Abstract
Different G1 cyclins confer functional specificity to the cyclin-dependent kinase (Cdk) Cdc28p in budding yeast. The Cln3p G1 cyclin is localized primarily to the nucleus, while Cln2p is localized primarily to the cytoplasm. Both binding to Cdc28p and Cdc28p-dependent phosphorylation in the C-terminal region of Cln2p are independently required for efficient nuclear depletion of Cln2p, suggesting that this process may be physiologically regulated. The accumulation of hypophosphorylated Cln2 in the nucleus is an energy-dependent process, but may not involve the RAN GTPase. Phosphorylation of Cln2p is inefficient in small newborn cells obtained by elutriation, and this lowered phosphorylation correlates with reduced Cln2p nuclear depletion in newborn cells. Thus, Cln2p may have a brief period of nuclear residence early in the cell cycle. In contrast, the nuclear localization pattern of Cln3p is not influenced by Cdk activity. Cln3p localization requires a bipartite nuclear localization signal (NLS) located at the C terminus of the protein. This sequence is required for nuclear localization of Cln3p and is sufficient to confer nuclear localization to green fluorescent protein in a RAN-dependent manner. Mislocalized Cln3p, lacking the NLS, is much less active in genetic assays specific for Cln3p, but more active in assays normally specific for Cln2p, consistent with the idea that Cln3p localization explains a significant part of Clnp functional specificity.
Cyclin-dependent kinases (Cdks) are serine/threonine protein kinases that are required for key events in the cell division cycle. Cdk activity depends on conformational changes induced by physical association with cyclin proteins and subsequent cyclin-dependent targeting of Cdk activity to potential substrates (3, 49, 50, 56). A single Cdk can carry out multiple functions, dependent on the identity of its cognate cyclin partner. In budding yeast, the Cdk Cdc28p is required for all key cell cycle events. When bound by any one of three G1 (Cln) cyclins, Cdc28p supports cell cycle entry (12). When bound by one of six B-type (Clb) cyclins, Cdc28p supports later cell cycle events, such as DNA replication and mitosis (54). For both G1 Cln cyclins and B-type Clb cyclins, specific cyclins provide functional specificity to the Cdk complex independent of expression patterns and abundance of associated kinase activity (14, 30, 38).
Deletion of all three G1-type cyclin CLN genes results in a cell cycle arrest before cell cycle initiation, as unbudded cells with 1C DNA content. CLN1, CLN2, and CLN3 are functionally redundant, since all double cln deletions result in viability. However, there are significant differences in the functional specificity of CLN2 (representative of the highly homologous CLN1 CLN2 gene pair) and CLN3. Functional analysis of CLN2 and CLN3 indicates that CLN3 is primarily required for the expression of genes during late G1, including CLN1 and CLN2. This Cln3p activity requires the SCB transcription complex, consisting of Swi4p and Swi6p (34). Once expressed, Cln2p is believed to trigger events required for cell cycle progression, such as bud emergence and activation of Clb-Cdc28p complexes (7, 12, 15, 40). Bck2p also triggers SCB-dependent transcription in G1, most likely working in parallel with Cln3p (17, 21). We have characterized the subcellular localization patterns of the G1 cyclins Cln2p and Cln3p and find strikingly distinct localization patterns. These patterns are consistent with the idea that Cln2p and Cln3p confer functional specificity to the Cdk Cdc28p and support cell cycle entry by distinct mechanisms. Cln3p, thought to be the physiologically relevant cyclin for activation of transcription (19, 38, 70), is localized primarily to the nucleus (45). Cln2, thought to trigger other events, such as bud emergence (7, 12, 15, 40), is localized primarily to the cytoplasm (45).
Differences in functional specificity between Cln2p and Cln3p can be assayed in specific genetic backgrounds. For example, CLN2 is able to support viability in the absence of Swi4p, while CLN3 is not (38, 55, 57). This suggests that the ability of CLN3 to rescue a cln1 cln2 strain requires Cln3p- and Swi4p-dependent transcription. The PCL1 and PCL2 gene pair (encoding cyclin homologues that can bind to and activate the Pho85p Cdk) is also essential for Cln3p-dependent viability in the absence of cln1 and cln2 (38, 43, 57). PCL1 and PCL2 are transcribed under the control of Swi4p (43, 57), suggesting that at least some of the Swi4p requirement in the cln1 cln2 background may be due to the Pcl1p, Pcl2p requirement. This requirement may involve promoting functions important for morphogenesis (37).
Studies of higher eukaryotic cyclins have established the importance of spatial control for regulation of cyclin and/or Cdk activity. The most extensively studied are the mitotic cyclin B1 and the G1 cyclin D1. When unphosphorylated, cyclin B1 localizes to the cytoplasm through physical association with the exportin CRM1. The physical association between cyclin B1 and CRM1 is abolished when cyclin B1 is phosphorylated, resulting in localization to the nucleus, possibly through interactions with the karyopherin α-β complex (46, 58, 82). Cyclin B1 may also enter the nucleus through physical interactions with cyclin F or independently through physical interactions with importin β (71). Phosphorylation of cyclin B1 has also been implicated in triggering nuclear import, independent of inhibition of binding to CRM1 (25, 83). Physical association of cyclin B1 with Cdk does not significantly contribute to nuclear import (46). In vitro studies show that both the cyclin B1-Cdk1 complex, mitogen-activated protein kinase, and casein kinase II are able to phosphorylate cyclin B1 in the region that associates with CRM1 (28, 42). The D-type cyclins function at early stages in the cell division cycle, responding to growth factor stimulation and facilitating the activation of cyclin-Cdk2 complexes required for entry into the phase of the cell cycle at which DNA replication occurs. The shift in localization of cyclin D1 from the nucleus to the cytoplasm at the onset of DNA replication is linked to the proteosomal degradation of cyclin D1 and requires phosphorylation by glycogen synthase kinase-3β on Thr-286. This shift in localization reflects phosphorylation-dependent export of cyclin D1 (18). Two different human B-type cyclins, B1 and B2, exhibit different subcellular localization (to microtubules and to the Golgi apparatus) (29). Interestingly, chimeras constructed that switch the localization signal of cyclin B1 and cyclin B2 also influence their ability to function, demonstrating that differences in subcellular localization influence the functional specificity of these cyclins (20).
In this paper, we analyze mechanisms that regulate localization of Cln2p and Cln3p. We address the role of Cdc28p in regulated localization of G1 cyclins in budding yeast and characterize the distinct mechanisms used to import different G1 cyclins into the nucleus.
MATERIALS AND METHODS
The relevant genotypes of the yeast strains used are shown in Table 1. All strains (except for the grr1 strain, MMY42 strain, ACY212, MMY103, MMY106, and controls) are congenic with BF264–15D (59). YPD (dextrose), YPG (galactose), and synthetic complete (SC) media for yeast growth were prepared as described previously (63). Yeast cells were transformed with plasmid DNA as described previously (24).
TABLE 1.
Strains used in this study
| Strain | Relevant genotypea |
|---|---|
| 1255-5C | MATaCLN1 CLN2 CLN3 |
| 1607-2D | MATacln1Δcln2Δcln3Δleu2::LEU2::GAL1::CLN3 |
| 1446-10B | cln1Δcln2Δcln3Δbck2::ARG4 leu2::LEU2::GAL1::CLN3 arg4 |
| 1591-10D | MATacln1Δcln2Δcln3Δpcl1::HIS3 pcl2::URA3 leu2::LEU2::GAL1::CLN1 his3 HIS2 |
| 1591-11A | cln1Δcln2ΔCLN3 leu2::LEU2::GAL1::CLN1 pcl1::HIS3 pcl2::URA3 his3 HIS2 |
| MMY6-7D | MATacln1Δcln2ΔCLN3 swi4::LEU2 pGAL1::CLN1/URA3 |
| 1559-3D-7 | MATα CLN1 CLN2 CLN3 CDC28HA::URA3 |
| MMY1 | MATaCdLN1 CLN2myc::LEU2 CLN3 |
| MMY4 | MATaCLN1 CLN2-4t3smyc::LEU2 CLN3 |
| cdc28 | cdc28-4 CLN1 CLN2 CLN3 |
| MMY5 | cdc28-4 CLN1 CLN2myc::LEU2 CLN3 |
| cdc34 | cdc34 CLN1 CLN2 CLN3 |
| MMY7 | cdc34 CLN1 CLN2myc::LEU2 CLN3 |
| MMY8 | MATaCLN1 CLN2s396, 427Amyc::LEU2 CLN3 |
| 1559-3B-7 | CDC28HACLN1 CLN2 CLN3 |
| YMT1871 | MATa grr1Δ |
| MMY13 | MATa grr1ΔCLN2myc::LEU2 |
| MMY32 | MATaCLN3myc::TRP1 |
| W303 | MATatrp1 leu2 his3 ura3 ade3 |
| W303d | MATa/MATa trp1/trp1 leu2/leu2 his3/his3 ura3/ura3 ade3/ade3 |
| MMY42 | MATa/MATa trp1/trp1 leu2/leu2 his3/his3 ura3/ura3 ade3/ade3 CLN2myc::LEU2/CLN2PA::HIS5 (S. pombe)b |
| ACY212 | gspΔpRAN/URA3 |
| MMY95 | CLN2myc::LEU2gspΔpGSP/URA3 |
| MMY97 | CLN2-4t3smyc::LEU2 gspΔpGSP/URA3 |
| MMY99 | gspΔpAC78 (GSP1/TRP1) |
| MMY100 | gspΔpAC607 (gsp1-1/TRP1) |
| MMY101 | gspΔpAC608 (gsp1-2/TRP1) |
| MMY102 | CLN2myc::LEU2 gspΔ pAC78 (gsp1-1/TRP1) |
| MMY103 | CLN2myc::LEU 2gspΔ pAC607 (gsp1-1/TRP1) |
| MMY104 | CLN2myc::LEU 2gspΔ pAC608 (gsp1-2/TRP1) |
| MMY105 | CLN2-4t3smyc::LEU2 gspΔpAC78 (GSP1/TRP1) |
| MMY106 | CLN2-4t3smyc::LEU2 gspΔpAC607 (gsp1-1/TRP1) |
| MMY107 | CLN2-4t3smyc::LEU2 gspΔpAC608 (gsp1-2/TRP1) |
All strains are congenic with BF264-15D: MATa trp1 leu2 ura3 ade1 his2, except W303, YMT1871, MMY13, and MMY42 (which are isogenic with each other); and ACY212 and MMY95-MMY106 (which are congenic with each other).
The HIS5 gene is from S. pombe (see Materials and Methods).
Plasmids.
The 9× myc-epitope-tagged CLN3 CEN/ARS plasmid pMM99, Cln2mycp integrating construct pMM56, and CEN/ARS plasmid pMM82 (CLN3::CLN2myc) have been described previously (45). The 9× myc tag is engineered at the COOH terminus of the cyclin proteins in these constructs.
The genomic copy of CLN2 was tagged by a COOH-terminal, in-frame integration of a PCR-derived DNA fragment encoding the immunoglobulin G (IgG) binding domain of protein A. This DNA fragment was amplified from the plasmid pProtA/HIS5, which carries the Schizosaccharomyces pombe HIS5 gene as its selectable marker (75), as described previously (1). This results in the expression of protein A (PA)-tagged CLN2 from the CLN2 promoter. Correct integration of the tag was confirmed by PCR analysis of genomic structure and immunoblotting. Initial integration was carried out in the diploid strain W303d, resulting in strain MMY17. This strain was sporulated, and haploid cells expressing PA-tagged CLN2 were obtained by tetrad dissection. The diploid strain MMY42, expressing both the PA- and 9× myc epitope-tagged versions of CLN2, was made by mating haploid MMY17 to haploid W303 strain expressing CLN2myc (constructed with integration vector pMM56 as described previously [45]).
The Cln3-Δ22mycp expression plasmid (pMM98) was constructed by PCR amplification of CLN3 coding sequences lacking the last 66 bp. The 3′ primer for the cln3-Δ22 amplifications was 5′-CTAGCGGCCGCTCAGTTGGGTGGG GGCAGAGCATGGGTG-3′, and the 5′ primer was 5′-CTTACATTCCATTGCATCTCCC-3′. The plasmid pMM99 (45) was used as the template for PCRs. PCR products were subsequently cloned into TOPOII vector with topoisomerase-based ligations (Invitrogen). EcoRI-NotI cassettes from the TOPOII clones were used to replace the EcoRI-NotI CLN3 sequences of pMM45 to give rise to pMM96. The 9× myc epitope tag NotI cassette from pAG4 (A. Gartner) was cloned into the NotI site following the CLN3 sequences in pMM96 to give rise to pMM98. The pMM98 plasmid was sequenced to confirm the absence of the last 66 bp of CLN3 coding sequence followed in frame by the 9× myc epitope tag. The nuclear localization signal (NLS), mutant nonfunctional NLS (mnls), nuclear export signal (NES), and mutant nonfunctional NES (mnes) Cln3-Δ22p-expressing plasmids were made by replacing the ClaI-EcoRI fragment of pMM98 with the ClaI-EcoRI fragments of pMM83 (NLS-CLN3-1), pMM84 (mnls-CLN3-1), pMM85 (NES-CLN3-1), and pMM86 (mnes-CLN3-1) to give rise to pMM104, pMM105, pMM106, and pMM107, respectively (45). The cln3 mutant cln3-A13 contains two amino acid substitutions of K106A and R108A. These mutations abolish detectable interactions with Cdc28p assayed by coimmunoprecipitation assay (unpublished data).
The CLN2-4T3S mutant integration vector pMM68 was made by replacing the 3× hemagglutinin (HA) tag of pMT480 (M. Tyers) with the NotI 9× myc epitope tag. The resulting clone was sequenced to ensure identity and that the Cln coding sequences were followed in frame by the myc epitope tags. The CLN3myc integration vector pMM162 was made by inserting the SalI-SacII cassette (containing the CLN3 promoter-driven 9× myc-tagged CLN3) of pMM99 into the integration vector pRS404. Strain MMY32 was produced by integration of this vector into strain 1255-5C.
The CLN3 promoter-driven 9× myc-tagged Cln2-4t3sp expression plasmid (pMM108) was constructed by replacing the SpeI-NotI carboxy-terminal CLN2 fragment of pMM82 (45) with the SpeI-NotI carboxy-terminal CLN2 fragment of pMM68 and subsequent addition of the NotI fragment containing the 9× myc tag. The CLN2 promoter-driven 9× myc-tagged Cln2p(pMM159) and Cln2-4t3sp(pMM151) expression plasmids were constructed by replacing the SalI-ClaI CLN3 promoter fragments of pMM82 and pMM108, respectively, with the SalI-ClaI CLN2 promoter fragment from pKL007 (38). The CLN3 promoter-driven 9× myc-tagged Cln2-KAEAp expression plasmid (pMM66) was constructed by replacing the NotI cassette containing the 3× HA tag of pKL005 (38) with the NotI cassette containing 9× myc of pMM99. The CLN2 promoter-driven 9× myc-tagged Cln2-KAEAp (pMM167) and Cln2-KAEA-4t3sp (pMM166) expression plasmids were constructed by replacing the ClaI-BclI N-terminal CLN2 fragments of pMM159 and pMM151, respectively, with the ClaI-BclI N-terminal CLN2 fragment of pMM66. Plasmids were sequenced to ensure the presence of KAEA mutations.
Plasmids expressing green fluorescent protein (GFP)-CLN fusions were constructed by using pKW431 (69). These plasmids encode fusion proteins consisting of the simian virus 40 (SV40) NLS (the nonfunctional mutant protein kinase inhibitor [PKI] NES) and two GFP molecules. The HindIII-EcoRI SV40 NLS fragment of pKW431 was replaced with the CLN3 NLS by homologous recombination in vivo, resulting in pMM131. The HindIII-CLN3 NLS-EcoRI fragment was produced by PCR amplification with the forward primer 5′ATACAATCT GCACAATATTTCAAGCTATACCAAGCATACAATAAGCTTATGAAAA AGAGATCTACTTCCTCTGTGGAT3′ and the reverse primer 5′TTGTTGA TATCAAGACCTGCTAATTTCAAGGCTAATTCATTGAATTCGCGAGTT TTCTTGAGGTTGCTACT3′. Nonfunctional Cln3 mnls-GFP fusion proteins were made by identical means, except the primers contain base changes that result in alanine substitution of the basic clusters at the beginning and end of the Cln3p NLS. Mutation of both clusters resulted in plasmid pMM133. Mutation of the first basic cluster resulted in pMM135, and mutation of the second basic cluster resulted in pMM137. The nonfunctional PKI NES in these constructs was presumably inert. Substitution of the functional PKI NES in the Cln3 NLS construct resulted in loss of nuclear accumulation of Cln3 NLS-PKI NES-GFP fusion (data not shown).
Indirect immunolocalization and GFP localization.
Exponentially growing cultures expressing the myc-tagged Cln proteins were fixed in growth medium with formaldehyde (1:10 dilution) for 60 min at 30°C with rotation. When indicated, cells were fixed by “fast fixation.” In this case, cells were harvested by filtration and fixed in buffer (0.1 M KH2PO4, 0.5 M MgCl2, 40 mM KOH) with formaldehyde (1:10 dilution) for 5 min. The extended fixation time (up to 2 h) did not change localization patterns with the fast fixation method, and fast fixation gave essentially identical results to standard fixation.
Cells were prepared for indirect immunofluorescence as described previously (45). For detection of the myc epitope, primary monoclonal antibody 9E10 (Santa Cruz Biotechnology) was diluted 1:200 in blocking solution, cleared for 2 min by centrifugation in a microcentrifuge, and incubated on the cells for 2 h. Secondary antibody (Cy3-conjugated antimouse IgG; Jackson Immunochemicals) was diluted 1:200 in blocking solution, cleared, and incubated on the wells for 2 h. The negative control for integrated myc-tagged Cln proteins was the wild-type strain 1255-5C. The negative controls for plasmid-based expression of myc-tagged Cln proteins were the vector controls pRS414 and pRS416 (66).
Cells expressing the GFP fusion proteins were grown to log phase, sonicated, placed on polylysine-coated slides, and visualized microscopically. To confirm the localization of Cln3 NLS-GFP signal in the nucleus, cells were sonicated briefly, washed in 1× phosphate-buffered saline (PBS), fixed for 12 min in 70% ethanol at room temperature, washed in 1× PBS, stained for 12 min with 1:1,000 dilution of 1-mg/ml 4′,6′-diamidino-2-phenylindole (DAPI) at room temperature, washed in 1× PBS, and assayed for fluorescence. This procedure allows simultaneous detection of DAPI and GFP signal.
Standard indirect immunofluorescence and GFP experiments were done on an Axoplan universal microscope (Carl Zeiss, Inc.) with a Hamamatsu digital camera. Images were collected with Openlab software version 1.2 (Improvision) and processed with Photoshop version 4.0 (Adobe systems). When noted, cells were analyzed with the DeltaVision restoration microscopy system (Applied Precision, Inc.) with a Roper CH350 full-frame KAF1401E 12-bit high-resolution charge-coupled device (CCD) camera. The DeltaVision system was mounted on an Olympus 1X-70 microscope, and the UplanApo100X oil 1.35NA objective was used. When using the DeltaVision system, images were collected, processed, and deconvolved by using softWoRx V2.5 software (Applied Precision, Inc.). All images presented in a single figure were captured and processed in parallel by identical means when noted.
Functional assays.
Cell viability and volume assays were carried out as described previously (45). Mutant strains (Table 1) were transformed with the following plasmids: pMM82, pMM99, pMM98, pMM104, pMM105, pMM106, pMM107, pMM60, pMM61, pMM92, pMM93 and pRS416. All plasmids are episomal centromeric low-copy plasmids and carry the TRP1 gene. The data presented are representative of data from at least two independent experiments.
Protein extraction, immunoprecipitations, cellular fractionation, and immunoblotting.
Total cellular protein lysates were obtained, immunoprecipitated, and immunoblotted as previously described (38, 45). Cellular fractionation was carried out exactly as previously described (45). The monoclonal antibody HA.11 was used for immunoprecipitation of HA-tagged Cdc28p. Polyclonal HA.11 antibody was used for Western blot detection of Cdc28hap present in immunoprecipitates. The polyclonal antibody α-myc A-14 (Santa Cruz Biotechnology) was used to detect the myc epitope in Western blot analysis. Detection was by enhanced chemiluminescence (ECL) with the Super Signal ECL kit (Pierce).
Elutriation.
The diploid strain MMY42 contains CLN2myc at one CLN2 locus and CLN2PA at the other CLN2 locus; both expressed from the CLN2 promoter. MMY42 was grown to log phase (optical density at 660 nm [OD660] ∼0.9) in YEPD at 30°C (1-liter culture), harvested by filtration, resuspended in icewater, sonicated, and separated on the basis of cell size by centrifugal elutriation with a Beckman J6-M elutriating rotor with a 40-ml chamber. Elutriation was carried out with 0°C water, and the rotor chamber was maintained at 4°C throughout the elutriation. The rotor was maintained at the speed of 2,400 rpm, and the first fraction was taken at the flow rate of 70 ml/min. Four-hundred-milliliter fractions were collected with 10% increments in flow rates between fractions. The first fraction was saved when the OD660 of the sample rose above 0.02. After collection of fractions, equivalent amounts (as determined by OD) of each fraction were concentrated by filtration, resuspended in ice-cold YPD medium, and immediately subjected to indirect immunofluorescence analysis and protein extraction. Unfixed samples were subjected to Coulter counter analysis to determine the size of the fractions and used to measure the OD660. Subsequent immunoblotting analysis was carried out as described above.
Metabolic poisoning.
Metabolic poisoning was carried out as described in reference 65. Exponential cultures (50 ml) were harvested by filtration, washed with an equal volume of water, and resuspended in 6 ml of metabolic poison (10 mM NaN3, 10 mM deoxyglucose in YEP medium lacking glucose). Cells were incubated at 30°C with rotation for 45 min. Cells were removed, collected by filtration, fast formaldehyde fixed (see above), and processed for indirect immunofluorescence (see above). The poison was washed out of the culture by filtration followed by washing with an equal volume of water. Cells were allowed to recover by resuspending the cells in YPD medium and incubation at 30°C with rotation. Samples were collected and fast formaldehyde fixed for indirect immunofluorescence after various times of recovery.
RESULTS
Localization of Cln2p is regulated by Cdc28p-dependent phosphorylation.
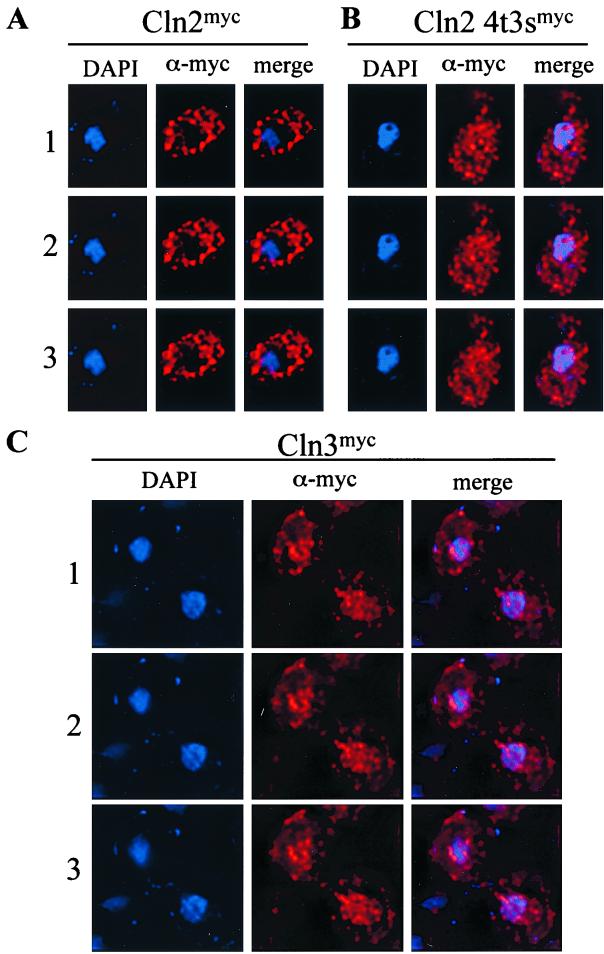
We showed previously that Cln2p was localized primarily to the cytoplasm (45) (Fig. 1). Since Cdc28p binds to Cln2p, phosphorylates Cln2p, and also localizes to the cytoplasm (78; our unpublished data), we investigated the possibility that Cdc28p activity might be important for Cln2p cytoplasmic localization. We assayed the localization pattern of a Cln2mycp mutant in which all seven potential Cdc28p phosphorylation sites have been mutated to alanine (Cln2-4t3smycp) (36, 62). Cln2-4t3sp binds Cdc28p, but the mutant Cln2p is not phosphorylated (36). myc-tagged CLN2-4T3S was integrated at the CLN2 locus in a wild-type strain. Wild-type Cln2p localizes primarily to the cytoplasm with clearly detectable “holes” of nuclear depletion, while the Cln2-4t3smycp mutant is no longer depleted in the nucleus and shows some nuclear accumulation (Fig. 1). We note that this nuclear accumulation is not complete, and a population of Cln2-4t3smycp remains in the cytoplasm (Fig. 1). It is unclear if the punctate nature of the Clnp staining in this assay results from fixation or reflects a relevant localization pattern in vivo. Consistent with these data, Cln2p also accumulates in the nucleus when CDC28 has been inactivated by using the temperature-sensitive allele cdc28-4 (Fig. 2); Cln2p phosphorylation is strongly reduced under these conditions (data not shown).
FIG. 1.
Indirect immunolocalization of Cln2mycp, Cln2-4t3smycp, and Cln3mycp. Strains expressing wild-type Cln2mycp (MMY1) (A), mutant Cln2-4t3smycp (MMY4) (B), or wild-type Cln3mycp (MMY32) (C) were assayed for indirect immunolocalization as described in Materials and Methods. Images were collected, processed, and deconvolved by using the DeltaVision restoration system (Applied Scientific) as described in Materials and Methods. Shown are three sections taken at 0.2-μm intervals labeled 1, 2, and 3. The DAPI signal indicates the position of the nucleus (blue), the α-myc signal indicates the position of the myc-epitope-tagged cyclin (red), and the merge combines these two images. The negative control (no myc-tagged cyclin) strain 1255-5C gave no detectable signal (data not shown).
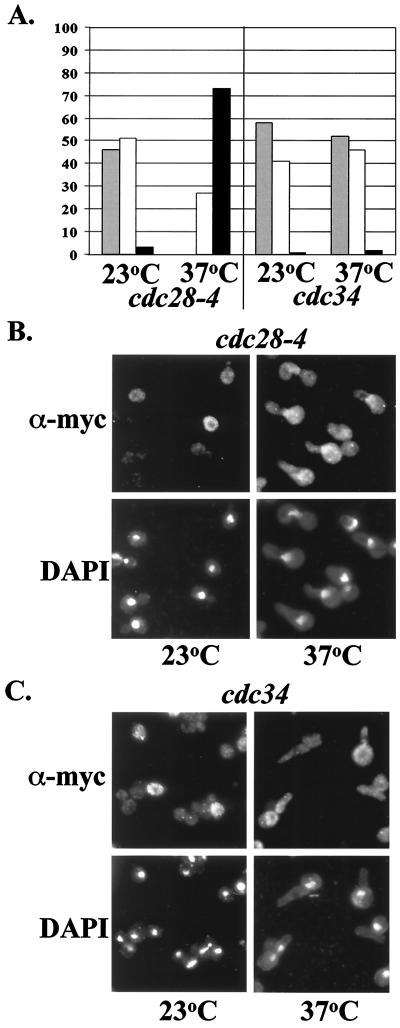
FIG. 2.
Indirect immunolocalization of Cln2mycp. cdc28-4 and cdc34 mutant cells expressing integrated myc-epitope-tagged Cln2p from the CLN2 promoter were assayed by indirect immunofluorescence as described in Materials and Methods. Cells were grown to log phase at 23°C. Cultures were split, and half was incubated at 23°C and half was incubated at 37°C for 2 h. (A) Quantitation of Cln2 localization. The percentage of cells showing nuclear depletion (gray bars), signal throughout the cell (white bars), or nuclear accumulation (black bars) is shown. A cell was scored positive for nuclear depletion when the position of the nucleus (DAPI signal) could be estimated correctly due to a reduction in Cln2mycp signal. A cell was scored positive for nuclear accumulation when the position of the nucleus (DAPI signal) could be estimated correctly due to an increase in Cln2mycp signal. When the position of the nuclei could not be determined based on Cln2mycp localization, the cells were scored as having a signal throughout the cell. Note that this procedure is unbiased in that the position of Cln2 depletion is determined from immunostaining before the position of the nucleus is determined from DAPI staining. Cln2 depletion from the vacuole will not be scored as nuclear depletion by this procedure. Data for Cln2 localization in the presence of mutant cdc28-4 and mutant cdc34 incubated at 23 and 37°C are shown. No fewer than 200 cells were counted with the DeltaVision microscopy restoration system. (B and C) Indirect immunolocalization of Cln2mycp expressed in mutant cdc28-4 (B) and mutant cdc34 (C) strains. The first row shows the indirect immunofluorescence with monoclonal anti-myc antibody 9E10 (α-myc), and the second row shows DAPI staining of DNA. Images were collected by standard microscopy.
Hypophosphorylated Cln2-4t3sp is stabilized because Cdc28p-dependent phosphorylation is required for Skp1–Cdc34–F-box (SCF)-dependent ubiquitination and subsequent degradation (36). We find that increased protein stability is not sufficient for the nuclear accumulation of Cln2p. CDC34 encodes the ubiquitin-conjugating component of SCF (6, 16, 77), and GRR1 is an SCF component that binds specifically to phosphorylated Cln2p, and this binding is required for SCF-dependent ubiquitination of Cln2p (6, 33, 41, 67, 68). Inactivation of CDC34 or GRR1, resulting in stabilization of Cln2p, does not alter Cln2p localization, since nuclear depletion is still observed, and no nuclear accumulation occurs (Fig. 2) (data not shown). Nuclear exclusion of Cln2p in the absence of CDC34 function is still dependent on CDC28-dependent phosphorylation, because Cln2p accumulates in the nucleus in a cdc28-4 cdc34 double mutant just as it does in a cdc28-4 single mutant (Fig. 2) (data not shown).
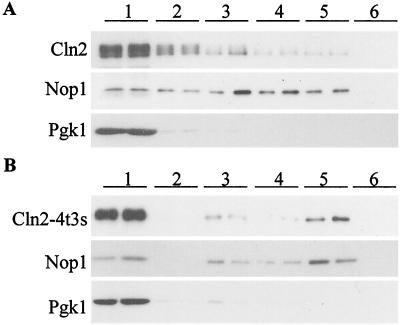
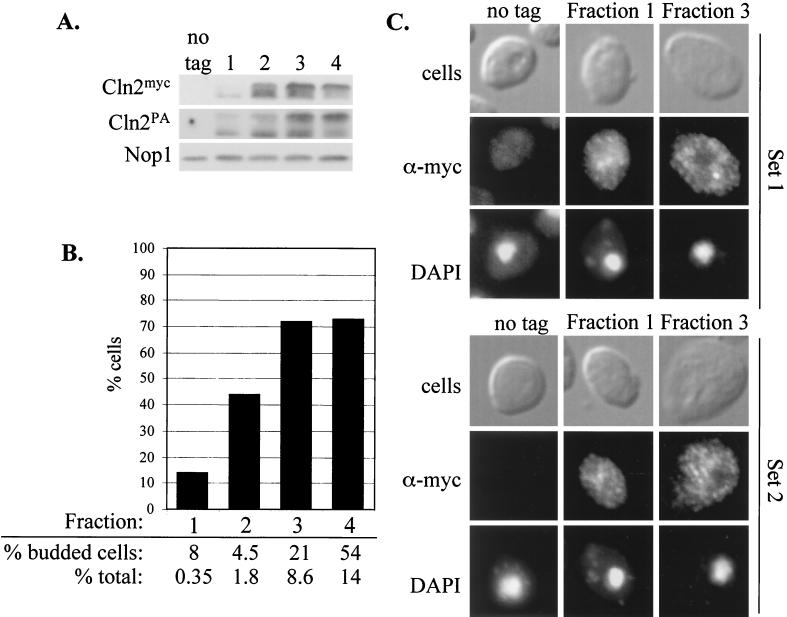
To confirm that hypophosphorylated Cln2p accumulates in the nucleus, nuclear and cytoplasmic components of the cell were separated and analyzed for Cln2mycp localization in cellular fractionation experiments. Wild-type Cln2mycp localizes primarily to the cytoplasm (Fig. 1) (45) and largely cofractionates with cytoplasmic marker Pgk1p (Fig. 3) (45). The Cln2-4t3smycp mutant shows an increase in nuclear localization (Fig. 1), and a subfraction cofractionates with nuclear marker Nop1p (Fig. 3). Consistent with the indirect immunofluorescence experiments, a significant amount of Cln2-4t3smycp cofractionates with the cytoplasmic Pgk1p (Fig. 3). Our ability to easily visualize the smaller amount of Cln2-4t3smycp accumulated in the nucleus is most likely due to the smaller size of the nuclear compartment compared to the cytoplasm, effectively concentrating the nuclear signal. (Note that the protein gels for the fractionation experiment in Fig. 3 are loaded in cell-equivalent amounts, not equal amounts of protein from the different fractions, to give a better representation of the overall distribution.) In both indirect immunofluorescence and cellular fractionation experiments, the Cln2-4t3smycp mutant shows an increase in nuclear localization compared with wild-type Cln2mycp. (Although these fractionation experiments were somewhat hampered by variable nuclear breakage, as indicated by recovery of the nuclear marker Nop1p in inappropriate fractions, concentration of Cln2-4t3smycp compared to that of wild-type Cln2mycp in the authentic nuclear fraction was reproducibly observed in multiple fractionations [data not shown].)
FIG. 3.
Localization of Cln2mycp and Cln2-4t3smycp by subcellular fractionation. Fractions were prepared as described in Materials and Methods. Duplicate samples of each fraction were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% polyacrylamide) and analyzed by Western blotting. Fractions are indicated at the top of each set of blots. Fraction 1 corresponds to the crude cytoplasmic fraction, fraction 2 corresponds to the surface of the sucrose gradient, fraction 3 corresponds to the surface-to-2.01 M sucrose interface, fraction 4 corresponds to the 2.01-to-2.1 M-sucrose interface, fraction 5 corresponds to the 2.1-to-2.3 M sucrose interface, and fraction 6 corresponds to the remaining pellet of the sucrose gradient. (A) Immunoblot analysis of fractions from a strain expressing Cln2mycp from the CLN2 promoter (MMY1). The position of Cln2mycp is indicated by “Cln2.” Corresponding immunoblot analysis of the Nop1p and Pgk1p fractionation is shown below the Cln2p blot. The nucleolar protein Nop1p detected with the monoclonal antibody D77 (4) indicates which fractions contain nuclear proteins. The cytoplasmic protein Pgk1p detected with the anti-Pgk1p polyclonal antibody (Molecular Probes) indicates which fractions contain cytoplasmic proteins. (B) Immunoblot analysis of fractions from a strain expressing Cln2-4t3smycp from the CLN2 promoter (MMY4). The Cln2-4t3smycp, Nop1p, and Pgk1p immunoblots are shown. A moderate level of nuclear breakage apparently occurred in both fractionation experiments, as indicated by the leakage of Nop1p into the intermediate and cytosolic fractions.
These data show a role of Cln2p phosphorylation by Cdc28p in regulated Cln2p localization.
Regulated subcellular localization of Cln2p requires binding to Cdc28p.
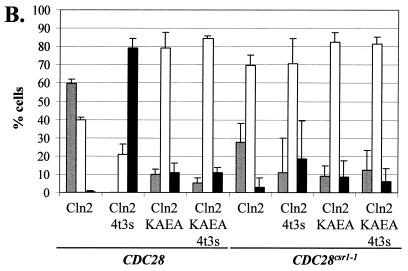
To ask if Cdc28p binding is necessary for Cln2p cytoplasmic localization, we made use of a Cln2p mutant (Cln2-KAEAp) with crippling mutations (K129A E183A) in the cyclin box region that mediates interaction with Cdc28p (27, 32, 38). The localization pattern of Cln2-KAEAmycp differs from that of wild-type Cln2mycp. Nuclear depletion is observed in approximately 60% of the cells expressing Cln2mycp, while it is observed in approximately 10% of the cells expressing Cln2-KAEAmycp (Fig. 4). Cln2p localization was also assayed in a strain expressing a mutant Cdc28p, Cdc28csr1-1p (cdc28-Q188P). Cdc28csr1-1p shows 100-fold reduced binding to wild-type Cln2p upon immunoprecipitation (39). Similar to the KA, EA cln2 mutation, the cdc28csr1-1 mutation causes a drop in the percentage of cells with nuclear depletion of Cln2mycp (Fig. 4). Residual binding between mutant Cdc28csr1-1p and wild-type Cln2p may occur, accounting for the remaining cells (approximately 30%) showing nuclear depletion. This residual binding may be lost in the case of the Cln2-KAEAmycp mutant (39). Using this mutant, we observed only a low level of nuclear depletion in the presence of either wild-type or mutant Cdc28csr1-1p. These data suggest that the physical association between Cln2p and Cdc28p is required for efficient nuclear exclusion of Cln2mycp.
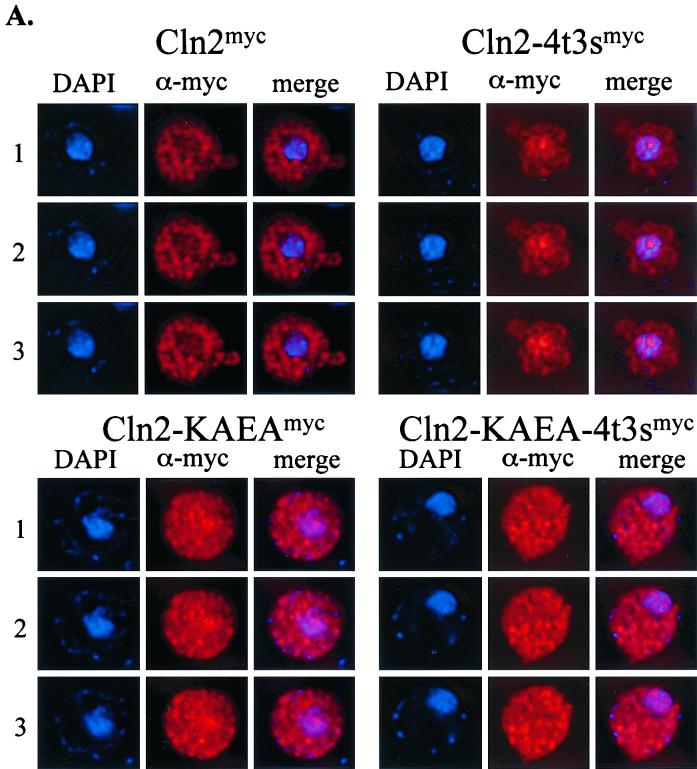
FIG. 4.
Requirement for binding to Cdc28p for regulated Cln2mycp localization. (A) Strain 1255-5C containing plasmids that express Cln2mycp (pMM159), mutant Cln2-4t3smycp (pMM151), mutant Cln2-KAEAmycp (pMM167), or mutant Cln2-KAEA-4t3smycp (pMM166) was assayed for indirect immunolocalization as described in Materials and Methods. Images were collected, processed, and deconvolved with the DeltaVision restoration system (Applied Scientific) as described in Materials and Methods. All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln2 protein from the CLN3 promoter. Shown are three sections taken at 0.2-μm intervals labeled 1, 2, and 3 for each set. The DAPI signal indicates the position of the nucleus (blue), the α-myc signal indicates the position of the myc epitope-tagged cyclin (red), and the merge combines these two images. The negative control (no myc-tagged cyclin) strain 1255-5C containing plasmid pRS416 gave no detectable signal (data not shown). (B) Quantitation of Cln2 localization. Percentages of cells showing nuclear depletion (gray bars), signal throughout the cell (white bars), or nuclear accumulation (black bars) were determined as described in the legend to Fig. 2. Data for Cln2 localization in the presence of wild-type Cdc28p (CDC28) and Cdc28csr1-1p (CDC28csr1-1) are shown. No fewer than 200 cells were counted for each experiment. The data shown are the average of three independent transformants with standard deviation indicated by the error bar. Images in this figure were captured and processed in parallel.
The hypophosphorylated Cln2-4t3smycp mutant shows nuclear accumulation in approximately 80% of the cells assayed. These data indicate that the physical interaction between Cln2p and Cdc28p is not sufficient for Cln2p cytoplasmic localization, since Cln2-4t3sp binds Cdc28p (36) and accumulates in the nucleus (Fig. 1, 3, and 4). Interestingly, the hypophosphorylated Cln2-4t3sp also requires binding to Cdc28p for regulated localization, since when the Cdc28p binding of the hypophosphorylated Cln2-4t3smycp is crippled (because of the cln2-KAEA mutation, the cdc28csr1-1 mutation, or both), we no longer observe nuclear accumulation of Cln2-4t3smycp (Fig. 4). This may explain the lack of strong nuclear accumulation of KAEAmycp (Fig. 4), which is hypophosphorylated like Cln2-4t3smycp, but unlike Cln2-4t3smycp binds to Cdc28p inefficiently (38; data not shown).
These results on nuclear exclusion of Cln2p can be accounted for by proposing that, first, only the localization of Cln2p-Cdc28p complex is regulated, but the Cln2p unbound to Cdc28p is distributed throughout the cell; and, second, that C-terminal phosphorylation of Cln2p in the Cln2p-Cdc28p complex is required for its efficient nuclear exclusion. The results with cdc28-4 (Fig. 2) are somewhat complicated to interpret in this framework because we don't know how much Cdc28-4p–Cln2p binding occurs in vivo.
We have been unable to identify an NLS or NES by inspection of the Cln2p sequence. Fusion of the wild-type or phosphorylation mutant Cln2p C terminus (residues 291 through 545, containing the seven Cdk phosphorylation sites) to GFP does not result in import of GFP to the nucleus (data not shown) and therefore gives no indication of an import activity encoded within this region of Cln2p. Also, no cytoplasmic export of GFP was observed upon fusing the Cln2p C terminus to an SV40 NLS-GFP fusion (data not shown). Thus, we have no evidence that this region of Cln2p provides signals for import or export. However, these regions of Cln2p do not include the Cdc28p binding domain, and these results might reflect a requirement for physical association with Cln2p for regulated localization, as suggested above.
Energy-dependent nuclear localization of Cln2p.
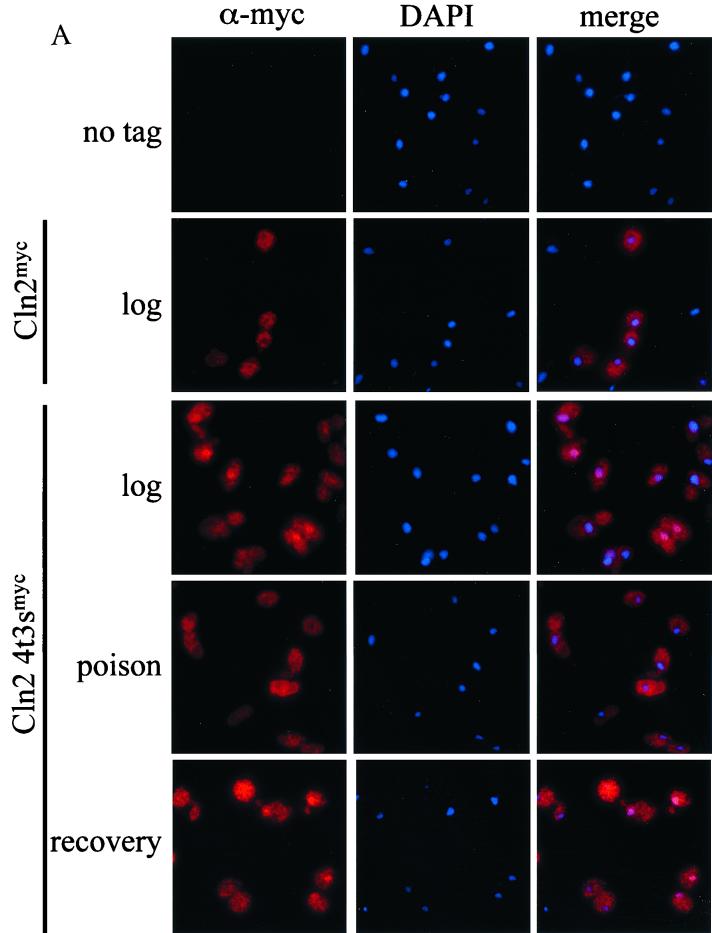
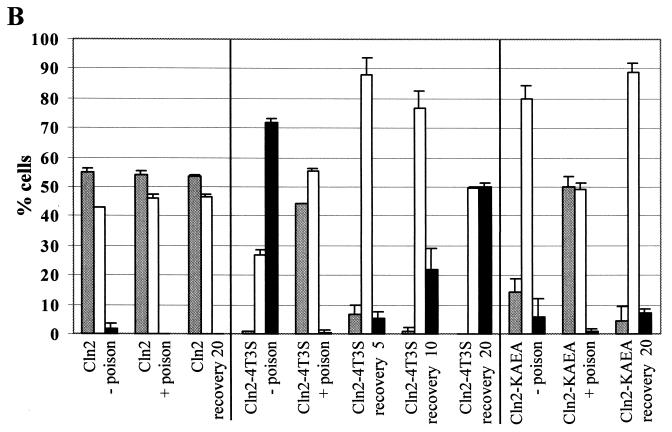
We assayed cells expressing wild-type or mutant Cln2mycp for energy-dependent localization by incubating the cells in metabolic poison and assaying Cln2p localization by indirect immunofluorescence. Exponential cultures were treated with metabolic poison as described previously (65). We find that after poison treatment, no difference in the localization pattern of wild-type Cln2mycp is observed (Fig. 5). In contrast, the Cln2-4t3smycp showed a dramatic change in localization, failing to accumulate in the nucleus in the presence of the poison (Fig. 5). Additionally, nearly 50% of the cells show some nuclear depletion of Cln2-4t3smycp (Fig. 5). We find that the maximal shift in localization occurred at 45 min of incubation with poison and did not change with longer incubations (up to 2 h). We were able to reconstitute nuclear accumulation by washing out the poison and incubating the cells in rich medium at 30°C. The nuclear accumulation of Cln2-4t3smycp after the poison was washed out was rapid and maximized between 15 and 20 min (Fig. 5). Steady-state protein levels of Cln2mycp did not change significantly when incubated with poison (data not shown), though the population of hypophosphorylated Cln2mycp increased. Upon recovery, there was some reduction in Cln2mycp levels combined with some recovery of Cln2mycp phosphorylation. Cln2-4t3smycp abundance did not change significantly when incubated with poison or during recovery (data not shown). Overall, poison effects on Cln2p abundance seem unlikely to account for the localization results.
FIG. 5.
Energy-dependent localization of Cln2mycp. Strains expressing wild-type untagged Cln2 (1255-5C), wild-type Cln2mycp (MMY1), Cln2-4t3smycp (MMY4), or Cln2-KAEAmycp (pMM167 in strain 1255-5C) were treated with metabolic poison and assayed for localization as described in Materials and Methods. (A) Indirect immunofluorescence of strains growing exponentially (log) after 45 min of treatment with metabolic poison (poison) and 20 min after removal of poison and incubation in rich medium at 30°C (recovery). Cells express untagged Cln2 (no tag), wild-type Cln2mycp (Cln2), or hypophosphorylated Cln2 mutant (Cln2-4t3smyc). Negative controls (no tag) incubated with poison and after recovery gave background signal identical to data shown for exponentially growing cells. (B) Quantitation of Cln2mycp localization during treatment with metabolic poison. Percentages of cells showing nuclear depletion (gray bars), signal throughout the cell (white bars), or nuclear accumulation (black bars) are shown as follows: exponentially growing cells expressing wild-type Cln2mycp without poison treatment (Cln2 − poison), cells expressing Cln2mycp after 45 min of treatment with poison (Cln2 + poison), cells expressing Cln2mycp after poison had been washed out and allowed to grow in rich medium for 20 min (Cln2 recovery 20), exponentially growing cells expressing mutant Cln2-4t3smycp without the addition of poison (Cln2-4t3s − poison), cells expressing Cln2-4t3smycp after 45 min of treatment with poison (Cln2-4t3s + poison), cells expressing Cln2-4t3smycp after poison had been washed out and allowed to grow in rich medium for 5, 10, and 20 min (Cln2-4t3s recovery 5, recovery 10, and recovery 20, respectively), cells expressing Cln2-KAEAmycp without addition of poison (Cln2-KAEA − poison), cells expressing Cln2-KAEAmycp after 45 min of treatment with poison (Cln2-KAEA + poison), and cells expressing Cln2-KAEAmycp after poison had been washed out and allowed to grow in rich medium for 20 min (Cln2-KAEA recovery 20). Quantitation was performed as described in the legend to Fig. 2. Essentially identical results were obtained with plasmid-based and genomic expression of Cln2p and Cln2-4t3sp. No fewer than 200 cells were counted for each experiment. The data shown are the average of two independent experiments with standard deviation indicated by the error bar. Images in this figure were captured and processed in parallel.
These data suggest that the nuclear accumulation of hypophosphorylated Cln2p occurs by energy-dependent active import. Nuclear depletion after incubation with poison introduces the possibility that nuclear export of Cln2-4t3smycp occurs in an energy independent manner, although we cannot exclude some contribution of nuclear degradation of Cln2p under these conditions. It is possible that once the Cln2-4t3smycp mutant moves into the cytoplasm, it is retained in the cytoplasm by an energy-independent mechanism, for example, by interaction with cytoplasmic proteins. Consistent with this, wild-type Cln2p continues to show nuclear depletion after up to 2 h of incubation in poison (data not shown).
Crippling the physical interaction between Cln2p and Cdc28p by introducing the cln2-KAEA or cdc28csr1-1 mutation results in apparent deregulation of Cln2p localization, with the majority of cells (approximately 80%) showing Cln2p signal throughout the cell (Fig. 4). To determine if the subpopulation of unbound Cln2p present in the nucleus entered by energy-dependent import, we assayed localization of the Cln2-KAEAmycp mutant after treatment with metabolic poison (Fig. 5). After poison treatment, Cln2-KAEAmycp showed nuclear depletion in approximately 50% of the cells assayed. Washing out the poison rapidly reconstituted localization of Cln2-KAEAp throughout the cell. Essentially identical results were obtained with Cln2-KAEA-4t3smycp and with the cdc28csr1-1 mutant (data not shown). These data suggest a constant energy-dependent nuclear import of Cln2p that occurs independent of Cln2p phosphorylation or Cln2p-Cdc28p complex formation. Nevertheless, interaction with Cdc28p appears to be required for efficient and phosphorylation-regulated Cln2p localization.
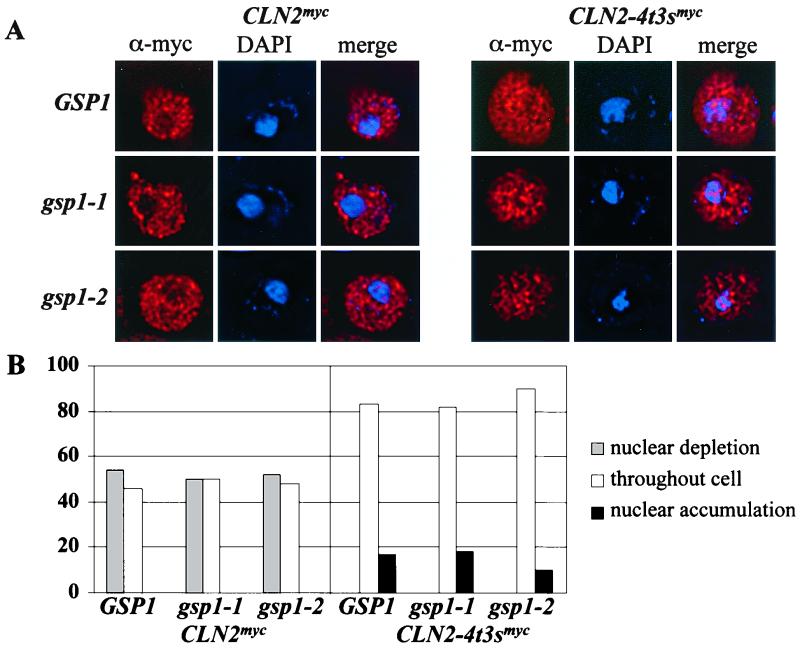
The metabolic energy requirement for nuclear accumulation of Cln2-4t3smycp could result from a requirement for GTP hydrolysis by the RAN GTPase (44, 47, 61). To test this possibility, we assayed wild-type Cln2mycp and mutant Cln2-4t3smycp localization in a temperature-sensitive ran mutant strain expressing the gsp1-1 and gsp1-2 alleles (79). No change in wild-type Cln2mycp localization was observed at permissive and nonpermissive temperatures (data not shown) (Fig. 6). In this strain, the Cln2-4t3smycp shows a localization pattern primarily throughout the cell, differing significantly from that observed with wild-type Cln2mycp. No RAN-dependent change in the localization pattern of Cln2-4t3sp was observed at permissive and nonpermissive temperatures (Fig. 6). Additionally, inactivation of the transport factors Crm1p, Msn5p, Los1p, Sxm1p, Mtr10p, Nmd5p, Kap104p, and Kap114p individually does not significantly alter wild-type or Cln2-4t3sp localization (data not shown). Localization of Cln2p might also be regulated by physical interactions with non-transport proteins, such as members of the 14-3-3 family (reviewed in reference 22). However, none of the potential phosphorylation sites mutated in this study clearly falls within described 14-3-3 binding motifs (52, 80).
FIG. 6.
Immunolocalization of Cln2mycp and Cln2-4t3smycp in the absence of Ran activity. GSP1 deletion strains carrying a plasmid that expresses either the wild-type GSP1, mutant gsp1-1, or mutant gsp1-2 alleles and integrated myc epitope-tagged Cln2p from the CLN2 promoter were assayed by indirect immunofluorescence as described in Materials and Methods. Cells were grown to log phase at 23°C. Cultures were split, and half was incubated at 23°C and half was incubated at 37°C for 2 h. (A) Indirect immunolocalization of Cln2mycp and Cln2-4t3smycp expressed in wild-type GSP1 strain (top row), gsp1-1 strain (second row) and gsp1-2 strain (third row) incubated at 37°C. The first column shows the indirect immunofluorescence with monoclonal anti-myc antibody 9E10 (red), the second column shows DAPI staining of DNA (blue), and the third column shows the merge of these two images. Images were collected with the DeltaVision microscopy restoration system. (B) Quantitation of Cln2 localization. The percentage of cells showing nuclear depletion (gray bars), signal throughout the cell (white bars), or nuclear accumulation (black bars) is shown. Quantitation was performed as described in the legend to Fig. 2. Data for Cln2mycp and Cln2-4t3smycp localization in the presence of wild- type and mutant GSP1 incubated at 37°C are shown. No fewer than 200 cells were counted with the DeltaVision microscopy restoration system.
These data suggest that the nuclear accumulation of hypophosphorylated Cln2p-Cdc28p occurs by a Ran-independent, yet energy-dependent form of active transport (Fig. 5 and 6). Export of Cln2p-Cdc28p from the nucleus may occur in an energy-independent manner (Fig. 5).
Cln2p nuclear depletion may be inefficient early in the cell cycle.
We analyzed Cln2p phosphorylation and subcellular localization in a population of rapidly growing cells separated by centrifugal elutriation into fractions of different cell size. The smallest (newborn) cells obtained from the elutriation contain Cln2p that is significantly underphosphorylated compared to larger unbudded cells (Fig. 7A). This is true for 9× myc-epitope-tagged Cln2p, as well as for protein A-tagged Cln2p (in both cases integrated at the CLN2 locus). Indirect immunofluorescence shows little or no detectable depletion of Cln2pmyc from the nucleus of the smallest unbudded cells (Fig. 7B and C), consistent with the idea that Cln2p shuttles between the cytoplasm and nucleus. These data also suggest the possibility that shuttling is differentially regulated, with more nuclear Cln2p present very early in the cell cycle.
FIG. 7.
Cell-cycle-regulated nuclear depletion of Cln2p. Rapidly growing cultures of diploid strain MMY42 were chilled and subjected to centrifugal elutriation to obtain fractions of cells of increasing size as described in Materials and Methods. (A) Fractions were subjected to protein extraction and immunoblot analysis to detect 9× myc epitope-tagged or PA epitope-tagged Cln2p, each expressed from a different allele from the CLN2 promoter. Nop1p levels were monitored as a loading control. (B) Fractions were assayed by indirect immunofluorescence to detect Cln2mycp localization. The bar graph shows cells that were scored for the presence of nuclear depletion as described in the legend to Fig. 2. No nuclear accumulation was observed in these fractions. The percentage of budded cells is the percentage of cells with buds in the population of cells in each fraction. The percentage of the total is the percentage of cell mass present in a fraction of the total amount of cell mass as determined by OD of samples. (C) Representative immunofluorescence results of two MMY42 elutriations (sets 1 and 2). Cells were assayed by indirect immunofluorescence as described in Materials and Methods. The first row shows differential interference contrast (DIC) images (cells), the second row shows the indirect immunofluorescence with monoclonal anti-myc antibody 9E10 (α-myc), and the third row shows DAPI staining of DNA. The untagged control from this experiment consisted of exponentially growing cells of strain W303d. The amounts of cells of fractions 1, 2, 3, and 4 were 66, 73, 78, and 89 fl, respectively, as determined by identification of peak channels by Coulter Counter analysis of cells.
We showed previously that addition of an NES to Cln2p significantly weakened its biological activity (45), implying that Cln2p might spend some time in the nucleus and that this nuclear residence might be important for its function. Our present results suggest that physical association with Cdc28p and phosphorylation may regulate nuclear accumulation of Cln2p; this binding or phosphorylation may be cell cycle regulated, leading to the potential for Cln2p to function in the nucleus specifically very early in the cell cycle.
Another hypothesis that could explain loss of nuclear exclusion of Cln2p in cells early in the cell cycle would be that Cln2p-Cdc28p complex formation is inefficient at this time. We do not yet have information on this point, because the extremely low yield of cells at this point in the cell cycle from the elutriating rotor (Fig. 7) makes a definitive analysis of Cln2p-Cdc28p complex formation in these cells technically challenging.
Hypophosphorylated Cln2p could occur early in the cell cycle because of low kinase activity or because of the activity of a phosphatase. We tested the idea that the Cdc14p phosphatase, which is known to reverse other Cdk phosphorylations (31, 73), might also dephosphorylate Cln2p. This idea seemed reasonable, since we detect hypophosphorylated Cln2p early in the cell cycle, not long after Cdc14p has been released from the nucleolus late in mitosis (5, 64). We found, though, that strong overexpression of Cdc14p from the GAL1 promoter sufficient to arrest the cell cycle (73) or deletion of the Cdc14p regulator CFI1 (74) has little or no effect on Cln2p phosphorylation or localization (data not shown).
Cln3p localization does not require Cdc28p activity or binding.
In contrast to Cln2p localization, the localization of Cln3p does not appear to be influenced by Cdc28p activity. Cln3p remains nuclear when expressed in the cdc28-4 strain at permissive and nonpermissive temperatures (data not shown). Additionally, mutations within the cyclin box of Cln3p that eliminate detectable interaction with Cdc28p (M. Miller, unpublished observations) do not alter Cln3p nuclear localization (data not shown). Nuclear localization of both wild-type and cyclin box-defective Cln3p was also observed in the presence of Cdc28csr1-1p mutant Cdc28p (data not shown); this Cdc28 mutation reduces immunoprecipitable Cln3p-Cdc28p complexes by about 10-fold (39). Thus, we were unable to detect any Cdc28p requirement for Cln3p localization.
Carboxy-terminal Cln3p bipartite NLS.
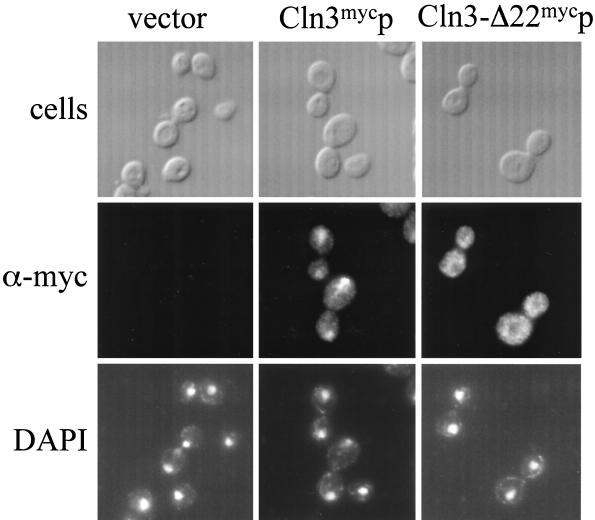
The CLN3-1 truncation lacks the C-terminal 177 amino acids of Cln3 and shows a partial increase of cytoplasmic localization when compared to full-length Cln3p (45). These data suggest that sequences important for the nuclear localization are present in the carboxy-terminal tail. The last 22 amino acids of Cln3p contain a potential bipartite NLS (this was proposed as a candidate NLS previously, although it was not tested functionally [81]). Deletion of the last 22 amino acids of Cln3p (Cln3-Δ22p) resulted in a primarily cytoplasmic localization, unlike the nuclear localization of Cln3mycp (Fig. 8). Thus, the last 22 amino acids are required for nuclear localization of Cln3p.
FIG. 8.
Indirect immunolocalization of Cln3-Δ22mycp. Wild-type (1255-5C) cells were transformed with plasmids pRS414 (vector), pMM99 (Cln3mycp), and pMM98 (Cln3-Δ22mycp). All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln3 protein from the CLN3 promoter. Transformants were assayed by indirect immunofluorescence as described in Materials and Methods. The first row shows DIC images (cells), the second row shows indirect immunofluorescence with monoclonal anti-myc antibody 9E10 (α-myc), and the third row shows DAPI staining of DNA. Images in this figure were captured and processed in parallel.
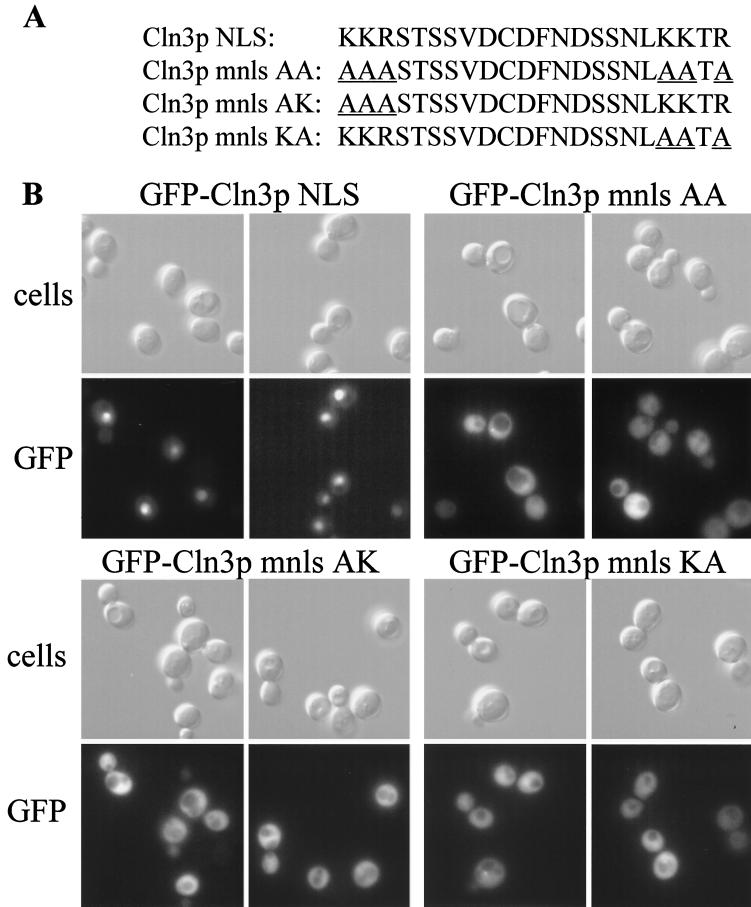
To determine if the putative Cln3p NLS is sufficient for nuclear localization, the last 22 amino acids of Cln3p were fused to the N terminus of the GFP expressed from the transport assay plasmid pKW431 (69). Since this sequence consists of two basic clusters separated by 15 amino acids (diagrammed in Fig. 9A), we also constructed mutant versions of the Cln3p NLS fused to GFP, in which the clusters of basic residues are mutated to alanine singly or together (Cln3p KA, AK, or AA NLS). The GFP proteins containing the Cln3p NLS show a staining pattern in living cells consistent with nuclear localization, while the GFP fusions containing the mutant Cln3p mnls sequences were cytoplasmic (Fig. 9B). To ensure that the Cln3p NLS shifts the localization of GFP into the nucleus, cells were fixed and assayed for both GFP localization and DAPI staining. The Cln3p-NLS-GFP fusion colocalized with the nuclear DAPI signal, although this procedure weakened the GFP signal significantly (data not shown).
FIG. 9.
Cln3-NLS-GFP localization. (A) Amino acid sequence of the bipartite-type Cln3 NLS. The mutant Cln3 nls AA substitutes the basic residues in the first and second basic clusters to alanine. The mutant Cln3 nls AK substitutes the first basic cluster with alanines, and the mutant Cln3 nls KA substitutes the second basic cluster with alanines. Mutated residues are underlined. (B) Immunofluorescent localization of the various Cln3 NLS-GFP in living cells. The top row shows DIC images (cells), and the bottom row shows GFP signal in an identical field of cells. Images in this figure were captured and processed in parallel.
Taken together, these data indicate that the CLN3p NLS is a bipartite NLS, which is necessary and sufficient for nuclear localization.
Cln3-Δ22p protein characterization.
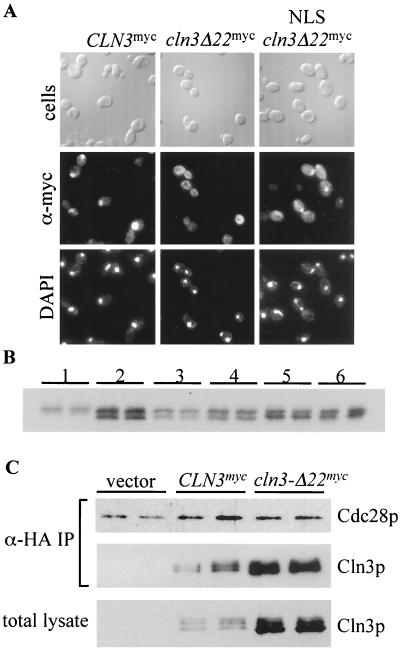
To determine if deletion of the Cln3 NLS alters steady-state expression levels, Western blot analyses were carried out on a wild-type strain expressing Cln3mycp and Cln3-Δ22mycp. Cln3-Δ22mycp was expressed at levels as high as wild-type Cln3mycp, and in 3 out of 5 experiments Cln3-Δ22mycp was expressed at higher levels than wild-type Cln3mycp (Fig. 10B, lanes 1 and 2 and data not shown).
FIG. 10.
Comparison of myc-tagged Cln3 proteins. (A) Indirect immunolocalization of NLS-Cln3-Δ22mycp. Wild-type (1255-5C) cells were transformed with plasmids pMM99 (Cln3mycp), pMM98 (Cln3-Δ22mycp), and pMM104 (NLS-Cln3-Δ22mycp). All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln3 protein from the CLN3 promoter. Transformants were assayed by indirect immunofluorescence as described in Materials and Methods. The first row shows DIC images (cells), the second row shows indirect immunofluorescence with monoclonal anti-myc antibody 9E10 (α-myc), and the third row shows DAPI staining of DNA. The vector control from this experiment (data not shown) gave no detectable signal. (B) Wild-type strain (1255-5c) transformed with plasmids pMM99 (Cln3myc, set 1), pMM98 (Cln3-Δ22myc, set 2), pMM104 (NLS-Cln3-Δ22myc, set 3), pMM105 (mnls-Cln3-Δ22myc, set 4), pMM106 (NES-Cln3-Δ22myc, set5), and pMM107 (mnes-Cln3-Δ22myc, set 6). All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln protein from the CLN3 promoter. Cellular lysates of two independent transformants for each clone were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide) and analyzed by Western blot analysis as described in Materials and Methods. (C) Coimmunoprecipitation assays were carried out as described in Materials and Methods. Strain 1559-3B-7 expressing HA-epitope-tagged Cdc28p was transformed with plasmids pRS414 (vector), pMM99 (CLN3myc), and pMM98 (cln3-Δ22myc). The top panel shows the anti-HA immunoblot of immunoprecipitated Cdc28hap. The middle panel shows the anti-myc immunoblot of Cln3myc protein present in the immunoprecipitation. The bottom panel shows the anti-myc immunoblot of Cln3myc protein present in the total cellular lysate used for the immunoprecipitation.
The NLS could overlap with functional PEST sequences in the Cln3p C-terminal third (13, 81); alternatively, altering Cln3p localization could directly affect Cln3p stability. To test this, exogenous localization signals (SV40 NLS or PKI NES) were added to Cln3-Δ22mycp to accentuate or correct the localization defect. The NLS resulted in a partial accumulation of Cln3-Δ22mycp in the nucleus (Fig. 10A). In repeated immunofluorescence experiments, the PKI NES appeared to result in a tighter exclusion of Cln3-Δ22mycp from the nucleus (data not shown), although the effect is subtle, since the bulk of Cln3-Δ22mycp is already cytoplasmic even without the NES. The mnls and mnes mutant sequences (containing single-amino-acid changes that disrupt the localization activity of the sequences) did not significantly alter the localization of Cln3-Δ22mycp (data not shown). No significant difference was observed between the steady-state levels of the NLS-Cln3-Δ22mycp (shifted to the nucleus [Fig. 10]) versus mnls-Cln3-Δ22mycp (cytoplasmic [data not shown]); therefore, mislocalization does not appear to cause the increase in steady-state levels of Cln3-Δ22mycp (Fig. 10B). The increase in steady-state expression of Cln3-Δ22mycp may be due to stabilization, since part of the final Cln3p PEST region is missing in this deletion (81). The effect of this mutation on Cln3mycp accumulation is significantly less than the effect of complete deletion of the C-terminal 177 amino acids of Cln3p (45), consistent with the presence of additional PEST sequences in Cln3p (81).
Differences in subcellular localization of Cln3p verses Cln3-Δ22p may influence Clnp-Cdc28p complex formation. To test this idea, a strain expressing HA-tagged Cdc28p was transformed with plasmids that express 9× myc-tagged Cln3p or Cln3-Δ22p. Immunoprecipitations of the HA-epitope-tagged Cdc28p results in coimmunoprecipitation of the myc-epitope-tagged Cln3p. We find that levels of Cln3p able to coimmunoprecipitate with Cdc28p correspond to the expression levels of these proteins found in total cellular lysates (Fig. 10C). These data indicate that the deletion of the last 22 amino acids of Cln3p does not significantly alter the ability of Cln3p to associate with Cdc28p.
Functional consequences of Cln3p mislocalization.
The last 22 amino acids of Cln3p encode an NLS that is important for the nuclear localization of Cln3p. Functional analysis was carried out to determine if this NLS was significant for Cln3p function in vivo. Previously, we demonstrated that shifts in the localization patterns of Cln3p correlate with changes in the functional profile of Cln3p. This study was carried out with the Cln3-1p mutant, which lacks the carboxy-terminal 177 amino acids of Cln3p. In addition to missing the C-terminal 22 amino acids (NLS characterized above), this mutant lacks several PEST domains and demonstrates a dramatic stabilization phenotype (13, 45, 81). In contrast, the Cln3-Δ22p mutant is a much smaller deletion, missing only the NLS and parts of a single PEST domain and causes significantly less stabilization of the protein than the deletion of the C-terminal 177 amino acids. In addition to determining if the C-terminal NLS defined in this study is a functionally significant NLS in vivo, we were also interested in whether the changes in the functional profile previously observed for the mislocalized Cln3-1p mutant (45) required hyperstabilization.
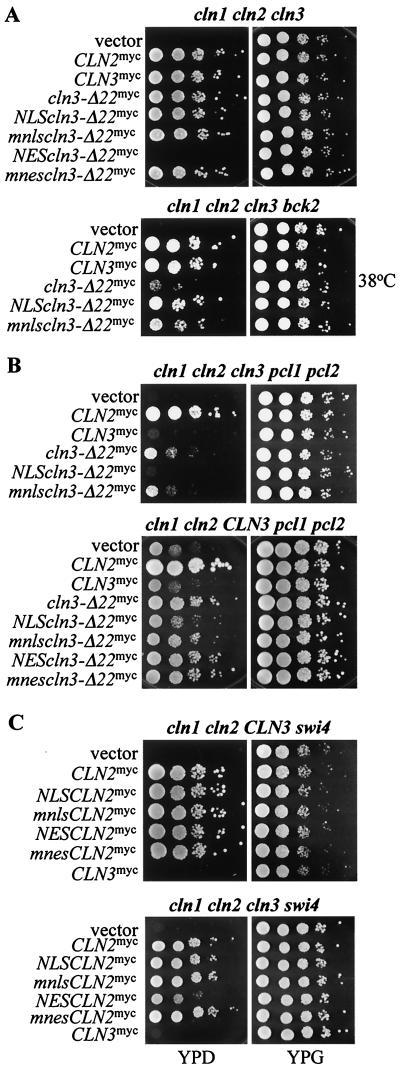
A cln1 cln2 cln3 GAL1::CLN1 strain is inviable on glucose medium (which turns off the GAL1::CLN1 cassette, resulting in cln1 cln2 cln3 inviability). We introduced plasmids encoding Cln3-Δ22p with or without mislocalization signals into this strain and compared the ability of these plasmid-bearing strains to form colonies on glucose-containing medium. The strain expressing NES-Cln3-Δ22p was inviable, while the strains expressing NLS-, mnls-, or mnes-Cln3-Δ22p and Cln3-Δ22p were viable (Fig. 11A). This result is different from that obtained with the Cln3-1p mutant and reveals an essential nuclear requirement for Cln3p not previously observed. The ability of NES-Cln3-1p to support viability in this strain may result from its overexpression. These data indicate that nuclear localization of Cln3p is required for efficient rescue of the triple cln deletion. The ability of Cln3-Δ22p to rescue the cln1 cln2 cln3 strain may reflect residual nuclear accumulation of Cln3-Δ22p, which may be eliminated by NES (but not mnes) addition.
FIG. 11.
Functional assays for Cln3-Δ22p. Mutant strains, each containing a GAL1::CLN for viability, were transformed with CLN plasmids. For each transformant, 10-fold serial dilutions were prepared from independent pools of transformants (5 to 10 colonies), and 3 μl of each dilution was plated onto both YPD (dextrose) and YPG (galactose) plates. Plates were incubated at 30°C, except where 38°C is indicated. The CLN plasmids used are pMM82 (CLN2myc), pMM99 (CLN3myc), pMM98 (cln3-Δ22myc), pMM104 (NLS-cln3-Δ22myc), pMM105 (mnls-cln3-Δ22myc), pMM106 (NES-cln3-Δ22myc), pMM107 (mnes-cln3-Δ22myc), pMM60 (NLS-CLN2myc), pMM61 (mnls-CLN2myc), pMM92 (NES-CLN2myc), and pMM93 (mnes-CLN2myc). All plasmids are episomal CEN (low copy number) and express the myc-tagged Cln protein from the CLN3 promoter. Strains are listed in Table 1. Genotypes are as follows (lowercase denotes deletion; uppercase denotes wild type, and GAL1:: CLN gene not indicated): cln1 cln2 cln3 and cln1 cln2 cln3 bck2 (A); cln1 cln2 cln3 pcl1 pcl2 and cln1 cln2 CLN3 pcl1 pcl2 (B); cln1 cln2 CLN3 swi4 and cln1 cln2 cln3 swi4 (C). (cl1 clz cl3 swi4 data were reproduced for comparison to the cln1 cln2 CLN3 swi4 strain from reference 45.)
The genetic requirement for Cln3p is increased in the absence of the BCK2 gene product, since Bck2p works in parallel with Cln3p to trigger transcription of SCF- and MBF-regulated genes in late G1 (see Introduction). Cln3-Δ22p has a reduced ability to rescue the bck2 cln1 cln2 cln3 strain, at least at elevated temperatures, compared to wild-type Cln3p (Fig. 11A). This defect was rescued by addition of the NLS to Cln3-Δ22p, consistent with the idea that this defect is specifically due to the largely cytoplasmic localization of Cln3-Δ22p. Addition of the mnls to Cln3-Δ22p was significantly less effective at restoring its activity (Fig. 9A). These data indicate that the C-terminal NLS of Cln3p is functionally significant, consistent with the idea that nuclear localization of Cln3p is important for function (45). These data also reveal an essential role of nuclear Cln3p in the cln strain that was not previously detected with Cln3-1p.
Deletion of pcl1 and pcl2 results in the inability of CLN3, but not CLN1 or CLN2, to provide the essential CLN function for cell cycle initiation (see Introduction). Thus a cln1 cln2 cln3 pcl1 pcl2 GAL1::CLN1 strain is rescued on glucose medium by CLN2, but not by CLN3 on plasmids. As observed for the CLN3-1 mutant (45), CLN3-Δ22 exhibits a weak ability to rescue this strain (Fig. 11B). This rescue was abolished by addition of the NLS (but not the mnls) to Cln3-Δ22p, suggesting that significant cytoplasmic localization of Cln3-Δ22p was required for rescue. As expected from the inability of NES-Cln3-Δ22p to rescue the cln1 cln2 cln3 strain (Fig. 11A), it was also unable to rescue the cln1 cln2 cln3 pcl1 pcl2 strain (data not shown). Taken together, these results could imply that rescue of the cln1 cln2 cln3 pcl1 pcl2 strain by Cln3-Δ22p required both cytoplasmic Cln3-Δ22p and some residual nuclear Cln3-Δ22p. If this were correct, then we would predict that the combination of wild-type nuclear Cln3p and the strongly cytoplasmic NES-Cln3-Δ22p might complement each other for rescue of a cln1 cln2 cln3 pcl1 pcl2 strain. Indeed, this was observed (Fig. 11B): provision of wild-type CLN3 in the chromosome and NES-CLN3-Δ22 on a plasmid resulted in significant viability in the absence of pcl1, pcl2, cln1, and cln2, while neither one alone was sufficient (Fig. 11B) (data not shown). Essentially similar results were obtained with a swi4 cln1 cln2 CLN3 GAL1::CLN1 deletion strain (data not shown). Therefore, as with pcl1 pcl2, deletion of swi4 appears to uncover a strong requirement for both cytoplasmic and nuclear CLN activity.
Consistent with this, we found previously that addition of an NES (but not an mnes) to Cln2p strongly reduced its ability to rescue a cln1 cln2 cln3 swi4 strain (45). According to the hypothesis that both nuclear and cytoplasmic CLN activities may be required in this context, NES-Cln2p should complement Cln3p for rescue of a cln1 cln2 cln3 swi4 strain, since Cln3p should provide the nuclear CLN function and NES-Cln2p should provide the cytoplasmic CLN function. This was observed (Fig. 11C): provision of wild-type CLN3 in the chromosome and NES-CLN2 on a plasmid resulted in significant viability in the absence of pcl1, pcl2, cln1, and cln2, while CLN3 alone was insufficient and NES-CLN2 alone was very inefficient (Fig. 11C).
Thus, wild-type Cln3p requires nuclear localization to carry out its normal roles, probably including activation of G1/S transcription (see Introduction), so that its mislocalization to the cytoplasm blocks its ability to rescue the cln1 cln2 cln3 strain. In contrast, mislocalization of Cln3p to the cytoplasm allows it to carry out some biological roles normally restricted to the cytoplasmic Cln2p. These results confirm and extend our previous results with mislocalized Cln3-1p, with much less experimental confounding of localization and protein abundance effects.
Localization-dependent cell size control by Cln3p.
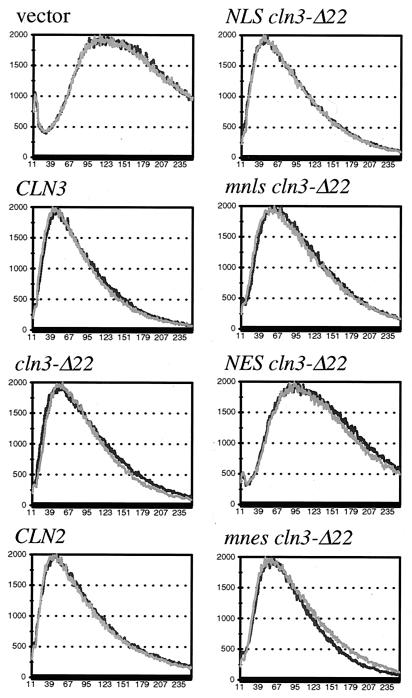
We also assayed for CLN activity by monitoring cell volume. Initiation of the cell cycle requires CLN activity for bud emergence and activation of Clbp-Cdc28p complexes. Cells with reduced CLN activity will have a delay in these events resulting in a population of relatively large cells due to a longer period of cell growth after cell division and before cell cycle initiation. Conversely, high CLN activity will result in a population of relatively small cells (11, 48, 53). Links between cell cycle progression and cellular growth may also exist in higher eukaryotic cells, suggesting that this process could be conserved (as reviewed in reference 10). Plasmids encoding the various mutant Cln3 proteins were introduced into a cln2 cln3 strain, and transformants were assayed for cell volume. (A cln2 cln3 strain was used to sensitize the assay to CLN function.) As expected, expression of Cln2mycp and Cln3mycp confers smaller cell volume than that of the vector control. Expression of NES-Cln3-Δ22mycp (but not the mnes controls) produces populations of cells with larger cell volumes, near the volume of control vector transformants (Fig. 12). These data show a strong reduction of CLN3 functional activity when Cln3p is exported out of the nucleus and into the cytoplasm through the combination of the Cln3p NLS deletion and addition of the PKI NES. Thus, as with the cln1 cln2 cln3 rescue assay, the cell volume assay for CLN activity leads to the conclusion that nuclear Cln3 may be required to efficiently drive cell cycle initiation.
FIG. 12.
Cell size assay for CLN gene function. CLN1 cln2 cln3 cells (1421-21D) were transformed with plasmids containing various CLN2 and CLN3 constructs (see the legend to Fig. 9). Cell size analysis was carried out as described in Materials and Methods. Data for two transformants are shown on each graph.
Although we expected the shift of Cln3-Δ22p to the cytoplasm (even without the NES) to decrease its activity in this assay, we find only a slight shift and widening of the peak distribution of cells expressing Cln3-Δ22p compared to wild-type Cln3p. This slight, but reproducible change is relieved by addition of the functional SV40 NLS, but not the mnls control (Fig. 12). These data are relevant to models describing the regulation of cell size in response to Clnp dosage resulting from Cln3p compartmentalization in the nucleus. Assuming that the nucleus maintains a constant volume through G1, as cells grow, the concentration of nuclear Cln3p could increase until some critical threshold is reached allowing cell cycle progression (as proposed in reference 23 and quantitatively modeled by reference 8). If so, the prediction would be that the redistribution of a significant amount of Cln3p to the cytoplasm (via deletion of the Cln3 NLS) should reduce the amount of Cln3p in the nucleus, thus delaying the ability of the nuclear Cln3p to reach this critical threshold. In this case, Cln3-Δ22p should give a delay in cell cycle progression compared to that of the wild type and a commensurate increase in cell size. This prediction is complicated in our experiment by the fact that Cln3-Δ22p is present at an elevated level compared to wild-type Cln3 (Fig. 10). Increased Cln3p protein levels are known to accelerate cell cycle progression (11, 53), consistent with the quantitative model (8). Hence, the lack of phenotype observed with Cln3-Δ22p may reflect a balance between accelerated cell cycle progression due to higher steady-state levels of Cln3p (at least some of which is likely to be nuclear) and slowed cell cycle progression due to the reduction of nuclear Cln3p. However, comparison of the NLS-Cln3-Δ22p and the mnls-Cln3-Δ22p provides us with a situation in which a clear difference in subcellular localization is observed by indirect immunofluorescence (Fig. 10), with no significant changes in expression levels (Fig. 10). In this case, we find only minor changes in cell size distribution dependent on subcellular localization, contrary to predictions made based on models in which nuclear compartmentalization of Cln3p is crucial for cell size control. It is possible that there is some compensating acceleration of cell cycle initiation by cytoplasmic Cln3p in this case.
Taken together, these data suggest that efficient nuclear localization of Cln3p is important for maintaining cell size, since NES-Cln3-Δ22p is almost unable to function in this assay. However, the relationship between Cln3p function, nuclear localization, and cell size control may be somewhat complex.
DISCUSSION
Different mechanisms control localization of the G1 cyclins Cln2p and Cln3p.
In addition to activating Cdks through conformational changes associated with cyclin-Cdk binding, the spatial context in which cyclin proteins function is an important component of Cdk regulation. In previous work, we found that the localization pattern of the G1cyclin Cln2p is primarily cytoplasmic and that of the G1 cyclin Cln3p is primarily nuclear (45). Here we have investigated the role that the Cdk Cdc28p plays in the control of distinct subcellular localization patterns of these cyclins. Using a mutant Cln2p in which all potential Cdc28p target sites have been mutated, we find a partial accumulation of Cln2p in the nucleus (Fig. 1). Inactivation of CDC28 also results in a partial accumulation of Cln2p in the nucleus as demonstrated by indirect immunofluorescence (Fig. 2). The nuclear accumulation of hypophosphorylated Cln2p does not reflect Cln2p stabilization, since stabilization of Cln2p via mutation of the CDC34 gene or deletion of the GRR1 gene does not alter Cln2p localization (Fig. 2 and data not shown). Taken together, these data are consistent with direct Cdc28p-dependent phosphorylation of Cln2 being required for nuclear depletion of Cln2p. Additionally, there is a requirement of Cln2p-Cdc28p complex formation in regulated Cln2p localization. Both nuclear depletion and nuclear accumulation are influenced by physical association with Cdc28p, as seen with mutations in Cln2p that are defective in binding Cdc28p and with mutations in Cdc28p that are defective for binding Cln2p (Fig. 4).
We also find a cell cycle-dependent shift in the phosphorylation state of Cln2p. Populations of small unbudded cells harvested by centrifugal elutriation from a log-phase culture (presumably the youngest cells in the population) contain largely hypophosphorylated Cln2p compared to populations of larger unbudded cells (presumably from later in the cell cycle). These newborn cells with hypophosphorylated Cln2p show little or no depletion of Cln2p from the nucleus. These data are consistent with regulated shuttling of Cln2p in and out of the nucleus, where nonphosphorylated Cln2p is enriched in the nucleus and phosphorylated Cln2p is enriched in the cytoplasm.
Phosphorylation of Cln2p also triggers Cln2p degradation. It remains unclear if the phosphorylation sites that target Cln2p for degradation are the same as the phosphorylation sites that regulate Cln2p localization. It is possible that the localization and stability of Cln2p are linked, such that the localization of Cln2p in the cytoplasm increases the instability of this protein. It also remains possible that multiple phosphorylation sites are involved in regulation of Cln2p localization. This might allow multiple cellular inputs to regulate Cln2p localization, as has been described for the higher eukaryotic cyclin B1 (83). We are currently investigating these possibilities.
Our findings on Cln2p localization can be explained by the model that Cln2p-Cdc28p complex can enter the nucleus in an energy-dependent manner and can probably leave the nucleus in an energy-independent manner. We do not yet know if Cln2p phosphorylation increases the rate of energy-independent nuclear export, decreases the rate of energy-dependent import, or influences potential cytoplasmic retention. All Cln2p traffic appears to be Ran independent. Some aspect of this regulation is interfered with in cells early in the cell cycle, which may allow a brief period of nuclear residence of Cln2p.
Unlike Cln2p, the localization of Cln3p does not require Cdc28p activity or binding (data not shown). Nuclear localization of Cln3p requires a C-terminal bipartite-type NLS. Deletion of this sequence results in a shift in localization of Cln3p to the cytoplasm, and fusion of this sequence to GFP results in nuclear localization of GFP; therefore, this sequence is necessary and sufficient for nuclear localization. The ability of this region to confer nuclear localization to GFP requires two basic clusters within the sequence, consistent with a bipartite-type structure of the Cln3p NLS. Additionally, we find that the Cln3p NLS-GFP fusion protein is defective for nuclear localization in a gsp1-1 Ran temperature-sensitive mutant strain (data not shown).
Our data indicate that the localization patterns of Cln2p and Cln3p are actively maintained in the cell and regulated by distinct mechanisms. The movement of Cln2p-Cdc28 complex into the nucleus is energy dependent and regulated by Cdc28p-dependent phosphorylation, yet appears to involve a nonclassical route, since Cln2p transport appears Ran independent. In contrast, Cln3p nuclear import appears Cdc28p independent and requires a bipartite-type localization signal that is RAN dependent. The existence of distinct mechanisms that regulate the subcellular localization patterns of the Cln2p and Cln3p cyclins may influence cyclin function and specificity (discussed below).
G1 cyclin localization is significant for cyclin function and specificity.
There are at least two ways in which subcellular localization of the G1 cyclins might be important for cyclin function. First, a cyclin might need to be in its usual location to carry out its normal function, so that mislocalization due to removal of localization signals and/or addition of mistargeting signals could reduce or eliminate its normal function. We see several examples of this in the present work. The strongly mistargeted NES-Cln3-Δ22p lacks biological activity in the cln1 cln2 cln3 rescue and cell volume assays (Fig. 11 and 12). (We assume that the function of Cln3-Δ22p is due to residual nuclear localization of Cln3-Δ22p.) Deletion of BCK2 sensitizes the cell to Cln3p activity (38, 45). In a bck2 cln1 cln2 cln3 background, Cln3-Δ22p is reduced in rescue activity compared to that of wild-type Cln3p (Fig. 11). This defect is rescued by addition of the SV40 NLS, which partially restores nuclear localization of this mutant. Interestingly, addition of an NES to Cln2p strongly reduces some activities of Cln2p that may specifically require transient Cln2p nuclear residence (45).
Previous attempts to detect functional defects of mislocalized Cln3p (Cln3-1p) in the absence of additional CLN genes (but in an otherwise wild-type strain) were unsuccessful (45). This is most likely due to additional effects of the larger CLN3-1 deletion compared to the 22-amino-acid C-terminal deletion in cln3-Δ22. Cln3-1p exhibits stronger nuclear localization than Cln3-Δ22p and also accumulates to a much higher level than Cln3-Δ22p. These effects are likely due to the presence of other sequences affecting Cln3p localization in the C-terminal 177 amino acids and to the presence of additional functional PEST sequences in this region (13, 72, 81). The present results obtained with the Cln3-Δ22p mutant are significantly more clear, because the deletion is much more precise, being restricted to the NLS rather than removing a large segment of protein with major effects on Cln3p stability.
A second way in which subcellular localization patterns of the G1 cyclins might be important for cyclin function is that localization could keep a cyclin from performing a task normally performed by a different cyclin. In this case, mislocalization due to removal of localization signals and/or addition of mistargeting signals could result in an alteration in the functional spectrum of a cyclin. Removal of Cln3p from the nucleus by deletion of its NLS results in Cln3p acquiring functional activities that are normally restricted to the cytoplasmic Cln2p, such as rescue of a cln1 cln2 CLN3 pcl1 pcl2 strain (Fig. 11). Mislocalized Cln3p is not as efficient in these assays as Cln2p, indicating that additional factors beyond simple nuclear versus cytoplasmic localization may contribute to cyclin functional specificity.
Complementation between nuclear Cln3p and artificially cytoplasmic Cln (either NES-Cln2p or NES-Cln3p-Δ22) (Fig. 11) provides strong evidence that cyclin functional specificity can be altered just by changing localization.
The Cln2 phosphorylation mutant, Cln2-4t3sp, has been reported to result in reduced mating factor sensitivity and accelerated cell cycle initiation (36). These defects may result from the stabilization of Cln2p, as suggested previously; but the defects could also result from the mislocalization of Cln2-4t3sp to the nucleus or from a combination of these factors. We showed previously that addition of an NES (but not an mnes) to Cln2p significantly reduced its biological activity (45). In this study, we find a transient population of Cln2p that is hypophosphorylated and that fails to exhibit nuclear depletion. These observations, combined with the observation that elimination of Cln2p phosphorylation resulted in a nuclear population of Cln2p (Fig. 1 and 2), suggest the hypothesis that Cln2p is able to reside in the nucleus when unphosphorylated, but not when phosphorylated, and that nuclear residence of Cln2p is important for its function.
The finding that hypophosphorylated Cln2p no longer enters the nucleus in the presence of metabolic poison indicates that the nuclear import of Cln2p is active. No evidence of active Cln2p export was observed in our studies. Our data establish a role for Cdk activity in the subcellular localization pattern of a G1 cyclin, Cln2p. Previous work with higher eukaryotic cells indicates that Cdk activity might regulate the localization of the mitotic cyclin B1 (28, 42). Phosphorylation of cyclin B1 at serine 113, resulting in nuclear localization, has the opposite effect on localization from phosphorylation of Cln2p, which results in cytoplasmic localization. In the case of cyclin B1, phosphorylation results in the inhibition of export (83). It has also been reported that nuclear import of cyclin B1 is positively regulated by phosphorylation at multiple sites in cyclin B1 (25, 83), again, having an opposite effect from that observed with Cln2p in our study. The phosphorylation-dependent localization of cyclin B1 also most likely occurs independent of physical interactions with cyclin F, since they involve phosphorylation events within the defined cytoplasmic retention sequence (CRS) domain of cyclin B1 (35). Interestingly, a Ran-independent mechanism for cyclin B1 nuclear import has been described. In this case, the Ran-independent transport of cyclin B1 required importin β (71), and seems to function independent of phosphorylation-dependent import (25, 83). We are currently addressing whether similar requirements exist for Cln2p nuclear import. Other examples of Ran-independent, energy-dependent transport have been characterized, including spliced mRNA (9), the U1A and U1B spliceosome proteins (26), the inhibitor of κBα (IκBα) (60), β-catenin (60), and histone 2b (76). The data with Cln2p are reminiscent of the data obtained with cyclin D1, in which glycogen synthase kinase 3β phosphorylation results in cytoplasmic localization and subsequent degradation of cyclin D1 (18). However, in the case of cyclin D1, phosphorylation results in the physical interaction with the Crm1 exportin, leading to active export (2). We do not believe that a similar mechanism exists for Cln2p localization, since we found no evidence of active export in our experiments, and the localization pattern of Cln2p is not changed in a mutant crm1 strain (Fig. 5) (data not shown). Additionally, in the case of Cln2p, the phosphorylation most likely occurs through the bound Cdk. Our data are consistent with the idea that phosphorylation of Cln2p forces the Cln2-Cdc28p complex into a regulated localization pattern, through a novel nonclassical pathway, altering the accessibility of critical substrates for active Cdk.
ACKNOWLEDGMENTS
We thank Mike Tyers, Stephan Lanker, Anita Corbett, Mike Rout, Karsten Weis, and Lucy Pemberton for generously providing plasmids and strains. We thank Alison North, Matt Jacobson, Yelena Fishilevich, Martha Klovstad, Caihong Li, Ben Timney, Ralph Waesch, and David Miller for helpful discussions and technical assistance.
This work was supported by NIH grant GM47238 to F.R.C. M.E.M. was supported by NRSA GM18782. This study was made possible in part by funds granted by the Charles H. Revson Foundation.
REFERENCES
- 1.Aitchison J D, Rout M P, Marelli M, Blobel G, Wozniak R W. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alt J R, Cleveland J L, Hannink M, Diehl J A. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- 4.Aris J P, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardin A J, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 6.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 7.Benton B K, Tinkelenberg A H, Jean D, Plump S D, Cross F R. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 1993;12:5267–5275. doi: 10.1002/j.1460-2075.1993.tb06222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K C, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson J J. Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell. 2000;11:369–391. doi: 10.1091/mbc.11.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clouse K N, Luo M, Zhou Z, Reed R. A Ran-independent pathway for export of spliced mRNA. Nat Cell Biol. 2000;3:97–99. doi: 10.1038/35050625. [DOI] [PubMed] [Google Scholar]
- 10.Coelho C M, Leevers S J. Do growth and cell division rates determine cell size in multicellular organisms? J Cell Sci. 2000;113:2927–2934. doi: 10.1242/jcs.113.17.2927. [DOI] [PubMed] [Google Scholar]
- 11.Cross F R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross F R. Starting the cell cycle: what's the point? Curr Opin Cell Biol. 1995;7:790–797. doi: 10.1016/0955-0674(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 13.Cross F R, Blake C M. The yeast Cln3 protein is an unstable activator of Cdc28. Mol Cell Biol. 1993;13:3266–3271. doi: 10.1128/mcb.13.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross F R, Yuste-Rojas M, Gray S, Jacobson M D. Specialization and targeting of B-type cyclins. Mol Cell. 1999;4:11–19. doi: 10.1016/s1097-2765(00)80183-5. [DOI] [PubMed] [Google Scholar]
- 15.Cvrcková F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deshaies R J, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Como C J, Chang H, Arndt K T. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diehl J A, Cheng M, Roussel M F, Sherr C J. Glycogen synthase kinase 3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;15:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dirick L, Böhm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Draviam V M, Orrechia S, Lowe M, Pardi R, Pines J. The localization of human cyclins B1 and B2 determines their substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J Cell Biol. 2001;152:1–15. doi: 10.1083/jcb.152.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein C B, Cross F R. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol Cell Biol. 1994;14:2041–2047. doi: 10.1128/mcb.14.3.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu H, Subramanian R, Masters S C. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 23.Futcher B. Cyclins and the wiring of the yeast cell cycle. Yeast. 1996;12:1635–1646. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1635::AID-YEA83%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 24.Gietz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 25.Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr Biol. 1999;9:680–689. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- 26.Hetzer M, Mattaj I W. An ATP-dependent, Ran-independent mechanism for nuclear import of the U1A and U2B" spliceosome proteins. J Cell Biol. 2000;148:293–303. doi: 10.1083/jcb.148.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang K N, Odinsky S A, Cross F R. Structure-function analysis of the Saccharomyces cerevisiae G1 cyclin Cln2. Mol Cell Biol. 1997;17:4654–4666. doi: 10.1128/mcb.17.8.4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izumi T, Maller J L. Phosphorylation of Xenopus cyclins B1 and B2 is not required for cell cycle transitions. Mol Cell Biol. 1991;11:3860–3867. doi: 10.1128/mcb.11.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackman M, Firth M, Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson M D, Gray S, Yuste-Rojas M, Cross F R. Testing cyclin specificity in the exit from mitosis. Mol Cell Biol. 2000;20:4483–4493. doi: 10.1128/mcb.20.13.4483-4493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaspersen S L, Charles J F, Morgan D O. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 32.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 33.Kishi T, Yamao F. An essential function of Grr1 for the degradation of Cln2 is to act as a binding core that links Cln2 to Skp1. J Cell Sci. 1998;111:3655–3661. doi: 10.1242/jcs.111.24.3655. [DOI] [PubMed] [Google Scholar]
- 34.Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 35.Kong M, Barnes E A, Ollendorff V, Donoghue D J. Cyclin F regulates the nuclear localization of cyclin B1 through a cyclin-cyclin interaction. EMBO J. 2000;19:1378–1388. doi: 10.1093/emboj/19.6.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 37.Lenburg M E, O'Shea E K. Evidence for a morphogenic function of the Saccharomyces cerevisiae Pho85 cyclin-dependent kinase. Genetics. 2001;157:39–51. doi: 10.1093/genetics/157.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine K, Huang K, Cross F R. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine K, Oehlen L J W M, Cross F R. Isolation and characterization of new alleles of the cyclin-dependent kinase gene CDC28 with cyclin-specific functional and biochemical defects. Mol Cell Biol. 1998;18:290–302. doi: 10.1128/mcb.18.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Meyer A N, Donoghue D. Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1997;94:502–507. doi: 10.1073/pnas.94.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- 44.Melchior F, Paschal B, Evans E, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller M E, Cross F R. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:542–555. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore J D, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 48.Moore S A. Cell size specific binding of the fluorescent dye calcofluor to budding yeast. Biochim Biophys Acta. 1990;1035:206–213. doi: 10.1016/0304-4165(90)90118-g. [DOI] [PubMed] [Google Scholar]
- 49.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 50.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 51.Moroianu J. Nuclear import and export: transport factors, mechanisms and regulation. Crit Rev Eukaryot Gene Expr. 1999;9:89–106. doi: 10.1615/critreveukargeneexpr.v9.i2.10. [DOI] [PubMed] [Google Scholar]
- 52.Muslin A J, Tanner J W, Allen P M. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 53.Nash R, Tokiwa G, Anand S, Erickson K, Futcher A B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 55.Nasmyth K, Dirick L. The role of SW14 and SW16 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 56.Nigg E A. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;16:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 57.Ogas J, Andrews B J, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 58.Pines J, Hunter T. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richardson H E, Wittenberg C, Cross F, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 60.Sachdev A, Bagchi S, Zhang D D, Mings A C, Hannink M. Nuclear import of IκBα is accomplished by a Ran-independent transport pathway. Mol Cell Biol. 2000;20:1571–1582. doi: 10.1128/mcb.20.5.1571-1582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schlendstedt G, Hurt E, Doyle V, Silver P A. Reconstitution of nuclear-protein transport with semi-intact yeast cells. J Cell Biol. 1993;123:785–798. doi: 10.1083/jcb.123.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider B L, Patton E E, Lanker S, Mendenhall M D, Wittenburg C, Futcher B, Tyers M. Yeast G1 cyclins are unstable in G1 phase. Nature. 1998;395:86–89. doi: 10.1038/25774. [DOI] [PubMed] [Google Scholar]
- 63.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 64.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W, Jang J, Shevchenko A, Charbonneau H, Deshaies R J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 65.Shulga N, Roberts P, Gu Z, Spitz L, Tabb M M, Nomura M, Goldfarb D. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sikorski R S, Hieter P. A system of shuttle vectors and yeast strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 68.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 69.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1(Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 70.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 71.Takizawa C G, Weis K, Morgan D. Ran-independent nuclear import of cyclin B1-Cdc2 by importin B. Proc Natl Acad Sci USA. 1999;96:7938–7943. doi: 10.1073/pnas.96.14.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Visintin R, Craig K, Hwang E S, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 74.Visintin R, Hwang E S, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 75.Wach A, Brachet C, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 76.Wiechens N, Fagotto F. CRM1- and Ran-independent nuclear export of β-catenin. Curr Biol. 2001;11:18–27. doi: 10.1016/s0960-9822(00)00045-2. [DOI] [PubMed] [Google Scholar]
- 77.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 78.Wittenberg C, Richardson S L, Reed S I. Subcellular localization of a protein kinase required for cell cycle initiation in Saccharomyces cerevisiae: evidence for an association between the CDC28 gene product and the insoluble cytoplasmic matrix. J Cell Biol. 1987;105:1527–1538. doi: 10.1083/jcb.105.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wong D H, Corbett A H, Kent H M, Stewart M, Silver P. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin S J, Smerdon S J, Cantely L C. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1996;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 81.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–740. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang J, Bardes E S, Moore J D, Brennan J, Powers M A, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang J, Song H, Walsh S, Bardes E S G, Kornbluth S. Combinatorial control of cyclin B1. Nuclear trafficking through phosphorylation at multiple sites. J Biol Chem. 2001;276:3604–3609. doi: 10.1074/jbc.M008151200. [DOI] [PubMed] [Google Scholar]