Abstract
Over the past two decades, it has become clear that the multiscale spatial and temporal organization of the genome has important implications for nuclear function. This review centers on insights gained from recent advances in light microscopy on our understanding of transcription. We discuss spatial and temporal aspects that shape nuclear order and their consequences on regulatory components, focusing on genomic scales most relevant to function. The emerging picture is that spatiotemporal constraints increase the complexity in transcriptional regulation, highlighting new challenges, such as uncertainty about how information travels from molecular factors through the genome and space to generate a functional output.
There is a growing appreciation that gene function is connected to the dynamic structure of chromosomes. In particular, spatiotemporal aspects of genome architecture are now appreciated as crucial to our understanding of eukaryotic gene expression, and thus to the main functional output of the nucleus (van Steensel and Furlong 2019). However, the mechanistic underpinnings and actual causal links between structure and function are scarce and present an obvious challenge for the next decade. Generation of vast atlases of Hi-C proximity maps over the past 10 years have provided a starting point for such studies (Dekker 2016), and they underscore the existence of DNA organization at the kb-Mb scales as an inherent, functionally important, component in gene activation. However, these atlases are limited in their ability to link single-cell, three-dimensional genome structures to specific transcriptional states. In addition, a characterization of how dynamic long-range interactions regulate single-cell transcriptional dynamics is currently missing, especially at the genomic scales, in the kilobase (kb) to megabase (Mb) range, where cis-regulatory elements (CREs) mediate functional interactions.
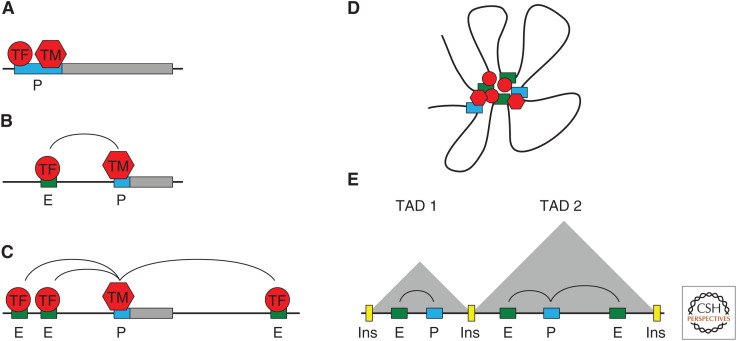
CREs are noncoding DNA regions that regulate transcription and include enhancers, chromatin insulators, silencers, and promoters (Wittkopp and Kalay 2012). Enhancers are short regulatory DNA sequences that control gene activity and contribute to the dynamic control of gene expression (de Laat and Duboule 2013; Bolt and Duboule 2020). The stochastic readout of regulators at promoter or enhancer sequences (Fig. 1A,B) gives rise to the time- and tissue-specific activation of subsets of genes to confer cell identity. Since their discovery four decades ago (Banerji et al. 1981, 1983; Moreau et al. 1981; Gillies et al. 1983; Mercola et al. 1983), enhancers have been largely regarded as autonomous, modular units, capable of activating transcription in a location- and orientation-independent manner. Enhancers are often at large distances, up to several Mb, from their respective target-gene promoters, most times with additional nontarget genes found within the intervening sequences (Furlong and Levine 2018; Schoenfelder and Fraser 2019). The canonical model is that enhancers regulate transcription by physically interacting with promoters over large genomic distances (Fig. 1C; Blackwood and Kadonaga 1998). Whole-genome methods have shown that the human genome is riddled with enhancers, with estimates ranging from hundreds of thousands to more than a million (Pennacchio et al. 2013; Schoenfelder and Fraser 2019; Xu et al. 2020). On average, a typical human gene is regulated by at least 10–20 different enhancers (Sanyal et al. 2012), raising the possibility that multiple enhancers may physically contact the same promoter (Fig. 1D,E).
Figure 1.
Increasing complexity in transcriptional control. (A) Transcription factors (TFs, red circle) and components of the transcription machinery (TM, red hexagon), such as RNA polymerase II and transcriptional activators like the Mediator complex, assemble at the promoter (P, blue), upstream of the gene body (gray). (B) TFs bind to enhancer elements (E, green), giving rise to temporal and tissue-specific gene regulation by interaction with a disjoint promoter element. (C) Multiple enhancers modulate the transcriptional activity of a promoter, often over large genomic distances. (D) CREs organized in nuclear space forming a three-dimensional gene locus, interacting with various factors (red). (E) Topologically associating domains (TADs) are thought to encapsulate such three-dimensional structures, forming a network of enhancer–promoter interactions. TADs are often demarcated by insulators (Ins, yellow).
Remarkably, this network of interactions may be modulated by other CREs, such as chromatin insulators. These are short, cis-regulatory sequences that block communication between promoters and enhancers (Reitman et al. 1990; Geyer and Corces 1992; Cai and Levine 1995). Genome-wide, thousands of sites are characterized as insulators, particularly enriched in intergenic and promoter regions. Insulators play a role in the formation of long-range interactions (Vogelmann et al. 2011, 2014; Yang and Corces 2012) and are involved in the global regulation of transcription (Bushey et al. 2009). Chromosome conformation capture (3C)-based assays (Lieberman-Aiden et al. 2009), which capture chromatin–chromatin interactions, revealed the existence of well-defined kb-Mb genomic regions displaying locally enhanced chromatin interactions (Dixon et al. 2012; Nora et al. 2012; Sexton et al. 2012). These genomic regions, called topologically associating domains (TADs), tend to be demarcated by insulators (Hou et al. 2012; Sexton et al. 2012; Rao et al. 2015) and often encapsulate enhancers and their target genes (Fig. 1E; Shen et al. 2012; Dowen et al. 2014; Symmons et al. 2014; Ji et al. 2016; Neems et al. 2016; Ron et al. 2017). Presently, the role of TADs in gene activity is still an open question (for a review, see Schoenfelder and Fraser 2019; Cavalheiro et al. 2021).
Disruption of TAD architecture, either by local duplications, deletions, or inversions that fuse adjacent TADs or form new TADs, were reported to trigger developmental defects in mammals by causing improper enhancer–promoter interactions (Northcott et al. 2014; Lupiáñez et al. 2015; Franke et al. 2016). In contrast, global disruption of TADs by depletion of CTCF or cohesin, two factors involved in formation of TADs, caused only mild changes in gene expression (Nora et al. 2017; Rao et al. 2017; Schwarzer et al. 2017), suggesting that TADs may be important for regulating only a subset of genes.
The reason for these apparent discrepancies may arise from the intrinsic dynamic properties of the chromatin fiber and transcription processes. Most studies assessing the transcriptional roles of TADs stem from a qualitative view and are based on ensemble, static measurements of TAD borders or contacts between CREs and promoters, combined with the assessment of bulk transcriptional outputs. This view entirely lacks the temporal and spatial aspects that are critical to understanding the genesis of these intrinsically dynamic processes, or to address key questions about the roles of TADs in transcriptional regulation: Are chromosome dynamics random and thus a key component to the stochasticity of gene expression? How can remote enhancers direct the correct spatial and temporal control of transcription? How do multiple enhancers interact dynamically to control promoter accessibility?
Renewed focus on spatial nuclear chromatin organization and its potential impact on gene regulation has recently brought a new twist to our understanding of eukaryotic transcription, broadening the potential angles evolution can use to interfere with the underlying mechanisms. Over the past 40 years, our view of transcription has gone from transcription factor (TFs) and machinery binding directly to promoters, to single or multiple enhancers mediating transcriptional control, to distal enhancer–promoter communication over megabase scales in cis and even in trans (Fig. 1). Now, a novel, spatial component comes into play, where loci are organized in three dimensions, and where organization can have functional consequences. In particular, recent imaging technologies have provided insightful tools to probe this organization and its dynamic impact on transcription. Here, we review how these new tools are starting to give new insights into otherwise inaccessible fundamental biological questions. Specifically, understanding how spatiotemporal changes in chromosome structure modulate genome function requires progress at four levels: (1) the relationship between genomic and physical scales, (2) the physical organization of regulatory elements, (3) the timescales involved in chromosome organization and transcription, and (4) the interplay between the spatiotemporal dynamics of the genome and its function. We discuss each in the following sections.
SPATIAL SCALES IN THE NUCLEUS
At the heart of the control of eukaryotic gene regulation is the organization and function of transcriptional enhancers, the major constituents of the noncoding genome that controls gene activity (Furlong and Levine 2018). A typical gene is regulated by multiple enhancers. Many of these enhancers possess overlapping regulatory activities, raising questions about proper cis-regulatory “trafficking,” whereby the correct enhancers interact with the appropriate target promoters (Bushey et al. 2008; Chopra et al. 2009). What is the influence of the exact position and affinity of DNA binding factors to CREs on transcriptional output? Answering these questions requires not only the relative genomic positioning of CREs in one dimension (Negre et al. 2011; ENCODE Project Consortium 2012), but also the direct detection of enhancer–promoter interactions in 3D to understand the impact of spatial chromatin architecture on function.
1D to 3D: Unintuitive Expectations
The physical properties of the chromatin fiber can be modeled using conventional polymer theory. In the simplest case, chromatin can be approximated by a polymer composed of a chain of identical monomers (de Gennes and Gennes 1979; Doi 1996; Rippe 2001; Nelson 2003; Wiggins et al. 2006; Doi 1996; Wiggins et al. 2006; Grosberg and Khokhlov 2011). The dynamics of the polymer chain are then governed by thermal fluctuations, excluded volume interactions, and rigidity parameters such as the effective persistence length. This simple model has been successfully used to predict the expected mean 3D distance between two DNA loci as a function of their genomic distance in sequence space (1D) (Rosa and Everaers 2008). Importantly, the conversion between 1D and 3D distances follows a power law (Mirny 2011) with a fractional exponent that varies between species, mainly due to changes in the physical properties of chromatin, such as the persistence length, the molecular composition of the chromatin fiber, genomic sizes, and nuclear volumes (Mirny 2011).
Importantly, the identified power–law relation between distances in sequence (1D) and in physical (3D) space is nonlinear. For instance, so-called distal enhancers in Drosophila are typically found at relatively short genomic distances (1–100 kb) (Ghavi-Helm et al. 2014) that can translate into relatively large average physical distances (200–400 nm) (Rosa and Everaers 2008; Cardozo Gizzi et al. 2019). Similarly, in mammals, enhancers found hundreds of kb from their target promoter are on average very far away (>1 μm [Rosa and Everaers 2008]). However, chromosomes are confined within the limited nuclear space, thus DNA loci located very far in genomic space (e.g., 2 Mb), or even on other chromosomes, are not proportionally separated in 3D space (e.g., 1 μm [Cattoni et al. 2017]).
The 1D sequence encodes information for proteins to bind to specific sites, which in turn can modulate the folding of chromatin in 3D. The binding of chromatin insulator proteins (e.g., CTCF), TFs, heterochromatin-associated complexes (e.g., Polycomb, HP1), or architectural proteins (e.g., cohesin, condensin) (Rowley and Corces 2016) promote 3D bridges between chromatin regions that can affect the frequencies of interactions expected for a homopolymer (Rowley and Corces 2016). For example, 3D loops between converging CTCF sites can produce specific 3D interactions between TAD borders (Rao et al. 2015) while cohesin-mediated loops can affect the contact frequencies within TADs (Nora et al. 2017; Rao et al. 2017). Alternatively, chromatin hubs linking multiple genomic loci can lead to the spatial clustering of regulatory elements (Beagrie et al. 2017; Allahyar et al. 2018; Oudelaar et al. 2018; Quinodoz et al. 2018; Espinola et al. 2021). Thus, both passive binding (e.g., CTCF, TFs) and active processes (e.g., cohesin/condensin looping, transcriptional elongation) can alter 3D chromatin organization at the kilobase to megabase scales, bringing into close spatial proximity loci that would be expected to reside far apart in a pure homopolymeric chromosome. Understanding these processes and their functional consequences holds potentially far-reaching insights into genome evolution, and requires methods able to dissect chromatin interactions in 3D.
3D Mapping: Sequencing and Imaging Approaches
Chromatin conformation can be measured by different types of methods. One class is sequencing-based, such as 3C and its derivatives, where chromatin contact frequencies measure the proximity between genomic loci averaged over a population of cells. The range over which genomic loci are cross-linked by ligation is much debated but computational models estimate the distance to be around 100 nm (Giorgetti et al. 2014).
An alternative class of approaches is based on microscopy techniques, especially fluorescence in situ hybridization (DNA-FISH), that directly detect pairwise distances in single cells. These are used to estimate imaging-derived contact frequencies by computing the proportion of cells displaying pairwise distances smaller than a critical radius RM (equivalent to integrating the spherical pairwise distance distribution up to RM) (Wang et al. 2016; Cattoni et al. 2017; Bintu et al. 2018; Cardozo Gizzi et al. 2019; Finn et al. 2019; Mateo et al. 2019).
It is worth noting that in neither case does the notion of “contact” imply physical interaction, but rather an estimate of whether two genomic loci are spatially close to each other. Thus, in the remainder of this review, we will refer to “proximity” rather than “contact frequency.” RM-values can be derived from control experiments where a single genomic locus is imaged in multiple colors/cycles (Cattoni et al. 2017; Cardozo Gizzi et al. 2019; Mateo et al. 2019) and RM-values between 150–500 nm have typically been used, producing good correlations between Hi-C and microscopy-based proximity frequencies (Wang et al. 2016; Bintu et al. 2018; Cardozo Gizzi et al. 2019; Mateo et al. 2019; Su et al. 2020). As such, the working definition of proximity is different for genomic-based and imaging-based methods, and its exact meaning will likely shift in future measurements with increasing genomic and optical resolutions. Determining the true measure for each method will likely involve a correlation analysis that reveals the scales at which proximity frequencies are correlated for both methods.
In the past, it has become common practice to assess the specificity of 3D interactions by using DNA-FISH to compare the mean 3D pairwise distance between two candidate genomic loci with that of a control that resides at the same genomic distance (Rao et al. 2015; Ogiyama et al. 2018; Benabdallah et al. 2019). However, relying purely on mean distances can be deceptive in cases where the full pairwise distance distribution departs from that of a single species or when specific 3D interactions are rare. For instance, while proximity frequency and mean spatial distance are inversely correlated for a homopolymer, this is not necessarily the case in the presence of sequence-dependent 3D interactions, where a 3D loop is rather expected to lead to a bimodal distance distribution (Giorgetti et al. 2014; Fudenberg and Imakaev 2017). In this case, the mean pairwise distance fails to capture sequence-specific looping interactions, particularly if these occur at low frequencies (e.g., <10%).
Even more counterintuitively, an increase in the frequency of sequence-specific interactions can lead to lower, equal, or even higher mean pairwise distances depending on the shape of the pairwise distance distribution (Giorgetti and Heard 2016; Fudenberg and Imakaev 2017). Thus, in a general case, a change in the mean pairwise distance between two genomic regions may not necessarily reflect changes in proximity. Instead, the proximity measured from the full pairwise distance distribution obtained from a DNA-FISH experiment should always be used to assess changes in 3D chromatin organization. These considerations are critical to derive models of enhancer function from DNA-FISH experiments.
Heterogeneity in chromosome organization is well documented by the observed width of pairwise distance distributions (Giorgetti et al. 2014; Cattoni et al. 2017; Bintu et al. 2018; Finn et al. 2019) and by large cell-to-cell variations in TAD volumes (Boettiger et al. 2016; Nir et al. 2018; Szabo et al. 2018; Luppino et al. 2020). These structural heterogeneities can originate from the intrinsic polymer dynamics of the chromatin fiber or from other extrinsic sources, such as active transcription acting on chromatin. Alternatively, DNA-binding proteins such as insulators (e.g., CTCF) (Hansen et al. 2017) or TFs (Izeddin et al. 2014; Normanno et al. 2015) can rapidly bind to and dissociate from DNA (seconds to minutes timescales) to shape the dynamics of local and global chromatin conformation. A final possibility is that the structure of the chromatin fiber is actually distinct in individual cells (Finn and Misteli 2019).
Bridging the Genomic and Molecular Scales
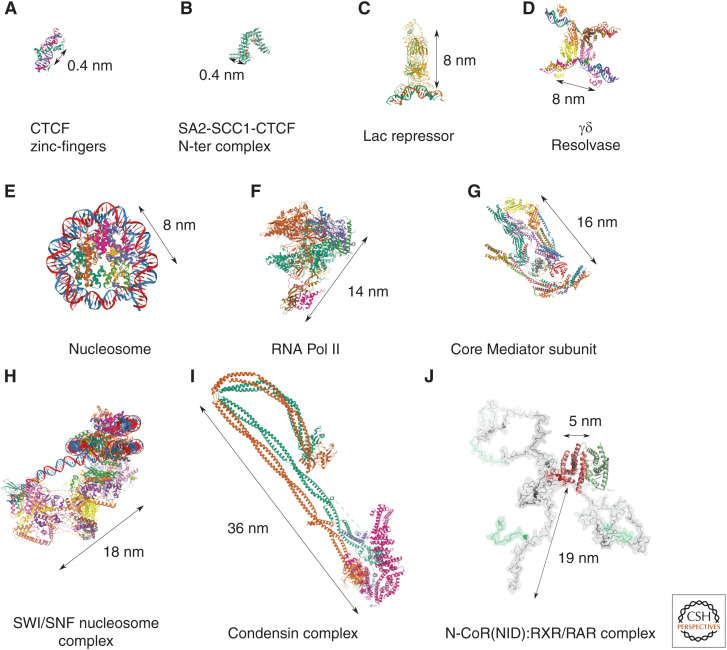
Genomic and imaging methods can map chromatin proximities in the 100–500 nm range; however, the factors mediating these interactions are typically much smaller. DNA-binding domains have typical sizes in the sub-nm range (e.g., CTCF zinc fingers 4–8) (Fig. 2A; Yin et al. 2017), similarly to protein–protein interaction domains (e.g., SA2-SCC1 cohesin subunit bound to CTCF amino-terminal domain) (Fig. 2B; Li et al. 2020b). DNA-bound TF sizes are commonly in the nanometer range (e.g., lactose operon repressor bound to operator DNA) (Fig. 2C; Lewis et al. 1996), while distances between DNA segments bridged by single-protein complexes can be ∼10 nm apart (e.g., γδ resolvase bound to site I DNA) (Fig. 2D; Li et al. 2005). Similarly, nucleosomes are ∼10 nm in size (Fig. 2E; Tachiwana et al. 2010). Multi-subunit machines, such as the Mediator complex, RNA polymerase II, or the SWI/SNF chromatin remodeling complex can reach sizes between 14 and 22 nm (Fig. 2F–H; Armache et al. 2005; Robinson et al. 2015; Nozawa et al. 2017; Wagner et al. 2020), while condensins can be up to 40 nm long (e.g., condensin complex from Saccharomyces cerevisiae) (Fig. 2I; Lee et al. 2020).
Figure 2.
Atomic-resolution structures and physical sizes of factors involved in chromosome organization and function. (A) CTCF zinc-fingers 4–8 bound to DNA (5YEG) (Yin et al. 2017), (B) SA2-SCC1 subunit of cohesin (green) bound to CTCF amino-terminal fragment (orange) (6QNX) (Li et al. 2020b), (C) Lac repressor bound to DNA (1LBG) (Lewis et al. 1996), (D) γδ resolvase in complex with site I DNA (1ZR4) (Li et al. 2005), (E) human nucleosome complex (3AFA) (Tachiwana et al. 2010), (F) complete 12-subunit RNA Pol II (5FJ8) (Armache et al. 2005), (G) crystal structure of the 15-subunit core Mediator complex from Schizosaccharomyces pombe (5N9J) (Nozawa et al. 2017), (H) SWI/SNF in complex with nucleosome (6TDA) (Wagner et al. 2020), (I) condensin complex from Saccharomyces cerevisiae (6YVU) (Lee et al. 2020), and (J) folded domains and three putative models of the asymmetric forms of the N-CoRNID:RXR/RAR complex are displayed (Cordeiro et al. 2019).
It is generally accepted that enhancer function requires looping to the promoter (for reviews, see Bulger and Groudine 2011; Schwarzer and Spitz 2014). Indeed, imaging-based studies recently reported that transcriptional activation can occur when enhancers get closer than 200–350 nm to promoters (Chen et al. 2018; Li et al. 2020a). Similarly, 3C data combined with modeling described the formation of 100–200 nm active “cages” (Di Stefano et al. 2020). Thus, there is a notable gap between genomic scales from in vivo enhancer–promoter distance measurements (200–350 nm) and molecular scales from atomic-resolution molecular models (1–50 nm).
Part of the explanation for this discrepancy may be the formation of higher-order molecular structures mediated by unstructured protein regions. In fact, multiple factors involved in transcriptional regulation and chromosome organization possess low-complexity/intrinsically disordered regions (IDRs) that may be able to bridge relatively large physical distances (Watson and Stott 2019). For instance, histone tails constitute sites of extensive posttranslational modifications that encode epigenetic information, and TFs have been long predicted to encode extensive IDRs (Liu et al. 2006). IDRs can increase the effective volumes occupied by protein complexes, as for the N-CoRNID:RXR/RAR nuclear receptor complex (Fig. 2J; Cordeiro et al. 2019). In addition, IDRs can fold by interacting with other factors, such as a region of the intrinsically disordered amino-terminal domain of CTCF when in interaction with the SA2-SCC1 subunit of cohesin (Li et al. 2020b). Notably, these interactions are necessary for loop formation (Li et al. 2020b; Pugacheva et al. 2020). Finally, IDRs can play many functions (Watson and Stott 2019), including formation of phase-separated compartments (Boija et al. 2018; Chong et al. 2018; Sabari et al. 2018). These alternative models will be explored in the following section.
SPATIAL CLUSTERING OF REGULATORY COMPONENTS
Gene expression is a highly regulated process that relies on a multitude of interactions between cis- and trans-acting factors. Whereas CREs are cis-regulatory genetic elements (such as enhancers, promoters, insulators, and silencers), trans-factors are molecular components, such as proteins, protein complexes, and noncoding RNAs that are involved in gene regulation in the form of TFs, coactivators (e.g., the Mediator complex), RNA Pol II, lncRNA, etc. (Schoenfelder and Fraser 2019). At present, very limited knowledge exists about the relative spatial organization and interactions of CREs and trans-factors in the nucleus. Which of these components, and how many, come together in the nuclear space? Do they display distinct spatial organizations? How are interactions between these elements mediated?
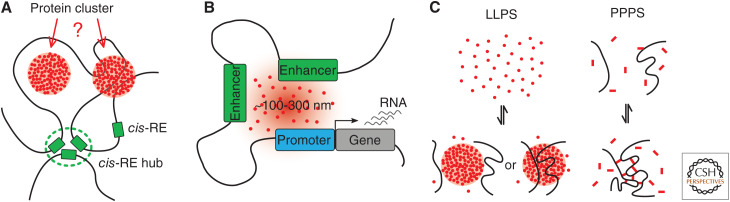
Recent findings revealed that both cis- and trans-regulatory components can come into close spatial proximity, and even cluster in spatially localized regions. No common terminology for the observed phenomena has been found as of now (Peng and Weber 2019). Thus, we made the choice to refer to clusters of chromatin regions containing CREs as CRE hubs, and nuclear aggregates of proteins or protein complexes as foci, without implying an underlying physical mechanism for either of them (Fig. 3A). The following sections will summarize evidence for the formation of CRE hubs, transcription-associated foci, and the physical mechanisms that may describe their formation.
Figure 3.
CRE (cis-regulatory element) hubs, protein foci, and phase-separated condensates. (A) Nuclear protein foci may or may not be DNA-associated/bound. The relationship between protein foci and CRE hubs is so far unknown. (B) Model for transcription-associated, foci-mediating enhancer–promoter interactions. (C) Two mechanisms of condensate formation via phase separation. Liquid–liquid phase separation (LLPS) is driven by weak, multivalent interactions between the constituents of the condensate (red dots) and does not require a scaffolding polymer. Conversely, polymer–polymer phase separation (PPPS) results from cross-linking the chromatin scaffold by a bridging factor (red rectangles).
Hubs of Cis-Regulatory Elements
In a canonical model, direct physical interaction between an enhancer and a promoter is required for enhancer action. The typical eukaryotic gene is regulated by multiple enhancers, particularly during development (Osterwalder et al. 2018; Fulco et al. 2019; Oudelaar and Higgs 2021), and many enhancers may be shared by multiple target genes (Ghavi-Helm et al. 2014), sometimes even simultaneously (Fukaya et al. 2016). These observations suggest that multiple enhancers and promoters come into close spatial proximity during transcription.
To directly probe the spatial clustering of multiple CREs, standard 3C-based techniques are not sufficient as they detect binary chromatin interactions. Consequently, sequencing and imaging-based technologies have been developed to detect multiway interactions. Among the former, “genome architecture mapping” (GAM) (Beagrie et al. 2017) and “split-pool recognition of interactions by tag extension” (SPRITE) (Quinodoz et al. 2018) were able to detect three-way interactions between super-enhancers in fixed mouse embryonic stem cells (mESCs). Alternatively, proximity-ligation methods were used to describe the existence of multiway enhancer interactions at the β- and α-globin locus during cell differentiation (Allahyar et al. 2018; Oudelaar et al. 2018). All in all, these results showed that multiple CREs can be captured in close proximity (Allahyar et al. 2018; Oudelaar et al. 2018). However, these methods are unable to detect multiple chromatin contacts and transcriptional status at once; thus, it was unclear whether clustering of CREs has a functional role.
Multiplexed imaging methods are ideally suited to tackle this question. Several complementary approaches rely on similar principles: “chromatin tracing” (Wang et al. 2016; Bintu et al. 2018), “Hi-M” (Cardozo Gizzi et al. 2019), “optical reconstruction of chromatin architecture” (ORCA) (Mateo et al. 2019), “chromosome walking” (Nir et al. 2018), as well as “seqFISH+” (Eng et al. 2019), and “MINA” (Liu et al. 2020). They combine microfluidics, wide-field microscopy, and Oligopaint FISH (Beliveau et al. 2012) to resolve the physical 3D position of tens to thousands of genomic loci in the nucleus, and can reach kilobase and nanometer precision.
These multiplexed imaging methods were recently used to simultaneously determine chromosome organization and transcription. ORCA revealed that the architecture of the Hox Polycomb TAD changes between different cell types in Drosophila embryos (Mateo et al. 2019). Hi-M was applied to detect CRE interactions and transcriptional status during early Drosophila development (Espinola et al. 2021) and revealed that CRE spatial clustering pre-dates gene activation and does not seem to depend on transcriptional state, with transcriptionally active and silent cells displaying similar CRE hubs (Espinola et al. 2021). These results are consistent with a concurrent Hi-C study demonstrating that chromatin conformation is independent of gene activity (Ing-Simmons et al. 2021), and with earlier reports showing that enhancer–promoter proximity can precede gene activation in mice or during Drosophila embryonic development (Montavon et al. 2011; Ghavi-Helm et al. 2014; Paliou et al. 2019). However, it is important to bear in mind that cell-specific changes in enhancer–promoter interaction networks during differentiation have also been well documented (Schoenfelder and Fraser 2019). Thus, we hypothesize that the roles of CRE hubs may be regulated either by altering their 3D structure and/or by modifying the cocktail of trans-acting factors they are bound by. These changes could be realized in different cell types by fine-tuning the abundance and binding of trans-acting factors.
Transcription-Associated Nuclear Foci
In eukaryotes, CREs are bound by TFs and coactivators (e.g., the histone acetyltransferase p300 or the Mediator complex). Interestingly, conventional and super-resolved imaging approaches revealed that many of these factors form nuclear foci (Cisse et al. 2013; Liu et al. 2014; Tsai et al. 2017; Boija et al. 2018; Cho et al. 2018; Chong et al. 2018; Dufourt et al. 2018; Mir et al. 2018; Sabari et al. 2018). This observation raised the possibility that trans-acting factors may form nuclear micro-environments where genes are coregulated. In some instances, transcription-associated nuclear foci have been reported to contain mRNAs (Boija et al. 2018; Sabari et al. 2018) and to associate with chromatin (Cho et al. 2018; Chong et al. 2018; Sabari et al. 2018), consistent with the observation of CRE hubs (Fig. 3A,B). However, it is not clear whether this is the norm or the exception.
The functional roles of transcription-associated nuclear foci in gene regulation are still unclear. For instance, are CREs always associated with nuclear foci? How many CREs take part in the formation of foci? Is a single CRE enough to form foci? Does the presence of multiple CREs increase transcriptional output? Answering these questions will require the ability to localize both trans-factors and multiple CREs simultaneously with high spatial resolution, and the use of perturbation methods (e.g., optogenetic manipulation of the low-complexity domains that mediate protein–protein interactions [Shin et al. 2017, 2018]). The next section describes the mechanisms that may be involved in the formation of transcription-associated nuclear foci and CRE hubs.
Phase Separation and Nuclear Structure
Phase separation has long been known as a process of self-organization in cells, and can explain the formation of membraneless compartments in the nucleus. Some of the most prominent examples include nuclear bodies (e.g., nucleoli, Cajal bodies, and DNA damage repair sites [Hyman et al. 2014; Banani et al. 2017; Shin and Brangwynne 2017; Boeynaems et al. 2018]) as well as chromatin itself (Gibson et al. 2019). For a current overview on the roles of phase separation in nuclear organization, we refer to Rippe (2021) and to recent reviews (Mir et al. 2019; Feric and Misteli 2021). Phase separation offers unique opportunities for controlling biochemical micro-environments by locally increasing the concentration of the constituents (and consequently chemical reaction rates), while still permitting dynamic exchange of reactants and products. Different mechanisms, involving distinct types of molecular interactions, have been proposed to lead to phase separation in the nucleus (Banani et al. 2017; Kato and McKnight 2017; Erdel and Rippe 2018). In addition, mechanisms not involving phase separation may also play a role in the formation of nuclear foci (McSwiggen et al. 2019).
Lately, liquid–liquid phase separation (LLPS) has received much attention as a possible mechanism of nuclear organization. LLPS compartments typically have spherical shapes, can fuse together, can deform under shear flow, are in exchange with the surrounding medium, and show a dynamic internal organization (Hyman et al. 2014). The key molecular driving force for the formation of liquid-like condensates are weak, multivalent interactions, typically involving proteins with IDRs (Banani et al. 2017; Shin and Brangwynne 2017).
Spatial clustering of super-enhancers often relies on the formation of condensates displaying typical properties of LLPS (Hnisz et al. 2017; Boija et al. 2018; Cho et al. 2018; Chong et al. 2018; Sabari et al. 2018). Consistently, the proteins involved in the formation of these condensates (TFs and coactivators) often contained IDRs that facilitated their nucleation. In this scenario, activating/repressive signals from CREs within a condensate may be transmitted over relatively large distances to the transcription machinery without requiring direct physical interactions. So far, it is unclear how long CREs remain associated with protein foci or whether promoters only need brief and transient encounters with these condensates for transcriptional activation (Cho et al. 2018). A more thorough discussion of the roles of LLPS in transcription can be found elsewhere (McSwiggen et al. 2019; Mir et al. 2019; Feric and Misteli 2021).
A second mechanism that can drive the formation of transcription-associated foci and CRE hubs is polymer–polymer phase separation (PPPS) (Erdel and Rippe 2018). This mechanism is based on the binding of soluble “bridging factors” to a long polymer chain, connecting two or more sites on the polymer. If the density of cross-links becomes sufficiently high, a polymer collapse takes place, resulting in a locally compact polymer globule. In this model, transient binding of cross-linking factors and their constant exchange with the surrounding nucleoplasm still leads to stable condensates, as long as a steady state with a sufficiently high density of bridging interactions is maintained (Erdel and Rippe 2018). Recently proposed models of phase separation for transcriptional control (Hnisz et al. 2017) are compatible with PPPS as they do not set any requirements on the chemical nature of the cross-links between chromatin chains. Counterintuitively, factors able to assemble liquid droplets in vitro may still form PPPS in vivo (Erdel et al. 2020). Despite their association to DNA, PPPS condensates are highly dynamic and, in some cases, can require the energy of ATP hydrolysis to ensure proper subnuclear positioning (Guilhas et al. 2020).
Perhaps the most important difference between LLPS and PPPS is that condensates formed via LLPS can persist independently of the chromatin scaffold, while condensates formed via PPPS strictly rely on such a scaffold and would disassemble in its absence (Fig. 3C). Future studies should experimentally test LLPS and PPPS models by examining concentration dependency and the need for a chromatin scaffold (Erdel and Rippe 2018; Erdel et al. 2020; Guilhas et al. 2020). These experiments would be particularly timely to further clarify whether the formation of CRE hubs requires transcription-associated foci and vice versa.
TIMESCALES OF NUCLEAR DYNAMICS
With the advent of high-resolution live imaging technologies, measurements of dynamic nuclear phenomena involving chromatin and binding factors have become a reality. These involve, on the one hand, measurements of dynamic parameters such as transition rates, binding/unbinding of factors, and more generally of kinetic rate constants that provide kinetic but not spatial information (Senecal et al. 2014; Zoller et al. 2018). On the other hand, it is now possible to go a step further to get real-time dynamic measurements with spatial information that, for example, follow the movement of chromatin or molecules in space and time (McCord et al. 2020; Shaban et al. 2020; Tortora et al. 2020). Here, we distinguish between cis-dynamics, that happen on the same chromosome, such as enhancer–promoter interactions or chromatin loop formation, and trans dynamics that involve interactions of trans-acting factors with chromatin, the formation of higher-order structures such as CRE hubs, or transvection.
Several studies, using both high-resolution and live-cell imaging but also single-cell 3C-based methods, have revealed a highly heterogeneous nature of genome organization in both space and time (Nagano et al. 2013; Cattoni et al. 2017; Flyamer et al. 2017; Stevens et al. 2017; Bintu et al. 2018; Hansen et al. 2018; Finn et al. 2019). However, few studies have in fact succeeded in quantifying actual chromosome dynamics in single cells, leaving a multitude of unanswered questions that are crucial for understanding genome organization and function. For example, the timescales over which the structure of chromosomes change remain largely unknown. How are single-cell chromosome topologies established during the cell cycle and during development? How does loop interaction frequency change across different timescales? How does the dynamic organization of the genome relate to gene expression and other nuclear processes, such as cell-type specification, DNA repair, or replication?
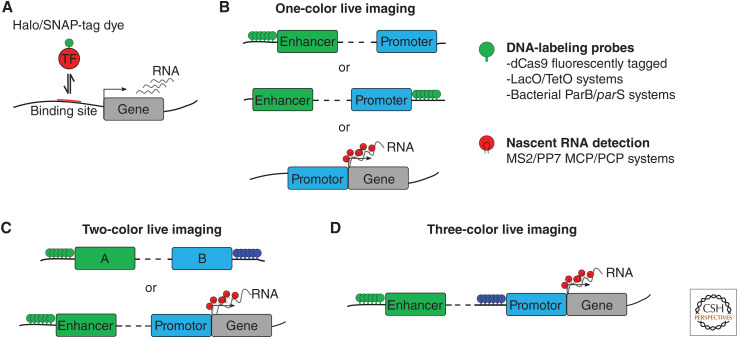
To directly address these and other questions, a number of imaging and labeling techniques have been developed (Fig. 4). Following chromatin dynamics in space and time not only requires development of state-of-the-art imaging technologies that often go beyond the diffraction limit (Lakadamyali and Cosma 2020; Brandão et al. 2021), but also development of highly sophisticated chromatin labeling capabilities that typically require a strenuous and time-consuming combination of molecular cloning, genome editing, and genetics (Sato et al. 2020; Shaban and Seeber 2020).
Figure 4.
Imaging genome and transcriptional dynamics in living cells. (A) Transcription factor dynamics monitored via two labeling strategies: fluorescent proteins or Halo/SNAP-tags coupled with dyes. (B) Strategies for fluorescent imaging of DNA and RNA in living cells. Several labeling approaches have facilitated imaging of chromosome dynamics in a sequence-specific manner: fluorescently tagged catalytically inactive cas9 enzymes (dCas9), fluorescently labeled operator binding protein (TetR, LacI), and the bacterial multimerizing ParB/parS system. (C) Two-color live imaging is used to simultaneously monitor the dynamics of two chromosomal regions or enhancer–promoter dynamics coupled with nascent transcription by using a combination of approaches shown in B. (D) Three-color live imaging allows probing for functional proximity: two colors for tagging enhancer–promoter pairs for example and one color for active transcription.
Several methods have been developed to visualize global chromatin dynamics at the nuclear scale (Shaban et al. 2020; Zidovska 2020). Imaging the dynamics of multiple genomic sites requires tools to fluorescently label specific sequences. Early methods for live imaging of specific genomic loci included the endogenous insertion of large binding site arrays for fluorescently tagged LacI or TetR repressors (Marshall et al. 1997; Heun et al. 2001; Chubb et al. 2002; Chuang et al. 2006; Kumaran and Spector 2008; Masui et al. 2011). Other approaches that require genome editing use the ParB/parS or ANCHOR DNA-labeling systems (Saad et al. 2014; Germier et al. 2017; Chen et al. 2018). In addition, catalytically inactive Cas9 enzymes (dCas9) tagged with green fluorescent protein (GFP) have been used to target specific genomic loci in living cells (Chen et al. 2013; Gu et al. 2018; Ma et al. 2018; Stanyte et al. 2018). These efforts led to the first direct measurements of CRE dynamics in living cells (Lucas et al. 2014; Germier et al. 2017; Herbert et al. 2017; Chen et al. 2018; Gu et al. 2018; Lim et al. 2018; Alexander et al. 2019; Khanna et al. 2019; Li et al. 2020a), assessments of large-scale chromatin dynamics (Zidovska et al. 2013; Nozaki et al. 2017; Shaban et al. 2018; Zidovska 2020), and quantification of DNA-binding factor interaction dynamics (Lionnet and Wu 2021).
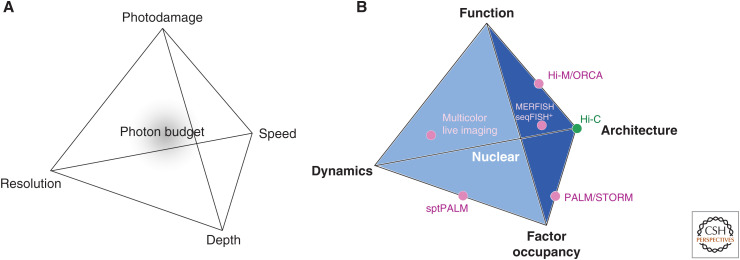
Many improvements in both microscopy and probe development are linked by their ability to control the photon budget (Planchon et al. 2011; Zhao et al. 2011; Lavis 2017): the number of detectable photons that a particular fluorophore contributes to the experiment, which is limited because of photochemistry and photobleaching (i.e., the permanent loss of fluorescence due to photo-induced chemical changes). However, further increases in localization precision, imaging rate, and imaging time, all require the detection of higher numbers of photons. This generates an optimization dilemma between resolution, speed, depth of view, and photodamage (Fig. 5A). Thus, the limiting factor is not the microscope; it is the photon budget. Close attention needs to be paid to the fluorophore choice because the photon budget is one of the most commonly neglected factors that significantly affects the feasibility and success of an experiment. A common strategy to sidestep this problem is to increase the number of fluorophores bound to individual molecules (Bertrand et al. 1998; Femino et al. 1998).
Figure 5.
Compromises in optimizing photon budget and measurements of nuclear properties. (A) The photon budget creates a tug-of-war between different desirable optimization strategies in photonic imaging. Whereas the ideal imaging experiment would push the limits of each property on the corners of the tetrahedron, the finite size of the photon budget sets a fundamental limit on the combined optimization of all four corners. Pushing one corner to its extreme limit implies giving up on the performance of the others. (B) The (epi)-genome contains all the information necessary to produce an entire organism. However, characterization of the functional output of the genome requires the ability to monitor multiple observables simultaneously. These include nuclear architecture, dynamics, occupancy by regulatory factors, and the biological output. Current technologies (showing here only a small number for simplicity) hardly accomplish a subset of these tasks in a single experiment.
Cis-Dynamics
The first pioneering work analyzing the dynamics of chromosomal loci was performed in lymphocyte B cells, where either VH or DHJH regions at the immunoglobulin gene loci were followed one-by-one by single-particle tracking (Lucas et al. 2014). These loci were shown to display a subdiffusive behavior with fractional Langevin motion and were mostly spatially confined. Similarly, in live mouse embryonic stem cells, the Fgf5 enhancer and promoter displayed subdiffusive behavior and their motility increased during differentiation to epiblast-like cells concomitant with transcriptional activation (Gu et al. 2018). The authors proposed that higher diffusivity of CREs increases stochastic encounters within TADs, potentially boosting successful enhancer–promoter interactions.
To partially avoid biases introduced by the inherent large-scale, three-dimensional motion of the entire nucleus, several efforts used two-color imaging to monitor the relative motion of two genomic loci on the same chromosome (Fig. 4C). This dual labeling strategy was used in lymphocyte B cells to characterize the relative movement of VH and DHJH regions (Khanna et al. 2019). The positions of VH and DHJH elements were found to undergo local, spatial fluctuations while their distance remains nearly constant over time, but abrupt changes in motion could be observed and suspected to originate from rapid temporal changes in large-scale chromatin conformations. These studies indicate that chromatin dynamics are largely subdiffusive in mammalian cells, with occasional abrupt changes in motion.
In mouse embryonic stem cells, live two-color imaging revealed that the Pou5f1 and Sox2 enhancers are frequently in proximity with their target gene transcription site (100–200 nm) (Li et al. 2020a). The authors argued that this 100–200-nm-sized cluster of enhancers could concentrate components of the transcription machinery and activate gene expression (Li et al. 2020a). Alexander et al. (2019) monitored the dynamics of the Sox2 gene in mouse embryonic stem cells and its essential endogenous SCR enhancer, positioned ∼100 kb away to show that the Sox2 enhancer–promoter spatial organization exhibited a high cell-to-cell variability. The two loci displayed sporadic sharp topological transitions within a tightly confined space in the ∼200 nm length scale. These observations suggest that enhancer–promoter pairs could be frequently and transiently interacting with each other, in contrast with the classical stable contact model (Deng et al. 2012, 2014; Bartman et al. 2016). However, it is unclear whether loop extrusion and chromatin conformation dynamics impact these transient enhancer–promoter interactions and what the consequences on transcription might be.
Trans-Dynamics
Another approach to decipher the time-dependent nature of the genome is to quantify the kinetics and dynamics of trans-acting factors, such as those required for loop extrusion, TAD formation, or stabilization of enhancer–promoter interactions. Their dynamics could potentially offer a route to study these processes and thus shed light on the underlying chromatin mechanics. For example, using single-molecule imaging approaches in live cells, the average residence time of cohesin (∼20 min) is about an order of magnitude longer than that of CTCF (∼1 min) (Hansen et al. 2017). These residence times are relatively stable compared to conventional TFs that can bind and dissociate from DNA on timescales of seconds (Mazza et al. 2012; Chen et al. 2014; Liu et al. 2014; Hansen et al. 2018) but highly dynamic compared to the length of the cell cycle (∼24 h), suggesting that chromatin loops dynamically form and break multiple times throughout the cell cycle (Hansen et al. 2018; Zhang et al. 2019). In addition, loop extrusion by the cohesin complex occurs at a maximum rate of ∼2 kb/sec in vitro (Davidson et al. 2019; Kim et al. 2019); thus, loops could likely play a role in the dynamics of enhancer–promoter interactions.
Several studies showed that components of the transcription machinery, such as RNA polymerase II, the Mediator complex, BRD4, and TFs, form transcription-associated nuclear phase-separated condensates (Cho et al. 2018; Chong et al. 2018; Sabari et al. 2018) (see also the section “Spatial Clustering of Regulatory Components” above). Using live-cell single-molecule imaging, Chong et al. showed that interactions between low-complexity regions of TFs are highly dynamic, typically on the second to minutes timescale. Consistently, Cho et al. used single-particle tracking to show that Mediator and RNA Pol II form large (>300 nm), long-lived (>100 sec), chromatin-associated clusters where they colocalize in a transcription-dependent manner. Mediator clusters display properties of phase-separated condensates and may mediate transient enhancer–promoter communication over large distances (a few hundred nanometers) (Fig. 3). Critically, in some instances, transcription-associated foci are highly dynamic and can form and dissociate in a matter of seconds (Liu et al. 2014; Cho et al. 2018; Dufourt et al. 2018). How these short-lived foci contribute to establishing long-range interactions between CREs remains an open question.
FUNCTIONAL CONSEQUENCES OF SPATIOTEMPORAL CHROMOSOME ORGANIZATION
After static and dynamic descriptions of nuclear organization, the next frontier is to provide a connection to biological function. What is the functional impact of nuclear order? Is there an evolved functional relationship between chromatin dynamics and transcription? To address these and similar questions, we need the ability to assess physical and genomic structure, chromatin dynamics and spatial distributions of regulatory factors, as well as biological function—ideally all simultaneously—which is a highly challenging task. Most likely, and similar to the dilemma of sharing a photon budget for imaging experiments, designing one experiment that can accomplish all these tasks simultaneously still lies somewhere in the future, and with current technologies one has to make choices about which of these features to optimize (Fig. 5B). Here we focus on initial progress to assess biological function in terms of measurements of transcription and its relationship to local genome organization.
Arguably one of the most functionally relevant outputs of the nucleus is its genetic program, which is executed via gene activity. Transcriptional dynamics can be monitored by in vivo visualization of nascent mRNA transcripts using bacteriophage-based reporter cassettes, as pioneered more than 20 years ago (Bertrand et al. 1998; Sato et al. 2020). MS2 and PP7 stem-loops are positioned in the gene body and detected with a fluorescently tagged coat protein to visualize nascent transcription in live cells (Janicki et al. 2004; Larson et al. 2011; Bothma et al. 2014). Measuring the signal intensity of the fluorescent signal at the site of gene activity was used to show that transcription is a stochastic process, subject to molecular noise, which exhibits transcriptional bursts with intervals of mRNA production followed by intervals of transcriptional inactivity (Rodriguez and Larson 2020). This stochasticity results in gene expression noise and cell-to-cell variability, the causes of which could possibly involve spatiotemporal fluctuations of the chromosome (Shah et al. 2018) or of CRE hubs (Shah et al. 2018).
A powerful application of this approach showed that enhancers control the frequency of transcriptional bursts, are able to coactivate linked genes, and yet can exhibit large spatial separation from their target genes even during transcriptional activation (Fukaya et al. 2016). These results contribute to the conflicting picture about the role of chromatin topology on genome function (Northcott et al. 2014; Lupiáñez et al. 2015; Franke et al. 2016; Nora et al. 2017; Rao et al. 2017; Schwarzer et al. 2017). While it is clear that enhancers and promoters can come into spatial proximity, and that physical proximity is somehow linked to transcriptional activation, such evidence is often correlative. Is enhancer–promoter proximity a consequence of transcriptional activation or is it needed for transcriptional activation? And if so, does transcription happen simultaneously with proximity or is it uncoupled? Even if CRE proximity is necessary for transcriptional activation, is it also necessary for sustained activity?
Answering these questions will require simultaneous detection of transcription and genome organization in the same cell. This is particularly important for enhancer–promoter interactions and the underlying mechanisms governing transcriptional control. Recent development of elaborate imaging-based methods—either via live-cell imaging or high-resolution fixed-tissue localization-based microscopy—have enabled the first direct visualization of long-range enhancer–promoter interactions coupled with transcriptional activity (Chen et al. 2018; Alexander et al. 2019; Mateo et al. 2019; Barinov et al. 2020; Li et al. 2020a; Espinola et al. 2021). Hi-M and ORCA have been used to visualize both chromatin structure and transcriptional activation in Drosophila to show that transcriptionally active and inactive cells display very similar enhancer–promoter proximity (Mateo et al. 2019; Espinola et al. 2021). Whereas these approaches enable the detection of transcription and the topology of multiple enhancers and promoters, they do not shed light on their dynamics.
To observe whether two distal chromosomal regions interact in a functionally significant manner, the notions of proximity or contact may no longer be sufficient. Rather, a more complex imaging assay is needed that involves simultaneous live image capture of three differently colored DNA tags: two to dynamically follow the motion of the distal chromosomal sites, such as an enhancer and a promoter, with the third tag serving as a reporter for functional proximity (i.e., it only lights up when specific events, such as transcription, occur). Here, the MS2/PP7-labeling systems are key in two ways: analysis of the fluorescence intensity signal for function and simultaneously tracking its location to gain insights into spatial relationships.
Using such an approach, progress toward a causal connection between dynamic enhancer–promoter communication and gene expression has recently been carried out. Live imaging experiments in Drosophila embryos visualized physical enhancer–promoter interactions and transcription at the eve locus in Drosophila embryos (Chen et al. 2018). Sustained physical proximity between the enhancer and the promoter of a distal reporter gene was shown to be necessary for transcriptional activity of the reporter gene. In addition, transcriptional activity also seems to stabilize this proximal conformation and have an impact on the physical size of the active gene locus, suggestive of a reciprocal interplay between enhancer–promoter dynamics and transcriptional activity (van Steensel and Furlong 2019).
In contrast, a similar study in mouse embryonic stem cells reported that enhancer–promoter proximity at the Sox2 locus is uncoupled from transcription (Alexander et al. 2019). In line with these results, 3D DNA-FISH and chromosome conformation capture in fixed cells revealed decreased spatial proximity between the Shh gene and its enhancers during the differentiation of mouse embryonic stem cells into neural progenitor cells (Benabdallah et al. 2019). It would be interesting to analyze enhancer–promoter interactions at the Shh locus using live imaging to determine the dynamics of these interactions at timescales shorter than days of cell differentiation, to see whether they are transient and unstable, as for the Sox2 locus. In many loci, several enhancers can activate transcription of a promoter, individually or conjointly. Thus, an observed lack of proximity may not be a good indicator for the need of enhancer–promoter proximity as some other enhancer in the neighborhood or within a CRE hub may be activating transcription.
It is still an open question what exactly “contact” or “proximity” mean in the context of an active enhancer–promoter pair, and how these notions relate to the emerging evidence of transcription-related CRE hubs and transcription-associated protein foci. Again, to discern between these models and to devise new ones, dynamic measurements would need to simultaneously track multiple CREs and factors in space and time. Testing current models of enhancer–promoter communication will also require imaging methods with high resolution to distinguish enhancer–promoter loops in physical interaction range (in and of itself an ill-defined length scale) from observed enhancer–promoter proximity (150–200 nm). Further progress toward this issue has been put forward using multicolor localization microscopy to achieve 1–2 kb resolution in an 18 kb gene locus in Drosophila, where the transcriptionally active enhancer–promoter pair was still more than 150 nm separated; thus, again not in physical interaction range (Barinov et al. 2020).
At the time of writing this review, there are still a number of unanswered questions, most prominently whether the observed spatial gaps between active enhancers and promoters prove to be a general feature of active transcription-associated loci. In that case, the most pressing question is to understand how information about activity spreads across large spatial distances (>100 nm) from the TF-bound active enhancer to the transcription-engaging promoter. It may take another generation of experiments and new technologies to make progress in the understanding of how the general architecture of the folded nuclear genome regulates genome function and vice versa. The observations above could suggest that gene regulation by stable enhancer–promoter looping is not a generality, and more elaborate models of long-range enhancer–promoter communication should take other spatial considerations or constraints into account.
CONCLUDING REMARKS
The last two decades have seen a revolution in optical imaging approaches with the advent of superresolution microscopy, live tracking of individual molecules, and multiplexed methods. Application of these approaches already made a strong impact on our understanding of the interplay between the structure and dynamics of the nucleus and its transcriptional output. However, understanding causality and gaining an integrated picture of how dynamic changes in chromosome organization control the timing and levels of transcription will require considerable further advances in several interdisciplinary areas. A crucial issue will be the necessity to measure in individual cells the multiple critical components of nuclear organization underlying gene activity: transcriptional output, spatial arrangement of CREs, dynamic long-range chromosomal interactions, and the epigenetic state of chromatin (Fig. 5B). Many of these quantities can already be measured in single cells, one-by-one or in pairs, but detecting them simultaneously presents a long-term challenge that will require significant advances in design and development of microscopy, as well as in labeling and genome-editing tools.
One area that is still largely underdeveloped is the single-cell study of the dynamics and genome occupancy of trans-regulatory factors and how they influence genome organization and function. For example, it will be crucial to monitor the dynamic binding of multiple TFs to CREs to understand how the latter turn into a state of activity that ultimately leads to a transcriptional output. Likewise, it will be critical to further integrate how signaling, environmental cues, or splicing shape the assembly and dynamics of CRE hubs and transcription-associated foci to regulate transcription.
There are still many open questions and new frontiers to explore. Progress in optical and electron microscopies have already achieved the spatial resolution necessary to start revealing the organization of chromatin in cells at the nanometer scale (Ricci et al. 2015; Jungmann et al. 2016; Ou et al. 2017). However, visualization of chromatin structure at that scale as inert/featureless beads may not be enough; to get a full understanding of the role of chromatin structure for biological function will require the detection of multiple types of molecules (e.g., proteins, RNA, DNA) with sequence specificity, nanometer precision, and dynamic probes. These developments have to go hand in hand with improvements in labeling and sample preparation procedures that conserve structures at the molecular scale. In addition, still largely lacking are perturbation techniques that can directly probe causal relationships between nuclear structure, the environment, and function. For example, optogenetic association or dissociation of molecular compounds or DNA loci should provide powerful handles for causality experiments. Excitingly, all these advances should bring a much needed facet to tracking single molecules interacting with DNA and asserting control over transcription programs, one factor at a time.
ACKNOWLEDGMENTS
We thank Pau Bernado for his critical reading of the manuscript and for help with Figure 2J. This work was supported in part by the European Union's Horizon 2020 Research and Innovation Program (Grant ID 724429) (M.N.), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, Project ID 431471305) (M.G.), the Laboratoire d'Excellence Revive (Investissement d'Avenir; ANR-10-LABX-73) (I.B.), the U.S. National Science Foundation, through the Center for the Physics of Biological Function (PHY–1734030), and by National Institutes of Health Grants R01GM097275, U01DA047730, and U01DK127429 (T.G.).
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, Huang B, Lomvardas S, Weiner OD. 2019. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 8: e41769. 10.7554/eLife.41769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahyar A, Vermeulen C, Bouwman BAM, Krijger PHL, Marjon JA, Geeven G, van Kranenburg M, Pieterse M, Straver R, Haarhuis JHI, et al. 2018. Enhancer hubs and loop collisions identified from single-allele topologies. Nat Genet 50: 1151–1160. 10.1038/s41588-018-0161-5 [DOI] [PubMed] [Google Scholar]
- Armache KJ, Mitterweger S, Meinhart A, Cramer P. 2005. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem 280: 7131–7134. 10.1074/jbc.M413038200 [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W. 1981. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27: 299–308. 10.1016/0092-8674(81)90413-X [DOI] [PubMed] [Google Scholar]
- Banerji J, Olson L, Schaffner W. 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33: 729–740. 10.1016/0092-8674(83)90015-6 [DOI] [PubMed] [Google Scholar]
- Barinov L, Ryabichko S, Bialek W, Gregor T. 2020. Transcription-dependent spatial organization of a gene locus. arXiv 2012.15819 [Google Scholar]
- Bartman CR, Hsu SC, Hsiung CCS, Raj A, Blobel GA. 2016. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol Cell 62: 237–247. 10.1016/j.molcel.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DCA, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543: 519–524. 10.1038/nature21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BR, et al. 2012. Versatile design and synthesis platform for visualizing genomes with oligopaint FISH probes. Proc Natl Acad Sci 109: 21301–21306. 10.1073/pnas.1213818110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, Bickmore WA. 2019. Decreased enhancer–promoter proximity accompanying enhancer activation. Mol Cell 76: 473–484.e7. 10.1016/j.molcel.2019.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. 1998. Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445. 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su JH, Sinnott-Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, Zhuang X. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362: eaau1783. 10.1126/science.aau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. 1998. Going the distance: a current view of enhancer action. Science 281: 60–63. 10.1126/science.281.5373.60 [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu CT, Zhuang X. 2016. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529: 418–422. 10.1038/nature16496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, et al. 2018. Protein phase separation: a new phase in cell biology. Trends Cell Biol 28: 420–435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. 2018. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175: 1842–1855.e16. 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt CC, Duboule D. 2020. The regulatory landscapes of developmental genes. Development 147: dev171736. 10.1242/dev.171736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M. 2014. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc Natl Acad Sci 111: 10598–10603. 10.1073/pnas.1410022111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandão HB, Gabriele M, Hansen AS. 2021. Tracking and interpreting long-range chromatin interactions with super-resolution live-cell imaging. Curr Opin Cell Biol 70: 18–26. 10.1016/j.ceb.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. 10.1016/j.cell.2011.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell 32: 1–9. 10.1016/j.molcel.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG. 2009. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev 23: 1338–1350. 10.1101/gad.1798209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Levine M. 1995. Modulation of enhancer–promoter interactions by insulators in the Drosophila embryo. Nature 376: 533–536. 10.1038/376533a0 [DOI] [PubMed] [Google Scholar]
- Cardozo Gizzi AM, Cattoni DI, Fiche JB, Espinola SM, Gurgo J, Messina O, Houbron C, Ogiyama Y, Papadopoulos GL, Cavalli G, et al. 2019. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol Cell 74: 212–222.e5. 10.1016/j.molcel.2019.01.011 [DOI] [PubMed] [Google Scholar]
- Cattoni DI, Cardozo Gizzi AM, Georgieva M, Di Stefano M, Valeri A, Chamousset D, Houbron C, Déjardin S, Fiche JB, González I, et al. 2017. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat Commun 8: 1753. 10.1038/s41467-017-01962-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalheiro GR, Pollex T, Furlong EE. 2021. To loop or not to loop: what is the role of TADs in enhancer function and gene regulation? Curr Opin Genet Dev 67: 119–129. 10.1016/j.gde.2020.12.015 [DOI] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, et al. 2013. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. 2014. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156: 1274–1285. 10.1016/j.cell.2014.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T. 2018. Dynamic interplay between enhancer–promoter topology and gene activity. Nat Genet 50: 1296–1303. 10.1038/s41588-018-0175-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse II. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361: 412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, et al. 2018. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361: ear2555. 10.1126/science.aar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Cande J, Hong JW, Levine M. 2009. Stalled Hox promoters as chromosomal boundaries. Genes Dev 23: 1505–1509. 10.1101/gad.1807309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831. 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol 12: 439–445. 10.1016/S0960-9822(02)00695-4 [DOI] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341: 664–667. 10.1126/science.1239053 [DOI] [PubMed] [Google Scholar]
- Cordeiro TN, Sibille N, Germain P, Barthe P, Boulahtouf A, Allemand F, Bailly R, Vivat V, Ebel C, Barducci A, et al. 2019. Interplay of protein disorder in retinoic acid receptor heterodimer and its corepressor regulates gene expression. Structure 27: 1270–1285.e6. 10.1016/j.str.2019.05.001 [DOI] [PubMed] [Google Scholar]
- Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters JM. 2019. DNA loop extrusion by human cohesin. Science 366: 1338–1345. 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- de Gennes PG, Gennes PG. 1979. Scaling concepts in polymer physics. Cornell University Press, Ithaca, NY. [Google Scholar]
- Dekker J. 2016. Mapping the 3D genome: aiming for consilience. Nat Rev Mol Cell Biol 17: 741–742. 10.1038/nrm.2016.151 [DOI] [PubMed] [Google Scholar]
- de Laat W, Duboule D. 2013. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature 502: 499–506. 10.1038/nature12753 [DOI] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. 2012. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149: 1233–1244. 10.1016/j.cell.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. 2014. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell 158: 849–860. 10.1016/j.cell.2014.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano M, Stadhouders R, Farabella I, Castillo D, Serra F, Graf T, Marti-Renom MA. 2020. Transcriptional activation during cell reprogramming correlates with the formation of 3D open chromatin hubs. Nat Commun 11. 10.1038/s41467-020-16396-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M. 1996. Introduction to polymer physics. Oxford University Press, Oxford, UK. [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schujiers J, Lee TI, Zhao K, et al. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159: 374–387. 10.1016/j.cell.2014.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourt J, Trullo A, Hunter J, Fernandez C, Lazaro J, Dejean M, Morales L, Nait-Amer S, Schulz KN, Harrison MM, et al. 2018. Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nat Commun 9: 5194. 10.1038/s41467-018-07613-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CHL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, et al. 2019. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568: 235–239. 10.1038/s41586-019-1049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rippe K. 2018. Formation of chromatin subcompartments by phase separation. Biophys J 114: 2262–2270. 10.1016/j.bpj.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rademacher A, Vlijm R, Tünnermann J, Frank L, Weinmann R, Schweigert E, Yserentant K, Hummert J, Bauer C, et al. 2020. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid–liquid phase separation. Mol Cell 78: 236–249.e7. 10.1016/j.molcel.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinola SM, Götz M, Bellec M, Messina O, Fiche JB, Houbron C, Dejean M, Reim I, Cardozo Gizzi AM, Lagha M, et al. 2021. Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat Genet 53: 477–486. 10.1038/s41588-021-00816-z [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. 1998. Visualization of single RNA transcripts in situ. Science 280: 585–590. 10.1126/science.280.5363.585 [DOI] [PubMed] [Google Scholar]
- Feric M, Misteli T. 2021. Phase separation in genome organization across evolution. Trends Cell Biol 23: S0962-8924(21)00047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Misteli T. 2019. Molecular basis and biological function of variability in spatial genome organization. Science 365: eaaw9498. 10.1126/science.aaw9498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn EH, Pegoraro G, Brandão HB, Valton AL, Oomen ME, Dekker J, Mirny L, Misteli T. 2019. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176: 1502–1515.e10. 10.1016/j.cell.2019.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana-Konwalski K. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544: 110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, Kraft K, Kempfer R, Jerković I, Chan WL, et al. 2016. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265–269. 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M. 2017. FISH-ing for captured contacts: towards reconciling FISH and 3C. Nat Methods 14: 673–678. 10.1038/nmeth.4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. 2016. Enhancer control of transcriptional bursting. Cell 166: 358–368. 10.1016/j.cell.2016.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Nasser J, Jones TR, Munson G, Bergman DT, Subramanian V, Grossman SR, Anyoha R, Doughty BR, Patwardhan TA, et al. 2019. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat Genet 51: 1664–1669. 10.1038/s41588-019-0538-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong EEM, Levine M. 2018. Developmental enhancers and chromosome topology. Science 361: 1341–1345. 10.1126/science.aau0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germier T, Kocanova S, Walther N, Bancaud A, Shaban HA, Sellou H, Politi AZ, Ellenberg J, Gallardo F, Bystricky K. 2017. Real-time imaging of a single gene reveals transcription-initiated local confinement. Biophys J 113: 1383–1394. 10.1016/j.bpj.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873. 10.1101/gad.6.10.1865 [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EEM. 2014. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512: 96–100. 10.1038/nature13417 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK. 2019. Organization of chromatin by intrinsic and regulated phase separation. Cell 179: 470–484.e21. 10.1016/j.cell.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies SD, Morrison SL, Oi VT, Tonegawa S. 1983. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33: 717–728. 10.1016/0092-8674(83)90014-4 [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Heard E. 2016. Closing the loop: 3C versus DNA FISH. Genome Biol 17: 215. 10.1186/s13059-016-1081-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. 2014. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157: 950–963. 10.1016/j.cell.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg A, Khokhlov AR. 2011. Giant molecules: here, there, and everywhere. World Scientific, Singapore. [Google Scholar]
- Gu B, Swigut T, Spencley A, Bauer MR, Chung M, Meyer T, Wysocka J. 2018. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359: 1050–1055. 10.1126/science.aao3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhas B, Walter JC, Rech J, David G, Walliser NO, Palmeri J, Mathieu-Demaziere C, Parmeggiani A, Bouet JY, Le Gall A, et al. 2020. ATP-driven separation of liquid phase condensates in bacteria. Mol Cell 79: 293–303.e4. 10.1016/j.molcel.2020.06.034 [DOI] [PubMed] [Google Scholar]
- Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. 2017. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6: e25776. 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Cattoglio C, Darzacq X, Tjian R. 2018. Recent evidence that TADs and chromatin loops are dynamic structures. Nucleus 9: 20–32. 10.1080/19491034.2017.1389365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S, Brion A, Arbona JM, Lelek M, Veillet A, Lelandais B, Parmar J, Fernández FG, Almayrac E, Khalil Y, et al. 2017. Chromatin stiffening underlies enhanced locus mobility after DNA damage in budding yeast. EMBO J 36: 2595–2608. 10.15252/embj.201695842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. 2001. Chromosome dynamics in the yeast interphase nucleus. Science 294: 2181–2186. 10.1126/science.1065366 [DOI] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169: 13–23. 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin Z, Corces V. 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell 48: 471–484. 10.1016/j.molcel.2012.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. 2014. Liquid–liquid phase separation in biology. Annu Rev Cell Dev Biol 30: 39–58. 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- Ing-Simmons E, Vaid R, Bing XY, Levine M, Mannervik M, Vaquerizas JM. 2021. Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nat Genet 53: 487–499. 10.1038/s41588-021-00799-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, Récamier V, Bosanac L, Cissé II, Boudarene L, Dugast-Darzacq C, Proux F, Bénichou O, Voituriez R, Bensaude O, et al. 2014. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife 3: e02230. 10.7554/eLife.02230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116: 683–698. 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, et al. 2016. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell 18: 262–275. 10.1016/j.stem.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R, Avendaño MS, Dai M, Woehrstein JB, Agasti SS, Feiger Z, Rodal A, Yin P. 2016. Quantitative super-resolution imaging with qPAINT. Nat Methods 13: 439–442. 10.1038/nmeth.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, McKnight SL. 2017. Cross-β polymerization of low complexity sequence domains. Cold Spring Harb Perspect Biol 9: a023598. 10.1101/cshperspect.a023598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N, Zhang Y, Lucas JS, Dudko OK, Murre C. 2019. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat Commun 10: 1–13. 10.1038/s41467-019-10628-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. 2019. Human cohesin compacts DNA by loop extrusion. Science 366: 1345–1349. 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180: 51–65. 10.1083/jcb.200706060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Cosma MP. 2020. Visualizing the genome in high resolution challenges our textbook understanding. Nat Methods 17: 371–379. 10.1038/s41592-020-0758-3 [DOI] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. 2011. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332: 475–478. 10.1126/science.1202142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis LD. 2017. Chemistry is dead. Long live chemistry! Biochemistry 56: 5165–5170. [DOI] [PubMed] [Google Scholar]
- Lee BG, Merkel F, Allegretti M, Hassler M, Cawood C, Lecomte L, O'Reilly FJ, Sinn LR, Gutierrez-Escribano P, Kschonsak M, et al. 2020. Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Nat Struct Mol Biol 27: 743–751. 10.1038/s41594-020-0457-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. 1996. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271: 1247–1254. 10.1126/science.271.5253.1247 [DOI] [PubMed] [Google Scholar]
- Li W, Kamtekar S, Xiong Y, Sarkis GJ, Grindley NDF, Steitz TA. 2005. Structure of a synaptic γδ resolvase tetramer covalently linked to two cleaved DNAs. Science 309: 1210–1215. 10.1126/science.1112064 [DOI] [PubMed] [Google Scholar]
- Li J, Hsu A, Hua Y, Wang G, Cheng L, Ochiai H, Yamamoto T, Pertsinidis A. 2020a. Single-gene imaging links genome topology, promoter–enhancer communication and transcription control. Nat Struct Mol Biol 27: 1032–1040. 10.1038/s41594-020-0493-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Haarhuis JHI, Sedeño Cacciatore Á, Oldenkamp R, van Ruiten MS, Willems L, Teunissen H, Muir KW, de Wit E, Rowland BD, et al. 2020b. The structural basis for cohesin-CTCF-anchored loops. Nature 578: 472–476. 10.1038/s41586-019-1910-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Heist T, Levine M, Fukaya T. 2018. Visualization of transvection in living Drosophila embryos. Mol Cell 70: 287–296.e6. 10.1016/j.molcel.2018.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Wu C. 2021. Single-molecule tracking of transcription protein dynamics in living cells: seeing is believing, but what are we seeing? Curr Opin Genet Dev 67: 94–102. 10.1016/j.gde.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. 2006. Intrinsic disorder in transcription factors. Biochemistry 45: 6873–6888. 10.1021/bi0602718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. 2014. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife 3: e04263. 10.7554/eLife.04236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Lu Y, Yang B, Chen Y, Radda JSD, Hu M, Katz SG, Wang S. 2020. Multiplexed imaging of nucleome architectures in single cells of mammalian tissue. Nat Commun 11: 2907. 10.1038/s41467-020-16732-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JS, Zhang Y, Dudko OK, Murre C. 2014. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell 158: 339–352. 10.1016/j.cell.2014.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppino JM, Park DS, Nguyen SC, Lan Y, Xu Z, Yunker R, Joyce EF. 2020. Cohesin promotes stochastic domain intermingling to ensure proper regulation of boundary-proximal genes. Nat Genet 52: 840–848. 10.1038/s41588-020-0647-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Tu LC, Naseri A, Chung YC, Grunwald D, Zhang S, Pederson T. 2018. CRISPR-Sirius: RNA scaffolds for signal amplification in genome imaging. Nat Methods 15: 928–931. 10.1038/s41592-018-0174-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 7: 930–939. 10.1016/S0960-9822(06)00412-X [DOI] [PubMed] [Google Scholar]
- Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, Hupé P, Barillot E, Belmont AS, Heard E. 2011. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell 145: 447–458. 10.1016/j.cell.2011.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568: 49–54. 10.1038/s41586-019-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza D, Abernathy A, Golob N, Morisaki T, McNally JG. 2012. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res 40: e119–e119. 10.1093/nar/gks701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RP, Kaplan N, Giorgetti L. 2020. Chromosome conformation capture and beyond: toward an integrative view of chromosome structure and function. Mol Cell 77: 688–708. 10.1016/j.molcel.2019.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, Tjian R. 2019. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 33: 1619–1634. 10.1101/gad.331520.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercola M, Wang XF, Olsen J, Calame K. 1983. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science 221: 663–665. 10.1126/science.6306772 [DOI] [PubMed] [Google Scholar]
- Mir M, Stadler MR, Ortiz SA, Hannon CE, Harrison MM, Darzacq X, Eisen MB. 2018. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 7: e40497. 10.7554/eLife.40497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M, Bickmore W, Furlong EEM, Narlikar G. 2019. Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase? Development 146: dev182766. 10.1242/dev.182766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA. 2011. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res 19: 37–51. 10.1007/s10577-010-9177-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147: 1132–1145. 10.1016/j.cell.2011.10.023 [DOI] [PubMed] [Google Scholar]
- Moreau P, Hen R, Wasylyk B, Everett R, Gaub MP, Chambon P. 1981. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res 9: 6047–6068. 10.1093/nar/9.22.6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neems DS, Garza-Gongora AG, Smith ED, Kosak ST. 2016. Topologically associated domains enriched for lineage-specific genes reveal expression-dependent nuclear topologies during myogenesis. Proc Natl Acad Sci 113: E1691–E1700. 10.1073/pnas.1521826113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531. 10.1038/nature09990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. 2003. Biological physics: energy, information, life. W. H. Freeman, New York. [Google Scholar]
- Nir G, Farabella I, Pérez Estrada C, Ebeling CG, Beliveau BJ, Sasaki HM, Lee SD, Nguyen SC, McCole RB, Chattoraj S, et al. 2018. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet 14: e1007872. 10.1371/journal.pgen.1007872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno D, Boudarène L, Dugast-Darzacq C, Chen J, Richter C, Proux F, Bénichou O, Voituriez R, Darzacq X, Dahan M. 2015. Probing the target search of DNA-binding proteins in mammalian cells using tetR as model searcher. Nat Commun 6: 7357. 10.1038/ncomms8357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, Shih DJH, Hovestadt V, Zapatka M, Sturm D, et al. 2014. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511: 428–434. 10.1038/nature13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, Tani T, Joti Y, Tomita M, Hibino K, Kanemaki MT, et al. 2017. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol Cell 67: 282–293.e7. 10.1016/j.molcel.2017.06.018 [DOI] [PubMed] [Google Scholar]
- Nozawa K, Schneider TR, Cramer P. 2017. Core Mediator structure at 3.4 Å extends model of transcription initiation complex. Nature 545: 248–251. 10.1038/nature22328 [DOI] [PubMed] [Google Scholar]
- Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang JM, Cavalli G. 2018. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol Cell 71: 73–88.e5. 10.1016/j.molcel.2018.05.032 [DOI] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, et al. 2018. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554: 239–243. 10.1038/nature25461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O'Shea CC. 2017. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357: eaag0025. 10.1126/science.aag0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudelaar MA, Higgs DR. 2021. The relationship between genome structure and function. Nat Rev Genet 22: 154–168. 10.1038/s41576-020-00303-x [DOI] [PubMed] [Google Scholar]
- Oudelaar MA, Davies JOJ, Hanssen LLP, Telenius JM, Schwessinger R, Liu Y, Brown JM, Downes DJ, Chiariello AM, Bianco S, et al. 2018. Single-allele chromatin interactions identify regulatory hubs in dynamic compartmentalized domains. Nat Genet 50: 1744–1751. 10.1038/s41588-018-0253-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliou C, Guckelberger P, Schöpflin R, Heinrich V, Esposito A, Chiariello AM, Bianco S, Annunziatella C, Helmuth J, Haas S, et al. 2019. Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc Natl Acad Sci 116: 12390–12399. 10.1073/pnas.1900672116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- A Peng, Weber SC. 2019. Evidence for and against liquid–liquid phase separation in the nucleus. Noncoding RNA 5: 50. 10.3390/ncrna5040050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. 2013. Enhancers: five essential questions. Nat Rev Genet 14: 288–295. 10.1038/nrg3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. 2011. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods 8: 417–423. 10.1038/nmeth.1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva EM, Kubo N, Loukinov D, Tajmul M, Kang S, Kovalchuk AL, Strunnikov AV, Zentner GE, Ren B, Lobanenkov VV. 2020. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc Natl Acad Sci 117: 2020–2031. 10.1073/pnas.1911708117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174: 744–757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2015. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 162: 687–688. 10.1016/j.cell.2015.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M, Lee E, Westphal H, Felsenfeld G. 1990. Site-independent expression of the chicken βA-globin gene in transgenic mice. Nature 348: 749–752. 10.1038/348749a0 [DOI] [PubMed] [Google Scholar]
- Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, Cosma MP. 2015. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160: 1145–1158. 10.1016/j.cell.2015.01.054 [DOI] [PubMed] [Google Scholar]
- Rippe K. 2001. Making contacts on a nucleic acid polymer. Trends Biochem Sci 26: 733–740. 10.1016/S0968-0004(01)01978-8 [DOI] [PubMed] [Google Scholar]
- Rippe K. 2021. Liquid–liquid phase separation in chromatin. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, Burlingame AL, Sali A, Kornberg RD. 2015. Molecular architecture of the yeast Mediator complex. eLife 4: e08719. 10.7554/eLife.08719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Larson DR. 2020. Transcription in living cells: molecular mechanisms of bursting. Annu Rev Biochem 89: 189–212. 10.1146/annurev-biochem-011520-105250 [DOI] [PubMed] [Google Scholar]
- Ron G, Globerson Y, Moran D, Kaplan T. 2017. Promoter–enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat Commun 8: 2237. 10.1038/s41467-017-02386-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Everaers R. 2008. Structure and dynamics of interphase chromosomes. PLoS Comput Biol 4: e1000153. 10.1371/journal.pcbi.1000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Corces VG. 2016. The three-dimensional genome: principles and roles of long-distance interactions. Curr Opin Cell Biol 40: 8–14. 10.1016/j.ceb.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad H, Gallardo F, Dalvai M, Tanguy-le-Gac N, Lane D, Bystricky K. 2014. DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet 10: e1004187. 10.1371/journal.pgen.1004187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361: eaar3958. 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113. 10.1038/nature11279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Das S, Singer RH, Vera M. 2020. Imaging of DNA and RNA in living eukaryotic cells to reveal spatiotemporal dynamics of gene expression. Annu Rev Biochem 89: 159–187. 10.1146/annurev-biochem-011520-104955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Fraser P. 2019. Long-range enhancer–promoter contacts in gene expression control. Nat Rev Genet 20: 437–455. 10.1038/s41576-019-0128-0 [DOI] [PubMed] [Google Scholar]
- Schwarzer W, Spitz F. 2014. The architecture of gene expression: integrating dispersed cis-regulatory modules into coherent regulatory domains. Curr Opin Genet Dev 27: 74–82. 10.1016/j.gde.2014.03.014 [DOI] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecal A, Munsky B, Proux F, Ly N, Braye FE, Zimmer C, Mueller F, Darzacq X. 2014. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep 8: 75–83. 10.1016/j.celrep.2014.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Shaban HA, Seeber A. 2020. Monitoring the spatio-temporal organization and dynamics of the genome. Nucleic Acids Res 48: 3423–3434. 10.1093/nar/gkaa135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban HA, Barth R, Bystricky K. 2018. Formation of correlated chromatin domains at nanoscale dynamic resolution during transcription. Nucleic Acids Res 46: e77. 10.1093/nar/gky269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban HA, Barth R, Bystricky K. 2020. Navigating the crowd: visualizing coordination between genome dynamics, structure, and transcription. Genome Biol 21: 278. 10.1186/s13059-020-02185-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Takei Y, Zhou W, Lubeck E, Yun J, Eng CHL, Koulena N, Cronin C, Karp C, Liaw EJ, et al. 2018. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174: 363–376.e16. 10.1016/j.cell.2018.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382. 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. 2017. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168: 159–171.e14. 10.1016/j.cell.2016.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP. 2018. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175: 1481–1491.e13. 10.1016/j.cell.2018.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyte R, Nuebler J, Blaukopf C, Hoefler R, Stocsits R, Peters JM, Gerlich DW. 2018. Dynamics of sister chromatid resolution during cell cycle progression. J Cell Biol 217: 1985–2004. 10.1083/jcb.201801157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O'Shaughnessy-Kirwan A, et al. 2017. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544: 59–64. 10.1038/nature21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JH, Zheng P, Kinrot SS, Bintu B, Zhuang X. 2020. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182: 1641–1659.e26. 10.1016/j.cell.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, Ettwiller L, Spitz F. 2014. Functional and topological characteristics of mammalian regulatory domains. Genome Res 24: 390–400. 10.1101/gr.163519.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Jost D, Chang JM, Cattoni DI, Papadopoulos GL, Bonev B, Sexton T, Gurgo J, Jacquier C, Nollmann M, et al. 2018. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci Adv 4: eaar8082. 10.1126/sciadv.aar8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi-Takanaka Y, Kimura H, Kurumizaka H. 2010. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc Natl Acad Sci 107: 10454–10459. 10.1073/pnas.1003064107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortora MM, Salari H, Jost D. 2020. Chromosome dynamics during interphase: a biophysical perspective. Curr Opin Genet Dev 61: 37–43. 10.1016/j.gde.2020.03.001 [DOI] [PubMed] [Google Scholar]
- Tsai A, Muthusamy AK, Alves MRP, Lavis LD, Singer RH, Stern DL, Crocker J. 2017. Nuclear microenvironments modulate transcription from low-affinity enhancers. eLife 6: e28975. 10.7554/eLife.28975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Furlong EEM. 2019. The role of transcription in shaping the spatial organization of the genome. Nat Rev Mol Cell Biol 20: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann J, Valeri A, Guillou E, Cuvier O, Nollmann M. 2011. Roles of chromatin insulator proteins in higher-order chromatin organization and transcription regulation. Nucleus 2: 358–369. 10.4161/nucl.2.5.17860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann J, Le Gall A, Dejardin S, Allemand F, Gamot A, Labesse G, Cuvier O, Nègre N, Cohen-Gonsaud M, Margeat E, et al. 2014. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet 10: e1004544. 10.1371/journal.pgen.1004544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner FR, Dienemann C, Wang H, Stützer A, Tegunov D, Urlaub H, Cramer P. 2020. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 579: 448–451. 10.1038/s41586-020-2088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Su JH, Beliveau BJ, Bintu B, Moffitt JR, Wu CT, Zhuang X. 2016. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353: 598–602. 10.1126/science.aaf8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M, Stott K. 2019. Disordered domains in chromatin-binding proteins. Essays Biochem 63: 147–156. 10.1042/EBC20180068 [DOI] [PubMed] [Google Scholar]
- Wiggins PA, van der Heijden T, Moreno-Herrero F, Spakowitz A, Phillips R, Widom J, Dekker C, Nelson PC. 2006. High flexibility of DNA on short length scales probed by atomic force microscopy. Nat Nanotechnol 1: 137–141. 10.1038/nnano.2006.63 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13: 59–69. 10.1038/nrg3095 [DOI] [PubMed] [Google Scholar]
- Xu H, Zhang S, Yi X, Plewczynski D, Li MJ. 2020. Exploring 3D chromatin contacts in gene regulation: the evolution of approaches for the identification of functional enhancer–promoter interaction. Comput Struct Biotechnol J 18: 558–570. 10.1016/j.csbj.2020.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Corces VG. 2012. Insulators, long-range interactions, and genome function. Curr Opin Genet Dev 22: 86–92. 10.1016/j.gde.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M, Wang J, Wang M, Li X, Zhang M, Wu Q, Wang Y. 2017. Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res 27: 1365–1377. 10.1038/cr.2017.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Emerson DJ, Gilgenast TG, Titus KR, Lan Y, Huang P, Zhang D, Wang H, Keller CA, Giardine B, et al. 2019. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576: 158–162. 10.1038/s41586-019-1778-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Young IT, de Jong JGS. 2011. Photon budget analysis for fluorescence lifetime imaging microscopy. J Biomed Opt 16: 086007. 10.1117/1.3608997 [DOI] [PubMed] [Google Scholar]
- Zidovska A. 2020. The self-stirred genome: large-scale chromatin dynamics, its biophysical origins and implications. Curr Opin Genet Dev 61: 83–90. 10.1016/j.gde.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovska A, Weitz DA, Mitchison TJ. 2013. Micron-scale coherence in interphase chromatin dynamics. Proc Natl Acad Sci 110: 15555–15560. 10.1073/pnas.1220313110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller B, Little SC, Gregor T. 2018. Diverse spatial expression patterns emerge from unified kinetics of transcriptional bursting. Cell 175: 835–847.e25. 10.1016/j.cell.2018.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]