Abstract
RAD24 has been identified as a gene essential for the DNA damage checkpoint in budding yeast. Rad24 is structurally related to subunits of the replication factor C (RFC) complex, and forms an RFC-related complex with Rfc2, Rfc3, Rfc4, and Rfc5. The rad24Δ mutation enhances the defect of rfc5-1 in the DNA replication block checkpoint, implicating RAD24 in this checkpoint. CHL12 (also called CTF18) encodes a protein that is structurally related to the Rad24 and RFC proteins. We show here that although neither chl12Δ nor rad24Δ single mutants are defective, chl12Δ rad24Δ double mutants become defective in the replication block checkpoint. We also show that Chl12 interacts physically with Rfc2, Rfc3, Rfc4, and Rfc5 and forms an RFC-related complex which is distinct from the RFC and RAD24 complexes. Our results suggest that Chl12 forms a novel RFC-related complex and functions redundantly with Rad24 in the DNA replication block checkpoint.
Eukaryotic cells employ a set of surveillance mechanisms to coordinate the onset of one event and the completion of the preceding event during the cell cycle. The mechanisms that ensure the proper ordering of cell cycle events have been termed checkpoint controls in eukaryotes (11). When DNA is damaged or DNA replication is blocked, the activation of checkpoint pathways arrests the cell cycle and induces the transcription of genes that facilitate DNA repair and/or replication (5, 33).
Checkpoint pathways are an evolutionarily conserved feature of eukaryotic cells. This feature is typified in the ATM and ATR family genes which encode phosphatidylinositol 3-kinase-related proteins possessing protein kinase activity (33). In the budding yeast Saccharomyces cerevisiae, MEC1 encodes an ATR-related protein and plays a critical role in checkpoint controls (14, 17, 32). Mec1 physically interacts with Pie1 (also called Lcd1 or Ddc2), a protein that exhibits limited homology to the fission yeast Rad26 protein (17, 19, 32). Likewise, in fission yeast the ATR family protein Rad3 forms a complex with Rad26 (4). DNA damage responses have been well characterized in budding yeast and consist of the G1-, S-, and G2/M-phase damage checkpoints (14). Both Mec1 and Pie1 are essential for all three DNA damage checkpoints, as well as the DNA replication block checkpoint.
In addition to MEC1 and PIE1, a number of genes that control the checkpoints in budding yeast have been identified. These include DDC1, MEC3, RAD9, RAD17, RAD24, and RAD53 (5, 14, 33). RAD53 encodes a protein kinase and functions downstream of MEC1 in the checkpoint pathway. Like Mec1, Rad53 plays an essential role in both the replication block and DNA damage checkpoints. Following DNA damage and replication block, the Rad53 protein is hyperphosphorylated and activated by a mechanism dependent on Mec1 (20, 26). Thus, Mec1 and Rad53 constitute a central checkpoint pathway in budding yeast. RAD9, RAD17, MEC3, DDC1, and RAD24 are also required for DNA damage checkpoints. Rad9 is hyperphosphorylated following DNA damage, and the phosphorylated Rad9 protein binds to Rad53, possibly to modulate its activity (6, 27, 30). Genetic evidence has suggested that RAD17, RAD24, MEC3, and DDC1 operate in a common checkpoint pathway. Indeed, Ddc1, Mec3, and Rad17 interact physically with each other and function in a complex to control the DNA damage checkpoints (12). It has been shown that Ddc1, Mec3, and Rad17 are structurally related to PCNA (1, 28, 29). RAD24 encodes a protein structurally related to the subunits of replication factor C (RFC) which is required for DNA replication and repair. RFC consists of one large subunit, Rfc1, and four small subunits, Rfc2, Rfc3, Rfc4, and Rfc5 (3). Rad24 also interacts with the four small RFC subunits, Rfc2, Rfc3, Rfc4, and Rfc5, to form an RFC-related complex (9, 16). Genetic evidence has indicated that Rad24 functions upstream of the Ddc1-Mec3-Rad17 complex in the checkpoint pathway (12). RFC loads PCNA onto the primer terminus of DNA, and then DNA polymerases δ and ɛ bind to the resulting DNA-RFC-PCNA complex to form a processive replication complex (31). By analogy, the RFC-related RAD24 complex is proposed to recruit a complex consisting of Ddc1, Mec3, and Rad17, each of which is related to PCNA, to damaged DNA (9, 16, 33).
We have shown that rfc5-1 mutants are defective not only in the DNA damage checkpoint but also in the DNA replication block checkpoint (24, 25). The observation that the rad24Δ mutation enhances the replication block checkpoint defect in rfc5-1 mutants suggests that Rad24 plays a role in the DNA replication block checkpoint (22). However, the rad24Δ mutation alone causes no obvious defect in the DNA replication block checkpoint (15, 22). These results suggest that RAD24 functions redundantly with other genes in this checkpoint pathway.
The CHL12 (also called CTF18) gene encodes a protein homologous to the Rad24 and RFC proteins (3, 13). CHL12 was identified in a screen for mutants exhibiting increased rates of mitotic loss of chromosomes and has been suggested to play a critical role in DNA metabolism (13). In this paper, we show that CHL12 and RAD24 function redundantly in the DNA replication block checkpoint: this checkpoint operates normally in the single chl12Δ and rad24Δ mutants but is defective in chl12Δ rad24Δ double mutants. We also show that Chl12 interacts physically with the four small RFC subunits to form a complex that is related to, but distinct from, the RFC and RAD24 complexes. Thus, Chl12 and Rad24 are both required for the DNA replication block checkpoint.
MATERIALS AND METHODS
Strains, media, and general methods.
The yeast strains used in this study are isogenic and are listed in Table 1. Standard genetic techniques were used for manipulating yeast strains. Synthetic complete (SC) medium containing 0.5% Casamino Acids and the appropriate supplements was used to maintain selection of URA3 plasmids.
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| KSC006 | MATaade1 his2 trp1 ura3 leu2 |
| KSC835 | MATarfc5-1::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1090 | MATarad24Δ::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1105 | MATarfc5-1::LEU2 rad24Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1148 | MATachl12Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1266 | MATachl12Δ::LEU2 rad24Δ::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1372 | MATaRFC1-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1373 | MATaRFC2-FLAG::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1374 | MATaRFC3-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1375 | MATaRFC4-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1376 | MATaRFC5-FLAG::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1377 | MATaRAD24-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1378 | MATaCHL12-HA::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1382 | MATaCHL12-HA::LEU2 RFC1-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1383 | MATaCHL12-HA::LEU2 RFC2-FLAG::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1384 | MATaCHL12-HA::LEU2 RFC3-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1385 | MATaCHL12-HA::LEU2 RFC4-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1386 | MATaCHL12-HA::LEU2 RFC5-FLAG::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1387 | MATaCHL12-HA::LEU2 RAD24-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1388 | MATaCHL12-HA::LEU2 RFC3-FLAG::URA3 rfc5-1::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1433 | MATaCHL12-HA::LEU2 RAD24-myc::TRP1 RFC1-FLAG::URA3 ade1 his2 trp1 ura3 leu2 |
| KSC1435 | MATaRFC1-FLAG::URA3 chl12Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
Plasmids and gene disruptions.
To create the CHL12 disruption plasmid, the amino- and carboxyl-terminal regions were amplified by PCR with the amino-terminal primers KS158 (5′-ACAGAACTATCTGCAGAATGTGAATATTGTC-3′) and KS162 (5′-CCAGAAGCTTGTTCTATATCACC-3′) or the carboxyl-terminal primers KS163 (5′-ACTGAATTCAATCCAAAATCGGTCAGAA-3′) and KS161 (5′-CGGAGCTGTGTCGACTACCGGCAGGATTGATTGCTAAT-3′). The amino- and carboxyl-terminal fragments were digested with the appropriate restriction enzymes (PstI-HindIII and EcoRI-SalI, respectively) and cloned into YIplac128 (8). The CHL12 disruption plasmid was cleaved by SalI and transformed into a diploid strain. The heterozygous diploid was sporulated, and the tetrads were dissected. Disruption of the CHL12 gene was confirmed by PCR. To construct a hemagglutinin (HA)-tagged version of CHL12, the carboxyl-terminal region of CHL12 was amplified by PCR with the primers KS160 (5′-CCATATCATGGTTTAAAATCGTGAACCAATT-3′) and KS171 (5′-CTCGGATCCCTTCCCACAGGTTATTCCAAGTC-3′). The SnaBI-BamHI-treated carboxyl-terminal fragment of CHL12 and a BamHI-SalI fragment containing sequence encoding HA epitopes were cloned into the SmaI-SalI-linearized YIplac128, creating YIp-CHL12-HA. The RAD24-FLAG integration plasmid YIpU-RAD24-FLAG was constructed as follows. The carboxyl-terminal region of RAD24 was amplified by PCR with the primers KS551 (5′-TCTGAGCTCGGGCGACATCAAGTGGAAGTT-3′) and KS552 (5′-CTCGGATCCGAGTATTTCCAGATCTGAATCTGAAAGGGA-3′). After treatment with SacI and BamHI, the fragment was cloned into SacI-BamHI-linearized pRS306FLAG (a gift from N. Lowndes). YCpRAD53-HA was described previously (24). To construct YIp-RAD24-myc, the EcoRI-HindIII fragment from YCpRAD24-myc (22) was cloned into YIplac204 (8). The CHL12-HA and RAD24-FLAG strains were generated by transforming YIp-CHL12-HA and YIp-RAD24-FLAG after treatment with XbaI. The RAD24-myc strains were obtained by transforming YIp-RAD24-myc after digestion with NcoI. The construction of strains carrying FLAG-tagged RFC1, RFC2, RFC3, RFC4, and RFC5 was described previously (9). Cells containing the tagged constructs expressed appropriate-sized proteins from their own promoters and showed no growth defect or increased sensitivity to DNA-damaging agents.
UV radiation and drug sensitivities.
The hydroxyurea (HU), UV radiation, or methyl methanesulfonate (MMS) sensitivity assay was performed as described previously (32).
MMS synchrony experiments.
To analyze cell cycle delay at the G2/M transition, log-phase cultures at 30°C were arrested with 15 μg of nocodazole/ml for 150 min to synchronize cells in G2/M. Cells arrested in G2/M were incubated with 0.25% MMS for 30 min and then washed to remove the nocodazole and MMS and released into fresh yeast extract-peptone-dextrose (YEPD). At timed intervals, cells were withdrawn and stained with 4′,6′-diamidino-2-phenylindole (DAPI) for microscopic examination. To examine regulation of the S-phase progression, cells were synchronized in G1 and released into MMS at 30°C as described previously (16). Briefly, cells were grown in YEPD and treated with 6 μg of α-factor/ml for 150 min. Cells synchronized with α-factor were released into YEPD containing 0.05% MMS. An experiment to analyze cell cycle delay at the G1/S transition following MMS treatment was carried out at 30°C (23). Cells were grown in YEPD and treated with 6 μg of α-factor/ml for 150 min. Cells arrested in G1/S were incubated with 0.25% MMS for 30 min and then washed to remove the α-factor and MMS and released into fresh YEPD. At timed intervals, cells were withdrawn for microscopic examination.
Immunofluorescence microscopic analysis.
To examine spindle elongation at 30°C, the culture was synchronized in the G1 phase by addition of 6 μg of α-factor/ml at 30°C for 150 min. The cells were then washed to remove the α-factor and released into YEPD with or without 10 mg of HU/ml at 30°C. Aliquots of cells were removed and processed for DNA flow cytometry analysis and indirect immunofluorescence microscopy as described previously (32).
Immunoblotting.
Protein extracts for immunoblotting were prepared and resolved by electrophoresis on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels as previously described (32). The proteins were then transferred to nylon membranes and subjected to immunoblotting analysis with the monoclonal anti-HA (3F10 or 16B12), anti-myc (9E10), and anti-FLAG (M2) antibodies, and antibody binding was detected using an ECL kit (Amersham Pharmacia Biotech).
Immunoprecipitation.
Cells were grown in preculture, diluted in YEPD, and allowed to grow for 3 h at 30°C. The cells were next harvested, washed, and resuspended in lysis buffer (24). An equal volume of glass beads was added, and the cells were lysed by vortexing. Extracts were prepared as described previously (24) and incubated at 4°C for 2 h with protein G-Sepharose beads bound with anti-HA (3F10) or anti-FLAG (M2) antibody. Immunoprecipitates were washed with lysis buffer containing 300 mM NaCl and subsequently with a wash buffer and boiled immediately in 1× SDS-PAGE sample buffer. The proteins were detected after immunoblotting was performed with the antibodies described above.
Sucrose density gradient centrifugation.
Extracts prepared in lysis buffer were separated by sucrose density gradient sedimentation in an SW60 rotor at 40,000 rpm for 16 h at 4°C as described previously (12). The gradients were fractionated from the top and subjected to immunoblotting with the antibodies described above.
RESULTS
CHL12 encodes a protein structurally related to Rad24.
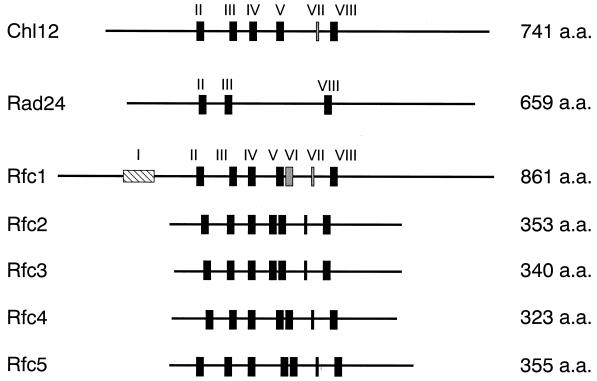
Rad24 has an essential role in the DNA damage checkpoint and forms an RFC-related complex with Rfc2, Rfc3, Rfc4, and Rfc5 (9, 16). Rad24 has been suggested to participate in the DNA replication block checkpoint (22). However, rad24Δ single mutants are not defective in this checkpoint (15, 22). One possible explanation is that Rad24 functions redundantly with other proteins in this checkpoint pathway. The CHL12 gene encodes a 741-amino-acid protein which is structurally related to the Rad24 and RFC proteins in S. cerevisiae (13). All of the RFC proteins contain seven conserved domains, termed RFC boxes II to VIII (3). Chl12 contains RFC boxes II to V, VII, and VIII (3), whereas the Rad24 protein contains RFC boxes II, III, and VIII (15) (Fig. 1). CHL12 and RAD24 encode proteins of similar sizes, although Chl12 shows more significant homology to the RFC proteins than to Rad24. Because of its similarities to Rad24, we investigated the possible redundant roles of Chl12 with Rad24 in the DNA replication block checkpoint.
FIG. 1.
Structures of the S. cerevisiae Chl12, Rad24, and RFC proteins. There are eight RFC boxes numbered consecutively from the amino terminus to the carboxyl terminus. All of the RFC proteins possess the RFC boxes II to VIII. Box I is present only in the largest RFC subunits. The solid and shaded boxes indicate high and moderate degrees of homology, respectively. a.a., amino acids.
Chl12 and Rad24 play redundant roles in the DNA replication checkpoint.
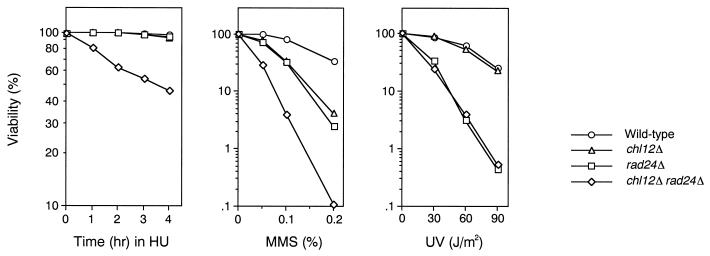
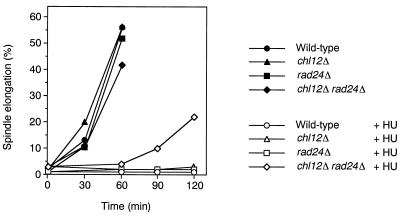
To address the potentially redundant roles of Chl12 and Rad24 in the DNA replication block checkpoint, we generated chl12Δ and rad24Δ single or double mutants and examined their sensitivities to HU treatment (Fig. 2). While neither chl12Δ nor rad24Δ single mutants were sensitive to HU, chl12Δ rad24Δ double mutants became sensitive. To test whether chl12Δ rad24Δ double mutants are defective in the replication block checkpoint, cells were synchronized with α-factor at G1 and released into medium with or without HU. Flow cytometric analysis showed that DNA replication was blocked by HU treatment in cells during the experiments (data not shown). In the absence of HU, half of the cells exhibited elongated spindles at 60 min after release from α-factor, indicating that cells of all strains entered into mitosis in similar manners (Fig. 3). In the presence of HU, wild-type, chl12Δ, and rad24Δ mutant cells were arrested as large budded cells with short spindles. Although chl12Δ rad24Δ double mutants delayed progression into mitosis, 20% of the cells displayed elongated spindles at 120 min after release into HU (Fig. 3). These results indicate that chl12Δ rad24Δ double mutants are partially defective in the DNA replication block checkpoint.
FIG. 2.
Viability of chl12Δ and chl12Δ rad24Δ mutants following exposure to HU, MMS, or UV light. Wild-type (KSC006), chl12Δ (KSC1148), rad24Δ (KSC1090), and chl12Δ rad24Δ (KSC1266) cells were grown in log phase at 30°C, treated with HU or MMS, and irradiated with UV light. The viability of the cells was estimated as described in Materials and Methods.
FIG. 3.
DNA replication block checkpoint in chl12Δ rad24Δ mutants. Cells were arrested at G1 with α-factor and then released in YEPD with or without 10 mg of HU/ml at 30°C. Aliquots of cells were collected at the indicated times and stained with anti-tubulin antibodies. The percentage of cells with elongated spindles was scored as described in Materials and Methods. The strains used were the wild type (KSC006), chl12Δ (KSC1148), rad24Δ (KSC1090), and chl12Δ rad24Δ (KSC1266).
Rad53 is phosphorylated in response to DNA replication block, and this phosphorylation has been shown to correlate with the activation of checkpoint pathways. To examine whether chl12Δ rad24Δ double mutants become defective in Rad53 phosphorylation following HU treatment, cells expressing Rad53-HA were arrested in G1 with α-factor and released into medium containing HU. Extracts were prepared from the cells after release and subjected to immunoblotting analysis. Rad53 was phosphorylated in chl12Δ and rad24Δ single mutants, similar to its phosphorylation in wild-type cells, whereas its phosphorylation was significantly reduced in chl12Δ rad24Δ double mutants (Fig. 4). Together, these results strongly suggest that Chl12 and Rad24 play redundant roles in the DNA replication block checkpoint.
FIG. 4.
Effect of chl12Δ rad24Δ mutation on Rad53 modification following replication block. Cells carrying YCpRAD53-HA were grown at 30°C, arrested in G1 with α-factor, and then released in YEPD containing 5 mg of HU/ml. Aliquots of cells were collected at the indicated times and subjected to immunoblotting analysis as described in Materials and Methods. The strains used were the wild-type (KSC006), chl12Δ (KSC1148), rad24Δ (KSC1090), and chl12Δ rad24Δ (KSC1266).
Effect of the chl12Δ mutation on response to DNA damage.
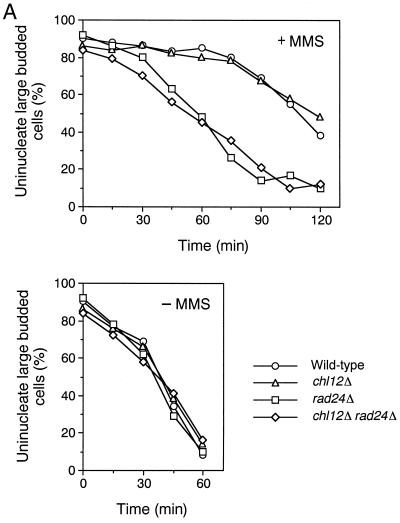
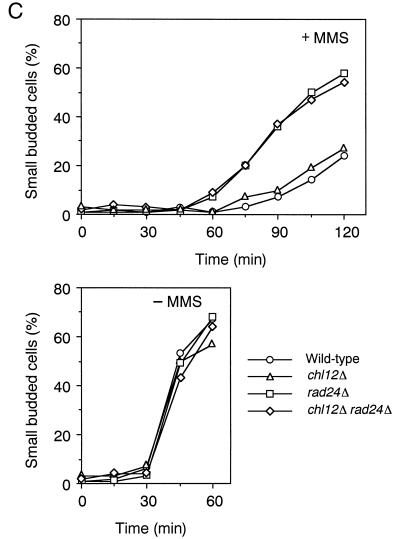
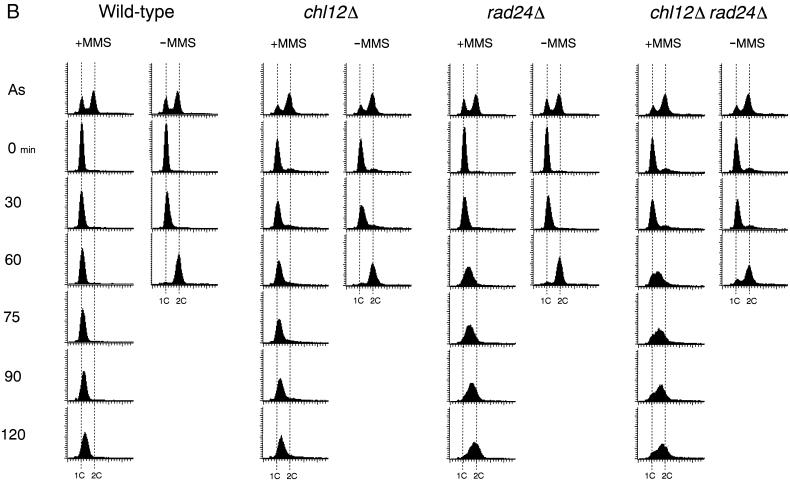
While the rad24Δ mutation affects the DNA replication checkpoint in combination with other mutations as described above, the rad24Δ mutation alone causes a defect in the DNA damage checkpoint. We therefore characterized the possible role of CHL12 in the DNA damage checkpoint. We first examined the sensitivity of chl12Δ and rad24Δ single or double mutants to UV irradiation and MMS treatment (Fig. 2). Both chl12Δ and rad24Δ single mutants showed sensitivity to MMS, and chl12Δ rad24Δ double mutants became more sensitive to MMS than either single mutant. In contrast to rad24Δ, the chl12Δ mutation had no apparent effect on sensitivity to UV irradiation. Moreover, chl12Δ rad24Δ double mutants were no more sensitive to UV irradiation than the single rad24Δ mutants. These results suggest that CHL12 is required for the proper response to DNA damage following MMS treatment but not following UV irradiation. It has been shown that RAD24 is required for the G1-, S-, and G2/M-phase DNA damage checkpoints following MMS treatment (7, 18). We therefore examined the checkpoint defect of chl12Δ single mutants and chl12Δ rad24Δ double mutants after MMS treatment. The G2/M-phase DNA damage checkpoint was examined by monitoring mitotic division following DNA damage (Fig. 5A). When cell cultures were released from nocodazole arrest after MMS treatment, wild-type cells showed delayed nuclear division while rad24Δ cells proceeded through mitosis faster than wild-type cells. In contrast to rad24Δ mutants, chl12Δ mutants underwent mitosis at the same rate as wild-type cells. Moreover, chl12Δ rad24Δ double mutants did not proceed through mitosis any faster than rad24Δ single mutant cells. The S-phase DNA damage checkpoint was analyzed by monitoring the DNA content of cells experiencing DNA damage after release from G1 block (Fig. 5B). When treated with MMS and released from α-factor arrest, wild-type cells exhibited lower rates of DNA synthesis. Although rad24Δ mutants progressed through the S phase faster than wild-type cells, chl12Δ mutants went through the S phase at the same rate as wild-type cells. Again, there was no difference between chl12Δ rad24Δ double mutants and rad24Δ single mutants with respect to the rate of DNA synthesis. The G1-phase DNA damage checkpoint was also examined (Fig. 5C). After MMS treatment, wild-type cells delayed the G1/S transition as judged by budding whereas rad24Δ mutants went through this transition faster. Once again, the chl12Δ mutation had no effect on the G1/S transition following DNA damage, either as a single mutation or in combination with the rad24Δ mutation. These results indicate that CHL12 is not required for the G1-, S- and G2/M-phase DNA damage checkpoints and does not have a role overlapping that of RAD24 in the DNA damage checkpoints.
FIG. 5.
DNA damage checkpoints in chl12Δ and chl12Δ rad24Δ mutants. (A) G2/M-phase DNA damage checkpoint in chl12Δ and chl12Δ rad24Δ mutants. Cells were grown at 30°C, arrested with nocodazole, and treated or not treated with MMS. At the indicated times after release of MMS-treated (+MMS) and untreated (−MMS) cultures from nocodazole, the percentage of uninucleate large budded cells was scored by DAPI staining. (B) S-phase DNA damage checkpoint in chl12Δ and chl12Δ rad24Δ mutants. Cells were synchronized with α-factor in G1 and released in either the presence or the absence of MMS at 30°C as described in Materials and Methods. Aliquots of cells were collected at the indicated times after release from α-factor treatment and examined for DNA content by flow cytometry. The dotted lines indicate the DNA content of 1C and 2C cells. The top panels represent asynchronous (As) cells not treated with MMS at 30°C and are included as a reference. (C) G1-phase DNA damage checkpoint in chl12Δ and chl12Δ rad24Δ mutants. Cells were synchronized with α-factor in G1 and treated with MMS (+MMS) or not treated (−MMS). At the indicated times after release from α-factor, the percentage of small budded cells was scored under microscopy. The strains used are the wild type (KSC006), chl12Δ (KSC1148), rad24Δ (KSC1090), and chl12Δ rad24Δ (KSC1266).
Chl12 forms a novel RFC-related complex with the RFC small subunits, Rfc2, Rfc3, Rfc4, and Rfc5.
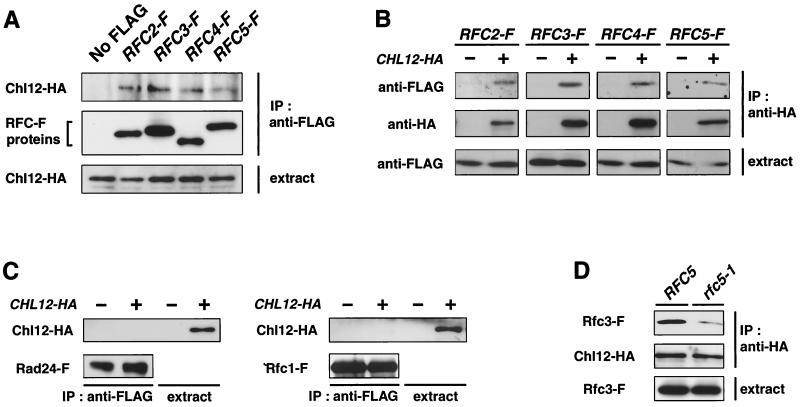
Rad24 forms an RFC-related complex with the four small RFC subunits, Rfc2, Rfc3, Rfc4, and Rfc5. Given the analogous functions of Chl12 and Rad24 in the replication block checkpoint, we examined whether Chl12 also interacts physically with Rfc2, Rfc3, Rfc4, and/or Rfc5 in vivo by immunoprecipitation experiments. We generated strains in which the corresponding genomic alleles were replaced with the HA-tagged CHL12 gene and/or one of each of the RFC genes tagged with FLAG. Extracts were prepared from cells and subjected to immunoprecipitation with anti-FLAG antibody. The immunoprecipitates were then analyzed by immunoblotting them with antibodies against the HA and FLAG epitopes. Chl12-HA was coprecipitated with Rfc2-FLAG, Rfc3-FLAG, Rfc4-FLAG, and Rfc5-FLAG in cells expressing both the tagged Chl12 and RFC proteins (Fig. 6A). In the reverse coimmunoprecipitation experiment, Chl12 was also found to interact physically with Rfc2, Rfc3, Rfc4, and Rfc5 (Fig. 6B). Rad24 and Rfc1 have been shown to form a complex with Rfc2, Rfc3, Rfc4, and Rfc5. We then tested whether Chl12 interacts physically with Rad24 or Rfc1. However, coprecipitation of Chl12 with Rfc1 or Rad24 was not detected (Fig. 6C). These observations demonstrate that Chl12 physically interacts in vivo with Rfc2, Rfc3, Rfc4, and Rfc5 but not with Rad24 or Rfc1.
FIG. 6.
Physical interaction of Chl12 with Rfc2, Rfc3, Rfc4, and Rfc5. (A and B) Interaction of Chl12 with Rfc2, Rfc3, Rfc4, and Rfc5. Extracts were prepared from CHL12-HA cells containing no FLAG construct or the indicated FLAG-tagged construct (A) or cells containing the indicated FLAG-tagged construct and no HA construct (−) or CHL12-HA (+) (B) and subjected to immunoprecipitation (IP) with anti-FLAG antibody (A) or anti-HA antibody (B). An F after a gene name indicates the addition of FLAG epitopes. The immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-FLAG or anti-HA antibody. Whole extracts were immunoblotted with anti-HA antibody (A) or anti-FLAG (B) antibody. (C) Interaction of Chl12 with Rad24 and Rfc1. Extracts were prepared from RAD24-FLAG and RFC1-FLAG cells containing no HA construct (−) or CHL12-HA (+) and subjected to IP and immunoblotting analysis as for panel A. (D) Interaction of Chl12 with Rfc3 in RFC5 and rfc5-1 mutant cells. Extracts were prepared from RFC5 or rfc5-1 cells containing CHL12-HA and RFC3-FLAG and were subjected to IP and immunoblotting analysis as for panel B.
In rfc5-1 mutants, the interaction among the small RFC subunits is defective, resulting in a decreased association of Rad24 with the small RFC subunits (16). Assuming that Chl12 forms an RFC-related complex similar to Rad24, we expected that the interaction of Chl12 with the small RFC subunits might also be defective in rfc5-1 mutants. We examined the interaction between Chl12 and Rfc3 in extracts prepared from wild-type or rfc5-1 cells expressing Chl12-HA and Rfc3-FLAG. Extracts were subjected to immunoprecipitation with anti-HA antibody, and the immunoprecipitates were analyzed by immunoblotting analysis with anti-Flag and anti-HA antibodies. The interaction of Chl12-HA with Rfc3-FLAG was decreased in rfc5-1 mutants compared to wild-type cells, whereas the expression of Chl12-HA and Rfc3-FLAG was similar in the two strains (Fig. 6D).
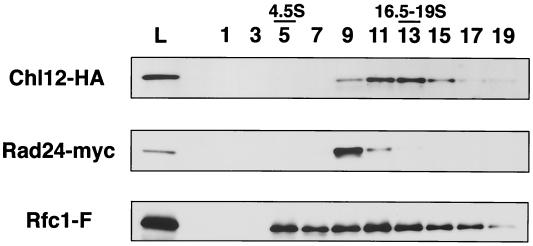
We further examined fractionation profiles of Chl12, Rad24, and Rfc1 in a sucrose density gradient centrifugation. Extracts prepared from cells coexpressing Chl12-HA, Rfc1-FLAG, and Rad24-myc were fractionated by sucrose density gradient centrifugation and subjected to immunoblotting analysis using anti-HA, anti-FLAG, and anti-myc antibodies. Chl12-HA sedimented as a 12S particle peaking at fractions 11 to 13 (Fig. 7). This is consistent with the idea that Chl12 is present as part of a larger complex. Rad24-myc sedimented as a 10S particle separately from Chl12-HA, peaking at fraction 9, whereas Rfc1-FLAG sedimented more broadly than Chl12-HA and was found in fractions 5 to 17 (Fig. 7). The chl12Δ mutation did not affect the sedimentation profile of Rfc1-FLAG (data not shown). Together, these results demonstrate that Chl12 forms a novel RFC-related complex which is distinct from the RFC and RAD24 complexes.
FIG. 7.
Sedimentation of Chl12, Rad24, and Rfc1 in sucrose density gradient centrifugation. Extracts were prepared from CHL12-HA RAD24-myc RFC1-FLAG (KSC1433) cells and separated by centrifugation in a 10 to 40% sucrose gradient for 16 h. The load on the gradient (L) and fractions (removed from the top of the gradient) were analyzed by immunoblotting using anti-FLAG, anti-HA, or anti-myc antibody. An F after a gene name indicates the addition of FLAG epitopes. Bovine serum albumin (4.5S) and thyroglobulin (16.5–19S) were separated simultaneously in an independent gradient as markers.
DISCUSSION
The RFC complex is required for DNA replication and repair and consists of one large and four small subunits (31). In S. cerevisiae, the large subunit of RFC is encoded by RFC1 (also called CDC44), and the four small subunits are encoded by RFC2, RFC3, RFC4, and RFC5. RFC is a structure-specific DNA-binding protein complex that recognizes a primer-template junction. Following its association with DNA at a primer end, RFC recruits PCNA onto DNA and then tethers DNA polymerase δ or ɛ to the primer junction (31). Rad24 shares a limited homology with the RFC subunits and forms an RFC-related complex with the four small RFC subunits (9, 16). RAD24 plays an essential role in the DNA damage checkpoint and functions upstream of Ddc1, Mec3, and Rad17. Ddc1, Mec3, and Rad17 possess PCNA-like structures and interact physically with each other. One model suggests that the RAD24 complex may recognize aberrant DNA structures and recruit the Ddc1-Mec3-Rad17 complex to damaged DNA.
It has been shown that the small RFC subunits Rfc2 and Rfc5 are involved in the DNA replication block checkpoint. This checkpoint response occurs normally in rad24Δ mutants, even though the rad24Δ mutation increases the severity of the checkpoint defect caused by the rfc5-1 mutation. This raises the possibility that the replication block checkpoint may be redundantly controlled by Rad24 and other RFC-related complexes. In this study, we have provided evidence indicating that Chl12, which is structurally related to the Rad24 and RFC proteins, forms an RFC-related complex and plays a role in the DNA replication block checkpoint. Although neither chl12Δ nor rad24Δ single mutants are defective, chl12Δ rad24Δ double mutants become defective in the DNA replication checkpoint. Consistent with this, phosphorylation of Rad53 following HU treatment is decreased in the chl12Δ rad24Δ double-mutant cells. Chl12 was found to interact physically with Rfc2, Rfc3, Rfc4, and Rfc5 but not with Rad24 or Rfc1. Moreover, Chl12 does not cosediment with Rad24 or Rfc1 in a sucrose density centrifugation. Thus, Chl12 forms a novel RFC-related complex and functions redundantly with Rad24 in the replication block checkpoint pathway. Because of their structural similarities to RFC, it is suggested that the Chl12- and Rad24-containing complexes function to recognize specific DNA structures and to recruit the proper apparatus to a stalled replication fork.
In rfc5-1 mutants, the interaction among the small RFC subunits is decreased, resulting in defective association between Rad24 and the small RFC subunits (16). We found that the association between Chl12 and the small RFC subunits was similarly affected by the rfc5-1 mutation. These observations suggest that the checkpoint defect observed in rfc5-1 mutants may be attributed, at least in part, to the decreased interaction of Chl12 and Rad24 with the small RFC subunits. However, these checkpoint defects could not be due entirely to the disruption of the CHL12 and RAD24 complexes, since the DNA replication block checkpoint is more defective in rfc5-1 single and rfc5-1 rad24Δ double mutants than in chl12Δ rad24Δ double mutants (data not shown). One possible explanation is that the remaining small RFC subunits are able to form a subcomplex retaining partial function. In fact, the human small RFC subunits form a subcomplex in vitro, although its in vivo function is not yet established (2). Alternatively, the replication block checkpoint might require the RFC complex containing Rfc1. Since disruption of RFC1 is lethal, this possibility would be best addressed if conditional rfc1 mutations were available. A nonlethal mutation allele of RFC1, cdc44-1, has no apparent effect on the DNA replication block checkpoint, either by itself or in combination with either chl12Δ or rad24Δ (data not shown). However, a more systematic analysis with rfc1 mutations would be required to argue against this possibility.
Although CHL12 plays a role in the DNA replication block checkpoint, CHL12 does not appear to be involved in the DNA damage checkpoints. In contrast, RAD24 plays a role in both the DNA replication block and damage checkpoints. If both the CHL12 and RAD24 complexes could bind to specific DNA structures, it is puzzling that CHL12 has no apparent role in the DNA damage checkpoints. One explanation could be that other checkpoint machineries might cooperate with the CHL12 or RAD24 complex to detect aberrant DNA structures and activate the checkpoint pathways. Following DNA damage, such checkpoint machineries might bind to the same DNA structures that the RAD24 complex could recognize but not ones that the CHL12 complex could. If this were the case, each of the CHL12 and RAD24 complexes would recognize distinct DNA structures resulting from DNA damage. Consistent with this model, chl12Δ and rad24Δ single mutants behave differently in response to DNA damage; rad24Δ mutants are sensitive to MMS and UV, whereas chl12Δ mutants are sensitive to MMS but not to UV. Moreover, an increase in the dosage of either complex failed to compensate for lack of the other complex in response to DNA damage; overexpression of CHL12 or RAD24 did not suppress MMS sensitivity of the other deletion mutation (data not shown). Finally, although our results indicate that Chl12 functions in a complex with the small RFC subunits, we cannot exclude the possibility that Chl12 could form a separate complex with other proteins. Such separate CHL12 complexes might be involved in repair of MMS-induced DNA lesions but not checkpoint response.
In fission yeast, the rad17+ gene, a homolog of RAD24, has been isolated and characterized (10), and the gene product has been shown to interact with the small RFC subunit (21). Thus, the fission yeast Rad17 protein might also form an RFC-related complex. Genetic evidence has suggested that the fission yeast rad17+ and budding yeast RAD24 genes might be functionally different; the rad24Δ mutation is defective only in the DNA damage checkpoint, whereas the rad17 mutation is defective in both the DNA damage and replication block checkpoints. However, given our observation that RAD24 plays a redundant role in the replication block checkpoint, the functions of these two related genes appear to be much more conserved than they initially appeared to be. Interestingly, a homolog of CHL12 has also been identified in the fission yeast genome database (data not shown), although it has not been characterized yet. It is possible that this Chl12 homolog plays an overlapping role with Rad17 in the fission yeast checkpoints. Homologs of the CHL12 and RAD24 genes have also been identified in humans (reference 33 and data not shown). The structural and functional conservation of the yeast genes suggests that these human homologs might control the DNA replication checkpoint in human cells.
ACKNOWLEDGMENTS
We thank C. Green, P. Hieter, N. Kouprina, N. Lowndes, and T. Shimomura for materials, M. Mayer and T. Tsurimoto for discussion, and M. Lamphier for critical readings of the manuscript.
T.N. and T.K. are recipients of a JSPS predoctoral fellowship. K.S. acknowledges support from the Naito Foundation. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas and General Research from the Ministry of Education, Science, Sports and Culture of Japan (K.M. and K.S.).
REFERENCES
- 1.Aravind L, Walker D R, Koonin E V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai J, Gibbs E, Uhlmann F, Phillips B, Yao N, O'Donnell M, Hurwitz J. A complex consisting of human replication factor C p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J Biol Chem. 1997;272:18974–18981. doi: 10.1074/jbc.272.30.18974. [DOI] [PubMed] [Google Scholar]
- 3.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards R J, Bentley N J, Carr A M. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 5.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 6.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 7.Frei C, Gasser S M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 8.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 9.Green C M, Erdjument-Bromage H, Tempst P, Lowndes N F. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths D J F, Barbet N C, McCready S, Lehmann A R, Carr A M. Fission yeast rad17; a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:229–234. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 12.Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouprina N, Kroll E, Kirillov A, Bannikov V, Zakharyev V, Larionov V. CHL12, a gene essential for the fidelity of chromosome transmission in the yeast Saccharomyces cerevisiae. Genetics. 1994;138:1067–1079. doi: 10.1093/genetics/138.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longhese M P, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 16.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paciotti V, Clerici M, Lucchini G, Longhese M P. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 18.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rouse J, Jackson S P. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5793–5800. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 21.Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai K K, Shimada C, Nojima H. Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol Biol Cell. 1999;12:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sidorova J M, Breeden L L. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Rfc5, replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugimoto K, Shimomura T, Hashimoto K, Araki H, Sugino A, Matsumoto K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc Natl Acad Sci USA. 1996;93:7048–7052. doi: 10.1073/pnas.93.14.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 27.Sun Z, Hsiao J, Fay D S, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 28.Thelen M P, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;19:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 29.Venclovas C, Thelen M P. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vialard J E, Gilbert C S, Green C M, Lowndes N F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 32.Wakayama T, Kondo T, Ando S, Matsumoto K, Sugimoto K. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:755–764. doi: 10.1128/MCB.21.3.755-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou B-B S, Elledge S J. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]