Abstract
Medication-related osteonecrosis of the jaw (MRONJ) is a rare but serious adverse drug reaction. Our previous whole-exome sequencing study found SIRT1 intronic region single-nucleotide polymorphism (SNP) rs7896005 to be associated with MRONJ in cancer patients treated with intravenous (iv) bisphosphonates (BPs). This study aimed to identify causal variants for this association. In silico analyses identified three SNPs (rs3758391, rs932658, and rs2394443) in the SIRT1 promoter region that are in high linkage disequilibrium (r2 > 0.8) with rs7896005. To validate the association between these SNPs and MRONJ, we genotyped these three SNPs on the germline DNA from 104 cancer patients of European ancestry treated with iv BPs (46 cases and 58 controls). Multivariable logistic regression analysis showed the minor alleles of these three SNPs were associated with lower odds for MRONJ. The odds ratios (95% confidence interval) and p values were 0.351 (0.164–0.751; p = 0.007) for rs3758391, 0.351 (0.164–0.751; p = 0.007) for rs932658, and 0.331 (0.157–0.697; p = 0.0036) for rs2394443, respectively. In the reporter gene assays, constructs containing rs932658 with variant allele A had higher luciferase activity than the reference allele, whereas constructs containing SNP rs3758391 and/or rs2394443 did not significantly affect activity. These results indicate that the promoter SNP rs932658 regulates the expression of SIRT1 and presumably lowers the risk of MRONJ by increasing SIRT1 expression.
Keywords: ANTIRESORPTIVES, BONE MODELING AND REMODELING, OSTEOBLASTS, OSTEONECROSIS OF THE JAW, PHARMACOGENOMICS
Introduction
Medication-related osteonecrosis of the jaw (MRONJ) is diagnosed as the exposure of the jaw bone (mandible, maxilla, or both) with slow healing for >8 weeks or no healing.(1) MRONJ is painful and affects the quality of life. MRONJ occurs as an infrequent adverse drug reaction of treatment with various therapeutic drugs, the most prominent being nitrogen-containing bisphosphonates (BPs) and the receptor activator of NF-κB ligand (RANKL) inhibitor denosumab.(2) BPs are widely used to treat or prevent osteoporosis or skeletal-related events (SREs) in cancer patients(3–5) and are the most common agents for osteoporosis treatment.(6) BPs have also shown high efficacy in preventing bone loss in patients with metastatic cancer, such as multiple myeloma (MM), breast cancer, and prostate cancer.(5) The first MRONJ case was reported by Marx RE in 2003.(7) Since then, the reported incidence of MRONJ ranges from 0.001% to 0.15% in osteoporosis patients(1,8–11) and 0.5% to 12% in patients with metastatic cancer.(12–18) Although many mechanisms for MRONJ have been proposed,(19,20) the pathogenesis of MRONJ remains largely undetermined. Many studies have suggested that there is a genetic component for MRONJ,(21–26) and the incidences of MRONJ vary in genetically diverse populations.(27)
Our previous whole-exome sequencing (WES) results identified a single-nucleotide polymorphism (SNP), rs7896005, within an intronic region of SIRT1 locus to be associated with MRONJ.(28) SIRT1 is a protein-coding gene, which encodes a member of the sirtuin family of conserved NAD+-dependent protein deacetylase. As a protein deacetylase, SIRT1 plays a vital role in multiple important biological processes, such as energy metabolism, inflammation, tumorigenesis, longevity, cell differentiation, and bone remodeling.(29–37) Based on the results of in silico analyses, there is no evidence indicating that previously identified WES index SNP rs7896005 was a functional SNP. The purpose of this study was to identify functional SNPs in high linkage disequilibrium (LD) with the index SNP that might be causal SNPs for this association in a retrospective case–control study.
Materials and Methods
In silico analysis
LDlink,(38) an in silico tool provided by the National Cancer Institute (NCI) Division of Cancer Epidemiology and Genetics, was used to identify all SNPs from phase 3 (version 5) of the 1000 Genomes Project that were in high LD (r2 > 0.8) with the previously discovered WES index SNP rs7896005 (including rs7896005). The Genotype-Tissue Expression (GTEx) portal(39) and single nucleotide polymorphism annotator (SNiPA)(40) were used to identify expression quantitative trait loci (eQTL) SNPs for SIRT1. UCSC Genome Browser,(41) SNPnexus,(42) SNiPA,(40) and Haploreg V4.1(43) were used to identify locus-related information for the SNPs. Based on the location of the SNPs, different in silico tools were used to predict the potential functional effects of the SNPs. Briefly, Encyclopedia of DNA Elements (ENCODE),(44,45) UCSC Genome Browser, Haploreg V4.1, SNPnexus, RegulomeDB,(46) and Ensembl(47) were used to identify SNPs in regulatory elements. miRWalk,(48) TargetScan,(49) PolymiRTS,(50) miRBase,(51) and miRDB(52,53) were used to predict SNPs that may potentially affect microRNA (miRNA) binding. Human Splicing Finder(54) and Alternative Splice Site Predictor (ASSP)(55) were used to predict the SNPs that potentially affect splicing (Fig. 1).
Fig 1.
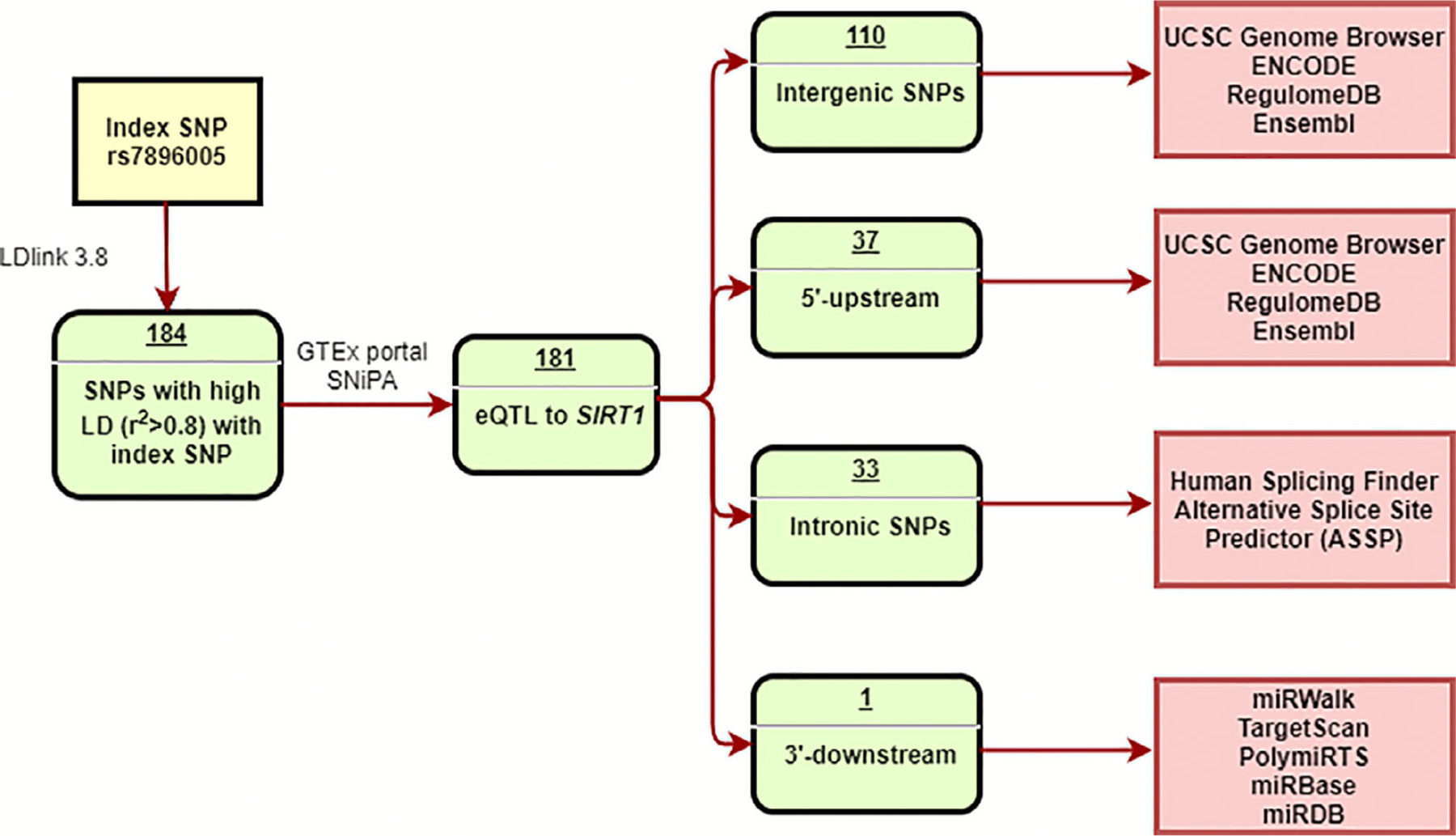
Flow chart for in silico analyses.
Patients
Genomic DNA samples were obtained from 104 cancer patients of European ancestry treated with intravenous (iv) BPs, including 46 patients who developed MRONJ (cases) and 58 who did not develop MRONJ (controls) from multiple institutions. All patients regardless of cancer stages or concomitant diseases who were treated with iv bisphosphonates (zolendronate or pamidronate) coming to Dr Moreb’s clinic or Dr Katz’s clinic at the University of Florida between 2016 and 2018 were approached for study participation. All patients who agreed to participate, signed consent, and provided genomic DNA were included in this study. Patients who developed MRONJ and confirmed by Dr Katz were included as MRONJ cases; those who did not develop MRONJ after at least 12 months of iv BP treatment were included as controls. Additional MRONJ cases were also obtained from Semmelweis University of Medical School and Dental School in Budapest, Hungary, and the University of Bologna in Bologna, Italy. All patients signed informed consent at each study site. This study was approved by the Institutional Review Board at the University of Florida (IRB nos. IRB201501186 and IRB201800934).
Genotyping
Seven SNPs identified from in silico analyses or our previous WES were genotyped in the 104 samples. Genomic DNA was isolated from buffy coat using the FlexiGene DNA kit (Qiagen, Valencia, CA, USA). SNPs rs7896005, rs3758391, rs3740051, and rs35706870 were genotyped using TaqMan Genotyping on QuantStudio 12 K Flex Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). All Taqman probes were ordered from Thermo Fisher Scientific (Winooski, VT, USA). SNPs with no exiting Taqman probed (rs12778366, rs932658, and rs2394443) were genotyped using pyrosequencing. Pyrosequencing assays were designed using pyrosequencing assay design software.(56) The PCR and sequencing primer information is shown in Supplemental Table S1. All primers for the pyrosequencing assay were ordered from Integrated DNA Technologies, Inc (Coralville, IA, USA).
Based on the genotyping results, SNPs were selected for further investigation. The potential function of promoter region SNPs was validated using the luciferase assay.
In vitro luciferase reporter assay
Cell culture:
The human osteoblast cell line (hFOB 1.19) purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA) was cultured in complete 1:1 mixture of Ham’s F12 Medium Dulbecco’s Modified Eagle’s Medium (DMEM) (without phenol red) (Thermo Fisher) supplemented with fetal bovine serum (Thermo Fisher) (10%), 2.5 mM L-glutamine, 0.3 mg/mL G418 (Thermo Fisher). Cells were grown at 34°C in a humidified atmosphere containing 5% CO2.
Plasmid constructs:
PCR and appropriate primers (IDT) were used to amplify two fragments of the SIRT1 promoter of different lengths (Promoter_short [1.13 kb] and Promoter_long [1.54 kb]) using human genomic DNA as a template. Briefly, PCR was performed in a maximum volume of 25 μL: 12.5 μL of CloneAmp HiFi PCR Premix (Takara Bio, Mountain View, CA, USA), 1 μL of forward primer, 1 μL of reverse primer, 8.5 μL of RNA free water, and 2 μL of DNA template. The parameters for the amplification reaction were the following: 95°C for 15 minutes, which was followed by 40 cycles of 98°C for 10 seconds, 60°C for 30 seconds, and 72°C for 1 to 2 minutes based on the length of DNA fragment. The primers were designed using the Takara In-Fusion primer design tool. All promoter fragments were purified using a DNA purification kit (Takara Bio). The primers are listed as 1.54 kb-NheI-forward-5′-ACCTGAGCTCGCTAGCAGGCTTCTAGGACTGGAGATG-3′, 1.13 kb-NheI-forward-5′-ACCTGAGCTCGCTAGCGGTAC CCCTCGTTTTACATCTGG-3′, and HinIII-reverse-5′-CCGGATTGC CAAGCTTTGCGGAGCGGCTCCCCGG-3′.
In-Fusion cloning:
The pGL4.10 [luc2] vector (Promega, Madison, WI, USA) was linearized by digestion with the NheI and HinIII restriction enzymes (New England BioLabs, Ipswich, MA, USA) and then purified using a DNA purification kit (Takara Bio). Two DNA fragments (Promoter_short and Promoter_long) from the SIRT1 promoter containing different combinations of candidate SNPs were then cloned into pGL4.10 vector using the In-Fusion cloning kit (Takara Bio). Clones were screened with restriction enzyme digestion, and DNA fragments were sequenced (Eton Bioscience, San Diego, CA, USA) to ensure no random mutations. All primers used for sequencing are listed in Supplemental Table S2. Plasmid DNA was prepared using the PureYield Plasmid Midiprep System (Promega).
Dual-luciferase reporter gene assay:
HFOB 1.19 cells were plated in 96-well plates with 10,000 cells in 100 μL per well. After 24 hours, 10 μL of FuGENE 6 reagent (Promega)/plasmid DNA (DNA/fuGENE6 ratio to 1:3) mixture was added to each well of cells for transfection. pGL4.74 (hRluc/TK) expressing Renilla luciferase was used as an internal control. Empty pGL4.10 [luc2] basic vector was used as a negative control. Forty-eight hours post-transfection, luciferase activities of the transfected cells were measured using the dual-luciferase reporter assay system (Promega) on the BioTek Synergy HT Reader (Thermo Fisher). All experiments were repeated independently in triplicate for three times. The firefly luciferase activity normalized by Renilla luciferase activity represents the transcriptional activities of the SIRT1 promoter. The luminescence ratio was represented by the ratio of the firefly luciferase activity and Renilla luciferase activity. The relative luciferase activity is the normalized ratio of the luminescence ratio, which normalized the results from each experimental sample to control samples repeated on each plate.
Statistical analysis
Multivariable logistic regression was performed to estimate the odds ratios (ORs) and 95% confidence interval (CI) of each potential candidate SNP with MRONJ adjusting for age and sex. To account for multiple comparisons, SNPs with p values <0.0125 (0.05/4) were considered statistically significant. Luciferase reporter assay results are represented as mean ± SE and analyzed with Student’s t test. A p value <0.0125 was considered statistically different. All statistical analyses were performed using SAS 9.4 (Cary, NC, USA).
Results
Three SIRT1 promoter SNPs were associated with MRONJ
A total of 184 SNPs in high LD (r2 > 0.8) with index SNP rs7896005 in the European population in the 1000 Genome Project were extracted using LDlink 3.8. After excluding SNPs that were not eQTLs for SIRT1 based on the GTEx database, 181 SNPs were classified into four groups: intergenic (110), 5′-upstream of SIRT1 (37), intronic (33), and 3′-downstream (1) (Fig. 1).
According to the UCSC Genome Browser, 20 of 181 SNPs were located at five potential enhancer/promoter elements of SIRT1 (Supplemental Fig. S1A; Supplemental Table S3). The RegulomeDB rank was represented in Supplemental Table S3. The rank of RegulomeDB refers to different available data types for a single coordinate, which gives a prediction of the potential function of these 20 SNPs. After evaluating Ensembl, ENCODE, and the Homo sapiens SIRT1 promoter region (LOC107832851) from National Center for Biotechnology Information (NCBI), SNPs rs3758391, rs932658, and rs2394443 were selected as candidate functional SNPs, all of which were located in the promoter region (GH10J067883) of SIRT1 gene (Supplemental Table S4; Supplemental Fig. S1B). The UCSC Genome Browser showed that rs932658 and rs2394443 were in a DNase I hypersensitivity signal peak, enriched for H3K27Ac and near CpG island, which mostly occurs in the promoter region (Supplemental Fig. S1B). The DNase I hypersensitivity signal peak identified the potential regulatory region, which is sensitive to cleavage by DNase H3K27Ac was defined as an enhancer mark, which was mostly found nearby the transcription start site. SNP rs3758391 was also in the promoter region and showed the perfect LD with rs932658. The UCSC Genome Browser also reported the interactions between this promoter region (GH10J067883) and other enhancer regions (GH10J067837, GH10J067848, GH10J067855). The 3D Genome Browser Hi-C data(57,58) predicted these SNPs are located in a topologically associated domain (TAD) in the ENCODE data using human mesenchymal stem cells line (H1-MSC) (Supplemental Fig. S1C). Based on the annotation by chromHMM, these SNPs are located at an active TSS (red). According to virtual 4C data, there is a potential interaction between this active TSS and the SIRT1 enhancer (orange). This linkage is also supported by DNase I hypertensive site (DHS) linkage. This region is also located at a H3K4me3 peak region, which is a promoter mark (Supplemental Fig. S1D). The allele frequency for all three promoter region candidate SNPs and the index SNP rs7896005 were shown in Supplemental Table S5.
Ensembl Genome Browser showed a total of six SIRT1 transcripts in the human genome (Supplemental Fig. S2), containing 11 exons and 10 introns. Based on the GTEx portal, there are 11 splice junctions. Intron 1–7 could potentially modify the SIRT1 transcript expression by affecting splicing. However, human splice finder (HSF) and alternative splice site predictor (ASSP) results showed that none of the intron SNPs affect splicing (Supplemental Table S6).
MicroRNAs (MiRNAs) are small non-coding RNAs, which affect mRNA stability or translation by base-pairing with complementary sequences within target mRNAs. miRNAs can also regulate protein expression via binding to the 3’ UTR region of mRNA. Studies have identified that miRNA can also bind to intron regions or promoter TATA-box motifs to affect mRNA stability and alternative expression.(59,60) We used miRWalk, TargetScan 7.2, and miRDB to extract all potential miRNA and their binding sites. None of the target SNPs are predicted to be located at a miRNA binding site.
After in silico analyses, a total of three SIRT1 promoter region SNPs (rs3758391, rs932658, and rs2394443) were identified as potential functional SNPs. The GTEx expression data for SNPs rs3758391, rs932658, and rs2394443 showed that the minor allele for each of these three SNPs in our study had increased SIRT1 mRNA expression in whole blood, with p values of 9.3 × 10−46, 9.3 × 10−46, and 4.2 × 10−33, respectively (Supplemental Fig. S3).
Genotyping validated three promoter SNPs associated with MRONJ
To further validate these three SNPs, we performed an independent genotyping study of 104 cancer patients of European ancestry treated with iv BPs, including 58 controls and 46 MRONJ cases. The demographics of the samples are shown in Table 1. The mean age of the patients was 64 years. For MRONJ cases, the mean duration of BP treatment was 26.55 months before the development of MRONJ. For controls, the mean duration of treatment was 33.06 months. There was no statistically significant difference in the duration of iv BP treatment between the two groups (p = 0.194). Multivariable logistic regression analysis found that the WES index SNP and the three candidate SNPs were associated with lower odds for MRONJ: OR = 0.331 and 95% CI 0.157–0.697 (p = 0.0036) for index SNP rs7896005, OR = 0.351 and 95% CI 0.164–0.751 (p = 0.007) for rs3758391, OR = 0.351 and 95% CI 0.164–0.751 (p = 0.007) for rs932658, and OR = 0.331 and 95% CI 0.157–0.697 (p = 0.0036) for rs2394443, respectively (Fig. 2).
Table 1.
Demographic Table of Patients Included in the Logistic Regression Analysis
| Characteristics | Total (n = 104) | MRONJ (n = 46) | Non-MRONJ (n = 58) | p Value |
|---|---|---|---|---|
| Age (years) (mean ± SD) | 63.92 ± 9.93 | 65.13 ± 10.47 | 62.97 ± 9.46 | 0.4 |
| Female (%) | 50.96% | 69.57% | 36.21% | 0.0008 |
| Duration of treatment (mean ± SD) | 29.94 ± 4.99 | 26.55 ± 7.36 | 33.06 ± 6.82 | 0.194 |
| Race | ||||
| European ancestry | 104 (100%) | 46 (100%) | 58 (100%) | |
| Cancer type | ||||
| Multiple myeloma | 68 (65.38%) | 15 (32.61) | 53 (91.38%) | 0.0001 |
| Breast cancer | 26 (25.00%) | 24 (52.17) | 2 (3.45%) | |
| Lung cancer | 1(0.96%) | 1 (2.17%) | 0 (0%) | |
| Prostate cancer | 6 (5.77%) | 3 (6.52) | 3 (5.17%) | |
| Renal cancer | 2 (1.92) | 2 (4.35) | 0 (0%) | |
| Cervix cancer | 1 (0.96%) | 1 (2.17%) | 0 (0%) |
MRONJ = medication-related osteonecrosis of the jaw.
Fig 2.
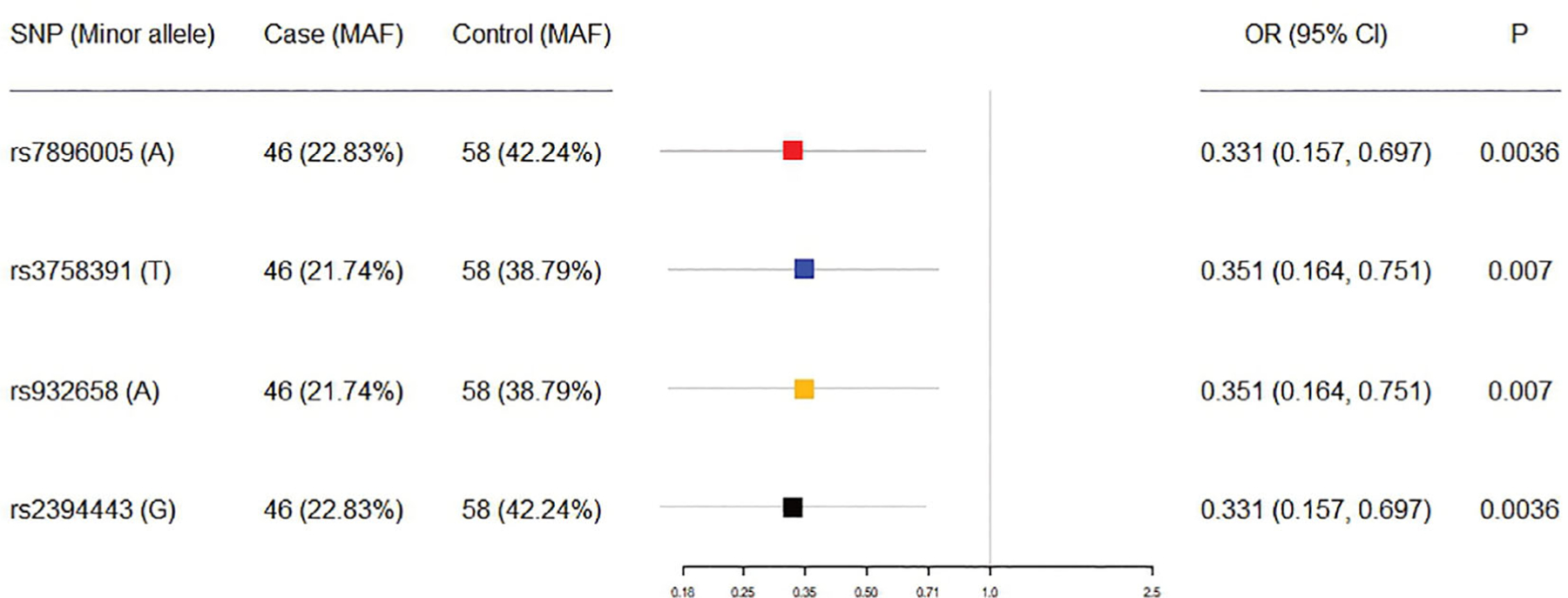
Odds ratio (OR) of three promoter region SNPs. MAF = minor allele frequency; CI = confidence interval.
SNP rs932658 affected promoter activity of SIRT1
To assess the effects of these SNPs on transcriptional activity, we performed luciferase reporter gene assays using DNA fragments derived from the promoter region of the SIRT1 gene. The Homo sapiens SIRT1 promoter region (LOC107832851) from NCBI is 2958 bp and includes 11 SNPs (Fig. 3A). We generated two DNA constructs: Promoter_short is a 1.13 kb DNA fragment, which included SNPs rs35706870, rs3740051, rs932658, rs3740053, and rs2394443; Promoter_long is a 1.54 kb DNA fragment, which included SNPs rs12778366, rs3758391, rs35706870, rs3740051, rs932658, rs3740053, and rs2394443 (Fig. 3B). The sequence information of the two DNA fragments is shown in Supplemental Table S7. After genotyping, samples with homozygous T/T, A/A, and A/A for rs12778366, rs35706870, and rs3740051 were selected for further studies. The LD plot showed complete LD between rs3740051 and rs3740053, so SNP rs3740053 homozygous A/A were also identified in these samples. From these samples, heterozygous T/C, A/C, G/C for SNPs rs3758391, rs932658, and rs2394443 were selected for cloning.
Fig 3.
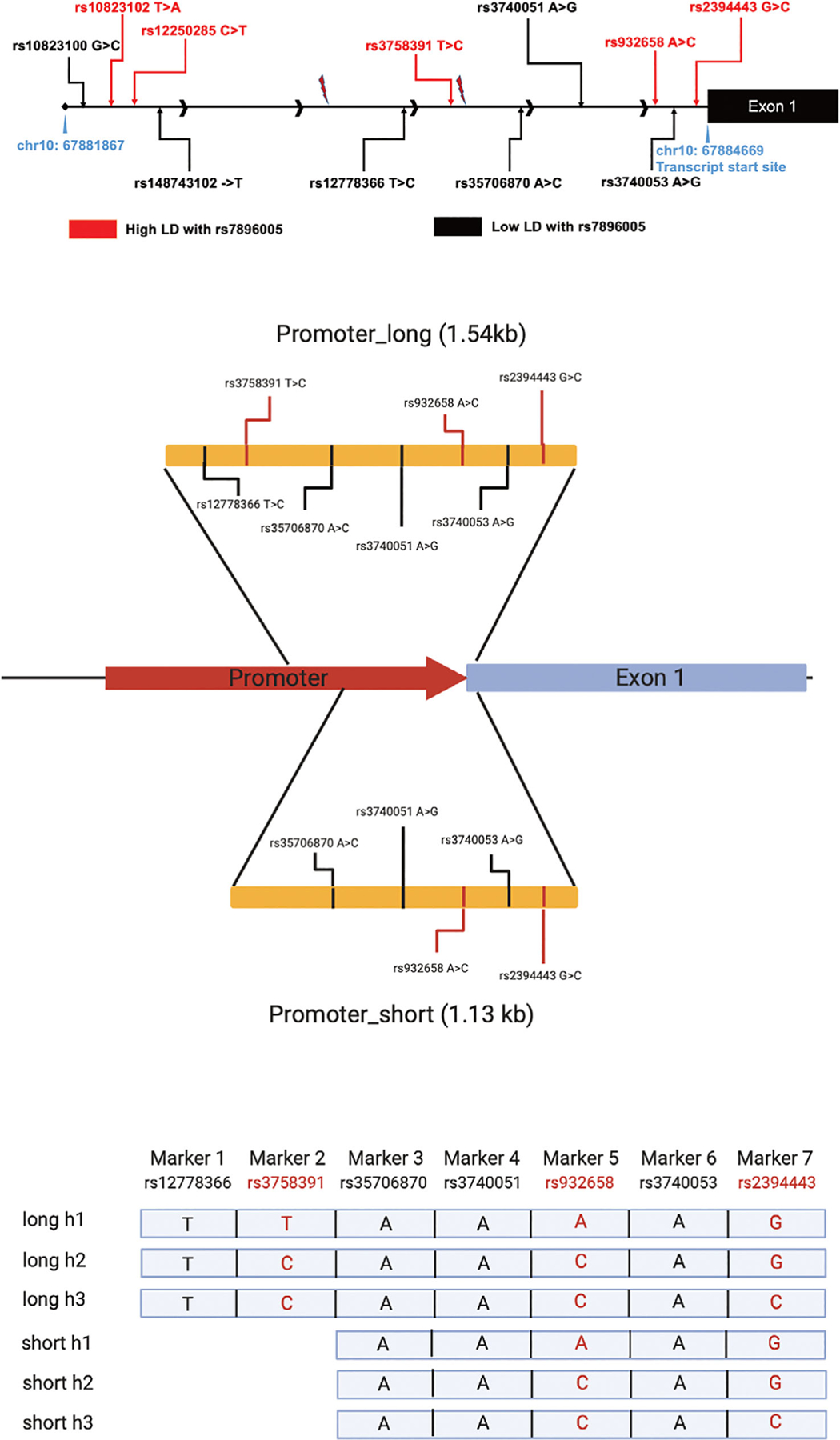
SNPs at SIRT1 promoter region. (A) Eleven SNPs are located in the SIRT1 promoter region according to the 1000 Genome Project. (B) We designed two promoter region constructs with different lengths: Promoter_short included SNPs rs35706870, rs3740051, rs932658, rs3740053, and rs2394443; Promoter_long included SNPs rs12778366, rs3758391, rs35706870, rs3740051, rs932658, rs3740053, and rs2394443. (C) The six possible haplotypes were used to perform luciferase reporter gene assay.
Three different haplotypes of the target SNPs are reported in human populations for both of the SIRT1 DNA fragments (Supplemental Fig. S2), all of which were identified in our samples (Fig. 3C). The construct Promoter_short containing the rs932659 variant A allele showed significantly higher luciferase activity compared with those containing the reference C allele (p = 0.0052), whereas SNP rs2394443 did not affect reporter gene activity (Fig. 4A, B). Similar results were obtained with the Promoter_long constructs, showing higher luciferase activity only when containing the variant A allele of rs932658 (Fig. 4C, D). Moreover, the two constructs with different lengths had similar luciferase activities (A-G versus T-A-G) (p = 0.93) (Fig. 4E, F). Therefore, the addition of SNP rs3758391 did not affect luciferase activity. These results demonstrate that the promoter SNP rs932658 increases transcriptional activity in reporter gene assays.
Fig 4.
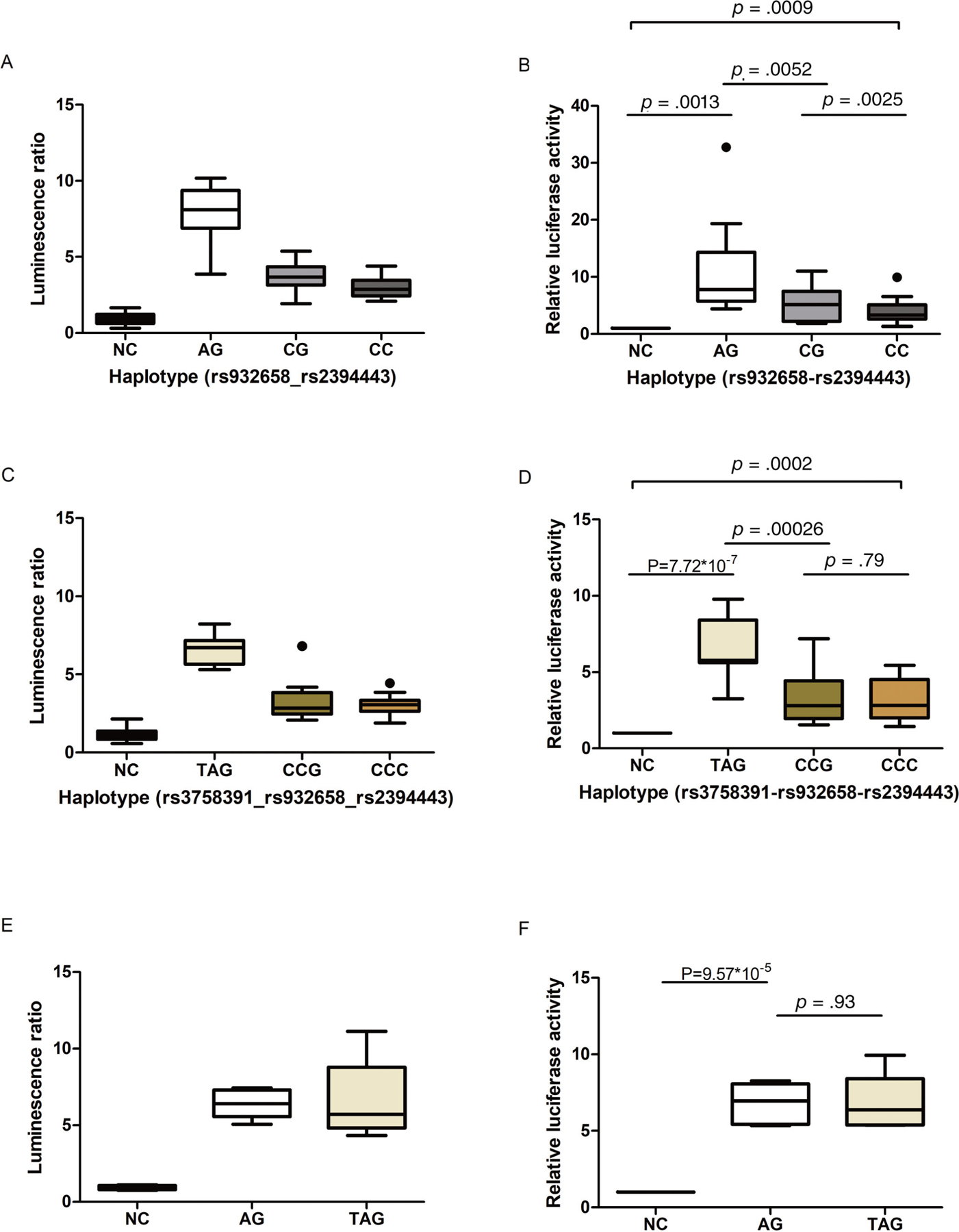
The effect of different haplotypes in luciferase reporter assays. Luminescence activity for different haplotypes is shown in a relative luminescence activity plot. (A, C, E) Luminescence ratio is represented by the ratio of the firefly luciferase activity and Renilla luciferase activity. (B, D, F) Box plots of the normalized ratios of the luminescence ratios, which normalized the results from each experimental sample to control samples repeated on each plate. (A–D) Polymorphism A of rs932658 increased promoter activity of SIRT1. rs2394443 did not affect promoter activity significantly. (E, F) There was no significant difference between haplotype A-G and haplotype T-A-G. Box plot key: line within box = median; upper boundary of box = third quartile (Q3); lower boundary of box = first quartile (Q1); vertical length of box = interquartile range (IQR); upper whisker = smaller value of the maximum and Q3 + 1.5*IQR; lower whisker = larger value of the minimum and Q1–1.5*IQR. Dots indicate outliers outside of the boundaries defined by whiskers.
To further confirm that rs932658 is the causal SNP, we performed a conditional logistic regression analysis in the 104 samples. After adjusting for rs932658, the index SNP was no longer associated with MRONJ (OR = 0.263 and 95% CI 0.026–2.674; p = 0.26).
Discussion
In this study, we identified three promoter region SNPs, rs3758391 (C > T), rs932658 (C > A), and rs2394443 (C > G) in the SIRT1 gene that were associated with lower odds of developing MRONJ among cancer patients who were treated with iv BPs for the prevention of SREs. All three SNPs were in high LD (r2 > 0.8) with our previous WES study index SNP rs7896005. In addition, in silico analyses using the GTEx portal provided evidence that the T allele of rs3758391, the A allele of rs932658, and the G allele of rs2394443 were associated with higher expression of SIRT1. Luciferase reporter gene assays showed that allele A of SNP rs932658 was associated with higher promoter activity of SIRT1, which leads to higher expression of SIRT1 mRNA. These results indicate that the promoter SNP rs932658 is the functional variant regulating the expression of SIRT1 and protects against MRONJ by increasing SIRT1 expression.
SIRT1 plays an important role in bone remodeling, especially by promoting bone formation.(6) SIRT1 could regulate the Wnt signaling pathway by deacetylating β-catenin,(61) causing β-catenin to accumulate in the nucleus and induce mesenchymal stem cell (MSC) differentiation to osteoblasts, rather than adipogenesis.(61,62) Low expression of SIRT1 also inhibits the RUNX2 pathway, which plays an important role in osteoblast differentiation.(63) SIRT1 also deacetylates the forkhead box proteins (FOX) O1, FOXO3, and FOXO4, which inhibit osteoclast differentiation and proliferation.(34) Based on these studies, SIRT1 may regulate bone remodeling by both promoting osteoblast differentiation while inhibiting osteoclast formation. in vivo experiments have shown that SIRT1 knockout (KO) animals develop defects in bone formation(64–66) and that deletion of SIRT1 in osteoblasts or osteoclasts induced loss of bone mass.(6,67) Furthermore, previous studies showed that SIRT1 stimulates MSC proliferation and differentiation into mature osteoblasts.(63,68) SIRT1 is associated with inflammation.(69) SIRT1 deacetylates the p65 protein, which is a subunit of the NF-κB complex and deacetylation of p65 leads to the inhibition of NF-κB signaling,(32) thereby reducing the immune response and inducing the inflammation process through the NF-κB pathway.(70) SIRT1 also promotes angiogenesis by regulating the MAPK pathway.(71) Inflammation and anti-angiogenesis are both risk factors of MRONJ based on the updated 2014 guidelines from the American Association of Oral and Maxillofacial Surgeons (AAOMS).(1) Therefore, SIRT1 may reduce the risk of MRONJ by inhibiting inflammation and inducing angiogenesis, improving healing of oral cavity wounds. Combined with the above results, the SIRT1 SNP rs932658 was associated with decreased risk of MRONJ induced by BPs by increasing the expression of SIRT1. Overall, increased expression of SIRT1 due to pharmacogenomic markers, such as polymorphism A of rs932658 identified in this study, may lower the risk of patients developing MRONJ when treated with BPs.
It is important to recognize that MRONJ is a complex disease that is likely driven by many genetic factors, environmental factors, and lifestyle. The genetic factors are just one factor. If a patient is homozygous or heterozygous for the risk allele, that does not mean that they will get MRONJ if they are treated with BPs. Rather it means they are at higher risk for MRONJ. There are other genetic factors that could contribute to the risk, as well as other environmental and lifestyle factors. Because MRONJ is rare, the only appropriate study design is a case–control study design. We found that the minor allele frequency of SNP rs932658 was 22% in MRONJ cases, but 39% in controls. So the appropriate interpretation of our result would be that patients with the minor allele A of the SNP rs932658 were at lower odds for MRONJ when treated with BPs than those who were homozygotes for major allele C of this SNP.
Our study had some limitations that need to be acknowledged. Because of the low rate of this drug-related adverse event, we used a case–control study design, which has some inherent limitations. One major limitation was that we could not ascertain that the controls who did not develop MRONJ at the time of analyses were truly controls. Nevertheless, in the literature, the median duration of treatment before the development of MRONJ was 15.6 months for zoledronate, 26.4 months for pamidronate, and 28.8 months for alendronate.(72) When we compared the duration of iv BP treatment between MRONJ cases and controls (26.55 versus 33.06 months), there were no statistical differences between the two groups. So the controls in our study were likely to be real controls. The other limitation was that because of the low event rate of MRONJ and therefore small sample size of our study, we could not match MRONJ cases and controls within each cancer type. This might not be an issue because the germline genetic polymorphisms associated with MRONJ is likely not cancer specific.
In conclusion, this study validated our previous WES results(28) and also suggested a causal variant for this association. These results indicate that the promoter SNP rs932658 regulates the expression of SIRT1 and presumably lowers the risk of MRONJ by increasing SIRT1 expression. SNPs rs932658 may be the potential pharmacogenomic marker for a personalized approach to regulate the use of BPs for the prevention of SREs. Based on our data, patients carrying the major allele C in rs932658 may be at higher risk of MRONJ and therefore may potentially benefit from lower exposure or less frequent administration of a BP agent. Further investigations are warranted before such findings can be translated into clinic settings.
Supplementary Material
Acknowledgments
This study was supported by the University of Florida Health Cancer Center. We also acknowledge the support of all the study participants.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures
All authors state that they have no conflicts of interest.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/jbmr.4185.
References
- 1.Ruggiero SL, Dodson TB, Fantasia J, et al. American Association of Oral and Maxillofacial Surgeons position paper on medication-related osteonecrosis of the jaw–2014 update. J Oral Maxillofac Surg 2014;72(10):1938–56. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Hamadeh IS, Song S, et al. Osteonecrosis of the jaw in the United States Food and Drug Administration’s adverse event reporting system (FAERS). J Bone Miner Res 2016;31(2):336–40. [DOI] [PubMed] [Google Scholar]
- 3.Hamadeh IS, Ngwa BA, Gong Y. Drug induced osteonecrosis of the jaw. Cancer Treat Rev 2015;41(5):455–64. [DOI] [PubMed] [Google Scholar]
- 4.Drake MT, Clarke BL, Khosla S. Bisphosphonates: mechanism of action and role in clinical practice. Mayo Clin Proc 2008;83(9): 1032–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gralow JR, Biermann JS, Farooki A, et al. NCCN task force report: bone health in cancer care. J Natl Compr Canc Netw 2009;7(Suppl 3): S1–32; quiz S3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zainabadi K, Liu CJ, Caldwell ALM, Guarente L. SIRT1 is a positive regulator of in vivo bone mass and a therapeutic target for osteoporosis. PLoS One 2017;12(9):e0185236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marx RE. Pamidronate (Aredia) and zoledronate (Zometa) induced avascular necrosis of the jaws: a growing epidemic. J Oral Maxillofac Surg 2003;61(9):1115–7. [DOI] [PubMed] [Google Scholar]
- 8.Khan AA, Morrison A, Hanley DA, et al. Diagnosis and management of osteonecrosis of the jaw: a systematic review and international consensus. J Bone Miner Res 2015;30(1):3–23. [DOI] [PubMed] [Google Scholar]
- 9.Lo JC, O’Ryan FS, Gordon NP, et al. Prevalence of osteonecrosis of the jaw in patients with oral bisphosphonate exposure. J Oral Maxillofac Surg 2010;68(2):243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malden N, Lopes V. An epidemiological study of alendronate-related osteonecrosis of the jaws. A case series from the south-east of Scotland with attention given to case definition and prevalence. J Bone Miner Metab 2012;30(2):171–82. [DOI] [PubMed] [Google Scholar]
- 11.Grbic JT, Black DM, Lyles KW, et al. The incidence of osteonecrosis of the jaw in patients receiving 5 milligrams of zoledronic acid: data from the health outcomes and reduced incidence with zoledronic acid once yearly clinical trials program. J Am Dent Assoc 2010;141 (11):1365–70. [DOI] [PubMed] [Google Scholar]
- 12.Stopeck AT, Fizazi K, Body JJ, et al. Safety of long-term denosumab therapy: results from the open label extension phase of two phase 3 studies in patients with metastatic breast and prostate cancer. Support Care Cancer 2016;24(1):447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saad F, Brown JE, Van Poznak C, et al. Incidence, risk factors, and outcomes of osteonecrosis of the jaw: integrated analysis from three blinded active-controlled phase III trials in cancer patients with bone metastases. Ann Oncol 2012;23(5):1341–7. [DOI] [PubMed] [Google Scholar]
- 14.Bamias A, Kastritis E, Bamia C, et al. Osteonecrosis of the jaw in cancer after treatment with bisphosphonates: incidence and risk factors. J Clin Oncol 2005;23(34):8580–7. [DOI] [PubMed] [Google Scholar]
- 15.Durie BG, Katz M, Crowley J. Osteonecrosis of the jaw and bisphosphonates. N Engl J Med 2005;353(1):99–102. [DOI] [PubMed] [Google Scholar]
- 16.Vahtsevanos K, Kyrgidis A, Verrou E, et al. Longitudinal cohort study of risk factors in cancer patients of bisphosphonate-related osteonecrosis of the jaw. J Clin Oncol 2009;27(32):5356–62. [DOI] [PubMed] [Google Scholar]
- 17.Qi WX, Tang LN, He AN, Yao Y, Shen Z. Risk of osteonecrosis of the jaw in cancer patients receiving denosumab: a meta-analysis of seven randomized controlled trials. Int J Clin Oncol 2014;19(2):403–10. [DOI] [PubMed] [Google Scholar]
- 18.Coleman R, Woodward E, Brown J, et al. Safety of zoledronic acid and incidence of osteonecrosis of the jaw (ONJ) during adjuvant therapy in a randomised phase III trial (AZURE: BIG 01–04) for women with stage II/III breast cancer. Breast Cancer Res Treat 2011;127(2):429–38. [DOI] [PubMed] [Google Scholar]
- 19.Ruggiero SL, Dodson TB, Assael LA, et al. American Association of Oral and Maxillofacial Surgeons position paper on bisphosphonate-related osteonecrosis of the jaws–2009 update. J Oral Maxillofac Surg 2009;67(5 Suppl):2–12. [DOI] [PubMed] [Google Scholar]
- 20.American Dental Association Council on Scientific Affairs. Dental management of patients receiving oral bisphosphonate therapy: expert panel recommendations. J Am Dent Assoc 2006;137(8): 1144–50. [DOI] [PubMed] [Google Scholar]
- 21.Di Martino MT, Arbitrio M, Guzzi PH, et al. A peroxisome proliferator-activated receptor gamma (PPARG) polymorphism is associated with zoledronic acid-related osteonecrosis of the jaw in multiple myeloma patients: analysis by DMET microarray profiling. Br J Haematol 2011; 154(4):529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz J, Gong Y, Salmasinia D, et al. Genetic polymorphisms and other risk factors associated with bisphosphonate induced osteonecrosis of the jaw. Int J Oral Maxillofac Surg 2011;40(6):605–11. [DOI] [PubMed] [Google Scholar]
- 23.Stockmann P, Nkenke E, Englbrecht M, et al. Major histocompatibility complex class II polymorphisms are associated with the development of anti-resorptive agent-induced osteonecrosis of the jaw. J Craniomaxillofac Surg 2013;41(1):71–5. [DOI] [PubMed] [Google Scholar]
- 24.La Ferla F, Paolicchi E, Crea F, et al. An aromatase polymorphism (g.132810C>T) predicts risk of bisphosphonate-related osteonecrosis of the jaw. Biomark Med 2012;6(2):201–9. [DOI] [PubMed] [Google Scholar]
- 25.Sarasquete ME, García-Sanz R, Marín L, et al. Bisphosphonate-related osteonecrosis of the jaw is associated with polymorphisms of the cytochrome P450 CYP2C8 in multiple myeloma: a genome-wide single nucleotide polymorphism analysis. Blood 2008;112(7):2709–12. [DOI] [PubMed] [Google Scholar]
- 26.Nicoletti P, Cartsos VM, Palaska PK, Shen Y, Floratos A, Zavras AI. Genomewide pharmacogenetics of bisphosphonate-induced osteonecrosis of the jaw: the role of RBMS3. Oncologist 2012;17(2):279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang G, Singh S, Chen Y, et al. Pharmacogenomics of osteonecrosis of the jaw. Bone 2019;124:75–82. [DOI] [PubMed] [Google Scholar]
- 28.Yang G, Hamadeh IS, Katz J, et al. SIRT1/HERC4 locus associated with bisphosphonate-induced osteonecrosis of the jaw: an exome-wide association analysis. J Bone Miner Res 2018;33(1):91–8. [DOI] [PubMed] [Google Scholar]
- 29.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000;403(6771):795–800. [DOI] [PubMed] [Google Scholar]
- 30.Imai SI, Guarente L. It takes two to tango: NAD. NPJ Aging Mech Dis 2016;2:16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X SIRT1 and energy metabolism. Acta Biochim Biophys Sin (Shanghai) 2013;45(1):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kauppinen A, Suuronen T, Ojala J, Kaarniranta K, Salminen A. Antagonistic crosstalk between NF-κB and SIRT1 in the regulation of inflammation and metabolic disorders. Cell Signal 2013;25(10): 1939–48. [DOI] [PubMed] [Google Scholar]
- 33.Yang H, Bi Y, Xue L, et al. Multifaceted modulation of SIRT1 in cancer and inflammation. Crit Rev Oncog 2015;20(1–2):49–64. [DOI] [PubMed] [Google Scholar]
- 34.Kim HN, Han L, Iyer S, et al. Sirtuin1 suppresses osteoclastogenesis by deacetylating FoxOs. Mol Endocrinol 2015;29(10):1498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duncan MT, DeLuca TA, Kuo HY, Yi M, Mrksich M, Miller WM. SIRT1 is a critical regulator of K562 cell growth, survival, and differentiation. Exp Cell Res 2016;344(1):40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zainabadi K Drugs targeting SIRT1, a new generation of therapeutics for osteoporosis and other bone related disorders? Pharmacol Res 2019;143:97–105. [DOI] [PubMed] [Google Scholar]
- 37.Yan S, Miao L, Lu Y, Wang L. Sirtuin 1 inhibits TNF-α-mediated osteoclastogenesis of bone marrow-derived macrophages through both ROS generation and TRPV1 activation. Mol Cell Biochem 2019;455 (1–2):135–45. [DOI] [PubMed] [Google Scholar]
- 38.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015;31(21): 3555–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium G The genotype-tissue expression (GTEx) project. Nat Genet 2013;45(6):580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 2015;31(8):1334–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 2014;24(4):697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dayem Ullah AZ, Oscanoa J, Wang J, Nagano A, Lemoine NR, Chelala C. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res 2018;46(W1):W109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40(Database issue):D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis CA, Hitz BC, Sloan CA, et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 2018;46(D1): D794–D01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22 (9):1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zerbino DR, Achuthan P, Akanni W, et al. Ensembl 2018. Nucleic Acids Res 2018;46(D1):D754–D61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One 2018; 13(10):e0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective micro-RNA target sites in mammalian mRNAs. Elife 2015;4(8):e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res 2014;42(Database issue):D86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from micro-RNA sequences to function. Nucleic Acids Res 2019;47(D1): D155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43(Database issue):D146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol 2019;20(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009;37(9):e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, Marín A. Characterization and prediction of alternative splice sites. Gene 2006;366(2):219–27. [DOI] [PubMed] [Google Scholar]
- 56.Langaee T, Ronaghi M. Genetic variation analyses by pyrosequencing. Mutat Res 2005;573(1–2):96–102. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Song F, Zhang B, et al. The 3D genome browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol 2018;19 (1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou X, Li D, Zhang B, et al. Epigenomic annotation of genetic variants using the roadmap epigenome browser. Nat Biotechnol 2015; 33(4):345–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin SL, Miller JD, Ying SY. Intronic microRNA (miRNA). J Biomed Biotechnol 2006;2006(4):26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Fan M, Zhang X, et al. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA 2014; 20(12):1878–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y, Song T, Peng J, et al. SIRT1 suppresses adipogenesis by activating Wnt/β-catenin signaling in vivo and in vitro. Oncotarget 2016; 7(47):77707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng G, Zheng K, Song D, et al. SIRT1 was involved in TNF-α-promoted osteogenic differentiation of human DPSCs through Wnt/β-catenin signal. In Vitro Cell Dev Biol Anim 2016;52(10): 1001–11. [DOI] [PubMed] [Google Scholar]
- 63.Zainabadi K, Liu CJ, Guarente L. SIRT1 is a positive regulator of the master osteoblast transcription factor, RUNX2. PLoS One 2017;12 (5):e0178520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemieux ME, Yang X, Jardine K, et al. The Sirt1 deacetylase modulates the insulin-like growth factor signaling pathway in mammals. Mech Ageing Dev 2005;126(10):1097–105. [DOI] [PubMed] [Google Scholar]
- 65.McBurney MW, Yang X, Jardine K, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol 2003;23(1):38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng HL, Mostoslavsky R, Saito S, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A 2003;100(19):10794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edwards JR, Perrien DS, Fleming N, et al. Silent information regulator (Sir)T1 inhibits NF-κB signaling to maintain normal skeletal remodeling. J Bone Miner Res 2013;28(4):960–9. [DOI] [PubMed] [Google Scholar]
- 68.Simic P, Zainabadi K, Bell E, et al. SIRT1 regulates differentiation of mesenchymal stem cells by deacetylating β-catenin. EMBO Mol Med 2013;5(3):430–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang H, Zhang W, Pan H, et al. SIRT1 activators suppress inflammatory responses through promotion of p65 deacetylation and inhibition of NF-κB activity. PLoS One 2012;7(9):e46364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 2009;27(4):693–733. [DOI] [PubMed] [Google Scholar]
- 71.Wahedi HM, Chae JK, Subedi L, et al. NED416, a novel synthetic Sirt1 activator, promotes cutaneous wound healing via the MAPK/Rho pathway. Int J Mol Med 2020;46(1):149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakamura M, Umetsu R, Abe J, et al. Analysis of the time-to-onset of osteonecrosis of jaw with bisphosphonate treatment using the data from a spontaneous reporting system of adverse drug events. J Pharm Health Care Sci 2015;1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


