Abstract
Prevalence of autism spectrum disorder (ASD) has been increasing in the United States in the past decades. The exact mechanisms remain enigmatic, and diagnosis of the disease still relies primarily on assessment of behavior. We first used a case–control design (75 idiopathic cases and 29 controls, enrolled at Boston Children’s Hospital from 2007-2012) to identify plasma biomarkers of ASD through a metabolome-wide association study approach. Then we leveraged a family-based design (31 families) to investigate the influence of shared genetic and environmental components on the autism-associated features. Using untargeted high-resolution mass spectrometry metabolomics platforms, we detected 19 184 features. Of these, 191 were associated with ASD (false discovery rate < 0.05). We putatively annotated 30 features that had an odds ratio (OR) between <0.01 and 5.84. An identified endogenous metabolite, O-phosphotyrosine, was associated with an extremely low autism odds (OR 0.17; 95% confidence interval 0.06-0.39). We also found that glutathione metabolism was associated with ASD (P = 0.048). Correlations of the significant features between proband and parents were low (median = 0.09). Of the 30 annotated features, the median correlations within families (proband–parents) were −0.15 and 0.24 for the endogenous and exogenous metabolites, respectively. We hypothesize that, without feature identification, family-based correlation analysis of autism-associated features can be an alternative way to assist the prioritization of potentially diagnostic features. A panel of ASD diagnostic metabolic markers with high specificity could be derived upon further studies.
Keywords: ASD, untargeted metabolomics, correlation globes, biomarker, shared environment, ASD diagnosis
Introduction
Autism spectrum disorder (ASD) is a group of neurodevelopmental disorders characterized by impairment in communication and social interaction, and repetitive behaviors.1 Over the past decades, the prevalence of ASD in the United States has increased to 1 in 42 boys and 1 in 189 girls 2, with earlier3,4 and inclusive3,5 diagnosis contributing 12% and 56%–60% to the rise in prevalence, respectively. Since the relative stability of genetic factors cannot explain such a dramatic change in prevalence over such a short period, the steady increase may be partially due to environmental factors or interplay between gene and environment.6 Recent twin studies have suggested that nongenetic contribution of ASD could be between 30% and 50%.7-10 At present, a diagnostic test for ASD is not available through any objective biofluid test but rely on assessing children's behavior and developmental delays by physicians and other health specialists.1,11
The metabolome includes all of the small molecules found within a biological sample.12 The majority of the molecules are endogenous metabolites that participate in the essential chemical reactions, while exogenous chemicals that originate from the external environment can also be identified.13 Untargeted high-resolution mass spectrometry (HRMS) provides broad-spectrum coverage of the metabolome and therefore is commonly used to profile the molecular signatures presenting physiological status of an individual. Data-driven studies of the plasma metabolome have been successfully used to answer biomedical questions, for example, characterizing the disrupted physiological processes and molecular fingerprint of disorders and/or discovering the exogenous exposures associated with health outcomes.14–18
Untargeted profiling of the metabolome has been applied to study ASD. These studies mostly employed a case–control design and aimed to identify biomarkers for early diagnosis, treatment, or prevent, and understand the pathophysiologic networks of ASD.19–30 To measure the blood or urine metabolome, analytical platforms such as gas chromatography–mass spectrometry, liquid chromatography–mass spectrometry, capillary electrophoresis–mass spectrometry, and nuclear magnetic resonance spectroscopy can be used. Each of these platforms has its own strengths and limitations in terms of speed, cost, sensitivity, and interpretability of the signals.31,32 and the final choice is dependent on the resources, sample types, and questions of interest in the studies. For exploratory analysis, employing multiple mass spectrometry platforms can maximize sensitivity and hence coverage of the metabolome, however, more than half of the published ASD metabolomics studies were using only a single platform.
Although powerful, untargeted metabolic profiling with HRMS has a major drawback—only a small fraction of the detected signals that have higher abundance in biofluids, such as glycolic acid, glycine, and arginine, can be routinely identified.33 Recent developments in mass spectral molecular networking and community-based spectral library have enhanced the interpretability of the metabolic profiles by providing correlation-based putative annotation, however, many of the features could still remain unknown.34 Furthermore, data-driven ASD studies typically generate a list of ASD-associated features and these potentially diagnostic features require structural confirmation and validation with independent cohorts. Since a majority of the pilot and exploratory studies employed a case–control design, associated features could be a mix of endogenous and exogenous molecules. Clearly, environmental chemicals have little biological relevance for reliable ASD prognosis or diagnosis, and the ability to distinguish the origins of features prior to tedious follow-up analyses would improve feature prioritization and accelerate pace of discovery.
We set two aims for this study. In Aim 1, we sought to discover new insights towards the molecular mechanism behind children with ASD using orthogonal liquid chromatography–mass spectrometry method to maximize the detection of plasma metabolome. In Aim 2, we posited that by studying the influence of shared genetic and environmental components in ASD families, we could deduce the origin and the role of the unknown ASD characterizing features. Specifically, we applied a metabolome-wide association study (MWAS) to an ASD-control comparison and conducted a series of family-based correlation analyses within and between probands and their parents. The overarching goal is to increase our understanding toward the etiology, physiology, and putative molecular biomarkers of ASD.
Methods
Study population
We recruited individuals with ASD and their parents, and unrelated neurotypical individuals as previously described.35,36 The MWAS used a case–control study design. We enrolled a total of 104 individuals (75 cases and 29 controls), who were part of a blood transcriptome study conducted at the Boston Children’s Hospital (BCH) in the years 2007-2012. We recruited unrelated controls from well-visit children to the Primary Care Center of BCH and from healthy individuals who visited the Division of Endocrinology of BCH for the evaluation of short stature. In our family-based correlation investigation, we enrolled parents of probands—40 mothers and 39 fathers. There were 31 families with at least one proband and at least one parent. BCH Institutional Review Board approved the study and we obtained informed consent from all participating subjects or their custodians prior to any data and sample collection.
Sample preparation and measurement
The whole blood was collected from participants in ethylenediaminetetraacetic acid-treated tubes and centrifuged at 2000 ×g for 10 min at room temperature to obtain plasma. These samples were then stored at −80°C until aliquoting and shipping. Plasma samples were thawed overnight in 4 °C refrigerator and then aliquoted into 0.5 mL screw-cap tubes prior to shipping in a dry iced box. Since blood samples were collected as part of standard clinical practice, time since last caloric intake varied from 5 min to 8 h. We prepared and analyzed the samples using established methods.37–39 We used dual-column chromatography (hydrophilic interaction liquid chromatography [HILIC; XBridge BEH Amide XP HILIC column; Waters, Waltham, MA, 50 × 2.1 mm, 2.5 μm] with positive electrospray ionization [ESI] and reversed-phase liquid chromatography [RPLC; C18 column; Higgins Analytical, Mountain View, CA, 50 × 2.1 mm, 2.6 μm] with negative ESI) coupled with HRMS for measuring the metabolome.
Data preprocessing
We converted raw data to open mzxml format using Proteowizard40 and conducted data preprocessing including peak detection, noise filtering, peak quantification and alignment, averaging signals of triplicates, peak matching and batch effect correction with apLCMS41 and xMSanalyzer.42 The pipeline generated a data table for each of HILIC and RPLC that comprises the relative signal intensities of 13 098 and 10 522 features, respectively.
We excluded features in the highest 20% of the coefficient of variations across quality control samples and those with greater than 80% zero intensity across study samples. The number of features available in our analyses was 10 173 (HILIC) and 8011 (RPLC). We presented the details about inclusion and exclusion criteria for ASD cases, controls and proband families, metabolome measurement, and analyses of quality control samples in the Supplementary material (Methods S1-S3).
Statistical analyses
Analytical overview
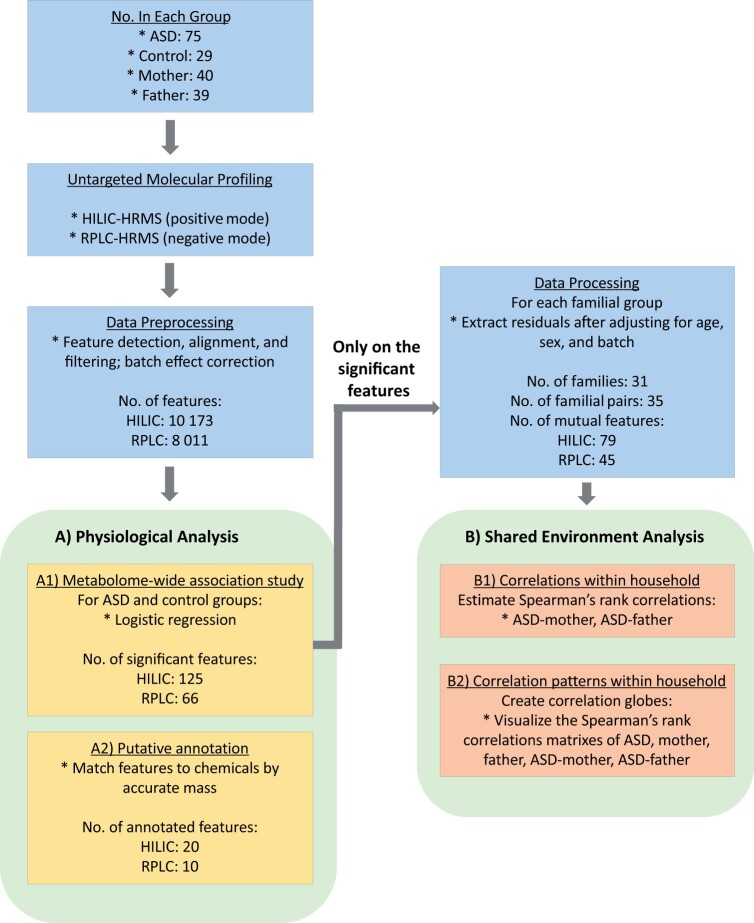
We divided the analysis into two parts as shown in Figure 1. (A) For Aim 1: metabolomic profiling of ASD and (B) for Aim 2: feature correlations in the shared environment. We conducted the analyses separately for HILIC and RPLC data. Using a case–control design, (A1) we executed an MWAS to unbiasedly discover features associated with ASD. Then, we conducted a series of analyses based on the MWAS selected features. (A2) We identified their chemical names through matching the measured mass-to-charge ratio (m/z) with the theoretical values. We leveraged a family-based design to understand the influence of shared genetic and environmental components on ASD. Specifically, we (B1) computed the correlation of the MWAS features in proband-mother and proband-father pairs and (B2) visualized the correlation patterns as correlation globes.43 Additionally, we provided complementary pathway enrichment for Aim 1 in the Supplementary material (Method S4).
Figure 1.
Overview of the data collection and statistical analyses conducted in this study. After collecting plasma samples of ASD cases, controls, and parents of ASD, we used two analytical platforms, HILIC and RPLC that were coupled with high-resolution mass spectrometry (HRMS) to conduct untargeted metabolic profiling. Raw data was processed, including feature detection, peak alignment, batch effect correction, and feature filtering before the data sets were sent to downstream analysis. For physiological analysis, we first run (A1) an MWAS on cases and controls to identify significant features associated with ASD (FDR = 0.05). Then we (A2) putatively named the compounds by matching their accurate masses to those unambiguously found in metabolite databases. Using the MWAS selected features, we conducted the shared environment analyses after adjusting for confounders (i.e., analyzed the extracted residuals from the adjusting model). We (B1) first investigated the Spearman’s rank correlations within households (i.e., in proband-mother, proband-father). Then we (B2) visualized the correlation patterns within households as correlation globes.
Metabolome-wide association study
To conduct an MWAS, we first extracted 75 cases and 29 controls from the data table. We filtered out features with zero variance in each group and analyzed the features that had greater than zero variance in both cases and controls. The number of available features was 10 143 (HILIC; 99.8% of the input features) and 7997 (RPLC; 99.8% of the input features). Then we scaled the log-transformed features and ran a generalized linear model as follows:
where ASD is a binary variable denoting the diagnosis of ASD; age is a continuous variable (months); sex is a binary variable; batch is a categorical variable denoting the analytical batch identifier of the samples. We used false discovery rate (FDR) to correct for multiplicity when determining statistical significance.
Annotation of features
We used xMSannotator to annotate detected features.44 For each feature, the accurate mass was searched against those chemicals listed in Human Metabolome Database (HMDB), Kyoto Encyclopedia of Genes and Genomes (KEGG), and LIPID MAPS45–47 at a 5 ppm error-tolerant window. To avoid generating excessive false annotations, we only used hydrogen adduct for m/z matching (HILIC: M + H; RPLC: M-H). A single feature could be unambiguously matched with unique chemical identity or matched with multiple chemicals in the same or across different databases. We only reported the MWAS significant features with unambiguous putative annotation in the main table and provided the full putative annotation results in the Supplementary material (Table S1: HILIC; Table S2: RPLC). We followed the identification confidence levels in high resolution mass spectrometric analysis by Schymanski et al. (level 1: confirmed structure, level 2: probable structure, level 3: tentative candidate(s), level 4: unequivocal molecular formula, and level 5: exact mass) and reported all the metabolite identification with a level 5 confidence level unless otherwise specified.48 For the ASD-associated features that are believed to be endogenous metabolites based on the putative annotation, we further validated their identities as follows: we identified spectral peaks in the in-house pooled plasma samples with sufficient intensity for tandem mass spectrometry. Tandem mass spectrometry (MS/MS) fragmentation was then performed on the matched parent m/z and the resulting fragmentation MS/MS spectra were compared to those found in two public databases—MetFrag49,50 and mzCloud (mzcloud.org)—to provide level 2 confidence (probable structure).48
Correlations within households
We studied the household correlations separately for mother and father instead of averaging their signals to a single value (i.e., proband-mother and proband-father pairs). In each of the proband, mother, and father groups, we first filtered MWAS features that had > 80% zero intensity and fit a model to subtract linear contributions of age, sex, and batch:
After matching with family ID, we had 35 pairs from 31 families and 79 mutual features in each group (HILIC; 45 mutual features for RPLC data). The model residuals were used as the input for all the familial correlation analyses. We estimated the Spearman’s rank correlations (rs) of the mutual features in proband-mother and proband-father and corrected for multiplicity with FDR. We used the proportion of significant correlations as a metric to gauge the influence of shared components.
Correlation patterns within household
We estimated five rs matrices—proband, mother, father, proband-mother, and proband-father—using the residuals estimated from the previous step. In addition, using Euclidean distance, we performed hierarchical clustering analysis on the features of ASD and arbitrarily set to create 9 feature groups to aid pattern inspection. The group membership was applied to mother and father and features were sorted in the same order to aid visual inspection of the correlation patterns within households. We created the correlation globes with the R package circlize (v 0.4.5).51
Computational settings
Our analyses took the following default settings unless otherwise specified. To control spurious findings due to multiple testing, we used a 5% FDR and reported the Q value, which is an FDR adjusted P-value. FDR was calculated separately for HILIC and RPLC analyses. We transformed, or scaled, each variable feaure to have mean zero and unit variance. We used a fudge factor of 1 (i.e., x + 1) to avoid directly log-transforming variables with zero intensity. Correlation refers to the estimation of Spearman’s rank correlations. We executed all analyses using the computing environment R (v 3.5.1). For reproducibility purposes, all analytic code is publicly available on GitHub via an MIT license (github.com/jakemkc/autism_shared_env).
Results
Study population
Demographic summary of the study population is shown in Table 1. The majority of the enrolled subjects were Caucasian and non-Hispanic with less than 5% Asian. We enrolled more individuals with ASD (75 versus 29) that were younger (98 versus 147 months) and with more male (83% versus 62%) when compared with neurotypical controls. Demographic characteristics of mother and father, including sample size (40 versus 39), age in months (493 versus 502), and ethnicity (98% versus 97% white) were similar.
Table 1.
Demographic characteristics of autism cases, parents of cases, and controls in this study
| Characteristic | Control | Autism | Mother | Father |
|---|---|---|---|---|
| Number | 29 | 75 | 40 | 39 |
| Age, median (interquartile range), months | 152 (34) | 83 (51) | 501.5 (94.7) | 504 (63) |
| Male, number (%) | 18 (62) | 62 (83) | 0 (0) | 39 (100) |
| Ethnicity, number (%) | ||||
| White | 27 (93) | 71 (95) | 39 (98) | 38 (97) |
| Asian | 1 (3) | 2 (3) | 0 (0) | 0 (0) |
| Unknown | 1 (3) | 2 (3) | 1 (3) | 1 (3) |
Molecular profile of autism spectrum disorder
Table 2 shows the putatively annotated chemicals together with their test statistics. Out of the MWAS significant features (HILIC: 125, RPLC: 66), we could annotate 20 (16%) and 10 (15.2%) of them by their accurate mass for HILIC and RPLC, respectively. Only a few of these were associated with an increased odd of ASD (HILIC: 5/20; RPLC: 0/10). ORs for HILIC data spanned from less than 0.01 to 5.84 whereas those for RPLC were from less than 0.01 to 0.29. Annotated chemicals came from diverse chemical classes, such as piperidines, flavonoids, carboxylic acids and derivatives for HILIC data, and carboxylic acids, naphthofurans, and glycerolipids for RPLC data. Using the MWAS significant features to conduct pathway enrichment analysis, we found that only glutathione metabolism was affected (Table S4; P-value = 0.048).
Table 2.
Putatively identified metabolites from the MWAS of autism cases and controls and the correlations of metabolites within the ASD families using two different analytical platforms
| Platform | Monoisotopic mass | OR (95% CI)a | Q value | dbIDb | Name | Source | Spearman correlationc |
||
|---|---|---|---|---|---|---|---|---|---|
| HILIC | ASD-Mother | ASD-Father | |||||||
| 129.1518 | < 0.01 | (< 0.01-0.03) | 0.04 | C01740 | Octylamine; N-Octylamine; Monoctylamine | Exogenous | −0.186 | 0.276 | |
| 155.1310 | <0.01 | (< 0.01-0.01) | 0.01 | C06184 | N-Methylpelletierine | Exogenous | 0.364 | 0.255 | |
| 149.9987 | 0.01 | (< 0.01-0.07) | 0.04 | HMDB42032 | Thiodiacetic acid | Endogenous | −0.282 | 0.178 | |
| 468.9511 | 0.04 | (0.01-0.15) | 0.01 | HMDB60640 | Lamivudine-triphosphate | Endogenous | NA | NA | |
| 85.0891 | 0.06 | (0.01-0.19) | 0.01 | HMDB34301 | Piperidine | Exogenous | −0.145 | −0.142 | |
| 323.3188 | 0.10 | (0.03-0.22) | 0.01 | HMDB34373 | N-(14-Methylhexadecanoyl)pyrrolidine | Exogenous | NA | NA | |
| 213.9879 | 0.10 | (0.03-0.25) | 0.01 | HMDB06801 | 2-Oxo-3-hydroxy-4-phosphobutanoic acid | Endogenous | NA | NA | |
| 141.0112 | 0.12 | (0.04-0.28) | 0.01 | HMDB60688 | Nornitrogen mustard | Endogenous | NA | NA | |
| 193.0600 | 0.13 | (0.04-0.31) | 0.02 | C16789 | Toxoflavine | Exogenous | NA | NA | |
| 350.0096 | 0.15 | (0.05-0.34) | 0.02 | HMDB37851 | Apigenin 7-sulfate | Exogenous | 0.346 | 0.299 | |
| 261.0402 | 0.17 | (0.06-0.39) | 0.02 | HMDB06049 | O-Phosphotyrosine | Endogenous | NA | NA | |
| 341.9739 | 0.19 | (0.08-0.39) | 0.01 | C18968 | Carbophenothion | Exogenous | 0.308 | 0.396 | |
| 276.0124 | 0.24 | (0.11-0.46) | 0.02 | C11570 | 2-(2-Chloro-phenyl)-5-(5-methylthiophen-2-yl)-134oxadiazole | NA | NA | NA | |
| 204.9810 | 0.26 | (0.13-0.49) | 0.02 | C12284 | Saccharin sodium anhydrous | Exogenous | NA | NA | |
| 378.1943 | 0.30 | (0.16-0.54) | 0.02 | HMDB61127 | 4R-Hydroxy solifenacin | Endogenous | NA | NA | |
| 355.9497 | 0.31 | (0.15-0.58) | 0.04 | HMDB15610 | Silver sulfadiazine | Exogenous | 0.204 | 0.331 | |
| 263.0844 | 2.89 | (1.66-5.73) | 0.05 | HMDB42056 | Tulobuterol | Exogenous | 0.023 | 0.236 | |
| 500.3866 | 3.14 | (1.74-6.37) | 0.04 | C15379 | 3beta-Hydroxylanostane-711-dione acetate | Exogenous | −0.020 | 0.078 | |
| 471.0923 | 3.49 | (1.82-7.79) | 0.05 | HMDB61083 | desbutyl-lumefantrine | Endogenous | −0.185 | −0.116 | |
| 650.2211 | 5.84 | (2.50-17.5) | 0.03 | LMPK12050358 | 5-Hydroxy-734-trimethoxy-8-methylisoflavone 5-O-neohesperidoside | Exogenous | 0.241 | 0.195 | |
| RPLP | |||||||||
| 326.2093 | <0.01 | (<0.01-<0.01) | 0.05 | HMDB31963 | (3b6b8a12a)-812-Epoxy-7(11)-eremophilene-6812-trimethoxy-3-ol | Exogenous | 0.003 | 0.312 | |
| 230.0579 | <0.01 | (<0.01-<0.01) | 0.02 | LMPK13110003 | Visnagin | Exogenous | −0.115 | −0.037 | |
| 410.1818 | 0.06 | (0.01-0.18) | 0.02 | C08093 | Oseltamivir phosphate | Exogenous | NA | NA | |
| 274.2144 | 0.11 | (0.03-0.28) | 0.02 | C13854 | 1-Dodecanoyl-sn-glycerol | NA | −0.155 | −0.074 | |
| 217.0773 | 0.17 | (0.06-0.36) | 0.02 | HMDB15328 | Captopril | Exogenous | 0.211 | 0.466 | |
| 216.0746 | 0.19 | (0.08-0.38) | 0.02 | C17359 | 8-Hydroxyalanylclavam | Exogenous | 0.281 | 0.498 | |
| 269.1991 | 0.21 | (0.09-0.40) | 0.02 | HMDB32255 | N-(Ethoxycarbonyl)methyl)-p-menthane-3-carboxamide | Exogenous | NA | NA | |
| 322.1780 | 0.24 | (0.11-0.48) | 0.03 | HMDB32702 | Zeranol | Exogenous | NA | NA | |
| 386.2305 | 0.25 | (0.11-0.48) | 0.03 | C11990 | Oleandolide | Exogenous | NA | NA | |
| 332.2563 | 0.29 | (0.15-0.53) | 0.03 | C19621 | Floionolic acid | Exogenous | NA | NA | |
Only features with level 5 identification confidence (exact mass) and unambiguous compound matching with Human Metabolome Database (HMDB), Kyoto Encyclopedia of Genes and Genomes (KEGG), and LIPID MAPS are shown. For silver sulfadiazine, it is now located in the Toxin and Toxin Target Database (T3DB) with an accession number T3D3068. All of the metabolites were matched by exact mass through comparison with records in reference databases. Only O-phosphotyrosine was identified to level 2 (probable structure) by matching the MS/MS spectrum. A putatively annotated metabolite by exact mass, 2-oxophytanic acid, was not shown because its tandem mass spectrometry (MS/MS) spectrum did not match with the records in databases. Metabolites are sorted by size of the OR.
Input features to the model were log-transformed and scaled to have mean zero and unit variance.
Unique ID for the corresponding chemicals found in HMD, KEGG or LIPID MAPS.
Spearman’s rank correlations of annotated metabolites within the ASD families (i.e., in proband-mother, proband-father). NA represents the metabolite not consistently detected within the families.
Correlations in the shared environment
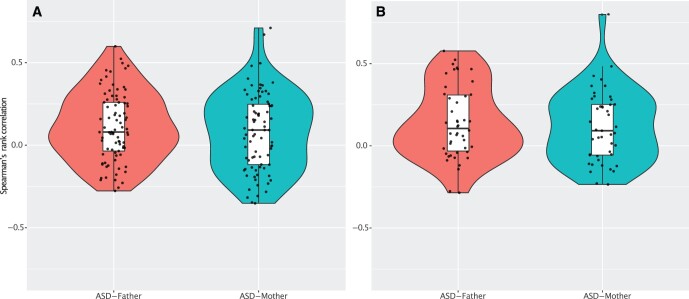
We show the distribution of the rss within households in Figure 2. For HILIC data, mean rs was 0.08 (range: −0.35 to 0.71) and 0.11 (range: −0.28 to 0.60) for proband-mother and proband-father, respectively. Only a few of the features had rs greater than 0.5. Proband-mother had a uniform-like distribution of rs, whereas that for proband-father is more resembled to normal. Of the 79 investigated features, 5% were significantly correlated—two were significant in proband-mother and proband-father, respectively, after FDR correction (i.e., four non-overlapping features). For RPLC data, mean rs was 0.12 (range: −0.23 to 0.80) and 0.13 (−0.29 to 0.58) for proband-mother and proband-father, respectively. Distribution of rss within households was similar to HILIC. Only five of all the features had rs greater than 0.5. For all 45 features investigated, 13% were significantly correlated—one was significant in proband-mother and five were significant in proband-father after FDR correction. All the significant correlations were positively correlated with a magnitude greater than 0.45.
Figure 2.
Violin plots showing the distributions of the correlations of ASD–associating features within the ASD families. (A) HILIC platform; (B) RPLC platform. Using the significant features found in the MWAS, we estimated the Spearman’s rank correlation for each feature between proband-mother, proband-father. The number of shared features for estimating correlations were 79 (HILIC) and 45 (RPLC) after data preprocessing for familial analysis. In each plot, we overlaid with a one-dimensional scatter plot and a boxplot showing median, interquartile ranges, and whiskers extending to the largest values within 1.5*interquartile range.
We show the within household rss of the MWAS significant metabolites in Table 2. Overall, only two out of seven (28.6%) endogenous and 13 out of 21 (61.9%) exogenous metabolites were consistently detected in the ASD families. The median rss within the families (across HILIC, RPLC, ASD-mother, and ASD-father) were −0.15 and 0.24 for endogenous and exogenous metabolites, respectively.
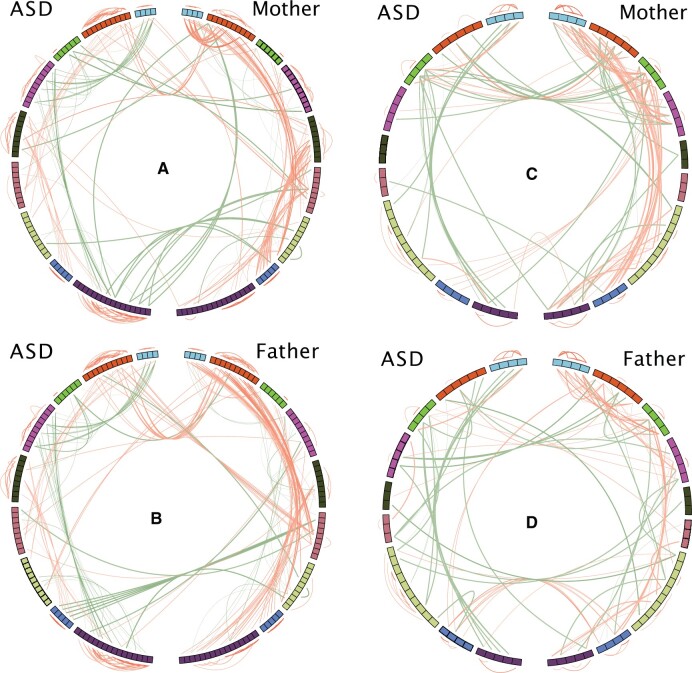
We show the within household correlation patterns in Figure 3. In the correlation globes depicting the patterns of 79 features from HILIC data, we did not observe strong similarity in the correlation patterns between proband and mother (Figure 3A) nor between proband and father (Figure 3B). Instead, we observed general similarity in patterns between mother (Figure 3A, right half) and father (Figure 3B, right half), and to a lesser extent, between proband-mother (Figure 3A, across the globe) and proband-father (Figure 3B, across the globe). These observations of correlation patterns were also found for the 45 features from RPLC data (Figure 3C and 3D).
Figure 3.
Correlation globe showing the correlation patterns in the shared environment. (A) and (B): HILIC platform; (C) and (D) RPLC platform. Using the HILIC platform as an example, we estimated the Spearman’s rank correlations of 79 ASD correlating features shared in ASD families (RPLC: 45 shared features). To facilitate visual inspection of the patterns, we assigned the features in ASD, mother, and father into nine feature groups based on the results from hierarchical clustering on ASD cases. Each correlation globe is showing the correlations within ASD, within parents (mother or father), and between ASD and parents (mother or father). Features are arranged as a circular track. Left and right halves of the globe represent features in ASD and parent (mother or father), respectively. Only Spearman’s rank correlations greater than 0.5 and smaller than −0.5 are shown as connections in the globe. Red line denotes positive correlation, and dark green line denotes a negative one. Color intensity and line width are proportional to the size of the correlation. Within-group and between-group correlations are shown outside and inside of the track, respectively. Correlations between ASD and parent are indicated by the lines linking across the vertical half of the globe.
Discussion
Molecular profiling of ASD
In this study, we employed orthogonal separation techniques coupled with HRMS to maximize coverage of the human plasma metabolome. Our comprehensive analyses have shown that a total of 191 chemical features were associated with ASD in our study cohort. These markers could be endogenous molecules acting as mediators in the causal pathways for ASD, noncausal indicators of ASD, or exogenous exposures with natural or anthropogenic origins. In the pathway enrichment analysis, we found that glutathione metabolism was perturbed in ASD.
In our initial annotation, we found three putative MWAS significant metabolites detected using HILIC could be endogenous compounds (Table 2). Upon further identification, we found that 2-oxophytanic acid was a false positive and concentration of 2-oxo-3-hydroxy-4-phosphobutanoic acid was too low for MS/MS comparison. Only O-phosphotyrosine (pTyr) was identified by a matched mass spectrum in the reference database.
pTyr (OR 0.17, 95% confidence interval [CI] 0.06-0.39) is a product of tyrosine phosphorylation, which is the addition of a phosphate group to tyrosine and is catalyzed by tyrosine kinases. Phosphorylation of tyrosine residues in proteins is an important post-translational modification that is implicated in various biological processes including cell–cell signaling and proliferation as well as neuronal maturation and synaptic plasticity.52 Concentration of pTyr in body fluid could be correlated with tyrosine kinase and pTyr phosphatase activities in tissue.53 In the current study, sorting out the proteins with tyrosine residue responsible for lower concentration of pTyr in plasma samples from ASD warrants further studies. Nonetheless, a receptor tyrosine kinase encoded by MET is implicated in pathophysiological changes of ASD. A non-coding promoter variant of MET, rs1858830 C allele is reported as a genetic risk factor of ASD. rs1858830 CC genotype decreases MET promoter activity, which results in down-regulation of MET gene expression.54
2-oxo-3-hydroxy-4-phosphobutanoic acid (OR 0.10, 95% CI 0.03-0.25) is part of the vitamin B6 pathway. Some have suggested that vitamin B6 can be used as a supplement in brain disorder. However, a randomized controlled study to investigate this relationship had shown negative results.55
We found a handful of exogenous chemicals including pharmaceuticals, natural dietary molecules, and food additives to be significantly associated with ASD (Table S4). The combination of case–control design and blood sampling of ASD cases after diagnosis make our findings merely suggestive about their potential roles in the cause and development of ASD. Nonetheless, these shortlisted chemicals could be good candidates for future etiological research.
Familial correlations
The metabolome of ASD cases is affected by genetics, the environment, and the disease. We found 191 features were associated with ASD in our case–control MWAS analysis. Many of these features were not putatively annotated and we hypothesize that these could be either (1) exogenous chemicals from confounders such as unique environment or (2) endogenous biomarkers that are pathophysiological signatures of ASD and possess diagnostic value.
We leveraged a family-based study design and sought to obtain hints about the origins of the ASD-associated features. Key findings of our correlation analyses include low rss (averaged across HILIC and RPLC data: proband-mother; median [interquartile range], 0.09 [0.35]; proband-father; median [interquartile range], 0.08 [0.3]) and low similarity in correlation patterns in proband-mother and proband-father. In a familial setting, assuming family members stay 12 h a day at home, proband share about 50% of the genetics and the environment with the parents. In other studies, the median rss of exogenous chemicals between couples were about 0.2143 (serum; polychlorinated biphenyls, organochlorine pesticides, polybrominated chemicals, per- and poly-fluoroalkyl substances, and metals) and 0.4156 (urine; triclosan, phenols, bisphenols, parabens, and phthalates). For child–parent pairs, the median rss were 0.31 and 0.68 for serum perfluorinated compounds and polybrominated diphenyl ethers, respectively.57,58 In addition, almost all of the exogenous chemicals were positively correlated in these studies.
Further, using the known origin of some of the putatively identified metabolites (Table 2), we found that only 28.6% of the ASD-associated endogenous metabolites were also detected in parents, which is much smaller than that for exogenous metabolites (61.9%). Also, the median rs within the families for endogenous metabolites was smaller than that of exogenous metabolites (−0.15 and 0.24, respectively). These comparisons and analyses suggest that familial correlation in the ASD families could be used to infer the origin of the ASD-associated features found in the case–control MWAS. Correlation has a range between −1 and +1 and we hypothesize that the smaller the familial correlation of an MWAS feature, the more likely that it is endogenous, and may rank higher in priority for follow-ups such as structural confirmation and validation of diagnostic value.
Several studies have shown the potential of using plasma metabolites as clinical biomarkers for diagnosing neurological diseases.28,59–62 For example, West et al. found a set of over 100 metabolites that were able to discriminate ASD subjects from typically developing children with a maximum accuracy (area under the curve) of 81%.28 In an Alzheimer disease study, Stamate et al. reported that the diagnostic performance of using plasma metabolites had the potential to match the well-established cerebrospinal fluid biomarkers.60
Limitations
We identified a few study limitations that pose a challenge to our findings. First, individuals with ASD were not matched with neurotypical controls by age and by sex; specifically, the control group was older with a higher proportion of female. To address this case–control imbalance, we adjusted the analyses by age and sex. Second, blood samples were not collected under an overnight fasting protocol. Our analysis may suffer from higher variability and hence reduced statistical power to detect associations. Third, for most of the putatively annotated metabolites, we did not conduct additional identification such as matching fragmentation patterns in a spectral library or comparison with authentic standards and thus they should be interpreted with caution. Fourth, autism is a complex and multifactorial disease and may require a larger sample size to capture a greater number of metabolites at smaller association sizes, that in totality, may ascribe more variation to autism. The sample size in this study (n = 104) is insufficient for a reliable subtype analysis. Last, our MWAS analysis could be confounded by unmeasured factors such as medication and dietary factors. The primary goal of this exploratory study is to investigate what deep metabolomics measurement could tell us about autism and the families with children who have autism.
Conclusions
Our case–control MWAS analysis revealed that a total of 191 features were associated with ASD. An identified metabolite, O-phosphotyrosine, was associated with a decreased risk of ASD. We also found glutathione metabolism was affected in ASD. Family-based correlation of ASD-associated features can assist the prioritization of potentially diagnostic features. Further studies are required to select a panel of reproducible metabolites, quantify their the diagnostic performance in different clinical settings of the metabolites, and identify the exogenous environmental factors associated with the metabolites.
Supplementary Material
Acknowledgments
We acknowledge the Precision Link Biobank at Boston Children’s Hospital for plasma sample storage and handling for metabolomics profiling. We thank Carles Hernandez-Ferrer and Vy Nguyen for providing feedback on the manuscript.
Supplementary material
Supplementary material is available at Exposome online.
Funding
The study was funded by research grants from the National Institute of Mental Health (R01MH107205) and National Institute of Allergy and Infectious Diseases (R01AI127250) and the National Institute of Environmental Health Sciences (R01ES032470)..
Conflict of interest statement
The authors declare no competing financial interests.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding authors.
References
- 1. Randall M, Egberts KJ, Samtani A. et al. Diagnostic tests for autism spectrum disorder (ASD) in preschool children. Cochrane Database Syst Rev. 2018. Jul 24;7(7):CD009044. doi: 10.1002/14651858.CD009044.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Christensen DL, Braun KVN, Baio J. et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 Sites, United States, 2012. MMWR Surveill Summ. 2018;65(13):1–23. doi: 10.15585/mmwr.ss6513a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hertz-Picciotto I, Delwiche L.. The rise in autism and the role of age at diagnosis. Epidemiology. 2009;20(1):84–90. doi: 10.1097/EDE.0b013e3181902d15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fountain C, King MD, Bearman PS.. Age of diagnosis for autism: individual and community factors across 10 birth cohorts. J Epidemiol Community Health. 2011;65(6):503–510. doi: 10.1136/jech.2009.104588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansen SN, Schendel DE, Parner ET.. Explaining the increase in the prevalence of autism spectrum disorders: the proportion attributable to changes in reporting practices. JAMA Pediatr. 2015;169(1):56–62. doi: 10.1001/jamapediatrics.2014.1893 [DOI] [PubMed] [Google Scholar]
- 6. Nevison CD. A comparison of temporal trends in United States autism prevalence to trends in suspected environmental factors. Environ Health. 2014;13(1):73. doi: 10.1186/1476-069X-13-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A.. The familial risk of autism. JAMA 2014;311(17):1770–1777. doi: 10.1001/jama.2014.4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hallmayer J, Cleveland S, Torres A. et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68(11):1095–1102. doi: 10.1001/archgenpsychiatry.2011.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaugler T, Klei L, Sanders SJ. et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46(8):881–885. doi: 10.1038/ng.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colvert E, Tick B, McEwen F. et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry. 2015;72(5):415–423. doi: 10.1001/jamapsychiatry.2014.3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lord C, Risi S, DiLavore PS, Shulman C, Thurm A, Pickles A.. Autism from 2 to 9 years of age. Arch Gen Psychiatry. 2006;63(6):694–701. doi: 10.1001/archpsyc.63.6.694 [DOI] [PubMed] [Google Scholar]
- 12. Nicholson JK, Lindon JC.. Metabonomics. Nature. 2008;455(7216):1054–1056. doi: 10.1038/4551054a [DOI] [PubMed] [Google Scholar]
- 13. Rappaport SM, Barupal DK, Wishart D, Vineis P, Scalbert A.. The blood exposome and its role in discovering causes of disease. Environ Health Perspect. 2014;122(8):769–774. doi: 10.1289/ehp.1308015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu R, Hong J, Xu X. et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23(7):859–868. doi: 10.1038/nm.4358 [DOI] [PubMed] [Google Scholar]
- 15. Ali SE, Farag MA, Holvoet P, Hanafi RS, Gad MZ.. A comparative metabolomics approach reveals early biomarkers for metabolic response to acute myocardial infarction. Sci Rep. 2016;6:36359. doi: 10.1038/srep36359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alvarez JA, Chong EY, Walker DI. et al. Plasma metabolomics in adults with cystic fibrosis during a pulmonary exacerbation: A pilot randomized study of high-dose vitamin D3 administration. Metabolism 2017;70:31–41. doi: 10.1016/j.metabol.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burgess LG, Uppal K, Walker DI. et al. Metabolome-wide association study of primary open angle glaucoma. Invest Ophthalmol Vis Sci. 2015;56(8):5020–5028. doi: 10.1167/iovs.15-16702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walker DI, Lane KJ, Liu K. et al. Metabolomic assessment of exposure to near-highway ultrafine particles. J Expo Sci Environ Epidemiol. 2019;29(4):469–483. doi: 10.1038/s41370-018-0102-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bent S, Lawton B, Warren T. et al. Identification of urinary metabolites that correlate with clinical improvements in children with autism treated with sulforaphane from broccoli. Mol Autism. 2018;9(1):35. doi: 10.1186/s13229-018-0218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diémé B, Mavel S, Blasco H. et al. Metabolomics study of urine in autism spectrum disorders using a multiplatform analytical methodology. J Proteome Res. 2015;14(12):5273–5282. doi: 10.1021/acs.jproteome.5b00699 [DOI] [PubMed] [Google Scholar]
- 21. Emond P, Mavel S, Aïdoud N. et al. GC-MS-based urine metabolic profiling of autism spectrum disorders. Anal Bioanal Chem. 2013;405(15):5291–5300. doi: 10.1007/s00216-013-6934-x [DOI] [PubMed] [Google Scholar]
- 22. Gevi F, Zolla L, Gabriele S, Persico AM.. Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Mol Autism. 2016;7(1):47. doi: 10.1186/s13229-016-0109-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuwabara H, Yamasue H, Koike S. et al. Altered metabolites in the plasma of autism spectrum disorder: a capillary electrophoresis time-of-flight mass spectroscopy study. PLoS One. 2013;8(9):e73814. doi: 10.1371/journal.pone.0073814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lussu M, Noto A, Masili A. et al. The urinary 1 H-NMR metabolomics profile of an Italian autistic children population and their unaffected siblings: Metabolomics profile of autistic children. Autism Res. 2017;10(6):1058–1066. doi: 10.1002/aur.1748 [DOI] [PubMed] [Google Scholar]
- 25. Ming X, Stein TP, Barnes V, Rhodes N, Guo L.. Metabolic perturbance in autism spectrum disorders: a metabolomics study. J Proteome Res. 2012;11(12):5856–5862. doi: 10.1021/pr300910n [DOI] [PubMed] [Google Scholar]
- 26. Noto A, Fanos V, Barberini L. et al. The urinary metabolomics profile of an Italian autistic children population and their unaffected siblings. J Matern Fetal Neonatal Med. 2014;27(sup2):46–52. doi: 10.3109/14767058.2014.954784 [DOI] [PubMed] [Google Scholar]
- 27. Wang H, Liang S, Wang M. et al. Potential serum biomarkers from a metabolomics study of autism. J Psychiatry Neurosci. 2016;41(1):27–37. doi: 10.1503/jpn.140009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. West PR, Amaral DG, Bais P. et al. Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS One. 2014;9(11):e112445. doi: 10.1371/journal.pone.0112445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ritz B, Yan Q, Uppal K. et al. Untargeted metabolomics screen of mid‐pregnancy maternal serum and autism in offspring. Autism Res. 2020;13(8):1258–1269. doi: 10.1002/aur.2311 [DOI] [PubMed] [Google Scholar]
- 30. Smith AM, Natowicz MR, Braas D. et al. A metabolomics approach to screening for autism risk in the children’s autism metabolome project. Autism Res. 2020;13(8):1270–1285. doi: 10.1002/aur.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ren J-L, Zhang A-H, Kong L, Wang X-J.. Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Adv. 2018;8(40):22335–22350. doi: 10.1039/C8RA01574K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dettmer K, Aronov PA, Hammock BD.. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26(1):51–78. doi: 10.1002/mas.20108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uppal K, Walker DI, Liu K, Li S, Go Y-M, Jones DP.. Computational metabolomics: a framework for the million metabolome. Chem Res Toxicol. 2016;29(12):1956–1975. doi: 10.1021/acs.chemrestox.6b00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li D, Gaquerel E.. Next-generation mass spectrometry metabolomics revives the functional analysis of plant metabolic diversity. Annu Rev Plant Biol. 2021;72(1):867–891. doi: 10.1146/annurev-arplant-071720-114836. [DOI] [PubMed] [Google Scholar]
- 35. Kong SW, Shimizu-Motohashi Y, Campbell MG. et al. Peripheral blood gene expression signature differentiates children with autism from unaffected siblings. Neurogenetics. 2013;14(2):143–152. doi: 10.1007/s10048-013-0363-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kong SW, Collins CD, Shimizu-Motohashi Y. et al. Characteristics and predictive value of blood transcriptome signature in males with autism spectrum disorders. PloS One. 2012;7(12):e49475. doi: 10.1371/journal.pone.0049475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walker DI, Perry-Walker K, Finnell RH. et al. Metabolome-wide association study of anti-epileptic drug treatment during pregnancy. Toxicol Appl Pharmacol. 2019;363:122–130. doi: 10.1016/j.taap.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith MR, Jarrell ZR, Orr M, Liu KH, Go Y-M, Jones DP.. Metabolome-wide association study of flavorant vanillin exposure in bronchial epithelial cells reveals disease-related perturbations in metabolism. Environ Int. 2021;147:106323. doi: 10.1016/j.envint.2020.106323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yan Q, Liew Z, Uppal K. et al. Maternal serum metabolome and traffic-related air pollution exposure in pregnancy. Environ Int. 2019;130:104872. doi: 10.1016/j.envint.2019.05.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kessner D, Chambers M, Burke R, Agus D, Mallick P.. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24(21):2534–2536. doi: 10.1093/bioinformatics/btn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu T, Park Y, Johnson JM, Jones DP.. apLCMS—adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25(15):1930–1936. doi: 10.1093/bioinformatics/btp291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Uppal K, Soltow QA, Strobel FH. et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics. 2013;14(1):15. doi: 10.1186/1471-2105-14-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chung MK, Kannan K, Louis GM, Patel CJ.. Toward capturing the exposome: exposure biomarker variability and coexposure patterns in the shared environment. Environ Sci Technol. 2018;52(15):8801–8810. doi: 10.1021/acs.est.8b01467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uppal K, Walker DI, Jones DP.. xMSannotator: an r package for network-based annotation of high-resolution metabolomics data. Anal Chem. 2017;89(2):1063–1067. doi: 10.1021/acs.analchem.6b01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanehisa M, Goto S.. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wishart DS, Feunang YD, Marcu A. et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fahy E, Sud M, Cotter D, Subramaniam S.. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35(Web Server issue):W606–W612. doi: 10.1093/nar/gkm324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schymanski EL, Jeon J, Gulde R. et al. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ Sci Technol. 2014;48(4):2097–2098. doi: 10.1021/es5002105 [DOI] [PubMed] [Google Scholar]
- 49. Ruttkies C, Schymanski EL, Wolf S, Hollender J, Neumann S.. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J Cheminform. 2016;8(1):3. doi: 10.1186/s13321-016-0115-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolf S, Schmidt S, Müller-Hannemann M, Neumann S.. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics. 2010;11(1):148. doi: 10.1186/1471-2105-11-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gu Z, Gu L, Eils R, Schlesner M, Brors B.. circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2812. doi: 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- 52. Bertling E, Englund J, Minkeviciene R. et al. Actin tyrosine-53-phosphorylation in neuronal maturation and synaptic plasticity. J Neurosci. 2016;36(19):5299–5313. doi: 10.1523/JNEUROSCI.2649-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kataoka H, Nakai K, Makita M.. Increase of phosphotyrosine levels in mouse urine and liver during liver regeneration after partial hepatectomy. Biochem Biophys Res Commun. 1994;201(2):909–916. doi: 10.1006/bbrc.1994.1788 [DOI] [PubMed] [Google Scholar]
- 54. Campbell DB, Sutcliffe JS, Ebert PJ. et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103(45):16834–16839. doi: 10.1073/pnas.0605296103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lerner V, Miodownik C, Kaptsan A, Cohen H, Loewenthal U, Kotler M.. Vitamin B6 as add-on treatment in chronic schizophrenic and schizoaffective patients: a double-blind, placebo-controlled study. J Clin Psychiatry. 2002;63(1):54–58. [DOI] [PubMed] [Google Scholar]
- 56. Nassan FL, Williams PL, Gaskins AJ. et al. Correlation and temporal variability of urinary biomarkers of chemicals among couples: Implications for reproductive epidemiological studies. Environ Int. 2019;123:181–188. doi: 10.1016/j.envint.2018.11.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu X(M), Bennett DH, Moran RE. et al. Polybrominated diphenyl ether serum concentrations in a Californian population of children, their parents, and older adults: an exposure assessment study. Environ Health. 2015;14(1):23. doi: 10.1186/s12940-015-0002-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wu X(M), Bennett DH, Calafat AM. et al. Serum concentrations of perfluorinated compounds (PFC) among selected populations of children and Adults in California. Environ Res. 2015;136:264–273. doi: 10.1016/j.envres.2014.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thijssen EH, La Joie R, Wolf A, Advancing Research and Treatment for Frontotemporal Lobar Degeneration (ARTFL) investigators et al. Diagnostic value of plasma phosphorylated tau181 in Alzheimer’s disease and frontotemporal lobar degeneration. Nat Med. 2020;26(3):387–397. Published online doi: 10.1038/s41591-020-0762-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stamate D, Kim M, Proitsi P. et al. A metabolite‐based machine learning approach to diagnose Alzheimer‐type dementia in blood: results from the European Medical Information Framework for Alzheimer disease biomarker discovery cohort. Alzheimers Dement (NY). 2019;5(1):933–938. doi: 10.1016/j.trci.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smith AM, King JJ, West PR. et al. Amino acid dysregulation metabotypes: potential biomarkers for diagnosis and individualized treatment for subtypes of autism spectrum disorder. Biol Psychiatry. 2019;85(4):345–354. doi: 10.1016/j.biopsych.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pan J-X, Xia J-J, Deng F-L. et al. Diagnosis of major depressive disorder based on changes in multiple plasma neurotransmitters: a targeted metabolomics study. Transl Psychiatry. 2018;8(1):130. doi: 10.1038/s41398-018-0183-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding authors.