Abstract
Nucleocytoplasmic trafficking of histone deacetylase 4 (HDAC4) plays an important role in regulating its function, and binding of 14-3-3 proteins is necessary for its cytoplasmic retention. Here, we report the identification of nuclear import and export sequences of HDAC4. While its N-terminal 118 residues modulate the nuclear localization, residues 244 to 279 constitute an authentic, strong nuclear localization signal. Mutational analysis of this signal revealed that three arginine-lysine clusters are necessary for its nuclear import activity. As for nuclear export, leucine-rich sequences located in the middle part of HDAC4 do not function as nuclear export signals. By contrast, a hydrophobic motif (MXXLXVXV) located at the C-terminal end serves as a nuclear export signal that is necessary for cytoplasmic retention of HDAC4. This motif is required for CRM1-mediated nuclear export of HDAC4. Furthermore, binding of 14-3-3 proteins promotes cytoplasmic localization of HDAC4 by both inhibiting its nuclear import and stimulating its nuclear export. Unlike wild-type HDAC4, a point mutant with abrogated MEF2-binding ability remains cytoplasmic upon exogenous expression of MEF2C, supporting the notion that direct MEF2 binding targets HDAC4 to the nucleus. Therefore, HDAC4 possesses intrinsic nuclear import and export signals for its dynamic nucleocytoplasmic shuttling, and association with 14-3-3 and MEF2 proteins affects such shuttling and thus directs HDAC4 to the cytoplasm and the nucleus, respectively.
How protein functions are regulated in vivo is a fundamental issue relevant to various biological processes. Lysine acetylation has recently emerged as a major form of posttranslational modification that regulates functions of histones, nonhistone chromosomal proteins, and transcription factors (8, 21, 29, 52, 54). Acetylation of histones and other chromosomal proteins regulates chromatin activities in transcription, replication, and recombination (3, 38, 42, 55, 62). Histone deacetylases (HDACs) are the enzymes responsible for reversing the acetylation of histones and other proteins. According to sequence homology and time of identification, mammalian HDACs can be divided into three classes. Class I HDACs (HDAC1, HDAC2, HDAC3, and HDAC8) show high similarity to the yeast deacetylase Rpd3 (4, 9, 12, 22, 56, 57, 65, 66). Class II HDACs (HDAC4, HDAC5, HDAC6, and HDAC7) possess catalytic domains significantly homologous to that of yeast Hda1 (13, 19, 27, 43, 48, 59, 60). Class III is comprised of proteins with catalytic domains similar to that of the yeast NAD+-dependent deacetylase Sir2 (15, 24, 31, 49).
Compared to class I deacetylases, much less is known about the second class (8). HDAC4, HDAC5, and HDAC7 are homologous, with their Hda1-related domains located in the C-terminal parts, whereas HDAC6 possesses tandem Hda1-related domains (13, 19, 27, 43, 59, 60). Like class I members, class II HDACs (except HDAC6) have been found to be corepressors recruited for transcriptional repression. The MEF2 transcription factors interact with HDAC4, HDAC5, HDAC7, and their related protein monocyte enhancer factor 2 (MEF2)-interacting transcription repressor (MITR) (also known as HDAC-related protein) to repress transcription (11, 32, 35, 43, 50, 60, 69). Moreover, this interaction is signal dependent and regulated during muscle differentiation (11, 35, 36, 67). HDAC4, HDAC5, and HDAC7 also interact with the nuclear receptor corepressors SMRT and N-CoR to repress transcription (23, 27).
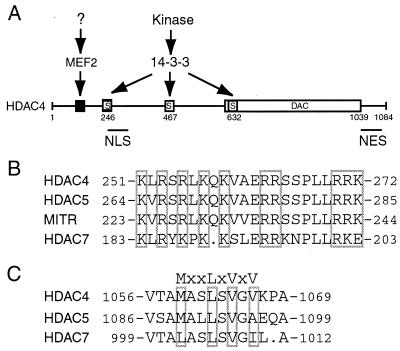
How are functions of different deacetylases regulated in vivo? Emerging evidence suggests that cellular compartmentalization is one major regulatory mechanism for class II HDACs (8, 28). Active nucleocytoplasmic shuttling has been shown for HDAC4 (20, 43, 61), HDAC5 (40, 41), HDAC6 (58), and HDAC7 (11). Moreover, such shuttling is tightly controlled. 14-3-3 proteins directly bind to HDAC4 and HDAC5 and negatively regulate their roles in transcriptional repression (20, 40, 61). 14-3-3 binding to HDAC5 and perhaps to its homologs (i.e., HDAC4 and HDAC7) plays an important role in regulating functions of MEF2 during muscle differentiation (11, 36, 40, 41, 53). Three serine residues of HDAC4 (i.e., S246, S467, and S632) mediate its binding to 14-3-3 proteins (20, 61). Unlike wild-type HDAC4, the triple mutant S246/467/632A is completely defective in 14-3-3 binding and is localized to the nucleus (20, 61), indicating that 14-3-3 binding is necessary for retaining HDAC4 in the cytoplasm. However, it remains unclear whether 14-3-3 binding alone is sufficient for cytoplasmic retention of HDAC4.
While characterizing the interesting link between HDAC4 and 14-3-3 proteins, we unexpectedly found that the mutant 118-1084/S246/467/632A, the triple mutant which lacks the N-terminal 118 residues of HDAC4, was mainly cytoplasmic or pancellular. To understand this intriguing finding, we engineered and analyzed various HDAC4 mutants, which has led to the identification of sequence elements that are important for nucleocytoplasmic trafficking of HDAC4. While the N-terminal 118 residues and MEF2-binding site of HDAC4 modulate its nuclear localization, residues 244 to 279 constitute an authentic, tripartite nuclear localization signal (NLS) and a C-terminal hydrophobic motif serves as a functional nuclear export signal (NES). This NES is required for CRM1-mediated nuclear export of HDAC4. Furthermore, both 14-3-3 binding and the NES-mediated nuclear export are required for cytoplasmic retention of HDAC4. We propose that subcellular distribution of HDAC4 is controlled by multiple mechanisms in vivo. Such a regulatory scheme may provide flexibility for fine-tuning biological functions of HDAC4.
MATERIALS AND METHODS
Molecular cloning.
Expression plasmids for HDAC4 and some deletion mutants have been described previously (60, 61). Additional HDAC4 mutants were generated by PCR with Expand (Roche) thermostable DNA polymerase or by site-directed mutagenesis with single-stranded uracil-containing templates and T7 DNA polymerase. DNA sequencing was performed with T7 Sequenase 2.0 (Amersham Pharmacia Biotech) for confirmation of mutations. Green fluorescent protein (GFP) constructs were derived from pEGFP-C2 (Clontech).
Green fluorescence microscopy.
NIH 3T3 and 293 cells were transfected with plasmids expressing GFP fusion proteins using SuperFect transfection reagent (Qiagen) (5, 60). At 16 h after transfection, living cells were analyzed by GFP fluorescence microscopy as described (61). Fluorescence images were collected using a charge-coupled device camera (Q-imaging, Inc.) linked to a computer running Northern Eclipse (version 5.0; Empix Imaging) and exported for further processing with Adobe Photoshop. Alternatively, cells were fixed with formaldehyde and counterstained with Hoechst 33528 to visualize the nuclei (61); Hoechst and green fluorescence images were subsequently collected.
Immunofluorescence microscopy.
To assess effects of MEF2 binding on subcellular localization of HDAC4 and its mutants, MEF2C expression plasmid was transfected into NIH 3T3 cells along with mammalian expression plasmids for HDAC4 or its mutants fused to GFP. To detect the expression of MEF2C, cells were fixed with formaldehyde 16 h after transfection, incubated with anti-MEF2C antibody, and stained with Cy3 anti-rabbit immunoglobulin G antibody (Jackson Immunoresearch) as previously described (37, 61). Cells were counterstained with Hoechst 33528 to visualize the nuclei. Expression of GFP fusion proteins was determined by green fluorescence microscopy. Similarly, effects of exogenous CRM1 on subcellular localization of HDAC4 mutants were determined.
Protein-protein interaction.
To analyze interaction of the MEF2C mutant M178 with HDAC4 mutants, in vitro maltose-binding protein (MBP) binding assays were carried out as described (60). For analysis of intra- or intermolecular interaction among HDAC4 molecules, its fragments were expressed as MBP fusion proteins in Escherichia coli, immobilized on amylose agarose (New England Biolabs), and incubated with HDAC4 or its fragments synthesized in vitro by use of a TNT-T7 coupled reticulocyte lysate system (Promega) in the presence of l-[35S]methionine (Amersham Pharmacia Biotech). Agarose beads were washed three times with buffer B (20 mM Tris-HCl [pH 8.0], 10% glycerol, 5 mM MgCl2, 0.1% NP-40, protease inhibitors) containing 0.15 M KCl and once with buffer B containing 0.5 M KCl. Bound proteins were then analyzed by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography as previously described (60).
Western blotting analysis.
Expression of GFP fusion proteins was also confirmed by Western blotting analysis of total cell extracts as previously described (6, 60). 293 cells were transfected with plasmids expressing GFP fusion proteins using SuperFect transfection reagent (Qiagen) (5, 60). At 16 h after transfection, cells were washed twice with ice-cold phosphate-buffered saline (PBS) and collected in ice-cold buffer B containing 0.15 M KCl or in buffer H (20 mM HEPES [pH 7.6], 20% glycerol, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 25 mM NaF, 10 mM β-glycerophosphate, 1 mM dithiothreitol, protease inhibitors). After being rotated at 4°C for 20 min, the cell lysates were cleared by high-speed centrifugation at 4°C, and the supernatants were collected as total cell extracts. For immunoblotting, the total cell extracts (∼10 μg/lane) were resolved by reducing SDS-PAGE, electro-transferred to nitrocellulose membrane, and subsequently immunoblotted with anti-GFP antibody (Santa Cruz Biotechnology; sc-8334). For blocking and antibody incubation, PBS containing 20% horse serum (GG free; Gibco BRL) and 0.15% Tween 20 (Sigma) was used. For washing, PBS with 0.15% Tween 20 was used. Blots were developed with Supersignal chemiluminescent substrate (Pierce).
BLAST search.
Amino acid sequence homology searches were performed at the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/) using PSI-BLAST with the matrix BLOSUM62 (1).
RESULTS
Compared to the yeast deacetylase Hda1, HDAC4 can be divided into three parts: an extended N-terminal region (residues 1 to 620), an Hda1-related deacetylase domain (residues 621 to 1039), and a small C-terminal module (residues 1040 to 1084) (Fig. 1A). The extended N-terminal region has been found to interact with MEF2 and 14-3-3 proteins (20, 43, 60, 61), whereas the function of the small C-terminal module remains elusive.
FIG. 1.
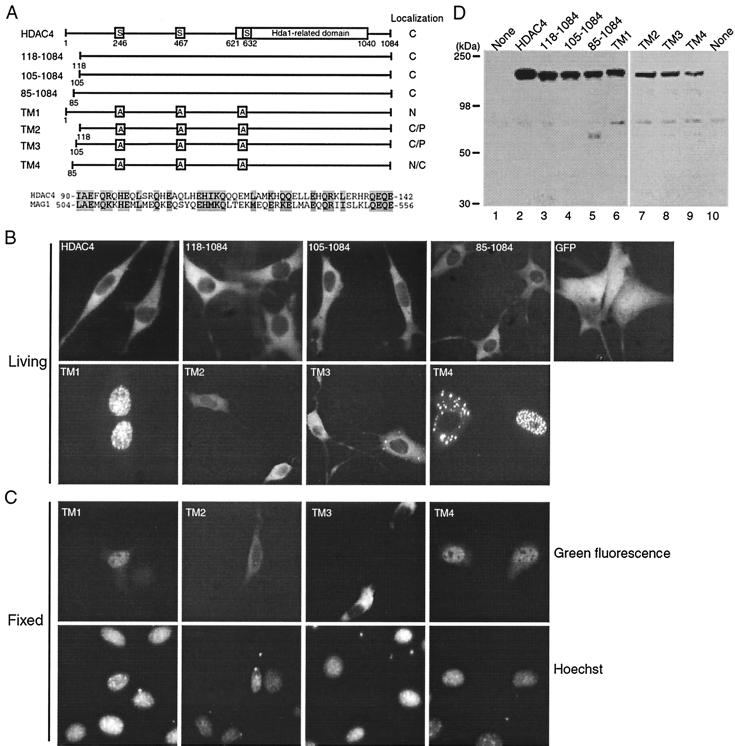
Role of the N-terminal 118 residues of HDAC4 in regulating its nuclear localization. (A) Schematic illustration of HDAC4 and mutants. For HDAC4, 14-3-3 binding sites (S246, S467, and S632) and the Hda1-homology domain are depicted by boxes. In the triple mutants TM1 to TM4, the three serine residues critical for 14-3-3 binding are changed to alanine. Subcellular localization of HDAC4 and mutants is summarized at right: C, predominantly cytoplasmic; N, predominantly nuclear; P, pancellular. Shown at the lower part of the panel is the sequence comparison between homologous regions of HDAC4 (residues 90 to 142) and MAG1 (residues 504 to 556), with identical or conserved residues shaded. (B and C) Representative green fluorescence images of living (B) or fixed (C) NIH 3T3 cells expressing GFP or its fusion proteins. Cells were transfected with expression plasmids for GFP or its fusion proteins and subsequently analyzed by fluorescence microscopy 16 h after transfection. (B) Living cells were directly used for microscopic analysis. (C) Transfected cells were fixed with formaldehyde, counterstained with Hoechst 33528 and analyzed by green fluorescence microscopy (top), with corresponding Hoechst fluorescence images also taken (bottom). (D) Expression of GFP fusion proteins. 293 cells were transfected with expression plasmids for indicated GFP fusion proteins, and total cell extracts were analyzed by immunoblotting with anti-GFP antibody.
Role of the N-terminal 118 residues of HDAC4 in modulating its nuclear localization.
We and others have previously shown that the HDAC4 triple mutant TM1 (Fig. 1A) is completely defective in 14-3-3 binding and thus predominantly nuclear (20, 61). As reported, GFP-HDAC4 and -TM1 were cytoplasmic and nuclear, respectively, whereas GFP itself was pancellular in NIH 3T3 cells (Fig. 1B and C). Also consistent with published reports (43, 61), the mutant 118-1084 was predominantly cytoplasmic (Fig. 1 and data not shown). Unexpectedly, we found that unlike GFP-TM1, GFP-TM2 was either cytoplasmic or pancellular (Fig. 1), suggesting that the N-terminal 118 residues are involved in regulating nuclear localization of HDAC4. BLAST searches revealed that residues 90 to 142 of HDAC4 show limited sequence similarity to the GTP-binding protein MAG1 (Fig. 1A) (7, 63). To address whether the MAG1-related region of HDAC4 is responsible for the observed difference between GFP-TM1 and -TM2, we engineered the mutants TM3 and TM4 fused to GFP (Fig. 1A). As shown in Fig. 1B and C, unlike GFP-TM3, GFP-TM4 was more similar to GFP-TM1, suggesting that the MAG1-homology region may be important for controlling nuclear localization of HDAC4. As previously reported (61), GFP-TM1 was nuclear in most cells. By contrast, GFP-TM4 was found to be nuclear in 40 to 50% of the cells expressing this fusion protein (data not shown), suggesting that the N-terminal 85 residues are also important for nuclear localization of HDAC4. To determine whether GFP fusion proteins are expressed as expected, we performed Western blotting analysis. As shown in Fig. 1D, GFP fusion proteins with expected sizes were detected. Taken together, these results indicate that the N-terminal 118 residues of HDAC4 play an important role in modulating its nuclear localization.
How do the N-terminal 118 residues of HDAC4 modulate its nuclear localization?
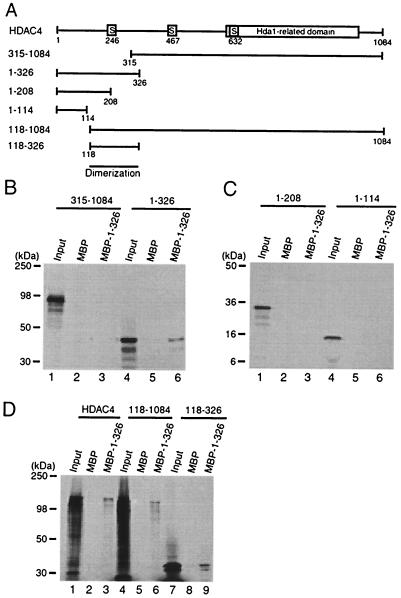
To promote its nuclear localization, the N-terminal 118 residues of HDAC4 may: (i) be involved in inter- or intramolecular interaction with HDAC4 itself (such interaction may affect the exposure of potential nuclear import or export sequences), (ii) interact with other nuclear proteins, or (iii) be (part of) an NLS. To distinguish among these possibilities, we first investigated whether the N-terminal part of HDAC4 mediates inter- or intramolecular interaction with HDAC4 itself. For this, we analyzed different HDAC4 deletion mutants (Fig. 2A) by in vitro binding assays using MBP or its fusion proteins immobilized on amylose agarose. As shown in Fig. 2B, no interaction was detectable between mutant 315-1084 and MBP–1-326 (lanes 1 to 3). By contrast, mutant 1-326 interacted with MBP–1-326 but not MBP itself (lanes 4 to 6). To test whether the N-terminal 118 residues are essential for this interaction, we tested the deletion mutants 1-208 and 1-114. As shown in Fig. 2C, neither mutant was retained by MBP–1-326, suggesting that residues 118 to 326 may be responsible for the interaction. In agreement with this, MBP–1-326 interacted with HDAC4, mutant 118-1084 and mutant 118-326 (Fig. 2D), suggesting that residues 118 to 326 of HDAC4 constitute a dimerization domain. Taken together, these results indicate that the N-terminal 118 residues of HDAC4 do not appear to be involved in inter- or intramolecular interaction with HDAC4 itself.
FIG. 2.
Mapping the dimerization domain of HDAC4. (A) Schematic representation of HDAC4 and deletion mutants. Motifs or domains are depicted by boxes as in Fig. 1A. (B to D) Interaction among HDAC4 proteins. HDAC4 deletion mutants were expressed as MBP fusion proteins in E. coli, immobilized on amylose agarose and incubated with HDAC4 or deletion mutants synthesized in vitro in the presence of l-[35S]methionine. Agarose beads were washed three times with buffer B–0.15 M KCl and once with buffer B–0.5 M KCl. Bound proteins were separated by SDS-PAGE and subsequently detected by autoradiography. Input represents 20% of the 35S-labeled protein used for each binding assay. Migrating positions of molecular markers are shown at the left of each panel.
As discussed above, the N-terminal 118 residues may interact with other nuclear proteins and thereby stimulate nuclear localization of HDAC4. Two proteins are known to have the potential to interact with the N-terminal part of HDAC4. Although the exact binding site has not been mapped, HDAC1 has been shown to be associated with MITR, a corepressor with sequence similarity to the N-terminal part of HDAC4 (50). Adenovirus E1A C-terminal-binding protein (CtBP) has also been shown to interact with HDAC4, and its N-terminal 118 residues possesses a putative CtBP-binding site (68). To test whether HDAC1 or CtBP modulates subcellular localization of HDAC4, we examined effects of their overexpression on intracellular distribution of GFP-HDAC4 by fluorescence microscopy. The results indicated that overexpression of HDAC1 or CtBP had minimal effects on the cytoplasmic localization of GFP-HDAC4 (data not shown), suggesting that neither HDAC1 nor CtBP is involved in modulating intracellular localization of HDAC4. It still remains possible, however, that an unidentified protein may interact with the N-terminal 118 residues of HDAC4 and thereby modulate its intracellular distribution.
To investigate whether the N-terminal 118 residues of HDAC4 constitute (or are part of) an NLS, we examined subcellular distribution of the HDAC4 deletion mutants 1-118 and 1-165 expressed as GFP fusion proteins (Fig. 3A). As shown in Fig. 3B, both mutants were partially enriched in the nucleus, suggesting that the N-terminal 118 residues of HDAC4 only possess weak nuclear targeting ability. This region does not show any sequence resemblance to classical arginine-lysine-rich nuclear import signals, raising the possibility that a yet-unknown protein interacts with this region of HDAC4 and regulates its subcellular localization (see Discussion).
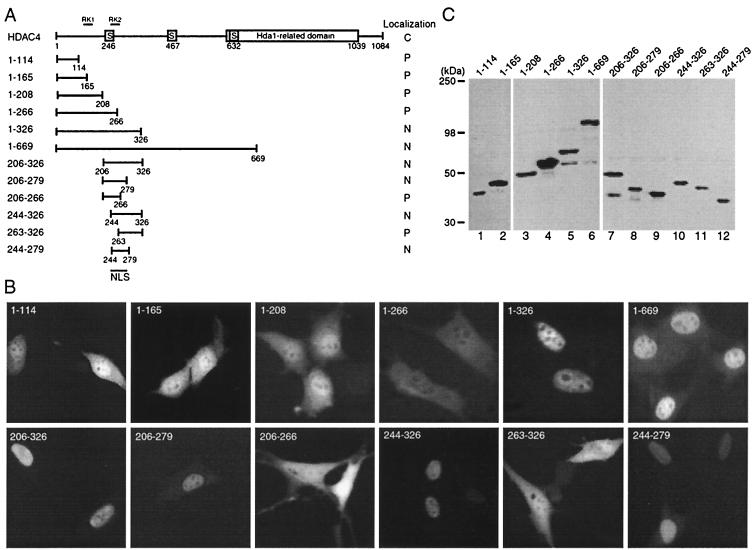
FIG. 3.
Mapping the NLS of HDAC4. (A) Schematic representation of HDAC4 and deletion mutants. Motifs or domains are depicted by boxes as in Fig. 1A. Also indicated are two arginine-lysine-rich regions: RK1 (residues 132 to 184) and RK2 (residues 242 to 283). (B) Representative green fluorescence images of living cells expressing HDAC4 mutants fused to GFP. NIH 3T3 cells were transfected with expression plasmids for indicated GFP fusion proteins and analyzed by live green fluorescence microscopy. (C) Expression of GFP fusion proteins. 293 cells were transfected with expression plasmids for indicated GFP fusion proteins, and total cell extracts were analyzed by immunoblotting with anti-GFP antibody.
Identification of an authentic HDAC4 NLS.
The distinct localization between GFP-TM1 (nuclear [Fig. 1]) and GFP-1-165 (partially enriched in the nucleus [Fig. 3]) further suggests that there is a strong NLS within residues 166 to 1084. Consistent with this, HDAC4 possesses two arginine-lysine-rich sequences (RK1 and RK2) (Fig. 3A). To further understand how nucleocytoplasmic distribution of HDAC4 is controlled, we decided to map its NLS. To take a systematic approach, we first analyzed the deletion mutants 1-208, 1-266, 1-326, and 1-669 expressed as GFP fusion proteins (Fig. 3A). As shown in Fig. 3B, the localization of mutant 1-208 was similar to that of mutants 1-114 and 1-165, suggesting that RK1 is not an NLS. Distinct from mutant 1-208, mutant 1-266 was pancellular (Fig. 3B). One explanation for this is that 14-3-3 binding to S246 of mutant 1-266 counteracts the weak nuclear targeting activity that the N-terminal 118 residues exhibit. Unlike mutant 1-266, the mutants 1-326 and 1-669 were exclusively or predominantly nuclear, indicating that residues 267 to 326 are important for the nuclear localization activity. Consistent with this, the mutant 206-326 was exclusively nuclear. Together, these results suggest that RK2 may possess an authentic NLS. To further map this NLS, we constructed and analyzed the mutants 206-279, 206-266, 244-326, and 263-326 (Fig. 3A). These mutants were designed according to the sequence of RK2 (residues 242 to 283, Fig. 4A). While mutants 206-279 and 244-326 were nuclear, mutants 206-266 and 263-326 were mainly pancellular (Fig. 3B), indicating that residues 244 to 279 are important for the nuclear localization activity. To test whether this region is sufficient, we examined the mutant 244-279 expressed as a GFP fusion protein (Fig. 3A). As shown in Fig. 3B, this mutant was exclusively nuclear, indicating that residues 244 to 279 of HDAC4 are capable of directing GFP to the nucleus. Western blotting analysis with anti-GFP antibody revealed that the deletion mutants used for mapping the NLS were correctly expressed (Fig. 3C). Taken together, these mapping data indicate that residues 244 to 279 of HDAC4 constitute an authentic NLS.
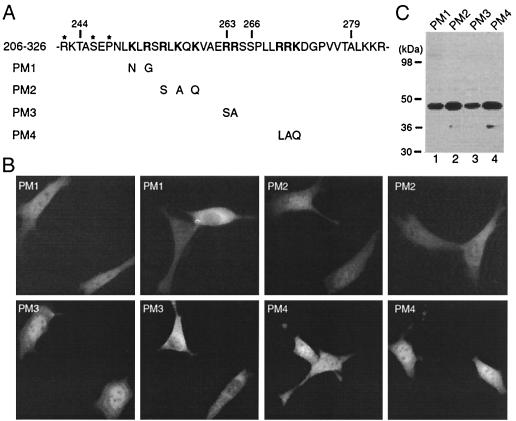
FIG. 4.
Mutational analysis of the NLS. (A) Illustration of mutant 206-326 and its point mutants. The amino acid sequence of residues 242 to 283 of HDAC4 is listed, with potentially important arginine-lysine residues shown in boldface type. Residues important for 14-3-3 binding are labeled with asterisks. The point mutants PM1 to PM4 were derived from the deletion mutant 206-326 by substitution of indicated arginine-lysine residues. (B) Representative green fluorescence images of living NIH 3T3 cells expressing PM1 to PM4 fused to GFP. Cells were transfected with expression plasmids for indicated GFP fusion proteins, and green fluorescence microscopy was performed with living cells. For each mutant, two images are shown to illustrate distinct localization in different cells. (C) Expression of GFP fusion proteins. 293 cells were transfected with expression plasmids for indicated GFP fusion proteins, and cell extracts were analyzed by immunoblotting with anti-GFP antibody.
Point mutational analysis of the HDAC4 NLS.
To further define the NLS, we sought to identify its critical residues. Residues 244 to 279 possess three clusters of arginine-lysine residues (Fig. 4A). To test whether these clusters are required for the nuclear targeting activity, we derived the point mutants PM1 to PM4 from the deletion mutant 206-326 by mutating arginine-lysine residues. GFP fusion proteins were expressed and analyzed by green fluorescence microscopy. As shown in Fig. 4B, unlike mutant 206-326, PM1 to PM4 were pancellular, cytoplasmic, or partially enriched in the nucleus. Western blotting analysis revealed that these point mutants were correctly expressed (Fig. 4C). Therefore, the three clusters of arginine-lysine residues are all necessary for the nuclear localization of mutant 206-326. This also implies that the NLS of HDAC4 is tripartite.
Nuclear export activity of leucine-rich sequences of HDAC4.
We wondered why GFP-TM2 (Fig. 1) was not localized to the nucleus, although it possesses the strong NLS just identified. Cytoplasmic localization of HDAC4 is sensitive to treatment with leptomycin B (LMB) (43, 61). LMB is a specific inhibitor of the nuclear export receptor CRM1 (14, 30, 46, 51), so HDAC4 is subject to active nuclear export. 14-3-3 binding to HDAC4 promotes its cytoplasmic retention (20, 43, 61). 14-3-3 proteins are dimeric, and each monomer is known to possess an active NES (34, 47). Therefore, one explanation for the active nuclear export of HDAC4 is that it binds to 14-3-3 proteins and is subsequently targeted to the cytoplasm through LMB-sensitive nuclear export. Alternatively, HDAC4 may possess an intrinsic NES that directs it to the cytoplasm. Since its 14-3-3 binding sites are impaired, the cytoplasmic localization of GFP-TM2 (Fig. 1) supports the latter possibility. However, this does not exclude the former. With this reasoning in mind, we dissected the underlying mechanisms by which HDAC4 is exported from the nucleus.
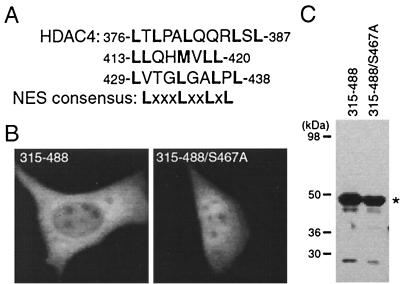
Some leucine-rich sequences are known export signals recognized by the nuclear export receptor CRM1 (45). HDAC4 possesses several leucine-rich sequences (Fig. 5A). In particular, residues 429 to 438 match exactly the NES consensus sequence derived from various known export signals (2). In light of this observation, we expressed and analyzed the HDAC4 mutant 315-488 as a GFP fusion protein. As shown in Fig. 5B, this mutant was partially enriched in the cytoplasm. Since this mutant contains a 14-3-3 binding site, we decided to investigate whether its partial enrichment in the cytoplasm is due to 14-3-3 binding. For this, S467 was substituted with alanine to generate the mutant 315-488/S467A. This mutant was found to be pancellular (Fig. 5B). Without active nuclear import and export, such a localization is expected since this mutant may be able to passively diffuse through nuclear pores (18). Western blotting analysis with anti-GFP antibody indicated that both 315-488 and 315-488/S467A were well expressed (Fig. 5C). Together, these results suggest that the enrichment of mutant 315-488 in the cytoplasm is due to 14-3-3 binding, implying that the leucine-rich sequences of HDAC4 do not have nuclear export activity.
FIG. 5.
Nuclear export activity of leucine-rich sequences of HDAC4. (A) Amino acid sequence of leucine-rich motifs of HDAC4, with leucine and methionine residues shown in boldface type. The consensus sequence of known leucine-rich export signals is also shown, with X denoting any amino acid residue. (B) Representative green fluorescence images of living cells expressing mutant 315-488 and its point mutant fused to GFP. NIH 3T3 cells were transfected with expression plasmids for the mutants, and green fluorescence microscopy was performed with living cells. (C) Expression of GFP fusion proteins. 293 cells were transfected with expression plasmids for GFP-315-488 and -315-488/S467A, and total cell extracts were analyzed by immunoblotting with anti-GFP antibody.
Mapping an NES to the C-terminal end of HDAC4.
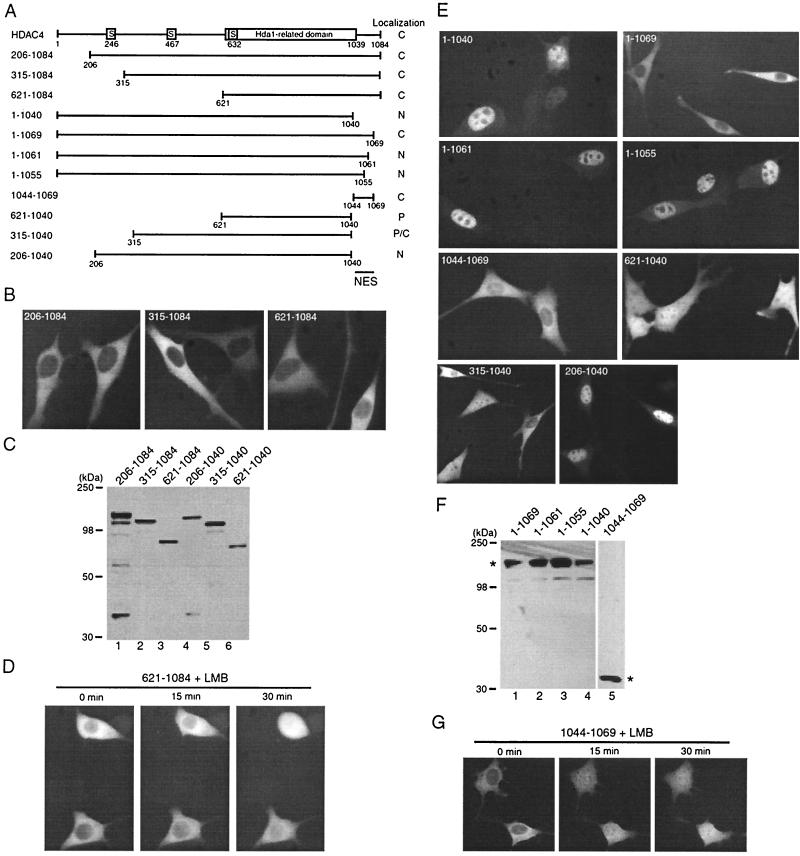
To investigate whether other sequences of HDAC4 may exhibit nuclear export activity, we examined subcellular localization of the deletion mutants 206-1084, 315-1084, and 621-1084 fused to GFP (Fig. 6A). As shown in Fig. 6B, these mutants were predominantly cytoplasmic. Western blotting analysis with anti-GFP antibody indicated that these deletion mutants were expressed as expected (Fig. 6C, lanes 1 to 3). To test whether the S632 14-3-3 binding site contributes to the cytoplasmic localization of mutant 621-1084, we analyzed the mutants 531-1084 and 531-1084/S632A expressed as GFP fusion proteins. Both mutants were found to be cytoplasmic (data not shown), suggesting that 14-3-3 binding is not the major mechanism by which mutant 621-1084 is sequestered to the cytoplasm. The cytoplasmic localization of mutant 621-1084 could be either that it is not imported to the nucleus or that it is subject to active nuclear export. To distinguish between these two possibilities, we utilized the nuclear export inhibitor LMB since HDAC4 is known to be exported in an LMB-sensitive manner (43, 61). As shown in Fig. 6D, LMB treatment inhibited the predominantly cytoplasmic localization of mutant 621-1084, indicating that mutant 621-1084 is subject to active nuclear export. The pancellular localization after LMB treatment is perhaps due to passive diffusion through nuclear pores (18). Therefore, residues 621 to 1084 possess an intrinsic NES.
FIG. 6.
Mapping intrinsic NES of HDAC4. (A) Schematic illustration of HDAC4 and its deletion mutants. Motifs or domains are depicted by boxes as in Fig. 1A. Subcellular localization of HDAC4 and mutants is summarized at right. (B and E) Representative green fluorescence images of living cells expressing HDAC4 and its deletion mutants fused to GFP. NIH 3T3 cells were transfected with expression plasmids for indicated GFP fusion proteins, and green fluorescence microscopy was performed with living cells. (C and F) Expression of GFP fusion proteins. 293 cells were transfected with expression plasmids for indicated GFP fusion proteins, and total cell extracts were analyzed by immunoblotting with anti-GFP antibody. (D and G) Effect of LMB on subcellular distribution of indicated GFP fusion proteins expressed in NIH 3T3 cells. After initial examination for green fluorescence, living cells expressing the indicated fusion proteins were treated with LMB (10 ng/ml), and their green fluorescence images were taken at indicated times. Under similar conditions, LMB had minimal effects on subcellular localization of GFP itself (data not shown).
To map the NES, we first considered whether it is located at the small C-terminal domain (residues 1040 to 1084) since this region is missing in Hda1 (Fig. 6A). This small domain was thus deleted to generate the mutant 1-1040 fused to GFP (Fig. 6A). This fusion protein was predominantly nuclear (Fig. 6E), indicating that the small C-terminal domain is indeed required for cytoplasmic localization of HDAC4. Since this mutant possesses intact 14-3-3 binding sites (20, 61), this exciting finding also indicates that 14-3-3 binding is not sufficient for retaining HDAC4 in the cytoplasm. To further define the small C-terminal domain, small deletions from the C-terminal end were engineered to express the mutants 1-1069, 1-1061, and 1-1055 as GFP fusion proteins (Fig. 6A). The mutant 1-1069 was predominantly cytoplasmic (Fig. 6E), suggesting that residues 1070 to 1084 of HDAC4 are dispensable for its cytoplasmic localization. The two mutants 1-1061 and 1-1055 were predominantly nuclear (Fig. 6E), indicating that residues 1062 to 1069 are essential for cytoplasmic retention of HDAC4. These results also imply that residues 1040 to 1069 of HDAC4 may constitute an NES. To test this hypothesis, we expressed the mutant 1044-1069 as a GFP fusion protein (Fig. 6A). As shown in Fig. 6E, this fusion protein was predominantly cytoplasmic. On the other hand, mutant 621-1040 was pancellular (Fig. 6E). Western blotting analysis with anti-GFP antibody indicated that these deletion mutants were correctly expressed (Fig. 6C, F). Together, these results indicate that residues 1044 to 1069 of HDAC4 function as an NES.
We also examined the mutants 315-1040 and 206-1040 expressed as GFP fusion proteins (Fig. 6A). Different from mutant 315-1084 (Fig. 3B), mutant 315-1040 was pancellular or cytoplasmic (Fig. 6D). Unlike mutant 206-1084 (Fig. 3B), mutant 206-1040 was predominantly nuclear (Fig. 6D). The distinct localization between the mutants 315-1040 and 206-1040 supports the aforementioned conclusion that residues 244 to 279 constitute an NLS (Fig. 3). The pancellular or cytoplasmic localization of mutant 315-1040 is expected since this mutant may be able to passively diffuse into the nucleus through nuclear pores and 14-3-3 binding to S467 and S632 of 315-1040 may promote its active nuclear export. Western blotting analysis with anti-GFP antibody indicated that mutants 206-1040 and 315-1040 were correctly expressed (Fig. 6C, lanes 4 to 5). Together, these results underscore the importance of residues 1044 to 1069 for cytoplasmic retention of HDAC4.
To determine whether residues 1044 to 1069 retain HDAC4 in the cytoplasm by nuclear export, we treated NIH 3T3 cells expressing GFP-1044-1069 with LMB. This fusion protein is small (∼28 kDa) and does not appear to contain an NLS, so it can passively diffuse through nuclear pores (18, 26). Therefore, this fusion protein would be expected to be pancellular if its nuclear export is inhibited by LMB. As shown in Fig. 6G, upon LMB treatment, mutant 1044-1069 became pancellular within 15 min, indicating that nuclear export of mutant 1044-1069 is sensitive to LMB. Therefore, residues 1044 to 1069 constitute an NES whose activity is LMB sensitive. Since LMB is a CRM1-specific inhibitor (14, 30, 46, 51), CRM1 may recognize this NES.
Point mutational analysis of the HDAC4 NES.
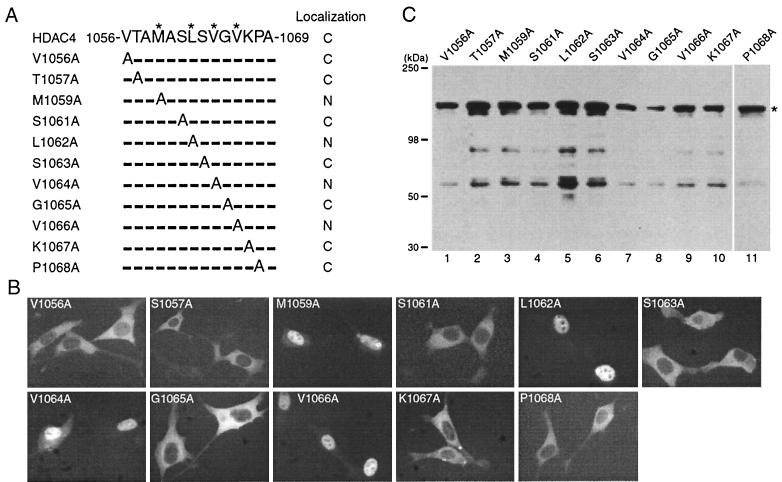
To further define the NES, we sought to identify its critical residues. Sequence inspection revealed that residues 1056 to 1069 constitute a highly hydrophobic motif (Fig. 7A). Since CRM1 is known to recognize leucine-rich or other hydrophobic motifs (45), residues 1056 to 1069 may constitute a functional NES. To determine which residues are important, we performed alanine scanning mutagenesis to generate mutants in which each nonalanine residue was replaced with alanine (Fig. 7A). Fluorescence microscopic analysis of these GFP fusion proteins revealed that substitution of M1059, L1062, V1064, or V1066 of HDAC4 led to nuclear accumulation of the resulting mutants (Fig. 7B). Western blotting analysis with anti-GFP antibody indicated that all point mutants were correctly expressed (Fig. 7C). Together, these results suggest that M1059, L1062, V1064, and V1066 are important for nuclear export of HDAC4. These residues constitute a hydrophobic motif, MXXLXVXV, where X represents any amino acid residue.
FIG. 7.
Mutational analysis of the NES. (A) Amino acid sequences of residues 1056 to 1069 of HDAC4 and mutants. M1059, L1062, V1064, and V1066 of HDAC4 are labeled with asterisks. For the point mutants, substituted and unchanged residues are indicated by the letter A (for alanine) and hyphens, respectively. (B) Representative green fluorescence images of living cells expressing HDAC4 mutants fused to GFP. NIH 3T3 cells were transfected with expression plasmids for indicated GFP fusion proteins, and green fluorescence microscopy was performed with living cells. (C) Expression of GFP fusion proteins. 293 cells were transfected with expression plasmids for indicated GFP fusion proteins, and total cell extracts were analyzed by immunoblotting with anti-GFP antibody. The asterisk at right marks the expected migrating position.
CRM1 directs NES-mediated nuclear export of HDAC4.
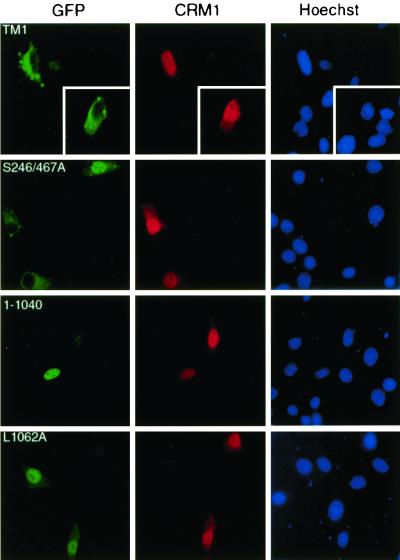
While the NES of HDAC4 is distinct from most known nuclear export sequences recognized by CRM1, its function appeared to be LMB sensitive (Fig. 6). This suggests that the NES may be regulated by CRM1. To substantiate this, we sought to examine directly whether CRM1 can mediate nuclear export of HDAC4 and how the NES is involved. It has been demonstrated that overexpression of CRM1 leads to nuclear exclusion of two transcription factors (17, 71). Since wild-type HDAC4 is mainly cytoplasmic, we tested whether overexpression of CRM1 can lead to nuclear exclusion of TM1 and S246/467A. While TM1 possesses no functional 14-3-3 binding sites, the double mutant S246/467A contains only one functional 14-3-3 binding site (S632 [Fig. 1A]). Both mutants have been found to be predominantly nuclear (61). As shown in Fig. 8, exogenous expression of CRM1 promoted cytoplasmic localization of both mutants, suggesting that CRM1 directs HDAC4 to the cytoplasm. To assess whether the NES of HDAC4 is involved, we examined the mutants 1-1040 and L1062A. 14-3-3 binding sites are intact in both mutants, but the NES is deleted in mutant 1-1040 (Fig. 6) and impaired by point mutation in L1062A (Fig. 7). As shown in Fig. 8, CRM1 overexpression had minimal effects on the nuclear localization of these two mutants, indicating that the NES of HDAC4 is required for its CRM1-mediated nuclear export.
FIG. 8.
Effects of overexpressed CRM1 on subcellular localization of HDAC4 mutants. An HA-CRM1 expression plasmid was transfected into NIH 3T3 cells along with mammalian expression plasmids for HDAC4 mutants fused to GFP. At 16 h after transfection, cells were fixed and stained with antihemagglutinin antibody to detect exogenous CRM1 (middle, red) by indirect immunofluorescence microscopy. Green fluorescence was used to determine subcellular localization of GFP fusion proteins (left) (green). The cells were counterstained with Hoechst 33528 to visualize nuclei (right) (blue). While endogenous CRM1 is enriched around the nuclear envelope, overexpressed CRM1 has been found to be pancellular or nuclear (17, 71).
Direct MEF2 binding targets HDAC4 to the nucleus.
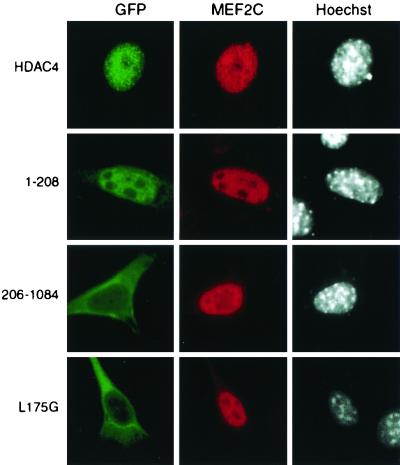
Identification of intrinsic nuclear import and export signals of HDAC4 further supports the notion that it is subject to dynamic nucleocytoplasmic shuttling. Binding of 14-3-3 proteins promotes cytoplasmic localization of HDAC4 by affecting such dynamic shuttling (20, 40, 61). This led us to ask whether association of other proteins also alters this shuttling. It has been shown that the HDAC4 mutant 118-1084 translocates to the nucleus upon exogenous expression of MEF2A in HeLa cells (43). It was not proven, however, whether direct MEF2 binding is required for this nuclear targeting. To further understand how MEF2 may affect intracellular localization of HDAC4, we first tested whether full-length HDAC4 is targeted to the nucleus upon exogenous expression of MEF2C in NIH 3T3 cells. As shown in Fig. 9, coexpression of MEF2C led to nuclear accumulation of GFP-HDAC4. We then asked whether the nuclear targeting of HDAC4 requires its NLS and/or MEF2-binding site. To address this, we assessed whether coexpression of MEF2C affects subcellular localization of the HDAC4 mutants 1-208 and 206-1084. While mutant 1-208 possesses the MEF2-binding site (32, 35, 43, 50, 60), mutant 206-1084 contains the NLS described above. In the absence of exogenous MEF2C, mutant 1-208 was partially enriched in the nucleus (Fig. 3), whereas mutant 206-1084 was predominantly cytoplasmic (Fig. 6). As shown in Fig. 9, expression of MEF2C promoted nuclear accumulation of mutant 1-208 but not mutant 206-1084, suggesting that MEF2 directs HDAC4 to the nucleus in a manner dependent on its MEF2-binding site but not NLS.
FIG. 9.
Effects of exogenous MEF2C on nuclear localization of HDAC4 and mutants. The MEF2C expression plasmid was transfected into NIH 3T3 cells along with mammalian expression plasmids for GFP fusion proteins of HDAC4 or its mutants. At 16 h after transfection, cells were fixed and stained with anti-MEF2C antibody to detect MEF2C by indirect immunofluorescence microscopy (middle) (red). Green fluorescence was used to determine subcellular distribution of GFP fusion proteins (left) (green). The cells were counterstained with Hoechst 33528 to visualize nuclei (right) (white).
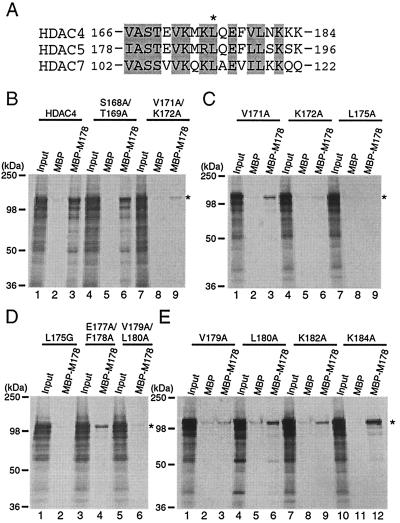
To further investigate whether direct MEF2 binding is essential for the nuclear targeting, we sought to analyze an HDAC4 point mutant that is completely defective in MEF2 binding. For this, we first conducted mutational analysis of the MEF2-binding site of HDAC4 to test whether point mutations can abrogate the MEF2 binding and to identify residues critical for such binding. We and others have located the MEF2-binding site to a small motif conserved among HDAC4, HDAC5, HDAC7, and MITR (Fig. 10A) (32, 35, 43, 50, 60). Mutagenesis was thus performed to substitute potentially important residues of this motif with alanine, and in vitro binding assays were utilized to assess how well each mutant binds to MEF2. For binding assays, M178, an MEF2C mutant containing its N-terminal 178 residues (60), was expressed as an MBP fusion protein. As reported, HDAC4 interacted with MBP-M178 but not MBP itself (Fig. 10B, lanes 1 to 3). Like wild-type HDAC4, the double mutant S168A/T169A interacted with M178 (lanes 4 to 6), indicating that S168 and T169 of HDAC4 are not critical for MEF2 binding. By contrast, the double mutant V171A/K172A was unable to interact with M178 (lanes 7 to 9), suggesting that V171 and/or K172 is important for MEF2 binding. Consistent with this, V171A weakly interacted with M178 (Fig. 10C, lanes 1 to 3), whereas K172A was completely defective in binding to M178 (lanes 7 to 9). Neither L175A (Fig. 10C, lanes 7 to 9) nor L175G (Fig. 10D, lanes 1 to 3) interacted with M178, indicating that L175 of HDAC4 is critical for MEF2 binding. In a similar fashion, V179, L180, and K182 of HDAC4 were found to be involved in interaction with MEF2 (Fig. 10D and E). Among the HDAC4 mutants analyzed, L175G is one whose MEF2-binding ability is completely abolished.
FIG. 10.
Mutational analysis of the MEF2-binding site of HDAC4. (A) Sequence comparison of residues 166 to 184 of HDAC4 with the corresponding regions of HDAC5 and HDAC7. Identical or conserved residues are shaded, and L175 of HDAC4 is indicated by an asterisk. (B to E) Interaction of the MEF2C mutant M178 with HDAC4 and point mutants. MBP or MBP-M178 was immobilized on amylose agarose and tested for interaction with HDAC4 or mutants synthesized in vitro in the presence of [35S]methionine. Bound proteins were separated by SDS-PAGE and subsequent autoradiography. Input represents 20% of the 35S-labeled protein used for each binding assay. Migrating positions of molecular markers are shown at the left of each panel, whereas the positions of HDAC4 and its mutants are indicated by asterisks at right.
We next analyzed GFP-L175G by fluorescence microscopy. Like GFP-HDAC4, GFP-L175G was cytoplasmic (data not shown). As shown in Fig. 9, exogenous expression of MEF2C failed to target this point mutant to the nucleus, supporting that direct MEF2 binding is indeed responsible for nuclear targeting of HDAC4 by MEF2. Along with published reports (20, 41, 43, 61), these results indicate that direct binding of 14-3-3 and MEF2 proteins to HDAC4 affects its dynamic nucleocytoplasmic shuttling and thereby targets it to the cytoplasm and the nucleus, respectively.
DISCUSSION
HDAC4 is known to function as a transcriptional corepressor (32, 35, 43, 50, 60, 69). Its corepressor function is subject to regulation by active nucleocytoplasmic trafficking (20, 43, 61). 14-3-3 proteins bind to HDAC4, sequester it to the cytoplasm, and thereby inhibit its corepressor function (20, 61). The results presented herein demonstrate that besides its 14-3-3 binding sites, HDAC4 possesses additional sequence elements that are also important for controlling its subcellular distribution (Fig. 11A).
FIG. 11.
(A) Model depicting how subcellular localization of HDAC4 is controlled. HDAC4 possesses intrinsic nuclear import and export signals important for its dynamic nucleocytoplasmic shuttling. Association with 14-3-3 or MFE2 proteins modulates the shuttling. 14-3-3 binding promotes cytoplasmic localization of HDAC4 by both inhibiting its nuclear import and stimulating its nuclear export, whereas MEF2 interacts with HDAC4 and targets it to the nucleus. While phosphorylation of S246, S467, and S632 of HDAC4 stimulates the binding of 14-3-3 proteins, it remains less clear how the interaction between MEF2 and HDAC4 is regulated. (B) Sequence comparison of the NLS of HDAC4 with the corresponding regions of HDAC5, MITR and HDAC7. Critical residues of the HDAC4 NLS are boxed with related residues of the other three proteins. (C) Sequence alignment of the NES of HDAC4 with the related regions of HDAC5 and HDAC7. Critical residues of the HDAC4 NES are boxed with corresponding residues of HDAC5 and HDAC7.
The N-terminal 118 residues of HDAC4 modulate its nuclear localization.
The N-terminal 118 residues play a contributing role in regulating nuclear localization of HDAC4 (Fig. 1). Residues 90 to 142 of HDAC4 show limited sequence similarity to MAG1 (Fig. 1A) (7, 63). The distinct subcellular localization between GFP-TM3 and -TM4 (Fig. 1) suggests that residues 85 to 105 are important for nuclear localization of HDAC4. GFP-TM4 was nuclear in 40 to 50% of expressing cells (data not shown); the N-terminal 85 residues also modulate subcellular localization of HDAC4.
How do the N-terminal 118 residues modulate subcellular localization of HDAC4? This region may not be involved in intra- or intermolecular interaction with HDAC4 molecules (Fig. 2). Moreover, neither HDAC1 nor CtBP appeared to modulate subcellular localization of HDAC4 (data not shown). Although the N-terminal 118 residues exhibited weak nuclear targeting activity (Fig. 3), this region does not appear to possess a classical arginine-lysine-rich NLS. This region may contain a novel NLS. Alternatively, this region may interact with a protein that awaits to be identified.
In agreement with the latter possibility, a transcriptional repression domain has been mapped to this region (50, 60, 69). When tethered, the N-terminal 208 residues of HDAC4 function as a strong, active transcriptional repression domain. However, neither the N-terminal 118 residues nor residues 118 to 620 are able to repress transcription (60), suggesting that the N-terminal 118 residues are necessary but not sufficient for transcriptional repression. These findings suggest that this repression domain of HDAC4 may interact with an unidentified nuclear protein. Involvement of the N-terminal 118 residues in such binding may explain the role in modulating subcellular localization of HDAC4. It will be interesting to identify this elusive protein and study its role in regulating subcellular localization and function of HDAC4.
Tripartite nuclear import signal of HDAC4.
Residues 244 to 279 of HDAC4 constitute a functional NLS (Fig. 11A). Mutational analysis of this NLS revealed that three clusters of arginine-lysine residues are necessary for its nuclear import activity (Fig. 4). Such a tripartite organization is distinct from known monopartite or bipartite nuclear import sequences (10, 26, 45). It is noteworthy that the HDAC4 mutant 206-326 was found to be nuclear, although it still possesses an intact 14-3-3 binding site (S246, Fig. 3). Therefore, this NLS is unique and strong. Since it is arginine-lysine rich, it can be recognized by importin α. Consistent with this, HDAC4 has been found to interact with importin α (20).
Shown in Fig. 11B is the sequence comparison of the HDAC4 NLS with the corresponding regions of HDAC5, MITR, and HDAC7. The NLS of HDAC4 is highly conserved among these three proteins, suggesting that their corresponding regions may constitute authentic nuclear import signals. Consistent with this, an HDAC5 fragment containing the putative NLS has been very recently shown to possess strong nuclear localization activity (40). Further experiments are needed to verify the putative import signals of MITR and HDAC7.
Hydrophobic nuclear export signal of HDAC4.
Deletion and point mutational analyses revealed that while leucine-rich sequences of HDAC4 do not exhibit nuclear export activity (Fig. 5), a hydrophobic motif (MXXLXVXV) located at its C-terminal end functions as an NES (Fig. 11A). Alanine substitution of the four critical residue led to nuclear accumulation of the resulting mutants (Fig. 7). These mutants possess all three 14-3-3 binding sites (20, 61), so they are presumably able to interact with 14-3-3 proteins. Therefore, besides the three 14-3-3 binding sites, the NES is also required for cytoplasmic retention of HDAC4.
This NES is different from most leucine-rich export signals identified in other proteins (45). However, the NES of cyclin B contains only one leucine: LXXXFXXVXI, where X represents any amino acid residue (39). Although most known export signals are binding sites of CRM1 (45), CRM1-independent protein nuclear export pathways have also been found (25, 33). Cytoplasmic localization of HDAC4 is sensitive to LMB, a known CRM1-specific inhibitor (14, 30, 46, 51), so CRM1 may be involved in its nuclear export. The nuclear export function of residues 1040 to 1069 is LMB sensitive (Fig. 6G), suggesting that CRM1 recognizes this NES. Consistent with this, we found that HDAC4 and CRM1 functionally interact in vivo and that such interaction requires the NES of HDAC4 (Fig. 8). We also conducted pulldown and coimmunoprecipitation assays to analyze the physical interaction between HDAC4 and CRM1 in vitro or in vivo. Various efforts failed to verify this (data not shown), suggesting that the physical interaction may be transient or too weak to be easily detected.
Illustrated in Fig. 11C is the sequence comparison of the HDAC4 NES with the corresponding regions of HDAC5 and HDAC7. While HDAC7 possesses LXXLXVXI, HDAC5 contains MXXLXVXA. It has been reported that HDAC5 is mainly nuclear in several cell lines (27, 32, 40). We also found that unlike GFP-HDAC4, GFP-HDAC5 was mainly nuclear in NIH 3T3 cells (data not shown). Interestingly, A1096 of HDAC5 corresponds to V1066 of HDAC4, and the point mutant V1066A of HDAC4 was predominantly nuclear (Fig. 4). Besides these distinctions, there are other differences between HDAC4 and HDAC5. First, HDAC5 has been very recently reported to possess an NES within the deacetylase domain (40). The corresponding region of HDAC4 does not appear to be an NES since the mutant 621-1040 was pancellular (Fig. 6E). Second, although HDAC4 and HDAC5 are homologous (overall amino acid sequence identity, 62%; similarity, 69%), their sequences are quite divergent in some regions (13, 19, 43, 59, 60). Third, unlike S632 of HDAC4, S661 of HDAC5 does not mediate 14-3-3 binding (20, 41, 61). Finally, while 14-3-3 binding to HDAC4 is constitutive in most cells tested, 14-3-3 binding to HDAC5 is dependent on activation of Ca2+/calmodulin-dependent kinases (20, 41, 61). Therefore, nuclear export of HDAC4 and HDAC5 seems to be differentially regulated.
Both 14-3-3 and MEF2 proteins regulate intracellular localization of HDAC4.
Mapping the NLS and NES of HDAC4 also shed light on how 14-3-3 proteins regulate its subcellular localization. First, the HDAC4 mutant 206-326 was exclusively nuclear, although it has an intact 14-3-3 binding site (S246, Fig. 3). Second, while mutant 315-488 was enriched in the cytoplasm, mutant 315-488/S467A was pancellular (Fig. 5). Third, deletion of residues 1040 to 1084 or alanine substitution of the critical residues of the NES led to nuclear accumulation of the resulting mutants, although these mutants still contain all three 14-3-3 binding sites (Fig. 6 and 7). Finally, the triple mutant TM1 was found to be mainly nuclear (Fig. 1) (20, 61), although its NES remains intact. Taken together, these findings indicate that both 14-3-3 binding and nuclear export mediated by the NES are required for cytoplasmic retention of HDAC4.
How does 14-3-3 binding promote cytoplasmic retention of HDAC4? The NLS of HDAC4 is only two residues away from the S246 14-3-3 binding site (Fig. 4A), so 14-3-3 binding to S246 may mask the NLS and thereby inhibit its nuclear targeting activity. Consistent with this, 14-3-3 binding has been found to interfere with the association of importin α with HDAC4 (20). 14-3-3 binding to S246 of HDAC4 may inhibit access of importin α to the NLS. Therefore, one mechanism by which 14-3-3 proteins negatively regulate nuclear localization of HDAC4 operates through direct inhibition of importin α binding to HDAC4. Substitution of S246 alone was found to be insufficient to alter cytoplasmic localization of HDAC4 (20, 61), so additional mechanisms may be involved. The distinct localization of mutants 315-488 and 315-488/S467A (Fig. 5) suggests that 14-3-3 binding also stimulates nuclear export of HDAC4. Since each 14-3-3 protein is known to contain an NES (34, 47), binding of dimeric 14-3-3 proteins to S467 of HDAC4 may provide an active NES. Therefore, as previously proposed (61), 14-3-3 binding may promote cytoplasmic retention of HDAC4 by both inhibiting its nuclear import and stimulating its nuclear export. Similar modes of action may also apply to some of the other 14-3-3 binding partners (16, 44, 64).
Besides 14-3-3 proteins, MEF2 binds to HDAC4 and affects its subcellular localization (Fig. 9) (43). MEF2 was able to direct the mutant 1-208 to the nucleus, although this mutant does not possess the NLS of HDAC4. Mutational analysis of the MEF2-binding site further supports that direct binding of MEF2 promotes nuclear import of HDAC4. Therefore, HDAC4 possesses multiple sequence elements controlling its subcellular localization (Fig. 11A). The intrinsic nuclear import and export signals of HDAC4 dictate its active shuttling between the cytoplasm and the nucleus. Such shuttling leads to a distribution equilibrium. Association of other proteins with HDAC4 then shifts this equilibrium towards the nucleus or the cytoplasm. Indeed, direct binding of 14-3-3 and MEF2 proteins to HDAC4 leads to its cytoplasmic and nuclear localization, respectively.
Cell signaling may regulate HDAC4 through controlling its interaction with 14-3-3 and MEF2 proteins. 14-3-3 binding motifs are putative phosphorylation sites of cyclic AMP- or Ca2+-calmodulin-dependent protein kinases, so these kinases may phosphorylate HDAC4, regulate its association with 14-3-3 proteins, and thereby affect its subcellular localization. Consistent with this, Ca2+-calmodulin-dependent kinases have been shown to phosphorylate HDAC5, stimulate binding of 14-3-3 proteins and regulate its nuclear export (40). Ca2+-calmodulin-dependent signaling has also been found to regulate MEF2 binding to HDAC4 (35, 40, 67). Since binding of MEF2 to HDAC4 leads to its nuclear localization (Fig. 9) (43), Ca2+-calmodulin-dependent signaling may regulate subcellular localization of HDAC4 through modulating its interaction with MEF2. The recent finding that oncogenic Ras stimulates localization of HDAC4 to the nucleus also supports that its subcellular distribution is regulated by cell signaling (70).
In summary, HDAC4 possesses an NLS and an NES for its dynamic shuttling between the cytoplasm and the nucleus. Direct binding of 14-3-3 and MEF2 proteins alters such shuttling and targets HDAC4 to the cytoplasm and the nucleus, respectively. The N-terminal 118 residues of HDAC4 affect its intracellular localization perhaps through interacting with an unidentified nuclear protein. Further investigation of multiple mechanisms through which cell signaling pathways modulate subcellular localization of HDAC4 shall shed light on how different deacetylases are differentially regulated in vivo.
ACKNOWLEDGMENTS
We thank S. Khochbin and E. N. Olson for sharing results about subcellular localization of HDAC4 and HDAC5, C. M. Grozinger and S. L. Schreiber for HDAC5 expression plasmids, M. Yoshida for leptomycin B, C. Dargemont and R. Lin for CRM1 cDNA, D. Gorlich for RanQ69L expression plasmid, J. Han for anti-MEF2C antibody, J. J. LeBrun for use of a FluoChem imaging system, and M. Park and S. Stifani for use of fluorescence microscopes. A.H.W. is the recipient of a Canadian Institutes of Health Research (CIHR) doctoral research award.
This work was supported by grants from the National Cancer Institute of Canada and a scholarship from the CIHR (to X.-J.Y.).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown C E, Lechner T, Howe L, Workman J L. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 4.Buggy J J, Sideris M L, Mak P, Lorimer D D, McIntosh B, Clark J M. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem J. 2000;350:199–205. [PMC free article] [PubMed] [Google Scholar]
- 5.Champagne N, Bertos N R, Pelletier N, Wang A H, Vezmar M, Yang Y, Heng H H, Yang X J. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J Biol Chem. 1999;274:28528–28536. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- 6.Champagne N, Pelletier N, Yang X J. The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene. 2001;20:404–409. doi: 10.1038/sj.onc.1204114. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y S, Patterson C E, Staeheli P. Interferon-induced guanylate-binding proteins lack an N(T)KXD consensus motif and bind GMP in addition to GDP and GTP. Mol Cell Biol. 1991;11:4717–4725. doi: 10.1128/mcb.11.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cress W D, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 9.Dangond F, Hafler D A, Tong J K, Randall J, Kojima R, Utku N, Gullans S R. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 10.Dingwall C, Laskey R A. Nuclear targeting sequences-a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 11.Dressel U, Bailey P J, Wang S C, Downes M, Evans R M, Muscat G E. A dynamic role for HDAC-7 in MEF2 mediated muscle differentiation. J Biol Chem. 2001;276:17361–17366. doi: 10.1074/jbc.M101508200. [DOI] [PubMed] [Google Scholar]
- 12.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischle W, Emiliani S, Hendzel M J, Nagase T, Nomura N, Voelter W, Verdin E. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 14.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 15.Frye R A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 16.Fu H, Subramanian R R, Masters S C. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 17.Gaubatz S, Lees J A, Lindeman G J, Livingston D M. E2F4 is exported from the nucleus in a CRM1-dependent manner. Mol Cell Biol. 2001;21:1384–1392. doi: 10.1128/MCB.21.4.1384-1392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorlich D. Transport into and out of the nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grozinger C M, Hassig C A, Schreiber S L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grozinger C M, Schreiber S L. Regulation of histone deacetylase 4 and 5 transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 22.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang G F, Johanson K, Sung C M, Liu R, Winkler J. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 23.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 24.Imai S I, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 25.Kaffman A, Rank N M, O'Neill E M, Huang L S, O'Shea E K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 26.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear localization. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 27.Kao H Y, Downes M, Ordentlich P, Evans R M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 28.Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D. Functional significance of histone deacetylase diversity. Curr Opin Genet Dev. 2001;11:162–166. doi: 10.1016/s0959-437x(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner E P, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemercier C, Verdel A, Galloo B, Curtet S, Brocard M P, Khochbin S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J Biol Chem. 2000;275:15594–15599. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- 33.Lipowsky G, Bischoff F R, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Gorlich D. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 2000;19:4362–4371. doi: 10.1093/emboj/19.16.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 35.Lu J, McKinsey T A, Nicol R L, Olson E N. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci USA. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J, McKinsey T A, Zhang C L, Olson E N. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 37.Maroun C R, Moscatello D K, Naujokas M A, Holgado-Madruga M, Wong A J, Park M. A conserved inositol phospholipid binding site within the pleckstrin homology domain of the Gab1 docking protein is required for epithelial morphogenesis. J Biol Chem. 1999;274:31719–31726. doi: 10.1074/jbc.274.44.31719. [DOI] [PubMed] [Google Scholar]
- 38.McBlane F, Boyes J. Stimulation of V(D)J recombination by histone acetylation. Curr Biol. 2000;10:483–486. doi: 10.1016/s0960-9822(00)00449-8. [DOI] [PubMed] [Google Scholar]
- 39.McBride K, McDonald C, Reich N C. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 2000;19:6196–6206. doi: 10.1093/emboj/19.22.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKinsey T A, Zhang C L, Lu J, Olson E N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinsey T A, Zhang C L, Olson E N. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci USA. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMurry M T, Krangel M S. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 43.Miska E A, Karlsson C, Langley E, Nielsen S J, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muslin A J, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 45.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 46.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 47.Rittinger K, Budman J, Xu J, Volinia S, Cantley L C, Smerdon S J, Gamblin S J, Yaffe M B. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 48.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sparrow D B, Miska E A, Langley E, Reynaud-Deonauth S, Kotecha S, Towers N, Spohr G, Kouzarides T, Mohun T J. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 52.Sterner D E, Berger S L. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart S, Crabtree G R. Regulation of the regulators. Nature. 2000;408:46–47. doi: 10.1038/35040690. [DOI] [PubMed] [Google Scholar]
- 54.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 55.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 56.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 57.Van de Wyngaert I, de Vries W, Kremer A, Neefs J, Verhasselt P, Luyten W H, Kass S U. Cloning and characterization of human histone deacetylase 8. FEBS Lett. 2000;478:77–83. doi: 10.1016/s0014-5793(00)01813-5. [DOI] [PubMed] [Google Scholar]
- 58.Verdel A, Curtet S, Brocard M P, Rousseaux S, Lemercier C, Yoshida M, Khochbin S. Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 59.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 60.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Th'ng J, Han J, Yang X-J. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang A H, Kruhlak M J, Wu J, Bertos N R, Vezmar M, Posner B I, Bazett-Jones D P, Yang X-J. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolffe A P, Matzke M A. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 63.Wynn T A, Nicolet C M, Paulnock D M. Identification and characterization of a new gene family induced during macrophage activation. J Immunol. 1991;147:4384–4392. [PubMed] [Google Scholar]
- 64.Yaffe M B, Elia A E H. Phosphoserine/threonine-binding domains. Curr Opin Cell Biol. 2001;13:131–138. doi: 10.1016/s0955-0674(00)00189-7. [DOI] [PubMed] [Google Scholar]
- 65.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 67.Youn H D, Grozinger C M, Liu J O. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J Biol Chem. 2000;275:22563–22567. doi: 10.1074/jbc.C000304200. [DOI] [PubMed] [Google Scholar]
- 68.Zhang C L, McKinsey T A, Lu J, Olson E N. Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J Biol Chem. 2000;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 69.Zhou X, Richon V M, Rifkind R A, Marks P A. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc Natl Acad Sci USA. 2000;97:1056–1061. doi: 10.1073/pnas.97.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou X, Richon V M, Wang A H, Yang X J, Rifkind R A, Marks P A. Histone deacetylase 4 associates with extracellular signal-regulated kinases 1 and 2, and its cellular localization is regulated by oncogenic Ras. Proc Natl Acad Sci USA. 2000;97:14329–14333. doi: 10.1073/pnas.250494697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu J, McKeon F. NF-AT activation requires suppression of Crm1-dependent export by calcineurin. Nature. 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]