Abstract
Objectives:
To characterize the clinical presentation, genomic alterations, pathologic phenotype and clinical management of microphthalmia-associated transcription factor (MITF) familial renal cell carcinoma (RCC), caused by a member of the TFE3, TFEB and MITF family of transcription factor genes.
Methods:
The clinical presentation, family history, tumor histopathology, and surgical management were evaluated and reported herein. DNA sequencing was performed on blood DNA, tumor DNA and DNA extracted from adjacent normal kidney tissue. Copy number and gene expression analyses on tumor and normal tissues were performed by Real-Time PCR. TCGA gene expression data were used for comparative analysis. Protein expression and subcellular localization were evaluated by immunohistochemistry.
Results:
Germline genomic analysis identified the MITF p.E318K variant in a patient with bilateral, multifocal type 1 papillary RCC and a family history of RCC. All tumors displayed the MITF variant and were characterized by amplification of chromosomes 7 and 17, hallmarks of type 1 papillary RCC. We demonstrated that MITF p.E318K variant results in altered transcriptional activity and that downstream targets of MiT family members, such as GPNMB, are dysregulated in the tumors.
Conclusions:
Association of the pathogenic MITF variant with bilateral and multifocal type 1 papillary RCC in this family supports its role as a risk allele for the development of RCC and emphasizes the importance of screening for MITF variants irrelevant of the RCC histologic subtype. This study identifies potential biomarkers for the disease, such as GPNMB expression, that may facilitate the development of targeted therapies for patients affected with MITF-associated RCC.
Keywords: Renal cell carcinoma, papillary renal cell carcinoma, MITF, germline mutation, GPNMB, Microphthalmia-Associated Transcription Factor
Graphical Abstract
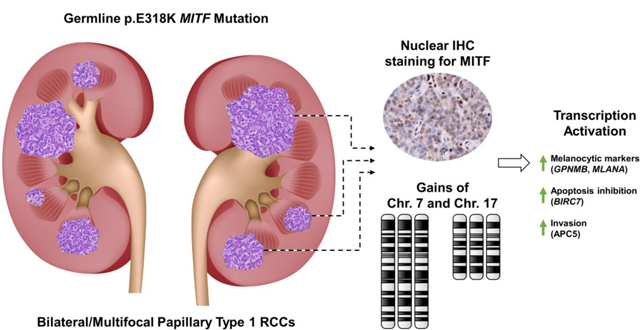
This report describes a case study of a patient with bilateral, multifocal type 1 papillary RCC and a family history of RCC that has the germline MITF p.E318K variant, known to predispose to kidney cancer. All tumors demonstrated a papillary type 1 histology and analyses of available tumors identified increased nuclear staining for the MITF transcription factor, somatic amplification of chromosomes 7 and 17, and altered expression of genes associated with tumorigenesis, including potential biomarkers, such as GPNMB.
INTRODUCTION
Familial forms of renal cell carcinoma (RCC) comprise approximately 5% of all cases of RCC, present with different histologic subtypes, and are associated with pathogenic mutations in several different genes, such as VHL, BAP1, MET, FLCN, FH, and SDHB/C/D.1, 2 Papillary RCC is the second most common RCC subtype (~15%) and is itself a heterogeneous disease that can be classified into two main histologically-defined subsets, termed type 1 and type 2 papillary RCC.3 A hereditary form of type 1 papillary RCC, Hereditary Papillary Renal Carcinoma (HPRC), is associated with activating mutations in the tyrosine kinase domain of the MET oncogene that results in bilateral, multifocal tumors.4 Whereas, a hereditary form of type 2 papillary RCC is associated with inactivating mutations of the fumarate hydratase (FH) tumor suppressor gene that result in aggressive type 2 tumors as a phenotypic feature of Hereditary Leiomyomatosis and Renal Cell Carcinoma (HLRCC).5 In addition, there are families with renal cell carcinoma in multiple generations for which no genetic basis has been identified.
In 1996, with Colin Cooper and his colleagues, we described a novel type of renal cell carcinoma characterized by a specific chromosomal translocation t(X;1)(p11.2; q21.1) in human papillary renal cell carcinoma and demonstrated a fusion of a gene at 1q21.2, which we designated PRCC, to the TFE3 gene at Xp11.2.6 TFE3 encodes a member of the microphthalmia transcription factor (MiT) basic helix-loop-helix family of transcription factors which includes TFE3, TFEB and MITF. Recently, a variant in the MITF gene, c.G1075A/p.E318K (rs149617956), has been recognized as a risk allele for the development of melanoma and RCC.7 While present at a low allele frequency in the normal population (0.0013 - ExAC and gnomAD databases8), enrichment for this variant is primarily associated with melanoma and has been reported at significantly higher allele frequencies in melanoma cohorts from Australia (0.017), the U.K. (0.018), France (0.016), Italy (0.009), and Spain, and within a large international cohort (0.014).7, 9–13 The MITF p.E318K variant is also associated with an increased risk of RCC with the majority of the patients presenting with a personal or family history of melanoma.7, 11, 14, 15
MITF, TFE3, TFEB and TFEC encode members of the microphthalmia-associated transcription factor (MiT) family that regulate cell growth and differentiation in response to nutrient availability within the cell and are controlled by subcellular localization.16 Somatic translocations involving either TFE3, TFEB, or MITF have been identified in cases of sporadic RCC that produce fusion proteins that constitutively localize to the nucleus, causing upregulation of a series of transcriptional targets that can drive RCC tumorigenesis.17–19 The MITF p.E318K variant represents the first germline alteration within this family of genes that associates with increased risk of cancer. Stability and function of MITF is normally tightly regulated by posttranslational modifications, such as ubiquitination, acetylation, phosphorylation, and SUMOylation.20 The MITF p.E318K variant occurs within one of the two consensus sequences (ΨKXE) for the addition of small ubiquitin-like-modifiers (SUMO; K182 and K316) in the MITF protein and results in loss of this SUMOylation site.7, 9 The SUMOylation of MITF represses transcriptional activity and loss of this SUMOylation site due to the MITF p.E318K variant has been shown to increase or dysregulate transcriptional activity by synergy control of genes with multiple MITF binding sites.7, 9, 21 In this study we describe a family with a history of RCC in which the proband presented with bilateral, multifocal type 1 papillary RCC and characterize the genotype and histologic and molecular features of these tumors.
MATERIALS AND METHODS
Patient:
The patient was evaluated and managed at the Hatfield Clinical Research Center, National Institutes of Health (NIH). Peripheral blood and tumor samples were obtained for DNA extraction and immunohistochemistry. This study was approved by the Institutional Review Board of the National Cancer Institute on the NCI-97-C-0147 protocol and the patient provided written informed consent.
DNA Sequencing:
DNA was extracted from patient’s blood, fresh frozen tumor samples from all three surgeries, and fresh frozen normal kidney tissue adjacent to tumors using Maxwell 16 Blood DNA or Tissue DNA purification kits. The MITF c.G1075A variant was validated by PCR using a Qiagen Taq PCR Core Kit (Qiagen, MD, USA) according to the manufacturer’s instructions, followed by bidirectional sequencing using the Big Dye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, CA, USA) according to the manufacturer’s specifications and run on an ABI 3130xl Genetic Analyzer (Applied Biosystems). Forward and reverse sequences were evaluated using Sequencher 5.2.4 (Genecodes, MI, USA). Primers used were MITF-Fw: AAGTCCTCTGTGCTCTGCCT and MITF-Rv: AGCTAAAGTCTGTGGTGAATTC.
Copy Number Assay:
Copy number was measured by Real-Time PCR using TaqMan® Copy Number Assays on a ViiA 7 Real-Time PCR system (Thermo Fisher Scientific) per manufacturer’s instructions and calculated as comparative CT (ΔΔCT) values. The following genes were assessed for chromosomal copy number: EGFR (Hs00711969_cn), HGF (Hs02939712_cn), MET (Hs01432482_cn), BRAF (Hs05008776_cn) on chromosome 7; FLCN (Hs00494676_cn) on chromosome 17p; VHL (Hs01945190_cn), MITF (Hs00391575_cn) on chromosome 3p. The TaqMan® Copy Number Reference Assay for RNAseP on chromosome 14 (Thermo Fisher Scientific) was used as reference gene and a normal control DNA was run in all experiments as reference for copy number analysis. 10ng of DNA were used for each reaction and samples were analyzed in triplicate.
Gene expression analysis:
RNA extraction from tissues and Real-Time PCR were performed as previously described.22 Expression levels were normalized to the control ACTB housekeeping gene (Hs99999903_m1) and calculated as comparative CT (ΔΔCT) values. A tumor sample was designated to represent the expression value of 1 to account for undetectable expression in normal kidney tissue. TaqMan® Gene Expression Assays (Thermo Fisher Scientific Inc.) were used to assess the expression levels of MITF (Hs01117294_m1) and several of its target genes GPNMB (Hs01095669_m1), BIRC7 (Hs01086675_m1), MLANA (Hs00194133_m1), CCND1 (Hs00765553_m1), LYST (Hs00915889_m1), ACP5 (Hs00356261_m1).
Immunohistochemistry:
Five-micron thick formalin-fixed paraffin-embedded sections were deparaffinized and blocked with methanol containing 30% H2O2. Antigen retrieval was performed with Citrate buffer at pH 6.0 in a steamer cooker. Slides were blocked in PBS with normal rabbit serum, incubated with primary antibodies overnight and with secondary antibodies conjugated with biotin for 40 min at room temperature. Signal was detected with streptavidin-HRP for 40 min at room temperature. DAB (3,3′-diaminobenzidine) counterstaining was performed by diluting DAB 1:40 in DAB buffer plus H2O2 for 2 min and Hematoxylin for 10 seconds (Vector Laboratories). Pictures were obtained with an AxioScan.Z1 Slide Scanner (Zeiss, Oberkochen, DE). The primary antibodies were used as follows: Human Osteoactivin/GPNMB anti goat (R&D Systems, Minneapolis, MN; 1:800), MITF anti mouse (EMD Millipore, Billerica, MA;1:50); TFE3 anti rabbit (Sigma Aldrich; 1:800), TFEB anti rabbit (Bethyl Laboratories, Montgomery, TX; 1:200).
Culturing of primary tumor cells:
Tumor was enzymatically dissociated with gentleMACS Dissociator (Miltenyi Biotec, Auburn, CA) according to manufacturer’s recommendations. Cells were cultured in humidified incubator at 5% O2 in DMEM culture media supplemented with 25mM glucose, 4mM glutamine and 1mM sodium pyruvate (Thermo Fisher Scientific) and seeded on 8-well chambered slides (ibidi, Martinsried, D) for immunofluorescence staining.
Immunofluorescence:
Cells were attached to chambered slides overnight. Cells were fixed in 10% formalin for 30 minutes, rinsed for 3 times with PBS, blocked and permeabilized with blocking buffer (PBS with 5% BSA, 1% TWEEN®20 and 0.1% Trito-X100, Sigma-Aldrich, St. Louis, MO) for 1 hour at room temperature and incubated for 1 hour with the primary antibody diluted in blocking buffer. Primary antibodies were used as follows: MITF anti mouse (EMD Millipore, Billerica, MA) 1:100; TFE3 anti rabbit (Sigma-Aldrich) 1:200; TFEB anti rabbit (Bethyl Laboratories, Montgomery, TX) 1:100. Following three 5-minute washes in PBST (PBS with 0.2% TWEEN®20) cells were incubated for 1 hour at room temperature with secondary antibodies Alexa Fluor 488 anti-mouse IgG (1:400) and Alexa Fluor 594 anti-rabbit IgG (1:400) (Thermo Fisher Scientific, Waltham, MA). Cell nuclei were counterstained with DAPI (0.1 μg/ml) for 15 minutes in PBST and cells were washed 3 times in PBST before mounting the slides with antifade mounting medium (Vector Laboratories, Burlingame, CA). Images were acquired with a fluorescence microscope (BZ-X700 Keyence, Osaka, JN).
RESULTS
A 43-year-old man of African American descent initially presented with bilateral, multifocal renal masses. A metastatic work up revealed no evidence of lymphadenopathy or disseminated disease. The father of the proband had previously undergone a partial nephrectomy at age 56 at an outside institution for the removal of a 2 cm renal tumor characterized as renal cell carcinoma showing “papillary growth with fairly well differentiated cells which showed clear cytoplasm in many areas” that was consistent with type 1 papillary RCC with clear cell features. The father died 13 years later with a diagnosis of arterionephrosclerosis. There was no additional history of RCC in other family members and no history of melanoma. Examination of the proband did not reveal any signs of melanoma (Fig. 1A). Pre-operative glomerular filtration rate (GFR) was 83 mL/min/1.73 m2.
Fig. 1: Clinical presentation of proband carrying the MITF p.E318K variant.
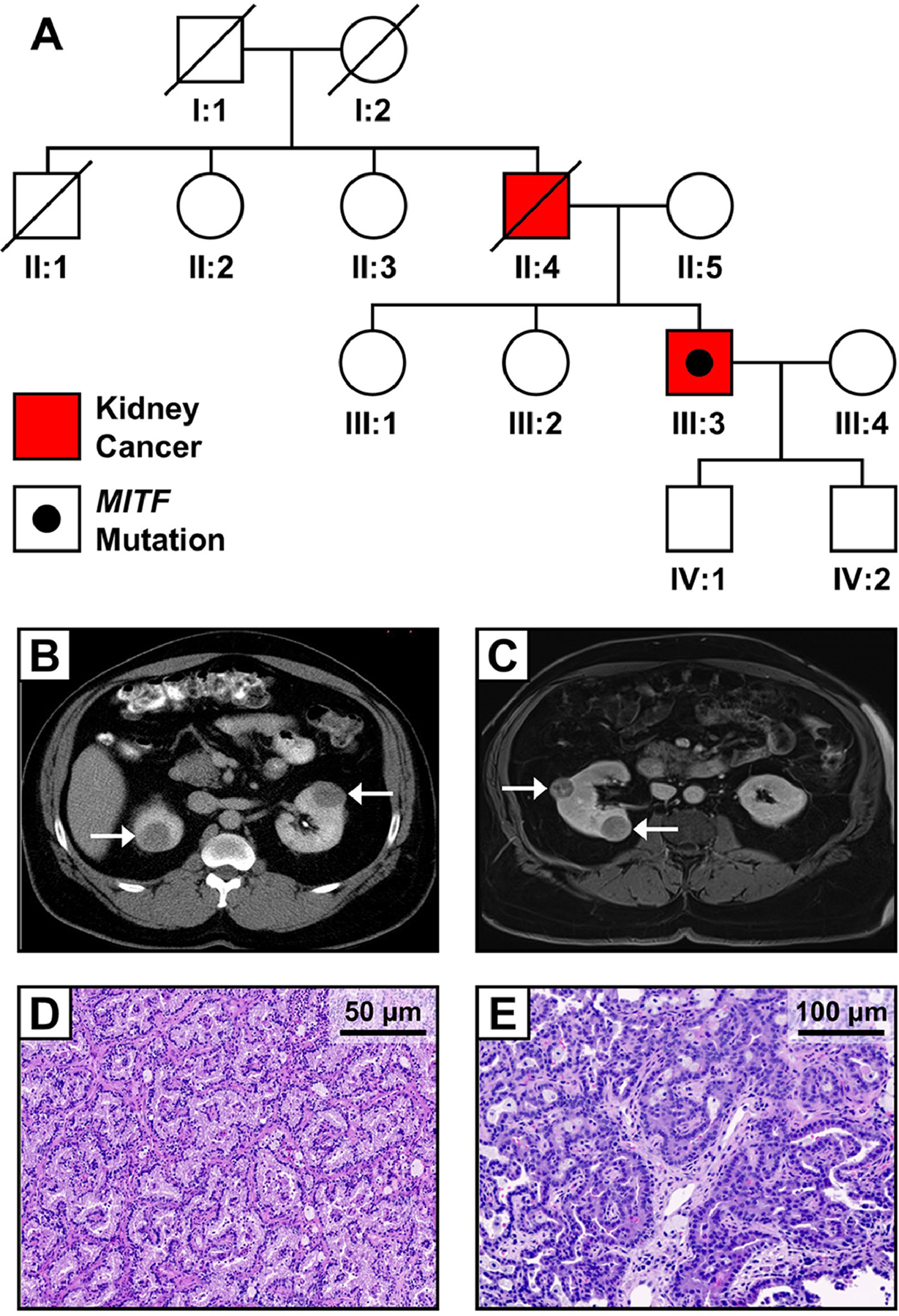
(A) Family pedigree for patient with germline MITF p.E318K variant and RCC. The proband (III:3) was diagnosed with bilateral and multifocal RCC at age 43. His father previously underwent surgery at an outside institution for RCC. Axial abdominal computed tomography (CT) scans from the proband before undergoing staged bilateral partial nephrectomies (B) and before second right-sided partial nephrectomy (C) reveal bilateral, multifocal kidney lesions, indicated by arrows. Histology of resected tumors was classified as type 1 papillary RCC, as shown by H&E stained sections of two different tumors from second partial nephrectomy (D, E).
The patient was managed with staged, bilateral partial nephrectomies. Surgical technique has been previously described.23 Briefly, Gerota’s fascia was incised and the kidney mobilized within Gerota’s fascia. Intraoperative ultrasound was used to define tumors. The renal hilum was clamped only for hilar and/or endophytic tumors. Tumor enucleation was performed by circumferentially incising the renal capsule. A plane was defined between the tumor pseudocapsule and renal parenchyma, and the underlying parenchyma was spared. Renorraphies were performed in the standard fashion, and Gerota’s fascia was reapproximated over the kidney at the end of the case.
The solitary right-sided renal tumor was resected via laparoscopic partial nephrectomy. Pathologic analysis revealed a 5.5 cm stage pT1b type 1 papillary RCC. Notably, a 0.5 cm “incipient” lesion was identified during the pathologic analysis of the adjacent resected “normal” tissue. Warm ischemia for the case was 29 minutes. Estimated blood loss (EBL) was 300 mL. GFR 3 months after surgery was 82 mL/min/1.73 m2. Eleven months later, the patient underwent a left-sided robotic-assisted partial nephrectomy during which four tumors, ranging from 2.1 cm to 6.9 cm were resected (Fig. 1B). Given the multifocal nature of these tumors, tumor enucleation was used. Two lesions were resected without clamping the kidney and two were removed during warm ischemia, with a total warm ischemia time of 44 minutes. EBL was 700 mL. Histologic evaluation of these lesions demonstrated three type 1 papillary RCCs and one type 1 papillary RCC with clear cell components. Incipient lesions were again identified in the “normal” tissue adjacent to the renal tumors. GFR 3 months after surgery was maintained at 82 mL/min/1.73 m2.
The patient was followed with annual imaging for 5 years, at which time a 1.5 cm lesion was found in the right kidney and several sub-centimeter indeterminate lesions were identified in the left kidney. Annual surveillance was recommended, but the patient was lost to follow-up. He returned four years later for surveillance imaging and four right-sided renal lesions were detected on imaging. The previously noted 1.5 cm mass had grown to ~3 cm, and three new lesions measuring 2.4, 2.3, and 0.9 cm were identified (Fig. 1C). Preoperative GFR was 78 GFR mL/min/1.73 m2. The now 53-year-old man underwent a reoperative right-sided robotic-assisted partial nephrectomy during which a total of nine tumors, a number of which were not apparent on imaging, were surgically removed. There was dense fibrosis around the kidney consistent with prior surgery and similar to our prior experience and description.24 All tumors were removed without renal ischemia, and EBL was 1700 mL. Pathologic evaluation identified eight type 1 papillary RCCs ranging in size from 1.0 to 3.2 cm, and a 3.9 cm type 1 papillary RCC with clear cell features (Fig. 1D and 1E). Currently, the patient has a 0.7 cm lesion in his left kidney on imaging which is being followed by regular surveillance. GFR at last follow-up was 71 mL/min/1.73 m2.
Due to the presence of bilateral, multifocal disease and a family history of RCC, the proband’s germline DNA was evaluated for mutation using a Clinical Laboratory Improvement Amendment (CLIA)-approved multi-gene Renal Cancer Panel of 18 RCC susceptibility genes that included the MET gene, which is associated with hereditary papillary renal carcinoma, as well as VHL, FH, FLCN, TSC1, TSC2, SDHB, SDHC, SDHD, MITF, BAP1, PTEN, TP53, EPCAM, MLH1, MSH2, MSH6, and PMS2. The germline mutation analysis revealed a heterozygous pathogenic variant in the MITF gene (p.E318K), while no additional germline variants were found in any of the other evaluated genes. This pathogenic variant was confirmed in the tumor and incipient lesion from the first surgery, in three analyzed tumors and the adjacent normal tissue sample from the second surgery, and in eight analyzed tumors from the third surgery (Fig. 2A). Notably, two of the tumors from the third surgery, including the tumor that was histologically classified as type 1 papillary RCC with clear cell features, showed MITF loss of heterozygosity (LOH) and only retained the pathogenic variant allele (Fig. 2A). However, copy number analysis of the MITF gene, located on chromosome 3p13, demonstrated that all evaluated tumors retained two copies of the gene and therefore both tumors demonstrated copy neutral LOH (Fig. 2B).
Fig. 2: Genetic analysis of MITF p.E318K variant in biological samples from proband.
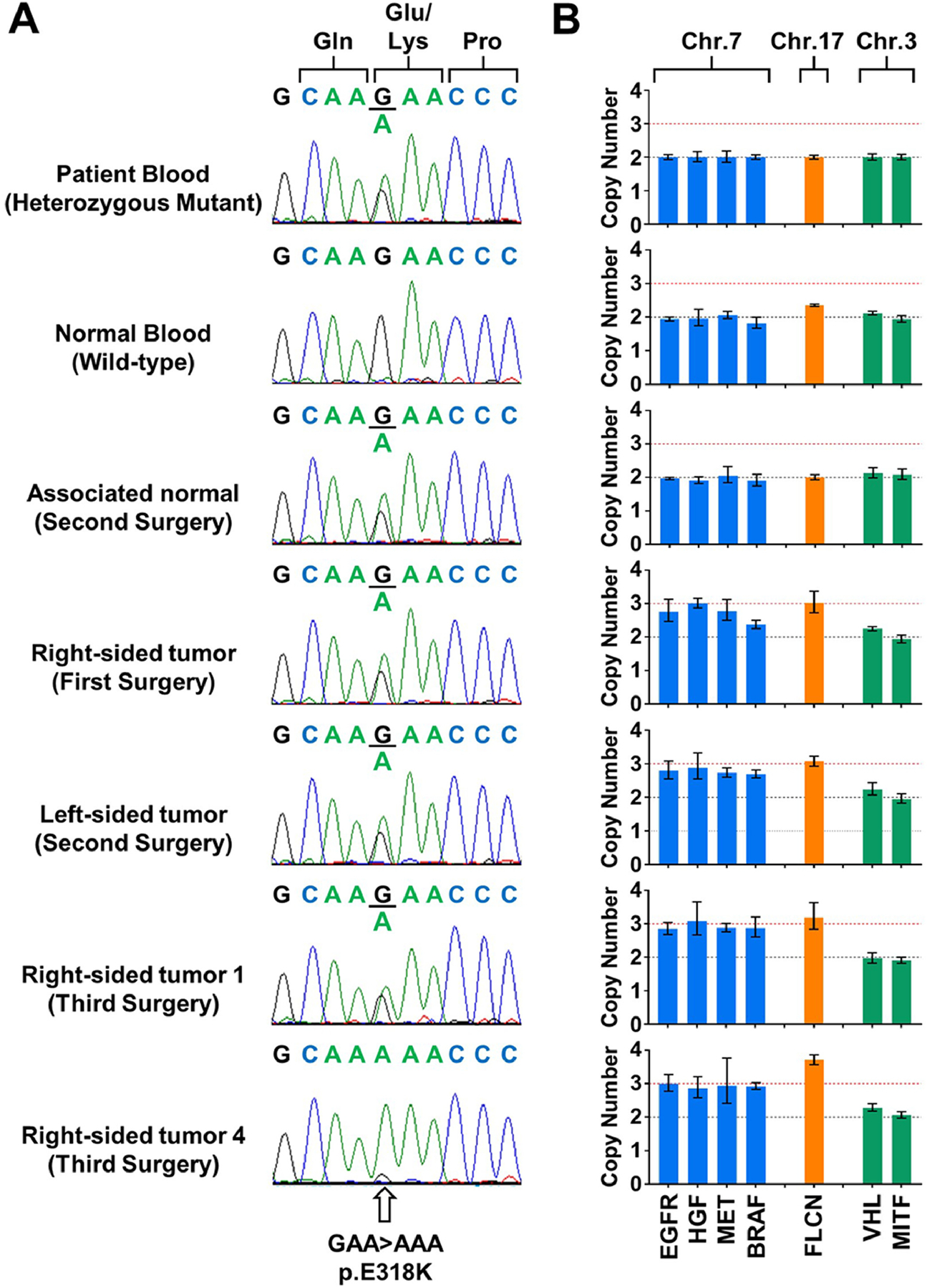
(A) Sequencing electropherograms demonstrate the MITF p.E318K variant in blood, normal kidney tissue and a selection of tumors from the proband. Right-sided tumor 4 shows loss of heterozygosity of the MITF p.E318K variant, while the remaining tumors were heterozygous for the variant. (B) Copy number analysis of chromosome 7 (assessed with EGFR, HGF, MET, BRAF), chromosome 17 (FLCN) and chromosome 3 (VHL and MITF) in these samples shows amplification of chromosomes 7 and 17 in all tumors, while retaining two copies of the short arm of chromosome 3. This demonstrated copy number neutral loss of heterozygosity in tumor 4.
Somatic gains of chromosomes 7 and 17 are hallmarks of sporadic type 1 papillary RCC.3 Real-Time PCR-based analysis was performed on four genes located on chromosome 7, namely EGFR (7p11.2), HGF (7p11.2), MET (7p11.2), BRAF (7p11.2), and one gene located on chromosome 17, FLCN (17p11.2). All evaluated tumors from all three surgeries showed amplification of both chromosomes 7 and 17 demonstrating between three or four copies of each chromosome, while only two copies of the chromosomes were detected in the patient’s blood DNA and normal kidney tissue (Fig. 2B). Somatic alterations of the MET tyrosine kinase domain were ruled out by sequencing all tumor samples. Due to presence of clear cell features, alterations of the VHL gene were evaluated since loss of this gene is characteristic of sporadic clear cell RCC.1 No mutations within the coding regions of VHL, promoter hypermethylation, or chromosomal loss were observed in any tumors with clear cell features.
Since altered transcriptional activity of MITF p.E318K variant protein was shown previously, we tested expression levels of transcriptional targets of MITF by Real-Time PCR in MITF-variant tumor tissue, adjacent normal kidney tissue and non-affected normal kidney tissue. Three MiT family target genes, GPNMB (glycoprotein NMB), the apoptosis inhibitor BIRC7 (Baculoviral IAP repeat-containing 7) and the melanocytic marker MLANA (Melan-A), were highly upregulated in tumor tissues as compared to normal kidney tissues (Fig. 3A). While ACP5 (Acid phosphatase 5, tartrate resistant), a gene involved in invasion, was mildly upregulated, the pigmentation-related gene LYST (lysosomal trafficking regulator) was not significantly different, and cyclin D1 (CCND1) was lower in tumors compared to normal kidney tissue (Fig. 3A). Gene expression of MITF itself was not significantly altered (Fig. 3A). To test whether these gene expression signals are common to papillary RCC or may be specific to MiT-associated tumors, we evaluated the same genes in papillary RCCs published by the TCGA project.3 Analysis showed that gene expression in MITF-variant tumors mostly corresponded to gene expression patterns observed between the TFE3- and TFEB-fusion RCC present in the TCGA cohort and the remaining non-fusion papillary RCC and normal kidney tissue, with upregulation of GPNMB, BIRC7, MLANA and ACP5 (Fig. 3B). These data confirm that the MITF p.E318K variant may alter its transcriptional activity and therefore contribute to tumorigenesis.
Fig. 3: Gene expression of MITF target genes.
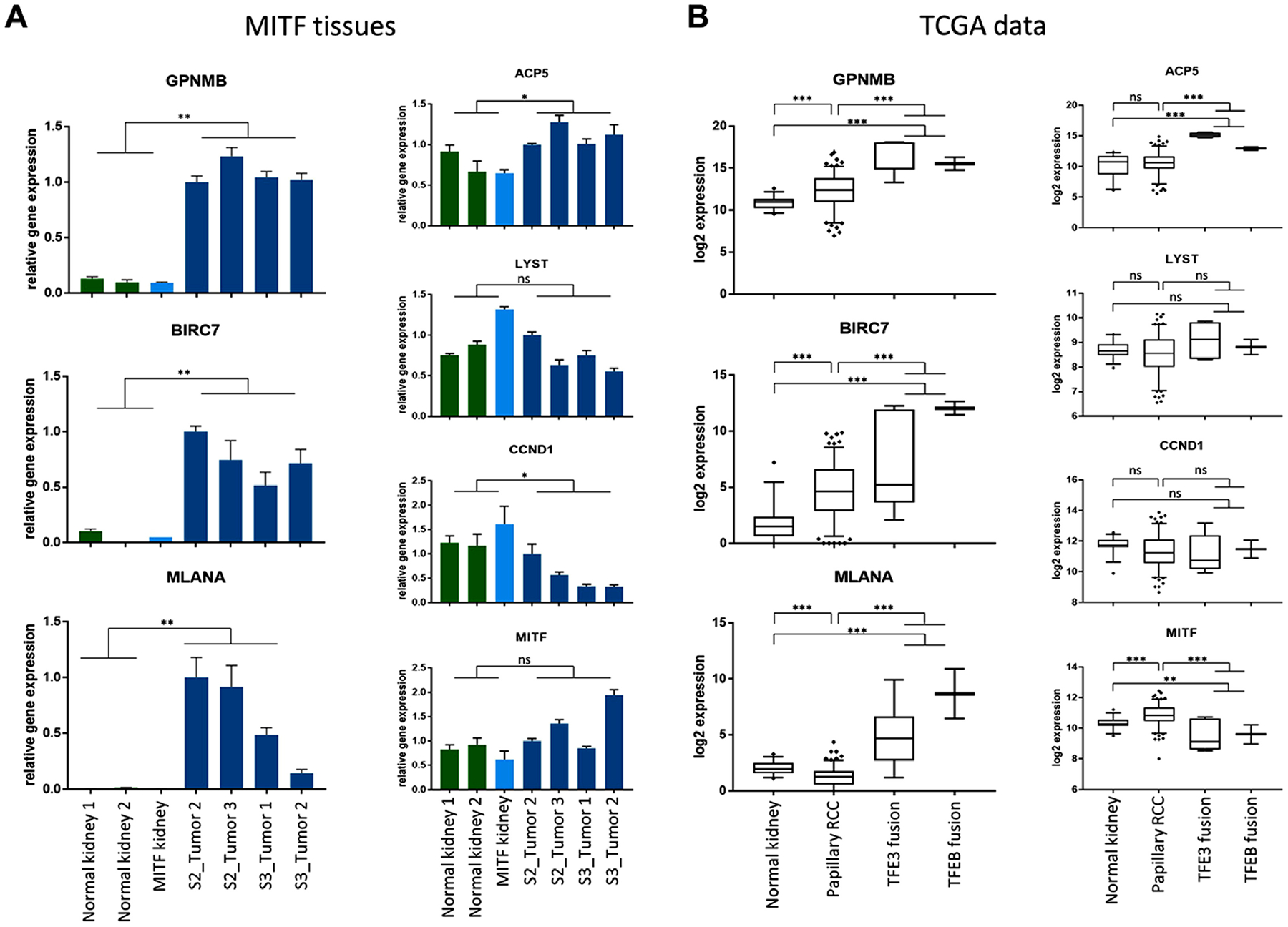
(A) Gene expression analysis by Real-Time PCR on RNA extracted from normal kidney (green bars), MITF p.E318K variant kidney (light blue bar) and MITF-variant tumors from second (S2) and third surgery (S3) (dark blue bars) shows overexpression of MITF target genes GPNMB, BIRC7 and MLANA. No significant differences were found in expression of MITF and LYST between normal and tumor tissues, decreased expression of CCND1 and increased expression of ACP5 in MITF p.E318K variant tumors. Comparison performed by Student t-test. (B) Evaluation of expression of MITF target genes among papillary RCC samples from TCGA project shows increased expression of GPNMB, BIRC7 and MLANA in TFE3-(n=6) and TFEB fusion (n=2) tumors as compared to other papillary RCC (n=153) and normal tissues (n=30). MITF expression was decreased and expression of ACP5 increased in TFE3-/TFEB-fusion RCC as compared to normal kidney tissue and papillary type 1 RCC samples. No significant difference in gene expression was found for CCND1 and LYST. Comparisons performed by one-way ANOVA. ns: not significant; ** p ≤ 0.01; *** p ≤ 0.001.
To confirm these results in tissues at the protein expression level, we performed IHC staining for MITF and GPNMB on unrelated normal kidney tissue, MITF-variant patient’s normal kidney tissue and tumors. While MITF expression was low in unrelated normal tissues, mildly increased expression that localized within the nucleus was seen in MITF-variant normal kidney tissue. Qualitative assessment of staining patterns indicated that MITF expression was substantially higher in the tumor tissue with predominantly nuclear localization. GPNMB expression was mostly absent from both unrelated and MITF-variant normal kidney tissues, while tumor tissues stained positive (Fig. 4A). IHC staining of other MiT family member proteins (TFE3 and TFEB) showed overall weak staining for both proteins, largely cytoplasmic localization of TFEB and nuclear localization of TFE3 (Fig. 4A). This result was replicated in early passage primary cells from MITF-variant tumors (Fig. 4B).
Fig. 4: Immunohistochemistry and immunofluorescence staining for MiT family member proteins and GPNMB.
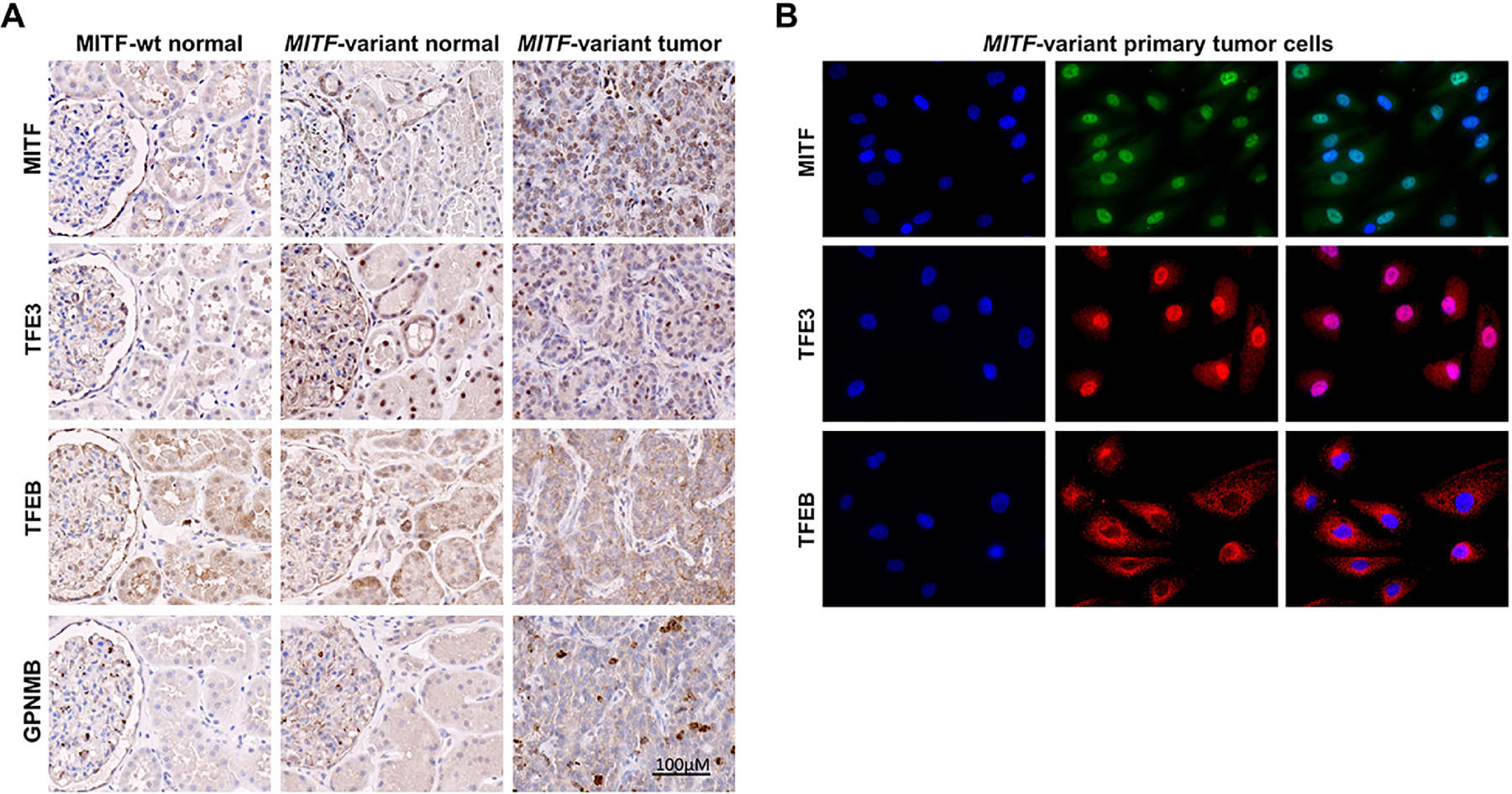
(A) Immunohistochemistry shows cytoplasmic staining of MITF in MITF wild type (wt) kidney tissue and predominantly nuclear localization of MITF in MITF p.E318K variant kidney and tumor tissues. TFE3 immunohistochemistry in MITF p.E318K variant kidney and tumor tissues shows predominantly nuclear staining. TFEB is localized to cytoplasm in MITF wild type (wt) and p.E318K variant tissues. While normal kidney expresses very low levels of GPNMB, tumor tissue carrying the MITF p.E318K variant displays areas of high GPNMB staining.
(B) Immunofluorescence staining for MiT family members (MITF, TFE3 and TFEB) in early passage primary tumor cells derived from the MITF p.E318K variant kidney tumor shows distinct nuclear localization of MITF and TFE3, and cytoplasmic localization of TFEB. Panels show DAPI stain (blue), protein of interest (green or red) and overlap.
DISCUSSION
MITF belongs to a family of microphthalmia-associated transcription factors that includes TFE3 and TFEB and alteration of these genes is strongly associated with RCC. Somatic translocations involving TFE3, TFEB, and, in rare cases, MITF that result in fusion proteins have been shown to be driver alterations in sporadic RCC.17–19 While, a specific germline activating variant of MITF, p.E318K, has been shown to be a risk factor for the development of melanoma and RCC.7, 11, 14 In this study we describe a family in which an individual presented with bilateral, multifocal type 1 papillary RCC with a germline MITF p.E318K pathogenic variant.
The histology of sporadic translocation RCCs is frequently characterized by clear cell and papillary features, but can be heterogeneous.25 Likewise, the histological subtype of RCC associated with the MITF p.E318K variant has shown a degree of variability in a small number of reports. Three studies have reported tumors classified as clear cell RCC, while two studies also classified tumors as papillary RCC, and one tumor as resembling a “juvenile RCC with translocation”, indicating the histopathology similar to that seen with somatic TFE3 translocations.7, 14, 15 The patient described in the current study presented with bilateral and multifocal type 1 papillary RCC that has not been previously described in association with the MITF p.E318K variant. Previously, bilateral and multifocal type 1 papillary RCC has only been associated with germline mutations of the MET oncogene in hereditary papillary renal carcinoma (HPRC).4 Furthermore, since this patient did not have a personal or family history of melanoma, these findings demonstrate that MITF mutations should be considered in the genetic evaluation of patients who present with this phenotype. The patient in this study presented with an early age of onset (43 years), that is consistent with other RCC predisposition syndromes. The average age of onset for RCC in reported patients with the MITF p.E318K variant was 52.5 years old with a range of onsets from 33 to 79 years old.7, 14, 15 To date, no imaging recommendations or clinical guidelines are available for carriers of the MITF p.E318K variant.26, 27 The early age of onset and potential for RCC to present as the sole feature suggests that germline MITF alteration should be considered in patients affected with or at risk for hereditary RCC.
Analysis of the tumors from the patient with the germline MITF p.E318K variant demonstrated further somatic changes of MITF in a small number of tumors. These tumors showed chromosomal copy number neutral loss of heterozygosity, and no tumors showed loss of chromosome 3. Sporadic type 1 papillary RCC is characterized by copy number gains of chromosomes 7 and 17 and such gains were observed in all of the tumors analyzed in this study.3 These genetic alterations resulted in amplification of the MET, EGFR, and BRAF genes, among others, and are likely to aid in tumorigenesis. The necessity for additional genetic alterations could account for the relatively low penetrance of RCC in MITF p.E318K variant carriers. In support of this hypothesis, sporadic TFE3/TFEB-translocation RCCs have been shown to frequently gain pathogenic mutations in chromatin remodeling genes.28
Under normal physiological conditions, the subcellular localization of members of the MiT transcription factor family is tightly regulated to control their transcriptional activity by permitting nuclear migration only in response to specific environmental cues.16 Sporadic TFE3-translocation RCCs are characterized by gene fusions that drive uncontrolled migration of the transcription factor into the nucleus producing a strong and specific IHC signal for nuclear TFE3 that can serve as a diagnostic marker.17, 18 MITF p.E318K variant tumors in this and a previous study demonstrated migration of MITF to the nucleus, where it could cooperate with other MiT family members in regulating gene expression of target genes.7 Physiological stimuli, such as reduced nutrient availability, induce nuclear translocation of MiT family members to stimulate autophagy, lysosomal biogenesis, cell cycle, cell survival, apoptosis, and protection against oxidative stress.7, 16, 19, 29, 30 While normal MITF protein responds to restore nutrient levels of the cells, p.E318K-variant MITF protein may activate an altered gene expression profile due to the alteration in SUMOylation-based control, which could contribute to tumorigenesis and tumor progression. This mechanism could explain the increased susceptibility of MITF p.E318K variant carriers to develop neoplasms. We showed that genes involved in apoptosis (BIRC7), invasion (ACP5) and cell cycle (CCND1) were differentially regulated in MITF p.E318K variant tumor tissues. In addition, melanocytic markers, such as GPNMB and MLANA, were highly upregulated in MITF p.E318K variant papillary RCC. GPNMB is upregulated in several tumors and has been shown to be a direct target of MITF and TFE3 transcription factors.31–33 The finding in the present study of increased expression of GPNMB, a highly glycosylated protein, may provide a tumor specific marker that could be exploited for therapy. An antibody-drug conjugate against GPNMB (Glembatumumab vedotin; CDX-011) has shown pharmacologic efficacy against breast cancer and melanoma.34–36 This antibody-drug conjugate could potentially provide a targeted therapy for MITF p.E318K variant patients with advanced disease.34–36
CONCLUSIONS
In summary, the present study has identified a patient carrying a germline MITF p.E318K variant from a family characterized by a history of RCC, who presented with bilateral, multifocal type 1 papillary RCC. To our knowledge, this is the first report characterizing the genotype, phenotype, pathologic features and genomic characteristics of MITF kidney cancer. It is notable that the here described renal phenotype in the context of MITF p.E318K variant consists of bilateral, multifocal papillary renal cell carcinoma. It is also notable that the MITF pathology is consistently papillary with clear cell features. This is precisely the pathology frequently described in TFE3 renal cell carcinoma. It is likely that MITF familial RCC is more common than is currently appreciated. When we first described TFE3 RCC in 1996,6 it was considered a novel and rare form of RCC. We now know that TFE3 and TFEB RCC make up 42 percent of RCC in children and young adults37 and are more common than previously appreciated in the adult population, where they may account for as much as 12 percent of type II papillary RCC38 and a surprising number of clear cell RCC’s.39 These findings illustrate that RCC in the context of a MITF p.E318K variant may present with a clear cell-papillary, non-clear cell, or unusual RCC histology, thus emphasizing the importance of screening for MITF variants in patients with familial renal cell carcinoma, regardless of their histopathologic subtype. Once identified, carriers of the MITF p.E318K variant should be screened for RCC and melanoma to allow for early detection and treatment of potential malignancies. Downstream targets of MITF and TFE3 transcription factors, such as GPNMB, provide opportunities for the development of targeted therapies in MITF p.E318K variant RCC.
Acknowledgements:
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, including grants ZIA BC011028, ZIA BC011038, Z01 BC011089, and ZIC BC011044. This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under Contract HHSN261200800001E. Sanger Sequencing was conducted at the CCR Genomics Core at the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Linehan WM. Genetic basis of kidney cancer: role of genomics for the development of disease-based therapeutics. Genome Res. 2012;22:2089–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farley MN, Schmidt LS, Mester JL, et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol Cancer Res. 2013;11:1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N, Linehan WM, Spellman PT, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med. 2016;374:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. [DOI] [PubMed] [Google Scholar]
- 5.Launonen V, Vierimaa O, Kiuru M, et al. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci U S A. 2001;98:3387–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidhar SK, Clark J, Gill S, et al. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses anovel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet. 1996;5:1333–1338. [DOI] [PubMed] [Google Scholar]
- 7.Bertolotto C, Lesueur F, Giuliano S, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. [DOI] [PubMed] [Google Scholar]
- 8.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoyama S, Woods SL, Boyle GM, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghiorzo P, Pastorino L, Queirolo P, et al. Prevalence of the E318K MITF germline mutation in Italian melanoma patients: associations with histological subtypes and family cancer history. Pigment Cell Melanoma Res. 2013;26:259–262. [DOI] [PubMed] [Google Scholar]
- 12.Potrony M, Puig-Butille JA, Aguilera P, et al. Prevalence of MITF p.E318K in Patients With Melanoma Independent of the Presence of CDKN2A Causative Mutations. JAMA Dermatol. 2016;152:405–412. [DOI] [PubMed] [Google Scholar]
- 13.Berwick M, MacArthur J, Orlow I, et al. MITF E318K’s effect on melanoma risk independent of, but modified by, other risk factors. Pigment Cell Melanoma Res. 2014;27:485–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoehr CG, Walter B, Denzinger S, et al. The Microphthalmia-Associated Transcription Factor p.E318K Mutation Does Not Play a Major Role in Sporadic Renal Cell Tumors from Caucasian Patients. Pathobiology. 2016;83:165–169. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen KA, Syed JS, Espenschied CR, et al. Advances in the diagnosis of hereditary kidney cancer: Initial results of a multigene panel test. Cancer. 2017;123:4363–4371. [DOI] [PubMed] [Google Scholar]
- 16.Martina JA, Diab HI, Lishu L, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kauffman EC, Ricketts CJ, Rais-Bahrami S, et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nat Rev Urol. 2014;11:465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Argani P MiT family translocation renal cell carcinoma. Semin Diagn Pathol. 2015;32:103–113. [DOI] [PubMed] [Google Scholar]
- 19.Durinck S, Stawiski EW, Pavia-Jimenez A, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2015;47:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellbrock C, Arozarena I. Microphthalmia-associated transcription factor in melanoma development and MAP-kinase pathway targeted therapy. Pigment Cell Melanoma Res. 2015;28:390–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem. 2005;280:146–155. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Vocke CD, Ricketts CJ, et al. Genomic and metabolic characterization of a chromophobe renal cell carcinoma cell line model (UOK276). Genes Chromosomes Cancer. 2017;56:719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baiocco JA, Ball MW, Pappajohn AK, et al. A comparison of outcomes for standard and multiplex partial nephrectomy in a solitary kidney: The National Cancer Institute experience. Urol Oncol. 2019;37:356 e351–356 e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuch B, Linehan WM, Bratslavsky G. Repeat partial nephrectomy: surgical, functional and oncological outcomes. Curr Opin Urol. 2011;21:368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magers MJ, Udager AM, Mehra R. MiT Family Translocation-Associated Renal Cell Carcinoma: A Contemporary Update With Emphasis on Morphologic, Immunophenotypic, and Molecular Mimics. Arch Pathol Lab Med. 2015;139:1224–1233. [DOI] [PubMed] [Google Scholar]
- 26.Freifeld Y, Ananthakrishnan L, Margulis V. Imaging for Screening and Surveillance of Patients with Hereditary Forms of Renal Cell Carcinoma. Curr Urol Rep. 2018;19:82. [DOI] [PubMed] [Google Scholar]
- 27.Carlo MI, Hakimi AA, Stewart GD, et al. Familial Kidney Cancer: Implications of New Syndromes and Molecular Insights. Eur Urol. 2019;76:754–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malouf GG, Su X, Yao H, et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res. 2014;20:4129–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartman ML, Czyz M. Pro-survival role of MITF in melanoma. J Invest Dermatol. 2015;135:352–358. [DOI] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas N Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One. 2010;5:e15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loftus SK, Antonellis A, Matera I, et al. Gpnmb is a melanoblast-expressed, MITF-dependent gene. Pigment Cell Melanoma Res. 2009;22:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tse KF, Jeffers M, Pollack VA, et al. CR011, a fully human monoclonal antibody-auristatin E conjugate, for the treatment of melanoma. Clin Cancer Res. 2006;12:1373–1382. [DOI] [PubMed] [Google Scholar]
- 34.Bendell J, Saleh M, Rose AA, et al. Phase I/II study of the antibody-drug conjugate glembatumumab vedotin in patients with locally advanced or metastatic breast cancer. J Clin Oncol. 2014;32:3619–3625. [DOI] [PubMed] [Google Scholar]
- 35.Ott PA, Hamid O, Pavlick AC, et al. Phase I/II study of the antibody-drug conjugate glembatumumab vedotin in patients with advanced melanoma. J Clin Oncol. 2014;32:3659–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yardley DA, Weaver R, Melisko ME, et al. EMERGE: A Randomized Phase II Study of the Antibody-Drug Conjugate Glembatumumab Vedotin in Advanced Glycoprotein NMB-Expressing Breast Cancer. J Clin Oncol. 2015;33:1609–1619. [DOI] [PubMed] [Google Scholar]
- 37.Cajaiba MM, Dyer LM, Geller JI, et al. The classification of pediatric and young adult renal cell carcinomas registered on the children’s oncology group (COG) protocol AREN03B2 after focused genetic testing. Cancer. 2018;124:3381–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linehan WM, Spellman PT, Ricketts CJ, et al. Comprehensive Molecular Characterization of Papillary Renal Cell Carcinoma. N Engl J Med. 2016;374:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas N Comprehensive Molecular Characterization of Clear Cell Renal Cell Carcinoma. Nature. 2013;499:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]


