Abstract
Background
In the nematode Caenorhabditis elegans, longevity in response to germline ablation, but not in response to reduced insulin/IGF1-like signaling, is strongly dependent on the conserved protein kinase minibrain-related kinase 1 (MBK-1). In humans, the MBK-1 ortholog DYRK1A is associated with a variety of disorders, most prominently with neurological defects observed in Down syndrome. To better understand mbk-1’s physiological roles and their dependence on genetic background, we analyzed the influence of mbk-1 loss on the transcriptomes of wildtype and long-lived, germline-deficient or insulin-receptor defective, C. elegans strains by RNA-sequencing.
Results
mbk-1 loss elicited global changes in transcription that were less pronounced in insulin-receptor mutant than in germline-deficient or wildtype C. elegans. Irrespective of genetic background, mbk-1 regulated genes were enriched for functions in biological processes related to organic acid metabolism and pathogen defense. qPCR-studies confirmed mbk-1 dependent induction of all three C. elegans Δ9-fatty acid desaturases, fat-5, fat-6 and fat-7, in wildtype, germline-deficient and insulin-receptor mutant strains. Conversely, mbk-1 dependent expression patterns of selected pathogen resistance genes, including asp-12, dod-24 and drd-50, differed across the genetic backgrounds examined. Finally, cth-1 and cysl-2, two genes which connect pathogen resistance to the metabolism of the gaseous messenger and lifespan regulator hydrogen sulfide (H2S), were commonly suppressed by mbk-1 loss only in wildtype and germline-deficient, but not in insulin-receptor mutant C. elegans.
Conclusion
Our work reveals previously unknown roles of C. elegans mbk-1 in the regulation of fatty acid desaturase- and H2S metabolic-genes. These roles are only partially dependent on genetic background. Considering the particular importance of fatty acid desaturation and H2S for longevity of germline-deficient C. elegans, we propose that these processes at least in part account for the previous observation that mbk-1 preferentially regulates lifespan in these worms.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-08176-y.
Keywords: Lifespan regulation, Aging, Insulin-like signaling, Germline stem cells, RNA-seq, Fatty acid desaturation, Pathogen defense, Hydrogen sulfide
Introduction
Caenorhabditis elegans mbk-1 (minibrain-related kinase 1) encodes an evolutionarily conserved dual-specificity tyrosine-regulated kinase (DYRK) orthologous to Drosophila melanogaster minibrain and human DYRK1A/B [1–4]. Human DYRK1A maps to chromosome 21q22.13 in the Down syndrome critical region, and its overexpression has been implicated in the neurological defects associated with this disorder [5, 6]. Inhibitory single nucleotide variants, small insertions/deletions or complete deletion of one DYRK1A-allele in patients or in mice also cause intellectual disability and other abnormalities, highlighting the sensitivity of DYRK1A-activity to gene dosage [7, 8]. DYRK1A is expressed in neuronal and non-neuronal tissues during development and adulthood [2, 9] and has been described to contribute to tumor development, both as an oncogene and as a tumor suppressor [2]. Indeed, individuals with Down syndrome display an elevated risk for certain types of childhood leukemia, but a lower risk for solid tumors across all ages [10, 11].
C. elegans mbk-1 differs from its related kinase mbk-2 (DYRK2) and from a more distant family member, hpk-1 (HIPK2) in its expression and subcellular localization patterns, as well as in its physiological role [3]. MBK-1 is expressed in all somatic cells during development and adulthood and localizes predominantly to the nucleus [3]. Overexpression of mbk-1 impairs olfactory behavior, paralleling the DYRK1A-overexpression induced neurological defects observed in other species [3]. On the other hand, mbk-1 inactivation does not cause obvious phenotypic alterations under standard culture conditions [3]. Yet, upon exposure to Pseudomonas aeruginosa strain PAO1, mbk-1 is required for protecting C. elegans from fast killing by this pathogen [12]. Moreover, mbk-1 loss strongly reduces longevity in response to germline ablation, while exerting smaller effects on longevity in response to reduced insulin-like signaling, or on wildtype lifespan [13]. The mechanisms by which mbk-1 promotes pathogen defense and longevity have not been fully elucidated.
Reduced activity of the conserved insulin/IGF1-(like) signaling pathway promotes longevity from C. elegans to mammals [14]. Similarly, loss of germline stem cells (GSCs) extends lifespan in C. elegans and in Drosophila melanogaster, and castration has been associated with longevity in men, suggesting that germline signals also constitute a wide-spread principle of lifespan regulation [15–18]. In C. elegans, longevity in response to daf-2 (insulin/IGF1-like receptor) inhibition and GSC ablation differ in their requirement for certain regulators, such as mbk-1, kri-1 and tcer-1 [13, 19, 20]. On the other hand, both pathways depend on a common set of conserved key transcription factors, including DAF-16 (FOXO), HLH-30 (TFEB), HSF-1 (HSF1) and SKN-1 (NRF2) [21]. In germline (GSC)-deficient, but not in daf-2 mutant C. elegans, mbk-1 contributes to full induction of daf-16 target genes, consistent with the model that mbk-1 exerts at least a part of its lifespan-modulatory activity through DAF-16 [13]. Of note, a DYRK1A phosphorylation site on FOXO1 appears to be conserved between C. elegans and humans [13, 22].
In this study, we aimed to better understand the mechanisms through which mbk-1 preferentially modulates C. elegans lifespan in response to GSC deficiency. Towards this goal, we examined the effect of mbk-1 loss on stress resistance and gene transcription of normal and long-lived, wildtype, germline-deficient or daf-2(−) mutant C. elegans strains, in functional assays and by RNA-sequencing.
Results
Loss of mbk-1 decreases heat and oxidative stress resistance in germline-deficient worms
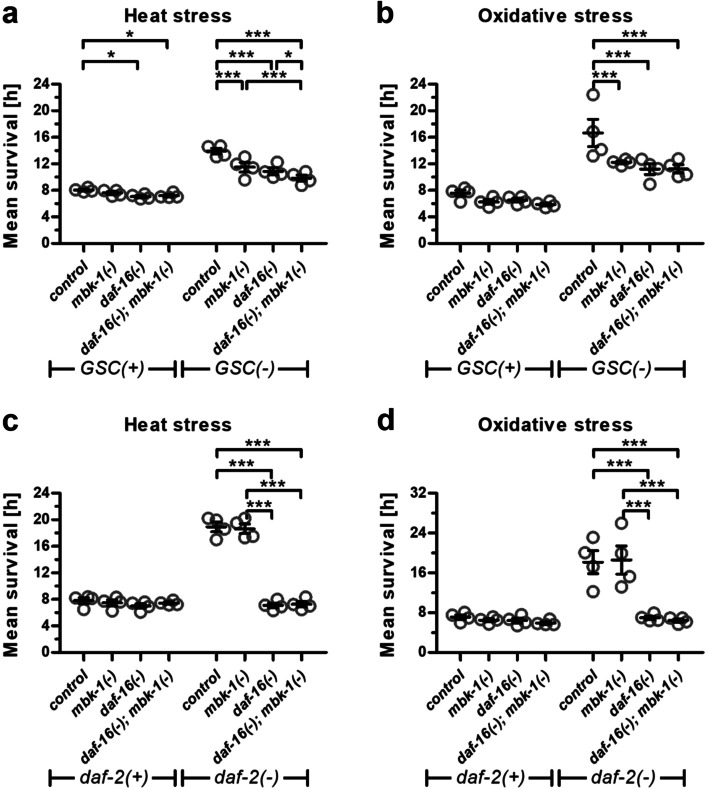
We previously reported that the longevity of GSC deficient C. elegans is strongly dependent on mbk-1 [13]. Conversely, long-lived daf-2(−) or wildtype worms do not, or only to a minor extent, require mbk-1 for lifespan regulation [13]. As changes in longevity are frequently accompanied by changes in resistance to environmental stress such as heat or oxidative stress [23], we examined how mbk-1 influences these properties in GSC(−), daf-2(−) and corresponding GSC(+) or daf-2(+) control worms (cf. Methods). Upon exposure to heat stress, the loss of function allele mbk-1(pk1398) (hereafter referred to as mbk-1(−); [3]), consistently reduced survival of GSC(−) worms, while a more moderate, or no reduction of survival was observed in daf-2(−) and in GSC(+)/daf-2(+) worms (Fig. 1a, c, Additional file 1: Table S1). Oxidative stress resistance on the other hand was consistently decreased in mbk-1(−) worms in GSC(−), GSC(+) and daf-2(+), but not in daf-2(−) background (Fig. 1b, d, Additional file 1: Table S1). Loss of daf-16 produced a statistically significant decrease in survival in response to each of the two stressors in all genetic backgrounds in all repetitions of the experiment, although effects were generally smaller in GSC(+)/daf-2(+) than in long-lived GSC(−) or daf-2(−) worms [24] (Additional file 1: Table S1). Yet, when daf-16 and mbk-1 loss were combined, further reduction of survival was consistently observed only in heat-stressed GSC(−) worms (Fig. 1; Additional file 1: Table S1). Collectively, these results are consistent with and expand the concept [13] that mbk-1 activity is particularly important for GSC(−) physiology and longevity. Moreover, our data rise the possibility that mbk-1 exerts its lifespan- and stress modulatory function in these worms through both, daf-16 dependent and –independent mechanisms.
Fig. 1.
Loss of mbk-1 decreases stress resistance of germline-deficient C. elegans. Strains of the genetic backgrounds indicated, carrying loss of function mutations in daf-16 and/or mbk-1 or no additional mutation (control) were subjected to heat (a, c) or oxidative stress (b, d) and survival was monitored. Data represent mean survival ± SEM from 4 biological replicates of the experiment. Statistical significance of survival differences between groups was determined by repeated measures two-way ANOVA with Bonferroni post tests (* p < 0.05, ** p < 0.01, *** p < 0.001). See Additional file 1: Table S1 for complete statistical analysis
Identification of differentially expressed genes upon loss of mbk-1
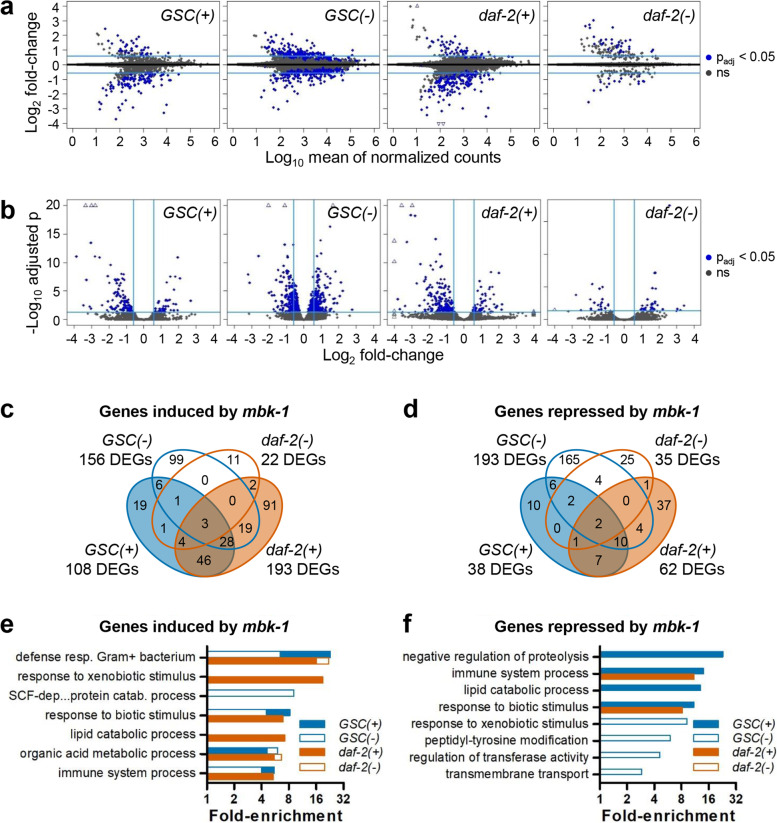
To identify genes whose expression is regulated in an mbk-1 dependent manner during adulthood, we performed whole transcriptome RNA-sequencing (RNA-seq) on mbk-1(+) and mbk-1(−) worms. To further account for the influence of genetic background on mbk-1 function, these experiments were conducted in GSC(−), daf-2(−) and in GSC(+) and daf-2(+) control strains. Downstream analyses focused on differentially expressed genes (DEGs) that displayed an expression fold-change (FC) difference > 1.5 between the strains compared. Only when comparing GSC(−) to GSC(+) worms, this FC-threshold was set to 4 (cf. Ref. [25]). Comparison of DEGs in daf-2(−) vs. daf-2(+), and in GSC(−) vs. GSC(+) worms revealed highly significant overlaps with published RNA-seq studies [25–27], thus validating our data (Additional file 1: Tables S2, 3; Additional file 2: Table S9; Additional file 3: Fig. S1). Globally, changes in gene expression upon mbk-1 loss were more limited in daf-2(−) than in GSC(−) or GSC(+)/daf-2(+) control worms. (Fig. 2a, b). Genes downregulated upon mbk-1 loss (mbk-1 induced genes) from GSC(+), GSC(−), daf-2(+) and daf-2(−) strains significantly overlapped with each other, and the same was observed for genes upregulated upon mbk-1 loss (mbk-1 repressed genes) (Fig. 2c, d; Additional file 1: Table S3, 4; Additional file 2: Table S9). In addition, irrespective of genetic background, mbk-1 induced DEGs significantly overlapped with most to all lists of daf-16 induced genes in long-lived mutants from six published studies [28–33] (Additional file 1: Table S5). On the other hand, significant overlap with most lists of daf-16 repressed genes was observed only for mbk-1 repressed genes from GSC(−) worms (Additional file 1: Table S5). Moreover, irrespective of genetic background, mbk-1 induced genes were commonly enriched for genes associated with the biological processes/gene ontology (GO) terms “organic acid metabolic process” and “defense response to gram-positive bacterium”, although this latter GOterm comprised only very few genes in long-lived mutant backgrounds (Fig. 2e; Additional file 1: Table S6). Conversely, enrichment analysis among mbk-1 repressed genes did not reveal any GO terms shared across all four backgrounds (Fig. 2f). Interestingly, in GSC(+)/daf-2(+) worms, both mbk-1 induced- and mbk-1 repressed genes, were enriched for functions in “response to biotic stimulus” and “immune system process” (Fig. 2e, f; Additional file 1: Table S6). In summary, our gene expression- and GO term enrichment analyses pointed towards a partially genetic background-dependent role for mbk-1 in immunity, reminiscent of a previous study [12], and towards a novel function of mbk-1 in the regulation of organic acid metabolism.
Fig. 2.
Global changes in gene expression upon mbk-1 loss in wildtype and long-lived C. elegans. a and b Differential gene expression between mbk-1(−) and mbk-1(+) worms in the backgrounds indicated, visualized in MA-plots (a) and Volcano plots (b). Light blue lines indicate |Log2FC| > 0.58 and padj < 0.05, open triangles indicate data points beyond axis limits. c and d Overlap of mbk-1 induced (c) and mbk-1 repressed genes (d) from the backgrounds indicated visualized by Venn-diagrams. See Table S4 for statistical analysis. e and f Biological processes enriched (q < 0.05) among mbk-1 induced (e) and mbk–1 repressed genes (f) in the backgrounds indicated. See Table S6 for complete analysis
Mbk-1 modulates the expression of fatty acid desaturase genes
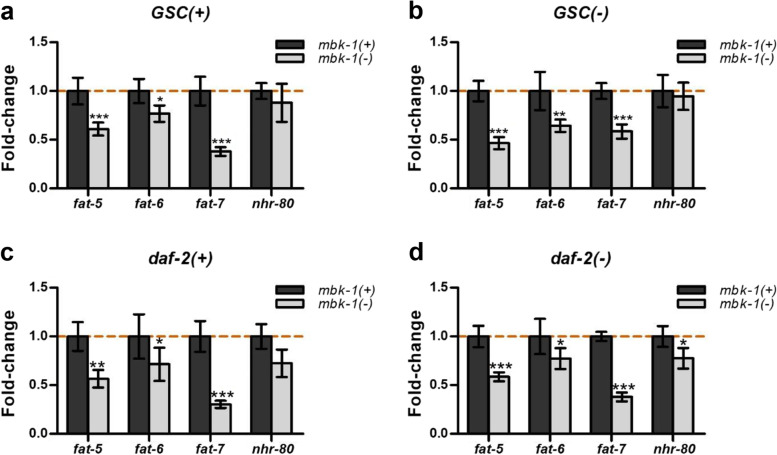
The GOterm “organic acid metabolic process” showed significant enrichment among mbk-1 induced genes irrespective of genetic background, and contained two of the three C. elegans Δ9-fatty acid desaturase genes [34], fat-5 and fat-6, in GSC(−) and in daf-2(+) worms (Fig. 2e, Additional file 1: Table S3). Differential expression of the third desaturase, fat-7, was not statistically significant in any of the four backgrounds examined (Additional file 2: Table S9). Previous reports indicate that fat-5; fat-6; fat-7 triple mutants are not viable, and that both, fat-6 and fat-7 together, are required for GSC(−) longevity, but not for normal wildtype lifespan [35, 36]. Similarly, the fat-5/fat-6/fat-7 transcriptional regulator nhr-80 is required for GSC(−), but not daf-2(−) longevity [35]. Given this preferential requirement of mbk-1 and Δ9-fatty acid desaturation for GSC(−) longevity, we examined the effect of mbk-1 loss on fat-5/fat-6/fat-7- and nhr-80 expression in GSC(−), daf-2(−) and corresponding GSC(+)/daf-2(+) control strains by qPCR. These experiments revealed repression of all three Δ9-desaturase genes in mbk-1(−) worms of all four backgrounds, while nhr-80 was suppressed to a statistically significant extent only in daf-2(−) worms (Fig. 3). Of note, in agreement with published studies [25–27, 35], GSC(−) and daf-2(−) worms displayed increased mRNA-levels of fat-5 and fat-6, but not fat-7 relative to corresponding GSC(+) or daf-2(+) animals (Additional file 2: Table S9, Additional file 3: Fig. S2). In summary, our qPCR analyses indicated that mbk-1 positively regulates Δ9-desaturase gene expression and that this regulatory role is independent of GSC and daf-2 status.
Fig. 3.
mbk-1 promotes the expression of fatty acid desaturase-genes. Gene expression levels were determined for the genes indicated in GSC(+) (a), GSC(−) (b), daf-2(+) (c) and daf-2(−) (d) by qPCR in day 2 adult worms. Data shown represent mean fold-changes ± SEM in mbk-1(−) relative to mbk-1(+) worms from four biological replicates. Statistical significance of expression differences was determined by two-way ANOVA with Bonferroni post tests (* p < 0.05, ** p < 0.01, *** p < 0.001)
Mbk-1 modulates the expression of genes implicated in pathogen defense and immunity
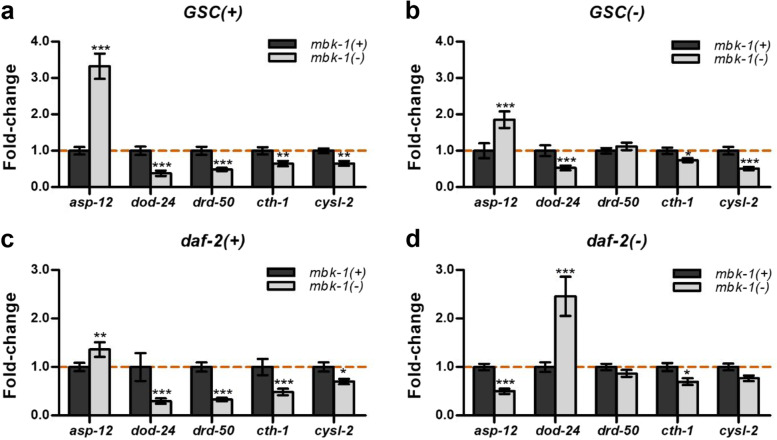
Our RNA-seq based gene expression- and GO term enrichment-analyses further suggested that mbk-1 loss disturbs the expression of genes related to pathogen defense/immunity, particularly in GSC(+)/daf-2(+) worms (Fig. 2e, f). To further validate this observation, we examined mRNA-levels of selected genes associated with both, “response to biotic stimulus” and the more specialized GOterm “immune system process” by qPCR. In agreement with our RNA-seq data, these qPCR-analyses indicated induction of asp-12 (a cathepsin E-ortholog) [37], as well as repression of dod-24 (an epoxide hydrolase 1 ortholog) [32] and drd-50 (nematode-specific) [37], upon mbk-1 loss in GSC(+)/daf-2(+) worms (Fig. 4a, c; Additional file 1: Table S3). Similarly, qPCR confirmed that mbk-1 loss causes asp-12 induction and dod-24 repression in GSC(−), but not in daf-2(−) worms (Fig. 4b, d). Moreover, drd-50 expression was consistently unaffected by mbk-1 loss in GSC(−) worms (Fig. 4b; Additional file 1: Table S3). Finally, our analyses confirmed previous studies [25–27] that had reported differential expression of asp-12, dod-24 and drd-50 between daf-2(−) and daf-2(+), and of asp-12 between GSC(−) and GSC(+) worms (Additional file 3: Fig. S3).
Fig. 4.
mbk-1 promotes the expression of pathogen defense- and H2S/HCN metabolism genes. Gene expression levels were determined for the genes indicated in GSC(+) (a), GSC(−) (b), daf-2(+) (c) and daf-2(−) (d) by qPCR in day 2 adult worms. Data shown represent mean fold-changes ± SEM in mbk-1(−) relative to mbk-1(+) worms from four biological replicates. Statistical significance of expression differences was determined by two-way ANOVA with Bonferroni post tests (* p < 0.05, ** p < 0.01, *** p < 0.001)
As none of the mbk-1 dependent DEGs validated above has been implicated in the same form of pathogen resistance than mbk-1 itself has been implicated in, we searched or RNA-seq data for genes that may mediate this known role of mbk-1. Specifically, mbk-1 protects C. elegans from fast-killing by a particular strain of the gram-negative bacterium P. aeruginosa, PAO1 [12]. Given that growing C. elegans in the presence of hydrogen sulfide (H2S) also confers PAO1-resistance [38], and that PAO1 kills C. elegans by hydrogen cyanide (HCN) poisoning [39], we focused on genes implicated in H2S- and/or HCN-metabolism. Indeed, qPCR-analysis indicated mbk-1 loss-induced downregulation of the cystathionine gamma lyase ortholog cth-1 [40] in all four backgrounds examined, confirming results or trends also observed in our RNA-seq data (Fig. 4; Additional file 1: Table S3). In addition, a known PAO1-resistance gene, the cyanoalanine synthase cysl-2 [38] was downregulated upon mbk-1 loss in GSC(−) and in GSC(+)/daf-2(+) C. elegans (Fig. 4). Collectively, these data indicate that mbk-1 modulates the expression of pathogen defense genes in a genetic background-dependent manner, and rise the possibility that mbk-1’s function in pathogen resistance/immunity is not limited to fast-killing by gram-negative P. aeruginosa PAO1.
Discussion
In this study, we show that the DYRK1-ortholog mbk-1, which is required for full longevity of germline-deficient C. elegans, also promotes stress resistance in these animals. Moreover, using a transcriptomics approach, we discovered that mbk-1 modulates the expression of genes implicated in pathogen defense and in fatty acid desaturation, a process that, as mbk-1, strongly contributes to GSC(−), but not to daf-2(−) longevity. Thus, our study helps explain mbk-1’s previously described activities in C. elegans lifespan regulation and pathogen resistance. To our knowledge, this report also is the first in any organism to profile the influence of a DYRK1A-family member on gene expression in the postmitotic adult stage.
Genes and pathways deregulated upon loss of mbk-1
Our RNA-seq profiling of normal and long-lived GSC(−) and daf-2(−) C. elegans revealed global changes in gene expression that were more limited in daf-2(−) than in GSC(−) or GSC(+)/daf-2(+) animals, but over-all similar across all backgrounds examined. Thus, it is not surprising that mbk-1 regulated genes were functionally enriched for some of the same biological processes. Remarkably, mbk-1 most consistently, i.e. across multiple or all backgrounds analyzed here, modulated the expression of genes implicated in organic acid metabolism and pathogen defense. Indeed, similar GO terms, such as “metabolic process” and “immune system process” were also detected in published gene expression studies on the mbk-1 ortholog in other systems, including zebrafish embryos [41], and HeLa cells [42]. Collectively, these data provide evidence for mbk-1/DYRK1A playing an evolutionarily conserved role in these processes.
Pathways that may account for mbk-1’s particular requirement in germline longevity
Loss of mbk-1 can decrease Δ9-fatty acid desaturase gene expression in GSC(−), daf-2(−) and in GSC(+)/daf-2(+) worms. All three C. elegans Δ9-fatty acid desaturases, fat-5, fat-6, and fat-7, are orthologous to the human Δ9-desaturases hSCD1 and hSCD5 and convert palmitoyl-CoA (fat-5) or stearoyl-CoA (fat-6/7) into palmitoyl- and oleoyl-CoA, respectively [43, 44]. While wildtype worms tolerate simultaneous inactivation of up to two desaturases with little to no lifespan decrease [35, 36], inactivation of fat-6, alone or in combination with fat-5, or concomitant inactivation of fat-6 and fat-7 results in a moderate to substantial reduction of GSC(−) longevity [35]. Therefore, full GSC(−) lifespan extension appears to be particularly dependent on fat-6, and reduced fat-6 expression in GSC(−); mbk-1(−) double-mutant worms may explain their reduced longevity. In daf-2(−) worms, the roles of fat-5, fat-6 and fat-7 for lifespan extension have not been explicitly examined, even though at least fat-5 and fat-6 are positively regulated by daf-16, a key factor for daf-2 longevity [30, 32, 33]. Yet, nhr-80, another transcriptional regulator of fat-5/fat-6/fat-7 [35, 36], is dispensable for daf-2(−) lifespan regulation [35]. Thus, it is unclear whether reduced fat-5/fat-6/fat-7 could also contribute to the slightly reduced longevity of daf-2(−); mbk-1(−) worms [13].
Role of mbk-1 in pathogen defense
Mbk-1 has been implicated in the resistance against a particular strain of the gram-negative bacterium P. aeruginosa, PAO1 [12], which produces HCN to rapidly kill C. elegans [39, 45]. More specifically, mbk-1 contributes to transcriptional activation of the key PAO1-resistance factor HIF-1, but the precise mechanisms remained unclear [12]. The results we present here suggest a potential solution for the so far “missing link” between mbk-1 and hif-1. Specifically, mbk-1 promotes the expression of cth-1, a cystathionine gamma lyase involved in the synthesis of H2S [40]. Moreover, mbk-1 induces the cyanoalanine synthase cysl-2, which directly detoxifies the PAO1 virulence factor HCN [38] in a reaction that also generates H2S [38]. H2S in turn was proposed as an endogenous activator of HIF-1 [46]. Beyond PAO1-resistance, it is interesting to note that GSC(−) worms display elevated levels of H2S relative to wildtype C. elegans and require transsulfuration pathway enzymes, and by extension: H2S, for their longevity [47]. Therefore, it is tempting to speculate that ensuring beneficial levels of H2S constitutes another mechanism via which mbk-1 ensures GSC(−) longevity.
Remarkably, we observed that mbk-1 in wildype C. elegans also promotes the expression of genes such as dod-24 and drd-50, which are upregulated upon exposure to another pathogenic P. aeruginosa strain, PA14 [37]. PA14, in contrast to PAO1, kills C. elegans slowly by gut colonization [12, 37]. Even though some PA14-induced genes such as asp-12 [37] are suppressed by mbk-1, our results provide evidence for a broader function of mbk-1 in pathogen defense that is not restricted to the previously reported protection from P. aeruginosa PAO1 [12]. Clearly, it will be interesting to examine how mbk-1 loss modulates C. elegans’ response to other pathogens in the future.
Conclusions
In summary, the data reported here suggest specific mechanisms underlying previously known activities of the C. elegans DYRK1-family kinase mbk-1 in the regulation of lifespan and pathogen defense. Moreover, they provide further evidence for genetic background-restricted, as well as for genetic background-independent functions of mbk-1. Specific novel regulatory influences of mbk-1 emerging from our studies include fatty acid desaturation, H2S-metabolism and P. aeruginosa PA14-resistance. As many components of these pathways, and mbk-1 itself, are evolutionarily conserved, our results may also be applicable to mbk-1 orthologs in postmitotic tissues of other species.
Methods
C. elegans strains and culture
Strains used in this study are listed in Additional file 1: Table S7. Worms were cultured following standard protocols on NG agar plates seeded with E. coli OP50 [48]. C. elegans carrying the glp-1(e2144ts)-mutation served as a genetic model for germline-deficiency [15, 47]. To eliminate germ cells, glp-1(ts) strains (referred to as GSC(−) in Results/Discussion), and corresponding glp-1(+) (i.e. GSC(+)) control strains, were incubated at 25 °C for the first 24 h of postembryonic development and subsequently, shifted to 20 °C for the remainder of the experiment. Daf-2(e1370) worms and corresponding daf-2(+) control worms were continuously cultured at 20 °C.
Stress resistance assays
Worms were synchronized by hypochlorite treatment and transferred to assay plates on day 2 of adulthood at a density of 20–30 worms per 3 cm plate for heat stress experiments and 50–60 worms per 3 cm plate for oxidative stress experiments. Survival was scored every 1–2 h. Heat stress was imposed by incubation at 35 °C. For oxidative stress experiments, assay plates contained 15.4 mM tert-butyl hydroperoxide (TBHP) and were prepared 12 h before starting the experiment.
Growing worms for RNA-extraction
To obtain synchronized populations, gravid adults were treated with hypochlorite and eggs were allowed to hatch in M9 overnight. ~ 700 L1 larvae per strain were plated on 10 cm NG agar plates seeded with concentrated E. coli OP50 and cultured at the required temperatures (cf. above). At the L4-stage, 500 worms per strain were transferred to two E. coli OP50-seeded 6 cm NG agar plates supplemented with 20 μM FUDR to inhibit germ cell proliferation and progeny production. At day 2 of adulthood, worms were harvested by washing them off their plates with M9. After additional washing with M9 and RNAse-free water, worms were suspended in 1 ml Trizol, snap-frozen in liquid nitrogen and stored at − 80 °C until RNA-extraction.
RNA-extraction
RNA was extracted using Trizol and cleaned up with the Monarch® Total RNA Miniprep Kit or RNA Cleanup Kit (New England Biolabs) according to the manufacturer’s instructions.
RNA-sequencing
RNA quality control, library preparation, and 50 bp single-end RNA-sequencing on the Illumina HiSeq4000 platform was performed at Eurofins Genomics, Ebersberg, Germany. All samples were analyzed in 3 biological replicates and a minimum of 33.2 * 106 reads (maximum 49.9 * 106, median 39.7 * 106 reads) per replicate were obtained, resulting in at least 16.5x genome coverage (maximum 24.9x, median 20x). A minimum of 91.4% of reads (maximum 96.9%, median 95.7%) could be mapped to the C. elegans reference genome (cf. below).
RNA-seq data analysis
Tools provided at https://usegalaxy.eu/ were used for initial analysis steps: FastQC (v0.72) and MultiQC (v1.7) for initial quality control; Trimmomatic (v0.36.5; ILLUMINACLIP with default settings, LEADING:20, TRAILING:20, SLIDINGWINDOW:5:20, MINLENGTH:20) for gentle trimming of reads; STAR (v2.7.2a) for alignment to the C. elegans reference genome (Wormbase release WS271); and featureCounts (v1.6.4) for read summarization [49–54]. One replicate of the daf-2(+); mbk-1(−) strain was excluded at this stage. Subsequent analyses were performed using Bioconductor (v3.8–3.12) packages and custom R scripts (v 3.5.3–4.0.4) [55]. Differential expression analysis with DESeq2 (v1.22.2–1.30.1) [56] was limited to genes detected with at least one count in each biological replicate of at least one of the strains analyzed. Genes with less than 10 counts across the samples compared were removed by pre-filtering. RNA-seq data was visualized with MA- and Volcano-plots using the apeglm-method in DESeq2 for shrinking log-fold-changes [57], and the R/Bioconductor-package EnhancedVolcano (v1.0.1–1.8.0). Differentially expressed genes (DEGs) were defined as padj (Benjamini-Hochberg adjusted p-value/FDR) < 0.05 in DESeq2-analysis and expression fold-change > 1.5/ < 0.6667 (|Log2FC| > 0.58).
Overlap of DEG-lists
Overlaps between lists of DEGs were visualized using the Venn and Euler Diagrams-App in Cytoscape (v3.8.2) [58]. Statistical significance of overlaps was calculated as the hypergeometric probability of detecting at least as many common genes as observed in the two lists, using the phyper-function in R. Representation factors were calculated as the number of overlaping genes divided by the expected number of overlapping genes in the two lists (http://nemates.org/MA/progs/overlap_stats.html; last accessed 12. April 2019). For all calculations, the number of genes in the genome was set to 18,980, i.e. the number of genes in Wormbase WS271 that passed low-count filtering during DESeq2-analysis (cf. above). DEG-lists from published studies were, if necessary, converted to current WBGene-IDs using WormMine (http://intermine.wormbase.org/tools/wormmine/begin.do; last accessed 04. September 2019) and adjusted to genes in Wormbase WS271 using custom R-scripts and manual curation.
GO term enrichment analysis of DEG lists
GO term enrichment analysis was performed using the enrichment tool provided by Wormbase (https://wormbase.org/tools/enrichment/tea/tea.cgi; last accessed: 31 March 2021) [59]. The q-value threshold was set to < 0.05 unless otherwise stated. GO term categories were retrieved using REViGO ([60] http://revigo.irb.hr/; last accessed 31 March 2021).
qPCR
One microgram total RNA was reverse-transcribed using LunaScript® RT SuperMix (New England Biolabs). qPCR-reactions were performed in 2–3 technical replicates in 20 μl reaction volume on an CFX Connect™ Real-Time PCR Detection System (Bio-Rad Laboratories) with iTaq™ Universal SYBR® Green Supermix (Bio-Rad Laboratories). The thermal cycling protocol comprised one activation step at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 10 s and combined annealing/extension at 60 °C for 30 s. Melting curve analysis was performed from 65 °C to 95 °C with 0.5 °C increments at 5 s per step. Data were analyzed by the ΔΔCt method and target gene expression levels were normalized to the geometric mean of cdc-42, tba-1 and Y45F10D.4 [61, 62]. Primer sequences are listed in Additional file 1: Table S8.
Statistical analysis
Statistical analysis was performed using Prism 5 (GraphPad Software, San Diego, CA, USA). Details on the particular tests used are specified in the figure legends.
Supplementary Information
Additional file 1: Table S1. Statistical analysis of stress resistance data. Accompanies Fig. 1. Worms of the strains indicated were subjected to heat or oxidative stress as specified in the individual table headlines. For each biological replicate of the respective experiment, the table lists mean and median survival times in h, standard deviations and standard errors of the mean, the number of worms scored, and p-values from Kaplan-Meier survival analysis with Mantel-Cox tests against the control-, daf-16(−) or mbk-1(−) strain of the same genetic background. In case of the GSC(−) and daf-2(−) control strain, survival analysis was performed relative to the GSC(+) or daf-2(+) control strain. Repeated measures two-way ANOVA with Bonferroni post tests was performed on the mean survival times determined in the individual biological replicates of the experiments. Statistically significant p-values (* p < 0.05, ** p < 0.01, *** p < 0.001) are highlighted in blue. Table S2. Genes regulated by daf-2(−) and GSC(−) in this work and in published studies. For each overlap, the number of genes shared between the two particular lists, the representation factor, and the p-value (in italics; hypergeometric probability) are indicated. Statistically significant overlaps (p < 0.05) are highlighted in blue. For these overlap analyses, lists of published studies were adjusted to WS271 and limited to genes that passed low-count filtering in our DESeq2-analyses. (cf. Methods). See main text for full references of the published studies included in the table. Table S3. Genes differentially expressed between mbk-1(−) and mbk-1(+) worms of the genetic backgrounds analyzed in this study. Differentially expressed genes (DEGs) were defined by padj < 0.05 in DESeq2-analysis and by the following expression fold-change thresholds: FC < 0.6667 (Log2FC < − 0.58) for genes downregulated upon mbk-1 loss. Expression of these genes is normally promoted by mbk-1 (mbk-1 induced DEGs). FC > 1.5 (Log2FC > 0.58) for genes upregulated upon mbk-1 loss. Expression of these genes is normally repressed by mbk-1 (mbk-1 repressed DEGs). In addition, the table lists DEGs detected in control comparisons between the following strains (all mbk-1(+)): downregulated < 0.6667-fold or upregulated > 1.5-fold in daf-2(−) relative to daf-2(+); downregulated < 0.25-fold (Log2FC < − 2) or upregulated > 4-fold (Log2FC > 2) in GSC(−) relative to GSC(+). See Additional file 2 for full DESeq2-data. Table S4. Overlaps of mbk-1 dependent DEGs in the genetic backgrounds analyzed in this study. Accompanies Fig. 2c, d. (a) Pairwise overlaps between DEG-lists. For each overlap, the number of genes shared between the two particular lists, the representation factor, and the p-value (in italics; hypergeometric probability) are indicated. Statistically significant overlaps are highlighted in blue. (b) Absolute numbers of mbk-1 dependent DEGs that are induced/repressed exclusively in the background indicated, or that are also induced/repressed in one or more of the three other backgrounds. (c) Percentages of mbk-1 dependent DEGs that are induced/repressed exclusively in the background indicated, or that are also induced/repressed in one or more of the three other backgrounds. Table S5. Overlap of genes regulated by mbk-1 in this work and by daf-16 in published studies. For each overlap, the number of genes shared between the two particular lists, the representation factor, and the p-value (in italics; hypergeometric probability) are indicated. Statistically significant overlaps (p < 0.05) are highlighted in blue. For these overlap analyses, lists of published studies were adjusted to WS271 and limited to genes that passed low-count filtering in our DESeq2-analyses. (cf. Methods). See main text for full references of the published studies included in the table. Table S6. GO term-enrichment among mbk-1 dependent DEGs in the genetic backgrounds analyzed in this study. Accompanies Fig. 2e, f. mbk-1 induced and -repressed DEGs from the backgrounds indicated were subjected to GO term enrichment analysis. q-values < 0.05 were considered statistically significant. The table covers all three GO term categories (biological process [bp], cellular component [cc] and molecular function [mf]), while only terms of the bp-category were plotted in Fig. 2e, f. Note that Fig 2e, f are further limited to GO terms of the bp-category with Nobserved ≥ 2 and enrichment fold-change ≥ 2 and that some GO terms were not plotted in these figures for the reasons specified in the Comment-column. Table S7. List of C. elegans strains used in this study. Table S8. List of qPCR-primers used in this study.
Additional file 2: Table S9. DESeq2-analysis of the RNA-seq data set generated in this study. Differential gene expression was determined between mbk-1(−) and mbk-1(+) worms (abbreviated as “-VS+” in column headers) in GSC(+), GSC(−), daf-2(+) and daf-2(−) background. In addition, control comparisons were made between the mbk-1(+) strains of the daf-2(−) and daf-2(+), and the GSC(−) and GSC(+) backgrounds. The table shows DESeq2-results for all genes that passed our filter criteria in at least one of the comparisons (cf. Methods).
Additional file 3: Fig. S1. Global changes in gene expression upon daf-2 or GSC loss. Accompanies Fig. 2. (a and b) Differential gene expression between the strains indicated was determined by RNA-seq and visualized in MA-plots (a) and Volcano plots (b). Light blue lines indicate |Log2FC| > 0.58 and padj < 0.05, open triangles indicate data points beyond axis limits. See Additional file 2: Table S9 for complete DESeq2-analysis results. Fig. S2. Influence of GSC and daf-2 status on fatty acid desaturase gene-expression. Accompanies Fig. 3. Expression levels of the genes indicated were determined in GSC(−) relative to GSC(+) (a), and in daf-2(−) relative to daf-2(+) worms (b) by qPCR in day 2 adults. Data shown represent mean fold-changes ± SEM from four biological replicates. Statistical significance of expression differences was determined by two-way ANOVA with Bonferroni post tests (* p < 0.05, ** p < 0.01, *** p < 0.001). Note that other authors [25] have proposed a fold-change of ~4 for detecting genes differentially expressed between GSC(+) and GSC(-) worms, as they differ in the presence of a germline. The following gene expression changes (or lack of change) relative to the corresponding GSC(+)/daf-2(+) strain were also observed in published studies: GSC(−): induction of fat-5/nhr-80 [25, 35], induction of fat-6 [35], no upregulation of fat-7 (our qPCR, [25]) or downregulation of fat-7 (our RNA-seq, [35]); daf-2(−): induction of fat-5 [26, 27] and fat-6 [26], repression of fat-7 [26, 27], no differential expression of nhr-80 [26, 27]. Fig. S3. Influence of GSC and daf-2 status on the expression of genes involved in pathogen defense and H2S/HCN metabolism. Accompanies Fig. 4. Expression levels of the genes indicated were determined in GSC(−) relative to GSC(+) (a), and in daf-2(−) relative to daf-2(+) worms (b) by qPCR in day 2 adults. Data shown represent mean fold-changes ± SEM from 3 to 4 biological replicates. Statistical significance of expression differences was determined by two-way ANOVA with Bonferroni post tests (* p < 0.05, ** p < 0.01, *** p < 0.001). Note that other authors [25] have proposed a fold-change of ~ 4 for detecting genes differentially expressed between GSC(+) and GSC(−) worms, as they differ in the presence of a germline. The following gene expression changes (or lack of change) relative to the corresponding GSC(+)/daf-2(+) strain were also observed in published studies: GSC(−): significant induction of asp-12 [25] (GSC(−) repressed genes not published in this study, thus it is unclear whether dod-24, drd-50, cth-1 and cysl-2 were not induced/induced < 4-fold, or even repressed by GSC loss); daf-2(−): significant induction of asp-12, significant repression of dod-24 and drd-50, no differential expression of cysl-2 [26, 27]; no differential expression of cth-1 in [27], but repression in [26].
Acknowledgements
The authors thank Jerome Mertens for helpful discussion and Werner Kapferer for technical help. Some C. elegans strains used in the present study were provided by the Caenorhabditis Genetic Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The Galaxy server that was used for some calculations is in part funded by Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and German Federal Ministry of Education and Research (BMBF grants 031 A538A/A538C RBC, 031L0101B/031L0101C de.NBI-epi, 031 L0106 de.STAIR (de.NBI)).
Abbreviations
- DEGs
Differentially expressed genes
- FC
Fold-change
- GO term
Gene ontology term
- GSCs
Germline stem cells
- RNA-seq
RNA-sequencing
- SEM
Standard error of the mean
- TBHP
Tert-butyl hydroperoxide
Authors’ contributions
H.I.D.M. conceived the study. H.I.D.M, J.K., and E.A. performed experiments. H.I.D.M., E.K.M.M. and P.J.D. analyzed data. H.I.D.M. drafted the manuscript. All authors revised the manuscript. The authors read and approved the final manuscript.
Funding
This study was funded by a grant from the Tiroler Wissenschaftsfonds to H.I.D.M. Research in E.K.M.M.’s lab is supported by Stiftung P.E. Kempkes and Deutsche José Carreras Leukämie-Stiftung. Funding for publication costs was provided by the University of Innsbruck. The funding bodies had no role in the design of the study, in collection, analysis, and interpretation of data, nor in writing the manuscript.
Availability of data and materials
The RNA sequencing data set generated during the present study has been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10898. Complete results from the DESeq2-analysis are also included in this article in Additional file 2: Table S9.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest relating to this manuscript.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hildegard I. D. Mack, Email: Hildegard.I.Mack@gmail.com
Jennifer Kremer, Email: kremerje@staff.uni-marburg.de.
Eva Albertini, Email: eva.albertini@uibk.ac.at.
Elisabeth K. M. Mack, Email: elisabeth.mack@staff.uni-marburg.de
Pidder Jansen-Dürr, Email: pidder.jansen-duerr@uibk.ac.at.
Reference
- 1.Aranda S, Laguna A, de la Luna S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011;25(2):449–462. doi: 10.1096/fj.10-165837. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez-Martinez P, Zahonero C, Sanchez-Gomez P. DYRK1A: the double-edged kinase as a protagonist in cell growth and tumorigenesis. Mol Cell Oncol. 2015;2(1):e970048. doi: 10.4161/23723548.2014.970048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raich WB, Moorman C, Lacefield CO, Lehrer J, Bartsch D, Plasterk RH, Kandel ER, Hobert O. Characterization of Caenorhabditis elegans homologs of the Down syndrome candidate gene DYRK1A. Genetics. 2003;163(2):571–580. doi: 10.1093/genetics/163.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tejedor F, Zhu XR, Kaltenbach E, Ackermann A, Baumann A, Canal I, Heisenberg M, Fischbach KF. Pongs O: minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;14(2):287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- 5.Dowjat WK, Adayev T, Kuchna I, Nowicki K, Palminiello S, Hwang YW, Wegiel J. Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett. 2007;413(1):77–81. doi: 10.1016/j.neulet.2006.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith DJ, Stevens ME, Sudanagunta SP, Bronson RT, Makhinson M, Watabe AM, O'Dell TJ, Fung J, Weier HU, Cheng JF, et al. Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat Genet. 1997;16(1):28–36. doi: 10.1038/ng0597-28. [DOI] [PubMed] [Google Scholar]
- 7.Fotaki V, Martinez De Lagran M, Estivill X, Arbones M, Dierssen M. Haploinsufficiency of Dyrk1A in mice leads to specific alterations in the development and regulation of motor activity. Behav Neurosci. 2004;118(4):815–821. doi: 10.1037/0735-7044.118.4.815. [DOI] [PubMed] [Google Scholar]
- 8.Ji J, Lee H, Argiropoulos B, Dorrani N, Mann J, Martinez-Agosto JA, Gomez-Ospina N, Gallant N, Bernstein JA, Hudgins L, et al. DYRK1A haploinsufficiency causes a new recognizable syndrome with microcephaly, intellectual disability, speech impairment, and distinct facies. Eur J Hum Genet. 2015;23(11):1473–1481. doi: 10.1038/ejhg.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 10.Lee P, Bhansali R, Izraeli S, Hijiya N, Crispino JD. The biology, pathogenesis and clinical aspects of acute lymphoblastic leukemia in children with Down syndrome. Leukemia. 2016;30(9):1816–1823. doi: 10.1038/leu.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet. 2000;355(9199):165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 12.Shao Z, Zhang Y, Ye Q, Saldanha JN, Powell-Coffman JA. C. elegans SWAN-1 Binds to EGL-9 and regulates HIF-1-mediated resistance to the bacterial pathogen Pseudomonas aeruginosa PAO1. PLoS Pathog. 2010;6(8):e1001075. doi: 10.1371/journal.ppat.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mack HID, Zhang P, Fonslow BR, Yates JR. The protein kinase MBK-1 contributes to lifespan extension in daf-2 mutant and germline-deficient Caenorhabditis elegans. Aging (Albany NY) 2017;9(5):1414–1432. doi: 10.18632/aging.101244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 15.Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295(5554):502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- 16.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399(6734):362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 17.Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones DL, Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci U S A. 2008;105(17):6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min KJ, Lee CK, Park HN. The lifespan of Korean eunuchs. Curr Biol. 2012;22(18):R792–R793. doi: 10.1016/j.cub.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 19.Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124(5):1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Ghazi A, Henis-Korenblit S, Kenyon C. A transcription elongation factor that links signals from the reproductive system to lifespan extension in Caenorhabditis elegans. PLoS Genet. 2009;5(9):e1000639. doi: 10.1371/journal.pgen.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denzel MS, Lapierre LR, Mack HID. Emerging topics in C. elegans aging research: transcriptional regulation, stress response and epigenetics. Mech Ageing Dev. 2019;177:4–21. doi: 10.1016/j.mad.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woods YL, Rena G, Morrice N, Barthel A, Becker W, Guo S, Unterman TG, Cohen P. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem J. 2001;355(Pt 3):597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou KI, Pincus Z, Slack FJ. Longevity and stress in Caenorhabditis elegans. Aging (Albany NY) 2011;3(8):733–753. doi: 10.18632/aging.100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sen I, Zhou X, Chernobrovkin A, Puerta-Cavanzo N, Kanno T, Salignon J, Stoehr A, Lin XX, Baskaner B, Brandenburg S, et al. DAF-16/FOXO requires protein phosphatase 4 to initiate transcription of stress resistance and longevity promoting genes. Nat Commun. 2020;11(1):138. doi: 10.1038/s41467-019-13931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinbaugh MJ, Narasimhan SD, Robida-Stubbs S, Moronetti Mazzeo LE, Dreyfuss JM, Hourihan JM, et al. Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife. 2015;4:e07836. [DOI] [PMC free article] [PubMed]
- 26.Senchuk MM, Dues DJ, Schaar CE, Johnson BK, Madaj ZB, Bowman MJ, Winn ME, Van Raamsdonk JM. Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLoS Genet. 2018;14(3):e1007268. doi: 10.1371/journal.pgen.1007268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo M, Seo K, Hwang W, Koo HJ, Hahm JH, Yang JS, Han SK, Hwang D, Kim S, Jang SK et al. RNA helicase HEL-1 promotes longevity by specifically activating DAF-16/FOXO transcription factor signaling in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2015;112(31):E4246–55. [DOI] [PMC free article] [PubMed]
- 28.Amrit FR, Steenkiste EM, Ratnappan R, Chen SW, McClendon TB, Kostka D, Yanowitz J, Olsen CP, Ghazi A. DAF-16 and TCER-1 Facilitate Adaptation to Germline Loss by Restoring Lipid Homeostasis and Repressing Reproductive Physiology in C. elegans. PLoS Genet. 2016;12(2):e1005788. doi: 10.1371/journal.pgen.1005788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen AT, Guo C, Itani OA, Budaitis BG, Williams TW, Hopkins CE, McEachin RC, Pande M, Grant AR, Yoshina S, et al. Longevity genes revealed by integrative analysis of isoform-specific daf-16/FoxO mutants of Caenorhabditis elegans. Genetics. 2015;201(2):613–629. doi: 10.1534/genetics.115.177998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaletsky R, Lakhina V, Arey R, Williams A, Landis J, Ashraf J, Murphy CT. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 2016;529(7584):92–96. doi: 10.1038/nature16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend Caenorhabditis elegans' lifespan in response to reproductive signals. Aging Cell. 2012;11(2):192–202. doi: 10.1111/j.1474-9726.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424(6946):277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 33.Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154(3):676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watts JL, Ristow M. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics. 2017;207(2):413–446. doi: 10.1534/genetics.117.300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goudeau J, Bellemin S, Toselli-Mollereau E, Shamalnasab M, Chen Y, Aguilaniu H. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 2011;9(3):e1000599. doi: 10.1371/journal.pbio.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brock TJ, Browse J, Watts JL. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet. 2006;2(7):e108. doi: 10.1371/journal.pgen.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A. 2006;103(38):14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budde MW, Roth MB. The response of Caenorhabditis elegans to hydrogen sulfide and hydrogen cyanide. Genetics. 2011;189(2):521–532. doi: 10.1534/genetics.111.129841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallagher LA, Manoil C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol. 2001;183(21):6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qabazard B, Li L, Gruber J, Peh MT, Ng LF, Kumar SD, Rose P, Tan CH, Dymock BW, Wei F, et al. Hydrogen sulfide is an endogenous regulator of aging in Caenorhabditis elegans. Antioxid Redox Signal. 2014;20(16):2621–2630. doi: 10.1089/ars.2013.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho HJ, Lee JG, Kim JH, Kim SY, Huh YH, Kim HJ, et al. Vascular defects of DYRK1A knockouts are ameliorated by modulating calcium signaling in zebrafish. Dis Model Mech. 2019;12(5):dmm037044. [DOI] [PMC free article] [PubMed]
- 42.Jang SM, Azebi S, Soubigou G, Muchardt C. DYRK1A phoshorylates histone H3 to differentially regulate the binding of HP1 isoforms and antagonize HP1-mediated transcriptional repression. EMBO Rep. 2014;15(6):686–694. doi: 10.15252/embr.201338356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim W, Underwood RS, Greenwald I, Shaye DD. OrthoList 2: a new comparative genomic analysis of human and Caenorhabditis elegans genes. Genetics. 2018;210(2):445–461. doi: 10.1534/genetics.118.301307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watts JL, Browse J. A palmitoyl-CoA-specific delta9 fatty acid desaturase from Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;272(1):263–269. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]
- 45.Darby C, Cosma CL, Thomas JH, Manoil C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1999;96(26):15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Budde MW, Roth MB. Hydrogen sulfide increases hypoxia-inducible factor-1 activity independently of von Hippel-Lindau tumor suppressor-1 in C. elegans. Mol Biol Cell. 2010;21(1):212–217. doi: 10.1091/mbc.E09-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei Y, Kenyon C. Roles for ROS and hydrogen sulfide in the longevity response to germline loss in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2016;113(20):E2832–E2841. doi: 10.1073/pnas.1524727113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK. Shi W: featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 50.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams CR, Baccarella A, Parrish JZ, Kim CC. Trimming of sequence reads alters RNA-Seq gene expression estimates. BMC Bioinformatics. 2016;17:103. doi: 10.1186/s12859-016-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018;46(W1):W537–W544. doi: 10.1093/nar/gky379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35(12):2084–2092. doi: 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angeles-Albores D, Lee RY, Chan J, Sternberg PW. Tissue enrichment analysis for C. elegans genomics. BMC Bioinformatics. 2016;17(1):366. doi: 10.1186/s12859-016-1229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C elegans. BMC Mol Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Chen D, Smith MA, Zhang B, Pan X. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS One. 2012;7(3):e31849. doi: 10.1371/journal.pone.0031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Statistical analysis of stress resistance data. Accompanies Fig. 1. Worms of the strains indicated were subjected to heat or oxidative stress as specified in the individual table headlines. For each biological replicate of the respective experiment, the table lists mean and median survival times in h, standard deviations and standard errors of the mean, the number of worms scored, and p-values from Kaplan-Meier survival analysis with Mantel-Cox tests against the control-, daf-16(−) or mbk-1(−) strain of the same genetic background. In case of the GSC(−) and daf-2(−) control strain, survival analysis was performed relative to the GSC(+) or daf-2(+) control strain. Repeated measures two-way ANOVA with Bonferroni post tests was performed on the mean survival times determined in the individual biological replicates of the experiments. Statistically significant p-values (* p < 0.05, ** p < 0.01, *** p < 0.001) are highlighted in blue. Table S2. Genes regulated by daf-2(−) and GSC(−) in this work and in published studies. For each overlap, the number of genes shared between the two particular lists, the representation factor, and the p-value (in italics; hypergeometric probability) are indicated. Statistically significant overlaps (p < 0.05) are highlighted in blue. For these overlap analyses, lists of published studies were adjusted to WS271 and limited to genes that passed low-count filtering in our DESeq2-analyses. (cf. Methods). See main text for full references of the published studies included in the table. Table S3. Genes differentially expressed between mbk-1(−) and mbk-1(+) worms of the genetic backgrounds analyzed in this study. Differentially expressed genes (DEGs) were defined by padj < 0.05 in DESeq2-analysis and by the following expression fold-change thresholds: FC < 0.6667 (Log2FC < − 0.58) for genes downregulated upon mbk-1 loss. Expression of these genes is normally promoted by mbk-1 (mbk-1 induced DEGs). FC > 1.5 (Log2FC > 0.58) for genes upregulated upon mbk-1 loss. Expression of these genes is normally repressed by mbk-1 (mbk-1 repressed DEGs). In addition, the table lists DEGs detected in control comparisons between the following strains (all mbk-1(+)): downregulated < 0.6667-fold or upregulated > 1.5-fold in daf-2(−) relative to daf-2(+); downregulated < 0.25-fold (Log2FC < − 2) or upregulated > 4-fold (Log2FC > 2) in GSC(−) relative to GSC(+). See Additional file 2 for full DESeq2-data. Table S4. Overlaps of mbk-1 dependent DEGs in the genetic backgrounds analyzed in this study. Accompanies Fig. 2c, d. (a) Pairwise overlaps between DEG-lists. For each overlap, the number of genes shared between the two particular lists, the representation factor, and the p-value (in italics; hypergeometric probability) are indicated. Statistically significant overlaps are highlighted in blue. (b) Absolute numbers of mbk-1 dependent DEGs that are induced/repressed exclusively in the background indicated, or that are also induced/repressed in one or more of the three other backgrounds. (c) Percentages of mbk-1 dependent DEGs that are induced/repressed exclusively in the background indicated, or that are also induced/repressed in one or more of the three other backgrounds. Table S5. Overlap of genes regulated by mbk-1 in this work and by daf-16 in published studies. For each overlap, the number of genes shared between the two particular lists, the representation factor, and the p-value (in italics; hypergeometric probability) are indicated. Statistically significant overlaps (p < 0.05) are highlighted in blue. For these overlap analyses, lists of published studies were adjusted to WS271 and limited to genes that passed low-count filtering in our DESeq2-analyses. (cf. Methods). See main text for full references of the published studies included in the table. Table S6. GO term-enrichment among mbk-1 dependent DEGs in the genetic backgrounds analyzed in this study. Accompanies Fig. 2e, f. mbk-1 induced and -repressed DEGs from the backgrounds indicated were subjected to GO term enrichment analysis. q-values < 0.05 were considered statistically significant. The table covers all three GO term categories (biological process [bp], cellular component [cc] and molecular function [mf]), while only terms of the bp-category were plotted in Fig. 2e, f. Note that Fig 2e, f are further limited to GO terms of the bp-category with Nobserved ≥ 2 and enrichment fold-change ≥ 2 and that some GO terms were not plotted in these figures for the reasons specified in the Comment-column. Table S7. List of C. elegans strains used in this study. Table S8. List of qPCR-primers used in this study.
Additional file 2: Table S9. DESeq2-analysis of the RNA-seq data set generated in this study. Differential gene expression was determined between mbk-1(−) and mbk-1(+) worms (abbreviated as “-VS+” in column headers) in GSC(+), GSC(−), daf-2(+) and daf-2(−) background. In addition, control comparisons were made between the mbk-1(+) strains of the daf-2(−) and daf-2(+), and the GSC(−) and GSC(+) backgrounds. The table shows DESeq2-results for all genes that passed our filter criteria in at least one of the comparisons (cf. Methods).
Additional file 3: Fig. S1. Global changes in gene expression upon daf-2 or GSC loss. Accompanies Fig. 2. (a and b) Differential gene expression between the strains indicated was determined by RNA-seq and visualized in MA-plots (a) and Volcano plots (b). Light blue lines indicate |Log2FC| > 0.58 and padj < 0.05, open triangles indicate data points beyond axis limits. See Additional file 2: Table S9 for complete DESeq2-analysis results. Fig. S2. Influence of GSC and daf-2 status on fatty acid desaturase gene-expression. Accompanies Fig. 3. Expression levels of the genes indicated were determined in GSC(−) relative to GSC(+) (a), and in daf-2(−) relative to daf-2(+) worms (b) by qPCR in day 2 adults. Data shown represent mean fold-changes ± SEM from four biological replicates. Statistical significance of expression differences was determined by two-way ANOVA with Bonferroni post tests (* p < 0.05, ** p < 0.01, *** p < 0.001). Note that other authors [25] have proposed a fold-change of ~4 for detecting genes differentially expressed between GSC(+) and GSC(-) worms, as they differ in the presence of a germline. The following gene expression changes (or lack of change) relative to the corresponding GSC(+)/daf-2(+) strain were also observed in published studies: GSC(−): induction of fat-5/nhr-80 [25, 35], induction of fat-6 [35], no upregulation of fat-7 (our qPCR, [25]) or downregulation of fat-7 (our RNA-seq, [35]); daf-2(−): induction of fat-5 [26, 27] and fat-6 [26], repression of fat-7 [26, 27], no differential expression of nhr-80 [26, 27]. Fig. S3. Influence of GSC and daf-2 status on the expression of genes involved in pathogen defense and H2S/HCN metabolism. Accompanies Fig. 4. Expression levels of the genes indicated were determined in GSC(−) relative to GSC(+) (a), and in daf-2(−) relative to daf-2(+) worms (b) by qPCR in day 2 adults. Data shown represent mean fold-changes ± SEM from 3 to 4 biological replicates. Statistical significance of expression differences was determined by two-way ANOVA with Bonferroni post tests (* p < 0.05, ** p < 0.01, *** p < 0.001). Note that other authors [25] have proposed a fold-change of ~ 4 for detecting genes differentially expressed between GSC(+) and GSC(−) worms, as they differ in the presence of a germline. The following gene expression changes (or lack of change) relative to the corresponding GSC(+)/daf-2(+) strain were also observed in published studies: GSC(−): significant induction of asp-12 [25] (GSC(−) repressed genes not published in this study, thus it is unclear whether dod-24, drd-50, cth-1 and cysl-2 were not induced/induced < 4-fold, or even repressed by GSC loss); daf-2(−): significant induction of asp-12, significant repression of dod-24 and drd-50, no differential expression of cysl-2 [26, 27]; no differential expression of cth-1 in [27], but repression in [26].
Data Availability Statement
The RNA sequencing data set generated during the present study has been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10898. Complete results from the DESeq2-analysis are also included in this article in Additional file 2: Table S9.