Abstract
SARS-CoV-2 mRNA vaccines confer robust protection against COVID-19, but the emergence of variants has generated concerns regarding the protective efficacy of the currently approved vaccines, which lose neutralizing potency against some variants. Emerging data suggest that antibody functions beyond neutralization may contribute to protection from the disease, but little is known about SARS-CoV-2 antibody effector functions. Here, we profiled the binding and functional capacity of convalescent antibodies and Moderna mRNA-1273 COVID-19 vaccine-induced antibodies across SARS-CoV-2 variants of concern (VOCs). Although the neutralizing responses to VOCs decreased in both groups, the Fc-mediated responses were distinct. In convalescent individuals, although antibodies exhibited robust binding to VOCs, they showed compromised interactions with Fc-receptors. Conversely, vaccine-induced antibodies also bound robustly to VOCs but continued to interact with Fc-receptors and mediate antibody effector functions. These data point to a resilience in the mRNA-vaccine-induced humoral immune response that may continue to offer protection from SARS-CoV-2 VOCs independent of neutralization.
Keywords: Fc effector function, SARS-CoV-2, COVID-19, vaccines, mRNA-1273 vaccination, variants of concern
SARS-CoV-2 mRNA vaccines provide cross-variant protection against COVID-19. Whether this is mediated strictly via neutralization or is linked to effector functions that may limit, rather than block, transmission remains unknown. Kaplonek et al. show that mRNA-1273-vaccination-induced antibodies preserve Fc effector responses across variants of concern, whereas antibodies induced following natural infection show compromised interactions with Fc-receptors.
Introduction
Remarkable progress in the battle against SARS-CoV-2 has been achieved with the approval and emergency use authorization of several COVID-19 vaccines globally. However, the emergence of variants of concern (VOCs), which are able to re-infect large numbers of previously immune individuals (Sabino et al., 2021; Zhou et al., 2021; Bergwerk et al., 2021; Lopez Bernal et al., 2021; Nasreen et al., 2021; Puranik et al., 2021), has reignited the concerns related to potential vaccine vulnerabilities and the prospective need for next-generation VOC-inspired vaccines. Yet, the persistence of protection by several COVID-19 vaccines, including mRNA-1273 Moderna (Chemaitelly et al., 2021), BNT162b2 Pfizer/BioNTech (Abu-Raddad et al., 2021; Kustin et al., 2021), and Johnson & Johnson (Sadoff et al., 2021) vaccines, in geographic regions of the world where VOCs evade neutralization (Garcia-Beltran et al., 2021a; Wall et al., 2021), argues for the importance of alternate immune mechanisms in the protection from COVID-19. Beyond the T cells that have been implicated in natural immunity to the virus (Weiskopf et al., 2020), antibody effector functions track with DNA, adenovirus serotype 26 vector-based (Ad26), and adjuvant protein-based vaccine-mediated protection in non-human primates (Mercado et al., 2020; Gorman et al., 2021). Moreover, the power of antibodies to leverage immune functions, including monocyte or neutrophil phagocytosis, and complement deposition has been implicated in the protection against many viral infections, including influenza virus, HIV, and Ebola (Lu et al., 2018).
Many mutations in VOCs have emerged in the receptor-binding domain (RBD) and at sites on the S1 that improve the stability and orientation of RBD, collectively improving attachment and infection and, thereby enhancing the virus infectivity (Dejnirattisai et al., 2021a). Neutralizing antibodies that strictly prevent infection must interfere with a limited surface area involved in the binding to the ACE2 receptor or prevent fusion; thus, it is not surprising that several mutations accumulating across the globe have been linked to the reduced sensitivity of VOCs to the neutralizing antibody activity (Zost et al., 2020). Conversely, vast numbers of antibodies generated during infection and following vaccination can bind outside of these footprints, across the entire surface of the spike antigen, and further recognize and potentially continue to confer protection against the diseases caused by VOCs. Importantly, although neutralizing antibodies are likely to be a key to preventing transmission, the antibodies that are able to bind outside RBD and leverage the antiviral activity of the innate immune system may confer protection against the disease, which is effectively capable of controlling and turning COVID-19 into a mild illness comparable to the common cold. However, whether the emerging SARS-CoV-2 VOCs affect these alternate humoral immune responses remains unclear.
Although neutralizing antibodies show a consistent decrease in function across VOCs when it comes to naturally acquired and vaccine-induced immunity (Dejnirattisai et al., 2021b; Garcia-Beltran et al., 2021b; Rees-Spear et al., 2021; Supasa et al., 2021; Barrett et al., 2021; Wall et al., 2021), emerging data point to antibody-dependent effector functions, such as neutrophil (antibody-dependent neutrophil phagocytosis ) and monocyte (antibody-dependent cellular phagocytosis ) phagocytosis as well as NK cytotoxicity (ADNK), in the resolution of natural infection (Mercado et al., 2020) and protection following vaccination and administration of monoclonal therapeutics in animal models (Gorman et al., 2021; Pinto et al., 2021; Yu et al., 2020). With the rise of VOCs that have begun to break through vaccine-induced immunity globally, a more profound understanding of the mediators of immunity, in addition to neutralization, is urgently needed. Thus, we aimed to define whether Fc effector functions were equally disrupted across VOCs. Given the ability of VOCs to re-infect previously naturally immune individuals (Andreano et al., 2020; Wibmer et al., 2021; Planas et al., 2021a), we probed the impact of the recently emerging SARS-CoV-2 VOCs on antibody binding and functional humoral immunity, induced by natural immunity or the mRNA-1273 vaccine, on both the spike (S) and RBD VOCs. Although naturally induced antibodies from convalescent patients bound robustly to the wild-type (WT) SARS-CoV-2 spike and to a slightly lesser degree to VOCs, the convalescent VOC-binding antibodies largely failed to interact with Fc-receptors. Conversely, although the mRNA-1273-induced antibodies bound similar to wild type and VOCs, they demonstrated negligible differences in the Fc-receptor engagement, maintaining largely conserved antibody effector functions. These data highlight a previously unappreciated functional gap in the immunity of convalescents and point to the resilience of functional mRNA-vaccine-induced antibodies that are likely to continue to bind and confer robust antibody effector functions against VOCs, even in the setting of compromised neutralization.
Results
mRNA-1273 vaccination induces a broader cross-isotype/subclass-binding antibody recognition of VOCs than natural infection
The emergence and rapid spread of multiple SARS-CoV-2 variants worldwide has raised concerns about the cross-protective activity of mRNA-induced vaccine immunity against these perpetually evolving VOCs. Decreased VOC neutralization has been observed across vaccines (Abdool Karim and de Oliveira, 2021). However, whether these VOCs also escape other humoral immune antiviral mechanisms remains unclear. To determine how VOCs affect humoral immunity, we profiled convalescent plasma and mRNA-vaccine-induced humoral immune responses across VOCs. Convalescent plasma samples (n = 305) were collected voluntarily from a community-based seroprevalence study at four sites across the US (California, Texas, North Carolina, and Florida), where the majority of participants experienced largely asymptomatic/mild symptoms following SARS-CoV-2 infection. The volunteers were followed on a monthly basis from May to August, 2020 prior to the release of vaccines; they were between the ages of 19 and 62 years, with a prevalence of 84% of males. These profiles were compared with peak immunogenicity time points (day 43, two weeks after boosting) from the phase 1 mRNA-1273 clinical trial (ClinicalTrials.gov number NCT04283461). Eligible vaccine participants (n = 45, with one patient being excluded from the analysis due to low sample volume ) included healthy adults, 18 to 55 years of age, with 49% of males and 51% of females (Jackson et al., 2020). Only limited differences were observed between vaccine doses (Figure S1); therefore, the analysis was performed for all doses combined.
Both natural infection and vaccination induce polyclonal pools of antibodies consisting of different isotypes/subclasses and specificities that act synergistically via the formation of immune complexes to elicit diverse effector functions (Jefferis et al., 1998). Several studies have shown differences in antibody titers and neutralization across natural infection and following mRNA vaccination to both the WT and VOC spike antigens (Assis et al., 2021; Jalkanen et al., 2021; Cho et al., 2021). However, whether the vaccines further shift the polyclonal humoral immune response remains unclear. Thus, here we aimed to deeply characterize and compare the humoral immune response following SARS-CoV-2 infection and COVID-19 mRNA-1273 vaccination across SARS-CoV-2 VOCs to better understand the underlying differences in immune responses and protection against potential infection. Collectively, we observed reduced IgG1, IgG3, IgM, and IgA binding to the B.1.1.7-like (N501YΔ69–70) and E484K (present in B.1.351 and P1) spike antigens compared with the WT D614G spike in convalescent responses (Figures 1A and 1B). Conversely, mRNA-1273-immunized individuals exhibited decreased isotype recognition only for B.1.1.7 VOCs compared with the D614G, with robustly conserved or even higher vaccine-induced IgG1, IgG3, IgA, and IgM binding across D614G E484K and D614G K417N spike variants (Figures 1C and 1D). No differences in the natural and vaccine-induced antibody responses were observed either between males and females, across age groups, or based on symptom severity (Figures S2–S6). These data point to potential convalescent vulnerabilities in IgG1, IgM, and IgA recognition of the VOCs but a more limited impact on mRNA vaccine-induced antibody binding to VOCs.
Figure 1.
SARS-CoV-2 natural infection and mRNA-1273 vaccination induce IgG1, IgG3, IgM, and IgA antibodies across SARS-CoV-2 VOCs
(A) The dot plots represent the relationship between WT D614G spike antibody binding (x axis) and N501YΔ69–70 (red) and D614G E484K (black) spike variants (y axis) across convalescent COVID-19 patients (n = 305).
(B) Line graphs represent the overall binding profile to the D614G S, N501YΔ69–70 S, and D614G E484K S plotted on the comparative line antigens in the same group of convalescent subjects.
(C) The correlation plot shows the relationship between WT D614G spike antibody binding (x axis) and the full B.1.1.7 (red), D614G E484K (black), and the D614G K417N (gray) spike variants (y axis) across mRNA-1273-vaccinated individuals at peak immunogenicity (n = 44).
(D) Line graphs represent the same data as the overall binding profile to the D614G S, B.1.17 S (red), D614G E484K S (black), and the D614G K417N S (gray) in the group of vaccinated subjects. A Pearson correlation was used to establish the strength of the relationship between WT and VOC antigen binding. Dots represent the mean value of replicates per serum sample. The fold change was calculated as a ratio of WT binding compared with each VOC, which is indicated in the bracket. Matched nonparametric Friedman test with Dunn’s multiple comparisons test was used to calculate the statistical significance for line graphs. Only statistically significant values are shown, and the asterisks represent the adjusted p values: (∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p <0.0001). See also Figures S1–S4.
mRNA-1273-induced VOC spike-specific antibodies show stable Fc-receptor binding
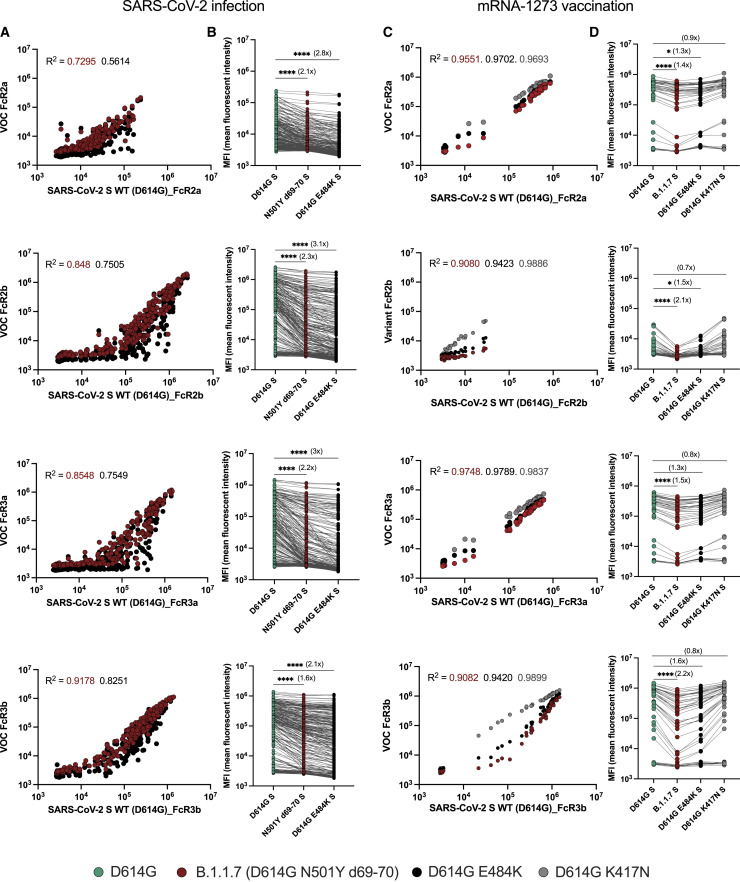
In addition to their ability to bind and block infection, antibodies leverage a wide array of antibody-pathogen functions via antibody constant domain (Fc-domain) interactions with the Fc-receptors found on innate immune cells (Lu et al., 2018). Importantly, Fc effector function has been linked to protection against several infections, including influenza (Boudreau and Alter, 2019), malaria (Suscovich et al., 2020), Ebola virus (Meyer et al., 2021), and SARS-CoV-2 (Gorman et al., 2021; Pinto et al., 2021; Yu et al., 2020). Given our emerging appreciation for the role of Fc effector function in the control/clearance of natural SARS-CoV-2 infection (Zohar et al., 2020; Atyeo et al., 2020), we explored whether antibodies that evolve in convalescent individuals or mRNA-1273 vaccinees have an equivalent capacity to interact with Fc-receptors. We profiled the binding to all low-affinity IgG-Fc-receptors (FcRs) involved in driving IgG effector function (Figure 2 ). A substantial variation was noted in the SARS-CoV-2-specific antibody binding to FcRs across the VOCs in convalescent plasma (Figures 2A and 2B). We observed an overall lower SARS-CoV-2-specific antibody binding to the activating FcγR2a in convalescent individuals compared with mRNA-vaccinated individuals. Conversely, the opposite pattern was observed for the inhibitory FcγR2b receptor, with enhanced binding to FcγR2b in convalescent individuals compared with vaccinees. Moreover, compared with the D614G spike FcR binding, the antibodies produced after natural infection exhibited a substantial reduction in the binding of FcγR2b, FcγR3a, and FcγR3b to the N501YΔ69–70 spike variant. Yet, a more prominent loss of FcR binding antibodies was seen for the D614G E484K binding antibodies in convalescent individuals. Importantly, reduced SARS-CoV-2-specific antibody FcR binding was notable, particularly at low convalescent antibody titers. By contrast, the convalescent individuals with robust D614G-FcR binding titers exhibited an equivalent binding to VOCs. These data point to compromised FcR interactions in convalescent immunity, particularly at low binding titers that may evolve due to mild infection or in the setting of waning immunity (Ibarrondo et al., 2020; Choe et al., 2021). Compromised FcR binding evolved despite largely preserved IgG binding to the VOCs, which may render a subset of previously immune individuals with low antibody titers vulnerable to re-infection with these new strains.
Figure 2.
More robust SARS-CoV-2 VOC-specific Fc-receptor binding is driven by mRNA-1273 vaccination compared with SARS-CoV-2 natural infection
(A) The dot plots show the overall FcγR2a, FcγR2b, FcγR3a, and FcγR3b binding to D614G (x axis) and to N501YΔ69–70 (red) and D614G E484K (black) spike variants (y axis) across convalescent COVID-19 patients (n = 305).
(B) The line graphs show the overall FcR binding profile to D614G S (green), N501YΔ69–70 S (red), and D614G E484K S (black) in the same group of convalescent subjects.
(C) The dot plots show the overall FcγR2a, FcγR2b, FcγR3a, and FcγR3b binding to D614G (x axis) and to B.1.1.7 (black), B.1.351 (red), and P.1 (gray) in mRNA-1273-vaccinated individuals (y axis).
(D) The line graph shows the overall FcR binding profile to the D614G, D614G E484K (black), and the D614G K417N (gray) spike variants across mRNA-1273-vaccinated individuals. A Pearson correlation was used to establish the strength of the relationship between wild-type and VOC antigen binding. Dots represent the mean value of replicates per serum sample. The fold change was calculated as a ratio of WT binding compared with each VOC, which is indicated in the bracket. Matched nonparametric Friedman test with Dunn’s multiple comparisons test was used to calculate the statistical significance for line graphs. Only statistically significant values are shown, and the asterisks represent the adjusted p values: (∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p <0.0001).
In contrast to convalescent plasma, mRNA-1273-vaccine-induced antibodies exhibited an overall stable binding to all the activating FcRs but lower binding to the inhibitory FcγR2b (Figures 2C and 2D). In comparison with D614G, a stronger decline was observed in FcγR2a, FcγR2b, Fcγ3a, and Fcγ3b binding to the B.1.1.7 spike, and a trend toward reduction was noted for the D614G E484K spike for FcγR2a and FcγR2b binding. However, reduced binding was proportional across binding titer levels and was not solely observed in individuals with low spike-specific binding titers. Additionally, no alteration in the SARS-CoV-2-specific antibody binding to FcRs was observed for D614G K417N spike. These data highlight the disconnect between the binding antibodies and FcR-engaging antibodies, where Fc-receptor binding was compromised in the SARS-CoV-2 spike-specific antibodies induced following natural infection but not following vaccination. Importantly, it is critical to note that compromised Fc-receptor binding was most clearly observed among the convalescent patients with lower antibody titers, whereas in the convalescent individuals with high titers, approximating levels observed in vaccinees, antibodies appeared to bind Fc-receptors robustly across the variants. These data point to a functional resilience in the mRNA-1273 vaccine-induced antibodies that are less affected by the VOCs compared with the low-titer convalescent antibodies.
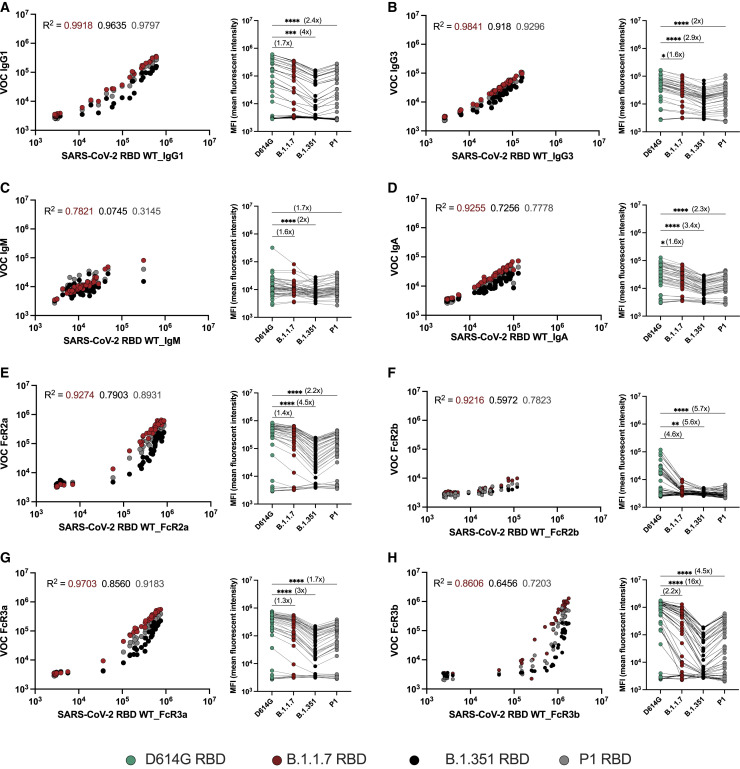
Both vaccine-induced and convalescent antibodies exhibit compromised VOC RBD-specific Fc-receptor binding
Despite the high efficacy of the mRNA vaccines against the original SARS-CoV-2 variant, waves of variants have emerged. These variants include amino acid substitutions that significantly diminish the neutralizing antibody activity (Garcia-Beltran et al., 2021a; Wall et al., 2021; Planas et al., 2021c) attributed to particular mutations in the RBD (Greaney et al., 2021). However, whether these changes in the RBD eliminate all RBD-specific antibody binding and antibody-mediated Fc-receptor interactions has yet to be defined. Here, we observed diminished binding of vaccine-induced RBD-specific IgG1 and IgG3 across the B.1.1.7 (alpha), B.1.351 (beta), and P.1 (gamma) RBDs (Figures 3A and 3B). Less substantial differences were noted for IgM binding across the variants, with a more pronounced loss of IgA binding (Figures 3C and 3D). Antibody-FcR binding was compromised across all VOC RBDs, most profoundly for B.1.351 (Figures 3E–3H). However, the FcR interactions with VOC RBDs were not completely eliminated but continued to bind in a correlated, albeit dampened, manner compared with the antibody binding to the WT RBD. These data point to a more profound reduction, but not a complete loss, of the vaccine-induced antibody FcR binding activity to VOC RBDs, despite largely preserved antibody-mediated Fc-activity to the full spike.
Figure 3.
mRNA-1273 vaccination induces RBD-specific antibody responses to SARS-CoV-2 VOCs
(A–H) The dot plots show the relationship of (A) IgG1, (B) IgG3, (C) IgM, and (D) IgA as well as (E) FcγR2a, (F) FcγR2b, (G) FcγR3a, and (H) FcγR3b binding profiles with the WT RBD (x axis) or RBD VOCs (y axis) across mRNA-1273-vaccinated individuals (n = 44). The line graphs show the response to each of the RBDs across the same set of mRNA-1273 vaccine samples. A Pearson correlation was used to establish the strength of the relationship between WT and VOC antigen binding. Dots represent the mean value of replicates per serum sample. The fold change was calculated as a ratio of WT binding compared with each VOC, which is indicated in the bracket. Matched nonparametric Friedman test with Dunn’s multiple comparisons test was used to calculate the statistical significance for line graphs. Only statistically significant values are shown, and the asterisks represent the adjusted p values: (∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001).
Fc effector functions are different in COVID-19 convalescents and vaccinees across VOCs
Given the differential FcR binding differences noted across the antibodies that are able to bind to the VOCs, we next sought to determine whether these differences also translated to changes in the antibody Fc effector function across full spike VOCs for SARS-CoV-2-infected and mRNA-1273-vaccinated individuals (Figure 4 ). Specifically, the antibodies from convalescent plasma displayed a prominent reduction in the antibody-dependent complement deposition (ADCD) for the B.1.351 (beta) variant and a similar, but non-significant, trend for the B.1.1.7 (alpha) variant spike compared with WT SARS-CoV-2 (Figure 4A). Natural antibody-mediated complement depositing functions for the P1 VOC spike antigen were higher than those for WT. As for the mRNA-1273 vaccine-induced SARS-CoV-2 spike-specific antibodies, only a minor reduction in ADCD was observed for the B.1.1.7 variant, with stable complement activity for the B.1.351 (beta) variant, and elevated complement fixation for the P1 (gamma) spike variant compared with WT SARS-CoV-2 (Figure 4B). Furthermore, a loss of ADNP was noted for the B.1.1.7 variant, with a more significant decline in the B.1.351-directed ADNP activity in the individuals with natural SARS-CoV-2 infection. Similar to ADCD, the ADNP activity against the P1 variants was higher than that against the WT SARS-CoV-2 (Figure 4C). Conversely, limited to no loss of ADNP activity was observed for the B.1.1.7 and P1 variants in mRNA-1273-vaccinated individuals compared with the ADNP activity for the D614G spike variant. However, a significant but not complete loss of ADNP was noted for the B.1.351 variant (Figure 4D). ADCP was reduced for all VOCs compared with the D614G variant across both convalescent individuals and vaccinated individuals (Figures 4E and 4F). Again, these data reinforce the disconnect between the quality and quantity of the humoral immune response to SARS-CoV-2, with equal or enhanced complement and neutrophil phagocytic functions following vaccination for the B.1.1.7 (alpha) and P.1 (gamma) variants but decreased activity for the B.1.351 (beta) variant. However, more substantial changes in the functional responses to the VOCs were seen following natural infection. Our study reveals some potential nuanced vulnerabilities to the emerging VOCs that may help refine our ability to strategically design next-generation vaccines or boosting regimens to provide maximal protection against the novel circulating variants of SARS-CoV-2.
Figure 4.
SARS-CoV-2-specific antibody-mediated effector functions to VOCs are different across COVID-19 convalescent individuals and mRNA-1273 vaccine recipients
(A–F) The functional vaccine-induced immune responses for antibody-mediated complement deposition (ADCD) (A and B), antibody-mediated neutrophil phagocytosis (ADNP) (C and D), and antibody-mediated monocyte phagocytosis (ADCP) (E and F) were analyzed in a cohort of SARS-CoV-2-infected (n = 305) and mRNA-1273-immunized individuals (n = 44). The dot plots represent the average functional activity in the cohort, with each dot representing the mean of two biological replicates. The fold change was calculated as a ratio of WT binding compared with each VOC, which is indicated in the bracket. Matched nonparametric Friedman test with Dunn’s multiple comparisons test was used to calculate the statistical significance for line graphs. Only statistically significant values are shown, and the asterisks represent the adjusted p values: (∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001).
Discussion
Neutralization represents a critical correlate of immunity for many clinically approved vaccines (Plotkin, 2010). However, protection from infection has been observed: (1) just weeks after primary mRNA and Ad26 immunization prior to the induction of neutralizing antibodies (Stephenson et al., 2021; Polack et al., 2020); (2) in regions of the world where the vaccine neutralizes the variants poorly (Alter et al., 2021); as well as (3) months after immunization despite declining neutralizing antibody titers (Thompson et al., 2021), collectively arguing for additional correlates of immunity against SARS-CoV-2. Antibodies may also contribute to protection via various additional mechanisms through their ability to interact with Fc-receptors or complement (Boudreau and Alter, 2019). Moreover, Fc effector functions are emerging as critical correlates of protection in natural SARS-CoV-2 infection (Zohar et al., 2020; Mercado et al., 2020) as well as in non-human primate vaccine studies (Gorman et al., 2021; Mercado et al., 2020). Yet, despite the early high efficacy of approved vaccines, re-infections are common in the setting of waning natural (Chen et al., 2021; Supasa et al., 2021) and vaccine immunity in the setting of a rise in VOCs (Supasa et al., 2021; Abdool Karim and de Oliveira, 2021). Yet, it is unclear how VOCs disrupt the wholistic natural or vaccine-induced humoral immune response and whether antibody functions beyond neutralization can compensate for a loss of neutralization.
Unlike neutralizing antibodies that must bind to the spike at precise sites, antibodies that are able to elicit Fc effector function can theoretically target the entire surface of the spike. Here, we demonstrated the disconnect between the quantity and quality of antibodies, measured by FcR binding to VOCs in convalescent plasma samples, marking unexpected vulnerabilities in the naturally induced humoral immune response to SARS-CoV-2 VOCs. Conversely, the mRNA-1273 vaccine-induced spike-specific antibodies exhibited a more robust FcR binding capacity and functionality, pointing to the vaccine-induced induction of more resilient antibodies that are able to target VOCs, bind FcRs, and drive function more flexibly than antibodies that evolve following natural infection. Thus, although the neutralizing antibody activity may be selectively compromised by particular variants, the ability of antibodies to continue to leverage the antiviral activity of Fc-effector functions may afford the humoral immune response the capability of providing persistent opsonophagocytic clearing functions that are able to attenuate the disease, should infection occur. Thus, although the ultimate goal of completely blocking SARS-CoV-2 transmission could profoundly accelerate the end of the pandemic, the ability of vaccines to provide protection against disease and death may transform SARS-CoV-2 into a less dangerous virus. The persistence of robust binding, FcR binding, and Fc-effector function inducing mRNA-vaccine-induced spike-specific antibodies may continue to provide a critical defense against the disease caused by emerging VOCs.
Natural resolution of SARS-CoV-2 infection is associated with the generation of a wide variety of different humoral immune titers that persist for several months (Wajnberg et al., 2020). However, data from Manaus, Brazil, illustrated the limited protective efficacy of naturally generated humoral immune responses against the emerging VOCs (Sabino et al., 2021). Incomplete natural immunity against VOCs may be related to lower quantities of antibodies induced in most convalescent individuals, which is substantially lower than those induced by mRNA vaccination. Moreover, highly heterogeneous antibody titers were observed among convalescent individuals, which exhibited linked heterogeneous binding capabilities to multiple VOCs, marked by concomitant differences in the FcR binding capabilities and Fc-effector functions across VOCs. Importantly, the disconnect between isotype-binding titers and FcR binding was largely observed at lower antibody titers, suggesting that above a particular threshold, even convalescent antibodies may confer a robust barrier against VOCs. Additionally, two doses of mRNA-1273 clearly drove high antibody titers, which were able to bind to FcRs effectively and drive highly resilient Fc-effector function across most VOCs. However, whether additional vaccine platforms will drive equivalent resilient antibody functions and whether additional heterologous prime-boosting strategies may drive robust and durable antibody effector functions remain unclear. Yet, it is likely that autologous or heterologous boosters drive robust increases not only in titers but also in FcR binding and function, thereby augmenting both Fab- (neutralization) and Fc-mediated antiviral mechanisms, providing the greatest level of protection against VOCs, even from those like Omicron that significantly evade neutralization (Planas et al., 2021b; Garcia-Beltran et al., 2021c; Gruell et al., 2021).
Thus, despite the loss of neutralization against some VOCs, continued recognition by non-neutralizing mechanisms of protective immunity may be sufficient to confer protection against the disease, eliminating the risk of mortality and morbidity previously associated with COVID-19. With emerging deeper phase 3 correlate analyses underway, the precise correlates of immunity will be defined across VOCs, providing critical insights to guide the future design and deployment of vaccines. The induction of broader, more flexible immune responses that maintain protection against disease and death may ultimately represent a more critical target for global immunity in the face of expected future viral evolution.
Limitations of the study
There are several limitations to this study. First, we do not know the precise dates of infection for the community-acquired COVID-19 convalescent individuals. However, based on repeat testing, the individuals were no more than three months from infection (Bartsch et al., 2021) following the resolution of acute infection, representing a time point of relative stability in the humoral immune response (Israel et al., 2021; Wajnberg et al., 2020). Unfortunately, we were not able to look at long-term vaccine or convalescent durability in this study. However, recent studies have pointed to differential stability in the FcR binding profiles across vaccine platforms, potentially accounting for the differences in real-world vaccine efficacy (Puranik et al., 2021; Chemaitelly et al., 2021). Second, convalescent individuals were recruited during the WT or D614G variant waves, and thus, we were unable to profile FcR binding profiles and function elicited by the VOC-mediated infections that may vary in their flexibility similar to the responses induced by mRNA-1273 vaccination. Third, the mRNA-1273 trial participants were healthy adults, 18 to 55 years of age. Conversely, the convalescent individuals were between the ages of 19- and 62 years, thus capturing several individuals a decade older than the vaccinees. Yet, no significant age-effects were observed across each cohort, which may be potentially related to the smaller sample size in these cohorts. Fourth, our community-acquired COVID-19 convalescent individuals experienced largely asymptomatic/mild symptoms following SARS-CoV-2 infection, and no differences were observed across symptom severities. Lastly, antibody-mediated Fc-effector functional assays were performed with cells from healthy donors rather than autologous cells from the convalescent or vaccinated individuals, which may differ and contribute to differential overall capabilities to respond and clear virus upon exposure (Kuri-Cervantes et al., 2020). Yet, despite these limitations, this study represents the first comprehensive analysis of the additional extra-neutralizing antibody properties of mRNA-1273-induced antibodies across VOCs that may provide persisting protection against severe disease and death even with a loss of neutralization.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Pacific Blue™ Mouse Anti-Human CD3 | BD Biosciences | CAT# 558117 RRID:AB 1595437 |
| Mouse Anti-Human IgG1-Fc PE | Southern Biotech | CAT# 9054-09 RRID:AB_2796628 |
| Mouse Anti-Human IgG2-Fc PE | Southern Biotech | CAT# 9060-09; RRID:AB_2796635 |
| Mouse Anti-Human IgG3-Fc PE | Southern Biotech | CAT# 9210-09; RRID:AB_2796701 |
| Mouse Anti-Human IgM-Fc PE | Southern Biotech | CAT# 9020-09 RRID:AB_2796577 |
| Mouse Anti-Human IgA1-Fc PE | Southern Biotech | CAT# 9130-09 RRID:AB_2796656 |
| Pacific Blue(TM) anti-human CD66b antibody | Biolegend | CAT# 305112 RRID:AB_2563294 |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 D614G WT Spike | Erica Saphire, La Jolla Institute for Immunology | N/A |
| SARS-CoV-2 D614G, N501Y, N501YΔ69-70, and P681H mutations Spike | Erica Saphire, La Jolla Institute for Immunology | N/A |
| SARS-CoV-2 D614G E484K Spike | Erica Saphire, La Jolla Institute for Immunology | N/A |
| SARS-CoV-2 D614G K417N Spike | Erica Saphire, La Jolla Institute for Immunology | N/A |
| SARS-CoV-2 N501YΔ69-70 Spike | Erica Saphire, La Jolla Institute for Immunology | N/A |
| SARS-CoV-2 WT RBD | Florian Krammer, Icahn School of Medicine at Mount Sinai | N/A |
| SARS-CoV-2 B.1.1.7 (N501Y) RBD | Florian Krammer, Icahn School of Medicine at Mount Sinai | N/A |
| SARS-CoV-2 B.1.351 (with N501Y, E484K, K417N mutations) RBD | Florian Krammer, Icahn School of Medicine at Mount | N/A |
| SARS-CoV-2 P1 (with N501Y, E484K, K417T mutations) RBD | Florian Krammer, Icahn School of Medicine at Mount | N/A |
| SARS-CoV-2 WT | LakePharma | N/A Custom order |
| SARS-CoV-2 B.1.1.7 | LakePharma | N/A Custom order |
| SARS-CoV-2 B.1.351 | LakePharma | N/A Custom order |
| SARS-CoV-2 P1 | LakePharma | N/A Custom order |
| HA A/Michigan/45/2015 (H1N1) | Immune Tech | IT-003-00105DTMp |
| HA A/Singapore/INFIMH-16-0019/2016 (H3N2) | Immune Tech | IT-003-00434DTMp |
| HA B/Phuket/3073/2013 | Immune Tech | IT-003-B11DTMp |
| Human Fc receptors | Produced at the Duke Human Vaccine Institute | N/A |
| Streptavidin-R-Phycoerythrin | Prozyme | CAT# PJ31S |
| FIX&Perm Cell Permeabilization Kit | Life Tech | CAT# GAS001S100 CAT# GAS002S100 |
| Brefeldin A | Sigma Aldrich | CAT# B7651 |
| GolgiStop | BD Biosciences | CAT# 554724 |
| EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) | Thermo Fisher | CAT# 77149 |
| Sulfo-NHS-LC-LC biotin | Thermo Fisher | CAT# A35358 |
| Software and algorithms | ||
| IntelliCyt ForeCyt (v8.1) | Sartorius | https://intellicyt.com/products/software/ |
| FlowJo (v10.7.1) | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo |
| Prism 9.2.0 (283) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| FluoSpheres™ NeutrAvidin™-Labeled Microspheres, yellow-green fluorescent (505/515), 1% solids | Invitrogen | N/A Custom order |
| FluoSpheres™ NeutrAvidin™-Labeled Microspheres, blue (fluorescent 350/440), 1% solids | Invitrogen | N/A Custom order |
| FluoSpheres™ NeutrAvidin™-Labeled Microspheres, crimson fluorescent (625/645),1% solids | Invitrogen | N/A Custom order |
| FluoSpheres™ NeutrAvidin™-Labeled Microspheres, red-orange fluorescent (565/580,)1% solids | Invitrogen | N/A Custom order |
| MagPlex microspheres | Luminex corporation | CAT# MC12001-01 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Galit Alter (galter@mgh.harvard.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Description of the cohorts
mRNA-vaccinated individuals
The phase 1, dose-escalation, open-label clinical trial was designed to determine the safety, reactogenicity, and immunogenicity of mRNA-1273 (mRNA-1273 ClinicalTrials.gov number, NCT04283461). Eligible participants (n = 45) were healthy adults 18 to 55 years of age who received two injections of trial vaccine 28 days apart at a dose of 25 μg (n = 15), 100 μg (n = 15), or 250 μg (n = 15) (Jackson et al., 2020). One person from a group receiving a dose of 100 μg was excluded from the analysis due to limited sample volume, therefore the analysis was performed with n = 44. The median age of the seropositive population was 32.6 years (range 18–53 years), the gender ratio was 49% male and 51% female, with 86% of non-Hispanic nor Latino and 14% of Hispanic or Latino participants. The study protocol was approved by the Advarra institutional review board. All participants provided written informed consent before enrollment. The trial was conducted under an Investigational New Drug application submitted to the Food and Drug Administration. The vaccine was co-developed by researchers at the National Institute of Allergy and Infectious Diseases (NIAID, the trial sponsor) and at Moderna (Cambridge, MA).
The mRNA-1273 vaccine, manufactured by Moderna, encodes the S-2P antigen, consisting of the SARS-CoV-2 glycoprotein with a transmembrane anchor and an intact S1–S2 cleavage site. S-2P was stabilized in its prefusion conformation by two consecutive proline substitutions at amino acid positions 986 and 987, at the top of the central helix in the S2 subunit (Jackson et al., 2020; Baden et al., 2021). The mRNA-LNP vaccine (mRNA-1273) was provided as a sterile liquid for injection at a concentration of 0.5 mg per milliliter. Normal saline was used as a diluent to prepare the doses administered. The vaccine was administered as a 0.5 ml injection in the deltoid muscle on days 1 and 29; follow-up visits were scheduled for 7 and 14 days after each vaccination and on days 57, 119, 209, and 394. Only the peak immunogenicity time point (day 43, two weeks following the boost) was included in this study.
Community-acquired COVID-19 individuals
Industry employees (Space Exploration Technologies Corp.) were volunteer tested for COVID-19, starting in April 2020. Participants completed a study survey including the collection of COVID-19 related symptoms, such as loss of smell/taste, fever, feverish/chills, cough, fatigue, headache, congestion, nausea/vomiting, diarrhea, sore throat, and body/muscle aches. The cohort included mild-symptomatic infections largely (Bartsch et al., 2021). Specifically, 68% of participant were asymptomatic (0 symptoms reported), 11%, 8%, 6%, 5% and 2% experienced respectively 1, 2, 3, 4 or 5 of mentioned symptoms. Upon obtaining informed consent, blood samples were collected (n = 305) and used for immune profiling. The median age of the seropositive population was 32 years (range 19–62 years), and 84% were males. The enrolled participants were 66% White, 8% Asian, 6% More than one race, 2% Black, 1% American Indian/Alaska Native, and 17% unknown. Volunteers were tested by PCR and for antibodies monthly. All antibody positive individuals were included in the study(Bartsch et al., 2021).
Primary Immune Cells
Fresh peripheral blood was collected by the MGH Blood bank from healthy human volunteers. All volunteers were over 18 years of age, provided written informed consent and all samples were de-identified before use. The studies were approved by the MGH (previously Partners Healthcare) Institutional Review Board. Human NK cells were isolated from fresh peripheral blood and maintained at 37°C, 5% CO2 in RPMI with 10% fetal bovine serum, L-glutamine, penicillin/streptomycin.
Cell Lines
THP-1 cells (ATCC) were grown in RPMI-1640 supplemented with 10% fetal bovine serum, L-glutamine, penicillin/streptomycin, HEPES, and beta-mercaptoethanol at 37°C, 5% CO2.
Method details
Luminex profiling
Antibody isotyping and Fcγ-receptor (FcγR) binding were conducted by multiplexed Luminex assay, as previously described (Brown et al., 2012; Brown et al., 2017). Briefly, SARS-CoV-2 D614G WT, B.1.1.7 (with D614G, N501Y, N501YΔ69-70, and P681H mutations), D614G E484K, D614G K417N, and N501YΔ69-70 Spike proteins (kindly provided by Erica Saphire, La Jolla Institute for Immunology) and SARS-CoV-2 WT, B.1.1.7 (N501Y), B.1.351 (with N501Y, E484K, K417N mutations), and P1 (with N501Y, E484K, K417T mutations) RBDs (kindly provided by Florian Krammer, Icahn School of Medicine at Mount Sinai) were used to profile specific humoral immune responses. Antigens were coupled to magnetic Luminex beads (Luminex Corp) by carbodiimide-NHS ester-coupling (Thermo Fisher). Antigen-coupled microspheres were washed and incubated with plasma samples at an appropriate sample dilution (1:100 for IgG3, IgA and IgM, 1:500 for IgG1 and, 1:1,000 for all low affinity Fcγ- receptors,) overnight at 4°C in 384-well plates (Greiner Bio-One). The high affinity FcR was not tested due to its minimal role in tuning antibody effector function (Nimmerjahn and Ravetch, 2008). Unbound antibodies were washed away, and antigen-bound antibodies were detected by using a PE-coupled detection antibody for each subclass and isotype (IgG1, IgG3, IgA1, and IgM; Southern Biotech), and Fcγ-receptors were fluorescently labeled with PE before addition to immune complexes (FcγR2a, FcγR2b, FcγR3a, and FcγR3b; Duke Protein Production facility). After one hour of incubation, plates were washed, and flow cytometry was performed with an IQue (Intellicyt), and analysis was performed on IntelliCyt ForeCyt (v8.1). PE median fluorescent intensity (MFI) is reported as a readout for antigen-specific antibody titers.
Effector functional assays
Antibody-dependent complement deposition (ADCD), antibody-dependent monocyte phagocytosis (ADCP), and antibody-dependent neutrophil phagocytosis (ADNP) were conducted as previously described (Butler et al., 2019; Karsten et al., 2019; Fischinger et al., 2019).
For ADCD, SARS-CoV-2 WT, B.1.1.7, B.1.351, and P1 Spike proteins (LakePharma) were coupled to magnetic Luminex beads (Luminex Corp) by carbodiimide-NHS ester-coupling (Thermo Fisher). Coupled beads were incubated for 2 hours at 37°C with serum samples (1:10 dilution) to form immune complexes and then washed to remove unbound immunoglobulins. In order to measure antibody-dependent deposition of C3, lyophilized guinea pig complement (Cedarlane) was diluted in gelatin veronal buffer with calcium and magnesium (GBV++) (Boston BioProducts) and added to immune complexes. Subsequently, C3 was detected with an anti-C3 fluorescein-conjugated goat IgG fraction detection antibody (Mpbio). The flow cytometry was performed with iQue (Intellicyt) and an S-Lab robot (PAA). ADCD was reported as the median of C3 deposition.
For ADCP and ADNP, SARS-CoV-2 WT, B.1.1.7, B.1.351, and P1 Spike proteins (LakePharma) were biotinylated using EDC (Thermo Fisher) and Sulfo-NHS-LC-LC biotin (Thermo Fisher) and coupled to blue (350/440), crimson (625/645), red-orange (565/580), and yellow-green (505/515) fluorescent Neutravidin-conjugated beads (Thermo Fisher), respectively. To form immune complexes, antigen-coupled beads were incubated for 2 hours at 37°C with 1:100 diluted serum samples and then washed to remove unbound immunoglobulins. For ADCP, the immune complexes were incubated for 16-18 hours with THP-1 cells (25,000 THP-1 cells per well at a concentration of 1.25×105 cells/ml in R10 cells/mL) and for ADNP for 1 hour with RBC-lyzed whole blood. Following the incubation, cells were fixed with 4% PFA. For ADNP, For ADNP granulocytes were isolated from whole blood by lysing RBC in ACK lysis buffer (1:10 blood in ACK lysis buffer) for 7 minutes before precipitation by centrifugation. Granulocytes were washed twice with cold PBS and resuspended at 2.5×105 cells/ml in R10. 50,000 cells per well were added to each well and incubated with immune complexes for 1 hour at 37°C, 5% CO2. Following the incubation, cells were fixed with 4% PFA. For ADNP, RBC-lyzed whole blood was washed, stained for CD66b+ (Biolegend) to identify neutrophils, and then fixed in 4% PFA. Flow cytometry was performed to identify the percentage of cells that had phagocytosed beads as well as the number of beads that had been phagocytosis (phagocytosis score = % positive cells × Median Fluorescent Intensity of positive cells/10000). The Flow cytometry was performed with 5 Laser LSR Fortessa Flow Cytometer, and analysis was performed using FlowJo V10.7.1.
Quantification and statistical analysis
All Luminex data were log-transformed, and all features were scaled and centered.
Statistical analyses were performed using GraphPad Prism 9.0 software. Dots represent the geometric mean fluorescent intensity (gMFI) value of replicates for each serum sample. The relationship of wildtype D614G (x-axis) to the recognition of the VOCs (y-axis) for each Fc-feature was assessed by Pearson correlation. The fold change was calculated as a ratio of wildtype binding compared to each VOC Line graphs show the gMFI per serum sample and the fold change was calculated as a ratio of wildtype binding compared to each VOC. Statistical differences were calculated using matched nonparametric Friedman test with Dunn’s multiple comparisons test. (∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p <0.0001).
Acknowledgment
We thank Nancy Zimmerman, Mark and Lisa Schwartz, an anonymous donor (financial support), Terry and Susan Ragon, and the SAMANA Kay MGH Research Scholars award for their support. We acknowledge the support from the Ragon Institute of MGH, MIT, and Harvard, the Massachusetts Consortium on Pathogen Readiness (MassCPR), the NIH (3R37AI080289-11S1, R01AI146785, U19AI42790-01, U19AI135995-02, U19AI42790-01, 1U01CA260476 – 01, and CIVIC75N93019C00052), and the Gates Foundation Global. This study was also supported by Health Vaccine Accelerator Platform funding (OPP1146996 and INV-001650), Translational Research Institute for Space Health through NASA Cooperative Agreement (NNX16AO69A), and the Musk Foundation. This work used samples from the phase 1 mRNA-1273 study (NCT04283461; doi: 10.1056/NEJMoa2022483). The mRNA-1273 phase 1 study was sponsored and primarily funded by the National Institute of Allergy and Infectious Diseases (NIAID) and the National Institutes of Health (NIH), Bethesda, MD. This trial has been partially funded with federal funds from the NIAID under grant awards UM1AI148373 to Kaiser Washington; UM1AI148576, UM1AI148684, and NIH P51 OD011132 to Emory University; and NIH AID AI149644 and contract award HHSN272201500002C to Emmes. Funding for the manufacture of mRNA-1273 phase 1 material was provided by the Coalition for Epidemic Preparedness Innovation.
Author contributions
P.K., B.J., and G.A. analyzed and interpreted the data. P.K., S.F., Y.C.B., J.K., J.S.B., and S.A.S. performed experiments. P.K., S.F., and D.C. performed the analysis. D.D., P.M., A.S.M., E.J.N., and E.R.M. managed samples and data collection for the community-acquired COVID-19 cohort. C.M., F.A., F.K., and E.O.S. produced SARS-CoV-2 S antigens. A.C. collected the samples and supervised and managed the clinical data for the Moderna mRNA-1273-vaccinated cohort. G.A. supervised the project. P.K. and G.A. drafted the manuscript. All authors critically reviewed the manuscript.
Declaration of interests
G.A. is the founder of Seromyx Systems Inc. A.C. is an employee of Moderna Inc. D.D., P.M., A.S.M., and E.R.M. are employees of Space Exploration Technologies Corp. All other authors have declared no competing interests.
Published: January 6, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2022.01.001.
Supplemental information
Data and code availability
-
•
The dataset generated for the study have been made available in the supplemental material.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants — clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Raddad L.J., Chemaitelly H., Butt A.A., National Study Group for COVID-19 Vaccination Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Dal Monego S., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis, R., Jain, A., Nakajima, R., Jasinskas, A., Khan, S., Palma, A., Parker, D.M., Chau, A., Specimen Collection Group, Obiero, J.M., et al. (2021) Distinct SARS-CoV-2 antibody reactivity patterns elicited by natural infection and mRNA vaccination. NPJ Vaccines 6, 132. [DOI] [PMC free article] [PubMed]
- Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J., et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532. doi: 10.1016/j.immuni.2020.07.020. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.R., Belij-Rammerstorfer S., Dold C., Ewer K.J., Folegatti P.M., Gilbride C., Halkerston R., Hill J., Jenkin D., Stockdale L., et al. Phase 1/2 trial of SARS-CoV-2 vaccine ChAdOx1 nCoV-19 with a booster dose induces multifunctional antibody responses. Nat. Med. 2021;27:279–288. doi: 10.1038/s41591-020-01179-4. [DOI] [PubMed] [Google Scholar]
- Bartsch Y.C., Fischinger S., Siddiqui S.M., Chen Z., Yu J., Gebre M., Atyeo C., Gorman M.J., Zhu A.L., Kang J., et al. Discrete SARS-CoV-2 antibody titers track with functional humoral stability. Nat. Commun. 2021;12:1018. doi: 10.1038/s41467-021-21336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., et al. Covid-19 breakthrough infections in vaccinated health care workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau C.M., Alter G. Extra-neutralizing FcR-mediated antibody functions for a universal influenza vaccine. Front. Immunol. 2019;10:440. doi: 10.3389/fimmu.2019.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.P., Dowell K.G., Boesch A.W., Normandin E., Mahan A.E., Chu T., Barouch D.H., Bailey-Kellogg C., Alter G., Ackerman M.E. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods. 2017;443:33–44. doi: 10.1016/j.jim.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.P., Licht A.F., Dugast A.-S., Choi I., Bailey-Kellogg C., Alter G., Ackerman M.E. High-throughput, multiplexed IgG subclassing of antigen-specific antibodies from clinical samples. J. Immunol. Methods. 2012;386:117–123. doi: 10.1016/j.jim.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A.L., Fallon J.K., Alter G. A sample-sparing multiplexed ADCP assay. Front. Immunol. 2019;10:1851. doi: 10.3389/fimmu.2019.01851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemaitelly H., Yassine H.M., Benslimane F.M., Al Khatib H.A., Tang P., Hasan M.R., Malek J.A., Coyle P., Ayoub H.H., Al Kanaani Z., et al. mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar. Nat. Med. 2021;27:1614–1621. doi: 10.1038/s41591-021-01446-y. [DOI] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe P.G., Kang C.K., Suh H.J., Jung J., Song K.H., Bang J.H., Kim E.S., Kim H.B., Park S.W., Kim N.J., et al. Waning antibody responses in asymptomatic and symptomatic SARS-CoV-2 infection. Emerg. Infect. Dis. 2021;27:327–329. doi: 10.3201/eid2701.203515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M.E., Supasa P., Case J.B., Zhao Y., Walter T.S., Mentzer A.J., Liu C., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200. doi: 10.1016/j.cell.2021.02.032. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954. doi: 10.1016/j.cell.2021.03.055. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischinger S., Fallon J.K., Michell A.R., Broge T., Suscovich T.J., Streeck H., Alter G. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods. 2019;473:112630. doi: 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Circulating SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. medRxiv. 2021 doi: 10.1101/2021.02.14.21251704. [DOI] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2021 doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M.J., Patel N., Guebre-Xabier M., Zhu A., Atyeo C., Pullen K.M., Loos C., Goez-Gazi Y., Carrion R., Tian J.H., et al. Collaboration between the Fab and Fc contribute to maximal protection against SARS-CoV-2 in nonhuman primates following NVX-CoV2373 subunit vaccine with Matrix-M™ vaccination. bioRxiv. 2021 doi: 10.1101/2021.02.05.429759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57. doi: 10.1016/j.chom.2020.11.007. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. medRxiv. 2021 doi: 10.1101/2021.12.14.21267769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel A., Shenhar Y., Green I., Merzon E., Golan-Cohen A., Schäffer A.A., Ruppin E., Vinker S., Magen E. Large-scale study of antibody titer decay following BNT162b2 mRNA vaccine or SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1101/2021.08.19.21262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalkanen P., Kolehmainen P., Häkkinen H.K., Huttunen M., Tähtinen P.A., Lundberg R., Maljanen S., Reinholm A., Tauriainen S., Pakkanen S.H., et al. COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat. Commun. 2021;12:3991. doi: 10.1038/s41467-021-24285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis R., Lund J., Pound J.D. IgG-Fc-mediated effector functions: molecular definition of interaction sites for effector ligands and the role of glycosylation. Immunol. Rev. 1998;163:59–76. doi: 10.1111/j.1600-065x.1998.tb01188.x. [DOI] [PubMed] [Google Scholar]
- Karsten C.B., Mehta N., Shin S.A., Diefenbach T.J., Slein M.D., Karpinski W., Irvine E.B., Broge T., Suscovich T.J., Alter G. A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J. Immunol. Methods. 2019;471:46–56. doi: 10.1016/j.jim.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes, L., Pampena, M.B., Meng, W., Rosenfeld, A.M., Ittner, C.A.G., Weisman, A.R., Agyekum, R., Mathew, D., Baxter, A.E., Vella, L., et al. (2020). Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection. bioRxiv. 2020.05.18.101717. 10.1101/2020.05.18.101717. [DOI]
- Kustin T., Harel N., Finkel U., Perchik S., Harari S., Tahor M., Caspi I., Levy R., Leschinsky M., Dror S.K., et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2 mRNA vaccinated individuals. Nat. Med. 2021;27:1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Gunn B.M., Malherbe D.C., Gangavarapu K., Yoshida A., Pietzsch C., Kuzmina N.A., Saphire E.O., Collins P.L., Crowe J.E., Jr., et al. Ebola vaccine-induced protection in nonhuman primates correlates with antibody specificity and Fc-mediated effects. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abg6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasreen S., Chung H., He S., Brown K.A., Gubbay J.B., Buchan S.A., Fell D.B., Austin P.C., Schwartz K.L., Sundaram M.E., et al. 2021. Effectiveness of COVID-19 vaccines against variants of concern in Ontario, Canada. medRxiv. [DOI] [Google Scholar]
- Nimmerjahn F., Ravetch J.V. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- Pinto D., Sauer M.M., Czudnochowski N., Low J.S., Tortorici M.A., Housley M.P., Noack J., Walls A.C., Bowen J.E., Guarino B., et al. Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization. 2021. bioRxiv. [DOI] [PubMed]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik A., Lenehan P.J., Silvert E., Niesen M.J.M., Corchado-Garcia J., O’Horo J.C., Virk A., Swift M.D., Halamka J., Badley A.D., et al. Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of alpha and delta variant prevalence. medRxiv. 2021 doi: 10.1101/2021.08.06.21261707. [DOI] [Google Scholar]
- Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., Thomas P., Graham C., Seow J., Lee N., et al. The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34:108890. doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Jr., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson K.E., Le Gars M., Sadoff J., de Groot A.M., Heerwegh D., Truyers C., Atyeo C., Loos C., Chandrashekar A., McMahan K., et al. Immunogenicity of the Ad26.COV2.S vaccine for COVID-19. JAMA. 2021;325:1535–1544. doi: 10.1001/jama.2021.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211. doi: 10.1016/j.cell.2021.02.033. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suscovich T.J., Fallon J.K., Das J., Demas A.R., Crain J., Linde C.H., Michell A., Natarajan H., Arevalo C., Broge T., et al. Mapping functional humoral correlates of protection against malaria challenge following RTS,S/AS01 vaccination. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb4757. [DOI] [PubMed] [Google Scholar]
- Thompson M.G., Burgess J.L., Naleway A.L., Tyner H., Yoon S.K., Meece J., Olsho L.E.W., Caban-Martinez A.J., Fowlkes A.L., Lutrick K., et al. Prevention and attenuation of Covid-19 with the BNT162b2 and mRNA-1273 vaccines. N. Engl. J. Med. 2021;385:320–329. doi: 10.1056/NEJMoa2107058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall E.C., Wu M., Harvey R., Kelly G., Warchal S., Sawyer C., Daniels R., Hobson P., Hatipoglu E., Ngai Y., Hussain S. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397:2331–2333. doi: 10.1016/S0140-6736(21)01290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519. doi: 10.1016/j.cell.2020.10.052. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The dataset generated for the study have been made available in the supplemental material.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.