Abstract
We report here on the identification and characterization of novel 2-enoyl thioester reductases of fatty acid metabolism, Etr1p from Candida tropicalis and its homolog Ybr026p (Mrf1′p) from Saccharomyces cerevisiae. Overexpression of these proteins in S. cerevisiae led to the development of significantly enlarged mitochondria, whereas deletion of the S. cerevisiae YBR026c gene resulted in rudimentary mitochondria with decreased contents of cytochromes and a respiration-deficient phenotype. Immunolocalization and in vivo targeting experiments showed these proteins to be predominantly mitochondrial. Mitochondrial targeting was essential for complementation of the mutant phenotype, since targeting of the reductases to other subcellular locations failed to reestablish respiratory growth. The mutant phenotype was also complemented by a mitochondrially targeted FabI protein from Escherichia coli. FabI represents a nonhomologous 2-enoyl-acyl carrier protein reductase that participates in the last step of the type II fatty acid synthesis. This indicated that 2-enoyl thioester reductase activity was critical for the mitochondrial function. We conclude that Etr1p and Ybr026p are novel 2-enoyl thioester reductases required for respiration and the maintenance of the mitochondrial compartment, putatively acting in mitochondrial synthesis of fatty acids.
Maintenance of functional mitochondria involves a large number of nuclear and a few mitochondrially encoded factors (13). These protein components have attracted considerable attention not only for the goal of understanding their role in the complex network of mitochondrial processes but also due to the existence of severe human respiratory disorders caused by the loss of some of these proteins (43). It is known that both mitochondrial protein and lipid deficiencies can compromise respiratory growth in Saccharomyces cerevisiae (9). Recently, a group of S. cerevisiae genes encoding mitochondrial proteins with similarities to the components of the prokaryotic type II fatty acid synthase (FAS) (38) have been identified that appear to be involved in the respiratory function of mitochondria (16, 41, 42). In contrast to the FAS type I system, which occurs as single-gene-encoded multifunctional enzyme complexes in eukaryotes (44), FAS type II systems consist of discrete monofunctional proteins (38). FAS II has been reported to exist in plant plastids and mitochondria in addition to prokaryotes (7, 14). As in plants, the putative FAS II in fungal mitochondria is assumed to generate precursors for the synthesis of lipoic acid (23, 50), in addition to which there exists fragmentary evidence for the involvement of the FAS II-generated fatty acids as constituents of mitochondrial membranes (31, 42, 52).
Here we report the identification and characterization of novel mitochondrial 2-enoyl thioester reductases, Etr1p from Candida tropicalis and its homolog Ybr026p from S. cerevisiae. The latter, also known as Mrf1′p, has been suggested to be necessary for the assembly of mitochondrial respiratory complexes by nuclear DNA binding function (51). Our data, however, demonstrate that the mitochondrial localization of both Etr1p and Ybr026p was required for restoring respiratory growth of S. cerevisiae cells devoid of Ybr026p, whereas localization of these proteins to other subcellular compartments failed to complement the respiration-deficient phenotype of the ybr026cΔ strain. Characterization of Etr1p and Ybr026p revealed that they catalyze the reaction trans-2-enoyl-(ACP/CoA) + NADPH + H+→acyl-(ACP/CoA) + NADP+, where ACP stands for acyl carrier protein and CoA stands for coenzyme A, which can contribute to the last step of FAS II. These results are discussed in terms of the biological function of novel proteins involved in mitochondrial synthesis of fatty acids.
MATERIALS AND METHODS
Primers, strains, media, and growth conditions.
The S. cerevisiae strains and primers used in the study are described in Table 1. S. cerevisiae strain BJ1991 (or alternatively BY4741) (4, 22) was maintained on rich YPD (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, and 2% [wt/vol] d-glucose) supplemented with 200 μg of Geneticin/ml for the selection of the ybr026cΔ strain(s). Transformed strains harboring URA3-marked plasmids were maintained on synthetic medium lacking uracil (SC-U) containing 2% (wt/vol) d-glucose (2). To examine complementation on nonfermentable carbon sources, transformants were grown on SC-U containing 3% (wt/vol) glycerol as the sole carbon source. For protein purification or immunoelectron microscopy (immuno-EM), the ybr026cΔ cells overexpressing Ybr026p or Etr1p were transferred from overnight cultures in SC-U to oleic acid medium (15) to an optical density at 600 nm of 0.2 and grown for 16 h at 30°C. C. tropicalis pK233 cells (American Type Culture Collection, Rockville, Md.) for purification or immuno-EM of the wild-type Etr1p were grown at 30°C for 20 h in oleic acid medium (12).
TABLE 1.
S. cerevisiae strains and primers used
| Strain or primer | Genotype or sequence (5′ → 3′) |
|---|---|
| Strains | |
| BJ1991a | MATα leu2 trp1 ura3-52 pep4-3 prb1-1122 |
| BY4741b | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| BJ1991 ybr026cΔ | ybr026c::kanMX4 MATα leu2 trp1 ura3-52 pep4-3 prb1-1122 |
| BY4741 ybr026cΔ | ybr026c::kanMX4 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| ybr026cΔ + Cta1p | ybr026c::kanMX4 pYE352::CTA1 |
| ybr026cΔ + full-length Etr1p | ybr026c::kanMX4 pYE352::ETR1 |
| ybr026cΔ + mature Etr1p | ybr026c::kanMX4 pYE352::ETR1mature |
| ybr026cΔ + full-length Ybr026p | ybr026c::kanMX4 pYE352::YBR026c |
| ybr026cΔ + mature Ybr026p | ybr026c::kanMX4 pYE352::YBR026mature |
| ybr026cΔ + nuclear Ybr026p | ybr026c::kanMX4 pYE352::nucYBR026 |
| ybr026cΔ + mtFabI | ybr026c::kanMX4 pYE352::mtFabI |
| BJ1991 + Ybr026p-GFP | MATα leu2 trp1 ura3-52 pep4-3 prb1-1122 pYE352::MLS1-YBR026c-GFP |
| Primers | |
| oligo(dT17) | GGTGACCCGGGAGATCTGAATTCTTTTTTTTTTTTTTTTT |
| YBR026CS1 | TAAGAAGGTTCATAATCTGCCAATTACATTTGATATAACAAGATGCGTACGCTGCAGGTCGAC |
| YBR026CS2 | TTTGGTGGTTGTCTAACTCTGAGCAATGCTAAGGCCCTTAATCGATGAATTCGAGCTCG |
| CTRED5′A | GAGCTCTAGAAGATGTATTCTGTATTGAAACAGTCAATTCG |
| CTRED5′B | GAGCTCTAGAAGATGATCACCGCCCAGGC |
| CTRED3′A | CGAGAAGCTTTTTAGTACGTAATCAACTGTTTACCATCC |
| CTRED3′B | CGAGCTCGAGTTTAGTACGTAATCAACTGTTC |
| SCMRF5′A | GAGCTCTAGAAGATGCTTCCCACATTCAAAC |
| SCMRF5′B | GAGCTCTAGAAGATGTCGTCCTCAGCTCATATT |
| SCMRF5′C | GAGCTCTAGAAGATGGCGCCGAAAAAGAAACGGAAGGTGATGTCGTCCTCAGCTCATCAGATT |
| SCMRF3′ | CGAGCTCGAGTTTACCATTCTAAACAACCATTT |
| FABI5′ | GTCCATGGGTTTTCTTTCCGGTAA |
| FABI3′ | GACTCGAGTTATTTCAGTTCGAGTTCGT |
| COQ3MT5′A | CTGCTAGCATGGGATTCATAATGTTGTT |
| COQ3MT3′A | GACCATGGCTTCAGAAGCATCTGTGCTC |
| SCMRFGFP5′ | GAGCGGTACCATGCTTCCCACATTCA |
| SCMRFGFP3′ | GAGCGGTACCCATCCATTCTAAAACAACCATTTTTTTC |
| ARS1 | BIOTIN-CAGATTTTATGTTTAGATCTTTTATGCTTG |
Protein purification and overexpression.
C. tropicalis cells (30 g [wet weight]) were washed with 50 mM KPi, pH 6.8, and resuspended in 100 ml of a solution containing 10 mM KPi (pH 7.0), 2 mM EDTA, 2 mM EGTA, 0.2 M KCl, 1.2 M (NH4)2SO4, 0.1 mM phenylmethylsulfonyl fluoride, 0.5 mM benzamidine hydrochloride (BA), and 0.5 mM dithiothreitol. Resuspended cells were disrupted with glass beads (0.5 mm in diameter) in a Bead-Beater homogenizer (Biospec Products, Bartlesville, Okla.). Cell debris was removed by centrifugation at 120,000 × g, and the supernatant was spun again at 6,000,000 × g. The supernatant was filtered through glass wool to remove lipids, and the sample was applied to a 3- by 8-cm phenyl-Sepharose 6 Fast Flow hydrophobic interaction column (Amersham Pharmacia Biotech, Uppsala, Sweden) equilibrated with 10 mM KPi (pH 7.0)–2 mM EDTA–2 mM EGTA–1.2 M (NH4)2SO4. After the column was washed with 300 ml of the equilibration buffer, the bound proteins were eluted with a decreasing linear (NH4)2SO4 gradient (total, 500 ml) from 1.2 to 0 M at a flow rate of 1 ml/min. The fractions of the first reductase activity peak were pooled and then equilibrated with and applied in 20 mM Tris-HCl (pH 8.0) to a Mono Q HR (5/5) anion-exchange column (Amersham Pharmacia Biotech). After the unbound proteins were washed off, the column was subjected to an increasing linear NaCl gradient from 0 to 0.5 M (total, 20 ml) at a flow rate of 1 ml/min. The fractions containing Etr1p were pooled, concentrated, and applied to a Superdex-200 HR (10/30) size-exclusion column (Amersham Pharmacia Biotech) equilibrated with 50 mM NaPi (pH 7.0)–0.15 M NaCl.
For purification of the Etr1p or Ybr026p produced in S. cerevisiae, oleic acid-induced cells were broken in a French press (Spectronic Instruments, Rochester, N.Y.). Purification was performed as described above for the wild-type Etr1p, except that the Mono Q HR (5/5) column was replaced by a 1- by 10-cm 2′5′ADP-Sepharose column (Amersham Pharmacia Biotech) equilibrated with 50 mM KPi (pH 7.0) containing 0.5 mM BA and dithiothreitol. The bound proteins were eluted with an increasing linear NaCl gradient from 0 to 2.0 M (total, 70 ml) at a flow rate of 1 ml/min. Also, unlike in the wild-type Etr1p purification, the Resource S ion-exchange column (Amersham Pharmacia Biotech) equilibrated with 25 mM morpholineethanesulfonic acid (MES)–NaCl (pH 5.5) was additionally used before the Superdex 200 column for purification of the overexpressed Etr1p.
Protein digestion and peptide sequencing.
A sample of the Etr1p purified from C. tropicalis (30 μg) was subjected to trypsin digestion according to the manufacturer's instructions (Promega Corp., Madison, Wis.), and the resulting peptides were separated on a μRPC C2/C18 SC 2.1/10 reverse-phase column (Amersham Pharmacia Biotech) in an acetonitrile gradient. Fractions were analyzed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Kompact Maldi III; Kratos Analytical, Manchester, United Kingdom), and samples with more than one peptide mass signal were further separated on a Sephasil C8, 5-μm SC 2.1/10 reverse-phase column (Amersham Pharmacia Biotech). A 72-μg sample of Etr1p was used for endoproteinase Glu-C (4 μg) (Promega) digestion, and the resulting peptides were separated as described above. Peptide sequencing was carried out by automated Edman degradation in an Applied Biosystems model 477A protein sequencer.
Cloning of genomic ETR1 fragments.
C. tropicalis genomic DNA (500 ng) was used as a template in a PCR with degenerate primers based on amino acid sequences obtained from tryptic digestions. The resulting 174-bp DNA fragment was ligated to pUC18 and sequenced. Genomic fragments flanking this DNA fragment were obtained with ligation-mediated PCR (26, 34) using PvuII-digested genomic DNA as a template. Primers nested within the previous PCR product were used for further amplifications. A 0.6-kb DNA fragment obtained was similarly subcloned into pUC18 and sequenced. To isolate additional genomic fragments a C. tropicalis genomic library was screened with a 32P-labeled probe consisting of the 0.6-kb amplification product using the cosmid library screening method (40). The library was prepared from HindIII-digested DNA ligated to pUC18 as described previously (2). A clone containing a 5-kb insert was detected and partially sequenced.
Isolation of ETR1 cDNA.
Reverse transcription (RT) was carried out using 200 ng of mRNA isolated from oleic acid-grown C. tropicalis cells as a template. An oligonucleotide termed oligo(dT17) was used as an antisense primer in an RT reaction with avian myeloblastosis virus reverse transcriptase (Promega). The primers used in the subsequent PCR corresponded to nucleotides 753 to 776 in the final sequence and to the sequence of oligo(dT17). The resulting PCR product of 0.54 kb was cloned into pUC18 and sequenced. Single-stranded cDNA was also prepared using 20 μg of total RNA isolated from oleic acid-grown C. tropicalis cells using oligo(dT17) as primer. The prepared cDNA served as a template for a PCR in which another primer was based on the sequence of the 5-kb genomic DNA library insert (see above) and designed to encode the first methionine from the N-terminal peptide sequence. The other primer was based on the 0.54-kb PCR product and was designed to include the first putative polyadenylation signal downstream of the stop codon. A DNA fragment of 1.2 kb was thus generated, cloned into pUC18, and sequenced.
Constructs for expression of Etr1p or Ybr026p variants.
The PCR amplifications were performed with Pfu polymerase (Stratagene, La Jolla, Calif.). The primers corresponding to the 5′ and 3′ ends of the different ETR1 and YBR026c variants contained an extra XbaI site at the 5′ end and a methionine start codon and an extra XhoI site (except CTRED3′A, which had a HindIII site) at the 5′ end and the stop codon, respectively. ETR1, corresponding to the full-length Etr1p, was amplified from the plasmid containing the 1.2-kb ETR1 fragment (see above) with the primer pair CTRED5′A/CTRED3′A. To generate ETR1mature, corresponding to the mature Etr1p lacking the N-terminal 22 amino acids (aa), PCR was performed with the primer pair CTRED5′B/CTRED3′B. YBR026c was amplified by PCR from BJ1991 genomic DNA with the primer pair SCMRF5′A-SCMRF3′, corresponding to the full-length Ybr026p, or SCMRF5′B-SCMRF3′, corresponding to the mature Ybr026p lacking the N-terminal 8 aa. For targeting Ybr026p to the nucleus, PCR was carried out with the primer pair SCMRF5′C-SCMRF3′, with the forward primer SCMRF5′C being synthesized on the basis of the simian virus 40 large T antigen nuclear targeting signal M-P-K-K-K-R-K-V (25). The resulting DNA fragments were purified by agarose gel electrophoresis and ligated in EcoRV-digested pBluescript SK(+) (pSK) (Stratagene). Following XbaI-XhoI (or -HindIII) double digestions of the pSK subclones, generated inserts encoding ETR1, ETR1mature, YBR026c, YBR026mature, and nucYBR026 were gel purified and ligated behind the S. cerevisiae catalase A (CTA1) promoter carried by pYE352 (12). The difference in codon usage between S. cerevisiae and C. tropicalis was compensated for by changing the universal Leu codon CUG (encoding Ser114 and -336 in Etr1p) to the Ser codon AGC by site-directed mutagenesis (Stratagene).
Construct for expression of FabI.
Escherichia coli fabI was PCR amplified from chromosomal DNA using the primer pair FABI5′-FABI3′, which introduced NcoI and XhoI sites at the 5′ and 3′ ends of the open reading frame, respectively. The mitochondrial targeting (MT) sequence of the S. cerevisiae COQ3 gene (21) was PCR amplified from S. cerevisiae genomic DNA with the primer pair COQ3MT5′-COQ3MT3′, which introduced NheI and NcoI sites at the 5′ and 3′ ends of the MT sequence, respectively. The resulting PCR products of 0.79 and 0.13 kb were digested with NcoI-XhoI and NheI-NcoI, gel purified, and ligated behind the CTA1 promoter in pYE352, resulting in pYE352::mtFabI. The COQ3 MT sequence used for the construct contained the region coding for aa 1 to 42 of COQ3 plus one alanine residue.
Ybr026p-GFP construct.
YBR026c was PCR amplified from pYE352::YBR026c (encoding full-length Ybr026p) with the primer pair SCMRFGFP5′-SCMRFGFP3′ and subcloned in pSK (see above). The plasmid generated was then digested with KpnI, resulting in a KpnI-KpnI fragment which was gel purified. An in-frame hybrid between the S. cerevisiae malate synthase (MLS1) promoter (17) and the cDNA encoding the Aequorea victoria green fluorescent protein (GFP) was generated by ligation of the YBR026c fragment in a KpnI site in pYE352::MLS1-GFP-SKL (5). The codons for the carboxy-terminal peroxisomal signal SKL (33) in GFP-SKL were removed from the GFP fusion by site-directed mutagenesis (Stratagene), changing the Ser codon TCC to a TAA stop codon, resulting in pYE352::MLS1-YBR026c-GFP.
Characterization of the reaction product.
trans-2,trans-4-Hexadienoyl-CoA or trans-2-hexenoyl-CoA (10 mM) was incubated with 0.2 ng of recombinant Etr1p or Ybr026p and 10 mM NADPH in 120 μl of 50 mM KPi (pH 7.4) at 37°C. After 0, 45, 90, and 180 min, 5 μl of 0.5 M KOH was added to aliquots of 20 μl from the reaction mixture, and free fatty acids were liberated by heating at 80°C for 3 h. After the addition of 80 μl of acetone, the mixture was evaporated under a nitrogen stream at 50°C. Twenty microliters of 1 M acetyl chloride in ethanol was added. Following a short sonication, the fatty acids were esterified overnight at 30°C. Ten microliters of saturated CuCl2 was added to the mixture and extracted with 20 μl of hexane. A 1-μl sample of the organic layer was subjected to gas-liquid chromatography using a gas chromatographic system (model 5890; Hewlett-Packard, Bad Homburg, Germany) with flame ionization detector and an FS-FFAP-CB-0.25 (20 m) column (Cs-Chromatographic Service GmbH, Langerwehe, Germany). The column head pressure was 35 kPa and the temperature program was 10 min at 60°C, followed by temperature gradients of 60 to 90°C with an increment of 4°C/min and 90 to 200°C with an increment of 20°C/min, and finally 200°C for 5 min.
Immuno-EM and fluorescence microscopy.
For immuno-EM, cells were fixed with 4% paraformaldehyde and 0.15% glutaraldehyde in Pi buffer (pH 6.8) and then subjected to further fixation by freeze substitution in absolute methanol at −80°C (3). Thin sections from cells embedded in LR White resin (Electron Microscopy Sciences, Fort Washington, Pa.) were incubated with anti-Etr1p antibodies diluted 1:150, followed by a protein A-gold complex. Counterstained sections were examined using a Philips EM 410 microscope (Philips Electron Optics, Eindhoven, The Netherlands). For fluorescence analysis, cells were spread on a polylysine-coated slide and left to dry for 15 min at 37°C. The slides were fixed for 1 min in cold acetone at −20°C and then stained for 30 s with 4′-6-diamidino-2-phenylindole (DAPI) at a final concentration of 25 ng/ml in phosphate-buffered saline. After drying at 37°C, the cells were examined for GFP and DAPI stains using an Olympus BX 60 microscope. By this rapid procedure using a low concentration of DAPI the mitochondrial staining is emphasized much more than that of the nucleus.
ARS1 protein binding analysis.
Binding interactions were analyzed by surface plasmon resonance (Biacore, Uppsala, Sweden). Biotinylated ARS1 oligonucleotide (200 μg/ml in 0.3 M NaCl, pH 7.0) was coupled to streptavidin on a sensorchip at a flow rate of 5 μl/min for 7 min, giving 2,200 response units (RU) in the sensorgram. Pure Ybr026p or Etr1p was injected over the immobilized ARS1 at a flow rate of 30 μl/min in 0.01 M HEPES (pH 7.4) containing 0.15 M NaCl, 3 mM EDTA, and 0.005% (vol/vol) surfactant P-20.
Other methods.
Enoyl reductase activity was measured using 60 μM trans-2-enoyl-CoA or trans-2,trans-4-hexadienoyl-CoA (synthesized via a mixed anhydride method [37]) at 22°C as described previously (11). The deletion strain ybr026cΔ::kanMX4 was generated using the short flanking homology procedure (49) with the primer pair YBR026CS1-YBR026CS2. Transformations were performed as previously described (8). Cytochrome spectra for the C. tropicalis and S. cerevisiae strains were obtained with a Shimadzu UV3000 spectrophotometer, following a published protocol (30). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described (29). Anti-Etr1p polyclonal antibodies were raised in rabbits. Immunoglobulin G was purified from antiserum by anion-exchange chromatography (2).
Nucleotide sequence accession number.
The sequence data for ETR1 have been submitted to the DDBJ, EMBL, and GenBank databases under accession number U94997.
RESULTS
A novel 2-enoyl thioester reductase, Etr1p from C. tropicalis
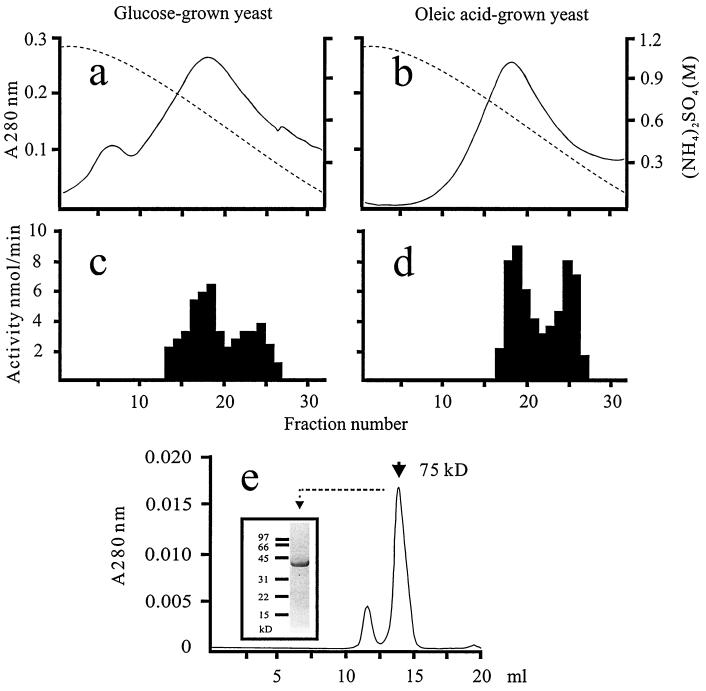
To obtain novel fatty enoyl thioester reductases from C. tropicalis, soluble protein extracts of cells grown on glucose or oleic acid were applied to a phenyl-Sepharose hydrophobic interaction column. When the bound proteins were eluted from the column (Fig. 1a and b), two reductase activity peaks (measured using trans-2,trans-4-hexadienoyl-CoA as the substrate) were observed (Fig. 1c and d). An immunoblot using antibodies against Sps19p, a peroxisomal 2,4-dienoyl-CoA reductase from S. cerevisiae (15), revealed a 34-kDa protein band in fractions 21 to 26 corresponding to an oleic acid-inducible reductase activity. We assumed the reductase activity in these fractions to represent the previously characterized 2,4-dienoyl-CoA reductase of the β-oxidation (11). This Sps19p-like protein was scarcely detectable in fractions 13 to 20, the first reductase activity peak. To study this first reductase, an enoyl thioester reductase, termed here Etr1p, was purified to apparent homogeneity by chromatography using phenyl-Sepharose, Mono Q, and Superdex 200 columns (Table 2). Superdex 200 size-exclusion chromatography indicated that Etr1p had a native molecular mass of 75 kDa, and SDS-PAGE resulted in a single protein band of 40 kDa (Fig. 1e). The latter value agrees with the 39,699 Da determined with matrix-assisted laser desorption ionization–time of flight mass spectroscopy, indicating that Etr1p was probably a homodimer.
FIG. 1.
Purification of Etr1p. Soluble extracts (5 mg) from glucose (a)- and oleic acid (b)-grown C. tropicalis were applied to a phenyl-Sepharose column. Bound proteins (solid line) were eluted with a decreasing linear (NH4)2SO4 gradient (dashed line). Respective elution profiles (c and d) of reductase activities determined with trans-2,trans-4-dienoyl-CoA as substrate are shown. (e) Etr1p (150 μg) from a Mono Q column was run on a Superdex 200 size-exclusion column and eluted at a volume of 13 to 16 ml. A sample from the pooled Etr1p was subjected to SDS-PAGE and Coomassie stained, as shown in the inset.
TABLE 2.
Purification of Etr1p from C. tropicalis
| Purification step | Total protein (mg) | Sp act (μmol/ min/mg) | Total units (μmol/min) | Yield (%) |
|---|---|---|---|---|
| Soluble extract | 140 | 0.21 | 29 | 100 |
| Phenyl-Sepharose column | 18 | 0.41 | 7.4 | 26 |
| Mono Q column | 1.1 | 2.7 | 2.9 | 10 |
| Superdex 200 column | 0.23 | 5.0 | 1.2 | 4a |
The low yield can be at least partially explained by the existence of several proteins (see Fig. 1) with reductase activity (measured with trans-2,trans-4-dienoyl-CoA as substrate) in the soluble extract.
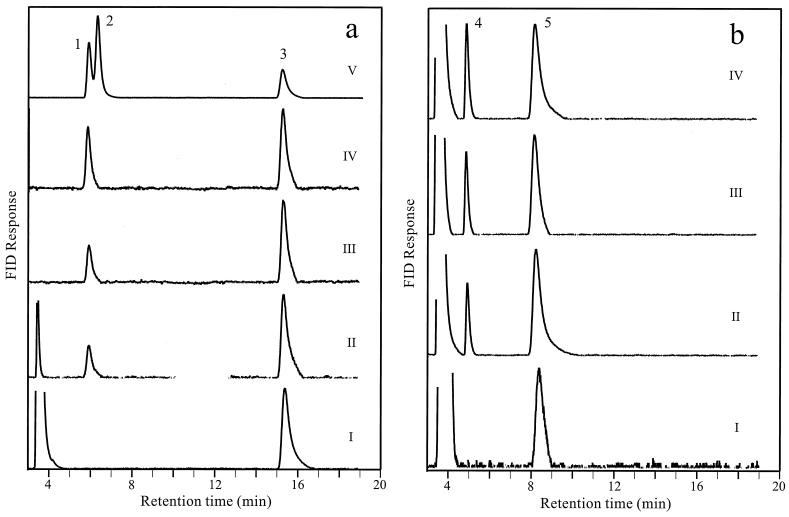
ETR1 was cloned (see below) and overexpressed in S. cerevisiae. Purified protein was incubated with trans-2,trans-4-hexadienoyl-CoA in the presence of NADPH. Gas chromatography performed on the hydrolyzed and esterified products of this incubation revealed the generation of 4-hexenoic acid ethyl ester in a time-dependent manner (Fig. 2a). No peaks corresponding to 2- or ethyl 3-hexenoic acid ethyl esters, which represent the respective end products of prokaryotic (32) or eukaryotic (28) 2,4-dienoyl-CoA reductases, were observed. Analysis of the reaction following replacement of trans-2,trans-4-hexadienoyl-CoA with trans-2-hexenoyl-CoA revealed the accumulation of hexanoic acid ethyl ester (Fig. 2b). This indicated that Etr1p was a 2-enoyl-CoA but not a 2,4-dienoyl-CoA reductase. The specific activity of Etr1p with trans-2-hexenoyl-CoA and trans-2,trans-4-hexadienoyl-CoA substrates was 20 and 10 μmol/min per mg of protein, respectively. In these enzyme assays NADH could not replace NADPH.
FIG. 2.
Gas chromatographic analysis of the reductase reaction products. Etr1p was incubated with trans-2,trans-4-hexadienoyl-CoA (a) or trans-2-hexenoyl-CoA (b) for 0 (I), 45 (II), 90 (III), and 180 (IV) min in the presence of NADPH, and the hydrolyzed and esterified reaction products were analyzed. After 180 min of incubation with trans-2,trans-4-hexadienoyl-CoA, trans-3-hexenoyl-CoA was added into the reaction mixture, shown in panel a (V). The peaks indicating different ethyl esters derived from CoA esters are trans-4-hexenoate (1), trans-3-hexenoate (2), trans-2,trans-4-hexadienoate (3), hexanoate (4), and trans-2-hexenoate (5). FID, flame ionization detector.
Molecular cloning of C. tropicalis ETR1
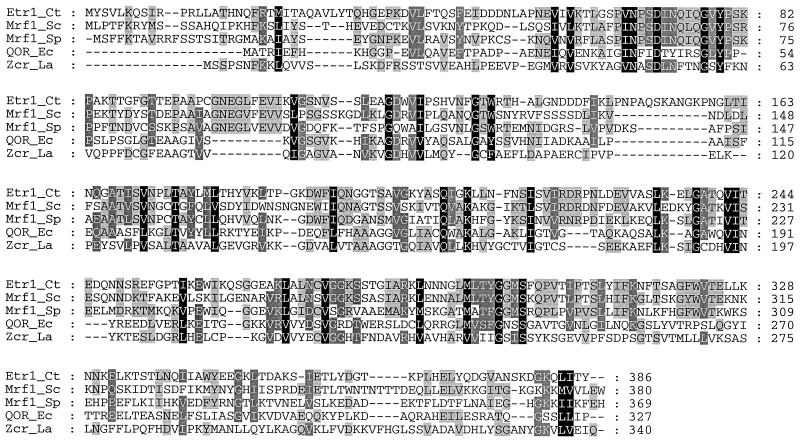
To obtain the ETR1 gene, purified Etr1p from C. tropicalis was subjected to trypsin and endoproteinase Glu-C digestions. Microsequencing of peptide fragments yielded a total number of 186 aa in partially overlapping sequences. As the sequence M-I-T-A-Q-A-V was obtained from undigested protein as well as from both of the protease digestion preparations, we assumed it to represent the N terminus of the mature protein. Based on the amino acid sequences obtained, degenerate primers were synthesized and PCR was performed using C. tropicalis genomic DNA as a template. Ligation-mediated PCR amplification and genomic library screening resulted in a DNA fragment containing an open reading frame of 1,158 nucleotides. The deduced protein product of ETR1 was a polypeptide of 386 aa that fully matched the sequences of the proteolytic peptides after adjustments were made to accommodate the nonuniversal codon usage of C. tropicalis (45). The predicted molecular mass for Etr1p was 42,167 Da, which included a presequence of 22 aa absent from the mature Etr1p (predicted molecular mass, 39,519 Da). RT-PCR with C. tropicalis mRNA and sequencing of the products verified that the ETR1 gene was intronless. A search of databases using BLAST (1) showed that the homolog of Etr1p with the highest amino acid identity (42%) was S. cerevisiae Ybr026p, also termed mitochondrial respiratory function protein (Mrf1p) (51), followed by the Ybr026p homolog in Schizosaccharomyces pombe (40% identity; accession number Q10488). Amino acid sequence alignment showed that Etr1p and Ybr026p belong to the medium-chain dehydrogenase/reductase (MDR) superfamily of proteins, which includes members with divergent functions, e.g., some quinone oxidoreductases and ζ-crystallins (Fig. 3) as well as glucose and alcohol dehydrogenases (24, 35).
FIG. 3.
Sequence alignment of Etr1p (Etr1_Ct) with mitochondrial respiratory function protein from S. cerevisiae (Mrf1_Sc; accession number D26606), a mitochondrial respiratory function protein homolog from S. pombe (Mrf1_Sp; accession number Q10488), quinone oxidoreductase from E. coli (QOR_Ec; accession number P28304), and ζ-crystallin from Leishmania amazonensis (Zcr_La; accession number L11705). The black and gray boxes indicate identity and similarity, respectively. The amino acid residues are numbered from the initial methionine (+1).
Comparison between Etr1p and Ybr026p.
Etr1p and Ybr026p were produced in a ybr026cΔ strain and purified as described above (from pYE352::ETR1 and pYE352::YBR026c; see Materials and Methods). Like Etr1p, Ybr026p was also found to carry out NADPH-dependent reduction of both trans-2-hexanoyl-CoA and trans-2,trans-4-hexadienoyl-CoA at the rates of 20 and 10 μmol/min per mg of protein, respectively. Further analysis using gas chromatography indicated that the respective products of the reactions were trans-4-hexenoyl-CoA and hexanoyl-CoA (data not shown). The similarity between the proteins raised the question of whether Etr1p might also bind an autonomously replicating sequence (ARS1), as has been shown for Ybr026p (51). Injection of purified Ybr026p over ARS1 coupled onto a plasmon resonance surface resulted in the observation of specific binding in a concentration-dependent manner. However, no response of specific binding was observed following injection of purified Etr1p.
C. tropicalis ETR1 restores respiration to the S. cerevisiae ybr026Δ strain.
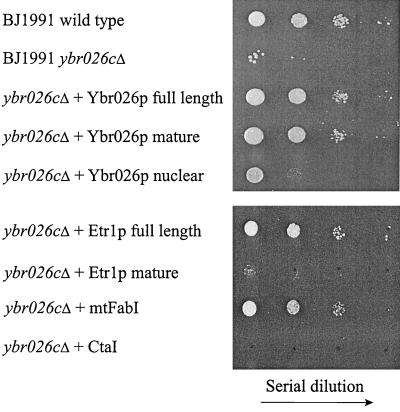
Ybr026p has been implicated as a factor needed to maintain respiratory competence in budding yeast. The ybr026cΔ strain is unable to grow on nonfermentable carbon sources and exhibits decreased contents of cytochromes (51). The similarity between the primary sequences of Etr1p and Ybr026p combined with the discovery that both represented 2-enoyl thioester reductases prompted us to examine whether this similarity extended to a common physiological function. Therefore, the ybr026cΔ strain was examined for heterologous complementation. Mutant ybr026cΔ cells were transformed with the plasmids pYE352::ETR1, pYE352::YBR026c, and control pYE352::CTA1 expressing Etr1p, Ybr026p, and catalase A (Cta1p), respectively. The ybr026cΔ strain expressing Cta1p was not able to grow on glycerol medium. The growth could be restored by expression of either Etr1p or Ybr026p (Fig. 4). To examine whether this restoration of respiratory growth was coincidental with the formation of mitochondrial cytochrome complexes, the cytochrome spectra of the strains were examined. The results showed that in cells lacking Ybr026p, cytochrome complexes were missing. In the ybr026cΔ strains expressing Etr1p or Ybr026p, formation of cytochromes was restored (data not shown). This indicated that both proteins share a common physiological function that involves mitochondrial cytochrome complexes and respiratory competence.
FIG. 4.
Comparison of the growth of S. cerevisiae strains on a nonfermentable carbon source. The S. cerevisiae strains BJ1991 wild type, BJ1991 ybr026cΔ, or BJ1991 ybr026cΔ transformed with plasmid DNA overexpressing either full-length Ybr026p, mature Ybr026p without 5′ leader sequence, Ybr026p extended with a nuclear targeting signal, full-length Etr1p, mature Etr1p without 5′ leader sequence, FabI extended with a mitochondrial targeting signal, or Cta1p were cultured on 3% synthetic complete glycerol medium for 5 days at 30°C.
Etr1p and Ybr026p are mitochondrial proteins that cause organelle enlargement upon their overexpression.
Etr1p is preceded by an N-terminal sequence (Fig. 3) that is similar to MT sequences (18). To examine the subcellular localization of Etr1p, immuno-EM was performed. C. tropicalis cells were grown to late log phase on oleic acid medium, fixed, and immunolabeled. Application of anti-Etr1p antibodies to the thin sections resulted in the decoration of mitochondrial structures (Fig. 5a). No specific labeling was observed in other cellular compartments. Further studies on the subcellular localization of Etr1p were carried out with spheroplast lysate prepared from C. tropicalis cells grown on oleic acid. The lysate was subjected to fractionation on a Nycodenz density gradient. In this gradient, comigration of a protein band of 40 kDa with antibodies to Etr1p was observed in cytochrome c oxidase (a marker for mitochondria) activity-containing fractions, thereby providing supporting evidence for a mitochondrial localization of Etr1p.
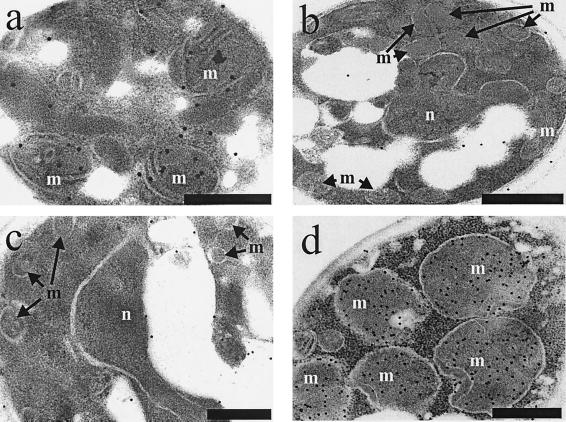
FIG. 5.
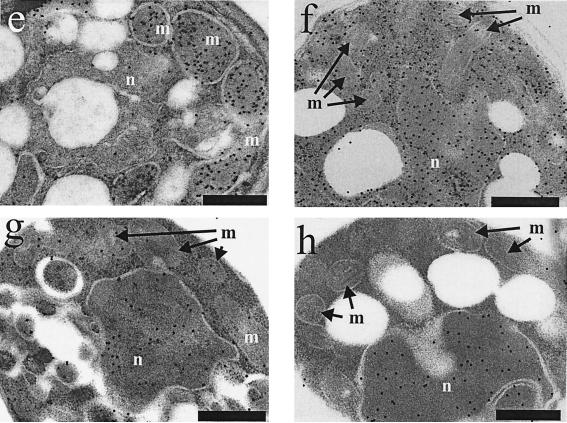
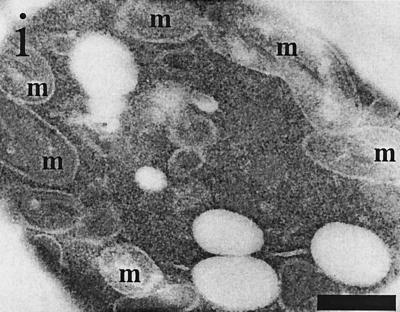
Immuno-EM of Etr1p, Ybr026p, and FabI. C. tropicalis wild type (a) and the S. cerevisiae strains BJ1991 (b), BJ1991 ybr026cΔ (c), and BJ1991 ybr026cΔ overexpressing either full-length Etr1p (d), full-length Ybr026p (e), mature Etr1p without 5′ leader sequence (f), matureYbr026p without 5′ leader sequence (g), Ybr026p extended with a nuclear targeting signal (h), or FabI extended with a mitochondrial targeting signal (i) were grown on oleic acid. Cells were treated with anti-Etr1p antibodies and gold particles conjugated to protein A. Mitochondria (m) and the nucleus (n) are indicated. The scale bars represent 250 nm.
The subcellular localization of Etr1p was also studied in a heterologous context. YBR026c-deficient S. cerevisiae cells overexpressing Etr1p were grown on the nonfermentable carbon source oleic acid. This ensured high-level transcription of ETR1 from the oleic acid-inducible CTA1 promoter, yielding a reductase activity of 0.26 μmol/min per mg of protein (measured with trans-2-hexenoyl-CoA as substrate) in the soluble extracts. Application of anti-Etr1p antibodies to thin sections decorated large globular structures in the immuno-EM (Fig. 5d). Since these contained cristae, we recognized these organelles as unusually enlarged mitochondria compared to those seen in the wild type (Fig. 5b). To test if this phenotype was due to the overexpression of Etr1p in the medium rich with oleic acid, we also induced overexpression of the protein on medium with galactose as the sole carbon source. Enlargement of mitochondria was also obvious in these cells (data not shown), indicating that the phenotype was not an effect of the carbon source.
The data described for Etr1p indicating the mitochondrial localization of the protein in both the native context and the heterologous context prompted a reinvestigation of Ybr026p, which was previously claimed to be a nuclear protein (51). Soluble protein extracts obtained from the ybr026cΔ strain overexpressing Ybr026p revealed a specific activity of 0.040 μmol/min per mg of protein. Immunoblotting experiments using anti-Etr1p antibodies revealed no visible band in the extracts of the ybr026cΔ strain, whereas a protein band of 40 kDa was identified in the strain overexpressing Ybr026p. This indicated that the anti-Etr1p antibodies cross-reacted with Ybr026p. Immuno-EM on YBR026c-deleted cells overexpressing Ybr026p showed mitochondrial decoration of the gold particles concurrent with the enlargement of mitochondria (Fig. 5e) which corresponded to the phenotype of the cells overexpressing Etr1p. Examination of the nucleus revealed only a few gold particles.
The deletion, complementation, and overexpression experiments described above were carried out in the BJ1991 ybr026cΔ strain, which carries the pep4-3 mutation. To rule out the possibility that the synergistic effect of the pep4-3 allele in conjuction with overexpression of MRF1 or ETR1 was causing the changes in mitochondrial morphology, we repeated the MRF1 overexpression experiment in the S. cerevisiae BY4741 ybr026cΔ strain. In these experiments we observed similar changes in mitochondrial morphology as in the BJ1991 strain background, hence ruling out a role of the pep4-3 allele in the MRF1 overexpression phenotype.
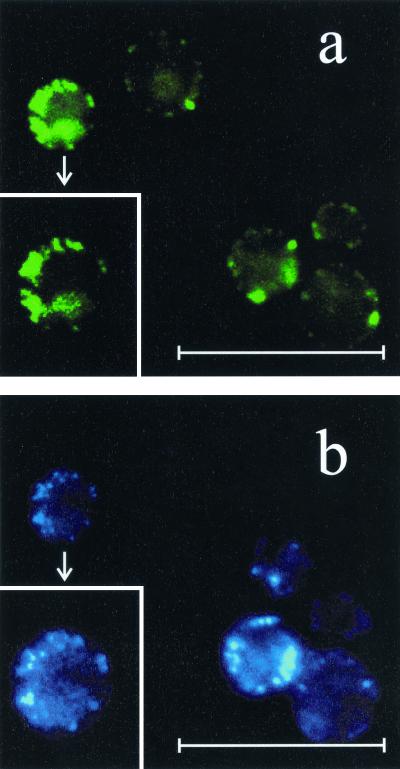
To provide further evidence for the mitochondrial localization for Ybr026p, we generated a Ybr026p-GFP fusion protein. Although the fusion protein was not as enzymatically active as wild-type Mrf1p and only very poorly if at all complemented the ybr026cΔ respiration-deficient phenotype, fluorescence microscopy of the BJ1991 cells expressing this reporter demonstrated a punctate pattern of green fluorescence coinciding with DAPI staining of mitochondrial DNA (Fig. 6). Hence, fusion with Ybr026p was sufficient to direct GFP to mitochondria in S. cerevisiae, thereby reiterating the discrepancy regarding its site of action in S. cerevisiae cells.
FIG. 6.
Fluorescent signals from Ybr026p-GFP and DAPI-stained DNA in S. cerevisiae BJ1991. (a) Cells overexpressing Ybr026p-GFP showed punctate green fluorescence. (b) DAPI staining (blue) of the same cells shown in panel a. Punctate staining of mitochondrial DNA was clearly visible and coincided with GFP fluorescence. Additionally, inserts of a single-cell magnification of the corresponding fields of Mrf1p-GFP and DAPI staining are shown. The scale bars represent 16.5 μm.
Complementation of the respiratory deficiency of the ybr026cΔ strain with Etr1p or Ybr026p requires their localization to mitochondria.
To address the possibility that despite their apparent mitochondrial localization both Ybr026p and Etr1p might act in the nucleus to restore respiration in the ybr026cΔ strain, different protein targeting variants were constructed. The ybr026cΔ strain was transformed with pYE352::ETR1mature expressing Etr1p that lacked the N-terminal 22 aa. Soluble protein extracts derived from these cells resulted in a reductase activity of 0.26 μmol/min per mg of protein, thereby confirming the expression and proper folding of this truncated protein. Immuno-EM showed that unlike wild-type Etr1p, this altered Etr1p was primarily extramitochondrial and was mostly detectable in the cytosol and nucleus (Fig. 5f). The mitochondrial compartment was not enlarged in these cells. Since these cells failed to grow on glycerol medium, on which the corresponding mutant cells expressing the wild-type Etr1p could grow abundantly (Fig. 4), this experiment indicated that the first 22 aa of C. tropicalis Etr1p were necessary for directing the protein to the mitochondria and that mitochondrial localization of Etr1p was essential for its function.
Our ultrastructural data pointed to a primarily mitochondrial localization for the S. cerevisiae protein (Fig. 5e), although it has been previously suggested that Ybr026p relies on the N-terminal 8 aa to be targeted to the nucleus. These 8 aa are missing from the mature protein (51), which we verified by N-terminal sequencing of the Ybr026p overexpressed in S. cerevisiae. To examine the role of these amino acids for intracellular targeting of Ybr026p, a strategy similar to the one described for Etr1p was applied to the S. cerevisiae protein. The ybr026cΔ strain was transformed with pYE352::YBR026mature, and enzyme assays demonstrated that overexpression of the truncated Ybr026p resulted in a reductase activity of 0.042 μmol/min per mg of protein in the soluble protein extract. Immuno-EM showed mitochondrial decoration of the gold particles, but unlike the complete protein (Fig. 5e), truncated Ybr026p was also present elsewhere in the cell (Fig. 5g). Since mutant cells expressing the truncation grew on glycerol (Fig. 4), the missing amino acids were dispensable for the protein function, probably because the truncation did not completely abolish mitochondrial localization.
To test the previous hypothesis that Ybr026p acts in the nucleus to maintain mitochondrial respiration (51), the protein was fused to the nuclear targeting signal of simian virus 40 protein large antigen (25). Enzyme assays corroborated that this fusion did not alter the activity of Ybr026p, since these resulted in a reductase activity of 0.039 μmol/min per mg of protein in the soluble protein extract. Immuno-EM confirmed that this protein variant was primarily localized in the nucleus and revealed that the mitochondria were not enlarged (Fig. 5h). However, despite the abundance of Ybr026p in the nucleus, the growth assay (Fig. 4) demonstrated that the respiration-deficient phenotype of the ybr026cΔ strain was only very poorly rescued. As the protein showed enzymatic activity comparable to the wild type, we take this result as an indication that the nuclear localization was not sufficient to complement the deletion. We cannot exclude the possibility that a small fraction of the overexpressed protein was still localized to the mitochondria despite the nuclear targeting signal. The combined data pointed to a model whereby both Ybr026p and Etr1p acted in the mitochondria to maintain respiration. However, it was by no means clear whether the mitochondrial function of the two homologs was enzymatic, i.e., as 2-enoyl thioester reductases, or whether they had structural roles in facilitating the formation of cytochrome complexes.
Complementation of the ybr026cΔ strain by E. coli fabI, encoding a FAS II enoyl-ACP reductase.
To discriminate between the two possibilities outlined above (enzymatic versus structural function), E. coli FabI, an NADPH-dependent 2-enoyl-ACP reductase, was expressed in the ybr026cΔ strain to test for complementation. FabI represents a nonhomologous protein participating with the prokaryotic FAS II (38). Mutant cells were transformed with pYE352::mtFabI encoding a FabI variant that was N-terminally extended with an MT signal. This transformation rescued the respiratory deficiency of the deletion strain, allowing for growth on glycerol (Fig. 4) as well as restoration of cytochrome formation (data not shown). Moreover, similar to overexpression of Etr1p or Ybr026p targeted to mitochondria, mtFabI overexpression in the ybr026cΔ strain also resulted in significant enlargement of these organelles (Fig. 5i).
DISCUSSION
C. tropicalis Etr1p and its S. cerevisiae homolog Ybr026p described here are novel NADPH-dependent 2-enoyl thioester reductases belonging to the functionally diverse MDR superfamily of proteins (24). Among the best-known 2-enoyl thioester reductases are enzymes in the multifunctional FAS I system from animals and fungi (27, 44) as well as monofunctional reductases, such as FabI from E. coli or InhA from Mycobacterium tuberculosis, participating with the prokaryotic FAS type II system (38). Additionally, enoyl reductases have been characterized from plant organelles (7) and from mammalian peroxisomes (10). Only recently other prokaryotic enoyl reductases (FabK and FabL) (19, 20) unrelated to FabI (or InhA) have also been discovered. However, any of these previously characterized enoyl reductases do not share significant amino acid sequence similarity with Ybr026p or Etr1p.
Thus far, a total of four nuclear genes from S. cerevisiae (ACP1, CEM1, MCT1, and OAR1) have been postulated to encode components involved in a mitochondrial FAS II (6, 16, 41). Inactivation of any one of these genes leads to a respiration-deficient phenotype, manifested by decreased contents of cytochromes and impaired growth on nonfermentable carbon sources, which is similar to the ybr026cΔ phenotype. Together with the observation that Ybr026p and Etr1p are 2-enoyl thioester reductases, this suggests that Ybr026p and Etr1p are fungal mitochondrial FAS II 2-enoyl-ACP reductases, components that have remained unidentified so far. The finding that YBR026c could be replaced by a well-characterized fabI (encoding 2-enoyl-ACP reductase) from E. coli lends substance to this hypothesis. This substitution by nonhomologous FabI indicates that the complementation was due to the reductase activity rather than to a structural function. Nevertheless, it has been shown that ACP associated with the mitochondrial FAS II also serves a structural role as an essential part of complex I in mitochondria of higher eukaryotes (39, 42, 46).
Our findings on the subcellular localization of Ybr026p and Etr1p support the hypothesis that these proteins can participate in mitochondrial fatty acid synthesis. The expression of differentially engineered YBR026c or ETR1 in the ybr026cΔ strain demonstrated that the mitochondrial, but not nuclear, localization of the gene products was necessary for the complementation. In agreement with previous observations (51), Ybr026p was able to bind to ARS1. However, Etr1p was not observed to bind to ARS1. Moreover, it is improbable that the heterologous complementation by mtFabI was due to an interaction with ARS1.
The molecular link between the mitochondrial FAS II and the assembly of the respiratory chain has remained unclear, but several mechanistic explanations have been put forward. Although elevations in the level of mitochondrial lipids are known to affect the integrity of the mitochondrial genome, it is not clear whether this can explain the impaired respiration in the FAS II-deficient strains (9). The complementation experiments with either YBR026c or ETR1 showed that either of these genes was sufficient for restoring respiratory growth to the ybr026cΔ strain (Fig. 4). This concurs with the previous findings, which indicated that intact mitochondrial DNA and apocytochromes are present in the ybr026cΔ cells (51), implying that neither the mitochondrial transcription nor translation is affected in the mutant cells.
Synthesis of an important mitochondrial enzyme cofactor, lipoic acid, is assumed to be dependent on octanoyl-ACP precursors generated in the mitochondria (23). Our preliminary data show that the ybr026cΔ strain lipoic acid level amounts to only 10% of the level of the wild type, which is similar to the previous results for the acp1Δ strain in S. cerevisiae (6). However, the data available on a collection of petite S. cerevisiae strains with low lipoic acid levels imply that the cellular lipoic acid per se cannot explain the petite phenotype of ybr026cΔ, as cytochromes are detectable in the S. cerevisiae lip1-5Δ strains (47). The incorporation of radioactivity from [2-14C]malonate into various acyl groups bound to ACP in Neurospora crassa (31, 52) as well as in plant mitochondria (14) indicates that mitochondrial fatty acid synthesis may also participate in the generation of lipids other than lipoic acid. Notably, an inner membrane-embedded mitochondrially encoded ND5 subunit of the NADH:ubiquinone oxidoreductase (complex I) and a core catalytic subunit 1 of cytochrome c oxidase (complex V) in N. crassa have been found to carry covalently linked myristoyl-(C14) groups. This kind of fatty acylation might not only provide the means for proper assembly but may also be required for the catalytic mechanism of the respiratory chain enzyme complexes (36, 48). There is also the possibility that a range of mitochondrial acyl-ACPs could contribute to the synthesis and repair of phospholipid membranes, as acyl-ACPs of the prokaryotic FAS II do (38, 42).
One striking morphological effect of the YBR026c (or ETR1) overexpression in S. cerevisiae is the significant enlargement of mitochondria that occupy over 30% of the cytoplasmic volume (Fig. 5d and e), while inactivation of the YBR026c results in the appearance of rudimentary organelles (Fig. 5c). Densitometry of Coomassie-stained SDS-PAGE gels of ybr026cΔ transformed with either pYE352::YBR026c or pYE352::ETR1 indicated that the overexpressed Ybr026p or Etr1p counted for approximately 3% of the soluble protein extracts. It appears more likely that the biological function of these proteins rather than an increase in the total protein concentration is responsible for the effects on the maintenance of mitochondria. Interestingly, E. coli FabI enoyl-ACP reductase of FAS II has been suggested to be the rate-limiting enzyme of the pathway (38). Analogously, this might also be the case in S. cerevisiae cells overexpressing Ybr026p, Etr1p, or FabI in mitochondria. It remains to be revealed which lipid species generated could explain the observed changes in the mitochondrial morphology and size.
ACKNOWLEDGMENTS
We thank Marika Kamps and Tanja Kokko for their technical assistance.
This work was supported by grants from the Academy of Finland and the Sigrid Jusélius Foundation.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 3.Baba M, Osumi M. Transmission and scanning electron microscopic examination of intracellular organelles in freeze-substituted Kloeckera and Saccharomyces cerevisiae. J Electron Microsc Tech. 1987;5:249–261. [Google Scholar]
- 4.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Brocard C, Lametschwandtner G, Koudelka R, Hartig A. Pex14p is a member of the protein linkage map of Pex5p. EMBO J. 1997;16:5491–5500. doi: 10.1093/emboj/16.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brody S, Oh C, Hoja U, Schweizer E. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Lett. 1997;408:217–220. doi: 10.1016/s0014-5793(97)00428-6. [DOI] [PubMed] [Google Scholar]
- 7.Caughey I, Kekwick R G. The characteristics of some components of the fatty acid synthetase system in the plastids from the mesocarp of avocado (Persea americana) fruit. Eur J Biochem. 1982;123:553–561. doi: 10.1111/j.1432-1033.1982.tb06568.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen D C, Yang B C, Kuo T T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 9.Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das A K, Uhler M D, Hajra A K. Molecular cloning and expression of mammalian peroxisomal trans-2-enoyl-coenzyme A reductase cDNAs. J Biol Chem. 2000;275:24333–24340. doi: 10.1074/jbc.M001168200. [DOI] [PubMed] [Google Scholar]
- 11.Dommes V, Kunau W H. 2,4-Dienoyl coenzyme A reductases from bovine liver and Escherichia coli. Comparison of properties. J Biol Chem. 1984;259:1781–1788. [PubMed] [Google Scholar]
- 12.Filppula S A, Sormunen R T, Hartig A, Kunau W H, Hiltunen J K. Changing stereochemistry for a metabolic pathway in vivo. Experiments with the peroxisomal beta-oxidation in yeast. J Biol Chem. 1995;270:27453–27457. doi: 10.1074/jbc.270.46.27453. [DOI] [PubMed] [Google Scholar]
- 13.Grivell L A, Artal-Sanz M, Hakkaart G, de Jong L, Nijtmans L G, van Oosterum K, Siep M, van der Spek H. Mitochondrial assembly in yeast. FEBS Lett. 1999;452:57–60. doi: 10.1016/s0014-5793(99)00532-3. [DOI] [PubMed] [Google Scholar]
- 14.Gueguen V, Macherel D, Jaquinod M, Douce R, Bourguignon J. Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J Biol Chem. 2000;275:5016–5025. doi: 10.1074/jbc.275.7.5016. [DOI] [PubMed] [Google Scholar]
- 15.Gurvitz A, Rottensteiner H, Kilpelainen S H, Hartig A, Hiltunen J K, Binder M, Dawes I W, Hamilton B. The Saccharomyces cerevisiae peroxisomal 2,4-dienoyl-CoA reductase is encoded by the oleate-inducible gene SPS19. J Biol Chem. 1997;272:22140–22147. doi: 10.1074/jbc.272.35.22140. [DOI] [PubMed] [Google Scholar]
- 16.Harington A, Schwarz E, Slonimski P P, Herbert C J. Subcellular relocalization of a long-chain fatty acid CoA ligase by a suppressor mutation alleviates a respiration deficiency in Saccharomyces cerevisiae. EMBO J. 1994;13:5531–5538. doi: 10.1002/j.1460-2075.1994.tb06890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartig A, Simon M M, Schuster T, Daugherty J R, Yoo H S, Cooper T G. Differentially regulated malate synthase genes participate in carbon and nitrogen metabolism of S. cerevisiae. Nucleic Acids Res. 1992;20:5677–5686. doi: 10.1093/nar/20.21.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haucke V, Schatz G. Reconstitution of the protein insertion machinery of the mitochondrial inner membrane. EMBO J. 1997;16:4560–4567. doi: 10.1093/emboj/16.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heath R J, Rock C O. A triclosan-resistant bacterial enzyme. Nature. 2000;406:145–146. doi: 10.1038/35018162. [DOI] [PubMed] [Google Scholar]
- 20.Heath R J, Su N, Murphy C K, Rock C O. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J Biol Chem. 2000;275:40128–40133. doi: 10.1074/jbc.M005611200. [DOI] [PubMed] [Google Scholar]
- 21.Hsu A Y, Poon W W, Shepherd J A, Myles D C, Clarke C F. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry. 1996;35:9797–9806. doi: 10.1021/bi9602932. [DOI] [PubMed] [Google Scholar]
- 22.Jones E W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977;85:23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan S W, Cronan J E., Jr A new metabolic link. The acyl carrier protein of lipid synthesis donates lipoic acid to the pyruvate dehydrogenase complex in Escherichia coli and mitochondria. J Biol Chem. 1997;272:17903–17906. doi: 10.1074/jbc.272.29.17903. [DOI] [PubMed] [Google Scholar]
- 24.Jornvall H, Hoog J O, Persson B. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 1999;445:261–264. doi: 10.1016/s0014-5793(99)00130-1. [DOI] [PubMed] [Google Scholar]
- 25.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 26.Kere J, Nagaraja R, Mumm S, Ciccodicola A, D'Urso M, Schlessinger D. Mapping human chromosomes by walking with sequence-tagged sites from end fragments of yeast artificial chromosome inserts. Genomics. 1992;14:241–248. doi: 10.1016/s0888-7543(05)80212-5. [DOI] [PubMed] [Google Scholar]
- 27.Kolodziej S J, Penczek P A, Schroeter J P, Stoops J K. Structure-function relationships of the Saccharomyces cerevisiae fatty acid synthase. Three-dimensional structure. J Biol Chem. 1996;271:28422–28429. doi: 10.1074/jbc.271.45.28422. [DOI] [PubMed] [Google Scholar]
- 28.Kunau W H, Dommes P. Degradation of unsaturated fatty acids. Identification of intermediates in the degradation of cis-4-decenoyl-CoA by extracts of beef-liver mitochondria. Eur J Biochem. 1978;91:533–544. doi: 10.1111/j.1432-1033.1978.tb12707.x. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lindenmayer A, Estabrook R W. Low-temperature spectral studies on the biosynthesis of cytochromes in baker's yeast. Arch Biochem Biophys. 1958;78:66–82. doi: 10.1016/0003-9861(58)90315-1. [DOI] [PubMed] [Google Scholar]
- 31.Mikolajczyk S, Brody S. De novo fatty acid synthesis mediated by acyl-carrier protein in Neurospora crassa mitochondria. Eur J Biochem. 1990;187:431–437. doi: 10.1111/j.1432-1033.1990.tb15322.x. [DOI] [PubMed] [Google Scholar]
- 32.Mizugaki M, Kimura C, Nishimaki T, Kawaguchi A, Okuda S, Yamanaka H. Studies on the metabolism of unsaturated fatty acids. XII. Reaction catalyzed by 2,4-dienoyl-CoA reductase of Escherichia coli. J Biochem (Tokyo) 1983;94:409–413. doi: 10.1093/oxfordjournals.jbchem.a134370. [DOI] [PubMed] [Google Scholar]
- 33.Monosov E Z, Wenzel T J, Luers G H, Heyman J A, Subramani S. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J Histochem Cytochem. 1996;44:581–589. doi: 10.1177/44.6.8666743. [DOI] [PubMed] [Google Scholar]
- 34.Mueller P R, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 35.Persson B, Zigler J S, Jr, Jornvall H. A super-family of medium-chain dehydrogenases/reductases (MDR). Sub-lines including zeta-crystallin, alcohol and polyol dehydrogenases, quinone oxidoreductase enoyl reductases, VAT-1 and other proteins. Eur J Biochem. 1994;226:15–22. doi: 10.1111/j.1432-1033.1994.tb20021.x. [DOI] [PubMed] [Google Scholar]
- 36.Plesofsky N, Gardner N, Videira A, Brambl R. NADH dehydrogenase in Neurospora crassa contains myristic acid covalently linked to the ND5 subunit peptide. Biochim Biophys Acta. 2000;1495:223–230. doi: 10.1016/s0167-4889(99)00170-6. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen J T, Borchers T, Knudsen J. Comparison of the binding affinities of acyl-CoA-binding protein and fatty-acid-binding protein for long-chain acyl-CoA esters. Biochem J. 1990;265:849–855. doi: 10.1042/bj2650849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rock C O, Cronan J E. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 39.Runswick M J, Fearnley I M, Skehel J M, Walker J E. Presence of an acyl carrier protein in NADH:ubiquinone oxidoreductase from bovine heart mitochondria. FEBS Lett. 1991;286:121–124. doi: 10.1016/0014-5793(91)80955-3. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schneider R, Brors B, Burger F, Camrath S, Weiss H. Two genes of the putative mitochondrial fatty acid synthase in the genome of Saccharomyces cerevisiae. Curr Genet. 1997;32:384–388. doi: 10.1007/s002940050292. [DOI] [PubMed] [Google Scholar]
- 42.Schneider R, Massow M, Lisowsky T, Weiss H. Different respiratory-defective phenotypes of Neurospora crassa and Saccharomyces cerevisiae after inactivation of the gene encoding the mitochondrial acyl carrier protein. Curr Genet. 1995;29:10–17. doi: 10.1007/BF00313188. [DOI] [PubMed] [Google Scholar]
- 43.Schon E A. Mitochondrial genetics and disease. Trends Biochem Sci. 2000;25:555–560. doi: 10.1016/s0968-0004(00)01688-1. [DOI] [PubMed] [Google Scholar]
- 44.Smith S. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. FASEB J. 1994;8:1248–1259. [PubMed] [Google Scholar]
- 45.Sugita T, Nakase T. Nonuniversal usage of the leucine CUG codon and the molecular phylogeny of the genus Candida. Syst Appl Microbiol. 1999;22:79–86. doi: 10.1016/S0723-2020(99)80030-7. [DOI] [PubMed] [Google Scholar]
- 46.Triepels R, Smeitink J, Loeffen J, Smeets R, Buskens C, Trijbels F, van den Heuvel L. The human nuclear-encoded acyl carrier subunit (NDUFAB1) of the mitochondrial complex I in human pathology. J Inherit Metab Dis. 1999;22:163–173. doi: 10.1023/a:1005402020569. [DOI] [PubMed] [Google Scholar]
- 47.Tzagoloff A, Dieckmann C L. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vassilev A O, Plesofsky-Vig N, Brambl R. Cytochrome c oxidase in Neurospora crassa contains myristic acid covalently linked to subunit 1. Proc Natl Acad Sci USA. 1995;92:8680–8684. doi: 10.1073/pnas.92.19.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 50.Wada H, Shintani D, Ohlrogge J. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc Natl Acad Sci USA. 1997;94:1591–1596. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamazoe M, Shirahige K, Rashid M B, Kaneko Y, Nakayama T, Ogasawara N, Yoshikawa H. A protein which binds preferentially to single-stranded core sequence of autonomously replicating sequence is essential for respiratory function in mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1994;269:15244–15252. [PubMed] [Google Scholar]
- 52.Zensen R, Husmann H, Schneider R, Peine T, Weiss H. De novo synthesis and desaturation of fatty acids at the mitochondrial acyl-carrier protein, a subunit of NADH:ubiquinone oxidoreductase in Neurospora crassa. FEBS Lett. 1992;310:179–181. doi: 10.1016/0014-5793(92)81324-f. [DOI] [PubMed] [Google Scholar]