Abstract
Glioblastoma (GBM) is an aggressive malignant brain tumor with median survival of 12 months and 5-year survival rate less than 5%. GBM is highly vascularized, and the interactions between tumor and endothelial cells play an important role in driving tumor growth. To study tumor-endothelial interactions, the gold standard co-culture model is transwell culture, which fails to recapitulate the biochemical or physical cues found in tumor niche. Recently, we reported the development of poly(ethylene-glycol)-based hydrogels as 3D niche that supported GBM proliferation and invasion. To further mimic the microanatomical architecture of tumor-endothelial interactions in vivo, here we developed a hydrogel-based co-culture model that mimics the spatial organization of tumor and endothelial cells. To increase the physiological relevance, patient-derived GBM cells and mouse brain endothelial cells were used as model cell types. Using hydrolytically-degradable alginate fibers as porogens, endothelial cells were deployed and patterned into vessel-like structures in 3D hydrogels with high cell viability and retention of endothelial phenotype. Co-culture led to a significant increase in GBM cell proliferation and decrease in endothelial cell expression of cell adhesion proteins. In summary, we have developed a novel 3D co-culture model that mimics the in vivo spatial organization of brain tumor and endothelial cells. Such model may provide a valuable tool for future mechanistic studies to elucidate the effects of tumor-endothelial interactions on tumor progression in a more physiologically-relevant manner.
Keywords: Tumor, Glioblastoma, Endothelial, Co-culture, Cancer model, Three-dimensional
1. Introduction
Tumor-vasculature interactions play an important role in driving tumor growth [1]. Nutrient deficiency in the growing tumor initiates signaling by tumor cells that induces phenotypic and genotypic changes in endothelial cells, the primary cell type of the vasculature [2]. These changes in endothelial cells result in extracellular matrix remodeling and sprouting of new blood vessels from existing ones to meet tumor metabolic needs [3]. In addition to responding to tumor cell signals, endothelial cells can also secrete paracrine factors and express surface ligands that act on tumor cells, promoting tumor progression [4–6]. Glioblastoma (GBM) is a highly vascularized, malignant brain tumor with median survival of 12 months and 5-year survival rate less than 5% [7–9]. During GBM tumor development, GBM cells migrate towards and organize into cuffs around normal vessels and induce migration and proliferation of endothelial cells for angiogenesis [1,10,11]. Characterization of GBM-endothelial interactions would further elucidate mechanisms underlying GBM progression and accelerate the development of novel therapeutics.
Given the importance of GBM-endothelial interactions, extensive efforts have been made to develop in vitro models for studying tumor and endothelial cell crosstalk. The most widely employed co-culture models include Transwell, direct monolayer, and spheroid co-culture. In Transwell co-culture, tumor and endothelial cells are cultured in a single well as monolayers, separated by a membrane. Transwell co-culture of GBM cells and human microvascular endothelial cells led to increased GBM invasiveness and increased endothelial cell proliferation [6]. In direct monolayer co-culture, tumor and endothelial cells are mixed and plated together in the same well. Direct monolayer co-culture of GBM and brain endothelial cells showed tumor co-localization with endothelial cells and increased tumor cell proliferation [4]. In a mixed co-culture spheroid, endothelial cells migrate throughout the tumor spheroid and form short tube-like formations that mimic vessels. Spheroid co-culture of breast tumor and endothelial cells demonstrated upregulation of proangiogenic factors and increased tumor growth, compared to tumor cell-only spheroids [12]. Although previous reports have yielded significant insight into tumor-endothelial interactions, previous co-culture models lack a 3D extracellular matrix, which is required for tumor cell invasion and endothelial cell sprouting. This limitation may result in differential cell responses compared to in vivo, highlighting the critical need for advanced 3D in vitro models that can recapitulate the brain tumor-vasculature microanatomical architecture found in vivo.
To this end, recent efforts have patterned endothelial cells into vessel-mimicking structures in 3D hydrogels using microfluidics or subtractive templating. Previous reports have developed endothelial cell-lined vascular networks using microfluidic circuits formed via soft lithography in collagen hydrogels [13]. Tumor cells can be introduced into the bulk collagen hydrogel for studying tumor-vasculature interactions. Microfluidic vascular models are advantageous in that they permit flow, which can mimic in vivo gradients, and allow for precise tuning of channel geometry and structure. However, using microfluidics to generate vascular networks requires complex experimental setups and facilities, as well as extensive attention to maintain channel fidelity, prevent air bubbles, and achieve homogeneous cell seeding. Alternatively, subtractive templating is a more facile method to pattern endothelial cells in which a template rod is inserted during hydrogel gelation and removed after crosslinking, forming a cylindrical channel. Following channel formation, a suspension of endothelial cells is flowed through the channel to allow for cell attachment [14]. Similar to microfluidics, tumor cells can be encapsulated in the bulk hydrogel prior to removal of the template rod. Although subtractive templating permits well-defined channel geometry and orientation, this process requires excessive numbers of endothelial cells for patterning and suffers from poor cell distribution. Given the limitations of current methodologies, there remains a critical need for a facile method to pattern endothelial cells into vessel-mimicking structures with homogeneous cell distribution at a tissue scale without special experimental setup.
To develop a physiologically-relevant co-culture model of GBM and endothelial interactions, it is important to choose appropriate model cell types that can retain in vivo cell phenotype. To model GBM tumor cells, most previous studies employed immortalized GBM cell lines, which poorly represent the phenotypes and genotypes of primary GBM cells [15]. In contrast, patient-derived GBM xenograft cells can retain phenotypes and genotypes of the original parental tumor [16]. For modeling endothelial cells, previous studies primarily employed human umbilical vein endothelial cells (HUVEC) or human microvascular endothelial cells (HMEC-1) [6,17]. However, brain endothelial cells are unique from peripheral endothelial cells. To regulate transport to substances into and out of the brain, brain endothelial cells have membrane transporter systems, lower number of endocytic vesicles, enzymes to degrade substances, and increased number of tight junctions [18]. Staining of tight junction proteins, such as occludin or ZO-1, revealed significant differences between commonly used endothelial cell lines (HUVEC, HMEC-1) and brain endothelial cells [19,20].
In the present study, we sought to engineer a 3D co-culture model that mimics the in vivo microanatomical architecture of brain tumorendothelial interactions using biomimetic hydrogels. Patient-derived glioblastoma xenograft (PDTX GBM) cells and mouse brain endothelial cells were used as model cell types to enable species-specific cell fate tracking in co-culture. Recently, we reported a poly(ethylene)-glycol (PEG)-based hydrogel system with brain-mimicking biochemical and mechanical properties that supported GBM tumor cell proliferation and invasion in 3D [21]. To pattern brain endothelial cells into luminal vessel-like structures in 3D hydrogels, we employed a technology we recently reported using hydrolytically degradable alginate microfibers as porogens [22,23]. To mimic in vivo vessel geometry, microchannel geometry was modulated by tuning the parameters involved in wet-spinning alginate microfibers. To recapitulate in vivo endothelial cellcell adhesions, endothelial cell density in alginate microfibers was modulated to achieve confluent endothelial cell-lined microchannels in 3D hydrogels. After optimization of microchannel geometry and endothelial cell density, the effects of co-culture on GBM and endothelial cell fates were analyzed, compared to GBM and endothelial mono-cultures. Cell behavior in co-culture was compared to mono-culture control by analyzing cell morphology using brightfield microscopy, proliferation using immunostaining, and gene expression using RT-PCR.
2. Materials and methods
2.1. Materials
8-arm PEG (MW ∼ 40 kDa) was purchased from JenKem Technology USA (Allen, TX, USA). Linear PEG (MW ∼ 1.5 kDa) was purchased from Sigma-Aldrich USA (St. Louis, MO, USA). RGD peptide (CRGDS) was purchased from Bio Basic, Inc (Amherst, NY, USA). Sodium hyaluronate (HA) (MW ∼ 20–40 kDa) was purchased from Lifecore Biomedical (Chaska, MN, USA). MMP-degradable peptide (KCGPQGIWGQCK) was purchased from GenScript (Piscataway, NJ, USA). 8-arm PEG-norbornene (PEG-NB) and linear PEG-dithiol (PEGSH) were synthesized as previously reported [24,25]. Thiolated sodium hyaluronate (HA-SH) was synthesized as previously reported [26]. Alginate was purchased from Sigma (St. Louis, MO, USA). Hydrolytically degradable alginate was synthesized as previously reported [22]. All other reagents were obtained from Fisher Scientific (Pittsburgh, PA, USA) unless otherwise noted.
2.2. Alginate microfiber formation
Alginate microfibers were formed by wet-spinning in calcium bath. Briefly, alginate was solubilized in tumor growth media at a concentration of 3% w/v. Alginate solution was loaded into a 1 mL syringe and capped with a 30-gauge needle. Using a syringe pump, alginate solution was injected into a stirring crosslinking bath of 1% (w/v) calcium chloride and 0.9% (w/v) sodium chloride. Alginate injection rate (mL/min) and calcium bath stir speed (rpm) were modulated to tune resultant microchannel diameter. After wet-spinning, alginate microfibers were transferred from the crosslinking bath into tumor growth media and chopped using a scalpel. Cut microfibers were then transferred into a solution of 20% 3 kDa linear PEG in tumor culture media for 1 h. This preincubation step prevents diffusion of hydrogel precursor solution into alginate microfibers.
2.3. Preparation of hydrogel precursor solution
Hydrogel precursor solution (6% PEG) was prepared from 8-arm PEG-NB, linear PEG-SH, and an MMP-cleavable peptide (KCGPQGIW-GQCK), which were mixed at a molar ratio of 2:3:3, respectively. To facilitate cell adhesion, RGD peptide (CRGDS) was added at a final concentration of 0.914 mM. To mimic brain ECM content, HA-SH was chemically incorporated at a final concentration of 0.004% (w/v), which was selected based on reported values in human brain tissue [27]. 20 kDa linear PEG (Sigma) was added at a final concentration of 16% (w/v) for additional modulation of hydrogel stiffness.
2.4. Encapsulation of alginate microfibers in 3D hydrogel
Preincubated alginate microfibers were mixed with hydrogel precursor solution at a 1 to 8 vol ratio. Microfiber-precursor solution was loaded into a 1 mL syringe, and the solution was crosslinked in the presence of photoinitiator lithium acylphosphinate (LAP) (0.05% w/v) under UV light (365 nm, 4 mW/cm2) for 5 min at RT with constant manual rotation during the 1st minute. The crosslinked hydrogel was then cut into individual 50 μL hydrogels.
2.5. Cell culture
Patient-derived adult GBM tumor xenograft cells (D-270 MG) were provided by the lab of Professor Gerald Grant at Stanford University. Tumor cells were maintained in growth media, consisting of improved minimal essential zinc option medium (Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum (Gibco, Life Technologies) and 0.1% gentamicin (Life Technologies) at 37 °C in 5% CO2 with media change every other day. Mouse brain microvascular endothelial cells (bEnd3) were obtained from ATCC (Manassas, VA, USA) and maintained in growth media, consisting of Dulbecco’s modified eagle medium high glucose with 20% fetal bovine serum (Gibco, Life Technologies) and 1% penicillin-streptmycin (Life Technologies) at 37 °C in 5% CO2 with media change every other day.
2.6. Encapsulation of endothelial cell-laden alginate microfibers
Trypsinized endothelial cells were homogeneously mixed in a 3% (w/v) solution of alginate at varying cell densities (1 M, 2 M, 3 M, 4 M, and 5 M cells/mL). Endothelial cell-laden alginate microfibers were prepared and encapsulated in hydrogels as above.
2.7. Cell viability
Cell viability was assessed using Live/Dead Cell Viability Assay kit (Life Technologies). Live/Dead reagent was prepared per manufacturer’s instructions. Hydrogels were immersed in Live/Dead reagent solution for 40 min and imaged using a Zeiss fluorescence microscope.
2.8. Confocal imaging of endothelial cells in microchannels
Hydrogels were fixed in 4% PFA for 1 h at RT and washed in PBS. Cell permeabilization was performed using 0.1% Triton X-100 in PBS. Heat-induced antigen retrieval was performed by equilibrating hydrogel in sodium citrate buffer for 1 h at RT, transferring hydrogel to heated buffer (90 °C) for 20 min, and replacing half the volume with PBS to cool for 20 min at RT. Nonspecific binding was blocked by incubating samples in blocking buffer for 1 h at RT. CD31 staining was performed by incubating samples in CD31 antibody (Abcam, Cambridge, United Kingdom) for 2 days at 4 °C. Samples were washed 3 times for 1 h each in PBS at RT. Secondary and nuclei staining was performed by incubating samples in secondary antibody (Life Technologies) and Hoechst 33342 dye (Cell Signaling Technologies, Danvers, MA, USA) for 2.5 h at RT. Samples were washed 3 times for 1 h each in PBS at RT. Samples were incubated in mounting media overnight and imaged the next day using Zeiss SP5 confocal microscope (Zeiss, Oberkochen, Germany). Maximal projections were obtained from 200 μm thick Z-stack with 2 μm step size.
2.9. Co-encapsulation of tumor and endothelial cells in hydrogels
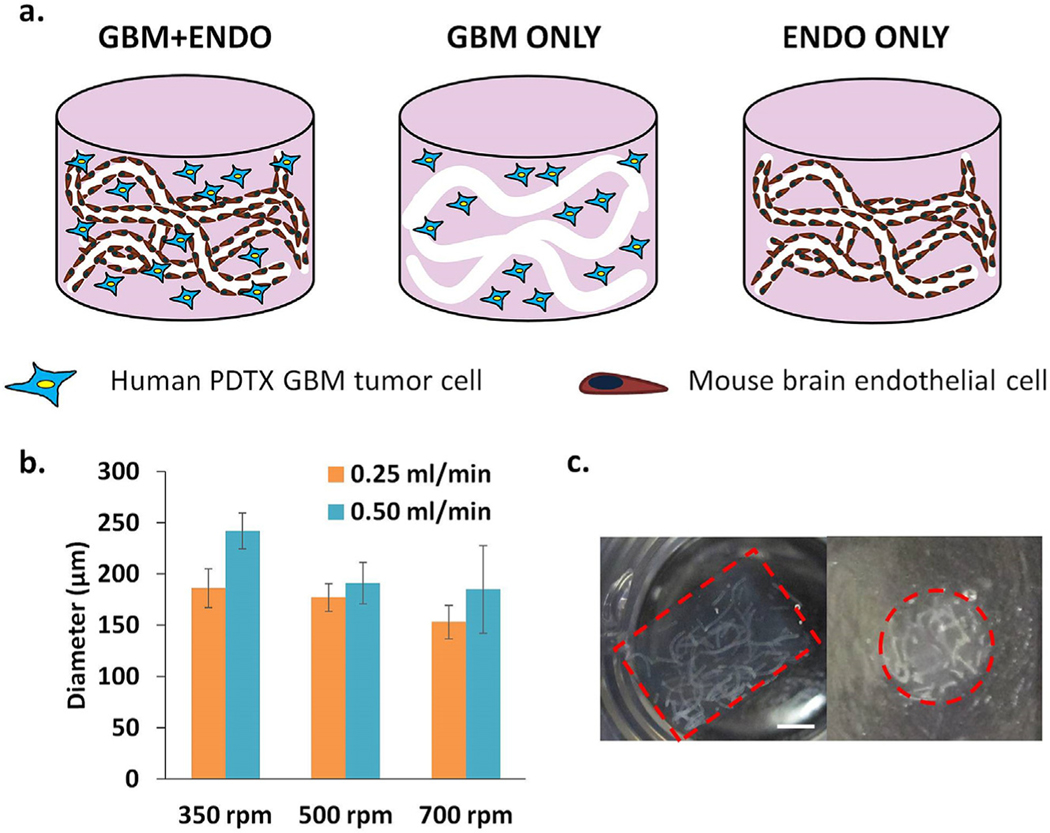
In the co-culture group (GBM + ENDO), endothelial and tumor cells were co-encapsulated in hydrogels (Fig. 1a). Trypsinized endothelial cells were homogeneously resuspended in a 3% (w/v) solution of alginate at a density of 4 M cells/ml. Endothelial cell-laden alginate microfibers were prepared as above. Trypsinized tumor cells were homogeneously resuspended in hydrogel precursor solution (0.5 M cells/mL). Endothelial cell-laden alginate microfibers and tumor cellladen hydrogel precursor solution were mixed and crosslinked into hydrogels as above. For endothelial only group (ENDO ONLY), endothelial cells were encapsulated in alginate microfibers and subsequently encapsulated in hydrogels, as described above (Fig. 1a). For tumor only group (GBM ONLY), tumor cells were co-encapsulated with acellular alginate microfibers, as described above (Fig. 1a). Hydrogels were then transferred to tumor growth media and maintained at 37 °C in 5% CO2 with media change every other day.
Fig. 1.
(a). Schematic of experimental group design. Endothelial cells (mouse brain) were patterned into vessellike structures in 3D biomimetic hydrogels using hydrolytically degradable alginate microfibers as porogens. In GBM + ENDO group, single patient-derived glioblastoma xenograft (PDTX GBM) cells were co-cultured with patterned endothelial cells. In GBM + ONLY group, single PDTX GBM cells were co-encapsulated with acellular alginate microfibers. In ENDO ONLY group, patterned endothelial cells were cultured alone. (b). Increasing calcium bath stir speed (350–700 rpm) led to decreased microchannel diameter. Increasing alginate injection rate (0.25, 0.50 mL/min) led to increased microchannel diameter. (c). Cross-sectional views of encapsulated alginate microfibers in 3D hydrogels. Left = side view. Right =top view. Red =hydrogel edge. Scale bar = 2 mm.
2.10. RT-PCR
Gene expression of endothelial cell adhesion proteins (Claudin-5, ZO-1, CD31), tumor CXCR4, and tumor/endothelial VEGFa were measured. To test primer specificity, total RNA was extracted from tumor and endothelial cells from 2D controls (Table S1). For 3D samples, total RNA was isolated from hydrogels using TRIzol (Life Technologies) (n = 3). cDNA was synthesized using SuperScript III First-Strand Synthesis kit (Life Technologies). RT-PCR was then performed using an Applied Biosystems 7900 Real-Time PCR system (Applied Biosystems, Life Technologies) using Power SYBR Green PCR Master Mix (Applied Biosystems, Life Technologies). Relative expression levels of target genes were determined using the comparative CT method. Target gene expression was first normalized to a housekeeping gene, GAPDH, followed by a second normalization to the gene expression level on day 1.
2.11. Immunohistochemical staining
To characterize the brain tumor cells(D-270 MG), cells were fixed with 4% PFA for 15 min at RT, followed by three PBS washes. Cells were then permeabilized in PBS containing 0.3% Triton X-100 for 10 min at RT, followed by blocking in PBS containing 10% BSA for 1 h at RT. Samples were then incubated with primary antibodies for Nestin (EMD Millipore, San Diego, CA, USA), β-Tubulin III (SigmaAldrich, St. Louis, MO, USA), or GFAP (Abcam, Cambridge, MA, USA) for 1 h at 37 °C, followed by secondary antibody for 1 h at RT. Cell nuclei were counterstained with Hoechst dye 33342 (Cell Signaling Technologies, Danvers, MA, USA) and Phalloidin-TRITC (SigmaAldrich, St. Louis, MO, USA) for 1 h at RT. Samples were mounted (Vector Laboratories, Burlingame, CA, USA), and images were taken using Zeiss fluorescence microscope (Zeiss, Oberkochen, Germany).
To analyze cell proliferation, tumor and endothelial cells were stained using anti-Ki67 primary antibody (Abcam, Cambridge, MA, USA). Tumor cells were identified using anti-human nuclear antigen primary antibody (Millipore, Billerica, MA, USA). To analyze endothelial cell adhesion proteins, endothelial cells were stained using primary antibodies for claudin-5 (Life Technologies), ZO-1 (Proteintech, Rosemont, IL, USA), and CD31 (Life Technologies). To analyze tight junctions of endothelial cells, endothelial cells were stained using primary antibody for VE-cadherin (R&D Systems, Minneapolis, MN, USA). Briefly, hydrogels were fixed in 4% PFA for 1 h at RT and washed in PBS. Samples were dehydrated overnight in 30% sucrose at 4 °C and cryopreserved in OCT (Sakura Finetek USA, Torrance, CA, USA) in liquid nitrogen. Samples were then cryosectioned at −20 °C (14 μm thickness). Sections were first permeabilized in PBS, pH 7.4 containing 0.4% Triton-X 100 for 1 h at RT. Heatmediated antigen retrieval was performed by incubating the sections in sodium citrate buffer (90 °C) for 2 min. Then, samples were blocked in PBS, pH 7.4 containing 2% goat serum, 1% BSA, and 0.1% Triton-X 100 for 1 h at RT. Samples were then incubated overnight with primary antibody at 4 °C, followed by secondary antibody for 1 h at RT. Cell nuclei were counterstained with Hoechst dye 33342 (Cell Signaling Technologies, Danvers, MA, USA) for 1 h at RT. Samples were mounted (Vector Laboratories, Burlingame, CA, USA), and images were taken using Zeiss fluorescence microscope (Zeiss, Oberkochen, Germany). Quantification of Ki67 positive and negative nuclei (n = 150 for each cell type and group) was performed manually using ImageJ software.
2.12. Characterization of VEGF release from GBM and GBM + ENDO
GBM and GBM + ENDO hydrogels (n = 3 for each group) were cultured in 500 μL of tumor growth media and maintained at 37 °C in 5% CO2 with media change every other day. Conditioned medium was collected every other day (Days 2, 4, and 6). Amount of VEGF in the collected media was measured using a commercially available VEGF ELISA kit (MyBioSource, San Diego, CA, USA) following manufacturer’s protocol.
2.13. Statistical analyses
GraphPad Prism (GraphPad Software, San Diego, CA, USA) was used to perform statistical analyses on hydrogel stiffness, cell proliferation, gene expression data, and VEGF release. Unpaired student’s t tests (assuming Gaussian distribution) and two-way analysis of variance (ANOVA) were used to determine statistical significance (p < 0.05). Error was reported as standard deviation unless otherwise noted.
3. Results
3.1. Characterization of D-270 MG cell line
To verify that D-270 MG cell lines maintained expression of genes characteristic of GBM in our study, we stained for neural markers Nestin, β-tubulin III, and GFAP [28,29]. D-270 MG cells showed uniform and strong staining for Nestin and β-tubulin III, which are markers for neural stem cells (NSCs) and early neurons [28], respectively (Figs. S1a and b). Our finding is consistent with previous reports of characterization of other GBM cells [30–32]. D-270 MG showed low signal for GFAP, an astrocytic marker (Fig. S1c). Given that GBM is a heterogeneous population of cells, our staining suggest D-270 MG are composed mostly of neural progenitor cells and low percentage of astrocytes [28,29].
3.2. Optimization of alginate microfiber formation and encapsulation in hydrogels
Sodium alginate was modified to be hydrolytically degradable using partial periodate oxidation, as reported in previous literature [33]. Microchannel diameter was modulated by tuning alginate wet-spinning parameters, calcium bath stir speed and alginate injection rate (Fig. 1b). Endothelial cells retained high cell viability when varying the wet-spinning parameters (Fig. S2). Calcium bath stir speed was selected to be 700 rpm, and alginate injection rate was selected to be 0.25 mL/min. Using these parameters, alginate fibers with diameter of 150 μm were able to be homogeneously encapsulated throughout 3D hydrogels (Fig. 1c).
3.3. Optimization of endothelial cell patterning in microchannels
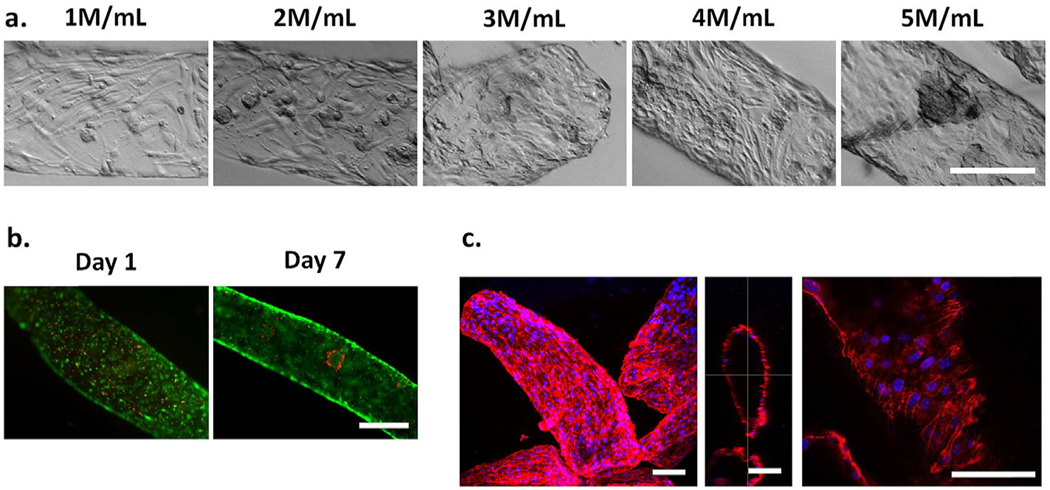
To recapitulate brain endothelial cell-cell adhesions in vitro, we hypothesized that a high endothelial cell density is required, based on observations in previous literature [34]. When plated in 2D monolayer at high cell density, bEnd3 cells formed confluent cell sheet within 24 h with high expression of CD31, an endothelial cell marker and cell adhesion protein (Fig. S3). Lowering cell density in 2D monolayer led to decreased CD31 expression (Fig. S3). To determine the optimal endothelial cell seeding density in microchannels, we modulated the concentration of endothelial cells in alginate solution from 1 M cells/ mL to 5 M cells/mL (Fig. 2a). As the density of endothelial cells increased, the confluency of the cells increased, as expected, and in the 5 M cells/ml group, cell aggregates were observed due to over-confluency. Therefore, 4 M cells/mL was selected, as this concentration permitted rapid formation of confluent monolayer in the microchannel within 24 h without significant cell aggregation. Endothelial cells in microchannels retained high cell viability over time (Fig. 2b). On day 7, as observed using confocal microscopy, endothelial cells displayed high expression of CD31, suggesting retention of endothelial cell phenotype in microchannel (Fig. 2c). Also, CD31 expression was found along the entire microchannel cross-section, demonstrating that bEnd3 cells can coat the entire microchannel surface. Taken together, by using hydrolytically degradable alginate fibers, bEnd3 cells can be patterned in microchannels in vessel-like structures with high cell viability and retention of endothelial cell phenotype.
Fig. 2. Optimization of endothelial cell density in hydrolytically degradable alginate microfibers for microchannel formation in 3D hydrogels.
(a). Increasing initial endothelial cell density in alginate solution led to increased cell confluency and eventual cell aggregation after 2 days in culture. Scale bar = 250 μm. (b). Endothelial cells (4 M cells/mL) retained high cell viability and proliferated over time in microchannels. Scale bar =500 μm. Green =live. Red =dead. (c). Endothelial cells (4 M/mL) expressed high levels of CD31, along microchannel length and diameter after 7 days in culture. Scale bar = 100 μm. Blue =nuclei. Red = CD31. Maximal projection (left), orthogonal view (middle), single slice (right).
3.4. Effects of co-culture on tumor and endothelial cell morphologies
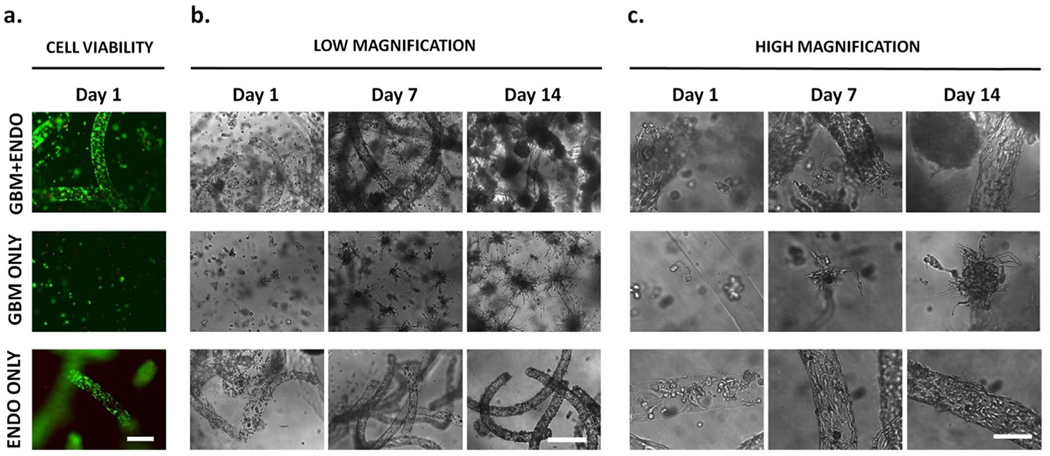
Following encapsulation, tumor and endothelial cells remained viable in all groups, indicating that the encapsulation process is cellfriendly (Fig. 3a). Endothelial and tumor cell morphologies in co-culture and mono-culture were monitored using brightfield microscopy (Fig. 3b and c). On day 1, in both GBM + ENDO and ENDO ONLY groups, most endothelial cells were rounded, while some cells were spreading, suggesting that the alginate fibers had degraded and released the cells within 24 h. By day 7, endothelial cells in ENDO ONLY group formed well organized, confluent cell monolayer with expected endothelial cell morphology. In contrast, in GBM + ENDO group, endothelial cell rounding was observed, a possible sign of monolayer disorganization and downregulation of endothelial cell adhesion proteins. By day 14, endothelial cells were poorly organized in GBM + ENDO group with rounded morphologies and cell protrusions extending beyond the microchannel border. In contrast, endothelial cells in ENDO ONLY group maintained expected endothelial cell morphologies and organized cell monolayer. For tumor cell morphology, tumor cells started with rounded morphologies on after encapsulation and spread with significant cell protrusions by day 7 with the same degree in GBM + ENDO and GBM ONLY group. By day 14, tumor cells in GBM + ENDO group that were directly adjacent to endothelial microchannels had more spherical morphology. In contrast, in GBM ONLY group, tumor cells formed large cell aggregates with radial protrusions.
Fig. 3. Effects of co-culture on cell morphology over time.
(a). High cell viability of tumor and endothelial cells was observed after encapsulation. Scale bar =500 μm. Green = live. Red = dead. (b),(c). Endothelial cells started with rounded morphology after release from alginate microfibers on day 1. In GBM + ENDO group, endothelial cells were more rounded and disorganized, compared to cells in ENDO ONLY group, after 14 days in culture. Tumor cells started with rounded morphology after encapsulation. In GBM ONLY group, tumor cells formed large cell aggregates with radial protrusions after 14 days in culture. When co-cultured with endothelial cells in GBM + ENDO group, tumor cells directly adjacent to endothelial microchannels had more spherical morphology. Scale bars = 500 μm (b), 125 μm (c).
3.5. Effects of co-culture on endothelial cell adhesions
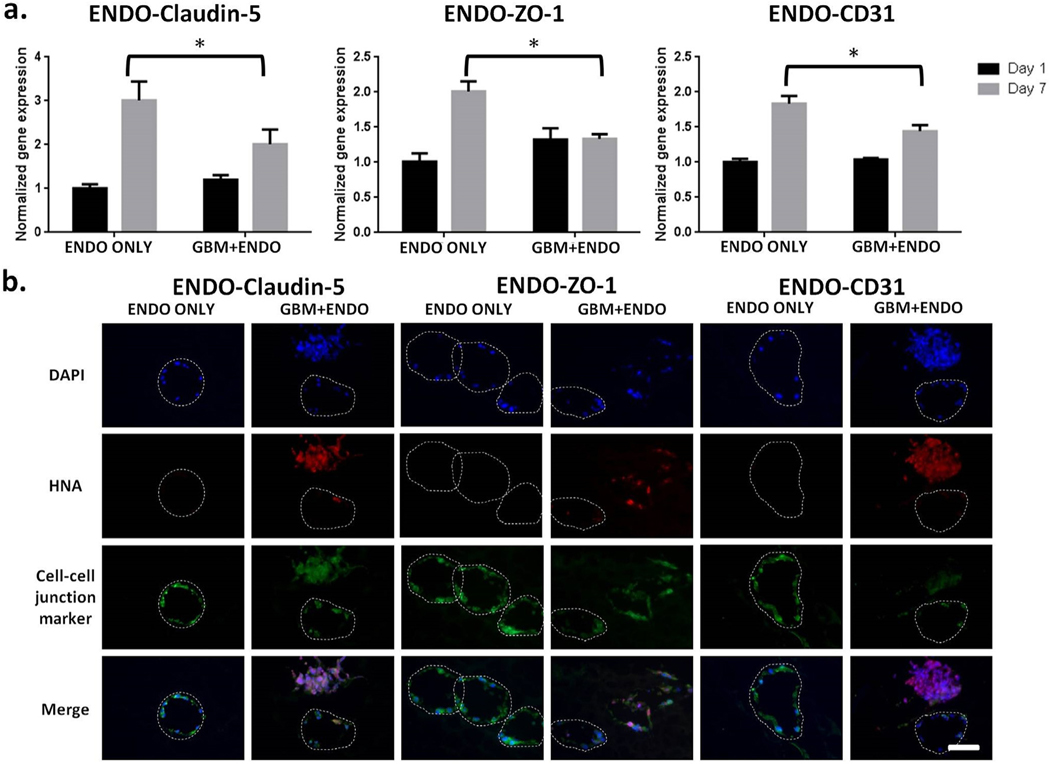
To determine the effects of co-culture on endothelial cell adhesion protein expression, gene expression and protein expression of claudin5, ZO-1, and CD31 were analyzed using RT-PCR and immunostaining, respectively. On day 7, gene expression levels of endothelial cell adhesion proteins (claudin-5, ZO-1, and CD31) were downregulated in GBM + ENDO group, as compared to ENDO ONLY group (Fig. 4a). On day 14, protein expression levels for claudin-5, ZO-1, and CD31, as evaluated using immunostaining, were decreased in GBM + ENDO group, as compared to ENDO ONLY group (Fig. 4b). To further characterize the tight junctions of endothelial cells, we also performed immunostaining of VE-cadherin. Both ENDO only and ENDO + GBM co-culture showed positive VE-Cadherin staining (Fig. S4).
Fig. 4. Co-culture led to downregulation of endothelial cell adhesion protein expression.
(a). Expression of cell adhesion proteins in endothelial cells on day 7, normalized to day 1. Mouse specific primers were used to measure gene expression levels of claudin-5, ZO-1, and CD31 using RT-PCR. * p < 0.05. (b). Immunostaining of cell-cell junction proteins on day 14. Blue = nuclei. Red = human nuclear antigen. Green = claudin-5, ZO-1, or CD31. White dashed line = microchannel edge. Scale bar =100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Effects of co-culture on tumor and endothelial cell proliferation
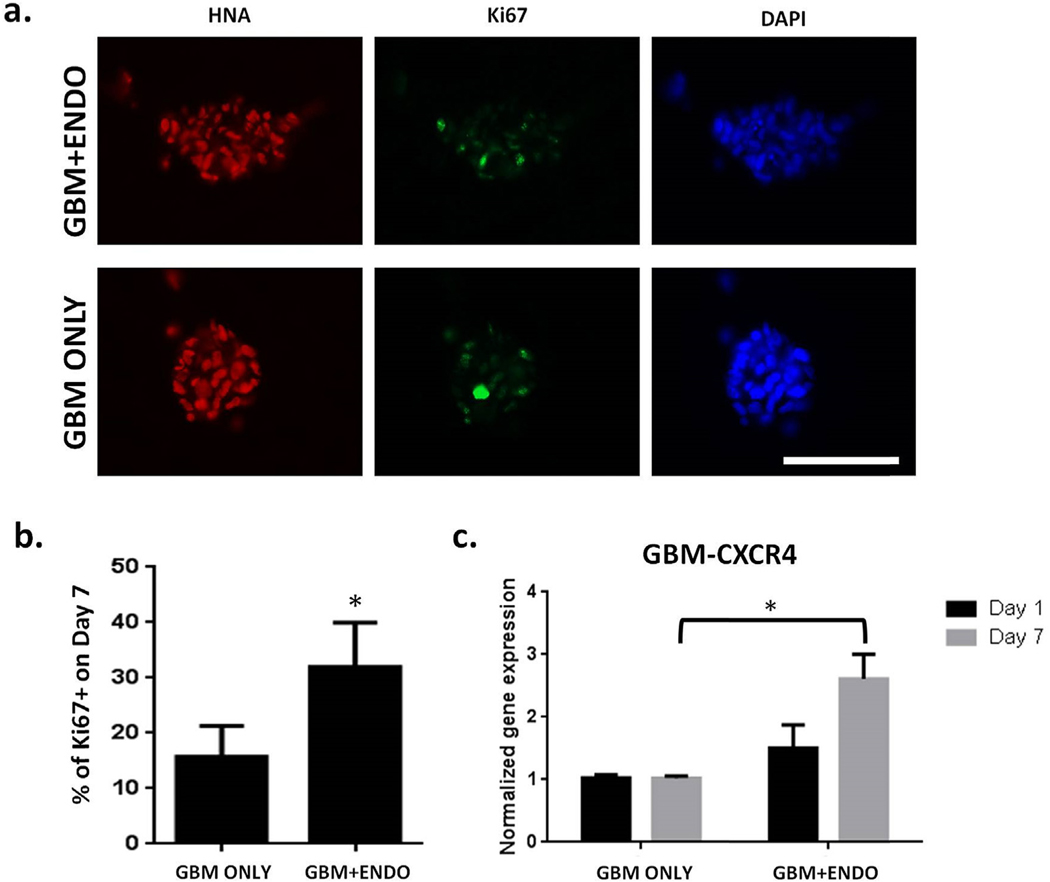
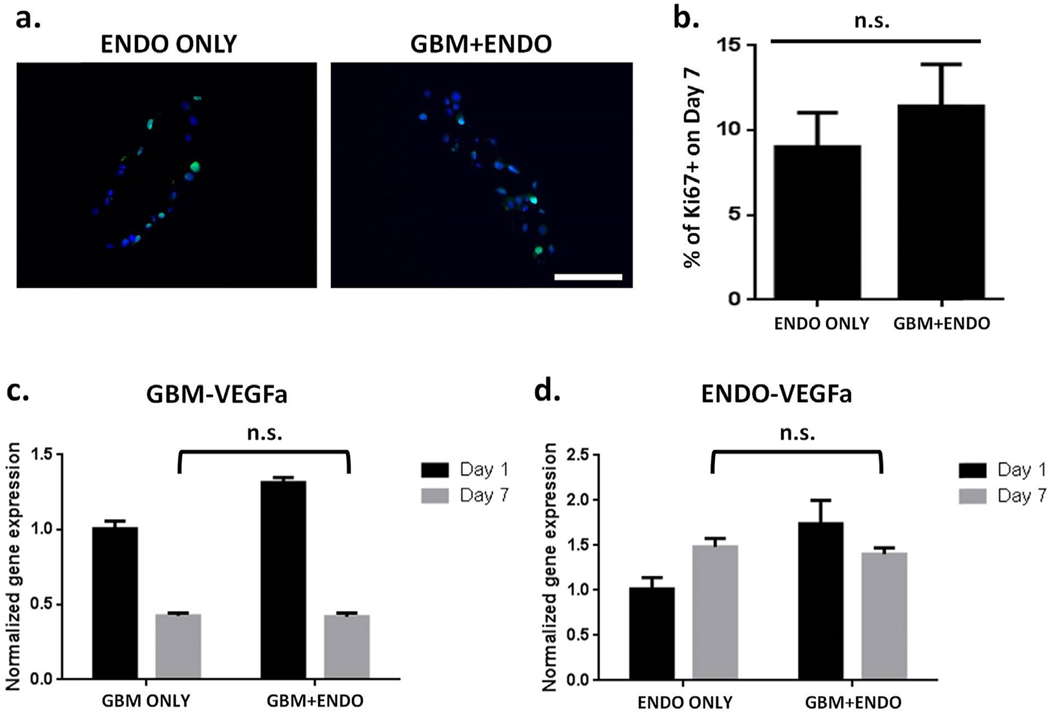
To analyze the effects of co-culture on cell proliferation, the percentage of Ki67 + cells were quantified on day 7 using immunostaining (Figs. 5 and 6). For tumor cells, the percentage of proliferating cells in GBM + ENDO group (32%) was significantly increased than in GBM ONLY group (16%) (Fig. 5b). CXCR4 is a chemokine receptor known to be involved in modulating tumor cell fates, including proliferation [35]. The gene expression levels of tumor CXCR4 was significantly upregulated in GBM + ENDO group by day 7, compared to GBM ONLY (Fig. 5c). For endothelial cells, comparable percentages of proliferating cells were observed in GBM + ENDO (11.4%) and ENDO (9.0%) groups (Fig. 6b). To determine if co-culture induced changes at the gene expression level, expression levels of VEGFa, a potent angiogenic growth factor for endothelial cell proliferation, were measured. Tumor and endothelial VEGFa gene expression levels were comparable in all groups on day 7 (Fig. 6c and d). In addition to gene expression analyses, we also conducted ELISA to further quantify secreted VEGF in medium from the GBM ONLY and GBM + ENDO groups. ELISA results showed co-culture led to significantly higher VEGF secretion by GBM at day 2 (Fig. S5). However, this difference was only transient. No significance difference in VEGF secretion was observed between the two groups at later time points (day 4 and day 6) (Fig. S5). The trend observed from ELISA is consistent with the trend we observed in gene expression data, which showed co-culture induced higher VEGF gene expression by GBM cells at day 1, but no differences were observed at day 7 (Fig. 6c).
Fig. 5. Co-culture with endothelial cells led to increased tumor cell proliferation.
(a). Immunostaining for proliferating (Ki67+) tumor cells on day 7. Scale bar = 100 μm. Red = human nuclear antigen. Green = Ki67. Blue = nuclei. (b). Percentage of proliferating (Ki67+) tumor cells on day 7 (n = 150). * p < 0.05. (c). Expression of CXCR4 in tumor cells on day 7, normalized to Day 1. Human specific primers were used to measure gene expression levels of CXCR4 using RT-PCR. * p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6. Endothelial cells in GBM + ENDO and ENDO ONLY groups had comparable levels of cell proliferation.
(a). Immunostaining for proliferating (Ki67 + cells) on day 7. Scale bar = 100 μm. Green = Ki67. Blue = nuclei. (b). Percentage of proliferating cells (Ki67+) on day 7 (n = 150). * p < 0.05. (c). Expression of VEGFa in tumor cells on day 7, normalized to day 1. Human specific primers were used to measure gene expression levels of VEGFa using RT-PCR. (d). Expression of VEGFa in endothelial cells on day 7, normalized to day 1. Mouse specific primers were used to measure gene expression levels of VEGFa using RT-PCR. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
To address the need for a physiologically relevant in vitro model for brain tumor-vasculature interactions, here we report a 3D co-culture model of GBM and brain endothelial cells with microanatomical architecture mimicry. In our model, endothelial cells were spatially patterned into vessel-like structures inside hydrogels with brain-mimicking biochemical and mechanical cues using hydrolytically-degradable alginate microfibers both as porogens and as cell-delivery vehicles. Our results demonstrate that this cell-patterning methodology was facile and efficient in generating microchannel porosity and deploying endothelial cells into vessel-like structures with high cell viability in 3D hydrogels. Patterned endothelial cells maintained endothelial cell phenotype, forming continuous cell sheet and vessel-like structures in 3D (Fig. 2). In co-culture, GBM cells exhibited significant increases in cell proliferation and gene expression levels of CXCR4, as compared to mono-culture (Fig. 5). Furthermore, in co-culture, endothelial cells significantly downregulated expression levels of cell adhesion proteins (Fig. 4), as compared to mono-culture.
To overcome the current limitations of conventional cell-patterning methods, we have engineered a facile, efficient methodology to generate vessel-like structures within 3D hydrogels using hydrolytically degradable alginate microfibers as porogens. Compared to microfluidics, which requires complex experimental setup and laborious protocols, our cell-patterning methodology does not involve special apparatus, providing ease of fabrication and convenience for scale up. Moreover, microfluidics also may use reagents that can be toxic to cells. In contrast, our methodology is cell-friendly, as shown by high cell viability on day 1 (Figs. 2b and 3a). Furthermore, uniform, confluent cell distribution is required to enable endothelial cells to form cell-cell adhesions [34]. Using microfluidics or subtractive templating to pattern endothelial cells often requires excessive amounts of cells and results in poor cell distribution along the microchannel length. Our results show that by using hydrolytically degradable alginate microfibers to deploy endothelial cells in microchannels, endothelial cells uniformly and confluently distribute along the microchannel lumen while maintaining endothelial cell phenotype (Fig. 2c). Compared to the advantages of conventional methods, limitations of our methodology include poor interconnectivity between microchannels and lack of flow. Also, the current alginate wet-spinning protocol can only synthesize microchannels with diameters ranging from 100 μm to millimeter scale, which is significantly larger the microvessels found in vivo, such as brain capillaries (7 μm) [36]. Future studies will focus on addressing these limitations to further enhance the applicability of our co-culture model.
It is worth pointing out that the model in this study does not provide functional blood vessels that form a complete and stable vascular structure. Instead, we sought to create a 3D co-culture model that mimics the spatial organization of just endothelial cells and tumor cells in 3D. As such, we chose only EC, but no stromal cells in the co-culture model. The advantage of using in vitro bioengineered model, rather than in vivo model, is that it allows us to ask one question at a time without multiple confounding factors. In this paper, we successfully achieved spatial patterning of endothelial cells in tubular structures to mimic the microanatomical structures, while co-encapsulating tumor cells around them. This is a substantial technological advance compared to simply mixing two cell types together in 3D, which cannot mimic spatial organization of the two cell types in vivo. To form stable and functional vessels, additional stromal cell types will likely be necessary such as pericytes and fibroblasts. Future studies can incorporate additional cell types to further elucidate the roles of additional cell types in the interactions of brain tumor and vasculature.
Tumor cell fates, such as proliferation and invasion, are known to be modulated by endothelial cell-secreted paracrine factors [4–6]. Co-culture of tumor and endothelial cells using direct monolayer co-culture has been shown to increase GBM cell proliferation [4]. When co-cultured with spatially patterned endothelial cells in 3D hydrogels, PDTX GBM cells exhibited significant increases in tumor cell proliferation, as shown by percentage of Ki67 + cells from 16% in mono-culture to 32% in co-culture (Fig. 4). One possible explanation for increased tumor cell proliferation in co-culture may be modulation of the SDF1a/CXCR4 pathway in tumor cells, as suggested in previous literature [4]. SDF1a is a chemokine found to be highly expressed by endothelial cells in tumor-associated blood vessels [4]. CXCR4 is a receptor for SDF1a, and previous work has shown that CXCR4 activation can induce survival and chemotaxis of brain tumor cells [37]. Inhibition of SDF1a/CXCR4 signaling between tumor and endothelial cells led to decreases in tumor cell proliferation [4]. Interestingly, in terms of tumor morphology, tumor aggregates in co-culture that were adjacent to endothelial microchannels were largely spherical with few cell protrusions (Fig. 3c). In the absence of endothelial cells, GBM tumor aggregates had radial protrusions extending from the aggregate surface. A possible explanation for the spherical morphology of tumor aggregates adjacent to endothelial microchannels may be that endothelial cells can create a local niche that supports the self-renewal of GBM stem cells, which typically grow as spheres [38]. Future experiments will investigate how co-culture influences the number and phenotype of GBM stem cells. Furthermore, previous literature has shown that, when co-cultured with endothelial tube networks, tumor cells migrated towards and co-localized with endothelial tubes, making direct contact with endothelial cells, a process mediated by endothelial cell-secreted SDF1a [4]. Also, in a Transwell invasion assay, endothelial cells promoted tumor cell invasion through SDF1a-mediated upregulation of MMP9 [6]. In the present study, no significant directional migration of tumor cells towards endothelial microchannels was observed. Discrepancies between our results and previous literature may be due to differences in the various endothelial cell types used. Previous reports used human brain microvascular endothelial cells and human microvascular endothelial cells. In the present study, mouse brain endothelial cells expressed very little SDF1a at both gene expression and protein levels (data not shown). The lack of directional migration of tumor cells towards endothelial cells suggests that additional stimulus may be needed in our co-culture model, such as hypoxia or other tumor resident cell types. Future studies will investigate how other microenvironmental factors can modulate tumor cell migration and invasion when co-cultured with endothelial cells.
With regards to the choice of model cell types, we have chosen a mouse endothelial cell line (bEnd3) to demonstrate the proof of concept in this study. We have chosen this cell line because previous reports have shown bEnd3 cells express tight junction protein claudin-5 in a similar way to those of primary mouse brain endothelial cells [39]. It is also characterized by the presence of a variety of transporters including P-gp, glucose transporter (GLUT1) and monocarboxylic acid transporter (MCT1) [40]. Therefore it provides a more physiologically relevant cell type than peripheral endothelial cells such as HUVECs. Recent studies by other groups have reported success to isolate and culture mouse or human derived brain microvascular endothelial cells [41,42]. It would be valuable to apply the model reported here for growing primarily isolated brain endothelial cells in the future, which will further increase the physiological relevance of the biological findings.
Tumor-associated endothelial cells have distinct morphologies, proliferation rates, and gene expression profiles, compared to normal brain endothelial cells [43]. In 3D hydrogels, endothelial cells co-cultured with tumor cells (GBM + ENDO group) became more rounded and disorganized, as compared to endothelial cells cultured alone (ENDO ONLY) (Fig. 3c). Disruption of endothelial cell morphology and monolayer organization in co-culture may be mediated by tumor-induced downregulation of cell adhesion proteins (claudin-5, ZO-1, and CD31) (Fig. 4a). Claudin-5 and ZO-1 participate in the formation of tight junctions between endothelial cells, which are essential for the low permeability and barrier properties of the blood-brain barrier (BBB) [44]. In addition to being a marker for endothelial cells, CD31, also known as PECAM, is highly enriched specifically at endothelial cell junctions, and is therefore used as an endothelial cell marker [45]. Brain tumors are characterized by breakdown of the BBB due to endothelial cell tight junction dysfunction. Significant decreases in tight junction proteins, claudin-1, claudin-5, and occludin, were correlated with the degree of tumor malignancy [46]. One possible explanation for tumor-induced BBB disruption is tumor-secreted paracrine factors that can modulate expression of proteins that can degrade tight junction proteins. For example, co-culture of GBM and endothelial cells led to decreased tight-junction protein expression due to TGF-β2 upregulation of MMP9, which can proteolytically degrade tight junction proteins [46]. Furthermore, tumor-secreted VEGF can also induce tight junction disassembly by changing the organization of tight-junction proteins [47]. Activation of VEGF receptors can lead to phosphorylation of cytoplasmic proteins involved in signal transduction, altering tight junction integrity [47]. VEGF can also change concentrations of cytosolic calcium in endothelial cells, modulating the actin cytoskeleton that regulates tight junction organization [47].
Throughout the process of tumor vascularization, GBM tumor cells are known to modulate endothelial cell proliferation via paracrine signals, such as VEGF, which promote migration and proliferation of endothelial cells, leading to vessel sprouting and angiogenesis [1]. Interestingly, in our study, gene expression levels of VEGF for tumor and endothelial cells was comparable in co-culture (GBM + ENDO) and mono-culture (GBM ONLY, ENDO ONLY) (Fig. 6c). These results may explain the comparable percentages of proliferating endothelial cells in co-culture (GBM + ENDO) and mono-culture (ENDO ONLY) (Fig. 6b). The lack of increased endothelial cell proliferation in co-culture suggests that additional stimulus may need to be incorporated in our co-culture system to model angiogenic interactions between tumor and endothelial cells. As an example, hypoxia is a potent regulator of tumor angiogenesis. In a low oxygen environment, tumor cells upregulate expression of HIF1α, an upstream transcription factor of VEGF [48]. Introduction of hypoxia into our co-culture system may induce tumor cells to upregulate VEGF expression and stimulate endothelial cell proliferation. Furthermore, other tumor resident cell types are known to participate in new blood vessel formation, such as pericytes, which can be co-encapsulated in our system. Pericytes are contractile cells that wrap around endothelial cells to support and maintain vessel growth and integrity. Pericytes can activate alternative pro-angiogenic pathways, such as PDGF receptor signaling [49].
5. Conclusion
GBM tumors are characterized by extensive vascularization, a phenomenon that critically depends on tumor-endothelial interactions. Here we report a 3D co-culture model that mimics in vivo spatial organization of tumor and endothelial cells using 3D biomimetic hydrogels and hydrolytically-degradable alginate fibers as porogens to deploy and pattern endothelial cells into vessel-like structures. Such cell-patterning methodology provide a facile, efficient method that can enable co-culture of tumor and endothelial cells at tissue scale without special experimental setup. Patterned endothelial cells retained high cell viability, proliferated, and maintained endothelial cell phenotype over time. Co-culture of brain endothelial cells and patient-derived glioblastoma cells recapitulated interactions observed in previous literature. Endothelial cells downregulated expression of cell adhesion proteins and adopted more rounded cell morphology. Co-culture also promoted increased tumor cell proliferation, and tumor aggregates that were directly adjacent to endothelial cells were largely spherical. Interestingly, endothelial cell proliferation was comparable in co-culture and mono-culture groups, suggesting that additional stimulus in the co-culture model may be required, such as hypoxia or additional cell types. In summary, we developed a 3D co-culture model that mimics the microanatomical architecture of tumor-endothelial interactions, which may provide a valuable tool for studying the role of tumorendothelial interactions on tumor progression and for evaluating novel drug targets.
Supplementary Material
Acknowledgements
This work was supported by the following grants: NIH R01DE024772 (F.Y.), NSF CAREER award (CBET-1351289) (F.Y.), and California Institute for Regenerative Medicine Tools and Technologies Award (RT3-07804) (F.Y.). The authors also acknowledge funding from the Stanford Child Health Research Institute Faculty Scholar Award (F.Y.), Stanford Bio-X IIP grant award (F. Y.), the Alliance for Cancer Gene Therapy Young Investigator award grant (F.Y.)., and the Stanford Chem-H Institute (F.Y.). C.W. would like to thank Stanford Graduate Fellowship and Stanford Interdisciplinary Graduate Fellowship for support. J. L. would like to thank Stanford Child Health Research Institute postdoctoral fellowship for support. The authors also appreciate technical assistance from Stanford Cell Sciences Imaging Facility for confocal imaging.
Footnotes
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time due to technical or time limitations.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2019.02.024.
References
- [1].Hardee ME, Zagzag D, Mechanisms of glioma-associated neovascularization, Am. J. Pathol. 181 (2012) 1126–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ruoslahti E, Specialization of tumour vasculature, Nat. Rev. Canc. 2 (2002) 83–90. [DOI] [PubMed] [Google Scholar]
- [3].Vaupel P, Harrison L, Tumor hypoxia: causative factors, compensatory mechanisms, and cellular response, Oncol. 9 (2004) 4–9. [DOI] [PubMed] [Google Scholar]
- [4].Rao S, Sengupta R, Choe EJ, Woerner BM, Jackson E, Sun T, et al. , CXCL12 mediates trophic interactions between endothelial and tumor cells in glioblastoma, PLoS One 7 (2012) e33005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhu TS, Costello MA, Talsma CE, Flack CG, Crowley JG, Hamm LL, et al. , Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells, Cancer Res. 71 (2011) 6061–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kenig S, Alonso MBD, Mueller MM, Lah TT, Glioblastoma and endothelial cells cross-talk, mediated by SDF-1, enhances tumour invasion and endothelial proliferation by increasing expression of cathepsins B, S, and MMP-9, Cancer Lett. 289 (2010) 53–61. [DOI] [PubMed] [Google Scholar]
- [7].Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, et al. , Longterm survival with glioblastoma multiforme, Brain 130 (2007) 2596–2606. [DOI] [PubMed] [Google Scholar]
- [8].McLendon RE, Halperin EC, Is the long-term survival of patients with intracranial glioblastoma multiforme overstated? Cancer 98 (2003) 1745–1748. [DOI] [PubMed] [Google Scholar]
- [9].Wen PY, Kesari S, Malignant gliomas in adults, N. Engl. J. Med. 359 (2008) 492–507. [DOI] [PubMed] [Google Scholar]
- [10].Holash J, Maisonpierre PC, Compton D, Boland P, Alexander CR, Zagzag D, et al. , Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF, Science 284 (1999) 1994–1998. [DOI] [PubMed] [Google Scholar]
- [11].Zagzag D, Amirnovin R, Greco M, Yee H, Holash J, Wiegand S, et al. , Vascular apoptosis and involution in gliomas precede neovascularization: a novel concept for glioma growth and angiogenesis, Lab. Invest. 80 (2000) 837–849. [DOI] [PubMed] [Google Scholar]
- [12].Upreti M, Abstract 5288: tumor-endothelial 3D spheroids for dynamic imaging of cancer progression, Cancer Res. 71 (2011) 5288. [Google Scholar]
- [13].Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, et al. , In vitro microvessels for the study of angiogenesis and thrombosis, Proceedings of the National Academy of Sciences of the United States of America, vol. 109, 2012, pp. 9342–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC, In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform, Frontiers in Bioengineering and Biotechnology 4 (2016) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li A, Walling J, Kotliarov Y, Center A, Steed ME, Ahn SJ, et al. , Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas, Mol. Canc. Res. MCR 6 (2008) 21–30. [DOI] [PubMed] [Google Scholar]
- [16].Joo KM, Kim J, Jin J, Kim M, Seol HJ, Muradov J, et al. , Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ, Cell Rep. 3 (2013) 260–273. [DOI] [PubMed] [Google Scholar]
- [17].Chen Z, Htay A, Dos Santos W, Gillies GT, Fillmore HL, Sholley MM, et al. , In vitro angiogenesis by human umbilical vein endothelial cells (HUVEC) induced by three-dimensional co-culture with glioblastoma cells, J. Neuro Oncol. 92 (2009) 121–128. [DOI] [PubMed] [Google Scholar]
- [18].Azad TD, Pan J, Connolly ID, Remington A, Wilson CM, Grant GA, Therapeutic strategies to improve drug delivery across the blood-brain barrier, Neurosurg. Focus 38 (2015) E9-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rüffer C, Strey A, Janning A, Kim KS, Gerke V, Cell−Cell junctions of dermal microvascular endothelial cells contain tight and adherens junction proteins in spatial proximity, Biochemistry 43 (2004) 5360–5369. [DOI] [PubMed] [Google Scholar]
- [20].Man S, Ubogu EE, Williams KA, Tucky B, Callahan MK, Ransohoff RM, Human brain microvascular endothelial cells and umbilical vein endothelial cells differentially facilitate leukocyte recruitment and utilize chemokines for T cell migration, Clin. Dev. Immunol. 2008 (2008) 384982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang C, Tong X, Yang F, Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using PEG-based hydrogels, Mol. Pharm. 11 (7) (2014. July 7) 2115–2125. [DOI] [PubMed] [Google Scholar]
- [22].Madl CM, Keeney M, Li X, Han LH, Yang F, Co-Release of Cells and Polymeric Nanoparticles from Sacrificial Microfibers Enhances Nonviral Gene Delivery inside 3D Hydrogels, Tissue Eng. Part C Methods (2014). [DOI] [PubMed] [Google Scholar]
- [23].Hammer J, Han LH, Tong X, Yang F, A facile method to fabricate hydrogels with microchannel-like porosity for tissue engineering. Tissue engineering Part C, Methods 20 (2014) 169–176. [DOI] [PubMed] [Google Scholar]
- [24].Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS, A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization, Adv. Mater. 21 (2009) 5005–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Anderson SB, Lin C-C, Kuntzler DV, Anseth KS, The performance of human mesenchymal stem cells encapsulated in cell-degradable polymer-peptide hydrogels, Biomaterials 32 (2011) 3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shu XZ, Liu Y, Luo Y, Roberts MC, Prestwich GD, Disulfide cross-linked hyaluronan hydrogels, Biomacromolecules 3 (2002) 1304–1311. [DOI] [PubMed] [Google Scholar]
- [27].Delpech B, Maingonnat C, Girard N, Chauzy C, Maunoury R, Olivier A, et al. , Hyaluronan and hyaluronectin in the extracellular matrix of human brain tumor stroma, Eur. J. Cancer 29 (1993) 1012–1017. [DOI] [PubMed] [Google Scholar]
- [28].Demirkan B. Gürsel, Shin Benjamin J., Burkhardt Jan-Karl, Kesavabhotla Kartik, Schlaff Cody D., Boockvar John A., Glioblastoma stem-like cells—biology and therapeutic implications, Cancers 3 (2) (2011) 2655–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yuan Xiangpeng, Curtin James, Xiong Yizhi, Liu Gentao, Sebastian Waschsmann-Hogiu Daniel L. Farkas, Black Keith L., John S. Yu, Isolation of cancer stem cells from adult glioblastoma multiforme, Oncogene 23 (58) (2004) 9392. [DOI] [PubMed] [Google Scholar]
- [30].Veselska Renata, Kuglik Petr, Cejpek Pavel, Svachova Hana, Neradil Jakub, Loja Tomas, Relichova Jirina, Nestin expression in the cell lines derived from glioblastoma multiforme, BMC Canc. 6 (no. 1) (2006) 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Palumbo Paola, Miconi Gianfranca, Cinque Benedetta, Lombardi Francesca, Cristina La Torre Soheila Raysi Dehcordi, Galzio Renato, Cimini Annamaria, Giordano Antonio, Cifone Maria Grazia, NOS2 expression in glioma cell lines and glioma primary cell cultures: correlation with neurosphere generation and SOX-2 expression, Oncotarget 8 (2017) 25582–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yan Tao, Kai Ove Skaftnesmo Lina Leiss, Sleire Linda, Wang Jian, Li Xingang, Per Øyvind Enger, Neuronal markers are expressed in human gliomas and NSE knockdown sensitizes glioblastoma cells to radiotherapy and temozolomide, BMC Canc. 11 (no. 1) (2011) 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bouhadir KH, Lee KY, Alsberg E, Damm KL, Anderson KW, Mooney DJ, Degradation of partially oxidized alginate and its potential application for tissue engineering, Biotechnol. Prog. 17 (2001) 945–950. [DOI] [PubMed] [Google Scholar]
- [34].Dejana E, Endothelial cell-cell junctions: happy together, Nat. Rev. Mol. Cell Biol. 5 (2004) 261–270. [DOI] [PubMed] [Google Scholar]
- [35].Ehtesham M, Min E, Issar NM, Kasl RA, Khan IS, Thompson RC, The role of the CXCR4 cell surface chemokine receptor in glioma biology, J. Neuro Oncol. 113 (2013) 153–162. [DOI] [PubMed] [Google Scholar]
- [36].Wong A, Ye M, Levy A, Rothstein J, Bergles D, Searson P, The blood-brain barrier: an engineering perspective, Front. Neuroeng. 6 (2013) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rubin JB, Kung AL, Klein RS, Chan JA, Sun Y, Schmidt K, et al. , A smallmolecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors, Proc. Natl. Acad. Sci. U. S. A 100 (2003) 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN, Cancer stem cells in glioblastoma, Genes Dev. 29 (2015) 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Watanabe T, Dohgu S, Takata F, Nishioku T, Nakashima A, Futagami K, et al. , Paracellular barrier and tightness junction protein expression in immortalized brain endothelial cell lines bEND.3, bEND.5 and mouse brain endothelial cell 4, Biol. Pharm. Bull. 36 (2013) 492–495. [DOI] [PubMed] [Google Scholar]
- [40].Omidi Y, Campbell L, Barar J, Connell D, Akhtar S, Gumbleton M, Evaluation of the immortalized mouse brain capillary endothelial line, b.End 3, as an in vitro blood-brain barrier model for drug uptake and transport studies, Brain Res. 990 (2003) 95–112. [DOI] [PubMed] [Google Scholar]
- [41].Ruck Tobias, Bittner Stefan, Epping Lisa, Herrmann Alexander M., Meuth Sven G., Isolation of primary murine brain microvascular endothelial cells, JoVE: JoVE 93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Navone Stefania E., Marfia Giovanni, Invernici Gloria, Cristini Silvia, Nava Sara, Balbi Sergio, Sangiorgi Simone, et al. , Isolation and expansion of human and mouse brain microvascular endothelial cells, Nat. Protoc. 8 (9) (2013) 1680. [DOI] [PubMed] [Google Scholar]
- [43].Charalambous C, Chen TC, Hofman FM, Characteristics of tumor-associated endothelial cells derived from glioblastoma multiforme, Neurosurg. Focus 20 (2006) E22. [DOI] [PubMed] [Google Scholar]
- [44].Bazzoni G, Dejana E, Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis, Physiol. Rev. 84 (2004) 869–901. [DOI] [PubMed] [Google Scholar]
- [45].Albelda SM, Muller WA, Buck CA, Newman PJ, Molecular and cellular properties of PECAM-1 (endoCAM/CD31): a novel vascular cell-cell adhesion molecule, J. Cell Biol. 114 (1991) 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ishihara H, Kubota H, Lindberg RLP, Leppert D, Gloor SM, Errede M, et al. , Endothelial cell barrier impairment induced by glioblastomas and transforming growth factor β2 involves matrix metalloproteinases and tight junction proteins, J. Neuropathol. Exp. Neurol. 67 (2008) 435–448. [DOI] [PubMed] [Google Scholar]
- [47].de Vries HE, Montagne L, Dijkstra CD, van der Valk P, Molecular and cellular biology of the blood-brain barrier and its characteristics in brain tumors, in: AliOsman F. (Ed.), Brain Tumors, Humana Press, Totowa, NJ, 2005, pp. 157–174. [Google Scholar]
- [48].Unruh A, Ressel A, Mohamed HG, Johnson RS, Nadrowitz R, Richter E, et al. , The hypoxia-inducible factor-1[alpha] is a negative factor for tumor therapy, Oncogene 22 (2003) 3213–3220. [DOI] [PubMed] [Google Scholar]
- [49].Liu Allan Y, Ouyang G, Tumor angiogenesis: a new source of pericytes, Curr. Biol. 23 (2013) R565–R568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.