Abstract
The CCAAT displacement protein (CDP), the homologue of the Drosophila melanogaster Cut protein, contains four DNA binding domains that function in pairs. Cooperation between Cut repeat 3 and the Cut homeodomain allows stable DNA binding to the ATCGAT motif, an activity previously shown to be upregulated in S phase. Here we showed that the full-length CDP/Cut protein is incapable of stable DNA binding and that the ATCGAT binding activity present in cells involves a 110-kDa carboxy-terminal peptide of CDP/Cut. A vector expressing CDP/Cut with Myc and hemagglutinin epitope tags at either end generated N- and C-terminal products of 90 and 110 kDa, suggesting that proteolytic cleavage was involved. In vivo pulse/chase labeling experiments confirmed that the 110-kDa protein was derived from the full-length CDP/Cut protein. Proteolytic processing was weak or not detectable in G0 and G1 but increased in populations of cells enriched in S phase, and the appearance of the 110-kDa protein coincided with the increase in ATCGAT DNA binding. Interestingly, the amino-truncated and the full-length CDP/Cut isoforms exhibited different transcriptional properties in a reporter assay. We conclude that proteolytic processing of CDP/Cut at the G1/S transition generates a CDP/Cut isoform with distinct DNA binding and transcriptional activities. These findings, together with the cleavage of the Scc1 protein at mitosis, suggest that site-specific proteolysis may play an important role in the regulation of cell cycle progression.
Genetic studies with Drosophila melanogaster indicated that cut plays an important role in determining cell-type specificity in several tissues (reviewed in reference 35). Defects caused by cut mutations appear to result from the fact that some cells have enrolled in the wrong developmental program (6, 8–10, 12, 13, 24, 29–31). In higher vertebrates, Cut proteins were originally characterized as CCAAT displacement proteins and have been termed CDP (CCAAT displacement protein), Clox (Cut-like homeobox), Cux-1 (Cut homeobox), and CDP-2 (2, 36, 43, 50).
CDP/Cut proteins are unique in that they contain four DNA binding domains: the Cut homeodomain (HD) and the three Cut repeats (CR1, CR2, and CR3), which are three regions of ∼70 amino acids that share from 52 to 63% amino acid identity with each other (1, 3, 7, 20, 21, 36). Using a panel of fusion proteins, we have shown that a Cut repeat cannot bind to DNA as a monomer but that certain combinations of domains exhibit high DNA binding affinity: CR1CR2, CR3HD, CR1HD, and CR2HD (33). CR1CR2 displayed rapid on and off rates and bound preferably to two CAAT or CGAT sites, organized as direct or inverted repeats. Accordingly, only CR1CR2 was able to bind to the CCAAT sequence, and its affinity was increased by the presence of a CA/GAT site at close proximity. Moreover, CR1CR2, but not CR3HD, was able to displace the NF-Y factor. Thus, the CCAAT-displacement activity involves Cut repeats 1 and 2 but not the Cut homeodomain. In contrast, various combinations involving the Cut homeodomain and one Cut repeat were found to make a stable interaction with ATNNAT motifs. In particular, CR3HD stably bound to DNA, preferably with the ATCGAT sequence. CDP/Cut proteins were generally found to function as transcriptional repressors (2, 17, 27, 28, 32, 38, 40, 43). In addition, CDP/Cut may also be able to participate in gene activation on specific promoters (45, 50). Indeed, CDP/Cut was found to be a component of the promoter complex HiNF-D, which is believed to contribute to the transcriptional induction of several histone genes at the G1/S phase transition of the cell cycle (5, 44, 45). In this instance, transcriptional activation could not be demonstrated in cotransfection assays, but it was proposed that the regulatory effect of CDP/Cut on transcription might vary depending on the proteins with which it interacts (45). In particular, supershift assays using antibodies against pRb-related proteins suggested that CDP/Cut might alternatively interact with pRb or p107 on different promoters (45).
We have recently obtained evidence that CDP/Cut may play an important role in cell cycle progression (16). Using the ATCGAT site as a probe, little CDP/Cut DNA binding was detected in G0 and early G1, unless cell extracts were previously treated with alkaline phosphatase. In contrast, strong DNA binding was observed in S phase. This was shown to result, at least in part, from dephosphorylation of the Cut HD by the Cdc25A phosphatase. Thus, apart from cdk2, CDP/Cut is the only known substrate for Cdc25A, a phosphatase that is required for G1/S transition and whose overexpression triggers S phase entrance (11, 25, 46). The increase in CDP/Cut activity coincided with a decrease in p21WAF1/CIP1/SDI1 mRNA. In cotransfection experiments, CDP/Cut repressed a reporter controlled by the p21 promoter, whereas an antisense CDP/Cut vector restored p21 expression in S phase. Moreover, p21 expression was repressed equally well by either Cdc25A or CDP/Cut. Altogether, these results led us to propose a model whereby Cdc25A activates the CDP/Cut repressor that subsequently down-regulates transcription of p21 in S phase.
Site-specific proteolysis has emerged as an important regulatory mechanism that plays a role in a number of cellular processes including transduction within the Notch and Hedgehog signaling pathways, sister-chromatid separation at anaphase, and generation of the amyloidogenic peptide in Alzheimer's disease (4, 15, 19, 42). In the field of transcription, proteolytic processing of transcription factors was shown to redirect the localization of these proteins or to generate specific isoforms with distinct biochemical properties (4, 14, 18, 22, 47, 49).
The present study was triggered by a surprising result. We found that a purified CDP/Cut protein exhibited DNA binding properties similar to that of CR1CR2, suggesting that CR3HD was weakly or not active in the context of the full-length protein. Moreover, in mammalian cells the ATCGAT binding activity that is upregulated in S phase could be supershifted with C-terminal-specific but not with N-terminal-specific anti-CDP/Cut antibodies. Further investigation led to the discovery that the full-length CDP/Cut protein is proteolytically cleaved to generate N-terminal and C-terminal peptides of 90 and 110 kDa and that processing is regulated during cell cycle progression. Interestingly, the full-length and 110-kDa CDP/Cut isoforms displayed different effects in a reporter assay, suggesting that processing of CDP/Cut serves to generate an isoform with distinct transcriptional properties.
MATERIALS AND METHODS
Plasmid construction.
For expression of the full-length CDP/Cut protein in SF9 insect cells, nucleotides (nt) 27 to 5101 from the human CDP/Cut cDNA (GenBank) accession no. M74099) was inserted into pBlueBac His2b (InVitrogen). The resulting plasmid was cotransfected with a helper plasmid to obtain baculoviruses expressing CDP/Cut with a histidine tag at the amino terminus. The DNA polymerase alpha-luciferase reporter construct was prepared as follows. PCR amplification was performed to obtain a fragment of genomic DNA containing nt 56 to 1657 of the human DNA polymerase alpha gene 5′ end (GenBank no. M64481). The primer at the 5′ and 3′ ends included EcoRI and BamHI sites, respectively, to allow cloning into the pBluescript KS vector (Stratagene). An EcoRV-NcoI fragment, including nt 56 to 1621 of the DNA polymerase alpha gene, was then subcloned into the corresponding sites of the luciferase reporter vector, pGL3 (Promega). The resulting plasmid contains sequences from −1517 to +49 relative to the transcription initiation site of the DNA polymerase alpha gene.
Expression and purification of CDP/Cut fusion proteins.
The his-tagged CR1CR2, CR3HD, and full-length CDP/Cut have been described previously (33). ET-15b-derived vector was introduced into the BL21(DE3) strain of Escherichia coli and induced with isopropyl-β-d-thiogalactopyranoside (IPTG). SF9 insect cells were infected with baculovirus encoding his-CDP/Cut and were incubated for 3 days. The fusion proteins were purified by affinity chromatography using procedures provided by the suppliers.
Cell culture and synchronization.
NIH 3T3, HeLa cells, and HS578T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 293 cells, in DMEM with 10% horse serum. To obtain cells in G0 phase using the serum starvation and stimulation method, NIH 3T3 cells were maintained for 3 days in DMEM plus 0.4% FBS (serum 0 h). The medium was then changed to DMEM plus 10% FBS and cells were harvested either 3 h later to obtain cells in early G1 (serum 3 h), 10 h later for mid-G1, or 18 h later for S phase. Synchronization in G1/S was performed using the double thymidine procedure (39). Cells were cultured overnight in DMEM plus 10% FBS supplemented with 2 mM thymidine, washed the next day, cultured for 10 h in DMEM plus 10% FBS, and finally further incubated overnight in the presence of 2 mM thymidine (thymidine 0 h). To allow cells to progress in the cell cycle, the medium was replaced with DMEM plus 10% FBS and cells were harvested 3 h later (thymidine 3 h). Fluorescence-activated cell sorter (FACS) analysis was performed as previously described (16)
Preparation of nuclear extracts.
Nuclear extracts were prepared according to the procedure of Lee et al., except that nuclei were obtained by submitting cells to 3 freeze-thaw cycles in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT] (26). Nuclei were then resuspended in buffer C (20 mM HEPES [pH 7.9], 25% glycerol, 1.5mM MgCl2, 420 mM NaCl2, 0.2 mM EDTA) and incubated at 4°C for 30 min. After 15 min of centrifugation, the supernatant was collected. Buffers A and C were supplemented with protease inhibitor mix tablets purchased from Roche. Total extracts were prepared by applying buffer X (50 mM HEPES [pH 7.9], 0.4 M KCl, 4 mM NaF, 4 mM Na3VO4, 0.2 mM EGTA, 0.2 mM EDTA, 0.1% NP-40, 10% glycerol, 0.5 mM DTT, protease inhibitor mix tablet from Roche) to a monolayer plate. After 10 min of incubation on ice, the resulting slurry was centrifuged for 15 min at 4°C and the supernatant was collected.
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed with 2 to 5 μg of nuclear extract from mammalian cells. The samples were incubated at room temperature for 5 min in a final volume of 30 μl of 25 mM NaCl, 10 mM Tris (pH 7.5), 1 mM MgCl2, 5 mM EDTA (pH 8.0), 5% of glycerol, and 1 mM DTT, with 30 ng of poly(dI-dC) and 30 μg of bovine serum albumin as nonspecific competitors. End-labeled double-stranded oligonucleotides (∼10 pg) were added and further incubated for 15 min at room temperature. Samples were loaded on a 5 or 4% polyacrylamide gel (30:1) and separated by electrophoresis at 8 V/cm in 0.5× Tris borate-EDTA. Gels were dried and visualized by autoradiography.
Oligonucleotides.
The sequences of oligonucleotides used in this study are as follows: CGATATCGAT (universal CDP/Cut consensus binding site), TCGAGACGATATCGATAAGCTTCTTTTC; ATCGAT, TCGAGACGGTATCGATAAGCTTCTTTTC. Underlining and boldface type indicate the recognition sequence within the oligonucleotides. CGAT...CGAT is the binding site for CR1CR2, while ATCGAT is the binding site for CR3HD.
CDP/Cut antibodies and Western blot analysis.
To generate polyclonal antibodies against various regions of CDP/Cut (1,505 amino acids [aa]), rabbits were injected with 500 μg of purified bacterial fusion protein containing various regions of CDP/Cut in Freund's complete adjuvant. αN and α861 were raised against aa 227 to 1003 and aa 861 to 936, respectively. The animals were boosted twice with 250 μg of protein, and serum was collected 10 days after the last boost. Polyclonal antibodies were purified by affinity chromatography. The serum was passed through two glutathione S-transferase (GST) affinity columns and the flowthrough was then applied to a GST-CDP/Cut affinity column to isolate antibodies against CDP/Cut. To generate N-term antibodies, the N serum was immunodepleted by being passed through a column containing GST-CDP/Cut 861-936 and GST-CDP/Cut 940-1036. For Western blot analysis, protein extracts were recovered as described above and separated by electrophoresis on 8% (see Fig. 2) or 6% polyacrylamide gels. Western blot analysis with αCut 861 and αCut N term was performed as previously described (16). Western blot analysis for Myc and hemagglutinin (HA) epitopes were done with minor modification. For the primary antibody incubation, αMyc or αHA antibodies were incubated with the membrane in TBST (10 mM Tris [pH 8] 150 mM NaCl, 0.1% Tween) for 1 h at room temperature. After four 10-min washes with TBST, secondary antibodies were added to membrane in TBST and incubated for 45 min at room temperature. Following four 10-min washes with TBST, proteins were visualized with the ECL system from Amersham.
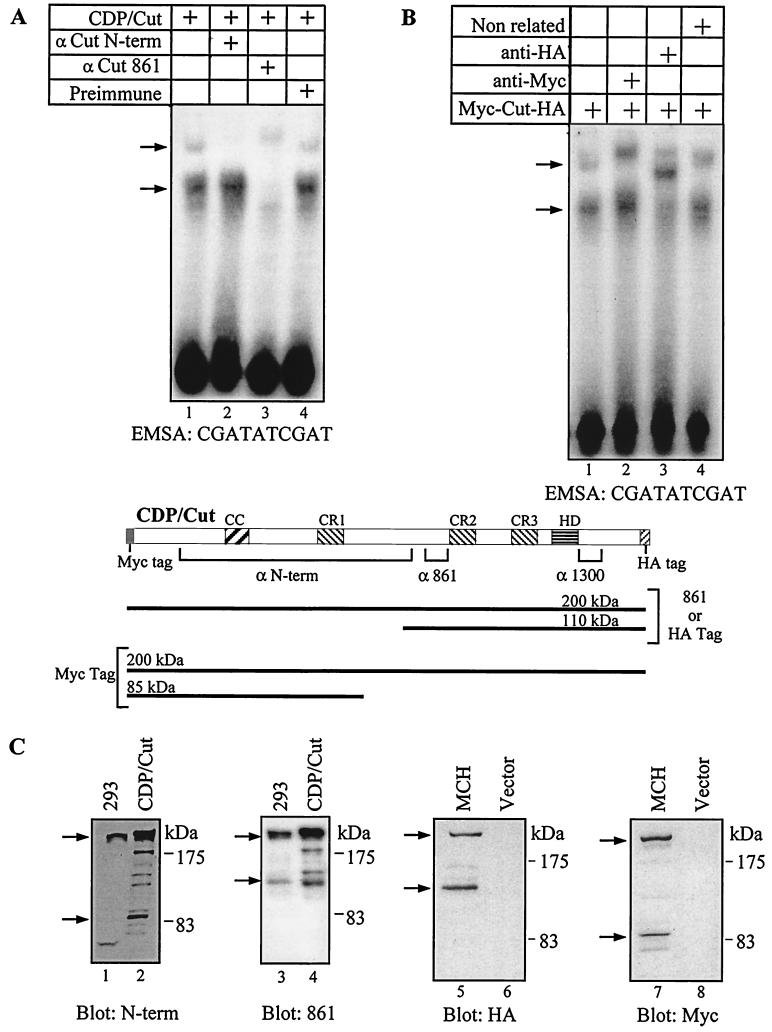
FIG. 2.
Two distinct retarded complexes are generated by CDP/Cut proteins, only one of which is supershifted by antibodies raised against the N-terminal region of the protein. (A and B) NIH 3T3 cells were transfected with a vector expressing the CDP/Cut protein (A) or Myc-Cut-HA, a CDP/Cut protein with Myc and HA epitope tags at its amino- and carboxy-termini, respectively (B). Nuclear extracts were prepared and analyzed in EMSA with oligonucleotides containing a universal CDP/Cut consensus-binding site (CGATATCGAT). The DNA and proteins were incubated either with no antibody (lane 1) or with the indicated antibodies (lanes 2 to 4). The preimmune serum is that for anti-861, the nonrelated antibody is a goat anti-rabbit immunoglobulin G. The arrows indicate the positions of the retarded complexes. Note that the lower retarded complex can be supershifted with anti-861 (A) and anti-HA (B) antibodies but not with anti-N-term (A) or anti-Myc (B) antibodies. A diagram of the protein is shown indicating the evolutionarily conserved domains and the regions recognized by the respective antibodies (CC, coiled-coil). (C) Nuclear extracts from untransfected 293 cells and transfected NIH 3T3 cells (CDP/Cut, MCH, Vector) were separated by electrophoresis on 8% polyacrylamide gels and analyzed in Western blots with CDP/Cut N-term, CDP/Cut 861, HA, and Myc antibodies. Note that the relative abundance of the full-length CDP/Cut protein appears less than that in subsequent blots using 6% polyacrylamide (Fig. 3, 4, and 5). This is because transfer of the 200-kDa protein to the membrane is less efficient in 8% polyacrylamide.
DNA affinity chromatography.
A 5′ biotinylated oligonucleotide containing an ATCGAT site was annealed with its unlabeled complementary oligonucleotide and bound to streptavidine agarose beads for 1 h in EMSA reaction buffer. HeLa nuclear extract (100 μg) was precleared with streptavidine agarose beads for 1 h and added to streptavidine-biotin-DNA beads and incubated for 1 h in EMSA buffer. The DNA-protein beads complex was washed three times for 5 min with EMSA buffer at 4°C, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Western blot analysis.
Luciferase assay.
HS578T cells were plated at 105 cells per 22.1 wells. The next day, a total amount of 1.5 μg of DNA was transfected, including 0.5 μg of reporter DNA and increasing amounts of effector DNA using ExGen500 (MBI; Fermentas). Each transfection was carried in 6 separate wells. Cells were harvested 40 h later. Cells from three wells served to prepare separate cytoplasmic extracts for luciferase assay, and cells from three other wells serve to prepare total extracts for Western blot analysis. Luciferase assays were performed as previously described, with minor modifications (16). Because the internal control plasmid is itself often repressed by CDP/Cut, as a control for transfection efficiency the purified β-galactosidase protein (Sigma) was included in the transfection mix, as previously described (23). The luciferase activity was then normalized based on β-galactosidase activity.
RESULTS
The full-length CDP/Cut protein exhibits DNA binding kinetics similar to these of CR1CR2.
The CDP/Cut protein contains four DNA binding domains: CR1, CR2, CR3, and HD. We have previously shown that two DNA binding domains are required for efficient DNA binding, and that the two most efficient combinations, CR1CR2 and CR3HD, exhibit different DNA binding specificities and kinetics (33). Whereas CR1CR2 only transiently bound to sequences containing direct or inverted repeats of the CGAT or CAAT motifs, CR3HD made a stable interaction with the ATCGAT sequence. To analyze DNA binding by the full length CDP/Cut protein, we designed oligonucleotides that can be recognized both by CR1CR2 and CR3HD. Oligonucleotides containing the CGATATCGAT sequence were recognized efficiently by CR1CR2, since two CGAT direct repeats are present (CGATATCGAT), and by CR3HD, since the ATCGAT sequence is present (CGATATCGAT) (Fig. 1A). These oligonucleotides can thus serve as universal CDP/Cut consensus binding sites.
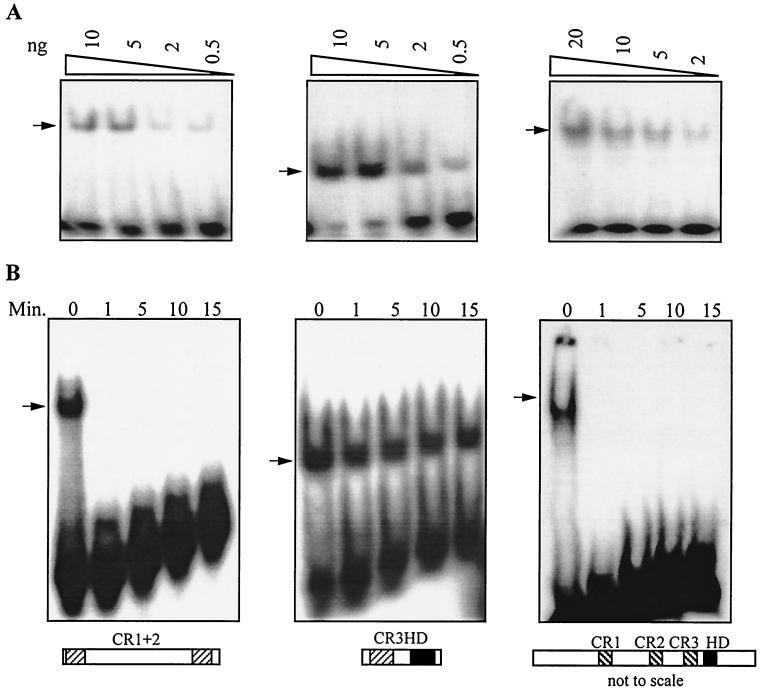
FIG. 1.
The full-length CDP/Cut protein exhibits DNA binding kinetics similar to that of CR1CR2. (A) DNA binding specificity of CR1CR2 (CR1+2), CR3HD, and CDP/Cut for CGATATCGAT. Decreasing amounts of the indicated fusion proteins were mixed with radiolabeled oligonucleotides containing the CGATATCGAT sequence at room temperature until equilibrium was reached. DNA-protein complexes were resolved on a nondenaturing polyacrylamide gel that was dried and autoradiographed. Note that oligonucleotides containing the CGATATCGAT sequence are recognized efficiently by CR1CR2, since two CGAT direct repeats are present (CGATATCGAT), and by CR3HD, since the ATCGAT sequence is present (CGATATCGAT). (B) DNA binding kinetics of CR1CR2, CR3HD, and CDP/Cut. One hundred nanograms of the indicated fusion protein was incubated with radiolabeled oligonucleotides containing the CGATATCGAT sequence at room temperature until the equilibrium was reached (15 min); 1,000-fold molar excess of unlabeled oligonucleotides was added, and at the indicated time points aliquots of the mixture were taken and analyzed by EMSA.
To investigate the DNA binding properties of the full length CDP/Cut protein, the protein was purified as a histidine-tagged fusion protein using a baculovirus expression system. When incubated with the CGATATCGAT probe, CDP/Cut generated one retarded complex (Fig. 1A). We next investigated the DNA binding kinetics of the full-length CDP/Cut protein. Like CR1CR2 but in contrast to CR3HD, CDP/Cut bound to DNA with a fast off rate (Fig. 1B). Thus, the full-length CDP/Cut protein binds only transiently to DNA, although it contains CR3HD that, on its own, makes a stable interaction with DNA. Altogether, these results suggest that the CR1CR2 bipartite DNA binding domain is active in the context of the full-length CDP/Cut protein, whereas CR3HD is weakly active or not active.
An amino-truncated 110-kDa CDP/Cut protein is responsible for the lower of two retarded complexes observed with the universal CDP/Cut consensus-binding site.
EMSAs were performed using the universal CDP/Cut consensus binding site, CGATATCGAT, and nuclear extracts from NIH 3T3 cells that had been transfected with a vector expressing the full-length CDP/Cut cDNA. Two main retarded complexes were observed (Fig. 2A, lane 1). Antibodies raised against different regions of the CDP/Cut protein were used in supershifting experiments in order to assess the nature of the protein(s) responsible for the retarded complexes. While the 861 antibodies supershifted both retarded complexes, the N-term antibodies supershifted only the highest complex, suggesting that the higher complex involved the full-length CDP/Cut protein and the lower complex, a CDP/Cut protein that is truncated at its amino-terminal end (Fig. 2A, lanes 2 and 3). In Western blot analyses with the N-term and 861 antibodies, several lower-molecular-weight species were observed in addition to the full-length 200-kDa CDP/Cut protein (Fig. 2C, lanes 1 to 4). Interestingly, a 110-kDa protein detected with the 861 antibody was also present in untransfected 293 cells, indicating that this smaller protein is normally expressed in untransfected human cells (Fig. 2C, lanes 3 and 4).
To confirm and extend the above results we prepared a vector, called MCH (Myc-Cut-HA), expressing the full-length CDP/Cut protein with Myc and HA epitope tags at its amino and carboxy termini, respectively. NIH 3T3 cells were transfected with the MCH vector and tested in EMSA with the universal CDP/Cut consensus-binding site. Again, two main retarded complexes were observed (Fig. 2B, lane 1). The Myc antibody supershifted only the highest retarded complex and the HA antibody supershifted both complexes, whereas an unrelated antibody had no effect (Fig. 2B, lanes 2 to 4). These results indicate that the protein responsible for the highest complex contains both epitope tags and therefore must be full length. In contrast, the protein generating the lower retarded complex contains an HA tag but not a Myc tag and must be truncated at its amino-terminal end. In Western blot analysis, the HA antibody revealed two main proteins: one with an apparent molecular size of 200 kDa corresponding to the full-length CDP/Cut and one of approximately 110 kDa. The Myc antibody also revealed two main proteins: one of 200 kDa and one of approximately 90 kDa. These results demonstrate that shorter CDP/Cut proteins can be generated from the full-length CDP/Cut coding sequences and that an amino-truncated CDP/Cut protein binds to the universal CDP/Cut binding site.
Two mechanisms could account for the generation of shorter CDP/Cut proteins. A proteolytic processing event may, at once, produce the 110-kDa protein seen with the HA antibody and produce the 90-kDa protein seen with the Myc antibody. Alternatively, the amino-truncated 110-kDa protein may be generated by translation at an internal start codon, while the carboxy-truncated 90-kDa protein would be generated by premature translation termination or proteolytic cleavage of the full-length protein. Experiments described below will address this issue. It is important to stress that several experiments were performed to ensure that shorter CDP/Cut isoforms were not simply generated as a result of proteolytic cleavage occurring postlysis. First, our lysis buffer contained an extensive cocktail of protease inhibitors (see Materials and Methods). Second, the shorter CDP/Cut isoforms were observed with a series of lysis buffers containing various concentrations of ionic and nonionic detergents (data not shown). Thirdly, treatment of cells with certain protease inhibitors for 4 h caused a reduction in the amount of short CDP/Cut isoforms (see Fig. 8 and data not shown). However, no reduction was observed when the same protease inhibitors were applied to cells only minutes prior to cell lysis (data not shown).
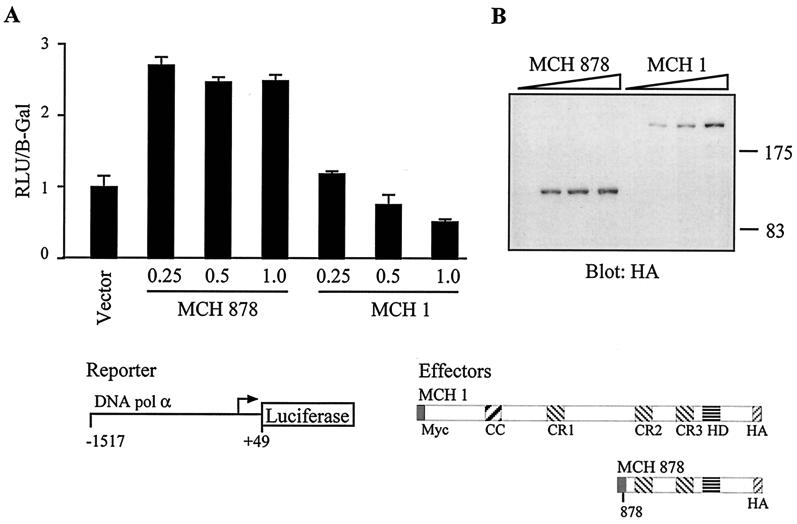
FIG. 8.
An amino-terminally truncated CDP/Cut isoform is capable of transcriptional activation. HS578T cells were cotransfected with the DNA polymerase alpha luciferase reporter construct and a vector expressing either nothing, full-length Myc-Cut-HA (MCH 1), or a MCH protein starting at aa 878 (MCH 878). (A) Cytoplasmic extracts were prepared and processed to measure luciferase activity. Means of 6 transfections are shown and the results are expressed as relative light units (RLU) normalized to β-galactosidase activity from an internal control. (B) Total extracts were prepared in parallel and analyzed in Western blots with anti-HA antibodies.
DNA affinity chromatography leads to the specific enrichment of a 110-kDa CDP/Cut protein.
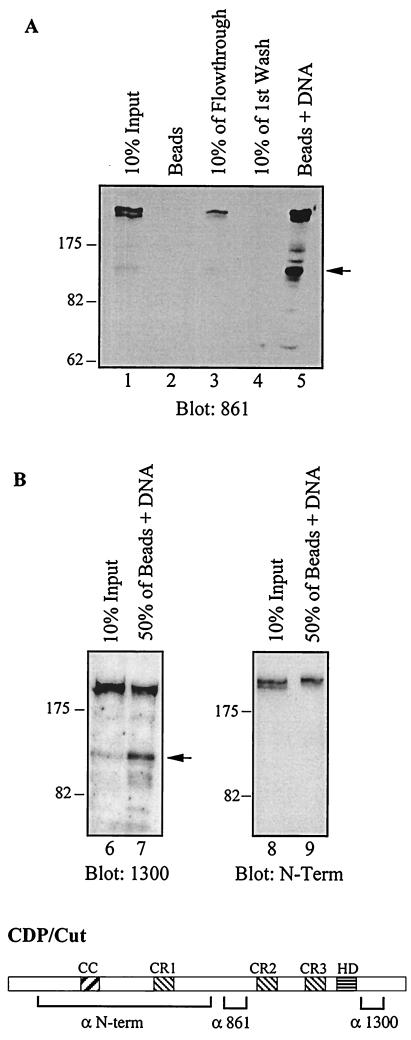
The results presented in Fig. 2 showed that the most abundant CDP/Cut protein species was the 200-kDa full-length protein. However, in EMSA the most abundant retarded complex was the fast-migrating one. Assuming that the fast-migrating complex involved a shorter CDP/Cut protein, these results could only be reconciled if the putative shorter species exhibited a higher DNA binding affinity or was able to bind more stably to DNA. To investigate this possibility, we performed DNA affinity chromatography and assessed the relative amounts of CDP/Cut protein species before and after the procedure (see Materials and Methods). In unfractionated nuclear extracts from HeLa cells, the 110-kDa species was clearly a minor species compared to the 200-kDa species (Fig. 3, lanes 1 and 6). However, DNA affinity chromatography led to the specific enrichment of the 110-kDa protein (Fig. 3, lanes 5 and 7). The 110-kDa Protein was recognized by the 861 and 1300 antibodies but not by the N-terminal CDP/Cut antibodies (see Fig. 2 for the position of their respective epitopes). These results clearly demonstrate the N-terminally truncated 110-kDa CDP/Cut protein can bind to DNA with high affinity.
FIG. 3.
DNA affinity chromatography with the ATCGAT sequence leads to the specific enrichment of a 110-kDa CDP/Cut protein. (A) Nuclear extracts from HeLa cells were subjected to affinity chroma- tography using as bait biotinylated oligonucleotides containing the ATCGAT sequence. Samples were then separated by electrophoresis on 6% polyacrylamide gels and analyzed by Western blotting with the anti-CDP/Cut 861 (lanes 1 to 5), 1300 (lanes 6 to 7), and N-term (lanes 8 and 9) antibodies.
S phase-specific proteolytic cleavage of CDP/Cut.
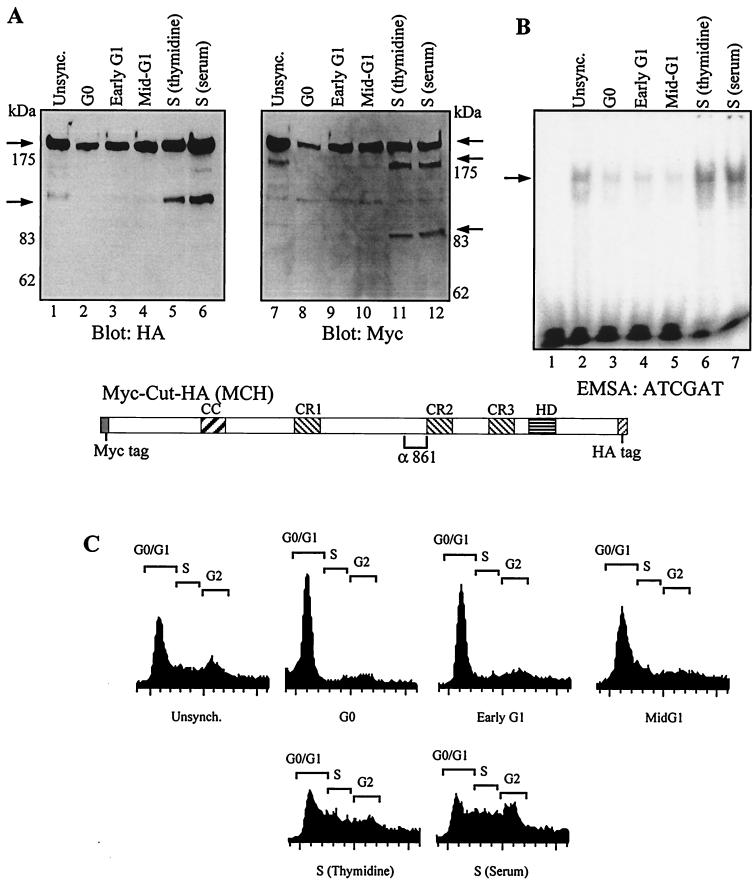
We have previously shown that CDP/Cut is regulated in a cell cycle-dependent manner and that DNA binding to the ATCGAT sequence increases as cells progress into S phase (16). We therefore verified whether expression of the shorter 110-kDa protein was also regulated during the cell cycle. NIH 3T3 cells were transfected with the MCH vector and then synchronized in G0, early G1, mid-G1, and S phases using serum starvation and restimulation or thymidine block (see Materials and Methods). Total extracts were prepared and analyzed by Western blot using the HA and Myc antibodies (Fig. 4A). The 110-kDa protein was not detected in the population of cells enriched in G0 and it was barely visible in early G1 and mid-G1, but it was strongly expressed in S phase using either method of synchronization. Interestingly, with the Myc antibody a protein of 90 kDa was detected in the two populations of cells enriched in S phase, suggesting that the 110-kDa HA-tagged protein and the 90-kDa Myc-tagged protein were generated at the same time, most likely by proteolytic cleavage. This hypothesis will be tested in pulse-chase labeling experiments (see Fig. 6). Interestingly, the Myc antibody also revealed a carboxy-terminally truncated protein that migrated with an apparent molecular size of 180 kDa. These findings raise the possibility that another processing event also takes place within the carboxy-terminal domain of the protein. This possibility is currently under investigation. EMSA were performed with the same extracts and oligonucleotides containing the ATCGAT consensus (Fig. 4B). These oligonucleotides were previously shown to be well recognized by CR3HD but weakly or not at all recognized by CR1CR2 (33). Indeed, only one retarded complex was detected, and its intensity was increased in S phase. Altogether these results demonstrate that the increase in ATCGAT binding activity at the G1/S transition coincides with the production of the N-terminally truncated 110-kDa CDP/Cut protein.
FIG. 4.
S phase-specific proteolytic cleavage of CDP/Cut. A cDNA encoding Myc-Cut-HA generates short proteins specifically in S phase. NIH 3T3 cells were transfected with a vector expressing Myc-Cut-HA, a CDP/Cut protein with Myc and HA epitope tags at its amino and carboxy termini, respectively. Cells were synchronized either by serum starvation and stimulation or by thymidine block, as described in Materials and Methods. Total extracts were prepared and analyzed in Western blots with anti-Myc and anti-HA antibodies (A) and in EMSA with an ATCGAT probe (B). Note that only a lower retarded complex is visible in EMSA with total extracts, whereas two complexes were observed with nuclear extracts (see Fig. 1). Cell cycle distribution was monitored by FACS analysis after staining of the DNA with propidium iodide (C). Unsync., unsynchronized.
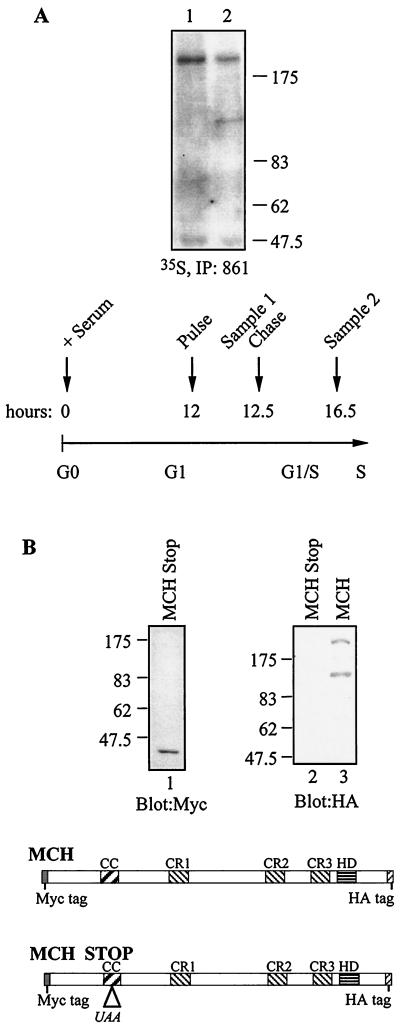
FIG. 6.
35S-pulse-chase labeling demonstrates that the 110-kDa amino-truncated protein derives from the 200-kDa full-length CDP/Cut protein. (A) NIH 3T3 cells were transfected with cDNA expressing Myc-Cut-HA in two 150 mm-diameter plates and synchronized in G0 by serum starvation for 72 h and then stimulated with fresh DMEM plus 10% FBS. The indicated times correspond to the time elapsed since release from the G0 block. At 12 h, the medium was replaced with complete labeling medium containing 35S-labeled methionine and cysteine. At 12 h and 30 min, total extracts were prepared from one plate of cells, while the medium was replaced with complete medium with cold methionine and cysteine in the second plate. At 16 h and 30 min, total extracts were prepared from the second plate. Samples 1 and 2 were immunoprecipitated with 861 antibody, resolved by PAGE, and revealed by autoradiography. (B) A cDNA expressing Myc-Cut-HA and Myc-Cut-HA with a stop codon inserted in the coiled-coil region were transfected in NIH 3T3 cells. Nuclear extracts were prepared and analyzed by Western blot with Myc and HA antibody.
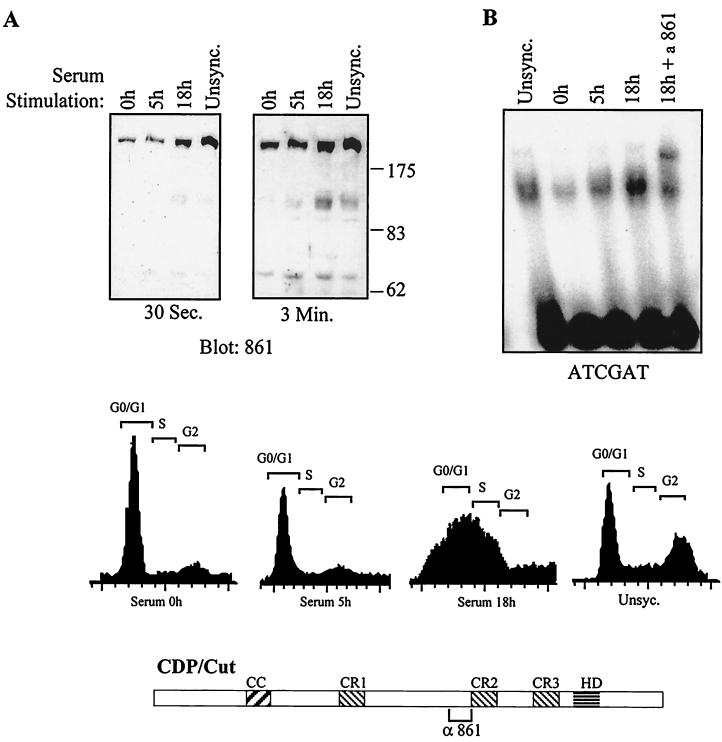
We then verified whether expression of the endogenous CDP/Cut protein was also regulated in a cell cycle-dependent manner. NIH 3T3 cells were synchronized by serum starvation and restimulation and were analyzed by Western blot using the 861 antibodies and in EMSA using the ATCGAT binding site. Interestingly, a doublet of bands at 110 kDa was observed at 18 h following serum stimulation in the population of cells enriched in S phase (Fig. 5A). Whether there are two processing events or one processing event followed by some posttranslational modification is not known at this point. It should be noted that only one 110-kDa band was detected in 293 cells using the same antibodies (see Fig. 2). Thus, there could be a difference in this respect between human and murine cells. Importantly, these novel bands correlate with the increase in the retarded complex seen in EMSA (Fig. 5B). In conclusion, amino-truncated CDP/Cut proteins are expressed predominantly in S phase, and the increase in their expression correlates with the increase in DNA binding to the ATCGAT sequence.
FIG. 5.
CDP/Cut proteins (110 kDa) are expressed in S phase. NIH 3T3 cells were synchronized in G0 by serum starvation for 72 h and then stimulated with fresh DMEM plus 10% FBS. The indicated times correspond to the time elapsed since release from the G0 block. Nuclear extracts were prepared and analyzed in Western blots with anti-Cut 861 antibodies (A) and in EMSA with the ATCGAT probe (B). Cell cycle distribution was monitored by FACS analysis after staining of the DNA with propidium iodide. Unsync., unsynchronized.
Pulse-chase labeling in vivo indicates that the 110-kDa protein derives from the 200-kDa full-length CDP/Cut protein.
The fact that the 110-kDa N-terminally truncated CDP/Cut protein was expressed concomitantly with a 90-kDa C-terminally truncated protein suggested that a proteolytic processing event was involved in the generation of the 110-kDa protein. To verify this hypothesis, we performed 35S-pulse-chase labeling in vivo and followed expression of the 200- and 110-kDa CDP/Cut proteins. NIH 3T3 cells were transfected with a vector expressing MCH and synchronized in G0 by serum starvation for 72 h. After serum starvation, cells were stimulated to reenter the cell cycle by replacing the medium for complete medium with 10% FBS. Twelve hours later, cells were incubated with complete labeling medium for 30 min. Total extracts were prepared from one plate (Fig. 6A, sample 1), while cells in the second plate were incubated with normal medium for an additional 4 h, at which time total extracts were prepared (Fig. 6A, sample 2). This scheme was chosen because previous cell synchronization experiments revealed that the 110-kDa protein was weakly expressed or not expressed in G1 phase but appeared later as cells progressed into S phase (Fig. 4 and 5). Following immunoprecipitation with the 861 antibody, the 110-kDa protein was not detected in sample 1, indicating that it is not synthesized in mid-G1 (Fig. 6A, lane 1). Yet the 110-kDa protein was clearly visible in cells that had been incubated an additional 4 h in the presence of cold medium (Fig. 6A, lane 2). Since radiolabeling took place from 12 h to 12.5 h and only full-length CDP/Cut was visible at 12.5 h, the presence of the 110-kDa protein at 16.5 h could only be accounted for by supposing that it was generated by cleavage of the full-length CDP/Cut protein. Moreover, as both samples were prepared in the same manner, the presence of the 110-kDa protein in one sample but not in the other further demonstrates that the proteolytic cleavage did not occur postlysis.
Another mechanism for the production of the N-terminally truncated 110-kDa protein would be translation initiation at an internal start site. To investigate this possibility, we modified the MCH vector by inserting a stop codon in the coiled-coil region upstream of CR1. The presence of a stop codon in the 5′ coding region should not affect expression of the 110-kDa protein if translation at a downstream start site is involved. The MCH-STOP vector produced a short, truncated protein that was easily detected with the Myc antibody, indicating that the stop codon was functional (Fig. 6B, lane 1). However, the 110-kDa protein could not be detected with the HA antibody (Fig. 6B, lane 2). We conclude that expression of the 110-kDa protein requires the prior production of the full-length 200-kDa CDP/Cut protein.
Altogether, these results exclude translation at an internal initiation site as the mechanism leading to the expression of the 110-kDa protein, and they are most consistent with the notion that this shorter protein is generated from the 200-kDa protein by proteolytic processing. Thus, translation in mid-G1 leads to the synthesis of a 200-kDa full-length CDP/Cut protein that is later processed into an amino-terminally truncated protein of 110 kDa with DNA binding properties similar to those of CR3HD.
Mapping of the sequences required for processing.
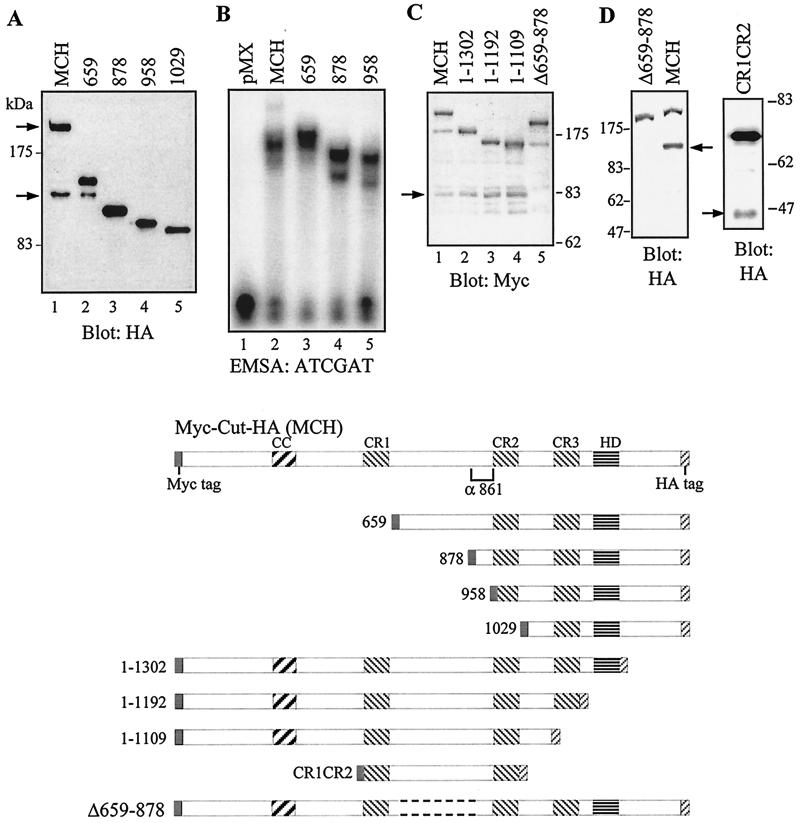
To map the sequences required for processing, we generated a series of vectors expressing amino-truncated Myc-Cut-HA proteins (Fig. 7A). The name of each vector corresponds to the first amino acid encoded by this vector. Only the full-length MCH and the 659 vectors produced the 110-kDa proteins (Fig. 7A). The 878, 958, and 1029 vectors generated shorter proteins. Thus, the site of cleavage is located between CR1 and CR2, more precisely between aa 659 and 878. In EMSA with the ATCGAT sequence, the 878 and 958 vectors generated retarded complexes that migrated faster than those produced by the full-length MCH vector. In contrast, 659 produced two retarded complexes, one comigrating with the MCH complex and one (more intense) that migrated more slowly. These results suggest that the full-length 659 protein, in contrast to the full-length MCH protein, is able to bind efficiently to the ATCGAT sequence. Since the full-length 659 protein is more abundant than its processed form (Fig. 7A, lane 2), it gives rise to the most abundant retarded complex in EMSA. We deduce from these results that the ATCGAT DNA binding activity is inhibited in the context of the full-length protein and that this inhibition can be released following amino-terminal truncation of the protein.
FIG. 7.
Proteolytic cleavage occurs downstream of CR1, between aa 659 and 878. NIH 3T3 cells were transfected with a series of vectors expressing amino- and carboxy-terminally truncated MCH proteins. Nuclear extracts were prepared and analyzed in Western blots with anti-HA or anti-Myc antibodies, as indicated, and in EMSA with the ATCGAT probe (B). A diagram of the proteins encoded by each construct is shown at the bottom. Note that only the full-length MCH and the 659 are processed into a shorter form. In EMSA, 659 generates two main retarded complexes, the higher of which migrates more slowly than that seen with MCH. This is because the nonprocessed 659 is more abundant than its processed derivative, and it is capable of stable binding to the ATCGAT probe.
We then generated a series of MCH vectors with progressive carboxy-terminal deletions. Since processing was observed with each construct, we conclude that the carboxy-terminal domain, the Cut HD, and the Cut repeat 3 can be removed without affecting processing (Fig. 7C). An internal deletion from aa 659 to 878 completely abolished processing (Fig. 7D), whereas a construct encoding only Cut repeats 1 and 2 and the linker between them generated a shorter protein of the expected molecular size (Fig. 7D). Altogether, our mapping data indicate that proteolytic cleavage occurs between aa 659 and 878 and does not require domains of the protein that are located far away from the cleavage site. Whether sequences between aa 659 and 878 are sufficient for cleavage is currently under investigation.
Processing changes the transcriptional properties of CDP/Cut.
Since the short isoform is capable of stable DNA binding, whereas the full-length protein only transiently binds to DNA, we envisaged that the two isoforms could exhibit different transcriptional properties. CDP/Cut has been characterized by us and other groups as a transcriptional repressor (2, 17, 27, 28, 32, 38, 40, 43). However, some results in the literature suggest that it may also participate in transcriptional activation (45, 50). This is also suggested by our own studies using a reporter construct containing the DNA polymerase alpha gene reporter (Truscott et al., in preparation). To compare the effect of the two CDP/Cut isoforms in a transient reporter assay, we needed to express the full-length protein in the absence of the processed isoform. In parallel studies, we found that little or no processing of CDP/Cut takes place in confluent cells. Therefore, in this experiment cells were plated at a higher cell density and, following transfection, were allowed to reach near confluence before harvesting them. As can be seen in Fig. 8B, no processed isoform was observed in cells transfected with the full-length Myc-Cut-Ha construct. In the reporter assay, we found that a recombinant protein corresponding to the p110 CDP/Cut isoform was able to stimulate expression from a DNA polymerase alpha gene reporter (Fig. 8A). In contrast, expression was decreased in the presence of full-length CDP/Cut (Fig. 8A). These results do not demonstrate that p110 directly activates the DNA polymerase alpha reporter. The results are also consistent with a model whereby the short CDP/Cut isoform would repress expression of a protein that, in turn, acts as a down-modulator of the DNA polymerase alpha gene promoter. Notwithstanding the exact mechanism-leading to an increase in reporter gene expression, these results suggest that an important consequence of CDP/Cut processing is to generate an isoform with distinct transcriptional properties.
DISCUSSION
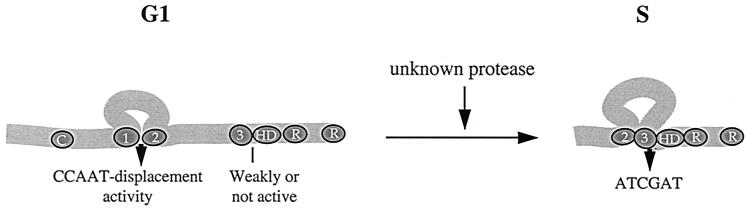
The CDP/Cut transcription factor contains two bipartite DNA binding domains, CR1CR2 and CR3HD, that exhibit distinct DNA binding activities. When expressed as fusion proteins, CR1CR2 bound with rapid on and off rates to DNA sequences containing direct or inverted repeats of the CAAT or CGAT motifs, whereas CR3HD made a stable interaction preferably with the ATCGAT sequence (33). To our surprise, we found that the full-length CDP/Cut protein could only make a transcient interaction with DNA (Fig. 1). In cells, the CDP/Cut protein that binds to the ATCGAT site was found to be an amino-terminally truncated 110-kDa protein (Fig. 2 and 3). Production of this protein was shown to involve the synthesis of the full-length protein followed by an S phase-specific proteolytic cleavage (Fig. 4 to 7). In reporter assays, an amino-terminally truncated CDP/Cut protein caused an increase, whereas the full-length CDP/Cut caused a decrease, in the expression from the DNA polymerase alpha gene promoter (Fig. 8). Altogether, our results demonstrated that proteolytic processing of CDP/Cut leads to the production of an isoform that displays distinct DNA binding and transcriptional activities (Fig. 9).
FIG. 9.
Modulation of CDP/Cut at the G1/S transition. In early G1, CDP/Cut exists as a full-length protein that cannot stably bind to the ATCGAT sequence. As cells reach S phase, CDP/Cut is proteolytically processed into an amino-truncated 110-kDa protein in which the CR3HD bipartite DNA binding domain is fully active.
The proteolytic processing event leading to the production of the 110-kDa CDP/Cut protein was found to occur at a much higher rate in populations of cells enriched in S phase, either by serum starvation and stimulation or by thymidine block, than in unsynchronized cells or in populations enriched in G0 or G1 (Fig. 4 and 5). This suggests that CDP/Cut processing begins as cells progress into S phase. It is not clear whether the triggering event at the end of G1 involves the protease or CDP/Cut. Two scenarios can be envisaged. The protease itself may be expressed or activated at the end of G1. Alternatively, we can envisage that the protease acts in a constitutive manner but that CDP/Cut needs to be posttranslationally modified in order to become a substrate of this protease.
Interestingly, S phase-specific proteolytic processing of the p27 cdk inhibitor has previously been reported (37). Processing was assumed to cause the inactivation of p27 by removing the cyclin interacting domain, CY. The cleavage of p27 and CDP/Cut suggests that proteolytic processing may be an important regulatory mechanism in the control of the G1/S transition. These results, along with the findings that sister-chromatid separation at anaphase is promoted by cleavage of the cohesin subunit Sec1, indicate that proteolytic processing may be involved in different phases of the cell cycle (42). Thus, two sorts of proteolysis appear to be needed to ensure cell cycle progression. The first is the destruction of several cell cycle regulators following ubiquitination by the Skp1–Cdc53/cullin–F-box protein complexes or the anaphase promoting complex/cyclosome (reviewed in reference 41). The latter is the precise cleavage of certain proteins to generate peptides with properties distinct from that of their precursors.
In most cases thus far reported the consequence of proteolytic processing was to redirect the localization of these proteins, in particular from a cytoplasmic site to the nucleus. For example, cleavage of SREBP and ATF6 in response to sterol deprivation and endoplasmic reticulum (ER) stress caused the release of their cleaved products from the ER membrane and their movement to the nucleus (22, 48). Cleavage of Cubitus interruptis in the absence of Hedgehog signaling enabled the translocation of the Cubitus interruptis 75-kDa isoform from microtubules in the cytoplasm to the nucleus where it functioned as a repressor (4). Processing of NF-κB p105 into NF-κB p50 was proposed to contribute to its nuclear translocation by separating the P50 N-terminal peptide from the ankyrin repeats which would retain it to a cytoplasmic anchor (18). In a few cases, specific proteolytic cleavage serves to generate novel isoforms with altered biochemical properties. Cleavage of full-length 38-kDa C/EBPβ leads to the production of a dominant-negative C/EBPβ isoform of 21 kDa, LIP (liver-enriched transcriptional inhibitory protein), that can bind to DNA but is devoid of transactivation potential (49). Autocatalytic processing of the C1 factor was found to generate a number of polypeptides that remained tightly associated together (47). In this case, processing resulted in the production of a protein complex with different properties, but whether processing also affected its localization remained unclear. In the case of CDP/Cut, processing did not seem to alter subcellular localization, as antibodies raised against N- or C-terminal peptides generated a strong signal primarily, if not exclusively, in the nucleus (data not shown). We cannot, of course, exclude the possibility that processing affects localization in a more subtle way within the nucleus itself. Clearly, however, processing of CDP/Cut generated an isoform with different DNA binding properties.
An amino-terminally truncated CDP/Cut protein, but not the full-length CDP/Cut protein, was able to stimulate expression from a reporter construct containing the DNA polymerase alpha gene promoter (Fig. 8). We do not know the mechanism of action of p110; it may directly activate transcription of the reporter, alternatively, it may repress expression of another repressor that binds to the DNA polymerase alpha promoter. Whatever its mechanism of action, we can speculate that the distinct transcriptional activities of the short and full-length CDP/Cut proteins may result from their differences in DNA binding activities. Whereas the full-length CDP/Cut protein could only make transient interaction with oligonucleotides containing the ATCGAT motif, shorter proteins containing CR3HD or CR2CR3HD were capable of stable DNA binding (Fig. 1 and reference 33). It is generally assumed that transcriptional activation requires stable interaction with the promoter. On the other hand, we and others have shown that CDP/Cut can repress by two mechanisms: active repression and competition for binding site occupancy (27, 28, 32). While transient DNA binding could cause repression by preventing the binding of an activator to an overlapping binding site, active repression would be expected to require stable DNA binding to the promoter in order to recruit a histone deacetylase. Future experiments should test the notion that stable DNA binding is required for active repression and transcriptional activation. Moreover, our results raise the interesting possibility that CDP/Cut isoforms may exhibit distinct biochemical activities on different promoters.
An important component of the transcriptional regulation at the G1/S transition involves the phosphorylation of retinoblastoma protein by G1 cyclin/CDKs and the concomitant release of E2F, which can then activate the transcription of genes whose products are required for DNA replication (reviewed in reference 34). However, some of the genes previously reported to be activated by E2F in cotransfection assays, including the DNA polymerase alpha gene, were recently found not to be primary targets of E2F, implying the involvement of other effectors acting downstream of, or in parallel with, E2F (46). Our results suggest that proteolytic cleavage of CDP/Cut in part may serve to activate the transactivation function of CDP/Cut (Fig. 8). These findings raise the possibility that the processed CDP/Cut isoform represents one of the G1/S effectors. Future experiments should aim to verify whether CDP/Cut function is required for the induction of S phase by E2F.
ACKNOWLEDGMENTS
A.N. is the recipient of a scholarship from the Fonds de la Recherche en Santé du Québec. N.M.S. is the recipient of a fellowship from Medical Research Council of Canada. This research was supported by grant no. 3497 from the National Cancer Institute of Canada and grant no. MT-11590 from the Canadian Institute of Health Research of Canada to A.N.
REFERENCES
- 1.Andres V, Chiara M D, Mahdavi V. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 1994;8:245–257. doi: 10.1101/gad.8.2.245. [DOI] [PubMed] [Google Scholar]
- 2.Andres V, Nadal-Ginard B, Mahdavi V. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development. 1992;116:321–334. doi: 10.1242/dev.116.2.321. [DOI] [PubMed] [Google Scholar]
- 3.Aufiero B, Neufeld E J, Orkin S H. Sequence-specific DNA binding of individual Cut repeats of the human CCAAT displacement/Cut homeodomain protein. Proc Natl Acad Sci USA. 1994;91:7757–7761. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aza-Blanc P, Ramirez-Weber F A, Laget M P, Schwartz C, Kornberg T B. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 5.Aziz F, Vanwijnen A J, Vanghan P S, Wu S J, Shakoori A R, Lian J B, Soprano K J, Stein J L, Stein G S. The integrated activities of Irf-2 (Hinf-M), Cdp/Cut (Hinf-D) and H4tf-2 (Hinf-P) regulate transcription of a cell cycle controlled human histone H4 gene - mechanistic differences between distinct H4 genes. Mol Biol Rep. 1998;25:1–12. doi: 10.1023/a:1006888731301. [DOI] [PubMed] [Google Scholar]
- 6.Blanc R. The production of wing scalloping in Drosophila melanogaster. Univ Calif Publ Zool. 1942;49:1–31. [Google Scholar]
- 7.Blochlinger K, Bodmer R, Jack J, Jan L Y, Jan Y N. Primary structure and expression of a product from cut, a locus involved in specifying sensory organ identity in Drosophila. Nature. 1988;333:629–635. doi: 10.1038/333629a0. [DOI] [PubMed] [Google Scholar]
- 8.Blochlinger K, Bodmer R, Jan L Y, Jan Y N. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 1990;4:1322–1331. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- 9.Blochlinger K, Jan L Y, Jan Y N. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development. 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- 10.Blochlinger K, Jan L Y, Jan Y N. Transformation of sensory organ identity by ectopic expression of Cut in Drosophila. Genes Dev. 1991;5:1124–1135. doi: 10.1101/gad.5.7.1124. [DOI] [PubMed] [Google Scholar]
- 11.Blomberg I, Hoffmann I. Ectopic expression of Cdc25A accelerates the G1/S transition and leads to premature activation of cyclin E- and cyclin A-dependent kinases. Mol Cell Biol. 1999;19:6183–6194. doi: 10.1128/mcb.19.9.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodmer R, Barbel S, Shepherd S, Jack J W, Jan L Y, Jan Y N. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell. 1987;51:293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 13.Braun W. The effect of puncture on the developing wing of several mutants of Drosophila melanogaster. J Exp Zool. 1942;84:325–350. [Google Scholar]
- 14.Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 15.Chan Y M, Jan Y N. Roles for proteolysis and trafficking in notch maturation and signal transduction. Cell. 1998;94:423–426. doi: 10.1016/s0092-8674(00)81583-4. [DOI] [PubMed] [Google Scholar]
- 16.Coqueret O, Berube G, Nepveu A. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates P21(Waf1/Cip1/Sdi1) in S phase. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dufort D, Nepveu A. The human cut homeodomain protein represses transcription from the c-myc promoter. Mol Cell Biol. 1994;14:4251–4257. doi: 10.1128/mcb.14.6.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan C M, Maniatis T. Generation of p50 subunit of NF-kappa B by processing of p105 through an ATP-dependent pathway. Nature. 1991;354:395–398. doi: 10.1038/354395a0. [DOI] [PubMed] [Google Scholar]
- 19.Gervais F G, Xu D, Robertson G S, Vaillancourt J P, Zhu Y, Huang J, LeBlanc A, Smith D, Rigby M, Shearman M S, Clarke E E, Zheng H, Van Der Ploeg L H, Ruffolo S C, Thornberry N A, Xanthoudakis S, Zamboni R J, Roy S, Nicholson D W. Involvement of caspases in proteolytic cleavage of Alzheimer's amyloidbeta precursor protein and amyloidogenic A beta peptide formation. Cell. 1999;97:395–406. doi: 10.1016/s0092-8674(00)80748-5. [DOI] [PubMed] [Google Scholar]
- 20.Harada R, Berube G, Tamplin O J, Denis-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol Cell Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada R, Dufort D, Denis-Larose C, Nepveu A. Conserved cut repeats in the human cut homeodomain protein function as DNA binding domains. J Biol Chem. 1994;269:2062–2067. [PubMed] [Google Scholar]
- 22.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Cell Biol. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howcroft T K, Kirshner S L, Singer D S. Measure of transient transfection efficiency using beta-galactosidase protein. Anal Biochem. 1997;244:22–27. doi: 10.1006/abio.1996.9868. [DOI] [PubMed] [Google Scholar]
- 24.Jack J, Dorsett D, Delotto Y, Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 1991;113:735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- 25.Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K A W, Bindereif A, Green M R. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Technol. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 27.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld E J, LeLeiko N S, Walsh M J. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. 1999;274:7803–7815. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 28.Lievens P M J, Donady J J, Tufarelli C, Neufeld E J. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J Biol Chem. 1995;270:12745–12750. doi: 10.1074/jbc.270.21.12745. [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Jack J. Regulatory interactions and role in cell type specification of the Malpighian tubules by the cut, kruppel, and caudal genes of Drosophila. Dev Biol. 1992;150:133–143. doi: 10.1016/0012-1606(92)90013-7. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, McLeod E, Jack J. Four distinct regulatory regions of the cut locus and their effect on cell type specification in Drosophila. Genetics. 1991;127:151–159. doi: 10.1093/genetics/127.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludlow C, Choy R, Blochlinger K. Functional analysis of Drosophila and mammalian Cut proteins in flies. Dev Biol. 1996;178:149–159. doi: 10.1006/dbio.1996.0205. [DOI] [PubMed] [Google Scholar]
- 32.Mailly F, Berube G, Harada R, Mao P L, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346–5357. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon N S, Berube G, Nepveu A. CCAAT displacement activity involves Cut repeats 1 and 2, not the Cut homeodomain. J Biol Chem. 2000;275:31325–31334. doi: 10.1074/jbc.M002912200. [DOI] [PubMed] [Google Scholar]
- 34.Muller H, Helin K. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta. 2000;1470:M1–M12. doi: 10.1016/s0304-419x(99)00030-x. [DOI] [PubMed] [Google Scholar]
- 35.Nepveu, A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene, in press. [DOI] [PubMed]
- 36.Neufeld E J, Skalnik D G, Lievens P M, Orkin S H. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 37.Shirane M, Harumiya Y, Ishida N, Hirai A, Miyamoto C, Hatakeyama S, Nakayama K, Kitagawa M. Down-regulation of p27(Kip1) by two mechanisms, ubiquitin-mediated degradation and proteolytic processing. J Biol Chem. 1999;274:13886–13893. doi: 10.1074/jbc.274.20.13886. [DOI] [PubMed] [Google Scholar]
- 38.Skalnik D G, Strauss E C, Orkin S H. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 39.Stein G S, Borun T W. The synthesis of acidic chromosomal proteins during the cell cycle of HeLa S-3 cells. I. The accelerated accumulation of acidic residual nuclear protein before the initiation of DNA replication. J Cell Biochem. 1972;52:292–307. doi: 10.1083/jcb.52.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Superti-Furga G, Barberis A, Schreiber E, Busslinger M. The protein CDP, but not CP1, Footprints on the CCAAT region of the g-globulin gene in unfractionated B-cell extracts. Biochim Biophys Acta. 1989;1007:237–242. doi: 10.1016/0167-4781(89)90046-8. [DOI] [PubMed] [Google Scholar]
- 41.Tyers M, Jorgensen P. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Gene Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 42.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 43.Valarche I, Tissier-Seta J P, Hirsch M R, Martinez S, Goridis C, Brunet J F. The mouse homeodomain protein Phox2 regulates Ncam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development. 1993;119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- 44.van Wijnen A J, Cooper C, Odgren P, Aziz F, De Luca A, Shakoori R A, Giordano A, Quesenberry P J, Lian J B, Stein G S, Stein J L. Cell cycle-dependent modifications in activities of pRb-related tumor suppressors and proliferation-specific CDP/cut homeodomain factors in murine hematopoietic progenitor cells. J Cell Biochem. 1997;66:512–523. doi: 10.1002/(sici)1097-4644(19970915)66:4<512::aid-jcb10>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 45.van Wijnen A J, van Gurp M F, de Ridder M C, Tufarelli C, Last T J, Birn-baum M, Vaughan P S, Giordano A, Krek W, Neufeld E J, Stein J L, Stein G S. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc Natl Acad Sci USA. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni M C, Helin K. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel J L, Kristie T M. Autocatalytic proteolysis of the transcription factor-coactivator Cl (HCF): a potential role for proteolytic regulation of coactivator function. Proc Natl Acad Sci USA. 2000;97:9425–9430. doi: 10.1073/pnas.160266697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. SREBP-1, a membrane-bound transcription factor released by sterol- regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 49.Welm A L, Timchenko N A, Darlington G J. C/EBPalpha regulates generation of C/EBP beta isoforms through activation of specific proteolytic cleavage. Mol Cell Biol. 1999;19:1695–704. doi: 10.1128/mcb.19.3.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoon S O, Chikaraishi D M. Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J Biol Chem. 1994;269:18453–18462. [PubMed] [Google Scholar]