Abstract
Repression of gene transcription is linked to regulation of chromatin structure through deacetylation of core histone amino-terminal tails. This action is mediated by histone deacetylases (HDACs) that function within active multiprotein complexes directed to the promoters of repressed genes. In vivo, HDAC3 forms a stable complex with the SMRT corepressor. The SMRT-HDAC3 complex exhibits histone deacetylase activity, whereas recombinant HDAC3 is an inactive enzyme. Here we report that SMRT functions as an activating cofactor of HDAC3. In contrast, SMRT does not activate the class II HDAC4, with which it also interacts. Activation of HDAC3 is mediated by a deacetylase activating domain (DAD) that includes one of two SANT motifs present in SMRT. A cognate DAD is present in the related corepressor N-CoR, which can also activate HDAC3. Mutations in the DAD that abolish HDAC3 interaction also eliminate reconstitution of HDAC activity. Using purified components, the SMRT DAD is shown to be necessary and sufficient for activation of HDAC3. Moreover, the DAD is required both for HDAC3 to function enzymatically and for the major repression function of SMRT. Thus, SMRT and N-CoR do not serve merely as platforms for HDAC recruitment but function as an integral component of an active cellular HDAC3 enzyme.
Reversible histone acetylation has been identified as a major regulator of eukaryotic gene transcription. Lysine residues in histone tails are acetylated by histone acetyltransferases (HATs) that function as transcriptional coactivators (5, 29, 49). The acetylation of histones results in a less restrictive chromatin structure that is generally associated with transcriptional activation (55). The HATs primarily function as part of large multiprotein coactivation complexes that are recruited to active genes by sequence-specific DNA binding proteins (11, 28). Although these complexes can contain multiple HATs, individual HATs are enzymatically active when purified from a variety of sources (38, 40, 48). These intrinsically active HATs can be inhibited or potentiated by various viral proteins and transcription factors (7, 8, 16, 43, 47).
Histone deacetylases (HDACs) reverse the enzyme reaction catalyzed by HATs, leading to a repressive chromatin structure (30, 42). Multiple HDACs of three major classes have been identified. Class I HDACs include HDACs 1, 2, 3, and 8 (10, 20, 50, 63, 64) and are homologous to the yeast Rpd3 deacetylase, whereas the class II deacetylases including HDACs 4, 5, 6, and 7 are more similar to yeast Hda1 (12, 27, 37, 57). A third class of HDACs include homologs of the yeast Sir2 silencing protein which was found to require a NAD cofactor for activity (24, 32, 46). HDACs have been found to function in vivo as large multiprotein complexes that are targeted by DNA binding proteins to actively repress gene transcription. Biochemically purified HDAC-containing complexes include NuRD-Mi2 complexes (52, 58, 62, 69), Sin3 complexes (17, 31, 68, 71), CoREST complexes (22, 65), and the core complexes containing nuclear hormone receptor corepressors N-CoR and SMRT (15, 34). Isolation of individual HDACs by recombinant methods without associated complex components or cofactors has generally yielded inactive forms of the deacetylases (24, 32, 33, 70). It has been shown that the deacetylase activity of HDAC1 is dependent upon MTA2 in the context of the Mi2-NuRD complex and upon CoREST in its cognate complex (70).
Two closely related corepressors, SMRT (silencing mediator for retinoid and thyroid receptors) (9) and N-CoR (nuclear receptor corepressor) (19), mediate repression functions for numerous transcription factors including unliganded nuclear hormone receptors (9, 19, 44, 66), the notch binding protein CBF-1 (26), and homeodomain proteins Rpx2, Pit1, and Pbx (3, 61). Studies with knockout mice have demonstrated that N-CoR is essential for viability, playing a critical role in the development of the brain and hematopoietic system (25). Both SMRT and N-CoR interact directly with multiple HDACs, including HDACs 3, 4, 5, and 7 (15, 21, 27, 34), and may associate with HDACs 1 and 2 via the Sin3 repressor (18, 39, 54). The current view of SMRT and N-CoR function holds that these corepressors passively recruit active HDACs to unliganded nuclear hormone receptors and other transcriptional repressors.
SMRT and N-CoR both exist in tight complexes containing TBL1 (a WD40 repeat-containing protein) and HDAC3 (15, 34). The functional interaction between N-CoR or SMRT and HDAC3 was also found in Xenopus oocytes, where HDAC3 is important for mediating repression by unliganded thyroid hormone receptor (34, 56). The purified SMRT-TBL1-HDAC3 complex possesses HDAC enzyme activity, whereas here we show that recombinant HDAC3 alone does not function as an HDAC. SMRT binding to HDAC3 is necessary and sufficient to form an active enzyme complex. This function of SMRT is mediated by a deacetylase activating domain (DAD) that is conserved in N-CoR, which also activates HDAC3. Thus, the role of SMRT and N-CoR is not only to deliver HDACs to target genes but to serve as critical cofactors in the formation of an active HDAC enzyme.
MATERIALS AND METHODS
Production of active HDAC complexes in vitro.
Myc-HDAC3 and cofactors were translated individually with the TNT T7 Quick coupled transcription-translation kit (Promega) according to the manufacturer's instructions. One hundred microliters of translated material was used as a source for each protein unless otherwise noted. After translation, the desired proteins were mixed and added to 2 volumes of buffer D-300 (300 mM KCl, 20 mM HEPES [pH 7.9], 0.25 mM EDTA, 10% glycerol, 0.1% Tween 20) containing protease inhibitors (Boehringer Mannheim Complete cocktail). Complexes were purified by incubation with anti-Myc agarose beads (Santa Cruz Biotechnology) for 16 to 18 h at 4°C. Beads were washed three times in buffer D-300 and once in HD buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% glycerol). The beads were then subjected to HDAC assay and immunoblot analysis.
HDAC assays.
3H-labeled acetylated histones were prepared essentially as described elsewhere (6). Deacetylase activities of immunoprecipitations were assayed by incubating the pelleted beads with 20,000 cpm of 3H-labeled acetylated HeLa histones in a total volume of 200 μl of HD buffer at 37°C for 1.5 h. The beads were collected and separated from the aqueous reaction mixtures. Reaction mixtures were quenched with 50 μl of stop solution (1 M HCl, 0.16 M HAc) and extracted with 600 μl of ethyl acetate. Released 3H-acetic acid was measured by scintillation counting of the organic phase.
Plasmid constructs.
All plasmids contained the T7 promoter for in vitro coupled transcription-translation. Myc-HDAC3 was produced by cloning HDAC3 cDNA (encoding amino acids 1 to 428) into pcDNA3.1myc/his (Invitrogen). Myc-HDAC4 was produced by cloning HDAC4 cDNA into pcDNA3.1myc/his and was described previously (16). Gal4-TBL1 was produced by cloning TBL1 cDNA (encoding amino acids 1 to 577) into pGal4-DBD (67). Full-length Flag-tagged mouse SMRT was a gift from Ronald Evans, full-length Flag-tagged mouse N-CoR was a gift from Michael G. Rosenfeld, and full-length rat MTA1 was a gift from Yasushi Toh. Full-length Gal4-SMRT was produced by inserting the EcoRV fragment from pCMX-SMRT (41) containing SMRT cDNA into pGal4-DBD, Gal4-mSMRT plasmids containing Gal4 DNA binding domain (DBD) fusions to mSMRT amino acids 1961 to 2473, 1531 to 1959, 1068 to 1530, 1 to 1028, 1 to 763, 305 to 489, 305 to 763, 305 to 427, 427 to 669, 427 to 489, 427 to 599, 305 to 455, and 395 to 489 were produced by PCR amplification of the corresponding DNA sequences followed by insertion into pGal4-DBD. Gal4-SMRT DAD mutants W432A, F440A, F451A, I454A, L458A, V463A, and Y470A were produced using the corresponding mutagenic oligonucleotides and the QuickChange site-directed mutagenesis kit (Stratagene). Gal4-SMRT 1-763Δ DAD (deleted amino acids 305 to 428) was produced by PCR amplification of mSMRT sequences encoding amino acids 1 to 304 and insertion in frame into pGal4-SMRT 1-763 with deletion of sequences encoding amino acids 1 to 428.
Immunoprecipitations.
Cells were washed in phosphate-buffered saline, lysed with lysis buffer A (150 mM NaCl, 40 mM Tris [pH 7.6], 10% glycerol, 0.3% NP-40) containing protease inhibitors (Boheringer Mannheim Complete cocktail) for 20 min, and pelleted at 20,000 × g. Supernatants were incubated with anti-Gal4-agarose-conjugated beads (Santa Cruz Biotechnology) at 4°C for 16 to 18 h. Beads were pelleted at 6,000 × g and washed successively in lysis buffer A, lysis buffer A containing 300 mM NaCl–0.1% NP-40, lysis buffer A containing 500 mM NaCl–0.1% NP-40, and HD buffer.
Immunoblot analysis and antibodies.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes with HMW transfer buffer (50 mM Tris, 380 mM glycine, 0.1% SDS, and 20% methanol). Blots were probed with primary antibodies in Tris-buffered saline containing 0.15% Tween 20 and 3.0% nonfat dry milk, followed by horseradish peroxidase (HRP)-conjugated anti-rabbit antibodies (Boehringer Mannheim). Alternatively, HRP-conjugated primary antibodies were directly used. All blots were visualized with the ECL kit (Amersham Pharmacia Biotech). Anti-Gal4 DBD rabbit polyclonal antibody and anti-MTA1 antibody were purchased from Santa Cruz Biotechnology. Anti-Myc–HRP was purchased from Invitrogen. Anti-Flag–HRP rabbit polyclonal antibody was purchased from Sigma Biochemicals.
Baculoviruses and purification of active HDAC3 complex.
Flag-HDAC3 and Myc-SMRT amino acids 1 to 587 were cloned into pBlueBac4.5, and recombinant baculoviruses were produced with the MaxBac 2.0 kit (Invitrogen). Sf9 cells were maintained in Sf-900 II SFm medium (Gibco BRL) containing Antibiotic-Antimycotic (Gibco BRL) and 0.1% Pluronic F-68 (Gibco BRL). Sf9 cells (7.5 × 108) were infected with Flag-HDAC3 baculovirus at a mutiplicity of infection of 5.0 in the absence or presence of Myc-SMRT 1-587 baculovirus at a multiplicity of infection of of 5.0. Control cells were mock infected with an equal volume of Sf-900 medium. Cells were lysed in lysis buffer A containing protease inhibitors (Boheringer Mannheim Complete cocktail), and the supernatant was incubated with 500 μl of anti-Flag agarose beads (Sigma) for 14 to 16 h at 4°C. Beads were washed successively in lysis buffer A, buffer D-300, buffer D-300 containing 500 mM KCl, buffer D-300 containing 700 mM KCl, and EB buffer (Tris-buffered saline containing 10% glycerol). Proteins were eluted with 3 × 500 μl of EB buffer containing 200 μg of FLAG peptide (Sigma) per ml.
Mammalian cell culture and transfection.
293T cells were maintained in DMEM (high glucose) supplemented with 10% fetal bovine serum and l-glutamine (all from GIBCO BRL). Cells were grown at 37°C in 5% CO2. 293T cells were transfected with FuGene6 (Boehringer Mannheim) according to the manufacturer's instructions. Cells were transfected for 48 h and washed with phosphate-buffered saline before being harvested for luciferase reporter assays and immunoprecipitations. In luciferase reporter assays, the Gal4 UAS × 5-SV40-luciferase reporter contains five copies of the Gal4 17-mer binding site. Luciferase assay kit (Promega) was used to determine relative levels of the luciferase gene product. Light units were normalized to a cotransfected β-galactosidase expression plasmid. Fold repression is relative to the Gal4 DBD, and results of duplicate samples are plotted.
RESULTS
HDAC3 enzyme activity requires SMRT.
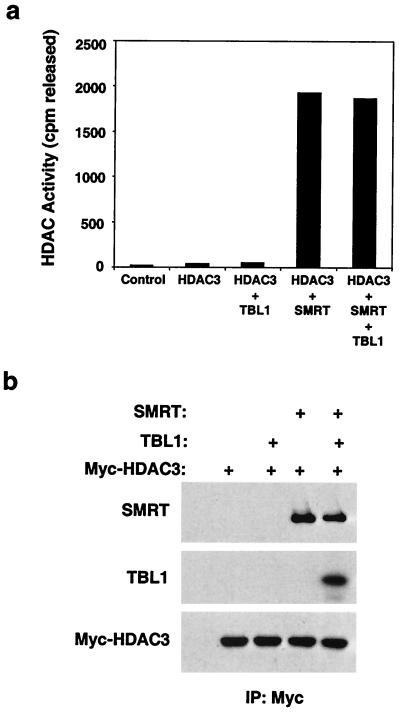
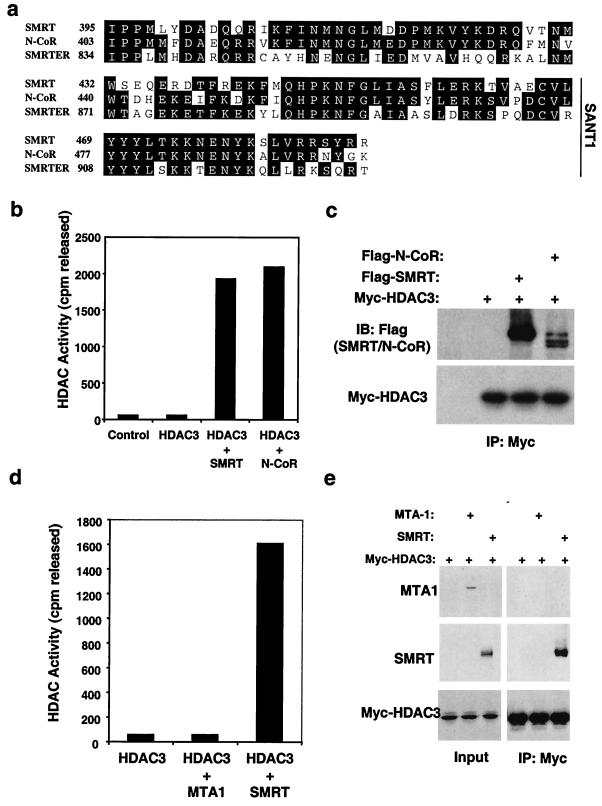
We found that recombinant HDAC3, produced by in vitro coupled transcription-translation in rabbit reticulocyte lysate, was devoid of enzyme activity (Fig. 1a). Since we had previously shown that the HDAC3-SMRT-TBL1 complex is an enzymatically active HDAC complex (15), we tested whether recombinant SMRT, TBL1, or both could reconstitute enzyme activity in an in vitro coupled transcription-translation system. HDAC3 and potential cofactors were combined, the complexes were captured on an affinity matrix, and the deacetylase activity was measured. Remarkably, addition of SMRT to HDAC3 created a potent HDAC enzyme (Fig. 1a). SMRT and HDAC3 physically associate under these conditions (Fig. 1b). TBL1, in contrast, did not activate HDAC3 and did not significantly alter the enzyme activity of HDAC3 in the presence of SMRT despite physically interacting with the HDAC3-SMRT complex (Fig. 1). Thus SMRT, and not TBL1, was required for enzymatic competency of HDAC3.
FIG. 1.
SMRT activates HDAC3 deacetylase activity in vitro. (a) SMRT core complex components were produced by T7-coupled transcription-translation. Myc-HDAC3 alone or mixed together with Gal4-TBL1 and/or Flag-SMRT was immunoprecipitated with anti-Myc agarose beads to isolate HDAC complexes, and the beads were assayed for HDAC activity. Immunoprecipitation of unprogrammed transcription-translation mix was used as a control. (b) Western blot analysis of the complexes in panel a. The upper panel was probed for SMRT (anti-Flag), the middle panel was probed for TBL1 (anti-Gal4), and the lower panel was probed for HDAC3 (anti-Myc). IP, immunoprecipitation.
SMRT binds but does not activate HDAC4.
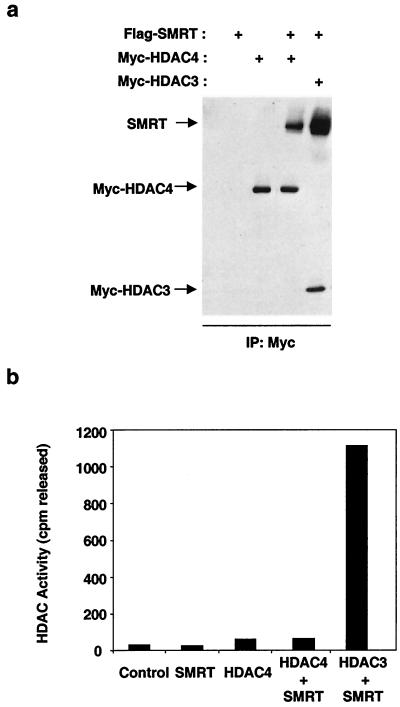
We next sought to determine whether association with SMRT activates HDACs other than HDAC3. Although HDAC3 is the sole HDAC found in stable endogenous SMRT and N-CoR complexes, we and others have previously shown that SMRT and N-CoR bind directly and specifically to class II HDACs, including HDAC4. This interaction was confirmed in a coimmunoprecipitation of Myc-HDAC4 and Flag-SMRT (Fig. 2a). However, whereas SMRT was associated with both HDAC3 and HDAC4 (Fig. 2a), only HDAC3 enzyme activity was reconstituted by this interaction (Fig. 2b). Thus, the ability of SMRT to activate HDAC activity is specific to HDAC3.
FIG. 2.
SMRT binds but does not activate HDAC4. (a) Western blot analysis of HDAC complexes formed in the presence or absence of SMRT. Proteins produced by T7-coupled transcription-translation were mixed and immunoprecipitated (IP) with anti-Myc agarose beads. All samples were resoved by SDS-PAGE and subjected to immunoblotting. Flag-SMRT (top) was probed with anti-Flag. Myc-HDAC4 and Myc-HDAC3 (middle and bottom) were probed with anti-Myc. (b) HDAC activity of the complexes in panel a. Note that 50 μl of translated HDAC3 and 100 μl of HDAC4 were used.
Multiple domains of SMRT interact with HDAC3, but only one functions as a DAD.
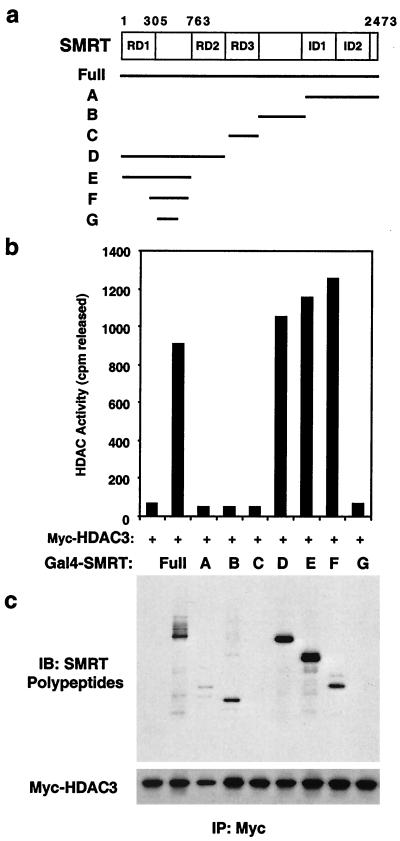
Polypeptides encompassing the full length of SMRT were produced in vitro and tested for the ability to render HDAC3 an active enzyme as well to interact with HDAC3. (Fig. 3). An HDAC4-interacting polypeptide corresponding to repression domain 3 (21) did not interact with or activate HDAC3 (fragment C in Fig. 3). Two C-terminal polypeptides containing regions previously implicated in HDAC3 interaction (34, 60) were confirmed to interact with HDAC3 but did not activate its enzyme activity (fragments A and B in Fig. 3). Thus, mere interaction with SMRT was not the sole determinant of HDAC3 activation. In contrast, the N terminus of SMRT interacted most strongly with HDAC3 and potently activated HDAC3 enzyme activity (fragment D in Fig. 3). This interaction could be localized to amino acids 305 to 763 (fragment F). Remarkably, these amino acids were sufficient for reconstitution of HDAC3 enzymatic activity (Fig. 3c).
FIG. 3.
SMRT contains a DAD. (a) Schematic representation of the SMRT corepressor and deletion mutants used to reconstitute HDAC3 activity. Gal4 DBD fusions to SMRT contained SMRT amino acids 1 to 2473 (full-length SMRT), SMRT amino acids 1961 to 2473 (fragment A), SMRT amino 1531 to 1959 (fragment B), SMRT amino acids 1068 to 1530 (fragment C), SMRT amino acids 1 to 1028 (fragment D), SMRT amino acids 1 to 763 (fragment E), SMRT amino acids 305 to 763 (fragment F), and SMRT amino acids 427 to 559 (fragment G). (b) HDAC activities of HDAC3-containing complexes. Myc-HDAC3 was incubated with equal amounts of Gal4-SMRT fusion proteins, immunoprecipitated with anti-Myc, and subjected to HDAC assay. (c) Immunoblot (IB) of anti-Myc agarose immunoprecipitated (IP) complexes. The upper panel was probed for SMRT polypeptides (anti-Gal4); the lower panel was probed for HDAC3 (anti-Myc).
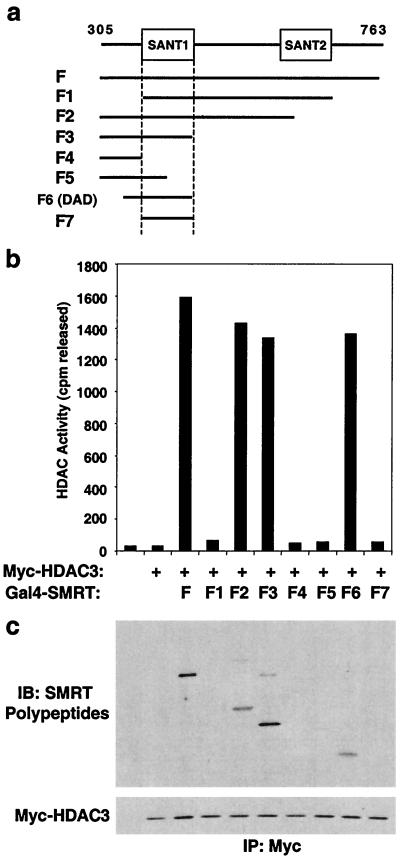
SMRT amino acids 305 to 763 contain two imperfect SANT repeats. (SANT1 and SANT2 [Fig. 4a]). The SANT domain is a region of homology between the SWI3, ADA2, N-CoR or SMRT, and TFIIIB proteins which has some similarity to the DBDs of Myb-related transcription factors (1). We therefore sought to determine if one or both SANT repeats were required for HDAC3 activation, using the series of SMRT-derived polypeptides shown in Fig. 4a. A region containing both SANT repeats and the intervening sequence (F1 in Fig. 4b) was unable to activate HDAC3. SANT2 was dispensable for efficient HDAC3 activation (F2 in Fig. 4b); however deletions of SANT1 (F4 and F5 in Fig. 4) resulted in abolition of both HDAC3 binding (F3 and F4 in Fig. 4b) and activity. SANT1 is necessary but not sufficient (compare F3 and F7 in Fig. 4b) for HDAC3 activation. Rather, SANT1 requires additional N-terminal flanking sequence (compare F6 and F7 in Fig. 4b) to form the intact DAD. Thus, the minimal SMRT DAD is approximately 95 amino acids (395 to 489 in SMRT). In all cases, interaction with the DAD-containing peptide was required for competency of the HDAC enzyme (Fig. 4c). The sufficiency of SANT1 was confirmed using a glutathione S-transferase fusion of SMRT amino acids 305 to 559, containing the DAD but not SANT2. Purified GST-SMRT 305-559 also activated HDAC3 activity (not shown). Thus, the two SANT sequences in SMRT are functionally distinct, with only SANT1 being required to activate HDAC3.
FIG. 4.
The SMRT DAD is comprised of SANT1 and its amino-terminal flank. (a) Schematic representation of SMRT amino acids 305 to 763 containing SANT1 and SANT2. Gal4 DBD–SMRT fusions contained amino acids 305 to 763 (F), amino acids 427 to 669 (F1), amino acids 305 to 559 (F2), amino acids 305 to 489 (F3), amino acids 305 to 427 (F4), amino acids 305 to 455 (F5), amino acids 395 to 489 (F6 [DAD]), and amino acids 427 to 489 (F7). (b) HDAC activities of HDAC3-containing complexes. Myc-HDAC3 was incubated with equal amounts of Gal4-SMRT fusions, immunoprecipitated with anti-Myc, and subjected to HDAC assay. (c) Immunoblot (IB) of anti-Myc-precipitated complexes. The upper panel was probed for SMRT polypeptides (anti-Gal4), and the lower panel was probed for HDAC3 (anti-Myc). IP, immunoprecipitation.
N-CoR also activates HDAC3 enzyme activity.
The DAD is highly conserved between SMRT and N-CoR (Fig. 5a). We and others have shown that N-CoR interacts with HDAC3 (15, 34, 60), and thus, we sought to determine if N-CoR could also create an active HDAC3 enzyme. N-CoR–HDAC3 as well as SMRT-HDAC3 complexes were formed in vitro and tested for HDAC activity (Fig. 5b). Recombinant N-CoR activated HDAC3 enzymatic activity as effectively as SMRT, a finding consistent with N-CoR's conserved DAD and ability to interact with HDAC3 (Fig. 5c). The DAD sequence is also conserved in the Drosophila protein SMRTER (Fig. 5a), which functions as a corepressor for the ecdysone receptor (53).
FIG. 5.
N-CoR corepressor binds and activates HDAC3. (a) Alignment of the SMRT DAD with homologous regions of the corepressors N-CoR and SMRTER. Residues conserved in at least two of the three proteins are shown on black background. (b) Myc-HDAC3 alone or mixed with either Flag-SMRT or Flag–N-CoR was immunoprecipitated with anti-Myc and subjected to HDAC assay. Immunoprecipitation of an unprogrammed transcription-translation mix was used as a control. (c) Immunoblot (IB) of HDAC3 complexes immunoprecipitated (IP) by anti-Myc. The upper panel was probed for SMRT or N-CoR (anti-Flag); the lower panel was probed for HDAC3 (anti-Myc). (d) Myc-HDAC3 alone or mixed with either MTA1 or Flag-SMRT was immunoprecipitated with anti-Myc and subjected to HDAC assay. (e) Immunoblot analysis of the Myc-HDAC3 immunoprecipitates in panel d. The upper panel was probed with anti-MTA1, the middle panel was probed for SMRT (anti-Flag), and the lower panel was probed for HDAC3 (anti-Myc). Five-percent inputs are shown on the left, and all proteins were produced by T7-coupled transcription-translation.
MTA1 is another SANT domain protein, known to be present in HDAC1 core complexes (22, 45, 51). In order to determine if the SANT-containing DADs of SMRT and N-CoR were specifically required for HDAC3 activation, MTA1 was tested for the ability to bind and activate HDAC3. Unlike N-CoR or SMRT, MTA1 neither activates (Fig. 5d) nor binds HDAC3 (Fig. 5e). This is most likely due to lack of conservation of the amino acids immediately N terminal to the SANT domain, which were shown earlier to be required for SMRT interaction and activation of HDAC3 (Fig. 4).
HDAC3 binding and activation by the SMRT DAD are strongly correlated.
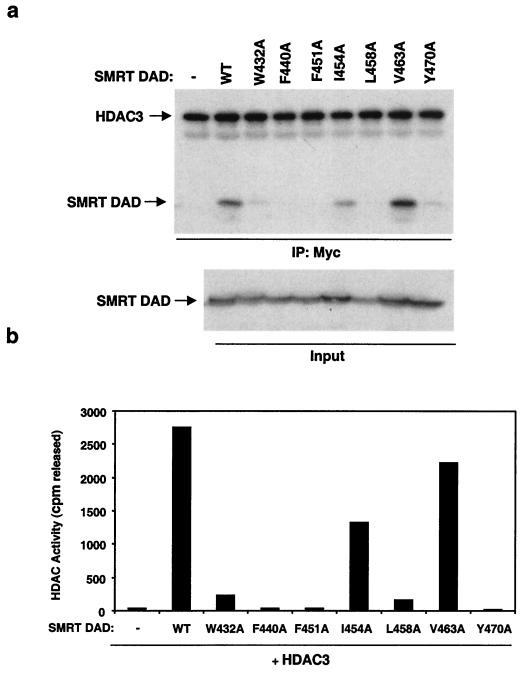
Having shown that the extended N-terminal SANT domain is required for HDAC3 binding and enzyme activation by SMRT, we further evaluated the role of the SANT domain by creating and studying seven point mutations across this region. All seven amino acids chosen for this analysis are identical in SMRT and N-CoR (Fig. 5a), and all proteins were expressed at similar levels (Fig. 6a). Figure 6 shows a remarkable correlation between the ability of these SMRT DAD point mutants to bind to HDAC3 and their ability to activate the enzyme. For example, the V463A mutant did not impair binding nor HDAC activity relative to the wild type, whereas the I454A mutant bound and activated HDAC3 about 50% as well as the wild type (Fig. 6). In contrast, mutations that dramatically reduced binding (W432A, F440A, F451A, V463A, and Y470A) also abrogated HDAC3 enzyme activation by the SMRT DAD. Thus, binding and activation of HDAC3 by the SMRT DAD are inseparable by this analysis.
FIG. 6.
HDAC3 activation correlates with SMRT DAD binding. Myc-HDAC3 was mixed with wild-type (WT) Gal4-DAD (Gal4 DBD fusion to SMRT amino acids 395 to 489) or the point mutant W432A, F440A, F451A, I454A, L458A, V463A, or Y470A. HDAC3 complexes were immunoprecipitated by anti-Myc agarose beads. (a) Immunoblot analysis of anti-Myc-precipitated complexes. The upper panel was probed for HDAC3 (anti-Myc), the middle panel was probed for associated SMRT DAD proteins (anti-Gal4), and the lower panel shows 5% inputs of SMRT DAD proteins. (b) HDAC-activating abilities of anti-Myc-precipitated complexes.
N and C termini of HDAC3 are required for interaction with the SMRT DAD.
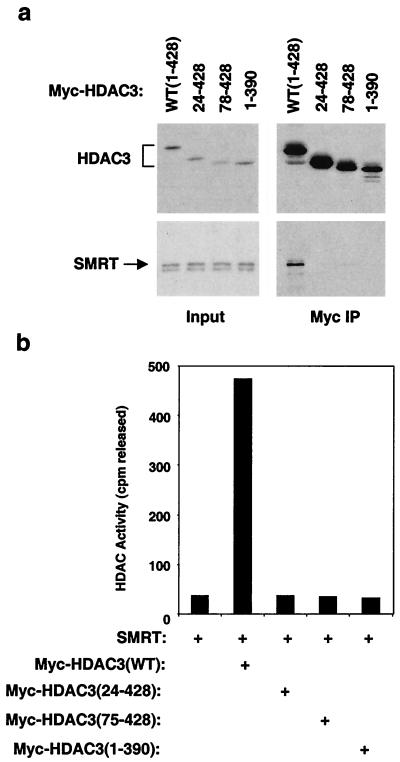
To ascertain which regions of HDAC3 mediate interaction with SMRT, we created HDAC3 deletion mutants and tested their deacetylase activity and ability to interact with a SMRT DAD-containing polypeptide. Deletion of the N-terminal 23 amino acids from HDAC3 abolished its interaction with SMRT (Fig. 7a) as well as its ability to be enzymatically activated by the SMRT DAD (Fig. 7b). Deletion of additional N-terminal amino acids led to the same result. Remarkably, deletion of 37 amino acids from the C terminus of HDAC3 also abrogated SMRT interaction (Fig. 7a) and activation of deacetylase activity (Fig. 7b). Thus, both the N and the C termini of HDAC3 are required for interaction with the SMRT DAD. This indicates that both ends of the HDAC3 polypeptide contribute to an intricately folded surface that interacts with the SMRT DAD.
FIG. 7.
HDAC3 interaction with SMRT. SMRT 1-763 fused to Gal4 DBD was mixed with wild-type (WT) or mutant Myc-HDAC3 (Myc-HDAC3 amino acids 24 to 428, 78 to 428, or 1 to 390), and HDAC3 complexes were immunoprecipitated (IP) by anti-Myc agarose beads. (a) Immunoblot analysis of HDAC3 complexes. HDAC3 deletion mutants were visualized by anti-Myc, and SMRT polypeptides were probed with anti-Gal4. Inputs are shown on the left. (b) HDAC assay of immunocomplexes. Note that 50 μl of full-length HDAC3 translation mix and 100 μl of all translated HDAC3 mutants were used to normalize protein levels.
Reconstitution of a pure, enzymatically active HDAC3-SMRT complex.
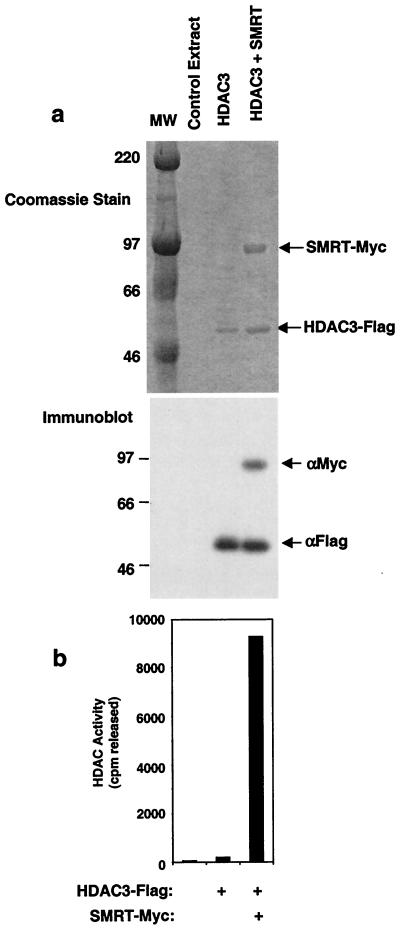
Having determined the necessity of the SMRT DAD for activation of recombinant HDAC3 enzyme in an in vitro transcription coupled translation system, we next utilized a baculoviral system to express and purify HDAC3 and a polypeptide containing the SMRT DAD. Full-length HDAC3 was expressed with a Flag epitope at its C terminus and purified to near homogeneity using Flag-agarose (Fig. 8a). This pure preparation of HDAC3 was enzymatically inactive (Fig. 8b), as previously observed for recombinant HDAC3 synthesized in reticulocyte lysate. HDAC3 was also purified after coinfection with a baculovirus expressing SMRT amino acids 1 to 587, containing the DAD. After anti-Flag affinity purification, SMRT was tightly and stoichiometrically associated with HDAC3 (Fig. 8). This nearly homogeneous SMRT-HDAC3 complex was extremely active as an HDAC (Fig. 8b); ∼50% of acetylated histone substrate had been deacetylated compared with ∼10 to 20% that we generally observe with the HDAC3 activity reconstituted with reticulocyte lysate-derived proteins. These experiments show that SMRT is sufficient as well as necessary for activation of HDAC3.
FIG. 8.
Purification of an enzymatically active HDAC3-SMRT complex. (a) Coomassie stain (top) of purified HDAC3 complex. Sf9 cells were mock infected (control) or infected with baculovirus expressing Flag-HDAC3, Myc-SMRT amino acids 1 to 587, or both. Sf9 lysates were purified by anti-Flag affinity chromatography and resolved by SDS-PAGE. Molecular weight markers (MW) and major polypeptides are indicated. Identities of complex components were verified by immunoblotting with anti-Myc and Flag-HDAC3 (below the Coomassie stain). (b) HDAC activities of the proteins in panel a.
Repressive function of the SMRT DAD.
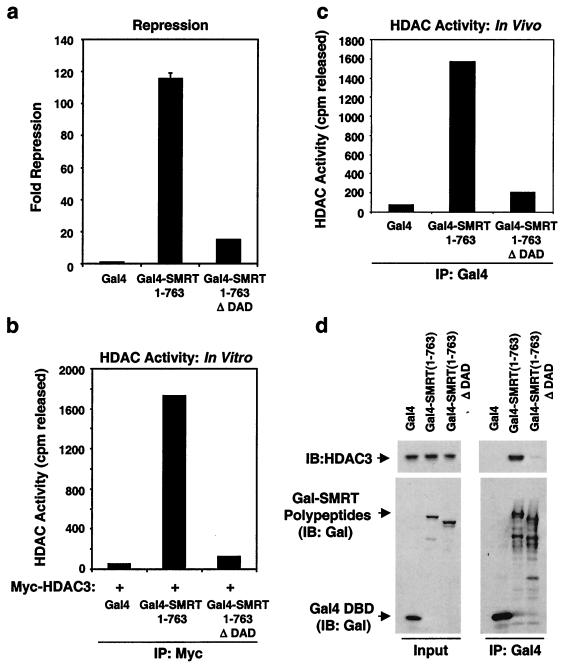
We next tested whether the SMRT DAD contributes to the repression function of SMRT. The N terminus of SMRT can be subdivided into at least three domains that mediate repression when fused to the Gal4 DBD. The Gal4 DBD fusion to SMRT amino acids 1 to 763 is a particularly powerful repressor (Fig. 9a). This region of SMRT contains the DAD but does not contain the region that interacts with the class II HDACs 4, 5, and 7 (53). Within the DAD region, we deleted amino acids 305 to 428 (immediately N terminal to SANT1), which are required for HDAC3 binding and activation. Deletion of this region within the DAD dramatically lowered the ability of Gal4 DBD–SMRT 1-763 to repress transcription in 293T cells (Fig. 9a). In vitro analysis confirmed the inability of this DAD deletion construct to activate HDAC3 (Fig. 9b). Moreover, Gal4–SMRT (1-763) coimmunoprecipitated with HDAC enzyme activity after transfection into 293T cells, unless the DAD domain was deleted (Fig. 9c). Indeed, endogenous HDAC3 coimmunoprecipitated with Gal4–SMRT (1-763) but not with the same polypeptide lacking the intact DAD (Fig. 9d). Thus, in vivo interaction and activation of HDAC3 by SMRT are as deduced from the in vitro studies. Residual repression activity of the DAD deletion construct may be mediated by the TBL1 interaction which we have previously determined to occur within amino acids 1 to 303 of SMRT. However, the main repression function of SMRT clearly tracked with HDAC3 interaction and reconstitution of deacetylase enzyme activity. These results are in agreement with previous studies where microinjection of anti-HDAC3 antibodies abrogated N-CoR-mediated repression in both Rat1 cells (25) and Xenopus oocytes (34).
FIG. 9.
The SMRT DAD is required for repression and deacetylase activity. (a) Repression of a Gal4-SV40-luciferase reporter by Gal4-SMRT 1-763 or Gal4-SMRT 1-763Δ DAD. Fold repression is expressed relative to Gal4 DBD. (b) In vitro HDAC activity of Myc-HDAC3 incubated with either Gal4 DBD, Gal4–SMRT 1-763, or Gal4–SMRT 1-763Δ DAD. HDAC3 complexes were immunoprecipitated (IP) with anti-Myc, and HDAC activity was expressed as the amount of acetate released. (c) HDAC activity of Gal4 DBD, Gal4–SMRT 1-763, or Gal4–SMRT 1-763Δ DAD immunoprecipitated by anti-Gal4 agarose after transfection into 293T cells. (d) Immunoblot (IB) of anti-Gal4 immunoprecipitates from 293T cells transfected with Gal4 DBD, Gal4–SMRT 1-763, or Gal4–SMRT 1-763Δ DAD. Inputs (2%) are shown on the left for comparison. The upper panels were probed for endogenous HDAC3 (anti-HDAC3), and the lower panels were probed for Gal4 DBD and Gal4-SMRT polypeptides (anti-Gal4).
DISCUSSION
Corepressors N-CoR and SMRT normally exist in tight and stoichiometric complexes with HDAC3. Here we have shown that recombinant HDAC3 is not an active deacetylase in and of itself but can be transformed into a competent enzyme by addition of SMRT or N-CoR. A previous study showing that HDAC3 has basal activity which is modestly potentiated by N-CoR is generally consistent with this finding; presumably, the basal activity of HDAC3 in that study reflected binding of endogenous N-CoR or SMRT homologs within Sf9 cells (60). In the present study, reconstitution of the SMRT-HDAC3 complex from a baculoviral system demonstrates that SMRT is sufficient to dramatically activate an otherwise inactive HDAC3 enzyme. On the basis of the present work, we suggest that these corepressors are absolutely required as activating cofactors for this HDAC enzyme. This is in stark contrast to the current model in which SMRT and N-CoR passively shepherd HDACs to transcriptional repressors.
SMRT contains a conserved short DAD that is necessary and sufficient for reconstituting deacetylase activity. The DAD contains the SANT1 domain which is necessary, though not sufficient, for DAD function. The DAD also requires the 37 amino acids N terminal to SANT1. HDAC3 enzyme activation by the SMRT DAD requires stable interaction as evidenced by the tight correlation between the abilities of SMRT DAD point mutants to bind and activate HDAC3 and is therefore likely to be stoichiometric rather than catalytic. Consistent with this, stoichiometric amounts of SMRT and HDAC3 are present in a purified, recombinant complex. While regions outside the SMRT DAD may bind HDAC3, they are dispensable for activation and may play a role in stabilizing the SMRT-HDAC3 complex in vivo.
Control of HDAC targeting, specificity, and activity remain central questions in determining how individual genes or gene sets are silenced. The regulatory mechanisms governing HDAC activity are beginning to be understood in the context of cofactor recruitment. It has previously been shown that the yeast silencing protein Sir2 requires NAD cofactor to become an active deacetylase (24, 32, 46). It has also been shown that recombinant HDAC1 is not a functional deacetylase without core components of the NuRD complex (70). MTA2, in particular, is critical for activating HDAC1 activity, but it is not known whether it is sufficient in the absence of other components of the NuRD complex. Although a specific DAD has not been sublocalized in MTA2, MTA2 also contains a single SANT motif. Interestingly, HDAC1 is also a component of the CoREST complex (22, 65). CoREST is a neural-specific repressor protein containing two SANT repeats similar to SMRT (2). Deletion of the N-terminal SANT domain of CoREST results in loss of HDAC1 interaction and of associated HDAC activity that is remarkably parallel to our finding that deletion of SANT1 of SMRT results in loss of HDAC3 interaction and associated HDAC activity (22, 65). The present studies in concert with the functional MTA2-HDAC1 and CoREST-HDAC1 interactions suggest a role of a single SANT motif in activating HDAC enzymes. Our finding that the SANT-containing protein MTA1 did not interact with or activate HDAC3 indicates that SANT-containing proteins are likely not interchangeable between specific complexes. It is of note that the 37 amino acids N terminal to SANT1 that are required for DAD function in SMRT are conserved in N-CoR and SMRTER but not in MTA1, MTA2, or CoREST. This suggests a code in which a single SANT motif combines with unique N-terminal amino acids to provide specificity of HDAC interaction and activation.
The requirement for protein partners such as SMRT, N-CoR, CoREST, and MTA2 to activate HDAC enzyme activity conceivably serves to restrict deacetylation to the chromatin environments of specific genes to which the HDACs have been targeted. Potential deacetylation of nonhistone substrates such as p53 and GATA (4, 14, 23, 35) may also be dependent on the formation of these active HDAC complexes. Although class II HDACs can be regulated by subcellular localization (13, 36, 37, 59), they are also inactive when expressed recombinantly and thus may require one or more activating subunits as an additional layer of regulatory control. Of note, although SMRT and N-CoR interact with the class II HDACs 4, 5, and 7 in vitro and in vivo (via a region that is C terminal to the DAD domain as well as SANT2 [21]), neither was sufficient to activate HDAC4 deacetylase activity in vitro. It is likely that other HDAC4-interacting proteins, alone or together with SMRT, are required for enzymatic competency of HDAC4 as a deacetylase.
ACKNOWLEDGMENTS
We are grateful to Eric Verdin and Wolfgang Fishle for HDAC3 cDNA and to Yasushi Toh for the MTA1 plasmid. We thank Xiao Hu, Clarice Chen, and Eric Huang for helpful discussions and the Wistar Baculovirus facility for Sf9 cell culture. We also thank Michael Pollack for DNA preparation.
This work was supported by grants DK45586 and DK43806 from the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH to M.A.L. DNA sequencing was supported by the Penn Digestive Disease Center.
REFERENCES
- 1.Aasland R, Steward A F, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR, and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- 2.Andres M E, Burger C, Peral-Rubio M J, Battaglioli E, Anderson M E, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahara H, Dutta S, Kao H Y, Evans R M, Montminy M. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol Cell Biol. 1999;19:8219–8225. doi: 10.1128/mcb.19.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 5.Brown C E, Lechner T, Howe L, Workman J L. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 6.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 8.Chen C J, Deng Z, Blobel G A, Lieberman P M. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol Cell Biol. 2001;21:476–487. doi: 10.1128/MCB.21.2.476-487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 10.Emiliani S, Fischle W, VanLint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 12.Grozinger C M, Hassig C A, Schreiber S L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grozinger C M, Schreiber S L. Regulation of histone deacetylase 4 and 5 transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 15.Guenther M G, Lane W S, Fischle W, Verdin E, Lazar M A, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and a WD40 repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 16.Hamamori Y, Sartorelli V, Ogryzko V, Puri P L, Wu H Y, Wang J Y, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 17.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–348. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 18.Heinzel T, Lavinsky R M, Mullen T-M, Soderstrom M, Laherty C D, Torchia J, Yuang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 19.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 20.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang G-F, Johanson K, Sung C-M, Liu R, Winkler J. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcriptional repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 21.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 22.Humphrey G W, Wang Y, Russanova V R, Hirai T, Qin J, Nakatani Y, Howard B H. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 23.Hung H L, Lau J, Kim A Y, Weiss M J, Blobel G A. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 25.Jepsen K, Harmanson O, Onami T M, Gleiberman A S, Lunyak V, McEvilly R J, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick S M, Mandel G, Glass C K, Rose D W, Rosenfeld M G. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 26.Kao H-Y, Ordentlich P, Koyano-Nakagawa K, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kao H Y, Downes M, Ordentlich P, Evans R M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 28.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 29.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 30.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 31.Laherty C E, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 32.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lechner T, Carrozza M J, Yu Y, Grant P A, Eberharter A, Vannier D, Brosch G, Stillman D J, Shore D, Workman J L. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3/Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J Biol Chem. 2000;275:40961–40966. doi: 10.1074/jbc.M005730200. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Wang J, Nawaz Z, Liu J M, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKinsey T A, Zhang C L, Lu J, Olson E N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miska E A, Karlsson C, Langley E, Nielsen S J, Pines J, Kouzarides T. HDAC4 associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 39.Nagy L, Kao H-Y, Chakvarkti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 40.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 41.Ordentlich P, Downes M, Xie W, Genin A, Spinner N B, Evans R M. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci USA. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 43.Reid J L, Bannister A J, Zegerman P, Martinez-Balbas M A, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sande S, Privalsky M L. Identification of TRACs, a family of co-factors that associate with and modulate the activity of nuclear hormone receptors. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt D R, Schreiber S L. Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry. 1999;38:14711–14717. doi: 10.1021/bi991614n. [DOI] [PubMed] [Google Scholar]
- 46.Smith J S, Brachmann C B, Celic I, Kena M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. A phylogenetically conserved NAD-dependent protein deacetylase activity. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soutoglou E, Viollet B, Vaxillaire M, Yaniv M, Pontoglio M, Talianidis I. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 2001;20:1984–1992. doi: 10.1093/emboj/20.8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 49.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 50.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 51.Toh Y, Kuninaka S, Endo K, Oshiro T, Ikeda Y, Nakashima H, Baba H, Kohnoe S, Okamura T, Nicolson G L, Sugimachi K. Molecular analysis of a candidate metastasis-associated gene, MTA1: possible interaction with histone deacetylase 1. J Exp Clin Cancer Res. 2000;19:105–111. [PubMed] [Google Scholar]
- 52.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 53.Tsai C-C, Kao H-Y, Yao T-P, McKeown M, Evans R M. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell. 1999;4:175–186. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- 54.Underhill C, Qutob M S, Yee S P, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 55.Urnov F D, Wolffe A P. A necessary good: nuclear hormone receptors and their chromatin templates. Mol Endocrinol. 2001;15:1–16. doi: 10.1210/mend.15.1.0589. [DOI] [PubMed] [Google Scholar]
- 56.Urnov F D, Yee J, Sachs L, Collingwood T N, Bauer A, Beug H, Shi Y-B, Wolffe A P. Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-ErbA yields a chromatin infrastructure-dependent transcription repression pathway. EMBO J. 2000;19:4074–4090. doi: 10.1093/emboj/19.15.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 58.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 59.Wang A H, Kruhlak M J, Wu J, Bertos N R, Vezmar M, Posner B I, Bazett-Jones D P, Yang X J. Regulation of histone deacetylase 4 by 14-3-3 proteins. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen Y D, Perissi V, Staszewski L M, Yang W M, Krones A, Glass C K, Rosenfeld M G, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu L, Lavinsky R M, Dasen J S, Flynn S E, McInerney E M, Mullen T M, Heinzel T, Szeto D, Korzus E, Durokawa R, Aggarwal A K, Rose D W, Glass C K, Rosenfeld M G. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 62.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 63.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global repressor RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang W-M, Yao Y-L, Sun J-M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 65.You A, Tong J K, Grozinger C M, Schreiber S L. CoREST is an integral component of the coREST-human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zamir I, Harding H P, Atkins G B, Horlein A, Glass C K, Rosenfeld M G, Lazar M A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with different repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zamir I, Zhang J, Lazar M A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Sun Z W, Iratni R, Erdjument B H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]