Summary
Background
Oogenesis is a fundamental process of human reproduction, and mitochondria play crucial roles in oocyte competence. Mitochondrial ATP-dependent Lon protease 1 (LONP1) functions as a critical protein in maintaining mitochondrial and cellular homeostasis in somatic cells. However, the essential role of LONP1 in maintaining mammalian oogenesis is far from elucidated.
Methods
Using conditional oocyte Lonp1-knockout mice, RNA sequencing (RNA-seq) and coimmunoprecipitation/liquid chromatography–mass spectrometry (Co-IP/LC–MS) technology, we analysed the functions of LONP1 in mammalian oogenesis.
Findings
Conditional knockout of Lonp1 in mouse oocytes in both the primordial and growing follicle stages impairs follicular development and causes progressive oocyte death, ovarian reserve loss, and infertility. LONP1 directly interacts with apoptosis inducing factor mitochondria-associated 1 (AIFM1), and LONP1 ablation leads to the translocation of AIFM1 from the cytoplasm to the nucleus, causing apoptosis in mouse oocytes. In addition, women with pathogenic variants of LONP1 lack large antral follicles (>10 mm) in the ovaries, are infertile and present premature ovarian insufficiency.
Interpretation
We demonstrated the function of LONP1 in regulating oocyte development and survival, and in-depth analysis of LONP1 will be crucial for elucidating the mechanisms underlying premature ovarian insufficiency.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2018YFC1004701), the National Nature Science Foundation of China (82001629, 81871128, 81571391, 81401166, 82030040), the Jiangsu Province Social Development Project (BE2018602), the Jiangsu Provincial Medical Youth Talent (QNRC2016006), the Youth Program of the Natural Science Foundation of Jiangsu Province (BK20200116) and Jiangsu Province Postdoctoral Research Funding (2021K277B).
Keywords: Mitochondria, LONP1, AIFM1, Oocyte development, Oocyte survival, Premature ovarian insufficiency
Research in Context.
Evidence before this study
As a classic mitochondrial protease, Mitochondrial ATP-dependent Lon protease 1 (LONP1) has been well demonstrated to regulate metabolism and perform mitochondrial quality control in vitro. A growing number of studies have indicated that LONP1 is involved in regulating the unfolded protein response and mitochondrial dynamics in vitro and mouse heart development. The ovarian reserve and oocyte quality determine the reproductive life of mammals. Oocytes contain the largest number of mitochondria among all cell types, and the mitochondrial quality and quantity determine the oocyte quality and survival. However, far less is known about the function of LONP1 in oocyte development.
Added value of this study
We found that LONP1 is associated with oocyte development and ovarian reserve decline upon ageing. Conditional knockout of Lonp1 in mouse oocytes in both the primordial and growing follicle stages causes defective development of oocytes from secondary follicles into antral follicles and infertility. Moreover, LONP1 ablation in mouse oocytes causes prolonged activation of the mitochondrial unfolded protein response (mtUPR) and defective mitochondrial biosynthesis and function and leads to the translocation of apoptosis-inducing factor mitochondria-associated 1 (AIFM1) from the cytoplasm to the nucleus, causing apoptosis in mouse oocytes. The AIFM1-induced apoptosis inhibitor partly improves the competence of Lonp1-knockout oocytes. Moreover, pathogenic variants of LONP1 are responsible for large-follicle abnormalities in some women with premature ovarian insufficiency.
Implications of all available evidence
Our data revealed a new role of LONP1 in oocyte development or ovarian ageing and provided new insights into the pathogenic causes of premature ovarian insufficiency.
Alt-text: Unlabelled box
Introduction
Female fertility is one of the first physiological functions adversely affected by ageing.1 The consistent live birth rate of pregnancies from oocyte donation in ageing women suggests that a decline in oocyte quality is the major contributing factor responsible for infertility upon ageing.2 Ageing and abnormal follicular activation or quiescence can lead to poor oocyte quality and premature ovarian insufficiency.3 Maternal ageing triggers a series of molecular alterations that may cause a decline in oocyte quality.4 However, only a few targets responsible for oocyte quality decline upon ageing have been identified.
Folliculogenesis is a lengthy, complex process in mammals that yields competent oocytes capable of being fertilized and supporting embryonic development. Follicle activation, growth, and maturation are accompanied by a series of dynamic changes, including granulosa cell proliferation, RNA and protein synthesis, and mitochondrial biogenesis in oocytes.5,6 When follicles develop to the secondary stage, oocytes shift their metabolic mode from glycolysis towards oxidative phosphorylation (OXPHOS) to meet the energy demands required for oocyte development.7 Unlike somatic cells, which contain ∼1000 mitochondria, a mature oocyte contains more than 100,000 mitochondria.1,8 However, it has been reported that mitochondrial quality may be more important than mitochondrial quantity in determining the oocyte quality and survival.4,9
Mitochondrial quality control (MQC) plays a vital role in the maintenance of mitochondrial function. MQC requires proper functioning of the mitochondrial chaperone system, the mitochondrial protease system, mitochondrial dynamics, and mitophagy.10 The accumulation of oxidatively damaged and unfolded proteins in mitochondria induces mitochondrial stresses, activating the mitochondrial unfolded protein response (mtUPR) pathway to rescue mitochondrial function.11 However, prolonged activation of the mtUPR leads to mitochondrial dysfunction and apoptosis.12,13
Mitochondrial ATP-dependent Lon protease 1 (LONP1) is a mitochondrial protease that regulates metabolism and mitochondrial function and responds to reactive oxygen species (ROS)-induced damage in somatic cells.14 LONP1 is involved in proteolysis, the degradation of misfolded proteins, and the cyclic uptake of specific mitochondrial enzymes.15 In addition, LONP1 regulates mitochondrial DNA (mtDNA) replication and mitochondrial biosynthesis and stabilizes the electron transport chain within mitochondria.16 LONP1 mutations or deficiencies are associated with various diseases of the nervous and motor systems, including cerebral, ocular, dental, auricular, and skeletal anomalies (CODAS) syndrome; Parkinson's disease; and amyotrophic lateral sclerosis.17 Despite significant progress in elucidating the roles of LONP1 in somatic cells, how LONP1-mediated MQC contributes to oocyte development and competence remains elusive.
Apoptosis-inducing factor mitochondria-associated 1 (AIFM1) is a mitochondrial inner-membrane-anchored protein with the N-terminus exposed to the mitochondrial matrix and the C-terminal portion exposed to the mitochondrial intermembrane space and is required for the maintenance of mitochondrial respiratory chain complex I in heathy cells.18 However, in response to cell death signalling stimuli, AIFM1 is cleaved by calpains and is then released from mitochondria into the cytosol and transported the nucleus to cause cell death by chromatin condensation and large-scale DNA fragmentation.19,20 HSP70 inhibits the nuclear import of AIFM by direct binding, and as a chaperone, HSP70 also binds LONP1 to facilitate mitochondrial protein folding and maturation.21,22
In the present study, we found that LONP1 is associated with oocyte development and ovarian reserve decline upon ageing. Conditional knockout of Lonp1 in mouse oocytes in both the primordial and growing follicle stages caused defective development of oocytes from secondary follicles into antral follicles and infertility. In addition, LONP1 ablation in mouse oocytes caused prolonged activation of the mtUPR, defective mitochondrial biosynthesis and function, and oocyte death. Mechanistically, LONP1 directly interacts with AIFM1, and LONP1 ablation led to the translocation of AIFM1 from the cytoplasm to the nucleus, causing apoptosis in mouse oocytes. Pathogenic variants of LONP1 are responsible for large-follicle abnormalities in some women with premature ovarian insufficiency. Hence, our findings suggest that LONP1 plays an essential role in oocyte development and is required for female fertility.
Methods
Clinical samples
In the present study, we evaluated two affected families with premature ovarian insufficiency who were recruited from the Center for Reproductive Medicine, Shandong University, China. The phenotypes and related numbers are shown in Table S1. The human studies were approved by the Ethics Review Board of the Center for Reproductive Medicine, Shandong University (IRB No. 2021-45), and all samples were donated voluntarily for scientific research after informed consent was obtained. Human oocytes (unfertilized oocytes after in vitro fertilization (IVF), young women: n=46; old women: n=20) were obtained from donors with approval from the Ethics Committee of Nanjing Drum Tower Hospital. All donors gave written informed consent.
Mice
Wild-type C57/BL6 mice were purchased from the Nanjing Biomedical Research Institute of Nanjing University, China, and Lonp1flox/flox, Gdf9-Cre, Zp3-Cre mice were generated previously.23,24 The mice were maintained under specific pathogen-free conditions in a controlled environment at a temperature of 20±2°C and a humidity of 50–70% on a 12:12 h light:dark cycle, and food and water were provided ad libitum. Mice lacking LONP1 in their oocytes were generated by crossing Lonp1flox/flox mice with Gdf9-Cre or Zp3-Cre mice. We named the mice with Gdf9-Cre-mediated conditional Lonp1 knockout in primordial oocytes “Lonp1Gdf9Cre cKO mice” and the mice with Zp3-Cre-mediated conditional Lonp1 knockout in growing oocytes “LonpZp3Cre cKO mice”. All mouse strains were bred with the C57BL/6 background. Animal care and experimental procedures were performed in accordance with the guidelines of the Experimental Animal Management Committee (Jiangsu, China) and were approved by the Ethics Review Board for Animal Experiments of the Affiliated Drum Tower Hospital of Nanjing University Medical School (No. 20171202). All applicable institutional and/or national guidelines for the care and use of animals were followed.
Controlled ovarian hyperstimulation and IVF
For controlled ovarian hyperstimulation, female mice were injected i.p. with 5 IU of equine chorionic gonadotropin (eCG) (Sansheng Pharmaceuticals, Ningbo, China), followed by 5 IU human chorionic gonadotropin (hCG) (Sansheng Pharmaceutical) 48 h later. After an additional 13 h, cumulus–oocyte complexes (COCs) were released from the oviducts (ampullae), and the number of oocytes was counted after digestion with 1% hyaluronidase (Sigma-Aldrich).9 To achieve IVF, in brief, COCs released from the oviducts of each mouse were placed in 100 µl of HTF medium and cultured in 5% CO2 at 37°C until insemination. After sperm capacitation, spermatozoa were added to droplets of HTF medium containing oocytes. After 4 h of incubation, the oocytes were recovered from the droplets, washed 3 times in M2 medium to remove the remaining spermatozoa and granulosa cells, and cultured in KSOM medium.
Fertility testing
For fertility testing, we continuously mated 6-week-old female Lonp1flox/flox or Lonp1 conditional knockout mice (n=6 for each genotype) with 12-week-old fertile males at a 1:1 ratio for 6 months. The numbers of pups and litters were recorded to determine the fertility rates.
Western blot analysis
For western blotting, we lysed ovary tissues/oocytes in RIPA lysis buffer containing protease inhibitor cocktail for 30 min at 4°C with rotation (100 oocytes/embryos per sample) and heated the lysates for 5 min at 95°C after adding 5 × SDS. The proteins were loaded onto 10% SDS-PAGE gels for electrophoresis, separated by SDS-PAGE, and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Darmstadt, Germany) using conventional methods.25 After immunoblotting with primary antibodies (Table S2), the membranes were washed in TBST and incubated with an HRP-linked goat anti-rabbit secondary antibody. Finally, the bands were detected using an Enhanced Chemiluminescence Detection Kit (Millipore). All the antibodies used in Western blot analysis and the following test were verified with positive control in our lab provided by the manufacturer.
Histological analysis and immunohistochemistry
Ovaries in the diestrous stage were removed and fixed in 10% formalin solution at room temperature overnight with rotation, dehydrated in an ascending series of ethanol solutions (70%, 80%, 90%, 95%, and 100%), and then embedded in paraffin. The ovaries were serially sectioned at a thickness of 5 μm and subjected to HE staining for follicle counting. The number of follicles in distinct stages containing oocytes with a clearly visible nucleus per ovary was counted according to our previous report.9 For immunochemistry, 3 μm-thick sections were deparaffinized and dehydrated with xylene and an ascending series of alcohol solutions. After antigen retrieval, the sections were incubated with primary antibodies (Table S2) at 4°C overnight, incubated with biotin-labelled secondary antibodies for 30 min at room temperature, and developed using a DAB peroxidase substrate kit (Zsbio, Beijing, China).
Real-time PCR (RT-PCR) analysis of oocytes
Oocyte RNA was amplified using a Single Cell Sequence Specific Amplification Kit (Vazyme, Nanjing, China) according to the manufacturer's instructions. For the RT-PCR analysis, we used ChamQ Universal SYBR qPCR Master Mix (Vazyme) and a qTOWER3 Instrument. The primers are listed in Table S2. Relative mRNA levels were determined by normalization to GAPDH mRNA levels (used as an internal control) using the 2−ΔΔt method in GraphPad software ((Version 7, GraphPad Software, San Diego, CA). All measurements were performed in triplicate for each experiment.
Immunofluorescence
Sections of ovaries were deparaffinized and rehydrated with xylene and an alcohol gradient, and heat-induced antigen retrieval was performed in 10 mM sodium citrate buffer (pH 6.0). After permeabilization with 1% Triton X-100 in PBS, the sections were blocked with 3% BSA in PBS for 60 min at room temperature. Oocytes or embryos were fixed in PBS-buffered 4% paraformaldehyde for 30 min at room temperature, followed by permeabilization with 0.5% Triton X-100 in PBS for 25 min. After blocking with 3% BSA in PBS, oocytes or embryos were incubated with primary antibodies (Table S2) diluted in blocking solution at 4°C overnight. We incubated the oocytes or embryos with secondary antibodies for 45 min and then counterstained them with DAPI (Life Technologies, Carlsbad, USA) for 10 min. The oocytes or embryos were mounted on glass slides using SlowFade® Gold Antifade Reagent (Life Technologies) and examined with a confocal laser scanning microscope (Leica, Wetzlar, Germany).
Analysis of the mitochondrial membrane potential
Superovulated oocytes were incubated in M2 medium containing JC-1 (0.6 µg/ml, Sigma Aldrich) or MitoTracker Red CMXROS (200 nM, Invitrogen) at 37°C for 30 min, followed by three washes with 1% BSA in PBS. The distribution and fluorescence intensity of JC-1 monomers and aggregates were captured by fluorescence microscopy (Leica Microsystems, Austria) and analysed using ImageJ software. The mitochondrial membrane potential was determined by measuring the ratio of red (JC-1 monomers) to green (JC-1 aggregates) fluorescence.
Evaluation of mitochondrial ROS levels
Oocytes were incubated in M2 medium containing MitoTracker Green (100 nM, Invitrogen) at 37°C for 30 min, followed by three washes with M2 medium. We then incubated the oocytes with M2 medium containing MitoSOX (0.25 µg/ml, Invitrogen) for an additional 15 min at 37°C. After washing, the distribution and fluorescence intensity of MitoTracker Green or MitoSOX were captured by fluorescence microscopy (Leica) and analysed using ImageJ software. Mitochondria-specific ROS levels were evaluated by measuring the ratio of red to green fluorescence.
Live imaging of oocyte mitochondria
Mitochondria were stained with MitoTracker Red CMXROS (200 nM, Invitrogen) for 30 min, and images were acquired immediately after washing. We collected all imaging data with a PerkinElmer UltraVIEW Vox spinning-disk microscope equipped with a Hamamatsu C9100-23B EMCCD camera and a CFI Apochromat TIRF 100 × objective (NA=1.49).26 For time-lapse imaging, we acquired stacked images containing 70 planes (1 μm/70 μm) every 30 seconds. All imaging experiments were performed at 37°C in the presence of 5% CO2. The data were analysed with Fiji ImageJ software.
Immunoprecipitation followed by liquid chromatography–tandem mass spectrometry (LC–MS/MS)
The 293T cell line was from ATCC. The cell line was verified by short tandem repeat (STR) analysis and routinely tested for mycoplasma contamination by PCR and confirmed negative in our lab. Isolated mitochondria from 293T cells were lysed in RIPA buffer containing protease inhibitor (PI) and protease inhibitor cocktail 2/3(PIC2/3). The lysates were centrifuged at 12,000 × g for 10 min, and the supernatant was subjected to immunoprecipitation with an anti-LONP1 antibody (Proteintech, Chicago, USA) and protein A/G beads. After incubation at 4°C overnight, the beads were washed 3 times with RIPA buffer. We then added 2 × SDS sample buffer to the beads at 95°C in a metal bath for 10 min, and the immunoprecipitated proteins were separated by SDS–PAGE analysis. The gel bands were digested for LC–MS/MS analyses.
Oocyte immunoprecipitation
Approximately 5,000 oocytes in the GV stage were lysed in RIPA buffer containing PI and PIC2/3. The lysates were centrifuged at 12,000 × g for 10 min, and the supernatant was subjected to immunoprecipitation with an anti-LONP1 antibody (Proteintech) and protein A/G beads. After incubation at 4°C overnight, the beads were washed three times with RIPA buffer. We then added 2 × SDS sample buffer to the beads at 95°C in a metal bath for 10 min, and the immunoprecipitated proteins were subjected to western blot analysis.
mtDNA analysis
The procedures used for quantitative RT-PCR (qRT-PCR) analysis of mtDNA were described previously.9 Briefly, one oocyte was placed into a PCR tube with 20 µl of lysis buffer, and after incubation at 55°C for 20 min, proteinase K was heat-inactivated at 90°C for 10 min. The samples were used directly for PCR analysis, and qRT-PCR was performed using a qTOWER3 instrument (Analytik, Jena, Germany). The human mtDNA copy number in blood cells from women with premature ovarian insufficiency was determined by comparing mitochondrial (mt ND5) and nuclear (g ACTIN) DNA levels using qRT-PCR.
TEM
Small pieces of ovarian cortex tissue and oocytes were fixed in 2.5% glutaraldehyde at 4°C for 24 h, and the oocytes were embedded in 2% low-melting agarose with a volume of 1.5 mm × 1.5 mm × 1.5 mm. After rinsing in ddH2O, the agarose containing the oocytes was fixed with 1% osmic acid, dehydrated in gradient ethanol solutions and embedded in resin. Ultrathin sections were double-stained with lead citrate and uranium acetate, and the samples were observed under a transmission electron microscope (JEOL JEM-1010, Japan).
Cell culture and transfection
293T cells were cultured with F12/DMEM containing 10% (v/v) foetal bovine serum (Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin (HyClone Laboratory, South Logan, Utah, USA) at 37°C and 5% CO2 in compressed air at high humidity. LONP1 siRNA (Ruibo, Guangzhou, China) was transfected into 293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), and the cells were collected 48 h after transfection for further investigation.
SMART RNA-seq analysis of oocytes
Total oocyte RNA was reverse-transcribed into cDNA and full-length amplified using a Discover-scTM WTA Kit V2 (#N711, Vazyme) according to the manufacturer's instructions. We purified the amplified cDNA products using Aliquot VAHTS DNA Clean Beads, and quality was assessed with an Agilent 2100 Bioanalyzer. We then used 1 ng of cDNA as starting material for library construction with a TruePrep DNA Library Prep Kit V2 for Illumina® (#TD503, Vazyme). Library sequencing and analysis were performed by GMINIX (Shanghai). GO enrichment analysis was performed with Metascape.27 Heatmaps were generated with TBtools.28
Whole-exome sequencing and variant analysis
Whole-exome sequencing was performed using peripheral blood from affected individuals. Functional prediction was performed using PolyPhen2 and the Mutation Taster program. The positions of mutated amino acids in the protein structure were analysed using the published structure of human LONP1.22 Variants were filtered using the following criteria: (1) variants with minor allelic frequencies less than 1% in the gnomAD, 1000 Genomes Project, and ExAC databases and not present in our control database and (2) variants functionally predicted to be damaging by at least two prediction software programs.
Statistics
Statistical analysis was performed with GraphPad Prism (version 7). Means were compared using an independent samples Student's t test or one-way ANOVA. The data are shown as the mean ± SEM. P values less than 0.05 were considered statistically significant.
Results
LONP1 is associated with oocyte development and ovarian ageing
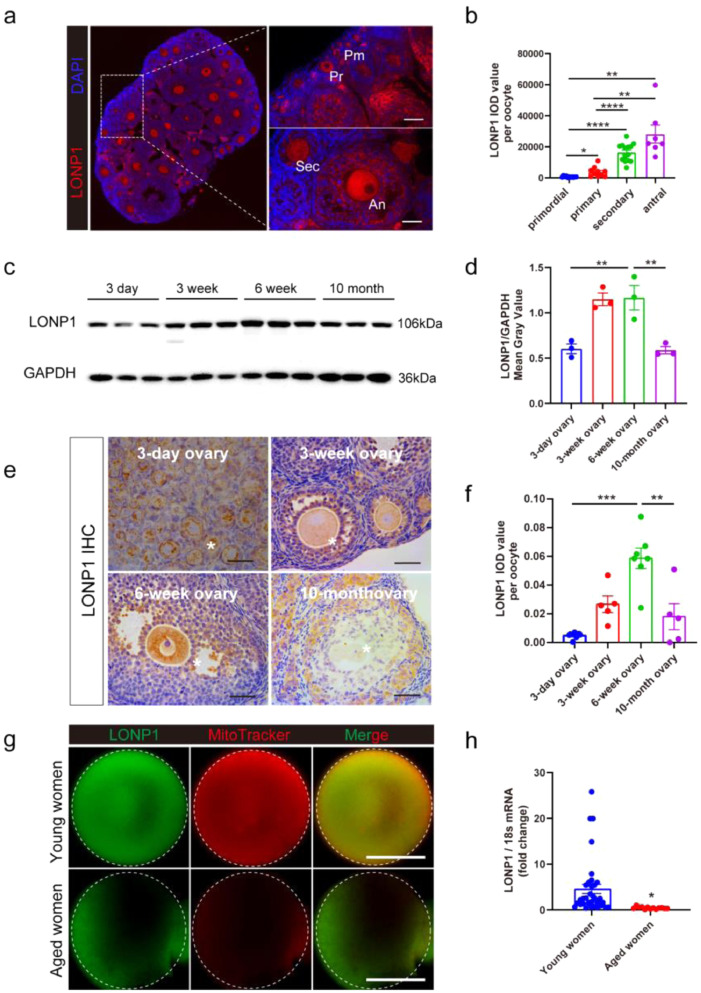
LONP1 expression during follicular development and ovarian ageing in mice and women was investigated. As shown in Figure. 1a, b, LONP1 was expressed in both oocytes and granulosa cells in all stages of follicular development, and the expression levels of LONP1 in oocytes gradually increased during follicular development (peaking in oocytes from secondary and antral follicles) in mice. Immunofluorescence analysis also revealed that LONP1 was highly expressed and mainly localized in the mitochondrial matrix in growing oocytes and preimplantation mouse embryos (Fig. S1).
Figure 1.
LONP1 is associated with oocyte development and ovarian ageing.
(a and b) Immunofluorescence staining of LONP1 in the ovaries of 3-week-old wild-type mice; Pm indicates oocytes from primordial follicles, Pr indicates oocytes from primary follicles, Sec indicates oocytes from secondary follicles, and An indicates oocytes from antral follicles. All oocytes from the ovaries of six 3-week wild-type mice were used for statistical analysis (*p < 0.05, **p < 0.01, ****p < 0.0001, one-way ANOVA). Scale bar = 50 μm. The integrated optical density (IOD) of each oocyte from follicles in different developmental stages was analysed using Image-Pro Plus 6.0. (c and d) western blot analysis of the expression of LONP1 in the ovaries of 3-day-old, 3-week-old, 6-week-old and 10-month-old mice (n=3, **p < 0.01, Student's t test). (e and f) Immunohistochemical analysis of the expression of LONP1 in oocytes in ovarian sections from 3-day-old, 3-week-old, 6-week-old and 10-month-old mice. The IOD of each oocyte from at least 5 mice was analysed (**p < 0.01, ***p < 0.001, one-way ANOVA). Scale bar = 50 μm. The asterisks indicate oocytes. (g) Immunofluorescence staining of LONP1 in the MII oocytes of young (<28 y) and aged (>38 y) women (n=10 vs. n=7). (h) qRT-PCR analysis of LONP1 in the MII oocytes of young (<28 y) and aged (>38 y) women (n=36 vs. n=13, p=0.0002).
In addition, the expression of LONP1 in oocytes was significantly elevated in the ovaries of 3-week-old and 6-week-old mice but markedly decreased in the ovaries of 10-month-old mice (Figure. 1c–f). Immunofluorescence analysis showed that the protein level of LONP1 in oocytes from aged women (ranging from 38 to 45 years old) was lower than those from young women (ranging from 21 to 28 years old) (Figure. 1g). Similarly, the mRNA expression level of LONP1 in oocytes from aged women was lower than that in oocytes from young women (Figure. 1h, 0.45 ± 0.07, n=13 vs. 4.60 ± 1.0, n=36; p = 0.0002). These results suggest that LONP1 is associated with oocyte development and ovarian ageing.
Lonp1 conditional knockout leads to infertility and compromised oocyte development
Because LONP1 is expressed in oocytes during all stages of follicular development, we then attempted to generate oocyte-specific and developmental stage-specific Lonp1-knockout mice (Lonp1Gdf9Cre cKO/LonpZp3Cre cKO mice) to assess the roles of LONP1 in oogenesis (Fig. S2).
As shown in Fig. S3a, b, and e, LONP1 was not detected in oocytes in Lonp1Gdf9Cre cKO mouse ovaries. In Lonp1Zp3Cre cKO mice, the expression of LONP1 appeared normal in primordial follicles but was significantly reduced in the oocytes of growing follicles (Fig. S3c–e). Six-month fertility testing was performed and indicated that unlike female Lonp1flox/flox mice, both Lonp1Gdf9Cre cKO and LonpZp3Cre cKO female mice were infertile (Fig. S3f, g). Collectively, these results indicate that LONP1 in oocytes is indispensable for oocyte development and female fertility.
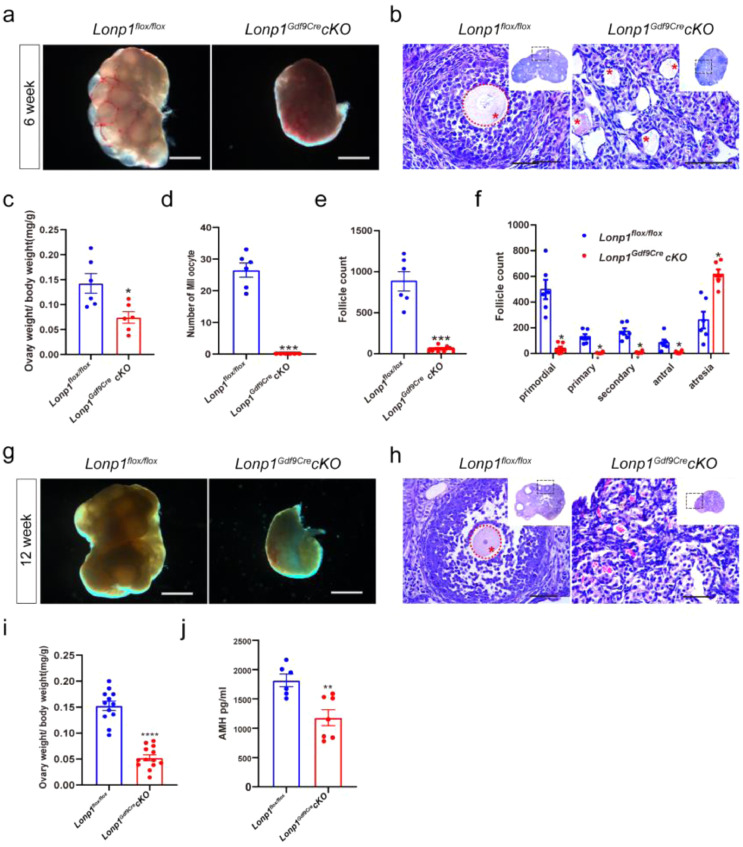
Lonp1Gdf9Cre cKO mice at 3 weeks of age have an indistinguishable ovary morphology and ovary index. However, the embryo development potential is significantly impaired after fertilization (Fig. S4; for details, see the supplemental text). In the 4th and 5th weeks, the cytoplasm of many oocytes in the ovarian medulla of Lonp1Gdf9Cre cKO mice shrunk significantly (Figs. S5, S6; for details, see the supplemental text). In the 6th week, the size of the ovaries was reduced significantly, and few antral follicles were present in the ovaries of Lonp1Gdf9Cre cKO mice (Figure. 2a, b). In addition, the ovary index of Lonp1Gdf9Cre cKO mice was significantly decreased (0.07 ± 0.01 vs. 0.14 ± 0.02, n=6, p=0.013, Figure. 2c), and Lonp1Gdf9Cre cKO mice no longer produced metaphase II (MII) oocytes after controlled ovarian hyperstimulation (0 vs. 26.50 ± 2.22, n=6, p<0.0001, Figure. 2d). As shown in Figure. 2e and f, the total number of follicles was significantly decreased (60.0 ± 16.38 vs. 882.5 ± 117.60, n=6, p=0.0008), as was the number of growing follicles, especially secondary and antral follicles (8.33 ± 3.58 vs. 171.70 ± 25.81, n=6, p=0.0013; 9.17 ± 3.96 vs. 83.33 ± 24.72, n=6, p=0.0296). Surprisingly, Lonp1 knockout caused the complete depletion of all follicles at the 12th week in Lonp1Gdf9Cre cKO mice (Figure. 2g–j). These results indicate that LONP1 plays a vital role in the development of growing follicles (especially secondary follicles).
Figure 2.
Oocyte-specific knockout of Lonp1 compromises oocyte development in mice. (a) Photographs of 6-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice. Scale bar = 500μm. (b) Micrographs of HE-stained ovarian sections from 6-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice. The asterisks indicate oocytes. Scale bar = 50 μm. (c) Ovary index values of 6-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=6, *p < 0.05, Student's t test). (d) Numbers of superovulated MII oocytes in 6-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=6, ***p < 0.001, Student's t test). (e and f) Total follicle numbers and numbers of follicles in different developmental stages per ovary in 6-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=6, *p < 0.05, ***p < 0.001, Student's t test and one-way ANOVA test). (g) Photographs of ovaries from 12-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice. Scale bar = 500 μm. (h) Micrographs of HE-stained ovarian sections from 12-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice. Scale bar = 500 μm. The asterisks indicate oocytes. (i) Ovary index values of 12-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (at least 6 mice per group were included, ****p < 0.0001, Student's t test). (j) Serum concentrations of anti-Mullerian hormone (AMH) in 12-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=6, **p < 0.01, Student's t test).
Furthermore, the ovaries of Lonp1Zp3Cre cKO mice were smaller than those of wild-type controls at 10 weeks of age. Growing oocytes of Lonp1Zp3Cre cKO mice exhibited abnormal morphology, and their embryo development potential was also significantly impaired after fertilization (Fig S7; for details, see the supplemental text).
Thus, the ablation of LONP1 in oocytes in both the primordial and growing follicular stages compromises oocyte development and results in premature ovarian insufficiency in mice.
Conditional Lonp1 knockout induces apoptosis-related gene expression in oocytes
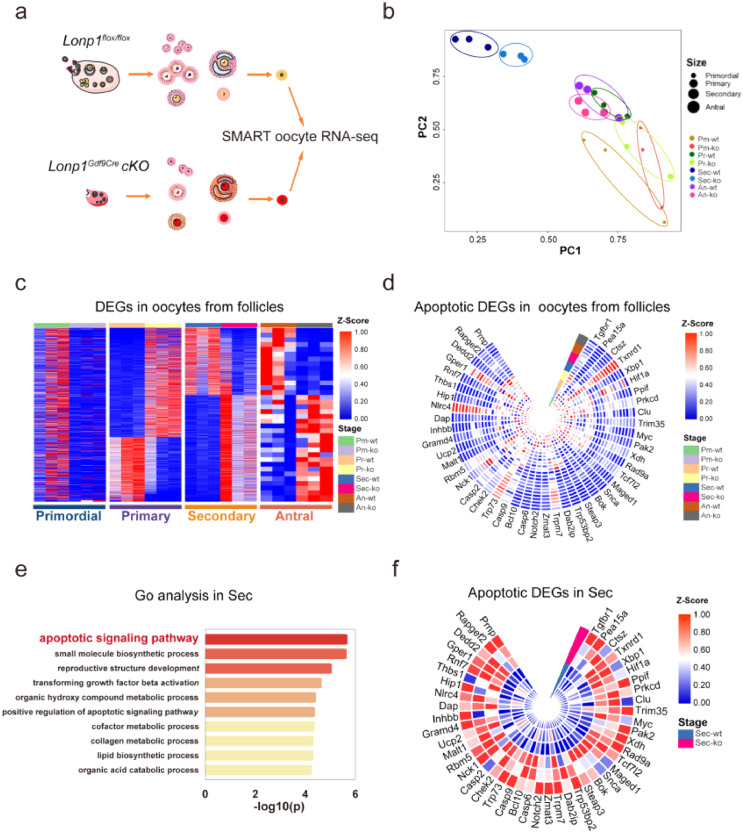
To explore the mechanisms underlying LONP1-dependent oogenesis, we performed SMART RNA-seq on oocytes from follicles in different developmental stages (the primordial, primary, secondary, and antral stages) and MII oocytes from 3-, 4-, and 5-week-old Lonp1Gdf9Cre cKO and Lonp1flox/flox mice (Figure. 3a). There were significant changes in gene expression between oocytes in secondary follicles from Lonp1Gdf9Cre cKO mice and those from Lonp1flox/flox mice, with 1929 differentially expressed genes (Figure. 3b, c). GO enrichment analysis revealed that Lonp1 knockout in mouse oocytes caused upregulation of the expression of genes involved in apoptotic signalling pathways and downregulation of the expression of genes involved in meiotic chromosomal segregation (Figure. 3d, e; Fig. S8a, b). Specifically, the expression levels of genes associated with apoptosis in oocytes from secondary follicles (i.e., Casp9, Casp6, Casp2, Ppif, Malt1, Dap, Dedd2, Maged1, Steap3, Myc, Bok, and Clu) were significantly increased (fold change > 1.5, p < 0.05) (Figure. 3f).
Figure 3.
Oocyte-specific knockout of Lonp1 induces apoptosis-related gene expression in oocytes from secondary follicles. (a) Representative pictures showing the flowchart of SMART RNA-seq analysis of Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mouse oocytes in different developmental stages and MII oocytes from Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice of different ages (n=3 for each genotype). (b) PCA of oocytes in different developmental stages from Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 for each genotype). (c) Analysis of differential gene expression in oocytes in different follicle developmental stages between Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 from each genotype). (d) GO enrichment analysis of apoptosis-related genes in oocytes from follicles in different developmental stages from Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 for each genotype). (e) GO enrichment analysis of the top-ten most enriched biological processes for upregulated genes in the secondary follicle stage between Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 for each genotype). (f) Representative genes related to the apoptotic signalling pathway expressed in oocytes from follicles in the secondary stage in Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 for each genotype). Pm indicates oocytes from primordial follicles, Pr indicates oocytes from primary follicles, Sec indicates oocytes from secondary follicles, and An indicates oocytes from antral follicles.
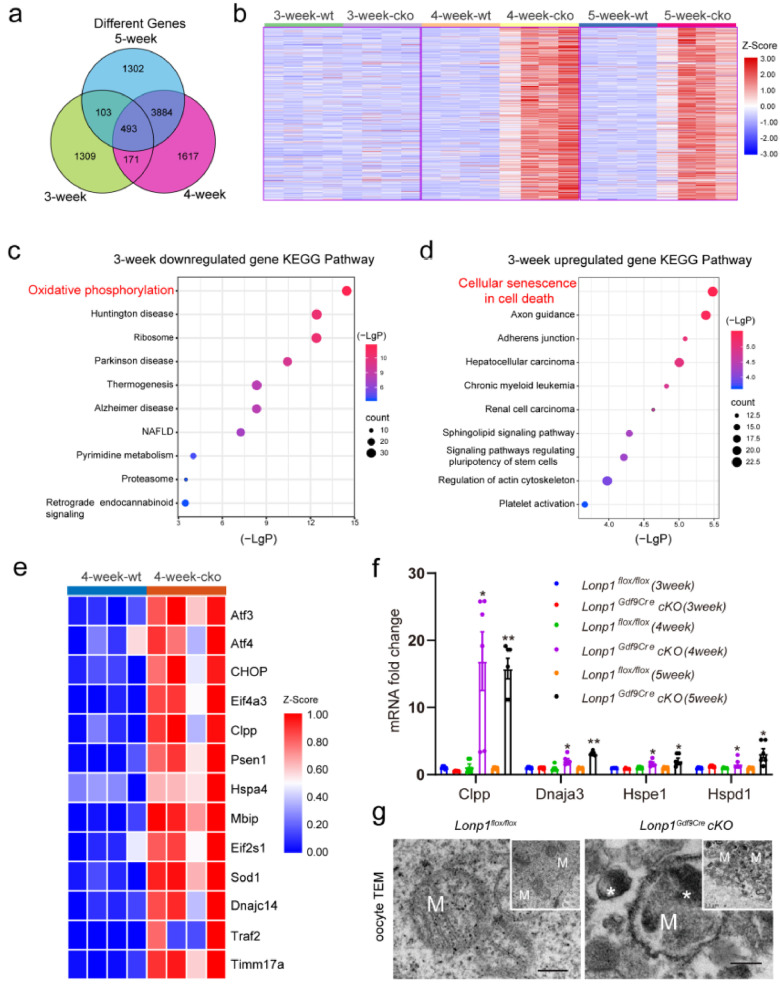
Moreover, transcriptomic analysis of MII oocytes from 3-, 4-, and 5-week-old Lonp1Gdf9Cre cKO and Lonp1flox/flox mice revealed 2076, 6165, and 5782 significantly differentially expressed genes, respectively (Figure. 4a). Further self-organizing map (SOM) analysis also showed that transcript levels were significantly increased in MII oocytes from 4- and 5-week-old Lonp1Gdf9Cre cKO mice (Figure. 4b). GO enrichment analysis showed that OXPHOS was compromised, the expression of genes related to cell death was activated in oocytes from 3-week-old Lonp1Gdf9Cre cKO mice (Figure. 4c, d) and ribosomal biogenesis was enhanced in oocytes from 4-week-old and 5-week-old Lonp1Gdf9Cre cKO mice (Fig. S8c, d). Specifically, these data show that the upregulated genes were related to persistent mitochondrial stress and mtUPR activation in Lonp1Gdf9Cre cKO mouse oocytes (Figure. 4e). The expression levels of the mtUPR markers Clpp, Dnaja3, Hspe1 and Hspd1 were also significantly elevated (fold change > 1.5, p < 0.05) (Figure. 4f, Fig. S9). Additionally, a large number of electron-dense aggregates related to the mtUPR were present in the mitochondrial matrix of oocytes from Lonp1Gdf9Cre cKO mice (Figure. 4g), suggesting prolonged activation of the mtUPR after LONP1 ablation.
Figure 4.
Bioinformatics analysis showing prolonged mtUPR in Lonp1 knockout MII oocytes. (a) Venn diagram illustrating the relationships among the differentially expressed genes identified by RNA-seq analysis of MII oocytes from 3-, 4-, and 5-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (b) SOM analysis of genes with altered transcript levels identified by RNA-seq analysis of MII oocytes from 3-, 4-, and 5-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (c) KEGG enrichment analysis of significantly downregulated genes between MII oocytes from 3-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (d) KEGG enrichment analysis of significantly upregulated genes between MII oocytes from 3-week-old Lonp1loxp/loxp mice and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (e) Differentially expressed genes involved in the mitochondrial stress response between MII oocytes from 4-week-old Lonp1loxp/loxp mice and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (f) qRT-PCR analysis of the expression of the mtUPR markers Clpp, Dnaja3, Hspe1 and Hspd1 in MII oocytes from Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=6, *p < 0.05, **p < 0.01). (g) Representative TEM images showing typical mitochondria in MII oocytes from Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 for each genotype). The asterisks indicate high-electron-density protein accumulation. Scale bar=0.5 μm.
LONP1 ablation impairs mitochondrial function and induces apoptosis in oocytes
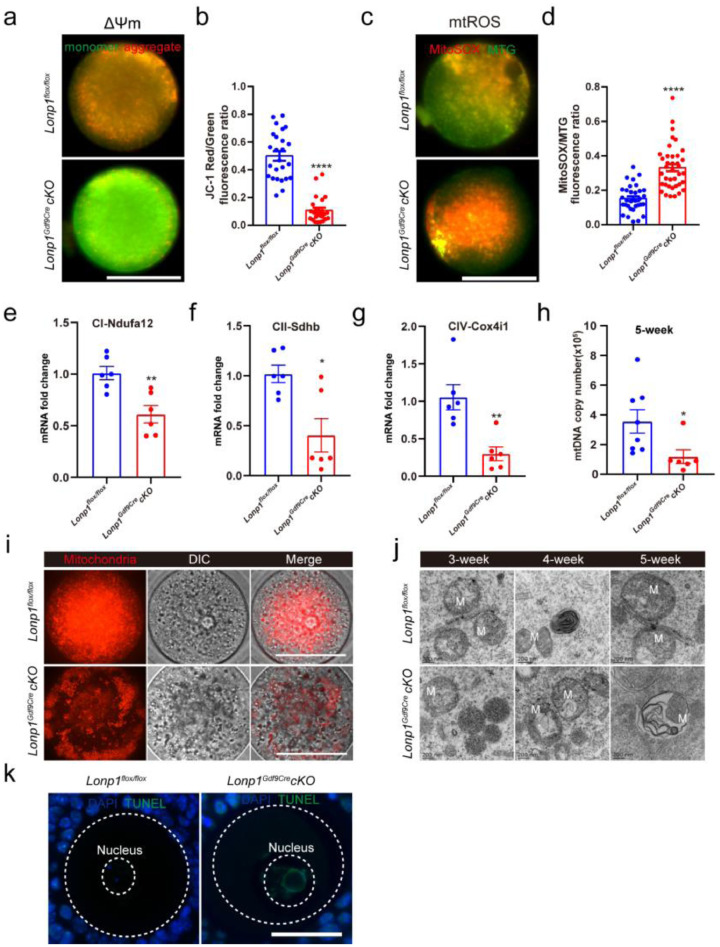
To assess the effects of Lonp1 knockout on the intracellular homeostasis of oocytes, mitochondrial functions were analysed in Lonp1Gdf9Cre cKO mice and Lonp1flox/flox mice. As shown in Figures. 5a, b and S10a, LONP1 ablation decreased the mitochondrial membrane potential but increased the levels of ROS in the mitochondria of MII oocytes (mitochondrial membrane potential, 0.11 ± 0.02, n=24 vs. 0.50 ± 0.03, n=25, p< 0.001; mitochondrial ROS levels, 0.33 ± 0.02, n=38 vs. 0.15 ± 0.01, n=32, p< 0.001) (Figure. 5c, d). Moreover, as shown in Figure. 5e–g, the expression levels of mitochondrial electron transport chain genes such as Ndufa12, Sdhb, and Cox4i-1 were significantly attenuated (Ndufa12, 0.61 ± 0.08 vs. 1.01 ± 0.07, n=6, p=0.0036; Sdhb, 0.30 ± 0.09 vs. 1.05 ± 0.17, n=6, p= 0.0026; Cox4i-1, 0.40 ± 0.17 vs. 1.02 ± 0.09, n=6, p= 0.0082). The mtDNA copy number was also diminished in the MII oocytes of 5-week-old Lonp1Gdf9Cre cKO mice (117614 ± 46427, n=6 vs. 355465 ± 78963, n=8, p=0.035, Figure. 5h). As shown in Figures. 5i and S10a and b, the absence of LONP1 caused abnormal mitochondrial distribution in oocytes. Consistently, transmission electron microscopy (TEM) analysis revealed the presence of abnormal mitochondrial structures in the GV oocytes of Lonp1Gdf9Cre cKO mice (Figure. 5j), indicating mitochondrial-related oocyte apoptosis and death.
Figure 5.
Oocyte-specific knockout of Lonp1 impairs mitochondrial biosynthesis and function. (a) Representative pictures of Lonp1loxp/loxp (upper) and Lonp1Gdf9Cre cKO (lower) mouse MII oocytes stained with JC‐1 (from 4-week-old mice of each genotype). Scale bar=50 μm. (b) Ratios of red:green JC‐1 fluorescence in Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mouse MII oocytes (n>24) from at least six mice (****p < 0.0001, Student's t test). (c) Representative pictures of Lonp1loxp/loxp (upper panel) and Lonp1Gdf9Cre cKO (lower panel) mouse MII oocytes stained with MitoTracker Green and MitoSOX (from 4-week-old mice of each genotype). Scale bar = 50 μm. (d) Ratios of MitoSOX:MitoTracker Green fluorescence in Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mouse MII oocytes (n>32) from at least six mice (****p < 0.0001, Student's t test). (e–g) qRT-PCR analysis of the expression of COX4i-1, Sdhb and Ndufa12 in Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mouse MII oocytes (n=6, *p < 0.05, **p < 0.01, Student's t test). (h) mtDNA copy numbers in oocytes from 5-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (at least six MII oocytes were included, *p < 0.05, Student's t test). (i) Representative immunofluorescence images showing the mitochondrial distributions in 4-week-old Lonp1loxp/loxp (upper) and Lonp1Gdf9Cre cKO (lower) mouse GV oocytes. Scale bar = 50 μm. (j) Representative TEM images showing mitochondria in GV oocytes from 3-, 4-, and 5-week-old Lonp1loxp/loxp (upper panel) and Lonp1Gdf9Cre cKO (lower panel) mice (n=3 for each genotype). M indicates mitochondria. Scale bar=0.2 μm. (k) TUNEL staining of oocytes from Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=6). Scale bar = 50 μm.
Specifically, in oocytes from growing follicles of 3-week-old Lonp1 knockout mice, the mitochondria were amorphous, and the mitochondrial membrane was slightly swollen (Figure. 5j). In Lonp1-knockout mouse oocytes, mitochondrial morphology was atypical, and the cristae structures were initially blurred at 4 and 5 weeks of age, showing vacuoles, irregular membrane structures and the disappearance of cristae structures (Figure. 5j). The disruption of mitochondria-related apoptosis was also confirmed by TUNEL staining of Lonp1-knockout oocytes (Figure. 5k). Furthermore, MII oocytes from Lonp1Gdf9Cre cKO mice exhibited spindle formation deficits and abnormal chromosomal segregation (Fig. S11a, b). Together, these results suggest that LONP1 is required for the normal functioning of mitochondria and antiapoptotic processes in oocytes.
LONP1 ablation causes AIFM1 nuclear translocation in oocytes
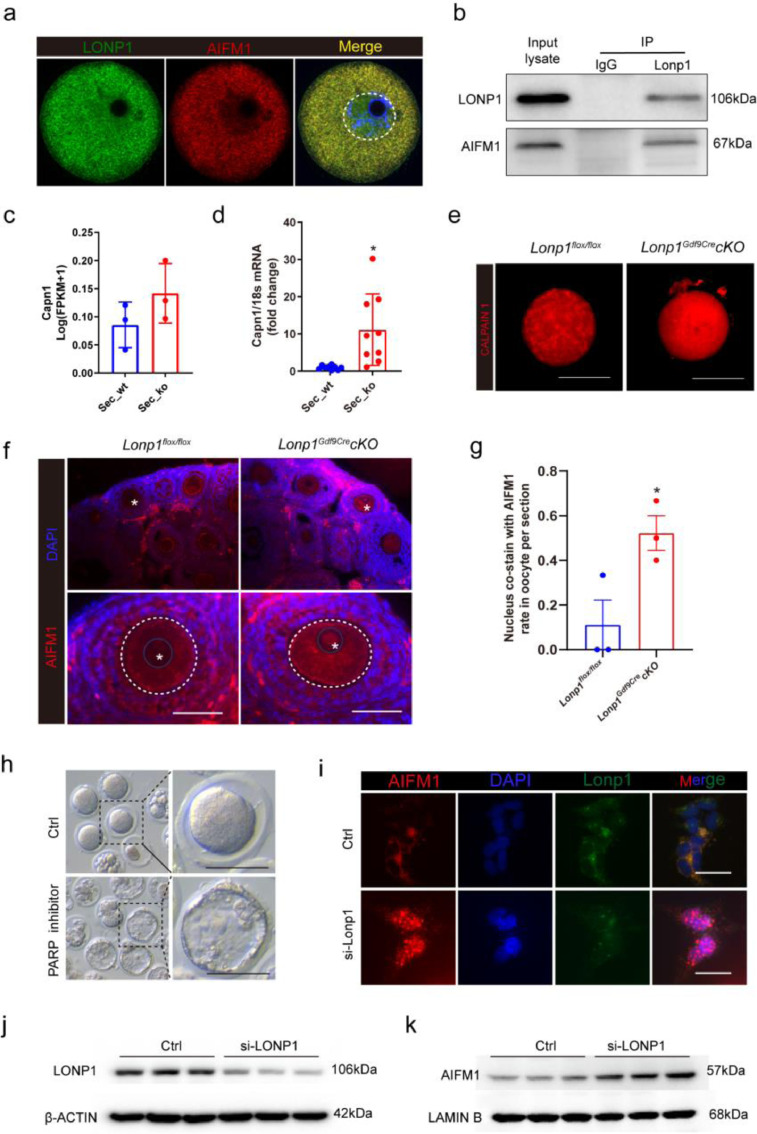
To determine the molecular association between LONP1 and apoptosis, endogenous coimmunoprecipitation (Co-IP) of cell lysates followed by MS was performed. The results revealed that AIFM1 associates with LONP1 (Table S3), which was confirmed by mitochondrial immunofluorescence and coimmunoprecipitation analysis of mouse GV oocytes (Figure. 6a, b). Calpain 1, which cleaves AIFM1, was significantly upregulated in GV oocytes from Lonp1Gdf9Cre cKO mice according to qRT-PCR and immunofluorescence analyses (Figure. 6c–e). Consistently, more AIFM1 translocated into the nucleus in oocytes from Lonp1Gdf9Cre cKO mice than in those from Lonp1flox/flox mice (0.52 ± 0.08 vs. 0.11 ± 0.11, n=3, p=0.044, Figure. 6f, g; Fig. S12). Pre-treatment with an AIFM1-induced apoptosis inhibitor (3-aminobenzamide) partly rescued Lonp1Gdf9Cre cKO mouse blastocyst formation (0% vs. 18.7%, p<0.01), suggesting an improvement in oocyte quality (Figure. 6h). Additionally, in somatic 293T cells, AIFM1 translocated from the cytoplasm to the nucleus after Lonp1 knockdown (Figure. 6i). Nuclear and cytoplasmic separation experiments after Lonp1 knockdown also showed that AIFM1 undergoes nuclear translocation (Figure. 6j and k). The above results indicate that apoptosis triggered by AIFM1 may be one of the important causes of premature ovarian insufficiency after LONP1 ablation in mouse oocytes.
Figure 6.
Oocyte-specific knockout of Lonp1 leads to AIFM1 nuclear translocation.
(a) LONP1 colocalized with AIFM1 in wild-type mouse oocytes, as indicated by immunofluorescence analysis (at least 30 GV oocytes were included). (b) Immunoprecipitation of LONP1 and AIFM1 in mouse oocytes (approximately 5000 wild-type GV oocytes were used). (c) FPKM values of Capn1 in oocytes from secondary follicles of Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 for each group). (d) qRT-PCR analysis of Capn1 in oocytes from secondary follicles of Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=9 for each group). (e) Immunofluorescence staining of CALPAIN1 in oocytes from secondary follicles of Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice. Scale bar = 50 μm. (f and g) Immunofluorescence staining of AIFM1 in the ovaries of 3-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 for each genotype). The asterisks indicate nuclei (*p < 0.05, Student's t test). Scale bar = 50 μm. (h) Blastocyst formation in oocytes from 3-week-old Lonp1GdfCre cKO mice after pretreatment with the PARP inhibitor 3-aminobenzamide to rescue AIFM1-induced apoptosis (n>30). (i) Immunofluorescence staining of AIFM1 in 293T cells after Lonp1 knockdown. Scale bar = 20 μm. Scale bar = 50 μm. (j and k) Immunoprecipitation of LONP1 and AIFM1 from the cytoplasmic and nuclear proteins of 293T cells after Lonp1 knockdown.
LONP1 mutation is associated with premature ovarian insufficiency in women
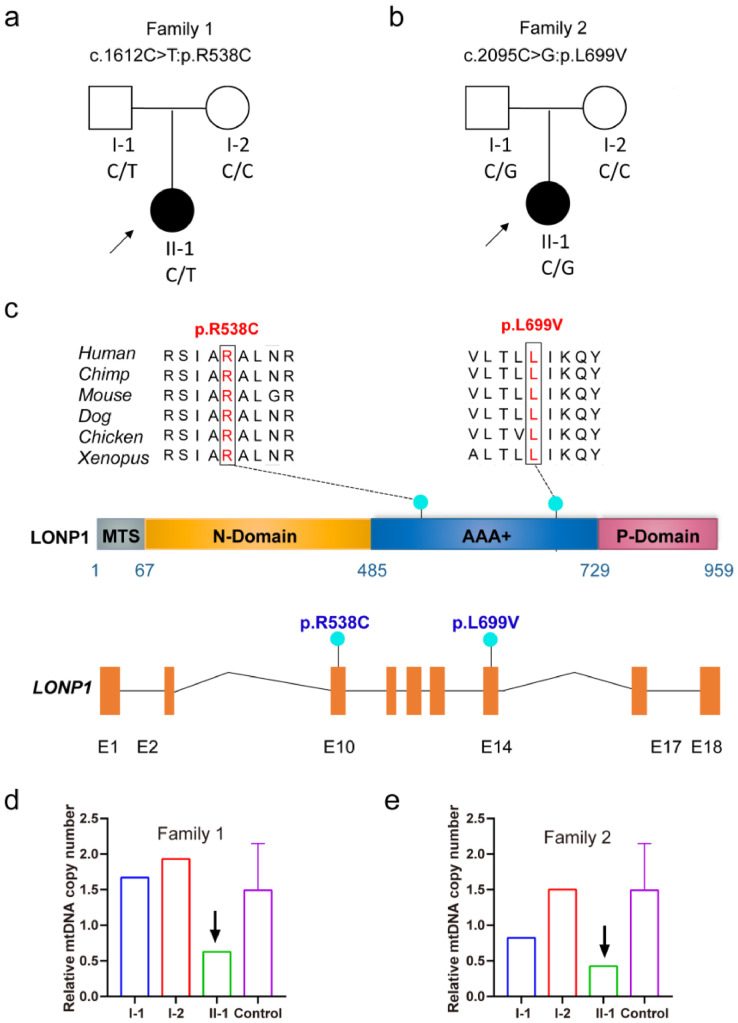
To investigate the association between LONP1 and the ovarian reserve in women, we recruited two unrelated families of women who had exhibited premature ovarian insufficiency and infertility for more than 3 years. The women had normal intelligence without other systemic abnormalities or a family history of other diseases. The proband of family 1 exhibited oligomenorrhea at 19 years old and menopause at 25 years old. No large antral follicles (>10 mm) were observed during controlled ovarian hyperstimulation (Figure. 7a). The proband of family 2 had exhibited a prolonged menstrual cycle since menarche, with a menstrual cycle of approximately 90–120 days. Only a few small antral follicles of approximately 1 mm were observed (Figure. 7b). Two potentially pathogenic heterozygous missense variants of LONP1 (c.1612C>T:p.R538C and c.2095C>G:p.L699V) with a minor allelic frequency <0.1% in three population databases (gnomAD, 1000 Genomes Project, and ExAC) and that may contribute to the occurrence and development of premature ovarian insufficiency were identified in the two families (Figure. 7c, Table S1).
Figure 7.
Identification of pathogenic variants of LONP1 associated with premature ovarian insufficiency. (a and b) Pedigrees of the two affected families. The black circles represent the affected individuals. (c) Mutation patterns and conservation of mutated amino acids in LONP1. The green dots indicate the mutations identified in this study. All mutations were missense mutations. (d and e) Relative mtDNA copy numbers in the two affected families.
Due to the importance of LONP1 function and constitutive expression in cells, many individuals with pathogenic homozygous LONP1 mutations seldom survive to reproductive age have children. Therefore, we were only able to recruit premature ovarian insufficiency patients with pathogenic heterozygous LONP1 variants. As predicted by PolyPhen-2 and Mutation Taster, the identified pathogenic variants are loss-of-function alleles and damaging missense variants of LONP1. The damaging missense variants are located in the ATPase domain on the surface of the LONP1 hexamer and thus may affect the proper functioning of LONP1 (Fig. S13). In addition, we also examined the mtDNA content in the blood cells of the two probands with LONP1 variations (Figure. 7d, e). The mtDNA/nDNA copy numbers in the probands of family 1 (629) and family 2 (425) were much lower than those in the probands of the control women (1491±643). This finding indicates that these LONP1 mutations reduce the mtDNA content in the cells of women with premature ovarian insufficiency.
Discussion
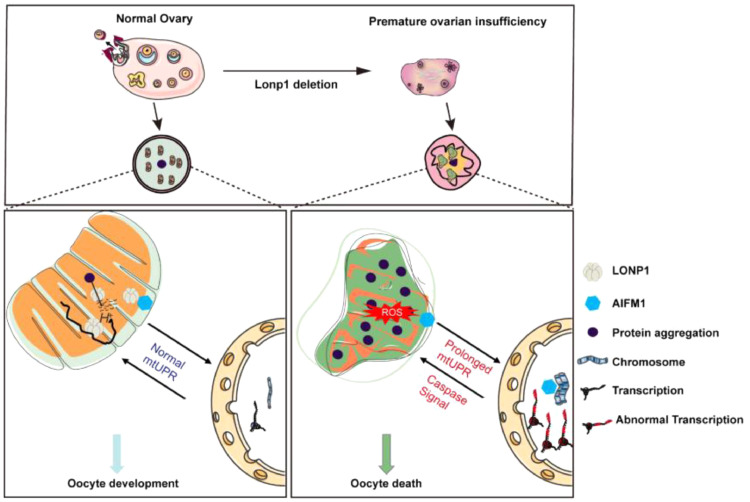
LONP1 is a protease responsible for degrading misfolded or damaged proteins within mitochondria and regulating numerous intramitochondrial or extramitochondrial processes in somatic cells.14,16 Although LONP1 mutations in humans have been found to be associated with severe diseases, the roles of LONP1 in mammalian reproduction remain elusive. In the present study, we report that LONP1 is associated with ovarian ageing and that several pathogenic LONP1 variants are associated with premature ovarian insufficiency. We further demonstrated that LONP1 is required for the proper functioning and biogenesis of mitochondria and the maintenance of mitochondrial morphology in oocytes. The absence of LONP1 appears to lead to excessive activation of the mtUPR in oocytes and subsequently to compromise of the oocyte quality and ovarian reserve, leading to infertility. The absence of LONP1 also causes the translocation of AIFM1 to the nucleus in the secondary follicle stage, promoting oocyte apoptosis and death (Figure. 8).
Figure 8.
Mechanism by which LONP1 maintains mammalian oocyte development and survival.
Because systemic knockout of Lonp1 is lethal to mouse embryos,29 we generated oocyte-specific knockout mouse models to study the roles of LONP1 during oocyte development and maturation. In mice with Gdf9-Cre-mediated oocyte-specific Lonp1 knockout, ovarian and early oocyte development appeared normal from birth until 3 weeks of age, as these mice showed a similar number of follicles to that of the controls. Surprisingly, in 6-week-old mice with oocyte-specific Lonp1 knockout, almost all oocytes in the secondary and antral follicular stages underwent death, and the ovarian reserve declined sharply. Similarly, women with pathological LONP1 variants showed abnormal large follicular development and premature ovarian insufficiency. These results indicate that LONP1 plays a vital role in regulating follicular development and survival.
Mitochondrial dysfunction is one of the primary causes of oocyte quality decline and death.30 The roles of LONP1 in mitochondrial function in cultured somatic cells have been reported previously.31,32 Mitochondrial replication takes place throughout the early process of oogenesis, but we found that LONP1 expression accompanies mitochondrial biogenesis in oocytes. Moreover, Lonp1 knockout in oocytes impairs mitochondrial function in the secondary follicle stage, when large-scale biogenesis of mitochondria begins. In addition, the expression of genes related to oxidative respiration and the electron transport chain was significantly downregulated in Lonp1Gdf9Cre cKO mice after the secondary follicle stage. These findings highlight that LONP1 in oocytes regulates mitochondrial function and OXPHOS, especially in the mammalian follicle growth stage. Interestingly, LONP1 appears to regulate the mitochondrial distribution and dynamics in oocytes. During meiosis in oocyte maturation, mitochondria appear to undergo fission more frequently than fusion.33 In contrast, LONP1 deletion in oocytes alters the balance of mitochondrial dynamics and leads to mitochondrial dysfunction. Similarly, Lonp1 knockout in the heart affects Opa1, as we reported previously, causing an imbalance between fission and fusion.23
Systemic knockout of mitochondrial proteases involved in MQC other than LONP1 (including CLPP, AFG3L2, and OMA1) does not cause embryonic lethality and allows embryos to develop normally.34, 35, 36 SMART RNA-seq analysis of oocytes after LONP1 ablation revealed that the expression of a large number of genes, especially mtUPR- and ribosome-related genes, was significantly upregulated in MII oocytes. Recent research has also shown that LONP1 is associated with RNA degradation and mitochondrial gene transcription.37 In mammalian somatic cells, LONP1 is one of the components of mitochondrial nucleoids, which are structures that pack mtDNA tightly into DNA-protein assemblies.15 Our transcriptomic analysis indicated that LONP1 may also play a role in ribosomal biogenesis and mRNA stabilization in oocytes.
LONP1 is involved in several cellular functions, including cellular metabolism and mitochondrial function, and LONP1 knockdown or inhibition leads to cell death in several somatic cell lines.38 Interestingly, we found that LONP1 interacts with AIFM1 to promote oocyte survival during follicular development. Only a few LONP1-interacting proteins have recently been identified, e.g., Hsp60/mtHsp70, p53, and COX4-1 subunits.22,39,40 The interaction of AIFM1 with LONP1 ensures the mitochondrial localization of AIFM1 within oocytes because Lonp1 knockout leads to nuclear AIFM1 translocation and induces apoptosis. And we also found that the Calpain 1, as a calcium-activated neutral protease, was significantly increase which may cleavage AIFM1 to soluble nonmembrane form and leads to nuclear translocation. Nuclear localization of AIFM1 affects chromosomal condensation and leads to chromosome fragmentation, inducing the release of the mitochondrial apoptogenic factors cytochrome c and caspase 9.41
During follicular growth, only a few follicles can grow and successfully develop to ovulation, and over 99% of follicles undergo atresia during development.42 Premature ovarian insufficiency is characterized by accelerated follicular atresia, and mitochondrial dysfunction may promote this process.43,44 However, the mechanisms underlying premature ovarian insufficiency remain unknown. In the present study, we identified pathogenic missense LONP1 variants associated with female infertility and premature ovarian insufficiency. Pathogenic LONP1 mutations and abnormal LONP1 activity cause multiple diseases in humans,17 and mutations have mainly been found in the AAA+ and protease domains, which are the Lon family protein regions that are conserved across species.45, 46, 47, 48 Similar to the LONP1 mutations found in some CODAS patients, LONP1 mutations in women with premature ovarian insufficiency are located in the AAA+ domain.49 In general, women with premature ovarian insufficiency who carry pathogenic heterozygous LONP1 mutations can undergo puberty. It is conceivable that different LONP1 mutations affect gene functions in a variety of ways and that pathogenic heterozygous LONP1 mutations in humans can lead to reproductive disorders earlier in puberty.15 However, how the identified pathogenic mutations affect LONP1 function and how to use LONP1 wild-type mRNA or proteins to rescue oocytes from women with premature ovarian insufficiency need further investigation.
In conclusion, we established that LONP1 plays a crucial role in oogenesis. LONP1 not only is involved in the regulation of mitochondrial functions but also plays a role in pathological processes involved in oocyte death by interacting with AIFM1. In-depth analysis of LONP1 will be crucial for elucidating the mechanisms underlying ovarian insufficiency.
Declaration of Competing Interest
The authors have declared that no conflicts of interest exist.
Acknowledgments
Author contributions
XS, LM, and GY contributed equally to this work. XS, LM, GY, YQ, HS, and LD were responsible for the experimental design and data interpretation. XS, LM, GL, JZ and LD performed the experiments. JL, YY, ZXL, XZ, YZ, ZD, YH, BY, YC, BL, and ZY contributed reagents and interpreted the data. XS, GY, CF, CL, SL, ZY, YQ, HS, and LD prepared the manuscript. YQ, HS, and LD verified the underlying data.
Funding source
This work was supported by grants from the National Key Research and Development Program of China (2018YFC1004701, LD), the National Nature Science Foundation of China (82001629, XS; 81871128, 81571391 and 81401166, LD; 82030040, HS), the Jiangsu Province Social Development Project (BE2018602, HS), Jiangsu Provincial Medical Youth Talent (QNRC2016006, LD), the Youth Program of Natural Science Foundation of Jiangsu Province (BK20200116, XS) and Jiangsu Province Postdoctoral Research Funding (2021K277B, XS).
Data sharing statement
Please contact Dr. Ding and Dr. Qin to request all data and reagents described in this article. SMART RNA-seq information and raw data can be found in NCBI Bioproject numbers PRJNA774941 and PRJNA774984 and Mendeley Data: http://dx.doi.org/10.17632/p4btc9z6hy.2.
Acknowledgment
We thank all researchers whose work was relevant to but not cited in this article due to limited space.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103790.
Contributor Information
Yingying Qin, Email: qinyingying1006@163.com.
Haixiang Sun, Email: stevensunz@163.com.
Lijun Ding, Email: dinglijun@nju.edu.cn.
Appendix. Supplementary materials
Reference
- 1.May-Panloup P, Boucret L, de la Barca JMC, Desquiret-Dumas V, Ferré-L'Hotellier1 V, Morinière C, et al. Ovarian ageing: the role of mitochondria in oocytes and follicles. Hum Reprod Update. 2016;22:725–743. doi: 10.1093/humupd/dmw028. [DOI] [PubMed] [Google Scholar]
- 2.Navot D, Bergh RA, Williams MA, Garrisi GJ, Guzman I, Sandler B, et al. Poor oocyte quality rather than implantation failure as a cause of age-related decline in female fertility. Lancet. 1991;337:1375–1377. doi: 10.1016/0140-6736(91)93060-M. [DOI] [PubMed] [Google Scholar]
- 3.Kawamura K, Kawamura N, Hsueh AJW. Activation of dormant follicles: a new treatment for premature ovarian failure? Curr Opin Obstet Gynecol. 2016;28:217–222. doi: 10.1097/GCO.0000000000000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilly JL, Sinclair DA. Germline energetics, aging, and female infertility. Cell Metab. 2013;17:838–850. doi: 10.1016/j.cmet.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babayev E, Seli E. Oocyte mitochondrial function and reproduction. Curr Opin Obstet Gynecol. 2015;27:175–181. doi: 10.1097/GCO.0000000000000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coticchio G, Dal Canto M, Mignini Renzini M, Guglielmo MC, Brambillasca F, Turchi D, et al. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum Reprod Update. 2015;21:427–454. doi: 10.1093/humupd/dmv011. [DOI] [PubMed] [Google Scholar]
- 7.Cinco R, Digman MA, Gratton E, Luderer U. Spatial characterization of bioenergetics and metabolism of primordial to preovulatory follicles in whole ex vivo murine ovary. Biol Reprod. 2016;95:129. doi: 10.1095/biolreprod.116.142141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- 9.Sheng X, Yang Y, Zhou J, Yan G, Liu M, Xu L, et al. Mitochondrial transfer from aged adipose-derived stem cells does not improve the quality of aged oocytes in C57BL/6 mice. Mol Reprod Dev. 2019;86:516–529. doi: 10.1002/mrd.23129. [DOI] [PubMed] [Google Scholar]
- 10.Vazquez-Calvo C, Suhm T, Büttner S, Ott M. The basic machineries for mitochondrial protein quality control. Mitochondrion. 2020;50:121–131. doi: 10.1016/j.mito.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian Y, Merkwirth C, Dillin A. Mitochondrial UPR: a double-edged sword. Trends Cell Biol. 2016;26:563–565. doi: 10.1016/j.tcb.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Melber A, Haynes CM. UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;28:281–295. doi: 10.1038/cr.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quiros PM, Langer T, Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 15.Gibellini L, De Gaetano A, Mandrioli M, Van Tongeren E, Bortolotti CA, Cossarizza A, et al. The biology of Lonp1: more than a mitochondrial protease. Int Rev Cell Mol Biol. 2020;354:1–61. doi: 10.1016/bs.ircmb.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Pinti M, Gibellini L, Liu Y, Xu S, Lu B, Cossarizza A. Mitochondrial Lon protease at the crossroads of oxidative stress, ageing and cancer. Cell Mol Life Sci. 2015;72:4807–4824. doi: 10.1007/s00018-015-2039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bota DA, Davies KJ. Mitochondrial Lon protease in human disease and aging: Including an etiologic classification of Lon-related diseases and disorders. Free Radic Biol Med. 2016;100:188–198. doi: 10.1016/j.freeradbiomed.2016.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joza N, Pospisilik JA, Hangen E, Hanada T, Modjtahedi N, Penninger JM, et al. AIF: not just an apoptosis-inducing factor. Ann N Y Acad Sci. 2009;1171:2–11. doi: 10.1111/j.1749-6632.2009.04681.x. [DOI] [PubMed] [Google Scholar]
- 19.Joza N, Susin SA, Daugas E, Stanford WL, Cho SK, Li CYJ, et al. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 20.Otera H, Ohsakaya S, Nagaura Z-I, Ishihara N, Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 2005;24:1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurbuxani S, Schmitt E, Cande C, Parcellier A, Hammann A, Daugas E, et al. Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene. 2003;22:6669–6678. doi: 10.1038/sj.onc.1206794. [DOI] [PubMed] [Google Scholar]
- 22.Shin CS, Meng S, Garbis SD, Moradian A, Taylor RW, Sweredoski MJ, et al. LONP1 and mtHSP70 cooperate to promote mitochondrial protein folding. Nat Commun. 2021;12:1–10. doi: 10.1038/s41467-020-20597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu B, Shangguan F, Huang D, Gong S, Shi Y, Song Z, et al. LonP1 orchestrates UPR mt and UPR ER and mitochondrial dynamics to regulate heart function. BioRxiv. 2019 doi: 10.1101/564492. [DOI] [Google Scholar]
- 24.Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian J, Zhang C, Kang N, Wang J, Kong N, Zhou J, et al. Attenuated monoamine oxidase a impairs endometrial receptivity in women with adenomyosis via downregulation of FOXO1. Biol Reprod. 2021:1–15. doi: 10.1093/biolre/ioab182. [DOI] [PubMed] [Google Scholar]
- 26.Zheng F, Jia B, Dong F, Liu L, Rasul F, He J, et al. Glucose starvation induces mitochondrial fragmentation depending on the dynamin GTPase Dnm1/Drp1 in fission yeast. J Biol Chem. 2019;294:17725–17734. doi: 10.1074/jbc.RA119.010185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Quiros PM, Espanol Y, Acin-Perez R, Rodriguez F, Barcena C, Watanabe K, et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8:542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 30.Labarta E, de los Santos MJ, Escribá MJ, Pellicer A, Herraiz S. Mitochondria as a tool for oocyte rejuvenation. Fertil Steril. 2019;111:219–226. doi: 10.1016/j.fertnstert.2018.10.036. [DOI] [PubMed] [Google Scholar]
- 31.Bayot A, Gareil M, Chavatte L, Hamon MP, L'Hermitte-Stead C, Beaumatin F, et al. Effect of Lon protease knockdown on mitochondrial function in HeLa cells. Biochimie. 2014;100:38–47. doi: 10.1016/j.biochi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Lee HJ, Chung K, Lee H, Lee K, Lim JH, Song J. Downregulation of mitochondrial lon protease impairs mitochondrial function and causes hepatic insulin resistance in human liver SK-HEP-1 cells. Diabetologia. 2011;54:1437–1446. doi: 10.1007/s00125-011-2074-z. [DOI] [PubMed] [Google Scholar]
- 33.Chiaratti MR, Garcia BM, Carvalho KF, Macabelli CH, Ribeiro FK da S, Zangirolamo AF, et al. Oocyte mitochondria: role on fertility and disease transmission. Anim Reprod. 2018;15:231–238. doi: 10.21451/1984-3143-AR2018-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, Babayev E, Jiang Z, Li G, Zhang M, Esencan E, et al. Mitochondrial unfolded protein response gene Clpp is required to maintain ovarian follicular reserve during aging, for oocyte competence, and development of pre-implantation embryos. Aging Cell. 2018;17:e12784. doi: 10.1111/acel.12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maltecca F, Aghaie A, Schroeder DG, Cassina L, Taylor BA, Phillips SJ, et al. The mitochondrial protease AFG3L2 is essential for axonal development. J Neurosci. 2008;28:2827–2836. doi: 10.1523/JNEUROSCI.4677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quirós PM, Ramsay AJ, Sala D, Fernández-Vizarra E, Rodríguez F, Peinado JR, et al. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 2012;31:2117–2133. doi: 10.1038/emboj.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeinert RD, Baniasadi H, Tu BP, Chien P. The Lon Protease Links Nucleotide Metabolism with Proteotoxic Stress. Mol Cell. 2020;79:758–767. doi: 10.1016/j.molcel.2020.07.011. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibellini L, Losi L, De Biasi S, Nasi M, Tartaro D.L., Pecorini S, et al. LonP1 differently modulates mitochondrial function and bioenergetics of primary versus metastatic colon cancer cells. Front Oncol. 2018;8 doi: 10.3389/fonc.2018.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao TY, Chiu YC, Fang WC, Cheng CW, Kuo CY, Juan HF, et al. Mitochondrial Lon regulates apoptosis through the association with Hsp60-mtHsp70 complex. Cell Death Dis. 2015;6:e1642. doi: 10.1038/cddis.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung YJ, Kao TY, Kuo CL, Fan CC, Cheng AN, Fang WC, et al. Mitochondrial Lon sequesters and stabilizes p53 in the matrix to restrain apoptosis under oxidative stress via its chaperone activity. Cell Death Dis. 2018;9:697. doi: 10.1038/s41419-018-0730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mierzewska H, Rydzanicz M, Biegański T, Kosinska J, Mierzewska-Schmidt M, Ługowska A, et al. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with AIFM1 mutation – a novel phenotype of the mitochondrial disease. Clin Genet. 2017;91:30–37. doi: 10.1111/cge.12792. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Peng X, Mei S. Autophagy in ovarian follicular development and atresia. Int J Biol Sci. 2019;15:726–737. doi: 10.7150/ijbs.30369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goswami D, Conway GS. Premature ovarian failure. Hum Reprod Update. 2005;11:391–410. doi: 10.1093/humupd/dmi012. [DOI] [PubMed] [Google Scholar]
- 44.Torrealday S, Kodaman P, Pal L. Premature ovarian insufficiency - an update on recent advances in understanding and management. F1000Research. 2017;6:2069. doi: 10.12688/f1000research.11948.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inui T, Anzai M, Takezawa Y, Endo W, Kakisaka Y, Kikuchi A, et al. A novel mutation in the proteolytic domain of LONP1 causes atypical CODAS syndrome. J Hum Genet. 2017;62:653–655. doi: 10.1038/jhg.2017.11. [DOI] [PubMed] [Google Scholar]
- 46.Strauss KA, Jinks RN, Puffenberger EG, Venkatesh S, Singh K, Cheng I, et al. CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ lon protease. Am J Hum Genet. 2015;96:121–135. doi: 10.1016/j.ajhg.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peter B, Waddington CL, Oláhová M, Sommerville EW, Hopton S, Pyle A, et al. Defective mitochondrial protease LonP1 can cause classical mitochondrial disease. Hum Mol Genet. 2018;27:1743–1753. doi: 10.1093/hmg/ddy080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinti M, Gibellini L, Nasi M, De Biasi S, Bortolotti CA, Iannone A, et al. Emerging role of Lon protease as a master regulator of mitochondrial functions. Biochim Biophys Acta. 2016;1857:1300–1306. doi: 10.1016/j.bbabio.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 49.Dikoglu E, Alfaiz A, Gorna M, Bertola D, Chae JH, Cho TJ, et al. Mutations in LONP1, a mitochondrial matrix protease, cause CODAS syndrome. Am J Med Genet Part A. 2015;167:1501–1509. doi: 10.1002/ajmg.a.37029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.