Abstract
Objectives
The acute respiratory distress syndrome (ARDS) is a heterogeneous condition, and identification of subphenotypes may help in better risk stratification. Our study objective is to identify ARDS subphenotypes using new simpler methodology and readily available clinical variables.
Setting
This is a retrospective Cohort Study of ARDS trials. Data from the US ARDSNet trials and from the international ART trial.
Participants
3763 patients from ARDSNet data sets and 1010 patients from the ART data set.
Primary and secondary outcome measures
The primary outcome was 60-day or 28-day mortality, depending on what was reported in the original trial. K-means cluster analysis was performed to identify subgroups. Sets of candidate variables were tested to assess their ability to produce different probabilities for mortality in each cluster. Clusters were compared with biomarker data, allowing identification of subphenotypes.
Results
Data from 4773 patients were analysed. Two subphenotypes (A and B) resulted in optimal separation in the final model, which included nine routinely collected clinical variables, namely heart rate, mean arterial pressure, respiratory rate, bilirubin, bicarbonate, creatinine, PaO2, arterial pH and FiO2. Participants in subphenotype B showed increased levels of proinflammatory markers, had consistently higher mortality, lower number of ventilator-free days at day 28 and longer duration of ventilation compared with patients in the subphenotype A.
Conclusions
Routinely available clinical data can successfully identify two distinct subphenotypes in adult ARDS patients. This work may facilitate implementation of precision therapy in ARDS clinical trials.
Keywords: adult intensive & critical care, adult intensive & critical care, respiratory medicine (see thoracic medicine)
Strengths and limitations of this study.
Largest cohort of patients used to identify subphenotypes of acute respiratory distress syndrome (ARDS) patients.
Subphenotypes were validated in the population of a large international ARDS randomised controlled trial.
Subphenotypes were identified by using only routinely collected clinical data.
Our use of data exclusively from randomised controlled trials does not prove generalisability to unselected ARDS populations.
The clinical utility of the subphenotypes has to be validated in a prospective study.
Introduction
The Berlin definition of acute respiratory distress syndrome (ARDS) encompasses acute hypoxemic respiratory failure due to a wide variety of etiologies.1 Due to this inclusion of heterogeneous conditions within the syndrome, there are significant clinical and biological differences that make ARDS challenging to treat.2 3 These differences among ARDS patients are associated with variation in risk of disease development and progression,3 4 potentially generating differential responses to treatments and interventions.5–10 Despite evidence, clinical risk stratification of ARDS patients still solely depends on PaO2/FiO2 ratios,11 12 possibly misleading the interpretation of results in clinical trials and clinicians when evaluating treatment options for patients.13
Therefore, identifying groups of patients who have similar clinical, physiologic or biomarker traits becomes relevant6 14 as it can help with stratification of patients producing better targeted therapies and interventions.15 These different groups can be defined as ARDS subphenotypes.4 14 Two ARDS subphenotypes have been consistently identified in previous studies.6–10 16–18 However, these models are complex, and significant barriers exist in their implementation and use in clinical practice. Existing models use up to 40 predictor variables, including biomarkers and other variables that are not readily available at the bedside.6–10 16–18 These limitations explain the current status quo of ARDS care, where clinicians must depend on the limited prognostic value of PaO2/FiO2 ratios instead of biologically distinct subphenotypes.
We hypothesised that the use of a simpler methodology and a small number of easily available clinical variables could identify new ARDS subphenotypes and thus provide the means to allow future implementation of bedside stratification.
Methods
Data source and participants
We performed a retrospective study using a deidentified data set pooling data from six randomised clinical trials in patients with ARDS, namely ARMA, ALVEOLI, FACTT, EDEN, SAILS and ART.19–24 Patients in ARMA, ALVEOLI, FACTT, EDENand SAILS trials were eligible if they met the American-European consensus for ARDS, including patients with a PaO2/FiO2 ratio <300 up to 48 hours before enrolment. From 1996 to 2013, these trials enrolled 902, 549, 1000, 1000 and 745 patients, respectively, and tested a variety of interventions.19–23 Between 2011 and 2017, the international ART study enrolled 1010 adult patients diagnosed with moderate to severe ARDS according to the Berlin definition (PaO2/FiO2 ratio <200) for less than 72 hours of duration and assessed two different ventilatory strategies.24 To avoid biases due to high mortality in the high tidal volume group of the ARMA study,19 which has not been standard of care since the beginning of 2000, only 473 patients receiving low tidal volume in that study were included.
Predictors
Six clinical trials were assessed to identify a set of clinical variables recorded closest to time of randomisation which were most commonly available across all data sets. The list of potential candidates was then further refined to include only those that are frequently observed in the routine care of ARDS patients at the time of its diagnosis according to judgement provided by intensive care unit physicians who participated in this study. To develop a clustering algorithm for potential rapid translation into clinical use, elements which would not be commonly found in the electronic health records (EHR) at the time of ARDS diagnosis, such as biomarker levels, ARDS risk factors, organ support apart from mechanical ventilation settings and severity scores, were excluded from model development. The treatment assignment in the original trials, and clinical outcomes were not considered in the model development.
After all assessment, 16 variables that are routinely collected as part of the usual care and which were uniformly present in all the trials were considered, including age, gender, arterial pH, PaO2, PaCO2, bicarbonate, creatinine, bilirubin, platelets, heart rate, respiratory rate, mean arterial pressure, positive end-expiratory pressure (PEEP), plateau pressure, FiO2 and tidal volume adjusted for predicted body weight (mL/kg PBW). The PBW was calculated as equal to 50+0.91 (centimetres of height, 152.4) in males and 45.5+0.91 (centimetres of height, 152.4) in females.18 These variables were grouped into five domains named demographics, arterial blood gases, laboratory values, vital signs and ventilatory variables. Plateau pressure was excluded due to a high rate of missingness across the trials included in the training set. Amount of missing data in the training data sets is reported in online supplemental eTable 1.
bmjopen-2021-053297supp001.pdf (405KB, pdf)
Outcomes
The primary outcome was 60-day mortality for all ARDSnet trials, and 28-day mortality for ART trial. Secondary outcomes included 90-day mortality, number of ventilator free days at day 2825 and the duration of mechanical ventilation in survivors within the first 28 days postenrolment.
Data preparation
Data preprocessing was performed before modelling, and the pooled data set was assessed for completeness and consistency. Patients with values out of the plausible physiological range for a specific variable were excluded from the final analysis (described in online supplemental eTable 2). The training data set was constructed using data from the two largest ARDSnet trials, EDEN and FACTT. The validation data set was sourced from the four remaining trials: ALVEOLI, ARMA, SAILS and ART. Means and SD for z-scoring variables were calculated from the training data set and subsequently applied to the validation data.
Statistical analysis
Baseline and outcome data were presented according to the assigned cluster. Continuous variables were presented as medians with their IQRs and categorical variables as total number and percentage. Proportions were compared using Fisher exact tests and continuous variables were compared using the Wilcoxon rank-sum test. Study outcomes were further compared using the median and mean absolute differences for continuous and categorical values, respectively.
Model development and validation
For the model development, the K-means clustering algorithm was used. K-means is one of the simplest and most used classes of clustering algorithms. In critical care research, unsupervised machine learning techniques have already been used in several studies, attempting to find homogeneous subgroups within a broad heterogeneous population.26 This specific algorithm identifies a K number of clusters in a data set by finding K centroids within the n-dimensional space of clinical features.26
For feature selection, different sets of candidate variables were tested to assess their ability to produce significantly different mortality probabilities in each cluster using the minimum amount of readily available clinical data. For each set of candidate variables, the optimal number of clusters was determined by comparing models with between 2 and 5 clusters, using the Elbow method27 and the Calinski-Harabasz index.28 Information about the methods for selecting the number of clusters are provided in the online supplemental material.
The following steps were performed for the final model selection: (1) all predictors were assessed for correlation (online supplemental eTable 3); and (2) 10 different combinations of the proposed variables were investigated. These combinations were developed based on the perceived clinical importance of each variable and its combinations. All 10 models were tested for the optimal number of clusters based on both the Elbow method and the Calinski-Harabasz index, as described above. The models were then compared, aiming for the minimum set of variables with high 60-day mortality separation. The description of each model is shown in online supplemental eTable 4.
Biological and clinical characteristics of the clusters were evaluated using clinical, laboratory and (when available) biomarker data to establish subphenotypes.4 All iterations in model development were done on the training set and the generalisability of the final model was assessed using the validation data set. K-means clustering analysis is structured to ignore cases with missing data. No assumption was made for missingness, and we therefore conducted a complete case analysis. Model development and evaluation was performed using Python V.3.8 and scikit-learn 0.23.1.
Patient and public involvement
There was no patient involvement in this study.
Data availability
Data from the ARDSnet studies (EDEN, FACTT, ARMA, ALVEOLI and SAILS) are publicly available from the NHLBI ARDS Network and data from the ART trial can be requested from study authors.
Results
Participants
Data from 4777 clinical trial patients were considered for inclusion. In total, four patients were excluded for having clinical measurements outside plausible range. The remaining 1998 patients from EDEN and FACTT trials were included in the training set, while the 2775 patients from ARMA, ALVEOLI, SAILS and ART were included in the validation cohort.
Baseline characteristics of the patients in the training and validation sets are presented in table 1. Pneumonia was the prevailing aetiology followed by sepsis and aspiration in all trials. Between 29.3% and 72.7% of the patients were receiving vasopressors at the time of randomisation. At randomisation, PaO2/FiO2 ratio ranged from 112 (75–158) to 134 (96–185) mm Hg, and PEEP from 8 (5–10) to 12 (10–14) cmH2O across trials. Mortality at 60 days for the ARDSnet trials ranged from 22.7% to 30.1%, while in the ART trial mortality at 28 days was 58.8%.
Table 1.
Baseline characteristics and clinical outcomes in the included trials
| Training set (n=1998) | Validation set (n=2775) | |||||
| EDEN (n=1000) |
FACTT (n=998) |
ALVEOLI (n=549) |
ARMA (n=472) |
ART (n=1010) |
SAILS (n=744) |
|
| Age, year | 52.0 (42.0–63.0) | 49.0 (38.0–60.8) | 50.0 (39.0–65.0) | 50.0 (37.8–65.0) | 52.0 (36.0–64.0) | 55.0 (42.0–66.0) |
| Male gender, no. (%) | 510 (51.0) | 533 (53.4) | 302 (55.0) | 285 (60.4) | 631 (62.5) | 365 (49.0) |
| Aetiology, no. (%) | ||||||
| Pneumonia | 650 (65.0) | 471 (47.2) | 221 (40.3) | 145 (30.7) | 555 (55.0) | 526 (70.7) |
| Sepsis | 147 (14.7) | 231 (23.1) | 120 (21.9) | 125 (26.5) | 196 (19.4) | 147 (19.8) |
| Aspiration | 96 (9.6) | 149 (14.9) | 84 (15.3) | 72 (15.3) | 58 (5.7) | 49 (6.6) |
| Trauma | 36 (3.6) | 74 (7.4) | 45 (8.2) | 59 (12.5) | 31 (3.1) | 6 (0.8) |
| Other | 71 (7.1) | 73 (7.3) | 79 (14.4) | 71 (15.0) | 170 (16.8) | 16 (2.2) |
| Severity of illness* | 73.0 (59.0–89.0) | 78.0 (62.0–94.0) | 78.0 (64.0–93.0) | 83.0 (70.0–97.0) | 63.0 (50.2–75.0) | 76.0 (61.0–92.0) |
| Vasopressors, no. (%) | 489 (48.9) | 397 (40.5) | 156 (29.3) | 147 (31.3) | 734 (72.7) | 395 (54.2) |
| Laboratory tests | ||||||
| White cell count, 109 /L | 12.0 (7.8–16.7) | 11.8 (7.2–17.1) | 11.6 (7.7–15.7) | 11.5 (7.5–16.2) | – | 13.9 (8.7–20.0) |
| Platelets, 109 /L | 169 (108–241) | 183 (106–258) | 157 (83–247) | 135 (80–211) | 175 (106–263) | 167 (96–247) |
| Creatinine, mg/dL | 1.2 (0.8–2.0) | 1.0 (0.7–1.5) | 1.0 (0.7–1.7) | 1.1 (0.8–1.7) | 1.3 (0.8–2.2) | 1.0 (0.7–1.7) |
| Bilirubin, mg/dL | 0.8 (0.5–1.4) | 0.8 (0.5–1.6) | 0.8 (0.5–1.5) | 1.0 (0.6–2.1) | 0.8 (0.4–1.5) | 0.8 (0.5–1.4) |
| Arterial blood gas | ||||||
| pH* | 7.36 (7.30–7.42) | 7.37 (7.30–7.43) | 7.40 (7.34–7.44) | 7.41 (7.35–7.45) | 7.28 (7.19–7.36) | 7.37 (7.31–7.42) |
| PaO2, mm Hg | 83 (68–108) | 79 (67–100) | 77 (67–93) | 76.5 (67–93) | 112 (81–155) | 83 (69–103) |
| PaO2/FiO2 | 125 (86–178) | 118 (80–163) | 134 (96–185) | 112 (75–158) | 112 (81–155) | 133 (89–178) |
| PaCO2, mm Hg | 38 (34–45) | 39 (34–45) | 38 (33–43) | 36 (31–41) | 50 (42–62) | 39 (34–45) |
| Bicarbonate, mmol/L | 21.0 (18.0–25.0) | 21.0 (17.4–25.0) | 22.0 (18.0–26.0) | 22.0 (18.0–25.0) | 22.9 (19.4–26.3) | 22.0 (18.0–25.0) |
| Ventilatory variables | ||||||
| Tidal volume, mL | 410 (360–470) | 450 (400–510) | 500 (420–600) | 700 (600–750) | 350 (308–400) | 400 (350–460) |
| Per PBW, mL/kg PBW | 6.3 (6.0–7.3) | 7.1 (6.1–8.1) | 7.9 (6.6–9.4) | 10.2 (9.0–11.3) | 5.9 (5.1–6.1) | 6.2 (6.0–7.1) |
| Plateau pressure, cmH2O | 24.0 (20.0–27.0) | 26.0 (22.0–30.0) | 26.0 (22.0–31.0) | 29.0 (24.8–34.0) | 26.0 (22.0–29.0) | 24.0 (19.0–28.0) |
| PEEP, cmH2O | 10 (5–12) | 10 (5–12) | 10 (5–12) | 8 (5–10) | 12 (10–14) | 10 (5–11) |
| FiO2 | 0.60 (0.50–0.80) | 0.60 (0.50–0.80) | 0.60 (0.50–0.80) | 0.60 (0.50–0.74) | 0.70 (0.60–1.00) | 0.60 (0.40–0.70) |
| Clinical outcomes | ||||||
| 28-day mortality, no. (%) | – | – | – | – | 594 (58.8) | – |
| 60-day mortality, no. (%) | 227 (22.7) | 268 (26.9) | 144 (26.2) | 141 (30.1) | – | 199 (26.7) |
| 90-day mortality, no. (%) | 233 (23.3) | 283 (28.6) | 148 (27.5) | 143 (30.8) | – | 204 (27.4) |
| Ventilator-free days, day 28 | 20.0 (0.0–24.0) | 17.0 (0.0–23.0) | 18.0 (0.0–24.0) | 13.0 (0.0–23.0) | 0.0 (0.0–13.0) | 20.0 (0.0–25.0) |
| Ventilator days in survivors | 7.0 (4.0–13.0) | 8.0 (5.0–16.0) | 8.0 (4.0–14.0) | 8.0 (4.0–15.0) | 13.0 (8.0–20.0) | 6.0 (4.0–11.0) |
Data are median (quartile 25th–quartile 75th) or N (%).
*Except for ART, that uses SAPS-3, all studies use APACHE-IV.
APACHE, Acute Physiology and Chronic Health Evaluation; PBW, predicted body weight; PEEP, positive end-expiratory pressure.
Predictor variables and model selection
The correlation between the 15 variables selected for clustering is shown in online supplemental eTable 3. The strongest correlation was between PEEP and FiO2 (r=0.49). The comparison of the 10 models regarding the optimal number of clusters based on both the Elbow method and the Calinski-Harabasz index is shown in online supplemental eFigure 1. In all models and methods, two clusters were a better fit than a higher number of clusters.
Across the 10 models, absolute mortality difference between cluster 1 and cluster 2 ranged from 3.9% to 13.1% for the FACTT study and between 0.1% and 8.1% for EDEN (online supplemental eTable 4). The models with the highest 60-day absolute mortality separation between the clusters for each of the two trials in the training set were then further evaluated. Models 6, 5 and 8 were consistently among the models with highest separation (online supplemental eTable 4). Model 8 was selected for further investigation, as it had the fewest variables (online supplemental eTable 5).
Clinical characteristics of each cluster
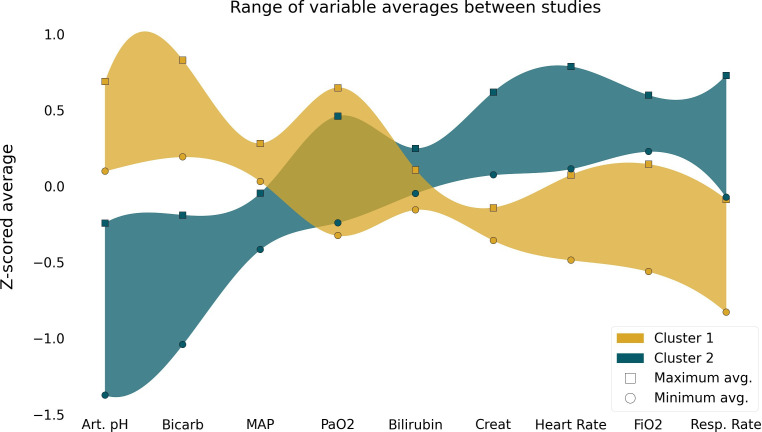
Based on model 8, only nine clinical and laboratory variables were needed to identify the two distinct clusters in ARDS patients, namely heart rate, mean arterial pressure, respiratory rate, bilirubin, bicarbonate, creatinine, PaO2, arterial pH and FiO2. For each variable in the model, opposing measurements could be observed for each cluster (figure 1 and online supplemental eFigure 2). For the ARDSnet trials, the incidence of cluster 1 patients varied from 57.8% (EDEN) to 73.6% (ARMA), and 41.5% of ART patients were part of cluster 1. Across all trials, patients in cluster 2 had higher severity of illness, rate of vasopressor, heart rate, respiratory rate, creatinine and bilirubin, as well as lower platelets, pH, blood urea nitrogen and bicarbonate compared with patients in cluster 1 (table 2, online supplemental eTables 6 and 7). In addition, 28-day, 60-day and 90-day mortality rate was higher in patients in cluster 2 in all trials (table 3). Likewise, for each trial, the number of ventilator-free days at day 28 was lower in patients in cluster 2 compared with cluster 1, and duration of ventilation in survivors was longer in cluster 1.
Figure 1.
Differences of the variables included in the cluster algorithm among clusters. Square symbols represent the study with the highest mean z score for each phenotype; circles represent the study with the lowest mean z score for each phenotype. The coloured bands are exclusively to help visualise the opposite trends of the variables on the different clusters; Art.pH, arterial pH; Bicarb, bicarbonate; MAP, mean arterial pressure; Creat, creatinine; Resp.Rate, respiratory rate.
Table 2.
Baseline characteristics and clinical outcomes according to the clusters and trials in the training set
| FACTT | EDEN | |||||
| Cluster 1 (n=407) |
Cluster 2 (n=294) |
P value | Cluster 1 (n=449) |
Cluster 2 (n=328) |
P value | |
| Age, year* | 50.0 (40.0–63.0) | 47.0 (36.0–58.0) | 0.002 | 53.0 (44.0–63.0) | 51.0 (41.0–62.2) | 0.183 |
| Male gender, no. (%) | 223 (54.8) | 151 (51.4) | 0.411 | 233 (51.9) | 168 (51.2) | 0.910 |
| Body mass index, kg/m2 | 27.5 (23.3–32.1) | 27.4 (23.0–32.7) | 0.938 | 29.1 (24.6–34.5) | 28.5 (23.4–35.1) | 0.476 |
| Caucasian, no. (%) | 269 (66.1) | 177 (60.2) | 0.129 | 349 (81.5) | 237 (75.7) | 0.067 |
| Aetiology, no. (%) | <0.001 | 0.003 | ||||
| Pneumonia | 201 (49.4) | 139 (47.3) | 296 (65.9) | 217 (66.2) | ||
| Sepsis | 78 (19.2) | 101 (34.4) | 50 (11.1) | 60 (18.3) | ||
| Aspiration | 67 (16.5) | 30 (10.2) | 45 (10.0) | 27 (8.2) | ||
| Trauma | 24 (5.9) | 8 (2.7) | 24 (5.3) | 5 (1.5) | ||
| Other | 37 (9.1) | 16 (5.4) | 34 (7.6) | 19 (5.8) | ||
| Prognostic scores | ||||||
| APACHE III | 69.0 (56.0–84.0) | 91 (76.0–105.0) | <0.001 | 66.0 (54.0–79.0) | 84.0 (71.0–100.2) | <0.001 |
| Use of vasopressor, no. (%) | 118 (29.5) | 189 (64.9) | <0.001 | 187 (41.6) | 209 (63.7) | <0.001 |
| Vital signs | ||||||
| Temperature, °C | 37.5 (36.8–38.2) | 37.6 (37.0–38.4) | 0.371 | 37.3 (36.8–37.8) | 37.3 (36.7–38.1) | 0.212 |
| Heart rate, bpm | 95.0 (81.0–110.0) | 114 (102–126) | <0.001 | 89 (77–102) | 101 (89–116) | <0.001 |
| Mean arterial pressure, mm Hg | 76.0 (68.0–88.0) | 71.0 (65.0–80.8) | <0.001 | 77.0 (68.0–84.0) | 71.0 (66.0–80.0) | <0.001 |
| SpO2, % | 96 (93–98) | 95 (92–97) | <0.001 | 96 (94–98) | 95 (92–98) | 0.032 |
| Urine output in 24 hours, mL | 1785 (1192–2853) | 1370 (842–2446) | <0.001 | 1505 (977–2250) | 1165 (566–1816) | <0.001 |
| Laboratory tests | ||||||
| Haematocrit, % | 30.0 (26.0–33.0) | 30.0 (24.2–35.0) | 0.272 | 30.0 (26.0–34.0) | 30.0 (26.0–35.0) | 0.919 |
| White cell count, 109 /L | 11.6 (7.3–16.3) | 11.7 (5.6–17.9) | 0.972 | 11.4 (7.7–15.5) | 12.7 (7.7–19.0) | 0.019 |
| Platelets, 109 /L | 195 (118.5–268) | 158 (87–237) | <0.001 | 163 (108–241) | 164 (103–227) | 0.552 |
| Creatinine, mg/dL | 0.9 (0.7–1.3) | 1.4 (1.0–2.0) | <0.001 | 1.0 (0.7–1.5) | 1.6 (1.0–2.8) | <0.001 |
| Bilirubin, mg/dL | 0.7 (0.5–1.3) | 0.9 (0.5–2.0) | 0.003 | 0.8 (0.5–1.3) | 0.8 (0.5–1.7) | 0.128 |
| Arterial blood gas | ||||||
| pH* | 7.41 (7.36–7.45) | 7.29 (7.23–7.35) | <0.001 | 7.40 (7.35–7.44) | 7.30 (7.24–7.35) | <0.001 |
| PaO2, mm Hg | 78 (68–100) | 78 (65–99) | 0.240 | 83 (70–107) | 81 (67–107) | 0.416 |
| PaO2/FiO2 | 132 (92–173) | 89 (65–126) | <0.001 | 133 (98–193) | 101 (73–162) | <0.001 |
| PaCO2, mm Hg | 39 (34–44) | 38.5 (33–47.9) | 0.877 | 38 (34–44) | 38 (33–46) | 0.55 |
| Bicarbonate, mmol/L | 24.0 (21.0–27.0) | 17.0 (14.0–20.0) | <0.001 | 23.0 (21.0–26.0) | 18.5 (15.0–21.0) | <0.001 |
| Ventilatory variables | ||||||
| Tidal volume, mL | 450 (400–530) | 450 (382–500) | 0.009 | 420 (356–487) | 400 (350–450) | 0.032 |
| Per PBW, mL/kg PBW | 7.1 (6.3–8.4) | 7.0 (6.0, 8.0) | 0.058 | 6.3 (6.0–7.5) | 6.1 (6.0–7.3) | 0.079 |
| Plateau pressure, cmH2O | 25.0 (20.0–29.0) | 28.0 (24.0–32.0) | <0.001 | 23.0 (19.0–27.0) | 24.0 (21.0–28.0) | 0.004 |
| PEEP, cmH2O | 8 (5–10) | 10 (8–14) | <0.001 | 10 (5–10) | 10 (8–14) | <0.001 |
| Respiratory rate, breaths/min | 22 (18–27) | 30 (24–35) | <0.001 | 22 (19–26) | 30 (25–35) | <0.001 |
| FiO2 | 0.50 (0.40–0.70) | 0.80 (0.60–1.00) | <0.001 | 0.60 (0.45–0.70) | 0.80 (0.60–1.00) | <0.001 |
Data are mean±SD, median (quartile 25th–quartile 75th) or N (%).
APACHE, Acute Physiology and Chronic Health Evaluation; PEEP, positive end-expiratory pressure; VT/PBW, tidal volume per predicted body weight.
Table 3.
Clinical outcomes according to clusters in each trial
| Cluster 1 | Cluster 2 | Difference (95% CI) | P value | |
| Training set | ||||
| FACTT | n=407 | n=294 | ||
| 60-day mortality, no. (%) | 94 (23.1) | 102 (34.7) | 11.6% (4.9% to 18.3%) | 0.001 |
| 90-day mortality, no. (%) | 103 (25.4) | 106 (36.3) | 10.9% (4.1% to 17.8%) | 0.002 |
| Ventilator-free days at day 28 | 19.0 (0.0–24.0) | 10.0 (0.0–21.0) | −9.0 (–11.9 to –6.1) | <0.001 |
| Duration of ventilation in survivors, days | 8.0 (4.0–13.0) | 10.0 (7.0–19.0) | 2.0 (0.5 to 3.5) | <0.001 |
| EDEN | n=449 | n=328 | ||
| 60-day mortality, no. (%) | 87 (19.4) | 90 (27.4) | 8.1% (2.1% to 14.0%) | 0.010 |
| 90-day mortality, no. (%) | 90 (20.0) | 93 (28.4) | 8.3% (2.3% to 14.3%) | 0.009 |
| Ventilator-free days at day 28 | 21.0 (0.0–25.0) | 15.0 (0.0–22.2) | −6.0 (–8.1 to –3.9) | <0.001 |
| Duration of ventilation in survivors, days | 6.0 (4.0–11.0) | 8.0 (6.0–18.0) | 2.0 (0.9 to 3.1) | <0.001 |
| Validation set | ||||
| ALVEOLI | n=336 | n=157 | ||
| 60-day mortality, no. (%) | 59 (17.6) | 68 (43.3) | 25.8% (17.7% to 33.8%) | <0.001 |
| 90-day mortality, no. (%) | 60 (18.1) | 70 (45.5) | 27.3% (19.2% to 35.5%) | <0.001 |
| Ventilator-free days at day 28 | 21.0 (4.8–25.0) | 2.0 (0.0–19.0) | −19.0 (–20.8 to –17.2) | <0.001 |
| Duration of ventilation in survivors, days | 7.0(4.0,13.0) | 11.0 (6.0–22.2) | 4.0 (2.1 to 5.9) | <0.001 |
| ARMA | n=279 | n=100 | ||
| 60-day mortality, no. (%) | 69 (24.8) | 42 (42.0) | 17.2% (6.9% to 27.5%) | 0.002 |
| 90-day mortality, no. (%) | 70 (25.5) | 42 (42.0) | 16.5% (6.0% to 26.9%) | 0.003 |
| Ventilator-free days at day 28 | 17.0 (0.0–24.0) | 2.0 (0.0–19.0) | −15.0 (–18.6 to –11.4) | <0.001 |
| Duration of ventilation in survivors, days | 7.0 (4.0–13.8) | 11.0 (5.0–18.0) | 4.0 (1.5 to 6.5) | 0.018 |
| SAILS | n=319 | n=188 | ||
| 60-day mortality, no. (%) | 80 (25.1) | 60 (31.9) | 6.8% (–1.2% to 14.9%) | 0.119 |
| 90-day mortality, no. (%) | 81 (25.4) | 63 (33.5) | 8.1% (0.0% to 16.3%) | 0.063 |
| Ventilator-free days at day 28 | 21.0 (0.0–25.0) | 16.0 (0.0–23.0) | −5.0 (–7.3 to –2.7) | <0.001 |
| Duration of ventilation in survivors, days | 6.0 (3.0–10.0) | 8.0 (5.0–14.0) | 2.0 (0.7 to 3.3) | <0.001 |
| ART | n=211 | n=298 | ||
| 28-day mortality, no. (%) | 81 (38.4) | 180 (60.4) | 22.0% (13.4% to 30.7%) | <0.001 |
| Ventilator-free days at day 28 | 0.0 (0.0–17.0) | 0.0 (0.0–7.8) | −0.0 (–1.0 to 1.0) | <0.001 |
| Duration of ventilation in survivors, days | 12.0 (8.0–20.0) | 13.5 (8.0–20.0) | 2.0 (–0.3 to 4.2) | 0.570 |
Data are median (quartile 25th–quartile 75th) or N (%). Difference is mean difference with (95% CI) for binomial variables and median difference with (95% CI) for continuous variables.
Identification of subphenotypes
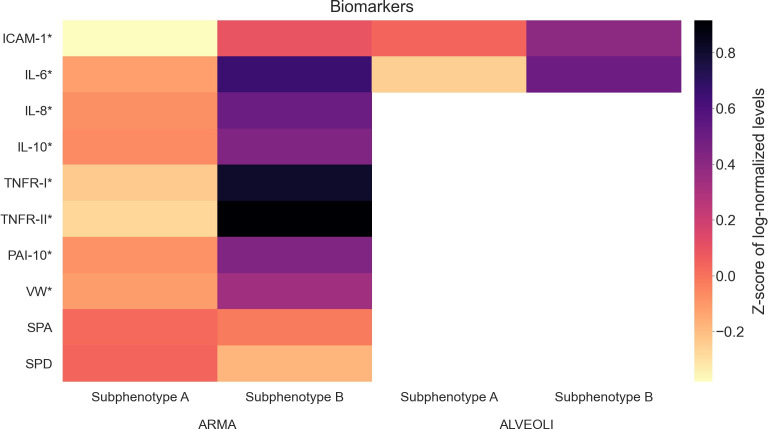
After comparing the clinical characteristics of the clusters, each cluster was assigned to represent a distinct subphenotype of ARDS, with patients in cluster 1 assigned to subphenotype A, and patients in cluster 2 assigned to subphenotype B. Using blood biomarker information available for a subset of patients from both ARMA and ALVEOLI, subphenotype B showed increased levels of proinflammatory markers when compared with subphenotype A (figure 2 and online supplemental eTables 8 and 9).
Figure 2.
Heat map of the biomarkers available for the ARMA and ALVEOLI trials. For better visualisation and due to difference in scales, the values were log-normalised and z-scored. Subphenotypes A and B are shown separately to highlight their differences.
Discussion
This study successfully demonstrated that nine easily obtainable clinical variables: arterial pH, partial O2 pressure, creatinine, bilirubin, bicarbonate, mean arterial pressure, heart rate, respiratory rate and FiO2 at the time of study enrolment can identify two distinct ARDS subphenotypes with different clinical and biologic characteristics as well as outcomes across the test and validation cohorts. There was good generalisability among diverse populations from multiple validation data sets with temporal and geographical differences.
It is understandable that researchers feel compelled to use as much information as possible to build robust models. This is supportable for two main reasons: (1) the well-known heterogeneity of complex syndromes such as ARDS and (2) the abundance of highly granular clinical data generated by EHRs. It is anticipated that analysing this vast amount of data will provide new knowledge regarding disease mechanisms by enabling researchers to find plausible hidden patterns within the data.29 However, this data-heavy approach has the potential drawback of using predictors which are not generally obtained in a time window prior to intervention, or worse yet, using variables that are not part of the routine standard of care for patients. The rationale of using fewer and easy to collect clinical variables is not new in the field of critical care. Prognostic models have already shown that it is indeed feasible to create meaningful models using fewer predictors.30 31
Unfortunately, unlike supervised algorithms (eg, regression analyses), unsupervised algorithms such as K-means clustering do not provide one straightforward and established metric to describe feature importance. In that sense, our approach of testing multiple sets of variables was also meant to select features that were most likely to be relevant, serving as surrogate for the feature selection step normally employed in supervised algorithms. While each individual variable by itself may not be significantly different across subphenotypes, their interaction in the nine-dimensional space of our model may be relevant.
Our initial choices to define variables commonly found in the EHR at ARDS diagnosis was inspired by a recent report from the WHO which showed an enormous discrepancy of medical devices availability in a survey across 135 countries.29 Recognising this inconsistency is essential for widespread implementation of machine learning models regardless of varying availability of resources across countries and health systems.29 The aim is to provide clinically relevant information within a defined and short period that might impact the delivery of effective interventions to the right patient population and to as many patients as possible.29
Recently, Sinha et al developed supervised-learning gradient boosted classifier models trained using 24 or 14 readily available clinical data elements to reproduce biomarker-derived subphenotypes which were previously identified by Calfee et al.17 Unlike Sinha et al, who predicted previously identified subphenotypes, our study has identified two subphenotypes de novo using a small set of clinical variables.
Although the subphenotypes that we have identified and those that have been previously published look similar, our work is distinct from previous studies in several ways. We employed different training and validation data sets as well as a different and well-established unsupervised learning technique. Moreover, we utilised a process for selecting predictors which is not comparable to previous studies. Acknowledging these differences is crucial. It would not be unexpected to assume that these deviations would be relevant enough to produce different subphenotypes.32 However, the clinical, laboratory characteristics and the clinical outcomes of our subphenotypes show that they are remarkably similar to subphenotypes found in previous papers, regardless of methodological differences.
At this point it is not possible to go beyond this comparative analysis, as there is no gold standard definition of ARDS subphenotypes.32 Nonetheless, our work does provide robust evidence that ARDS does indeed have two subphenotypes that can be systematically identified, despite major differences in population assessed and methodological approach used compared with previous studies. It also reinforces that we should continue to explore the underlying biological pathways of such subphenotypes to find responders to new or previously tested therapies.
Our study has several strengths. First, it is the largest cohort of patients that has been studied to develop distinct subphenotypes of ARDS patients. Moreover, our validation cohort included patients from the ART trial, allowing us to validate our model in the contemporaneous population of a large international randomised clinical trial in addition to the ARDSnet studies used in other subphenotyping studies. Second, our subphenotyping model was developed exclusively on the training set and then validated across multiple separate data sets. Nevertheless, similar separation in mortality was seen between the two subphenotypes across all trials. Third, we used the K-means algorithm to identify our subphenotypes, and the results obtained with this technique can be easily interpreted by clinicians and implemented in clinical practice. Finally, this is the first phenotyping study that has used easily available clinical variables to identify ARDS phenotypes de novo, which allows for early identification of these patients in the clinical care at the bedside. Using this algorithm with a small number of routinely collected variables could enable our model to be applied in trials that either retrospectively or prospectively assess interventions targeted to each subphenotype.
This study also has limitations. First, we have developed our models exclusively on patients enrolled in clinical trials. Due to the strict inclusion and exclusion criteria of these clinical trials, the generalisability of these results needs to be evaluated in unselected ARDS populations. Although there are clear clinical and biomarker differences between the identified subphenotypes, the model’s clinical utility needs to be prospectively validated and further investigated. Additionally, our biomarker analysis is limited to those patients in which the data were made publicly available by the study authors, but future collection of biomarker data in a prospective study will allow more robust understanding of the underlying biology and validation of the subphenotype model. Also, K-means clustering does not handle missing data, and no approach was used to impute missing values. However, the extremely low rate of missingness in our study makes this issue less relevant. Finally, future work should analyse previous trials to identify possible differential treatment responses for the subphenotypes of ARDS patients identified in this study.
Conclusions
This study confirms the existence of two distinct subphenotypes in ARDS patients using a novel clustering model on routinely collected clinical data. This work may allow for easier identification of ARDS subphenotypes to facilitate implementation of precision clinical trial enrolment and development of targeted therapies in a variety of settings without the added burdens of biomarker evaluation.
Supplementary Material
Footnotes
Twitter: @abhiduggalMD
AD, RK, EVA and LB contributed equally.
Contributors: AD, RK, EVA and LB participated in study design and analysis, drafted and revised the manuscript and are the guarantor of the document. MTS, DAR, JO, FGZ, ABC, IM, DMP, LNL and ASN participated in interpretation of data analysis, drafted the manuscript and revised it for critically important intellectual content. ROD participated in the study design, analysis, interpretation of data analysis and final revision of the manuscript content.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: RK, EVA, LB, JO, DR and ROD are employees of Endpoint Health and ASN reported receiving personal fees from Dräger unrelated to the submitted work.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information. Data from the ARDSnet studies (EDEN, FACTT, ARMA, ALVEOLI) may be requested from the NIH-NHLBI BioLINCC repository. These are available by application through the BioLINCC website (https://biolincc.nhlbi.nih.gov/resource_overview/), subject to established review processes for their distribution to qualified investigators and data from the ART trial can be requested from study authors.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All patients provided informed consent in the original trials. This secondary analysis study was exempt from IRB review because it does not meet the definition of human subject as defined in 45 CFR 46.102. WIRB Work Order #1-1228617-1.
References
- 1.ARDS Definition Task Force, Ranieri VM, Rubenfeld GD, et al. Acute respiratory distress syndrome: the Berlin definition. JAMA 2012;307:2526–33. 10.1001/jama.2012.5669 [DOI] [PubMed] [Google Scholar]
- 2.Thille AW, Esteban A, Fernández-Segoviano P, et al. Comparison of the Berlin definition for acute respiratory distress syndrome with autopsy. Am J Respir Crit Care Med 2013;187:761–7. 10.1164/rccm.201211-1981OC [DOI] [PubMed] [Google Scholar]
- 3.Reilly J, Calfee C, Christie J. Acute respiratory distress syndrome phenotypes. Semin Respir Crit Care Med 2019;40:019–30. 10.1055/s-0039-1684049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy K, Sinha P, O'Kane CM, et al. Subphenotypes in critical care: translation into clinical practice. Lancet Respir Med 2020;8:631–43. 10.1016/S2213-2600(20)30124-7 [DOI] [PubMed] [Google Scholar]
- 5.Shankar-Hari M, Fan E, Ferguson ND. Acute respiratory distress syndrome (ARDS) phenotyping. Intensive Care Med 2019;45:516–9. 10.1007/s00134-018-5480-6 [DOI] [PubMed] [Google Scholar]
- 6.Calfee CS, Delucchi K, Parsons PE, et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2014;2:611–20. 10.1016/S2213-2600(14)70097-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Famous KR, Delucchi K, Ware LB, et al. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med 2017;195:331–8. 10.1164/rccm.201603-0645OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calfee CS, Delucchi KL, Sinha P, et al. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med 2018;6:691–8. 10.1016/S2213-2600(18)30177-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinha P, Delucchi KL, Thompson BT, et al. Latent class analysis of ARDS subphenotypes: a secondary analysis of the statins for acutely injured lungs from sepsis (sails) study. Intensive Care Med 2018;44:1859–69. 10.1007/s00134-018-5378-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos LD, Schouten LR, van Vught LA, et al. Identification and validation of distinct biological phenotypes in patients with acute respiratory distress syndrome by cluster analysis. Thorax 2017;72:876–83. 10.1136/thoraxjnl-2016-209719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson ND, Fan E, Camporota L, et al. The Berlin definition of ARDS: an expanded rationale, justification, and supplementary material. Intensive Care Med 2012;38:1573–82. 10.1007/s00134-012-2682-1 [DOI] [PubMed] [Google Scholar]
- 12.Bellani G, Laffey JG, Pham T, et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA 2016;315:788. 10.1001/jama.2016.0291 [DOI] [PubMed] [Google Scholar]
- 13.Gattinoni L, Vassalli F, Romitti F. Benefits and risks of the P/F approach. Intensive Care Med 2018;44:2245–7. 10.1007/s00134-018-5413-4 [DOI] [PubMed] [Google Scholar]
- 14.Matthay MA, Arabi YM, Siegel ER, et al. Phenotypes and personalized medicine in the acute respiratory distress syndrome. Intensive Care Med 2020;46:2136–52. 10.1007/s00134-020-06296-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shankar-Hari M, Rubenfeld GD. Population enrichment for critical care trials: phenotypes and differential outcomes. Curr Opin Crit Care 2019;25:489–97. 10.1097/MCC.0000000000000641 [DOI] [PubMed] [Google Scholar]
- 16.Sinha P, Delucchi KL, McAuley DF, et al. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir Med 2020;8:247–57. 10.1016/S2213-2600(19)30369-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinha P, Churpek MM, Calfee CS. Machine learning classifier models can identify acute respiratory distress syndrome phenotypes using readily available clinical data. Am J Respir Crit Care Med 2020;202:996–1004. 10.1164/rccm.202002-0347OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitsios GD, Yang L, Manatakis DV, et al. Host-Response subphenotypes offer prognostic enrichment in patients with or at risk for acute respiratory distress syndrome. Crit Care Med 2019;47:1724–34. 10.1097/CCM.0000000000004018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acute Respiratory Distress Syndrome Network, Brower RG, Matthay MA, et al. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med 2000;342:1301–8. 10.1056/NEJM200005043421801 [DOI] [PubMed] [Google Scholar]
- 20.Brower RG, Lanken PN, MacIntyre N, et al. Higher versus lower positive end-expiratory pressures in patients with the acute respiratory distress syndrome. N Engl J Med 2004;351:327–36. 10.1056/NEJMoa032193 [DOI] [PubMed] [Google Scholar]
- 21.National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome (ARDS) Clinical Trials Network, Wheeler AP, Bernard GR, et al. Pulmonary-Artery versus central venous catheter to guide treatment of acute lung injury. N Engl J Med 2006;354:2213–24. 10.1056/NEJMoa061895 [DOI] [PubMed] [Google Scholar]
- 22.National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome (ARDS) Clinical Trials Network, Rice TW, Wheeler AP, et al. Initial trophic vs full enteral feeding in patients with acute lung injury: the EDEN randomized trial. JAMA 2012;307:795–803. 10.1001/jama.2012.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Heart, Lung, and Blood Institute ARDS Clinical Trials Network, Truwit JD, Bernard GR, et al. Rosuvastatin for sepsis-associated acute respiratory distress syndrome. N Engl J Med 2014;370:2191–200. 10.1056/NEJMoa1401520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Writing Group for the Alveolar Recruitment for Acute Respiratory Distress Syndrome Trial (ART) Investigators, Cavalcanti AB, Suzumura Érica Aranha, et al. Effect of lung recruitment and titrated positive end-expiratory pressure (PEEP) vs low PEEP on mortality in patients with acute respiratory distress syndrome: a randomized clinical trial. JAMA 2017;318:1335. 10.1001/jama.2017.14171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yehya N, Harhay MO, Curley MAQ, et al. Reappraisal of Ventilator-Free days in critical care research. Am J Respir Crit Care Med 2019;200:828–36. 10.1164/rccm.201810-2050CP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castela Forte J, Perner A, van der Horst ICC. The use of clustering algorithms in critical care research to unravel patient heterogeneity. Intensive Care Med 2019;45:1025–8. 10.1007/s00134-019-05631-z [DOI] [PubMed] [Google Scholar]
- 27.KETCHEN Jr. DJ, Shook CL. The application of cluster analysis in strategic management research: an analysis and critique. Strateg Manag J 1996;17:441–58. [DOI] [Google Scholar]
- 28.Caliński T, Harabasz J. A dendrite method for cluster analysis. Communications in Statistics 1974;3:1–27. [Google Scholar]
- 29.Bulgarelli L, Deliberato RO, Johnson AEW. Prediction on critically ill patients: The role of "big data". J Crit Care 2020;60:64–8. 10.1016/j.jcrc.2020.07.017 [DOI] [PubMed] [Google Scholar]
- 30.Johnson AEW, Kramer AA, Clifford GD. A new severity of illness scale using a subset of acute physiology and chronic health evaluation data elements shows comparable predictive accuracy. Crit Care Med 2013;41:1711–8. 10.1097/CCM.0b013e31828a24fe [DOI] [PubMed] [Google Scholar]
- 31.Deliberato RO, Escudero GG, Bulgarelli L, et al. SEVERITAS: an externally validated mortality prediction for critically ill patients in low and middle-income countries. Int J Med Inform 2019;131:103959. 10.1016/j.ijmedinf.2019.103959 [DOI] [PubMed] [Google Scholar]
- 32.DeMerle KM, Angus DC, Baillie JK, et al. Sepsis subclasses: a framework for development and interpretation. Crit Care Med 2021;49:748–59. 10.1097/CCM.0000000000004842 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2021-053297supp001.pdf (405KB, pdf)
Data Availability Statement
Data from the ARDSnet studies (EDEN, FACTT, ARMA, ALVEOLI and SAILS) are publicly available from the NHLBI ARDS Network and data from the ART trial can be requested from study authors.
Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information. Data from the ARDSnet studies (EDEN, FACTT, ARMA, ALVEOLI) may be requested from the NIH-NHLBI BioLINCC repository. These are available by application through the BioLINCC website (https://biolincc.nhlbi.nih.gov/resource_overview/), subject to established review processes for their distribution to qualified investigators and data from the ART trial can be requested from study authors.