Abstract
The punc gene, encoding a member of the neural cell adhesion molecule family expressed in the developing central nervous system, limbs, and inner ear, was identified. To extend studies of the normal expression pattern of punc and to determine its function, a mouse strain bearing a lacZ/neo insertion in a 5′ coding exon was created. The complex pattern of punc expression in embryos from embryonic day 9.5 (E9.5) to E11.5 was mimicked accurately by β-galactosidase (β-Gal) activity. As development proceeded, the distribution of β-Gal activity was increasingly restricted, finally becoming confined to the brain and inner ear by E15.5. In the adult, β-Gal activity was detected in several regions of the inner ear and brain and was particularly strong in the cerebellar Bergmann glia. Genetic analysis of this null allele demonstrated that punc is not required for normal embryogenesis. Interestingly, comparisons of β-Gal activity and punc transcripts in heterozygous and homozygous mutant individuals demonstrated that punc is negatively autoregulated in some tissues. Adult punc-deficient mice were overtly normal and had normal hearing. Compared with control littermates, however, homozygous mutants had significantly reduced retention times on the Rotarod, suggesting a role for Bergmann glia-expressed Punc in the cerebellar control of motor coordination.
Neural cell adhesion molecules (NCAMs) play roles in both the developing and the adult central nervous system (CNS) NCAMs have a single-pass transmembrane domain or are anchored to the cell surface via a glycosylphosphatidyl inositol linkage. Their extracellular domains contain variable numbers of immunoglobulin (Ig) and fibronectin type III (FNIII) repeats, and their intracellular domains are noncatalytic, distinguishing them from the receptor protein tyrosine kinase and phosphatase subfamilies. NCAMs participate in homophilic and/or heterophilic binding interactions and, in some cases, soluble or matrix-bound ligands have been identified. Despite the fact that NCAMs do not have catalytic activity, they are thought to participate in signaling either by extracellular interactions with catalytic subfamily members or by intracellular interactions with signal adaptors (39, 41).
NCAMs have complex, dynamic, and overlapping expression patterns during nervous system development. Many family members are also expressed widely in the adult. Thus, the expression and adhesive interaction data for a given family member can only provide clues to its function. Characterization of the phenotypes of mutants has been invaluable in determining the functions of these genes. To date, the phenotypes of individuals bearing mutations in the genes encoding four different NCAMs (NCAM, L1, DCC, and contactin) have been reported in vertebrates (1, 5, 6, 9, 10, 15, 17, 27, 37, 38, 45). Targeted ablation of these molecules substantiated other evidence that they function during development in axon guidance, fasciculation, and cell migration and linked specific developmental events to particular NCAMs. In contrast to the L1, DCC, and contactin mutants and despite widespread NCAM expression, NCAM-deficient mice have relatively mild abnormalities. This outcome allowed studies of its roles in nervous system function in the adult, which suggested that NCAM is required for one form of long-term potentiation in the hippocampus (4).
The punc gene, which encodes a new NCAM, was identified in a differential display screen of mRNA isolated from transgenic mice that overexpress the Islet-1 transcription factor. punc transcripts were found predominantly in the developing limbs and spinal cord, where its expression level was inversely correlated with that of islet-1. With four extracellular Ig domains and two FNIII repeats, Punc defined a new class within the neural Ig superfamily (31).
Here we describe studies aimed at determining the function of punc. We identified an embryonic stem (ES) cell line with a gene-trap insertion in punc. Expression of punc in the developing inner ear and localization of the mouse and human genes to syntenic regions of chromosomes 9(40) and 15q22, respectively prompted evaluation of PUNC as a candidate for the recessive deafness locus, DFNB16. Radiation hybrid mapping of PUNC relative to DFNB16 flanking markers showed that PUNC lies outside of the DFNB16 critical region and is unlikely to be responsible for this disorder. To facilitate studies of punc expression and determine its function, we generated a mouse strain with a lacZ insertion in punc. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining revealed that punc expression is more complex than previously appreciated during mid-gestation. punc expression becomes progressively restricted to the brain and inner ear during late gestation and is maintained in these tissues in the adult. Comparisons of β-Galactosidase (β-Gal) activity and punc transcript levels in heterozygous and homozygous mutant individuals showed that Punc has a tissue-specific role in negatively regulating the level of its own transcript. punc-deficient mice were viable and fertile but had subtly impaired motor coordination that could be correlated with expression of punc in the Bergmann glia of the adult cerebellum.
MATERIALS AND METHODS
Gene trapping and 5′-RACE (rapid amplification of cDNA ends).
The gene trap vector, pGTV1 (44), was modified by inserting an additional 51 bp of the adenovirus 2 splice acceptor upstream of the minimal acceptor sequence present in pGTV1 and by deleting pBluescript KS(+) sequences between the NotI and the proximal SspI sites to create pGTV2. The gene trap cell line, 24-B9, was isolated following electroporation of 106 R1 ES cells (26) with 25 μg of pGTV2 and selection in medium containing 350 μg of G-418 (Life Technologies) per ml. Aliquots of 24-B9 cells were tested for β-Gal activity before and after the application of thyroid hormone, nerve growth factor, and retinoic acid as described previously (44).
Total RNA was isolated from undifferentiated 24-B9 ES cells using TRIzol (Life Technologies) according to the manufacturer's protocol. One microgram of 24-B9 RNA was used as template for reverse transcription (RT) and PCR amplification of the 5′ ends of lacZ-containing RNAs (43). The PCR products were cloned and sequenced by standard methods (21). To isolate additional sequence from the trapped gene, a primer (SLM111, 5′-ACTATGAATGTGTGGCTCAG-3′) was designed from the novel 5′-RACE sequence and used together with a λgt11 primer (SLM103, 5′-AGACCAACTGGTAATGGTAGC-3′) to amplify a 1.2-kb cDNA fragment from an embryonic day 11.5 (E11.5) mouse cDNA library (Clontech). This DNA fragment was cloned into pBluescript KS(−), and sequence analysis showed no differences relative to the coding sequence of punc found in GenBank accession no. AF026465, nucleotides 605 to 1748 (31). An alternatively spliced exon, located at the 5′ end of punc, was identified by additional screening of the E11.5 cDNA library (GenBank accession no. AF236125). Signal peptides were predicted using the program PSORT (http://psort.nibb.ac.jp/).
Mapping of punc. (i) Mouse.
A 460-bp NcoI-to-SmaI DNA fragment (Identical to nucleotides 748 to 1203 of GenBank accession no. AF026465) isolated from the punc open reading frame was hybridized with nylon filters carrying TaqI-digested DNA samples from the Jackson Laboratory BSS Backcross panel (29). This probe detected a DNA fragment of 7.5 kbp in C57BL/6J DNA and one of 7.8 kbp in M. spretus DNA. The resulting strain distribution data were analyzed using MapMaker software (http://www.jax.org/resources/documents/cmdata).
(ii) Human.
A human expressed sequence tag (EST) with 77% sequence identity to the 3′ untranslated region (UTR) of mouse punc was identified (GenBank accession no. AA860555). To confirm that this EST was derived from human PUNC, 5′-RACE was performed on human fetal brain cDNA (kindly provided by Mark Keating, Howard Hughes Medical Institute) using a primer designed according to the EST sequence (SLM218, 5′-GAAGAAGATAGAATTTGTGGTG-3′). Sequence analysis of the largest 5′-RACE clone showed that the human 3′ UTR was preceded by 81 bp encoding 27 amino acids. This coding region had 89% nucleotide sequence identity with mouse punc. Two primers were designed according to the human PUNC sequence (forward, 5′-GAAGAAGATAGAATTTGTGGTG-3′; reverse, 5′-AGTCAAAATGAGCAGAGTGTG-3′). A 260-bp fragment was produced by PCR amplification of human DNA as the template but was not detected with hamster DNA as the template. Chromosomal assignment was determined using the NIGMS human-rodent somatic cell hybrid mapping panel 1 (kindly provided by Mark Keating). Concordance-discordance analysis of the PCR results placed PUNC on chromosome 15. Finer mapping of PUNC was obtained by screening the Stanford G3 Radiation Hybrid Mapping Panel (Research Genetics). DNAs from 83 hybrid clones were analyzed by PCR to detect those that contained the human PUNC sequence. Two DFNB16-flanking markers, D15S1039 and D15S155 (2), were also mapped to the same panel using PCR conditions suggested by Research Genetics. The markers were localized by two-point maximum likelihood analysis (http://wwwshgc.stanford.edu/Mapping/rh/search.html).
Gene targeting.
A 19-kbp DNA fragment containing exon 2 of the punc gene was isolated from a λFixII library prepared from mouse strain 129/Sv genomic DNA (Stratagene). The coding sequence of punc was disrupted by insertion into the MscI site of exon 2 of a lacZ gene that carried a consensus Kozak translation initiation codon and nuclear localization signal and was followed by a PGKneobpA cassette (35). LoxP sites flanked the PGKneobpA cassette. A short stretch of synthetic oligonucleotides (5′-GGCCGCTAAGTGAGTAAGCCGCCCGCC-3′) was placed in front of the lacZ sequence to ensure the closure of all three possible reading frames and to prevent production of a signal sequence–β-Gal fusion protein that might not have enzymatic activity (34). The final targeting vector contained 5.5 kbp of punc DNA upstream of the disruption in exon 2 and 3 kbp of punc DNA downstream of the disruption and was flanked by two thymidine kinase (TK) expression cassettes (kindly provided by Kirk Thomas, University of Utah). A total of 25 μg of the linearized vector was introduced into 106 R1-45 mouse ES cells, which were grown in medium containing 380 μg of G418 per ml and 2 μM ganciclovir (23). Correctly targeted cell lines were identified using Southern blot hybridization analysis of DNA. The 5′-flanking probe was a 450-bp DNA fragment that was PCR amplified from genomic DNA using the primers SLM185 (5′-CTAGGAAACCTCTCCCTATG-3′) and SLM207 (5′-GATAATCGAGCAAGATGACATG-3′). The 3′-flanking probe was a 500-bp DNA fragment that was amplified from genomic DNA using primers SLM186 (5′-GCAATGTAAGGAATTGAGCTG-3′) and SLM208 (5′-TCAACCCTCACACTATGAGC-3′). The neo probe was a 630-bp PstI-to-XbaI fragment of pPGKneobpA. Thirty-three percent of the drug-resistant clones carried the targeted allele. Cells from three correctly targeted lines were diluted to 0.5 × 105/ml and used to generate germ line chimeras by the one-step coculture method (19). The PGKneobpA cassette was removed from the targeted allele by mating heterozygous puncLN mice with a strain that expresses CRE in the germ line (33). The absence of PGKneobpA sequences in the resulting offspring was confirmed by PCR analysis using internal primers that amplify a 295-bp fragment in heterozygous DNA samples (SLM10, 5′-GCCTGCTTGCCGAATATCATGG-3′; SLM74, 5′-AAACAACAGATGGCTGGCAAC-3′). In addition, carriers of the CRE transgene were identified by PCR screening with CRE-specific primers (tCreF1, 5′-GGATTTCCGTCTCTGGTGTAGC-3′; tCreR1, 5′-ACCATTGCCCCTGTTTCACTATC-3′) and were not used in propagating puncL mice.
Genotyping of mice.
A PCR assay was used to genotype offspring of germ line chimeras and of intercrosses between heterozygotes. The primers (SLM24 and SLM25) that amplified a 333-bp fragment of the lacZ allele have been described previously (43). The forward primer for the wild-type punc allele was SLM136 (5′-AAATGATGATATTGCCAACCC-3′). The reverse primer for the wild-type allele was SLM137 (5′-CTACCTCCTTGCTCCGCC-3′). These punc primers amplified a fragment of 180 bp.
Northern blotting.
Total RNA was isolated from tissues or staged embryos as described above. mRNA was purified using an Oligotex mRNA isolation kit (Qiagen). Five micrograms of each mRNA sample was separated in a formaldehyde-agarose gel, transferred to nylon membranes, and hybridized with 32P-labeled probes as described earlier (43). The 3′ punc probe was the 1.2-kb cDNA fragment described above, and the 5′ punc probe was a 164-bp cDNA fragment obtained by PCR using forward primer SLM179 (5′-GGTCTGGGCCATTCTGCTG-3′) and reverse primer SLM180 (5′-GTGGTGTGGGTGCCCTCTG-3′). The simian virus 40 (SV40) probe was prepared from viral DNA. Hybridization with a chicken β-actin cDNA fragment served as a loading control. punc transcript levels were quantified by densitometry (Personal Densitometer SI; Amersham Pharmacia Biotech) of the hybridization signals on the X-ray films and normalized to β-actin hybridization signals.
In situ hybridization.
A 540-bp fragment of punc cDNA (nucleotides 603 to 1203 of GenBank accession no. AF026465) was cloned into pBluescript vectors KS(−) and SK(−). Sense and antisense digoxigenin-labeled riboprobes were synthesized using T7 RNA polymerase from the respective vectors. Whole E9.5 and E10.5 embryos were processed for in situ hybridization as described elsewhere (14) and photographed. Stained embryos were then embedded in gelatin and cryostat sectioned at 10 μm as described earlier (36).
X-Gal staining and immunohistochemistry.
To localize β-Gal activity in embryos, heterozygotes were intercrossed, and all of the littermates from each pregnant female were fixed and stained with X-Gal under identical conditions and photographed as described previously (43). All embryos at E14.5 and older were manually hemisected halfway through the fixation step, and all staining was terminated before any background signal was detected in wild-type embryos. For the puncLN strain, we examined two to four homozygous mutants and four to eight heterozygotes at each day of development between E8.5 and P0, except E16.5, which was omitted from the study. For the puncL heterozygous intercrosses, we examined two to four homozygotes and four to eight heterozygous embryos at E11.5, E13.3, and E15.5 only.
To detect β-Gal activity in the adult animals were anesthetized and perfused with the same fixative used for embryos. Most tissues were then dissected and stained as for the embryos. To permit access of stain to all areas of the brain, this tissue was cut into 1-mm slices using a mouse brain mold (Braintree Scientific) and then incubated with X-Gal as described for the embryos (43). To ensure the access of all reagents to the dissected inner ear, this tissue was manually perfused through the round window using a tuberculin syringe. X-Gal staining of adult tissues was performed on two individuals of each genotype for each punc strain. To examine expression patterns in more detail, embroys, brain slices, and inner ears were postfixed, dehydrated, embedded in paraffin, sectioned at 10 μm, and counterstained with eosin as described previously (44).
For glial fibrillary acidic protein (GFAP) immunohistochemistry, X-Gal-stained puncL brain slices were washed with phosphate-buffered saline (PBS), equilibrated in 15% sucrose at 4°C overnight, and embedded with OCT compound. Then, 10-μm cryosections were collected onto slides (ProbeOn Plus; Fisher Scientific), fixed in cold methanol for 10 min, and incubated in cold ethanol for another 10 min. The slides were then washed with PBS and blocked with 4% horse serum for 1 h at room temperature before the addition of antibodies directed against GFAP (Boehringer-Mannheim) diluted 1:100 in PBS with 4% horse serum. The sections were incubated with antibody for 3 h at room temperature and washed with PBS prior to incubation for 1 h at room temperature with peroxidase-conjugated goat anti-mouse IgG (Jackson ImmunoResearch) diluted 1:500 in PBS. Finally, the sections were washed three times in PBS and developed in diaminobenzidene (DAB) (50 μg/ml in PBS) with 0.0015% H2O2.
Behavioral phenotyping. (i) General.
Wild-type, heterozygous, and homozygous mutant mice used for behavioral tests were obtained by intercrossing heterozygotes. The parental puncLN heterozygotes were F1 offspring of the original germ line chimeric mice. The parental puncL heterozygotes were offspring of the F1 animals crossed to the CRE-deleter strain (33). Animals were tested at approximately 6 to 8 weeks of age in a procedure room separate from their home cage room.
(ii) Motor coordination.
Mice were placed on a Rotarod (diameter, 3.2 cm; Accelerating Rota-Rod for Mice 7650; Biological Research Apparatus), while it was turning at a fixed speed. Female mice were tested at 27 rpm, while males were tested at 31 rpm (maximum speed). These speeds were chosen based on preliminary testing of wild-type mice. Use of the accelerating mode (0 to 31 rpm, maximum acceleration setting) for the Rotarod was unsatisfactory because wild-type mice could stay on the rod for the full test period at the first trial. Thus, we sought to determine a fixed speed at which a few mice could remain on the rod for only a few seconds at the first trial. This condition allowed discrimination of differing abilities over time. Retention time on the rod was recorded in seconds, up to a maximum of 300 during two trials per day for five consecutive days (20). The test population was composed of 15 female and 19 male wild types, 17 female and 19 male heterozygotes, and 17 female and 19 male homozygous mutants.
The main objective of the statistical data analysis was to determine whether genotype had any effect on the retention time. Since the response variable was a time to an event, the problem fell into the category of lifetime data analysis. To adjust for multiple factors (trail number and gender), data were analyzed by the proportional hazards regression model. The analysis was stratified by gender because the experimental conditions were different for males and females. Following the terminology of lifetime data analysis, the probability S(t) that an animal was still on the rod at time t was called the retention survival function. Kaplan-Meier estimates of the cumulative probability of retention are shown (see Fig. 7). The effects of genotype were adjusted for the effect of the trial using Cox's proportional hazards model (Statistica; Statsoft).
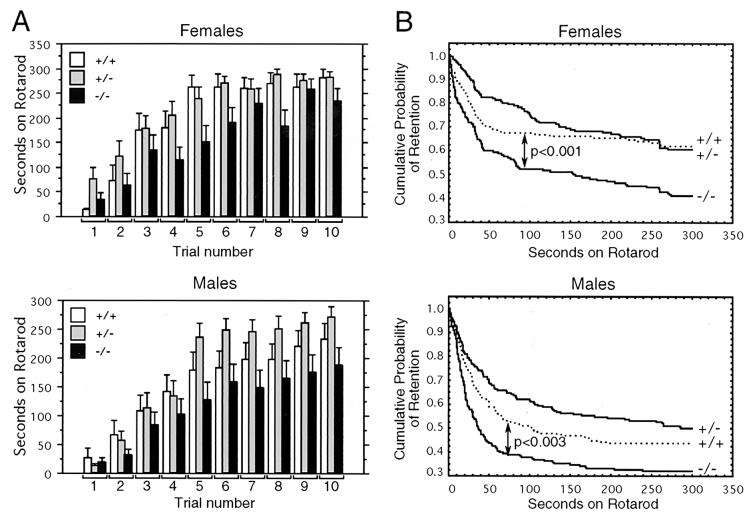
FIG. 7.
punc mutant mice have impaired motor coordination. (A) The average retention times on the Rotarod for males and females are shown for each trial and genotype. Wild-type (female, n = 15; male, n = 19), heterozygous (female, n = 17; male, n = 19), and homozygous mutant (female, n = 17; male, n = 19) groups are indicated by open, gray, and black boxes, respectively. Bars above each box indicate one standard error of the mean. (B) Statistical analysis of the Rotarod data separated by sex. The graphs show the probability of retention on the rod with respect to time for animals of each genotype: wild type (+/+, dotted line), heterozygotes (+/−, upper solid line), and homozygous mutants (−/−, lower solid line). P values shown are for the comparison of wild-type to homozygous mutant animals. The difference between the wild-type and heterozygous curves was not statistically significant for either sex.
Other tests.
Auditory brainstem response thresholds for click and 8-, 16-, and 32-kHz tone pip stimuli were measured by using hardware and software from Intelligent Hearing Systems according to the methods described by Zheng et al. (46). Swimming behavior was tested according to the method of Marshall and Berrios (24). Sensitivity to temperature was tested according to the method of Dahme et al. (7). Olfaction was tested according to the method of Cremer et al. (6).
RESULTS
Isolation of a gene trap cell line with a lacZ insertion in punc.
A mouse ES cell line, designated 24-B9, was isolated from an ongoing gene trap screen that has been described previously (44). β-Gal activity was detected in undifferentiated 24-B9 cells and was insensitive to the application of a variety of different growth factors and hormones. Chimeric embryos prepared using 24-B9 cells and analyzed at E11.5 and E13.5 exhibited β-Gal activity in a variety of locations, including the otic epithelium, facial mesenchyme, ventral neural tube, and limb mesenchyme (data not shown). Southern blot hybridization analysis showed that a single copy of the gene trap vector was integrated into the genome of 24-B9 cells (data not shown), demonstrating that the pattern of β-Gal activity observed in chimeric embryos reflected the activity of a single gene. 5′-RACE was used to isolate 68 bp of endogenous trapped gene sequences located at the 5′ end of lacZ mRNA isolated from 24-B9 cells (Fig. 1A). The trapped sequences were not in frame with respect to the lacZ gene, suggesting that translation of the hybrid lacZ mRNA was initiated from the Kozak consensus ATG that is present on the gene trap vector (44).
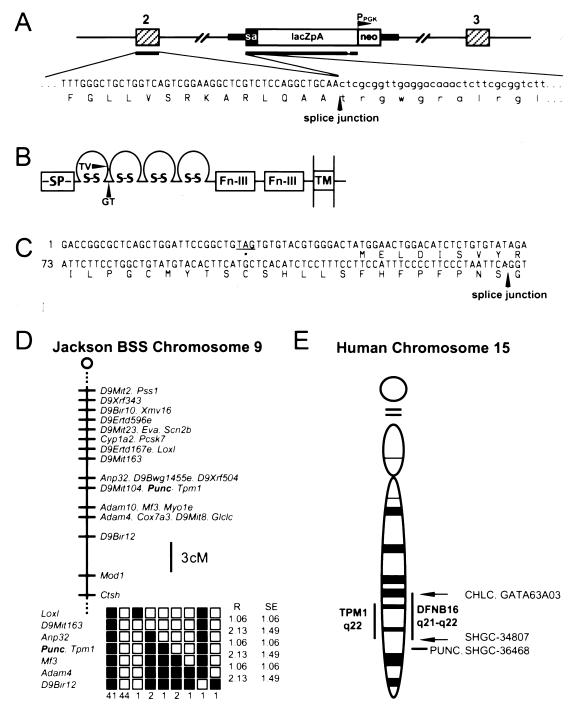
FIG. 1.
Characterization and mapping of punc gene, which encodes a member of the neural cell adhesion molecule family. (A) Diagram of the gene trap vector insertion in 24-B9 ES cells. The gene trap vector, composed of a splice acceptor (sa), lacZ gene (lacZpA), and PGKneobpA cassette (neo) inserted between exons 2 and 3 (hatched boxes) of punc. The nucleotide (upper line) and the corresponding protein sequence (lower line) near the splice junction joining punc exon 2 to the vector sequences is indicated below the diagram. Capitalized letters in the nucleotide sequence indicate punc sequence, whereas lowercase letters indicate the vector sequence. The arrow indicates the position of the splice junction. Only 43 of the 68 bp of punc sequence identified by 5′-RACE are shown. (B) Predicted domain structure of Punc protein. Ig and FNIII domains are indicated by loops and boxes, respectively. The sites at which Punc was disrupted by the gene trap vector (GT) or the targeting vector (TV) are marked by arrows. SP, signal peptide; TM, transmembrane domain. (C) Nucleotide sequence (upper line) and corresponding amino acid sequence (lower line) of the alternative 5′ punc exon (GenBank accession no. AF236125). An in-frame stop codon (star) preceding the open reading frame is underlined. An arrow indicates the splice junction between the alternative exon and exon 2. (D) At the top of the panel is a map figure from the Jackson BSS backcross showing part of mouse chromosome 9. The map is depicted with the centromere toward the top. A 3-cM scale bar is shown to the right of the figure. Loci mapping to the same position are listed in alphabetical order. Missing typings were inferred from surrounding data where the assignment was unambiguous. Raw data were from the Jackson Laboratory (http://www.jax.org/resources/documents/cmdata). At the bottom of the panel is a haplotype figure from the Jackson Laboratory BSS backcross showing part of chromosome 9 with loci linked to punc. Loci are listed in order with the most proximal at the top. The black boxes represent the C57BL6/JEi allele, and the white boxes represent the SPRET/Ei allele. The number of animals with each haplotype is given at the bottom of each column of boxes. The percent recombination (R) between adjacent loci is given to the right of the figure, along with the standard error (SE) for each value. Missing typings were inferred from surrounding data where the assignment was unambiguous. (E) Map figure of human chromosome 15 showing the localization of PUNC. Vertical bars on the left and right indicate the regions where TPM1 and DFNB16, respectively, are located. Arrows indicate the location of loci linked to the two DFNB16 flanking markers. The horizontal bar indicates the region where PUNC and its linked loci were mapped.
An additional 1.2 kbp of the trapped gene was isolated from an E11.5 cDNA library. Sequence analysis of this DNA fragment indicated that the gene trap vector had integrated into an intron of a gene encoding a 793-amino-acid single-pass transmembrane protein that is a new member of the Ig superfamily (Fig. 1B). The predicted protein comprises a signal peptide, an extracellular domain composed of four Ig repeats and two FNIII repeats, and an intracellular domain that is very small and has no recognizable motifs. The gene trap insertion was located in a position predicted to disrupt translation of the encoded protein between the first and second Ig repeats (Fig. 1B). In some cDNAs, an additional exon of 60 bp encoding 20 amino acids was located between the sequences encoding the second FNIII repeat and those of the transmembrane domain. While this work was in progress, the same gene (including the additional internal exon) was identified by Salbaum (31), who named it punc.
An additional alternatively spliced exon located at the 5′ end of the punc gene was identified in E11.5 cDNA (Fig. 1C). This exon contained an in-frame ATG initiation codon preceded by a stop codon and was spliced directly to sequences encoding the first Ig domain. Since cDNAs carrying this alternative 5′ exon did not appear to encode a signal peptide, it is possible that there exists an intracellular form of Punc. RT-PCR analysis demonstrated that the alternatively spliced 5′ exon was present in E9.5 to newborn embryos, although its levels were lower than those of the signal peptide-encoding form (data not shown).
Chromosomal localization of punc and elimination of PUNC as a candidate for DFNB16.
Mouse punc was mapped by hybridizing a punc cDNA fragment to the TaqI-digested Jackson Laboratories BSS interspecific backcross mapping panel (29). Analysis of the strain distribution pattern of a TaqI polymorphism showed that punc is located in the middle of chromosome 9, approximately 40 centimorgans (cM) from the centromere, in the vicinity of Tpm1 (Fig 1D). This result is consistent with the mapping of punc to the D-E1 region of chromosome 9 by fluorescence in situ hybridization (30) but provides higher resolution.
Since punc expression was found in the otic epithelium of 24-B9 chimeric embryos and the gene mapped near Tpm1, the human homologue of which maps to 15q22, we considered the possibility that human PUNC might be a candidate for the nonsyndromic recessive deafness locus, DFNB16, which had been mapped to 15q21-q22 (2). To address this possibility, a human EST from the PUNC locus was identified (GenBank accession no. AA860555). To localize human PUNC relative to DFNB16, two flanking markers for DFNB16, D15S1039 and D15S155 (2), were mapped to the Stanford G3 radiation hybrid mapping panel, along with the PUNC-containing EST (Fig. 1E). The PUNC EST was most closely linked to SHGC-36468, with an LOD score of 7 at 28 cR (Bin 34). This localization is consistent with the localization of PUNC to 15q22.3-23 by in situ hybridization (30). The distal marker for DFNB16, D15S1039, was most closely linked to CHLC.GATA63A03, with an LOD score of 9 at cR (Bin 14). The proximal marker, D15S155, was most closely linked to SHGC-34807 with an LOD score of 13 at 8 cR (Bin 31). These data suggest that human PUNC is located distal to the critical region for DFNB16 and is therefore not a candidate gene. Recent mapping data for DFNB16 are consistent with this result and suggest that the responsible gene may be even more proximally located than originally reported (40).
Expression of punc mRNA.
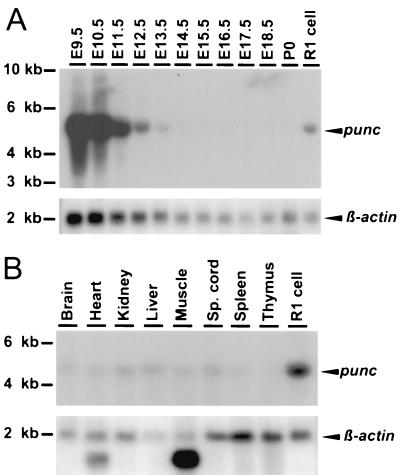
To determine the relative levels of punc expression during development, a stage-specific Northern blot was prepared from wild-type E9.5 to newborn embryo mRNA and hybridized with the 1.2-kbp punc cDNA fragment (Fig. 2). Strong expression of a 5-kb transcript was detected in E9.5 and E10.5 embryos. Levels of this transcript decreased gradually from E11.5 and were almost extinguished by E15.5, suggesting that punc could function during mid-gestation (Fig. 2A). The quantity of punc transcripts present in several adult mouse tissues was also determined. Northern blot hybridization analysis of mRNA derived from brain, heart, kidney, liver, muscle, spinal cord, spleen, and thymus showed similarly low levels of the 5-kb punc transcript (Fig. 2B).
FIG. 2.
Northern blot analysis of punc expression at embryonic stages and in adult tissues. (A) Stage-specific Northern blot of whole-embryo mRNA. (B) Northern blot of adult tissue mRNA. Each lane contains 5 μg of the indicated mRNA. A 1.2-kb punc cDNA was used as a probe. The lower panels show the results of subsequent hybridization with a β-actin probe. The positions of size markers are indicated on the left.
Gene targeting of punc.
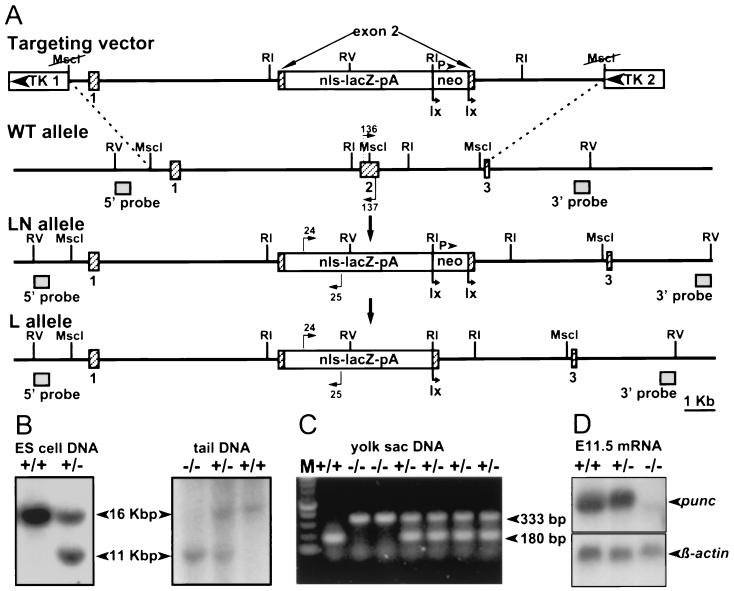
To determine the role of punc, a mouse strain carrying a mutation in the gene was required. Since fluorescence in situ hybridization analysis revealed that a high proportion of 24-B9 ES cells had an abnormal chromosome number accompanied by loss of the Y chromosome, that cell line was not a suitable candidate for germ line transmission of the gene trap punc allele (data not shown). Instead, the punc gene was disrupted by homologous recombination (Fig. 3). A targeting vector was designed to disrupt punc expression by insertion of a reporter gene (nls-lacZ) and a loxP-flanked PGKneobpA expression cassette into the Msc1 site of exon 2 (Fig. 3A). This site is located within sequences that encode the first Ig domain (Fig. 1B). To ensure activity of the β-Gal enzyme, production of a protein with both a signal sequence and a nuclear localization signal was prevented by placing a short stretch of synthetic oligonucleotides containing stop codons in all three reading frames immediately upstream of nls-lacZ. Thus, β-Gal activity from this construct was only expected to reflect punc transcription and was expected to be similar to the gene trap insertion except that β-Gal would be located in the nucleus rather than the cytoplasm. Two thymidine kinase expression cassettes were also included in the final construct for negative selection. The vector was introduced into R1-45 ES cells, which were cultured under standard selective conditions. Clones of targeted cells were identified by Southern blot hybridization analysis. As predicted, a 5′-flanking probe detected a 16-kb EcoRV fragment in DNA isolated from R1-45 cells, whereas DNA isolated from targeted clones contained both the 16-kb EcoRV fragment and an 11-kb EcoRV fragment (Fig. 3B). This result was confirmed by using a 3′-flanking probe (data not shown). In addition, hybridization with a neo probe confirmed that the new sequences were present at a single site in the genome (data not shown).
FIG. 3.
Gene targeting of punc. (A) The targeting vector is depicted on the first line. Exon 2 of punc was disrupted by insertion of a nuclear localized lacZ (nls-lacZpA), followed by a PGKneobpA expression cassette flanked by two loxP sites (lx). Thymidine kinase expression cassettes (TK1 and TK2) were placed at either end of the punc DNA (horizontal line). Hatched boxes indicate punc exons. The wild-type allele is shown on the second line. Shaded boxes indicate the probes used in Southern blot analysis. The locations of primers used to detect the wild-type allele (positions 136 and 137) are indicated with arrows. The third line shows the lacZ/neo targeted allele (LN). The locations of primers used to detect the mutant allele (24, 25) are indicated by arrows. The fourth line shows the lacZ targeted allele (L) after Cre-mediated removal of the PGKneobpA cassette. Key restriction enzyme recognition sites are indicated (RI and RV, EcoRI and EcoRV). (B) Southern blot hybridization analysis of DNA extracted from ES cell clones or from mouse tails. DNA was digested with EcoRV and probed with the 5′ probe. Genotypes (+/+, wild type; +/−, heterozygous; −/−, homozygous mutant) are indicated above each lane. The sizes of the bands are indicated. (C) PCR analysis of DNA extracted from yolk sacs. Each DNA sample was amplified with the wild-type and mutant primer pairs, which gave rise to bands of 180 and 330 bp, respectively. (D) Northern blot analysis of mRNA extracted from E11.5 mouse embryos of each punc genotype and hybridized sequentially with a 3′ punc probe and a β-actin probe.
Cells from three targeted clones were used to generate germ line chimeras, establishing the puncLN strain. A PCR-based assay was developed to distinguish the three possible punc genotypes (Fig. 3C). In addition, a puncL strain, which lacked the neo expression cassette, was established by mating F1 puncLN heterozygotes to a Cre-expressing mouse (33). No phenotypic differences between these two strains were detected, and the majority of the data presented here are derived from analyses of the original puncLN strain.
To determine whether the puncLN allele was a null, Northern blot hybridization was used to detect punc transcripts in E11.5 embryos of each genotype. A punc cDNA probe located 3′ of the lacZ insertion site detected the expected 5-kb transcript in mRNA samples derived from wild-type and heterozygous embryos. This transcript was not detected in homozygous mutant mRNA (Fig. 3D). However, a very small quantity of a shorter punc-hybridizing transcript was seen in the mutant mRNA sample. This transcript was present at similarly low levels in mRNA samples extracted from homozygous puncL embryos, suggesting that it did not initiate from the PGKneobpA sequences present in the puncLN allele but absent from the puncL allele (data not shown). As expected from the Northern blot results, it was possible to use RT-PCR to amplify small amounts of product from samples of homozygous mutant RNA derived from either punc allele when the primers were both located in punc exons downstream of the lacZ insertion site. However, RT-PCR amplification of homozygous mutant RNA samples invariably failed when one punc primer was located in an exon upstream of the lacZ insertion site and the other was located in an exon downstream of the lacZ insertion site (data not shown). These results support the conclusion that the rare transcript detected in mutant mRNA by hybridization with the 3′ punc probe was not formed by a splice that simply deleted the disrupted exon. Rather, they suggest that this aberrant transcript may initiate from somewhere within the lacZ sequences and is likely to be formed by splicing these sequences to downstream punc sequences. The punc exons located downstream of lacZ do not include an in-frame consensus translation initiation codon. Even if, however, the truncated transcript could be translated starting with the first non-consensus AUG, the resulting protein would lack the signal peptide and half of the first Ig domain of Punc. It is likely, therefore, that the targeted alleles are functionally null.
Mice homozygous for puncLN or puncL are viable and fertile.
Heterozygous puncLN or puncL animals did not exhibit any overt abnormalities when compared with their wild-type littermates. To assess whether punc has an essential embryonic function, heterozygous puncLN mice were intercrossed, and the offspring were genotyped. Of 131 E8.5-to-E17.5 embryos, 32 (24%) were wild type, 71 (54%) were heterozygous, and 28 (21%) were homozygous mutants, consistent with a normal Mendelian distribution. Of 109 adult offspring of the intercross, 23 (21%) were wild type, 55 (50%) were heterozygous, and 31 (28%) were homozygous mutants, again suggesting a normal Mendelian distribution. The homozygous mutant offspring on a mixed 129/Sv and C57BL/6 genetic backgound were overtly normal; they gained weight normally, and both sexes were fertile. Similar results were obtained with the puncL strain (data not shown). Therefore, punc does not have an essential embryonic or postnatal function.
Localization of punc in embryos and adults.
To determine the tissues that express punc, which might provide clues to its function, we examined the patterns of β-Gal activity in heterozygous and homozygous puncLN embryos using X-Gal staining (Fig. 4). No β-Gal activity was detected in embryos younger than E9.5. Since Salbaum reported punc expression in the neuroectoderm at E7.5 and E8.5 (31), and E8.5 puncLN heterozygotes and homozygotes did not exhibit any β-Gal activity, RT-PCR analysis of E8.5 heterozygous RNA was performed. Both punc and lacZ transcripts were readily detected and whole-mount in situ hybridization using a punc probe supported the RT-PCR results (data not shown). Thus, it is possible that the absence of X-Gal staining at E8.5 was caused by a delay in the translation of lacZ mRNA and/or in the assembly of active β-Gal tetramers.
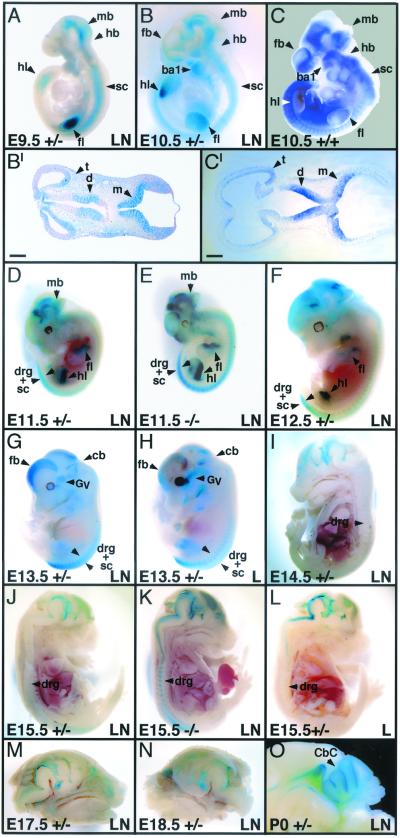
FIG. 4.
Sites of β-Gal activity as revealed by X-Gal staining of embryos heterozygous or homozygous mutant for targeted punc alleles. (A, B, and D to O) X-Gal-stained whole embryos. Embryos in panels I to O were hemisected prior to staining. The age, genotype, and punc strain for each embryo is shown at the bottom of the panel. Heterozygotes are indicated by +/−, and homozygous mutants are indicated by −/−. The lacZ/neo insertion is indicated by LN, and the lacZ insertion is indicated by L. (C) In situ hybridization of a punc probe with an E10.5 wild-type embryo. (B′) Transverse section (10 μm) through the brain of the embryo shown in panel B. (C′) Transverse section (10 μm) through the brain of the embryo shown in panel C. Structures mentioned in the text are indicated with arrows and are abbreviated as follows: forebrain (fb), midbrain (mb), hindbrain (hb), spinal cord (sc), forelimb (fl), hindlimb (hl), DRG (drg), branchial arch (ba), telencephalon (t), diencephalon (d), metenecphalon (m), trigeminal ganglion (Gv), cerebellum (cb) and cerebellar cortex (Cbc). Objective magnifications used for photography were as follows: A, ×4; B, ×2.5; C, ×2.5; and B′ and C′, ×6.6 (scale bar indicates 0.2 mm) and D to F, ×1.6; G and H, ×1.2; I to L, ×1; M, ×1.2; N, ×1; and O, ×3.2.
In E9.5 embryos, very strong β-Gal activity was detected in the CNS, including the midbrain, hindbrain, and spinal cord, as well as in the limb buds (Fig. 4A). At E10.5, expression of β-Gal in the CNS, now including the forebrain, was still strong and was confined to the ventral domain of expressing areas. Expression of β-Gal in the limbs began to show some restriction to the anterior domain and expression was initiated in the branchial arches (Fig. 4B). To confirm that the pattern of β-Gal activity reflected punc mRNA localization, RNA in situ hybridization analysis using a punc probe was performed on whole embryos at E9.5 (data not shown) and E10.5 (Fig. 4C). As expected, the localization of punc mRNA in normal embryos was very similar to the localization of β-Gal activity in similarly staged puncLN embryos (Fig. 4B and C and data not shown). This finding was confirmed by comparing sections of the X-Gal-stained embryos with those of the embryos processed for in situ hybridization. Transverse sections taken through the brain showed comparable localization of the X-Gal and alkaline phosphate precipitates in the telencephalon, diencephalon, and metencephalon (Fig. 4B′ and C′). In E11.5 (Fig. 4D) and E12.5 (Fig. 4F) embryos, β-Gal activity in the midbrain persisted, but spinal cord expression began to retreat posteriorly. Staining of the dorsal root ganglia (DRG) was also observed. X-Gal staining of the limb buds decreased and was concentrated anteriorly and moved proximally. Furthermore, the pattern of β-Gal activity in E11.5 puncL heterozygous embryos, from which the PGKneobpA cassette had been removed, was the same as that seen in puncLN heterozygotes, demonstrating that expression of the punc/lacZ transcript was not affected by the presence of the PGK promoter (data not shown). By E13.5, β-Gal activity in the spinal cord and DRG had retracted posteriorly (Fig. 4G). Strong β-Gal expression could also be detected in the developing cerebellum, the trigeminal ganglion and in the dorsal forebrain. Again, the same pattern of β-Gal activity was observed in puncL embryos at this stage (Fig. 4H). In E14.5 and E15.5 embryos, in addition to the brain staining, a low level of β-Gal activity was variably detected in the DRG of heterozygotes (Fig. 4I, J, and L). In contrast, β-Gal activity was uniformly very strong in all DRG of homozygous mutant embryos (Fig. 4K). In E16.5 and older embryos, β-Gal activity in both heterozygotes (Figs. 4M and N) and homozygotes (data not shown) was only detected in the brain and inner ear (not shown). In newborn mice, β-Gal activity was strongest in the cerebellum, especially in its anterior domain (Fig. 4O).
To localize punc expression in adults, animals were perfused with fixative, and various organs were stained with X-Gal. No β-Gal activity was detected above background levels in heart, kidney, liver, spleen, spinal cord, DRG, testis, thymus, or lung (data not shown). However, relatively strong β-Gal activity was localized to distinct regions of the brain, most notably the cerebellum (Fig. 5A) and the CA2 region of the hippocampus (Fig. 5B). Weaker β-Gal activity was also detected in the olfactory bulb (Fig. 5B). As was the case for embryos, no differences in the distribution of β-Gal activity between the adult brains from puncLN and puncL strains were noted (Fig. 5C and D).
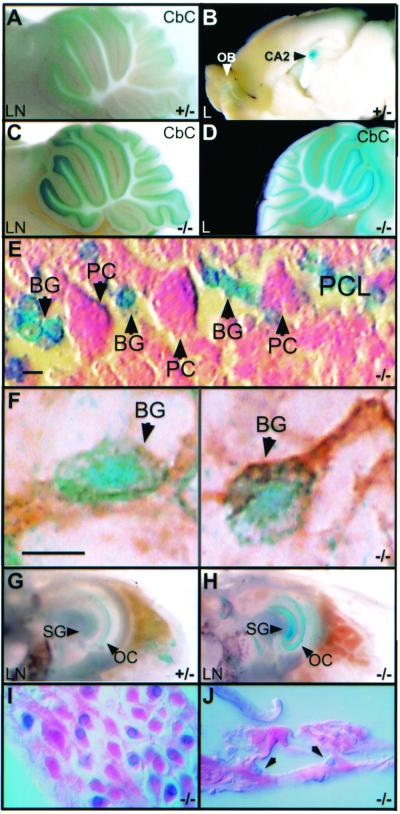
FIG. 5.
X-Gal staining of adult tissues showing that punc is expressed in the adult brain and inner ear. The genotype and punc strain for each sample is indicated at the bottom of the panels (heterozygotes, +/−; homozygous mutants, −/−). The lacZ/neo insertion is indicated by LN, and the lacZ insertion is indicated by L. (A to D) X-Gal-stained sagittal brain slices (1 mm) showing punc expression in the cerebellar cortex (Cbc), the CA2 region of the hippocampus (CA2), and the olfactory bulb (OB). A, C, and D, objective magnification of ×2.0; B, objective magnification of ×0.8. (E) Section (10 μm) of slice in panel D showing small β-Gal-expressing cells in the position of Bergmann glia (BG) between the Purkinje cell bodies (PC). The Purkinje cell layer (PCL) is indicated. Scale bar, 5 μm. (F) Cryosection (10 μm) of X-Gal-stained puncL/L cerebellum immunostained with antibody against GFAP (brown) showing colocalization of β-Gal activity with this marker of Bergmann glia (BG). Scale bar, 5 μm. (G and H) Medial view of whole X-Gal-stained inner ears showing β-Gal activity in the spiral ganglion (SG) and support cells of the Organ of Corti (OC). Objective magnification of ×3 .2. (I) Section (10 μm) of inner ear in panel H showing β-Gal-expressing neurons in the spiral ganglion. Objective magnification of ×40. (J) Section (10 μm) of inner ear in panel H showing β-Gal activity in the pillar cells, which are indicated with arrows. Objective magnification of ×40.
Sections taken through the cerebellum revealed that the neclei of small cells found in the same layer as the large Purkinje cell bodies expressed β-Gal (Fig. 5E). To determine whether these cells might be Bergmann glia, cerebellar sections that had been stained with X-Gal were processed for immunohistochemical detection of the intermediate filament protein, GFAP, a marker of glial cells (Fig. 5F). Indeed, the X-Gal precipitate was found in cells that were also GFAP positive, suggesting that punc mRNA is expressed by Bergmann glia.
The only other adult organ in which β-Gal activity could be detected was the inner ear (Fig. 5G and H). Moderate β-Gal activity was detected in the neurons of the spiral ganglion (Fig. 5I), and weak activity was evident in supporting cells, particularly the pillar cells of the Organ of Corti (Fig. 5J).
punc expression is negatively autoregulated.
Visual comparisons of the quantity of X-Gal precipitate produced by puncLN heterozygous and homozygous mutant littermate embryos processed and stained under identical conditions suggested that in some tissues of homozygotes, β-Gal activity was more than twofold higher than the levels found in heterozygotes. For example, β-Gal activity in the DRG anterior to the hindlimb was relatively strong in both E11.5 and E15.5 in puncLN/LN embryos, yet its expression level in puncLN/+ embryos varied from very weak to undetectable (compare Fig. 4E and K with Fig. 4D and J). This difference in DRG β-Gal activity between genotypes was also seen in E12.5 to E14.5 embryos (data not shown). Moreover, striking differences in the quantity of X-Gal staining intensity were also observed between puncLN/LN and puncLN/+ brain and inner ear samples (compare Fig. 5A with C and Fig. 5G with H). Similar observations were made when the X-Gal precipitate produced by puncL heterozygotes was compared with that of puncL homozygous littermates (data not shown). In contrast, many of the X-Gal-stained regions, particularly those in young embryos, had the expected twofold difference in staining intensity between heterozygous and homozygous samples (compare the limbs in Figs. 4D and E). These observations suggested that punc transcription or message stability might normally be negatively autoregulated in some tissues.
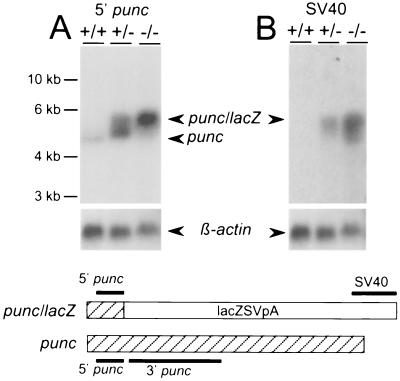
To address this issue, Northern blot hybridization was carried out on E11.5 mRNA extracted from whole wild-type, heterozygous, and homozygous puncLN embryos. This stage was chosen because the wild-type transcript was abundant enough to be detected by very short punc probes. The same Northern blot that had been previously hybridized with the 1.2-kb 3′ punc cDNA probe (Fig. 3D) was stripped and hybridized sequentially with a 164-bp 5′ punc cDNA probe to detect all transcripts initiated from the punc promoter and an SV40 probe to detect the 3′ end of lacZ-containing mRNA (Fig. 6). As expected, the 5′ punc cDNA probe detected a 5-kb punc transcript in both wild-type and heterozygous mRNA, and it revealed a larger punc/lacZ hybrid transcript in both heterozygous and homozygous mutant mRNA (Fig. 6A). Interestingly, the amount of the wild-type transcript detected in mRNA derived from heterozygotes, which have only one wild-type allele, was fourfold higher than that detected in mRNA derived from wild-type controls, which have two wild-type alleles. The amount of the hybrid transcript was only about twofold higher in homozygous mutant mRNA than in heterozygous mRNA. This observation is consistent with the finding that only a region of the spinal cord in homozygous mutant E11.5 embryos had excess β-Gal activity (Fig. 4D and E) and that mRNA was extracted from whole embryos, which also included many regions with the expected levels of X-Gal staining. Significantly, the amount of the hybrid transcript detected in homozygous mutant mRNA with the 5′ probe was much higher (about eightfold) than the levels of the wild-type punc transcript detected with the 5′ probe in wild-type mRNA (Fig. 6A). These results are consistent with the idea that punc transcription or mRNA stability is negatively autoregulated in wild-type cells. Hybridization with the SV40 probe (Fig. 6B) confirmed the identity and relative quantities of the hybrid mRNA and also revealed the presence of an mRNA species in the homozygous mutant sample that corresponded in size with that seen after hybridization with the 3′ punc probe (Fig. 3D). Since this transcript was found only in mRNA samples isolated from homozygous mutant embryos, Punc may also regulate its initiation or stability. Similar hybridization results were obtained using mRNA isolated from puncL strain embryos at E11.5 (data not shown).
FIG. 6.
punc transcription is negatively autoregulated. Northern blot analysis of 5 μg of mRNA isolated from E11.5 embryos of the indicated genotypes (+/+, wild type; +/−, heterozygous; −/−, homozygous mutant). (A) Hybridization of the blot with the 5′ punc probe. (B) Hybridization of the blot with the SV40 probe. The lower panels show the results of a subsequent hybridization with β-actin. The positions and the relative sizes of the probes (thick lines) are indicated below the Northern blots in schematic diagrams of the wild-type and punc/lacZ transcripts. The hatched boxes represent punc sequences, and the open boxes represent lacZ sequences.
Mice homozygous for puncLN performed poorly in a test of motor coordination.
Further characterization of the punc mutant strains focused on adult behaviors potentially affected by punc/lacZ-expressing cells. To assess inner ear function, animals of all three genotypes were tested for threshold shifts in the auditory brainstem response, for the righting reflex, and for swimming posture. To assess olfaction, behavior in the presence of attractive versus repellant bedding was observed. No differences in performance between animals of different genotypes were found in the results of any of these tests (data not shown). Furthermore, comparisons of the gross morphology and innervation of the inner ear in heterozygous and homozygous mutants at P0 revealed no differences (data not shown).
Since punc was expressed in the cerebellum, we assessed the mutant strain for motor coordination. Observation of the mutant animals did not reveal overt signs of ataxia. To assess more-subtle coordination deficits, the animals were placed for 10 trails on a fixed-speed Rotarod. Preliminary experiments with wild-type naive mice suggested that while males could stay on the rod for a few seconds at the highest speed (31 rpm), females could not. Therefore, females were tested at 27 rpm, the highest speed at which they could stay on for a few seconds, and males were tested at 31 rpm. For both sexes, retention time on the rod was measured up to 300 s. Animals that remained on the rod for the full test period were given a score of 300 that was treated in the statistical analysis as censored. The average retention time increased with trial number for all three genotypes (Fig. 7A). Overall, the effects of trial number and genotype were highly significant (P < 0.0001). Also, the learning ability of male and female mice (effect of trial number) was similar. In addition, survival analysis showed that the cumulative probability of retention on the rod for homozygous mutant individuals was significantly lower than that of wild-type animals (Fig. 7B, P < 0.001 for females and P < 0.003 for males). Although heterozygotes had average retention times that were somewhat longer than those of wild-type animals, the differences were not statistically significant for either sex (P < 0.25 for females, P < 0.19 for males).
The motor coordination defect was not correlated with any obvious structural abnormality of the mutant cerebellae. Their size, foliation pattern, and laminar structure appeared normal (Fig. 5A, C, and D and data not shown). In addition, inspection of hematoxylin-and-eosin-stained paraffin sections revealed normal numbers and positions of Purkinje cells and Bergmann glia. Furthermore, the number of interneurons in the molecular layer was also normal (data not shown). The motor coordination defect could not be attributed to sensory deficits. Homozygous mutant mice behaved similarly to wild-type and heterozygous mice when placed on a 55°C platform. They also responded normally to gentle pinching of the footpad. Finally, motor neuron development appeared normal as revealed by Islet-1 immunohistochemistry at E11.5 (data not shown).
DISCUSSION
Characterization of the gene disrupted by the 24-B9 gene trap insertion led to the identification of mouse punc, which encodes a member of the Ig superfamily. Punc protein, with an extracellular domain composed of four Ig repeats and two FNIII repeats and a single transmembrane domain is predicted to have structural properties similar to NCAMs. To investigate punc function, we generated mouse strains with either a lacZ/neo or a lacZ insertion exon 2 of the punc gene. Experimental evidence and theoretical considerations argue that the punc alleles are functionally null. First, the inserted DNA abolished expression of any normally sized punc mRNA in homozygous mutant embryos from both stains. The aberrantly short punc-containing mRNA found in mutant embryos was expressed at very low levels, was likely to have initiated from within the lacZ sequences, and did not contain a consensus in-frame translation initiation codon. Thus, even if this 5′-truncated punc transcript could be translated starting with a nonconsensus AUG, the resulting protein would lack a signal sequence and the entire first Ig domain, which are encoded upstream of the inserted DNA. Therefore, it is unlikely that the mutant punc alleles produce functional Punc protein.
Northern blot analysis showed that punc mRNA is expressed strongly between E9.5 and E11.5. After this time, the proportion of punc transcripts in whole-embryo mRNA decreases precipitously and punc transcripts are detected at very low levels in older embryos and in a variety of adult tissues. Construction of the transcriptional fusion between punc and lacZ permitted an extensive survey of the pattern of punc expression, as revealed by β-Gal activity, during embryogenesis and in the adult. Between E9.5 and E11.5, β-Gal activity in whole embryos coincided with the punc RNA in situ analysis of tissue sections published by Salbaum (31). We found very strong β-Gal activity in the CNS, limbs, and branchial arches. Sections taken through the heads of X-Gal-stained embryos at E11.5 revealed the otic epithelium to be an additional site of β-Gal activity (data not shown). Although punc mRNA was difficult to detect in samples isolated from whole embryos older than E11.5, X-Gal staining of puncLN embryos revealed that punc continues to be expressed throughout development. Its expression domain is progressively restricted, becoming confined to the brain and inner ear by E16.5.
Despite the strong and dynamically changing expression of punc in early embryos, no morphologic abnormalities were found in homozygous mutant embryos. This suggests that its embryonic function is subtle and/or redundant. Analysis of appropriate double-mutant combinations could reveal an embryonic role for Punc. Other noncatalytic Ig superfamily members, such as DCC (18), are particularly good candidates. Since netrin-1, a ligand for DCC, is required for normal inner ear development (32), it is possible that DCC plays a role in ear development. Thus, the double mutant combination of dcc with punc might reveal a role for punc in inner ear development or in other tissues that express both genes.
Notably, we found that punc gene expression is negatively autoregulated. In some regions of homozygous mutant embryos, particularly the spinal cord and dorsal root ganglia of embryos at E11.5 and older, the β-Gal activity increased more than twofold compared with the same region in heterozygous littermates stained under identical conditions. This increased β-Gal activity was also noted in the spiral ganglion and the cerebellum of adult homozygous mutants. The results of Northern hybridization of E11.5 mRNA with a variety of probes support this observation. The 5′ punc probe, which detects all transcripts initiated from the punc promoter, revealed that, relative to wild-type embryos, punc heterozygotes have fourfold-increased levels of the wild-type punc transcript and that punc homozygotes have eightfold-increased levels of the hybrid punc/lacZ transcript. It is also notable that the 3′ punc probe, which can only detect wild-type transcripts, revealed that wild-type and heterozygous punc embryos have similar amounts of punc mRNA. In the absence of autoregulation, heterozygotes would be expected to have half as much punc as do wild-type animals. Taken together, these results suggest that in some tissues Punc may play a dose-dependent role in negatively regulating its own transcription or mRNA stability. The difference between the ratio of wild-type transcript detected in heterozygous versus wild-type mRNA using the 5′ and 3′ probes (fourfold versus even) may reflect differences in the stability of wild-type punc mRNA in tissues that are or are not subject to autoregulation.
Since the major form of Punc is predicted to be found at the cell surface, it is likely that the proposed autoregulation is indirect, as illustrated in the model (Fig. 8). Although the intracellular domain of Punc does not include any recognizable motifs that would suggest a link to a known signaling pathway, it is possible that Punc could interact directly with one of those pathways. Alternatively, Punc may interact with another transmembrane protein that has signaling activity as, for example, some Ig superfamily members are thought to initiate signaling to the nucleus by interacting with fibroblast growth factor receptors (25, 41). As the increased β-Gal activity in homozygous mutants is restricted to certain locations, it is possible that this autoregulatory phenomenon is dependent on the reception of an as-yet-unidentified signal and/or ligand (Fig. 8B). In this scenario, the proposed Punc ligand is not expected to be expressed at all sites of Punc expression and, at those sites, punc expression is not autoregulated (Fig. 8A). The physiological significance of punc autoregulation is unclear. Mice with a lacZ insertion in ncam were not reported to have this property (15), but it will be interesting to learn whether any other members of neural Ig superfamily display similar regulation.
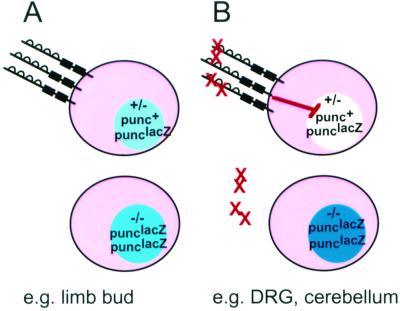
FIG. 8.
Model for negative autoregulation of punc gene expression. Punc protein is symbolized by a line with four Ig domains (half circles) and two FNIII domains (boxes) located outside the cell body (pink). For each cell, the intesity of color shading the nucleus (shades of blue or plain white) represents the relative level of β-Gal activity. (A) In most tissues and in all sites of punc expression prior to E11.5, the punc RNA level, as detected by the nuclear β-Gal activity, is proportional to the number of copies of the targeted punc/lacZ allele. (B) In Punc-expressing cells that are exposed to a Punc ligand (red X), a signal (red line with a bar at the end) is sent to the cell to reduce the level of punc RNA. In this diagram, the effect of the Punc-mediated signal is depicted as acting in the nucleus at the level of transcription initiation, but posttranscriptional mechanisms are not excluded.
punc expression was observed in only two adult tissues: the inner ear and the brain. Mice that were homozygous for either of the punc mutant alleles had normal inner ear structure as well as normal balance, posture, and auditory brainstem response thresholds, suggesting that if punc plays a role in the adult inner ear, its function has not been discovered or is redundant. Expression of punc in the cerebellum prompted testing of motor coordination in the homozygous mutants. We found that their performance on the Rotarod was significantly impaired relative to their wild-type or heterozygous littermates. Since the mutants increased their retention times on the Rotarod with experience, it seems that they were capable of learning the task, but that they did not do so as effectively as the controls. This suggests a role for Punc in the control of motor coordination, although a role in learning or memory cannot be rigorously excluded.
How might punc play a role in motor coordination? The punc mutant is quite different from the classic mouse mutants that have impaired motor coordination (12) in that the punc mutant cerebellum is grossly normal with respect to size, cytoarchitecture, and foliation. Double-labeling of X-Gal-stained nuclei in the Purkinje cell layer with antibodies directed against GFAP showed that punc is expressed by Bergmann glia. During cerebellar development, Bergmann glia play a role in neuronal migration. In the adult, Bergmann glia are intimately associated with Purkinje cells, which provide the major cerebellar output. The cell bodies of Bergmann glia are located among the Purkinje cell bodies and the Bergmann glia fibers extend between the Purkinje cell dendrites (8, 28). Therefore, Bergmann glia are in a position to interact directly with and potentially modulate the function of Purkinje cells in the adult. In addition, Bergmann glia fibers surround the excitatory synapses made by granule cell climbing and parallel fibers with Purkinje cell dendrites (28) and electrical stimulation of parallel fibers affects the intracellular calcium concentration in microdomains of Bergmann glia fibers (13). Thus, the loss of Bergmann glia-expressed Punc from the cell surface might disrupt interactions between Bergmann glia and Purkinje cell bodies and/or dendrites or granule cell axons and account for the observed defects in motor coordination in puncLN homozygotes. The relevance of adhesive contacts between neurons and glia to neuronal function is underscored by the reduction in hippocampal long-term potentiation seen in transgenic mice that ectopically express L1 in astrocytes (22).
Two other genes expressed in Bergmann glia, namely, glast, which encodes a glutamate transporter, and vimentin, which encodes an intermediate filament protein, also cause mild motor discoordination when mutated. The motor discoordination of glast mutant homozygotes could not be correlated with any structural abnormalities of the cerebellum but was associated with persistent multiple climbing fiber innervation of Purkinje cells (42). Ultrastructural studies of the cerebellum of vimentin-negative mice revealed subtle abnormalities of the Bergmann glia and Purkinje cells (3, 11). Although we did not detect any morphologic abnormalities of the cerebellum in the punc mutants by light microscopy, it is possible that an ultrastructural study focused on the relationships between Purkinje and granule neurons and Bergmann glia would be revealing. Finally, the intriguing similarity between the punc and vimentin mutant phenotypes could be manifest simply because these genes act in the same cell type. However, given that the many cell adhesion molecules of the integrin and cadherin classes function through interactions with the cytoskeleton and that some CAMS may function similarly (16), it would be interesting to determine whether Punc provides a link to the Bergmann glia intermediate filament cytoskeleton via vimentin.
ACKNOWLEDGMENTS
We are very grateful for materials and advice provided by colleagues. Shannon Odelberg added loxP sites to our gene trap vector. Mary Barter and Lucy Rowe of the Jackson Laboratory performed the analysis of the backcross mapping data. Mario Capecchi provided stocks of R1-45 ES cells and Cre mice. The DNA Sequencing and Transgenic/Knockout Cores at the University of Utah carried out DNA sequencing and assisted with germ line transmission, respectively. Scott Rogers provided antibodies against GFAP and expertise in neuroanatomy. Bernd Fritzsch checked inner ear innervation of punc mutants. Jeanne Wehner advised us on Rotarod testing. Alexander Tsodikov (Huntsman Cancer Institute, Cancer Center Support Grant 2P30 CA42A14) performed statistical analyses of the Rotarod data. The manuscript was improved by critical comments from Scott Rogers, Mario Capecchi, Gary Schoenwolf, Tracy Wright, and Carl Thummel.
This work was supported by grants from the NIDCD (R01-DC02043) and from the Huntsman Cancer Institute (HCI Pilot Project).
REFERENCES
- 1.Berglund E O, Murai K K, Fredette B, Sekerkova G, Marturano B, Weber L, Mugnaini E, Ranscht B. Ataxia and abnormal cerebellar microorganization in mice with ablated contactin gene expression. Neuron. 1999;24:739–750. doi: 10.1016/s0896-6273(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 2.Campbell D A, McHale D P, Brown K A, Moynihan L M, Houseman M, Karbani G, Parry G, Janjua A H, Newton V, al-Gazali L, Markham A F, Lench N J, Mueller R F. A new locus for non-syndromal, autosomal recessive, sensorineural hearing loss (DFNB16) maps to human chromosome 15q21–q22. J Med Genet. 1997;34:1015–1017. doi: 10.1136/jmg.34.12.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colucci-Guyon E, Gimenez Y R M, Maurice T, Babinet C, Privat A. Cerebellar defect and impaired motor coordination in mice lacking vimentin. Glia. 1999;25:33–43. [PubMed] [Google Scholar]
- 4.Cremer H, Chazal G, Carleton A, Goridis C, Vincent J D, Lledo P M. Long-term but not short-term plasticity at mossy fiber synapses is impaired in neural cell adhesion molecule-deficient mice. Proc Natl Acad Sci USA. 1998;95:13242–13247. doi: 10.1073/pnas.95.22.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cremer H, Chazal G, Goridis C, Represa A. NCAM is essential for axonal growth and fasciculation in the hippocampus. Mol Cell Neurosci. 1997;8:323–335. doi: 10.1006/mcne.1996.0588. [DOI] [PubMed] [Google Scholar]
- 6.Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, et al. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- 7.Dahme M, Bartsch U, Martini R, Anliker B, Schachner M, Mantei N. Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat Genet. 1997;17:346–349. doi: 10.1038/ng1197-346. [DOI] [PubMed] [Google Scholar]
- 8.de Blas A L. Monoclonal antibodies to specific astroglial and neuronal antigens reveal the cytoarchitecture of the Bergmann glia fibers in the cerebellum. J Neurosci. 1984;4:265–273. doi: 10.1523/JNEUROSCI.04-01-00265.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deiner M S, Kennedy T E, Fazeli A, Serafini T, Tessier-Lavigne M, Sretavan D W. Netrin-1 and DCC mediate axon guidance locally at the optic disc: loss of function leads to optic nerve hypoplasia. Neuron. 1997;19:575–589. doi: 10.1016/s0896-6273(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 10.Fazeli A, Dickinson S L, Hermiston M L, Tighe R V, Steen R G, Small C G, Stoeckli E T, Keino-Masu K, Masu M, Rayburn H, Simons J, Bronson R T, Gordon J I, Tessier-Lavigne M, Weinberg R A. Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature. 1997;386:796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- 11.Gimenez Y R M, Langa F, Menet V, Privat A. Comparative anatomy of the cerebellar cortex in mice lacking vimentin, GFAP, and both vimentin and GFAP. Glia. 2000;31:69–83. doi: 10.1002/(sici)1098-1136(200007)31:1<69::aid-glia70>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 12.Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 13.Grosche J, Matyash V, Moller T, Verkhratsky A, Reichenbach A, Kettenmann H. Microdomains for neuron-glia interaction: parallel fiber signaling to Bergmann glial cells. Nat Neurosci. 1999;2:139–143. doi: 10.1038/5692. [DOI] [PubMed] [Google Scholar]
- 14.Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 15.Holst B D, Vanderklish P W, Krushel L A, Zhou W, Langdon R B, McWhirter J R, Edelman G M, Crossin K L. Allosteric modulation of AMPA-type glutamate receptors increases activity of the promoter for the neural cell adhesion molecule, N-CAM. Proc Natl Acad Sci USA. 1998;95:2597–2602. doi: 10.1073/pnas.95.5.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes R O. Cell adhesion: old and new questions. Trends Cell Biol. 1999;9:M33–M37. [PubMed] [Google Scholar]
- 17.Kamiguchi H, Hlavin M L, Lemmon V. Role of L1 in neural development: what the knockouts tell us. Mol Cell Neurosci. 1998;12:48–55. doi: 10.1006/mcne.1998.0702. [DOI] [PubMed] [Google Scholar]
- 18.Keino-Masu K, Masu M, Hinck L, Leonardo E D, Chan S S, Culotti J G, Tessier-Lavigne M. Deleted in colorectal cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- 19.Khillan J S, Bao Y. Preparation of animals with a high degree of chimerism by one-step coculture of embryonic stem cells and preimplantation embryos. BioTechniques. 1997;22:544–549. doi: 10.2144/97223rr06. [DOI] [PubMed] [Google Scholar]
- 20.Lalonde R, Bensoula A N, Filali M. Rotorod sensorimotor learning in cerebellar mutant mice. Neurosci Res. 1995;22:423–426. doi: 10.1016/0168-0102(95)00916-h. [DOI] [PubMed] [Google Scholar]
- 21.Levis R. Strategies for Cloning PCR products. In: Dieffenbach C W, Dveksler G S, editors. PCR primer: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 539–554. [Google Scholar]
- 22.Luthi A, Mohajeri H, Schachner M, Laurent J P. Reduction of hippocampal long-term potentiation in transgenic mice ectopically expressing the neural cell adhesion molecule L1 in astrocytes. J Neurosci Res. 1996;46:1–6. doi: 10.1002/(SICI)1097-4547(19961001)46:1<1::AID-JNR1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 23.Mansour S L, Thomas K R, Capecchi M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 24.Marshall J F, Berrios N. Movement disorders of aged rats: reversal by dopamine receptor stimulation. Science. 1979;206:477–479. doi: 10.1126/science.504992. [DOI] [PubMed] [Google Scholar]
- 25.Mason I. Cell signalling. Do adhesion molecules signal via FGF receptors? Curr Biol. 1994;4:1158–1161. doi: 10.1016/s0960-9822(00)00263-3. [DOI] [PubMed] [Google Scholar]
- 26.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ono K, Tomasiewicz H, Magnuson T, Rutishauser U. N-CAM mutation inhibits tangential neuronal migration and is phenocopied by enzymatic removal of polysialic acid. Neuron. 1994;13:595–609. doi: 10.1016/0896-6273(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 28.Palay S L, Chan-Palay V. Cerebellar cortex. Heidelberg, Germany: Springer-Verlag Berlin; 1974. [Google Scholar]
- 29.Rowe L B, Nadeau J H, Turner R, Frankel W N, Letts V A, Eppig J T, Ko M S, Thurston S J, Birkenmeier E H. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm Genome. 1994;5:253–274. doi: 10.1007/BF00389540. . (Erratum, 5:463.) [DOI] [PubMed] [Google Scholar]
- 30.Salbaum J M. Genomic structure and chromosomal localization of the mouse gene punc. Mamm Genome. 1999;10:107–111. doi: 10.1007/s003359900953. [DOI] [PubMed] [Google Scholar]
- 31.Salbaum J M. Punc, a novel mouse gene of the immunoglobulin superfamily, is expressed predominantly in the developing nervous system. Mech Dev. 1998;71:201–204. doi: 10.1016/s0925-4773(98)00005-7. [DOI] [PubMed] [Google Scholar]
- 32.Salminen M, Meyer B I, Bober E, Gruss P. Netrin 1 is required for semicircular canal formation in the mouse inner ear. Development. 2000;127:13–22. doi: 10.1242/dev.127.1.13. [DOI] [PubMed] [Google Scholar]
- 33.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skarnes W C, Moss J E, Hurtley S M, Beddington R S P. Capturing genes encoding membrane and secreted proteins important for mouse development. Proc Natl Acad Sci USA. 1995;92:6592–6596. doi: 10.1073/pnas.92.14.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 36.Stark M R, Sechrist J, Bronner-Fraser M, Marcelle C. Neural tube-ectoderm interactions are required for trigeminal placode formation. Development. 1997;124:4287–4295. doi: 10.1242/dev.124.21.4287. [DOI] [PubMed] [Google Scholar]
- 37.Tomasiewicz H, Ono K, Yee D, Thompson C, Goridis C, Rutishauser U, Magnuson T. Genetic deletion of a neural cell adhesion molecule variant (N-CAM-180) produces distinct defects in the central nervous system. Neuron. 1993;11:1163–1174. doi: 10.1016/0896-6273(93)90228-j. [DOI] [PubMed] [Google Scholar]
- 38.Treloar H, Tomasiewicz H, Magnuson T, Key B. The central pathway of primary olfactory axons is abnormal in mice lacking the N-CAM-180 isoform. J Neurobiol. 1997;32:643–658. doi: 10.1002/(sici)1097-4695(19970620)32:7<643::aid-neu1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Vaughn D E, Bjorkman P J. The (Greek) key to structures of neural adhesion molecules. Neuron. 1996;16:261–273. doi: 10.1016/s0896-6273(00)80045-8. [DOI] [PubMed] [Google Scholar]
- 40.Villamar M, del Castillo I, Valle N, Romero L, Moreno F. Deafness locus DFNB16 is located on chromosome 15q13–q21 within a 5-cM interval flanked by markers D15S994 and D15S132. Am J Hum Genet. 1999;64:1238–1241. doi: 10.1086/302321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh F S, Doherty P. Neural cell adhesion molecules of the immunoglobulin superfamily: role in axon growth and guidance. Annu Rev Cell Dev Biol. 1997;13:425–456. doi: 10.1146/annurev.cellbio.13.1.425. [DOI] [PubMed] [Google Scholar]
- 42.Watase K, Hashimoto K, Kano M, Yamada K, Watanabe M, Inoue Y, Okuyama S, Sakagawa T, Ogawa S, Kawashima N, Hori S, Takimoto M, Wada K, Tanaka K. Motor discoordination and increased susceptibility to cerebellar injury in GLAST mutant mice. Eur J Neurosci. 1998;10:976–988. doi: 10.1046/j.1460-9568.1998.00108.x. [DOI] [PubMed] [Google Scholar]
- 43.Yang W, Mansour S L. Expression and genetic analysis of prtb, a gene that encodes a highly conserved proline-rich protein expressed in the brain. Dev Dyn. 1999;215:108–116. doi: 10.1002/(SICI)1097-0177(199906)215:2<108::AID-DVDY3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 44.Yang W, Musci T S, Mansour S L. Trapping genes expressed in the developing mouse inner ear. Hear Res. 1997;114:53–61. doi: 10.1016/s0378-5955(97)00146-9. [DOI] [PubMed] [Google Scholar]
- 45.Yee K T, Simon H H, Tessier-Lavigne M, O'Leary D M. Extension of long leading processes and neuronal migration in the mammalian brain directed by the chemoattractant netrin-1. Neuron. 1999;24:607–622. doi: 10.1016/s0896-6273(00)81116-2. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Q Y, Johnson K R, Erway L C. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]