Abstract
The CSL family protein CBF1 is a nuclear mediator of Notch signaling and has been predicted to contain an N-terminal nuclear localization signal in exon 4. Surprisingly, we found that CBF1 carrying mutations at codon 233 or 249 within exon 7 was restricted to the cytoplasm. In mammalian and yeast two-hybrid assays, these mutations were also associated with a loss of CBF1-mediated transcriptional repression and a severely impaired interaction with the corepressors SMRT and CIR. Overexpression of SMRT rescued the ability of mutant CBF1 to target to the nucleus of transfected cells and similarly rescued nuclear targeting of enhanced green fluorescent protein (EGFP)-CBF1 exons 6 to 9 CBF1(6-9)carrying the codon 233 or 249 mutations. Carboxy-terminally truncated SMRT with amino acids (aa) 1291 to 1495 deleted was unable to rescue the nuclear targeting of mutant EGFP-CBF1(6-9). In yeast two-hybrid assays, the SMRT aa 1291 to 1495 domain interacted with SKIP and SMRT aa 1291 to 1495 colocalized with SKIP within the nuclei of cotransfected cells. Comparison of the intracellular localization of CBF1(6-9) with that of CBF1(5-9) further supported the suggestion that nuclear targeting of CBF1 is dependent on the formation of a CBF1-SMRT-SKIP corepressor complex. These observations suggest that nuclear targeting of CBF1 is itself a component of CBF1-mediated gene regulation and that in the absence of signaling, CBF1 enters the nucleus precommitted to a transcriptional repression function. The activators NotchIC (the intracellular domain of Notch) and Epstein-Barr virus EBNA2 also mediated nuclear targeting of mutant CBF1, consistent with the competition model for activator versus corepressor binding to CBF1.
Ligand-induced activation of the Notch transmembrane receptor initiates a signaling pathway that is highly conserved through species from worms to humans. Notch signaling regulates cell fate decisions and affects cellular proliferation, differentiation, and programmed cell death (2, 12, 34). A current model suggests that ligand binding induces an intramembrane proteolytic cleavage event that is dependent on presenilins and results in the release of the intracellular domain of Notch, NotchIC, which then enters the nucleus to effect transcriptional reprogramming (35, 54). A proteolytic cleavage site between G1743 and V1744 that released NotchIC was identified in cells overexpressing a constitutively activated membrane-bound form of Notch1 (46), and when this cleavage site was mutated within the mouse Notch1 gene, the homozygous mutation was embryo lethal, with a phenotype similar to that seen in Notch1-null embryos (18, 25, 51). Genetic screens in Caenorhabditis elegans identified a presenilin homologue, sel-12, as a modifier of Notch (Lin-12) function, and disruption of both the sel-12 and hop-1 presenilin homologues results in a developmental phenotype that resembles that generated by loss of activity of the Notch homologues lin-12 and glp-1 (28, 29, 30, 55). The embryonic defects in mice lacking both presenilin 1 and presenilin 2 are also consistent with a conserved role for presenilins in Notch function in mammalian cells (7).
Constitutive expression of NotchIC recapitulates many of the features of ligand-mediated Notch signaling (24, 31, 42, 49). Exogenously expressed NotchIC is detected in the nucleus (10, 31, 49), but the presence of endogenously generated NotchIC in the nucleus has been difficult to visualize directly other than in neuronal cells (3, 43). However, assays in which nuclear transcriptional reporters were used to assess Notch function have clearly demonstrated a linkage between the generation of an endogenous NotchIC species and nuclear reporter activity (46, 48).
The best-characterized nuclear mediators of Notch signaling are the CSL family of DNA binding proteins [CBF1/RBPJ-κ in mammalian cells, Su(H) in drosophila, and LAG-1 in C. elegans]. These proteins were initially recognized as downstream effectors of Notch in genetic analyses and subsequently shown to directly interact with NotchIC (5, 15, 16, 19, 27). Human CBF1 recognizes the core DNA sequence GTGGGAA (32, 52), and DNA-bound CBF1 acts as a transcriptional repressor by bringing to the promoter a corepressor complex whose known components include SMRT, SKIP, SAP30, Sin3A, CIR, HDAC1, and HDAC2 (17, 22, 58, 59). The presence of histone deacetylases in this complex implies that transcriptional repression is mediated in part through chromatin remodeling (50). Negative regulation through direct contacts with the basal transcription machinery has also been described (8). Activation of the promoter by NotchIC appears to involve displacement of the corepressor complex from CBF1 by NotchIC (15, 16, 59) along with the recruitment of coactivators such as Mastermind and the GCN4 and PCAF histone acetylases, which interact with NotchIC (13, 26, 40, 57). The Epstein-Barr virus immortalizing protein EBNA2 mimics NotchIC by targeting CBF1 and similarly displacing the corepressor complex and bringing the coactivators p300, PCAF, and CBP to the promoter (14, 20, 53, 58).
SKIP was found to play an interesting role in the conversion from CBF1-mediated transcriptional repression to activation. SKIP was originally identified as an interacting partner of the avian retroviral oncogene v-Ski, whose cellular homolog, c-Ski, has recently been recognized as a component of the HDAC-corepressor complex that associates with the Mad and thyroid hormone receptor complexes, and we identified SKIP as a CBF1-interacting protein in a yeast two-hybrid screen (6, 38, 59). When expressed as a Gal4-fusion, SKIP represses expression from a reporter containing Gal4 binding sites and SKIP interacts with SMRT and CIR in the corepressor complex (58, 59). On the other hand, SKIP is also present in the activation complex, where it makes key tethering contacts with the NotchIC and EBNA2 activators. Mutations in NotchIC or EBNA2 that cause loss of SKIP interaction also impair the ability of NotchIC and EBNA2 to activate reporters containing CBF1 binding sites. Further, the ability of NotchIC to prevent differentiation of C2C12 myoblasts is ablated in cells in which SKIP protein levels are severely reduced by the expression of antisense SKIP mRNA. Thus, SKIP and CBF1 each make contacts with the corepressor complex and also each contact the activator complex. The corepressor and activator contacts are mutually exclusive, leading to a competition model for the switch from repression to activation (59).
CBF1 is present in the nuclei of transfected cells, and a group of positively charged amino acids between codons 81 and 84 has been assumed to represent an N-terminal nuclear localization signal (NLS) (1). The intracellular localization of CBF1 has not, therefore, been thought of as a component of CBF1-mediated gene regulation. We now provide evidence that nuclear localization of CBF1 is a regulated process that is mediated by interactions with the SMRT-SKIP corepressor complex. Entry of CBF1 into the nucleus preassociated with corepressor proteins would ensure transcriptional repression as the default setting for CBF1-bound promoters. Experiments examining the intracellular localization of CBF1 also provided evidence strengthening the concept of competition between the corepressor complex and the EBNA2 activator for interaction with CBF1.
MATERIALS AND METHODS
Plasmids.
Enhanced green fluorescent protein (EGFP)-CBF1(exons 6 to 9 [6-9]; EEF233AAA) (pSZ64) and EGFP-CBF1(6-9; KLV249AAA) (pSZ65) were generated by PCR amplification using SG5-Flag-CBF1(EEF233AAA) (pJH278) or SG5-Flag-CBF1(KLV249AAA) (pJH319), respectively, as the PCR template, followed by ligation of the BglII-cleaved fragments into the BglII site of pEGFP-C2 (Clontech). SG5-Myc-CBF1(5-9) (pSZ82) was made by ligating the CBF1(5-9) fragment (obtained through PCR using Gal4-CBF1 as the template and LGH4066 and LGH3373 as primers) into the BamHI-cut SG5-Myc (pDY48) vector. SG5-Flag-CBF1(6-9) (pJH337) was generated by ligating the CBF1(6-9) fragment (obtained by PCR using LGH1423 and LGH1450 as primers and Gal4-CBF1 as the template) into the BglII-cut SG5-Flag (pJH272) vector. CMX-SMRT(1-1290) (pSZ69) was generated from pCMX-SMRT by BglII cleavage, followed by Klenow filling in and religation. This removed amino acid (aa) residues 1291 to 1495 of SMRT. A BglII/BamHI fragment from pCMX-SMRT was ligated into pSG5-Flag (pJH272) cut with BglII or into pSG5-Myc (pDY48) cut with BamHI to make pSG5-Flag-SMRT(1290-1495) (pSZ77) or SG5-Myc-SMRT(1290-1495) (pSZ78), respectively. The yeast expression clone pACTII-SMRT(1290-1495) (pSZ75) was made by moving SMRT(1290-1495) as a BglII/BamHI fragment from pCMX-SMRT into the BamHI site of the vector pACTII. The same fragment was also ligated into the BamHI site of the vector pAS1-CYH2 to generate pAS1-SMRT(1290-1495) (pSZ76). SG5-Flag-CBF1(RLI261) (pJH320) was generated by moving a CBF1(RLI261AAA) fragment into the BglII site in the SG5-Flag vector (pJH253). 5xGal4TK-CAT, TK-luciferase, SG5-CBF1 (pJH156), SG5-CBF1(EEF233) (pJH157), SG5-hemagglutinin (HA)-SKIP (pJH277), SG5-Flag-CBF1(EEF233) (pJH278), SG5-Flag-Notch1IC (pJH279), SG5-Flag-CBF1 (pJH282), SG5-Flag-CBF1(KLV249) (pJH319), SG5-CIR-Myc (pJH402), SG5-CIR-Flag (pJH518), SG5-EBNA2 (pPDL151), and SG5-EBNA2(1-415) (pPDL179) and the yeast clones Gal4 DNA binding domain (DBD)-CBF1 (pJH137), Gal4DBD-SKIP (pJH313), Gal4DBD-CIR (pJH491), Gal4ACT-CBF1 (pJH346), Gal4ACT-SKIP (pJH177), and Gal4ACT-CIR (pJH178) have been previously described (17, 58, 59). CMX-SMRT, CMX-SMRT-Flag, yeast Gal4ACT-SMRT, and yeast Gal4DBD-SMRT(649-811) were generous gifts from R. Evans.
Immunofluorescence assays.
Plasmid DNA (0.5 μg) was transfected by the calcium phosphate procedure into Vero cells seeded at 0.8 × 105 cells per well in two-well LabTek slides (Nunc) and grown in Dulbecco modified Eagle medium plus 10% fetal calf serum. Two days after transfection, cells were washed and fixed in 1% paraformaldehyde in phosphate buffered saline for 10 min at room temperature. Fixed cells were washed and permeabilized in 0.2% Triton X-100 in phosphate-buffered saline for 20 min on ice. After washing, the cells were incubated with primary antibodies for 1 h at 37°C. Primary antibodies were diluted as follows: mouse anti-EBNA2 (Dako), 1:1,000; mouse anti-Flag (Sigma), 1:1,000; mouse anti-Myc (Sigma), 1:1,000; goat anti-SMRT (N-20; Santa Cruz), 1:400; rabbit anti-CBF1, 1:500; rabbit anti-SKIP, 1:500; rabbit anti-CIR, 1:500. The secondary antibodies fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit immunoglobulin (Ig; 1:200) and rhodamine-conjugated goat anti-mouse Ig or donkey anti-goat Ig (Chemicon; 1:200) were incubated for 0.5 h at 37°C. The slides were then washed and mounted in MOWIOL solution (Calbiochem). Images were captured using a Leitz fluorescence microscope and ImagePro software (Media Cybernetics).
Yeast two-hybrid assays.
Yeast two-hybrid assays were performed as previously described, by using yeast strain Y190 (58, 59). β-Galactosidase activity was measured from three independent cotransformants by using 2-nitrophenyl β-d-galactopyranoside as the substrate. The amount of 2-nitrophenol liberated after 2 to 4 h of incubation was measured by determining the A420.
Reporter assays.
HeLa cells were maintained in Dulbecco modified Eagle medium plus 10% fetal calf serum and plated in six-well plates (Nunc) at 1.2 × 105 cells per well 1 day prior to transfection. Transfections were performed essentially as previously described (58, 59), by using 0.5 μg of each plasmid DNA. Vector DNA was used to keep the total amount of transfected DNA constant. Chloramphenicol acetyltransferase (CAT) and luciferase assays were performed as previously described (58, 59), and each experiment was performed in triplicate.
RESULTS
Mutant forms of CBF1 with changes in exon 7 that affect SMRT interaction also affect CBF1 nuclear localization.
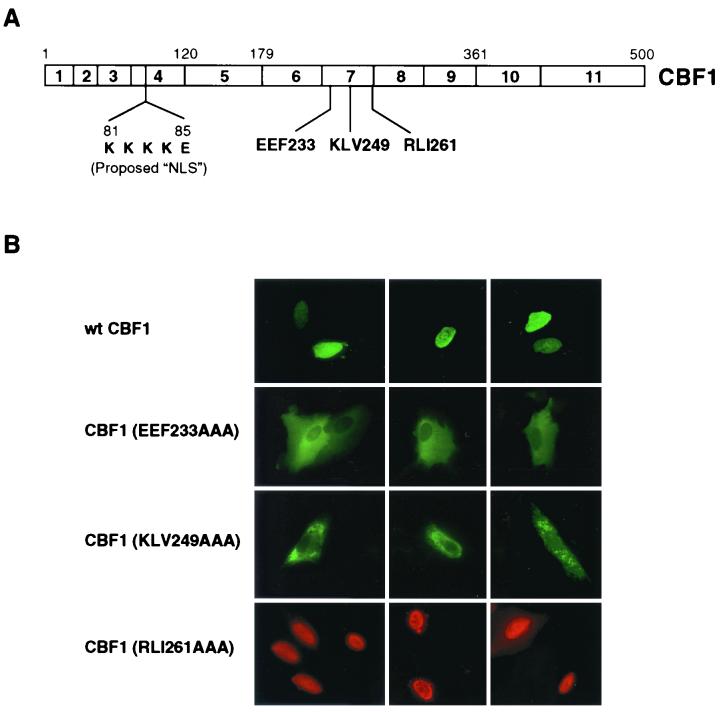
CBF1 localizes to the nuclei of transfected cells as illustrated in Fig. 1, in which Flag-tagged CBF1 was transfected into Vero cells and visualized in an indirect immunofluorescence assay using anti-Flag antibody. An NLS has been proposed to exist in exon 4 of CBF1 (Fig. 1) (1, 23). However, we observed that mutation of two separate positions in exon 7, EEF233AAA and KLV249AAA, resulted in Flag-CBF1 variants that were retained in the cytoplasm of transfected Vero cells (Fig. 1B). In contrast, an adjacent but further downstream mutation at codon 261 (RLI261AAA) did not affect the normal pattern of nuclear staining (Fig. 1B).
FIG. 1.
Mutations in CBF1 exon 7 abolish nuclear localization. (A) Schematic structure of CBF1 showing the exon structure, the proposed NLS (aa 81 to 85 [KKKKE]) (1), and three mutations in exon 7. (B) Immunofluorescence assay showing the intracellular localization of wt Flag-CBF1, the mutant forms CBF1(EEF233) and CBF1(KLV249), and a control mutant CBF1(RLI261) in transfected Vero cells. Flag-CBF1 was detected by using mouse anti-Flag as the primary antibody and either FITC-conjugated (green) or rhodamine-conjugated (red) goat anti-mouse Ig as the secondary antibody.
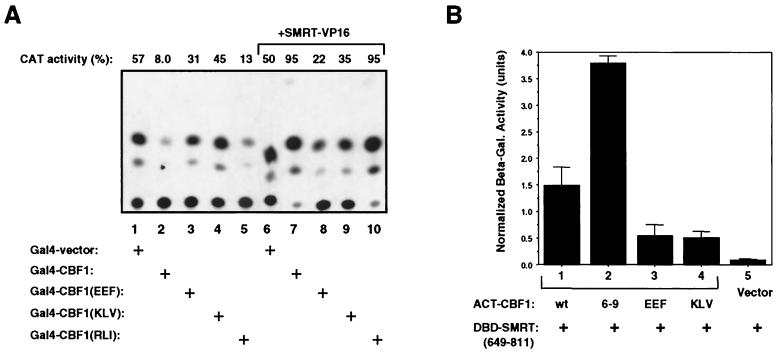
We had previously recognized that the CBF1 EEF233 and KLV249 mutant forms were associated with a loss of CBF1-mediated transcriptional repression (14, 16). This is illustrated in the transient-expression assay whose results are shown in Fig. 2A, where the behavior of Gal4-CBF1 fusion proteins containing the EEF233, KLV249, and RLI261 mutants are compared. Transfection of a 5xGal4BS-CAT reporter with Gal4-CBF1 resulted in the expected repression of CAT reporter expression (Fig. 2A, lane 2). Gal4-CBF1 fusions carrying the EEF233 and KLV249 mutations were ineffective at mediating transcriptional repression (Fig. 2A, lanes 3 and 4), whereas the Gal4-CBF1(RLI261) mutant still repressed expression of the CAT reporter (Fig. 2A, lane 5). Loss of CBF1-mediated repressive activity has been associated with loss of interaction with the corepressor SMRT (22). This is illustrated in the mammalian two-hybrid assay whose results are shown in Fig. 2A, lanes 6 to 10, where SMRT is expressed as a fusion with the transcriptional activation domain of the herpes simplex virus VP16 protein (SMRT-VP16). In cotransfected HeLa cells, interaction between Gal4-CBF1 and SMRT-VP16 leads to activation of the 5xGal4BS-CAT reporter through tethering of the VP16 activation domain to promoter-bound Gal4-CBF1. In this assay, both the wild-type and the RLI261 mutant Gal4-CBF1 proteins interacted with SMRT-VP16, as indicated by activation of the CAT reporter (Fig. 2B, lanes 7 and 10), whereas cotransfection of SMRT-VP16 with the Gal4-CBF1 proteins carrying the EEF233 and KLV249 mutations did not result in reporter activation (Fig. 2A, lanes 8 and 9).
FIG. 2.
CBF1 mutant proteins with changes in exons 6 to 9 that do not target to the nucleus are also impaired for SMRT interaction. (A) CBF1(EEF233) and CBF1(KLV249) do not mediate transcriptional repression, nor do they interact with SMRT in a mammalian two-hybrid assay. HeLa cells were transfected with the 5xGal4TK-CAT reporter and Gal4DBD-CBF1 or the indicated Gal4DBD-CBF1 mutant forms. Cotransfected Gal4-CBF1 or Gal4-CBF1(RLI261) repressed expression of the CAT reporter (lanes 2 and 5), whereas the Gal4-CBF1(EEF233) and Gal4-CBF1(KLV249) fusion proteins did not mediate significant repression (lanes 3 and 4). Interaction between CBF1 and SMRT was tested by cotransfection of SMRT fused with the activation domain of herpes simplex virus VP16 (lanes 6 to 10). SMRT-VP16 activated expression in the presence of Gal4-CBF1 (lane 7) and Gal4-CBF1(RLI261) (lane 10), indicating an interaction between SMRT and these two proteins. Gal4 vector (lane 6), Gal4-CBF1(EEF233) (lane 8), and Gal4-CBF1(KLV249) (lane 9) did not respond to the addition of SMRT-VP16, indicating a lack of interaction. (B) In yeast two-hybrid assays, interaction between SMRT and CBF1(EEF233) and CBF1(KLV249) was severely impaired but not completely abolished. The relative strength of the protein-protein interactions was quantified by the measurement of β-galactosidase induction in yeast cotransformed with the indicated Gal4DBD and Gal4ACT fusion proteins. CBF1(EEF233) and CBF1(KLV249) retained a weak interaction with SMRT (aa 649 to 811), as indicated by the measured β-galactosidase activity and by the pale blue color observed in a yeast colony lift assay (data not shown).
These reporter assays suggested that there was a correlation between the intracellular localization of CBF1 and its ability to interact with SMRT. The CBF1(EEF233) and CBF1(KLV249) mutant forms localized to the cytoplasm and had lost the ability to interact with SMRT, while the wild-type and RLI261 mutant proteins maintained SMRT interaction and were found in the nuclei of transfected cells. [It should be noted that the loss of activity shown by the Gal4-CBF1(EEF233) and Gal4-CBF1(KLV249) proteins in these assays is not due to inappropriate localization. The Gal4 DBD (aa 1 to 147) contains an NLS (44), and consequently, the Gal4-CBF1 fusion proteins are transported efficiently to the nucleus.] To further verify the effects of the EEF233 and KLV249 mutations on interaction with SMRT, a yeast two-hybrid assay was performed using a DBD-SMRT (aa 649 to 811) construction that contains the domain of SMRT known to interact with CBF1 (22). Interaction in cotransformed yeast between DBD-SMRT (aa 649 to 811) and ACT-CBF1 plasmids was measured by the induction of β-galactosidase activity (Fig. 2B). As expected, SMRT (aa 649 to 811) interacted with both full-length CBF1 and CBF1(6-9) (Fig. 2C, lanes 1 and 2). Interaction between SMRT(649-811) and the EEF233 and KLV249 mutant forms was significantly impaired but not completely abolished in this assay (Fig. 2B, lanes 3 and 4).
Overexpression of SMRT rescues nuclear localization of mutant CBF1.
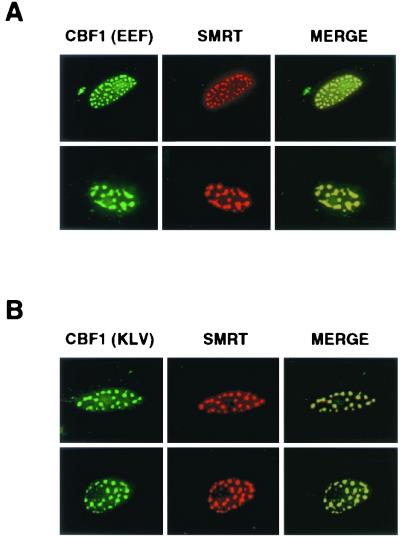
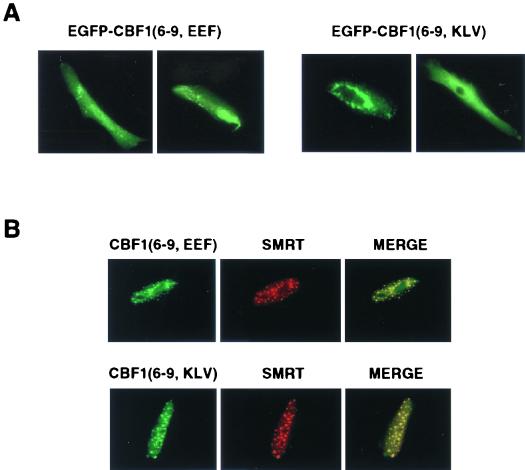
The yeast two-hybrid assay suggested that there remained some residual interaction between SMRT and the mutant CBF1 proteins. We wondered if overexpression of SMRT would compensate for the reduced affinity of binding. The intracellular localization of the CBF1(EFF233) and CBF1(KLV249) proteins was therefore examined in Vero cells cotransfected with an SMRT expression vector. In the cotransfected cells, the CBF1 mutant proteins were detected in the nucleus, where they colocalized with SMRT in punctate spots (Fig. 3A and B). The CBF1(EEF233) and CBF1(KLV249) proteins contain the predicted NLS in exon 4. To ensure that the observed results were independent of any contribution from this region or other regions of CBF1, we also tested EEF233 and KLV249 mutants that were in a background of CBF1(6-9). Cotransfection of SMRT also rescued nuclear localization of the CBF1(EEF233) and CBF1(KLV249) mutant proteins in the background of EGFP-CBF1(6-9) (Fig. 4A and B). The EGFP-CBF1(6-9) mutant protein again colocalized with SMRT in the nucleus, although the intranuclear distribution pattern was less distinctly punctate, suggesting that regions of CBF1 outside of exons 6 to 9 may have an impact on the intranuclear destination.
FIG. 3.
Overexpression of the corepressor SMRT rescues nuclear targeting of mutant CBF1. Results of indirect immunofluorescence assays show Vero cells cotransfected with SMRT and CBF1(EEF233) (A) or CBF1(KLV249) (B). CBF1(EEF233) (A, left, green), CBF1(KLV249) (B, left, green), and SMRT-Flag (middle, red) each showed nuclear punctate staining. In the merged images (right, yellow), SMRT-Flag colocalized with the mutant CBF1 proteins. Rabbit anti-CBF1 and mouse anti-Flag antibodies were used as primary antibodies. Secondary antibodies were FITC-conjugated donkey anti-rabbit Ig (green) and rhodamine-conjugated goat anti-mouse Ig (red).
FIG. 4.
Overexpressed SMRT also rescued nuclear targeting of mutant EGFP-CBF1(6-9). (A) Immunofluorescence assay in transfected Vero cells showing that the EEF233 and KLV249 mutations also disrupt nuclear localization of EGFP-tagged CBF1(6-9). (B) Indirect immunofluorescence assay in Vero cells cotransfected with SMRT and EGFP-CBF1(6-9, EEF) or EGFP-CBF1(6-9, KLV). EGFP-CBF1(6-9, EEF) (upper panel, green) or EGFP-CBF1(6-9, KLV) (lower panel, green) entered the nucleus in the presence of SMRT-Flag (red) and colocalized with SMRT (merge, yellow). Mouse anti-Flag antibody and rhodamine-conjugated goat anti-mouse Ig were used as primary and secondary antibodies, respectively.
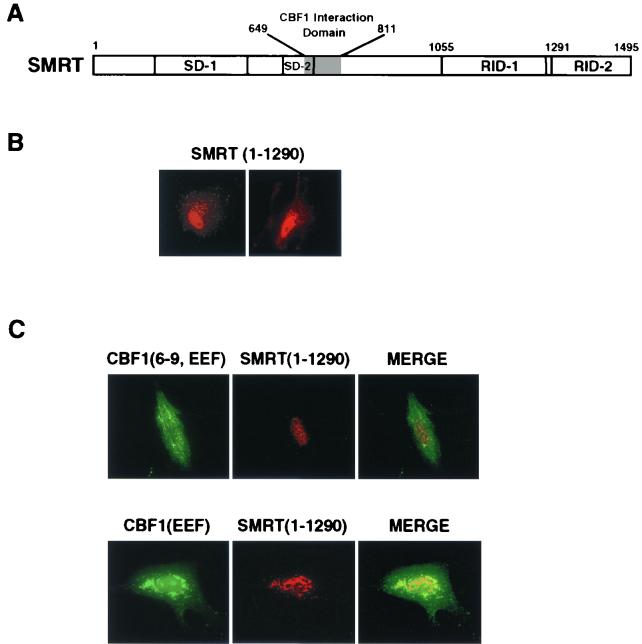
The SMRT RID-2 domain (aa 1291 to 1495) is required for nuclear translocation of CBF1.
The CBF1 interaction domain is located between aa 649 and 811 of SMRT (Fig. 5A) (22). We wished to determine whether any other domains of SMRT are required for CBF1 nuclear transport. One of the SMRT deletion mutant proteins that were created to address this question proved to have an interesting phenotype. Deletion of the carboxy-terminal RID-2 domain generated SMRT(1-1290), which, in transfected cells, localized predominantly to the nucleus, with some additional weak cytoplasmic staining (Fig. 5B). Cotransfected SMRT(1-1290) was unable to mediate nuclear localization of either EGFP-CBF1(6-9, EEF) or full-length CBF1(EEF233) (Fig. 5C), suggesting that the CBF1 interaction domain of SMRT was insufficient for effective CBF1 nuclear targeting and that sequences in the carboxy terminus of SMRT are also important for this function.
FIG. 5.
A C-terminally truncated form of SMRT is unable to rescue nuclear localization of mutant CBF1. (A) Schematic drawing of SMRT showing the CBF1 interaction domain (shaded), silencing domains (SD), and nuclear receptor interaction domains (RID). (B) Immunofluorescence assay in transfected Vero cells illustrating that SMRT(1-1290), in which the RID-2 domain (aa 1291 to 1495) is deleted, retains a predominantly nuclear localization. Goat anti-SMRT and rhodamine-conjugated donkey anti-goat Ig were used as the staining antibodies. (C) SMRT(1-1290) could not relocate EGFP-CBF1(6-9, EEF233) or full-length CBF1(EEF233) into the nucleus. Immunofluorescence assay show the intracellular distribution of EGFP-CBF1(6-9, EEF), CBF1(EEF233), and SMRT(1-1290) in cotransfected Vero cells. Goat anti-SMRT antibody and rhodamine-conjugated donkey anti-goat Ig were used to stain SMRT (upper and lower panels). Rabbit anti-CBF1 antibody and FITC-conjugated donkey anti-rabbit Ig were used to stain full-length CBF1(EEF233) (lower panel).
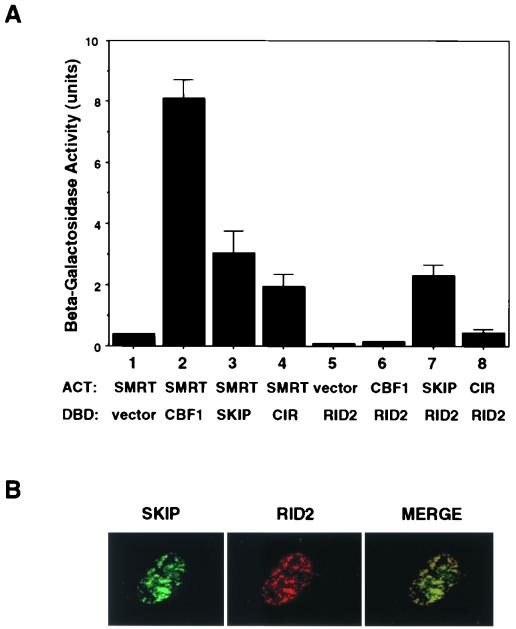
SMRT(1291-1495) interacts with SKIP.
A yeast two-hybrid assay was performed to determine the identities of proteins interacting with the SMRT(1291-1495) RID-2 domain. As illustrated in Fig. 6A (lanes 1 to 4), by the induction of β-galactosidase activity in cotransformed yeast, intact SMRT interacts with CBF1 and with two of the members of the associated corepressor complex, SKIP and CIR. When the SMRT RID-2 domain was tested in this assay, (Fig. 6A, lanes 5 to 8), the strongest interaction was with SKIP. The level of β-galactosidase activity induced in cells cotransformed with SKIP and the SMRT RID-2 domain was very similar to that observed in cells cotransformed with SKIP and intact SMRT. Consistent with previous mapping data (22), there was no interaction between CBF1 and this region of SMRT.
FIG. 6.
The RID-2 domain of SMRT interacts with SKIP. (A) Yeast two-hybrid assay in which β-galactosidase induction is used as a measure of protein-protein interaction. CBF1, SKIP, and CIR all interact with full-length SMRT (lanes 2, 3, and 4). However, only SKIP interacts significantly with SMRT(RID-2) (lane 7). (B) Immunofluorescence assay showing colocalization of HA-SKIP (left, green) and Flag-SMRT(RID2) (middle, red) in cotransfected Vero cells. The primary antibodies were rabbit anti-SKIP IgG and mouse anti-Flag antibody. The secondary antibodies were FITC-conjugated donkey anti-rabbit Ig and rhodamine-conjugated goat anti-mouse Ig.
To provide additional support for an association between SKIP and the SMRT RID-2 domain, a Flag-SMRT(1291-1495) expression vector was generated and cotransfected into Vero cells with HA-SKIP. Indirect immunofluorescence assays showed complete colocalization of HA-SKIP with Flag-SMRT(1291-1495) in the nuclei of cotransfected cells (Fig. 6B).
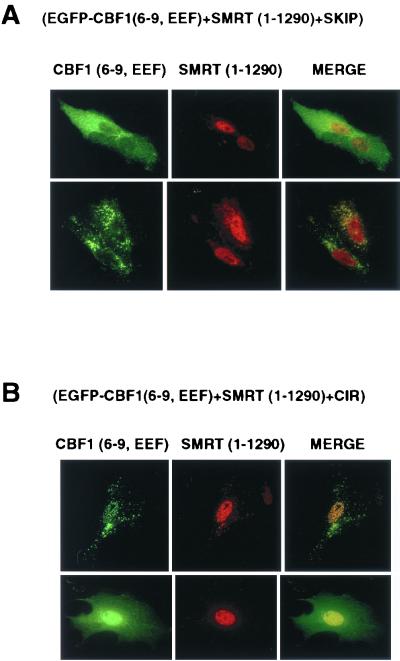
Efficient nuclear targeting may involve an assembled SMRT-SKIP-CIR corepressor complex.
We demonstrated in Fig. 5C that SMRT(1-1290) with the RID-2 domain deleted is unable to rescue nuclear targeting of EGFP-CBF1(6-9, EEF233) and in Fig. 6 that the RID-2 domain interacts with SKIP. Consistent with these observations, increasing the concentration of SKIP by cotransfection had no effect on the cytoplasmic distribution of EGFP-CBF1(6-9, EEF233) in the presence of SMRT(1-1290) (Fig. 7A), presumably because SKIP is unable to interact with SMRT with RID-2 deleted. However, increasing the concentration of the corepressor CIR in this same assay led to partial recovery of nuclear targeting by EGFP-CBF1(6-9, EEF) (Fig. 7B). CIR interacts with SMRT and also interacts strongly with SKIP (59), and we believe that CIR may tether SKIP to SMRT(1-1290) to reform a SKIP-CIR-SMRT complex and fulfill the requirements for CBF1 nuclear transport.
FIG. 7.
SKIP and CIR participate in SMRT-mediated nuclear targeting of CBF1. (A) Cotransfected SKIP did not rescue EGFP-CBF1(6-9, EEF) nuclear localization in the presence of SMRT(1-1290). An immunofluorescence assay was performed on Vero cells cotransfected with EGFP-CBF1(6-9, EEF), SMRT(1-1290), and HA-SKIP. (B) In cotransfected Vero cells CIR-Myc partially restored EGFP-CBF1(6-9, EEF) nuclear localization in the presence of SMRT(1-1290). Cells were stained for SMRT(1-1290) with goat anti-SMRT antibody.
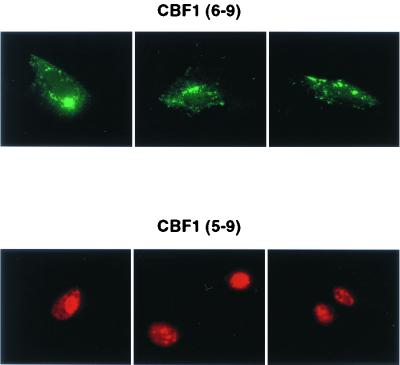
Further reinforcement of this model came from a comparison of the intracellular localization of Flag-CBF1(wt [wild-type] 6-9) and Myc-CBF1(wt 5-9) (Fig. 8). Whereas Flag-CBF1(wt 6-9) gave a cytoplasmic signal in transfected cells, Myc-CBF1(wt 5-9) was completely nuclear. SKIP interaction with CBF1 requires the presence of exon 5 (Zhou, unpublished data), and hence, a major difference between these two constructions is the ability to interact with SKIP.
FIG. 8.
The SKIP interaction domain, CBF1 exon 5, facilitates nuclear targeting. Immunofluorescence assay comparing the intracellular localizations of Flag-CBF1(6-9) (upper panel) and Myc-CBF1(5-9) (lower panel) in transfected Vero cells. Secondary antibodies were either FITC conjugated (upper panel) or rhodamine conjugated (lower panel).
The activators NotchIC and EBNA2 also mediate CBF1 nuclear localization.
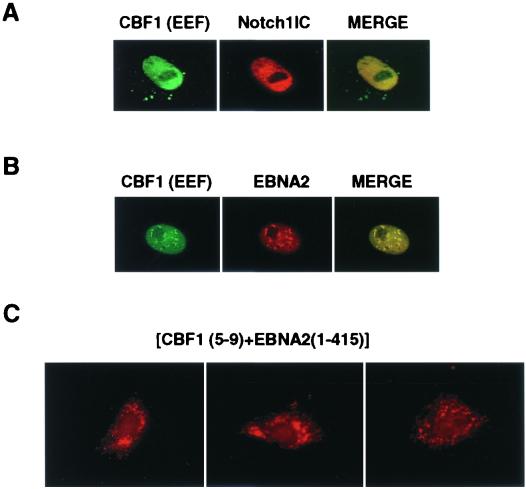
Evidence has been presented that nuclear localization of CBF1 is mediated through association with an SMRT-corepressor complex. The question arises as to the effect of the activators NotchIC and EBNA2. To address this point, Flag-CBF1(EEF233) was cotransfected into Vero cells with expression vectors for NotchIC or EBNA2. Both NotchIC and EBNA2 mediated nuclear entry of Flag-CBF1(EEF233) (Fig. 9A and B). As expected, truncated EBNA2(1-415) with the region containing the EBNA2 NLS deleted did not mediate nuclear translocation of Flag-CBF1(EEF233) (data not shown). More interestingly, EBNA2(1-415) was able to retain the normally nuclear Myc-CBF1(5-9) protein in the cytoplasm (Fig. 9C). This observation suggests that EBNA2 can outcompete the SMRT-SKIP-CIR complex for interaction with CBF1 and provides supporting evidence for the competition model for conversion of CBF1 from a mediator of repression to a mediator of activation.
FIG. 9.
NotchIC and EBNA2 can translocate CBF1(EEF233) into the nucleus. In indirect immunofluorescence assays in Vero cells, cotransfection of NotchIC (A) or EBNA2 (B) led to the detection of nuclear CBF1(EEF233) (left, green). CBF1 was detected by using rabbit anti-CBF1 and FITC-conjugated donkey anti-rabbit Ig; Flag-NotchIC (red) was detected by using rhodamine-conjugated goat anti-mouse Ig, and EBNA2 (red) was stained with anti-EBNA2 monoclonal antibody and rhodamine-conjugated anti-mouse Ig. (C) EBNA2 (aa 1 to 415), a truncated mutant protein that lacks the major NLS, retained normally nuclear Flag-CBF1(5-9) (red) in the cytoplasm.
A summary of the experimental data is provided in Fig. 10.
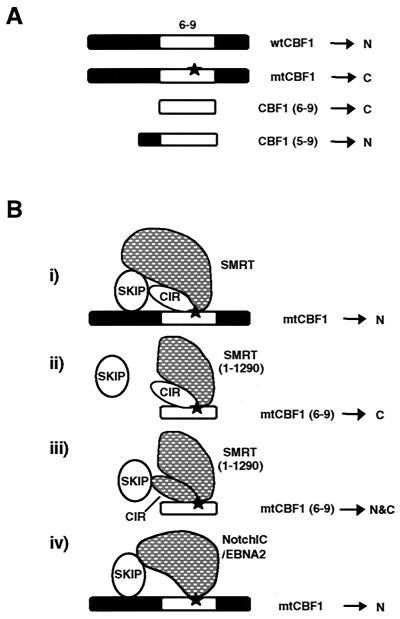
FIG. 10.
Summary of protein-protein interactions that affect CBF1 nuclear targeting. (A) Schematic of the wt and mutant (mt) CBF1 proteins showing the relative locations of exons 6 to 9 (white box) and the EEF233 and KLV249 mutations (star) and noting their intracellular localization, i.e., nuclear (N) or cytoplasmic (C). (B) Summary of experimental data. Transfected proteins are indicated by shading, and endogenous proteins are unshaded. (i) Increasing the concentration of SMRT overcomes the destabilized interaction brought about by the EEF233 or KLV249 mutation and restores nuclear targeting of mutant CBF1. (ii) Truncated SMRT(1-1290) with the RID2 domain deleted is unable to mediate this activity. The RID2 domain deletion abolishes the interaction between SKIP and SMRT. This result suggests that the presence of SKIP in the SMRT-corepressor complex is important for nuclear targeting of CBF1. (iii) The addition of transfected CIR to the situation shown in scheme ii allows partial recovery of nuclear localization. CIR interacts with SKIP and can bring endogenous SKIP to the SMRT(1-1290) complex. (iv) NotchIC and EBNA2 also mediate nuclear localization of mutant CBF1 in transfected cells. This NotchIC-EBNA2-CBF1 interaction recapitulates displacement of the SMRT-corepressor complex, an event that normally takes place in the nucleus.
DISCUSSION
The mechanisms involved in the intranuclear transduction of Notch signaling are incompletely understood. Both CSL protein dependent (35) and CSL-independent (33, 37, 39, 47) pathways have been described. CSL-partnered transcriptional activation by Notch is currently the better characterized mechanistically. In mammalian cells, domains within NotchIC and CBF1 that mediate interaction have been mapped, as have some of the regions within CBF1 that mediate other protein and DNA contacts. The region of CBF1 encoded by exons 6 to 9 (aa 179 to 361) binds to the corepressors SMRT and CIR, and a triple alanine mutation at CBF1 aa 233 to 235 has previously been shown to result in loss of both SMRT and CIR interactions (17, 22). The corepressor binding domain has been further defined in the present study with the demonstration that alanine substitution at aa 249 to 251 also results in loss of both CIR (17) and SMRT interactions (Fig. 2), whereas substitution at aa 261 to 263 does not affect either (Fig. 2 and unpublished results). Loss of SMRT and CIR interactions correlates with loss of CBF1 transcriptional repression activity (14, 16, 17), consistent with the known interactions between these proteins and components of the Sin3-Sap30-HDAC complex that mediates chromatin remodeling and repression. We now find that there is an additional correlation between the EEF233 and KLV249 mutations and the intracellular localization of CBF1. Whereas wt CBF1 localizes to the nuclei of transfected cells, loss of SMRT and CIR interactions resulted in a cytoplasmic CBF1 localization.
All of the data obtained in pursuing the relationship between SMRT-corepressor binding and nuclear localization of CBF1 are consistent with a core dependence on SMRT-SKIP contacts for CBF1 to become nuclear. The EEF233 and KLV249 mutations appear to destabilize, but not completely eliminate, the ability of CBF1 to bind to SMRT, and increasing the concentration of SMRT by introducing transfected SMRT was able to overcome the effects of the mutations and rescue the nuclear localization of the CBF1 EEF233 and KLV249 mutants. However, a truncated SMRT that was deleted for the C-terminal RID2 domain was unable to mediate nuclear rescue. The RID2 domain proved to interact with SKIP. Overexpression of CIR could partially compensate for loss of the SMRT RID2 domain, presumably because CIR interacts strongly with SKIP and could indirectly tether SKIP back onto the CBF1-SMRT-CIR complex. Finally, CBF1(6-9), which contains the SMRT-CIR interaction domain, was cytoplasmic in transfected cells, while the addition of exon 5 in CBF1(5-9) was sufficient to convert the protein to a nuclear localization. Exon 5 (aa 120 to 179) contains sequences required for SKIP binding to CBF1.
Nuclear targeting is commonly associated with the presence within the protein of either a short stretch of basic amino acids (NLS) or two basic motifs separated by a 10-aa spacer (bipartite NLS) that bind to importins and lead to association with, and transport through, the nuclear pore complex (21). However, other mechanisms also exist, including localization that is regulated through intermolecular interactions and complex formation. For example, interaction with 14–3-3 proteins modulates subcellular localization by masking export or docking signal sequences and nuclear localization of the catalytic subunit of telomerase (TERT) is regulated by 14–3-3 proteins (36). Nuclear localization by association with other NLS-containing proteins that participate in the same functional complex has been described, for example, for proteins involved in the assembly of virus capsids and in viral DNA replication (4, 11, 41, 56). The short stretch of positive amino acids in CBF1 exon 4 (aa 81 to 85) originally identified as a potential NLS has not been directly tested for NLS function. Our experiments suggest that, if active, this KKKKE signal is insufficient for nuclear transport of CBF1 since the EEF233 and KLV249 mutations, which are located some distance away from this sequence, result in a cytoplasmic phenotype. Based on the mutagenesis data, the nuclear localization of CBF1(5-9) and the protein-protein interactions that mapped to this region of CBF1, we conclude that CBF1 nuclear localization is a regulated process that is dependent on intermolecular interactions with the SMRT and SKIP binding partners.
A dependence on SMRT-corepressor interactions for CBF1 nuclear entry has implications for CBF1-mediated gene regulation. If CBF1 enters the nucleus preassembled in a complex with corepressors, then promoter-bound CBF1 would constitutively confer transcriptional repression. This scenario has the corollary that activation would then require competitive dissociation of the repression complex by the activators NotchIC and viral EBNA2. We previously noted that both EBNA2 and NotchIC interacted with the same region of CBF1 (aa 179 to 361; exons 6 to 9) that was required for repression activity (14, 15) and that the EEF233 and KLV249 CBF1 mutations that ablate SMRT-CIR interaction also impaired NotchIC interaction, as measured in mammalian and yeast two-hybrid assays, with the KLV249 mutation being the more severely debilitated (16). Sakai et al. (45) found that mutations in both the DBD of CBF1 (RBPJ-κ aa 212 to 227, CBF1 aa 186 to 201) and in a second region that was bounded by the aa 249 mutation (RBPJ-κ aa 275 to 323, CBF1 aa 249 to 297) affected NotchIC binding. This evidence for overlap of the repressor and activator interacting domains has been supported by competition experiments in which overexpression of SMRT led to displacement of the NotchIC and EBNA2 activators (22, 58, 59). Evidence has also been presented for competition between the EBNA2 and NotchIC proteins for CBF1 binding (45). Epstein-Barr virus EBNA2 is a nuclear protein, and it seems most likely that competition between EBNA2 and the repression complex for CBF1 binding would occur in the nucleus. An initial model for Notch signal transduction in drosophila suggested that the drosophila CSL protein Su(H) was bound to full-length Notch at the plasma membrane and entered the nucleus with NotchIC on ligand activation of Notch. This model was based on the observation that Su(H) was relocated to the cytoplasm in dually Notch- and Su(H)-transfected S2 cells (9). We found that transfected NotchIC could relocalize CBF1(EEF233) into the nucleus in transfected cells (Fig. 9) and that mutant cytoplasmic CBF1 redistributes within the cytoplasm to colocalize with Notch in dually transfected cells (Zhou, unpublished data), further indicating that interaction between these proteins can occur in the cytoplasm. However, we have been unable to obtain evidence for detectable levels of transfected wt CBF1 associated with membrane-bound Notch in GFP-Notch-overexpressing cells. Thus, at least to date, the evidence favors a nuclear location for NotchIC interactions with wt CBF1 and corepressor displacement.
Published experiments showing that binding between the corepressor complex and the NotchIC-EBNA2 activator is mutually exclusive have used overexpression of SMRT to compete away activation by NotchIC-EBNA2. More biologically relevant for the conversion from a constitutive repressive state to a state of activated gene expression would be a demonstration that the NotchIC-EBNA2 activator can outcompete binding by the SMRT-corepressor complex. Such a demonstration could be seen in Fig. 9C, in which normally nuclear CBF1(5-9) was held in the cytoplasm by an EBNA2 derivative with the region containing the EBNA2 NLS deleted. The experimental data have indicated that association with the SMRT-SKIP corepressor is required for CBF1 nuclear localization. If this corepressor complex is displaced by a competing protein that is itself unable to enter the nucleus, as is the case for the EBNA2(1-415) variant, then the outcome would be as observed, with CBF1(5-9) being retained in the cytoplasm. Thus, the intracellular relocalization of CBF1(5-9) also provided additional evidence in support of the competition-corepressor displacement model for NotchIC-EBNA2 transcriptional activation.
ACKNOWLEDGMENTS
We are grateful to R. Evans for providing SMRT plasmids, and we thank Feng Chang for manuscript preparation.
This work was funded by Public Health Service grant R01 CA42245 to S.D.H.
REFERENCES
- 1.Amakawa R, Jing W, Ozawa K, Matsunami N, Amaguchi Y, Matsuda F, Kawaichi M, Honjo T. Human Jk recombination signal binding protein gene (IGKJRB): comparison with its mouse homologue. Genomics. 1993;17:306–315. doi: 10.1006/geno.1993.1326. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Berezovska O, Jack C, McLean P, Aster J C, Hicks C, Xia W, Wolfe M S, Weinmaster G, Selkoe D J, Hyman B T. Rapid Notch1 nuclear translocation after ligand binding depends on presenilin-associated gamma-secretase activity. Ann N Y Acad Sci. 2000;920:223–226. doi: 10.1111/j.1749-6632.2000.tb06926.x. [DOI] [PubMed] [Google Scholar]
- 4.Calder J M, Stow E C, Stow N D. On the cellular localization of the components of the herpes simplex virus type 1 helicase-primase complex and the viral origin-binding protein. J Gen Virol. 1992;73:531–538. doi: 10.1099/0022-1317-73-3-531. [DOI] [PubMed] [Google Scholar]
- 5.Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H) Development. 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 6.Dahl R, Wani B, Hayman M J. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene. 1998;16:1579–1586. doi: 10.1038/sj.onc.1201687. [DOI] [PubMed] [Google Scholar]
- 7.Donoviel D B, Hadjantonakis A K, Ikeda M, Zheng H, Hyslop P S, Bernstein A. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 1999;13:2801–2810. doi: 10.1101/gad.13.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou S, Zeng X, Cortes P, Erdjument-Bromage H, Tempst P, Honjo T, Vales L D. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol Cell Biol. 1994;14:3310–3319. doi: 10.1128/mcb.14.5.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortini M E, Artavanis-Tsakonas S. The Suppressor of Hairless protein participates in Notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 10.Fortini M E, Rebay I, Caron L A, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 11.Gao Z, Krithivas A, Finan J E, Semmes O J, Zhou S, Wang Y, Hayward S D. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication complex. J Virol. 1998;72:8559–8567. doi: 10.1128/jvi.72.11.8559-8567.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 13.Helms W, Lee H, Ammerman M, Parks A L, Muskavitch M A, Yedvobnick B. Engineered truncations in the Drosophila Mastermind protein disrupt Notch pathway function. Dev Biol. 1999;215:358–374. doi: 10.1006/dbio.1999.9477. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh J J-D, Hayward S D. Masking of the CBF1/RBPJk transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh J J-D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh J J-D, Nofziger D E, Weinmaster G, Hayward S D. EBV Immortalization: Notch2 interacts with CBF1 and blocks differentiation. J Virol. 1997;71:1938–1945. doi: 10.1128/jvi.71.3.1938-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsieh J J-D, Zhou S, Chen L, Young D B, Hayward S D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppert S S, Le A, Schroeter E H, Mumm J S, Saxena M T, Milner L A, Kopan R. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 19.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 20.Jayachandra S, Low K G, Thlick A E, Yu J, Ling P D, Chang Y, Moore P S. Three unrelated viral transforming proteins (vIRF, EBNA2, and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci USA. 1999;96:11566–11571. doi: 10.1073/pnas.96.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Kaffman A, O'Shea E K. Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 22.Kao H-Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaichi M, Oka C, Shibayama S, Koromilas A E, Matsunami N, Hamaguchi Y, Honjo T. Genomic organization of mouse J kappa recombination signal binding protein (RBP-J kappa) gene. J Biol Chem. 1992;267:4016–4022. [PubMed] [Google Scholar]
- 24.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 25.Krebs L T, Xue Y, Norton C R, Shutter J R, Maguire M, Sundberg J P, Gallahan D, Closson V, Kitajewski J, Callahan R, Smith G H, Stark K L, Gridley T. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 26.Kurooka H, Honjo T. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 27.Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 28.Levitan D, Greenwald I. Effects of SEL-12 presenilin on LIN-12 localization and function in Caenorhabditis elegans. Development. 1998;125:3599–3606. doi: 10.1242/dev.125.18.3599. [DOI] [PubMed] [Google Scholar]
- 29.Levitan D, Greenwald I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature. 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Greenwald I. HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc Natl Acad Sci USA. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 32.Ling P D, Hsieh J J-D, Ruf I K, Rawlins D R, Hayward S D. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J Virol. 1994;68:5375–5383. doi: 10.1128/jvi.68.9.5375-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuno K, Go M J, Sun X, Eastman D S, Artavanis-Tsakonas S. Suppressor of Hairless-independent events in Notch signaling imply novel pathway elements. Development. 1997;124:4265–4273. doi: 10.1242/dev.124.21.4265. [DOI] [PubMed] [Google Scholar]
- 34.Miele L, Osborne B. Arbiter of differentiation and death: Notch signaling meets apoptosis. J Cell Physiol. 1999;181:393–409. doi: 10.1002/(SICI)1097-4652(199912)181:3<393::AID-JCP3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Mumm J S, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 36.Muslin A J, Xing H. 14–3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 37.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 38.Nomura T, Khan M M, Kaul S C, Dong H-D, Wadhwa R, Colmenares C, Kohno I, Ishii S. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ordentlich P, Lin A, Shen C P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petcherski A G, Kimble J. Mastermind is a putative activator for Notch. Curr Biol. 2000;10:R471–R473. doi: 10.1016/s0960-9822(00)00577-7. [DOI] [PubMed] [Google Scholar]
- 41.Plafker S M, Gibson W. Cytomegalovirus assembly protein precursor and proteinase precursor contain two nuclear localization signals that mediate their own nuclear translocation and that of the major capsid protein. J Virol. 1998;72:7722–7732. doi: 10.1128/jvi.72.10.7722-7732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebay I, Fehon R G, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 43.Redmond L, Oh S R, Hicks C, Weinmaster G, Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- 44.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 45.Sakai T, Taniguchi Y, Tamura K, Minoguchi S, Fukuhara T, Strobl L J, Zimber-Strobl U, Bornkamm G W, Honjo T. Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2. J Virol. 1998;72:6034–6039. doi: 10.1128/jvi.72.7.6034-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 47.Shawber C, Nofziger D, Hsieh J J-D, Lindsell C, Bogler O, Hayward S D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 48.Struhl G, Adachi A. Nuclear access and action of Notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 49.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 50.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 51.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 52.Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinmaster G. Notch signal transduction: a real rip and more. Curr Opin Genet Dev. 2000;10:363–369. doi: 10.1016/s0959-437x(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 55.Westlund B, Parry D, Clover R, Basson M, Johnson C D. Reverse genetic analysis of Caenorhabditis elegans presenilins reveals redundant but unequal roles for sel-12 and hop-1 in Notch-pathway signaling. Proc Natl Acad Sci USA. 1999;96:2497–2502. doi: 10.1073/pnas.96.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu F Y, Ahn J-H, Alcendor D J, Jang W-J, Xiao J, Hayward S D, Hayward G S. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J Virol. 2001;75:1487–1506. doi: 10.1128/JVI.75.3.1487-1506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu L, Aster J C, Blacklow S C, Lake R, Artavanis-Tsakonas S, Griffin J D. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 58.Zhou S, Fujimuro M, Hsieh J J-D, Chen L, Hayward S D. A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J Virol. 2000;74:1939–1947. doi: 10.1128/jvi.74.4.1939-1947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou S, Fujimuro M, Hsieh J J-D, Chen L, Miyamoto A, Weinmaster G, Hayward S D. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]