsenic trioxide treatment. Talanta 1(184):446–451. 10.1016/j.talanta.2018.03.021 [DOI] [PubMed] [Google Scholar]
- Chen J, Yoshinaga M, Rosen BP (2019) The antibiotic action of methylarsenite is an emergent property of microbial communities. Mol Microbiol 111:487–494. 10.1111/mmi.14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SC, Sun GX, Yan Y, Konstantinidis KT, Zhang SY, Deng Y, Li XM, Cui HL, Musat F, Popp D, Rosen BP, Zhu YG (2020) The Great Oxidation Event expanded the genetic repertoire of arsenic metabolism and cycling. Proc Natl Acad Sci USA 117:10414–10421. 10.1073/pnas.2001063117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H, Hou Y, Li G, Zhang Y, Coulon F, Cai C (2020) In vitro model insights into the role of human gut microbiota on arsenic bioaccessibility and its speciation in soils. Environ Pollut 263:114580. 10.1016/j.envpol.2020.114580 [DOI] [PubMed] [Google Scholar]
- Chouchane S, Snow ET (2001) In vitro effect of arsenical compounds on glutathione-related enzymes. Chem Res Toxicol 14(5):517–522. 10.1021/tx000123x [DOI] [PubMed] [Google Scholar]
- Cohen SM, Arnold LL, Uzvolgyi E, Cano M, St John M, Yamamoto S, Lu X, Le XC (2002) Possible role of dimethylarsinous acid in dimethylarsinic acid-induced urothelial toxicity and regeneration in the rat. Chem Res Toxicol 15:1150–1157. 10.1021/tx020026z [DOI] [PubMed] [Google Scholar]
- Colognato R, Coppede F, Ponti J, Sabbioni E, Migliore L (2007) Genotoxicity induced by arsenic compounds in peripheral human lymphocytes analysed by cytokinesis-block micronucleus assay. Mutagenesis 22(4):255–261. 10.1093/mutage/gem010 [DOI] [PubMed] [Google Scholar]
- Costa M (2019) Review of arsenic toxicity, speciation and polyadenylation of canonical histones. Toxicol Appl Pharmacol 15(375):1–4. 10.1016/j.taap.2019.05.006 [DOI] [PubMed] [Google Scholar]
- Crecelius EA (1977) Changes in the chemical speciation of arsenic following ingestion by man. Environ Health Perspect 19:147–150. 10.1289/ehp.7719147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen WR (2008) Is arsenic an aphrodisiac? The sociochemistry of an element? RSC Publishing, Cambridge [Google Scholar]
- Cullen WR (2014) Chemical mechanism of arsenic biomethylation. Chem Res Toxicol 27:457–461. 10.1021/tx400441h [DOI] [PubMed] [Google Scholar]
- Currier JM, Svoboda M, de Moraes DP, Matousek T, Dĕdina J, Stýblo M (2011a) Direct analysis of methylated trivalent arsenicals in mouse liver by hydride generation-cryotrapping-atomic absorption spectrometry. Chem Res Toxicol 24:478–480. 10.1021/tx200060c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier JM, Svoboda M, Matoušek T, Dědina J, Stýblo M (2011b) Direct analysis and stability of methylated trivalent arsenic metabolites in cells and tissues. Metallomics 3:1347–1354. 10.1039/c1mt00095k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier J, Saunders RJ, Ding L, Bodnar W, Cable P, Matoušek T, Creed JT, Stýblo M (2013) Comparative oxidation state specific analysis of arsenic species by high-performance liquid chromatography-inductively coupled plasma-mass spectrometry and hydride generation-cryotrapping-atomic absorption spectrometry. Anal At Spectrom 28(6):843–852. 10.1039/C3JA30380BJ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier JM, Ishida MC, González-Horta C, Sánchez-Ramírez B, Ballinas-Casarrubias L, Gutiérrez-Torres DS, Cerón RH, Morales DV, Terrazas FA, Del Razo LM, García-Vargas GG, Saunders RJ, Drobná Z, Fry RC, Matoušek T, Buse JB, Mendez MA, Loomis D, Stýblo M (2014) Associations between arsenic species in exfoliated urothelial cells and prevalence of diabetes among residents of Chihuahua, Mexico. Environ Health Perspect 122(10):1088–1094. 10.1289/ehp.1307756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier JM, Douillet C, Drobná Z, Stýblo M (2016) Oxidation state specific analysis of arsenic species in tissues of wild-type and arsenic (+3 oxidation state) methyltransferase-knockout mice. J Environ Sci (China) 49:104–112. 10.1016/j.jes.2016.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Razo LM, Styblo M, Cullen WR, Thomas DJ (2001) Determination of trivalent methylated arsenicals in biological matrices. Toxicol Appl Pharmacol 174(3):282–293. 10.1006/taap.2001.9226 [DOI] [PubMed] [Google Scholar]
- Del Razo LM, García-Vargas GG, Valenzuela OL, Castellanos EH, Sánchez-Peña LC, Currier JM, Drobná Z, Loomis D, Stýblo M (2011) Exposure to arsenic in drinking water is associated with increased prevalence of diabetes: a cross-sectional study in the Zimapan and Lagunera regions in Mexico. Environ Health 24(10):73. 10.1186/1476-069X-10-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker DA, Geter DR, Roop BC, Ward WO, Ahlborn GJ, Allen JW, Nelson GM, Ouyang M, Welsh W, Chen Y, O’Brien T, Kitchin KT (2009) Oncogene expression profiles in K6/ODC mouse skin and papillomas following a chronic exposure to monomethylarsonous acid. J Biochem Mol Toxicol 23(6):406–418. 10.1002/jbt.20304 [DOI] [PubMed] [Google Scholar]
- Delnomdedieu M, Basti MM, Otvos JD, Thomas DJ (1993) Transfer of arsenite from glutathione to dithiols: a model of interaction. Chem Res Toxicol 6(5):598–602. 10.1021/tx00035a002 [DOI] [PubMed] [Google Scholar]
- Delnomdedieu M, Basti MM, Otvos JD, Thomas DJ (1994) Reduction and binding of arsenate and dimethylarsinate by glutathione: a magnetic resonance study. Chem Biol Interact 90(2):139–155. 10.1016/0009-2797(94)90099-x [DOI] [PubMed] [Google Scholar]
- Delnomdedieu M, Styblo M, Thomas DJ (1995) Time dependence of accumulation and binding of inorganic and organic arsenic species in rabbit erythrocytes. Chem Biol Interact 98:69–83. 10.1016/0009-2797(95)03636-z [DOI] [PubMed] [Google Scholar]
- Devesa V, Del Razo LM, Blakely A, Drobna Z, Waters SB, Hughes MF, Styblo M, Thomas DJ (2004) Comprehensive analysis of arsenic metabolites by pH-specific hydride generation atomic absorption spectrometry. J Anal At Spectrom 19:1460–1467 [Google Scholar]
- Dheeman DS, Packianathan C, Pillai JK, Rosen BP (2014) Pathway of human AS3MT arsenic methylation. Chem Res Toxicol 27:1979–1989. 10.1021/tx500313k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders RJ, Drobná Z, Walton FS, Xun P, Thomas DJ, Stýblo M (2012) Methylation of arsenic by recombinant human wild-type arsenic (+3 oxidation state) methyltransferase and its methionine 287 threonine (M287T) polymorph: Role of glutathione. Toxicol Appl Pharmacol 264(1):121–130. 10.1016/j.taap.2012.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodmane PR, Arnold LL, Pennington KL, Thomas DJ, Cohen SM (2013) Effect of dietary treatment with dimethylarsinous acid (DMA(III)) on the urinary bladder epithelium of arsenic (+3 oxidation state) methyltransferase (As3mt) knockout and C57BL/6 wild type female mice. Toxicology 8(305):130–135. 10.1016/j.tox.2013.01.015 [DOI] [PubMed] [Google Scholar]
- Dong LQ, Zhang RB, Langlais P, He H, Clark M, Zhu L, Liu F (1999) Primary structure, tissue distribution, and expression of mouse phosphoinositide-dependent protein kinase-1, a protein kinase that phosphorylates and activates protein kinase Czeta. J Biol Chem 274(12):8117–8122. 10.1074/jbc.274.12.8117 [DOI] [PubMed] [Google Scholar]
- Dopp E, Hartmann LM, Florea AM et al. (2004) Uptake of inorganic and organic derivatives of arsenic associated with induced cytotoxic and genotoxic effects in Chinese hamster ovary (CHO) cells. Toxicol Appl Pharmacol 201(2):156–165. 10.1016/j.taap.2004.05.017 [DOI] [PubMed] [Google Scholar]
- Dopp E, von Recklinghausen U, Hartmann LM, Stueckradt I, Pollok I, Rabieh S, Hao L, Nussler A, Katier C, Hirner AV, Rettenmeier AW (2008) Subcellular distribution of inorganic and methylated arsenic compounds in human urothelial cells and human hepatocytes. Drug Metab Dispos 36(5):971–979. 10.1124/dmd.107.019034 [DOI] [PubMed] [Google Scholar]
- Dopp E, von Recklinghausen U, Diaz-Bone R, Hirner AV, Rettenmeier AW (2010) Cellular uptake, subcellular distribution and toxicity of arsenic compounds in methylating and non-methylating cells. Environ Res 110(5):435–442. 10.1016/j.envres.2009.08.012 [DOI] [PubMed] [Google Scholar]
- Douillet C, Currier J, Saunders J, Bodnar WM, Matousek T, Styblo M (2013) Methylated trivalent arsenicals are potent inhibitors of glucose-stimulated insulin secretion by murine pancreatic islets. Toxicol Appl Pharmacol 267(1):11–15. 10.1016/j.taap.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillet C, Huang MC, Saunders RJ, Dover EN, Zhang C, Stýblo M (2017) Knockout of arsenic (+3 oxidation state) methyltransferase is associated with adverse metabolic phenotype in mice: the role of sex and arsenic exposure. Arch Toxicol 91(7):2617–2627. 10.1007/s00204-016-1890-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover EN, Beck R, Huang MC et al. (2018) Arsenite and methylarsonite inhibit mitochondrial metabolism and glucose-stimulated insulin secretion in INS-1 832/13 beta cells. Arch Toxicol 92(2):693–704. 10.1007/s00204-017-2074-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z, Jaspers I, Thomas DJ, Styblo M (2003) Differential activation of AP-1 in human bladder epithelial cells by inorganic and methylated arsenicals. FASEB J 17(1):67–69. 10.1096/fj.02-0287fje [DOI] [PubMed] [Google Scholar]
- Drobna Z, Xing W, Thomas DJ, Stýblo M (2006) shRNA silencing of AS3MT expression minimizes arsenic methylation capacity of HepG2 cells. Chem Res Toxicol 19(7):894–898. 10.1021/tx060076u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z, Naranmandura H, Kubachka KM, Edwards BC, Herbin-Davis K, Styblo M, Le XC, Creed JT, Maeda N, Hughes MF, Thomas DJ (2009) Disruption of the arsenic (+3 oxidation state) methyltransferase gene in the mouse alters the phenotype for methylation of arsenic and affects distribution and retention of orally administered arsenate. Chem Res Toxicol 22:1713–1720. 10.1021/tx900179r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobná Z, Walton FS, Paul DS, Xing W, Thomas DJ, Stýblo M (2010) Metabolism of arsenic in human liver: the role of membrane transporters. Arch Toxicol 84(1):3–16. 10.1007/s00204-009-0499-7 [DOI] [PubMed] [Google Scholar]
- Drobná Z, Del Razo LM, García-Vargas GG, Sánchez-Peña LC, Barrera-Hernández A, Stýblo M, Loomis D (2013) Environmental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico. J Expo Sci Environ Epidemiol 23(2):151–155. 10.1038/jes.2012.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunivin TK, Yeh SY, Shade A (2019) A global survey of arsenic-related genes in soil microbiomes. BMC Biol 17:45. 10.1186/s12915-019-0661-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein M, Eleazer R, Rea M, Fondufe-Mittendorf Y (2017) Epigenomic reprogramming in inorganic arsenic-mediated gene expression patterns during carcinogenesis. Rev Environ Health 32(1–2):93–103. 10.1515/reveh-2016-0025 [DOI] [PubMed] [Google Scholar]
- Elshenawy OH, Abdelhamid G, Soshilov AA, Denison MS, El-Kadi AO (2017) Down-regulation of cytochrome P450 1A1 by monomethylarsonous acid in human HepG2 cells. Toxicol Lett 15(270):34–50. 10.1016/j.toxlet.2017.02.012 [DOI] [PubMed] [Google Scholar]
- Ezeh PC, Xu H, Lauer FT, Liu KJ, Hudson LG, Burchiel SW (2016) Monomethylarsonous acid (MMA+3) inhibits IL-7 signaling in mouse pre-B cells. Toxicol Sci 149(2):289–299. 10.1093/toxsci/kfv233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann J, Lai VWM, Cullen WR, Ma M, Lu X (1999) XC Le sample preparation and storage can change arsenic speciation in human urine. Clin Chem 45(11):1988–1997 [PubMed] [Google Scholar]
- Ferragut Cardoso AP, Udoh KT, States JC (2020) Arsenic-induced changes in miRNA expression in cancer and other diseases. Toxicol Appl Pharmacol 15(409):115306. 10.1016/j.taap.2020.115306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AB, Buchet JP, Lauwerys RR (1985) Arsenic uptake, cytotoxicity and detoxification studied in mammalian cells in culture. Arch Toxicol 57:168–172. 10.1007/BF00290882 [DOI] [PubMed] [Google Scholar]
- Fischer AB, Buchet JP, Lauwerys RR (1989) Cellular metabolism of arsenic studied in mammalian cells in vitro. Environ Geochem Health 11:87–92. 10.1007/BF01758656 [DOI] [PubMed] [Google Scholar]
- Francesconi KA, Kuehnelt D (2004) Determination of arsenic species: a critical review of methods and applications, 2000–2003. Analyst 129(5):373–395. 10.1039/b401321m [DOI] [PubMed] [Google Scholar]
- Fu Z, Gilbert ER, Liu D (2013) Regulation of insulin synthesis and secretion and pancreatic beta-cell dysfunction in diabetes. Curr Diabetes Rev 9:25–53 [PMC free article] [PubMed] [Google Scholar]
- Gamboa-Loira B, Cebrián ME, Franco-Marina F, López-Carrillo L (2017) Arsenic metabolism and cancer risk: a meta-analysis. Environ Res 156:551–558. 10.1016/j.envres.2017.04.016 [DOI] [PubMed] [Google Scholar]
- Garbinski LD, Rosen BP, Chen J (2019) Pathways of arsenic uptake and efflux. Environ Int 126:585–597. 10.1016/j.envint.2019.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbinski LD, Rosen BP, Yoshinaga M (2020) Organoarsenicals inhibit bacterial peptidoglycan biosynthesis by targeting the essential enzyme MurA. Chemosphere 254:126911. 10.1016/j.chemosphere.2020.126911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Montalvo EA, Valenzuela OL, Sánchez-Peña LC, Albores A, Del Razo LM (2011) Dose-dependent urinary phenotype of inorganic arsenic methylation in mice with a focus on trivalent methylated metabolites. Toxicol Mech Methods 21(9):649–655. 10.3109/15376516.2011.603765 [DOI] [PubMed] [Google Scholar]
- Gong Z, Lu X, Cullen WR, Le XC (2001) Unstable trivalent arsenic metabolites, monomethylarsonous acid and dimethylarsinous acid. J Anal At Spectrom 6:1409–1413 [Google Scholar]
- Gong Z, Lu X, Ma M, Watt C, Le XC (2002a) Arsenic speciation analysis. Talanta 58(1):77–96. 10.1016/s0039-9140(02)00258-8 [DOI] [PubMed] [Google Scholar]
- Gong Z, Jiang G, Cullen WR, Aposhian HV, Le XC (2002b) Determination of arsenic metabolic complex excreted in human urine after administration of sodium 2,3-dimercapto-1-propane sulfonate. Chem Res Toxicol 15(10):1318–1323. 10.1021/tx020058m [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Kobayashi Y, Cui X, Hirano S (2005) A new metabolic pathway of arsenite: arsenic-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol 79:183–191. 10.1007/s00204-004-0620-x [DOI] [PubMed] [Google Scholar]
- Hernández-Zavala A, Matoušek T, Drobná Z, Paul DS, Walton F, Adair BM, Jiří D, Thomas DJ, Stýblo M (2008) Speciation analysis of arsenic in biological matrices by automated hydride generation-cryotrapping-atomic absorption spectrometry with multiple microflame quartz tube atomizer (multiatomizer). J Anal At Spectrom 23:342–351. 10.1039/b706144g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler J, Zdrenka R, Reichel RAD, Weber DG, Rozynek P, Johnen G, Dopp E, Hirner AV (2011) Intracellular, time-resolved speciation and quantification of arsenic compounds in human urothelial and hepatoma cells. J Anal At Spectrom 26(12):2396–2403. 10.1039/C1JA10150A [DOI] [Google Scholar]
- Hirano S, Kobayashi Y, Cui X, Kanno S, Hayakawa T, Shraim A (2004) The accumulation and toxicity of methylated arsenicals in endothelial cells: important roles of thiol compounds. Toxicol Appl Pharmacol 198:458–467. 10.1016/j.taap.2003.10.023 [DOI] [PubMed] [Google Scholar]
- Hirata M, Mohri T, Hisanaga A, Ishinishi N (1989) Conversion of arsenite and arsenate to methylarsenic and dimethylarsenic compounds by homogenates prepared from livers and kidneys of rats and mice. Appl Organometal Chem 3:335–341. 10.1002/aoc.590030406 [DOI] [Google Scholar]
- Hosseini MJ, Shaki F, Ghazi-Khansari M, Pourahmad J (2013) Toxicity of Arsenic (III) on isolated liver mitochondria: a new mechanistic approach. Iran J Pharm Res 12(Suppl):121–138 [PMC free article] [PubMed] [Google Scholar]
- Hsu KC, Sun CC, Huang YL (2011) Arsenic speciation in biomedical sciences: recent advances and applications. Kaohsiung J Med Sci 27(9):382–389. 10.1016/j.kjms.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Ke Q, Costa M, Shi X (2004) Molecular mechanisms of arsenic carcinogenesis. Mol Cell Biochem 255(1–2):57–66. 10.1023/b:mcbi.0000007261.04684.78 [DOI] [PubMed] [Google Scholar]
- Huang K, Chen C, Zhang J, Tang Z, Shen Q, Rosen BP, Zhao FJ (2016) Efficient arsenic methylation and volatilization mediated by a novel bacterium from an arsenic-contaminated paddy soil. Environ Sci Technol 50:6389–6396. 10.1021/acs.est.6b01974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Xu Y, Packianathan C, Gao F, Chen C, Zhang J, Shen Q, Rosen BP, Zhao FJ (2018a) Arsenic methylation by a novel ArsM As(III) S-adenosylmethionine methyltransferase that requires only two conserved cysteine residues. Mol Microbiol 107:265–276. 10.1111/mmi.13882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MC, Douillet C, Dover EN, Zhang C, Beck R, Tejan-Sie A, Krupenko SA, Stýblo M (2018b) Metabolic phenotype of wild-type and As3mt-knockout C57BL/6J mice exposed to inorganic arsenic: the role of dietary fat and folate intake. Environ Health Perspect 126(12):127003. 10.1289/EHP3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Douillet C, Styblo M (2019) Arsenite and its trivalent methylated metabolites inhibit glucose-stimulated calcium influx and insulin secretion in murine pancreatic islets. Arch Toxicol 93(9):2525–2533. 10.1007/s00204-019-02526-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MF, Devesa V, Adair BM, Styblo M, Kenyon EM, Thomas DJ (2005) Tissue dosimetry, metabolism and excretion of pentavalent and trivalent monomethylated arsenic in mice after oral administration. Toxicol Appl Pharmacol 208(2):186–197. 10.1016/j.taap.2005.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MF, Devesa V, Adair BM, Conklin SD, Creed JT, Styblo M, Kenyon EM, Thomas DJ (2008) Tissue dosimetry, metabolism and excretion of pentavalent and trivalent dimethylated arsenic in mice after oral administration. Toxicol Appl Pharmacol 227:26–35. 10.1016/j.taap.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MF, Edwards BC, Herbin-Davis KM, Saunders J, Styblo M, Thomas DJ (2010) Arsenic (+3 oxidation state) methyltransferase genotype affects steady-state distribution and clearance of arsenic in arsenate-treated mice. Toxicol Appl Pharmacol 249:217–223. 10.1016/j.taap.2010.09.017 [DOI] [PubMed] [Google Scholar]
- Kala SV, Kala G, Prater CI, Sartorelli AC, Lieberman MW (2004) Formation and urinary excretion of arsenic triglutathione and methylarsenic diglutathione. Chem Res Toxicol 17(2):243–249. 10.1021/tx0342060 [DOI] [PubMed] [Google Scholar]
- Kitchin KT, Wallace K (2008) The role of protein binding of trivalent arsenicals in arsenic carcinogenesis and toxicity. J Inorg Biochem 102(3):532–539. 10.1016/j.jinorgbio.2007.10.021 [DOI] [PubMed] [Google Scholar]
- Kligerman AD, Doerr CL, Tennant AH et al. (2003) Methylated trivalent arsenicals as candidate ultimate genotoxic forms of arsenic: induction of chromosomal mutations but not gene mutations. Environ Mol Mutagen 42(3):192–205. 10.1002/em.10192 [DOI] [PubMed] [Google Scholar]
- Kuo CC, Moon KA, Wang SL, Silbergeld E, Navas-Acien A (2017) The association of arsenic metabolism with cancer, cardiovascular disease, and diabetes: a systematic review of the epidemiological evidence. Environ Health Perspect 125(8):087001. 10.1289/EHP577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz RC, Hays AM (2006) Role of oxidative stress in arsenic-induced toxicity. Drug Metab Rev 38(4):791–804. 10.1080/03602530600980108 [DOI] [PubMed] [Google Scholar]
- Le XC, Ma M, Cullen WR, Aposhian HV, Lu X, Zheng B (2000a) Determination of monomethylarsonous acid, a key arsenic methylation intermediate, in human urine. Environ Health Perspect 108:1015–1018. 10.1289/ehp.001081015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le XC, Lu X, Ma M, Cullen WR, Aposhian HV, Zheng B (2000b) Speciation of key arsenic metabolic intermediates in human urine. Anal Chem 72:5172–5127. 10.1021/ac000527u [DOI] [PubMed] [Google Scholar]
- Leslie EM (2012) Arsenic-glutathione conjugate transport by the human multidrug resistance proteins (MRPs/ABCCs). J Inorg Biochem 108:141–149. 10.1016/j.jinorgbio.2011.11.009 [DOI] [PubMed] [Google Scholar]
- Li J, Waters SB, Drobna Z, Devesa V, Styblo M, Thomas DJ (2005) Arsenic (+3 oxidation state) methyltransferase and the inorganic arsenic methylation phenotype. Toxicol Appl Pharmacol 204:164–169. 10.1016/j.taap.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Li X, Geng Z, Chang J, Wang S, Song X, Hu X, Wang Z (2013a) Identification of the third binding site of arsenic in human arsenic (III) methyltransferase. PLoS ONE 8:e84231. 10.1371/journal.pone.0084231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cao J, Wang S, Geng Z, Song X, Hu X, Wang Z (2013b) Residues in human arsenic (+3 oxidation state) methyltransferase forming potential hydrogen bond network around S-adenosylmethionine. PLoS ONE 8:e76709. 10.1371/journal.pone.0076709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pawitwar SS, Rosen BP (2016) The organoarsenical biocycle and the primordial antibiotic methylarsenite. Metallomics 8:1047–1055. 10.1039/c6mt00168h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Cullen WR, Thomas DJ (1999) Methylarsenicals and arsinothiols are potent inhibitors of mouse liver thioredoxin reductase. Chem Res Toxicol 12:924–930. 10.1021/tx9900775 [DOI] [PubMed] [Google Scholar]
- Lin S, Del Razo LM, Styblo M, Wang C, Cullen WR, Thomas DJ (2001) Arsenicals inhibit thioredoxin reductase in cultured rat hepatocytes. Chem Res Toxicol 14:305–311. 10.1021/tx0001878 [DOI] [PubMed] [Google Scholar]
- Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, Hall LL, Simeonsson JB, Thomas DJ (2002) A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J Biol Chem 277:10795–10803. 10.1074/jbc.M110246200 [DOI] [PubMed] [Google Scholar]
- Liu Z (2010) Roles of vertebrate aquaglyceroporins in arsenic transport and detoxification. Adv Exp Med Biol 679:71–81. 10.1007/978-1-4419-6315-4_6 [DOI] [PubMed] [Google Scholar]
- Lu X, Arnold LL, Cohen SM, Cullen WR, Le XC (2003) Speciation of dimethylarsinous acid and trimethylarsine oxide in urine from rats fed with dimethylarsinic acid and dimercaptopropane sulfonate. Anal Chem 75(23):6463–6468. 10.1021/ac034868u [DOI] [PubMed] [Google Scholar]
- Lu M, Wang H, Li XF, Lu X, Cullen WR, Arnold LL, Cohen SM, Le XC (2004) Evidence of hemoglobin binding to arsenic as a basis for the accumulation of arsenic in rat blood. Chem Res Toxicol 17(12):1733–1742. 10.1021/tx049756s [DOI] [PubMed] [Google Scholar]
- Lu M, Wang H, Li XF, Arnold LL, Cohen SM, Le XC (2007) Binding of dimethylarsinous acid to cys-13alpha of rat hemoglobin is responsible for the retention of arsenic in rat blood. Chem Res Toxicol 20(1):27–37. 10.1021/tx060195+ [DOI] [PubMed] [Google Scholar]
- Luvonga C, Rimmer CA, Yu LL, Lee SB (2020) Analytical methodologies for the determination of organoarsenicals in edible marine species: a review. J Agric Food Chem 68(7):1910–1934. 10.1021/acs.jafc.9b04525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciaszczyk-Dziubinska E, Wawrzycka D, Wysocki R (2012) Arsenic and antimony transporters in eukaryotes. Int J Mol Sci 13(3):3527–3548. 10.3390/ijms13033527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal BK, Ogra Y, Suzuki KT (2001) Identification of dimethylarsinous and monomethylarsonous acids in human urine of the arsenic-affected areas in West Bengal, India. Chem Res Toxicol 14(4):371–8. 10.1021/tx000246h [DOI] [PubMed] [Google Scholar]
- Mandal BK, Ogra Y, Suzuki KT (2003) Speciation of arsenic in human nail and hair from arsenic-affected area by HPLC-inductively coupled argon plasma mass spectrometry. Toxicol Appl Pharmacol 189(2):73–83. 10.1016/s0041-008x(03)00088-7 [DOI] [PubMed] [Google Scholar]
- Mandal BK, Ogra Y, Anzai K, Suzuki KT (2004) Speciation of arsenic in biological samples. Toxicol Appl Pharmacol 198(3):307–318. 10.1016/j.taap.2003.10.030 [DOI] [PubMed] [Google Scholar]
- Mandal BK, Suzuki KT, Anzai K (2007) Impact of arsenic in foodstuffs on the people living in the arsenic-affected areas of West Bengal, India. J Environ Sci Health A Tox Hazard Subst Environ Eng 42(12):1741–1752. 10.1080/10934520701564244 [DOI] [PubMed] [Google Scholar]
- Marapakala K, Qin J, Rosen BP (2012) Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry 51:944–951. 10.1021/bi201500c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marapakala K, Packianathan C, Ajees AA, Dheeman DS, Sankaran B, Kandavelu P, Rosen BP (2015) A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase. Acta Crystallogr D Biol Crystallogr 71(Pt 3):505–515. 10.1107/S1399004714027552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner K, Musil S, Mikšík I, Dědina J (2018) Investigation of hydride generation from arsenosugars—is it feasible for speciation analysis? Anal Chim Acta 30(1008):8–17. 10.1016/j.aca.2018.01.009 [DOI] [PubMed] [Google Scholar]
- Martin EM, Stýblo M, Fry RC (2017) Genetic and epigenetic mechanisms underlying arsenic-associated diabetes mellitus: a perspective of the current evidence. Epigenomics 9(5):701–710. 10.2217/epi-2016-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass MJ, Tennant A, Roop BC et al. (2001) Methylated trivalent arsenic species are genotoxic. Chem Res Toxicol 14(4):355–361. 10.1021/tx000251l [DOI] [PubMed] [Google Scholar]
- Matoušek T, Hernández-Zavala A, Svoboda M, Langrová L, Adair BM, Drobná Z, Thomas DJ, Stýblo M, Dědina J (2008) Oxidation state specific generation of arsines from methylated arsenicals based on L-cysteine treatment in buffered media for speciation analysis by hydride generation—automated cryotrapping—gas chromatography-atomic absorption spectrometry with the multiatomizer. Spectrochim Acta Part B At Spectrosc 63(3):396–406. 10.1016/j.sab.2007.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoušek T, Currier JM, Trojánková N, Saunders RJ, Ishida MC, González-Horta C, Musil S, Mester Z, Stýblo M, Dědina J (2013) Selective hydride generation- cryotrapping—ICP-MS for arsenic speciation analysis at picogram levels: analysis of river and sea water reference materials and human bladder epithelial cells. J Anal At Spectrom 28(9):1456–1465. 10.1039/C3JA50021G [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maull EA, Ahsan H, Edwards J, Longnecker MP, Navas-Acien A, Pi J, Silbergeld EK, Styblo M, Tseng CH, Thayer KA, Loomis D (2012) Evaluation of the association between arsenic and diabetes: a National Toxicology Program workshop review. Environ Health Perspect 120(12):1658–1670. 10.1289/ehp.1104579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meakin CJ, Szilagyi JT, Avula V, Fry RC (2020) Inorganic arsenic and its methylated metabolites as endocrine disruptors in the placenta: mechanisms underpinning glucocorticoid receptor (GR) pathway perturbations. Toxicol Appl Pharmacol 15(409):115305. 10.1016/j.taap.2020.115305 [DOI] [PubMed] [Google Scholar]
- Musil S, Matoušek T, Currier JM, Stýblo M, Dědina J (2014) Speciation analysis of arsenic by selective hydride generation-cryotrapping-atomic fluorescence spectrometry with flame-in-gas-shield atomizer: achieving extremely low detection limits with inexpensive instrumentation. Anal Chem 86(20):10422–10428. 10.1021/ac502931k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng JC, Johnson D, Imray P, Chiswell B, Moore MR (1998) Speciation of arsenic metabolites in the urine of occupational workers and experimental rats using an optimised hydride cold-trapping method. Analyst 123(5):929–933. 10.1039/a707726b [DOI] [PubMed] [Google Scholar]
- Nollen M, Ebert F, Moser J, Mullenders LH, Hartwig A, Schwerdtle T (2009) Impact of arsenic on nucleotide excision repair: XPC function, protein level, and gene expression. Mol Nutr Food Res 53(5):572–582. 10.1002/mnfr.200800480 [DOI] [PubMed] [Google Scholar]
- Notch EG, Goodale BC, Barnaby R, Coutermarsh B, Berwin B, Taylor VF, Jackson BP, Stanton BA (2015) Monomethylarsonous acid (MMAIII) has an adverse effect on the innate immune response of human bronchial epithelial cells to Pseudomonas aeruginosa. PLoS ONE 10:e0142392. 10.1371/journal.pone.0142392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okina M, Yoshida K, Kuroda K, Wanibuchi H, Fukushima S, Endo G (2004) Determination of trivalent methylated arsenicals in rat urine by liquid chromatography-inductively coupled plasma mass spectrometry after solvent extraction. J Chromatogr B Analyt Technol Biomed Life Sci 799(2):209–215. 10.1016/j.jchromb.2003.10.028 [DOI] [PubMed] [Google Scholar]
- Oliva-González C, Uresti-Rivera EE, Galicia-Cruz OG, Jasso-Robles FI, Gandolfi AJ, Escudero-Lourdes C (2017) The tumor suppressor phosphatase and tensin homolog protein (PTEN) is negatively regulated by NF-κb p50 homodimers and involves histone 3 methylation/deacetylation in UROtsa cells chronically exposed to monomethylarsonous acid. Toxicol Lett 5(280):92–98. 10.1016/j.toxlet.2017.08.013 [DOI] [PubMed] [Google Scholar]
- Pace C, Banerjee TD, Welch B, Khalili R, Dagda RK, Angermann J (2016) Monomethylarsonous acid, but not inorganic arsenic, is a mitochondria-specific toxicant in vascular smooth muscle cells. Toxicol In Vitro 35:188–201. 10.1016/j.tiv.2016.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packianathan C, Kandavelu P, Rosen BP (2018a) The structure of an As(III) S-adenosylmethionine methyltransferase with 3-coordinately bound As(III) depicts the first step in catalysis. Biochemistry 57:4083–4092. 10.1021/acs.biochem.8b00457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packianathan C, Li J, Kandavelu P, Sankaran B, Rosen BP (2018b) Reorientation of the methyl group in MAs(III) is the rate-limiting step in the ArsM As(III) S-adenosylmethionine methyltransferase reaction. ACS Omega 3:3104–3112. 10.1021/acsomega.8b00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren M, Engström K, Hallström BM, Wahlberg K, Søndergaard DA, Säll T, Vahter M, Broberg K (2017) AS3MT-mediated tolerance to arsenic evolved by multiple independent horizontal gene transfers from bacteria to eukaryotes. PLoS ONE 12(4):e0175422. 10.1371/journal.pone.0175422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul DS, Harmon AW, Devesa V, Thomas DJ, Styblo M (2007a) Molecular mechanisms of the diabetogenic effects of arsenic: inhibition of insulin signaling by arsenite and methylarsonous acid. Environ Health Perspect 115(5):734–742. 10.1289/ehp.9867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul DS, Hernández-Zavala A, Walton FS, Adair BM, Dedina J, Matousek T, Stýblo M (2007b) Examination of the effects of arsenic on glucose homeostasis in cell culture and animal studies: development of a mouse model for arsenic-induced diabetes. Toxicol Appl Pharmacol 222(3):305–314. 10.1016/j.taap.2007.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul DS, Devesa V, Hernandez-Zavala A, Adair BM, Walton FS, Drobnâ Z, Thomas DJ, Styblo M (2008) Environmental arsenic as a disruptor of insulin signaling. Met Ions Biol Med 10:1–7 [PMC free article] [PubMed] [Google Scholar]
- Petrick JS, Ayala-Fierro F, Cullen WR, Carter DE, Vasken AH (2000) Monomethylarsonous acid (MMA(III)) is more toxic than arsenite in Chang human hepatocytes. Toxicol Appl Pharmacol 163:203–207. 10.1006/taap.1999.8872 [DOI] [PubMed] [Google Scholar]
- Petrick JS, Jagadish B, Mash EA, Aposhian HV (2001) Monomethylarsonous acid (MMA(III)) and arsenite: LD(50) in hamsters and in vitro inhibition of pyruvate dehydrogenase. Chem Res Toxicol 14(6):651–656. 10.1021/tx000264z [DOI] [PubMed] [Google Scholar]
- Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C (2006) Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA 103:2075–2080. 10.1073/pnas.0506836103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab A, Meharg AA, Jaspars M, Genney DR, Feldmann J (2004) Arsenic–glutathione complexes—their stability in solution and during separation by different HPLC modes. J Anal At Spectrom 19:183–190. 10.1039/B307945G [DOI] [Google Scholar]
- Rabieh R, Hirner HV, Matschullat J (2008) Determination of arsenic species in human urine using high performance liquid chromatography (HPLC) coupled with inductively coupled plasma mass spectrometry (ICP-MS). J Anal At Spectrom 23:544–549. 10.1039/b718840d [DOI] [Google Scholar]
- Reid MS, Hoy KS, Schofield JRM, Uppal JS, Lin Y, Lu X, Peng H, Le XC (2020) Arsenic speciation analysis: a review with an emphasis on chromatographic separations. Trends Anal Chem 123:115770. 10.1016/j.trac.2019.115770 [DOI] [Google Scholar]
- Riedmann C, Ma Y, Melikishvili M, Godfrey SG, Zhang Z, Chen KC, Rouchka EC, Fondufe-Mittendorf YN (2015) Inorganic Arsenic-induced cellular transformation is coupled with genome wide changes in chromatin structure, transcriptome and splicing patterns. BMC Genomics 16(1):212. 10.1186/s12864-015-1295-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggenbeck BA, Banerjee M, Leslie EM (2016) Cellular arsenic transport pathways in mammals. J Environ Sci (China) 49:38–58. 10.1016/j.jes.2016.10.001 [DOI] [PubMed] [Google Scholar]
- Rosen BP, Liu Z (2009) Transport pathways for arsenic and selenium: a minireview. Environ Int 35(3):512–515. 10.1016/j.envint.2008.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Paul B (2016) The global menace of arsenic and its conventional remediation—a critical review. Chemosphere 158:37–49. 10.1016/j.chemosphere.2016.05.043 [DOI] [PubMed] [Google Scholar]
- Schwerdtle T, Walter I, Mackiw I, Hartwig A (2003) Induction of oxidative DNA damage by arsenite and its trivalent and pentavalent methylated metabolites in cultured human cells and isolated DNA. Carcinogenesis 24(5):967–974. 10.1093/carcin/bgg018 [DOI] [PubMed] [Google Scholar]
- Shen S, Li XF, Cullen WR, Weinfeld M, Le XC (2013) Arsenic binding to proteins. Chem Rev 113(10):7769–7792. 10.1021/cr300015c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiobara Y, Ogra Y, Suzuki KT (2001) Animal species difference in the uptake of dimethylarsinous acid (DMA(III)) by red blood cells. Chem Res Toxicol 14(10):1446–1452. 10.1021/tx015537k [DOI] [PubMed] [Google Scholar]
- Smeester L, Rager JE, Bailey KA, Guan X, Smith N, García-Vargas G, Del Razo LM, Drobná Z, Kelkar H, Stýblo M, Fry RC (2011) Epigenetic changes in individuals with arsenicosis. Chem Res Toxicol 24(2):165–167. 10.1021/tx1004419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Crecelius EA, Reading JC (1977) Airborne arsenic exposure and excretion of methylated arsenic compounds. Environ Health Perspect 19:89–93. 10.1289/ehp.771989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollome J, Martin E, Sethupathy P, Fry RC (2016) Environmental contaminants and microRNA regulation: transcription factors as regulators of toxicant-altered microRNA expression. Toxicol Appl Pharmacol 1(312):61–66. 10.1016/j.taap.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Geng Z, Zhu J, Li C, Hu X, Bian N, Zhang X, Wang Z (2009) Structure-function roles of four cysteine residues in the human arsenic (+3 oxidation state) methyltransferase (hAS3MT) by site-directed mutagenesis. Chem Biol Interact 179:321–328. 10.1016/j.cbi.2008.12.018 [DOI] [PubMed] [Google Scholar]
- Song X, Geng Z, Li X, Zhao Q, Hu X, Zhang X, Wang Z (2011) Functional and structural evaluation of cysteine residues in the human arsenic (+3 oxidation state) methyltransferase (hAS3MT). Biochimie 93:369–375. 10.1016/j.biochi.2010.10.010 [DOI] [PubMed] [Google Scholar]
- Spuches AM, Wilcox DE (2008) Monomethylarsenite competes with Zn2+ for binding sites in the glucocorticoid receptor. J Am Chem Soc 130(26):8148–8149. 10.1021/ja802179p [DOI] [PubMed] [Google Scholar]
- States JC, Singh AV, Knudsen TB, Rouchka EC, Ngalame NO, Arteel GE, Piao Y, Ko MS (2012) Prenatal arsenic exposure alters gene expression in the adult liver to a proinflammatory state contributing to accelerated atherosclerosis. PLoS ONE 7(6):e38713. 10.1371/journal.pone.0038713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice S, Liu G, Matulis S, Boise LH, Cai Y (2016) Determination of multiple human arsenic metabolites employing high performance liquid chromatography inductively coupled plasma mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 15(1009–1010):55–65. 10.1016/j.jchromb.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styblo M, Delnomdedieu M, Thomas DJ (1996) Mono- and dimethylation of arsenic in rat liver cytosol in vitro. Chem Biol Interact 99:147–164. 10.1016/0009-2797(95)03666-0 [DOI] [PubMed] [Google Scholar]
- Styblo M, Serves SV, Cullen WR, Thomas DJ (1997) Comparative inhibition of yeast glutathione reductase by arsenicals and arsenothiols. Chem Res Toxicol 10:27–33. 10.1021/tx960139g [DOI] [PubMed] [Google Scholar]
- Styblo M, Del Razo LM, Vega L, Germolec DR, LeCluyse EL, Hamilton GA, Reed W, Wang C, Cullen WR, Thomas DJ (2000) Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Arch Toxicol 74:289–299. 10.1007/s002040000134 [DOI] [PubMed] [Google Scholar]
- Stýblo M, Drobná Z, Jaspers I, Lin S, Thomas DJ (2002) The role of biomethylation in toxicity and carcinogenicity of arsenic: a research update. Environ Health Perspect 110(Suppl 5):767–71. 10.1289/ehp.110-1241242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PF, Hu YJ, Ho IC, Cheng YM, Lee TC (2006) Distinct gene expression profiles in immortalized human urothelial cells exposed to inorganic arsenite and its methylated trivalent metabolites. Environ Health Perspect 114(3):394–403. 10.1289/ehp.8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki KT, Tomita T, Ogra Y, Ohmichi M (2001) Glutathione-conjugated arsenics in the potential hepato-enteric circulation in rats. Chem Res Toxicol 14(12):1604–1611. 10.1021/tx0155496 [DOI] [PubMed] [Google Scholar]
- Suzuki KT, Mandal BK, Katagiri A, Sakuma Y, Kawakami A, Ogra Y, Yamaguchi K, Sei Y, Yamanaka K, Anzai K, Ohmichi M, Takayama H, Aimi N (2004) Dimethylthioarsenicals as arsenic metabolites and their chemical preparations. Chem Res Toxicol 17(7):914–921. 10.1021/tx049963s [DOI] [PubMed] [Google Scholar]
- Thomas DJ (2007) Molecular processes in cellular arsenic metabolism. Toxicol Appl Pharmacol. 10.1016/j.taap.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Rosen BP (2013) Arsenic methyltransferases. In: Kretsinger RH, Uversky VN, Permyakov EA(eds) Encyclopedia of metalloproteins, vol 222(3). Springer, New York, pp 365–73 [Google Scholar]
- Thomas DJ, Styblo M, Lin S (2001) The cellular metabolism and systemic toxicity of arsenic. Toxicol Appl Pharmacol 176(2):127–144. 10.1006/taap.2001.9258 [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, Devesa V, Styblo M (2007) Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp Biol Med (Maywood) 232:3–13) [PMC free article] [PubMed] [Google Scholar]
- Tokar EJ, Diwan BA, Thomas DJ, Waalkes MP (2012) Tumors and proliferative lesions in adult offspring after maternal exposure to methylarsonous acid during gestation in CD1 mice. Arch Toxicol 86:975–982. 10.1007/s00204-012-0820-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CH (2007) Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 25(1):1–22. 10.1080/10590500701201695 [DOI] [PubMed] [Google Scholar]
- Twaddle NC, Vanlandingham M, Beland FA, Doerge DR (2018a) Metabolism and disposition of arsenic species after repeated oral dosing with sodium arsenite in drinking water. II. Measurements in pregnant and fetal CD-1 mice. Food Chem Toxicol 115:178–184. 10.1016/j.fct.2018.03.010 [DOI] [PubMed] [Google Scholar]
- Twaddle NC, Vanlandingham M, Fisher JW, Doerge DR (2018b) Metabolism and disposition of arsenic species from controlled dosing with sodium arsenite in adult female CD-1 mice. III. Toxicokinetic studies following oral and intravenous administration. Food Chem Toxicol 121:676–686. 10.1016/j.fct.2018.09.068 [DOI] [PubMed] [Google Scholar]
- Twaddle NC, Vanlandingham M, Beland FA, Doerge DR (2019) Metabolism and disposition of arsenic species from controlled dosing with dimethylarsinic acid (DMAV) in adult female CD-1 mice. V. Toxicokinetic studies following oral and intravenous administration. Food Chem Toxicol 130:22–31. 10.1016/j.fct.2019.04.045 [DOI] [PubMed] [Google Scholar]
- Vahter M (2002) Mechanisms of arsenic biotransformation. Toxicology 27(181–182):211–217. 10.1016/s0300-483x(02)00285-8 [DOI] [PubMed] [Google Scholar]
- Vahter M, Concha G (2001) Role of metabolism in arsenic toxicity. Pharmacol Toxicol 89(1):1–5. 10.1034/j.1600-0773.2001.d01-128.x [DOI] [PubMed] [Google Scholar]
- Valenzuela OL, Borja-Aburto VH, Garcia-Vargas GG, Cruz-Gonzalez MB, Garcia-Montalvo EA, Calderon-Aranda ES, Del Razo LM (2005) Urinary trivalent methylated arsenic species in a population chronically exposed to inorganic arsenic. Environ Health Perspect 113(3):250–254. 10.1289/ehp.7519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela OL, Drobná Z, Hernández-Castellanos E, Sánchez-Peña LC, García-Vargas GG, Borja-Aburto VH, Stýblo M, Del Razo LM (2009) Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions. Toxicol Appl Pharmacol 239(2):200–207. 10.1016/j.taap.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M (2006) Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact 160(1):1–40. 10.1016/j.cbi.2005.12.009 [DOI] [PubMed] [Google Scholar]
- Vega L, Styblo M, Patterson R, Cullen W, Wang C, Germolec D (2001) Differential effects of trivalent and pentavalent arsenicals on cell proliferation and cytokine secretion in normal human epidermal keratinocytes. Toxicol Appl Pharmacol 172(3):225–232. 10.1006/taap.2001.9152 [DOI] [PubMed] [Google Scholar]
- Venkatratnam A, Douillet C, Topping BC, Shi Q, Addo KA, Ideraabdullah FY, Fry RC, Styblo M (2021) Sex-dependent effects of preconception exposure to arsenite on gene transcription in parental germ cells and on transcriptomic profiles and diabetic phenotype of offspring. Arch Toxicol 95(2):473–488. 10.1007/s00204-020-02941-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalkes MP, Liu J, Diwan BA (2007) Transplacental arsenic carcinogenesis in mice. Toxicol Appl Pharmacol 222:271–280. 10.1016/j.taap.2006.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton FS, Waters SB, Jolley SL, LeCluyse EL, Thomas DJ, Styblo M (2003) Selenium compounds modulate the activity of recombinant rat AsIII-methyltransferase and the methylation of arsenite by rat and human hepatocytes. Chem Res Toxicol 16:261–265. 10.1021/tx025649r [DOI] [PubMed] [Google Scholar]
- Walton FS, Harmon AW, Paul DS, Drobna Z, Patel YM, Styblo M (2004) Inhibition of insulin-dependent glucose uptake by trivalent arsenicals: possible mechanism of arsenic-induced diabetes. Toxicol Appl Pharmacol 198(3):424–433. 10.1016/j.taap.2003.10.026 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhou J, Lu X, Gong Z, Le XC (2004) Arsenic speciation in urine from acute promyelocytic leukemia patients undergoing arsenic trioxide treatment. Chem Res Toxicol 17(1):95–103. 10.1021/tx0341714 [DOI] [PubMed] [Google Scholar]
- Wang S, Li X, Song X, Geng Z, Hu X (2012) Wang Z Rapid equilibrium kinetic analysis of arsenite methylation catalyzed by recombinant human arsenic (+3 oxidation state) methyltransferase (hAS3MT). J Biol Chem 287:38790–38799. 10.1074/jbc.M112.368050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Geng Z, Shi N, Li X, Wang Z (2014) The functions of crucial cysteine residues in the arsenite methylation catalyzed by recombinant human arsenic (III) methyltransferase. PLoS ONE 9:e110924. 10.1371/journal.pone.0110924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters SB, Devesa V, Del Razo LM, Styblo M, Thomas DJ (2004) Endogenous reductants support the catalytic function of recombinant rat cyt19, an arsenic methyltransferase. Chem Res Toxicol 17:404–409. 10.1021/tx0342161 [DOI] [PubMed] [Google Scholar]
- Wnek SM, Jensen TJ, Severson PL, Futscher BW, Gandolfi AJ (2010) Monomethylarsonous acid produces irreversible events resulting in malignant transformation of a human bladder cell line following 12 weeks of low-level exposure. Toxicol Sci 116:44–57. 10.1093/toxsci/kfq106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Johnson W, Spayd S, Hall GS, Buckley B (2006) Arsenic speciation analysis of human urine using ion exchange chromatography coupled to inductively coupled plasma mass spectrometry. Anal Chim Acta 578(2):186–194. 10.1016/j.aca.2006.06.076 [DOI] [PubMed] [Google Scholar]
- Yamauchi H, Fowler BA (1994) Toxicity and metabolism of inorganic and methylated arsenicals in arsenic in the environment, Part II: human health and ecosystem effects. In: Nriagu JO (ed) Wiley, pp 35–53 [Google Scholar]
- Yan H, Wang N, Weinfeld M, Cullen WR, Le XC (2009) Identification of arsenic-binding proteins in human cells by affinity chromatography and mass spectrometry. Anal Chem 81(10):4144–4152. 10.1021/ac900352k [DOI] [PubMed] [Google Scholar]
- Yan Y, Ye J, Xue XM, Zhu YG (2015) Arsenic demethylation by a C·as lyase in Cyanobacterium Nostoc sp. PCC 7120. Environ Sci Technol 49(24):14350–8. 10.1021/acs.est.5b03357 [DOI] [PubMed] [Google Scholar]
- Yan X, Li J, Liu Q, Peng H, Popowich A, Wang Z, Li XF, Le XC (2016) p-Azidophenylarsenoxide: an arsenical “Bait” for the in situ capture and identification of cellular arsenic-binding proteins. Angew Chem Int Ed Engl 55(45):14051–14056. 10.1002/anie.201608006 [DOI] [PubMed] [Google Scholar]
- Yan Y, Xue XM, Guo YQ, Zhu YG, Ye J (2017) Co-expression of cyanobacterial genes for arsenic methylation and demethylation in Escherichia coli offers insights into arsenic resistance. Front Microbiol 24(8):60. 10.3389/fmicb.2017.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Frenkel K (2002) Arsenic-mediated cellular signal transduction, transcription factor activation, and aberrant gene expression: implications in carcinogenesis. J Environ Pathol Toxicol Oncol 21(4):331–342 [PubMed] [Google Scholar]
- Yokohira M, Arnold LL, Pennington KL, Suzuki S, Kakiuchi-Kiyota S, Herbin-Davis K, Thomas DJ, Cohen SM (2010) Severe systemic toxicity and urinary bladder cytotoxicity and regenerative hyperplasia induced by arsenite in arsenic (+3 oxidation state) methyltransferase knockout mice. A preliminary report. Toxicol Appl Pharmacol 246:1–7. 10.1016/j.taap.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Yokohira M, Arnold LL, Pennington KL, Suzuki S, Kakiuchi-Kiyota S, Herbin-Davis K, Thomas DJ, Cohen SM (2011) Effect of sodium arsenite dose administered in the drinking water on the urinary bladder epithelium of female arsenic (+3 oxidation state) methyltransferase knockout mice. Toxicol Sci 121:257–266. 10.1093/toxsci/kfr051 [DOI] [PubMed] [Google Scholar]
- Yoshinaga-Sakurai K, Shinde R, Rodriguez M, Rosen BP, El-Hage N (2020) Comparative cytotoxicity of inorganic arsenite and methylarsenite in human brain cells. ACS Chem Neurosci 11(5):743–751. 10.1021/acschemneuro.9b00653 [DOI] [PubMed] [Google Scholar]
- Zakharyan RA, Aposhian HV (1999) Enzymatic reduction of arsenic compounds in mammalian systems: the rate-limiting enzyme of rabbit liver arsenic biotransformation is MMA(V) reductase. Chem Res Toxicol 12:1278–1283. 10.1021/tx9901231 [DOI] [PubMed] [Google Scholar]
- Zakharyan RA, Sampayo-Reyes A, Healy SM, Tsaprailis G, Board PG, Liebler DC, Aposhian HV (2001) Human monomethylarsonic acid (MMA(V)) reductase is a member of the glutathione-S-transferase superfamily. Chem Res Toxicol 14:1051–1057. 10.1021/tx010052h [DOI] [PubMed] [Google Scholar]
- Zangi R, Filella M (2012) Transport routes of metalloids into and out of the cell: a review of the current knowledge. Chem Biol Interact 197(1):47–57. 10.1016/j.cbi.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Zhang HN, Yang L, Ling JY, Czajkowsky DM, Wang JF, Zhang XW, Zhou YM, Ge F, Yang MK, Xiong Q, Guo SJ, Le HY, Wu SF, Yan W, Liu B, Zhu H, Chen Z, Tao SC (2015) Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc Natl Acad Sci USA 112(49):15084–15089. 10.1073/pnas.1521316112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Fennel EMJ, Douillet C, Stýblo M (2017) Exposures to arsenite and methylarsonite produce insulin resistance and impair insulin-dependent glycogen metabolism in hepatocytes. Arch Toxicol 91(12):3811–3821. 10.1007/s00204-017-2076-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YG, Yoshinaga M, Zhao FJ, Rosen BP (2014) Earth abides arsenic biotransformations. Ann Rev Earth Planet Sci 42:443–467 [DOI] [PMC free article] [PubMed] [Google Scholar]


