Abstract
As a natural environment for human-microbiota interactions, healthy mucus houses a remarkably stable and diverse microbial community. Maintaining this microbiota is essential to human health, both to support the commensal bacteria that perform a wide array of beneficial functions and to prevent the outgrowth of pathogens. However, how the host selects and maintains a specialized microbiota remains largely unknown. In this viewpoint, we propose several strategies by which mucus may regulate the composition and function of the human microbiota and discuss how compromised mucus barriers in disease can give rise to microbial dysbiosis.
Keywords: microbial dysbiosis, microbiota, mucin glycans, mucins, mucus
Introduction
The human body is estimated to host 38 trillion bacterial cells [1] in large and diverse microbial communities on external surfaces covered by skin and on internal surfaces lined with mucus. Although microbes in these complex communities encounter numerous potential competitors for nutrients and space, a healthy microbiota remains relatively stable [2,3] and diverse [4] (Fig. 1A). Commensal microbes promote human health in a variety of ways, including by resisting invasion by potential pathogens [5], metabolizing otherwise inaccessible carbohydrates [6], synthesizing vitamins [6], and priming the adaptive immune system [7]. On the other hand, shifts in microbiota composition and function cause an imbalance—dysbiosis—that is implicated in a wide range of inflammatory, cardiovascular, and autoimmune diseases [8–12] (Fig. 1B). It is therefore imperative that the host effectively manages its microbial inhabitants, but how the body selects and maintains complex microbial communities is not fully understood.
Fig. 1.
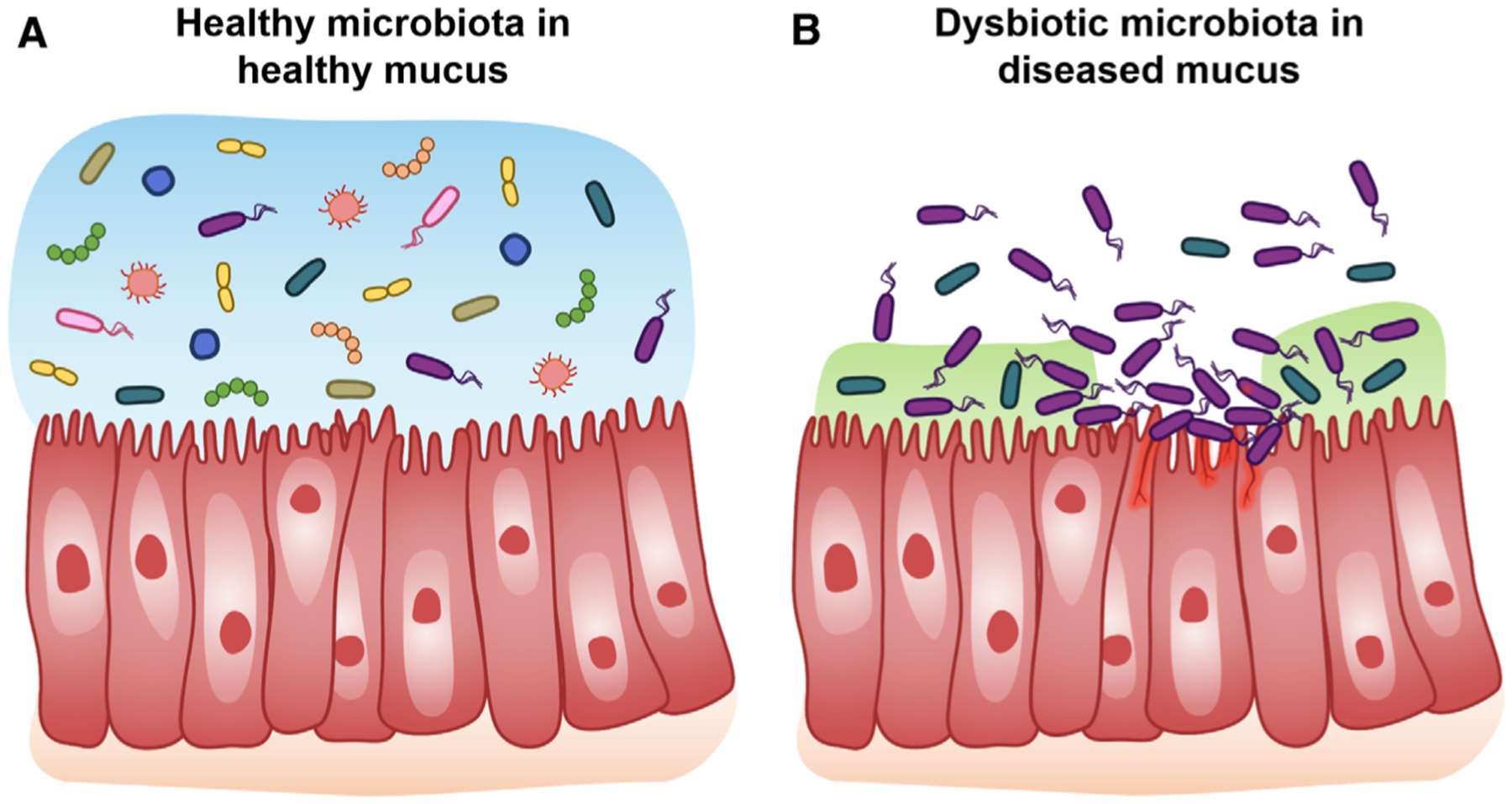
Overview schematic of microbial communities in healthy and diseased mucus. (A) Healthy mucus selects for and maintains a diverse, yet specific, microbial community. (B) Diseases with compromised mucus barriers are often associated with microbial dysbiosis.
The majority of the human microbiota resides in the mucus of the digestive tract, but mucus also harbors distinct microbial communities along the oral cavity and the respiratory and urogenital tracts. Mucus is a complex viscoelastic matrix containing water, gel-forming mucin glycoproteins, lipids, and many proteins, including antimicrobial immune factors [13]. Critical to the structural and biological activity of mucus are mucins, extraordinarily large glycoproteins (>2 MDa) consisting of approximately 50–80% carbohydrate by mass [13] (Fig. 2). The mucus barrier is dynamic, as mucins are constantly being synthesized, secreted, post-translationally modified, degraded, and cleared. The mucus gel varies in thickness, viscoelasticity, and composition to perform various protective functions across the body that is essential for health. For example, dysfunctional mucus barriers play major roles in ailments including cystic fibrosis (CF) [14], inflammatory bowel disease [15], Sjogren’s syndrome [16], and preterm delivery [17], which are also associated with microbial dysbiosis, underscoring the integral role of mucus in regulating the microbiota.
Fig. 2.
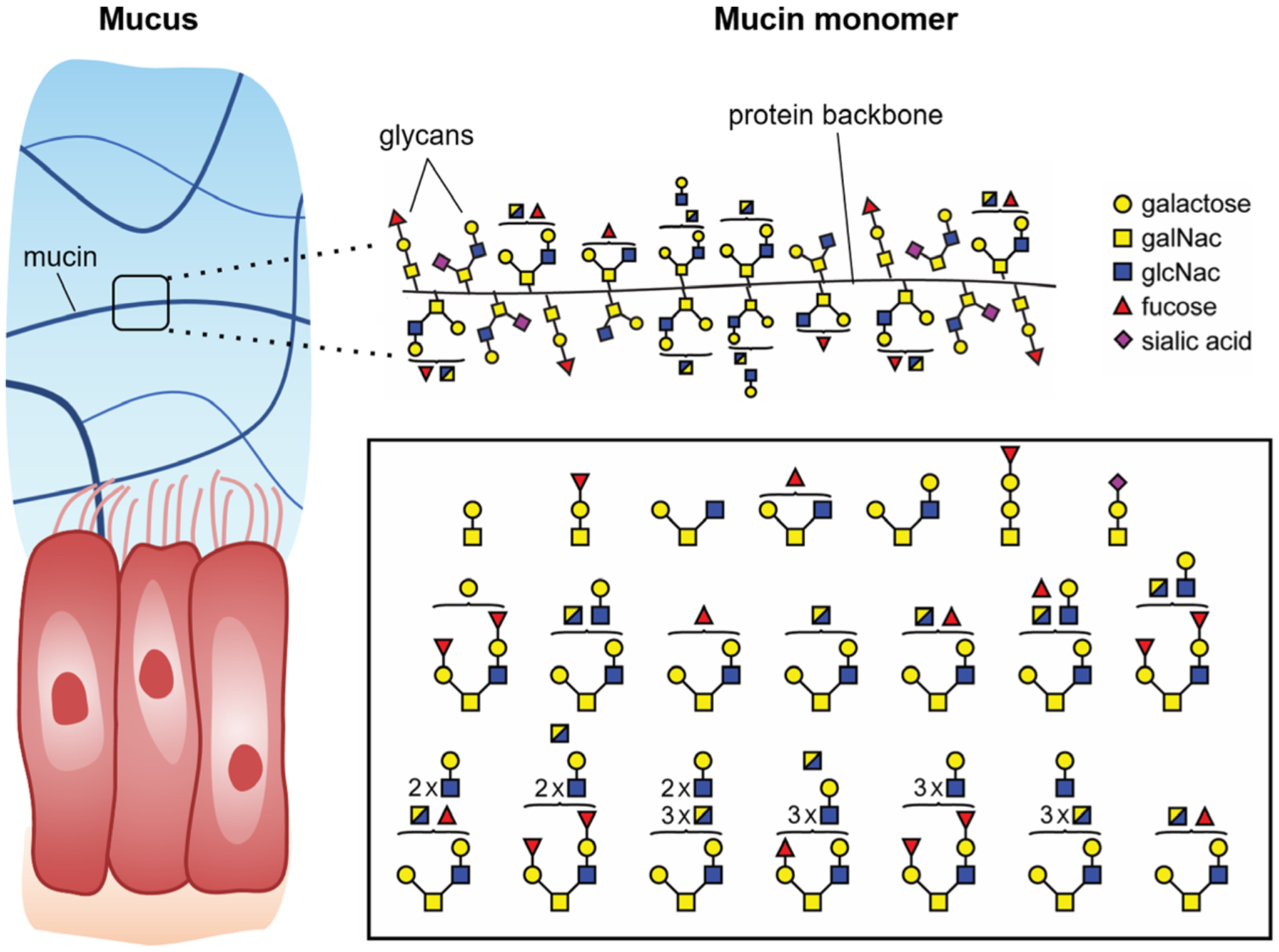
Mucin glycoproteins are the primary structural component of mucus gels. Mucin monomers are densely grafted with diverse and complex glycans. Shown are representative glycan structures isolated from MUC5AC, as identified by mass spectrometry.
Considered the first line of defense against infection, the host employs mucus in several ways to protect the inner epithelia of the body from invading microbes. For example, mucus traps or aggregates bacteria, enabling their clearance from the body [18,19]; this function is critical in delicate tissues such as the lungs and oral cavity. Mucus also acts as a physical barrier between bacteria and the epithelial surface, as exemplified by the dense inner layer of mucus that lines the colon and is generally impenetrable to bacteria. While important, these classical models of mucus’ protective roles are incomplete, as they fail to explain how mucus accommodates trillions of commensal microbes and enables the coexistence of diverse community members.
To close this gap, we hypothesize that mucus is a bioactive environment that not only excludes intruders, but also selects for and stabilizes a healthy microbiota. Here, we consider three strategies by which mucus may organize microbiota: (1) by providing a rich nutrient source to select and retain specific microbes, (2) by spatially dispersing microbial communities, and (3) by providing a source of signals that can directly impact microbial gene expression and behavior. The focus of this Perspective is not the role of mucus as a physical barrier, which has been more thoroughly studied and evaluated elsewhere [18,20,21]. Instead, we highlight several mechanisms by which mucus may actively regulate the microbiota.
A feast for all: mucin glycoproteins support a metabolically diverse microbial community
Mucus provides nutrition for mucus-dwelling microbes and may contribute to the selection of various commensal microbes that make up our microbiota (Fig. 3A). This nutritive role largely stems from mucin glycoproteins, which are the major structural and functional units of the mucus barrier.
Fig. 3.
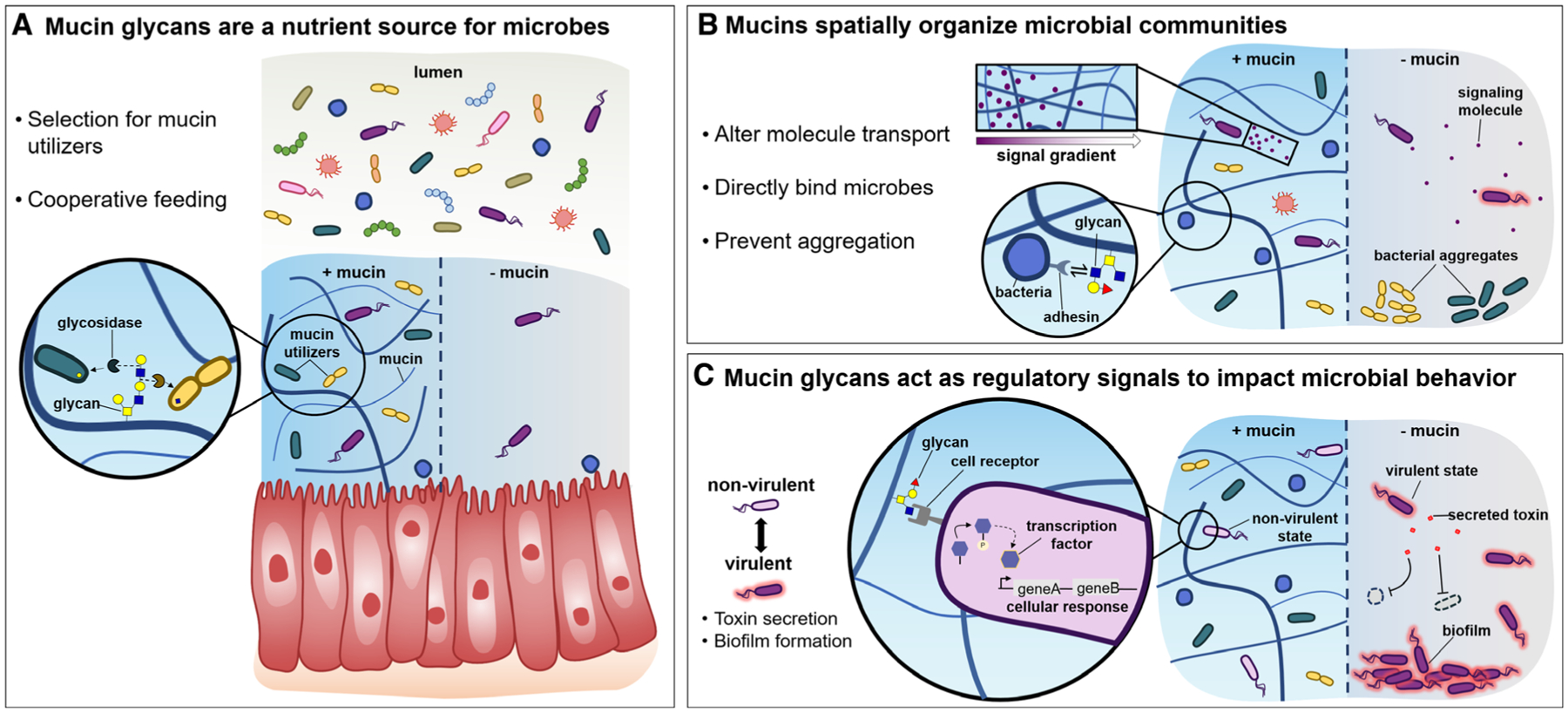
Proposed mechanisms of influence of mucins on microbial communities. A. Mucin glycans are a source of diverse and complex nutrients, which can metabolically shape the microbiota. Glycans may select for beneficial microbes which produce specific glycosidases, as well as facilitate cooperation across species which produce complementary degradative enzymes. B. Mucin networks may spatially organize bacterial communities in several ways, including by directly binding microbes, by altering group behaviors such as aggregation, or by impacting the transport of nutrients, host immune factors, and/or signaling molecules which may shape the assembly of microbial communities. C. Mucin glycans act through regulatory signaling pathways to attenuate virulent behavior. In the presence of mucins, potentially pathogenic microbes may sense and respond to mucin glycans, which enables their transition a host-compatible state within a healthy community. Without mucin regulation, aggressive microbes may overtake the community, forming a dysbiotic microbiota.
Several commensal species are known to degrade mucins [22–25]. For example, Bacteroides possess an extensive set of carbohydrate-utilization genes and have been shown to degrade mucins in vitro [24]. In turn, Bacteroides benefit the host by providing metabolic products that are readsorbed through the large intestine [26], which may modulate the host immune system to more effectively manage inflammation [27]. Further, characterization of a model microbiota of 177 reference genomes present in the human gut revealed that bacteria may harbor > 9000 carbohydrate-degrading enzymes, although significant functional redundancy is expected [28]. For context, the human genome contains coding sequences for at most 17 enzymes to digest glycans [28]. This complex set of mucin glycans may enable the host to support the growth of commensal microbes with different metabolic capabilities; in contrast, if only a few nutrients were provided by the host, aggressive microbial competition and eventual outgrowth could ensue. Host mucins may also buffer the microbial metabolic landscape against large variations in host diet.
Cooperation and cross-feeding between microbial strains are also important in a heterogeneous environment. Mucins present hundreds of unique glycan structures that require the action of linkage-specific glycosidases to be effectively broken down and utilized, which could require cooperation between strains (Fig. 3A). Different bacterial species often exhibit specialized functions, and their interactions can lead to biologically useful community dynamics and structures. Although the challenges of isolating natural mucin glycans [29] have limited experimental characterization of their bioactivity, there are several examples of how glycans in other contexts influence microbial community assembly, two of which we expand upon below.
A recent study found that polysaccharide composition can mediate community assembly of marine microbes specializing in distinct metabolic functions: Initial colonization on each substrate was achieved by specialized primary degraders, while successional dynamics were driven by metabolic cross-feeding interactions with a diverse group of broad-range taxa [30]. Extrapolating these principles of community assembly to the human microbiota suggests that mucin glycans may determine species composition by selecting for primary degraders and their associated broader communities. Ultimately, this strategy allows the host to bolster a beneficial and diverse microbiota; modulating mucin glycosylation levels and profiles could establish niche-specific communities.
Another example is found in human milk oligosaccharides (HMOs), which are thought to function as prebiotics, selectively cultivating a desirable gut microbiota [31]. HMOs present an analogous pool of hundreds of complex sugars which are similar in structure to mucin glycans and have fascinating bioactive properties. Because human infants lack the enzymatic capability to process HMOs [31], undigested oligosaccharides become the primary carbon source in the intestines [31]. As a result, bacterial strains involved in initial colonization of the gut must utilize HMOs. Accordingly, breastfed infants have higher proportions of Lactobacillus and Bifidobacterium [32], which are known to metabolize HMOs [33], than infants fed formula, which contains a lower abundance and diversity of oligosaccharides [34]. Similarly, early colonization of mucosal niches may be guided by a bacterial strain’s ability to forage mucin glycans, which include structures shared with HMOs as well as structures distinct from those in HMOs.
Do mucin glycans support microbial coexistence in ways other than as a source of nutrition? Recent work suggests that this is likely. Mucin glycans are not a preferred carbon source for many host commensals, including the well-studied Bacteroides thetaiotaomicron [35]. It is now known that these commensals prefer to eat saccharides that are available through human consumption of food, as well as surface glycans on the sloughed-off surfaces of epithelial cells, and that they only turn to mucin glycans when these other carbon sources are depleted [36]. Therefore, while mucin glycans likely play an integral role in the initial establishment of diverse communities, their importance may decrease when dietary polysaccharides are abundant. Further, offering a wide array of nutrients does not uniquely support commensals—several bacterial pathogens have developed strategies to benefit from host glycan metabolism and to disrupt healthy microbial communities. For example, the opportunistic pathogen Pseudomonas aeruginosa can grow on short-chain fatty acids in the CF lung that are generated during the consumption of mucin glycans by host commensals [37]. Clostridium perfingens SM101, an opportunistic pathogen in the gut, has also been shown to grow on intestinal mucins [38]. Furthermore, a set of genes involved in mucus and sugar utilization was exclusively identified in a clade of Ruminococcus gnavus enriched in irritable bowel disease patients [39]. Since preventing the outgrowth of these potentially harmful bacteria is essential to maintaining a healthy microbiota, mucus environments likely contribute to the selection and maintenance of healthy microbes beyond serving as a food source.
I want my own space! How mucus spatially organizes bacterial populations
Another feature of mucus is its ability to spatially organize populations of microbes. Microbial ecologists have long postulated [40,41] that a variety of different bacteria can thrive when allowed to form individual niches protected from competition, and indeed, various theoretical and experimental models have demonstrated that heterogeneous spatial structure supports diverse bacterial populations [42,43]. The importance of spatial structure has been demonstrated in stabilizing in vitro bacterial communities [44], as well as maintaining diversity within different mucus layers of the gut [45]. Consequently, by providing a three-dimensional scaffold for colonization, mucus may play a pivotal role in shaping the microbiota.
Mucus networks of varying pore size and adhesiveness may mediate spatial organization of bacterial communities in a variety of ways (Fig. 3B). Bacterial adhesion to mucus has been shown to influence bacterial colonization [46,47], and in vitro mucin binding assays with commensals such as Bacteroides fragilis [48] and Lactobacillus fermentum [49] substantiate the hypothesis that adhesion to mucosal surfaces allows beneficial bacteria to protect the host from invasion by potential pathogens. This hypothesis has been further supported by simulations of bacterial communities in the host epithelium, which suggest that host modulation of bacterial adhesion can be an important positive selection strategy [50], as adherent cells better resist displacement by nonadherent cells that are otherwise more competitive. Bacteria that bind mucin directly can further shape the environment by acting as sites of attachment for other bacteria, as well as point sources and sinks for diffusible metabolites and other factors. For example, the complex community structure in dental plaque is thought to form through initial attachment of Streptococcus and Actinomyces to the salivary pellicle, followed by attachment by other species including Fusobacterium nucleatum, which physically binds early and late colonizers [51].
In addition to binding directly to microbes, the spatial properties of mucus may also affect microbial group behaviors such as bacterial motility and aggregation [52]. For example, the heterogeneous glycan presentation of mucins may contribute to microbial movement, as glycans on mucins may act as chemoattractants to bacteria including P. aeruginosa [53] and Campylobacter jejuni [54]. Further, recent work has shown that mucins can prevent certain pathogens from aggregating and forming biofilms, and can also disperse cells from preformed aggregates and biofilms [55–58]. The inability of these pathogens to aggregate in mucus could affect microbial communication. For instance, cells in large aggregates have been reported to exhibit stronger quorum sensing activity than cells that are more uniformly distributed [59–61]. It is conceivable that this mucin-derived spatial separation could impact cell-cell communication among pathogens and perhaps even commensal species, although this hypothesis remains to be rigorously tested.
Mucins may also spatially distribute chemical signals that can influence the development of bacterial communities. The distribution of small molecules through mucus can be generally affected by bulk flow, as molecules are transported through advection [62]. This principle likely extends to microbially produced small molecules; for example, quorum sensing has been observed to change under different flow conditions, with a higher flow rate generally reducing quorum sensing [63–65]. In mucus, flow via mucociliary clearance is determined by coordinated ciliary activity and mucus viscosity [66]. Since mucins are the primary factor underlying mucus viscosity, alterations in mucin content can significantly impact flow [18,21]. These observations suggest that mucus may impact the advection of microbial signaling molecules and, consequently, the assembly of microbial communities.
Mucins could also affect the diffusion of small molecules through specific interactions. Selective transport of small molecules through mucus is a complex process influenced by electrostatic, hydrophobic, and specific binding interactions that together determine a particle’s adhesiveness to mucins [67]. Such interaction filtering may establish gradients in nutrients and other factors, which have been shown to shape the assembly of microbial communities [68]. Although there have yet to be systematic studies of the movement of many small molecules through mucus, mucin has empirically been shown to bind small-molecule drugs including polymyxin and fluoroquinolone antibiotics [69], protecting P. aeruginosa from killing by these antibiotics [70]. Oligopeptides have also exhibited modulated transport through mucus due to subtle differences in charge distribution [71], and mucins inhibit the diffusion of pyocyanin, a small molecule produced by P. aeruginosa [60]. As technical advances facilitate the measurement of small-molecule diffusion and binding to mucins, future studies may illuminate how mucins impact the distribution of diffusible molecules, thereby regulating microbial behavior.
Mucus can “talk” to microbes: Mucins present host-derived signals that can directly impact microbial gene expression and behavior
Thus far, we have discussed how mucin glycans can serve as food sources to support beneficial microbes, and we have highlighted how the spatial structure afforded by mucus can impact microbial communities. However, emerging evidence suggests that mucus can contribute to microbial coexistence through a third mechanism: by providing a source of signals that can directly trigger changes in bacterial gene expression and phenotypes.
To date, the signaling potential of mucus has been primarily investigated in terms of its striking ability to attenuate virulence in a variety of pathogens. Despite the traditional view of mucus as a simple physical barrier, multiple studies from our laboratory suggest that mucins can directly suppress the virulence of potential pathogens by changing their identity from harmful pathogens to host-compatible commensals [55–58] (Fig. 3C). In turn, the virulence-attenuating properties of mucin may contribute to the coexistence of microbes in mucus environments.
The ability of mucins to suppress microbial virulence is remarkably broad, as various serotypes of mucins (MUC5AC, MUC5B, MUC2) in sites across the human body (mouth, lungs, gut) attenuate virulence in evolutionarily distant pathogens, including Gram-negative microbes like P. aeruginosa [58,72], Gram-positive bacteria like Streptococcus mutans [56], and fungal species like Candida albicans [57]. Notably, mucins suppress biofilm formation across all three of these species [56–58] and also promote the dispersal of pre-formed P. aeruginosa biofilms [58]. Further, mucins directly trigger changes in the expression of virulence genes. For example, RNA sequencing revealed that mucins globally downregulate virulence pathways in P. aeruginosa [72], including the type I, II, III, and VI secretion systems, quorum sensing, phenazine production, and iron acquisition. Similarly, incubation of C. albicans with mucin downregulated multiple virulence genes, including those involved in biofilm formation, proteinase secretion, and filamentation [73]. Mucin-induced downregulation of virulence genes also prevents these pathogens from killing host cells and other microbes. For instance, P. aeruginosa and C. albicans were unable to kill human epithelial cells effectively in vitro in the presence of mucin [57,72], and P. aeruginosa was less virulent on an in vivo porcine burn wound model of infection when mucin was added [72]. Further, S. mutans was unable to outcompete the oral commensal Streptococcus sanguinis when incubated with mucin [74], while P. aeruginosa could not utilize its type VI secretion system (T6SS) to kill Escherichia coli or Burkholderia cepaciain the presence of mucin glycans [75]. As> 200 structures of mucin glycans have already been discovered [76], we propose that a “glycan code” differentially impacts virulence pathways in distinct pathogens (Fig. 3C).
By what mechanisms do mucin glycans serve as signaling molecules? Glycans can be directly sensed by receptors, such as the membrane boundhistidine kinases that are widely distributed across bacteria. For example, the RetS sensor kinase in P. aeruginosa has structural homology to other carbohydrate-binding proteins [77] and is considered a master regulator of virulence [78]. We have recently demonstrated that mucin glycans act as a signal for RetS [75], which triggers the downregulation of virulence traits associated with a chronic infection state, including theT6SS [75]. Another possibility is that mucin glycans may act through dedicated sugar sensing and utilization pathways by mimicking nutrient signals. To identify other receptors involved in sensing mucin glycans across species, we believe that in vivo screens with systematic knockouts of receptors in a particular microbe as well as in vitro screens to look for binding between purified receptor domains and mucin glycans will yield valuable mechanistic insights. Many microbes also encode other sugar-binding proteins, such as adhesins, which could interact with mucin glycans.
Glycans may also signal in indirect ways. For instance, RNA sequencing of P. aeruginosa revealed that dozens of putative metabolic genes are differentially expressed in response to mucins and mucin glycans [72,75], suggesting that glycans may induce dramatic changes in the metabolic states of these microbes. A growing body of work suggests that changes in metabolic state are associated with the expression of virulence pathways [79]. A recent modeling study identified dozens of genes involved in both virulence-factor production and primary metabolism in P. aeruginosa [80], and fluctuations in central metabolic pathways such as the citric acid cycle can lead to changes in the activity of antimicrobial systems such as the T6SS [81–83]. Many of the annotated metabolic genes identified in the above RNA-seq experiments have not been characterized, but a deeper understanding of how mucin glycans alter the metabolic state of pathogens may yield novel insights into how these signals indirectly affect virulence-associated behaviors across microbial species.
Interestingly, the virulence-attenuating effects of mucin glycans are also evident with the aforementioned HMOs, a separate class of human-produced glycans that are the third most abundant solid in human breastmilk [84]. Although traditionally viewed as food sources for commensals, recent work suggests that HMOs also play important roles in protection against pathogens, which could in turn promote microbial coexistence. For example, the HMOs 2-fucosyllactose and 6’-sialyllactose block the adhesion of Gram-negative E. coli and Gram-positive Salmonella fyris to epithelial cell surfaces [85], a necessary step prior to invasion. It has also been reported that nonsialylated HMOs inhibit the growth of pathogenic Streptococcus species [86]. Although the exact mechanism of this bacteriostatic activity is unclear, transposon mutagenesis identified a putative glycosyltransferase that may play a role in the response to HMOs [86]. Further, incubation of C. albicans with HMOs delays the yeast-to-hyphae transition of this fungal pathogen [87], which is reminiscent of the virulence suppression effects of mucin [57].
Virulence suppression by HMOs can also be indirect. For example, various host commensals such as Bifidobacterium species can utilize HMOs as sole carbon sources [88]. Interestingly, it has been reported that incubation of pathogenic E. coli O157:H7 and Salmonella typhimurium with spent media taken from cultures of Bifidobacterium species in which HMOs were supplied as a carbon source led to the downregulation of various virulence genes [89]. This observation suggests that metabolic by-products of HMOs generated by host commensals may serve as virulence-suppressing signals, although the actual signals in spent media have not yet been identified at the molecular level.
Overall, a clearer picture is emerging in which mucins and their associated glycans serve as virulence-attenuating signals for various opportunistic pathogens. However, the effects of mucin on other residents of the mucus environment—such as the trillions of commensal microbes that reside in mucus, as well as host immune cells—have not been well-studied. Commensal microbes are generally regarded as ‘host-compatible’, but the process by which mucus is populated by commensals is not fully understood. Whether mucins accomplish this selection by serving as food sources, establishing spatial structure and/or by acting as signals that directly impact commensal gene expression and physiology will be a fascinating area of future research. Similarly, it is currently unclear if and how mucin glycans alter the behavior of host immune cells. Intriguingly, immune cells encode various sugar-binding receptors, such as Siglecs [90,91], which raises the possibility that these host cells sense and respond to mucin glycans [92]. Ultimately, further investigation of how mucins and their associated glycans affect both commensals and host cells will provide further insights into how mucus manages the microbiota.
Compromised mucus gives rise to microbial dysbiosis
While healthy mucus houses a stable microbiota, various mucosal diseases including ulcerative colitis and cystic fibrosis are associated with microbial dysbiosis. In the gastrointestinal tract, degradation of mucus has been linked to the pathogenesis of inflammatory conditions [93,94]. Mucus which has undergone glycosidic and proteolytic degradation by enteric bacteria is less viscous [95] and more permeable to toxins and microbes [96] which can induce damage and inflammatory responses. This, in turn, may lead to the widespread killing of host commensals, which will decrease microbial diversity. Furthermore, mucin oligosaccharides in inflammatory bowel disease exhibit drastically shorter chain lengths, decreased sulfation, and increased sialylation [97]. Such modifications may weaken the ability of mucus to maintain microbial coexistence through the mechanisms discussed above, as glycans potentially play key roles in the nutrient presentation, structural arrangement, and virulence suppression functions of mucus.
At the other end of the spectrum, abnormally thick and viscous mucus is more susceptible to microbial infection than healthy mucus. CF is a genetic disorder in which the structure and function of the CF transmembrane conductance regulator protein are disrupted, leading to abnormally thick lung epithelial mucus [14]. In addition to the higher mucin content of CF mucus, inflammatory cell necrosis produces large quantities of extracellular polymers like DNA and F-actin [14,98,99], which likely alter mucus structure. This diseased mucus is more susceptible to colonization by various opportunistic pathogens such as P. aeruginosa, which can form dense bacterial communities in this niche that are resistant to both clearance by the immune system and to antibiotic treatment [100]. Changes to the mucus environment in CF may therefore influence biofilm-forming behavior, a key virulence determinant for many pathogens [101–103].
Importantly, colonization by P. aeruginosa is one of the largest sources of morbidity and mortality in CF patients [104], highlighting the clinical importance of clarifying the links between mucins and the microbiota of the lung. Thanks to several decades-spanning studies [105–107], we now know that both microbial diversity and lung function are highest in younger (<10 years of age) CF patients [108]. Strikingly, the increased prevalence over time of opportunistic pathogens including P. aeruginosa and Burkholderia correlates with decreased trends in both lung function and microbial diversity in the lung microbiota [108]. However, we still do not fully understand why the diseased CF lung allows certain pathogens to dominate, for example due to a malfunction in providing food for microbes, a change in the spatial structure of mucus or of the microbial community, a loss of virulence-attenuating signals, or a combination of these factors. Further research is warranted to better characterize the critical distinctions between healthy and diseased mucus environments, and to understand how these differences trigger changes in microbial communities and ultimately in host health.
Final perspectives and conclusions
Our understanding of mucus has dramatically evolved over the years. However, the current textbook description of mucus as a simple protective barrier is woefully incomplete. In this Perspective, we have highlighted three major roles that mucus plays in regulating the microbiota. First, heavily glycosylated mucins serve as food sources for diverse host commensals. Second, spatial structure provided by mucin gel networks helps microbes carve out specific niches. Third, mucins directly serve as virulence-attenuating signals to transition potential pathogens into a host-compatible state. Together, these three factors likely help select for and accommodate a diverse, yet specialized microbial community in our mucus environments, which is critical to human health. However, many questions remain. Is the ability to utilize mucin glycans widespread among bacteria, or exclusive to mucus-dwelling commensals? Are mucin gel networks able to impede the movement of microbial signaling molecules such as autoinducers? What are the actual signals in mucus environments that directly suppress the expression of virulence genes, and what are the receptors that sense them? Do mucin glycans alter microbial and/or host metabolism? How do mucins signal host cells, such as immune cells? What are the key differences between healthy and diseased mucus environments, such as the CF lung? We envision that answers to these critical questions and others will enable the development of therapeutics for diseases of the mucus environment, and that deeper understanding of the mechanisms that drive microbial coexistence in mucus environments will empower us to design novel treatments to promote mucosal and microbial health.
Acknowledgements
The authors thank the National Institutes of Health (R01-EB017755-041745302) and the National Science Foundation (NSFCareerPHY-1454673) for financial support. BXW and CMW are also supported by a National Science Foundation Graduate Research Fellowship (1745302). The authors also thank Otto Cordero for thoughtful input on the Viewpoint.
Abbreviations
- CF
cystic fibrosis
- HMOs
human milk oligosaccharides
- MUC2
mucin 2
- MUC5AC
mucin 5AC
- MUC5B
mucin 5B
- T6SS
type VI secretion system
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Sender R, Fuchs S & Milo R (2016) Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14, e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajilić-Stojanović M, Heilig HGHJ, Tims S, Zoetendal EG & de Vos WM (2013) Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol 15, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 3.Schloissnig S, Arumugam M, Sunagawa S et al. (2013) Genomic variation landscape of the human gut microbiome. Nature 493, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huttenhower C, Gevers D, Knight R et al. (2012) Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickard JM, Zeng MY, Caruso R & Núñez G (2017) Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 279, 70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowland I, Gibson G, Heinken A et al. (2018) Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr 57, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belkaid Y & Harrison OJ (2017) Homeostatic immunity and the microbiota. Immunity 46, 562–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carding S, Verbeke K, Vipond DT, Corfe BM & Owen LJ (2015) Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 26, 26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho I & Blaser MJ (2012) The human microbiome: at the interface of health and disease. Nat Rev Genet 13, 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan XC, Tickle TL, Sokol H et al. (2012) Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wade WG (2013) The oral microbiome in health and disease. Pharmacol Res 69, 137–143. [DOI] [PubMed] [Google Scholar]
- 12.Ma B, Forney LJ & Ravel J (2012) Vaginal Microbiome: Rethinking health and disease. Annu Rev Microbiol 66, 371–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bansil R & Turner BS (2018) The biology of mucus: composition, synthesis and organization. Adv Drug Deliv Rev 124, 3–15. [DOI] [PubMed] [Google Scholar]
- 14.Hill DB, Vasquez PA, Mellnik J et al. (2014) A biophysical basis for mucus solids concentration as a candidate biomarker for airways disease. Waigh T, ed. PLoS One 9, e87681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Shen X, Li Y et al. (2016) Therapeutic potential to modify the mucus barrier in inflammatory bowel disease. Nutrients. 8, 10.3390/nu8010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhury NMA, Proctor GB, Karlsson NG, Carpenter GH & Flowers SA. Reduced Mucin-7 (Muc7) sialylation and altered saliva rheology in Sjögren’s Syndrome associated oral dryness. Mol Cell Prot 15, 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Critchfield AS, Yao G, Jaishankar A et al. (2013) Cervical mucus properties stratify risk for preterm birth. PLoS One 8, 10.1371/journal.pone.0069528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cone RA (2009) Barrier properties of mucus. Adv Drug Deliv Rev 61, 75–85. [DOI] [PubMed] [Google Scholar]
- 19.Lynge Pedersen AM & Belstrøm D (2019) The role of natural salivary defences in maintaining a healthy oral microbiota. J Dent 80, S3–S12. [DOI] [PubMed] [Google Scholar]
- 20.Wagner CE, Wheeler KM & Ribbeck K (2018) Mucins and their role in shaping the functions of mucus barriers. Annu Rev Cell Dev Biol 34, 189–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai SK, Wang Y-Y, Wirtz D & Hanes J (2009) Micro- and macrorheology of mucus. Adv Drug Deliv Rev 61, 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derrien M, Vaughan EE, Plugge CM & de Vos WM (2004) Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54, 1469–1476. [DOI] [PubMed] [Google Scholar]
- 23.Martens EC, Lowe EC, Chiang H et al. (2011) Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. Eisen JA, ed. PLoS Biol 9, e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertont AM & Stanley RA (1982) In vitro utilization of mucin by Bacteroides Fragilis. Appl Environ Micrbiol 43, 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crost EH, Tailford LE, Monestier M et al. (2016) The mucin-degradation strategy of Ruminococcus gnavus : The importance of intramolecular trans-sialidases. Gut Microbes 7, 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooper LV, Midtvedt T & Gordon JI (2002) How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22, 283–307. [DOI] [PubMed] [Google Scholar]
- 27.Troy EB & Kasper DL (2010) Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci 15, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Kaoutari A, Armougom F, Gordon JI, Raoult D & Henrissat B (2013) The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11, 497–504. [DOI] [PubMed] [Google Scholar]
- 29.Li Z & Chai W (2019) Mucin O-glycan microarrays. Curr Opin Struct Biol.. 10.1016/j.sbi.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 30.Enke TN, Datta MS, Schwartzman J, Barrere J, Pascual-García A & Cordero OX (2019) Modular Assembly of Polysaccharide-Degrading Marine Microbial Communities. Curr Biol. 29, 10.1016/j.cub.2019.03.047. [DOI] [PubMed] [Google Scholar]
- 31.Bier DM, German JB & Lönnerdal B. Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria. Vol 62. Nestec Ltd., Vevey/S. Karger AG; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rinne M, Kalliomaki M, Arvilommi H, Salminen S & Isolauri E (2005) Effect of probiotics and breastfeeding on the bifidobacterium and lactobacillus/Enterococcus microbiota and humoral immune responses. J Pediatr 147, 186–191. [DOI] [PubMed] [Google Scholar]
- 33.Zúñiga M, Monedero V & Yebra MJ (2018) Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Front Microbiol 9, 1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward RE, Niñonuevo M, Mills DA, Lebrilla CB & German JB (2006) In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 72 (6), 4497–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwalm ND, Townsend GE & Groisman EA (2017) Prioritization of polysaccharide utilization and control of regulator activation in Bacteroides thetaiotaomicron. Mol Microbiol 104, 32–45. [DOI] [PubMed] [Google Scholar]
- 36.Sonnenburg JL, Xu J, Leip DD et al. (2005) Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959. [DOI] [PubMed] [Google Scholar]
- 37.Flynn JM, Niccum D, Dunitz JM & Hunter RC (2016) Evidence and role for bacterial mucin degradation in cystic fibrosis airway disease. PLoS Pathog 12, e1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacMillan JL, Vicaretti SD, Noyovitz B et al. (2019) Structural analysis of broiler chicken small intestinal mucin O-glycan modification by Clostridium perfringens. Mol Cell Biol 98, 5074–5088. [DOI] [PubMed] [Google Scholar]
- 39.Hall AB, Yassour M, Sauk J et al. (2017) A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 9, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kneitel JM & Chase JM (2004) Trade-offs in community ecology: linking spatial scales and species coexistence. Ecol Lett 7, 69–80. [Google Scholar]
- 41.Ghoul M & Mitri S (2016) The ecology and evolution of microbial competition. Trends Microbiol 24, 833–845. [DOI] [PubMed] [Google Scholar]
- 42.Kerr B, Riley MA, Feldman MW & Bohannan BJM (2002) Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418, 171–174. [DOI] [PubMed] [Google Scholar]
- 43.Portell X, Pot V, Garnier P, Otten W & Baveye PC (2018) Microscale heterogeneity of the spatial distribution of organic matter can promote bacterial biodiversity in soils: insights from computer simulations. Front Microbiol 9, 1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HJ, Boedicker JQ, Choi JW & Ismagilov RF (2008) Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A 105, 18188–18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu H-P, Lai Y-C, Huang S-W, Chen H-C, Hsieh C & Yu H-T (2015) Spatial heterogeneity of gut microbiota reveals multiple bacterial communities with distinct characteristics. Sci Rep 4, 6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tasteyre A, Barc M-C, Collignon A, Boureau H & Karjalainen T (2001) Role of FliC and FliD Flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun 69, 7937–7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danne C, Robbe-Masselot C, Aymeric L et al. (2015) Streptococcus gallolyticus Pil3 pilus is required for adhesion to colonic mucus and for colonization of mouse distal colon. J. Infect 212, 1646–1655. [DOI] [PubMed] [Google Scholar]
- 48.Huang JY, Lee SM & Mazmanian SK (2011) The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe 17, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas M, Ascencio F & Conway PL (2002) Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl Environ Microbiol 68, 2330–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mcloughlin K, Schluter J, Rakoff-Nahoum S, Smith AL & Foster KR (2016) Host selection of microbiota via differential adhesion. Cell Host Microbe 19, 550–559. [DOI] [PubMed] [Google Scholar]
- 51.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE & Borisy GG (2016) Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A 113, E791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Figueroa-Morales N, Dominguez-Rubio L, Ott TL & Aranson IS (2019) Mechanical shear controls bacterial penetration in mucus. Sci Rep. 9, 10.1038/s41598-019-46085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nelson JW, Tredgett MW, Sheehan JK, Thornton DJ, Notman D & Govan JR (1990) Mucinophilic and chemotactic properties of Pseudomonas aeruginosa in relation to pulmonary colonization in cystic fibrosis. Infect Immun 58, 1489–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hugdahl MB, Beery JT & Doyle MP (1988) Chemotactic behavior of Campylobacter jejuni. Infect Immun 56, 1560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caldara M, Friedlander RS, Kavanaugh NL, Aizenberg J, Foster KR & Ribbeck K (2012) Mucin biopolymers prevent bacterial aggregation by retaining cells in the free-swimming state. Curr Biol 22, 2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frenkel ES & Ribbeck K (2015) Salivary mucins protect surfaces from colonization by cariogenic bacteria. Appl Environ Microbiol 81, 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kavanaugh NL, Zhang AQ, Nobile CJ, Johnson AD & Ribbeck K (2014) Mucins suppress virulence traits of Candida albicans. MBio 5, 10.1128/mBio.01911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Co JY, Cárcamo-Oyarce G, Billings N et al. (2018) Mucins trigger dispersal of Pseudomonas aeruginosa biofilms. npj Biofilms Microbiomes 4, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao M, Zheng H, Ren Y et al. (2016) A crucial role for spatial distribution in bacterial quorum sensing. Sci Rep. 6, 10.1038/srep34695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darch SE, Simoska O, Fitzpatrick M et al. (2018) Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc Natl Acad Sci U S A 115, 4779–4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Connell JL, Kim J, Shear JB, Bard AJ & Whiteley M (2014) Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc Natl Acad Sci U S A 111, 18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karimi A, Karig D, Kumar A & Ardekani AM (2015) Interplay of physical mechanisms and biofilm processes: review of microfluidic methods. Lab Chip 15, 23–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim MK, Ingremeau F, Zhao A, Bassler BL & Stone HA (2016) Local and global consequences of flow on bacterial quorum sensing. Nat Microbiol 1, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo X, Wu HC, Tsao CY et al. (2012) Biofabrication of stratified biofilm mimics for observation and control of bacterial signaling. Biomaterials 33, 5136–5143. [DOI] [PubMed] [Google Scholar]
- 65.Meyer A, Megerle J, Kuttler C et al. (2012) Dynamics of AHL mediated quorum sensing under flow and non-flow conditions. Phys Biol 9, 10.1088/1478-3975. [DOI] [PubMed] [Google Scholar]
- 66.Quraishi MS, Jones NS & Mason J (1998) The rheology of nasal mucus: a review. Clin Otolaryngol Allied Sci 23, 403–413. [DOI] [PubMed] [Google Scholar]
- 67.Witten J & Ribbeck K (2017) The particle in the spider’s web: transport through biological hydrogels. Nanoscale 9, 8080–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J, Hanke A, Tegetmeyer HE et al. (2017) Impacts of chemical gradients on microbial community structure. ISME J 11, 920–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Witten J, Samad T & Ribbeck K (2019) Molecular characterization of mucus binding. Biomacromol 20, 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samad T, Co JY, Witten J & Ribbeck K (2019) Mucus and mucin environments reduce the efficacy of polymyxin and fluoroquinolone antibiotics against Pseudomonas aeruginosa. ACS Biomater Sci Eng 5, 1189–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li LD, Crouzier T, Sarkar A, Dunphy L, Han J & Ribbeck K (2013) Spatial configuration and composition of charge modulates transport into a mucin hydrogel barrier. Biophys J 105, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wheeler KM, Cárcamo-Oyarce G, Turner BS et al. Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nat Microbiol 4 2146–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chin V, Lee T, Rusliza B & Chong P (2016) Dissecting Candida albicans infection from the perspective of C. albicans virulence and omics approaches on host-pathogen interaction: a review. Int J Mol Sci 17, 1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frenkel ES & Ribbeck K (2017) Salivary mucins promote the coexistence of competing oral bacterial species. ISME J 11, 1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang BX, Wheeler KM, Cady KCet al.Mucin glycans signal through the sensor kinase RetS to inhibit virulence-associated traits in Pseudomonas aeruginosa. bioRxiv April 2020:2020.03.31.018614. doi: 10.1101/2020.03.31.018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin C, Kenny DT, Skoog EC et al. (2017) Structural diversity of human gastric mucin glycans. Mol Cell Proteomics 16, 743–758. [DOI] [PubMed] [Google Scholar]
- 77.Anantharaman V & Aravind L (2003) Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genom 4, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lapouge K, Schubert M, Allain FH-T & Haas D (2007) Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67, 241–253. [DOI] [PubMed] [Google Scholar]
- 79.Richardson AR, Somerville GA & Sonenshein AL (2015) Regulating the intersection of metabolism and pathogenesis in gram-positive bacteria. Microbiol Spectr 3, 10.1128/microbiolspec.MBP-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartell JA, Blazier AS, Yen P et al. (2017) Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nat Commun 8, 14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Septer AN, Bose JL, Lipzen A, Martin J, Whistler C & Stabb EV (2015) Bright luminescence of Vibrio fischeri aconitase mutants reveals a connection between citrate and the Gac/Csr regulatory system. Mol Microbiol 95, 283–296. [DOI] [PubMed] [Google Scholar]
- 82.Minato Y, Fassio SR, Wolfe AJ & Häse CC (2013) Central metabolism controls transcription of a virulence gene regulator in Vibrio cholerae. Microbiology 159 (Pt 4), 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harper L, Balasubramanian D, Ohneck EA et al. (2018) Staphylococcus aureus responds to the central metabolite pyruvate to regulate virulence. MBio 9, e02272–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Plaza-Díaz J, Fontana L & Gil A (2018) Human milk oligosaccharides and immune system development. Nutrients 10, 1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Facinelli B, Marini E, Magi G et al. (2019) Breast milk oligosaccharides: effects of 2′-fucosyllactose and 6′-sialyllactose on the adhesion of Escherichia coli and Salmonella fyris to Caco-2 cells. J Matern Neonatal Med 32, 2950–2952. [DOI] [PubMed] [Google Scholar]
- 86.Lin AE, Autran CA, Szyszka A et al. (2017) Human milk oligosaccharides inhibit growth of group B Streptococcus. J Biol Chem 292, 11243–11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gonia S, Tuepker M, Heisel T, Autran C, Bode L & Gale CA (2015) Human milk oligosaccharides inhibit Candida albicans invasion of human premature intestinal epithelial cells. J Nutr 145, 1992–1998. [DOI] [PubMed] [Google Scholar]
- 88.Özcan E & Sela DA (2018) Inefficient metabolism of the human milk oligosaccharides lacto-N-tetraose and lacto-N-neotetraose shifts Bifidobacterium longum subsp. infantis physiology. Front Nutr 5, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bondue P, Crèvecoeur S, Brose F et al. (2016) Cell-free spent media obtained from Bifidobacterium bifidum and Bifidobacterium crudilactis grown in media supplemented with 3′-Sialyllactose modulate virulence gene expression in Escherichia coli O157:H7 and Salmonella Typhimurium. Front Microbiol 7, 1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Crocker PR. Siglecs in innate immunity. Curr Opin Pharmacol 2005;5(4 SPEC. ISS):431–437. [DOI] [PubMed] [Google Scholar]
- 91.MacAuley MS, Crocker PR & Paulson JC (2014) Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol 14, 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Belisle JA, Horibata S, Jennifer GAA et al. (2010) Identification of Siglec-9 as the receptor for MUC16 on human NK cells, B cells, and monocytes. Mol Cancer 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roberton AM & Corfield AP. Mucin degradation and its significance in inflammatory conditions of the gastrointestinal tract. Medical Importance of the Normal Microflora. Boston, MA: Springer, US; 1999:222–261. [Google Scholar]
- 94.Corfield AP, Williams AJ, Clamp JR, Wagner SA & Mountford RA (1988) Degradation by bacterial enzymes of colonic mucus from normal subjects and patients with inflammatory bowel disease: the role of sialic acid metabolism and the detection of a novel O-acetylsialic acid esterase. Clin Sci (Lond) 74, 71–78. [DOI] [PubMed] [Google Scholar]
- 95.Crowther RS, Roomi NW, Fahim REF & Forstner JF (1987) Vibrio cholerae metalloproteinase degrades intestinal mucin and facilitates enterotoxin-induced secretion from rat intestine. Biochim Biophys Acta – Gen Subj 924, 393–402. [DOI] [PubMed] [Google Scholar]
- 96.Gibold L, Garenaux E, Dalmasso G et al. (2016) The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn’s disease-associated Escherichia coli. Cell Microbiol 18, 617–631. [DOI] [PubMed] [Google Scholar]
- 97.Shirazi T, Longman RJ, Corfield AP, Probert CSJ & Shirazi T (2011) Mucins and inflammatory bowel disease. J Gastroenterol Hepatol 27, 28–38. [Google Scholar]
- 98.Voynow JA & Mengr BKR (2009) Mucins, mucus, and sputum. Chest 135, 505–512. [DOI] [PubMed] [Google Scholar]
- 99.Porto BN & Stein RT (2016) Neutrophil extracellular traps in pulmonary diseases: too much of a good thing? Front Immunol 7, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matsui H, Wagner VE, Hill DB et al. (2006) A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A 103, 18131–18136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hall-Stoodley L & Stoodley P (2005) Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol 13, 7–10. [DOI] [PubMed] [Google Scholar]
- 102.Krzyściak W, Jurczak A, Kościelniak D, Bystrowska B & Skalniak A (2014) The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis 33, 499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hasan F, Xess I, Wang X, Jain N & Fries BC (2009) Biofilm formation in clinical Candida isolates and its association with virulence. Microbes Infect 11, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Emerson J, Rosenfeld M, McNamara S, Ramsey B & Gibson RL (2002) Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 34, 91–100. [DOI] [PubMed] [Google Scholar]
- 105.Renwick J, McNally P, John B et al. The microbial community of the cystic fibrosis airway is disrupted in early life. Williams S, ed. PLoS One 2014;9:e109798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paganin P, Fiscarelli EV, Tuccio V et al. Changes in cystic fibrosis airway microbial community associated with a severe decline in lung function. Hartl D, ed. PLoS One 2015;10:e0124348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao J, Schloss PD, Kalikin LM et al. (2012) Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109, 5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Coburn B, Wang PW, Diaz Caballero J et al. (2015) Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 5, 10241. [DOI] [PMC free article] [PubMed] [Google Scholar]


