Abstract
The experiment was conducted to compare the differences of gut microbiota and metabolic status of sows with different litter sizes on days 30 and 110 of gestation, and uncover the relationship between the composition of maternal gut microbiota during gestation and sow reproductive performance. Twenty-six Large White × Landrace crossbred multiparous sows (2nd parity) with similar back fat thickness and body weight were assigned to two groups [high-reproductive performance group (HP group) and low-reproductive performance group (LP group)] according to their litter sizes and fed a common gestation diet. Results showed that compared with LP sows, HP sows had significantly lower plasma levels of triglyceride (TG) on gestation d 30 (P < 0.05), but had significantly higher plasma levels of TG, non-esterified fatty acid, tumor necrosis factor-α, and immunoglobulin M on gestation d 110 (P < 0.05). Consistently, HP sows revealed increased alpha diversity and butyrate-producing genera, as well as fecal butyrate concentration, on gestation d 30; HP sows showed significantly different microbiota community structure with LP sows (P < 0.05) and had markedly higher abundance of Firmicutes (genera Christensenellaceae_R-7_group and Terrisporobacter) which were positively related with litter size on gestation d 110 than LP sows (P < 0.05). In addition, plasma biochemical parameters, plasma cytokines, and fecal microbiota shifted dramatically from gestation d 30 to d 110. Therefore, our findings demonstrated that microbial abundances and community structures differed significantly between sows with different litter sizes and gestation stages, which was associated with changes in plasma biochemical parameters, inflammatory factors, and immunoglobulin. Moreover, these findings revealed that there was a significant correlation between litter size and gut microbiota of sows, and provided a microbial perspective to improve sow reproductive performance in pig production.
Keywords: gestation stage, gut microbiota, litter size, metabolic status, reproductive performance, sow
Introduction
Diverse microbial communities reside at various sites within a mammalian body (1, 2). Gut microbiota makes up the vast majority of body's microbes and with an estimated number of several trillion most probably outnumber human body cells (3). The gut microbiota is shaped by many environmental factors, such as host genetics (4), diet (5), and the immune system (6), and has been reported to play a vital role in inflammation, metabolic syndrome (7), energy metabolism (8), and immunity (9).
Previous study in humans showed that the body experiences extensive hormonal, metabolic, and immunological changes over the course of normal and healthy pregnancy (10), accompanied by dramatic changes in maternal gut microbiota (11). Koren et al. (10) showed normal pregnancy to be accompanied by a profound change of gut microbiota from the first to the third trimester with an increase in the Proteobacteria and Actinobacteria abundances which might be connected with the maternal metabolic profile. Uryu et al. (12) demonstrated that sow productivity on different farms was likely related to changes in fecal microbe composition. Besides, research showed that dietary probiotic supplementation in gestating sow diet could increase the number of piglets total born (13, 14). Further, Al-Asmakh et al. (15) found that maternal microbiota could regulate placental development and then might affect the development of the growing offspring in mice. This research suggests that maternal gut microbiota during gestation is affecting sow reproductive performance. However, there is little literature available about whether the composition of gut microbiota during gestation is associated with improved sow reproductive performance.
The early and late pregnancy are two critical stages for embryonic survival and development (16, 17). In the present study, we aimed to explore the relationship between reproductive performance and maternal gut microbiota during gestation through comparing the fecal microbiota characteristics and metabolic status of sows with high (>12 piglets per litter) and low litter size (≤ 12 piglets per litter) on day 30 of gestation (G30) and on day 110 of gestation (G110).
Materials and Methods
Ethical Approval
This study was conducted at the pig breeding farm in Shandong Province. The animal use protocol for this research was approved by the Animal Care and Use Committee of Shandong Agricultural University (Approval Number: SDAUA-2019-019).
Animals and Experimental Design
Twenty-six Large White × Landrace crossbred multiparous sows (2nd parity) with similar back fat thickness (BF, 15.28 ± 0.45 mm) and body weight (174.34 ± 2.72 kg) were used in this study. The BF at the last rib was measured using a HG 9300 digital diagnostic ultrasound device (Caresono Technology Co. Ltd., Nanjing, China). After artificial insemination, the individual sow was housed individually in a gestation stall (2.37 × 0.65 × 1.13 m) kept at 21 ± 1°C. All the sows were mated within 3 days and fed a common fortified corn–soybean meal gestation diet (Supplementary Table 1) which was formulated to meet or exceed National Research Council (18) nutrient requirements. All sows received a daily meal at 0900 h and were fed the same amount of feed (days 1 to 89 of gestation 2.46 kg/d; days 90 of gestation to farrowing, 2.89 kg/d) during the entire gestation. On day 110 of gestation, sows were moved from gestation to farrowing rooms and kept in individual farrowing crates measuring 2.40 × 1.80 × 0.90 m thereafter. Backfat thickness and body weight of individual sow were measured at breeding and within 24 h of farrowing. At farrowing, the numbers of total born piglets, live born piglets, and dead born piglets per litter, as well as litter weight, were recorded, and the averages were calculated. Thus, two groups were generated (Table 1): 13 sows with litter size lower than the average in this trial (12.7 piglets) were classified as the low-reproductive performance group (LP group), while 13 sows with litter size higher than the average in this trial (12.7 piglets) labeled as the high-reproductive performance group (HP group). Sows had free access to water throughout the experiment and did not receive vaccine, antibiotics, or other medication in the feed or for any therapeutic purposes after insemination.
Table 1.
Litter size and litter weight in low- and high-reproductive performance groups.
| Items | Groupa | P -value | |
|---|---|---|---|
| LP | HP | ||
| No. of sows | 13 | 13 | - |
| Backfat thickness, mm | |||
| Breeding | 15.33 ± 0.74 | 14.84 ± 0.53 | 0.599 |
| Farrowing | 19.16 ± 0.72 | 17.82 ± 0.77 | 0.218 |
| Body weight, kg | |||
| Breeding | 179.32 ± 4.34 | 175.31 ± 4.18 | 0.631 |
| Farrowing | 238.77 ± 3.50 | 231.84 ± 3.91 | 0.199 |
| Litter size | |||
| Total born | 9.77 ± 0.53 | 15.54 ± 0.66 | <0.001 |
| Born alive | 9.46 ± 0.57 | 14.31 ± 0.61 | <0.001 |
| Dead | 0.31 ± 0.13 | 1.23 ± 0.30 | 0.013 |
| Litter weight, kg | |||
| Total born | 16.55 ± 0.64 | 22.16 ± 0.84 | <0.001 |
| Born alive | 16.03 ± 0.66 | 21.21 ± 0.83 | <0.001 |
LP, Sows in low-Reproductive Performance Group; HP, Sows in High-Reproductive Performance Group.
Values are mean ± standard error (n = 13).
Sample Collection
Fasting blood samples (12 h overnight) and fresh fecal samples from all healthy sows were collected on day 30 and day 110 of gestation before feeding in the morning. Samples were grouped as follows: LP30 and LP110: sows with low-reproductive performance on day 30 and day 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on day 30 and day 110 of gestation, respectively. Blood samples (5 mL) from the ear veins were collected into a tube containing heparin sodium and centrifuged at 3,000× g for 15 min. Plasma samples was transferred to 200 μL centrifuge tubes and stored at −20°C until analysis. Fecal samples (about 5 g) were collected from the rectum by a sterilized fecal collection tube and then stored at −80°C immediately for the detection of short-chain fatty acids (SCFAs) and analysis of microbiota.
Plasma Biochemical Parameters Analysis
Plasma biochemical parameters, including glucose (GLU), cholesterol (CHOL), triglyceride (TG), high density lipoprotein cholesterol (HDL-C), low density lipoprotein cholesterol (LDL-C), and non-esterified fatty acid (NEFA), were determined with commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) using standard spectrophotometric methods on an Autolab-PM4000 Automatic Analyzer (AMS Co., Rome, Italy) as previously described (19).
Analysis of Inflammatory Factors, Immunoglobulins, and Reproductive Hormones
Concentrations of interleukin-2 (IL-2), interleukin-6 (IL-6), interleukin-10 (IL-10), tumor necrosis factor-α (TNF-α), immunoglobulin A (IgA), immunoglobulin G (IgG), immunoglobulin M (IgM), progesterone, estrogen, lutropin, and prolactin in the plasma of sows were determined with commercial ELISA kits (Feiya Biotechnology Co. Ltd., Yancheng, China) as described in Supplementary Methods.
Determination of Fecal SCFAs
The fecal SCFAs of sows were measured by a Varian CP-3800 gas chromatograph (Palo Alto, CA, USA) equipped with a micro-injector, a flame ionization detector, and a capillary chromatographic column as described in Supplementary Methods.
Microbial Analysis
Microbial composition and diversity were analyzed as previously described in Li et al. (20). Briefly, bacterial genomic DNA was extracted from frozen fecal samples with an E.Z.N.A.TM Stool DNA kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer's protocol. After DNA concentration and purity monitoring, DNA was diluted to 1 ng/μL using sterile water, and the V4 hypervariable region of 16S rDNA was amplified with 515F and 806R primer (5′-GTGCCAGCMGCCGCGGTAA-3′ and 5′-GGACTACHVGGGTWTCTAAT-3′, respectively), on the Illumina HiSeq PE2500 platform by Novogene (Beijing, China). Filtered, non-chimeric high-quality sequences (tags) sharing over 97% sequence similarity were clustered into the same operational taxonomic units (OTUs) by Uparse software (21), and then classified to different taxonomic levels with SILVA database (22) based on Mothur algorithm to annotate taxonomic information. Operational taxonomic units abundance information were normalized using a standard of sequence number corresponding to the sample with the least sequences for subsequent analysis of alpha diversity and beta diversity. Shannon, Simpson, Chao 1, and ACE indexes were chosen to ascertain differences in alpha diversity based on different groups (23, 24), and Bray-Curtis distances were calculated and visualized using Principal Coordinate Analysis (PCoA) (25). The statistical differences in alpha and beta diversity of bacterial communities between the two groups were examined using the Wilcoxon rank-sum test. Significant difference among the microbial communities was accessed with the analysis of similarity (ANOSIM) test.
Statistical Analysis
The individual sow was regarded as the experimental unit for all variables. Differences in the data including plasma biochemical parameters, inflammatory factors, and fecal SCFAs were evaluated using the independent t-test (LP vs. HP) or paired t-test (G30 vs. G110) procedure of SAS 9.0 (Institute Inc., Cary, NC, USA) following normal distribution assessment using a Shapiro-Wilk's statistic (W > 0.05). Multiple testing was corrected by using the Benjamini-Hochberg false discovery rate. Spearman's correlations were used to assess the associations between bacterial abundance and litter size, as well as plasma biochemical indices. Treatment differences were considered statistically significant at P < 0.05, and 0.05 ≤ P < 0.10 was considered a statistical trend. Values are expressed as mean ± standard error in tables and figures.
Results
Changes of Fecal Microbial Diversity
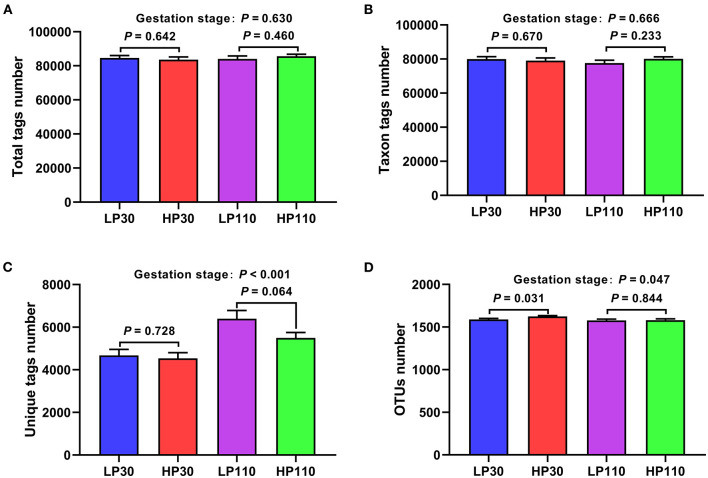
A total of 4,392,562 total tags, 4,118,486 taxon tags, and 274,447 unique tags were obtained from 52 sow fecal samples, with an average of 84,472 ± 730, 79,202 ± 720 and 5,278 ± 179 per sample, respectively (Figures 1A–C). Based on 97% sequence similarity, a total of 21,114 OTUs were found in the HP group on day 30 of gestation, with an significantly higher average of 1,624 ± 10 OUT per sample compared to an average of 1,589 ± 12 OUT per sample in the LP group, where 20,657 OTUs were found in total (Figure 1D); the HP group tended had lower unique tags than the LP group on day 110 of gestation (5,500 ± 248 vs. 6,397 ± 389; P = 0.064; Figure 1C). In addition, from gestation d 30 to d 110, the unique tags number was significantly increased on average (P < 0.05), but OTUs number was decreased (P = 0.047).
Figure 1.
Operational taxonomic unit (OUT) clustering and annotation of sow fecal samples on d 30 and d 110 of gestation. (A) Total tags number; (B) taxon tags number; (C) unique tags number; (D) OTUs number. LP30 and LP110: sows with low-reproductive performance on d 30 and d 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and d 110 of gestation, respectively. Gestation stage: difference in the variations between gestation d 30 and d 110. Values are mean ± standard error (n = 13).
To determine whether the sample size was sufficient for OUT testing, the species accumulation curves (SAC) was used in the present study. The SAC (Supplementary Figure 1) tended to flatten as the sample number of analyzed sequences increased up to 52, suggesting that the sample size was enough for OTUs testing and could estimate the species richness of the habitat.
The results of fecal microbial community structures assessment are shown in Figure 2. On d 30 of gestation, the HP group had significantly higher Shannon index (P < 0.05) and tended to have a higher Chao 1 index compared with LP group (P = 0.069); on d 110 of gestation, no significant differences were observed in alpha diversity between the two groups (P > 0.05).
Figure 2.
Difference on bacteria community diversity and richness among different groups on d 30 and 110 of gestation. (A) Shannon index; (B) Simpson index; (C) Chao 1 index; (D) ACE index. LP30 and LP110: sows with low-reproductive performance on d 30 and 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and 110 of gestation, respectively. Gestation stage: difference in the variations between gestation d 30 and 110. Values are mean ± standard error (n = 13).
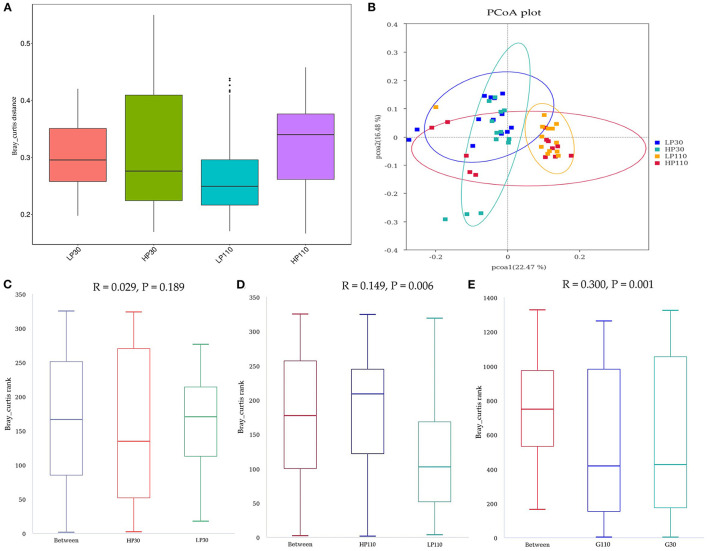
In addition, to measure the evolutionary distance between microbiotas (beta diversity), the PCoA profile for sow fecal samples based on the Bray-Curtis distance was used in the present study, and ANOSIM test was used to assess significant differences among the microbial communities (Figures 3A,B). The results suggested that LP and HP groups had close distance on gestation d 30 which showed that the two groups had no significant difference in the microbial community (P = 0.189, Figure 3C); an obvious separation was observed in PCoA between samples from HP group and LP group on d 110 of gestation. The ANOSIM test also indicated that the two groups had notably different microbiota structures on gestation d 110 (P = 0.006, Figure 3D), and sows in HP group had greater beta diversity compared to sows in LP group at 110 days of gestation (P < 0.05). Besides, the Bray-Curtis distance analysis showed a global shift in microbial community composition from gestation d 30 to d 110 (P = 0.001, Figures 3B,E).
Figure 3.
The beta diversity of microbial communities in the groups on d 30 and d 110 of gestation. (A) The Bray-Curtis distance within each group. (B) The principal coordinate analysis (PCoA) profile of the two groups displayed with the Bray-Curtis distance. Each dot represents one sample from each group. The percent variation explained by each principal coordinate is indicated on the X and Y axis. (C–E) Analysis of similarity (ANOSIM). R value is scaled to lie between −1 and +l. Generally, 0 < R < 1 and P < 0.05 represents that there were significant differences between the groups. LP30 and LP110: sows with low-reproductive performance on d 30 and d 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and d 110 of gestation, respectively. n = 13 for each group.
Changes in Relative Abundance of Phyla and Genera
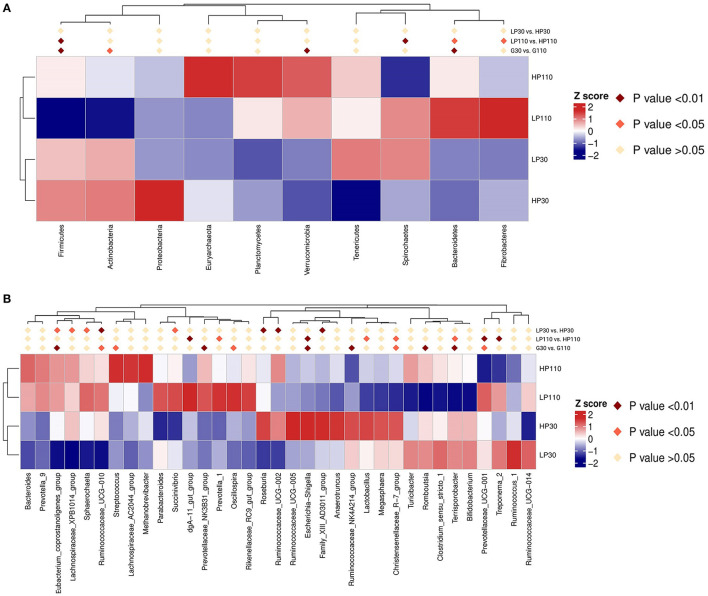
As shown in Figure 4, Firmicutes and Bacteroidetes were the most predominant phyla which accounted for more than 75%, followed by Spirochaetes and Tenericutes, in both groups during gestation (Figure 4A). No significant differences were observed in the top 10 phyla which accounted for more than 99.5% of the total bacteria population between the LP and HP groups on d 30 of gestation (P > 0.05, Figure 5A). However, the relative abundance of Firmicutes in the HP groups was significantly higher (P < 0.05) than that in the LP group, while the relative abundances of Bacteroidetes, Spirochaetes, and Fibrobacteres were significantly lower (P < 0.05) than that in the LP group on d 110 of gestation. In addition, the relative abundances of Firmicutes and Actinobacteria were significantly decreased (P < 0.05), while Bacteroidetes and Verrucomicrobia were significantly increased from gestation d 30 to d 110 (P < 0.05).
Figure 4.
Changes of the relative abundance at phylum and genus levels. (A) Unweighted pair-group method with arithmetic mean (UPGMA) clustering analysis with the Bray-Curtis distance. The left panel shows the phylogenic tree, and the right panel displays the relative abundance of each group at the phylum level. (B) The phylogenetic tree constructed based on the sequence of the top 35 genera. The branches with different colors in the inner circle represent their corresponding phylum, and the stacked column chart in the outer circle indicates the relative abundance of each genus in different treatments. LP30 and LP110: sows with low-reproductive performance on d 30 and 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and 110 of gestation, respectively. n = 13 for each group.
Figure 5.
Heatmap distribution of OTUs in sow feces for all groups. (A) Comparison of the relative abundances of sow fecal microbiota in the top 10 at the phylum level. (B) Comparison of the relative abundances of sow fecal microbiota in the top 35 at the genus level. Different colors show the relative abundance of taxa. Fecal bacterial abundances were standardized with Z-score prior to the analyses. Positive z-scores reflect abundances above the average, whereas a negative z-score indicates abundance below the average. LP30 and LP110: sows with low-reproductive performance on d 30 and d 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and d 110 of gestation, respectively. n = 13 for each group.
The species phylogenetic tree evolution was constructed by multiple sequences alignments to obtain the representative sequence of the top 35 genera. As shown in Figure 4B, the relative abundance of Firmicutes was contributed by Clostridium_sensu_stricto_1, Streptococcus, Lactobacillus, Christensenellaceae_R-7_group, Ruminococcaceae_UCG-002, Ruminococcaceae_NK4A214_group, Ruminococcaceae_UCG-005, Ruminococcaceae_UCG-014, Ruminococcus_1, and Lachnospiraceae_XPB1014_group; Bacteroidetes mainly distributed with Rikenellaceae_RC9_gut_group, Prevotellaceae_UCG-001, and Prevotellaceae_NK3B31_group; Spirochaetes was dominated by Treponema_2. In the LP and HP groups, Treponema_2 and Clostridium_sensu_stricto_1 were the top two genera on d 30 of gestation, and Treponema_2 and Streptococcus were the most dominant on d 110 of gestation. Of the top 35 genera, compared with sows in the LP group, sows in the HP group had significantly higher (P < 0.05) relative abundances of Eubacterium_coprostanoligenes_group, Lachnospiraceae_XPB1014_group, Sphaerochaeta, Ruminococcaceae_UCG-010, Roseburia, Ruminococcaceae_UCG-002, and Family_XIII_AD3011_group, and significantly lower (P < 0.05) Succinvibrio on d 30 of gestation; sows in the HP group had significantly lower (P < 0.05) Treponema_2, Prevotellaceae_UCG-001, Prevotella_1 and dgA-11_gut_group, as well as significantly higher (P < 0.05) Lactobacillus, Christensenellaceae_R-7_group, Terrisporobacter, and Escherichia-Shigella on d 110 of gestation (Figure 5B). Besides, sow fecal samples from gestation d 110 had significantly higher abundances of Streptococcus, Prevotellaceae_NNK3B31_group, Oscillospira, Eubacterium_coprostanoligenes_group, and Ruminococcaceae_UCG-010, but significantly lower Prevotellaceae_UCG-001, Terrisporobacter, Escherichia-Shigella, Ruminococcaceae_NK4A214_group, Christensenellaceae_R-7_group, and Romboutsia than those of sow fecal samples from gestation d 30 (P < 0.05).
Correlation Analysis Between Sow Reproductive Performance and Fecal Microbiota
On day 30 of gestation, at the phylum level, the relative abundances of Firmicutes and Actinobacteria tended to be positively correlated with litter size (P < 0.10), while the relative abundance of Proteobacteria tended to be negatively correlated with litter size (P < 0.10); at the genus level, the relative abundances of Turicibacter and Ruminococcaceae_UCG-014 had significant positive correlations with litter size (P < 0.05), and the relative abundances of Clostridium_sensu_stricto_1 and Romboutsia tended to be positively correlated with litter size (P < 0.10, Table 2).
Table 2.
The Spearman's correlation test between the sow fecal microbiota and litter size.
| Phase of gestation | Phylum | Genus |
|---|---|---|
| D 30 of gestation | Firmicutes (0.351+) | Clostridium_sensu_stricto_1 (0.376+) |
| Turicibacter (0.479*) | ||
| Ruminococcaceae_UCG-014 (0.533**) | ||
| Romboutsia (0.346+) | ||
| Proteobacteria (−0.331+) | - | |
| Actinobacteria (0.369+) | - | |
| D110 of gestation | Firmicutes (0.492*) | Lactobacillus (0.365+) |
| Clostridium_sensu_stricto_1 (0.434*) | ||
| Turicibacter (0.445*) | ||
| Terrisporobacter (0.466*) | ||
| Christensenellaceae_R-7_group (0.406*) | ||
| Bacteroidetes (−0.402*) | Rikenellaceae_RC9_gut_group (−0.495*) | |
| Spirochaetes (−0.526**) | Treponema_2 (−0.490*) | |
| Sphaerochaeta (−0.347+) | ||
| Proteobacteria (0.365+) | Escherichia-Shigella (0.578**) | |
| Actinobacteria (−0.627**) | - |
The correlation tends to be significant at a level of 0.10;
the correlation is significant at a level of 0.05;
the correlation is significant at a level of 0.01.
On d 110 of gestation, at the phylum level, significant positive correlation between the relative abundance of Firmicutes and litter size was observed (P < 0.05), and the relative abundances of Bacteroidetes, Spirochaetes, and Actinobacteria were all significantly negatively correlated with litter size (P < 0.05); at the genus level, Clostridium_sensu_stricto_1, Turicibacter, Terrisporobacter, Christensenellaceae_R-7_group, and Escherchia-Shigella exhibited the significantly positive correlations with litter size (P < 0.05), while Rikenellaceae_RC9_gut_group, Treponema_2, and Sphaerochaeta had significant negative correlations with litter size (P < 0.05). In addition, the phylum Proteobacteria and genus Lactobacillus displayed a tendency to be positively correlated with litter size (P < 0.10), and the genus Sphaerochaeta tended to be negatively correlated with litter size (P < 0.10).
Changes of Fecal SCFAs Concentrations During Gestation
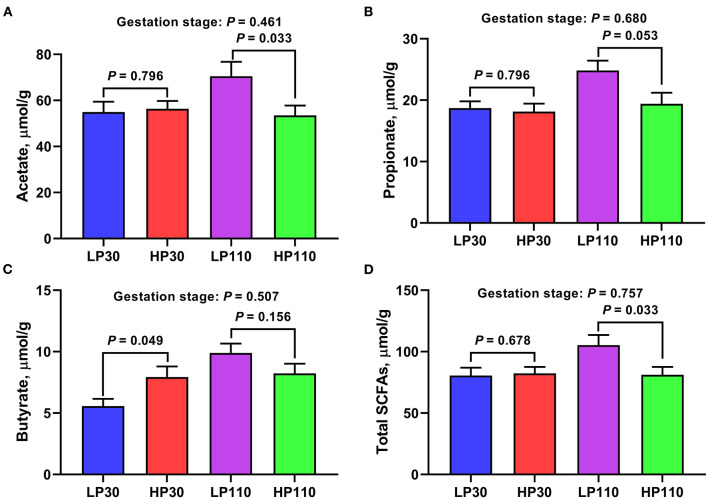
The concentrations of fecal short-chain fatty acids on d 30 and d 110 of gestation in the two groups are listed in Figure 6. On d 30 of gestation, there were no significant differences in fecal acetate, propionate, and total SCFAs concentrations between the LP group and HP group (P > 0.05), but sows from HP group showed significantly higher butyrate concentration than those of sows from LP group (P < 0.05). On d 110 of gestation, sows in the HP group had significantly lower acetate and total SCFAs concentrations than sows in the LP group (P < 0.05), and the propionate concentration in the HP group tended to be lower than that in the LP group (P = 0.053). The fecal SCFAs concentrations of sows on d 110 of gestation did not differ with that of sows on d 30 of gestation (P > 0.05).
Figure 6.
Fecal short-chain fatty acids (SCFAs) concentrations in low- and high-reproductive performance groups on d 30 and 110 of gestation. (A) Acetate. (B) Propionate. (C) Butyrate. (D) Total SCFAs. Total SCFAs, the sum of acetate, propionate, and butyrate. LP30 and LP110: sows with low-reproductive performance on d 30 and 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and 110 of gestation, respectively. Gestation stage: difference in the variations between gestation d 30 and d 110. Values are mean ± standard error (n = 13).
Changes in Plasma Metabolites During Gestation
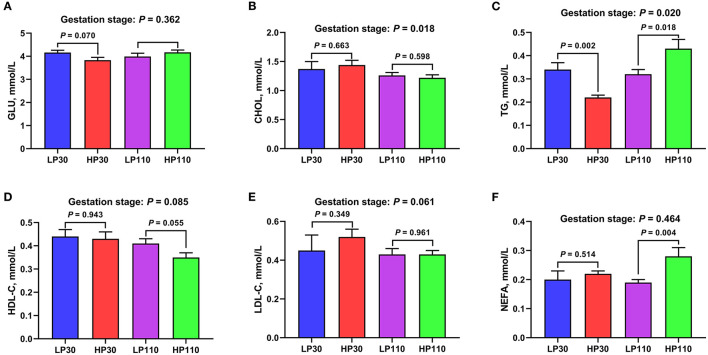
As shown in Figure 7, on day 30 of gestation, significantly lower plasma TG levels were observed in the HP group compared with those in the LP group (P < 0.05); sows in the HP group tended to have a lower plasma GLU concentration than sows in the LP group (P = 0.070). On d 110 of gestation, sows in the HP group had significantly higher plasma TG and NEFA levels (P < 0.05) and tended to have lower plasma HDL-C concentration compared with those of sows in the LP group (P = 0.055). Besides, the concentrations of CHOL (P = 0.018), HDL-C (P = 0.085), and LDL-C (P = 0.061) were decreased, and the levels of TG (P = 0.020) were increased from gestation d 30 to d 110.
Figure 7.
The levels of plasma metabolites in low- and high-reproductive performance groups on d 30 and 110 of gestation. (A) Glucose (GLU); (B) Cholesterol (CHOL); (C) Triglyceride (TG); (D) High density lipoprotein cholesterol (HDL-C); (E) Low density lipoprotein cholesterol (LDL-C); (F) Non-esterified fatty acid (NEFA). LP30 and LP110: sows with low-reproductive performance on d 30 and 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and 110 of gestation, respectively. Gestation stage: difference in the variations between gestation d 30 and d 110. Values are mean ± standard error (n = 13).
Changes of Plasma Inflammatory Factors and Immunoglobulins During Gestation
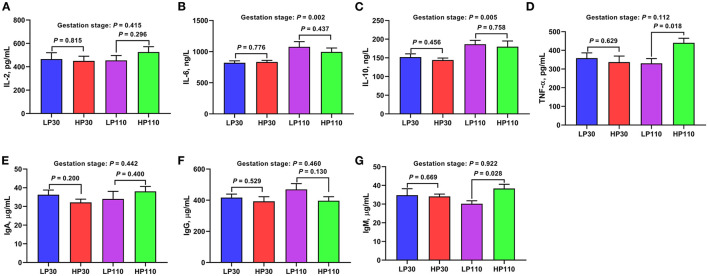
The levels of plasma inflammatory factors and immunoglobulins are shown in Figure 8. There were no significant differences in the plasma concentrations of inflammatory factors and immunoglobulins on d 30 of gestation between the LP and HP groups (P > 0.05). However, significantly higher TNF-α and IgM concentrations were observed in the HP group compared with those in the LP group on d 110 of gestation (P < 0.05). In addition, the plasma IL-6 and IL-10 levels were significantly increased (P < 0.05) from gestation d 30 to d 110.
Figure 8.
The levels of inflammatory factors and immunoglobulins in low- and high-reproductive performance groups on d 30 and 110 of gestation. (A) Interleukin-2 (IL-2); (B) Interleukin-6 (IL-6); (C) Interleukin-10 (IL-10); (D) Tumor necrosis factor-α (TNF-α); (E) Immunoglobulin A (IgA); (F) Immunoglobulin G (IgG); (G) Immunoglobulin M (IgM). LP30 and LP110: sows with low-reproductive performance on d 30 and 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and 110 of gestation, respectively. Gestation stage: difference in the variations between gestation d 30 and d 110. Values are mean ± standard error (n = 13).
Changes of Plasma Hormone Contents During Gestation
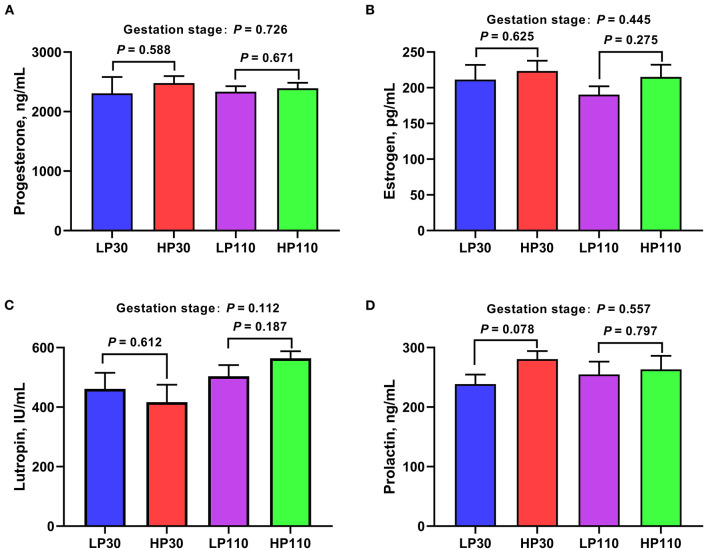
The plasma hormone contents are shown in Figure 9. There were no significant differences in the plasma hormone concentrations on d 30 and d 110 of gestation between the LP and HP groups (P > 0.05). The plasma hormone contents of sows on gestation d 110 did not differ from those of sows on gestation d 30 (P > 0.05).
Figure 9.
The plasma concentrations of reproductive hormones in low- and high-reproductive performance groups on d 30 and 110 of gestation. (A) Progesterone; (B) Estrogen; (C) Lutropin; (D) Prolactin. LP30 and LP110: sows with low-reproductive performance on d 30 and 110 of gestation, respectively; HP30 and HP110: sows with high-reproductive performance on d 30 and 110 of gestation, respectively. Gestation stage: difference in the variations between gestation d 30 and d 110. Values are mean ± standard error (n = 13).
Correlation Analysis Between Fecal Microbial Abundance and Plasma Biochemical Indices
At the phylum level (Figure 10A), the plasma levels of CHOL, HDL-C, and LDL-C showed positive correlations with the abundances of Firmicutes and Actinobacteria (P < 0.05) and negative correlations with the abundances of Bacteroidetes and Fibrobacteres (P < 0.05); the plasma TG level had significant positive correlation with Euryarchaeota abundance (P < 0.05); the plasma IL-2 concentration was significantly positively correlated with Proteobacteria abundance (P < 0.05); the plasma IL-6 concentration was significantly positively correlated with the abundance of Bacteroidetes (P < 0.05); the plasma IgA concentration had significant negative correlation with Verrucomicrobia (P < 0.05). In addition, the IL-10 concentration tended to be negatively correlated with the abundance of Tenericutes (P < 0.10), and the IgM concentration tended to be negatively correlated with the abundance of Euryarchaeota (P < 0.10).
Figure 10.
Correlation analysis between the plasma biochemical indices and sow fecal microbiota. (A) At phylum level; (B) At genus level. GLU, Glucose; CHOL, cholesterol; TG, triglyceride; HDL-C, high density lipoprotein cholesterol, LDL-C, low density lipoprotein cholesterol; NEFA, non-esterified fatty acid; IL-2, interleukin-2; IL-6, interleukin-6; IL-10, interleukin-10; TNF-α, tumor necrosis factor-α; IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M. +The correlation tends to be significant at a level of 0.10; *the correlation is significant at a level of 0.05; ** the correlation is significant at a level of 0.01.
At the genus level (Figure 10B), the plasma level of CHOL showed positive correlations with the abundances of Clostridium_sensu_stricto_1, Christensenellaceae_R-7_group, and Terrisporobacter (P < 0.05) and negative correlations with the abundances of Eubacterium_coprostanoligenes_group, dgA-11_gut_group, Ruminococcaceae_UCG-010, and Sphaerochaeta (P < 0.05). The plasma HDL-C concentration was significantly positively correlated with the abundance of Christensenellaceae_R-7_group (P < 0.05) and was significantly negatively correlated with the abundances of Eubacterium_coprostanoligenes_group, dgA-11_gut_group, and Sphaerochaeta (P < 0.05). The plasma LDL-C level had significant positive correlations with the abundances of Clostridium_sensu_stricto_1 and Christensenellaceae_R-7_group (P < 0.05) and had significant negative correlations with the abundances of dgA-11_gut_group, Ruminococcaceae_UCG-010, and Sphaerochaeta (P < 0.05). The plasma NEFA concentration displayed positive correlation with the abundance of Eubacterium_coprostanoligenes_group and negative correlations with the abundances of Turicibacter, Terrisporobacter, and Romboutsia (P < 0.05). The plasma IL-2 concentration was significantly positively correlated with the abundances of Clostridium_sensu_stricto_1 and Succinivibrio, and significantly negatively correlated with the abundance of Ruminococcaceae_UCG-010 (P < 0.05). The plasma IL-6 level showed positive correlation with the Prevotellaceae_NNK3B31_group abundance (P < 0.05). The plasma IL-10 concentration had significant positive correlations with the abundances of Prevotellaceae_NNK3B31_group and Sphaerochaeta (P < 0.05). The plasma TNF-α concentration was significantly positively correlated with the abundances of Anaerotruncus (P < 0.05). The plasma level of IgA showed positive correlation with the Lachnospiraceae_XPB1014_group abundance (P < 0.05). The plasma level of IgG indicated negative correlation with the Christensenellaceae_R-7_group abundance (P < 0.05). The plasma IgM concentration was significantly positively correlated with the abundances of Ruminococcaceae_UCG-014 and Lachnospiraceae_AC2044_group (P < 0.05), and was significantly negatively correlated with the abundances of Methanobrevibacter and dgA-11_gut_group (P < 0.05).
Discussion
Maternal metabolism changes dramatically during the gestation period. Especially, maternal glucose and lipid metabolism plays a vital role in the initiation and development of gestation (26). The early stage of gestation can be regarded as an anabolic state to meet the fetal-placental and maternal demands of late gestation and lactation, with an increase in maternal fat stores and small increases in insulin sensitivity (27). The present study showed that the sows in the HP group had lower plasma levels of GLU and TG than those in LP group on d 30 of gestation. Plasma levels of GLU and TG are important indicators of glycolipid metabolism. Plasma lipid profiles at early pregnancy may predict the incidence and severity of pre-eclampsia in humans (28). The previous study in humans showed that higher plasma GLU concentration in the first trimester of pregnancy was a risk factor for adverse perinatal and neonatal outcomes, such as diabetes-related complications, gestational hypertension, and obesity (29). Similarly, a study in dairy cows demonstrated that high glucose levels at early gestation had an adverse impact on early embryonic development (30). The reason might be related to high nutritional level that increased the metabolic clearance rate of progesterone (31). The results might suggest that higher glucose level was not conducive to the development of embryos. Besides, higher plasma TG concentration is usually associated with abnormal lipid metabolism and causally related to an increased risk of cardiovascular disease in the clinic (32). Previous research indicated that higher plasma TG concentration demonstrated a poor health status of a gestating sow (33). Therefore, the sows in HP group is in a better physical state than those in LP group on d 30 of gestation.
In contrast, the sows in HP group showed higher plasma levels of NEFA and TG on d 110 of gestation. Late pregnancy is characterized as a catabolic state with increased insulin resistance which leads to increases in concentrations of maternal glucose and NEFA in plasma, allowing for greater substrate availability for rapid fetal development (27, 34). Serum NEFA, one of the most important biomarkers of energy balance status, is the product of lipolysis of storage fat, such as TG. Elevated plasma NEFA level mediates many adverse metabolic effects, including obesity, insulin resistance, hypertension, and chronic inflammation (35–38). Consistently, increased plasma TNF-α concentration was found in the sows of HP group on d 110 of gestation. Tumor necrosis factor-alpha is a highly pleiotropic cytokine and is thought of as a vital mediator of inflammatory responses, metabolic activation, and cell death (39). The results of the present study demonstrated that HP sows might be in a more dramatic catabolic status to ensure the normal growth and development of the fetus during late gestation, leading to greater inflammation than LP sows, which was in accord with previous results in Shao et al. (40).
It is well-known that the dramatic changes of the microbial community can usually affect the health status of the host. In the present study, higher observed species, Shannon index, and Chao 1 index, as well as OTUs number, which was used to assess fecal microbial community richness and diversity, were observed in the HP group compared with the LP group on d 30 of gestation. Gut microbial diversity has been regarded as a new biomarker of health and metabolic capacity and low microbial diversity was often associated with poor health status such as inflammatory response, oxidative stress, and obesity (41, 42). In addition, sows in the HP group had the higher abundances of Eubacterium_coprostanoligenes_group, Lachnospiraceae_XPB1014_group, Ruminococcaceae_UCG-010, Roseburia, and Ruminococcaceae_UCG-002 on d 30 of gestation. Eubacterium coprostanoligenes is a cholesterol-reducing bacterium and inversely correlated with the inflammatory response (43, 44). Li et al. (45) found that feeding Eubacterium coprostanoligenes to germ-free mice decreased blood CHOL concentration. Consistently, the correlation analysis in the present study also demonstrated that the relative abundance of Eubacterium_coprostanoligenes_group was negatively correlated with plasma CHOL concentration. Lachnospiraceae family are abundant in healthy humans (46) and can impact their hosts by producing SCFAs, converting primary to secondary bile acids, and competitively inhibiting colonization of intestinal pathogens (47, 48). Ruminococcaceae, which has carbohydrate-active enzymes, sugar transport mechanisms, and metabolic pathways for the degradation of complex plant materials (49), is a common digestive tract microbe. Fomenky et al. (50) showed that Ruminococcaceae might enhance mucus production and benefit to improve inflammatory responses in calves. In the present study, Ruminococcaceae_UCG-010 was shown to be negatively associated with the plasma concentration of proinflammatory factor IL-2. Roseburia is a prominent gut-associated butyrate-producing genus (51) and inversely correlated with many diseases, such as inflammatory bowel disease (52) and atherosclerotic lesion (53). Consistently, increased fecal butyrate concentration was found in sows in the HP group. Microbial-driven butyrate has been shown to exhibit protective effects toward inflammatory diseases (54). Previous study has shown that butyrate oxidation can make up around 70 and 60% of the oxygen consumption in human descending colon and ascending colon, and inhibit the proliferation of aerobic pathogens (55). These findings might partly explain the better health status of HP sows at early gestation.
Interestingly, the significant difference in alpha diversity disappeared, but significant difference was observed in beta diversity between HP and LP groups on d 110 of gestation. This was in keeping with the results in Uryu et al. (12) who explored the relationship between sow productive capacities and the fecal microbiota in different farms. However, Shao et al. (40) reported that alpha diversity and beta diversity both differed between sows with high- and low-reproductive performance during late gestation. It might suggest that the beta diversity, not alpha diversity, was a critical factor to evaluate the effect of gut microbiota on sow reproductive performance (12).
In addition, compared with sows in the LP group, sows in the HP group had the lower abundances of Bacteroidetes (including Prevotellaceae_UCG-001, Prevotella_1, and dgA-11_gut_group) and Spirochaetes (Treponema_2) which were negatively correlated with litter size, but the higher abundance of Firmicutes (containing Lactobacillus, Christensenellaceae_R-7_group, and Terrisporobacter) and genus Escherichia-Shigella exhibited positive correlations with litter size on day 110 of gestation. In the present study, Firmicutes and Bacteroidetes were the most predominant phyla, regardless of the stage of gestation, which were in accordance with previous studies on sows (40, 56, 57). A previous study in obese children showed that the abundance of Firmicutes had the positive association with plasma TNF-α level (58). Bacteroidetes, as well as Treponema_2, includes a large number of cellulases, glycoside hydrolases, glycosyl transferases, and have the capacity to degrade polymers such as cellulose, hemicellulose, and lignin (59, 60), which might be the reason for the decreases in fecal concentrations of acetate, propionate, and total SCFAs. Previous studies indicated that a changed gut microbiota characterized by increased levels of Firmicutes and depleted Bacteroidetes was associated with chronic or low-grade inflammation (11). Escherichia-Shigella, belonging to phylum Proteobacteria, is generally taken as non-pathogenic bacteria and can become pathogenic bacteria when stimulated by stress (61). Shao et al. (40) also reported that predicted metabolic functions related to lipopolysaccharide biosynthesis significantly higher in HP sows than in LP sows during late gestation. The greater production of total SCFAs and propionate on d 110 of gestation in the LP group may be a compensatory mechanism in order to ensure the survival of fetuses and try to reduce pathogenic microorganisms, which need to be further studied. Moreover, Koren et al. (10) showed that dramatical alterations of species and abundance of gut microbiota contributed to the metabolic changes during gestation which was characterized by greater adiposity and insulin resistance to meet the needs of the rapid growth of fetuses during late gestation in human. Therefore, it might suggest that the alteration in gut microbiota during late gestation, associated with the increases in plasma TG and NEFA, in sows with high-reproductive performance might be more conducive to the growth and development of the fetus.
Interestingly, we also found increased abundance of Terrisporobacter that had significant negative correlations with the plasma NEFA concentration, which might be helpful to decrease the plasma NEFA from the HP sow and resist inflammatory response during late gestation. We also found increased plasma IgM concentration in HP sows on d 110 of gestation, which might be related to the increased abundance of Lactobacillus and the decreased abundance of dgA-11_gut_group. Wang et al. (62) reported that Lactobacillus supplementation in weanling piglets could increase plasma level of IgM. Immunoglobulin M, serving as the first line of host defense against infections, is the first antibody isotype to appear during immune responses and plays a vital role in immune regulation and immunological tolerance (63). The abundance of dgA-11_gut_group was negatively correlated with the plasma IgM concentration in the present study. This might be an important reason that the microecological balance of the intestinal tract of HP sows could restore during lactation (40).
In addition, we explored the shifts in plasma parameters, fecal metabolites, and microbiota from gestation d 30 to d 110 in the present study. The results indicated that plasma level of TG was increased, but levels of CHOL, HDL-C, and LDL-C were reduced on d 110 of gestation. Ji et al. (56) also showed that plasma concentrations of total CHOL and HDL-C were reduced from gestation d 60 to d 110. It suggested that lipid metabolism in the hepatic and adipose tissues of sows were activated to maintain the nutritional needs of the fetus in late gestation. Dramatic switches in lipid catabolism were often associated with inflammatory responses (64). Consistently, plasma concentrations of IL-6 and IL-10 were both elevated on d 110 of pregnancy. Interleukin-6 is a pleiotropic pro-inflammatory cytokine and involved in chronic inflammation and immune regulatory cascades (65). Interleukin-10, a prototypical anti-inflammatory cytokine produced by CD4 (+) cells, plays an important role in inhibiting inflammatory reaction by suppressing the upstream activities of antigen presenting cells and T cell functions (66). Increased serum concentration of IL-6 frequently accompanied an increased level of IL-10 in serum under inflammatory conditions (67). The alteration of abundances of phyla Firmicutes, Bacteroidetes, and Verrucomicrobia was in keeping with the results in Zhou et al. (11) that Firmicutes was significantly decreased while Bacteroidetes and Verrucomicrobia increased from d 30 to d 110 of gestation. However, Zhou et al. (11) observed an increase in Actinobacteria at late gestation, which was in line with Liu et al. (68). It suggested that the changes of abundance of Actinobacteria might be not associated with the progress of gestation. In terms of the genus level, the relative abundances of fecal Streptococcus, Oscillospira, and Ruminococcaceae_UCG-010 were increased, and that of fecal Terrisporobacter was decreased with progression of pregnancy. Zhou et al. (11) also showed increased Oscillospira and decreased Terrisporobacter in sow feces from gestation d 30 to d 110. The changes of abundances of Terrisporobacter and Ruminococcaceae_UCG-010 were in accord with alteration of plasma CHOL concentration. Streptococcus, including Gram-positive organisms shaped in cocci and organized in chains, are commensals, pathogens, and opportunistic pathogens for humans and animals (69). Previous study in humans also reported that Streptococcus was enriched in late gestation compared to in early gestation (10). However, Zhou et al. (11) found a reduction in fecal Streptococcus from gestation d 30 to d 110. Therefore, further study is required to indentify which microbiome is involved in the progress of pregnancy. Interestingly, the SCFAs were not significantly altered during gestation although significant microbiota compositions occurred, which was consistent with Liu et al. (68) and Zhou et al. (11). Above all, the sows underwent dramatic metabolic changes over the course of a normal pregnancy, which was associated with the profound alteration of the gut microbiota.
Conclusion
In summary, our findings demonstrated that microbial abundances and community structures differed significantly between sows with different litter sizes during gestation, which was associated with changes in plasma biochemical parameters, inflammatory factors, and immunoglobulin, as well as fecal metabolites. Besides, plasma biochemical parameters and cytokines shifted dramatically from gestation d 30 to d 110, which were associated with the alterations in microbial composition and diversity. These findings revealed that sow reproductive performance might be associated with the changes of maternal gut microbiota during gestation and provided a microbial perspective to improve sow reproductive performance in pig production.
Data Availability Statement
The assembled HiSeq sequences obtained in the present study were submitted to National Center of Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession PRJNA721963 (Illumina sequences).
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Shandong Agricultural University.
Author Contributions
YL: conceptualization, investigation, supervision, and writing—review and editing. JC: data curation, project administration, and writing—original draft. JC and FL: formal analysis. WY: funding acquisition. SJ and YL: methodology. JC and YL: software. FL, WY, and YL: validation. JC and SJ: visualization. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Starting Research Fund from the Shandong Agricultural University (040/72185) and the Shandong Province Pig Industry Technology System (SDAIT-08-04).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.793174/full#supplementary-material
References
- 1.Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. (2009) 136:1989–2002. 10.1053/j.gastro.2009.02.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letarov A, Kulikov E. The bacteriophages in human- and animal body-associated microbial communities. J Appl Microbiol. (2009) 107:1–13. 10.1111/j.1365-2672.2009.04143.x [DOI] [PubMed] [Google Scholar]
- 3.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. (2016) 14:e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. (2011) 9:279–90. 10.1038/nrmicro2540 [DOI] [PubMed] [Google Scholar]
- 5.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. (2011) 334:105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. (2009) 325:617–20. 10.1126/science.1172747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chassaing B, Ley RE, Gewirtz AT. Intestinal epithelial cell toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology. (2014) 147:1363–77.e1317. 10.1053/j.gastro.2014.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. (2011) 13:517–26. 10.1016/j.cmet.2011.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng J, Tang Y, Huang Y. Gut health: The results of microbial and mucosal immune interactions in pigs. Anim Nutr. (2021) 7:282–94. 10.1016/j.aninu.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Bäckhed HK, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. (2012) 150:470–80. 10.1016/j.cell.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P, Zhao Y, Zhang P, Li Y, Gui T, Wang J, et al. Microbial mechanistic insight into the role of inulin in improving maternal health in a pregnant sow model. Front Microbiol. (2017) 8:2242. 10.3389/fmicb.2017.02242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uryu H, Tsukahara T, Ishikawa H, Oi M, Otake S, Yamane I, et al. Comparison of productivity and fecal microbiotas of sows in commercial farms. Microorganisms. (2020) 8:1469. 10.3390/microorganisms8101469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Böhmer BM, Kramer W, Roth-Maier DA. Dietary probiotic supplementation and resulting effects on performance, health status, and microbial characteristics of primiparous sows. J Anim Physiol Anim Nutr. (2006) 90:309–15. 10.1111/j.1439-0396.2005.00601.x [DOI] [PubMed] [Google Scholar]
- 14.Zhang Q, Li J, Cao M, Li Y, Zhuo Y, Fang Z, et al. Dietary supplementation of Bacillus subtilis PB6 improves sow reproductive performance and reduces piglet birth intervals. Anim Nutr. (2020) 6:278–87. 10.1016/j.aninu.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Asmakh M, Hedin L, Pettersson S. Maternal microbiota regulate glucocorticoids levels and placental development in mice. Endocr Abstr. (2015) 37:1. 10.1530/endoabs.37.OC5.111032665 [DOI] [Google Scholar]
- 16.Che L, Yang Z, Xu M, Zhang Z, Liu P, Xu S, et al. Dietary energy intake affects fetal survival and development during early and middle pregnancy in Large White and Meishan gilts. Anim Nutr. (2015) 1:152–9. 10.1016/j.aninu.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Che L, Xu M, Yang Z, Xu S, Che L, Lin Y, et al. Detection of placental proteomes at different uterine positions in Large White and Meishan Gilts on gestational day 90. PLoS ONE. (2016) 11:e0167799. 10.1371/journal.pone.0167799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Research Council . Nutrient Requirements of Swine, 11th rev.ed. Washington, DC: National Academy Press; (2012). [Google Scholar]
- 19.Li Y, He J, Zhang L, Liu H, Cao M, Lin Y, et al. Effects of dietary fiber supplementation in gestation diets on sow performance, physiology and milk composition for successive three parities. Anim Feed Sci Tech. (2021) 276:114945. 10.1016/j.anifeedsci.2021.114945 [DOI] [Google Scholar]
- 20.Li Y, Zhao X, Zhang L, Zhan X, Liu Z, Zhuo Y, et al. Effects of a diet supplemented with exogenous catalase from penicillium notatum on intestinal development and microbiota in weaned piglets. Microorganisms. (2020) 8:30391. 10.3390/microorganisms8030391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 22.Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol. (2017) 261:169–76. 10.1016/j.jbiotec.2017.06.1198 [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Li S, Zheng J, Li W, Jiang X, Zhao X, et al. Effects of dietary Clostridium butyricum supplementation on growth performance, intestinal development, and immune response of weaned piglets challenged with lipopolysaccharide. J Anim Sci Biotechnol. (2018) 9:62. 10.1186/s40104-018-0275-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallego S, Esbrí JM, Campos JA, Peco JD, Martin-Laurent F, Higueras P. Microbial diversity and activity assessment in a 100-year-old lead mine. J Hazard Mater. (2021) 410:124618. 10.1016/j.jhazmat.2020.124618 [DOI] [PubMed] [Google Scholar]
- 25.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. (2005) 71:8228–35. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto J, Barros AS, Domingues MR, Goodfellow BJ, Galhano E, Pita C, et al. Following healthy pregnancy by NMR metabolomics of plasma and correlation to urine. J Proteome Res. (2015) 14:1263–74. 10.1021/pr5011982 [DOI] [PubMed] [Google Scholar]
- 27.Lain KY, Catalano PM. Metabolic changes in pregnancy. Clin Obstet Gynecol. (2007) 50:938–48. 10.1097/GRF.0b013e31815a5494 [DOI] [PubMed] [Google Scholar]
- 28.Ekhator CN, Ebomoyi MI. Blood glucose and serum lipid profiles during pregnancy. Afr J Diabetes Med. (2012) 20:e15320. 10.1097/MD.0000000000015320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salman L, Arbib N, Borovich A, Shmueli A, Chen R, Wiznitzer A, et al. The impact of first trimester fasting glucose level on adverse perinatal outcome. J Perinatol. (2018) 38:451–5. 10.1038/s41372-018-0045-7 [DOI] [PubMed] [Google Scholar]
- 30.Leane S, Herlihy MM, Curran F, Kenneally J, Forde N, Simintiras CA, et al. The effect of exogenous glucose infusion on early embryonic development in lactating dairy cows. J Dairy Sci. (2018) 101:11285–96. 10.3168/jds.2018-14894 [DOI] [PubMed] [Google Scholar]
- 31.Jindal R, Cosgrove JR, Foxcroft GR. Progesterone mediates nutritionally induced effects on embryonic survival in gilts. J Anim Sci. (1997) 75:1063–70. 10.2527/1997.7541063x [DOI] [PubMed] [Google Scholar]
- 32.Kockx M, Kritharides L. Triglyceride-rich lipoproteins. Cardiol Clin. (2018) 36:265–75. 10.1016/j.ccl.2017.12.008 [DOI] [PubMed] [Google Scholar]
- 33.Bin P, Azad MAK, Liu G, Zhu D, Kim SW, Yin Y. Effects of different levels of methionine on sow health and plasma metabolomics during late gestation. Food Funct. (2018) 9:4979–88. 10.1039/C8FO01477A [DOI] [PubMed] [Google Scholar]
- 34.Père MC, Etienne M. Insulin sensitivity during pregnancy, lactation, and postweaning in primiparous gilts. J Anim Sci. (2007) 85:101–10. 10.2527/jas.2006-130 [DOI] [PubMed] [Google Scholar]
- 35.Shi X, Li D, Deng Q, Li Y, Sun G, Yuan X, et al. NEFAs activate the oxidative stress-mediated NF-κB signaling pathway to induce inflammatory response in calf hepatocytes. J Steroid Biochem Mol Biol. (2015) 145:103–12. 10.1016/j.jsbmb.2014.10.014 [DOI] [PubMed] [Google Scholar]
- 36.Spiller S, Blüher M, Hoffmann R. Plasma levels of free fatty acids correlate with type 2 diabetes mellitus. Diabetes Obes Metab. (2018) 20:2661–9. 10.1111/dom.13449 [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Li C, Ali I, Li L, Wang G. N-acetylcysteine modulates non-esterified fatty acid-induced pyroptosis and inflammation in granulosa cells. Mol Immunol. (2020) 127:157–63. 10.1016/j.molimm.2020.09.011 [DOI] [PubMed] [Google Scholar]
- 38.Yang WS, Li R, Shen YQ, Wang XC, Liu QJ, Wang HY, et al. Importance of lipid ratios for predicting intracranial atherosclerotic stenosis. Lipids Health Dis. (2020) 19:160. 10.1186/s12944-020-01336-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-α-induced apoptosis by NF-κB. Science. (1996) 274:787–9. 10.1126/science.274.5288.787 [DOI] [PubMed] [Google Scholar]
- 40.Shao Y, Zhou J, Xiong X, Zou L, Kong X, Tan B, et al. Differences in gut microbial and serum biochemical indices between sows with different productive capacities during perinatal period. Front Microbiol. (2020) 10:3047. 10.3389/fmicb.2019.03047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiao Y, Sun J, Ding Y, Le G, Shi Y. Alterations of the gut microbiota in high-fat diet mice is strongly linked to oxidative stress. Appl Microbiol Biotechnol. (2013) 97:1689–97. 10.1007/s00253-012-4323-6 [DOI] [PubMed] [Google Scholar]
- 42.Gong D, Gong X, Wang L, Yu X, Dong Q. Involvement of reduced microbial diversity in inflammatory bowel disease. Gastroent Res Pract. (2016) 2016:1–7. 10.1155/2016/6951091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye JZ, Li YT, Wu WR, Shi D, Fang DQ, Yang LY, et al. Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J Gastroenterol. (2018) 24:2468–81. 10.3748/wjg.v24.i23.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vlacil AK, Schuett J, Ruppert V, Soufi M, Oberoi R, Shahin K, et al. Deficiency of Nucleotide-binding oligomerization domain-containing proteins (NOD) 1 and 2 reduces atherosclerosis. Basic Res Cardiol. (2020) 115:47. 10.1007/s00395-020-0806-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Batt SM, Wannemuehler M, Dispirito A, Beitz DC. Effect of feeding of a cholesterol-reducing bacterium, Eubacterium coprostanoligenes, to germ-free mice. Lab Anim Sci. (1998) 48:253–5. [PubMed] [Google Scholar]
- 46.Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol. (2002) 9:33–9. 10.1111/j.1574-6941.2002.tb00904.x [DOI] [PubMed] [Google Scholar]
- 47.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. (2015) 517:205–8. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. (2016) 19:443–54. 10.1016/j.chom.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. (2013) 5:627–40. 10.3390/d5030627 [DOI] [Google Scholar]
- 50.Fomenky BE, Do DN, Talbot G, Chiquette J, Bissonnette N, Chouinard YP, et al. Direct-fed microbial supplementation influences the bacteria community composition of the gastrointestinal tract of pre- and post-weaned calves. Sci Rep. (2018) 8:14147. 10.1038/s41598-018-32375-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson H, Barrett H, Gomez-Arango L, McIntyre HD, Callaway L, Dekker Nitert M. Ketonuria is associated with changes to the abundance of Roseburia in the gut microbiota of overweight and obese women at 16 weeks gestation: a cross-sectional observational study. Nutrients. (2019) 11:1836. 10.3390/nu11081836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. (2020) 51:102590. 10.1016/j.ebiom.2019.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasahara K, Krautkramer KA, Org E, Romano KA, Kerby RL, Vivas EI, et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat Microbiol. (2018) 3:1461–71. 10.1038/s41564-018-0272-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong X, Zhang Z, Wang S, Cao L, Zhou L, Sun A, et al. Microbial-driven butyrate regulates jejunal homeostasis in piglets during the weaning stage. Front Microbiol. (2019) 9:3335. 10.3389/fmicb.2018.03335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu J, Lin S, Zheng B, Cheung PC. Short-chain fatty acids in control of energy metabolism. Crit Rev Food Sci Nutr. (2018) 58:1243–49. 10.1080/10408398.2016.1245650 [DOI] [PubMed] [Google Scholar]
- 56.Ji YJ, Li H, Xie PF, Li ZH, Li HW, Yin YL, et al. Stages of pregnancy and weaning influence the gut microbiota diversity and function in sows. J Appl Microbiol. (2019) 127:867–79. 10.1111/jam.14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Liu H, Zhang L, Yang Y, Lin Y, Zhuo Y, et al. Maternal dietary fiber composition during gestation induces changes in offspring antioxidative capacity, inflammatory response, and gut microbiota in a sow model. Int J Mol Sci. (2020) 21:31. 10.3390/ijms21010031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orbe-Orihuela YC, Lagunas-Martínez A, Bahena-Román M, Madrid-Marina V, Torres-Poveda K, Flores-Alfaro E, et al. High relative abundance of firmicutes and increased TNF-α levels correlate with obesity in children. Salud Publica Mex. (2018) 60:5–11. 10.21149/8133 [DOI] [PubMed] [Google Scholar]
- 59.Rosewarne CP, Cheung JL, Smith WJ, Evans PN, Tomkins NW, Denman SE, et al. Draft genome sequence of Treponema sp. strain JC4, a novel spirochete isolated from the bovine rumen. J Bacteriol. (2012) 194:4130. 10.1128/JB.00754-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, et al. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. (2013) 7:1026–37. 10.1038/ismej.2012.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lutful Kabir SM. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int J Environ Res Public Health. (2010) 7:89–114. 10.3390/ijerph7010089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang T, Teng K, Liu Y, Shi W, Zhang J, Dong E, et al. Lactobacillus plantarum PFM 105 promotes intestinal development through modulation of gut microbiota in weaning piglets. Front Microbiol. (2019) 10:90. 10.3389/fmicb.2019.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu J, Wang Y, Xiong E, Hong R, Lu Q, Ohno H, et al. Role of the IgM Fc receptor in immunity and tolerance. Front Immunol. (2019) 10:529. 10.3389/fimmu.2019.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim SW, Weaver AC, Shen YB, Zhao Y. Improving efficiency of sow productivity: nutrition and health. J Anim Sci Biotechnol. (2013) 4:26. 10.1186/2049-1891-4-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jordan SC, Choi J, Kim I, Wu G, Toyoda M, Shin B, et al. Interleukin-6, a cytokine critical to mediation of inflammation, autoimmunity and allograft rejection: therapeutic implications of il-6 receptor blockade. Transplantation. (2017) 101:32–44. 10.1097/TP.0000000000001452 [DOI] [PubMed] [Google Scholar]
- 66.Kumar R, Ng S, Engwerda C. The role of IL-10 in malaria: a double edged sword. Front Immunol. (2019) 10:229. 10.3389/fimmu.2019.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jndoyan ZT, Bablumyan AY, Ginosyan KV, Shekoyan SV. Correlations between indicators of interleukin-10 and interleukin-6 in patients with periodic disease. Ter Arkh. (2018) 90:38–41. 10.26442/terarkh201890338-41 [DOI] [PubMed] [Google Scholar]
- 68.Liu H, Hou C, Li N, Zhang X, Zhang G, Yang F, et al. Microbial and metabolic alterations in gut microbiota of sows during pregnancy and lactation. FASEB J. 33:4490–501. 10.1096/fj.201801221RR [DOI] [PubMed] [Google Scholar]
- 69.Haenni M, Lupo A, Madec JY. Antimicrobial resistance in Streptococcus spp. Microbiol Spectr. (2018) 6:9804. 10.1128/9781555819804.ch8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled HiSeq sequences obtained in the present study were submitted to National Center of Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession PRJNA721963 (Illumina sequences).