Significance
Here we present an unprecedented multiscale simulation platform that enables modeling, hypothesis generation, and discovery across biologically relevant length and time scales to predict mechanisms that can be tested experimentally. We demonstrate that our predictive simulation-experimental validation loop generates accurate insights into RAS-membrane biology. Evaluating over 100,000 correlated simulations, we show that RAS–lipid interactions are dynamic and evolving, resulting in: 1) a reordering and selection of lipid domains in realistic eight-lipid bilayers, 2) clustering of RAS into multimers correlating with specific lipid fingerprints, 3) changes in the orientation of the RAS G-domain impacting its ability to interact with effectors, and 4) demonstration that RAS–RAS G-domain interfaces are nonspecific in these putative signaling domains.
Keywords: RAS dynamics, RAS-membrane biology, multiscale modeling, multiscale infrastructure, massive parallel simulations
Abstract
RAS is a signaling protein associated with the cell membrane that is mutated in up to 30% of human cancers. RAS signaling has been proposed to be regulated by dynamic heterogeneity of the cell membrane. Investigating such a mechanism requires near-atomistic detail at macroscopic temporal and spatial scales, which is not possible with conventional computational or experimental techniques. We demonstrate here a multiscale simulation infrastructure that uses machine learning to create a scale-bridging ensemble of over 100,000 simulations of active wild-type KRAS on a complex, asymmetric membrane. Initialized and validated with experimental data (including a new structure of active wild-type KRAS), these simulations represent a substantial advance in the ability to characterize RAS-membrane biology. We report distinctive patterns of local lipid composition that correlate with interfacially promiscuous RAS multimerization. These lipid fingerprints are coupled to RAS dynamics, predicted to influence effector binding, and therefore may be a mechanism for regulating cell signaling cascades.
RAS driven cancers are common (1), difficult to treat (2), and a major cause of death worldwide (3). KRAS, the isoform most frequently associated with disease, is mutated in nearly all pancreatic cancers and often in lung and colorectal cancers (4, 5). Only recently, with the development of covalent inhibitors of the G12C mutant (6), has direct targeting of RAS been successful, and more broadly applicable inhibitors are needed.
The RAS subfamily comprises peripheral membrane proteins with a conserved globular GTP-binding domain (G-domain) (7) that is tethered to the cell membrane by a prenylated ∼20-residue C-terminal domain called the hypervariable region (HVR) (8, 9). RAS proteins function as molecular switches whose active conformations, stabilized by GTP binding, interact with several protein effectors to control cell growth, proliferation, differentiation, and migration (10). Constitutive activation of oncogenic RAS perturbs several cellular signaling cascades, including the MAPK pathway, which RAS accesses via activation of RAF kinase at the plasma membrane (PM).
There is substantial interest in assessing the ability of RAS molecules to dimerize (11–13) or colocalize (14–17) at the membrane, because RAS-dependent RAF activation requires dimerization of RAF (18–20). Although wild-type KRAS4b, a common splice variant of KRAS (hereafter referred to as RAS), does not dimerize on two-component supported lipid bilayers (21), it preferentially colocalizes with anionic lipids in the liquid-disordered domains of giant unilamellar vesicles (22) and clusters on the scale of tens of nanometers in extracted PM sheets (14, 23, 24).
Preferential interaction of RAS with anionic lipids is mediated by 11 positively charged lysines in its HVR (25, 26). However, charge complementarity is insufficient to fully describe RAS nanoclustering, which is exquisitely sensitive to lipid composition (27–29). Even less is known about the influence of RAS–lipid coupling on RAS self-assembly and effector activation. While several feasible dimer interfaces have been reported (11–13, 15, 30–37), how RAS forms dimers, if at all, remains a major area of interest.
The fundamental challenge of investigating RAS activation events is that many of the proposed mechanisms involve time and lengths scales currently not accessible. For example, functional events in RAS dynamics that may preferentially depend on local depletion or enrichment of specific lipids are extremely difficult to observe directly, either in computational or biological experiments. Experimentally, we use single-particle tracking (SPT) to follow HaloTag-conjugated RAS via total internal reflection fluorescence microscopy in live HeLa cells. While the broad heterogeneity in lateral diffusion observed by SPT (Fig. 1A) is indicative of multiple RAS subpopulations that have distinct patterns of interaction with lipids and other cellular components, it provides little insight on local lipid environments and their effect on RAS behavior. Similarly, we can use detailed molecular dynamics (MD) simulations to probe specific lipid environments, but systems large enough to support substantial fluctuations in lipid and protein concentrations cannot be practically simulated long enough to observe relevant fluctuations. Instead, we present a multiscale infrastructure that directly couples a macroscale continuum simulation capable of observing RAS clustering and long-range lipid rearrangement, with a massive ensemble of microscale MD simulations that provides detailed insights into local dynamics. Both scales are connected through a machine learning (ML) informed dynamic sampling process (38), which enables mapping of findings from the MD simulations onto the continuum simulation, resulting in microscale insights that are observable over macrospatial and temporal scales.
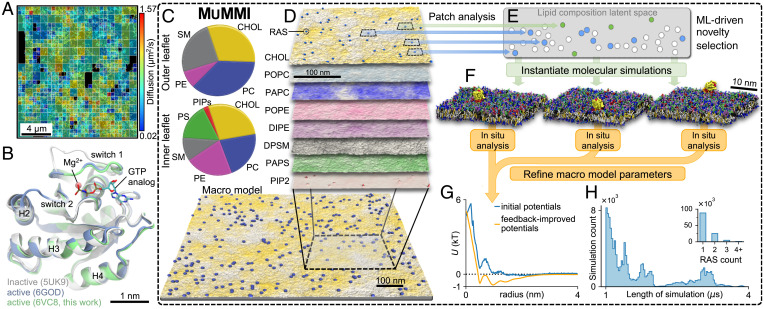
Fig. 1.
Experimental input and computational approach. (A) Diffusion mapping of single molecules of KRAS4b tethered to or within 100 nm of the PM in a 16 × 16-μm2 region of a live HeLa cell accumulated over 10 s. (B) Crystal structures of wild-type KRAS in active (green and blue; GppNHp-bound) and inactive (gray; GppCH2p-bound configurations). (C) Average macro model lipid composition. (D–F) The Multiscale Machine-learned Modeling Infrastructure (MuMMI). (D) Representative snapshots of each of the different lipid distributions in the inner leaflet of a 0.3 × 0.3-μm2 region of the full 1 × 1-μm2 macro simulation; color saturation indicates local lipid density. (E) Schematic illustrating latent space encoding of lipid composition in 30 × 30-nm2 membrane patches. From the candidate patches (blue and green), those that are most dissimilar (green) to existing (white) CG simulations are selected and used to spawn new CG simulations. (F) Representative CG simulation systems (water not shown). (G) Improvement of macro model parameter inputs from feedback iteration. (H) Distribution of CG simulation duration and (Inset) number of RAS per patch.
More specifically, to enable simulations of active wild-type RAS, we solve its crystal structure bound to the GTP analog GppNHp at a resolution of 2.5 Å (Fig. 1B and SI Appendix, S1.2.2). We choose to focus the effort on wild-type GTP-loaded KRAS4b because GTP hydrolysis will not occur on the time scale of the simulations, and because available structures of wild-type or oncogenic KRAS4b bound to the RAS binding domain (RBD) of RAF1 are similar. This present work will serve as the foundation for understanding activation of RAF by GTP-loaded RAS. We mimic the composition of a biologically relevant PM (39) by employing an asymmetric eight-lipid mixture (40), here called the average RAS lipid composition (ARC) and is composed of cholesterol, phosphatidylcholines (PC), phosphatidylethanolamines (PE), phosphatidylserine (PS), phosphatidylinositol bisphosphate (PIP2), and sphingomyelin (SM), with varying acyl chain length and saturation (Fig. 1C and SI Appendix, S1.2.1).
Multiscale Simulation Framework
As discussed above, the behavior of RAS is mediated by the local lipid environment, which is not readily accessible experimentally. In principle, computational models can provide the necessary details for small neighborhoods around RAS, but they require a priori knowledge of which protein confirmations and local lipid environments are of interest. Traditional computational studies typically choose an average lipid composition and focus on providing many replicates and long-running trajectories. However, in vivo, proteins experience a wide range of different lipid configurations and, especially considering effects such as charged lipids, a protein is in fact unlikely to experience an average lipid environment. Instead, we expect the local lipid composition to be heavily influenced by the proteins and the codependence of the lipid and proteins is one of the key properties of interest. Manually selecting protein conformations and sets of lipid compositions can provide valuable insights, but requires a prior knowledge of what amplitudes and along what dimensions one should adjust the simulations. Unfortunately, running an MD simulation large enough and long enough to naturally express a much wider range of lipid environments and protein conformations is practically not feasible. Here, we introduce MuMMI, the Multiscale Machine-learned Modeling Infrastructure (Fig. 1 C–H and SI Appendix, S1.1), which combines the relatively low computational cost of MD simulations in local neighborhoods around proteins with a diverse sampling of environments equivalent to running an otherwise infeasible MD simulation. As discussed in more details below, the resulting system efficiently samples local environments from a macroscale model, such that the resulting ensemble of MD simulations can be used to study the entire macroscale model at what amounts to MD resolution. For a full description of MuMMI, see SI Appendix, S1.1, and for the description of the model generation and parameterization, see SI Appendix, S1.2.
MuMMI employs a macro model (Fig. 1D) to explore interactions between lipids and proteins over large length and time scales. The macro model represents a lipid bilayer as a set of density fields describing the density of each lipid type as a function of space and evolves them using dynamic density functional theory. Proteins, RAS in our case, are represented as single, rotationally averaged beads, which interact with the lipids and other proteins through defined interaction potentials evolved using MD equations of motion. In this work, the macro model consists of a 1 × 1-µm2 PM, eight-lipid type (ARC) bilayer that is resolved at roughly nanometer resolution and with 300 RAS molecules on the inner leaflet. The RAS proteins exist in one of two distinct states representing different orientations with respect to the membrane, which they can transition between. During the MuMMI multiscale simulation, the macro model ran for 150 µs. The theory behind the macro model is described in Stanton et al. (41) and summarized in SI Appendix, S1.1.3. The macro model parameterization is described in SI Appendix, S1.2.6, while the MD simulations used to derive the initial parameters and the initial RAS state definition are described in SI Appendix, S1.2.4 and S1.2.5, respectively.
While not providing sufficient resolution to probe molecular processes, the macro model faithfully models the overall lipid dynamics and naturally creates local fluctuations of the average membrane composition in which certain lipid types are enriched or depleted, driven by membrane mechanics and protein–membrane interactions. In particular, the macro model explores a wide range of lipid environments beneath the RAS proteins and protein stoichiometries mimicking the diverse configurations encountered in a cell PM (SI Appendix, S2.2.1). More specifically, MuMMI focuses on local, here 30 × 30 nm2, subregions (hereafter, patches) and their corresponding proteins at both the macro- and at near-atomistic scales. One potential solution to explore the phase space of patches would be to randomly sample from the macro model, creating the corresponding coarse-grained (CG) Martini (42) MD simulations (Fig. 1 C–F) and ultimately assembling a rough approximation of the expected behavior at the macroscale. However, the patch distribution is highly nonuniform with some configurations occurring orders-of-magnitude more frequently than others. Nevertheless, compared to the relevant biological time scales even rare events, occurring at a frequency of 150 μs in the macro model, should not be considered outliers. In fact, rare lipid compositions might trigger signaling events that drastically amplify their overall impact and it is these cases we are most interested in studying. Therefore, MuMMI implements a ML-driven dynamic-importance (DynIm) sampling (38) to select a maximally diverse subset of patches to evaluate with CG simulations.
MuMMI uses a ML-driven selection process, which first encodes local lipid concentrations (patches) into a 15-dimensional latent space using a variational autoencoder that learns to identify similarity among macro configurations and then uses a novelty-sampling scheme to select the patches explored by the macro model that are most dissimilar to the ones already simulated at the CG scale (Fig. 1 C–F and SI Appendix, S1.1.5 and S1.2.7). The ML model was trained prior to running the multiscale simulation using 302,000 patches generated from a separate run of the macro model; it was used for inference to dynamically select novel patches generated during the MuMMI multiscale simulation. (Computational limitations prohibited retraining the autoencoder on-the-fly; therefore, the pretrained ML model was used throughout.) Selected patches are used to spawn new CG simulations creating a maximally diverse set of CG simulations expected to explore a much wider range of RAS behavior. Simultaneously, MuMMI maintains sufficient information about the nonselected patches to accurately reconstruct the true, unbiased distribution of patches in latent space (SI Appendix, S1.1.5, S2.2.3, and S2.3.2). Note that the reconstructed distribution has a significantly wider support than the one that could have been recovered from a traditional random sampling (SI Appendix, Fig. S15) of comparable size. In other words, by deliberately including rare patches in a correctly weighted manner, our distribution is equivalent to a substantially longer macro simulation and thus is able to explore more lipid environments and protein configurations than otherwise possible. Ultimately, the weighted distribution allows computation of accurate, population-wide averages of any property of interest computed from the CG simulations. Furthermore, as more computing resources become available, the sampling of patches with corresponding CG data becomes denser, ultimately converging to a state at which, for every patch created by the macro model, MuMMI has either the corresponding CG simulation or one for a patch so similar as to be considered equivalent. At this point, the ensemble of CG simulations, together with the macro model data, jointly represent a simulation at macrolevel time and length scales but effectively at MD precision.
At the microscale, CG MD simulations are used to capture interactions of proteins and lipids. Martini CG simulations provide over two orders-of-magnitude speedup compared to all-atom simulations and are well suited to capture overall lipid–lipid, protein–lipid, and protein–protein interactions, especially at longer length and time scales (42–45). The reduced degrees-of-freedom come with a price, and model limitations, such as lack of directional hydrogen bonds, need to be considered when interpreting the results, especially for protein–protein interactions (46, 47) (SI Appendix, S1.2). In MuMMI, each selected macro model patch is converted into a CG simulation containing ∼140K particles. Membranes are built with subgrid resolution and statespecific placement of one or more RAS molecules sampled from a library (Fig. 1F and SI Appendix, S1.1.4), resulting in a large sampling of lipid compositions and RAS configurations (SI Appendix, S2.2 and S2.3.2). These CG simulations are performed on GPUs using ddcMD (48) (SI Appendix, S1.1.2) and analyzed in situ (SI Appendix, S1.1.6). The on-the-fly analysis of each CG simulation is weighted based on its representative patch in macro model example (SI Appendix, S2.2.3) and used to continuously refine the macro model protein–lipid parameters for each protein state (Fig. 1G and SI Appendix, S2.2.4), incrementally improving the accuracy of the macro model with improved sampling.
MuMMI represents an emerging new capability that delivers a macroscale model at experimentally relevant time and length scales with effective microscale resolution. The simulations presented in this study represent a massive ensemble comprising 120K, 1- to 4-μs-long, unique but correlated CG simulations, simulated over several weeks using one of the world’s largest supercomputers, Sierra, and consuming ∼5.6 million GPU hours (SI Appendix, S2.1). These CG simulations capture and quantify RAS–lipid dynamics across local compositions observed in the much larger macro simulation. A new massively parallel workflow (49) was developed to orchestrate MuMMI (SI Appendix, S1.1.1) and MuMMI is capable of utilizing some of the largest supercomputers in the world; its functionality is also equally applicable to modest and more-commonly available compute resources.
RAS–Lipid Colocalization
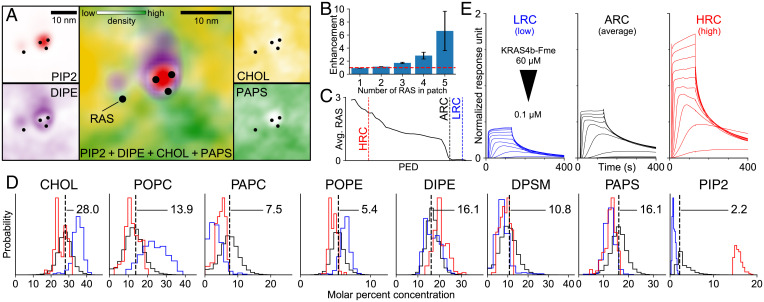
Lipid fingerprints are spatially resolved lipid concentration signatures around proteins (50) and can be clearly observed for RAS in the PM both at the macro and CG resolution (see below). The macro simulation reveals lipid redistribution around RAS (Fig. 2A), indicative of discrete lipid fingerprints, and a propensity for RAS to self-associate (Fig. 2B). The latter is consistent with experimental RAS clustering as seen in atomic force microscopy experiments with simple lipid mixtures (22, 51).
Fig. 2.
Lipid-dependence of RAS colocalization. (A) Representative macro model inner leaflet lipid densities around RAS, shown separately (small boxes) and together (large central box). Color saturation indicates local lipid density. (B) Population ratio of RAS multimer sizes observed in the macro simulation vs. a random uniform distribution. (C) Average number of RAS in macro model regions (radius 5 nm) along the primary embedding dimension (PED) from function preserving projection analysis (SI Appendix, S2.8.2). Vertical lines denote thresholds used to define high-RAS (HRC), initial average (ARC), and low-RAS (LRC) lipid compositions. (D) Distributions of inner leaflet lipid concentrations for all patches with RAS (black), HRC (red), and LRC (blue). The ARC is represented by a dashed vertical line. (E) Surface plasmon resonance sensorgrams of RAS adhesion to liposomes with the LRC, ARC, and HRC lipid compositions. Each subplot contains multiple traces representing distinct RAS concentrations (twofold dilutions, 60 to 0.1 μM).
To explore the connection between the local lipid environment and RAS clustering we use function preserving projections (52), which uses ML to identify two principal directions that are linear combinations of local lipid concentrations and maximally correlate these concentrations with the number of RAS in a patch (Fig. 2C and SI Appendix, S2.8.2 and S2.8.3). A subselection within the high/low RAS regions of the embedding is used to find unimodal lipid compositions that are either RAS depleted (low RAS colocalization, LRC) or enriched (high RAS colocalization, HRC) as classified in Fig. 2D and SI Appendix, Table S4. In particular, the HRC is enriched in PIP2 and depleted in cholesterol and polyunsaturated lipids, whereas the LRC exhibits opposite trends. These findings on our in silico membrane corroborate RAS’s preferential association with PIP2 on experimental artificial membranes (53, 54). However, a discrepancy still exists between reconstituted systems and the observed enrichment of PS and phosphatidic acids in RAS nanoclusters on intact-cell PM sheets (14, 24). While some lipid concentrations, such as 15.5% PIP2 in the HRC, are extreme for the global PM, our macroscale model indicates their presence in spatially localized regions due to concentration fluctuations and protein-mediated enrichment. Surface plasmon resonance confirms that, compared to the initial average lipid composition (ARC), RAS binding is weaker to the low LRC and stronger to the high HRC compositions (Fig. 2E), consistent with specific lipid concentrations influencing RAS recruitment and colocalization.
RAS Diffusion and Preferential Interaction with PIP2
Peripheral membrane proteins display dynamic diffusional properties as they interact with the hierarchical architecture of the PM (55). SPT of RAS proteins identified three discrete populations with diffusion coefficients represented here by values acquired from HeLa cells: one dominant fast mobile component (0.96 ± 0.03 μm2/s), an intermediate component (0.26 ± 0.03 μm2/s), and a slow component (0.05 ± 0.01 μm2/s) (56). Defining the disposition of RAS in these discrete diffusional populations is challenging, and subdiffraction microscopic techniques do not provide sufficient resolution to accurately measure the size of these RAS complexes. Coupling conformational fluctuations of protein structure with dynamic diffusional properties of RAS on the membrane can only be bridged using simulations.
It is well documented that experimentally measured RAS has a preference for liquid-disordered membrane patches enriched in anionic lipids. A variety of different experimental techniques, using different lipid mimetics (supported lipid bilayers, bilayers, nanodiscs, or liposomes) composed of simple two-lipid mixtures (21, 25) to those composed of five or more lipids (36, 57), all indicate that KRAS has a preference for PS (22, 57) and phosphatidylinositol. Lysine residues within the HVR of RAS (27) are known to interact with PS in mammalian PMs and, as a consequence, impact the propensity of RAS to form nanoclusters. Previous MD simulations have identified that RAS can exist in multiple membrane-bound orientational states (58–61) while bound to two component lipid mixtures. These simulations are limited in their complexity of the lipid mixtures represented, even though the complexity of the lipid composition in the PM is well established (62). In addition, the length of the simulations is not long enough to explore even the shortest lifetime of RAS molecules identified in mammalian cells (56). Therefore, to represent more biologically relevant models, extended time scales and increased complexity of the lipid bilayer are needed in such simulations.
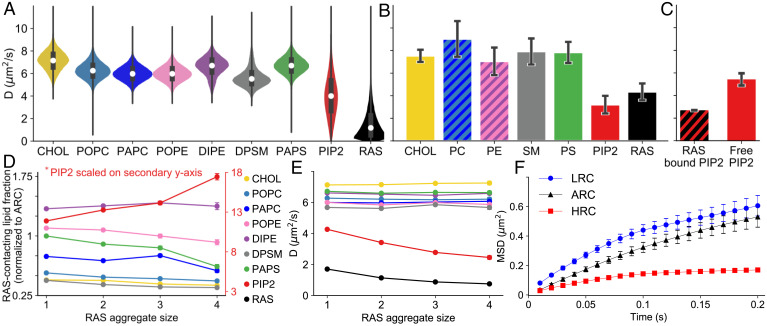
In this study, the diffusion properties of the lipids and RAS on ARC were evaluated in simulation as well as in experiments using fluorescence lifetime correlation spectroscopy (FLCS). All lipid types show a broad distribution of diffusion coefficients among the 88,392 unique CG simulations with one RAS molecule (Fig. 3A and SI Appendix, S2.7). This variation reflects the dependence of lipid diffusion on membrane composition. For example, a 10% change in cholesterol content slows the diffusion of all lipids except PIP2 by a factor of 2 (SI Appendix, Fig. S37). For a more complete discussion of the impacts of membrane composition, see SI Appendix, S2.3.2. The ARC diffusion rates calculated from FLCS experiments are shown in Fig. 3B. All lipids except PIP2 diffuse at similar average rates in both FLCS and in the ensemble of simulations, whereas RAS and PIP2 diffuse two to three times slower (Fig. 3 A and B). Note, absolute diffusion numbers can vary significantly depending on experimental method used (63), as well as simulation force field, size, and thermostat (64, 65) (SI Appendix, S.2.7 and Table S3) and should only be considered as semiqualitative. Here, all simulation diffusion numbers are scaled by the Martini canonical factor of 4 (42). Interestingly, in the simulation a direct interaction with RAS slows the diffusion of PIP2 (Fig. 3C). In CG simulations with multiple RAS molecules, increased aggregation of RAS is associated with local enrichment of PIP2 and depletion of PAPS (1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphatidylserine) (Fig. 3D), emphasizing the potential importance of charge density. RAS aggregation also slows the diffusion of both RAS and PIP2 (Fig. 3E), while the diffusion of other lipids remain unaltered.
Fig. 3.
Diffusion of RAS and lipids. (A and B) Distributions of lateral diffusion rates, D, for lipids and RAS in (A) CG simulations and (B) FLCS on the ARC. (C) Values of D for PIP2 conditioned on interaction with monomeric RAS in the CG simulations. (D) Proportion of each lipid type in RAS’s first solvation shell (within 0.55 nm), normalized by its molar fraction in the initial average lipid mixture (ARC), as a function of RAS aggregate size. (E) Values of D as a function of RAS aggregate size. (F) Mean square displacement (MSD) curves of JF646-labeled RAS from SPT on supported lipid bilayers with the low (LRC), average (ARC), and high (HRC) RAS lipid compositions.
To further evaluate how the lipid composition affects RAS diffusion, we use SPT to study RAS on supported lipid bilayers with low (LRC), average (ARC) and high (HRC) RAS colocalization compositions. Mean square displacement plots from 50,000 to 100,000 tracks obtained on each bilayer show that RAS undergoes anomalous diffusion that is slower and more confined on HRC compared to LRC and ARC (Fig. 3F). A more rigorous hidden Markov modeling (HMM) analysis of the SPT data on HRC and LRC identifies three interconverting states in which RAS diffusion is slow (∼0.1 μm2/s), medium (∼0.5 μm2/s), and fast (∼4 μm2/s) (SI Appendix, S2.8.4). It also reveals that the lipid-dependence of RAS diffusion (Fig. 3F) results from a decrease in slow and medium diffusion rates and an increased population of slowly diffusing RAS in the HRC (SI Appendix, S2.8.4). One interpretation of these data is that the high PIP2 concentration in HRC directly correlates to large RAS aggregate that diffuses much slower compared to less PIP2 and a smaller RAS cluster on LRC, although other factors can also slow diffusion.
Taken together, simulation and experiments identify a lipid composition (HRC) that recruits more RAS (Fig. 2 C–E) and favors RAS multimerization or confinement (Fig. 3C), slowing its diffusion (Fig. 3F). Lipid-driven RAS clustering provides an attractive hypothesis as an initial organizational step in nucleating a molecular signaling platform for efficient recruitment of RAS effectors.
RAS–RAS Association
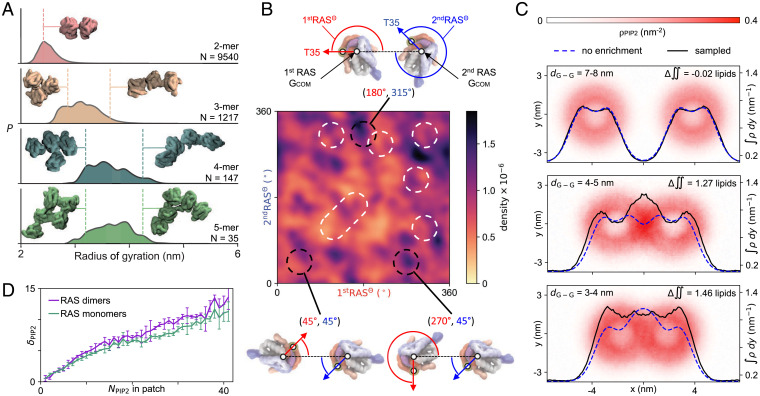
RAS multimers are hypothesized to function as signaling hotspots, and defining RAS–RAS interaction is critical to understanding the role of colocalization in signaling competence (14). Indeed, specific RAS–RAS interfaces have been identified in previously published works (11, 13, 15, 30, 31). However, analysis of 10,939 MuMMI simulations with RAS–RAS contacts reveals a broad distribution of interfacial arrangements (Fig. 4A), including published interfaces (13, 31, 32, 66) (Fig. 4B). The resulting modest (approximately twofold) interfacial preferences are inconsistent with the existence of a distinct dimer interface (Fig. 4B). As such, distinct lipid fingerprints may facilitate initial RAS association through preorientation of the protein. Due to these multiple potential interfaces and heterogeneity seen in RAS–RAS association (Fig. 4B), it is problematic to deconvolute a nonrotationally averaged lipid fingerprint for RAS multimers. Nevertheless, the local lipid environments of RAS aggregates are enriched/depleted in different lipid species, including enriched in PIP2 and depleted of ordered lipids in comparison to RAS monomers (Figs. 3D and 4 C and D, and SI Appendix, Figs. S43 and S44). This lipid–RAS codependence is consistent with a signaling mechanism in which lipid–RAS interactions concentrate RAS prior to RAF binding, and in which subsequent RAF dimerization is facilitated by this RAF-independent RAS colocalization.
Fig. 4.
RAS–RAS interactions. (A) Distributions of radii of gyration based on RAS aggregation number. (B) Sampling of the protein–protein interfaces projected as a density map of two angles. The first angle (1stRASΘ, in red) is between the vectors center of mass (COM) of the first RAS G-domain to COM of the second RAS G-domain (1stRAS GCOM→2ndRAS GCOM) and 1stRAS GCOM→1stRAS T35. The second angle (2ndRASΘ, in blue) is between the vectors 1stRAS GCOM→2ndRAS GCOM and 2ndRAS GCOM→2ndRAS T35. Illustrative examples of the interfaces defined by the angles are shown. White dashed regions indicate various interfaces presented in the literature (13, 31, 32, 66). Data are only plotted for 1stRAS GCOM→2ndRAS GCOM < 6.0 nm from simulations where the RAS proteins start apart (SI Appendix, Fig. S54). (C) PIP2 remodeling based on RAS–RAS G-domain separation, dG-G, shown with G-domain centers of mass on the x axis. Data show PIP2 density (red heatmaps), PIP2 density integrated over −4 nm < y < 4 nm (black lines), and a model reflecting translation of static PIP2 density distributions (blue dashed lines). Differences between the integrated density and the static translation model indicates regions of enrichment and depletion of PIP2 lipids during RAS–RAS association. (D) Preferential binding coefficients showing the enrichment of PIP2 among lipids in contact with RAS, dPIP2, as a function of the number of PIP2 per CG patch, shown separately for RAS dimers and monomers. The larger dPIP2 values for dimers indicate that dimers formation is favored by higher PIP2 concentration.
Lipid Dependence of RAS Orientation and Competence for Effector Binding
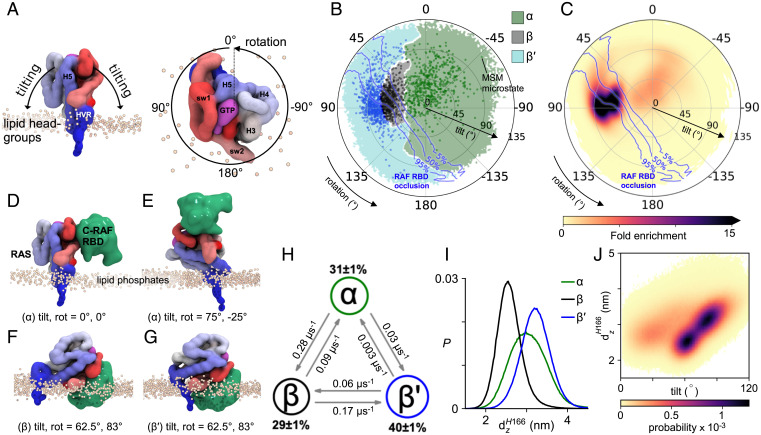
RAS is a closely tethered peripheral membrane protein, and the orientation of the G-domain controls access to its effector binding site through reversible membrane-based occlusion (67, 68). We define the orientation of the RAS G-domain with respect to the membrane surface (69). Briefly, a tilt angle defines the deflection of the long axis of helix 5 from the bilayer normal, and a rotation angle defines the direction in which that tilt occurs (Fig. 5A). CG simulations with one RAS reveal statistical confinement of RAS to specific orientations; moreover, Markov-state modeling of RAS orientation reveals three kinetic states of RAS orientation in which tilting brings G-domain helices 3 to 5 (α-state) or β-sheets 1 to 3 and switch I (β and β′ states) toward the membrane (Fig. 5 B and G and SI Appendix, S2.4 and S2.5).
Fig. 5.
RAS orientations. (A) Definition of tilt and rotation angles based on a reference orientation with the long axis of G-domain helix 5 perpendicular to the membrane surface. (B) Kinetic states of G-domain orientation. Dots represent Markov state modeling (MSM) microstates and color associates sampled orientations with the nearest microstate. Blue isocontours define the likelihood of membrane-based occlusion of RAF binding. (C) G-domain disposition in simulations with one RAS. (D–G) Representative configurations of α, β, and β′ states illustrating orientation-dependent competence for effector binding. (H) MSM state populations and transition rates. (I) State-specific distributions of the displacement of the H166 backbone bead from the bilayer center, dzH166. (J) G-domain tilt vs. dzH166.
The most populated RAS orientations are incompatible with effector binding (Fig. 5C). We use the RBD of RAF (PDB ID code 4G0N) as the relevant effector and observe that α-states project the effector binding interface of RAS away from the membrane (Fig. 5 B–E), possibly positioning RAS to bind its effectors (59, 70). Conversely, effector binding is occluded in most of β′- and half of the β-state orientations (Fig. 5 B, C, F, and G). Previously described exposed and occluded orientations (59, 70, 71) are included in the α and β/β′ states, respectively, and an experimentally driven model of RAS–RAF association straddles the dividing line between the α and β states (70). HMM analysis indicates that direct transitions between β′ and α are relatively rare (Fig. 5H and SI Appendix, S2.4) and interconversion between these states typically occurs via the β-state intermediate.
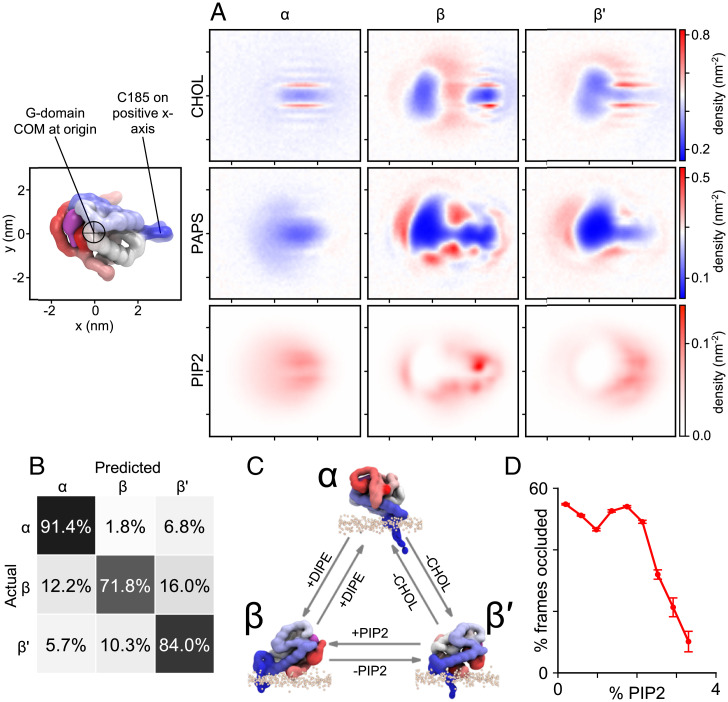
Consistent with the macro simulation, CG simulations reveal distinct RAS lipid fingerprints, which exhibit spatially complex patterns of lipid enrichment and depletion around the G-domain, HVR, and farnesyl anchor (SI Appendix, S2.6, and Fig. S30). A variety of protein–lipid interactions contribute to the modulation of G-domain orientation by PM lipids, and distinct lipid fingerprints observed for the different RAS orientational states (Fig. 6A) are specific enough that a convolutional neural network can predict RAS states with an accuracy of 82% based on inner leaflet spatial lipid distributions (Fig. 6B and SI Appendix, S2.6.2). Among the complex lipid signatures, regression analysis shows that enrichment/depletion of different lipids species affects the likelihood of transitions between states (Fig. 6C and SI Appendix, S2.6.3). For example, changes in the number of PIP2 lipids interacting with the G-domain emerge as a good predictor of transitions between β and β′ states, an orientational change that can modulate effector occlusion (Fig. 5B).
Fig. 6.
RAS–lipid interactions. (A) Relative lipid densities around monomeric RAS. The center of mass of the G-domain is at the origin and the backbone bead of C-terminal HVR residue C185 is on the positive x axis. (B) The confusion matrix shows the accuracy of ML predictions of the RAS state based on local lipid compositions. Each row corresponds to an actual state (computed using tilt/rotation) and each column to a predicted state; off-diagonal elements are the errors made by the prediction. (C) Local lipid composition is predictive for transitions between states. Transition probabilities increase when noted lipid concentrations (+) increase and (−) decrease (see SI Appendix, S2.6.3). (D) RAF-occlusion vs. PIP2 content.
Significantly, occlusion of the RAF-binding interface by the membrane is relieved by PIP2 at molar concentrations above 2% (Fig. 6D). The kinetic barrier between β and β′ states is related to a specific PIP2 binding site at the junction between the G-domain and the HVR, near H166 (Fig. 6A). The exchange between metastable β and β′ states (Fig. 5 B and H), appears to be controlled by reversible membrane adhesion of this region of the HVR (Fig. 5I), which must disengage to permit extensive G-domain tilting and transition from β to β′ (Fig. 5J). In this work, we identify molecular details by which lipid–RAS interactions regulate occlusion, revealing a plausible strategy for inhibition of RAS function. Indeed, the identification of a small molecule that stabilizes KRAS in the β-state and prevents RAF binding validates this approach and motivates the search for more potent molecules (71).
Discussion
The unique capabilities of MuMMI to operate and provide insights at two vastly different temporal and spatial scales in a coordinated manner represent a fundamentally unique technology in computational biology. Furthermore, this study highlights the necessity of such a capability by demonstrating the importance of considering membrane proteins and lipids collectively, as a single unit with two highly interdependent components. Only through the combination of the macroscale sampling of diverse local lipid environments with the microscale dynamics at the molecular level can we observe these distinct RAS–lipid interaction patterns, which were subsequently used to design the supporting experiments.
The combination of simulation and experiment reveals that RAS not only remodels its local lipid environment, but that it is itself regulated by these distinct lipid patterns. For example, an increased concentration of GTP-loaded KRAS4b is associated with PIP2 enrichment and cholesterol depletion, while PIP2 drives functionally relevant orientational changes in RAS. The differential influence of lipid fingerprints on interactions of RAS with regulators, such as SOS (72), and effectors such as RAF, PI3K, and RALGDS, are unknown, but there is clear potential for lipid patterning to define recruitment sites for discrete effectors and thereby segregate RAS-mediated signal transduction pathways. For membrane proteins in general, there is mounting evidence from both simulation and experiment for these types of “symbiotic” relationships between proteins and their surrounding membrane environment (45, 73, 74), establishing another mechanism for the cell to impose fine control over molecular function.
The current work finds RAS–RAS interactions to be interfacially nonspecific and validates all previously proposed interfaces as part of a broad ensemble of possible interactions. This interfacial promiscuity suggests that RAS multimer formation is mediated by lipids rather than specific interfacial contacts, and leads us to conclude that nonenzymatically relevant mutations, some of which impact tumor growth (11), may impact the dynamics of RAS multimer formation through changes in protein–lipid interactions.
MuMMI uses cohesive multiscale coupling, which in this application reveals both the broad scope and fine details of membrane remodeling that underlies functionally relevant RAS–lipid dynamics. More importantly, MuMMI has been designed to be generalizable and, as such, opens new avenues for potential research and expands the range of questions that can be addressed using biomolecular simulation. With a preinvestment in macro model development or model tuning and significant computational resources, a MuMMI-style campaign could be run instead of nearly any simulation ensemble, resulting in a queryable broad ensemble over a large set of relevant conditions, instead of predefined sampling along a given reaction coordinate.
Materials and Methods
An overview of MuMMI is given in Multiscale Simulation Framework and a detailed description of MuMMI is provided in SI Appendix, S1.1. The model generation and parameterization are described in SI Appendix, S1.2. Simulation analysis is described in SI Appendix, S1.2 and experimental methods in SI Appendix, S1.4. MuMMI is composed of numerus subcomponents, both freely available third-party applications and custom codes that all have been made available. Custom codes are: ddcMD (https://github.com/LLNL/ddcMD), ddcMDconverter (https://github.com/LLNL/ddcMDconverter), Maestro (https://github.com/LLNL/maestrowf), Flux (https://github.com/flux-framework), PyTarIdx (https://github.com/LLNL/pytaridx), DynIm (github.com/LLNL/dynim), MuMMI-core (that contains the workflow manager, https://github.com/mummi-framework/mummi-core), and MuMMI-ras (the remaining application components, https://github.com/mummi-framework/mummi-ras).
Supplementary Material
Acknowledgments
We thank Siewert-Jan Marrink and Carsten F. E. Schroer from the University of Groningen for advice on Martini and GTP parameters. From Frederick National Laboratory for Cancer Research (FNLCR) we thank Matt Drew, Shelley Perkins, and Vanessa Wall from the National Cancer Institute (NCI) RAS Initiative for cloning, expression, and protein purification; Jeff Clogston from Nanotechnology Characterization Laboratory for quality control of the liposome lipid composition; and Will Heinz from Optical Microscopy Analysis Laboratory for assistance with atomic force microscopy. This work was performed under the auspices of the US Department of Energy (DOE) by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344; Los Alamos National Laboratory (LANL) under Contract DE-AC5206NA25396; Oak Ridge National Laboratory under Contract DE-AC05-00OR22725; Argonne National Laboratory under Contract DE-AC02-06-CH11357; and under the auspices of the NCI by FNLCR under Contract HHSN261200800001E. Release: LA-UR-19-32061, LLNL-JRNL-799963. This work has been supported in part by the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) program established by the US DOE and the NCI of the NIH. We thank the Livermore Institutional Grand Challenge and LANL Institutional computing for computing time. The LANL Institutional Computing Program is supported by the US DOE National Nuclear Security Administration under Contract DE-AC52-06NA25396.
Footnotes
Reviewers: J.G., Georgia Institute of Technology; L.M., Centre National de la Recherche Scientifique; and M.S., University of Oxford.
Competing interest statement: F.M. is a consultant for Amgen, Daiichi Ltd., Frontiers Med, Exuma Biotech, Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, Inc., PellePharm, Pfizer Inc., PMV Pharma, and Quanta Therapeutics. F.M. is consultant and cofounder for (with ownership interest including stock options) BridgeBio, Olema Pharmaceuticals, Inc., and Quartz. F.M. is the scientific director of the National Cancer Institute RAS Initiative at the Frederick National Laboratory for Cancer Research/Leidos Biomedical Research, Inc. F.M. has been recipient of research grants from Daiichi Sankyo and Gilead Sciences and has a current grant from Boehringer-Ingelheim. H.I.I. and M.S. are coauthors on a 2019 review article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113297119/-/DCSupplemental.
Data Availability
All macro and micro simulation input and parameter files with an example micro simulation are available at Biochemical and Biophysical Systems Group, https://bbs.llnl.gov/RAS-lipid-dependent-dynamics-data.html. Aggregated saved simulation data is ~220 TB and is openly available on the NIH MoDaC server (https://modac.cancer.gov/). The atomic coordinates and structure factors of the GMPPNP-bound KRAS have been deposited in the Protein Data Bank (PDB ID 6VC8).
Change History
January 21, 2022: The byline has been updated.
References
- 1.Bailey M. H., et al. ; MC3 Working Group; Cancer Genome Atlas Research Network, Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeton A. B., Salter E. A., Piazza G. A., The RAS-effector interaction as a drug target. Cancer Res. 77, 221–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCormick F., K-Ras protein as a drug target. J. Mol. Med. (Berl.) 94, 253–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephen A. G., Esposito D., Bagni R. K., McCormick F., Dragging Ras back in the ring. Cancer Cell 25, 272–281 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Hobbs G. A., Der C. J., Rossman K. L., RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129, 1287–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lito P., Solomon M., Li L.-S., Hansen R., Rosen N., Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 351, 604–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valencia A., Chardin P., Wittinghofer A., Sander C., The RAS protein family: Evolutionary tree and role of conserved amino acids. Biochemistry 30, 4637–4648 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Willumsen B. M., Christensen A., Hubbert N. L., Papageorge A. G., Lowy D. R., The p21 RAS C-terminus is required for transformation and membrane association. Nature 310, 583–586 (1984). [DOI] [PubMed] [Google Scholar]
- 9.Neale C., García A. E., Methionine 170 is an environmentally sensitive membrane anchor in the disordered HVR of K-RAS4B. J. Phys. Chem. B 122, 10086–10096 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Bourne H. R., Sanders D. A., McCormick F., The GTPase superfamily: A conserved switch for diverse cell functions. Nature 348, 125–132 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Ambrogio C., et al. , KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell 172, 857–868 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Nan X., et al. , RAS-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 7996–8001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muratcioglu S., et al. , GTP-dependent K-Ras dimerization. Structure 23, 1325–1335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y., Hancock J. F., Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim. Biophys. Acta 1853, 841–849 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Sarkar-Banerjee S., et al. , Spatiotemporal analysis of K-Ras plasma membrane interactions reveals multiple high order homo-oligomeric complexes. J. Am. Chem. Soc. 139, 13466–13475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abankwa D., Gorfe A. A., Mechanisms of Ras membrane organization and signaling: Ras rocks again. Biomolecules 10, 1522 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Q. N., et al. , RAS nanoclusters: Dynamic signaling platforms amenable to therapeutic intervention. Biomolecules 11, 377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber C. K., Slupsky J. R., Kalmes H. A., Rapp U. R., Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 61, 3595–3598 (2001). [PubMed] [Google Scholar]
- 19.Rajakulendran T., Sahmi M., Lefrançois M., Sicheri F., Therrien M., A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Lavoie H., Therrien M., Regulation of RAF protein kinases in ERK signalling. Nat. Rev. Mol. Cell Biol. 16, 281–298 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Chung J. K., et al. , K-Ras4B remains monomeric on membranes over a wide range of surface densities and lipid compositions. Biophys. J. 114, 137–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weise K., et al. , Membrane-mediated induction and sorting of K-Ras microdomain signaling platforms. J. Am. Chem. Soc. 133, 880–887 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Prior I. A., Muncke C., Parton R. G., Hancock J. F., Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160, 165–170 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y., et al. , Signal integration by lipid-mediated spatial cross talk between Ras nanoclusters. Mol. Cell. Biol. 34, 862–876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillette W. K., et al. , Farnesylated and methylated KRAS4b: High yield production of protein suitable for biophysical studies of prenylated protein-lipid interactions. Sci. Rep. 5, 15916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadwallader K. A., Paterson H., Macdonald S. G., Hancock J. F., N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol. Cell. Biol. 14, 4722–4730 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y., et al. , Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 168, 239–251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y., Prakash P. S., Liang H., Gorfe A. A., Hancock J. F., The KRAS and other prenylated polybasic domain membrane anchors recognize phosphatidylserine acyl chain structure. Proc. Natl. Acad. Sci. U.S.A. 118, e2014605118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y., Gorfe A. A., Hancock J. F., RAS nanoclusters selectively sort distinct lipid headgroups and acyl chains. Front. Mol. Biosci. 8, 686338 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R., Membrane-associated RAS dimers are isoform-specific: K-Ras dimers differ from H-Ras dimers. Biochem. J. 473, 1719–1732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakash P., et al. , Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers. Sci. Rep. 7, 40109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K.-Y., et al. , Two distinct structures of membrane-associated homodimers of GTP- and GDP-bound KRAS4B revealed by paramagnetic relaxation enhancement. Angew. Chem. Int. Ed. Engl. 59, 11037–11045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sayyed-Ahmad A., Cho K.-J., Hancock J. F., Gorfe A. A., Computational equilibrium thermodynamic and kinetic analysis of K-Ras dimerization through an effector binding surface suggests limited functional role. J. Phys. Chem. B 120, 8547–8556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Güldenhaupt J., et al. , N-Ras forms dimers at POPC membranes. Biophys. J. 103, 1585–1593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inouye K., Mizutani S., Koide H., Kaziro Y., Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 275, 3737–3740 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Erwin N., et al. , Lipoprotein insertion into membranes of various complexity: Lipid sorting, interfacial adsorption and protein clustering. Phys. Chem. Chem. Phys. 18, 8954–8962 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Kapoor S., et al. , The role of G-domain orientation and nucleotide state on the RAS isoform-specific membrane interaction. Eur. Biophys. J. 41, 801–813 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Bhatia H., et al. , Machine learning based dynamic-importance sampling for adaptive multiscale simulations. Nat. Mach. Intell. 3, 401–409 (2021). [Google Scholar]
- 39.Ingólfsson H. I., et al. , Lipid organization of the plasma membrane. J. Am. Chem. Soc. 136, 14554–14559 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Ingólfsson H. I., et al. , Capturing biologically complex tissue-specific membranes at different levels of compositional complexity. J. Phys. Chem. B 124, 7819–7829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanton L., et al. , Dynamic density functional theory of multicomponent cellular membranes. arXiv [Preprint] (2021). https://arxiv.org/abs/2112.08651 (Accessed 22 December 2021). [Google Scholar]
- 42.Marrink S. J., Risselada H. J., Yefimov S., Tieleman D. P., de Vries A. H., The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 111, 7812–7824 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Ingólfsson H. I., et al. , The power of coarse graining in biomolecular simulations. Wiley Interdiscip. Rev. Comput. Mol. Sci. 4, 225–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marrink S. J., et al. , Computational modeling of realistic cell membranes. Chem. Rev. 119, 6184–6226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corradi V., et al. , Emerging diversity in lipid-protein interactions. Chem. Rev. 119, 5775–5848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alessandri R., et al. , Pitfalls of the Martini model. J. Chem. Theory Comput. 15, 5448–5460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Souza P. C. T., et al. , Martini 3: A general purpose force field for coarse-grained molecular dynamics. Nat. Methods 18, 382–388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X., et al. , ddcMD: A fully GPU-accelerated molecular dynamics program for the Martini force field. J. Chem. Phys. 153, 045103 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Di Natale F., et al. , A massively parallel infrastructure for adaptive multiscale simulations: Modeling RAS initiation pathway for cancer. in Proceedings of SC’19: The International Conference for High Performance Computing, Networking, Storage and Analysis (ACM, Denver, CO, 2019), Article no. 57, pp. 1–16.
- 50.Corradi V., et al. , Lipid-protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 4, 709–717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weise K., et al. , Dissociation of the K-RAS4B/PDEδ complex upon contact with lipid membranes: Membrane delivery instead of extraction. J. Am. Chem. Soc. 134, 11503–11510 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Liu S., Anirudh R., Thiagarajan J. J., Bremer P.-T., Uncovering interpretable relationships in high-dimensional scientific data through function preserving projections. Machine Learning: Sci. Technol. 1, 045016 (2020). [Google Scholar]
- 53.McLean M. A., Stephen A. G., Sligar S. G., PIP2 influences the conformational dynamics of membrane-bound KRAS4b. Biochemistry 58, 3537–3545 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Cao S., et al. , K-Ras G-domain binding with signaling lipid phosphatidylinositol (4,5)-phosphate (PIP2): Membrane association, protein orientation, and function. J. Biol. Chem. 294, 7068–7084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kusumi A., Suzuki K. G. N., Kasai R. S., Ritchie K., Fujiwara T. K., Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 36, 604–615 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Goswami D., et al. , Membrane interactions of the globular domain and the hypervariable region of KRAS4b define its unique diffusion behavior. eLife 9, e47654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lakshman B., et al. , Quantitative biophysical analysis defines key components modulating recruitment of the GTPase KRAS to the plasma membrane. J. Biol. Chem. 294, 2193–2207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prakash P., Gorfe A. A., Membrane orientation dynamics of lipid-modified small GTPases. Small GTPases 8, 129–138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prakash P., Zhou Y., Liang H., Hancock J. F., Gorfe A. A., Oncogenic K-RAS binds to an anionic membrane in two distinct orientations: A molecular dynamics analysis. Biophys. J. 110, 1125–1138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z.-L., Buck M., Computational modeling reveals that signaling lipids modulate the orientation of K-Ras4A at the membrane reflecting protein topology. Structure 25, 679–689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gorfe A. A., Hanzal-Bayer M., Abankwa D., Hancock J. F., McCammon J. A., Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J. Med. Chem. 50, 674–684 (2007). [DOI] [PubMed] [Google Scholar]
- 62.van Meer G., Voelker D. R., Feigenson G. W., Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L., et al. , Molecular diffusion measurement in lipid bilayers over wide concentration ranges: A comparative study. ChemPhysChem 9, 721–728 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Basconi J. E., Shirts M. R., Effects of temperature control algorithms on transport properties and kinetics in molecular dynamics simulations. J. Chem. Theory Comput. 9, 2887–2899 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Venable R. M., et al. , Lipid and peptide diffusion in bilayers: The Saffman-Delbrück Model and periodic boundary conditions. J. Phys. Chem. B 121, 3443–3457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee K.-Y., et al. , Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: A model for KRAS oligomerization. Chem. Sci. (Camb.) 12, 12827–12837 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abankwa D., Gorfe A. A., Inder K., Hancock J. F., Ras membrane orientation and nanodomain localization generate isoform diversity. Proc. Natl. Acad. Sci. U.S.A. 107, 1130–1135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abankwa D., et al. , A novel switch region regulates H-Ras membrane orientation and signal output. EMBO J. 27, 727–735 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Travers T., et al. , Molecular recognition of RAS/RAF complex at the membrane: Role of RAF cysteine-rich domain. Sci. Rep. 8, 8461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazhab-Jafari M. T., et al. , Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc. Natl. Acad. Sci. U.S.A. 112, 6625–6630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang Z., et al. , Inhibition of K-RAS4B by a unique mechanism of action: Stabilizing membrane-dependent occlusion of the effector-binding site. Cell Chem. Biol. 25, 1327–1336 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Gureasko J., et al. , Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 15, 452–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van den Bogaart G., et al. , Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muller M. P., et al. , Characterization of lipid-protein interactions and lipid-mediated modulation of membrane protein function through molecular simulation. Chem. Rev. 119, 6086–6161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All macro and micro simulation input and parameter files with an example micro simulation are available at Biochemical and Biophysical Systems Group, https://bbs.llnl.gov/RAS-lipid-dependent-dynamics-data.html. Aggregated saved simulation data is ~220 TB and is openly available on the NIH MoDaC server (https://modac.cancer.gov/). The atomic coordinates and structure factors of the GMPPNP-bound KRAS have been deposited in the Protein Data Bank (PDB ID 6VC8).