Researchers show how the antidepressant drug fluoxetine targets a viral protein and inhibits viral replication.
Abstract
Enteroviruses are globally prevalent human pathogens responsible for many diseases. The nonstructural protein 2C is a AAA+ helicase and plays a key role in enterovirus replication. Drug repurposing screens identified 2C-targeting compounds such as fluoxetine and dibucaine, but how they inhibit 2C is unknown. Here, we present a crystal structure of the soluble and monomeric fragment of coxsackievirus B3 2C protein in complex with (S)-fluoxetine (SFX), revealing an allosteric binding site. To study the functional consequences of SFX binding, we engineered an adenosine triphosphatase (ATPase)–competent, hexameric 2C protein. Using this system, we show that SFX, dibucaine, HBB [2-(α-hydroxybenzyl)-benzimidazole], and guanidine hydrochloride inhibit 2C ATPase activity. Moreover, cryo–electron microscopy analysis demonstrated that SFX and dibucaine lock 2C in a defined hexameric state, rationalizing their mode of inhibition. Collectively, these results provide important insights into 2C inhibition and a robust engineering strategy for structural, functional, and drug-screening analysis of 2C proteins.
INTRODUCTION
Viruses belonging to the genus Enterovirus, within the family Picornaviridae, are responsible for a broad range of diseases. This genus contains four clinically relevant enterovirus (EV) species (EV-A to EV-D) and three rhinovirus (RV) species (RV-A to RV-C). Notable members of these species include polioviruses (PVs), coxsackieviruses, echoviruses, and numbered EVs (e.g., EV-A71 and EV-D68). EV-associated diseases include respiratory infections, hand-foot-and-mouth disease, conjunctivitis, viral myocarditis, pancreatitis, aseptic meningitis, encephalitis, and acute flaccid paralysis (1). RVs cause the common cold but can also trigger the exacerbation of asthma and chronic obstructive pulmonary disease. While most EV infections are mild and self-limiting, fatal complications may arise in immunocompromised patients and young children (2). Outbreaks of EV-A71 and EV-D68 represent major public health concerns because they are associated with severe neurological complications (3, 4). Vaccines are only available for PV and EV-A71, with the latter only approved in China (5, 6). Treatment of other clinically important EVs is currently limited to supportive care. Given the large numbers of serotypes (>100 EVs and >200 RVs), the development of a pan-EV and RV vaccine seems unfeasible. This underscores the urgent clinical need for the development of broad-spectrum antivirals to treat EV-associated diseases.
The EV genome encodes seven nonstructural proteins (2A to 2C and 3A to 3D), which are involved in replication, and four structural proteins (VP1 to VP4), which form the capsid. Several direct-acting agents (DAAs) targeting either the capsid, the viral polymerase (3Dpol), or the viral protease (3Cpro) have been identified. While some of these compounds reached clinical trials, their development was stopped because of limited efficacy, poor bioavailability, or toxicity issues (7). An alternative target for antiviral drug discovery is the nonstructural protein 2C, which exhibits several important hallmarks of a promising DAA target. 2C is functionally indispensable and plays an essential role in several steps of the EV life cycle, e.g., viral RNA (vRNA) replication and encapsidation (8). Moreover, 2C is highly conserved (9), raising the possibility for broad-spectrum DAA development.
2C belongs to the helicase superfamily 3 (SF3) and acts as an adenosine triphosphate (ATP)–dependent RNA helicase and ATP-independent RNA remodeler (10). The role of this enzyme in viral replication likely requires an oligomeric organization, which has already been observed for recombinant 2C proteins (11–13). Previous studies suggested that the functional oligomerization state of 2C is hexameric, consistent with other members of the SF3 helicase family (14, 15). Structural and functional characterization of the full-length oligomeric 2C protein remains challenging due to the N-terminal amphipathic helix that renders the protein poorly soluble (11–13, 16, 17). Recent structures of a soluble, monomeric fragment of 2C from EV-A71 and PV provided molecular details of the adenosine triphosphatase (ATPase) domain, a cysteine-rich zinc finger, and a C-terminal helical domain (14, 15). Nevertheless, a structure of the oligomeric 2C remains elusive.
Given the central role of 2C during viral replication, it is not unexpected that several compounds targeting 2C, such as guanidine hydrochloride (GuaHCl), 2-(α-hydroxybenzyl)-benzimidazole (HBB), MRL-1237, and TBZE-029, have been identified (16, 18–20). In addition, several drug-repurposing screens have uncovered Food and Drug Administration (FDA)–approved drugs such as fluoxetine, dibucaine, pirlindole, and zuclopenthixol as inhibitors of EV-B and EV-D species, but their mode of action is not yet understood (21, 22). Fluoxetine (Prozac) is a selective serotonin reuptake inhibitor that is used clinically for the treatment of major depression and anxiety disorders. Mode-of-action studies revealed that only the S-enantiomer of fluoxetine (SFX) potently binds the 2C and inhibits EV-B and EV-D replication (23, 24). Despite the growing number of identified 2C-targeting compounds, the molecular basis for their antiviral activity remains unknown (25, 26). Such information would provide an invaluable resource for structure-based design of potent, pan-EV antivirals with low toxicity.
Here, we present the 1.5-Å crystal structure of the soluble fragment of CV-B3 2C in complex with SFX. The structure reveals a highly conserved hydrophobic pocket, distal to the ATP-binding site, into which the SFX trifluoro-phenoxy moiety inserts. To functionally validate our structure and study the inhibitory mechanism of SFX and other 2C-targeting compounds, we engineered a CV-B3 2C protein fused to a heterologous hexamerization domain. The chimeric protein recovered ATPase activity, allowing us to investigate the inhibitory effect of several antivirals targeting EV 2C, each of which displayed a dose-dependent ATPase inhibition. Moreover, incubation of our engineered 2C protein with SFX or dibucaine demonstrated that these drugs stabilize a ring-shaped hexameric complex. This allowed us to capture the first three-dimensional structure of the 2C hexamer by cryo–electron microscopy (cryo-EM) and suggests an inhibitory mechanism for these drugs. Together, these data provide new mechanistic insights into the mode of action of 2C-targeting compounds and offer unique tools for the design and validation of 2C inhibitors.
RESULTS
Crystal structure of CV-B3 Δ116-2C in complex with SFX
Inspired by recent structural reports of 2C protein fragments from EV-A and EV-C species (14, 15), we attempted a similar truncation strategy to obtain crystals of a fluoxetine-sensitive EV-B species member, namely, CV-B3. Thermal denaturation experiments showed that SFX increased the melting temperatures (Tm + 1.51°C) of the CV-B3 Δ116-2C, confirming that this truncated protein can be bound by the drug (fig. S1). To understand how SFX binds to 2C, we attempted to crystallize the truncated 2C construct in the presence or absence of the drug. While we were able to obtain a high-resolution (1.5 Å) structure of the complex (table S1), no crystals were obtained for the apo 2C protein. This suggests that SFX stabilizes the 2C protein in a conformation prone to crystallization.
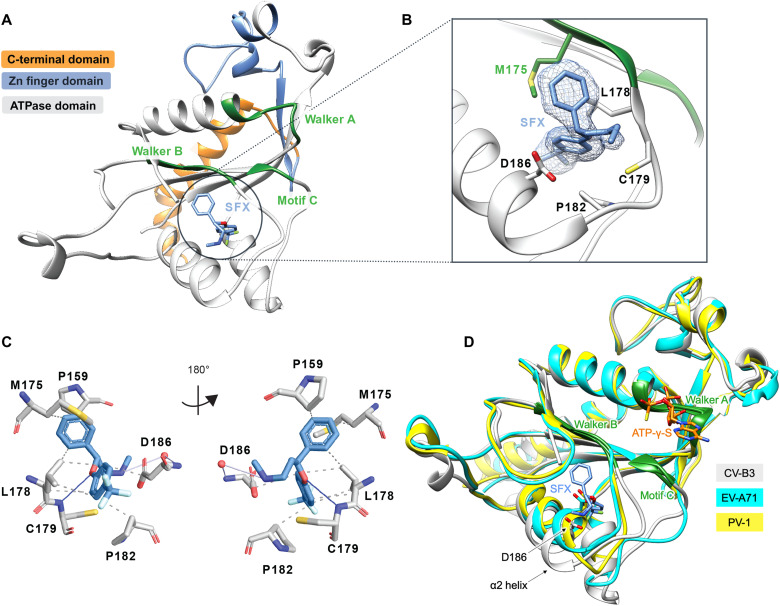
The overall fold of CV-B3 Δ116-2C is similar to the analogous structure from EV-A71 and PV, comprising a Rossmann fold core domain, a zinc finger domain, and a C-terminal helix (Fig. 1A and fig. S2). The Walker A motif is located on the loop connecting β1 and α1, forming a putative phosphate-binding loop (P-loop). The Walker B motif is found between β3 and α2, while the Walker C motif, containing N223, is found at the tip of β4 (27). Two large flexible loops located between strands β2-β3 and α2-β4 shield one side of the central sheet. Together, these loops form a hydrophobic cavity into which the SFX molecule binds (Fig. 1, A and B, and fig. S2). The interaction between SFX and the CV-B3 2C is mainly mediated by hydrophobic interactions involving the residues L157, P159, M175, D176, L178, C179, P182, and D186 (Fig. 1, B and C). The complex is also stabilized by a hydrogen bond between the amide group of the C179 main chain and the hydroxyl group of SFX. Last, in one of our structures, SFX interacts via its amino group with the main chain of D186 (Fig. 1C).
Fig. 1. Crystal structure of CV-B3 Δ116-2C in complex with SFX.
(A) The C-terminal part of 2C is highlighted in orange. The zinc finger domain is highlighted in blue, and the ATPase domain is highlighted in gray. Within the ATPase domain, the catalytic center, comprising Walker A, Walker B, and motif C, is highlighted in green. SFX is located close to the Walker B domain and is colored blue. (B) Zoomed-in view of the SFX binding pocket showing the interacting residues and electron density for the bound drug. Green residues are part of the Walker B motif. (C) Protein-ligand interaction profile of SFX with the 2C residues. Hydrophobic interactions are shown as dashed lines (P159, M175, L178, P182, and D186). Hydrogen bonds are shown as blue lines (C179), and water-mediated hydrogen bonds are colored light blue (D186). Figure was generated using PLIP (60). (D) Overlay of the 2C crystal structure of CV-B3 (gray) with the 2C crystal structure of PV-1 (PDB: 5Z3Q, chain A) (yellow) and EV-A71 (PDB: 5GRB, chain A) (cyan) in complex with ATP-γ-S.
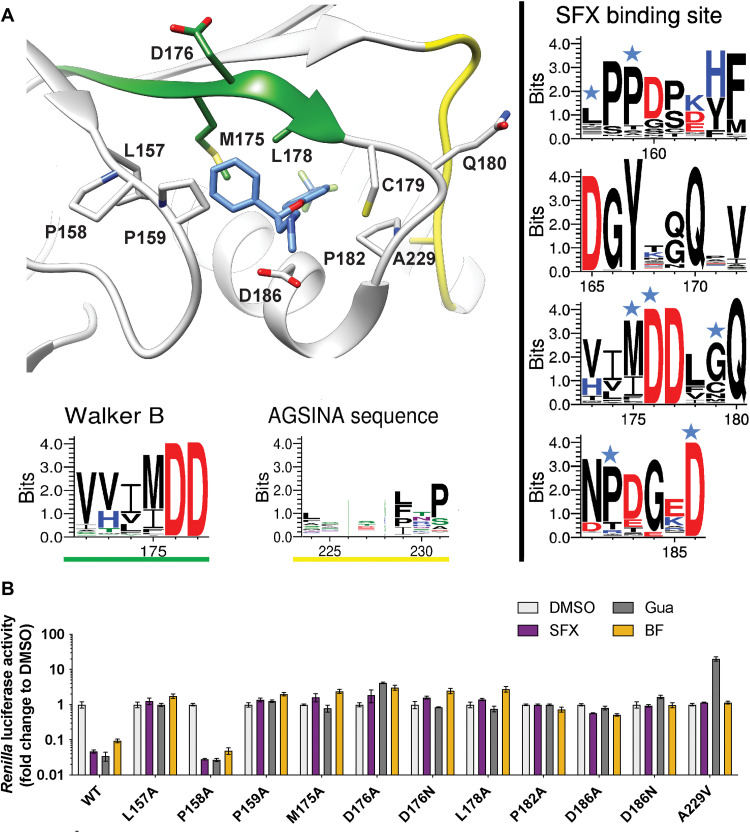
The accessibility of SFX can be associated with two main conformational differences between the CV-B3 2C structure and those of EV-A71 and PV-1. The first is a 10° tilt of the α2 helix (Fig. 1D), enlarging the binding site of SFX by moving D186 away from the methylamine group of SFX. Together with this, the Walker B–carrying loop is positioned away from the binding pocket in the CV-B3 Δ116-2C, as illustrated by the 210° rotation of the side chain of Q180, making the cavity accessible to SFX. In addition, residues 180 to 182, located downstream of the Walker B motif, participate in main-chain hydrogen bonding with the 224-AGSINA-229 loop, located downstream of motif C (Fig. 2A). This provides direct evidence for cross-talk between the SFX binding site and the AGSINA loop, which is a known hotspot for compound resistance and dependence mutations (16).
Fig. 2. Mutational analysis of the SFX binding site in CV-B3 2C.
(A) Schematic structure of CV-B3 2C highlighting the residues involved in SFX binding. The Weblogo represents the conservation of amino acids based on an alignment of the 2C protein from EVs that cause disease in humans. The blue stars on top of the amino acid residues in the right panels indicate an interaction with the ligand SFX. (B) The residues that interact with SFX were introduced into an infectious CV-B3 cDNA clone containing the Renilla luciferase reporter gene upstream of the capsid-coding region (Rluc-CV-B3). In vitro transcribed RNA was transfected into cells, and Renilla luciferase was used as a sensitive and quantitative readout for vRNA replication. The experiment was performed three times independently in biological triplicates. One representative experiment is shown.
Mutational analysis of the SFX binding site
Sequence analysis of the SFX binding site revealed that all residues, except cysteine at position 179, are conserved in SFX-resistant and SFX-sensitive EV species (Fig. 2A and fig. S3). To assess the functional consequences of alterations in the SFX binding residues, we performed mutagenesis of amino acids within, or adjacent to, the drug-binding pocket. L157A, P158A, P159A, M175A, D176A, D176N, L178A, P182A, D186A, and D186N were reverse-engineered into a CV-B3 infectious complementary DNA (cDNA) clone containing a Renilla luciferase (CV-B3-Rluc) reporter gene or in an infectious clone of CV-B3 (table S2). The replication efficiency of these mutants was then assessed in the presence or absence of 2C inhibitors. With the wild type (WT) virus, infected cells treated with SFX, Gua, and BF738735 should result in reduced Renilla luciferase activity compared to dimethyl sulfoxide (DMSO), which serves as a positive control for viral replication. BF738735 was included because it inhibits virus replication independently of 2C. In the special case of the A229V mutant, replication was only detected in the presence of the inhibitor Gua. Nearly all mutated viruses were not able to replicate their vRNA. Only the P158A mutant, which is not in direct contact with SFX, was able to replicate. This mutation did not alter the sensitivity of 2C to SFX or GuaHCl (Fig. 2B). We next tested the impact of introducing less stringent mutations into the SFX binding site. These mutations were selected using sequence information from other picornaviruses such as hepatitis A virus and Aichivirus (fig. S3). Unexpectedly, even the introduction of chemically similar amino acids (i.e., L178I and D186E) resulted in no detectable vRNA replication (fig. S4). Consistently, upon introduction of mutations into a CV-B3 infectious clone without a reporter gene, no viable viruses were recovered, except for mutation P158A (table S2). Notably, after independent passages of cells transfected with the P159A mutant, viruses were recovered carrying a second-site compensatory mutation, namely, A229V, which lies in proximity to P159. This substitution was previously shown to confer dependence to several compounds, including Gua, HBB, and MRL-1237 (16, 18, 20, 28). Virus carrying the P159A/A229V double mutation showed a small decrease in sensitivity to SFX (fig. S5). Together, these data demonstrate that residues in the SFX binding pocket are highly mutationally constrained, presumably to preserve the function of 2C. To further test this hypothesis, we introduced the M175A mutation into the recombinant Δ116-2C construct and assessed its stability in a thermal shift assay (TSA). The Tm of the M175A mutant was ~5°C lower than that of the WT and tended to precipitate during purification or upon incubation with SFX (fig. S6A). This suggests that the M175A mutation causes unfavorable structural rearrangement, resulting in the destabilization of 2C, which might perturb its proper function, providing a plausible explanation for its replication deficiency. On the other hand, when P158, which is not in direct contact with SFX, was substituted by an alanine, the protein exhibited only a ~1°C reduction in Tm and could still be bound by SFX (fig. S6B), consistent with the replication data (Fig. 2B).
To gain more insight into the binding pocket of SFX, we raised resistant CV-B3 via a clonal selection procedure as described previously (26). Several SFX-resistant viruses were obtained, containing single (I227V), double (I227V/A229V), or triple (A224V/I227V/A229V) mutations in the 224-AGSINA-229 loop (fig. S7). Mutations in this loop have been previously identified in CV-B3 resistant to 2C-targeting compounds (16, 29). In addition, we previously demonstrated that substitutions C179Y, C179F, and F190L also provide resistance to SFX (26). Notably, these latter mutations, as well as 224-AGSINA-229 loop mutations, provide cross-resistance against dibucaine (fig. S8). Our crystal structure of CV-B3 Δ116-2C in complex with SFX now allows us to cluster these mutations into two groups, those directly involved in SFX binding site (i.e., C179F, C179Y, and F190L) and those in the 224-AGSINA-229 loop located downstream of the Walker C motif, which do not directly interact with SFX (Fig. 2). The exact role of the 224-AGSINA-229 loop in conferring drug resistance, or dependence, is difficult to assess. However, its contribution to the stabilization of the Walker B–containing loop in an “open conformation,” suitable for SFX binding, corroborates this motif as a credible hotspot for resistance mutations. To further demonstrate that this loop plays an important role in fluoxetine sensitivity, we transplanted the 224-AGSINA-229 loop from CV-B3 2C into the 2C protein of the EV-A71 BrCr strain, which contains 224-ASNIIV-229 and is insensitive to SFX, despite the SFX binding residues being conserved. EV-A71 harboring the 224-AGSINA-229 loop gained sensitivity toward SFX, providing evidence that the 224-AGSINA-229 loop contributes to drug sensitivity (fig. S9).
Engineering an enzymatically active 2C hexamer to study the mechanism of action of SFX and other inhibitors
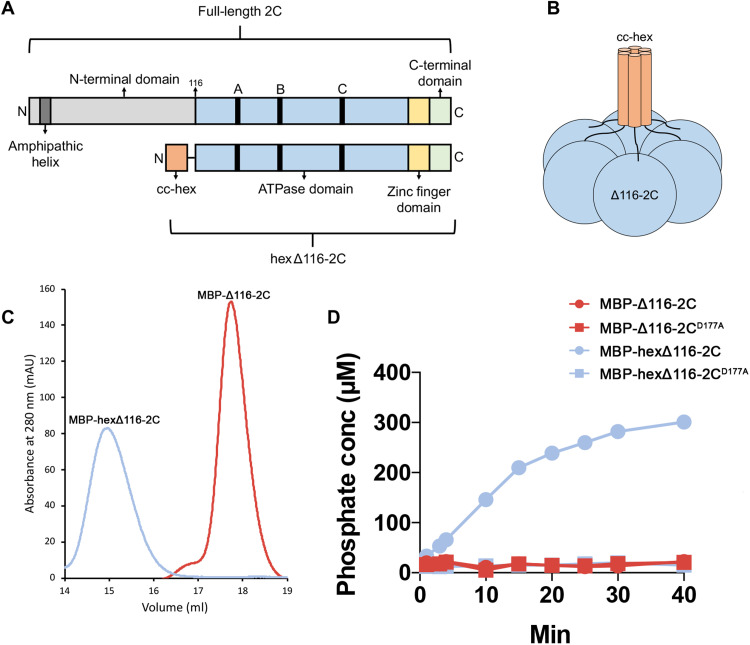
The 2C protein contains three canonical motifs required for nucleotide binding and hydrolysis, Walker A, Walker B, and motif C (9). Previous experiments demonstrated that SFX has little effect on the ATPase activity of the truncated, monomeric 2C protein of CV-B3 (29). However, the optimal activity of SF3 helicases requires their ATPase domains to be arranged in a hexameric complex (27, 30, 31). Therefore, studies of truncated 2C proteins should be complemented with biochemical analysis of the enzymatically active hexameric complex. It has been demonstrated previously that the N-terminal domain of 2C is not only important for oligomerization and ATPase activity but also renders the protein poorly soluble (11, 12, 17). With this in mind, we attempted to purify the maltose binding protein (MBP)–tagged full-length CV-B3 2C, which resulted in a heterogeneous protein preparation as shown by the size exclusion chromatography (SEC) profile. This indicated that a mixture of high–molecular weight 2C complexes were present (fig. S10). As an alternative strategy, we sought to uncouple CV-B3 2C from the unfavorable biochemical properties of the N-terminal amphipathic helix but retain the ability to form hexamers. Fusion of N- or C-terminal “assistant hexamer” proteins is an established method to study AAA+ ATPases (32–34). On the basis of the proposed hexamer model described for EV-A71 and PV 2C (14, 15), we hypothesized that we could fuse a hexameric parallel coiled coil, CC-hex-D24 (35), to the N terminus of CV-B3 Δ116-2C via a 10–amino acid linker sequence (Fig. 3, A and B). This would, in principle, enhance the local concentration of the 2C protein and correctly orientate the molecules in a manner reminiscent of membrane attachment, normally facilitated by the N-terminal amphipathic helix (36). The small size of the CC-hex coiled coil (32 amino acids) also permits the additional attachment of a cleavable His-MBP tag to promote solubility and facilitate affinity purification of the protein (fig. S11). As expected, the hexΔ116-2C elutes as a single peak that corresponds to a hexamer, whereas Δ116-2C is exclusively monomeric (Fig. 3C). In addition, mass photometry was further used to verify the molecular weights of the aforementioned proteins. The experiments revealed peaks at average masses of 56 ± 18 and 388 ± 74 kDa (average ± SD) corresponding to 2C monomer and hexamer within 20 and 8% of the predicted masses, respectively (fig. S12). We next used the malachite green assay to assess the ATPase activity of our hexΔ116-2C, which demonstrated that the engineered protein is enzymatically active. In contrast, the monomeric protein and catalytic dead D177A mutant showed little to no ATPase activity (Fig. 3D and fig. S13). Together, the data show that the hexamerization is necessary to promote the 2C-dependent ATPase activity, offering a technological platform to screen or validate 2C ATPase inhibitors.
Fig. 3. Engineering a soluble, enzymatically active 2C hexamer.
(A) Linear representation of the full-length CV-B3 2C protein (top) and the engineered hexΔ116-2C construct (bottom). The full-length 2C protein comprises an N-terminal domain (light gray); ATPase domain (blue); Walker A, B, and C motifs (black); zinc finger domain (yellow); and C-terminal extension (green). In hexΔ116-2C (residues 116 to 329), the N-terminal domain, which contains the amphipathic helix (dark gray), is replaced by the 32-residue cc-hex coiled-coil sequence (orange). (B) Schematic representation of the hexΔ116-2C construct. (C) Migration profile of the MBP-tagged monomeric Δ116-2C and hexΔ116-2C by SEC. mAU, milli-absorbance unit at 280 nm. (D) Comparison of the ATPase activity for the WT hexΔ116-2C, the hexΔ116-2C D177A mutant, WT Δ116-2C, and Δ116-2C D177A mutant. A representative result of three independent experiments is shown.
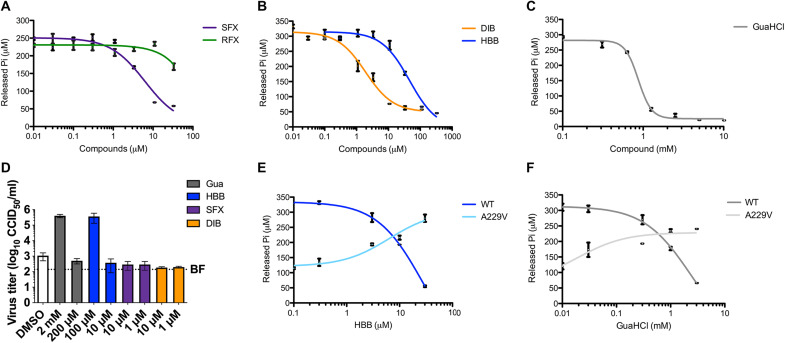
Next, we investigated the effect of the fluoxetine enantiomers—SFX and RFX—on 2C-dependent ATP hydrolysis (Fig. 4A). We showed that SFX inhibits ATPase activity in a dose-dependent manner with an EC50 (median effective concentration) value of ~5 μM, in the same concentration range as the dissociation equilibrium constant (Kd of 9.5 μM) obtained in a binding assay (23). Consistent with the binding assay, RFX was markedly less efficient at inhibiting ATPase activity. We also tested the inhibitory effect of dibucaine, HBB, and GuaHCl, known or suspected to target the 2C protein (17, 19, 22). All of the selected compounds reduced the ATPase activity of 2C in a dose-dependent manner, underscoring the link between inhibition of ATP hydrolysis and antiviral effect (Fig. 4, B and C). We then used hexΔ116-2C construct to evaluate the effect of mutations in or adjacent to the SFX binding site on the activity of the enzyme. The M175A mutant had little, if any, ATPase activity, but the P158A mutant was comparable to the WT and remained susceptible to SFX (fig. S14). These results are in line with our vRNA replication data and thermal shift experiments.
Fig. 4. 2C inhibitors reduce the ATPase activity of 2C and have antiviral effect.
(A to C) Effects of SFX and RFX (A), dibucaine (DIB) and [2-(alpha-hydroxybenzyl)-benzimidazole] (HBB) (B), or guanidium hydrochloride (GuaHCl) (C) on 2C-dependent ATPase activity. (D) Replication of CV-B3 containing the A229V 2C mutation in the presence of GuaHCL, HBB, SFX, or DIB (or a nonrelated replication inhibitor, BF738735, which targets a host lipid kinase). At 8 hours after infection, cells were freeze-thawed, and virus titers of lysates were determined. (E and F) Comparison of ATPase activity of WT 2C and the A229V mutant in the presence of HBB (E) and GuaHCl (F). Experiments shown in (A) to (C), (E), and (F) were performed twice, each time in technical duplicates. The experiment in (D) was performed in biological triplicates.
It is known that besides drug resistance, 2C-targeting compounds can induce drug dependence mutations. In the latter scenario, the presence of the compound is required for efficient EV replication (16, 18, 28, 31). The 2C mutant, A229V, has been shown to confer dependence on GuaHCl for efficient CV-B3 replication (16), but the underlying mechanism is unknown. Upon testing replication of CV-B3 carrying the A229V 2C mutation, we observed efficient replication in the presence of GuaHCl and HBB but not SFX and dibucaine (Fig. 4D). When the A229V mutant was introduced into our hexΔ116-2C construct, we observed reduced specific ATPase activity compared to the WT protein (Fig. 4E). The level of ATPase activity observed with the A229V 2C protein is fully and partly recovered in the presence of HBB and GuaHCl, respectively (Fig. 4, E and F). These data reveal that the A229V mutation negatively affects the ATPase activity of 2C and that GuaHCl and HBB can restore the enzymatic activity.
SFX and dibucaine stabilize a ring-shaped 2C hexamer
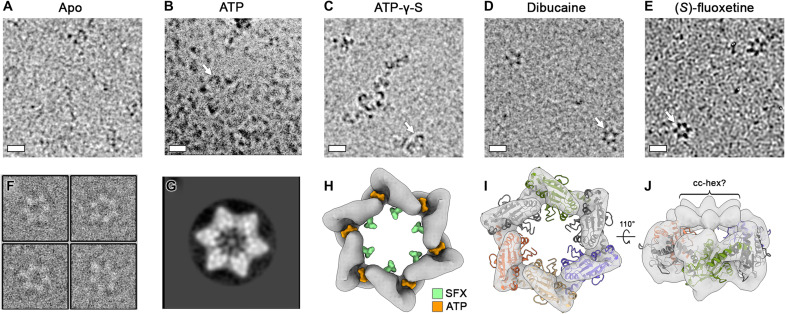
Having demonstrated that SFX and dibucaine can inhibit the 2C-dependent ATPase activity, we next wanted to determine how these compounds affect the quaternary structure of 2C. Cryo-EM analysis of our untagged engineered protein (fig. S16), in the absence of any nucleotide or inhibitor, did not reveal any distinct oligomeric structures (Fig. 5A). Upon incubation of hexΔ116-2C with ATP, a small number of “hook-shaped” oligomers were observed (Fig. 5B). These also formed in the presence of the ATP analog, adenosine 5′-O-(3-thiotriphosphate) (ATP-γ-S), but at a higher abundance (Fig. 5C). In contrast, when hexΔ116-2C was incubated with SFX or dibucaine, well-defined hexamers were discernable (Fig. 5, D to F). Single-particle analysis of SFX-bound hexamers produced distinct star-shaped two-dimensional (2D) class averages, some of which contained an electron dense feature in the central channel (Fig. 5G). Efforts to determine the cryo-EM structure of the SFX-bound 2C hexamer were hampered by the strong preferred orientation, low abundance, and instability of the particles. Nevertheless, a ~12-Å-resolution reconstruction of the complex was determined by using a 30° stage tilt during data collection (Fig. 5H and figs. S16 and S17). The cryo-EM map corresponds to ~140 kDa of ordered molecular weight and has a diameter of ~110 Å, consistent with the dimensions of the EV-A71 2C hexamer model proposed previously (14). Moreover, a cleft is present between the 2C protomers, which correlates with the expected location of the ATP-binding pocket (Fig. 5H). We next generated a hexameric model of the SFX-bound CV-B3 2C, based on the crystal structure of the JC polyomavirus large T antigen (37), as described by Guan et al. (14), and fitted this as a rigid body into our cryo-EM map. The overall agreement between the cryo-EM density and the hexameric model of CV-B3 2C supports the proposed mechanism of C terminus–mediated oligomerization (Fig. 5, I and J). In our crystal structures, the C-terminal helix (α6 and α7) appears to be broken in a “doorknob” fashion into two helices α6 and α7, due to crystal packing, and α7 helix is in intensive contact with the zinc-binding domain of a symmetrical chain (fig. S18).
Fig. 5. SFX and dibucaine stabilize a ring-shaped 2C hexamer.
(A) Cryo-electron micrographs of apo, (B) ATP-incubated, (C) ATP-γ-S–incubated, (D) dibucaine-incubated, or (E) SFX-incubated hexΔ116-2C. Scale bars, 10 nm. (F) Representative extracted particles and (G) 2D class average from untilted micrographs of SFX-incubated hexΔ116-2C. Weak density can be observed within the center of the 2C hexamer. (H) Surface representation of the 12-Å-resolution cryo-EM map of hexΔ116-2C in complex with SFX, generated from 30° tilted cryo-EM data. The map is shown at a contour level of 0.0661, and the binding positions of SFX (green) and ATP (orange) are superposed. (I) As shown in (H) with a hexameric 2C model, generated using the crystal structure of the monomer, overlaid. For clarity, pore-loop residues 85 to 100 are omitted. (J) A 110° rotated view of the model shown in (I), with the map shown at a contour level of 0.059. Putative density for the linker/cc-hex is indicated.
Together, these results indicate that SFX or dibucaine binding locks 2C in a defined hexameric state that does not permit ATP hydrolysis and, by extension, the associated functions such as helicase activity. Consistent with our cryo-EM observations, when SEC is performed on hexΔ116-2C in the presence of SFX, the complex exhibits an increased retention time, indicative of a reduced hydrodynamic radius (fig. S19A). In contrast, the presence of SFX did not greatly alter the elution profile of the monomeric Δ116-2C protein, except for a slightly more pronounced left shoulder peak, indicative of low-level oligomerization (fig. S19B). Therefore, it is likely that the SFX-mediated stabilization of the ring-shaped hexamer requires not only a high local concentration of protein but also their correct orientation with respect to one another.
DISCUSSION
EVs are globally prevalent pathogens responsible for many diseases. There is an unmet need for broad-spectrum therapeutics to treat EV infections. The EV 2C protein is an attractive target for direct-acting antivirals, given that it is a highly conserved and functionally indispensable protein, undertaking several pleiotropic functions during the viral life cycle including membrane rearrangement, RNA unwinding, and genome encapsidation (8). Over the past decades, several structurally disparate compounds targeting 2C, such as the FDA-approved drugs fluoxetine and dibucaine, among others, were identified (21, 22). Subsequently, a number of resistance mutations were obtained to gain insights into their mode of action (29). However, the binding sites and the mode of action remained elusive, hampering the rational-based improvement of the compounds. It has often been hypothesized that 2C inhibitors bind directly in the catalytic site. One study suggested that dibucaine analogs target the ATP-binding site (38), but no direct evidence supporting this hypothesis was presented. Recently, we obtained evidence for an allosteric binding site based on CV-B3, EV-A71, and EV-D68 mutants that were raised against a highly potent 2C inhibitor (26). Viruses resistant to this inhibitor contained mutations in, or adjacent to the α2 helix, which is distal to the catalytic site. Here, we provide a high-resolution crystal structure of the CV-B3 2C protein in complex with SFX. This crystal structure revealed a hydrophobic binding pocket that accommodated the ligand between the α2 helix and the Walker B domain. Our data demonstrate that SFX targets an allosteric site on 2C that is clearly distinct from the catalytic site. The obtained structure not only provides valuable insights into the mode of action of SFX but also represents a novel tool for the structure-based drug design of highly potent 2C inhibitors.
Several structural features are associated with the accessibility of SFX to bind the 2C protein of CV-B3 but not other EVs. One structural key determinant allowing SFX to access the binding pocket is the 224-AGSINA-229 loop, which is also a hotspot of resistance mutations. This is supported by our finding that exchange of the 224-ASNIIV-229 loop of the SFX-insensitive EV-A71 BrCr with the 224-AGSINA-229 loop implemented an SFX-sensitive phenotype to the resulting chimeric EV-A71 virus. The 224-229 loop is not the only determinant for SFX sensitivity, since we found previously that several clinical isolates of EV-A71 without changes in the 224-ASNIIV-229 are sensitive to SFX. Despite extensive efforts, we were not able to obtain crystals of the CV-B3 apo-2C and characterize the conformational changes that occur upon SFX binding. However, comparison of the SFX-bound CV-B3 2C to that of EV-A71 (apo and ATP-γ-S–bound) and PV (apo) shows that the α2 helix is tilted 10° away from the SFX binding site in CV-B3 2C, which creates the solvent-accessible volume required for drug binding. Similarly, the Walker B loop of CV-B3 2C is positioned further away from the SFX binding site compared to EV-A71 and PV 2C, which further contributes to the formation of the hydrophobic pocket required to accommodate SFX. This suggests that, in solution, 2C exhibits a high degree of conformational plasticity and that SFX binds to and stabilizes an open conformation of 2C.
The crystal structure of the 2C monomer in complex with SFX is not sufficient to decipher its mode of action as this should take into account the expected hexameric nature of AAA+ proteins. Several lines of evidence suggest that the catalytically active form of 2C is composed of higher oligomeric structures (10, 11, 31). Production of 2C without its N-terminal helical domain yields monomeric protein that has little, if any, ATPase activity (29). To overcome the biochemical limitations of 2C and to study the mode of action of 2C-targeting antivirals, we engineered an ATP-hydrolyzing, hexameric form of the 2C ATPase domain. The resulting construct produced homogeneous oligomers that displayed robust ATPase activity compared to the inactive monomeric form of 2C (Fig. 3). Using the engineered protein, we demonstrate that the FDA-approved drugs fluoxetine and dibucaine as well as GuaHCl and HBB inhibit the ATPase activity in a dose-dependent manner. Thus, our hexameric 2C construct represents a novel tool for in vitro screening of 2C-targeting ATPase inhibitors. This tool might be of particular interest to identify potential 2C inhibitors for difficult-to-culture EVs such as the RV-C species members (39). Besides this, this platform can provide mechanistic insights into the mode of action of specific mutations, as shown here for the drug-dependent A229V mutation, which we revealed to impair ATPase activity in the absence of inhibitors, but to stimulate ATPase activity in the presence of GuaHCl and HBB. Given that resistance mutations to a single 2C inhibitor are readily generated, the use of an antiviral cocktail would be a logical strategy. This could include another 2C inhibitor with a different mechanism of action or a compound that targets another viral protein.
A hexameric structure of 2C has long been sought after in the picornavirus field but has remained elusive because of the unfavorable biochemical properties of the full-length protein, especially its amphipathic N-terminal α helix. Therefore, cryo-EM analysis of CV-B3 2C protein, incubated with SFX, provides the first three-dimensional reconstruction of a hexameric complex and represents a substantial step toward understanding this enigmatic protein. Although artificially generated by the addition of a hexamerization domain, the recovery of the ATPase activity and the binding of SFX unambiguously confirm the relevance of the construct. Previous studies have shown that the 2C protein of foot-and-mouth disease virus (FMDV), a member of the genus Aphthovirus within the family Picornaviridae, uses a coordinated ATP hydrolysis mechanism. Using negative stain analysis, the same study demonstrated that FMDV 2C (34–318), containing the motif C mutation N207A (which interferes with ATP hydrolysis but not binding), could form hexamers in the presence of ATP and RNA (12). It is likely that EV 2C proteins must undergo conformation changes during ATP hydrolysis, RNA binding, and unwinding to drive their helicase function. Conceivably, 2C hexamers could transition between an open and closed state as they carry out their biological activities, as seen in phage genome packaging motors (40). Consistent with this, we observed open hook-shaped 2C oligomers in the presence of an ATP analog and closed, ring-like structures in the presence of SFX and dibucaine. Collectively, our biochemical and cryo-EM analysis suggests that drug binding locks 2C in a defined hexameric conformation and prevents propagation of conformational changes required for ATP hydrolysis, resulting in inhibition of viral replication. A similar mechanism was shown for a small-molecule inhibitor of the human AAA+ ATPase, p97 (41). The allosteric p97 inhibitor binds at the interface of two adjacent AAA domains and prevents the conformational changes that are required for ATPase activity of p97. Because SFX does not bind to a 2C-2C interface, it is likely that drug binding induces conformation changes within the monomer that translate to stabilization of the hexameric ring. As shown for several AAA+ ATPases, cryo-EM provides insights that are not available from crystallographic analysis alone (41–43). In the case of 2C, the observed stabilizing effect of SFX, observed in solution, complements the high-resolution information obtained by crystallography and provides insights into the allosteric inhibitory mechanism of 2C-targeting compounds. Our findings emphasize the importance of using an integrative structural biology approach to fully understand the function and mechanisms of inhibition of 2C. Previous studies have shown that purified, full-length PV 2C cannot be uncoupled from its solubility tag and contains a mixture of oligomeric species, both of which would hamper structural analysis (11). Similarly, an N-terminally extended 2C protein of echovirus 30 was shown to form heterogeneous ring-shaped structures (13). Here, we demonstrate that replacement of the N-terminal region of 2C with a hexamerization domain can produce a homogeneous complex amenable to cryo-EM. It remains possible that 2C can adopt multiple stoichiometries, as seen for the adeno-associated virus nonstructural Rep proteins (44). Nevertheless, the modular nature of our construct means that the cc-hex component can, in principle, be replaced with any other parallel coiled coil (45). Going forward, our engineering strategy should facilitate further structural and functional studies of oligomeric 2C proteins and their interaction with inhibitors, vRNA, and other viral/host factors.
MATERIALS AND METHODS
Cells and reagents
Buffalo green monkey (BGM) cells (purchased from the European Cell Culture Collection) and HeLa R19 cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium (Lonza, Switzerland) supplemented with 10% (v/v) fetal calf serum (Lonza). All cell lines were grown at 37°C in 5% CO2. Medium was refreshed every 2 to 4 days, and cells were passaged at >80% confluence with the use of phosphate-buffered saline and trypsin-EDTA (0.05%) for up to 10 passages. The cell lines were routinely tested for mycoplasma contamination. SFX, RFX, GuaHCl, and dibucaine were purchased from Sigma-Aldrich. HBB was provided by A.B. (Cardiff University). GuaHCl and ATP-γ-S (Sigma-Aldrich) were dissolved in water at 2 M and 20 mM stock concentration, respectively. All other compounds were dissolved in DMSO at 10 mM stock concentration.
Cloning and construct design
For crystallography experiments, the coding sequence of CV-B3 2C (amino acids 117 to 329) was cloned into the expression vector pmCox 20A as previously described (46). For ATPase assays and cryo-EM experiments, the coding sequence of CV-B3 2C (amino acids 116 to 329) was inserted into pET-28b plasmid. The hexΔ116-2C construct was produced by inserting a 32-residue codon-optimized cc-hex coding sequence followed by a linker (resulting peptide: GELKAIAQELKAIAKELKAIAWELKAIAQGAG; linker: GSGSYFQSNA; Genscript) at the 5′end of the CV-B3 2C coding sequence. For both constructs, a 3C protease–cleavable N-terminal His6-MBP tag was included to facilitate expression and purification. An overview of the hexΔ116-2C construct is shown in fig. S11. The P158A, M175A, D177A, and A229V variants were produced by site-directed mutagenesis using the Δ116-2C and hexΔ116-2C plasmids as a template.
Protein expression and purification
The CV-B3 Δ116-2C protein used for crystallogenesis was produced in Escherichia coli T7 Iq Express pLysS (New England Biolabs). Cells were grown in TB (Terrific Broth) medium containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) at 37°C. Expression was induced overnight at 17°C with 0.5 mM isopropyl-thiogalactopyranoside (IPTG). The cells were harvested by centrifugation and resuspended in lysis buffer (50 mM tris, 300 mM NaCl, 10 mM imidazole, 5% glycerol, and 0.1% Triton X-100) complemented with lysozyme (0.25 mg/ml), deoxyribonuclease (DNase; 10 μg/ml), and 1 mM phenylmethylsulfonyl fluoride. The bacterial sample was sonicated and centrifuged at 18,000g for 30 min at 4°C. Soluble CV-B3 Δ116-2C was purified from the supernatant by immobilized metal affinity chromatography (IMAC) on a HisTrap column (GE Healthcare) and eluted with a buffer consisting of 50 mM tris, 300 mM NaCl, and 500 mM imidazole (pH 8). The eluted protein was subjected to dialysis in 50 mM tris (pH 8), 400 mM NaCl, 10 mM imidazole, and 1 mM DTT (dithiothreitol), followed by an overnight cleavage by Tobacco Etch Virus (TEV) protease (molar ratio, 1/20), followed by a second IMAC to remove the tag and uncleaved protein. The recombinant protein was lastly purified by SEC on a HiLoad 16/60 Superdex 75 column (GE Healthcare) in 10 mM Hepes (pH 7.5) and 300 mM NaCl.
For EM and functional assays, E. coli Rosetta 2 (DE3) cells (Sigma-Aldrich) were transformed with the plasmids coding for the WT, D177A, and A229V CV-B3 2C proteins and grown in 2× YT medium containing kanamycin (50 μg/ml) and chloramphenicol (30 μg/ml) at 37°C until the optical density at 600 nm (OD600nm) reached 0.3. The temperature was then reduced to 18°C, and when the OD600nm reached 0.5, protein expression was induced with 0.5 mM IPTG. Following expression for 16 hours, the cells were harvested by centrifugation and resuspended in lysis buffer (50 mM tris, 300 mM NaCl, 10 mM imidazole, 5% glycerol, 0.1% Triton X-100, and 1 mM MgCl2) complemented with lysozyme (0.25 mg/ml), DNase I (10 μg/ml), and 1 tablet of EDTA-free protease inhibitor cocktail. The bacterial sample was sonicated and centrifuged at 25,000g for 45 min at 4°C. The supernatant was passed through a 0.45-μm filter and incubated with 1 ml of Ni-NTA (nitrilotriacetic acid) resin at 4°C for 1 hour on a roller. The beads were washed with wash I buffer (50 mM tris, 300 mM NaCl, 10 mM imidazole, and 1 mM MgCl2) and wash II buffer (50 mM tris, 300 mM NaCl, 10 mM imidazole, and 1 mM MgCl2) and then eluted with a buffer consisting of 50 mM tris, 300 mM NaCl, and 500 mM imidazole (pH 8). For ATPase assays, the eluted proteins were concentrated to ~200 μl and purified by SEC on a Superose 6 10/300 GL column (GE Healthcare) in 10 mM Hepes (pH 7.5), 150 mM NaCl, and 10 mM MgCl2. The protein-containing fractions were pooled, concentrated to ~4 mg/ml, and flash-frozen in 20-μl aliquots. To remove the N-terminal His6-MBP tag for cryo-EM analysis, the Ni-NTA–eluted hexΔ116-2C protein was concentrated to 200 μl, combined with 2 μg of HRV-3C protease (Sigma-Aldrich), and dialyzed overnight in buffer containing 50 mM tris (pH 8), 200 mM NaCl, and 0.5 mM DTT. The cleaved protein was lastly purified by SEC on a Superose 6 10/300 GL column (GE Healthcare) in 25 mM tris (pH 8), 300 mM NaCl, and 1 mM MgCl2. The fractions corresponding to hexΔ116-2C were pooled, concentrated to ~4 mg/ml, and flash-frozen in 10-μl aliquots. The Superose 6 10/300 GL column was calibrated using a high–molecular weight (43 to 699 kDa) gel filtration calibration kit (Cytiva).
Crystallogenesis, data collection, and structure determination
Structure Screens 1 and 2 and Stura Footprint Screen (Molecular Dimensions Ltd.) were used for initial screening. Trials were assessed in SWISSCI 3 Lens Crystallization Plates with three wells per reservoir using 400-, 300-, and 200-nl drops. The drops contained increasing volumes (100, 200, and 300 nl) of protein solution at a concentration of 8 to 9 mg/ml in complex with or without 2.5 mM SFX (from 50 mM stock in 100% DMSO) and 100 nl of mother liquor. Crystal hits were observed only in the presence of SFX, in 0.1 M MES (pH 6.5) and polyethylene glycol (PEG) 20K 12%. Optimization of crystal growth conditions led to the following condition: 0.1 M MES (pH 5.7 to 6.7) and PEG 20K 7.5/15%. Crystals appeared after 24 hours. They were soaked in 25% (v/v) PEG 200 added to the mother liquor and cooled in liquid nitrogen. X-ray diffraction data were collected at the European Synchrotron Radiation Facility, Grenoble. The structure was solved by molecular replacement using Phaser with PV-2C-ΔN-3Mut structure [Protein Data Bank (PDB) code: 5Z3Q, D chain] as the search model and was refined to a resolution of 1.4 Å. The crystal belongs to the space group of P212121, with one copy of CV-B3 Δ116-2C in the asymmetric unit. In addition, crystals were also obtained in another crystallization solution containing 0.1 M MES (pH 6.3 to 6.7), PEG 5000 MME 23/33%, and 0.2 M ammonium sulfate. Crystals in mother liquor were soaked with 0.3 μl of 20 mM ATP overnight at 20°C. The datasets were collected at Proxima-2A-Synchrotron SOLEIL. The structure was solved by molecular replacement using Phaser with the structure of CV-B3 Δ116-2C in complex with SFX as a search model and refined to a resolution of 1.5 Å. All of the datasets were processed using XDS (47) and scaled with SCALA (Collaborative Computational Project, number 4, 1994). The model building was performed in Coot (48), and the structures were refined using phenix.refine (49). The statistics of data collection, structure refinement, and structure validation are summarized in table S1. Figures of CV-B3 Δ116-2C structures and structural alignments were drawn by Chimera (50). 2C sequence alignments were performed with ClustalOMEGA (51), and secondary structure element was annotated using the ESPript 3.0 server (52).
Protein stability assay
To assess the folding of CV-B3 Δ116-2C and its ability to bind SFX, the thermal stability of recombinant CV-B3 Δ116-2C in the presence of SFX was monitored by fluorescence-based TSA using a Bio-Rad CFX Connect. TSA plates were prepared by dispensing into each well the 2C protein [final concentration of 15 μM in 10 mM Hepes (pH 7.5) and 300 mM NaCl], which was mixed with 1 μl of SFX (from 20 mM stock in 100% DMSO and 1 mM final concentration in 4% DMSO) and a SYPRO orange solution in concentrations recommended by the manufacturer in a final volume of 25 μl.
Reverse engineering of SFX interaction residues
The CV-B3 mutations in 2C L157A, P158A, P159A, M175A, M175I, M175G, D176A, D176N, L178A, L178I, P182A, D186A, D186E, and D186N were first introduced into the Rluc-CV-B3 reporter viruses with a Q5 site-directed mutagenesis kit (New England Biolabs, Bioké, Leiden, The Netherlands) (53). The Rluc-CV-B3 reporter viruses contain a Renilla luciferase gene upstream of the capsid-coding region, which allows a fast and quantitative readout for virus replication. Next, a 705–base pair fragment containing the desired mutation was isolated using the enzymes Bss HII and Xba I and reintroduced into the original nonmutagenized Rluc-CV-B3 backbone. After site-directed mutagenesis, the newly generated plasmids were subjected to Sanger sequencing to confirm the existence of the 2C mutation. The corresponding primers that were used for site-directed mutagenesis can be found in table S3. vRNA was transcribed in vitro using the T7 RiboMAX Express Large Scale RNA production system (Promega, Leiden, The Netherlands) according to the manufacturer’s protocol. In a 96-well plate, HeLa R19 cells were transfected with 7.5 ng of vRNA. One hour after transfection, the medium was replaced by fresh medium and/or compound-containing medium. After 8 hours, cells were lysed, and luciferase activity was determined using the Renilla luciferase Assay System (Promega, Leiden, The Netherlands). Since most of the mutations abrogated virus replication, we cloned the abovementioned 2C mutations into the p53CB3/T7 infectious clone using Bss HII and Xba I (53). Similar to the Rluc-CV-B3 plasmids, the infectious clones were linearized with Mlu I, RNA was transcribed in vitro, and 150 ng of vRNA was transfected into six wells containing either HeLa R19 or BGM cells. All transfections were performed in triplicates. Transfection of CV-B3-2C[A229V] was performed in the presence of 1 mM GuaHCl. Three days after transfection, the transfected cells were subjected to three freeze-thaw cycles, and the lysates were passaged three times on either HelaR19 or BGM cells, respectively. If cytopathic effect was observed during passaging of the viruses, vRNA was isolated with the NucleoSpin RNA Virus kit (Macherey-Nagel, Leiden, The Netherlands) according to the manufacturer’s protocol. The vRNA was reverse-transcribed into cDNA with random hexamer primers using the TaqMan Reverse Transcription Reagents (Applied Biosystems), and polymerase chain reaction (PCR) was performed to isolate the 2C regions with the forward primer binding in 2B 5′-CTAACCAAATATGTGAGC-3′ and the reverse primer binding in 3A 5′-CTCACTGTCTACCGATTTGAG-3′. The same primers were used for Sanger sequencing of the PCR product. If virus was obtained, virus titers were determined by endpoint dilution titration and calculated according to the method of Reed and Muench (54) and expressed as 50% cell culture infective dose (CCID50). The obtained CV-B3-2C[A229V] virus was titrated in the presence of 2 mM GuaHCl.
Single-cycle infection
Confluent HeLa R19 (25,000 cells per well in a 96-well plate) cells were infected with virus at a multiplicity of infection of 0.1 or 1 at 37°C for 30 min. Next, the medium was removed, and fresh (compound-containing) medium was added to the cells. At the indicated time points, the medium was discarded, and cells were subjected to three times freeze-thawing to determine the virus titers by endpoint dilution using the methods of Reed and Muench. In the case of RLuc-CV-B3 infection, cells were lysed 8 hours after infection to determine the luciferase activity with the Renilla luciferase Assay System (Promega). The CV-B3-2C[A229V] mutant was titrated in the presence of 2 mM GuaHCl.
ATPase assay
The release of inorganic phosphate during 2C-mediated ATPase hydrolysis was measured using the Malachite Green Phosphate Assay Kit (Sigma-Aldrich, Zwijndrecht, The Netherlands). Each 50-μl reaction comprised 500 nM recombinant 2C protein and 1 mM ATP in reaction buffer [10 mM Hepes (pH 7.5), 150 mM NaCl, and 10 mM MgCl2]. The reaction buffer was always prepared fresh, and recombinant 2C proteins were thawed immediately before use. For testing the inhibitory effect of 2C-targeting compounds, the final DMSO concentration in each reaction was 1%. Samples were incubated for 30 min at 37°C and then diluted fourfold with water to give a final ATP concentration of 0.25 mM, as per the manufacturer’s instructions. To terminate the enzyme reaction, 80 μl of the diluted sample was mixed with 20 μl of solution AB in a 96-well plate. The plates were incubated for 30 min at room temperature, and absorbance was measured using a microplate reader at OD630. To determine the phosphate concentration in each sample, the OD630 values were plotted against a standard curve using GraphPad Prism version 8.
Mass photometry
Mass photometry experiments were performed on a Refeyn OneMP mass photometer (Refeyn). Borosilicate microscope coverslips (24 mm by 50 mm 1.5H, Marienfeld) were cleaned in four sequential sonication rounds of 5 min alternating between the first isopropanol followed by Milli-Q water. Silicone cell culture gaskets (50 wells; 3 mm diameter × 1 mm depth; Grace Bio-Labs) were cut into sets of four wells, which were placed onto clean coverslips. Before data acquisition, coverslip wells were loaded with approximately 15 μl of buffer containing 10 mM Hepes (pH 7.5), 150 mM NaCl, and 10 mM MgCl2, followed by focusing the instrument on the glass-liquid interface. Samples of the His6-MBP–tagged Δ116-2C and hexΔ116-2C constructs were diluted into the buffer-loaded well to an estimated final concentration of approximately 10 nM, followed by recording for 60 s (6000 frames). Recordings were processed in DiscoverMP (Refeyn) and calibrated using NativeMark Unstained Protein Marker (Thermo Fisher Scientific).
Cryo–electron microscopy
HexΔ116-2C (1 mg/ml) was either used directly for grid preparation or first incubated with 5 mM ATP, 5 mM ATP-γ-S, 100 μM SFX, or 100 μM dibucaine (final DMSO concentration of 0.5%), at 4°C for 30 min. Three microliters of each sample was dispensed on Quantifoil R1.2/1.3 200-mesh grids (Quantifoil Micro Tools GmbH) that had been freshly glow-discharged for 30 s at 20 mA using a PELCO easyGLow Glow Discharge Cleaning System (Tedpella). Grids were blotted for 5 s using Whatman no. 1 filter paper and immediately plunge-frozen into liquid ethane cooled by liquid nitrogen using a Vitrobot Mark IV plunger (Thermo Fisher Scientific) equilibrated to ~95% relative humidity at 4°C. Screening of the apo, ATP-γ-S–incubated, and SFX- or dibucaine-incubated samples was performed using a 200-kV Talos Arctica (Thermo Fisher Scientific) equipped with a Gatan K2 Summit direct detector. Subsequently, two datasets were collected on the SFX-incubated sample using a Titan Krios cryo-TEM (Thermo Fisher Scientific) operating at 300 keV. For the first dataset, movies were collected using a K2 direct electron detector operating in electron counting mode, at ×165,000 magnification corresponding to a calibrated pixel size of 0.842 Å/pixel over a defocus range of −1.5 to −2.5 μm. A total of 1527 movies were collected using a dose rate of 8.8 e−/pixel per second for a total of 4 s (40 fractions), resulting in a total exposure of ~50 e−/Å2 (1.25 e−/Å2 per fraction). To account for the preferred orientation exhibited by the 2C hexamers, an alpha tilt of +30° was used for the second data collection. Movies were collected using a K3 direct electron detector operating in super-resolution mode, at ×64,000 magnification corresponding to a super-resolution pixel size of 0.69 Å/pixel over a defocus range of −2 to −4 μm. A total of 6119 movies were collected using a dose rate of 12.6 e−/pixel per second for a total of 2 s (37 fractions), resulting in a total exposure of ~54 e−/Å2 (1.45 e−/Å2 per fraction). All cryo-EM data were acquired using the EPU 2 software (Thermo Fisher Scientific).
Image processing
For the untilted data, collected movie stacks were manually inspected and then imported in Relion version 3.0.1 (55). Drift and gain correction were performed with MotionCor2 (56), and GCTF was used to estimate the contrast transfer function for each micrograph (57). Approximately 200 particles were picked manually and 2D classified. The best resulting class was then used as a template for autopicking in Relion, resulting in 42,206 particles. Fourier-binned (2 × 2) particles were extracted in a 240-pixel box and subjected to a round of 2D classification, after which 3616 particles were retained. Using the “molmap” command in UCSF Chimera, residues 358 to 628 of the JC polyomavirus helicase [PDB ID: 5J40 (37)] were used to generate a 50-Å-resolution starting model. Particles selected from 2D classification were 3D autorefined (with C6 symmetry), which produced a highly anisotropic map. Following unsuccessful attempts to solve the preferred particle orientation with buffer additives, tilted data collection was used. Tilted movies were Fourier-binned (2 × 2) during motion correction with MotionCor2 (56), and goCTF was used to estimate the defocus gradient of each micrograph (58). Approximately 1000 particles were picked manually and 2D classified. The best resulting class was then used as a template for autopicking in Relion, resulting in 180,069 particles. Fourier-binned (2 × 2) particles were extracted in an 80-pixel box and subjected to 3D classification, after which 53,854 particles were selected. After a second round of 3D classification, 11,024 particles were retained. A final round of no-alignment 3D classification was performed, yielding a final stack of 6856 particles. Subsequent 3D autorefinement (with C6 symmetry) and postprocessing yielded a map with an estimated resolution of 12 Å, based on the gold-standard Fourier shell correlation = 0.143 criterion. An ad hoc negative B-factor of −200 Å2 was applied during the final postprocessing step. An overview of the data processing pipeline is shown in fig. S16. Figures of the CV-B3 hexΔ116-2C reconstruction were made in UCSF Chimera X (59).
Acknowledgments
We thank S. Attoumani and M. den Boer for technical assistance and M. Vanevic for computational support.
Funding: D.L.H. is funded by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement (no. 842333) and holds an EMBO nonstipendiary long-term fellowship (ALTF 1172-2018). P.E.K. is the recipient of a scholarship of the Foundation “Méditerranée Infection,” Marseille. This work was supported by the European Union (Horizon 2020 Marie Skłodowska-Curie ETN “ANTIVIRALS,” grant agreement number 642434 to A.B., F.J.M.v.K., and B.Co.). This work is also supported by the Netherlands Organization for Scientific Research (NWO-ECHO-711.017.002 and NWO-VICI-91812628 to F.J.M.v.K.) and by the Life Science Research Network Wales (grant no. NRNPGSep14008 to A.B., an initiative funded through the Welsh Government's Ser Cymru program). This work was supported by the European Research Council under the European Union’s Horizon2020 Programme (ERC Consolidator grant agreement 724425–BENDER). T.M.S. and J.S. are supported by the Dutch Research Council NWO Gravitation 2013 BOO, Institute for Chemical Immunology (ICI, 024.002.009). This work benefited from access to the Netherlands Centre for Electron Nanoscopy (NeCEN) at Leiden University, an Instruct-ERIC center with assistance from R. Dillard.
Author contributions: Conceptualization: D.L.H., P.E.K., L.B., F.J.M.v.K., and B.Co. Investigation: D.L.H., P.E.K., L.B., N.P., F.P.F., T.D., A.L.W.v.V., T.M.S., and J.S. Formal analysis: D.L.H., P.E.K., L.B., N.P., F.P.F., and J.S. Validation: D.L.H., P.E.K., L.B., F.J.M.v.K., and B.Co. Resources: J.S., B.Ca., T.Z.-B.-M., F.F., F.J.M.v.K., and B.Co. Data curation: D.L.H., P.E.K., and L.B. Writing—original draft: D.L.H., P.E.K., L.B., F.J.M.v.K., and B.Co. Writing—review and editing: D.L.H., P.E.K., L.B., N.P., F.P.F., T.D., A.L.W.v.V., T.M.S., J.S., B.Ca., E.D., A.B., T.Z.-B.-M., F.F., F.J.M.v.K., and B.Co. Visualization: D.L.H., P.E.K., and L.B. Supervision: D.L.H., J.S., B.Ca., E.D., T.Z.-B.-M., F.F., F.J.M.v.K., and B.Co. Project administration: D.L.H., P.E.K., F.J.M.v.K., and B.Co. Funding acquisition: D.L.H., J.S., A.B., F.F., F.J.M.v.K., and B.Co.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Atomic coordinates and structure factors for CV-B3 Δ116-2C in complex with SFX are deposited in the PDB under accession codes 6S3A and 6T3W. The EM density map for the SFX-incubated hexΔ116-2C has been deposited to the Electron Microscopy Data Bank under the accession EMD-12798. Expression plasmid of CVB3 2C used for protein crystallization can be provided by Aix Marseille Université pending scientific review and a completed material transfer agreement. The expression plasmid can be requested on the EVA repository (https://european-virus-archive.com/). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S19
Tables S1 to S4
REFERENCES AND NOTES
- 1.Tapparel C., Siegrist F., Petty T. J., Kaiser L., Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 14, 282–293 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Chapman N. M., Kim K. S., Persistent coxsackievirus infection: Enterovirus persistence in chronic myocarditis and dilated cardiomyopathy. Curr. Top. Microbiol. Immunol. 323, 275–292 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Puenpa J., Wanlapakorn N., Vongpunsawad S., Poovorawan Y., The history of enterovirus A71 outbreaks and molecular epidemiology in the asia-pacific region. J. Biomed. Sci. 26, 75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messacar K., Asturias E. J., Hixon A. M., Van Leer-Buter C., Niesters H. G. M., Tyler K. L., Abzug M. J., Dominguez S. R., Enterovirus D68 and acute flaccid myelitis—Evaluating the evidence for causality. Lancet Infect. Dis. 18, e239–e247 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aw-Yong K. L., NikNadia N. M. N., Tan C. W., Sam I. C., Chan Y. F., Immune responses against enterovirus A71 infection: Implications for vaccine success. Rev. Med. Virol. 29, e2073 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Lin J.-Y., Kung Y.-A., Shih S.-R., Antivirals and vaccines for enterovirus A71. J. Biomed. Sci. 26, 65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baggen J., Thibaut H. J., Strating J. R. P. M., Van Kuppeveld F. J. M., The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 16, 368–381 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Wang S. H., Wang K., Zhao K., Hua S. C., Du J., The structure, function, and mechanisms of action of enterovirus non-structural protein 2C. Front. Microbiol. 11, 615965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya A. E., Koonin E. V., Wolf Y. I., A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262, 145–148 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Xia H., Wang P., Wang G. C., Yang J., Sun X., Wu W., Qiu Y., Shu T., Zhao X., Yin L., Qin C. F., Hu Y., Zhou X., Human enterovirus nonstructural protein 2CATPase functions as both an RNA helicase and ATP-independent RNA chaperone. PLOS Pathog. 11, e1005067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams P., Kandiah E., Effantin G., Steven A. C., Ehrenfeld E., Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J. Biol. Chem. 284, 22012–22021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweeney T. R., Cisnetto V., Bose D., Bailey M., Wilson J. R., Zhang X., Belsham G. J., Curry S., Foot-and-mouth disease virus 2C is a hexameric AAA+ protein with a coordinated ATP hydrolysis mechanism. J. Biol. Chem. 285, 24347–24359 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papageorgiou N., Coutard B., Lantez V., Gautron E., Chauvet O., Baronti C., Norder H., De Lamballerie X., Heresanu V., Ferté N., Veesler S., Gorbalenya A. E., Canard B., The 2C putative helicase of echovirus 30 adopts a hexameric ring-shaped structure. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 1116–1120 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Guan H., Tian J., Qin B., Wojdyla J. A., Wang B., Zhao Z., Wang M., Cui S., Crystal structure of 2C helicase from enterovirus 71. Sci. Adv. 3, e1602573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan H., Tian J., Zhang C., Qin B., Cui S., Crystal structure of a soluble fragment of poliovirus 2CATPase. PLOS Pathog. 14, e1007304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Palma A. M., Heggermont W., Lanke K., Coutard B., Bergmann M., Monforte A.-M., Canard B., De Clercq E., Chimirri A., Pürstinger G., Rohayem J., van Kuppeveld F., Neyts J., The thiazolobenzimidazole TBZE-029 inhibits enterovirus replication by targeting a short region immediately downstream from motif c in the nonstructural protein 2C. J. Virol. 82, 4720–4730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfister T., Wimmer E., Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274, 6992–7001 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Hadaschik D., Klein M., Zimmermann H., Eggers H. J., Nelsen-Salz B., Dependence of echovirus 9 on the enterovirus RNA replication inhibitor 2-(α-hydroxybenzyl)-benzimidazole maps to nonstructural protein 2C. J. Virol. 73, 10536–10539 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein M., Hadaschik D., Zimmermann H., Eggers H. J., Nelsen-Salz B., Picornavirus replication inhibitors HBB and guanidine in the echovirus-9 system: The significance of viral protein 2C. J. Gen. Virol. 81, 895–901 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Shimizu H., Agoh M., Agoh Y., Yoshida H., Yoshii K., Yoneyama T., Hagiwara A., Miyamura T., Mutations in the 2C region of poliovirus responsible for altered sensitivity to benzimidazole derivatives. J. Virol. 74, 4146–4154 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo J., Quinn K. K., Kye S., Cooper P., Damoiseaux R., Krogstad P., Fluoxetine is a potent inhibitor of coxsackievirus replication. Antimicrob. Agents Chemother. 56, 4838–4844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulferts R., De Boer S. M., Van Der Linden L., Bauer L., Lyoo H. R., Maté M. J., Lichière J., Canard B., Lelieveld D., Omta W., Egan D., Coutard B., Van Kuppeveld F. J. M., Screening of a library of FDA-approved drugs identifies several enterovirus replication inhibitors that target viral protein 2C. Antimicrob. Agents Chemother. 60, 2627–2638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer L., Manganaro R., Zonsics B., Strating J. R. P. M., El Kazzi P., Lorenzo Lopez M., Ulferts R., Van Hoey C., Maté M. J., Langer T., Coutard B., Brancale A., Van Kuppeveld F. J. M., Fluoxetine inhibits enterovirus replication by targeting the viral 2C protein in a stereospecific manner. ACS Infect. Dis. 5, 1609–1623 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manganaro R., Zonsics B., Bauer L., Lorenzo Lopez M., Donselaar T., Zwaagstra M., Saporito F., Ferla S., Strating J. R. P. M., Coutard B., Hurdiss D. L., van Kuppeveld F. J. M., Brancale A., Synthesis and antiviral effect of novel fluoxetine analogues as enterovirus 2C inhibitors. Antiviral Res. 178, 104781 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Musharrafieh R., Zhang J., Tuohy P., Kitamura N., Bellampalli S. S., Hu Y., Khanna R., Wang J., Discovery of quinoline analogues as potent antivirals against enterovirus D68 (EV-D68). J. Med. Chem. 62, 4074–4090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauer L., Manganaro R., Zonsics B., Hurdiss D. L., Zwaagstra M., Donselaar T., Welter N. G. E., Van Kleef R. G. D. M., Lopez M. L., Bevilacqua F., Raman T., Ferla S., Bassetto M., Neyts J., Strating J. R. P. M., Westerink R. H. S., Brancale A., Van Kuppeveld F. J. M., Rational design of highly potent broadspectrum enterovirus inhibitors targeting the nonstructural protein 2C. PLoS Biol. 18, e3000904 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickman A. B., Dyda F., Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15, 77–85 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Eggers H. J., Tamm I., Drug dependence of enteroviruses: Variants of Coxsackie A9 and ECHO 13 viruses that require 2-(α-hydroxybenzyl)-benzimidazole for growth. Virology 20, 10536–10539 (1963). [Google Scholar]
- 29.Ulferts R., Van Der Linden L., Thibaut H. J., Lanke K. H. W., Leyssen P., Coutard B., De Palma A. M., Canard B., Neyts J., Van Kuppeveld F. J. M., Selective serotonin reuptake inhibitor fluoxetine inhibits replication of human enteroviruses B and D by targeting viral protein 2C. Antimicrob. Agents Chemother. 57, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gai D., Zhao R., Li D., Finkielstein C. V., Chen X. S., Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, (2004). [DOI] [PubMed] [Google Scholar]
- 31.Tolskaya E. A., Romanova L. I., Kolesnikova M. S., Gmyl A. P., Gorbalenya A. E., Agol V. I., Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J. Mol. Biol. 236, 1310–1323 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Lu C., Turley S., Marionni S. T., Park Y. J., Lee K. K., Patrick M., Shah R., Sandkvist M., Bush M. F., Hol W. G. J., Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with Increased ATPase activity. Structure 21, 1707–1717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monroe N., Han H., Shen P. S., Sundquist W. I., Hill C. P., Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase. eLife 6, e24487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi H., Rampello A. J., Glynn S. E., Engineered AAA+ proteases reveal principles of proteolysis at the mitochondrial inner membrane. Nat. Commun. 7, 13301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaccai N. R., Chi B., Thomson A. R., Boyle A. L., Bartlett G. J., Bruning M., Linden N., Sessions R. B., Booth P. J., Brady R. L., Woolfson D. N., A de novo peptide hexamer with a mutable channel. Nat. Chem. Biol. 7, 935–941 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Echeverri A. C., Dasgupta A., Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208, 540–553 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Bonafoux D., Nanthakumar S., Bandarage U. K., Memmott C., Lowe D., Aronov A. M., Bhisetti G. R., Bonanno K. C., Coll J., Leeman J., Lepre C. A., Lu F., Perola E., Rijnbrand R., Taylor W. P., Wilson D., Zhou Y., Zwahlen J., Ter Haar E., Fragment-based discovery of dual JC Virus and BK virus helicase inhibitors. J. Med. Chem. 59, 7138–7151 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Tang Q., Xu Z., Jin M., Shu T., Chen Y., Feng L., Zhang Q., Lan K., Wu S., Zhou H. B., Identification of dibucaine derivatives as novel potent enterovirus 2C helicase inhibitors: In vitro, in vivo, and combination therapy study. Eur. J. Med. Chem. 202, 112310 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Bochkov Y. A., Palmenberg A. C., Lee W. M., Rathe J. A., Amineva S. P., Sun X., Pasic T. R., Jarjour N. N., Liggett S. B., Gern J. E., Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat. Med. 17, 627–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodson M., Pajak J., Mahler B. P., Zhao W., Zhang W., Arya G., White M. A., Jardine P. J., Morais M. C., A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA. Sci. Adv. 7, eabc1955 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banerjee S., Bartesaghi A., Merk A., Rao P., Bulfer S. L., Yan Y., Green N., Mroczkowski B., Neitz R. J., Wipf P., Falconieri V., Deshaies R. J., Milne J. L. S., Huryn D., Arkin M., Subramaniam S., 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science 351, 871–875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su M., Guo E. Z., Ding X., Li Y., Tarrasch J. T., Brooks C. L., Xu Z., Skiniotis G., Mechanism of Vps4 hexamer function revealed by cryo-EM. Sci. Adv. 3, e1700325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao M., Brunger A. T., Recent advances in deciphering the structure and molecular mechanism of the AAA + ATPase N-ethylmaleimide-sensitive factor (NSF). J. Mol. Biol. 428, 1912–1926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santosh V., Musayev F. N., Jaiswal R., Zárate-Pérez F., Vandewinkel B., Dierckx C., Endicott M., Sharifi K., Dryden K., Henckaerts E., Escalante C. R., The cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states. Nucleic Acids Res. 48, 12983–12999 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woolfson D. N., Coiled-coil design: Updated and upgraded. Subcell. Biochem. 82, 12983–12999 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Lantez V., Dalle K., Charrel R., Baronti C., Canard B., Coutard B., Comparative production analysis of three phlebovirus nucleoproteins under denaturing or non-denaturing conditions for crystallographic studies. PLoS Negl. Trop. Dis. 5, e936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabsch W., XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lanke K. H. W., van der Schaar H. M., Belov G. A., Feng Q., Duijsings D., Jackson C. L., Ehrenfeld E., van Kuppeveld F. J. M., GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J. Virol. 83, 11940–11949 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reed L. J., Muench H., A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497 (1938). [Google Scholar]
- 55.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su M., goCTF: Geometrically optimized CTF determination for single-particle cryo-EM. J. Struct. Biol. 205, 22–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salentin S., Schreiber S., Haupt V. J., Adasme M. F., Schroeder M., PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 43, W443–W447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S19
Tables S1 to S4