Abstract
DNA variants that alter gene expression in trans are important sources of phenotypic variation. Nevertheless, the identity of trans-acting variants remains poorly understood. Single causal variants in several genes have been reported to affect the expression of numerous distant genes in trans. Whether these simple molecular architectures are representative of trans-acting variation is unknown. Here, we studied the large RAS signaling regulator gene IRA2, which contains variants with extensive trans-acting effects on gene expression in the yeast Saccharomyces cerevisiae. We used systematic CRISPR-based genome engineering and a sensitive phenotyping strategy to dissect causal variants to the nucleotide level. In contrast to the simple molecular architectures known so far, IRA2 contained at least seven causal nonsynonymous variants. The effects of these variants were modulated by nonadditive, epistatic interactions. Two variants at the 5′-end affected gene expression and growth only when combined with a third variant that also had no effect in isolation. Our findings indicate that the molecular basis of trans-acting genetic variation may be considerably more complex than previously appreciated.
Keywords: eQTL, trans eQTL, IRA2, yeast, regulatory variation, causal variant, single-nucleotide variants
Introduction
Gene expression variation is a key bridge between DNA variation and organismal traits (Albert and Kruglyak 2015). In particular, trans-acting genetic effects profoundly shape gene expression in yeast crosses (Brem et al. 2002; Albert et al. 2018) and human populations (Grundberg et al. 2012; Wright et al. 2014) and are especially important contributors to phenotypic variation (The GTEx Consortium 2020). Trans-acting DNA variants change the activity or abundance of a regulatory factor. This factor influences the expression of other genes, which can be located anywhere in the genome. In yeast crosses, trans-acting variation arises almost exclusively from “hotspot” regions that contain one or potentially more DNA variants that alter the expression of dozens to thousands of genes (Albert et al. 2018). Due to their profound effects, these hotspots serve as models for understanding trans-acting regulatory variation.
A critical unsolved question is the nature of the individual DNA variants that cause trans-acting variation. Genetic effects on gene expression are commonly identified as expression quantitative trait loci (eQTLs). Like QTLs affecting organismal traits (Mackay et al. 2009), most eQTLs span wide genomic regions, often with multiple genes and dozens of DNA variants. Several yeast hotspots have been dissected to causal genes (Smith and Kruglyak 2008; Lewis and Gasch 2012; Brion et al. 2013) and single causal variants (Brem et al. 2002; Yvert et al. 2003; Brown et al. 2008; Zhu et al. 2008; Kim et al. 2009; Fehrmann et al. 2013; Sudarsanam and Cohen 2014; Lutz et al. 2019). However, experimental biases favor the identification of single variants with strong effects, compared to multiple variants with smaller effects (Rockman 2012). Careful dissection of QTLs for organismal traits has shown that QTLs need not arise from single causal variants (Steinmetz et al. 2002; Fidalgo et al. 2006; Gerke et al. 2009; Flint and Mackay 2009; Rat Genome Sequencing and Mapping Consortium 2013; Fay 2013). The complexity of the genetic architectures underlying the profound trans-acting effects on gene expression is unknown.
A particularly prominent trans-eQTL hotspot is caused by variation in the protein-coding region of the IRA2 gene (Smith and Kruglyak 2008). IRA2 encodes a key regulator of the conserved RAS signaling pathway. Together with IRA1, IRA2 is one of two paralogous genes that encode the yeast homologs of the human Neurofibromin (NF1) gene. NF1 mutations can cause neurofibromatosis type 1, a disease characterized by uncontrolled cell growth (Ballester et al. 1990; Ratner and Miller 2015). In a cross between the laboratory strain “BY” and the vineyard isolate “RM”, IRA2 resides in a trans-eQTL hotspot that affects the abundance of up to 1240 mRNAs (Albert et al. 2018), numerous proteins (Albert et al. 2014; Großbach et al. 2019), and cell growth in diverse environmental conditions (Bloom et al. 2013; Breunig et al. 2014; Wang and Kruglyak 2014). Variation in IRA2 may also underlie QTLs for a variety of traits in strains other than BY and RM (Parts et al. 2011; Wang et al. 2019; Stojiljkovic et al. 2020). In spite of these important effects on many traits, the causal variant or variants in IRA2 remain unknown. In part, this is due to the large size of IRA2. At 9,240 base pairs (bp), the IRA2 open reading frame (ORF) is one of the 10 longest ORFs in the yeast genome and contains many variants (Cherry et al. 2012).
Here, we leveraged recent advances in genome engineering and phenotyping (Lutz et al. 2019) to dissect the molecular basis of the IRA2 hotspot on trans gene expression. We show that IRA2 contains at least seven causal nonsynonymous variants whose effects are modulated by epistatic interactions among neighboring variants and along the length of the gene. In contrast to the simpler cases known so far, IRA2 shows that trans-eQTL hotspots can have a complex molecular genetic architecture, even in a single gene.
Materials and methods
Yeast strains and media
The Saccharomyces cerevisiae strains used here are derived from S288C [BY4741 (MATa, his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), referred to as “BY” in the text] and RM-11a [RM HO(BY) (MATa, his3Δ1::CloNAT, leu2Δ0, ura3Δ0 HO(BY allele)) AMN1(BY allele), referred to as “RM”]. A complete listing of the strains used in this study can be found in Supplementary Table S1. All primers are listed in Supplementary Table S2. We used the following media (recipes are for 1 l):
For general yeast strain growth and storage:
YPD (10 g yeast extract, 20 g peptone, 20 g glucose);
For selection of the GFP-HIS3MX6 cassette in GPH1 tagging:
SDC-His (1.66 g SC-His-Leu-Ura, 100 mg leucine, 200 mg uracil, 20 g glucose);
For selection of the KanMX4 cassette in IRA2 gene deletions and verifying allele exchanges by CRISPR-Swap:
YPD + G418 [G418 sulfate (Fisher Scientific cat # BP6731); 200 μg/ml];
For selection of the CRISPR-Swap plasmid in cells after transformation:
SDC-Leu (1.66 g SC -His -Leu -Ura (Sunrise Science; cat # 1327-030), 50 mg histidine, 200 mg uracil, 20 g glucose);
For phenotyping Gph1-GFP expression:
YNB LowFlo (6.7 g yeast nitrogen base-folic acid-riboflavin with ammonium sulfate and without amino acids (Sunrise Science; cat # 1536-050), 20 g glucose, 50 mg histidine, 100 mg leucine, 50 mg methionine, 200 mg uracil). Sterilized by filtration; and
For iodine staining:
YNB [6.7 g yeast nitrogen base with ammonium sulfate and without amino acids (BD Biosciences cat# 291940), 20 g glucose, 50 mg histidine, 100 mg leucine, 50 mg methionine, 200 mg uracil].
For solid media, 20 g/l agar was added prior to autoclaving. Yeast was grown at 30°C.
GPH1-GFP tagging and IRA2 gene deletions
Insertions of cassettes for genome modification were performed using a standard PCR-based one-step method (Longtine et al. 1998). Yeast transformations were performed using a standard LiAc procedure (Gietz and Schiestl 2007). Transformants expressing the selectable marker were single colony purified, and insertion of the cassette into the correct location and absence of the unmodified wild type allele were verified by colony PCR.
Strain BY GPH1-GFP:HIS3MX6 (YFA0644) was obtained from the GFP collection (Huh et al. 2003). RM HO(BY) GPH1-GFP:HIS3MX6 (YFA0649) was created by integration of the GPH1-GFP allele, which was PCR amplified from YFA0644 genomic DNA using primers OFA0471 and OFA0472, into RM HO(BY) (YFA0254).
Strains BY GPH1-GFP ira2Δ::KanMX (YFA0650 and YFA01430) and RM GPH1-GFP ira2::KanMX (YFA0654) were created by integration of the ira2Δ::kanMX allele, which was PCR amplified from YFA0666 genomic DNA using primers OFA0468 and OFA0469. BY GPH1-GFP ira2Δ::KanMX was created twice because glycerol stock storage at −80°C of YFA0650 resulted in a stark reduction in CRISPR-Swap efficiency.
Strain BY GPH1-GFP ira2_block1Δ::KanMX (YFA1443) was created by replacing IRA2 block 1 sequence with the KanMX cassette PCR amplified from nej1Δ::KanMX (YFA0007) genomic DNA using primers OFA1079 and OFA1080.
Creation of IRA2 allele fragments used as repair templates for CRISPR-Swap
All IRA2 allele fragments were PCR amplified from genomic DNA or commercially synthesized DNA (Twist Bioscience) using Phusion Hot Start Flex DNA polymerase (NEB). Genomic DNA was isolated using the 10-min preparation (Hoffman and Winston 1987). All PCR fragments were analyzed by agarose gel electrophoresis, excised, purified using Monarch DNA Gel Extraction Kit (NEB), and quantified using Qubit fluorometric quantification (Thermo Fisher Scientific).
The IRA2(BY) allele was amplified from BY4741 or YLK1879 genomic DNA and the IRA2(RM) allele (sometimes referred to as “full-RM”) from YFA0254 or YLK1950 genomic DNA using primers OFA0468 and OFA0469. These primers amplify a 9,431 bp IRA2 fragment with termini (123 bp at the 5′ end and 68 bp at the 3′ end) that have a sequence identical to the region flanking the ira2Δ::KanMX cassette.
The IRA2 block alleles were created using gene splicing by overlap extension (SOEing; Horton et al. 1989). Each of the 4 block regions was PCR amplified from BY (BY4741 for blocks 1 and 2 and YLK1879 for blocks 3 and 4) and RM (YFA0254) genomic DNA. Primers used were: block 1, OFA0467, and OFA0455; block 2, OFA0456 and OFA0457; block 3, OFA0458 and OFA0459; and block 4, OFA0460 and OFA0544. Pairwise fusions of block 1 and block 2 (four combinations) were amplified with primers OFA0467 and OFA0457 and block 2 and block 3 (four combinations) were amplified with primers OFA0458 and OFA0544. Finally, all pairwise fusions of blocks 1 and 2, and blocks 3 and 4 (16 combinations) were fused by amplification with primers OFA0468 and OFA0469. The IRA2 block 1 RM allele was created in the same manner as the IRA2 block allele RBBB.
Single-variant alleles were created by PCR SOEing using complementary primers containing the desired RM variant (see Supplementary Table S3). In the first step, two PCR amplifications were performed using BY (YLK1879) genomic DNA as a template. The first amplification used the variant-specific reverse primer and OFA0467, and the second amplification used the variant-specific forward primer and OFA0544. In the second step, the two PCR fragments were fused using primers OFA0468 and OFA0469.
The IRA2 block 1 synonymous and nonsynonymous variant alleles were PCR amplified using primers OFA0468 and OFA1124. Amplification with these primers creates a 1,978 bp IRA2 fragment with termini (123 bp at the 5′ end and 54 bp at the 3′ end) that are identical to the region flanking the ira2_block1Δ::KanMX cassette. The synBYnonsynBY and synRMnonsynRM alleles were amplified from BY and RM genomic DNA (YLK1879 and YLK1950, respectively), and the synBYnonsynRM and synRMnonsynBY alleles were amplified from synthetic DNA (Twist Bioscience).
The block 1 variant replacement alleles were created in two steps. In the first step, two fragments were PCR amplified using BY genomic DNA (YLK1879) or synBYnonsynRM synthetic DNA (Twist Bioscience) as a template. In the second step, the two fragments were fused by PCR SOEing using primers OFA0468 and OFA1124 (see Supplementary Table S3 for further details).
The alleles of all combinations of variants 3, 4, and 6 were created by PCR SOEing. The individual variant alleles were created first and then used as templates for the creation of the remaining combinations (Supplementary Table S3).
CRISPR-Swap
We followed the protocol of Lutz et al. (2019). For each CRISPR-Swap transformation, the amount of the IRA2 allele repair fragments ranged from 1000 to 1500 ng, and the amount of plasmid pFA0055 (Addgene #131774) was ∼300 ng. pFA0055 expresses Cas9 as well as the guide RNA (gCASS5a) that directs Cas9 to the 5′-end of the KanMX cassette. We recovered between 10 and 320 transformants when engineering in BY using the 9,431 bp IRA2 repair template and 504 to more than 900 transformants when using the 1,978 bp IRA2 block 1 repair template. Of the single colony-streaked transformants, 98–100% had lost G418 resistance, indicating a successful allele exchange.
The 16 chimeric block strains were engineered in one batch, as were the strains with the four arrangements of synonymous and nonsynonymous variants in block 1. The BY and RM background strains were engineered in two batches: one created the BY GPH1-GFP IRA2(BY) and RM GPH1-GFP IRA2(RM) strains and the other the BY GPH1-GFP IRA2(RM) and RM GPH1-GFP IRA2(BY) strains. The IRA2 single-variant strains were engineered in four batches, corresponding to the four IRA2 blocks. In each batch, a new IRA2(BY) strain was also engineered, and in batches 1, 2, and 4 a new IRA2(RM) strain was engineered. The IRA2(BY) strains from these four batches were similar in their effect on Gph1-GFP expression and therefore were grouped into one IRA2(BY) genotype that served as the baseline for determining the effects of the other alleles. The IRA2(RM) strains were similarly grouped. The IRA2 block 1 RM strain was created in the same batch (block 1) as variants 1–8. The 16 block 1 variant replacement alleles were created in two batches. In the first batch were alleles, A, D, E, F, G, H, I, J, K, L, M, N along with block 1 full-RM (allele P) and full-BY (allele O), and the second batch had alleles b, c, g, and h, with letter definitions given in Supplementary Table S3. The strains carrying all combinations of variants 3, 4, and 6 as RM were created in two batches. The first batch had all alleles except for ff (variants 4 and 6 RM). In the second batch with the ff allele, a new BY allele was also created.
Verification of IRA2 alleles after CRISPR-Swap
All G418 sensitive transformants after CRISPR-Swap were assumed to have exchanged the KanMX allele for the provided IRA2 allele. To verify the presence of the correct IRA2 allele and that strain mix-ups did not occur during the experimental procedures, some of the strains were partially genotyped at different stages (see below for details). Verified strains are designated in Supplementary Table S1.
The IRA2 alleles in the BY GPH1-GFP IRA2(BY) (YFA0658 and YFA0659) and RM GPH1-GFP IRA2(RM) (YFA0662 and YFA0663) strains were genotyped as BY or RM based on an EcoRI site present in the RM but not the BY allele. Genomic DNA isolated from these strains was PCR-amplified with OFA0068 and OFA0070. The PCR product was then digested with EcoRI and analyzed by agarose gel electrophoresis.
For each of the 16 IRA2 block alleles, a representative strain, either transformant #1 or #2, was confirmed to have the expected arrangement of blocks by verification of the expected variants at the block borders after Sanger sequencing. Genomic DNA was isolated from cultures that were started from glycerol stock plates (YPFA010 and YPFA011) and used for PCR amplification. For each strain, the junction between blocks 1 and 2 was amplified using primers OFA0585 and OFA0905 and sequenced with OFA0904, and the junction between blocks 3 and 4 was amplified using primers OFA0906 and OFA0907 and sequenced with OFA0903. Prior to sequencing, the fragments were analyzed by gel electrophoresis and purified using a Monarch DNA Gel Extraction Kit (NEB). All strains had the expected variants in the sequenced fragment. One strain, RRBR #1 had a de novo variant in the sequenced region resulting in an aGc to aTc (S1860I) amino acid change. This strain was not removed from phenotyping and its Gph1-GFP expression was similar to that of the other RRBR transformants. None of the RBRR transformants were sequenced.
To identify potential ira2 loss of function alleles among the transformants of the 16 different IRA2 block strains, cells were spotted onto YNB agar plates and stained with iodine vapors. Iodine stains wild type yeast cells containing glycogen a dark reddish brown. Cells with elevated RAS-GTP levels, as in the case of an ira2Δ, are unable to store glycogen and therefore stain pale yellow in the presence of iodine (Gil and Seeling 1999). For each of the 16 strains, 7–8 transformants were stained with iodine and 7/127 stained pale yellow, possibly due to errors created during the multiple-round PCR amplification of the IRA2 alleles. Transformants that stained pale yellow were excluded from phenotyping.
In the IRA2 single-variant strains, the expected amino acid change was verified for transformants of N148H, S149N, N201S, Y302H, H306D, A345T, N467D, V567I, Q757H, and 1507V. Genomic DNA was isolated from cells taken from the randomly arrayed starter culture plates and PCR amplified using primers OFA1053 and OFA1054. The PCR fragment was then Sanger sequenced in the region of the expected variant using the following primers: OFA0592 for N148H and S149N; OFA0589 for N201S; OFA0591 for Y302H, H306D, and A345T; OFA0536 for N467D; OFA0904 for V567I; OFA0539 for Q757H; and OFA0540 for I1507V.
Each of the block 1 variant replacement alleles and the combinations of variants 3, 4, and 6 as RM alleles were Sanger-sequenced prior to the CRISPR-Swap procedure to verify that the majority of the repair template molecules had the correct sequence. After CRISPR-Swap, genomic DNA was isolated from select variant 3, 4, and 6 transformants and PCR-amplified with OFA0467 and OFA0905. The resulting fragments were Sanger-sequenced with OFA0467, OFA1117, OFA0536, and OFA1122 to verify the entire block 1 sequence. See Supplementary Table S1 for more information.
Phenotyping of Gph1-GFP expression and growth rates in the plate reader
Starter cultures were inoculated with cells from glycerol stocks and grown overnight at 30°C in 800–1000 μl of YPD medium in a 2-ml deep-96-well-plate. The plates were sealed with a Breathe Easy membrane (Diversified Biotech) and placed on an Eppendorf MixMate set at 1100 rpm. After overnight growth, the starter culture plates were sealed with aluminum foil and stored at 4°C for the duration of phenotyping.
For each phenotyping plate run, 10 μl of resuspended cells from the starter culture plate were used to inoculate 600–800 μl of YNB LowFlo medium in a 2-ml deep-well plate. These pre-cultures were grown overnight as described for the starter cultures. After overnight growth, the pre-cultures were diluted to an OD600 = 0.05 in 100 μl of YNB LowFlo medium in a 96-well flat-bottom plate (Costar) and the plates were sealed with a Breathe Easy membrane (Diversified Biotech).
The strains were grown for ∼24 h in a Synergy H1 (BioTek Instruments) plate reader at 30°C with readings taken every 15 min for 97 cycles with 10 s of orbital shaking between reads and 13 min between cycles. Cell growth was characterized using absorbance readings at 600 nm and Gph1-GFP expression was measured from the bottom of the plate using excitation at 488 nm and emission at 520 nm.
Data were processed as described in Lutz et al. (2019). Briefly, we fit growth curves to each well to identify the inflection point at which the yeast culture begins to exit exponential growth. We extracted the fluorescence and OD600 values at this point as well as the two-time points flanking it, took their respective averages, and calculated the log2 of the ratio between the average fluorescence value and the average OD600 value as the Gph1-GFP expression level for the given well.
Growth rates were extracted from the same plate reader data, defined as the number of doublings per hour at the inflection point.
Designation of IRA2 domains
Conserved regions between Ira2 and NF1 were determined by multiple sequence alignment of Ira2/Ira1/NF1 sequences from Candida glabrata (KTB00138.1), Saccharomyces paradoxus (translated from CP020290.1) Kluyveromyces lactis (translated from CR382125.1), Homo sapiens (P21359.2), and S. cerevisiae (AAA34710.1 and AAA34709.1) using Clustal Omega (Sievers et al. 2011), as provided through the EMBL-EBI analysis tool API (Madeira et al. 2019).
The CHD (Neurofibromin CTD-homology domain) and Sec-PH (Sec14 homologous and pleckstrin homology-like domain) were defined by previous studies (D’angelo et al. 2006; Luo et al. 2014). We defined the Threonine-Serine Rich Domain (TSRD) as a region that extends from amino acid 399 to 1021 and is enriched for serines and threonines, but not enriched for cysteines. This region has 23/24 of the detected phosphorylation sites in Ira2 (Holt et al. 2009; Swaney et al. 2013; Lanz et al. 2021). We chose the start of the TSRD at a cluster of serines and the end at the last detected phosphorylated residue. The TSRD is 16.5% serine and 10.6% threonine, while the flanking amino acids 1–398 and 1022–1645 have 13.1% and 6.3% serine and 8.5% and 5.3% threonine residues, respectively.
Ancestral alleles and BY and RM allele frequencies
Ancestral alleles were determined by comparison with two different evolutionary outgroups to BY and RM. First, we determined the nucleotide allele present in the closely related species S. paradoxus after an alignment of IRA2(BY) to GenBank #AABY01000044.1. Second, we obtained the nucleotide allele present in the Taiwanese S. cerevisiae isolate EN14S01 (“standardized name”: “AMH”) from Peter et al. (2018). EN14S01 is a member of the highly diverged clade 17 thought to have branched early from all other S. cerevisiae isolates. Ancestral alleles defined by these two outgroups are in good agreement (Supplementary Table S4). The population allele frequencies of the BY and RM variants across 1011 S. cerevisiae species were obtained from Peter et al. (2018). Predictions of variant effects were obtained using PROVEAN (Choi and Chan 2015; http://provean.jcvi.org) and Mutfunc (Wagih et al. 2018; http://mutfunc.com) and are based on amino acid conservation and properties. Variant changes resulting in PROVEAN scores of less than −2.5 are predicted to be deleterious.
Statistical analyses of allele effects
All statistical analyses were conducted in R (https://www.r-project.org) version 4.0.4. The effect of RM alleles on the expression of GPH1-GFP and the growth rate (y) was estimated using the “lmer” function in the lme4 package (Bates et al. 2015) for fitting mixed-effects linear models. In each case, one RM allele (such as a block or a single variant, see below) was compared to strains with the BY allele that were measured during the same runs in the plate reader. The models included the genotype (BY vs the given RM allele) as a fixed effect, plate and transformant as random effects (denoted in parentheses), and the residual error (ε):
The effects of genotype were estimated as slopes in the linear models. Significance was calculated by using type I Analysis of variance (ANOVA), as provided by the lmerTest package (Kuznetsova et al. 2017). These linear models and ANOVAs were performed to test the following effects:
The RM vs BY allele in both the BY or RM background.
The various BY/RM combinations of the four blocks.
The effect of each individual nonsynonymous variant compared to BY.
Testing for epistasis using ANOVA
We used linear models implemented in the nlme package (Pinheiro et al. 2021) to test for epistasis between the BY or RM background genotype and full BY or RM IRA2 allele, among the four blocks, and among the single variants 3, 4, and 6 in block 1. The models fitted the effects of all respective alleles (full BY/RM, the four blocks, or the three variants) simultaneously, along with all two-way, three-way (blocks and the three variants in block 1), and four-way interactions (blocks):
Background-allele interaction model:
Block model:
Three-variant model:
We tested the significance of each term using a type I ANOVA through the sequential addition of terms to the model above.
Testing for epistasis between single-nucleotide variant effects by bootstrapping
Except for variants 3, 4, and 6, the single-nucleotide variants were not engineered in all combinations, precluding the use of interaction terms in a linear model. Instead, we asked whether the effects of single variants in block 1 could sum to the observed effect of block 1. To properly take into account measurement error across our experimental design, in which individual transformants were run multiple times in several different runs of a plate reader, we used bootstrapping. We performed stratified sampling with replacement across measurement plates, ensuring that each genotype was represented at the same sample size as in the full data. For each bootstrapped experiment, we fit linear models as described above and extracted the effect estimate. Single-variant effect estimates were summed to calculate the effect they would be expected to have together under an additive model. We performed 10,000 of these bootstraps to generate two distributions. The first distribution represented the difference between BY and the full block 1 RM allele, and the second distribution represented the sum of the effects of the single variants in block 1. Significance testing was performed by calculating the overlap between the two distributions. If the central 95% quantile range of the two distributions did not overlap, we considered it significantly unlikely that the additive effects of the single variants are able to account for the observed effect of the multivariant allele. The less the two bootstrapped distributions overlap, the stronger the evidence for epistatic interactions among the given set of single variants.
Population genetic analyses
DNA variation data from 1011 yeast isolates (Peter et al. 2018) was obtained from the “1011Matrix.gvcf.gz” file available at http://1002genomes.u-strasbg.fr/files/ and handled using the vcfR R package (Knaus and Grünwald 2017). We used the popgenome R package (Pfeifer et al. 2014) to compute diversity parameters and the dNdScv package (Martincorena et al. 2017) to compute dN/dS. Sequences for the fixed substitutions between species were extracted from a local alignment of IRA2 sequences from BY and S. paradoxus (GenBank: AABY01000044.1) using SNP-sites (Page et al. 2016). Integrated haplotype score (iHS) statistics were computed using the rehh package in R (Gautier and Vitalis 2012), using a freqbin parameter of 10.
Code availability
All analysis code is available at: https://github.com/Krivand/Multiple-causal-DNA-variants-in-a-single-gene-affect-gene-expression-in-trans
Results
Protein-coding variation in IRA2 affects gene expression in trans
Between BY and RM, the IRA2 coding region differs at 26 nonsynonymous and 61 synonymous variants (Figure 1A). Comparison of the nonsynonymous variants to those in the ancestral Taiwanese isolate shows that 19 are derived in RM and 7 are derived in BY (Peter et al. 2018; Supplementary Table S4). Two nonsynonymous variants are relatively rare in the population, with a derived allele frequency of 10% or less; four other variants are predicted to be deleterious by the PROVEAN tool (Choi and Chan 2015), and five variants were predicted to be “impactful” by the mutfunc tool (Wagih et al. 2018; Supplementary Table S4). Together, this information did not suggest an individual variant as the obvious source of the effects arising from IRA2.
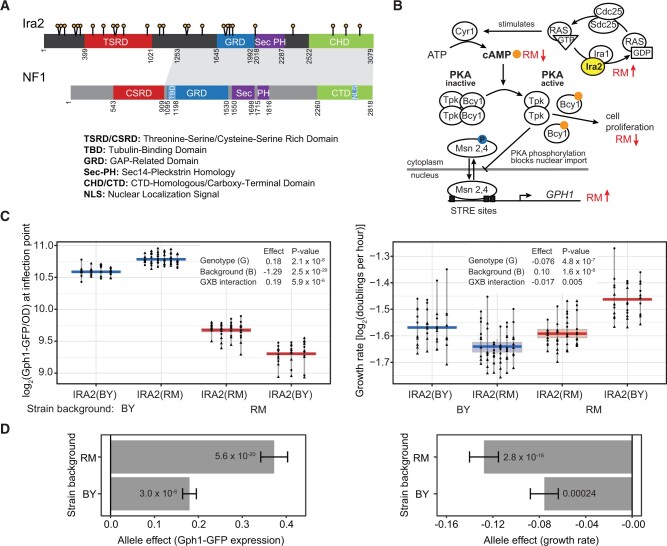
Figure 1.
RM variants within the coding region of IRA2 increase Gph1-GFP expression in trans. (A) Schematic showing nonsynonymous variants (lollipops) and domains (colored boxes) in the Ira2 protein. The NF1 schematic is modified from Bergoug et al. (2020). Amino acid positions of domain and conservation boundaries are indicated. The shaded region connects the extended regions of amino acid conservation between Ira2 (positions 1253–3079) and NF1 (positions 909–2818). (B) The role of IRA2 in the cAMP-PKA pathway including GPH1. The anticipated change in the levels or activity in the presence of the RM vs the BY allele is indicated by red arrows. Activation of PKA occurs upon binding of cAMP (orange circle) to the regulator Bcy1, resulting in its dissociation from the catalytic subunits (Tpk1, 2 or 3). See main text for more details. Figure is adapted from Santangelo (2006). (C) Effect of the IRA2 RM coding variants on Gph1-GFP expression and growth rate in the BY and RM backgrounds. For each strain, three to six transformants were phenotyped at least 10 times during two plate reader runs. Different shapes represent different plate reader runs. Lines connect measurements of the same transformant. The boxplots show the median as thicker central lines and the first and third quartiles computed on averaged phenotypes per transformant. (D) The IRA2 RM allele effect in the BY and RM backgrounds. The allele effect is the log2 of the average Gph1-GFP expression or growth rate in the presence of the RM allele minus that in the presence of the BY allele. Error bars are the standard error of the estimate. P-values are for a comparison between the RM allele and the BY allele in the designated background.
The Ira2 protein negatively regulates the cAMP-PKA signaling pathway by stimulating conversion of RAS GTPase (Ras1,2) from the active GTP-bound form to the inactive GDP-bound form (Figure 1B). RAS-GTP activates adenylate cyclase (Cyr1) to produce cAMP. High cAMP results in activation of Protein Kinase A (PKA). Phosphorylation of proteins by PKA leads to an increase in cell proliferation and a decrease in stress tolerance (Thevelein and de Winde 1999). One target of PKA is the transcription factor Msn2,4, which upon phosphorylation is retained in the cytoplasm where it is unable to activate expression of genes that contain stress-response elements (Martínez-Pastor et al. 1996; Görner et al. 1998). One of these genes is GPH1, which encodes glycogen phosphorylase, an enzyme required for the breakdown of glycogen (Wohler Sunnarborg et al. 2001). The expression of GPH1 mRNA and Gph1 protein is strongly affected by the IRA2 locus, with the RM allele resulting in higher expression (eQTL LOD = 21, protein QTL LOD = 60; Albert et al. 2014, 2018). Therefore, we chose Gph1 tagged with green fluorescent protein (GFP) as a phenotypic readout of trans gene expression in strains with engineered IRA2 alleles (Material and methods).
The presence of the RM allele compared to the BY allele of the IRA2 ORF resulted in a significantly higher Gph1-GFP expression and lower growth rate, confirming that coding variants in IRA2 contribute to the effects of this locus (Figure 1C). These effects were present in both the BY and RM strain backgrounds. The effects were larger in RM than in BY (ANOVA; Figure 1, C and D, and Supplementary Tables S5 and S6), suggesting the presence of epistatic interactions between variation in the IRA2 coding region and other variants elsewhere in the genome.
Increased expression of Gph1-GFP in the presence of the IRA2-RM allele is consistent with the direction of known effects of this locus (Albert et al. 2014, 2018). Based on the function of Ira2 (Figure 1B), increased GPH1 expression and decreased growth are expected in the presence of a more active IRA2 allele. Thus, given that RM carries more derived nonsynonymous alleles than BY, our findings agree with earlier results (Smith and Kruglyak 2008) that the RM allele of IRA2 has evolved higher activity than the more ancestral BY allele.
IRA2 harbors multiple causal variants with extensive epistatic interactions
Due to the large number of variants in the IRA2 ORF, we divided it into four blocks balancing size and number of nonsynonymous variants (Figure 2A). We then created 16 chimeric alleles representing all combinations of the four blocks to narrow in on the causal variant(s) and allow testing for potential nonadditive interactions between the blocks. These and all following allele replacements were performed in BY due to the higher efficiency of genome engineering in this background (Lutz et al. 2019).
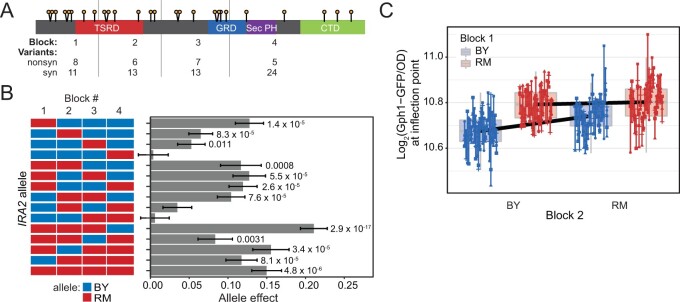
Figure 2.
Effect of blocks of IRA2 variants on Gph1-GFP expression. (A) Schematic of Ira2 showing the nonsynonymous variants (lollipops), the block 1–4 junctions (grey vertical lines) and the number of nonsynonymous (nonsyn) and synonymous (syn) variants in each block. (B) Allele effect of each of the 16 block combinations. Each allele was represented by five or six transformants that were each phenotyped five times. The allele effect is the log2 of the average Gph1-GFP expression in the presence of the indicated allele minus that in the presence of the BY allele. Error bars are the standard error of the estimate. P-values are for a comparison between the given allele and the BY allele. Only significant values are displayed. P-values were not corrected for multiple testing. See Supplementary Table S7 for all P-values. (C) Epistasis between blocks 1 and 2. The figure shows residual Gph1-GFP expression after removal of random effects of plate and transformant identity (Materials and methods). Measurements are grouped by their alleles at blocks 1 and 2. The alleles of blocks 3 and 4 are any combination of BY or RM. The black lines connect the average allele effect for the indicated genotype. Epistasis is highlighted by the steeper slope of the line when block 1 is BY rather than RM.
We measured the effect of the alleles on Gph1-GFP expression and growth rate (Supplementary Tables S7 and S8). At each of the first three blocks, the IRA2 RM sequence significantly increased Gph1-GFP expression, with the largest effect resulting from block 1 (Figure 2B, Table 1, Supplementary Figure S1, and Supplementary Table S7). Thus, there must be at least three causal variants in the IRA2 RM allele, one in each of blocks 1, 2, and 3.
Table 1.
ANOVA of the effects of IRA2 blocks 1–4 as RM
| Block | Effect | Std. error | t-Value | P-Value |
|---|---|---|---|---|
| 1 | 0.13 | 0.02 | 7.05 | 7.0 × 10-12 |
| 2 | 0.06 | 0.02 | 3.58 | 0.00038 |
| 3 | 0.05 | 0.02 | 2.91 | 0.0038 |
| 4 | 0 | 0.02 | 0.21 | 0.84 |
| 1 and 2 | −0.08 | 0.03 | −2.97 | 0.0032 |
| 1 and 3 | −0.05 | 0.03 | −2.07 | 0.039 |
| 1 and 4 | −0.01 | 0.03 | −0.47 | 0.64 |
| 2 and 3 | −0.01 | 0.03 | −0.53 | 0.59 |
| 2 and 4 | −0.03 | 0.03 | −1.29 | 0.20 |
| 3 and 4 | −0.05 | 0.03 | −1.98 | 0.049 |
| 1, 2, and 3 | 0.11 | 0.04 | 2.98 | 0.0030 |
| 1, 2, and 4 | 0.01 | 0.04 | 0.25 | 0.81 |
| 1, 3, and 4 | 0.09 | 0.04 | 2.36 | 0.019 |
| 2, 3, and 4 | 0.09 | 0.04 | 2.57 | 0.011 |
| 1, 2, 3, and 4 | −0.16 | 0.05 | −3.03 | 0.0026 |
Effect estimates are polarized such that positive values correspond to higher expression linked to the respective RM allele. Effects were computed using a mixed-effects linear model and P-values computed by type I ANOVA. Effect size confidence intervals were estimated using a bootstrap procedure. The displayed P-values were not corrected for multiple testing. P-values less than 0.05 are shown in bold.
One combination of blocks (with the RM allele at the first three blocks and the BY allele at the last block, “RRRB”) resulted in significantly higher Gph1-GFP expression than the allele carrying RM alleles at all four blocks (One-way ANOVA: P = 0.035, Figure 2B). Thus, higher Gph1-GFP levels than those driven by the full RM allele can be achieved by combinations of BY and RM variants. Considering that block 4 had no effect when swapped in isolation (Figure 2B, Table 1, and Supplementary Table S7), this difference between the RRRB construct and the full RM allele suggests that there are epistatic interactions between variants in block 4 and variants in the other blocks.
Comprehensive ANOVA on our fully crossed set of block chimeras revealed widespread nonadditive epistatic interactions along the length of IRA2. These included interactions between three pairs of blocks, with the strongest effect occurring between blocks 1 and 2 (Table 1 and Figure 2C). Higher-order interactions also existed among combinations of three blocks. They were further modulated by alleles in the remaining block, forming a significant four-way interaction (Table 1). In summary, IRA2 harbors multiple variants that affect Gph1-GFP expression in trans, with effects that are shaped by extensive nonadditive interactions.
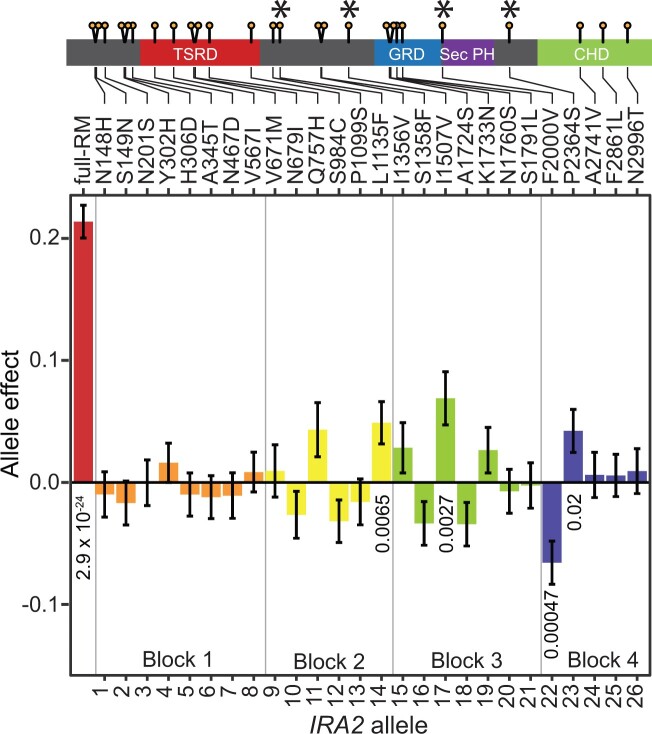
Minor effects of individual nonsynonymous RM variants
To examine the effects of single variants in IRA2, we engineered and phenotyped 26 IRA2 strains that each carried the RM allele at a single nonsynonymous variant (Figure 3A, Supplementary Figure S2, and Supplementary Tables S9 and S10). Four single-variant RM alleles (L1135F, I1507V, F2000V, and P2364S) significantly altered Gph1-GFP expression. Of these four identified causal variants, only P2364S was predicted to be deleterious by PROVEAN, and only L1135F (but not P2364S) was predicted to be “impactful” by mutfunc. None were rare among yeast isolates, with derived allele frequencies ranging from 0.36 to 0.51 (Peter et al. 2018; Supplementary Table S4). The causal variants are located in three different blocks: L1135F is in block 2, I1507V is in block 3, and F2000V and P2364S are in block 4 (Figure 1).
Figure 3.
Effect of each nonsynonymous RM variant in IRA2 on Gph1-GFP expression. Each allele was represented by nine transformants that were each measured five times. The first eight variants (N148H through V567I) were measured three additional times in an additional plate configuration. Significant P-values less than 0.05 for a comparison between the given allele and the BY allele are shown below the bars. These P-values were not corrected for multiple testing. See Supplementary Table S9 for all P-values. In the gene model, single variants with a significant effect are marked with an asterisk.
While three of the single RM variants increased Gph1-GFP expression, the F2000V variant reduced it. This direction of effect is in the opposite direction of that of the full IRA2-RM allele, revealing transgressive segregation of causal variants in a single gene. Variant F2000V lies within block 4 along with the variant P2364S, at which the RM allele significantly increased Gph1-GFP expression. Block 4 as a whole did not significantly alter Gph1-GFP expression, suggesting that the effects of variants F2000V and P2364S cancel each other out (Figure 2B). These results show that no single variant underlies the trans effect of IRA2-RM on Gph1-GFP expression. Instead, a combination of multiple variants must give rise to the overall effect of the full-RM allele at IRA2.
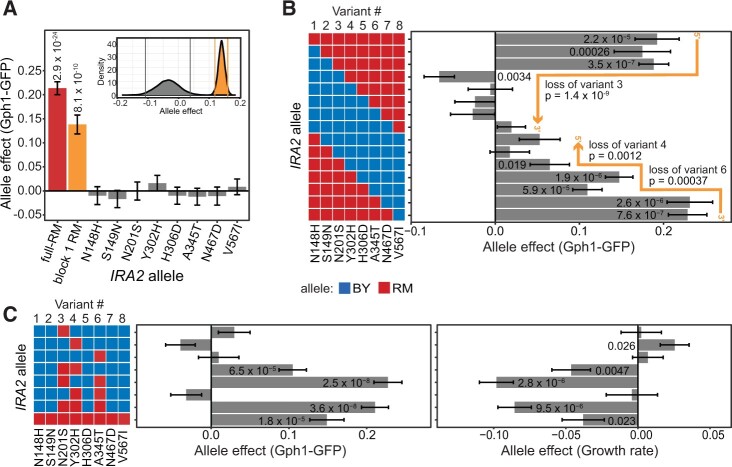
Epistatic effects among three variants near the 5′ end of IRA2
While the block 1 region at the 5′-end of IRA2 had a large effect on Gph1-GFP expression, none of the eight single variants in this block had a significant effect on their own (Figure 4A, Supplementary Figure S3, and Supplementary Table S11). A bootstrap analysis showed that the additive effects of the single variants cannot account for the effect of block 1 (Figure 4A, inset). In these comparisons, the block 1 RM allele also contains synonymous variants that are not present in the single nonsynonymous variant alleles. Although synonymous variants do not change protein sequences, they have been reported to affect complex traits (She and Jarosz 2018; Sharon et al. 2018). For example, their effects could arise from changes in translation that could affect Ira2 expression or function, especially for variants close to the 5′-end of the gene (Tuller et al. 2010; Plotkin and Kudla 2011). To test the combined effect of the synonymous variants, we engineered alleles of block 1 that carried each of the four combinations of synonymous and nonsynonymous variants. These alleles showed that while the nonsynonymous variants in block 1 had a strong effect (ANOVA P = 8 × 10−7), the synonymous variants had no discernible effect (P = 0.20; Supplementary Figure S4). Thus, a nonadditive interaction between at least two of the eight nonsynonymous variants in block 1 must underlie the effect of this region.
Figure 4.
Epistatic interactions between nonsynonymous variants in block 1. (A) Effects of each of the eight nonsynonymous RM variant alleles in block 1 and the block 1 RM and full-RM alleles. P-values are for a comparison between the given allele and the BY allele. Only significant P-values less than 0.05 are displayed. See Supplementary Table S11 for all P-values. Each allele was represented by 9 or 10 transformants that were each phenotyped three (block 1 RM) or eight (variants 1–8) times. Inset plot shows the bootstrap analysis of the effect of the block 1 RM allele compared to the summed effects of the eight variants in block 1. Same-color vertical lines mark the central 95% quantile range for each distribution. (B) Effect of progressive RM-to-BY allele replacement within block 1 on Gph1-GFP expression. Each allele was represented by five or six transformants, each measured five times. The P-values on the orange lines are for a comparison between the last allele to contain the indicated RM variant and the first allele without this RM variant. See Supplementary Tables S12 and S13 for all P-values. (D) Effect of all combinations of nonsynonymous RM variant alleles 3, 4, and 6 on Gph1-GFP expression (left panel) and growth rate (right panel), see Supplementary Tables S14 and S15, respectively, for all P-values. Each allele was represented by five transformants that were each measured eight times. (B,C) Significant differences (uncorrected P < 0.05) are shown for a comparison between the given allele and the BY allele. All P-values are available in Supplementary Tables S6 and S8, respectively.
To narrow in on the epistatic variants, we progressively replaced RM alleles with BY alleles from each end of block 1. We anticipated that if the replaced RM allele is engaged in an epistatic interaction, we would observe a drop in Gph1-GFP expression. Indeed, replacement of RM alleles with BY alleles from the 5′ end toward the 3′ end of block 1 resulted in a significant drop in Gph1-GFP expression at variant 3 (N201S). From the 3′ end toward the 5′ end, drops resulted at variants 6 (A345T) and 4 (Y302H) (Figure 4B, Supplementary Figure S5, and Supplementary Tables S12 and S13). These experiments highlighted variants 3, 4, and 6 as potential epistatic interaction partners. All three variants were among the five predicted to be impactful by mutfunc, and variant 4 was also predicted to be deleterious by PROVEAN.
We directly tested for epistatic interactions between variants 3, 4, and 6 by engineering and phenotyping all combinations of these variants (Figure 4C, Supplementary Figure S6, Table 2, and Supplementary Tables S14–S16). As above, none of these three variants increased Gph1-GFP expression in isolation. In sharp contrast to these weak single variant effects, combinations of RM alleles at variants 3 and 4 (P = 6.5 × 10−5), and especially at variants 3 and 6 (P = 2.5 × 10−8) significantly increased Gph1-GFP expression (Figure 4C). These combined variant effects exceeded those expected from their individual effects (interaction P-values: 3 and 4: P = 3.7 × 10−7; 3 and 6: P = 4.8 × 10−15). The combination of RM alleles at variants 3 and 6 exceeded the effect of the entire block 1 (P = 0.0034). A combination of RM alleles at variants 4 and 6 did not increase Gph1-GFP expression (P = 0.13). No further increase in expression was caused by the addition of the RM allele at variant 4 to the RM alleles at variants 3 and 6 (P = 0.24). The effect of these variants on growth rate mirrored their effect on Gph1-GFP (Figure 4C and Supplementary Table S15). Thus, the effect of block 1 on trans gene expression and growth rate is caused by epistatic interactions between two pairs of variants: variants 3 and 4, and especially variants 3 and 6.
Table 2.
ANOVA of the effects of IRA2-RM variants 3, 4 and 6 on Gph1-GFP
| Variant | Effect | Std. error | t-Value | P-Value |
|---|---|---|---|---|
| 3 | 0.030 | 0.015 | 2.03 | 0.044 |
| 4 | −0.039 | 0.015 | −2.67 | 0.0084 |
| 6 | 0.010 | 0.015 | 0.68 | 0.50 |
| 3 and 4 | 0.11 | 0.022 | 5.31 | 3.7 × 10-7 |
| 3 and 6 | 0.19 | 0.022 | 8.70 | 4.8 × 10-15 |
| 4 and 6 | −0.0026 | 0.022 | −0.18 | 0.91 |
| 3, 4 and 6 | −0.089 | 0.031 | −2.84 | 0.0051 |
P-values less than 0.05 are shown in bold.
Population genetic analyses
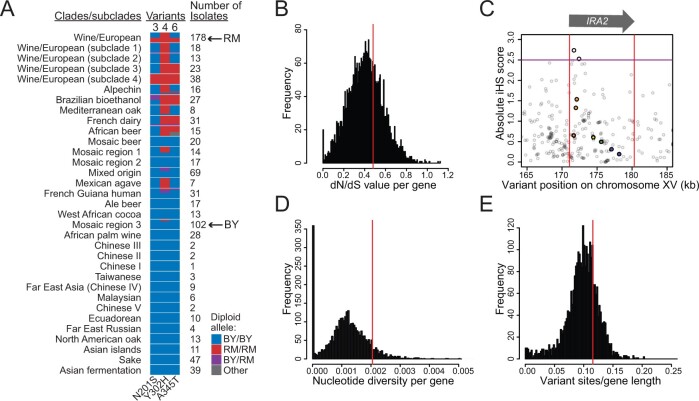
We examined the distribution of the derived RM alleles at variants 3, 4, and 6 across a population of 1011 S. cerevisiae isolates (Figure 5 and Supplementary Table S17; Peter et al. 2018). Variant 4 (Y302H) is the most common of the three variants. Its RM allele is nearly fixed among isolates from the European wine clade, of which RM is a member. The RM allele of variant 6 (A345T) occurs in a subset of isolates that also carry the RM allele at variant 4. Variant 3 (N201S) is the rarest of the three variants. Its RM allele is found exclusively in isolates that also carry the RM alleles at variant 4, most of which also carry variant 6. Thus, variant 3 likely arose on a haplotype that already carried variants 4 and 6. In our allele engineering, strains carrying only the RM alleles at variants 4 and 6 had Gph1-GFP expression similar to the ancestral BY haplotype, suggesting that these two variants did not convey increased Ira2 activity until the subsequent appearance of variant 3. Thus, the emergence of increased Ira2 activity in RM was shaped by the evolutionary sequence in which the three epistatic variants arose.
Figure 5.
Population genetic analysis of IRA2. (A) Haplotypes formed by variants 3 (N201S), 4 (Y302H), and 6 (A345T). The clades and subclades are as defined in Peter et al. (2018) and are listed in order of relatedness. Clade/subclade haplotypes with greater than a 10% frequency are shown proportional to their frequency within each clade/subclade. (B,D,E) Distribution of population genetic parameter estimates for all yeast ORFs. IRA2 is indicated by vertical red lines. (C) Absolute iHS statistics centered on IRA2 (position indicated by the arrow and by red vertical lines), calculated in the clade of European wine strains. The seven causal variants identified here are indicated by larger colored dots. Two variants with iHS > 2.5 [purple line, following (Voight et al. 2006)] are indicated. These two variants segregate among wine strains, but do not differ between BY and RM. kb: thousand base pairs.
To ask whether the multiple derived alleles that increase Ira2 activity in RM could have been subject to positive selection we computed the normalized ratio of nonsynonymous and synonymous variants (dN/dS). An excess of synonymous variants (dN/dS < 1) is an indication of negative selection purging amino acid-changing variants from the population, while an excess of nonsynonymous variants (dN/dS > 1) can indicate positive selection driving adaptive change of the protein sequence (Kimura 1977). Across all yeast isolates, IRA2 had a dN/dS of 0.48 (Figure 5B). Similarly, the European wine clade isolates showed a dN/dS of 0.45, providing no evidence that positive selection is acting on the protein sequence of this gene, unless positive selection were extremely strong (Kryazhimskiy and Plotkin 2008), which we do not consider likely in the case of IRA2. In accordance with previous results using a smaller population sample (Smith and Kruglyak 2008), the fraction of nonsynonymous polymorphisms in the S. cerevisiae population was significantly higher than the fraction of nonsynonymous substitutions that have become fixed between S. cerevisiae and the closely related species S. paradoxus, irrespective of whether polymorphisms at low frequency were removed from the analyses (Charlesworth and Eyre-Walker 2008), or whether the entire S. cerevisiae population or only the European wine clade was analyzed (Fisher’s exact test P ≤ 0.009, Supplementary Table S18). This excess of nonsynonymous polymorphisms indicates relaxed selective constraint or, potentially, positive selection on the IRA2 sequence within S. cerevisiae (Rand and Kann 1996). To further probe whether recent positive selection may have acted on the causal IRA2 variants in the clade of European wine strains, we computed iHS (Voight et al. 2006) among this clade. IRA2 contained two variants with elevated absolute iHS scores of more than 2.5 in the wine strains (Figure 5C), but none of the seven causal variants had elevated iHS scores. Thus, while we cannot rule out that the IRA2 sequence may have experienced positive selection, a more parsimonious interpretation of these results is that the causal variants identified here likely evolved under neutral genetic drift.
NF1, the human homolog of IRA2 and IRA1, is known as a hypermutable gene (Philpott et al. 2017), as reflected in a large fraction of neurofibromatosis cases that are caused by independent mutations (Clementi et al. 1990; Friedman 1999). In S. paradoxus, loss-of-function mutations commonly occur in IRA2 as well as IRA1, resulting in altered morphology and growth (Roop and Brem 2013). In experimental evolution studies of S. cerevisiae, adaptive inactivating mutations frequently arise in IRA2 and IRA1 (Lang et al. 2013; Kryazhimskiy et al. 2014; Venkataram et al. 2016; Li et al. 2019). These observations raise the question whether multiple (as opposed to a single) causal variants may have arisen in IRA2 due to a high mutation rate in this gene. To test this idea, we analyzed sequence diversity at IRA2 compared to other genes. IRA2 had a nucleotide diversity of 0.2%, placing it in the 91st percentile of all yeast genes (Figure 5D and Supplementary Table S19). Although IRA2 carries 1,068 variants across the yeast population, this value was not unusually high after accounting for the large size of the IRA2 ORF (76th percentile of all genes, Supplementary Figure 5E). These two metrics do not suggest an unusually high amount of variation and mutation at IRA2.
Discussion
Our dissection of a trans-eQTL hotspot revealed a complex molecular genetic architecture with at least seven causal variants and extensive epistatic interactions in a single gene. While we focused on protein-coding variants in the IRA2 ORF, variants in the IRA2 promoter, terminator, or even other genes in this region may further add to the overall effect of this hotspot. All seven variants we detected were missense variants that altered the sequence of the Ira2 protein. Synonymous variants had no detectable effect, at least not in the 5′ region of IRA2.
We explored why IRA2 harbors multiple causal variants, in contrast to the single causal variants found in other trans-eQTL hotspots. Loss-of-function mutations in IRA2 and its paralog IRA1 are frequently observed in experimental evolution studies in S. cerevisiae (Lang et al. 2013; Kryazhimskiy et al. 2014; Venkataram et al. 2016; Li et al. 2019) and lead to altered growth and morphology in several natural isolates of S. paradoxus (Roop and Brem 2013). NF1, the human homolog of IRA1 and IRA2, has a high deleterious mutation rate (Philpott et al. 2017) in part due to frequent recombination between repetitive sequences in the NF1 region (Upadhyaya et al. 2008). These sequences do not exist in yeast IRA2. In line with the absence of this mutagenic mechanism, IRA2 did not show elevated levels of genetic diversity compared to other yeast genes, suggesting that a high mutation rate does not explain why IRA2 carries multiple causal variants. Instead, IRA2 may simply present a prominent mutational target due to its large size. In addition, detection bias toward single variants with strong effects (Rockman 2012) may have obscured additional genes with multiple causal variants at other hotspots. If so, our results on IRA2 may foreshadow future discoveries of genes with complex molecular genetic architectures. We were able to detect the small effects of the causal variants in IRA2 because together they create a large effect that manifests as a trans-eQTL hotspot. It is interesting to speculate how many variants of similar effect may go unnoticed by QTL mapping because they fail to co-occur in a fashion that creates a detectable QTL (Kroymann and Mitchell-Olds 2005; Metzger and Wittkopp 2019; Bernstein et al. 2019).
Most of the causal variants we identified were derived in RM, where they increased the activity of the Ira2 protein. This gain of Ira2 function is in contrast to the mutations often found in studies of experimental evolution. In those studies, loss-of-function mutations of IRA2, IRA1, and other inhibitors of RAS signaling are beneficial because they confer faster growth (Lang et al. 2013; Kryazhimskiy et al. 2014; Venkataram et al. 2016; Li et al. 2019), as do experimental deletions of these genes (Sezmis et al. 2018). While the evolutionary characteristics of the gain-of-function variants in RM are different from the loss-of-function mutations seen in experimental evolution, both sets of variants act through the same cellular mechanisms. The effects of our causal variants on GPH1 expression were anticorrelated with their effects on growth, even if our growth rate measurements were less precise than those of gene expression. In particular, the increases in Gph1-GFP abundance caused by combinations of the three epistatic variants in block 1 were nearly perfectly mirrored by slower growth caused by the same combinations (Figure 4C). While our attention was drawn to IRA2 due to its widespread effects on gene expression (Albert et al. 2018), these concordant effects on gene expression and growth—an independent trait—suggest that it is likely that the same causal variants influence the numerous traits affected by the IRA2 locus in the BY/RM cross (Nguyen Ba et al. 2021).
Although the causal variants result in increased Ira2 activity, we did not find convincing evidence that these variants have evolved under positive selection. This result may be unsurprising given that these variants tend to reduce growth. However, loss-of-function mutations of IRA2 specifically affect only certain stages of yeast growth (Li et al. 2019), whose relative importance may vary with specific environmental niches. Likewise, EMS-induced mutations in IRA2 can affect gene expression from the TDH3 promoter in trans in a manner that depends on the precise media conditions (Duveau et al. 2021), and the effects of the BY and RM IRA2 alleles on mRNA abundance differ between media containing ethanol and glucose (Smith and Kruglyak 2008). These results suggest that the evolutionary fate of a new mutation in IRA2 may be highly dependent on the environment an isolate occupies. Further, the epistatic interactions we discovered among causal variants show that the effects, and therefore the fate, of new variants can be strongly dependent on the genetic background on which they occur.
The causal variants in IRA2 highlight the promise and challenge in predicting causal variants. No simple annotation or effect prediction was able to correctly and specifically pick all seven causal variants that we identified. While the PROVEAN tool missed most of the causal variants, mutfunc was able to predict four out of seven variants with high specificity (four of the five predicted variants were in fact causal). Interestingly, these four variants included the three epistatic variants in block 1, although these variants have at most modest effects in isolation when occurring in the BY reference strain. Future work will be needed to investigate why neither tool was able to correctly identify all seven causal variants.
Our results support an emerging view that trans-acting genetic effects on gene expression are as complex as those on organismal traits. Quantitative traits and gene expression can be highly polygenic (Flint and Mackay 2009; Bloom et al. 2013; Boyle et al. 2017; Visscher et al. 2017), such that the expression of a given gene is shaped by many loci across the genome (Albert et al. 2018; Metzger and Wittkopp 2019; Brion et al. 2020). Each of these loci can harbor multiple causal genes (Steinmetz et al. 2002; Metzger and Wittkopp 2019; Bernstein et al. 2019). In turn, as we showed here, a single causal trans-acting gene can have multiple causal variants. Our findings extend the pattern of multifactorial complexity of regulatory variation to the finest possible scale of individual nucleotides in a single gene. The molecular basis of trans-acting regulatory variation in humans and other multicellular organisms will likely prove to be at least as complex.
Data availability
All data needed to reproduce the analyses presented in this paper are available on github at https://github.com/Krivand/Multiple-causal-DNA-variants-in-a-single-gene-affect-gene-expression-in-trans. All strains constructed in this work are available on request.
Supplementary material is available at GENETICS online.
Supplementary Material
Acknowledgments
We thank Laura Sherer, Kaushik Renganaath, Laura Johnson, and Christian Brion. for technical contributions and discussions.
Funding
This work was supported by NIH grant R35GM124676 to F.W.A.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Albert FW, Bloom JS, Siegel J, Day L, Kruglyak L. 2018. Genetics of trans-regulatory variation in gene expression. eLife. 7:e35471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert FW, Kruglyak L. 2015. The role of regulatory variation in complex traits and disease. Nature Rev Genet. 16:197–212. [DOI] [PubMed] [Google Scholar]
- Albert FW, Treusch S, Shockley AH, Bloom JS, Kruglyak L. 2014. Genetics of single-cell protein abundance variation in large yeast populations. Nature. 506:494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, et al. 1990. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 63:851–859. doi:10.1016/0092–8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw. 67:48.doi:10.18637/jss.v067.i01. [Google Scholar]
- Bergoug M, Doudeau M, Godin F, Mosrin C, Vallée B, et al. 2020. Neurofibromin structure, functions and regulation. Cells. 9:2365.doi:10.3390/cells9112365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein MR, Zdraljevic S, Andersen EC, Rockman MV. 2019. Tightly linked antagonistic‐effect loci underlie polygenic phenotypic variation in C. elegans. Evol Lett. 3:462–473. doi:10.1002/evl3.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Lite T-LV, Kruglyak L. 2013. Finding the sources of missing heritability in a yeast cross. Nature. 494:234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell. 169:1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science. 296:752–755. [DOI] [PubMed] [Google Scholar]
- Breunig JS, Hackett SR, Rabinowitz JD, Kruglyak L. 2014. Genetic basis of metabolome variation in yeast. PLoS Genet. 10:e1004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion C, Ambroset C, Sanchez I, Legras J-L, Blondin B. 2013. Differential adaptation to multi-stressed conditions of wine fermentation revealed by variations in yeast regulatory networks. BMC Genomics. 14:681.doi:10.1186/1471–2164-14–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion C, Lutz SM, Albert FW. 2020. Simultaneous quantification of mRNA and protein in single cells reveals post-transcriptional effects of genetic variation. eLife. 9:e60645.doi:10.7554/eLife.60645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Landry CR, Hartl DL, Cavalieri D. 2008. Cascading transcriptional effects of a naturally occurring frameshift mutation in Saccharomyces cerevisiae. Mol Ecol. 17:2985–2997. [DOI] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A. 2008. The McDonald-Kreitman test and slightly deleterious mutations. Mol Biol Evol. 25:1007–1015. doi:10.1093/molbev/msn005. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, et al. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40: D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Chan AP. 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 31:2745–2747. doi:10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi M, Barbujani G, Turolla L, Tenconi R. 1990. Neurofibromatosis-1: a maximum likelihood estimation of mutation rate. Hum Genet. 84: 116–118. doi:10.1007/BF00208923. [DOI] [PubMed] [Google Scholar]
- D’angelo I, Welti S, Bonneau F, Scheffzek K. 2006. A novel bipartite phospholipid‐binding module in the neurofibromatosis type 1 protein. EMBO Rep. 7:174–179. doi:10.1038/sj.embor.7400602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duveau F, Vande Zande P, Metzger BP, Diaz CJ, Walker EA, et al. 2021. Mutational sources of trans-regulatory variation affecting gene expression in Saccharomyces cerevisiae. eLife. 10:e67806.doi:10.7554/eLife.67806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC. 2013. The molecular basis of phenotypic variation in yeast. Curr Opin Genet Dev. 23:672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann S, Bottin-Duplus H, Leonidou A, Mollereau E, Barthelaix A, et al. 2013. Natural sequence variants of yeast environmental sensors confer cell-to-cell expression variability. Mol Syst Biol. 9:695–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Barrales RR, Ibeas JI, Jimenez J. 2006. Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci U S A. 103:11228–11233. doi:10.1073/pnas.0601713103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Mackay TFC. 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19:723–733. doi:10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM. 1999. Epidemiology of neurofibromatosis type 1. Am J Med Genet. 89:1–6. [PubMed] [Google Scholar]
- Gautier M, Vitalis R. 2012. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 28:1176–1177. doi:10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
- Gerke J, Lorenz K, Cohen B. 2009. Genetic interactions between transcription factors cause natural variation in yeast. Science. 323:498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2:31–34. [DOI] [PubMed] [Google Scholar]
- Gil R, Seeling JM. 1999. Characterization of Saccharomyces cerevisiae strains expressing ira1 mutant alleles modeled after disease-causing mutations in NF1. Mol Cell Biochem. 202:109–118. doi:10.1023/A:1007058427880. [DOI] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, et al. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586–597. doi:10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großbach J, Gillet L, Clément-Ziza M, Schmalohr CL, Schubert OT, et al. 2019. Integration of transcriptome, proteome and phosphoproteome data elucidates the genetic control of molecular networks. bioRxiv. doi:10.1101/703140. [Google Scholar]
- Grundberg E, Small KS, Hedman ÅK, Nica AC, Buil A, et al. 2012. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 44:1084–1089. doi:10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 57:267–272. doi:10.1016/0378–1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villén J, Johnson AD, Gygi SP, et al. 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 325:1682–1686. doi:10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 77:61–68. doi:10.1016/0378–1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. doi:10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kim HS, Huh J, Fay JC. 2009. Dissecting the pleiotropic consequences of a quantitative trait nucleotide. FEMS Yeast Res. 9:713–722. doi:10.1111/j.1567-1364.2009.00516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1977. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 267:275–276. doi:10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Knaus BJ, Grünwald NJ. 2017. vcfr: a package to manipulate and visualize variant call format data in R. Mol Ecol Resour. 17:44–53. doi:10.1111/1755-0998.12549. [DOI] [PubMed] [Google Scholar]
- Kroymann J, Mitchell-Olds T. 2005. Epistasis and balanced polymorphism influencing complex trait variation. Nature. 435:95–98. doi:10.1038/nature03480. [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S, Plotkin JB. 2008. The population genetics of dN/dS. PLoS Genet. 4:e1000304.doi:10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science. 344:1519–1522. doi:10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB. 2017. lmerTest Package: tests in linear mixed effects models. J Stat Softw. 82: doi:10.18637/jss.v082.i13. [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, et al. 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 500:571–574. doi:10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz MC, Yugandhar K, Gupta S, Sanford EJ, Faça VM, et al. 2021. In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 22:e51121.doi:10.15252/embr.202051121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Gasch AP. 2012. Natural variation in the yeast glucose-signaling network reveals a new role for the Mig3p transcription factor. G3 (Bethesda). 2:1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Petrov DA, Sherlock G. 2019. Single nucleotide mapping of trait space reveals Pareto fronts that constrain adaptation. Nat Ecol Evol. 3:1539–1551. doi:10.1038/s41559-019–0993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Iii AM, Demarini DJ, Shah NG, Wach A, et al. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. doi:10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luo G, Kim J, Song K. 2014. The C-terminal domains of human neurofibromin and its budding yeast homologs Ira1 and Ira2 regulate the metaphase to anaphase transition. Cell Cycle. 13:2780–2789. doi:10.4161/15384101.2015.945870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S, Brion C, Kliebhan M, Albert FW. 2019. DNA variants affecting the expression of numerous genes in trans have diverse mechanisms of action and evolutionary histories. PLoS Genet. 15:e1008375.doi:10.1371/journal.pgen.1008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Stone EA, Ayroles JF. 2009. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 10:565–577. [DOI] [PubMed] [Google Scholar]
- Madeira F, Mi Park Y, Lee J, Buso N, Gur T, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47: W636–W641. doi:10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, et al. 2017. Universal patterns of selection in cancer and somatic tissues. Cell. 171:1029–1041.e21. doi:10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, et al. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227–2235. doi:10.1002/j.1460–2075.1996.tb00576.x. [PMC free article] [PubMed] [Google Scholar]
- Metzger BPH, Wittkopp PJ. 2019. Compensatory trans-regulatory alleles minimizing variation in TDH3 expression are common within Saccharomyces cerevisiae. Evol Lett. 3:448–461. doi:10.1002/evl3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Lawrence KR, Rego-Costa A, Gopalakrishnan S, Temko D, et al. 2021. Barcoded bulk QTL mapping reveals highly polygenic and epistatic architecture of complex traits in yeast. Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, et al. 2016. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2:e000056. doi:10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parts L, Cubillos FA, Warringer J, Jain K, Salinas F, et al. 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 21:1131–1138. doi:10.1101/gr.116731.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, Chiara MD, Friedrich A, Yue J-X, Pflieger D, et al. 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 556:339–344. doi:10.1038/s41586-018–0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol. 31:1929–1936. doi:10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott C, Tovell H, Frayling IM, Cooper DN, Upadhyaya M. 2017. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 11: doi:10.1186/s40246-017–0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2021. Nlme: linear and nonlinear mixed effects models. R package version 3.1-153. https://CRAN.R-project.org/package=nlme [Google Scholar]
- Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 12:32–42. doi:10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Kann LM. 1996. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 13:735–748. doi:10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Rat Genome Sequencing and Mapping Consortium. 2013. Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat Genet. 45:767–775. doi:10.1038/ng.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner N, Miller SJ. 2015. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 15:290–301. doi:10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV. 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution. 66:1–17. doi:10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roop JI, Brem RB. 2013. Rare variants in hypermutable genes underlie common morphology and growth traits in wild Saccharomyces paradoxus. Genetics. 195:513–525. doi:10.1534/genetics.113.155341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo GM. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 70:253–282. doi:10.1128/MMBR.70.1.253–282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezmis AL, Malerba ME, Marshall DJ, McDonald MJ. 2018. Beneficial mutations from evolution experiments increase rates of growth and fermentation. J Mol Evol. 86:111–117. doi:10.1007/s00239-018–9829-9. [DOI] [PubMed] [Google Scholar]
- Sharon E, Chen S-AA, Khosla NM, Smith JD, Pritchard JK, et al. 2018. Functional genetic variants revealed by massively parallel precise genome editing. Cell. 175:544–557. e16. doi:10.1016/j.cell.2018.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She R, Jarosz DF. 2018. Mapping causal variants with single-nucleotide resolution reveals biochemical drivers of phenotypic change. Cell. 172:478–490.e15. doi:10.1016/j.cell.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.doi:10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Kruglyak L. 2008. Gene–environment interaction in yeast gene expression. PLoS Biol. 6:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, et al. 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 416:326–330. [DOI] [PubMed] [Google Scholar]
- Stojiljkovic M, Foulquié-Moreno MR, Thevelein JM. 2020. Polygenic analysis of very high acetic acid tolerance in the yeast Saccharomyces cerevisiae reveals a complex genetic background and several new causative alleles. Biotechnol Biofuels. 13:126.doi:10.1186/s13068-020–01761-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P, Cohen BA. 2014. Single nucleotide variants in transcription factors associate more tightly with phenotype than with gene expression. PLoS Genet. 10:e1004325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, et al. 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods. 10:676–682. doi:10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2020. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 369:1318–1330. doi:10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 33:904–918. doi:10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, et al. 2010. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 141:344–354. doi:10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Kluwe L, Spurlock G, Monem B, Majounie E, et al. 2008. Germline and somaticNF1 gene mutation spectrum in NF1-associated malignant peripheral nerve sheath tumors (MPNSTs). Hum Mutat. 29:74–82. doi:10.1002/humu.20601. [DOI] [PubMed] [Google Scholar]
- Venkataram S, Dunn B, Li Y, Agarwala A, Chang J, et al. 2016. Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell. 166:1585–1596.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, et al. 2017. 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72.doi:10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O, Galardini M, Busby BP, Memon D, Typas A, et al. 2018. A resource of variant effect predictions of single nucleotide variants in model organisms. Mol Syst Biol. 14:e8430.doi:10.15252/msb.20188430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kruglyak L. 2014. Genetic basis of haloperidol resistance in Saccharomyces cerevisiae is complex and dose dependent. PLoS Genet. 10:e1004894.doi:10.1371/journal.pgen.1004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Qi Q, Lin Y, Guo Y, Liu Y, et al. 2019. QTL analysis reveals genomic variants linked to high-temperature fermentation performance in the industrial yeast. Biotechnol Biofuels. 12:59.doi:10.1186/s13068-019-1398-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohler Sunnarborg S, Miller SP, Unnikrishnan I, LaPorte DC. 2001. Expression of the yeast glycogen phosphorylase gene is regulated by stress-response elements and by the HOG MAP kinase pathway. Yeast. 18:1505–1514. doi:10.1002/yea.752. [DOI] [PubMed] [Google Scholar]
- Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, et al. 2014. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 46:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, et al. 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 35:57–64. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang B, Smith EN, Drees B, Brem RB, et al. 2008. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 40:854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to reproduce the analyses presented in this paper are available on github at https://github.com/Krivand/Multiple-causal-DNA-variants-in-a-single-gene-affect-gene-expression-in-trans. All strains constructed in this work are available on request.
Supplementary material is available at GENETICS online.