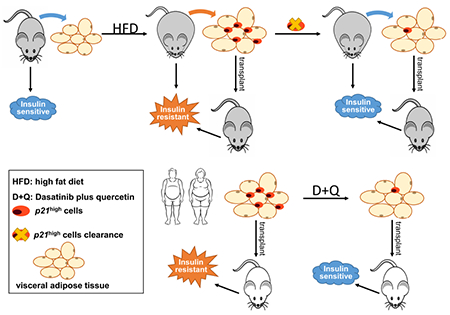
Summary
Insulin resistance is a pathological state often associated with obesity, representing a major risk factor for type 2 diabetes. Limited mechanism-based strategies exist to alleviate insulin resistance. Here, using single-cell transcriptomics, we identify a small, critically important, but previously unexamined cell population, p21Cip1-highly-expressing (p21high) cells, which accumulate in adipose tissue with obesity. By leveraging a p21-Cre mouse model, we demonstrate that intermittent clearance of p21high cells can both prevent and alleviate insulin resistance in obese mice. Exclusive inactivation of the NF-κB pathway within p21high cells, without killing them, attenuates insulin resistance. Moreover, fat transplantation experiments establish that p21high cells within fat are sufficient to cause insulin resistance in vivo. Importantly, a senolytic cocktail, dasatinib plus quercetin, eliminates p21high cells in human fat ex vivo, and mitigates insulin resistance following xenotransplantation into immuno-deficient mice. Our findings lay the foundation for pursuing the targeting of p21high cells as a new therapy to alleviate insulin resistance.
Keywords: cellular senescence, diabetes, senolytics, fat transplantation, NF-κB
eTOC
Wang et al. identify a previously unexamined senescent cell population (p21high cells) in fat and demonstrate their causal role in metabolic dysfunction with obesity, as well as the underlying mechanism involved. Treatment with a senolytic cocktail, dasatinib plus quercetin, reduces p21high cells in human fat and alleviates their harmful metabolic effects following transplantation into immuno-deficient mice in vivo.
Graphical Abstract
Introduction
More than 93 million US adults are obese (Centers for Disease Control and Prevention, 2018). Obesity accelerates the aging processes (Tchkonia et al., 2010), and is among the most significant risk factors for the development of type 2 diabetes (T2D), one of the leading causes of death for older adults (Palmer et al., 2015). Insulin resistance (IR) is the hallmark and the earliest detectable abnormality for prediabetes (Defronzo, 2009), a precursor of T2D (DeFronzo et al., 2015). IR refers to impaired tissue responses to insulin signaling, such as in the fat, liver and muscle (Muoio and Newgard, 2008), which eventually leads to pancreatic β-cell dysfunction, and results in abnormal insulin secretion. Due to its complexity, mechanisms by which IR and metabolic dysfunction develop are not fully understood. Besides lifestyle changes, such as exercise and healthy diet, common medications for metabolic dysfunction and T2D include metformin, insulin and insulin secretion inducers, insulin sensitizers, and incretin hormone modulators (DeFronzo et al., 2015). Novel therapeutic targets for IR and metabolic dysfunction are needed to optimize and improve current treatments.
Cellular senescence, refers to the cell fate that occurs in response to a range of cellular stresses and that involves, in essence, an irreversible proliferative arrest (Gorgoulis et al., 2019). Senescent cells accumulate in multiple tissues with obesity and aging (Munoz-Espin and Serrano, 2014; Palmer et al., 2019; Schafer et al., 2016), and they secrete a variety of pro-inflammatory cytokines, chemokines and proteases, termed the senescence-associated secretory phenotype (SASP) (Coppé et al., 2008; Xu et al., 2015b). A role for cellular senescence in promoting metabolic dysfunction has been recently identified (Aguayo-Mazzucato et al., 2019; Palmer et al., 2019; Xu et al., 2015a). However, several major questions remain unanswered. First, the transgenic mouse models used in these previous studies only targeted p16INK4a-highly-expressing (p16high) cells, but senescent cells are highly heterogeneous (Gasek et al., 2021; Gorgoulis et al., 2019; Tripathi et al., 2021). The understanding of cellular senescence has been evolving and it has been recognized that high p16 expression might not be specific or sufficient to define senescent cells because not all p16high cells are senescent (Frescas et al., 2017; Hall et al., 2017), and not all senescent cells express high levels of p16 (Gorgoulis et al., 2019). Thus, it is important to examine the role of other senescent cells in metabolic dysfunction and other conditions. Second, the mechanisms by which senescent cells cause metabolic dysfunction are largely unknown in vivo due to the limitations of current models. Third, other than a limited number of studies (Yoshimoto et al., 2013), it is largely unknown whether it is feasible to target senescent cells only in specific tissues, rather than systemically to alleviate metabolic dysfunction.
In this study, we leveraged a single-cell transcriptomic (SCT) analysis to identify a previously unexplored senescent cell population, p21Cip1-highly-expressing (p21high) cells. While p21 is one of the two major regulators and cellular markers for senescent cells (Gorgoulis et al., 2019), the role of p21high cells remains largely unknown in various pathological conditions in vivo. Recently, we generated and validated a new p21-CreERT2 transgenic mouse model containing a p21 promoter driving a bicistronic message consisting of Cre recombinase (Cre) fused to a tamoxifen-inducible estrogen receptor (ER) element (Wang et al., 2021). p21high cells targeted by this model have a number of typical senescence-related features including enlarged cell size, higher senescence-associated beta-galactosidase (SA-β-gal) activity, inhibited cell proliferation, lower Lamin B1 expression, and a SASP (Wang et al., 2021). By crossing this mouse model with a number of floxed mouse models, we managed to monitor, kill or modulate p21high cells in vivo without affecting other cells (Wang et al., 2021). Here, we used these mouse models along with fat transplantation to investigate the role and underlying mechanisms of p21high cells in metabolic dysfunction with obesity in vivo. Our findings demonstrate that p21high cells in gonadal visceral adipose tissue (gVAT) play a causal role in obesity-induced metabolic dysfunction, and they provide new insights into novel therapeutic targets to alleviate this condition.
Results
p21high cells accumulate in adipose tissue with obesity
To gain a deeper and comprehensive understanding of p21high cells in adipose tissue with obesity, we fed 2-month-old male C57BL/6 mice with a regular chow diet (RCD) or high-fat diet (HFD) for 2 months, and performed SCT analysis on the stromal vascular fraction (SVF) of gVAT from 3 RCD-fed lean and 3 HFD-fed obese mice (labelled by different hashtags allowing us to distinguish cells from different donors). After quality control, we obtained SCT information from 11,401 and 7,283 cells from lean and obese mice, respectively. On average, more than 2,800 genes and 11,000 polyadenylated RNA transcripts were detected per cell. Using unsupervised clustering and the expression of cell-type specific markers (Figure S1A), we identified a total of 14 cell clusters covering 9 different cell types (Figure 1A). We found that the gVAT from obese mice contained more p21high cells than the gVAT from lean mice, and that these p21high cells were mostly preadipocytes, endothelial cells and macrophages (Figure 1B–1D). Most sub-clusters in each cell type express a higher level of p21 in SVF from obese mice compared to SVF from lean mice, indicating that p21 alone might not be sufficient and specific enough to define a diet-induced sub-cluster of cells. Interestingly, at the time point of 1.5 to 2 months after HFD feeding, p16high cell accumulation is not evident in gVAT from obese mice (Figure S1B), which was further confirmed by RNA in situ hybridization assay (RNAScope, Figure S1C–1E) and flow cytometry analysis (Figure S1F). After 10 months of HFD feeding, we observed accumulation of both p21high and p16high cells in the gVAT (Figure S1G), and these 2 populations are distinguishable in spite of some overlap (Figure S1H).
Figure 1. Single cell analysis of p21high cells in gVAT from lean and obese mice.
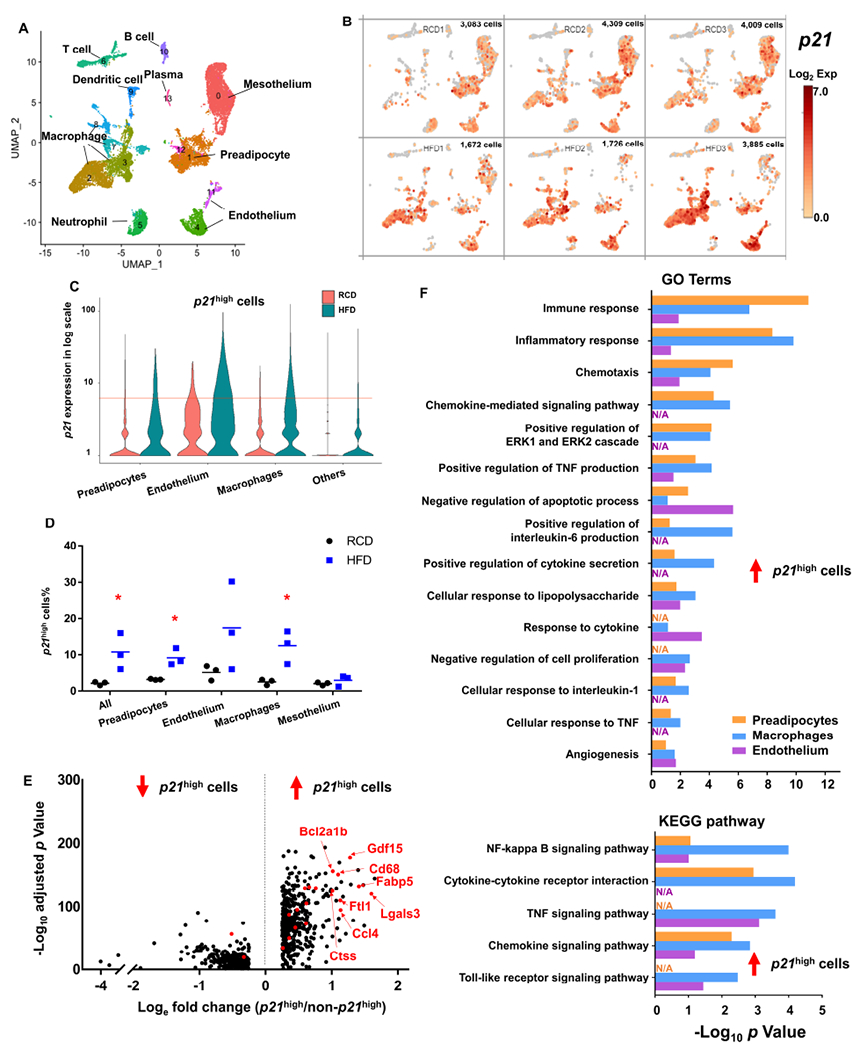
(A) Uniform manifold approximation and projection (UMAP) plot for unsupervised clustering of the single cells from SVF from RCD and HFD gVAT identified a total of 14 cell populations.
(B) UMAP plots showing log2 (normalized expression of p21) in RCD (upper) and HFD (bottom) mice. Each panel represented one mouse, and the number of single cells was shown on the top right of each panel.
(C) Violin plot of p21 expression in preadipocytes, endothelium, macrophages and others from RCD and HFD mice. Cells with p21 expression level > 6 (red line) were considered to be p21high.
(D) Proportion of p21high cells (p21 expression level > 6) in all cells, preadipocytes, endothelium, macrophages and mesothelium. n = 3 for both groups. n represents the number of biological replicates with 1 technical replicate. Results were shown as means ± s.e.m. *P < 0.05; two-tailed Welch’s t-test.
(E) Volcano plot of the down (left) and up-regulated (right) DEGs in p21high cells in all cell types. Overlapped DEGs among p21high preadipocytes, endothelium and macrophages are highlighted in red.
(F) Gene ontology (upper) and KEGG (bottom) analysis (https://david.ncifcrf.gov/) of the up-regulated DEGs in p21high cells of 3 cell types.
See also Figure S1.
We next compared the transcriptomes between p21high cells (1,143 cells) and all other cells (17,541 cells), and acquired differentially expressed genes (DEGs). We found 463 genes were up-regulated (logefold change > 0.25) and 302 were down-regulated (logefold change < −0.25) in the p21high cells (adjusted p value < 0.05; Figure 1E). To account for differences in cell type composition, we also acquired the DEGs of p21high preadipocytes, macrophages and endothelial cells. We found 21 genes were overlapped in all 3 DEG datasets (highlighted by red in Figure 1E). Using gene ontology (GO) analysis, we found p21high cells showed higher activity of inflammatory response, chemotaxis, negative regulation of apoptosis and angiogenesis than the other cells among the three cell types (Figure 1F). In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that NF-kappa B and chemokine signaling pathways were enriched in up-regulated genes from p21high cells for all three cell types (Figure 1F).
Next, we leveraged our newly generated p21-Cre mouse model to validate these findings. This model carries a transgene (Figure S2A) containing 3,225bp of the p21 promoter driving a CreERT2 as well as an internal ribosome entry site (IRES) followed by an enhanced green fluorescence protein (GFP), which allows us to detect p21high cells. Indeed, GFP+ cells expressed higher levels of p21 protein than GFP− cells (Figure S2B). We isolated SVF from obese or lean mice, and analyzed GFP+ cells in various cell populations using flow cytometry (Figure S2C). Based on SCT results (Figure S1A), we defined CD45−; CD31−; SCA1+ cells as preadipocytes, CD45−; CD31+ cells as endothelial cells and CD45+; F4/80+; CD11b+ cells as macrophages. We found all these cell types contained more GFP+ p21high cells in SVF from obese mice than lean mice (Figure S2D), consistent with our SCT analysis. Moreover, we crossed the p21-Cre mice with floxed knock-in tdTomato (Tom) (driven by CMV early enhancer/chicken β actin [CAG] promoter) mice (Madisen et al., 2010), and generated p21-Cre/+; Tom/+ (PT) mice (Figure S3A), which allow us to detect p21high cells by tdTomato. Similar to GFP, Tom+ cells express higher levels of p21 protein than Tom− cells (Figure S3B), and Tom+ p21high cells in SVF from obese mice were mainly preadipocytes, endothelial cells and macrophages (Figure S3C). In addition to gVAT, we examined Tom+ p21high cells in another 4 major metabolism-related tissues (liver, muscle, pancreas, and brown fat). We detected very few Tom+ p21high cells in all 5 tissues in RCD-fed lean mice. After 3 months of HFD feeding, Tom+ p21high cells were only observed in gVAT. After 6 months of HFD feeding, the Tom+ cells percentage in gVAT was 10-20%, and we started to see small numbers of Tom+ cells (~1-2%) in the liver (which is consistent with a previous study (Yoshimoto et al., 2013)), while Tom+ cells remained undetectable in the pancreas, muscle and brown fat (Figure S3D). These findings demonstrate the existence of a unique p21high cell population, which accumulates in gVAT with obesity.
Intermittent clearance of p21high cells alleviates metabolic dysfunction in obese mice
We next examined whether p21high cells play a causal role in metabolic dysfunction induced by obesity. We first crossed the p21-Cre mice with floxed knock-in firefly luciferase (LUC) mice (Safran et al., 2003), which contain a loxP-flanked STOP fragment between the Gt(ROSA)26Sor promoter and LUC. We generated p21-Cre/p21-Cre; LUC/LUC mice, and further crossed these mice with floxed diphtheria toxin A (DTA) mice (Figure 2A), which contain a floxed-STOP cassette followed by DTA driven by ROSA promoter (Voehringer et al., 2008). We generated p21-Cre/+; LUC/+ (PL) mice, allowing us to detect p21high cells in living mice through bioluminescent imaging (BLI) in a temporal manner and p21-Cre/+; LUC/DTA (PLD) mice (Figure 2A), enabling us to specifically kill (by DTA) as well as monitor p21high cells (by LUC) in vivo. This breeding strategy allowed us to use PL and PLD littermates for metabolic testing to minimize potential confounding effects from the litter size (Parra-Vargas et al., 2020). We fed both PL and PLD male mice with RCD or HFD for 2 months, and then treated them with 1 course (daily for 2 days) of tamoxifen (TAM) to activate Cre. Using BLI, we found that PL mice under RCD had little BLI signal, while a much higher BLI signal was observed in HFD-fed PL mice (Figure S4A and S4B), demonstrating the accumulation of p21high cells. The BLI signals in HFD-fed PLD mice were significantly lower compared to HFD-fed PL mice (Figure S4A and S4B), indicative of successful clearance of p21high cells by DTA. Using flow cytometry, we further confirmed that fewer GFP+ p21high preadipocytes, macrophages and leukocytes were in gVAT from PLD mice compared to their PL littermates, while the number of p21high endothelial cells was not altered (Figure 2B). Clearance of p21high cells resulted in lower SA-β-gal activity in the gVAT (Figure S4C). As SA-β-gal activity is not specific to senescent cells (Hall et al., 2017), we further assessed the senescent cell burden in the gVAT using cell proliferation and telomere-associated foci (TAF) assays, two relatively reliable and specific senescence markers. We found that gVAT from PLD mice contain fewer TAF-positive cells and have more EdU-positive cells than PL gVAT (Figure 2C–2E), indicating that clearance of p21high cells can reduce senescent cells. Similar to Figure S3D, p21high cells indicated by the BLI signals were primarily located in visceral fat in PL mice (Figure S4D). These data demonstrates that p21high cells are induced in visceral fat with obesity, and can be eliminated in PLD mice.
Figure 2. Clearance of p21high cells alleviates metabolic dysfunction from 2 months of HFD.
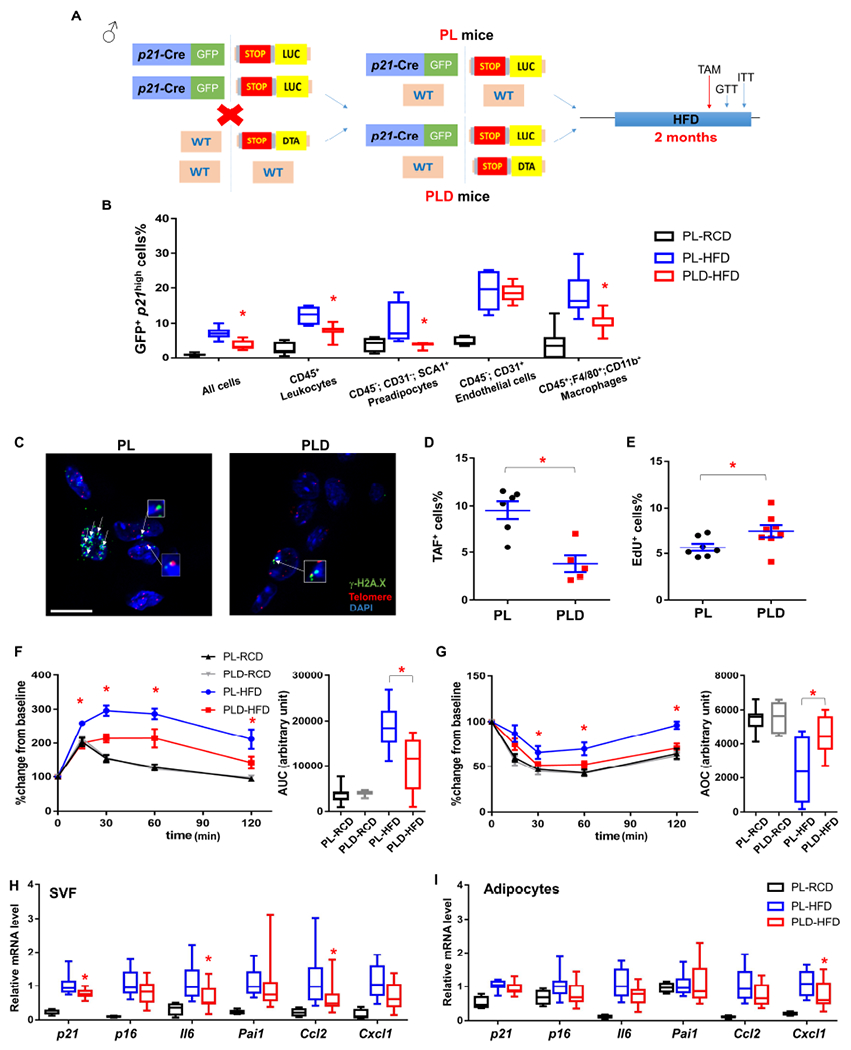
(A) Genetic crosses and experimental design.
(B) Percent of cells that are GFP+ across different cell types within gVAT.
(C) Representative TAF images (4-5 images per biological replicate). A single layer obtained from z-stacks with 20+ layers is shown. TAFs are indicated by white arrows. Gamma H2A.X (green), Telomere probe (red), DAPI (blue). Cells are considered to be TAF+ if they contain no less than 3 TAFs from the entire z-stacks. Scale bar = 10 μm.
(D) Proportion of TAF+ cells.
(E) Proportion of EdU+ cells.
(F) GTT curve (mean ± s.e.m.) and area under curve (AUC), (G) ITT curve (mean ± s.e.m.) and area over curve (AOC) in PL and PLD mice.
(H) Relative mRNA expression in SVF.
(I) Relative mRNA expression in adipocytes.
For B, n = 7 for all groups. For D, n= 6 for PL, n = 5 for PLD. For E, n = 7 for PL, n = 8 for PLD. For F and G, n = 7 for PL-RCD and PLD-RCD; n = 8 for PL-HFD and PLD-HFD. For H and I, n = 4 for PL-RCD, n = 11 for PL-HFD and PLD-HFD. For B, E-I, n represents the number of biological replicates with 1 technical replicate. For D, n represents the number of biological replicates with 2-5 technical replicates. Results were shown as mean ± s.e.m. or box-and-whisker plots, where a box extends from the 25th to 75th percentile with the median shown as a line in the middle, and whiskers indicate the smallest and largest values. *P < 0.05 vs PL-HFD by one-way ANOVA (B, H-I), by two-way ANOVA (F-G), or by two-tailed Welch’s t-test (D-E).
We next investigated whether clearance of p21high senescent cells impacts metabolic function. We found PL male mice had impaired glucose homeostasis (by glucose tolerance test; GTT) and insulin sensitivity (by insulin tolerance test; ITT) after 2 months of HFD feeding while elimination of p21high cells in HFD-fed PLD littermates alleviated this metabolic dysfunction (Figure 2F and 2G). These findings were repeated in another independent cohort of mice using a similar experimental design (Figure S4E). No difference in GTT or ITT was found between PL and PLD mice fed with RCD (Figure 2F and 2G), consistent with the fact that few p21high cells were observed in these mice (Figure S3D and S4A). Although tamoxifen can impact insulin sensitivity (Guillaume et al., 2017), we administrated tamoxifen (also only 2 doses) to all groups, making it a less likely contributor to the metabolic function improvement observed in PLD mice. HFD-fed PL and PLD mice had similar GTT and ITT results without TAM administration (Figure S4F), suggesting the “leaking” of Cre or STOP cassette without TAM in these mice is minimal.
PL and PLD mice had similar body weight, fat mass and lean mass before and after TAM treatment (Figure S5A), indicating that clearance of p21high cells did not alter these parameters. As certain behavioral changes, such as activity and calorie restriction, are well known to impact insulin sensitivity, we measured food intake and daily activity using the Comprehensive Laboratory Animal Monitoring System (CLAMS) in these mice, and no difference was found (Figure S5B and S5C), suggesting that improved insulin sensitivity is likely due to tissue function improvement rather than behavioral changes. Pancreatic β-cells can be another major contributor to improved glucose tolerance, even if no p21high cells were observed in pancreas (Figure S3D). To test this possibility, we performed glucose stimulated insulin secretion (GSIS) tests to assess β-cell function, and quantified the size and number of pancreatic islets by insulin staining. No difference was observed between PL and PLD mice (Figure S5D–S5F), suggesting pancreatic β-cells might not be a major contributor to the metabolic benefits we observed in PLD mice.
To gain mechanistic insight, we isolated SVF and adipocytes from gVAT from obese PL and PLD mice along with lean PL mice. Clearance of p21high cells resulted in a modest (~25%) but statistically significant lower p21 expression in PLD SVF than the PL SVF (Figure 2H), consistent with the fact that our p21-Cre model only targets a small percentage (~10%, Figure S2D and S3C) of SVF cells (p21high cells) without affecting p21 expression in other cells. Another key senescence marker, p16, was not changed in PLD SVF (Figure 2H). p21high cells highly express several SASP genes (Wang et al., 2021), which are closely associated with insulin resistance (Roden and Shulman, 2019). Clearance of p21high cells showed less expression of most of these SASP genes in the whole PLD SVF than the PL SVF (Figure 2H). For adipocytes, p21 expression level was similar between PL and PLD adipocytes (Figure 2I), suggesting that adipocytes might not be a major cell type in the p21high cell population targeted by our model. Despite this, expression of one SASP gene (Cxcl1) was lower in PLD adipocytes than PL adipocytes (Figure 2I), consistent with our previous finding that the SASP can induce SASP gene expression in nearby non-senescent cells (Xu et al., 2018; Xu et al., 2015b). Impairment of adipogenesis can lead to insulin resistance with aging or obesity (Palmer et al., 2019; Xu et al., 2015a). However, for both SVF and adipocyte populations, the expression levels of several key adipogenesis markers were not different between PL and PLD groups (Figure S5G and S5H), indicating that adipogenesis might not be a major mechanism for the metabolic benefits observed in PLD mice. Elimination of p21high cells did not statistically alter leukocyte (CD45+ cells) numbers including CD45+;F4/80+;CD11b+ macrophages (Figure S5I and S5J). Notably, we found that the expression levels of several key markers of M2-like macrophages are higher in PLD mice than PL mice (Figure S5K), suggesting that M2-polarized macrophages could be a potential contributor to the improved metabolic function.
Intermittent clearance of p21high cells has long-term benefits protecting against metabolic dysfunction in obese mice
We next investigated if long-term clearance of p21high cells could be beneficial for metabolic function in obesity. Although HFD-fed male PLD mice had better glucose tolerance and insulin sensitivity than PL mice after 1 course of p21high cell clearance (Figure 2F and 2G), the benefits disappeared after 1 month without TAM administration (Figure 3A–3C), but became evident again shortly after 1 more round of p21high cell clearance (Figure 3D–3F). Monthly clearance of p21high cells (2 doses of TAM per month) was sufficient to protect PLD mice from HFD-induced metabolic dysfunction for at least 8 months (Figure S6A).
Figure 3. Intermittent clearance of p21high cells alleviates insulin resistance with obesity over the long term.
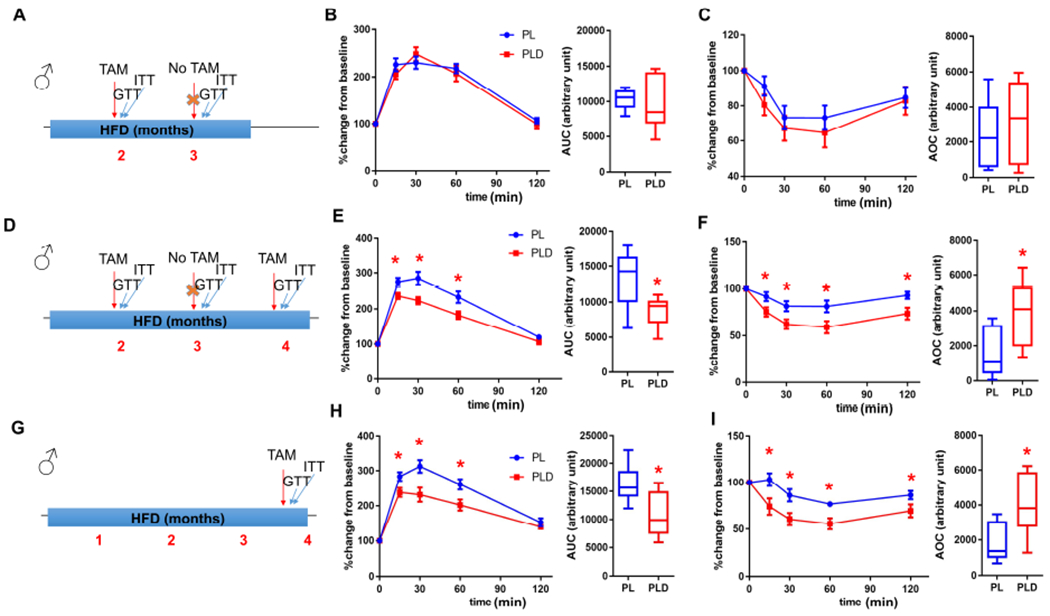
(A) Experimental timeline for B and C.
(B-C) GTT curve (mean ± s.e.m.) and AUC (B), ITT curve (mean ± s.e.m.) and AOC (C) in PL and PLD mice 3 months after initiating HFD.
(D) Experimental timeline for E and F.
(E-F) GTT curve (mean ± s.e.m.) and AUC (E), ITT curve (mean ± s.e.m.) and AOC (F) in PL and PLD mice 4 months after initiating HFD.
(G) Experimental timeline for H and I.
(H-I) GTT curve (mean ± s.e.m.) and AUC (H), ITT curve (mean ± s.e.m.) and AOC (I) in PL and PLD mice 4 months after initiating HFD.
For B, C, E, F, H and I, n = 8 for all groups. n represents the number of biological replicates with 1 technical replicate. Results were shown as box-and-whisker plots, where a box extends from the 25th to 75th percentile with the median shown as a line in the middle, and whiskers indicate the smallest and largest values. *P < 0.05 vs PL by two-way ANOVA (GTT and ITT curves), or by two-tailed Welch’s t-test (AUC and AOC).
See also Figure S6.
From a translational perspective, it is critical to determine whether targeting p21high cells is still beneficial once insulin resistance is at an advanced stage. To test this, we fed both PL and PLD male mice with a HFD for 4 months, and then treated them with 1 course of TAM. Both glucose homeostasis and insulin sensitivity was improved in PLD compared to PL mice (Figure 3G–3I), indicating that eliminating p21high cells can alleviate metabolic dysfunction even at later stages. Notably, although most of the mouse experiments were performed at UConn Health (Farmington, CT), we had a cohort of mice tested at Mayo Clinic (Rochester, MN) with consistent findings. We treated PL and PLD male mice with TAM for 2 courses, 1 month and 5 months after HFD feeding respectively. HFD-induced glucose intolerance and insulin resistance was largely alleviated in PLD mice after only 4 doses of TAM (2 doses per course) within 5 months (Figure S6B), similar to our findings at UConn Health. Thus, local environmental effects, including differences in the microbiome, are unlikely to be major contributors to these findings. All these data provide evidence that intermittent clearance of p21high cells can confer long-term protection against obesity-induced metabolic dysfunction, and the timing of clearance can be quite flexible, enhancing potential clinical applicability.
Clearance of p21high cells has less metabolic benefit in female than male mice
Male C57BL/6 mice are known to have higher susceptibility to HFD-induced metabolic dysfunction compared to female mice (Champy et al., 2008; Yeadon, 2015). Although we mainly used male mice for our study because of this, we also examined the effect of p21high cells clearance in female mice (Figure 4A). Unlike male mice, 2 months of HFD feeding did not induce p21high cell accumulation in PL female mice (Figure 4B and 4C), which might partially explain lower vulnerability to metabolic stress in female mice. After 1 course of TAM administration, we found no difference in GTT results between HFD-fed PL and PLD female mice, although both of these groups showed mild glucose tolerance impairment and higher body weight compared to RCD-fed mice (Figure 4D and 4E). Improvements in glucose tolerance and insulin sensitivity became evident in PLD female mice after 7 monthly courses of TAM treatment on a HFD (Figure 4F and 4G). Additionally, body weight did not differ between groups (Figure 4H). Thus, p21high cells play a causal role in metabolic dysfunction in both male and female mice.
Figure 4. Clearance of p21high cells has less metabolic benefit in female than male mice.
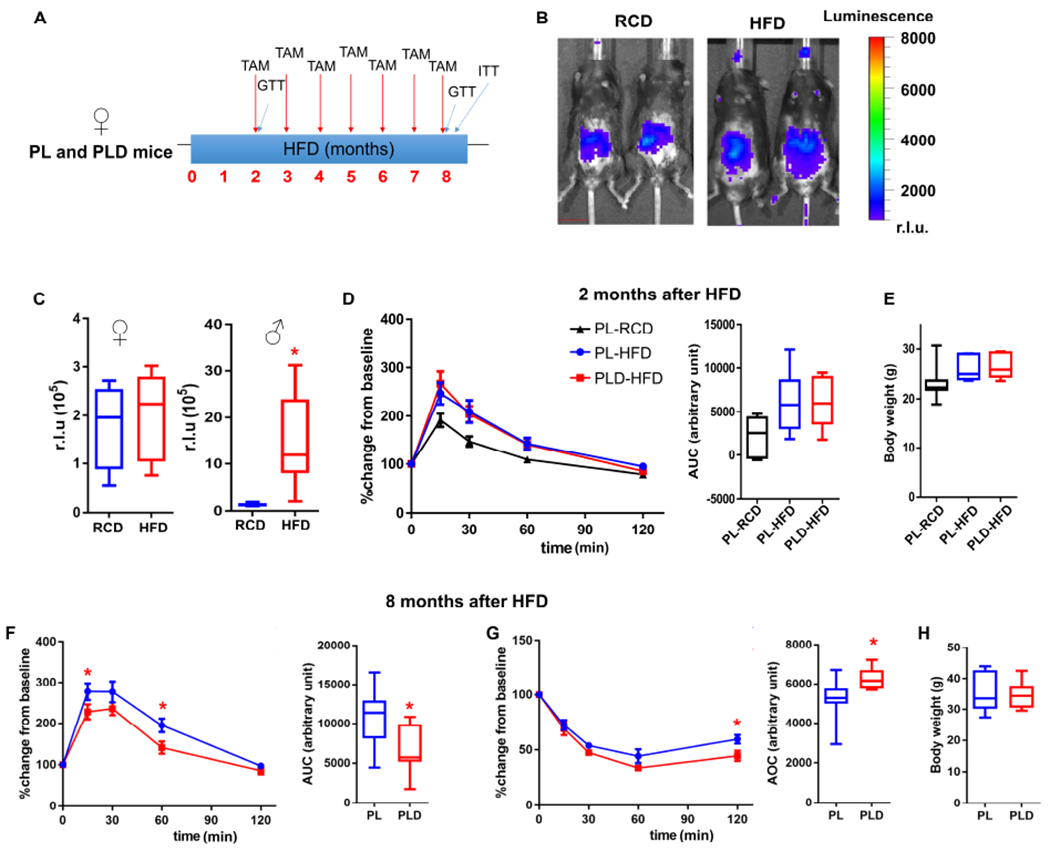
(A) Experimental design.
(B) Representative images (1 image per 4 - 7 biological replicates) of LUC activity in female PL mice fed with RCD (n = 4) or HFD (n = 7). Scale bar = 15 mm.
(C) Quantification of LUC activity.
(D-E) GTT curve (mean ± s.e.m.) and AUC (D), body weight (E) in female RCD-fed PL, HFD-fed PL, and HFD-fed PLD mice after 2 months of HFD feeding.
(F-H) GTT curve (mean ± s.e.m.) and AUC (F), ITT curve (mean ± s.e.m.), and AOC (G), body weight (H) in female PL and PLD mice after 8 months of HFD feeding.
For C, n = 4 for female RCD, n = 7 for female HFD, n = 6 for male RCD, n = 8 for male HFD. For D-H, n = 7 for all groups. For C-H, n represents the number of biological replicates with 1 technical replicate. Results were shown as box-and-whisker plots, where a box extends from the 25th to 75th percentile with the median shown as a line in the middle, and whiskers indicate the smallest and largest values. *P < 0.05 vs RCD by two-tailed Welch’s t-test (C); *P < 0.05 vs PL by two-tailed Welch’s t-test (F and G) or by two-way ANOVA (GTT and ITT curves).
Inactivation of the NF-κB pathway specifically in p21high cells alleviates obesity-induced metabolic dysfunction
In our SCT results, we found the NF-κB pathway is activated in p21high cells in vivo both with obesity (Figure 1F) and aging (Wang et al., 2020). Activation of NF-κB signaling is tightly linked to insulin resistance and early stages of diabetes (Patel and Santani, 2009). Moreover, the NF-κB pathway plays a crucial role in the SASP (Chien et al., 2011) and a number of SASP genes could induce insulin resistance (Palmer et al., 2015). Based on these points, we hypothesized that inhibition of NF-κB pathway in p21high cells might alleviate IR in obese mice. To test this, we obtained floxed Rela mice (Heise et al., 2014), which have a loxP-flanked exon 1 of the Rela gene (v-rel reticuloendotheliosis viral oncogene homolog A, or p65, a subunit crucial for NF-κB activation (Chen and Greene, 2004)). Upon Cre activation, Rela is mutated, leading to inactivation of the NF-κB pathway (Heise et al., 2014). By using p21-Cre/+; Tom/+; +/+ (PT) and p21-Cre/+; Tom/+; Relafl/fl (PT-Rela) mice (Figure 5A), we found that Rela mutation in p21high cells did not alter the p21high cell number (Figure 5B) after TAM administration. Rela protein level was significantly lower in p21high cells (but not in non-p21high cells) from PT-Rela gVAT vs. PT gVAT (Figure 5C), demonstrating the efficiency and specificity of Rela mutation in our model. We next crossed p21-Cre/+; Relafl/fl mice with +/+; Relafl/fl mice to generate +/+; Relafl/fl (Rela) and p21-Cre/+; Relafl/fl (P-Rela) littermates. We fed both Rela and P-Rela mice with a HFD for 2 months, and then treated them with TAM for 1 course (Figure 5D). Although Rela was only mutated in p21high cells in the P-Rela mice, it resulted in lower expression levels of a number of SASP genes in the whole gVAT (Figure 5E). p21 and p16 expression levels were not statistically different between Rela and P-Rela gVAT (Figure 5E), consistent with the fact that inactivation of Rela does not affect survival of senescent cells (Chien et al., 2011). Importantly, P-Rela mice had better glucose tolerance and alleviated insulin resistance compared to Rela mice after HFD feeding (Figure 5F and 5G). Similar to clearance of p21high cells, inactivation of Rela in p21high cells did not affect body composition, food intake, or daily activity (Figure 5H–5J). These findings indicate that the NF-κB pathway is one of the key mechanisms responsible for p21high cells causing IR in vivo.
Figure 5. Inactivation of the NF-κB pathway specifically in p21high cells alleviates obesity-induced metabolic dysfunction.
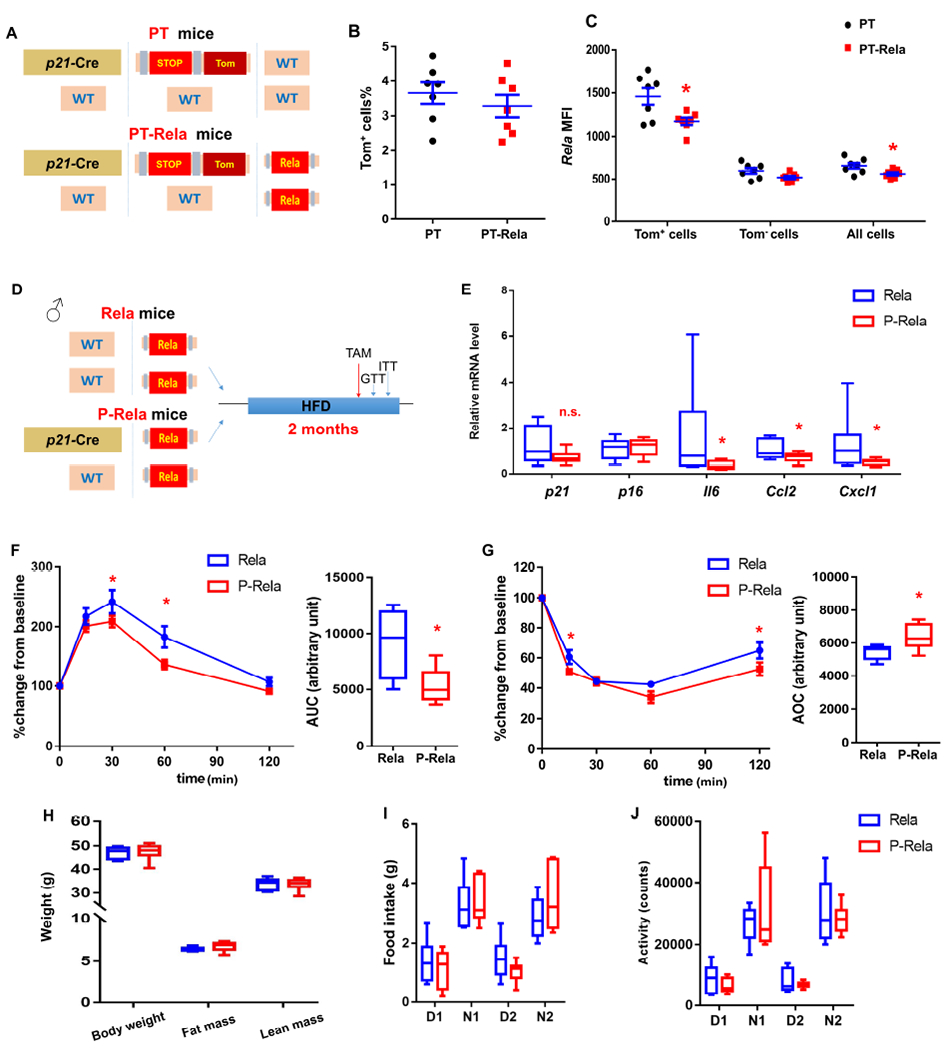
(A) Transgenic schematic of PT and PT-Rela mice.
(B) Proportion of Tom+ p21high cells in SVF from PT and PT-Rela mice fed with HFD.
(C) Mean fluorescence intensity (MFI) of Rela staining by flow cytometry in Tom+, Tom− and all SVF cells.
(D) Experimental design.
(E) Relative mRNA expression in gVAT.
(F) GTT curve (mean ± s.e.m.) and AUC, (G) ITT curve (mean ± s.e.m.) and AOC in HFD-fed Rela and P-Rela mice.
(H) Body composition.
(I-J) Food intake (I) and activity (J) during daytime (D) and night (N) for 2 days in HFD-fed Rela and P-Rela mice.
For B and C, n = 7 for both groups. Results were shown as means ± s.e.m. For E, n = 8 for Rela, n = 9 for P-Rela. For F and G, n = 7 for Rela, n = 6 for P-Rela. For H-J, n = 6 for both groups. For B-C, E-J, n represents the number of biological replicates with 1 technical replicate. Results were shown as box-and-whisker plots, where a box extends from the 25th to 75th percentile with the median shown as a line in the middle, and whiskers indicate the smallest and largest values. n.s, no significance vs Rela by two-tailed Welch’s t-test (E). *P < 0.05 vs PT (C) or Rela (E, F, G) by two-tailed Welch’s t-test, or by two-way ANOVA (GTT and ITT curves).
Elimination of p21high cells in gVAT is sufficient to alleviate obesity-induced metabolic dysfunction
Our p21-Cre mouse model targets p21high cells in all tissues. To explore the translational potential of targeting p21high cells further, we next examined whether eliminating p21high cells in a specific tissue could be feasible to achieve metabolic benefits. Based on histological analysis (Figure S3D) and BLI results (Figure S4D), we found that p21high cells mainly reside in gVAT. To test whether gVAT could be the primary targeting tissue with respect to p21high cells and metabolic dysfunction, we performed gVAT transplantation experiments. We first transplanted gVAT isolated from LUC transgenic mice into wild-type (WT) recipient mice (Figure S7A). Seven days after transplantation, we detected a strong BLI signal in the recipient mice (Figure S7B), and this signal remained detectable for at least 3 months after transplantation. As only donor gVAT carries the LUC transgene, this demonstrates that gVAT grafts remain biologically active in the recipients for months after transplantation. Consistent with this, we observed that the gVAT grafts were connected to the recipient fat 2 months after transplantation (Figure S7C), and had normal morphology by hematoxylin and eosin (H&E) staining (Figure S7D). These results validated our gVAT transplantation method. Next, we fed both PL and PLD male donor mice with a HFD for 3 months, and administrated TAM to them for 2 courses, each at 2 months and 3 months after HFD feeding respectively (Figure 6A). This experimental design allows accumulation of p21high cells in gVAT from PL mice as well as clearance of these cells in gVAT from PLD mice (Figure 2B and S4D). We then transplanted 0.7 g gVAT from either PL or PLD donor mice into the peritoneal cavity of 2-month-old lean recipient male mice. Transplantation of PL or PLD gVAT grafts did not affect the body composition or total weight in the recipient mice (Figure 6B). The weight (Figure 6C) and morphology (Figure S7D) of PL and PLD gVAT grafts were also similar in the recipient mice 2 months after transplantation, excluding the possibility of differential graft survival between PL and PLD donors. Importantly, compared to the sham group (mice with surgery but without gVAT transplantation), mice transplanted with gVAT from obese PL mice had impaired glucose tolerance and insulin sensitivity while elimination of p21high cells in PLD gVAT largely prevented these deleterious effects (Figure 6D and 6E). PL gVAT and PLD gVAT did not change daily activity, food intake or GSIS in the recipient mice (Figure 6F–6H). These results indicate that p21high cells in gVAT are sufficient to induce metabolic dysfunction in obese mice.
Figure 6. Elimination of p21high cells in gVAT is sufficient to alleviate obesity-induced metabolic dysfunction.
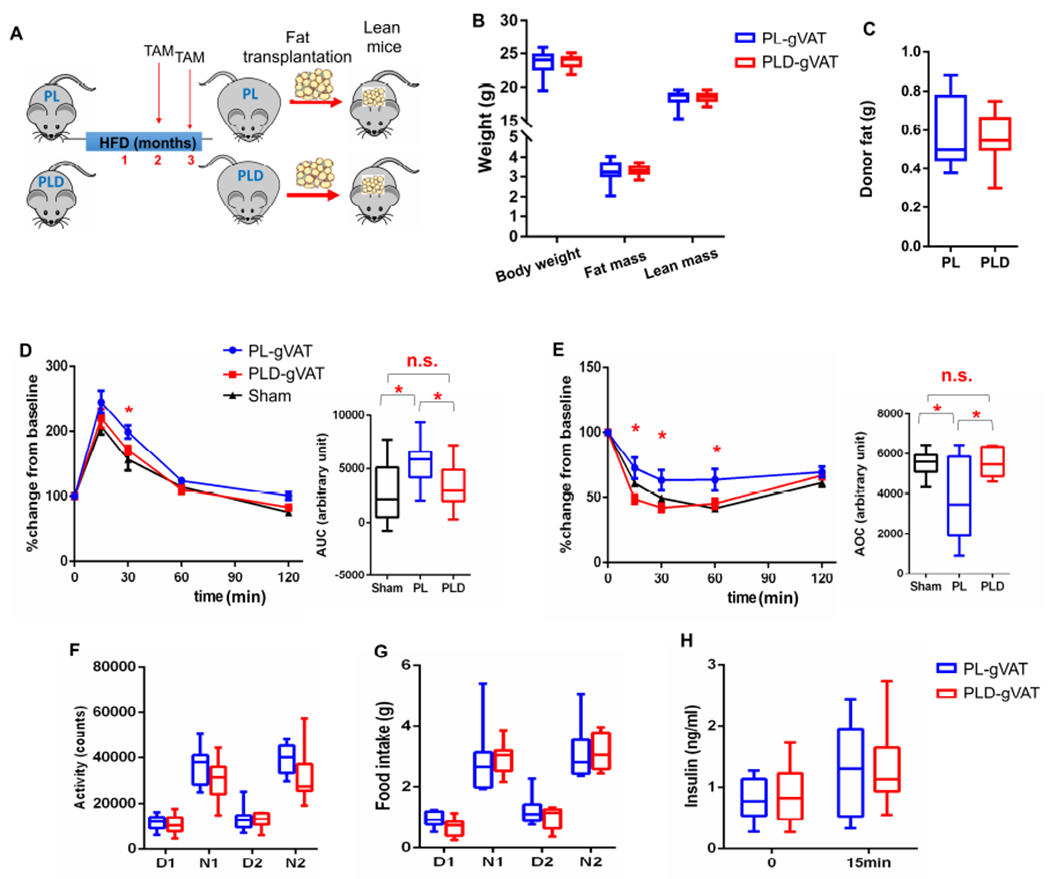
(A) Schematic of mouse gVAT transplantation experiments.
(B) Body composition.
(C) Donor fat weight 2 months after transplantation.
(D-E) GTT curve (mean ± s.e.m.) and AUC (D), ITT curve (mean ± s.e.m.) and AOC (E) in lean mice transplanted with no tissue (Sham), gVAT from PL mice (PL-gVAT), and gVAT from PLD mice (PLD-gVAT).
(F-G) Activity (F) and food intake (G) during daytime (D) and night (N) for 2 days.
(H) Plasma insulin at baseline and 15 minutes after glucose injection.
For B, C, H, n = 10 for both groups. For D and E, n = 8 for Sham, n = 10 for PL-gVAT, n = 10 for PLD-gVAT. For F, G, n = 8 for both groups. For B-H, n represents the number of biological replicates with 1 technical replicate. Results were shown as box-and-whisker plots, where a box extends from the 25th to 75th percentile with the median shown as a line in the middle, and whiskers indicate the smallest and largest values, n.s, no significance vs Sham by one-way ANOVA (D, E). *P < 0.05 vs PL by two-way ANOVA (GTT and ITT curves), or by one-way ANOVA (AUC and AOC).
See also Figure S7.
Pharmacological elimination of p21high cells in human VAT from individuals with obesity alleviates its harmful effect on metabolic function
We explored whether we could pharmacologically target p21high cells. Dasatinib plus quercetin (D+Q) has been shown to specifically kill senescent cells (Xu et al., 2018). By treating HFD-fed PT mice with D+Q, we found that D+Q-treated mice had significantly lower numbers of Tom+ p21high cells compared to control group, especially the p21high preadipocytes (Figure 7A).
Figure 7. Pharmacological elimination of p21high cells in VAT from humans with obesity alleviates its harmful effect on metabolic function in mice.
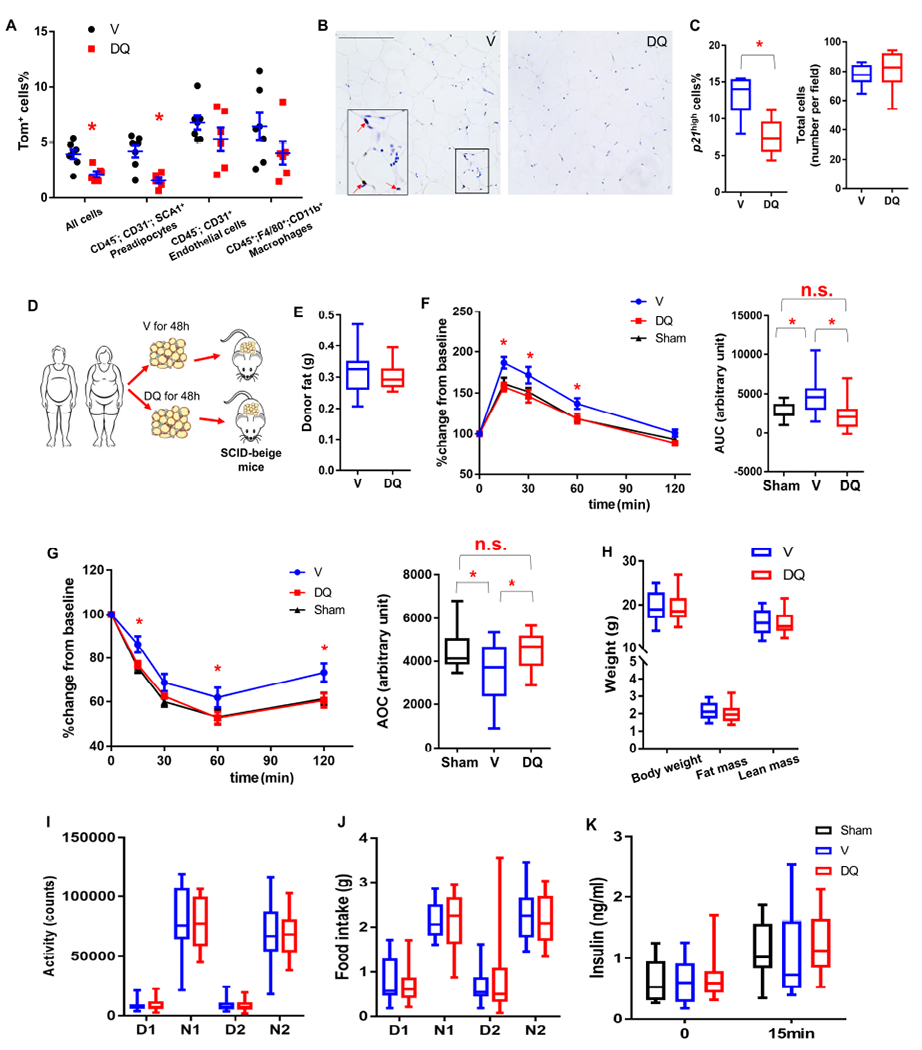
(A) Proportion of Tom+ cells in different cell types in gVAT from HFD-fed PT mice treated with D+Q (DQ) or vehicle (V).
(B) Representative p21 staining micrographs of human VAT explants treated with DMSO (V) or D+Q (DQ) (3 images per 8 biological replicates in each group). Red arrows indicate p21high cells. Scale bar = 250 μm.
(C) Percent of cells that are p21high cells among all cells and total cell number/field.
(D) Experimental design for human VAT transplantation.
(E) Human donor fat weight 2 months after transplantation.
(F-G) GTT curve (mean ± s.e.m.) and AUC (F), ITT curve (mean ± s.e.m.) and AOC (G) in SCID-Beige mice transplanted with no tissue (Sham), VAT explants from humans with obesity treated with DMSO (V) or D+Q (DQ).
(H) Body composition.
(I-J) Activity (I) and food intake (J) during daytime (D) and night (N) for 2 days.
(K) Plasma insulin at baseline and 15 minutes after glucose injection.
For A, n = 7 for V, n = 6 for DQ. Results were shown as means ± s.e.m. For C, n = 8 for both groups. For E-K, n = 12 for Sham, n = 13 for V, n = 14 for DQ. For A, C, E-H, n represents the number of biological replicates with 1 technical replicate. Results were shown as box-and-whisker plots, where a box extends from the 25th to 75th percentile with the median shown as a line in the middle, and whiskers indicate the smallest and largest values. n.s, no significance vs Sham by one-way ANOVA (F-AUC, G-AOC). *P < 0.05 vs V by two-tailed Welch’s t-test (A, C), by one-way ANOVA (F, G), or by two-way ANOVA (GTT and ITT curves).
We next examined whether D+Q could eliminate p21high cells in VAT samples from humans with obesity and whether such treated tissues could achieve in vivo metabolic benefits when transplanted into mice. We previously showed that D+Q could reduce senescent cells (using p16, telomere-associated foci [TAF], and senescence-associated β -galactosidase [SA-β gal] as markers) ex vivo in VAT explants from humans with obesity (Xu et al., 2018). Here, we examined p21high cell in VAT from humans with obesity. After 48h of D+Q (1 μM+20 μM) treatment, D+Q- treated VAT from human with obesity contained fewer p21high cells compared to vehicle- (V-) treated VAT from the same donor, while total cell numbers were not changed (Figure 7B and 7C), indicating that D+Q indeed could kill naturally occurring p21high cells in human tissue. D+Q also reduced pro-inflammatory SASP cytokine secretion by VAT from humans with obesity, including IL-6, IL-8 and MCP-1, without affecting adipokine secretion (Xu et al., 2018).
As a number of these cytokines are tightly associated with insulin resistance (Palmer et al., 2015), we next investigated whether D+Q can alleviate harmful metabolic effects of VAT from humans with obesity in vivo after transplantation into immune-deficient mice. We transplanted 0.5g of D+Q- or V-treated human VAT into the peritoneal cavity of SCID-beige (severe combined immunodeficiency-beige) mice (Figure 7D), which have impaired T cell, B cell, and natural killer cell function (Mosier et al., 1993). Similar to the mouse VAT transplantation experiments above, D+Q- or V- treated human VAT grafts were of similar weight in the recipient mice one month after transplantation (Figure 7E). Importantly, 0.5g V-treated VAT from humans with obesity induced glucose intolerance and insulin resistance in the SCID-beige recipient mice when compared to the Sham mice while D+Q treatment in these VAT samples fully mitigated the harmful metabolic effect of the transplanted VAT (Figure 7F and 7G). Body composition, daily activity, food intake and pancreatic β-cell function were not different between SCID-beige mice transplanted with V-treated and D+Q-treated human VAT (Figure 7H–7K). Thus, D+Q had similar metabolic effects on VAT from humans with obesity to those of clearing p21high cells in VAT from obese mice. These findings demonstrate that p21high cells in VAT are a key therapeutic target and that senolytic drugs could be a therapeutic strategy for alleviating obesity-induced metabolic dysfunction.
Discussion
Although it is postulated that senescent cells are highly heterogeneous, most of the studies using transgenic mouse models have been primarily focused on p16high senescent cells in various conditions. Here, we identify and characterize a novel senescent cell population, p21high cells. By sequencing more than 18,000 cells using SCT analysis in gVAT from obese and lean mice, we demonstrated that p21high cells and p21high cells are indeed two distinct cell populations, at least in the context of obesity. They differ in a number of ways: (1) Kinetics. We showed that p21high cells accumulate in adipose tissue 1.5-2 months after HFD feeding, whereas p16high cells accumulation is not evident. This was demonstrated by SCT analysis, RNAScope, and flow cytometry. After 10 months of HFD feeding, we observed accumulation of both p21high cells and p16high cells in the gVAT. Therefore, p21high cells seem to accumulate in adipose tissue earlier than p16high cells with obesity; (2) Cell distribution. In adipose tissue from mice fed a HFD for 10 months, p21high cells and p16high cells are distinguishable in spite of some overlap. Moreover, p16high cells are reported to accumulate in the pancreas from obese mice (Aguayo-Mazzucato et al., 2019), where we found very few p21high cells. Thus, p21high and p16high cells are indeed two distinct cell populations with different cell numbers, cell distributions and tissue locations; (3) Physiological role in metabolic dysfunction. Previously, we and others showed that genetic clearance of p16high cells improved glucose tolerance in obese mice, while insulin sensitivity was not improved significantly after 2-3 months of HFD feeding (Aguayo-Mazzucato et al., 2019; Palmer et al., 2019). Our study establishes that clearance of p21high cells could improve both glucose tolerance and insulin sensitivity, which is evident as early as 2 month after initiating high fat feeding. Moreover, clearance of p16high cells can improve pancreatic beta cell function with obesity (Aguayo-Mazzucato et al., 2019) while clearance of p21high cells has little effect. Thus, the role of p21high cells in vivo could be quite different from that of p16high cells at least in obesity, and quite possibly in other conditions.
Although p16 expression keeps unchanged in most p21high cells, a number of key senescence-related features are observed in p21high cells, including bigger cell size, higher SA-β-gal activity, reduced cell proliferation, and decreased Lamin B1 expression (Wang et al., 2021). Moreover, clearance of p21high cells inhibits TAF+ cells and improves cellular proliferation in gVAT. These findings further support the notion that high expression of p16 might not be enough to define senescent cells.
To the best of our knowledge, little evidence to date has demonstrated tissue-specific roles for senescent cells in vivo, since all drugs were administrated systemically to clear senescent cells in all tissues. For example, one study indicated that clearance of p16high cells in pancreatic β-cells contributed to improved glucose tolerance (Aguayo-Mazzucato et al., 2019). However, such studies cannot exclude the contribution from other tissues such as liver, adipose tissue, and muscle. In this study, using gVAT transplantation, we provide evidence that clearance of p21high cells only in adipose tissue is sufficient to prevent obesity-induced metabolic dysfunction. This could represent essential knowledge for developing gVAT-specific interventions to treat metabolic dysfunction, while minimizing the potential undesirable side effects from clearance of senescent cells in other tissues.
The role of senescent cells in various conditions has been extensively investigated in recent years. However, the underlying mechanisms by which senescent cells cause tissue dysfunction are rarely examined in vivo. In this study, by leveraging p21-Cre mice, we managed to mutate Rela to inhibit the NF-κB pathway only in p21high cells, and demonstrated that the NF-κB pathway is responsible for metabolic dysfunction induced by p21high cells in vivo with obesity. Notably, although the NF-κB pathway was only inhibited in a small subset of cells (p21high cells), it decreases inflammation in the whole gVAT, which is consistent with the fact that SASP can induce inflammation in nearby tissues (Xu et al., 2018; Xu et al., 2015b). Systemic inactivation of the NF-κB pathway could lead to serious side effects. Our findings indicate that targeting the NF-κB pathway only in p21high cells might be sufficient to improve insulin sensitivity in individuals with obesity, and minimize unintended off-target effects.
In this study, we leverage both GFP and Tom to target p21high cells. GFP is one part of the p21-Cre transgene, and is directly driven by the p21 promoter, theoretically representing ongoing p21 expression. For PT mice, p21high cells can be targeted by Tom after tamoxifen administration, which indicates the history of Cre recombination and high p21 expression. We found both GFP+ and Tom+ cells (shortly after tamoxifen treatment) expressed higher levels of p21 protein than the negative cells, validating our model for assessing and examining p21high cells in vivo. In future studies, by administrating tamoxifen at different times, PT mice also offer an opportunity to examine cells in which p21 expression level is high at certain time points and then decreases.
Our study implicates p21high cells as a new and potentially translatable target for T2D interventions. Monthly clearance of p21high cells can provide long-term protective effects on IR in obese mice. Importantly, at two time points of p21high cell clearance (2 months and 4 months after HFD feeding), the metabolic benefits are comparable, which further enhances the translational potential of this approach. Of note, clearance of p21high cells is completely distinct from interfering with p21 gene expression and function. p21 plays an essential role in a wide range of cellular events, and inactivation or suppression of the p21 gene in vivo can lead to tumorigenesis (Martin-Caballero et al., 2001). Our strategy here is to eliminate a small portion (2-15%) of cells which highly express p21, while not significantly reducing p21 gene expression in other cells. This is supported by the fact that clearance of p21high cells results in only 25% reduction of p21 expression in the whole population of SVF cells. In addition, we show that p21high cells can be pharmacologically eliminated by D+Q in human VAT. Recently, intermittent administration of D+Q has been proven to be relatively safe in older human individuals (Justice et al., 2019), and can effectively reduce p21high cells in adipose tissue in vivo in human participants (Hickson et al., 2019). Our findings here fill an important knowledge gap by demonstrating the in vivo benefits of D+Q on human fat tissues, and provide support for clinical trials testing D+Q in IR and T2D.
In summary, by using a powerful p21-Cre mouse model, we provide the proof-of-concept evidence that p21high cells play a causal role in IR through the NF-κB pathway, and intermittent clearance of these cells in gVAT can improve glucose tolerance and insulin sensitivity in individuals with obesity. We also acquired the in vivo transcriptomic signatures of p21high cells by SCT analysis, which could allow us and others to develop new pharmacological interventions to specifically target these cells. Moreover, the human adipose tissue transplantation model we developed here can be leveraged to screen a range of senolytic drugs or other agents on human individuals’ adipose tissues to alleviate IR in vivo, which has potential to be an invaluable tool for future precision medicine.
Limitations of Study
One limitation of our model is that we are not able to investigate the cell-type-specific effects of p21high cells, since all p21high cells are targeted by the model. In addition, because p21 alone might not be sufficient to define senescence, p21high cells most likely do not represent all the senescent cells and not all the p21high cells might be senescent. New animal models are needed to further explore these issues in future studies. Further, our study can only demonstrate that the NF-κB pathway is one of the responsible mechanisms underlying p21high cell mediated phenotypes. However, it is still unknown what downstream targets of this pathway are key for these effects. More investigation is needed to further examine this and other potential mechanisms, such as M2 macrophage polarization. As p21 is involved in the regulation of the hair cycle (Ohtani et al., 2007), it would also be important and clinically significant to examine the role of p21high cells in the hair cycle in states of obesity using the p21-Cre model in the future. We have previously shown that D+Q can reduce p16high cells in human and mouse VAT (Palmer et al., 2019; Xu et al., 2018). It is currently not feasible for us to discriminate the effects of D+Q on p21high cells from p16high cells and possibly other cells in human tissues. Moreover, our SCID-beige mouse model indicates that immune cell-independent mechanisms in the recipient mice might be responsible for the improvement of metabolic function in D+Q-treated human VAT. We previously showed that VAT from humans with obesity secreted a number of cytokines, including IL-6, IL-8, and MCP-1, which can directly impair insulin sensitivity (Palmer et al., 2015). As D+Q can effectively reduce secretion of these cytokines from VAT from humans with obesity (Xu et al., 2018), we speculate that this could be one mechanism for the insulin resistance observed in the recipient mice. Future studies are needed to examine other potential mechanisms.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ming Xu (mixu@uchc.edu).
Materials availability
p21-Cre mice used in this work are available from the lead contact upon request. The investigators are happy to share the p21-Cre mouse model with the research community. The mice will be shipped after a Material Transfer Agreement has been approved by Mayo Clinic. For nonprofit research purpose, the requestor will be responsible only for the shipping cost.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
Original data for creating all graphs in the paper are provided in Data S1
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD16/CD32 antibody | eBioscience | 14-0161-86 |
| APC anti-mouse CD45 antibody | eBioscience | 17-0451-83 |
| PE-Cyanine7 anti-mouse Sca-1 antibody | eBioscience | 25-5981-82 |
| PerCP-Cyanine5.5 anti-mouse CD31 antibody | Biolegend | 102420 |
| PE-Cyanine7 anti-mouse F4/80 antibody | eBioscience | 25-4801-82 |
| Brilliant Violet 421™ anti-mouse F4/80 Antibody | BioLegend | 123132 |
| PerCP-Cyanine5.5 anti-mouse CD11b antibody | eBioscience | 45-0112-82 |
| Brilliant Violet 711™ anti-mouse/human CD11b Antibody | BioLegend | 101242 |
| Anti-CDKN2A/p16INK4a antibody [EPR20418] | Abcam | ab211542 |
| p21 (WAF1, Cip1) polyclonal antibody | eBioscience | 14-6715-81 |
| Anti-NF-kB p65 antibody | Abcam | ab16502 |
| Goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 568 | Invitrogen | A-11011 |
| Goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 647 | Invitrogen | A-21244 |
| Phospho-histone H2A.X (Ser139) (20E3) rabbit mAb | Cell Signaling Technology | 9718 |
| Goat anti-rabbit IgG antibody (H+L), biotinylated | Vector Lab | BA-1000 |
| Single cell hashing antibody | BioLegend | TotalSeq anti-mouse Hashtag #3-5 |
| Kapa Library Quantification Kit | Roche | KK4824 |
| Insulin (C27C9) rabbit mAb | Cell Signaling Technology | 9008S |
| Human p21 antibody | Sigma-Aldrich | 421M-17 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Teklad Global 18% Protein Rodent Diet | ENVIGO | 2918 |
| Rodent diet with 60 kcal% Fat | Research Diets | D12492 |
| Tamoxifen | Sigma-Aldrich | T5648 |
| Corn oil | Sigma-Aldrich | C8267 |
| Collagenase type II | Sigma-Aldrich | C6885 |
| PBS (10X) | Fisher bioreagents | BP6651 |
| DNase I | Sigma-Aldrich | D5025 |
| ACK Lysing Buffer | Gibco | A1049201 |
| BSA | Sigma-Aldrich | A9647 |
| Kapa biosystems qPCR reagents and kits | Kapa Biosystems qPCR Reagents and Kits | Kapa Biosystems qPCR Reagents and Kits |
| TRIzol Reagent | Invitrogen | 15596018 |
| Chloroform | Sigma-Aldrich | 366927 |
| Isopropanol | Sigma-Aldrich | 34863 |
| Ethanol | Sigma-Aldrich | E7023 |
| UltraPure™ distilled water | Invitrogen | 10977 |
| M-MLV Reverse Transcriptase | Invitrogen | 28-025-013 |
| RNaseOUT™ Ribonuclease Inhibitor | Invitrogen | 10-777-019 |
| Random primers | Invitrogen | 48190-011 |
| dNTP set (100 mM) | Invitrogen | 10297-018 |
| PerfeCTa® FastMix® II | Quantabio | 95118 |
| Isoflurane, USP | Piramal Critical Care | 66794-013-25 |
| D-Luciferin, Potassium Salt | Gold Biotechnology | LUCK-1g |
| Glucose | Acros organics | 41095-5000 |
| Insulin (U-100) | Humulin® R | 0002-8215-17 |
| Ketoprofen (100mg/ml) | Zoetis | N/A |
| Dasatinib | LC Laboratories | D3307 |
| Quercetin | Sigma-Aldrich | Q4951 |
| Phosal 50 PG | Lipoid LLC | 368315 |
| PEG-400 | Sigma-Aldrich | 91893 |
| Fetal Bovine Serum | Corning | 35-011-CV |
| RPMI medium 1640 | Gibco | 11835-030 |
| Sodium Pyruvate (100 mM) | Gibco | 11360-070 |
| MEM Non-Essential Amino Acids (100 X) | Gibco | 11140-050 |
| MEM Vitamins (100 X) | Gibco | 11120-052 |
| Penicillin-Streptomycin | Gibco | 15140-122 |
| Paraformaldehyde solution, 4% in PBS | Thermo Fisher Scientific | AAJ19943K2 |
| Sucrose | Sigma-Aldrich | 84097 |
| Tissue-Plus™ O.C.T. Compound | Thermo Fisher Scientific | 23-730-571 |
| Diamond antifade mountant with DAPI | Life Technologies | P36962 |
| Triton X-100 | Sigma-Aldrich | T9284 |
| Harris Hematoxylin | Leica biosystems | 3801560 |
| Eosin | Leica biosystems | 3801600 |
| Toluene | Avantor Performance Materials | JTB/9351-03 |
| BOND IHC Polymer Detection Kit | Leica biosystems | DS9800 |
| RNAscope™ 2.5 LSx Reagent Kit-Brown | ACD | 322700 |
| RNAscope® 2.5 LS Positive Control Probe_Mm-PPIB | ACD | 313918 |
| RNAscope® 2.5 LS Negative Control Probe_dapB | ACD | 312038 |
| RNAscope® LS 2.5 Probe- Mm-Cdkn1a | ACD | 408558 |
| RNAscope® LS 2.5 Probe- Mm-Cdkn2a | ACD | 411018 |
| Avidin/Biotin blocking kit | Vector Lab | SP-2001 |
| Streptavidin, DyLight® 649 | Vector Lab | SA-5649-1 |
| TelC-Cy3 PNA FISH probe | Panagene | F1002 |
| Goat serum | Sigma-Aldrich | G9023 |
| EdU | Cayman Chemical | 20518 |
| Ascorbic acid | Alfa Aesar™ | A1561322 |
| Alexa Fluor 647 azide | Thermo Fisher Scientific | A10277 |
| X-gal | Teknova | X1220 |
| Probes/primers for QPCR | ||
| Tbp | IDT | Mm.PT.39a.22214839 |
| p21 | IDT | Mm.PT.58.17125846 |
| p16 | IDT | Mm.PT.58.8388138 |
| Il6 | IDT | Mm.PT.58.10005566 |
| Pai1 | IDT | Mm.PT.58.6413525 |
| Ccl2 | IDT | Mm.PT.58.42151692 |
| Cxcl1 | IDT | Mm.PT.58.42076891 |
| Pparg | IDT | Mm.PT.58.31161924 |
| Fabp4 | IDT | Mm.PT.58.43866459 |
| Adipoq | IDT | Mm.PT.58.9719546 |
| Mrc1 | IDT | Mm.PT.58.42560062 |
| Mgl2 | IDT | Mm.PT.58.21616779 |
| Cd163 | IDT | Mm.PT.58.15897116 |
| Critical Commercial Assays | ||
| Insulin Mouse Ultra Sensitive ELISA | Crystal Chem | 90080 |
| Deposited Data | ||
| Single-cell RNA sequence data Raw data | Gene Expression Omnibus Data S1 | GSE161872 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 | Jackson Laboratory | 000664 |
| floxed knock-in tdTomato mice | Jackson Laboratory | 007914 |
| floxed knock-in LUC mice | Jackson Laboratory | 005125 |
| floxed DTA mice | Jackson Laboratory | 009669 |
| floxed Rela mice | Jackson Laboratory | 024342 |
| SCID-beige mice | ENVIGO | N/A |
| p21-Cre mice | This study | N/A |
| Software and Algorithms | ||
| FlowJo software V10.7 | Becton, Dickinson & Company | https://www.flowjo.com/solutions/flowjo/downloads |
| minispec Plus software V7.0 | Bruker | N/A |
| Living Image 4.5.5 | PerkinElmer | https://www.perkinelmer.com.cn/lab-products-and-services/resources/in-vivo-imaging-software-downloads.html |
| Oxymax®-CLAMS V5.51 | Columbus | N/A |
| CLAMS examination tool (CLAX) V2.2.15 | Columbus | N/A |
| Image J 1.42q | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Zen Blue 2.3 | Zeiss | N/A |
| ZEN lite 3.2 | Zeiss | N/A |
| Other | ||
| AimStrip Plus blood glucose testing system | Germaine Laboratories | 37321 |
| Glucose test strips | Germaine Laboratories | 37350 |
| Countess II automated cell counter | Thermo Fisher Scientific | AMQAX1000 |
| 4200 Tapestation system | Agilent Technologies | N/A |
| NovaSeq™ 6000 sequencing system | Illumina | Novogene |
| 10X Chromium platform | 10x Genomics | N/A |
| BD LSR II flow cytometer | BD Biosciences | N/A |
| BD FACS Aria II flow cytometer | BD Biosciences | N/A |
| CFX96 Touch Real-Time PCR detection system | Bio-Rad Laboratories | 1855196 |
| minispec mq7.5 TD-NMR analyzer | Bruker | LF50 |
| Comprehensive laboratory animal monitoring system (CLAMS) | Columbus | N/A |
| EVOS XL Core imaging system | Thermo Fisher Scientific | AMEX1000 |
| ZOE™ Fluorescet cell imager | Bio-Rad Laboratories | 1450031 |
| Axio Observer Z1 | Zeiss | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse model
p21-Cre mice were generated on C57BL/6 background mice and genotyped as previously described (Wang et al., 2021). p21-Cre/+; Tom/+ (PT) mice were generated by crossing p21-Cre mice with floxed knock-in tdTomato mice (Jackson’s Laboratory). The p21-Cre mice were crossed with floxed knock-in firefly luciferase (LUC) mice (Jackson’s Laboratory) to get the p21-Cre/p21-Cre; LUC/LUC mice, which were further crossed with floxed diphtheria toxin A (DTA) mice (Jackson’s Laboratory) to generate the p21-Cre/+; LUC/+ (PL) and p21-Cre/+; LUC/DTA (PLD) littermates. Similarly, p21-Cre mice were crossed with Relafl/fl mice (Jackson’s Laboratory) to get p21-Cre/+; Relafl/fl mice, which were further crossed with Relafl/fl mice to obtain +/+; Relafl/fl (Rela) and p21-Cre/+; Relafl/fl (P-Rela) littermates. p21-Cre/p21-Cre; Tom/Tom mice and Rela mice were bred to generate p21-Cre/+; Tom/+; Relafl/fl (PT-Rela) mice. All mice were used as the result of in-house matings and maintained under 12:12 light/dark cycle (07:00 to 19:00 light on) at 25 ± 2°C with free access to water and regular chow diet (R CD) containing 18 kcal% fat, 58 kcal% carbohydrate and 24 kcal% protein (Teklad global 18% protein rodent diet, ENVIGO, Indianapolis, IN). All the mice experiments were conducted with age- and gender-matched mice with their health status checked daily by the investigators and technicians from UConn Health. The sex, genotype, and age information for the mice was indicated in the Results. Littermate controls were used if possible. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at UConn health.
HFD models and drug treatments
For the high-fat diet (HFD) mouse model challenge, the 2-3-month-old male or female mice were fed with a HFD containing 60 kcal% fat, 20 kcal% carbohydrate and 20 kcal% protein (Research Diets, New Brunswick, NJ). Then mice received intraperitoneal (i.p.) injection of 2 mg tamoxifen/corn oil solution (Sigma-Aldrich, St. Louis, MO) or equal volume of corn oil for two consecutive days at the designated time points. For in vivo D+Q administration, dasatinib (5 mg/kg; LC Laboratories, Woburn, MA) and quercetin (50 mg/kg; Sigma Aldrich) were dissolved in vehicle containing 60% Phosal 50 PG (Lipoid LLC, Ludwigshafen, Germany), 30% PEG-400 (Sigma Aldrich), and 10% ethanol. D+Q or vehicle was given by oral gavage for five consecutive days before subsequent experiments.
Mouse gVAT transplantation
For the mouse gVAT transplantation, gVAT from the donor mice was immediately dissociated, cut into approximately 0.1g pieces, and kept in saline until transplantation. The recipient mice were anesthetized under isoflurane and a total of 0.7 g gVAT was transplanted into their intraperitoneal cavity near to the gVAT location. The abdominal wall was then closed with 6-0 Vicryl suture and the skin incision was closed using BD Autoclip™ wound clips. The recipient mice were subcutaneously injected with 4 mg/kg of ketoprofen solution (Zoetis, Parsippany-Troy Hills, NJ) to relieve pain. Mice in the Sham group received same surgery but without gVAT transplantation. Body composition was measured for the recipient mice 3 days before the surgery and 6 days after the surgery, then the mice were detected for behavior and feed intake for another 3 days. GTT and ITT were performed at day 10 and day 17 after the surgery, respectively. GSIS were tested 1.5 months after the surgery.
Human VAT transplantation
The protocol was approved by the Mayo Clinic Foundation Institutional Review Board for Human Research. Informed written consent was obtained from all individuals. Human omental adipose tissues were donated by 3 female and 1 male Caucasian individuals with obesity (Table S1) with an average BMI and range of 43.42 ± 6.50 kg/m2 (mean ± s.d.; range, 37-52) and an average age at 51.75 ± 17.71 years ( mean ± s.d.; range, 28-68). Individual VAT sample was cut into small slices, separated into two groups treated with Dasatinib (1 μM) + Quercetin (20 μM) or vehicle solution (DMSO) at 37°C under 5% CO2 atmosphere for 48 h. Human VAT samples were then washed twice using saline. A total of 0.5 g vehicle or DQ-treated VAT slices (from the same individual donor) were transplanted into 3-month-old SCID beige recipient mice using the same surgery procedures as mouse VAT transplantation. For each human donor, the fat tissues were transplanted into 3-4 recipient mice per group. Body composition was measured for the recipient mice 3 days before the surgery and 6 days after the surgery. Because the fat transplantation procedure and all the following tests are stressful to the mice, especially for the immune-deficient mice, we let these mice rest for at least 1 week between tests to minimize the complication from these stressors. GTT and ITT were performed at day 9 and day 16 after the surgery, respectively. CLAMS were tested 1 month after the surgery, and GSIS were measured 1.5 months after the surgery.
METHOD DETAILS
SCT sample preparation and sequencing
The gVAT samples were cut into small pieces and then digested with 1 mg/ml type II collagenase/10 μg/ml DNase I/PBS (Sigma-Aldrich) at 37°C for 1 h. After spinning for 10 min at 1200 rpm, SVF cells were collected, filtered through 100 μm cell strainer and then lysed by ACK buffer (Gibco™, Thermo Fisher Scientific, Waltham, MA) at room temperature for 10 min to get rid of blood cells. The purified SVF cells were blocked with a FcR antibody (CD16/32, eBioscience, San Diego, CA) on ice for 10 min. SVF cells from each mouse were labeled with 1 μg cell hashtag oligo (HTO) conjugated antibodies (TotalSeq anti-mouse Hashtag #3-5, BioLegend, , San Diego, CA) according to the manufacturer’s protocol (BioLegend protocol #5009). Cells were washed and suspended in PBS containing 0.04% BSA and immediately processed as follows. Cells were counted on Countess II automated cell counter (Thermo Fisher Scientific), and were loaded onto one lane of a 10X Chromium microfluidic chip. Single cell capture, barcoding, and library preparation were performed using the 10X Chromium platform version 3 chemistry (10X Genomics, Pleasanton, CA) and according to the manufacturer’s protocol (#CG00052) with modifications for generating the hashtag library (BioLegend protocol #5009) (Stoeckius et al., 2018). cDNA and libraries were checked for quality on Agilent 4200 Tapestation (Agilent Technologies, Santa Clara, CA), quantified by using Kapa Library Quantification Kit (Roche, Basel, Switzerland), and pooled using a ratio of 95% gene expression library and 5% hashtag library before sequencing. Both sets of libraries were sequenced via the NovaSeq™ 6000 sequencing system with flow cell type S2 (Illumina, San Diego, CA).
SCT data processing, quality control and analysis
Illumina base call files for all libraries were converted to FASTQs using bcl2fastq v2.22.0.422 (Illumina). Gene expression FASTQ files were aligned to the mm10 (GRCh38.93, 10X Genomics mm10 reference 3.3.0) genome using the version 3.1.0 Cell Ranger count pipeline (10X Genomics), resulting in two cell-by-gene digital count matrices. Hashtag library FASTQs were processed using CITE-Seq-Count (version 1.4.2) which produces a cell-by-hashtag digital count matrix for each hash tag library. Downstream analysis was performed using the Seurat R toolkit (version 4.0.4) (Stuart et al., 2019). Cells were assigned labels corresponding to each input mouse using the HTODemux function. Barcodes identified as HTO doublets and negatives were first excluded. Then cells were further excluded from downstream analysis if they did not meet the following criteria: (1) more than 300 genes detected per cell; (2) less than 20% mitochondrial transcripts; (3) less than 1% hemoglobin transcripts. After quality control filtering, 11,401 and 7,283 cells remained in the RCD and HFD mice groups, respectively.
The gene expression matrices from the RCD and HFD batches were normalized by the total UMI counts in each cell, multiplied by 10,000, log-transformed, and the top 2,000 most variable genes were selected using the variance stabilizing transformation method. The two batches were then integrated using the standard workflow in Seurat toolkit, resulting in an integrated gene expression matrix. Using the integrated matrix, the mean expression of each gene in the cells was scaled to 0, and a linear dimensional reduction (principal component analysis, PCA) was performed on the scaled data. To cluster the cells, the first 40 PCs, which were determined by manual inspection of the elbow plot, were used to construct a K-nearest neighbor (KNN) graph. The clusters were identified through the Louvain algorithm on the KNN graph, where the resolution parameter was set as 0.2, yielding 14 cell clusters. The single cells were visualized in a 2D UMAP embedding that was computed from the KNN graph. To define the cell clusters, marker genes, which (1) detected at a minimum of 25% of cells in the cluster and (2) the mean expression in the cluster compared to all other cells is higher than 0.25 (log scale), were generated by the FindAllMarkers function using Wilcoxon Rank Sum test under the “RNA” assay. The cell clusters were assigned to specific cell populations based on the expression of canonical markers of these cell populations. We defined cells to be p21high if their p21 (Cdkn1a) counts were greater than six. Differentially expressed genes were generated by comparing the interested cell groups using the FindMarkers function. A Loupe file was generated by combining the two libraries using CellRanger aggr and subsequently CellRanger reanalyze using the final set of cell barcodes exported from the finalized Seurat object. Categorical cell annotations and UMAP coordinates were exported from the Seurat object and imported into the Loupe file, and the single-cell data were visualized by feature and violin plots using the Loupe Brower.
Flow cytometry analysis
Purified SVF cells were incubated with FcR blocking antibody, followed by staining with fluorescence-labeled CD45, Sca-1, F4/80, CD11b (eBioscience) or CD31 (Biolegend) for 30 min. Cells were then washed with 2% FBS/PBS buffer and suspended in 0.1 μg/ml of DAPI staining solution before GFP or tdTomato detection. For p21, p16 and NF-κB assessments, SVF cells were fixed in 4% paraformaldehyde (PFA) solution (Thermo Fisher Scientific) for 15 min, permeabilized in 0.2% Triton X-100 for 10 min, and blocked in 1% BSA/PBS solution at room temperature for 1 h. After overnight incubation with anti-p21 primary antibody (1:40, Thermo Fisher Scientific), anti-p16 primary antibody (1:200, abcam, Cambridge, UK) or anti-NF-κB p65 primary antibody (1:200, Abcam) in 1% BSA/PBS at 4°C, cells were rinsed in PBS and stained with Alexa Fluor 647-conjugated or Alexa Fluor 568-conjugated anti-rabbit secondary antibody (1:200, Thermo Fisher Scientific) for 1 h at room temperature. Cells were detected using the BD LSR II flow cytometer (BD Biosciences, San Jose, CA). Data analysis was performed on the FlowJo software V10.7 (Becton, Dickinson & Company, Franklin Lakes, NJ).
RNA extraction and quantitative real-time PCR analysis
gVAT pieces derived from RCD or HFD-fed male mice treated as indicated in figure legends were digested and centrifuged by using same methods as the single cell sample preparation. SVF and adipocyte fractions were then separated and collected, respectively. Total RNA was extracted using TRIzol reagent (Invitrogen, Thermo Fisher Scientific), purified with chloroform, isopropanol and 75% ice cold ethanol (Sigma-Aldrich), dissolved in RNase free water, and then reverse transcribed to cDNA with M-MLV reverse transcriptase kit (Invitrogen, Thermo Fisher Scientific). Quantitative real-time PCR (QPCR) was conducted using PerfeCTa® FastMix® II (Quantabio, Beverly, MA) on CFX96 Real-Time PCR detection system (Bio-Rad Laboratories, Hercules, CA). After pre-incubation at 95 °C for 3 min, the QPCR was performed as 39 cycles of 95 °C for 5 s and 60 °C for 30 s. At the end of QPCR, the threshold cycle (CT) values were obtained. The relative transcriptional level of target genes was calculated by the 2−ΔΔCT method with TATA-binding protein (Tbp) as an internal control. ΔΔCT values were used for statistical analysis. Probes and primers for Tbp, p21, p16, Il6, Pai1, Ccl2, Cxcl1, Pparg, Fabp4, Adipoq, Mrc1, Mgl2, and Cd163 were purchased from Integrated DNA Technologies (IDT, Coralville, Iowa).
RNAscope chromogenic in situ hybridization assay
gVAT was collected from mice fed with RCD or 2-month of HFD, and fixed with 4% PFA at 4°C before dehydration in 70% alcohol reagent and embedding in paraffin, which were further cut into 10-μm sections. The p21 and p16 mRNA CISH assays were performed on gVAT using RNAscope 2.5 LSx Reagent Kit-Brown (Advanced Cell Diagnostics (ACD), Newark, CA) on the Leica Bond RX stainer (Leica Biosystems, Wetzlar, Germany) according to the manufacturer’s instructions and previously reported methods (Thangaiah et al., 2021). In brief, slices were dewaxed and baked prior to been exposed to the retrieval reagent (BOND Epitope Retrieval Solution 2, Leica) for 15 min at 95°C and the Protease III pretreatment for 10 min at 40°C. Slides were then incubated for 2 h at 40°C with either the Mm-Cdkn1a (p21), Mm-Cdkn2a (p16), Mm-PPIB, or dapB probe (ACD) followed by signal amplification. Subsequently, samples were incubated in DAB for 10 minutes, hematoxylin for 5 minutes, and bluing reagent for 2 minutes. After water rinse and dehydration, slides were scanned using an Aperio ScanScope (Leica) at 40X magnification. The mRNA expression levels of p21 and p16 were evaluated according to an ACD scoring system (https://acdbio.com/dataanalysisguide.) by counting number of dots per cell, which correlate with RNA copy number. All experiments were confirmed that the positive control Mm-PPIB stain scored at ≥ 2 and the negative control dapB score at < 1.
Telomere-associated foci (TAF)-immunofluorescence in situ hybridization staining
The TAF immuno-FISH was performed on PL and PLD gVAT samples as described previously (Xu et al., 2018). Briefly, the PFA-fixed, paraffin-embedded gVAT sections were dewaxed rehydrated and heated in 0.01 M citrate buffer at pH 6.0 for 10 min at 95°C. After antigen retrieval, slices were blocked in goat serum (1:60, Sigma-Aldrich) for 1 h and avidin/biotin (Vector Lab, Burlingame, CA) for 15 min at room temperature, followed by overnight incubation with anti-γH2A.X primary antibody (1:200, Cell Signaling Technology, Danvers, MA) at 4°C. The next day, slides were stained with biotinylated, anti-rabbit secondary antibody (Vector Lab) for 30 min and additional DyLight 649 conjugated streptavidin (Vector Lab) for 20 min. For telomeres FISH counterstaining, slides were further fixed, dehydrated, and then denatured for 10 min at 80°C in hybridization buffer (70% formamide (Sigma-Aldrich), 25 mM MgCl2, 1 M Tris pH 7.2, 5% blocking reagent (Roche) with a 2.5 μg/ml Cy-3-labeled telomere-specific (CCCTAA) peptide nucleic acid (PNA) probe (Panagene, Daejeon, Korea) for 2 h at room temperature in the dark. Sections were stained with DAPI before imaging on an Axio Observer.Z1/7 equipped with an Apotome.2 (Zeiss, Oberkochen, Germany). In-depth Z-stacking (a minimum of 20 optical slices with 63X/1.40 oil objective) was achieved for Apotome 3D deconvolution using Zen Blue 2.3 and further TAFs counting on ZEN lite 3.2 (Zeiss). Cells are considered to be TAF+ if they contain no less than 3 TAFs from the entire z-stacks. For each sample, a minimum of 150 cells were counted.
Immunofluorescence staining
gVAT, liver, muscle, pancreas and brown fat were obtained from lean PT control or obese PT mice by 3- or 6-month HFD feeding. Tissues were all fixed with 4% PFA solution, dehydrated by using 30% sucrose/PBS (Sigma-Aldrich), and then embedded into O.C.T compound (Thermo Fisher Scientific). Cryosections at 6 μm of thickness were prepared for liver, muscle and pancreas and 10 μm cryosections were for gVAT and brown fat samples. For detecting islets of obese PL and PLD mice feeding with HFD for 3 months, pancreas cryosections were permeabilized with 0.3% Triton X-100 (Sigma-Aldrich) in 1% BSA for 10 min, blocked with 1% BSA for 30 min, and incubated with Alexa Fluor 647-conjugated rabbit anti-insulin monoclonal antibody (Cell Signaling Technology) for 1 h at room temperature. Tissue sections were covered with DAPI-contained mountant (ProLong, Life Technologies, Carlsbad, CA) prior to being imaged on a ZOE™ Fluorescet cell imager (Bio-Rad Laboratories). Islet area, defined as insulin staining positive area, and total pancreas area were quantified using Image J V1.42q (National Institutes of Health, Bethesda, Maryland).
Bioluminescence imaging (BLI)
Two days after tamoxifen treatment, PLD mice were subjected to a partial abdomen shave under isoflurane (Piramal Critical Care, Bethlehem, PA) anesthesia and then i.p. injected with 3 mg luciferin (Gold Biotechnology, Olivette, MO) in 200 μL PBS 5 min before imaging. Luminescent images were collected with a 3-minute exposure to IVIS® spectrum in vivo imaging system (PerkinElmer, Waltham, MA). Luminescent signal in the region of interest (ROI) was quantified using the Living Image 4.5.5 software (PerkinElmer).
To map the luminescence signal distribution, mice were i.p. injected with 200 μL luciferin solution 5 min before being sacrificed by cervical spine dissociation. Tissues including visceral fat, inguinal fat, brown fat, liver, muscle, pancreas, kidney, perirenal fat, intestine, lung, heart, brain and spleen were dissected for the luminescence imaging in the same way as in vivo test.
Cellular senescence-associated beta-galactosidase (SA-β-gal) activity assay
Cellular SA-β-gal activity was measured as previously described with slight modifications (Xu et al., 2015b). Briefly, a small piece of gVAT fat was acquired and fixed for 10 min in 2% formaldehyde/0.2% glutaraldehyde (Sigma-Aldrich)/PBS at room temperature. Fat trunks were then immersed in a rotated tubes full of freshly prepared SA-β-gal activity solution (1mg/ml of X-gal, 40 mM citricacid/sodium phosphate at pH 6.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2) at 37°C in dark. Photographs of gVAT samples among different groups were taken after 6 h of incubation and PBS rinse.
In vivo EdU incorporation assessment
Mice were intraperitoneally injected with 10 mg/kg of 5- ethynyl- 2′- deoxyuridine (EdU; Cayman Chemical, Ann Arbor, MI) in PBS 20 h before sacrifice. SVF cells were fixed and permeabilized as described above. For EdU-incorporatd DNA detection, cells were stained with a fluorescent solution consisting of 2 μM Alexa Fluor 647 azide (Thermo Fisher Scientific), 2 mM CuSO4, 10 mM ascorbic acid, and 100 mM Tris-HCl (pH 7.5) for 30 min at room temperature. After that, cells were washed with PBS and analyzed via BD LSR II flow cytometer (BD Biosciences).
Metabolic function tests
Mice were fasted for 6 h (GTT) or 4 h (ITT) followed by being administrated with 1.5 g/kg glucose (Acros organics, Fair Lawn, NJ) or 0.8 mU/kg insulin (Humulin® R U100, Eli Lilly and Company, Indianapolis, IN) via intraperitoneal injection, respectively. Tail vein blood at baseline and indicated time points after the injection was collected and measured for the glucose level by using handheld glucometer (Germaine™ Laboratories, San Antonio, TX).
For the GSIS assay, mice were fasted for 4 h, and then intraperitoneally injected with 1.5 g/kg glucose. Blood was collected via cheek punch at baseline and 15 minutes post glucose administration. Plasma samples were then isolated and tested for the insulin concentration by applying an ultra-sensitive mouse insulin ELISA kit (Crystal Chem, Elk Grove Village, IL).
Body composition measurement
Mice were placed in a plunger-contained restraining cylinder to keep immobile, and subsequently transferred into minispec mq7.5 TD-NMR analyzer (the “LF50”, Bruker, Billerica, MA). The fat and lean mass would be automatically detected at the same time. Data were then analyzed by minispec Plus software V7.0 (Bruker). C57BL/6 mice were adopted for calibration.
Comprehensive laboratory animal monitoring system (CLAMS)
The CLAMS (Columbus Instruments, Columbus, OH) instrument was adopted to quantitate individual mice on their daily activity and feed intake. Mice were weighed and individually housed in chambers under 12 h light/dark cycle (07:00 to 19:00 light on) at 25 ± 2°C and were with free access to water and food for three consecutive days. Activity, food consumption and water intake were monitored, and related date were recorded by Oxymax®-CLAMS software V5.51 (Columbus Instruments). Data were further analyzed with the CLAMS Examination Tool (CLAX) V2.2.15 (Columbus Instruments).
Immunohistochemical (IHC) staining
The PFA-fixed, paraffin-embedded blocks of human gVAT explants after DQ or vehicle treatment were cut into 10 μm sections, and then incubated with p21 primary antibody (1:200, Sigma-Aldrich) at room temperature for 15 min, followed by staining with a BOND IHC Polymer Detection Kit (Leica Biosystems). Images were acquired using EVOS XL Core imaging system (Thermo Fisher Scientific).
Hematoxylin and eosin staining
Ten days after the mice fat transplantation, the recipient mice were sacrificed and the donor fats were obtained. The PFA-fixed, paraffin-embedded blocks were cut into 10 μm sections. Slices were deparaffinized with toluene (J.T.Baker®, Avantor Performance Materials, Center Valley, PA), hydrated using an ethanol gradient, and then stained with hematoxylin solution (Leica biosystems). After rinse with water, slices were further stained with eosin solution (Leica biosystems), dehydrated by applying absolute alcohol and then cleared in toluene. Images were acquired using EVOS XL Core imaging system.
QUANTIFICATION AND STATISTICAL ANALYSIS
Results were shown as means ± s.e.m or box-and-whisker plots, where a box extends from the 25th to 75th percentile with the median shown as a line in the middle, and whiskers indicate the smallest and largest values, with n representing the number of biological replicates. GraphPad Prism was used for statistical analysis and graphs. The normality of data distribution was determined by Kolmogorov-Smirnov test. Mean values were compared by two-tailed Welch’s t-test with unequal variance or one-way ANOVA or two-way ANOVA with Fisher’s LSD test as indicated in figure legends. P < 0.05 was considered to be statistically significant. The exact statistical parameters were indicated in the figure legends. For the fat transplantation experiments, recipient mice were randomly distributed into different groups according to their bodyweight. For other experiments, transgenic mice were assigned to experimental groups based on their genotypes. Sample sizes were determined based on the means and variation of previous pilot or published experiments. No data was excluded for statistical analysis. All the key findings were reliably reproduced in several independent cohorts with large biological replicates. Investigators were blinded to allocation during experiments and outcome assessments.
Supplementary Material
Inclusion and diversity.
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure sex balance in the selection of non-human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Highlights.
p21high cells, distinct from p16high cells, accumulate in fat with obesity
Intermittent p21high cell clearance both prevents and alleviates insulin resistance
Exclusive inactivation of NF-κB in p21high cells improves insulin sensitivity
A senolytic reduces p21high cells in human fat and alleviates its metabolic harm in vivo
Acknowledgements
The authors are grateful to colleagues in the UConn Center on Aging for helpful and constructive discussion, Zhifang Hao for histology service, and Stefanie Farkas for administrative assistance. This work was supported in part by a ISG seed grant from UConn Health (M.X.), the Regenerative Medicine Initiative for Diabetes- Career Development Award from Mayo Clinic (M.X.), Glenn Foundation for Medical Research and AFAR Grant for Junior Faculty (M.X.), the Esperance Fellowship in Personalized Nutrition (N.S.G), The Kenneth and Paula Munson Family Fund for Student Support in Health Sciences Fellowship (N.S.G and T.K.), the Diana Jacobs Kalman/AFAR Scholarship for Research in the Biology of Aging (N.S.G.), The UConn/JAX-GM Training Program in Genomic Science T32HG010463 (R.L.C), Robert and Arlene Kogod (J.L.K.), the Connor Group (J.L.K.), Robert J. and Theresa W. Ryan (J.L.K.), the Noaber Foundation (J.L.K.), Travelers Chair in Geriatrics and Gerontology (G.A.K.), and NIH grants R37AG013925 (J.L.K.), K08CA215105 (A.M.), P01AG062413 (J.L.K.), R33AG061456 (J.L.K., T.T., G.A.K.), P30AG067988 (G.A.K.), AG063528 (M.X.), AG066679 (M.X.), and AG068860 (M.X.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
M.X., A.K.P., T.T., and J.L.K, have a financial interest related to this research. Patents on senolytic drugs (including PCT/US2016/041646, filed at the US Patent Office) are held by Mayo Clinic. These studies were conducted in compliance with Mayo Clinic conflict of interest policies.
References
- Aguayo-Mazzucato C, Andle J, Lee TB Jr., Midha A, Talemal L, Chipashvili V, Hollister-Lock J, van Deursen J, Weir G, and Bonner-Weir S (2019). Acceleration of beta Cell Aging Determines Diabetes and Senolysis Improves Disease Outcomes. Cell Metab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2018). Adult Obesity Facts. [Google Scholar]
- Champy MF, Selloum M, Zeitler V, Caradec C, Jung B, Rousseau S, Pouilly L, Sorg T, and Auwerx J (2008). Genetic background determines metabolic phenotypes in the mouse. Mamm Genome 19, 318–331. [DOI] [PubMed] [Google Scholar]
- Chen LF, and Greene WC (2004). Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol 5, 392–401. [DOI] [PubMed] [Google Scholar]
- Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, et al. (2011). Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev 25, 2125–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé JP, Patil C, Rodier F, Sun Y, Muñoz DP, Goldstein J, Nelson PS, Desprez PY, and Campisi J (2008). Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6, 2853–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defronzo RA (2009). Banting Lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes 58, 773–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo RA, Ferrannini E, Groop L, Henry RR, Herman WH, Holst JJ, Hu FB, Kahn CR, Raz I, Shulman GI, et al. (2015). Type 2 diabetes mellitus. Nat Rev Dis Primers 1, 15019. [DOI] [PubMed] [Google Scholar]
- Frescas D, Hall BM, Strom E, Virtuoso LP, Gupta M, Gleiberman AS, Rydkina E, Balan V, Vujcic S, Chernova OB, et al. (2017). Murine mesenchymal cells that express elevated levels of the CDK inhibitor p16(Ink4a) in vivo are not necessarily senescent. Cell Cycle 16, 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasek NS, Kuchel GA, Kirkland JL, and Xu M (2021). Strategies for targeting senescent cells in human disease. Nature Aging 1, 870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K, Ferbeyre G, et al. (2019). Cellular Senescence: Defining a Path Forward. Cell 179, 813–827. [DOI] [PubMed] [Google Scholar]
- Guillaume M, Handgraaf S, Fabre A, Raymond-Letron I, Riant E, Montagner A, Vinel A, Buscato M, Smirnova N, Fontaine C, et al. (2017). Selective Activation of Estrogen Receptor alpha Activation Function-1 Is Sufficient to Prevent Obesity, Steatosis, and Insulin Resistance in Mouse. The American journal of pathology 187, 1273–1287. [DOI] [PubMed] [Google Scholar]
- Hall BM, Balan V, Gleiberman AS, Strom E, Krasnov P, Virtuoso LP, Rydkina E, Vujcic S, Balan K, Gitlin II, et al. (2017). p16(Ink4a) and senescence-associated beta-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging (Albany NY) 9, 1867–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise N, De Silva NS, Silva K, Carette A, Simonetti G, Pasparakis M, and Klein U (2014). Germinal center B cell maintenance and differentiation are controlled by distinct NF-kappaB transcription factor subunits. J Exp Med 211, 2103–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson LJ, Langhi Prata LGP, Bobart SA, Evans TK, Giorgadze N, Hashmi SK, Herrmann SM, Jensen MD, Jia Q, Jordan KL, et al. (2019). Senolytics decrease senescent cells in humans: Preliminary report from a clinical trial of Dasatinib plus Quercetin in individuals with diabetic kidney disease. EBioMedicine 47, 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice JN, Nambiar AM, Tchkonia T, LeBrasseur NK, Pascual R, Hashmi SK, Prata L, Masternak MM, Kritchevsky SB, Musi N, et al. (2019). Senolytics in idiopathic pulmonary fibrosis: Results from a first-in-human, open-label, pilot study. EBioMedicine 40, 554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Caballero J, Flores JM, Garcia-Palencia P, and Serrano M (2001). Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res 61, 6234–6238. [PubMed] [Google Scholar]
- Mosier DE, Stell KL, Gulizia RJ, Torbett BE, and Gilmore GL (1993). Homozygous scid/scid;beige/beige mice have low levels of spontaneous or neonatal T cell-induced B cell generation. J Exp Med 177, 191–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Espin D, and Serrano M (2014). Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 15, 482–496. [DOI] [PubMed] [Google Scholar]
- Muoio DM, and Newgard CB (2008). Mechanisms of disease:Molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol 9, 193–205. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Imamura Y, Yamakoshi K, Hirota F, Nakayama R, Kubo Y, Ishimaru N, Takahashi A, Hirao A, Shimizu T, et al. (2007). Visualizing the dynamics of p21(Waf1/Cip1) cyclin-dependent kinase inhibitor expression in living animals. Proceedings of the National Academy of Sciences of the United States of America 104, 15034–15039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AK, Tchkonia T, LeBrasseur NK, Chini EN, Xu M, and Kirkland JL (2015). Cellular Senescence in Type 2 Diabetes: A Therapeutic Opportunity. Diabetes 64, 2289–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AK, Xu M, Zhu Y, Pirtskhalava T, Weivoda MM, Hachfeld CM, Prata LG, van Dijk TH, Verkade E, Casaclang-Verzosa G, et al. (2019). Targeting senescent cells alleviates obesity-induced metabolic dysfunction. Aging Cell 18, e12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Vargas M, Ramon-Krauel M, Lerin C, and Jimenez-Chillaron JC (2020). Size Does Matter: Litter Size Strongly Determines Adult Metabolism in Rodents. Cell Metab 32, 334–340. [DOI] [PubMed] [Google Scholar]
- Patel S, and Santani D (2009). Role of NF-kappa B in the pathogenesis of diabetes and its associated complications. Pharmacological reports : PR 61, 595–603. [DOI] [PubMed] [Google Scholar]
- Roden M, and Shulman GI (2019). The integrative biology of type 2 diabetes. Nature 576, 51–60. [DOI] [PubMed] [Google Scholar]
- Safran M, Kim WY, Kung AL, Horner JW, DePinho RA, and Kaelin WG Jr. (2003). Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging 2, 297–302. [DOI] [PubMed] [Google Scholar]
- Schafer MJ, White TA, Evans G, Tonne JM, Verzosa GC, Stout MB, Mazula DL, Palmer AK, Baker DJ, Jensen MD, et al. (2016). Exercise Prevents Diet-Induced Cellular Senescence in Adipose Tissue. Diabetes 65, 1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM 3rd, Smibert P, and Satija R (2018). Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T, Morbeck DE, Von Zglinicki T, Van Deursen J, Lustgarten J, Scrable H, Khosla S, Jensen MD, and Kirkland JL (2010). Fat tissue, aging, and cellular senescence. Aging Cell 9, 667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaiah JJ, Koepplin JW, and Folpe AL (2021). RNAscope CSF1 chromogenic in situ hybridization: a potentially useful tool in the differential diagnosis of tenosynovial giant cell tumors. Hum Pathol 115, 1–9. [DOI] [PubMed] [Google Scholar]
- Tripathi U, Misra A, Tchkonia T, and Kirkland JL (2021). Impact of Senescent Cell Subtypes on Tissue Dysfunction and Repair: Importance and Research Questions. Mech Ageing Dev 198, 111548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D, Liang HE, and Locksley RM (2008). Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J Immunol 180, 4742–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Liu Z, Chen VP, Wang L, Inman CL, Zhou Y, Guo C, Tchkonia T, Rowe DW, Kuchel GA, et al. (2020). Transplanting cells from old but not young donors causes physical dysfunction in older recipients. Aging Cell, e13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wang L, Gasek NS, Zhou Y, Kim T, Guo C, Jellison ER, Haynes L, Yadav S, Tchkonia T, et al. (2021). An inducible p21-Cre mouse model to monitor and manipulate p21-highly-expressing senescent cells in vivo. Nature Aging 1, 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Palmer AK, Ding H, Weivoda MM, Pirtskhalava T, White TA, Sepe A, Johnson KO, Stout MB, Giorgadze N, et al. (2015a). Targeting senescent cells enhances adipogenesis and metabolic function in old age. Elife 4, e12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, Inman CL, Ogrodnik MB, Hachfeld CM, Fraser DG, et al. (2018). Senolytics improve physical function and increase lifespan in old age. Nature medicine 24, 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Tchkonia T, Ding H, Ogrodnik M, Lubbers ER, Pirtskhalava T, White TA, Johnson KO, Stout MB, Mezera V, et al. (2015b). JAK inhibition alleviates the cellular senescence-associated secretory phenotype and frailty in old age. Proceedings of the National Academy of Sciences of the United States of America 112, E6301–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeadon J (2015). CHOOSING AMONG TYPE II DIABETES MOUSE MODELS. [Google Scholar]
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H , Hattori M, et al. (2013). Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
Original data for creating all graphs in the paper are provided in Data S1
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD16/CD32 antibody | eBioscience | 14-0161-86 |
| APC anti-mouse CD45 antibody | eBioscience | 17-0451-83 |
| PE-Cyanine7 anti-mouse Sca-1 antibody | eBioscience | 25-5981-82 |
| PerCP-Cyanine5.5 anti-mouse CD31 antibody | Biolegend | 102420 |
| PE-Cyanine7 anti-mouse F4/80 antibody | eBioscience | 25-4801-82 |
| Brilliant Violet 421™ anti-mouse F4/80 Antibody | BioLegend | 123132 |
| PerCP-Cyanine5.5 anti-mouse CD11b antibody | eBioscience | 45-0112-82 |
| Brilliant Violet 711™ anti-mouse/human CD11b Antibody | BioLegend | 101242 |
| Anti-CDKN2A/p16INK4a antibody [EPR20418] | Abcam | ab211542 |
| p21 (WAF1, Cip1) polyclonal antibody | eBioscience | 14-6715-81 |
| Anti-NF-kB p65 antibody | Abcam | ab16502 |
| Goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 568 | Invitrogen | A-11011 |
| Goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 647 | Invitrogen | A-21244 |
| Phospho-histone H2A.X (Ser139) (20E3) rabbit mAb | Cell Signaling Technology | 9718 |
| Goat anti-rabbit IgG antibody (H+L), biotinylated | Vector Lab | BA-1000 |
| Single cell hashing antibody | BioLegend | TotalSeq anti-mouse Hashtag #3-5 |
| Kapa Library Quantification Kit | Roche | KK4824 |
| Insulin (C27C9) rabbit mAb | Cell Signaling Technology | 9008S |
| Human p21 antibody | Sigma-Aldrich | 421M-17 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Teklad Global 18% Protein Rodent Diet | ENVIGO | 2918 |
| Rodent diet with 60 kcal% Fat | Research Diets | D12492 |
| Tamoxifen | Sigma-Aldrich | T5648 |
| Corn oil | Sigma-Aldrich | C8267 |
| Collagenase type II | Sigma-Aldrich | C6885 |
| PBS (10X) | Fisher bioreagents | BP6651 |
| DNase I | Sigma-Aldrich | D5025 |
| ACK Lysing Buffer | Gibco | A1049201 |
| BSA | Sigma-Aldrich | A9647 |
| Kapa biosystems qPCR reagents and kits | Kapa Biosystems qPCR Reagents and Kits | Kapa Biosystems qPCR Reagents and Kits |
| TRIzol Reagent | Invitrogen | 15596018 |
| Chloroform | Sigma-Aldrich | 366927 |
| Isopropanol | Sigma-Aldrich | 34863 |
| Ethanol | Sigma-Aldrich | E7023 |
| UltraPure™ distilled water | Invitrogen | 10977 |
| M-MLV Reverse Transcriptase | Invitrogen | 28-025-013 |
| RNaseOUT™ Ribonuclease Inhibitor | Invitrogen | 10-777-019 |
| Random primers | Invitrogen | 48190-011 |
| dNTP set (100 mM) | Invitrogen | 10297-018 |
| PerfeCTa® FastMix® II | Quantabio | 95118 |
| Isoflurane, USP | Piramal Critical Care | 66794-013-25 |
| D-Luciferin, Potassium Salt | Gold Biotechnology | LUCK-1g |
| Glucose | Acros organics | 41095-5000 |
| Insulin (U-100) | Humulin® R | 0002-8215-17 |
| Ketoprofen (100mg/ml) | Zoetis | N/A |
| Dasatinib | LC Laboratories | D3307 |
| Quercetin | Sigma-Aldrich | Q4951 |
| Phosal 50 PG | Lipoid LLC | 368315 |
| PEG-400 | Sigma-Aldrich | 91893 |
| Fetal Bovine Serum | Corning | 35-011-CV |
| RPMI medium 1640 | Gibco | 11835-030 |
| Sodium Pyruvate (100 mM) | Gibco | 11360-070 |
| MEM Non-Essential Amino Acids (100 X) | Gibco | 11140-050 |
| MEM Vitamins (100 X) | Gibco | 11120-052 |
| Penicillin-Streptomycin | Gibco | 15140-122 |
| Paraformaldehyde solution, 4% in PBS | Thermo Fisher Scientific | AAJ19943K2 |
| Sucrose | Sigma-Aldrich | 84097 |
| Tissue-Plus™ O.C.T. Compound | Thermo Fisher Scientific | 23-730-571 |
| Diamond antifade mountant with DAPI | Life Technologies | P36962 |
| Triton X-100 | Sigma-Aldrich | T9284 |
| Harris Hematoxylin | Leica biosystems | 3801560 |
| Eosin | Leica biosystems | 3801600 |
| Toluene | Avantor Performance Materials | JTB/9351-03 |
| BOND IHC Polymer Detection Kit | Leica biosystems | DS9800 |
| RNAscope™ 2.5 LSx Reagent Kit-Brown | ACD | 322700 |
| RNAscope® 2.5 LS Positive Control Probe_Mm-PPIB | ACD | 313918 |
| RNAscope® 2.5 LS Negative Control Probe_dapB | ACD | 312038 |
| RNAscope® LS 2.5 Probe- Mm-Cdkn1a | ACD | 408558 |
| RNAscope® LS 2.5 Probe- Mm-Cdkn2a | ACD | 411018 |
| Avidin/Biotin blocking kit | Vector Lab | SP-2001 |
| Streptavidin, DyLight® 649 | Vector Lab | SA-5649-1 |
| TelC-Cy3 PNA FISH probe | Panagene | F1002 |
| Goat serum | Sigma-Aldrich | G9023 |
| EdU | Cayman Chemical | 20518 |
| Ascorbic acid | Alfa Aesar™ | A1561322 |
| Alexa Fluor 647 azide | Thermo Fisher Scientific | A10277 |
| X-gal | Teknova | X1220 |
| Probes/primers for QPCR | ||
| Tbp | IDT | Mm.PT.39a.22214839 |
| p21 | IDT | Mm.PT.58.17125846 |
| p16 | IDT | Mm.PT.58.8388138 |
| Il6 | IDT | Mm.PT.58.10005566 |
| Pai1 | IDT | Mm.PT.58.6413525 |
| Ccl2 | IDT | Mm.PT.58.42151692 |
| Cxcl1 | IDT | Mm.PT.58.42076891 |
| Pparg | IDT | Mm.PT.58.31161924 |
| Fabp4 | IDT | Mm.PT.58.43866459 |
| Adipoq | IDT | Mm.PT.58.9719546 |
| Mrc1 | IDT | Mm.PT.58.42560062 |
| Mgl2 | IDT | Mm.PT.58.21616779 |
| Cd163 | IDT | Mm.PT.58.15897116 |
| Critical Commercial Assays | ||
| Insulin Mouse Ultra Sensitive ELISA | Crystal Chem | 90080 |
| Deposited Data | ||
| Single-cell RNA sequence data Raw data | Gene Expression Omnibus Data S1 | GSE161872 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 | Jackson Laboratory | 000664 |
| floxed knock-in tdTomato mice | Jackson Laboratory | 007914 |
| floxed knock-in LUC mice | Jackson Laboratory | 005125 |
| floxed DTA mice | Jackson Laboratory | 009669 |
| floxed Rela mice | Jackson Laboratory | 024342 |
| SCID-beige mice | ENVIGO | N/A |
| p21-Cre mice | This study | N/A |
| Software and Algorithms | ||
| FlowJo software V10.7 | Becton, Dickinson & Company | https://www.flowjo.com/solutions/flowjo/downloads |
| minispec Plus software V7.0 | Bruker | N/A |
| Living Image 4.5.5 | PerkinElmer | https://www.perkinelmer.com.cn/lab-products-and-services/resources/in-vivo-imaging-software-downloads.html |
| Oxymax®-CLAMS V5.51 | Columbus | N/A |
| CLAMS examination tool (CLAX) V2.2.15 | Columbus | N/A |
| Image J 1.42q | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Zen Blue 2.3 | Zeiss | N/A |
| ZEN lite 3.2 | Zeiss | N/A |
| Other | ||
| AimStrip Plus blood glucose testing system | Germaine Laboratories | 37321 |
| Glucose test strips | Germaine Laboratories | 37350 |
| Countess II automated cell counter | Thermo Fisher Scientific | AMQAX1000 |
| 4200 Tapestation system | Agilent Technologies | N/A |
| NovaSeq™ 6000 sequencing system | Illumina | Novogene |
| 10X Chromium platform | 10x Genomics | N/A |
| BD LSR II flow cytometer | BD Biosciences | N/A |
| BD FACS Aria II flow cytometer | BD Biosciences | N/A |
| CFX96 Touch Real-Time PCR detection system | Bio-Rad Laboratories | 1855196 |
| minispec mq7.5 TD-NMR analyzer | Bruker | LF50 |
| Comprehensive laboratory animal monitoring system (CLAMS) | Columbus | N/A |
| EVOS XL Core imaging system | Thermo Fisher Scientific | AMEX1000 |
| ZOE™ Fluorescet cell imager | Bio-Rad Laboratories | 1450031 |
| Axio Observer Z1 | Zeiss | N/A |


